
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Genet. , 12 October 2021
Sec. Evolutionary and Population Genetics
Volume 12 - 2021 | https://doi.org/10.3389/fgene.2021.770853
This article is part of the Research Topic The Origination of Genetic Novelties: New Genes, New Regulations, and New Cell Types View all 8 articles
Notch signaling pathway plays crucial roles in animal development. Protein ubiquitination contributes to Notch signaling regulation by governing the stability and activity of major signaling components. Studies in Drosophila have identified multiple ubiquitin ligases and deubiquitinating enzymes that modify Notch ligand and receptor proteins. The fate of ubiquitinated substrates depend on topologies of the attached ubiquitin chains, which are determined by the ubiquitin conjugating enzymes (E2 enzymes). However, which E2 enzymes participate in Notch signal transduction remain elusive. Here, we report that the E2 enzyme UbcD1 is required for Notch signaling activation during Drosophila wing development. Mutations of UbcD1 lead to marginal nicks in the adult wing and reduction of Notch signaling targets expression in the wing imaginal disc. Genetic analysis reveal that UbcD1 functions in the signaling receiving cells prior to cleavage of the Notch protein. We provide further evidence suggesting that UbcD1 is likely involved in endocytic trafficking of Notch protein. Our results demonstrate that UbcD1 positively regulates Notch signaling and thus reveal a novel role of UbcD1 in development.
Notch signaling pathway plays crucial roles in developmental processes such as tissue patterning, cell proliferation and cell fate determination (Bray, 2016). Malfunction of Notch signaling results in various malignant diseases in human, including neuropsychiatric diseases, metabolic disorders and multiple types of cancer (Salazar and Yamamoto, 2018). The core components and signal transduction routes of Notch signaling are highly conserved among the animal kingdom (Fortini, 2009). Named after the wing margin nicking phenotype observed in the Drosophila mutant, the Notch gene encodes a transmembrane protein which functions as signal receptor (Bray, 2016). Binding of Notch with ligand proteins Delta or Serrate, which are presented at the membrane of signal sending cells leads to a series of proteolytic cleavage of the Notch protein (Fortini, 2009). As a consequence, the Notch intracellular domain (NICD) is released and translocates into nucleus in the signal receiving cells (Kopan and Ilagan, 2009). NICD interacts with the transcription factor Suppressor of Hairless [Su(H)] and the co-activator Mastermind (Mam) to form a ternary complex. The Su(H)/NICD/Mam complex recognizes specific cis-regulatory regions and activates transcription of Notch target genes. In the absence of signal input, Su(H) recruits co-repressors and inhibits the expression of Notch targets (Kopan and Ilagan, 2009).
The Notch signaling is tightly controlled by auxiliary factors that modulate the expression, stability and activity of the core components (Fortini, 2009). Recent studies have revealed that protein ubiquitination is extensively involved in the regulation of Notch signaling pathway (Le Bras et al., 2011; Weinmaster and Fischer 2011). Protein ubiquitination is a reversible post-translational modification catalyzed by four distinct enzymes. The E1 (Ub-activating) and E2 (Ub-conjugating) enzymes are responsible for activating and conjugating the ubiquitin (Ub) moiety, respectively. The E3 (Ub ligases) enzyme recognizes specific substrates and transfers Ub from E2 onto them. The deubiquitinating enzyme (DUB) removes Ub from substrate proteins to counteract the ubiquitination process (Grabbe et al., 2011). E2 enzymes are now considered as the main determinant for the topology of ubiquitin chains, which directs the ubiquitinated substrates towards distinct fates (Ye and Rape, 2009).
Multiple E3s and DUBs have been demonstrated to regulate Notch signaling during fly development (Moretti and Brou, 2013). In the signal sending cells, E3 ligases Neuralized (Neur) and Mind bomb (Mib1) promote mono-ubiquitination of the ligand proteins Delta and Serrate to facilitate their endocytosis (Yeh et al., 2000; Lai et al., 2001; Pavlopoulos et al., 2001; Itoh et al., 2003; Le Borgne and Schweisguth, 2003; Li and Baker, 2004; Lai et al., 2005). Ubiquitination and endocytosis of ligand proteins are required for initiation of signal transduction in various tissues (Yeh et al., 2001; Le Borgne et al., 2005; Pitsouli and Delidakis, 2005; Wang and Struhl, 2005; Skwarek et al., 2007; Miller and Posakony, 2018). In the signal receiving cells, Notch molecules are ubiquitinated by E3 ligases Nedd4 and Suppressor of deltex [Su(dx)] and targeted for lysosomal degradation to avoid ligand independent activation (Cornell et al., 1999; Mazaleyrat et al., 2003; Sakata et al., 2004; Wilkin et al., 2004; Dalton et al., 2011). The E3 ubiquitin ligase Deltex (Dx) was isolated as a positive regulator of Notch signaling which genetically and physically interacts with Notch (Xu and Artavanis-Tsakonas, 1990; Diederich et al., 1994; Matsuno et al., 1995; Matsuno et al., 2002). Subsequent studies reveal that Dx promotes ubiquitination and ligand independent activation of Notch through the endocytic machinery (Hori et al., 2004; Wilkin et al., 2008; Hori et al., 2011; Yamada et al., 2011). Interestingly, Dx is also capable of inhibiting Notch activation in certain developmental contexts (Mukherjee et al., 2005; Fuwa et al., 2006; Dutta et al., 2017). The E3 ligase cbl is found to target both Dl (Wang et al., 2010) and Notch (Bala Tannan et al., 2018) for degradation. The DUB enzyme Fat facets (Faf) enhances Delta endocytosis to promote Notch signaling during fly eye development (Cadavid et al., 2000; Chen and Fischer, 2000; Chen et al., 2002; Overstreet et al., 2004), while another DUB enzyme USP5 negatively regulates Notch signaling in the same tissue (Ling et al., 2017). Several other DUBs have been implicated in Notch signaling regulation during wing development, but their substrates are still elusive (Zhang et al., 2012).
To date, very little is known about the roles of E2 enzymes in Notch signaling. Here we report that the E2 enzyme UbcD1 (also known as effete) positively regulates Notch signaling activity in the signal receiving cells during Drosophila wing development. UbcD1 is a highly conserved class I E2 enzyme (Treier et al., 1992), which plays important roles in a broad spectrum of cellular and developmental events. UbcD1 participates in regulation of telomere behavior (Cenci et al., 1997; Cipressa et al., 2013), apoptosis (Ryoo et al., 2002; Yeh and Bratton, 2013), innate immunity (Chen et al., 2017), dendrite pruning (Kuo et al., 2006), oogenesis (Ohlmeyer and Schupbach, 2003; Chen et al., 2009), neuroblast proliferation (Li et al., 2014) as well as Hedgehog (Hh) signaling and fly wing patterning (Pan et al., 2017). Our study represents the first analysis for the role of UbcD1 in Notch signaling pathway, which will help to understand the functional complexity and diversity of UbcD1.
All fly stocks and crosses were maintained at 25°C on standard media. The stocks used in this study are: FRT82B,UbcD1s1782/TM6B (#111415; Kyoto Stock Center); FRT82B,UbcD18/TM6B (Chen et al., 2009); FRT82B,UbcD1mer1/TM6B (Pan et al., 2017); NRE-EGFP (#30728; Bloomington Drosophila Stock Center, BDSC); dpp-Gal4, UAS-mCD8-GFP/TM3, dpp-Gal4, UAS-mRFP/TM3, C5-Gal4,UAS-GFP/TM6B and C96-Gal4,UAS-GFP/TM6B (Zhang et al., 2012; Li et al., 2019); UbcD1 RNAi (#26011; Vienna Drosophila Resource Center); UAS-Dl (#26694; BDSC); UAS-NFL (#52309; BDSC); UAS-NICD (Xie et al., 2014); UAS-UbcD1WT and UAS-UbcD1C85A (Pan et al., 2017); tub-GFP-LAMP1 (Akbar et al., 2009). The Ubx-Flp; FRT82B, Ubi-RFP/TM6B and Ubx-Flp; FRT82B, Ubi-GFP/TM6B stock were used to induce somatic clones in wing disc. The hsFlp; Tub-Gal4, UAS-GFP/Cyo; FRT82B, Tub-Gal80 stock was used to generate MARCM clones as previously described (Chang et al., 2021).
The genotypes in the experiments are listed below:
Figure 1A: Ubx-Flp; FRT82B, Ubi-RFP.
Figure 1B–D: FRT82B, UbcD1s1782 × Ubx-Flp; FRT82B, Ubi-RFP.
Figure 1E: NRE-GFP; FRT82B, UbcD1s1782 × Ubx-Flp; FRT82B, Ubi-RFP.
