
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Genet. , 13 December 2021
Sec. Livestock Genomics
Volume 12 - 2021 | https://doi.org/10.3389/fgene.2021.772557
This article is part of the Research Topic Omics Technologies in Livestock Improvement: From Selection to Breeding Decisions View all 30 articles
 Chun Huang1
Chun Huang1 Fei Ge1
Fei Ge1 Xiaoming Ma1
Xiaoming Ma1 Rongfeng Dai1
Rongfeng Dai1 Renqing Dingkao2
Renqing Dingkao2 Zhuoma Zhaxi3
Zhuoma Zhaxi3 Getu Burenchao3
Getu Burenchao3 Pengjia Bao1
Pengjia Bao1 Xiaoyun Wu1
Xiaoyun Wu1 Xian Guo1
Xian Guo1 Min Chu1
Min Chu1 Ping Yan1*
Ping Yan1* Chunnian Liang1*
Chunnian Liang1*Cattle-yak, as the hybrid offspring of cattle (Bos taurus) and yak (Bos grunniens), demonstrates obvious heterosis in production performance. Male hybrid sterility has been focused on for a long time; however, the mRNAs and non-coding RNAs related to muscle development as well as their regulatory networks remain unclear. The phenotypic data showed that the production performance (i.e., body weight, withers height, body length, and chest girth) of cattle-yak was significantly better than that of the yak, and the economic benefits of the cattle-yak were higher under the same feeding conditions. Then, we detected the expression profiles of the longissimus dorsi muscle of cattle-yak and yak to systematically reveal the molecular basis using the high-throughput sequencing technology. Here, 7,126 mRNAs, 791 lncRNAs, and 1,057 circRNAs were identified to be differentially expressed between cattle-yaks and yaks in the longissimus dorsi muscle. These mRNAs, lncRNA targeted genes, and circRNA host genes were significantly enriched in myoblast differentiation and some signaling pathways related to muscle development (such as HIF-1 signaling pathway and PI3K-Akt signaling pathway). We constructed a competing endogenous RNA (ceRNA) network and found that some non-coding RNAs differentially expressed may be involved in the regulation of muscle traits. Taken together, this study may be used as a reference tool to provide the molecular basis for studying muscle development.
Yak, a special germplasm resource mainly inhabiting the Qinghai-Tibet Plateau, has been optimized for living and a source of living for the local herdsmen. The cattle-yak constitutes the hybrid of cattle (Bos taurus) and yak (Bos grunniens) exhibiting outstanding hybrid vigor in growth rate, meat performance, plateau adaptability, etc. The meat of cattle-yak is highly enriched in protein but has lower fat than yak, meeting the requirements of a popular healthy and high-quality diet (Song et al., 2019; Wang et al., 2021b). Inhabiting the high-altitude environment, cattle-yak provides a natural green food favored by the consumers. The crossbreeding technology has been widely applied in animal breeding, as well as these hybrid individuals have been farmed for a long time forming gradually new indigenous breeds. Therefore, it is highly significant to understand the mechanisms regulating muscle growth and development of the generated crossbreed. The longissimus dorsi (LD) muscle is one of the important representatives of muscle tissues in animals, which is closely related to the individual skeletal muscle growth and development, as well as with the intramuscular fat content, and tenderness. A series of reported myogenic regulatory molecules regulate the myoblast’s proliferation and myophagism, further influencing the growth process, including the identified genes MYOG (Rudnicki and Jaenisch, 1995) and MYF5 (Xu et al., 2019), which are involved in regulating the myoblast differentiation, muscle growth, and meat quality traits in livestock. Previous studies have indicated that MYOD as a key regulator of myotube formation promotes the myotube’s differentiation (Tapscott, 2005; Wang et al., 2017). The CAPN family are important candidate genes in the growth and degradation of the muscle fibers, and are specifically expressed in the skeletal muscle (Gandolfi et al., 2011).
In fact, the studies indicated that the biological processes were not only regulated by protein-coding RNA—mRNA, as the sequencing technology developed. Non-coding RNAs (ncRNAs), including long non-coding RNA (lncRNA), circular RNA (circRNA), and microRNA (miRNA), are profoundly involved in diverse biological processes and are regulating them by various mechanisms. The lncRNAs universally acknowledged participating in chromatin transcriptional/epigenetic regulation by interacting with the chromatin regulators as “molecular scaffold” or decoys to activate or repress transcription (Caretti et al., 2006; Korostowski et al., 2012). Many lncRNAs have been proven to play a vital role in skeletal muscle development; for example, the lncRNA MAR1 positively correlates with muscle differentiation and growth in vitro and in vivo (Zhang et al., 2018). The lnc-smad7/miR-125b/Smad7 (SMAD family member 7) and IGF2 axes are instrumental in myoblast differentiation and regeneration of muscle in two different pig breeds (Song et al., 2018). In addition, continuously growing discoveries have reported the role of novel circRNA in skeletal muscle. CircZfp609 derived from Zinc Finger Protein 609, can inhibit the myogenic differentiation via the sponge miR-194-5p in the mouse myoblast cell line C2C12 (Wang et al., 2019b). The circFGFR4 was generated from the fibroblast growth factor receptor 4 (FGFR4) and could simulate the bovine primary myoblast’s differentiation through the circFGFR4-miR-107-WNT3A axis in cattle (Li et al., 2018). MicroRNA response elements are considered to be “talking mediators” of mRNAs, lncRNAs, and transcribed pseudogenes (Ji et al., 2019), and these response elements have important roles in various biological processes by forming a large number of complex regulatory networks. Numerous reports on the conjoint effect of miRNA and mRNA conjointly on developing skeletal muscle have been proved. FGFR1, which could prevent muscle fibrogenesis, is a functional target of miR-214-3p (Arrighi et al., 2021). The miR-183 and miR-96 were found to negatively regulate fat usage in the skeletal muscle via targeting FoxO1 and PDK4 (Wang et al., 2021a). Zhang et al. (2021) experimentally confirmed that miR-22-3p regulated the WFIKKN2 gene in adipocyte differentiation in muscle fat metabolism of Yanbian cattle. A targeted relationship between the oar-miR-655-3p and oar-miR-381-5p with ACSM3 and ABAT has been found to have crucial roles in sheep muscle organogenesis, myoblast migration (Sun et al., 2019).
In recent years, with the deepening of the research on the function of miRNAs, a new theory named competing endogenous RNA (ceRNA) has emerged. At the same time, some studies reported that mRNAs, lncRNAs, and circRNAs might regulate the gene function via miRNA and act as ceRNAs in various biological processes (Salmena et al., 2011; Yu et al., 2019). In the whole transcriptome, a comprehensive post-transcriptional regulatory network formed by ceRNA activity has greatly widened the cognition of functional genetic information in the genome. There are effective interactions among the lncRNA, circRNA, and mRNA with miRNA, and they can take significant effect in various processes of regulation in animals. LncRNAs act as molecular sponges for the miRNAs that specifically inhibit the target mRNAs so that they can give play to the protection of mRNAs (Li et al., 2019). For instance, a previous study reported that MAML1 and MEF2C, as transcription factors, activate the late-differentiation muscle genes, and linc-MD1 can regulate their expression as a ceRNA by sponging miR-133 and miR-135 (Cesana et al., 2011). LncRNA H19 can regulate muscle differentiation as a molecular sponge for the LET7 family in the developing embryo and adult muscles (Kallen et al., 2013).
Until now, most studies have been based on focusing on the cattle-yak for exploring the male sterility mechanism and barely referred to the superiority in the growth mechanism. Here, we have measured the growth traits of cattle-yaks and domestic Ashidan yaks under the same feeding and management, and systematically explored the differences of the longissimus dorsi muscles for the first time using the whole-transcriptome sequencing. Furthermore, the ceRNA network was constructed to identify the key factors involved in muscle growth and development. This study will thus help in improving yak breeding and provide new ideas for studying the genetic mechanism of muscle growth.
All the animal experiments were approved by Lanzhou Institute of Husbandry and Pharmaceutical Sciences of the Chinese Academy of Agricultural Sciences (CAAS) with the grant number: No. 2019-002. All the slaughter as well as sampling procedures strictly complied with the Guidelines on the Ethical Treatment of Experimental Animals of China.
Thirty cattle-yaks (Aberdeen Angus ♂ × Yak ♀) and 30 yaks were tracked to measure the production performance indices (withers height, body weight, chest girth, and body length) at three stages of growth (i.e., birth, 3 months, and 6 months). Cattle-yaks (n = 3, 6 months old) and yaks (n = 3, 6 months old) were selected randomly to be slaughtered for longissimus dorsi muscle. These samples were collected for transcriptome sequencing and Real-time quantitative polymerase chain reaction analysis (RT-qPCR). All the samples were stored in liquid nitrogen (−80°C) for the subsequent tests.
The total RNA was isolated with TRIzol (Invitrogen, Carlsbad, CA, United States) following the manufacturer’s instructions, and the concentration and quality of RNA were assessed by 1.5% agarose gel electrophoresis and Thermo Scientific NanoDrop 2000c (ThermoFisher Scientific Inc., Waltham, MA, United States).
