- 1Department of Internal Medicine, Shunde Women and Children’s Hospital (Maternity and Child Healthcare Hospital of Shunde Foshan), Guangdong Medical University, Foshan, China
- 2Key Laboratory of Research in Maternal and Child Medicine and Birth Defects, Guangdong Medical University, Foshan, China
- 3Shunde Women and Children’s Hospital (Maternity and Child Healthcare Hospital of Shunde Foshan), Matenal and Child Research Institute, Guangdong Medical University, Foshan, China
- 4State Key Laboratory for Quality Research of Chinese Medicines, Macau University of Science and Technology, Taipa, Macau (SAR) China
- 5Department of Clinical Laboratory, People’s Hospital of Haiyuan County, Zhongwei, China
- 6Department of Endocrinology, Affiliated Hospital of Guangdong Medical University, Zhanjiang, China
- 7Department of Ultrasound, Shunde Women and Children’s Hospital (Maternity and Child Healthcare Hospital of Shunde Foshan), Guangdong Medical University, Foshan, China
Background: Insulin-like growth factor-1 (IGF-1) has been demonstrated to increase fatty acid β oxidation during fasting, and play an important role in regulating lipid metabolism and type 2 diabetes mellitus (T2DM). The rs35767 (T > C) polymorphism, a functional SNP was found in IGF-1 promoter, which may directly affect IGF-1 expression. However, the inconsistent findings showed on the IGF-1 rs35767 polymorphism and T2DM risk.
Methods: We performed a comprehensive meta-analysis to estimate the association between the IGF-1 rs35767 and T2DM risk among four genetic models (the allele, additive, recessive and dominant models).
Results: A total 49,587 T2DM cases and 97,906 NDM controls were included in the allele model, a total 2256 T2DM cases and 2228 NDM controls were included in the other three genetic models (the additive; recessive and dominant models). In overall analysis, the IGF-1 rs35767 was shown to be significantly associated with increased T2DM risk for the allele model (T vs. C: OR = 1.251, 95% CI: 1.082–1.447, p = 0.002), additive model (homozygote comparisons: TT vs. CC: OR = 2.433, 95% CI: 1.095–5.405, p = 0.029; heterozygote comparisons: TC vs. CC: OR = 1.623, 95% CI: 1.055–2.495, p = 0.027) and dominant model (TT + CT vs. CC: OR = 1.934, 95% CI: 1.148–3.257, p = 0.013) with random effects model. After omitting Gouda’s study could reduce the heterogeneity, especially in the recessive model (TT vs. CC + CT: I2 = 38.7%, p = 0.163), the fixed effects model for recessive effect of the T allele (TT vs. CC + CT) produce results that were of borderline statistical significance (OR = 1.206, 95% CI: 1.004–1.448, p = 0.045). And increasing the risk of T2DM in Uyghur population of subgroup for the allele model.
Conclusion: The initial analyses that included all studies showed statistically significant associations between the rs35767 SNP and type 2 diabetes, but after removing the Gouda et al. study produced results that were mostly not statistically significant. Therefore, there is not enough evidence from the results of the meta-analysis to indicate that the rs35767 SNP has a statistically significant association with type 2 diabetes.
1 Introduction
Diabetes is one of the most common chronic metabolic disorder diseases in the worldwide, over 90% of the diabetes patients are type 2 diabetes mellitus (T2DM), which is characterized by insulin resistance in peripheral tissues and dysregulated insulin secretion by pancreatic beta (β) cells (Banerjee and Vats, 2014; Song et al., 2015). Substantial evidence suggests that insulin resistance, an inherited genetic defect, is the basis and major feature of T2DM (DeFronzo and Tripathy, 2009; Cai et al., 2019; Li et al., 2021). Insulin resistance is attributable to excess fatty acids and proinflammatory cytokines, which leads to impaired glucose transport and increases fat breakdown. Since there is an inadequate response or production of insulin, the body responds by inappropriately increasing glucagon, thus further contributing to hyperglycemia. Accumulated data have revealed that lipid abnormalities are associated with insulin resistance and contribute to T2DM (Johnson and Olefsky, 2013; Perry et al., 2014; Li et al., 2021). Studies have also revealed lipid metabolism-related genes and their single-nucleotide polymorphisms (SNPs) associated with insulin resistance and the development of T2DM (Ruchat et al., 2009; Dupuis et al., 2010; Chistiakov et al., 2012; Langberg et al., 2012; Mannino et al., 2013; Li et al., 2014; Thankamony et al., 2014; Yuan et al., 2015; Li et al., 2021).
Insulin-like growth factor-1 (IGF-1) is a circulating growth factor which structure is highly homologous with pro-insulin. IGF-1 is expresses in insulin-resistant tissue, it downregulates free fatty acid and increases fatty acid β oxidation during fasting (Thankamony et al., 2014; Li et al., 2021). It plays an important role in regulating lipid metabolism and insulin sensitivity (Seppä et al., 2015; Gouda et al., 2019), since it effects on glucose homeostasis and associated with insulin resistance (Li et al., 2011; Li et al., 2014; Dai et al., 2015; Ming et al., 2015; Yuan et al., 2015; Wei et al., 2018; Liao et al., 2019; Regué et al., 2019; Li et al., 2021). Previous studies have been reported that people with a low IGF-1 level are prone to have diabetes mellitus (Chen et al., 2013; Colao et al., 2013; Shankar and Li, 2013). Polymorphisms in the IGF-1 gene can directly affect IGF-1 expression. The rs35767 (T > C) polymorphism, a functional SNP was found in IGF-1 promoter, in which the promoter with C allele showed a higher transcriptional activity than promoter with T allele (Telgmann et al., 2009). Therefore, rs35767 may contribute to insulin resistance involving lipid metabolism in T2DM.
A significant association of IGF-1 rs35767 with T2DM has been reported in several case-control studies (Gouda et al., 2019; Gulixiati·Maimaitituersun. 2020; Wang. 2019; Zhang et al., 2017; Song et al., 2015). However, seven studies failed to replicate the results (Dupuis et al., 2010; Hu et al., 2010; Fujita et al., 2012; Liu et al., 2012; Zhao et al., 2016; Li et al., 2019; Li et al., 2021). In veiw of the inconsistent results, whether IGF-1 rs35767 is associated with T2DM remains to be determined. In this meta-analysis, we estimate the association of IGF-1 rs35767 with T2DM among four different genetic models.