Figure 2A, C, D: FRT82B, UbcD1mer1 × Ubx-Flp; FRT82B, Ubi-RFP.
Figure 2B, F, G: FRT82B, UbcD18 × Ubx-Flp; FRT82B, Ubi-RFP.
Figure 2E: NRE-GFP; FRT82B, UbcD1mer1 × Ubx-Flp; FRT82B, Ubi-RFP.
Figure 3A: dpp-Gal4, UAS-mCD8-GFP.
Figure 3B: dpp-Gal4, UAS-mCD8-GFP × UbcD1 RNAi.
Figure 3C: C5-Gal4, UAS-GFP.
Figure 3D: C5-Gal4, UAS-GFP × UbcD1 RNAi.
Figure 3E: C96-Gal4, UAS-GFP.
Figure 3F: C96-Gal4, UAS-GFP × UbcD1 RNAi.
Figure 4A: hsFlp; Tub-Gal4, UAS-GFP; FRT82B, Tub-Gal80 × FRT82B, UbcD1mer1.
Figure 4B: hsFlp; Tub-Gal4, UAS-GFP; FRT82B, Tub-Gal80 × UAS-Dl; FRT82B.
Figure 4C: hsFlp; Tub-Gal4, UAS-GFP; FRT82B, Tub-Gal80 × UAS-Dl; FRT82B, UbcD1mer1.
Figure 4D: hsFlp; Tub-Gal4, UAS-GFP; FRT82B, Tub-Gal80 × UAS-NFL; FRT82B, UbcD1mer1.
Figure 4E: hsFlp; Tub-Gal4, UAS-GFP; FRT82B, Tub-Gal80 × UAS-NICD; FRT82B, UbcD1mer1.
Figure 5A, B: FRT82B, UbcD1mer1 × Ubx-Flp; FRT82B, Ubi-GFP.
Figure 5C, D: dpp-Gal4, UAS-mCD8-GFP × UbcD1 RNAi.
Figure 6A, B: FRT82B, UbcD1mer1 × Ubx-Flp; FRT82B, Ubi-GFP.
Figure 6C: dpp-Gal4, UAS-mCD8-GFP × UbcD1 RNAi.
Figure 6D: dpp-Gal4, UAS-mRFP × tub-GFP-LAMP1; UbcD1 RNAi.
Figure 7A: hsFlp; Tub-Gal4, UAS-GFP; FRT82B, Tub-Gal80 × UAS-UbcD1WT; FRT82B, UbcD1mer1.
Figure 7B: hsFlp; Tub-Gal4, UAS-GFP; FRT82B, Tub-Gal80 × UAS-UbcD1C85A; FRT82B, UbcD1mer1.
Figure 7C, F: dpp-Gal4, UAS-mCD8-GFP × UAS-UbcD1WT; UbcD1 RNAi.
Figure 7D, G: dpp-Gal4, UAS-mCD8-GFP × UAS- UbcD1C85A; UbcD1 RNAi.
Figure 7E: dpp-Gal4, UAS-mCD8-GFP × UbcD1 RNAi.
Third-instar larvae were dissected in cold PBS and fixed with 4% paraformaldehyde for 15 min at room temperature. The wing discs were washed with 0.1% Triton X-100 in PBS (PBST) and blocked in 0.2% BSA in PBST for 1 h before incubating with primary antibodies overnight at 4°C. The primary antibodies used in this study are: mouse anti-Cut (1:200; 2B10; Developmental Studies Hybridoma Bank, DSHB), mouse anti-Wg (1:200; 4D4; DSHB), mouse anti-NICD (1:200; C17.9C6; DSHB), mouse anti-NECD (1:200; C458.2H; DSHB), mouse anti-Dl (1:200; C594.9B; DSHB), mouse anti-Rab7 (1:200; Rab7; DSHB), mouse anti-Hrs (1:200; Hrs8-2; DSHB). After washing with PBST, wing discs were immersed in second antibodies conjugated with Alexa Fluor 488 (1:200; Invitrogen) or Alexa Fluor 568 (1:200; Invitrogen) for 1 h at room temperature. After washing with PBST for three times, wing discs were dissected and mounted in the VECTASHIELD mounting medium (Vector Laboratories). For LysoTracker staining, wing discs were dissected in Schneider’s Drosophila medium (#21720024, Thermo Fisher) and incubated in medium containing LysoTracker (1:20000; L7528; Invitrogen) for 5 min at room temperature. After washed by fresh medium, the wing discs were mounted and imaged. The fluorescence images were acquired with Leica SP8 confocal microscope and assembled in Photoshop and ImageJ.
Adult wings were dissected from flies after fixed in isopropanol for at least 24 h and mounted in 50% glycerol. Images of adult wings were captured using a Leica DMIL inverted microscope equipped with a QImaging QICAM Fast 1394 digital camera.
Using a somatic mosaic screen strategy (Ren et al., 2018), we isolated an UbcD1 allele that impairs Notch signaling during fly wing development. Marginal nicks were observed in fly wings bearing homozygous UbcD1s1782 clones (Figures 1A,B), a typical phenotype caused by Notch loss-of-function (LOF) (Blair, 2007; Bray, 2016). Notch activates the expression of target genes such as cut and wingless (wg) in cells located at the dorsal-ventral (D/V) boundary in the wing imaginal disc (Supplementary Figures S1A,B). The expression of Cut and Wg were abolished in UbcD1s1782 homozygous cells (Figures 1C,D). The transcriptional activity of Notch signaling could be visualized by the NRE-GFP reporter (Saj et al., 2010), and the expression of NRE-GFP was also dampened in UbcD1s1782 homozygous clones (Figure 1E and Supplementary Figure S1C). These observations suggest that Notch signaling activity is disrupted in UbcD1s1782 mutant wing disc cells.
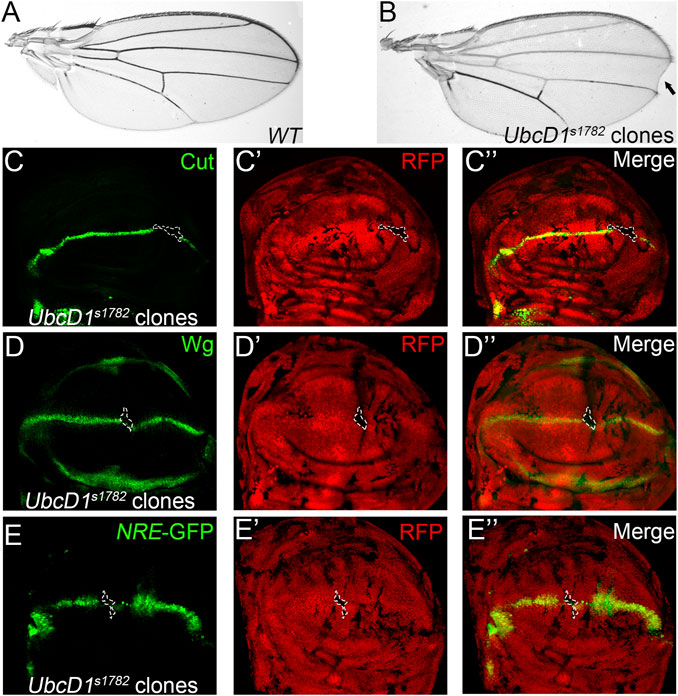
FIGURE 1. UbcD1s1782 shows phenotypes that resemble Notch LOF in Drosophila wing. (A–B) Wing of the parental Ubx-Flp; FRT82B, Ubi-RFP stock is shown as wild type control (A). Wing margin loss (black arrow) is observed in fly wings bearing UbcD1s1782 homozygous clones (B). (C–E) Expression of Notch signaling targets Cut (C), Wg (D) and the reporter NRE-GFP (E) are abolished in UbcD1s1782 homozygous mutant clones. Mutant clones are marked by absence of RFP. Representative mutant clones are circled by dashed lines.
To further establish a role of UbcD1 in Notch signaling transduction, two additional UbcD1 alleles were tested. Both UbcD1mer1 and UbcD18 are LOF alleles that have been shown to cause developmental defects in various fly tissues (Chen et al., 2009; Pan et al., 2017). Upon induction of somatic mosaic clones, both alleles led to wing margin nicks (Figures 2A,B). Expression of Cut (Figure 2C), Wg (Figure 2D) as well as the NRE-GFP reporter (Figure 2E) were reduced in UbcD1mer1 homozygous cells. Similarly, UbcD18 mutant cells were also deficient of Cut (Figure 2F) and Wg (Figure 2G) expression. Taken together, we conclude that UbcD1 positively regulates Notch signaling during fly wing development.