Equal quantities of RNA were pooled from each sample. Then, the TruSeq Stranded Total RNA with Ribo-Zero Gold Kit (Illumina, San Diego, CA, United States) was used for digesting the ribosomal RNA (rRNA) in the DNA-free RNA. According to the manufacturer’s instructions, we performed the construction of library preparation with NEB Next Ultra Directional RNA LibraryPrep Kit for Illumina (NEB, Ipswich, MA, United States). The size and purity of libraries were validated by Agilent Technologies 2100 Bioanalyzer (Agilent, Santa Clara, CA). Finally, the samples were sequenced using Illumina HiSeq 2500 Technology (LC Sciences, Houston, TX, United States) with a 150-bp paired-end run.
Raw reads generated during high-throughput sequencing were in fastq format. The raw sequencing dataset supporting the results of cattle-yaks in this study was deposited at NCBI’s Gene Expression Omnibus database (http://www.ncbi.nlm.nih.gov/geo/, accession number: PRJNA753699). The raw sequencing dataset of yaks has been uploaded to the NCBI Database in the previous study (Ma et al., 2020). To get high-quality clean reads that could be used for later analysis, the Trimmomatic software (Bolger et al., 2014) was applied to remove adaptors, low-quality bases, and N-bases.
The remaining high-quality cleaned reads of each sample were aligned to the yak reference genome (BosGru_v2.0) using the HiSAT2 software (Kim et al., 2015). All the samples were assessed by genomic and gene alignment. After obtaining the comparison result bam file, the reads on the comparison gene were assembled using the StringTie software (Pertea et al., 2015), and every single transcript assembled by each sample was fused and spliced into a merged transcript.
We performed the following steps to obtain the potential lncRNA candidates for subsequent analysis: (1) The Cuffcompare software (Ghosh and Chan, 2016) was used to compare the merged transcripts with the reference transcripts one by one, and the transcripts marked with “i,” “u,” “x,” and “o” were retained after clarifying the position class of the remaining transcripts. (2) Transcripts with length >200 nt and exon number ≥2 were obtained. (3) The above transcripts were analyzed about the coding ability by CPC2 (Kang et al., 2017), CNCI (Sun et al., 2013), PLEK (Li et al., 2014), and Pfam (Sonnhammer et al., 1998) software to remove the transcripts with coding potential and obtain potential lncRNA candidates.
Cis-acting and trans-acting modes are two main ways to predict the targets of lncRNAs (Mercer et al., 2009). We calculated the locations of the paired lncRNAs and mRNAs for the cis-acting prediction. The lncRNA with no nearest protein-coding gene within 100 kb upstream or downstream was excluded in subsequent analysis. For trans-acting regulatory mode, the LncTar was used to calculate the free energy between them to predict the regulatory targets, since the expression of lncRNA is determined to be independent of the location of mRNA.
We used the CIRI software and predicted the circRNA based on the BWA software (Li and Durbin, 2009; Gao et al., 2015). Since it is an authoritative software, it has the characteristics of high sensitivity and multiple screening for reducing false positives. Firstly, we aligned the clean reads to the reference genome to obtain the SAM file using the BWA software. Then, the CIRI software was used to scan for PCC signals (paired chiastic clipping signals), and circRNA sequences were predicted based on junction reads and GT-AG cleavage signals. The expressional levels of circRNAs were quantified by the RPM algorithm.
The expression levels of the mRNAs and lncRNAs were calculated through the fragments per kilobase of transcript per million reads mapped (FPKM) value (Liu et al., 2018) using the Cuffdiff program. The DESeq software (Anders and Huber, 2010) was used to standardize the counts of each sample and calculate the fold change (FC). A negative binomial distribution test (NB) was used to test the difference significance of counts. The differentially expressed mRNAs (DEMs), lncRNAs (DELs), and circRNAs (DECs) were screened according to the results of |log 2 FC| >1 and p < 0.05 eventually.
To further understand gene function, Gene Ontology (GO, http://www.geneontology.org) terms and the Kyoto Encyclopedia of Genes and Genomes (KEGG, https://www.genome.jp/kegg/) were used for enrichment analysis of functional pathways. Each GO and KEGG enrichment term was confirmed by Hypergeometric Distribution Test. Then, the p-value was corrected by Benjamini and Hochberg multiple tests. The enrichment with p-value lower than 0.05 was considered significant.
To acquire a better understanding of the interactions of the mRNAs, lncRNAs, circRNAs, and miRNAs, a lncRNA–circRNA–miRNA–mRNA regulatory network was constructed based on the ceRNA hypothesis (Salmena et al., 2011). MiRanda (John et al., 2004) was used to predict the pairs of miRNA–lncRNA, miRNA–mRNA, and miRNA–circRNA. The Spearman correlation coefficient (SCC) was used to evaluate the pairwise correlations of miRNA–lncRNA, miRNA–mRNA, and miRNA–circRNA; the value greater than 0.8 was considered relevant for constructing the network and p < 0.05 was regarded as being statistically significant. The Cytoscape software (version 3.5.1) was used to display the results visually.
The RNA for verifying the gene expression was the same as RNA used in the above Illumina sequencing. The RNA was reverse transcribed into cDNA by HiScript® II 1st Strand cDNA Synthesis Kit (Vazyme, Nanjing, Jiangsu, China). Glyceraldehyde-3-phosphate dehydrogenase (GAPDH), as endogenous control, was used to normalize target gene expression. The total 20-μl reaction system of qPCR included 10 ng of cDNA, 10 μl of SYBR Premix Ex Taq II (TaKaRa, Dalian, China), each 1 μl of forward primer and reverse primer, and 7 μl of ddH2O. The RT-qPCR conditions included the preincubation at 95°C for the 30 s, 45 cycles of 10 s at 95°C and 60 s at 59°C, then ended at 72°C for 30 s. Each experiment was repeated three times, and the results of relative RNA expression were calculated according to the cycle threshold (Ct) value 2–ΔΔCt (Livak and Schmittgen, 2001).
Analysis of the measurement results showed that the cattle-yaks had a significant improvement in production performance compared to the yaks in each age stage (p < 0.001). As can be seen from Table 1, cattle-yaks were stronger and taller at birth than yaks. In terms of body weight, at birth, the cattle-yak increased by about 22.60% compared to the yaks, and the body length, height, and chest girth of cattle-yaks were also increased by 17.62%, 11.06%, and 8.92%, respectively. From birth to 6 months old, the cattle-yak showed obvious heterosis with varying degrees of improvement in various indicators.
After filtering out the low-quality and supernumerary reads, we obtained a total of 310,089,232 and 308,742,564 clean reads with greater than 93.77% of Q30 from the LD muscles of cattle-yaks and yaks, respectively. As shown in Supplementary Table S1, the successful alignment of reads to the reference genome was approximately over 94.55%.
After rigorous screening and filtration, we detected a large number of lncRNAs and cirRNAs (Figures 1A,E). Among them, 1,817 lncRNAs with an average length of 1,449 bp were discovered as novel lncRNAs (Figure 1B). The largest proportion of lncRNAs was over 2,000 bp (19.37%) and the proportion of lncRNA containing two exons was about 70.61% (Figure 1C). The total lncRNAs of 135 exonic antisense, 219 intronic antisense, 105 intergenic downstream antisense, 202 intergenic upstream, 204 exonic sense, 380 intronic sense, 141 intergenic downstream sense and 140 intergenic up-stream sense were identified in our results (Figure 1D). The average length of detected circRNAs was 3,250 bp and the length of the most circRNAs was over 2,000 bp (16.92%) (Figure 1F). As evident in Figure 1E, the circRNAs of sense-overlapping accounts for 89% of the total. The number of circRNAs located in the exonic and intronic was 332 and 137, respectively, while the antisense-overlapping circRNAs occupied about 1%.