2 Materials and Methods
2.1 Literature Search
The Google Scholar, PubMed and Chinese National Knowledge Infrastructure were comprehensively searched for related studies published before July 31, 2021, using the key terms: “insulin-like growth factor 1 or IGF-1 or IGF1,” “rs35767 or rs35767 (T > C) or rs35767 (A > G),” “polymorphism or SNP or mutation or variant” and “diabetes or type 2 diabetes or T2DM.” All searches had no language limitations. Eligible studies were estimated by reading full texts, and excluded substandard studies.
2.2 Inclusion and Exclusion Criteria
The following inclusion criteria: 1) case-control or cohort studies that relate to the IGF-1 rs35767 and T2DM risk; 2) sufficient raw data or adequate data for assessing odds ratios (ORs) with corresponding 95% confidence intervals (CIs); and 3) The diagnostic standard of T2DM conformed to the World Health Organization.
The following exclusion criteria: 1) not a case-control study; 2) irrelevant to IGF-1 rs35767 and T2DM risk; 3) lacking detailed data; and 4) control subjects is not in Hardy-Weinberg equilibrium (HWE).
2.3 Data Extraction
Data were independently extracted by two authors from the eligible studies and collected the following data: first author, year of publication, origin, the numbers of T2DM cases and NDM controls, gender and age, BMI (kg/m2), the distributions number of genotype and alleles, ORs with 95% CI, or ability to calculate the OR and 95% CI. p-value for the HWE of NDM controls.
2.4 Statistical Analysis
Statistical analyses using the STATA v.14.0 software (Stata Corporation, TX, United States). Four genetic models were evaluated in this meta-analysis: the allele model (T vs. C); the additive model (homozygote comparisons: TT vs. CC; heterozygote comparisons: TC vs. CC); the recessive model (TT vs. CC + CT) and the dominant model (TT + CT vs. CC). Using Q-test and I2 test to estimate the genetic heterogeneity. OR with corresponding 95% CIs were calculated by the random effectss model when p < 0.01 and I2 > 50% (He et al., 2015; Zhang et al., 2016). Otherwise, the fixed effectss model were used. Sensitivity analyses were implemented to evaluate the stability of the overall effect by excluding a study at a time. The Hardy-Weinberg equilibrium for the NDM controls was assessed using Pearson’s Chi-squared test. Using Bgger’s test to evaluate publication bias (Shen et al., 2015; Li et al., 2016; Liu et al., 2017; Han et al., 2019).
3 Results
3.1 Study Inclusion and Characteristics
A total of 182 potential articles obtained through initial search. 51 duplicates were excluded. Then 131 studies were screened on title and abstract, 84 of them were excluded. The left 47 articles were evaluated by full-text reading, 35 of them were excluded cause that 22 were not case-control researchs, 10 were not related to rs35767 or T2DM, three did not provided sufficient data. 12 articles were included that there are six articles including five in English and one in Chinese just of the allele model data, and other six articles including four in English and two in Chinese of four genetic models data (the allele, additive, recessive and dominant models). Flow chart of researches selection in the meta-analysis was shown in Figure 1. A total 49,587 T2DM cases and 97,906 NDM controls were included in the allele model, a total 2256 T2DM cases and 2228 NDM controls were included in the other three genetic models (the additive; recessive and dominant models). The characteristics of each study are shown in Table 1 and Supplementary Table S1.
3.2 Meta-Analysis
The association between the IGF-1 rs35767 polymorphism and T2DM were evaluated using ORs and 95% CI in the allele model (12 studies, 49587 T2DM cases and 97906 NDM controls) and the additive; recessive and dominant models (6 studies, 2256 T2DM cases and 2228 NDM controls).
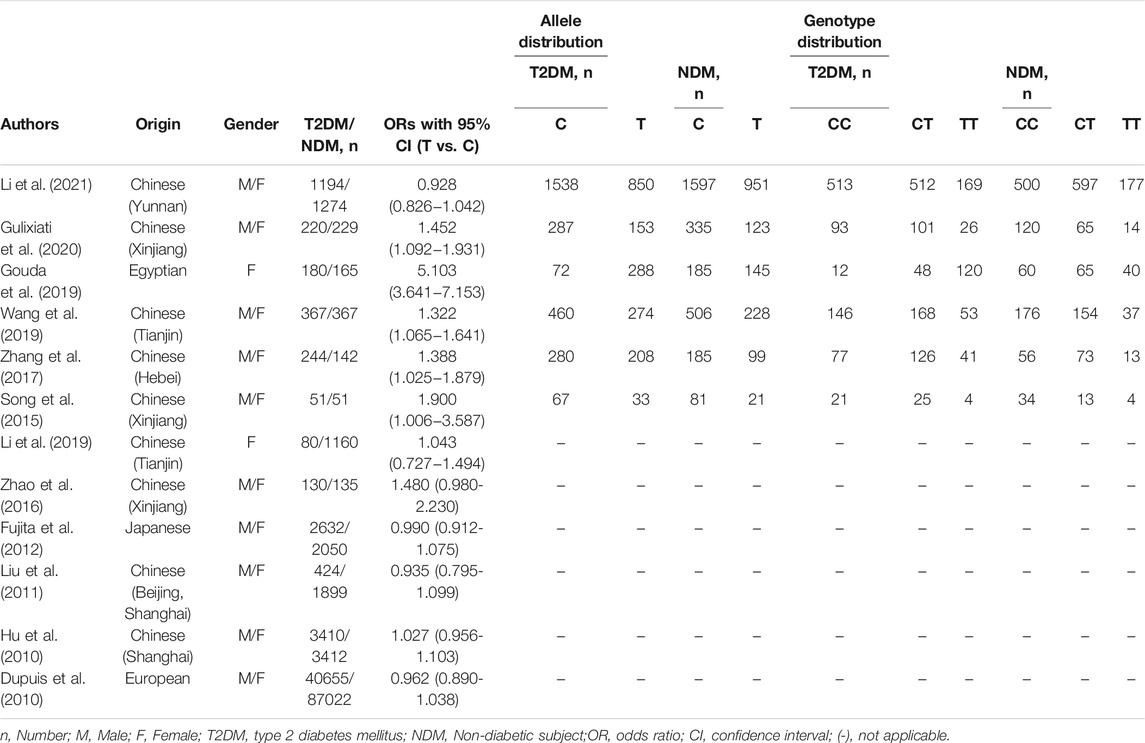
In overall analysis, A random effects model were used to analyze the allele, additive, recessive and dominant models. The IGF-1 rs35767 was shown to be significantly associated with increased T2DM risk for the allele model (T vs. C: OR = 1.251, 95% CI: 1.082–1.447,p = 0.002), additive model (homozygote comparisons: TT vs. CC: OR = 2.433, 95% CI: 1.095–5.405, p = 0.029; heterozygote comparisons: TC vs. CC: OR = 1.623, 95% CI: 1.055–2.495, p = 0.027) and dominant model (TT + CT vs. CC: OR = 1.934, 95% CI: 1.148–3.257, p = 0.013). The results showed no significant difference for the recessive model (TT vs. CC + CT: OR = 1.876, 95% CI: 0.989–3.559, p = 0.054) (Figure 2).