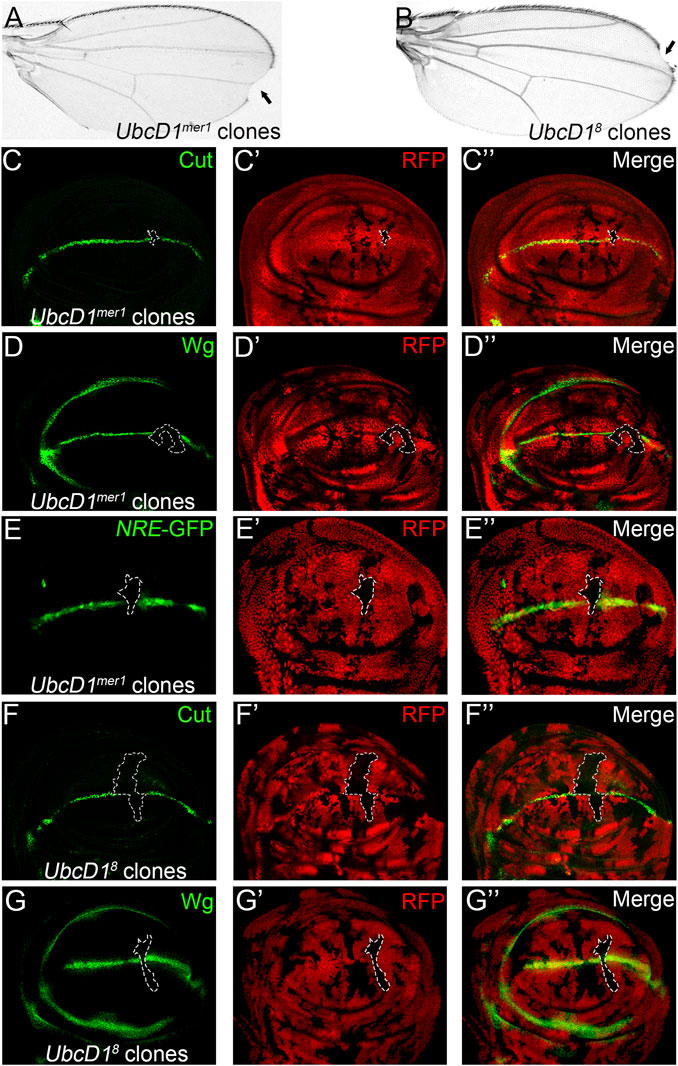
FIGURE 2. UbcD1 mutants inhibit Notch signaling activity in Drosophila wing. (A–-B) Wing margin nicks (black arrow) are observed in fly wings bearing UbcD1mer1 (A) and UbcD18 (B) homozygous clones. (C–E) Expression of Notch signaling targets Cut (C), Wg (D) and the reporter NRE-GFP (E) are abolished in UbcD1mer1 homozygous mutant cells. (F–G) Expression of Cut (F) and Wg (G) are abolished in a subset of UbcD18 homozygous cells. Mutant clones are marked by absence of RFP. Representative mutant clones are circled by dashed lines. The expression pattern of Cut, Wg and NRE-GFP in wild type wing discs are shown in Supplementary Figure S1.
Notch signaling operates among two group of cells, UbcD1 might function in either signal sending or receiving cells in the process of signal transduction. Cell type specific RNAi experiments were performed to further distinguish in which group of cells Ubcd1 are required. Knock-down of UbcD1 by a transgenic RNAi construct resulted in significant inhibition of Cut expression in the wing disc (Figures 3A,B). The C5-Gal4 (Hall et al., 2017; Li et al., 2019) was used to drive UbcD1 RNAi in the signal-sending cells. Knock-down of UbcD1 in the signal sending cells showed little impact on Cut expression (Figures 3C,D). When the C96-Gal4 was used to drive UbcD1 RNAi in the signal-receiving cells (Zhang et al., 2012), reduction of Cut was observed (Figures 3E,F). Therefore, UbcD1 likely functions in the signal-receiving cells to regulate Notch signaling activity.
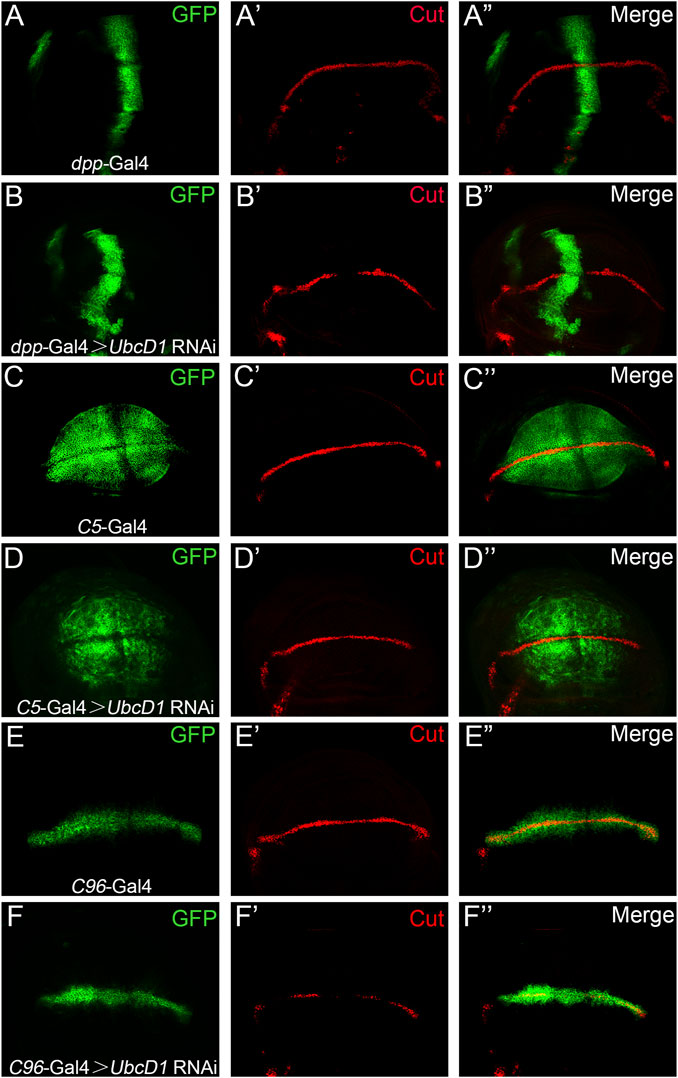
FIGURE 3. UbcD1 knock-down in the signaling receiving cells inhibit Notch activity. (A–B) When driven by dpp-Gal4 in the anterior-posterior border region (A), UbcD1 RNAi leads to reduction of Cut expression (B). (C–D) The C5-Gal4 drives GFP expression in the signal-sending cells (C). Expression of Cut are not affected when UbcD1 RNAi are driven by the C5-Gal4 (D). (E–F) The C96-Gal4 expression domain is restricted within the signal-receiving cells (E). Knocking-down UbcD1 in the signal-receiving cells by C96-Gal4 disrupts Cut expression (F). The Gal4 expression domain are marked by GFP.
To dissect how UbcD1 regulates Notch signal transduction, we used the MARCM system (Lee and Luo, 2001) to overexpress Dl and Notch proteins in UbcD1mer1 mutant cells. In UbcD1mer1 MARCM clones which are positively marked by GFP, the expression of Cut was abolished (Figure 4A). Expression of Dl in wild type cells led to induction of Cut in cells surrounding the MARCM clones (Figure 4B), as they received excessive signal inputs from cells inside the clone. In UbcD1mer1 mutant cells, overexpression of Dl was still capable of inducing Cut expression in the surrounding cells (Figure 4C). These results confirm that UbcD1 is dispensable in the signal sending cells.