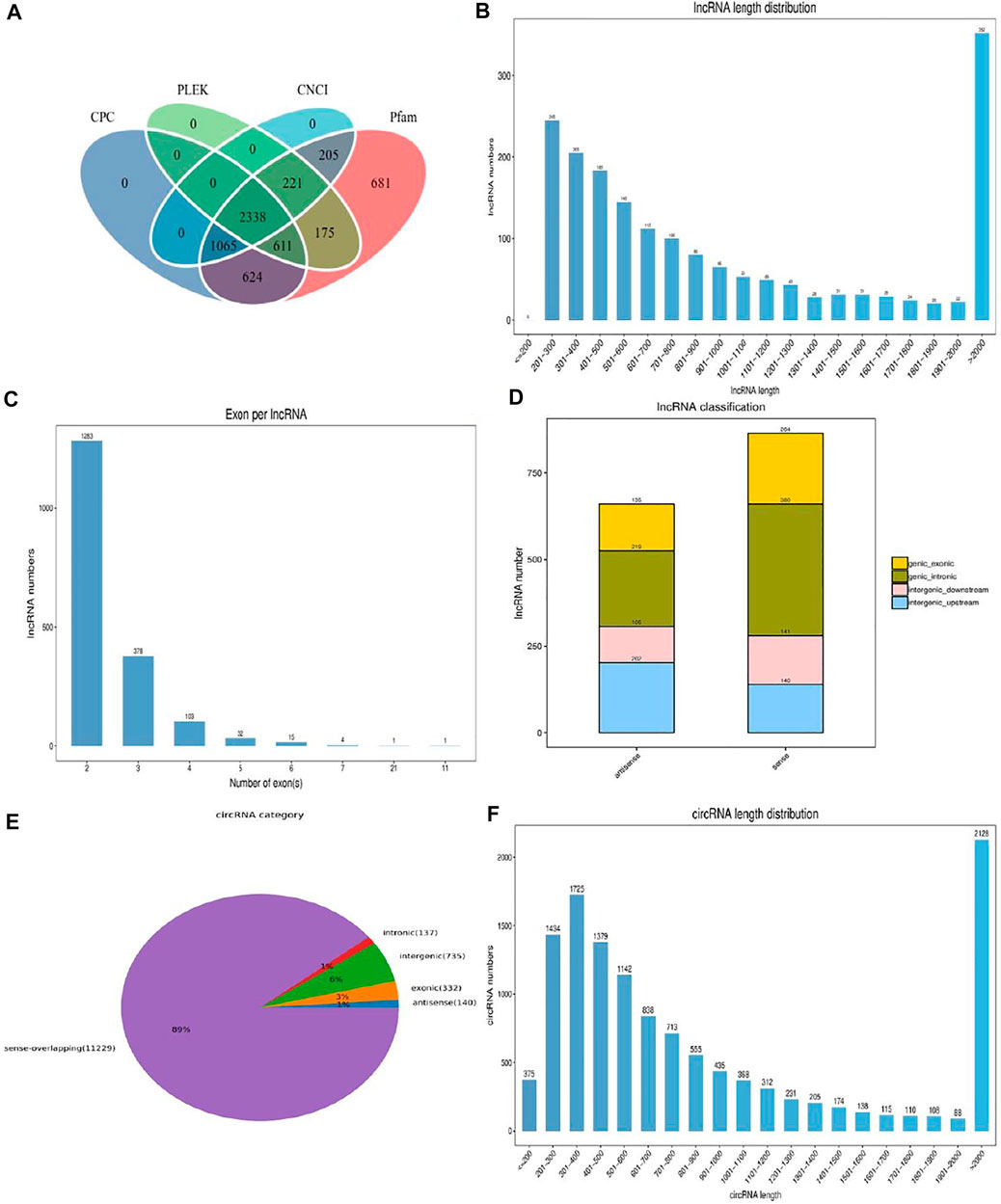
FIGURE 1. The description of identified lncRNAs and circRNAs. (A) Screening of the candidate lncRNAs in longissimus dorsi muscle. (B) The length distribution of the novel lncRNAs. (C) Exon number distribution of novel lncRNAs. (D) The classification of novel lncRNAs. (E) The structure type pie chart of circRNAs. (F) The length distribution of circRNAs.
The study identified 7,126 mRNAs, 791 lncRNAs, and 1,057 circRNAs to have significant differential expressions (Supplementary Tables S2–S4). There were 6902 DE mRNAs, 742 DE lncRNAs, and 273 DE circRNAs, respectively, which were detected in the cattle-yaks and yaks (Figures 2A–C). However, 119 mRNAs, 41 lncRNAs, and 232 circRNAs were detected to express only in the CY group. Other 105 mRNAs, 8 lncRNAs, and 552 circRNAs were identified only in the Y group. To find the overall distribution of differential expression, the volcano plot was drawn based on the results of the differential expression. Compared with the yaks of the control group, we found 3,563 upregulated mRNAs, 455 upregulated lncRNAs, and 353 upregulated circRNAs in the cattle-yaks. Meanwhile, there were 3,563 downregulated mRNAs, 336 downregulated lncRNAs, and 704 downregulated circRNAs, respectively, in the cattle-yaks (Figures 2D–F).
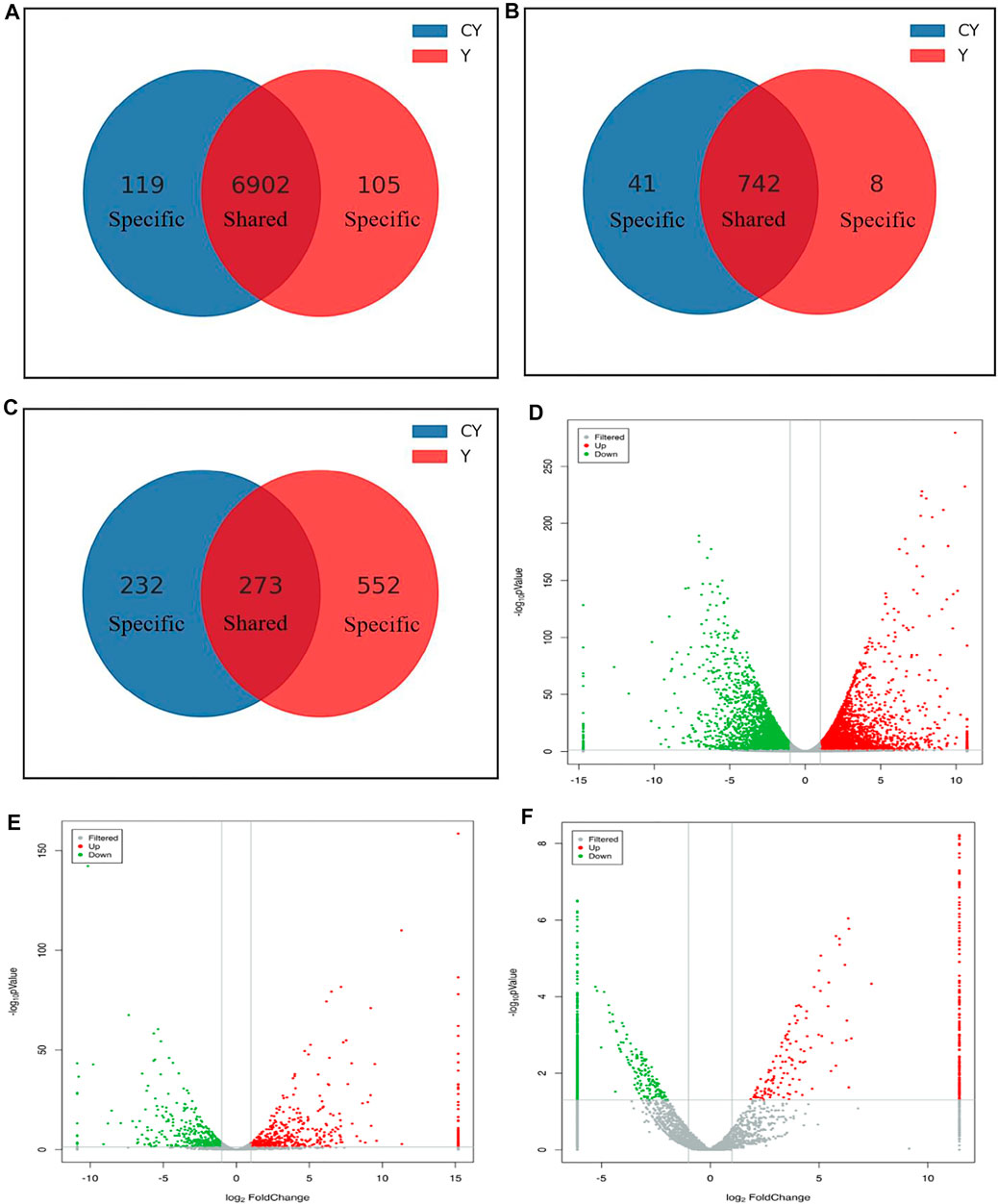
FIGURE 2. Comparative analysis of the differentially expressed mRNAs and ncRNAs between cattle-yaks and yaks. The specific and shared mRNAs (A), lncRNAs (B), and circRNAs (C) between the two groups. The analysis of differentially expressed mRNAs (D), lncRNAs (E), and circRNAs (F) between two groups. Note: The red and green points represented upregulated and downregulated mRNAs, lncRNAs, and circRNAs, respectively. The gray points represented no significant differences. The gray vertical lines showed |log2FC| = 1, and the gray horizontal lines showed p = 0.05.
GO analysis described the molecular functions performed by the DE ncRNAs, the cellular environment in which they were located, and the biological processes involved. We respectively selected the top 10 biological processes with the most significant enrichment of mRNAs, lncRNAs, and circRNAs (Figure 3A). Obviously, the DEMs were mainly enriched in regulation of osteoblast proliferation, negative regulation of fat cell proliferation, and regulation of cellular response to hypoxia. The targeted genes of DELs were most enriched in terms involving regulation of fat cell differentiation, ephrin receptor signaling pathway, and muscle contraction. Protein autophosphorylation, fatty acid catabolic process, and regulation of Rho protein signal transduction were the most significant enrichment terms for the sourced genes for DECs. As evident from Figures 3B–D, we conducted the KEGG analysis using KEGG public pathway database and draw the augmented scatter diagram of the selected target genes. Between cattle-yaks and yaks, the DEMs were enriched in the PI3K−Akt signaling pathway, MAPK signaling pathway, Fatty acid metabolism, Citrate cycle, etc. (Figure 3B). The DEL adjacent genes were significantly related to the protein digestion and absorption, GnRH signaling pathway, alanine, aspartate and glutamate metabolism, and so on (Figure 3C). The host genes of DECs were significantly associated with some pathways, such as those related to vitamin digestion, ABC transporters, cGMP−PKG signaling pathway, etc. (Figure 3D). Interestingly, some DEMs and the host genes of DECs were found to be enriched in the same pathways (i.e., hippo signaling pathway, cell cycle-caulobacter, and calcium signaling pathway), and some DEM and DEL adjacent genes were enriched in the same oxytocin signaling pathways.