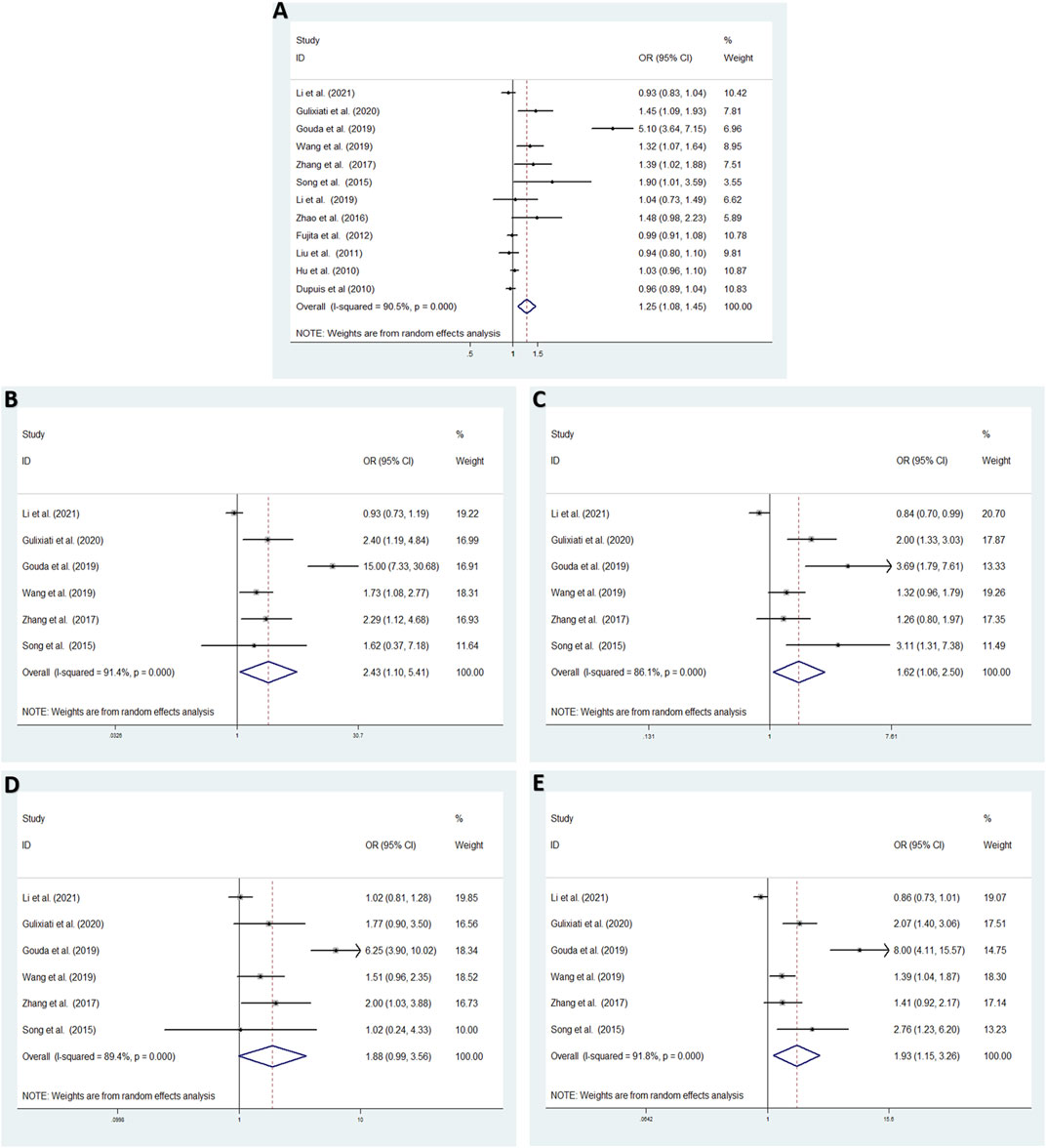
FIGURE 2. Meta-analysis with a random effects model for the association between the IGF-1 rs35767 and T2DM susceptibility. (A) Allele model, T vs. C. (B) Additive model (homozygote comparisons): TT vs. CC.(C) Additive model (heterozygote comparisons): TC vs. CC. (D) Recessive model, TT vs. CC + CT.(E) Dominant model, TT + CT vs. CC. OR: odds ratio, CI: confidence interval, I-squared: measure to quantify the degree of heterogeneity in meta-analyses.
3.3 Sensitivity Analysis
Aiming to estimate the influence of each study on the overall OR below four genetic models and analysis the sources of high heterogeneity, sensitivity meta-analysis was performed with random effects model. The results were showed in Figure3, omitting Gouda’s study could reduce the heterogeneity, especially in the recessive model (TT vs. CC + CT: I2 = 38.7%, p = 0.163), the fixed effects model for recessive effect of the T allele (TT vs. CC + CT) produce results that were of borderline statistical significance (OR = 1.206, 95% CI: 1.004–1.448, p = 0.045); In addtion, other three modal show moderate degree of heterogeneity (T vs. C: I2 = 64.9%, p = 0.002; TT vs. CC: I2 = 70.3%, p = 0.009; TC vs. CC: I2 = 84.0%, p = 0.000; TT + CT vs. CC: I2 = 85.8%, p = 0.000, respectively), the result showed no significant association between IGF-1 rs35767 and T2DM risk with random effects model (T vs. C: OR = 1.065, 95% CI: 0.983–1.153, p = 0.126; TT vs. CC: OR = 1.603, 95% CI: 0.996–2.578, p = 0.052; TC vs. CC: OR = 1.407, 95% CI: 0.937–2.112, p = 0.099; TT + CT vs. CC: OR = 1.469, 95% CI: 0.978–2.207, p = 0.064, respectively) (Figure 4). Since there are not sufficient evidence to draw the conclusion that the rs35767 SNP is associated with T2DM.