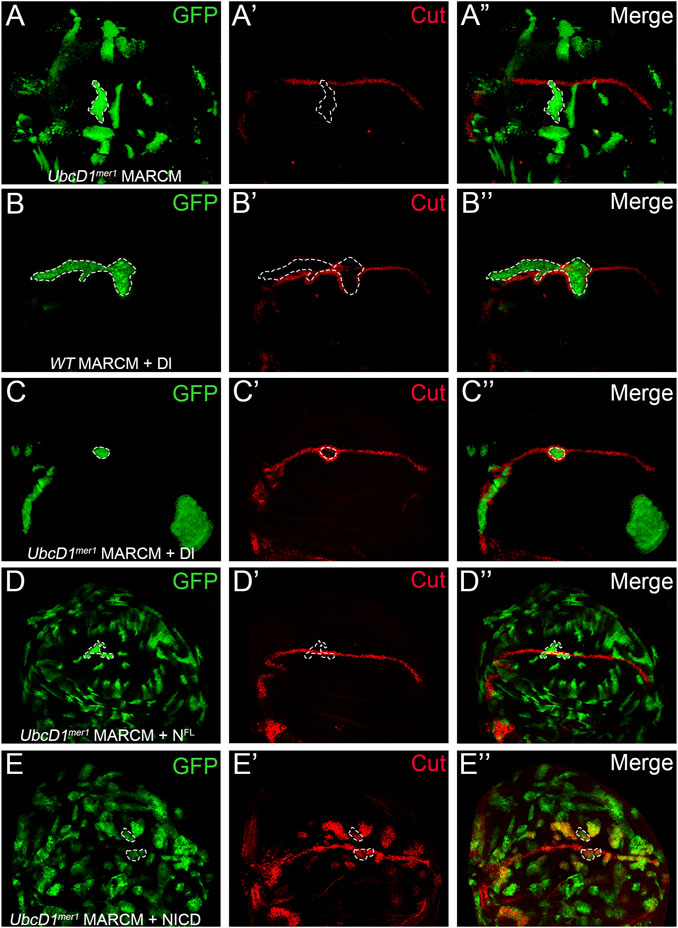
FIGURE 4. UbcD1 functions upstream of Notch protein processing. (A) In UbcD1mer1 MARCM clones, the expression of Cut are reduced. (B–C) Over-expression of Dl in both wild type (B) and UbcD1mer1 mutant cells (C) result in induction of Cut in cells surrounding the MARCM clone. (D) The full-length Notch protein restores Cut expression in UbcD1mer1 homozygous cells. (E) NICD robustly induces Cut expression in UbcD1mer1 mutant cells. MARCM clones are marked by GFP. Representative clones are circled by dashed lines.
In clones located at the D/V boundary, over-expression of Dl was insufficient to rescue Cut expression (Figure 4C). In contrast, the full-length Notch protein was able to restore the expression of Cut in UbcD1mer1 homozygous cells (Figure 4D). When NICD was introduced into UbcD1mer1 mutant cells, ectopic expression of Cut was robustly induced (Figure 4E). These genetics analysis suggests that UbcD1 functions in the signal receiving cells, presumably at early steps before the cleavage of full-length Notch protein.
Giving that UbcD1 functions up-stream of Notch protein processing, the potential effects on Notch protein were further examined. In UbcD1mer1 homozygous mutant cells, Notch proteins accumulated as puncta when labeled by an antibody recognizing the intracellular domain (Figure 5A and Supplementary Figure S2A). Similar distribution defect was observed using a second antibody raised against the extracellular domain of Notch protein (Figure 5B and Supplementary Figure S2B). Consistently, RNAi knock-down of UbcD1 also resulted in aggregation of Notch proteins (Figures 5C,D and Supplementary Figures S2C,D).
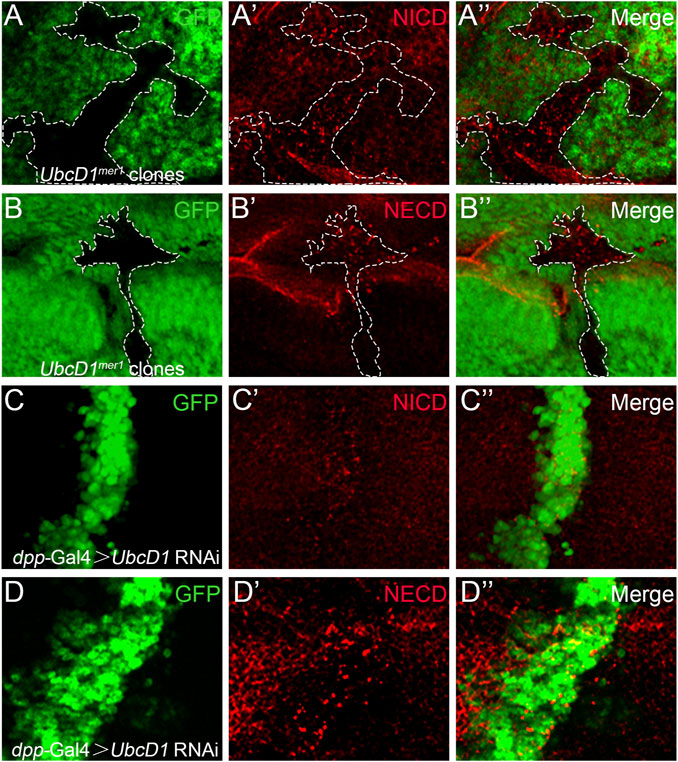
FIGURE 5. UbcD1 affects Notch distribution. (A–B) In UbcD1mer1 mutant cells, Notch proteins form puncta when labeled by antibodies against NICD (A) and NECD (B). Mutant clones are marked by absence of GFP. Representative mutant clones are circled by dashed lines. (C–D) In UbcD1 RNAi cells, Notch proteins form puncta when labeled by antibodies against NICD (C) and NECD (D). The RNAi expressing cells are marked by GFP. Panels (A–D) are magnification of a portion of Supplementary Figure S2.
Accumulation of Notch proteins accompanied with reduction of signaling activity have been found in mutations of the endolysosomal pathway components (Vaccari et al., 2008; Vaccari et al., 2010; Ren et al., 2018). Therefore, whether UbcD1 is involved in the endolysosomal machinery was investigated. In UbcD1mer1 mutant cells, early endosomes as labeled by Hrs were not significantly affected (Figure 6A and Supplementary Figure S3A), but formation of Rab7-positive late endosomes was inhibited (Figure 6B and Supplementary Figure S3B). Lacking of Rab7 associated late endosomes might disrupt subsequent events such as endolysosome acidification and cargo degradation. Interestingly, when applied to live wing discs, strong accumulation of the acidotrophic fluorescent dye LysoTracker was observed in UbcD1 RNAi cells (Figure 6C and Supplementary Figure S3C). This result indicates that despite the reduction of late endosome maturation, acidification of endocytic organelles are enhanced in UbcD1 RNAi cells. We further examined the lysosomal activity using a GFP-Lamp1 fusion protein that undergoes rapid lysosomal degradation in physiological context (Akbar et al., 2009). GFP-Lamp1 was hardly detectable in wild type wing imaginal disc cells, while knock-down of UbcD1 by RNAi caused a significant accumulation of GFP-Lamp1 (Figure 6D and Supplementary Figure S3D). These results indicate that UbcD1 might safeguard the integrity of the endolysosomal machinery to promote Notch signal transduction.
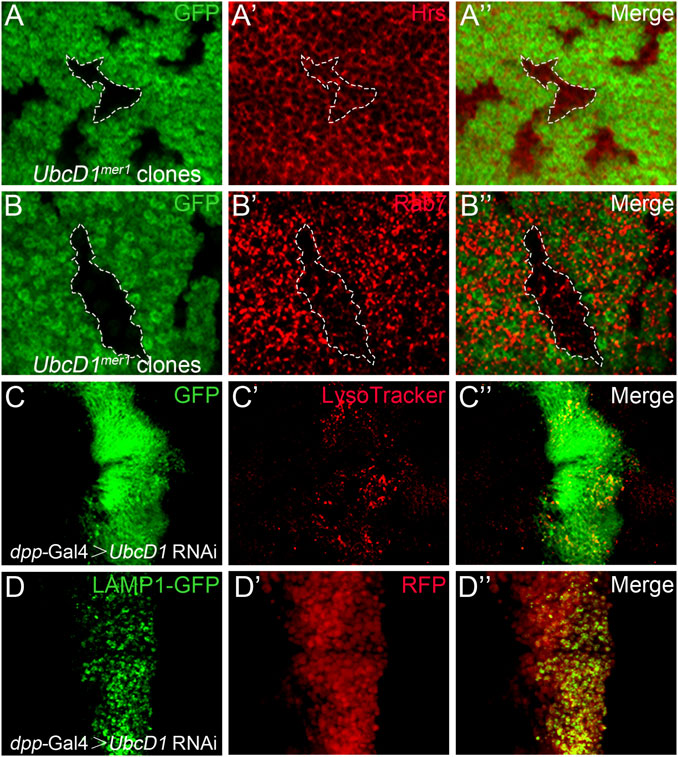
FIGURE 6. UbcD1 affects endolysosomal machinery. (A–B) In UbcD1mer1 mutant cells, Hrs positive early endosomes are not affected (A) while Rab7 association with endosomes are reduced (B). Mutant clones are marked by absence of GFP. Representative mutant clones are circled by dashed lines. (C–D) In UbcD1 RNAi cells, accumulation of LysoTracker (C) and GFP-LAMP1 (D) are evident. The RNAi expressing cells are marked by GFP (C) or RFP (D). Panels (A–D) are magnification of a portion of Supplementary Figure S3.