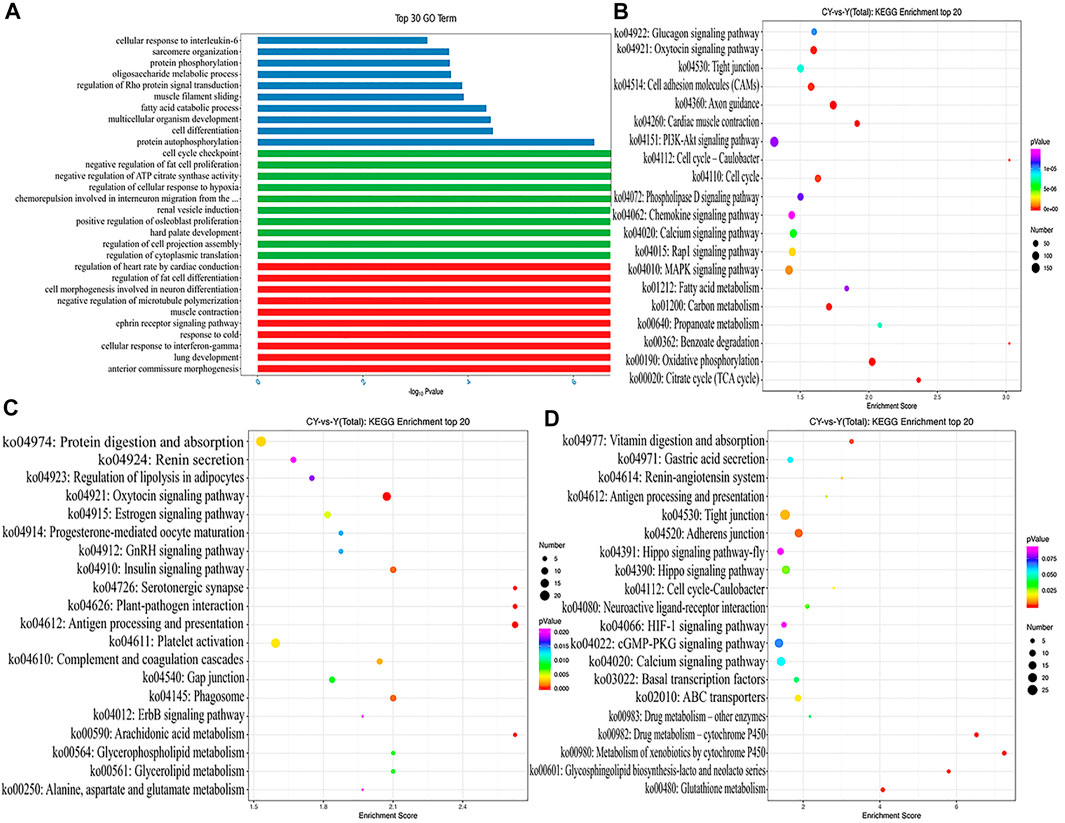
FIGURE 3. (A) GO analysis with the top 10 enrichment biological processes for DEMs, DELs targets, and DECs host genes between cattle-yaks and yaks. KEGG analysis with the top 20 KEGG enrichment pathways for DEMs (B), DELs target genes (C), and DECs host genes (D) between the cattle-yaks and yaks.
In order to further explore the DEMs related to muscle growth and fatness, we screened out the related genes. These 117 DEMs related to muscle development and fat deposition are shown in Supplementary Table S5. The functional predictions of GO in DEMs (Figure 4A) mainly focused on some terms of skeletal muscle development, muscle cell differentiation, and regulation of canonical Wnt signaling pathway, including positive regulation of myoblast differentiation and skeletal muscle fiber development. According to the KEGG pathway analysis (Figure 4B), some pathways were enriched significantly by these DEMs, such as Hippo signaling pathway, regulating many biological processes involved in proliferation, survival and differentiation of cell, organ size, and tissue homeostasis. In addition, some DEMs enriched significantly in Notch signaling pathway that can regulate the differentiation and development of cells, tissues, and organs through the interaction between the adjacent cells.
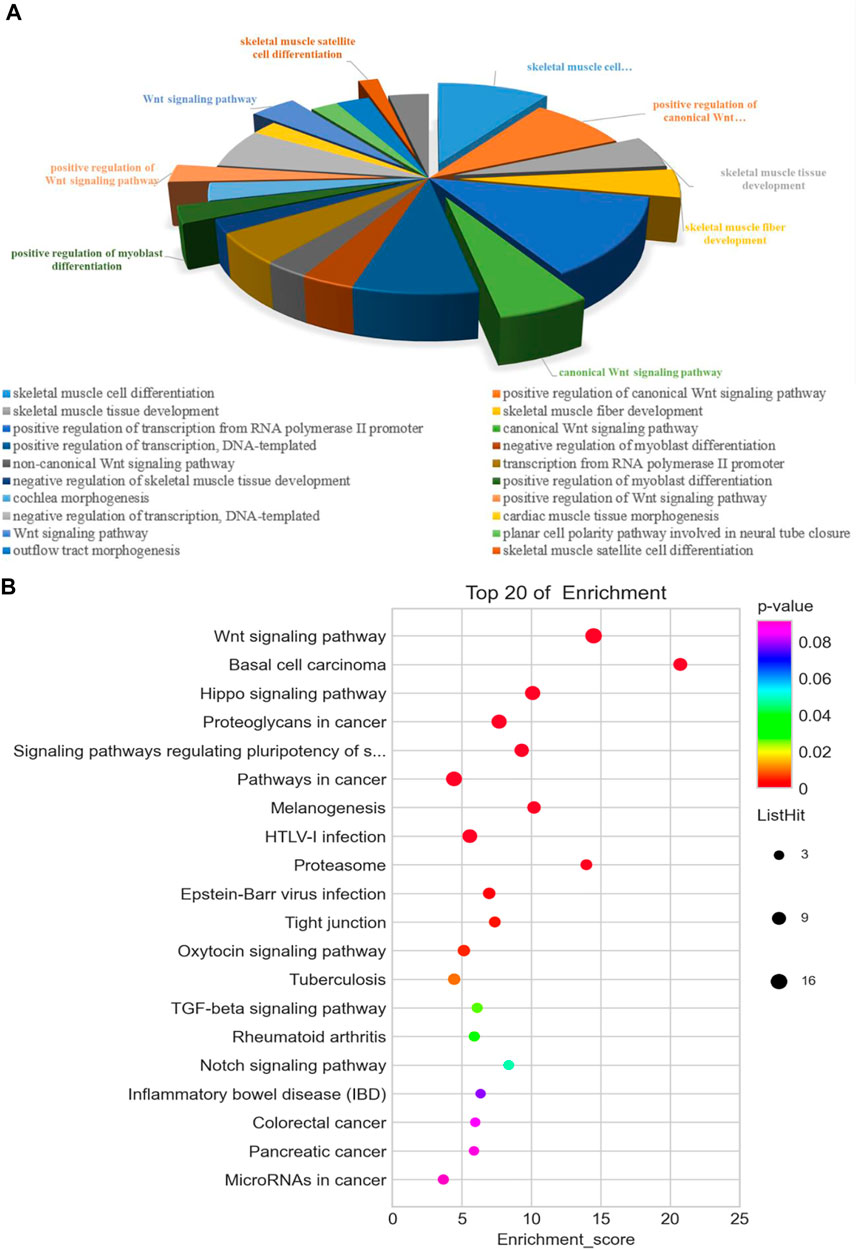
FIGURE 4. Functional analysis of 117 differentially expressed genes associated with muscle development and fatness between the cattle-yak and yak. GO (A) and KEGG (B) analysis.
Previous studies have shown that mRNAs, lncRNAs, and circRNAs may regulate gene function through miRNAs as ceRNAs in different processes (Salmena et al., 2011; Yu et al., 2019), indicating that ceRNAs and their miRNAs may work in concert with each other. Combined with the DEMs, DELs, and DECs related to muscle development and co-differentially expressed, we constructed the integrated ceRNA network. This ceRNA network contained 11 DEMs, 6 DELs, 8 DECs, and 33 relationships (Figure 5). Tcons-00034903 and bta-miR-2039 have a shared target gene WNT4. Similarly, we also found the same results in bta-miR-2316-Tcons-00029868-MYH4 and bta-miR-1777a-Tcons-00027748-SIX5. This ceRNA network might provide valuable information for the development of the longissimus dorsi in cattle-yaks and yaks.