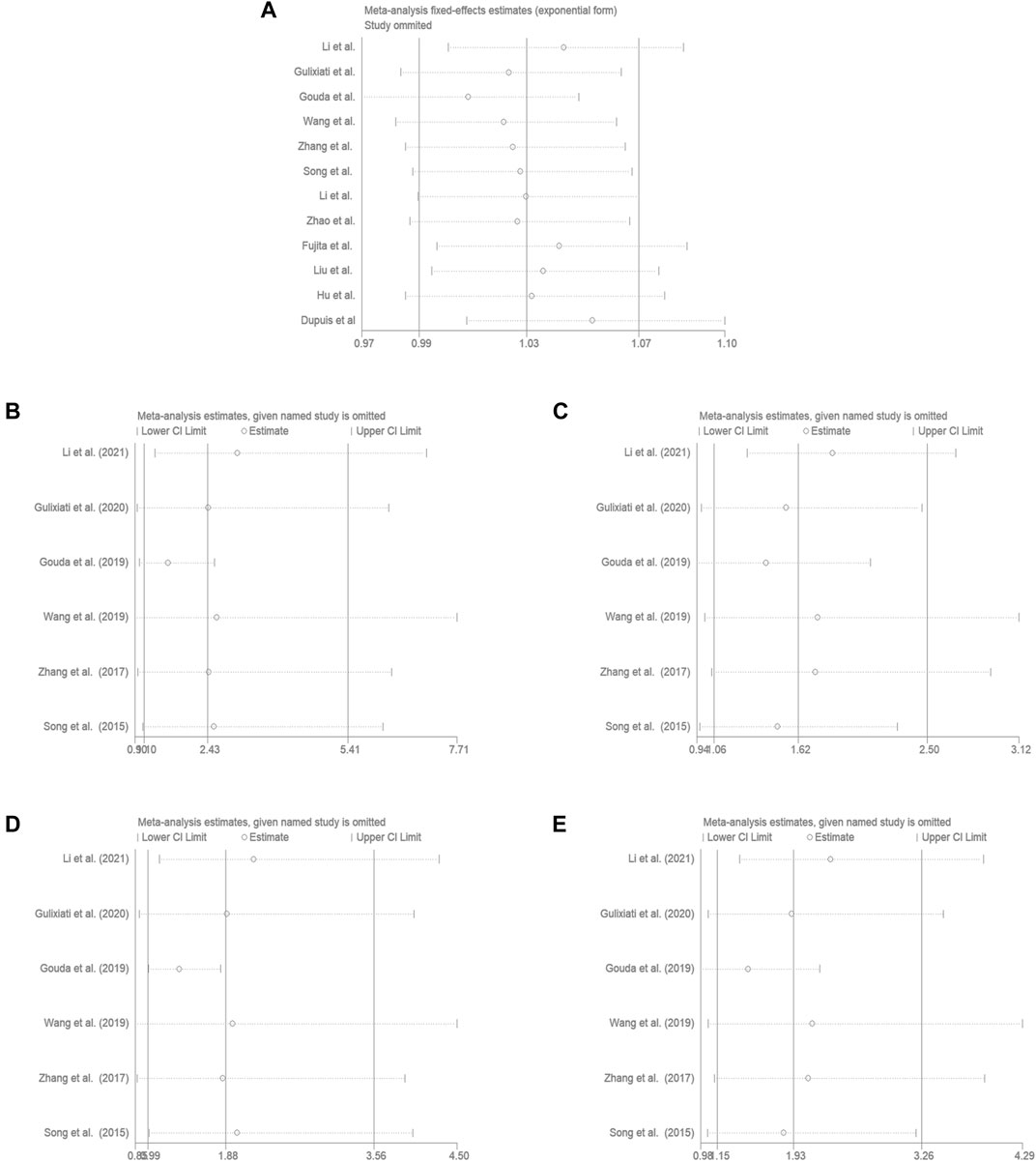
FIGURE 3. Sensitivity analysis by iteratively removing one study at a time. (A) Allele model, T vs. C. (B) Additive model (homozygote comparisons): TT vs. CC. (C) Additive model (heterozygote comparisons): TC vs. CC. (D) Recessive model, TT vs. CC + CT. (E) Dominant model, TT + CT vs. CC.
FIGURE 4. Meta-analysis for the association between the IGF-1 rs35767 and T2DM susceptibility after omitting Gouda’s study. (A) Allele model, T vs. C (random effects model). (B) Additive model (homozygote comparisons): TT vs. CC. (random effects model). (C) Additive model (heterozygote comparisons): TC vs. CC (random effects model) (D) Recessive model, TT vs. CC + CT (fixed effects model) (E) Dominant model, TT + CT vs. CC (random effects model). OR: odds ratio, CI: confidence interval, I-squared: measure to quantify the degree of heterogeneity in meta-analyses.
3.4 Subgroup-Analyses
As high heterogeneity was observed, we performed subgroup-analysis according to origin to evaluate the association between rs35767 and T2DM susceptibility in the allele model. The results suggested that rs35767 was significantly related to the risk of T2DM in XinJiang, China subgroup (T vs. C: OR = 1.508, 95% CI: 1.210–1.878, p = 0.000) with fixed effects model; a random effects model were used to analyze the other provinces, China, rs35767 was shown no significant association with T2DM risk (T vs. C: OR = 1.051, 95% CI: 0.943–1.173, p = 0.369); and not a significantly associatied in the other countries subgroup (excluding Gouda et al. Literature) (T vs. C: OR = 0.975, 95% CI: 0.922–1.031, p = 0.376) with fixed effects model (Figure 5).