A conserved Cystine residue at position 85 (C85) is required for the Ub conjugating activity of UbcD1 (Pan et al., 2017). Using the MARCM system, we found that reduction of Cut expression in UbcD1mer1 homozygous mutant cells was rescued by over-expression of UbcD1WT (Figure 7A), but not the “catalytic dead” form UbcD1C85A (Figure 7B). Similarly, only UbcD1WT (Figure 7C) but not UbcD1C85A (Figure 7D) was capable of restoring Cut expression in UbcD1 RNAi cells. Expression of another Notch target, Wg, was also rescued by UbcD1WT (Figures 7E,F) but not UbcD1C85A (Figure 7G) in UbcD1 RNAi cells. These results demonstrate that the Ub conjugating activity is essential for UbcD1 to ensure Notch activation.
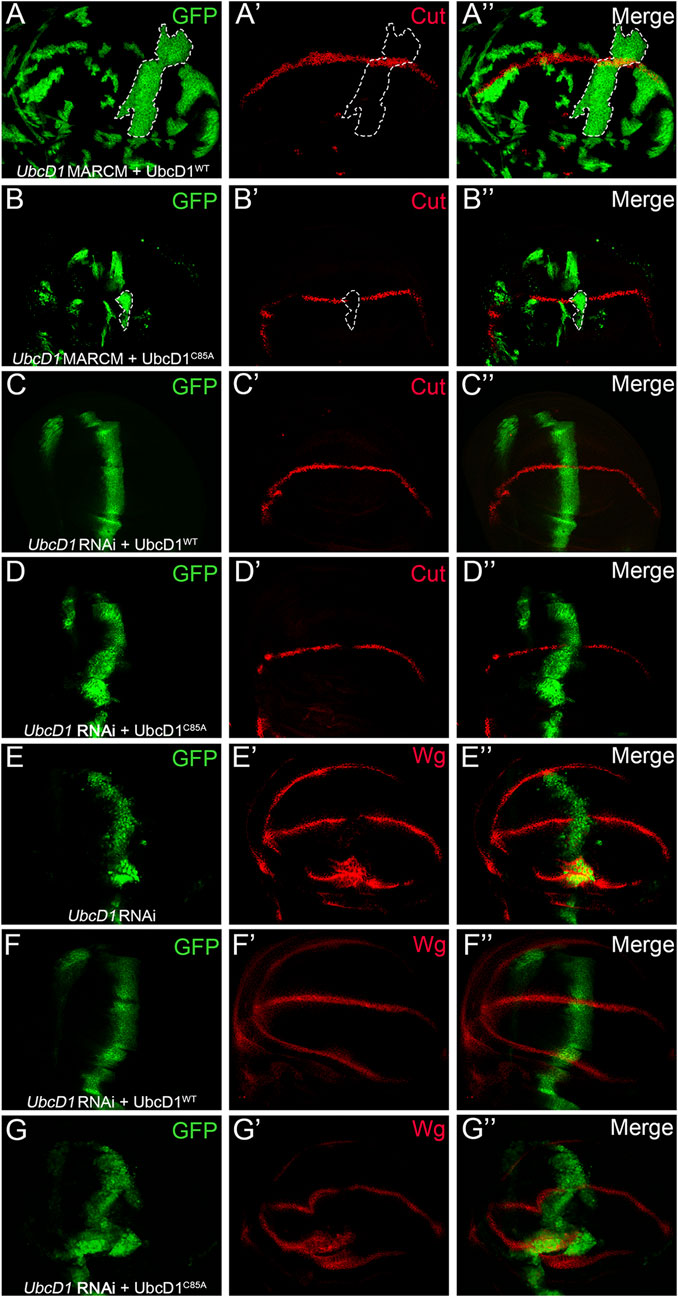
FIGURE 7. UbcD1 functions as an E2 enzyme. (A–B) In UbcD1mer1 mutant cells, UbcD1WT (A) but not UbcD1C85A (B) is sufficient to restore the expression of Cut. MARCM clones are marked by GFP. Representative clones are circled by dashed lines. (C–D) UbcD1WT (C) but not UbcD1C85A (D) is able to rescue the reduction of Cut expression caused by UbcD1 RNAi. (E–G) RNAi knock-down of UbcD1 leads to inhibition of Wg expression (E), which is rescued by UbcD1WT (F) but not UbcD1C85A (G). Note that Wg are accumulated in cells at the edge of wing pouch upon UbcD1 RNAi (E, G). The RNAi expressing cells are marked by GFP.
Formation of wings made insects the first group of animals that gained the ability to fly during evolution (Shimmi et al., 2014). The shape, size and venation patterns of insect wings are highly stereotyped and species specific, and these traits are widely used in biology researches ranging from species identification, organ development to evolutionary modelling (Parchem et al., 2007). Our knowledge of the genetic and molecular basis of insect wing development largely originates from studies in the model insect, Drosophila melanogaster (De Celis and Diaz-Benjumea, 2003). The Notch mutant likely represents one of the first recorded Drosophila mutations that affect wing development. Later studies demonstrate that the Notch signal pathway is highly conserved among the insects and regulates various developmental processes across different species. Notch signaling regulates wing margin formation in Drosophila hydei (Van Breugel and Langhout, 1983) and sheep blowfly (Davies et al., 1996; Chen et al., 1998), wing morphogenesis in silkworm (Sato et al., 2008; Ling et al., 2015) and pigment patterns in the butterfly wing (Reed, 2004; Reed and Serfas, 2004). Further studies indicate that Notch signaling is required for oogenesis in Blattella germanica (Irles et al., 2016) and locust (Song et al., 2019), reproductive constraint in the adult worker honeybee (Duncan et al., 2016), appendage development in silk worm (Liu, 2012) and camouflage patterns in caterpillars (Jin et al., 2020). Recent studies reveal crucial role of Notch signaling during body segmentation in insect species such as cockroaches (Pueyo et al., 2008; Chesebro et al., 2013) and silkworm (Liu, 2013). Whether Notch signaling regulates segmentation in cricket is still under debate (Kainz et al., 2011; Mito et al., 2011), but segmentation in Drosophila (Liao and Oates, 2017) and grasshopper (Dearden and Akam, 2000) is likely independent of Notch signaling. These studies highlight the important and diverse roles of Notch signaling, identification of new factors involved in Notch signal transduction will help us to better understand how it operates to control insect development.
Our data presented here suggests a novel role for UbcD1 as a positive regulator of the Notch signaling pathway during fly wing development. Previous studies have found that UbcD1 genetically interacts with the DUB Faf (Cadavid et al., 2000) and E3 ligase Neur (Lai et al., 2001), both of which regulate Dl endocytic trafficking during fly eye development. However, whether and how Dl protein and Notch signaling are affected in UbcD1 mutant eye disc cells have not been investigated (Cadavid et al., 2000; Lai et al., 2001). Furthermore, Faf is dispensable for fly wing development (Fischer-Vize et al., 1992). The E3 ligase Neur is essential for sensory precursors specification but not wing margin formation and other Notch signaling dependent processes during wing development (Yeh et al., 2000; Lai and Rubin, 2001). Therefore, UbcD1 is likely involved in Notch signaling regulation in multiple tissues and developmental contexts, targeting distinct signal molecules and transduction steps. Our genetic analysis suggests that UbcD1 functions presumably at early steps before the cleavage of full-length Notch protein, but also impacts later transduction events such as Notch trafficking and distribution in the developing wing. The molecular targets of UbcD1 and the exact mechanisms that how UbcD1 impacts Notch signaling still remains elusive.
Alternatively, UbcD1 might regulate Notch signaling indirectly through cellular processes such as endolysosomal trafficking. Our results indicate that UbcD1 is likely required for maturation of late endosomes and following steps towards lysosomal degradation. A crucial event during endosome maturation is Rab conversion, during which the early organizer Rab5 is replaced by the late organizer Rab7. Recent work identifies Dmon1, a member of the Sand1/Mon1 protein family, as a crucial factor for Rab conversion during fly wing development (Yousefian et al., 2013). In fly wing disc cells, LOF of Dmon1 results in reduced association of Rab7 with endosomes, enhancement of endolysosomal acidification and accumulation of Notch proteins (Yousefian et al., 2013). The high similarity of these LOF phenotypes indicate that UbcD1 might be involved in Rab conversion. The exact role of UbcD1 in the endolysosomal machinery remains an open question.