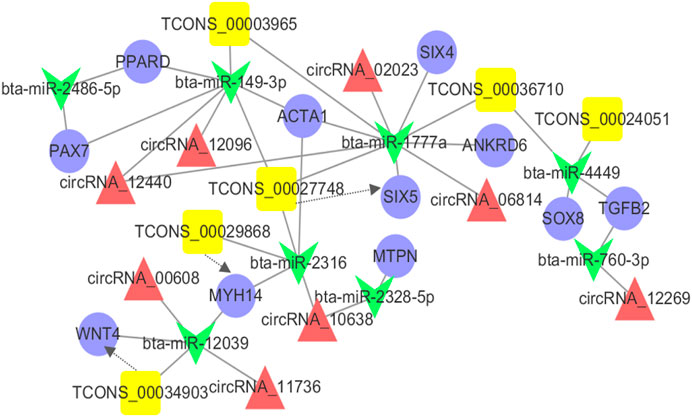
FIGURE 5. The ceRNA co-regulation network. The blue circle, yellow box, red triangle and green V-type represented the differentially expressed mRNAs, lncRNAs, circRNAs, and miRNA, respectively. The solid line indicated the co-regulation between miRNAs and other transcripts. The dotted line indicated the co-regulation between the lncRNAs and mRNAs.
Four mRNAs (MYH14, LOC106700760, PIK3R2, and FGFR4), 2 lncRNAs (Tcons-00034903 and Tcons-00004303), and 2 circRNAs (circ00012096 and circ00012564) were selected randomly for verifying the sequencing results through real-time quantitative PCR (Figure 6). The primers were designed using the Primer-BLAST web tool from the National Center for Biotechnology Information (NCBI) (Supplementary Table S6). Their expression patterns were highly consistent with the sequencing results, indicating that the gene expression profiles obtained in this study had high repeatability and reliability.
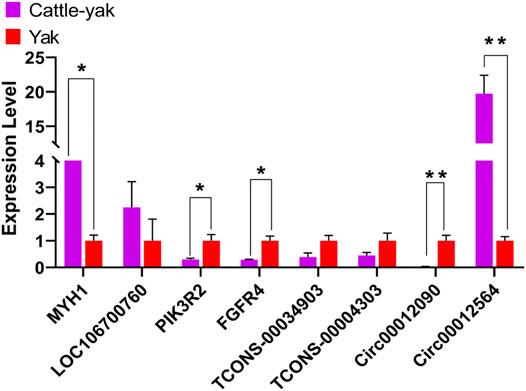
FIGURE 6. The results of the real-time quantitative PCR validation of the expression level. ** indicates p < 0.01, * indicates p < 0.05. The data represented the mean ± SEM from three biological replicates, and each measurement was repeated 3 times.
Muscle growth is a complex economic trait owing to various physiological and biochemical indices. It concomitantly involves many gene expressions and regulations. To date, people have a great demand for high-quality meat and nutrition with improving living standards; it is an urgent problem for improving the performance of yaks. Cattle-yak, as hybrid offspring of yak (♀) and cattle (♂), has been found to have significant improvement in the production performance (Guo et al., 2019; Dingkao et al., 2020; Luo et al., 2020; Jiang et al., 2021). However, the economic benefit of cattle-yaks has been ignored for a long time. The variations in the genes and proteins in the muscle structure are presumed generally to be affected by production performance (Joo et al., 1999); this study is the first time to systematically explore the differences of growth and development between cattle-yaks and yaks based on transcriptomics. To study the regulatory mechanism of growth and muscle development of cattle-yaks and yaks in a better way, we performed transcriptome analysis of the longissimus dorsi muscle. It was also the first time to compare the differences in the expression profiles of mRNA, lncRNA, and circRNA between the cattle-yaks and yaks, so as to determine the key factors involved in muscle growth and development. We measured the phenotypic data strictly of 30 cattle-yaks and yaks at three age groups of growth using the standard method of measurement, and the results obtained were similar to the previous studies; in each period of cattle-yaks with the same feeding conditions, production performance indexes of cattle-yaks were significantly higher than those of yaks (p < 0.001). We speculated that these differences may be due to genetic factors rather than the effects of feeding management on these traits. Therefore, the study on the mechanism regulating muscle growth and development in hybrid yaks and yak breeds can better help in enhancing the production performance of the yaks, providing help for exploring the regulatory mechanism of muscle development in mammals.
Typically, the growth and development of muscles are regulated by the core genes and signal transduction pathways (Myers et al., 2006; Ayuso et al., 2015). Compared to the yak group, a total of 7,126 DEMs, 791 DELs, and 1,057 DECs were identified in the cattle-yak group. Subsequently, the GO and KEGG pathway enrichment analysis revealed some important DEMs related to muscle growth and fat deposition, which was consistent with the results obtained after measuring the production performance, indicating that the production performance of the cattle-yak was better than that of the yak (Tumennasan et al., 1997). In addition, some DEMs, DELs, and DECs related to the immune system have reflected the adaptation of the yak to the high-altitude environment, and the adaptation of the yaks to the cold and high-altitude environment was well-preserved by the cattle-yaks. In succession, we focused on some DEMs related to the production performance of cattle-yaks and yaks. A total of 117 DEMs were identified that were related to the differentiation and proliferation of myoblasts, AMPK signaling pathway, MAPK signaling pathway, PI3K–Akt signaling pathway, etc. (Neri et al., 2002; Anderson, 2006; Gehart et al., 2010; Thomson, 2018). Among these DEMs, some have been found to have known functions in muscle growth and development. For example, myostatin (MSTN), as a member of the TGF-β superfamily, has a proven role as a growth differentiation factor (Kollias and McDermott, 2008; Li et al., 2008) playing an important role in mice (McPherron et al., 1997), cattle (McPherron and Lee, 1997), and humans (Schuelke et al., 2004) by negatively regulating the growth and differentiation of myoblasts, as well as other mammals via controlling both the activation and proliferation of the satellite cells (skeletal muscle stem cells) (McCroskery et al., 2003). The expression level of the MSTN gene in our study was found to be downregulated in the cattle-yaks; hence, the expression of this gene was considered to be consistent with its function. This also provided new evidence for the function of the MSTN gene in longissimus dorsi of cattle-yaks and the high conservation across species. Myogenin (MYOG), including myogenic factor 6 (MYF6), is a regulatory factor in the family of Myogenic regulatory factors (MRFs), which is mainly involved in the fusion and differentiation of myoblasts (CHARGÉ and RUDNICKI, 2004; Buckingham and Vincent, 2009). Interestingly, the paired box transcription factor 7 (PAX7) gene and the MYOG gene were found to be downregulated, but the MYF6 expression was upregulated in the cattle-yaks. Previous studies have shown that overexpression of gene PAX7 can induce MYOG expression to inhibit myogenesis and prevent the differentiation of the muscle cell (Chen et al., 2010; Dey et al., 2011). Therefore, this also justified the downregulation of both PAX7 and MYOG gene expression in cattle-yaks with better production performance than yaks. Additionally, some genes, such as actin alpha 1 skeletal muscle (ACTA1) and actin alpha cardiac muscle 1 (ACTC1), also play a key role in the differentiation and fusion of muscle cells, and have a positive impact on the myogenesis of the skeletal muscles (Ciecierska et al., 2020). Although these studies have helped in predicting the functions and accuracy of the key genes, the other functional DEMs related to muscle growth and development still need to be further studied on their expression regulation in longissimus dorsi muscle.
The main non-coding RNAs, the lncRNAs and circRNAs, are receiving more and more attention, as they can participate in the regulation of various biological processes in different ways (Liu et al., 2017; Marchese et al., 2017; Wang et al., 2019a). The identified DELs and DECs in this study were involved in regulating the promotion of muscle growth and development. LncRNA can exert its important action through various biological and pathological processes by demonstrating trans, cis, and antisense effects. Our results showed abundant differentially expressed lncRNAs in the skeletal muscle of cattle-yak and yak, suggesting that the lncRNAs may not be the exclusive by-products of mRNA in the cattle-yak but have specific roles. The long non-coding RNA acts differently in the nucleus and cytoplasm due to their different locations, where they are involved in the muscle development regulation on both embryonic and growing stages. According to the KEGG analysis, the differentially expressed lncRNAs and circRNAs were identified to functionally relate to some hormone regulation, myoblast proliferation, and metabolic pathways. CircRNAs being another type of non-coding RNA also act as an important regulatory role in skeletal muscle growth and development (Greco et al., 2018). Multiple studies have confirmed that abundant circRNAs exist in the skeletal muscle and their expression levels change dynamically during the process of myoblast differentiation (Legnini et al., 2017). Notably, we found that many DECs host genes are enriched in the HIF-1 signaling pathway in the KEGG analysis. HIF-1 signaling pathway is the core signaling pathway induced by hypoxia involved in regulating the proliferation and differentiation of myoblasts upon hypoxic conditions (Ogilvie et al., 2000). HIF-1 pathway can upregulate its target genes, some of which involve the regulation of proliferation and differentiation of myoblasts. These DEC host genes were found to be significantly enriched in the HIF-1 signaling pathway, indicating a close relationship with the regulation of muscle growth and development under high-altitude and hypoxic environments. The regulatory mechanism of these circRNAs and their host genes on muscle development hence deserves further studies. Furthermore, our studies also indicated some lncRNAs and circRNAs to be specifically or mainly expressed in the longissimus dorsi muscle of cattle-yaks (e.g., Tcons-00005361, Tcons-00037918, and circRNA-02529), indicating that these ncRNAs were generated on purpose to have specific effects in the muscle development of the cattle-yaks.