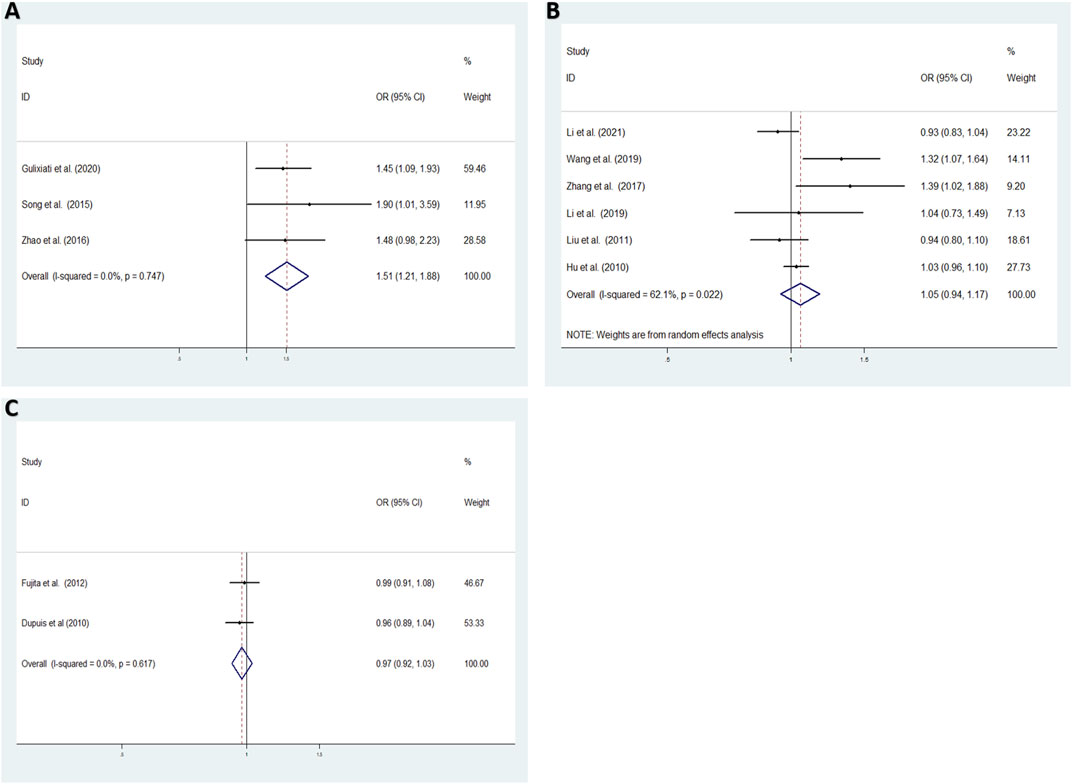
FIGURE 5. The association between the IGF-1 rs35767 and T2DM susceptibility in the subgroup for the allele model (T vs. C) (A) XinJiang, China (fixed effects model) (B) Other provinces, China (random effects model) (C) Other countries (fixed effects model). OR: odds ratio, CI: confidence interval, I-squared: measure to quantify the degree of heterogeneity in meta-analyses.
3.5 Publication Bias
The funnel plot was showed to be visually symmetrical (Supplementary Figure S1, Supplementary Figure S2, Supplementary Figure S3). Begg’s and Egger’s tests were performed to detect publication bias. There was no significant publication bias appeared in all genetic models in overall analysis via Begg’s test (all p > 0.05, Supplementary Table S2), but for Egger’s test, there was publication bias in the additive model (heterozygote comparisons) (Egger’s test, p = 0.006, Supplementary Table S2). We did not determine publication bias for Begg’s test after omitting Gouda’s study and subgroup analysis in all genetic models (all p > 0.05, Supplementary Table S3, Supplementary Table S4). However, for Egger’s test, there were publication bias in the allele model (Egger’s test, p = 0.039, Supplementary Table S3) and additive model (heterozygote comparisons) (Egger’s test, p = 0.031, Supplementary Table S4).
4 Discussion
Previous studies have showed the inconsistent findings of association between IGF-1 rs35767 and the risk of T2DM. Gouda et al. revealed that the TT, TT + CT genotypes of rs35767 were associated with an increased risk of T2DM in pregnant Egyptian women respectively (Gouda et al., 2019). This results were successfully replicated in a Chinese Han population (Wang. 2019). Zhang et al. found that the A allele of rs35767 contributed to the risk of developing T2DM in a Chinese Han population (Zhang.et al., 2017). More recently, two studies documented that the association of the rs35767 in IGF-1 was associated with T2DM in a Uyghur population in China (GulixiatiMaimaitituersun. 2020; Song et al., 2015). However, some studies did not find evidence of an association between rs35767 and T2DM (Dupuis et al., 2010; Hu et al., 2010; Fujita et al., 2012; Liu et al., 2012; Zhao et al., 2016; Li et al., 2019; Li et al., 2021). It is worth noting that the four largest studies with the most statistical power (Dupuis et al., Hu et al., Fujita et al., and Li et al., 2021) did not report statistically significant associations between the rs35767 SNP and T2DM, whereas five small studies (Song et al., Zhang et al., Wang et al., Gouda et al., and Gulixiati et al.) all reported statistically significant associations. Compared the subjects selected for the studies between the larger and smaller studies, we found that in the Gouda et al. study, the mean body mass index (BMI) of subjects with T2DM was 34.26 ± 5.7, which is very different from a mean BMI of 26.96 ± 4.57 for control subjects without T2DM. There were a significant difference concerning BMI between T2DM and control groups. This study included only pregnant women, was observed to be very influential on the initial analyses.
Previous studies have been reported that people with a low IGF-1 level are prone to have diabetes mellitus (Colao et al., 2013; Shankar and Li, 2013). The functional SNP rs35767 (T > C) in IGF-1 promoter, with C allele showed a higher transcriptional activity than promoter with T allele (Telgmann et al., 2009). In terms of mechanism, the higher transcriptional activity of the C allele IGF-1 promoter was contributed by the C/EBPD transcription activator, which bound exclusively to the C allele, but not to the T allele (Chen et al., 2013; Telgmann et al., 2009). Therefore, there may have low IGF-1 expression level when promoter with rs35767 T allele, which contribute to the development of T2DM. In our meta-analysis, we found that T allele, TT genotype, TT + CT genotype of rs35767 increased T2DM risk in overall analysis, as well as increasing the risk of T2DM in Uyghur population. After omitting Gouda’s study, the result showed TT genotype were of borderline statistical significance.