Given the broad cellular activities of UbcD1, it is not surprising to find that UbcD1 might regulate multiple signaling pathways during wing development. It has been shown that UbcD1 negatively regulates Hh signaling activation in the wing (Pan et al., 2017). When UbcD1 expression was inhibited by RNAi, down-regulation of Wg was observed in cells located at the D/V boundary due to disruption of Notch signaling transduction. In contrast, accumulation of Wg were found in UbcD1 RNAi cells at the edge of wing pouch (Figure 7E). The expression of Wg is regulated by signaling pathways other than Notch at this region. For example, in response to cell apoptosis, another cellular event that involves UbcD1 (Ryoo et al., 2002), the JNK pathway is sufficient to induce Wg expression in these cells (Ryoo et al., 2004). Whether and how UbcD1 is involved in these pathways during wing development awaits further investigation.
The raw data supporting the conclusion of this article will be made available by the authors, without undue reservation.
FZ and JZ conceived and designed the study. FZ and YC developed the methodology and performed the experiments. FZ, YC, JS, and JZ analyzed and interpreted the data. FZ and YC wrote the manuscript. JS and JZ reviewed and revised the manuscript. JS and JZ supervised the project. All authors contributed to the article and approved the submitted version.
This work was supported by the National Natural Science Foundation of China (31772526 and 31970478 to JZ and 32030012 to JS).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
We thank Drs. Dahua Chen, Yun Zhao, Zhouhua Li, Renjie Jiao, Wei Song, Alan Jian Zhu, Wei Wu, the Bloomington Stock Center, the Kyoto Fly Stock Center, the Vienna Drosophila Resource Center and the Developmental Studies Hybridoma Bank for fly stocks and antibodies. We thank the Core Facility of Drosophila Resource and Technology, Center for Excellence in Molecular Cell Science, Chinese Academy of Sciences for providing fly stocks and reagents.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2021.770853/full#supplementary-material
Akbar, M. A., Ray, S., and Krämer, H. (2009). The SM Protein Car/Vps33A Regulates SNARE-Mediated Trafficking to Lysosomes and Lysosome-Related Organelles. MBoC 20, 1705–1714. doi:10.1091/mbc.e08-03-0282
Bala Tannan, N., Collu, G., Humphries, A. C., Serysheva, E., Weber, U., and Mlodzik, M. (2018). AKAP200 Promotes Notch Stability by Protecting it from Cbl/lysosome-Mediated Degradation in Drosophila melanogaster. Plos Genet. 14, e1007153. doi:10.1371/journal.pgen.1007153
Blair, S. S. (2007). Wing Vein Patterning inDrosophilaand the Analysis of Intercellular Signaling. Annu. Rev. Cel Dev. Biol. 23, 293–319. doi:10.1146/annurev.cellbio.23.090506.123606
Bray, S. J. (2016). Notch Signalling in Context. Nat. Rev. Mol. Cel Biol 17, 722–735. doi:10.1038/nrm.2016.94
Cadavid, A. L., Ginzel, A., and Fischer, J. A. (2000). The Function of the Drosophila Fat Facets Deubiquitinating Enzyme in Limiting Photoreceptor Cell Number Is Intimately Associated with Endocytosis. Development 127, 1727–1736. doi:10.1242/dev.127.8.1727
Cenci, G., Rawson, R. B., Belloni, G., Castrillon, D. H., Tudor, M., Petrucci, R., et al. (1997). UbcD1, a Drosophila Ubiquitin-Conjugating Enzyme Required for Proper Telomere Behavior. Genes Dev. 11, 863–875. doi:10.1101/gad.11.7.863
Chang, X., Zhang, F., Li, H., Mo, D., Shen, J., and Zhang, J. (2021). Characterization of a New Mastermind Allele Identified from Somatic Mosaic Screen. Cell Dev. 165, 203664. doi:10.1016/j.cdev.2021.203664
Chen, D., Wang, Q., Huang, H., Xia, L., Jiang, X., Kan, L., et al. (2009). Effete-mediated Degradation of Cyclin a Is Essential for the Maintenance of Germline Stem Cells in Drosophila. Development 136, 4133–4142. doi:10.1242/dev.039032
Chen, L., Paquette, N., Mamoor, S., Rus, F., Nandy, A., Leszyk, J., et al. (2017). Innate Immune Signaling in Drosophila Is Regulated by Transforming Growth Factor β (TGFβ)-Activated Kinase (Tak1)-Triggered Ubiquitin Editing. J. Biol. Chem. 292, 8738–8749. doi:10.1074/jbc.M117.788158
Chen, X., and Fischer, J. A. (2000). In Vivo Structure/Function Analysis of the Drosophila Fat Facets Deubiquitinating Enzyme Gene. Genetics 156 (4), 1829–1836. doi:10.1093/genetics/156.4.1829
Chen, X., Zhang, B., and Fischer, J. A. (2002). A Specific Protein Substrate for a Deubiquitinating Enzyme: Liquid Facets Is the Substrate of Fat Facets. Genes Dev. 16 (3), 289–294. doi:10.1101/gad.961502
Chen, Z., Newsome, T., McKenzie, J. A., and Batterham, P. (1998). Molecular Characterization of the Notch Homologue from the Australian Sheep Blowfly, Lucilia cuprina. Insect Biochem. Mol. Biol. 28, 601–612. doi:10.1016/s0965-1748(98)00032-0
Chesebro, J. E., Pueyo, J. I., and Couso, J. P. (2013). Interplay between a Wnt-dependent Organiser and the Notch Segmentation Clock Regulates Posterior Development in Periplaneta americana. Biol. Open 2, 227–237. doi:10.1242/bio.20123699
Cipressa, F., Romano, S., Centonze, S., zur Lage, P. I., Vernì, F., Dimitri, P., et al. (2013). Effete, a Drosophila Chromatin-Associated Ubiquitin-Conjugating Enzyme that Affects Telomeric and Heterochromatic Position Effect Variegation. Genetics 195, 147–158. doi:10.1534/genetics.113.153320
Cornell, M., Evans, D. A. P., Mann, R., Fostier, M., Flasza, M., Monthatong, M., et al. (1999). The Drosophila melanogaster Suppressor of Deltex Gene, a Regulator of the Notch Receptor Signaling Pathway, Is an E3 Class Ubiquitin Ligase. Genetics 152, 567–576. doi:10.1093/genetics/152.2.567
Dalton, H. E., Denton, D., Foot, N. J., Ho, K., Mills, K., Brou, C., et al. (2011). Drosophila Ndfip Is a Novel Regulator of Notch Signaling. Cell Death Differ 18, 1150–1160. doi:10.1038/cdd.2010.130
Davies, A. G., Game, A. Y., Chen, Z., Williams, T. J., Goodall, S., Yen, J. L., et al. (1996). Scalloped Wings Is the Lucilia cuprina Notch Homologue and a Candidate for the Modifier of Fitness and Asymmetry of Diazinon Resistance. Genetics 143, 1321–1337. doi:10.1093/genetics/143.3.1321
De Celis, J. F., and Diaz-Benjumea, F. J. (2003). Developmental Basis for Vein Pattern Variations in Insect Wings. Int. J. Dev. Biol. 47, 653–663.