In recent years, extensive studies on the function of miRNAs have provided a new theory named competing for endogenous RNA (ceRNA). To understand the process of development of the skeletal muscle, a ceRNA regulatory network was constructed based on the combination with the DEMs, DELs, and DECs related to muscle development and were expressed co-differentially. The results showed that 11 DEMs, 6 DELs, and 8 DECs cross-talked with another through 8 differential expressions of the microRNAs. This also indicated that the development of the longissimus dorsi muscle in cattle-yak was a complex regulation process of a balanced level of gene expression under a high-altitude and hypoxic environment. As reported, peroxisome proliferator-activated receptor delta (PPARD) acted as a vital regulator in adipogenesis and lipid metabolism (Kim et al., 2006; Chen and Yang, 2014). Ankyrin repeat domain 6 (ANKRD6) also played a role in regulating crucial events in developing vertebrates and invertebrates (Schwarz-Romond et al., 2002; Moeller et al., 2006). Therefore, we speculated that these ncRNAs might also contribute to muscle development by indirectly regulating the gene expression of PPARD and ANKRD6. Not only that, we observed three important ceRNA subnetworks from the ceRNA network, showing that TCONS-00024051 and its target SOX8 “talked” to each other through the same bta-miR-1777a response element, while TCONS-00034903 and its target WNT4, TCONS-00029868 and its target ACTA1 “talked” to each other through bta-miR-12039 and bta-miR-4449 response elements, respectively. Therefore, we speculated that these three subnetworks may be crucially associated and function in regulating muscle development. Notably, as a member of the “unconventional” non-muscle myosin II family of molecular motors, myosin heavy chain 14 (MYH14) gene has been identified as a key regulator of muscle fiber type (van Rooij et al., 2009; Bell et al., 2010). In our results, MYH14 and Tcons-00029868 were both upregulated in longissimus dorsi muscle, which indicates that this lncRNA may have a cis-regulatory relationship with MYH14 gene. From our results, these DEMs, DELs, and DECs were not only involved in the process of muscle development, but might also be involved in lipogenesis as ceRNAs.
In conclusion, we compared the production performance of cattle-yaks and yaks, it was also the first time to compare the expressional features of mRNAs, lncRNAs, and circRNAs in the longissimus dorsi tissue of cattle-yaks and yaks. In our results, the abundant mRNAs and ncRNAs were identified, some of which were specifically expressed in cattle-yaks. According to the bioinformatics analyses, the ncRNAs were found to be not only connected with the myoblast differentiation and proliferation, skeletal development, and signaling pathway of muscle growth, but be also useful as ceRNAs of important transcription factors (such as SOX8 and PPARD). In addition, the ceRNA network (11 DEMs, 6 DELs, and 8 DECs) that we constructed may have the considerable effects on regulation of muscle growth. This study provided new insights into the genetic basis of muscle growth and laid the foundation for further study of the role of these ncRNAs in regulating muscle growth.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/, PRJNA753699.
The animal study was reviewed and approved by the Lanzhou Institute of Husbandry and Pharmaceutical Sciences Chinese Academy of Agricultural Sciences. Written informed consent was obtained from the owners for the participation of their animals in this study.
CL, PY, and CH contributed to conception and design of the study. CH and FG organized the database. FG and XM performed the statistical analysis. CH, FG, and XM wrote the first draft of the manuscript. RFD, RQD, ZZ, and GB collected the production data. XG, PB, XM, XW, and MC wrote sections of the manuscript. All authors contributed to manuscript revision, read, and approved the submitted version.
Work presented in this paper was supported by the Project of Innovative Research on Yak Molecular Breeding Technology under the Innovation Population of Basic Research in Gansu Province, grant number 20JR5RA580, the Agricultural Science and Technology Innovation Program, grant number CAAS-ASTIP-2014-LIHPS-01, the National Beef Cattle Industry Technology and System, grant number CARS-37, and the Herbivorous Livestock Industry and Technical System of Gansu Province, grant number GARS-08.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors, and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2021.772557/full#supplementary-material
Anders, S., and Huber, W. (2010). Differential Expression Analysis for Sequence Count Data. Genome Biol. 11 (10), R106. doi:10.1186/gb-2010-11-10-r106
Anderson, D. H. (2006). Role of Lipids in the MAPK Signaling Pathway. Prog. Lipid Res. 45 (2), 102–119. doi:10.1016/j.plipres.2005.12.003
Arrighi, N., Moratal, C., Savary, G., Fassy, J., Nottet, N., Pons, N., et al. (2021). The FibromiR miR-214-3p Is Upregulated in Duchenne Muscular Dystrophy and Promotes Differentiation of Human Fibro-Adipogenic Muscle Progenitors. Cells 10 (7), 1832. doi:10.3390/cells10071832
Ayuso, M., Fernández, A., Núñez, Y., Benítez, R., Isabel, B., Barragán, C., et al. (2015). Comparative Analysis of Muscle Transcriptome between Pig Genotypes Identifies Genes and Regulatory Mechanisms Associated to Growth, Fatness and Metabolism. PLoS One 10 (12), e0145162. doi:10.1371/journal.pone.0145162
Bell, M. L., Buvoli, M., and Leinwand, L. A. (2010). Uncoupling of Expression of an Intronic microRNA and its Myosin Host Gene by Exon Skipping. Mol. Cel Biol. 30 (8), 1937–1945. doi:10.1128/mcb.01370-09
Bolger, A. M., Lohse, M., and Usadel, B. (2014). Trimmomatic: a Flexible Trimmer for Illumina Sequence Data. Bioinformatics 30 (15), 2114–2120. doi:10.1093/bioinformatics/btu170
Buckingham, M., and Vincent, S. D. (2009). Distinct and Dynamic Myogenic Populations in the Vertebrate Embryo. Curr. Opin. Genet. Dev. 19 (5), 444–453. doi:10.1016/j.gde.2009.08.001
Caretti, G., Schiltz, R. L., Dilworth, F. J., Di Padova, M., Zhao, P., Ogryzko, V., et al. (2006). The RNA Helicases P68/p72 and the Noncoding RNA SRA Are Coregulators of MyoD and Skeletal Muscle Differentiation. Dev. Cel 11 (4), 547–560. doi:10.1016/j.devcel.2006.08.003
Cesana, M., Cacchiarelli, D., Legnini, I., Santini, T., Sthandier, O., Chinappi, M., et al. (2011). A Long Noncoding RNA Controls Muscle Differentiation by Functioning as a Competing Endogenous RNA. Cell 147 (2), 358–369. doi:10.1016/j.cell.2011.09.028
Chargé, S. B. P., and Rudnicki, M. A. (2004). Cellular and Molecular Regulation of Muscle Regeneration. Physiol. Rev. 84 (1), 209–238. doi:10.1152/physrev.00019.2003
Chen, J.-F., Tao, Y., Li, J., Deng, Z., Yan, Z., Xiao, X., et al. (2010). microRNA-1 and microRNA-206 Regulate Skeletal Muscle Satellite Cell Proliferation and Differentiation by Repressing Pax7. J. Cel Biol. 190 (5), 867–879. doi:10.1083/jcb.200911036
Chen, L., and Yang, G. (2014). PPARs Integrate the Mammalian Clock and Energy Metabolism. PPAR Res. 2014, 1–6. doi:10.1155/2014/653017
Ciecierska, A., Motyl, T., and Sadkowski, T. (2020). Transcriptomic Profile of Primary Culture of Skeletal Muscle Cells Isolated from Semitendinosus Muscle of Beef and Dairy Bulls. Ijms 21 (13), 4794. doi:10.3390/ijms21134794
Dey, B. K., Gagan, J., and Dutta, A. (2011). miR-206 and -486 Induce Myoblast Differentiation by Downregulating Pax7. Mol. Cel Biol. 31(1), 203–214. doi:10.1128/MCB.01009-10
Dingkao, R. Q., Shi, H. M., Li, P. X., Cairang, N. R., Ma, D. L., Niu, X. L., et al. (2020). Application of Early Pregnancy Diagnostic Technique in Xianan Cattle Production. China Cattle Sci. 46 (5), 11–14.