In overall analysis, high heterogeneity among studies were detected in four genetic models, which might be a result of the difference in ethnicity, country, genetic background and environmental factors (e.g., dietary, life style, climates) (Qin et al., 2010). Then omittied Gouda’s study, the heterogeneity was reduced. We found that the subjects of Gouda’s study were pregnant Egyptian women, but participants of other studies were both male and female. Thus gender ratio may also had a certain impact on heterogeneity. The subgroup-analyses were detected by origin in allele model, the subgroup of Uyghur in Xinjiang, China have no heterogeneity, but other subgroups still had high heterogeneity. It is noteworthy that a previous study in Xinjiang found that 19.6% of Uyghur had diabetes, exceptionally higher than that in Kazakh (7.3%) and Han Chinese (9.1%) (Li et al., 2012; Song et al., 2015). The marriage pattern and unique life style might be responsible for the observation. One hand, the practice of endogamy in Uyghur population might also be a reason (Wang et al., 2003; Mamet et al., 2005). On the other hand, The most Uyghurs have different dietary habits from Han Chinese. They have more meat, high carbohydrate diets with a higher salt (more than 20 g per day) and less unsaturated fatty acids compared with Han Chinese (Zhai et al., 2007).
There still have several limitations in our meta-analysis. Firstly, there were limited studies which estimated IGF-1 rs35767 and T2DM risk, only six articles had four gene models data, and the other six articles had only one allele model data. Secondly, the results did not adjustment the potential risk factors, including gender, body mass index, age, drinking and smoking status, and environmental factors. Thirdly, some results showed relatively obvious heterogeneity, but research the source of heterogeneity needs to more larger sample. Finally, some groups results existed potential publication bias in Egger’s test. Therefore, the results of the article should be interpreted carefully.
5 Conclusion
This is the first time to perform a meta-analysis to systematically summarize the association between the IGF-1 rs35767 and T2DM susceptibility. Overall, there is not enough evidence from the results of the meta-analysis to indicate that the rs35767 SNP has a statistically significant association with T2DM. Further more studies are necessary to verify the results.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material further inquiries can be directed to the corresponding authors.
Author Contributions
QZ, DZ, and QdD were suitable for the study design, literature searches, statistical analysis, and manuscript preparation. The study was supervised by XC, YW, and RG.
Funding
Support for this work includes funding from the National Natural Science Foundation of China (81873649); Doctoral scientific research Initiate funding project of Shunde Women and Children’s Hospital of Guangdong Medical University (Maternity and Child Healthcare Hospital of Shunde Foshan) (2020BSQD007); Guangdong Medical University Research Foundation (GDMUM2020008 and GDMUM2020012); Medical Research Project of Foshan Health Bureau (20210188 and 20210289).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2021.774489/full#supplementary-material
References
Banerjee, M., and Vats, P. (2014). Reactive Metabolites and Antioxidant Gene Polymorphisms in Type 2 Diabetes Mellitus. Indian J. Hum. Genet. 20, 10–19. doi:10.4103/0971-6866.132747
Cai, X., Xia, L., Pan, Y., He, D., Zhu, H., Wei, T., et al. (2019). Differential Role of Insulin Resistance and β-cell Function in the Development of Prediabetes and Diabetes in Middle-Aged and Elderly Chinese Population. Diabetol. Metab. Syndr. 11, 24. doi:10.1186/s13098-019-0418-x
Chen, H. Y., Huang, W., Leung, V. H. K., Fung, S. L. M., Ma, S. L., Jiang, H., et al. (2013). Functional Interaction between SNPs and Microsatellite in the Transcriptional Regulation of Insulin-like Growth Factor 1. Hum. Mutat. 34, 1289–1297. doi:10.1002/humu.22363
Chistiakov, D. A., Nikitin, A. G., Smetanina, S. A., Bel'chikova, L. N., Suplotova, L. A., Shestakova, M. V., et al. (2012). The Rs11705701 G>A Polymorphism of IGF2BP2 Is Associated with IGF2BP2 mRNA and Protein Levels in the Visceral Adipose Tissue - A Link to Type 2 Diabetes Susceptibility. Rev. Diabet Stud. 9, 112–122. doi:10.1900/RDS.2012.9.112
Colao, A., Di Somma, C., Cascella, T., Pivonello, R., Vitale, G., Grasso, L. F. S., et al. (2008). Relationships between Serum IGF1 Levels, Blood Pressure, and Glucose Tolerance: an Observational, Exploratory Study in 404 Subjects. Eur. J. Endocrinol. 159, 389–397. doi:10.1530/EJE-08-0201
Dai, N., Zhao, L., Wrighting, D., Krämer, D., Majithia, A., Wang, Y., et al. (2015). IGF2BP2/IMP2-Deficient Mice Resist Obesity through Enhanced Translation of Ucp1 mRNA and Other mRNAs Encoding Mitochondrial Proteins. Cel Metab. 21, 609–621. doi:10.1016/j.cmet.2015.03.006
DeFronzo, R. A., and Tripathy, D. (2009). Skeletal Muscle Insulin Resistance Is the Primary Defect in Type 2 Diabetes. Diabetes Care 32 (Suppl. 2), S157–S163. doi:10.2337/dc09-S302
Dupuis, J., Langenberg, C., Prokopenko, I., Saxena, R., Soranzo, N., Jackson, A. U., et al. (2010). New Genetic Loci Implicated in Fasting Glucose Homeostasis and Their Impact on Type 2 Diabetes Risk. Nat. Genet. 42, 105–116. doi:10.1038/ng.520
Fujita, H., Hara, K., Shojima, N., Horikoshi, M., Iwata, M., Hirota, Y., et al. (2012). Variations with Modest Effects Have an Important Role in the Genetic Background of Type 2 Diabetes and Diabetes-Related Traits. J. Hum. Genet. 57, 776–779. doi:10.1038/jhg.2012.110
Gouda, W., Mageed, L., AzmyOkasha, O. A., Okasha, A., Shaker, Y., and Ashour, E. (2019). Association of Genetic Variants in IGF-1 Gene with Susceptibility to Gestational and Type 2 Diabetes Mellitus. Meta Gene 21, 100588. doi:10.1016/j.mgene.2019.100588
Gulixiati·Maimaitituersun, (2020). “IGF-1Gene Polymorphism and Hyperuricemia in Xinjiang Uyghur Population and Type 2 Diabetes,” in A Dissertation Submitted to Xinjiang Medical University in Partial Fullfillment of the Requirements for the Degree of Master of Public Health. Xinjiang, China: Xinjiang Medical University. (in Chinese).