Dearden, P., and Akam, M. (2000). A Role for Fringe in Segment Morphogenesis but Not Segment Formation in the Grasshopper, Schistocerca gregaria. Dev. Genes Evol. 210, 329–336. doi:10.1007/s004270000072
Diederich, R. J., Matsuno, K., Hing, H., and Artavanis-Tsakonas, S. (1994). Cytosolic Interaction between Deltex and Notch Ankyrin Repeats Implicates Deltex in the Notch Signaling Pathway. Development 120, 473–481. doi:10.1242/dev.120.3.473
Duncan, E. J., Hyink, O., and Dearden, P. K. (2016). Notch Signalling Mediates Reproductive Constraint in the Adult Worker Honeybee. Nat. Commun. 7, 12427. doi:10.1038/ncomms12427
Dutta, D., Paul, M. S., Singh, A., Mutsuddi, M., and Mukherjee, A. (2017). Regulation of Notch Signaling by the Heterogeneous Nuclear Ribonucleoprotein Hrp48 and Deltex in Drosophila melanogaster. Genetics 206 (2), 905–918. doi:10.1534/genetics.116.198879
Fischer-Vize, J. A., Rubin, G. M., and Lehmann, R. (1992). The Fat Facets Gene Is Required for Drosophila Eye and Embryo Development. Development 116, 985–1000. doi:10.1242/dev.116.4.985
Fortini, M. E. (2009). Notch Signaling: The Core Pathway and its Posttranslational Regulation. Dev. Cel 16, 633–647. doi:10.1016/j.devcel.2009.03.010
Fuwa, T. J., Hori, K., Sasamura, T., Higgs, J., Baron, M., and Matsuno, K. (2006). The First Deltex Null Mutant Indicates Tissue-specific Deltex-dependent Notch Signaling in Drosophila. Mol. Genet. Genomics. 275, 251–263. doi:10.1007/s00438-005-0087-3
Grabbe, C., Husnjak, K., and Dikic, I. (2011). The Spatial and Temporal Organization of Ubiquitin Networks. Nat. Rev. Mol. Cel Biol 12, 295–307. doi:10.1038/nrm3099
Hall, E. T., Pradhan-Sundd, T., Samnani, F., and Verheyen, E. M. (2017). The Protein Phosphatase 4 Complex Promotes the Notch Pathway and Wingless Transcription. Biol. Open 6, 1165–1173. doi:10.1242/bio.025221
Hori, K., Fostier, M., Ito, M., Fuwa, T. J., Go, M. J., Okano, H., et al. (2004). DrosophilaDeltex Mediates Suppressor of Hairless-independent and Late-Endosomal Activation of Notch Signaling. Development 131, 5527–5537. doi:10.1242/dev.01448
Hori, K., Sen, A., Kirchhausen, T., and Artavanis-Tsakonas, S. (2011). Synergy between the ESCRT-III Complex and Deltex Defines a Ligand-independent Notch Signal. J. Cel Biol. 195, 1005–1015. doi:10.1083/jcb.201104146
Irles, P., Elshaer, N., and Piulachs, M.-D. (2016). The Notch Pathway Regulates Both the Proliferation and Differentiation of Follicular Cells in the Panoistic Ovary of Blattella germanica. Open Biol. 6, 150197. doi:10.1098/rsob.150197
Itoh, M., Kim, C.-H., Palardy, G., Oda, T., Jiang, Y.-J., Maust, D., et al. (2003). Mind Bomb Is a Ubiquitin Ligase that Is Essential for Efficient Activation of Notch Signaling by Delta. Dev. Cel 4, 67–82. doi:10.1016/s1534-5807(02)00409-4
Jin, H., Yoda, S., Liu, L., Kojima, T., and Fujiwara, H. (2020). Notch and Delta Control the Switch and Formation of Camouflage Patterns in Caterpillars. iScience 23, 101315. doi:10.1016/j.isci.2020.101315
Kainz, F., Ewen-Campen, B., Akam, M., and Extavour, C. G. (2011). Notch/Delta Signalling Is Not Required for Segment Generation in the Basally Branching Insect Gryllus Bimaculatus. Development 138, 5015–5026. doi:10.1242/dev.073395
Kopan, R., and Ilagan, M. X. G. (2009). The Canonical Notch Signaling Pathway: Unfolding the Activation Mechanism. Cell 137, 216–233. doi:10.1016/j.cell.2009.03.045
Kuo, C. T., Zhu, S., Younger, S., Jan, L. Y., and Jan, Y. N. (2006). Identification of E2/E3 Ubiquitinating Enzymes and Caspase Activity Regulating Drosophila Sensory Neuron Dendrite Pruning. Neuron 51, 283–290. doi:10.1016/j.neuron.2006.07.014
Lai, E. C., Deblandre, G. A., Kintner, C., and Rubin, G. M. (2001). Drosophila Neuralized Is a Ubiquitin Ligase that Promotes the Internalization and Degradation of delta. Dev. Cel 1, 783–794. doi:10.1016/s1534-5807(01)00092-2
Lai, E. C., Roegiers, F., Qin, X., Jan, Y. N., and Rubin, G. M. (2005). The Ubiquitin ligaseDrosophilaMind Bomb Promotes Notch Signaling by Regulating the Localization and Activity of Serrate and Delta. Development 132, 2319–2332. doi:10.1242/dev.01825
Lai, E. C., and Rubin, G. M. (2001). Neuralized Functions Cell-Autonomously to Regulate a Subset of Notch-dependent Processes during Adult Drosophila Development. Dev. Biol. 231, 217–233. doi:10.1006/dbio.2000.0124
Le Borgne, R., Remaud, S., Hamel, S., and Schweisguth, F. (2005). Two Distinct E3 Ubiquitin Ligases Have Complementary Functions in the Regulation of delta and Serrate Signaling in Drosophila. Plos Biol. 3, e96. doi:10.1371/journal.pbio.0030096
Le Borgne, R., and Schweisguth, F. (2003). Unequal Segregation of Neuralized Biases Notch Activation during Asymmetric Cell Division. Dev. Cel 5, 139–148. doi:10.1016/s1534-5807(03)00187-4
Le Bras, S., Loyer, N., and Le Borgne, R. (2011). The Multiple Facets of Ubiquitination in the Regulation of Notch Signaling Pathway. Traffic 12, 149–161. doi:10.1111/j.1600-0854.2010.01126.x
Lee, T., and Luo, L. (2001). Mosaic Analysis with a Repressible Cell Marker (MARCM) for Drosophila Neural Development. Trends Neurosciences 24, 251–254. doi:10.1016/s0166-2236(00)01791-4
Li, S., Wang, C., Sandanaraj, E., Aw, S. S. Y., Koe, C. T., Wong, J. J. L., et al. (2014). The SCF Slimb E3 Ligase Complex Regulates Asymmetric Division to Inhibit Neuroblast Overgrowth. EMBO Rep. 15, 165–174. doi:10.1002/embr.201337966
Li, Y., and Baker, N. E. (2004). The Roles of Cis-Inactivation by Notch Ligands and of Neuralized during Eye and Bristle Patterning in Drosophila. BMC Dev. Biol. 4, 5. doi:10.1186/1471-213X-4-5
Li, Y., Liu, T., and Zhang, J. (2019). The ATPase TER94 Regulates Notch Signaling during Drosophila wing Development. Biol. Open 8, bio038984. doi:10.1242/bio.038984
Liao, B.-K., and Oates, A. C. (2017). Delta-notch Signalling in Segmentation. Arthropod Struct. Dev. 46, 429–447. doi:10.1016/j.asd.2016.11.007
Ling, L., Ge, X., Li, Z., Zeng, B., Xu, J., Chen, X., et al. (2015). MiR-2 Family Targetsawdandfngto Regulate wing Morphogenesis inBombyx Mori. RNA Biol. 12, 742–748. doi:10.1080/15476286.2015.1048957
Ling, X., Huang, Q., Xu, Y., Jin, Y., Feng, Y., Shi, W., et al. (2017). The Deubiquitinating Enzyme Usp5 Regulates Notch and RTK Signaling duringDrosophilaeye Development. FEBS Lett. 591, 875–888. doi:10.1002/1873-3468.12580
Liu, W. (2013). Bmdelta Phenotype Implies Involvement of Notch Signaling in Body Segmentation and Appendage Development of Silkworm, Bombyx mori. Arthropod Struct. Dev. 42, 143–151. doi:10.1016/j.asd.2012.10.002
Liu, W. (2012). Functional Analyses in the Silkworm,Bombyx mori, Support a Role for Notch Signaling in Appendage Development but Not theGroucho-dependent Pair-Rule Process. J. Exp. Zool. (Mol. Dev. Evol. 318, 651–662. doi:10.1002/jez.b.22470
Matsuno, K., Diederich, R. J., Go, M. J., Blaumueller, C. M., and Artavanis-Tsakonas, S. (1995). Deltex Acts as a Positive Regulator of Notch Signaling through Interactions with the Notch Ankyrin Repeats. Development 121, 2633–2644. doi:10.1242/dev.121.8.2633
Matsuno, K., Ito, M., Hori, K., Miyashita, F., Suzuki, S., Kishi, N., et al. (2002). Involvement of a Proline-Rich Motif and RING-H2 finger of Deltex in the Regulation of Notch Signaling. Development 129, 1049–1059. doi:10.1242/dev.129.4.1049