Gandolfi, G., Pomponio, L., Ertbjerg, P., Karlsson, A. H., Nanni Costa, L., Lametsch, R., et al. (2011). Investigation on CAST, CAPN1 and CAPN3 Porcine Gene Polymorphisms and Expression in Relation to post-mortem Calpain Activity in Muscle and Meat Quality. Meat Sci. 88 (4), 694–700. doi:10.1016/j.meatsci.2011.02.031
Gao, Y., Wang, J., and Zhao, F. (2015). CIRI: an Efficient and Unbiased Algorithm for De Novo Circular RNA Identification. Genome Biol. 16 (1), 4. doi:10.1186/s13059-014-0571-3
Gehart, H., Kumpf, S., Ittner, A., and Ricci, R. (2010). MAPK Signalling in Cellular Metabolism: Stress or Wellness? EMBO Rep. 11 (11), 834–840. doi:10.1038/embor.2010.160
Ghosh, S., and Chan, C.-K. K. (2016). “Analysis of RNA-Seq Data Using TopHat and Cufflinks,” in Plant Bioinformatics: Methods and Protocols. Editor D. Edwards (New York, NY: Springer New York), 339–361. doi:10.1007/978-1-4939-3167-5_18
Greco, S., Cardinali, B., Falcone, G., and Martelli, F. (2018). Circular RNAs in Muscle Function and Disease. Ijms 19 (11), 3454. doi:10.3390/ijms19113454
Guo, S., Ma, D., Li, B., Bao, Z., Zhang, T., and Xu, G. (2019). Determination of Growth and Development Indexes of Cattle-Yak in Gannan alpine Pasture Area. China Herbivore Sci. 039 (002), 73–75. doi:10.3969/j.issn.2095-3887.2019.02.021
Ji, Q., Cai, G., Liu, X., Zhang, Y., Wang, Y., Zhou, L., et al. (2019). MALAT1 Regulates the Transcriptional and Translational Levels of Proto-Oncogene RUNX2 in Colorectal Cancer Metastasis. Cell Death Dis. 10 (6), 378. doi:10.1038/s41419-019-1598-x
Jiang, H., Zhang, C. F., Zhang, Q., Sun, G. M., Cao, H. W., CiJi, Y. D., et al. (2021). Study on Yak Economic Crossbreeding and Growth Performance of F1 Generation. Tibet J. Agric. Sci. 43 (02), 22–24.
John, B., Enright, A. J., Aravin, A., Tuschl, T., Sander, C., and Marks, D. S. (2004). Human MicroRNA Targets. Plos Biol. 2 (11), e363. doi:10.1371/journal.pbio.0020363
Joo, S. T., Kauffman, R. G., Kim, B. C., and Park, G. B. (1999). The Relationship of Sarcoplasmic and Myofibrillar Protein Solubility to Colour and Water-Holding Capacity in Porcine Longissimus Muscle. Meat Sci. 52 (3), 291–297. doi:10.1016/S0309-1740(99)00005-4
Kallen, A. N., Zhou, X.-B., Xu, J., Qiao, C., Ma, J., Yan, L., et al. (2013). The Imprinted H19 lncRNA Antagonizes Let-7 microRNAs. Mol. Cel 52 (1), 101–112. doi:10.1016/j.molcel.2013.08.027
Kang, Y.-J., Yang, D.-C., Kong, L., Hou, M., Meng, Y.-Q., Wei, L., et al. (2017). CPC2: a Fast and Accurate Coding Potential Calculator Based on Sequence Intrinsic Features. Nucleic Acids Res. 45 (W1), W12–W16. doi:10.1093/nar/gkx428
Kim, D. J., Bility, M. T., Billin, A. N., Willson, T. M., Gonzalez, F. J., and Peters, J. M. (2006). Pparβ/δ Selectively Induces Differentiation and Inhibits Cell Proliferation. Cell Death Differ 13 (1), 53–60. doi:10.1038/sj.cdd.4401713
Kim, D., Langmead, B., and Salzberg, S. L. (2015). HISAT: a Fast Spliced Aligner with Low Memory Requirements. Nat. Methods 12 (4), 357–360. doi:10.1038/nmeth.3317
Kollias, H. D., and McDermott, J. C. (2008). Transforming Growth Factor-β and Myostatin Signaling in Skeletal Muscle. J. Appl. Physiol. 104 (3), 579–587. doi:10.1152/japplphysiol.01091.2007
Korostowski, L., Sedlak, N., and Engel, N. (2012). The Kcnq1ot1 Long Non-coding RNA Affects Chromatin Conformation and Expression of Kcnq1, but Does Not Regulate its Imprinting in the Developing Heart. Plos Genet. 8 (9), e1002956. doi:10.1371/journal.pgen.1002956
Legnini, I., Di Timoteo, G., Rossi, F., Morlando, M., Briganti, F., Sthandier, O., et al. (2017). Circ-ZNF609 Is a Circular RNA that Can Be Translated and Functions in Myogenesis. Mol. Cel 66 (1), 22–37. e29. doi:10.1016/j.molcel.2017.02.017
Li, A., Zhang, J., and Zhou, Z. (2014). PLEK: a Tool for Predicting Long Non-coding RNAs and Messenger RNAs Based on an Improved K-Mer Scheme. BMC Bioinformatics 15 (1), 311. doi:10.1186/1471-2105-15-311
Li, H., and Durbin, R. (2009). Fast and Accurate Short Read Alignment with Burrows-Wheeler Transform. Bioinformatics 25 (14), 1754–1760. doi:10.1093/bioinformatics/btp324
Li, H., Wei, X., Yang, J., Dong, D., Hao, D., Huang, Y., et al. (2018). circFGFR4 Promotes Differentiation of Myoblasts via Binding miR-107 to Relieve its Inhibition of Wnt3a. Mol. Ther. - Nucleic Acids 11, 272–283. doi:10.1016/j.omtn.2018.02.012
Li, Z. B., Kollias, H. D., and Wagner, K. R. (2008). Myostatin Directly Regulates Skeletal Muscle Fibrosis. J. Biol. Chem. 283 (28), 19371–19378. doi:10.1074/jbc.M802585200
Li, Z., Cai, B., Abdalla, B. A., Zhu, X., Zheng, M., Han, P., et al. (2019). LncIRS1 Controls Muscle Atrophy via Sponging miR‐15 Family to Activate IGF1‐PI3K/AKT Pathway. J. Cachexia, Sarcopenia Muscle 10 (2), 391–410. doi:10.1002/jcsm.12374
Liu, J., Liu, T., Wang, X., and He, A. (2017). Circles Reshaping the RNA World: from Waste to Treasure. Mol. Cancer 16 (1), 58. doi:10.1186/s12943-017-0630-y
Liu, X., Liu, K., Shan, B., Wei, S., Li, D., Han, H., et al. (2018). A Genome-wide Landscape of mRNAs, lncRNAs, and circRNAs during Subcutaneous Adipogenesis in Pigs. J. Anim. Sci Biotechnol 9 (1), 76. doi:10.1186/s40104-018-0292-7
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2−ΔΔCT Method. Methods 25 (4), 402–408. doi:10.1006/meth.2001.1262
Luo, Q. B., Buren, C. G. T., Si, Q., Zhaxi, Z. M., Zhang, S., Chang, L., et al. (2020). Study on Crossbreeding Effect between Angus Beef Cattle and Yak. China Cattle Sci. 46 (5), 8–10.