Han, Z., Wang, T., Tian, R., Zhou, W., Wang, P., Ren, P., et al. (2019). BIN1 Rs744373 Variant Shows Different Association with Alzheimer's Disease in Caucasian and Asian Populations. BMC bioinformatics 20, 691. doi:10.1186/s12859-019-3264-9
He, D., Ma, L., Feng, R., Zhang, L., Jiang, Y., Zhang, Y., et al. (2015). Analyzing Large-Scale Samples Highlights Significant Association between Rs10411210 Polymorphism and Colorectal Cancer. Biomed. Pharmacother. 74, 164–168. doi:10.1016/j.biopha.2015.08.023
Hu, C., Zhang, R., Wang, C., Wang, J., Ma, X., Hou, X., et al. (2010). Variants from GIPR, TCF7L2, DGKB, MADD, CRY2, GLIS3, PROX1, SLC30A8 and IGF1 Are Associated with Glucose Metabolism in the Chinese. PLoS One 5, e15542. doi:10.1371/journal.pone.0015542
Johnson, A. M. F., and Olefsky, J. M. (2013). The Origins and Drivers of Insulin Resistance. Cell 152, 673–684. doi:10.1016/j.cell.2013.01.041
Langberg, K. A., Ma, L., Ma, L., Sharma, N. K., Hanis, C. L., Elbein, S. C., et al. (2012). Single Nucleotide Polymorphisms in JAZF1 and BCL11A Gene Are Nominally Associated with Type 2 Diabetes in African-American Families from the GENNID Study. J. Hum. Genet. 57, 57–61. doi:10.1038/jhg.2011.133
Li, L., Yang, Y., Yang, G., Lu, C., Yang, M., Liu, H., et al. (2011). The Role of JAZF1 on Lipid Metabolism and Related Genes In Vitro. Metabolism 60, 523–530. doi:10.1016/j.metabol.2010.04.021
Li, N., Wang, H., Yan, Z., Yao, X., Hong, J., and Zhou, L. (2012). Ethnic Disparities in the Clustering of Risk Factors for Cardiovascular Disease Among the Kazakh, Uygur, Mongolian and Han Populations of Xinjiang: a Cross-Sectional Study. BMC Public Health 12, 499. doi:10.1186/1471-2458-12-499
Li, W., Huang, Y., Liu, H., Zhang, S., Wang, L., Li, N., et al. (2019). Significance of SNPs from Previous Genome-wide Association Study in Prediction of Postpartum Diabetes Among Pregnant Women with Gestational Diabetes Mellitus. Chin. J. Public Health 35, 6. (in Chinese). doi:10.11847/zgggws1117960
Li, X., Yang, M., Wang, H., Jia, Y., Yan, P., Boden, G., et al. (2014). Overexpression of JAZF1 Protected ApoE-Deficient Mice from Atherosclerosis by Inhibiting Hepatic Cholesterol Synthesis via CREB-dependent Mechanisms. Int. J. Cardiol. 177, 100–110. doi:10.1016/j.ijcard.2014.09.007
Li, Y., He, S., Li, C., Shen, K., Yang, M., Tao, W., et al. (2021). Evidence of Association between Single-Nucleotide Polymorphisms in Lipid Metabolism-Related Genes and Type 2 Diabetes Mellitus in a Chinese Population. Int. J. Med. Sci. 18, 356–363. doi:10.7150/ijms.53004
Li, Y., Song, D., Jiang, Y., Wang, J., Feng, R., Zhang, L., et al. (2016). CR1 Rs3818361 Polymorphism Contributes to Alzheimer's Disease Susceptibility in Chinese Population. Mol. Neurobiol. 53, 4054–4059. doi:10.1007/s12035-015-9343-7
Liao, Z. Z., Wang, Y. D., Qi, X. Y., and Xiao, X. H. (2019). JAZF1, a Relevant Metabolic Regulator in Type 2 Diabetes. Diabetes Metab. Res. Rev. 35, e3148. doi:10.1002/dmrr.3148
Liu, C., Li, H., Qi, L., Loos, R. J. F., Qi, Q., Lu, L., et al. (2012). Variants in GLIS3 and CRY2 Are Associated with Type 2 Diabetes and Impaired Fasting Glucose in Chinese Hans. PLoS One 6, e21464. doi:10.1371/journal.pone.0021464
Liu, G., Xu, Y., Jiang, Y., Zhang, L., Feng, R., and Jiang, Q. (2017). PICALM Rs3851179 Variant Confers Susceptibility to Alzheimer's Disease in Chinese Population. Mol. Neurobiol. 54, 3131–3136. doi:10.1007/s12035-016-9886-2
Mamet, R., Jacobson, C. K., and Heaton, T. B. (2005). Ethnic Intermarriage in Beijing and Xinjiang, China, 1990. J. Comp. Fam. Stud. 36, 187–204. doi:10.3138/jcfs.36.2.187
Mannino, G. C., Greco, A., De Lorenzo, C., Andreozzi, F., Marini, M. A., Perticone, F., et al. (2013). A Fasting Insulin-Raising Allele at IGF1 Locus Is Associated with Circulating Levels of IGF-1 and Insulin Sensitivity. PLoS One 8, e85483. doi:10.1371/journal.pone.0085483
Ming, G.-f., Xiao, D., Gong, W.-j., Liu, H.-x., Liu, J., Zhou, H.-h., et al. (2014). JAZF1 Can Regulate the Expression of Lipid Metabolic Genes and Inhibit Lipid Accumulation in Adipocytes. Biochem. Biophysical Res. Commun. 445, 673–680. doi:10.1016/j.bbrc.2014.02.088
Perry, R. J., Samuel, V. T., Petersen, K. F., and Shulman, G. I. (2014). The Role of Hepatic Lipids in Hepatic Insulin Resistance and Type 2 Diabetes. Nature 510, 84–91. doi:10.1038/nature13478
Qin, L., Zhao, J., Wu, Y., Zhao, Y., Chen, C., Xu, M., et al. (2010). Association between Insulin-like Growth Factor 1 Gene Rs35767 Polymorphisms and Cancer Risk. Medicine (Baltimore) 98, e18017. doi:10.1097/MD.0000000000018017