Ma, X., Fu, D., Chu, M., Ding, X., Wu, X., Guo, X., et al. (2020). Genome-Wide Analysis Reveals Changes in Polled Yak Long Non-coding RNAs in Skeletal Muscle Development. Front. Genet. 11 (365). doi:10.3389/fgene.2020.00365
Marchese, F. P., Raimondi, I., and Huarte, M. (2017). The Multidimensional Mechanisms of Long Noncoding RNA Function. Genome Biol. 18 (1), 206. doi:10.1186/s13059-017-1348-2
McCroskery, S., Thomas, M., Maxwell, L., Sharma, M., and Kambadur, R. (2003). Myostatin Negatively Regulates Satellite Cell Activation and Self-Renewal. J. Cel Biol. 162 (6), 1135–1147. doi:10.1083/jcb.200207056
McPherron, A. C., Lawler, A. M., and Lee, S.-J. (1997). Regulation of Skeletal Muscle Mass in Mice by a New TGF-P Superfamily Member. Nature 387 (6628), 83–90. doi:10.1038/387083a0
McPherron, A. C., and Lee, S.-J. (1997). Double Muscling in Cattle Due to Mutations in the Myostatin Gene. Proc. Natl. Acad. Sci. 94 (23), 12457–12461. doi:10.1073/pnas.94.23.12457
Mercer, T. R., Dinger, M. E., and Mattick, J. S. (2009). Long Non-coding RNAs: Insights into Functions. Nat. Rev. Genet. 10 (3), 155–159. doi:10.1038/nrg2521
Moeller, H., Jenny, A., Schaeffer, H.-J., Schwarz-Romond, T., Mlodzik, M., Hammerschmidt, M., et al. (2006). Diversin Regulates Heart Formation and Gastrulation Movements in Development. Proc. Natl. Acad. Sci. 103 (43), 15900–15905. doi:10.1073/pnas.0603808103
Myers, S. A., Wang, S.-C. M., and Muscat, G. E. O. (2006). The Chicken Ovalbumin Upstream Promoter-Transcription Factors Modulate Genes and Pathways Involved in Skeletal Muscle Cell Metabolism. J. Biol. Chem. 281 (34), 24149–24160. doi:10.1074/jbc.M601941200
Neri, L. M., Borgatti, P., Capitani, S., and Martelli, A. M. (2002). The Nuclear Phosphoinositide 3-kinase/AKT Pathway: a New Second Messenger System. Biochim. Biophys. Acta 1584 (2), 73–80. doi:10.1016/S1388-1981(02)00300-1
Ogilvie, M., Yu, X., Nicolas-Metral, V., Pulido, S. M., Liu, C., Ruegg, U. T., et al. (2000). Erythropoietin Stimulates Proliferation and Interferes with Differentiation of Myoblasts. J. Biol. Chem. 275 (50), 39754–39761. doi:10.1074/jbc.M004999200
Pertea, M., Pertea, G. M., Antonescu, C. M., Chang, T.-C., Mendell, J. T., and Salzberg, S. L. (2015). StringTie Enables Improved Reconstruction of a Transcriptome from RNA-Seq Reads. Nat. Biotechnol. 33 (3), 290–295. doi:10.1038/nbt.3122
Rudnicki, M. A., and Jaenisch, R. (1995). The MyoD Family of Transcription Factors and Skeletal Myogenesis. Bioessays 17 (3), 203–209. doi:10.1002/bies.950170306
Salmena, L., Poliseno, L., Tay, Y., Kats, L., and Pandolfi, P. P. (2011). A ceRNA Hypothesis: The Rosetta Stone of a Hidden RNA Language? Cell 146 (3), 353–358. doi:10.1016/j.cell.2011.07.014
Schuelke, M., Wagner, K. R., Stolz, L. E., Hübner, C., Riebel, T., Kömen, W., et al. (2004). Myostatin Mutation Associated with Gross Muscle Hypertrophy in a Child. N. Engl. J. Med. 350 (26), 2682–2688. doi:10.1056/NEJMoa040933
Schwarz-Romond, T., Asbrand, C., Bakkers, J., Kühl, M., Schaeffer, H.-J., Huelsken, J., et al. (2002). The Ankyrin Repeat Protein Diversin Recruits Casein Kinase Iε to the β-catenin Degradation Complex and Acts in Both Canonical Wnt and Wnt/JNK Signaling. Genes Dev. 16 (16), 2073–2084. doi:10.1101/gad.230402
Song, C., Huang, Y., Yang, Z., Ma, Y., Chaogetu, B., Zhuoma, Z., et al. (2019). RNA-seq Analysis Identifies Differentially Expressed Genes in Subcutaneous Adipose Tissue in Qaidaford Cattle, Cattle-Yak, and Angus Cattle. Animals 9 (12), 1077. doi:10.3390/ani9121077
Song, C., Wang, J., Ma, Y., Yang, Z., Dong, D., Li, H., et al. (2018). Linc-smad7 Promotes Myoblast Differentiation and Muscle Regeneration via Sponging miR-125b. Epigenetics 13 (6), 591–604. doi:10.1080/15592294.2018.1481705
Sonnhammer, E., Eddy, S. R., Birney, E., Bateman, A., and Durbin, R. (1998). Pfam: Multiple Sequence Alignments and HMM-Profiles of Protein Domains. Nucleic Acids Res. 26 (1), 320–322. doi:10.1093/nar/26.1.320
Sun, L., Lu, S., Bai, M., Xiang, L., Li, J., Jia, C., et al. (2019). Integrative microRNA-mRNA Analysis of Muscle Tissues in Qianhua Mutton Merino and Small Tail Han Sheep Reveals Key Roles for Oar-miR-655-3p and Oar-miR-381-5p. DNA Cel Biol. 38 (5), 423–435. doi:10.1089/dna.2018.4408
Sun, L., Luo, H., Bu, D., Zhao, G., Yu, K., Zhang, C., et al. (2013). Utilizing Sequence Intrinsic Composition to Classify Protein-Coding and Long Non-coding Transcripts. Nucleic Acids Res. 41 (17), e166. doi:10.1093/nar/gkt646
Tapscott, S. J. (2005). The Circuitry of a Master Switch: Myod and the Regulation of Skeletal Muscle Gene Transcription. Development 132 (12), 2685–2695. doi:10.1242/dev.01874
Thomson, D. (2018). The Role of AMPK in the Regulation of Skeletal Muscle Size, Hypertrophy, and Regeneration. Ijms 19 (10), 3125. doi:10.3390/ijms19103125
Tumennasan, K., Tuya, T., Hotta, Y., Takase, H., Speed, R. M., and Chandley, A. C. (1997). Fertility Investigations in the F1 Hybrid and Backcross Progeny of Cattle (Bos taurus) Andyak (B. Grunniens) in Mongolia. Cytogenet. Cel Genet 78 (1), 69–73. doi:10.1159/000134633
van Rooij, E., Quiat, D., Johnson, B. A., Sutherland, L. B., Qi, X., Richardson, J. A., et al. (2009). A Family of microRNAs Encoded by Myosin Genes Governs Myosin Expression and Muscle Performance. Dev. Cel 17 (5), 662–673. doi:10.1016/j.devcel.2009.10.013
Wang, C., Liu, W., Nie, Y., Qaher, M., Horton, H. E., Yue, F., et al. (2017). Loss of MyoD Promotes Fate Transdifferentiation of Myoblasts into Brown Adipocytes. EBioMedicine 16, 212–223. doi:10.1016/j.ebiom.2017.01.015
Wang, H., Ma, M., Li, Y., Liu, J., Sun, C., Liu, S., et al. (2021a). miR‐183 and miR‐96 Orchestrate Both Glucose and Fat Utilization in Skeletal Muscle. EMBO Rep. 22, e52247. doi:10.15252/embr.202052247
Wang, J., Ren, Q., Hua, L., Chen, J., Zhang, J., Bai, H., et al. (2019a). Comprehensive Analysis of Differentially Expressed mRNA, lncRNA and circRNA and Their ceRNA Networks in the Longissimus Dorsi Muscle of Two Different Pig Breeds. Ijms 20 (5), 1107. doi:10.3390/ijms20051107
Wang, Y., Li, M., Wang, Y., Liu, J., Zhang, M., Fang, X., et al. (2019b). A Zfp609 Circular RNA Regulates Myoblast Differentiation by Sponging miR-194-5p. Int. J. Biol. Macromolecules 121, 1308–1313. doi:10.1016/j.ijbiomac.2018.09.039
Wang, Y., Wang, Z., Hu, R., Peng, Q., Xue, B., and Wang, L. (2021b). Comparison of Carcass Characteristics and Meat Quality between Simmental Crossbred Cattle, Cattle‐yaks and Xuanhan Yellow Cattle. J. Sci. Food Agric. 101 (9), 3927–3932. doi:10.1002/jsfa.11032
Xu, X., Mishra, B., Qin, N., Sun, X., Zhang, S., Yang, J., et al. (2019). Differential Transcriptome Analysis of Early Postnatal Developing Longissimus Dorsi Muscle from Two Pig Breeds Characterized in Divergent Myofiber Traits and Fatness. Anim. Biotechnol. 30 (1), 63–74. doi:10.1080/10495398.2018.1437045
Yu, C., Longfei, L., Long, W., Feng, Z., Chen, J., Chao, L., et al. (2019). LncRNA PVT1 Regulates VEGFC through Inhibiting miR‐128 in Bladder Cancer Cells. J. Cel Physiol. 234 (2), 1346–1353. doi:10.1002/jcp.26929
Zhang, J. S., Xu, H. Y., Fang, J. C., Yin, B. Z., Wang, B. B., Pang, Z., et al. (2021). Integrated microRNA-mRNA Analysis Reveals the Roles of microRNAs in the Muscle Fat Metabolism of Yanbian Cattle. Anim. Genet. 52, 598–607. doi:10.1111/age.13126
Keywords: cattle-yak, Bos grunniens, transcriptome, ceRNA, lncRNA, circRNA, skeletal muscle
Citation: Huang C, Ge F, Ma X, Dai R, Dingkao R, Zhaxi Z, Burenchao G, Bao P, Wu X, Guo X, Chu M, Yan P and Liang C (2021) Comprehensive Analysis of mRNA, lncRNA, circRNA, and miRNA Expression Profiles and Their ceRNA Networks in the Longissimus Dorsi Muscle of Cattle-Yak and Yak. Front. Genet. 12:772557. doi: 10.3389/fgene.2021.772557
Received: 08 September 2021; Accepted: 15 November 2021;
Published: 13 December 2021.
Edited by:
Marcos De Donato, Instituto de Tecnología y Educación Superior de Monterrey (ITESM), MexicoReviewed by:
Ran Di, Institute of Animal Sciences (CAAS), ChinaCopyright © 2021 Huang, Ge, Ma, Dai, Dingkao, Zhaxi, Burenchao, Bao, Wu, Guo, Chu, Yan and Liang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ping Yan, cGluZ3lhbmx6QDE2My5jb20=; Chunnian Liang, Y2h1bm5pYW4yMDA2QDE2My5jb20=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.