Regué, L., Minichiello, L., Avruch, J., and Dai, N. (2019). Liver-specific Deletion of IGF2 mRNA Binding protein-2/IMP2 Reduces Hepatic Fatty Acid Oxidation and Increases Hepatic Triglyceride Accumulation. J. Biol. Chem. 294, 11944–11951. doi:10.1074/jbc.RA119.008778
Ruchat, S.-M., Elks, C. E., Loos, R. J. F., Vohl, M.-C., Weisnagel, S. J., Rankinen, T., et al. (2009). Association between Insulin Secretion, Insulin Sensitivity and Type 2 Diabetes Susceptibility Variants Identified in Genome-wide Association Studies. Acta Diabetol. 46, 217–226. doi:10.1007/s00592-008-0080-5
Seppä, S., Voutilainen, R., and Tenhola, S. (2015). Markers of Insulin Sensitivity in 12-Year-Old Children Born from Preeclamptic Pregnancies. J. Pediatr. 167, 125–130. doi:10.1016/j.jpeds.2015.04.015
Shankar, A., and Li, J. (2013). Positive Association between High-Sensitivity C-Reactive Protein Level and Diabetes Mellitus Among US Non-hispanic Black Adults. Exp. Clin. Endocrinol. Diabetes 116, 455–460. doi:10.1055/s-2007-1004563
Shen, N., Chen, B., Jiang, Y., Feng, R., Liao, M., Zhang, L., et al. (2015). An Updated Analysis with 85,939 Samples Confirms the Association between CR1 Rs6656401 Polymorphism and Alzheimer's Disease. Mol. Neurobiol. 51, 1017–1023. doi:10.1007/s12035-014-8761-2
Song, M., Zhao, F., Ran, L., Dolikun, M., Wu, L., Ge, S., et al. (2015). The Uyghur Population and Genetic Susceptibility to Type 2 Diabetes: Potential Role for Variants inCDKAL1,JAZF1, andIGF1Genes. OMICS: A J. Integr. Biol. 19, 230–237. doi:10.1089/omi.2014.0162
Telgmann, R., Dördelmann, C., Brand, E., Nicaud, V., Hagedorn, C., Pavenstädt, H., et al. (2009). Molecular Genetic Analysis of a Human Insulin‐like Growth Factor 1 Promoter P1 Variation. FASEB j. 23, 1303–1313. doi:10.1096/fj.08-116863
Thankamony, A., Capalbo, D., Marcovecchio, M. L., Sleigh, A., Jørgensen, S. W., Hill, N. R., et al. (2014). Low Circulating Levels of IGF-1 in Healthy Adults Are Associated with Reduced β-Cell Function, Increased Intramyocellular Lipid, and Enhanced Fat Utilization during Fasting. J. Clin. Endocrinol. Metab. 99, 2198–2207. doi:10.1210/jc.2013-4542
Wang, l. (2019). Relation between SNPs in IGF1 Promoter Region and Colorectal Cancer Risk in Type 2 Diabetes Patients. MS Dissertation. Tianjin: Tianjin Medical University. (in Chinese).
Wang, W., Wise, C., Baric, T., Black, M. L., and Bittles, A. H. (2003). The Origins and Genetic Structure of Three Co-resident Chinese Muslim Populations: the Salar, Bo'an and Dongxiang. Hum. Genet. 113, 244–252. doi:10.1007/s00439-003-0948-y
Wei, Q., Zhou, B., Yang, G., Hu, W., Zhang, L., Liu, R., et al. (2018). JAZF1 Ameliorates Age and Diet-Associated Hepatic Steatosis through SREBP-1c -dependent Mechanism. Cell Death Dis 9 (9), 859. doi:10.1038/s41419-018-0923-0
Yuan, L., Luo, X., Zeng, M., Zhang, Y., Yang, M., Zhang, L., et al. (2015). Transcription Factor TIP27 Regulates Glucose Homeostasis and Insulin Sensitivity in a PI3-kinase/Akt-dependent Manner in Mice. Int. J. Obes. 39, 949–958. doi:10.1038/ijo.2015.5
Zhai, F., He, Y., Wang, Z., and Hu, Y. (2007). [Status and Characteristic of Dietary Intake of 12 minority Nationalities in China]. Wei Sheng Yan Jiu 36, 539–541. doi:10.3969/j.issn.1000-8020.2007.05.004 (in Chinese).
Zhang, J., Chen, X., Zhang, L., and Peng, Y. (2017). IGF1 Gene Polymorphisms Associated with Diabetic Retinopathy Risk in Chinese Han Population. Oncotarget 8, 88034–88042. doi:10.18632/oncotarget.21366
Zhang, S., Li, X., Ma, G., Jiang, Y., Liao, M., Feng, R., et al. (2016). CLU Rs9331888 Polymorphism Contributes to Alzheimer's Disease Susceptibility in Caucasian but Not East Asian Populations. Mol. Neurobiol. 53, 1446–1451. doi:10.1007/s00702-014-1260
Keywords: type 2 diabete mellitus, insulin-like growth factor-1, rs35767, susceptibility, meta-analysis
Citation: Zeng Q, Zou D, Zeng Q, Chen X, Wei Y and Guo R (2021) Association Between Insulin-like Growth Factor-1 rs35767 Polymorphism and Type 2 Diabetes Mellitus Susceptibility: A Meta-Analysis. Front. Genet. 12:774489. doi: 10.3389/fgene.2021.774489
Received: 12 September 2021; Accepted: 25 October 2021;
Published: 22 November 2021.
Edited by:
Liangcai Zhang, Janssen Research and Development, United StatesReviewed by:
Tonia Carter, Marshfield Clinic, United StatesYanfang Chen, Wright State University, United States
Copyright © 2021 Zeng, Zou, Zeng, Chen, Wei and Guo. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiaoming Chen, Y2hlbnhpYW9taW5nQGdkbXUuZWR1LmNu; Yue Wei, d2VpeXVlMTM4QDE2My5jb20=; Runmin Guo, cnVubWluLmd1b0BnZG11LmVkdS5jbg==
†These authors have contributed equally to this work
 Qiaoli Zeng
Qiaoli Zeng Dehua Zou
Dehua Zou Qiaodi Zeng5†
Qiaodi Zeng5† Runmin Guo
Runmin Guo