Mazaleyrat, S. L., Fostier, M., Wilkin, M. B., Aslam, H., Evans, D. A. P., Cornell, M., et al. (2003). Down-regulation of Notch Target Gene Expression by Suppressor of Deltex. Dev. Biol. 255, 363–372. doi:10.1016/s0012-1606(02)00086-6
Miller, S. W., and Posakony, J. W. (2018). Lateral Inhibition: Two Modes of Non-autonomous Negative Autoregulation by Neuralized. Plos Genet. 14, e1007528. doi:10.1371/journal.pgen.1007528
Mito, T., Shinmyo, Y., Kurita, K., Nakamura, T., Ohuchi, H., and Noji, S. (2011). Ancestral Functions of Delta/Notch Signaling in the Formation of Body and Leg Segments in the Cricket Gryllus Bimaculatus. Development 138, 3823–3833. doi:10.1242/dev.060681
Moretti, J., and Brou, C. (2013). Ubiquitinations in the Notch Signaling Pathway. Ijms 14, 6359–6381. doi:10.3390/ijms14036359
Mukherjee, A., Veraksa, A., Bauer, A., Rosse, C., Camonis, J., and Artavanis-Tsakonas, S. (2005). Regulation of Notch Signalling by Non-visual β-arrestin. Nat. Cel Biol. 7, 1191–1201. doi:10.1038/ncb1327
Ohlmeyer, J. T., and Schüpbach, T. (2003). Encore Facilitates SCF-ubiquitin-proteasome-dependent Proteolysis during Drosophila Oogenesis. Development 130, 6339–6349. doi:10.1242/dev.00855
Overstreet, E., Fitch, E., and Fischer, J. A. (2004). Fat Facets and Liquid Facets Promote Delta Endocytosis and Delta Signaling in the Signaling Cells. Development 131, 5355–5366. doi:10.1242/dev.01434
Pan, C., Xiong, Y., Lv, X., Xia, Y., Zhang, S., Chen, H., et al. (2017). UbcD1 Regulates Hedgehog Signaling by Directly Modulating Ci Ubiquitination and Processing. EMBO Rep. 18, 1922–1934. doi:10.15252/embr.201643289
Parchem, R. J., Perry, M. W., and Patel, N. H. (2007). Patterns on the Insect wing. Curr. Opin. Genet. Dev. 17, 300–308. doi:10.1016/j.gde.2007.05.006
Pavlopoulos, E., Pitsouli, C., Klueg, K. M., Muskavitch, M. A. T., Moschonas, N. K., and Delidakis, C. (2001). Neuralized Encodes a Peripheral Membrane Protein Involved in delta Signaling and Endocytosis. Dev. Cel 1, 807–816. doi:10.1016/s1534-5807(01)00093-4
Pitsouli, C., and Delidakis, C. (2005). The Interplay between DSL Proteins and Ubiquitin Ligases in Notch Signaling. Development 132, 4041–4050. doi:10.1242/dev.01979
Pueyo, J. I., Lanfear, R., and Couso, J. P. (2008). Ancestral Notch-Mediated Segmentation Revealed in the Cockroach Periplaneta americana. Proc. Natl. Acad. Sci. 105, 16614–16619. doi:10.1073/pnas.0804093105
Reed, R. D. (2004). Evidence for Notch-Mediated Lateral Inhibition in Organizing Butterfly wing Scales. Dev. Genes Evol. 214, 43–46. doi:10.1007/s00427-003-0366-0
Reed, R. D., and Serfas, M. S. (2004). Butterfly wing Pattern Evolution Is Associated with Changes in a Notch/Distal-Less Temporal Pattern Formation Process. Curr. Biol. 14, 1159–1166. doi:10.1016/j.cub.2004.06.046
Ren, L., Mo, D., Li, Y., Liu, T., Yin, H., Jiang, N., et al. (2018). A Genetic Mosaic Screen Identifies Genes Modulating Notch Signaling in Drosophila. PLoS One 13, e0203781. doi:10.1371/journal.pone.0203781
Ryoo, H. D., Bergmann, A., Gonen, H., Ciechanover, A., and Steller, H. (2002). Regulation of Drosophila IAP1 Degradation and Apoptosis by Reaper and ubcD1. Nat. Cel Biol. 4, 432–438. doi:10.1038/ncb795
Ryoo, H. D., Gorenc, T., and Steller, H. (2004). Apoptotic Cells Can Induce Compensatory Cell Proliferation through the JNK and the Wingless Signaling Pathways. Dev. Cel 7, 491–501. doi:10.1016/j.devcel.2004.08.019
Saj, A., Arziman, Z., Stempfle, D., van Belle, W., Sauder, U., Horn, T., et al. (2010). A Combined Ex Vivo and In Vivo RNAi Screen for Notch Regulators in Drosophila Reveals an Extensive Notch Interaction Network. Dev. Cel 18, 862–876. doi:10.1016/j.devcel.2010.03.013
Sakata, T., Sakaguchi, H., Tsuda, L., Higashitani, A., Aigaki, T., Matsuno, K., et al. (2004). Drosophila Nedd4 Regulates Endocytosis of Notch and Suppresses its Ligand-independent Activation. Curr. Biol. 14, 2228–2236. doi:10.1016/j.cub.2004.12.028
Salazar, J. L., and Yamamoto, S. (2018). Integration of drosophila and Human Genetics to Understand Notch Signaling Related Diseases. Adv. Exp. Med. Biol. 1066, 141–185. doi:10.1007/978-3-319-89512-3_8
Sato, K., Matsunaga, T. M., Futahashi, R., Kojima, T., Mita, K., Banno, Y., et al. (2008). Positional Cloning of a Bombyx Wingless Locus Flügellos (Fl) Reveals a Crucial Role for Fringe that Is Specific for Wing Morphogenesis. Genetics 179, 875–885. doi:10.1534/genetics.107.082784
Shimmi, O., Matsuda, S., and Hatakeyama, M. (2014). Insights into the Molecular Mechanisms Underlying Diversified wing Venation Among Insects. Proc. R. Soc. B. 281, 20140264. doi:10.1098/rspb.2014.0264
Skwarek, L. C., Garroni, M. K., Commisso, C., and Boulianne, G. L. (2007). Neuralized Contains a Phosphoinositide-Binding Motif Required Downstream of Ubiquitination for delta Endocytosis and Notch Signaling. Dev. Cel 13, 783–795. doi:10.1016/j.devcel.2007.10.020
Song, J., Li, W., Zhao, H., and Zhou, S. (2019). Clustered miR-2, miR-13a, miR-13b and miR-71 Coordinately Target Notch Gene to Regulate Oogenesis of the Migratory Locust Locusta migratoria. Insect Biochem. Mol. Biol. 106, 39–46. doi:10.1016/j.ibmb.2018.11.004
Treier, M., Seufert, W., and Jentsch, S. (1992). Drosophila UbcD1 Encodes a Highly Conserved Ubiquitin-Conjugating Enzyme Involved in Selective Protein Degradation. EMBO J. 11 (1), 367–372. doi:10.1002/j.1460-2075.1992.tb05059.x
Vaccari, T., Duchi, S., Cortese, K., Tacchetti, C., and Bilder, D. (2010). The Vacuolar ATPase Is Required for Physiological as Well as Pathological Activation of the Notch Receptor. Development 137, 1825–1832. doi:10.1242/dev.045484
Vaccari, T., Lu, H., Kanwar, R., Fortini, M. E., and Bilder, D. (2008). Endosomal Entry Regulates Notch Receptor Activation in Drosophila melanogaster. J. Cel Biol 180, 755–762. doi:10.1083/jcb.200708127
Van Breugel, F. M. A., and Langhout, B. V. Z. (1983). The NOTCH Locus of DROSOPHILA HYDEI: Alleles, Phenotypes and Functional Organization. Genetics 103, 197–217. doi:10.1093/genetics/103.2.197
Wang, W., and Struhl, G. (2005). Distinct Roles for Mind Bomb, Neuralized and Epsin in Mediating DSL Endocytosis and Signaling in Drosophila. Development 132, 2883–2894. doi:10.1242/dev.01860
Wang, Y., Chen, Z., and Bergmann, A. (2010). Regulation of EGFR and Notch Signaling by Distinct Isoforms of D-Cbl during Drosophila Development. Dev. Biol. 342, 1–10. doi:10.1016/j.ydbio.2010.03.005
Weinmaster, G., and Fischer, J. A. (2011). Notch Ligand Ubiquitylation: What Is it Good for?. Dev. Cel 21, 134–144. doi:10.1016/j.devcel.2011.06.006
Wilkin, M. B., Carbery, A.-M., Fostier, M., Aslam, H., Mazaleyrat, S. L., Higgs, J., et al. (2004). Regulation of Notch Endosomal Sorting and Signaling by Drosophila Nedd4 Family Proteins. Curr. Biol. 14, 2237–2244. doi:10.1016/j.cub.2004.11.030
Wilkin, M., Tongngok, P., Gensch, N., Clemence, S., Motoki, M., Yamada, K., et al. (2008). Drosophila HOPS and AP-3 Complex Genes Are Required for a Deltex-Regulated Activation of Notch in the Endosomal Trafficking Pathway. Dev. Cel 15, 762–772. doi:10.1016/j.devcel.2008.09.002
Xie, G., Yu, Z., Jia, D., Jiao, R., and Deng, W. M. (2014). E(y)1/TAF9 Mediates the Transcriptional Output of Notch Signaling in Drosophila. J. Cel Sci 127, 3830–3839. doi:10.1242/jcs.154583
Xu, T., and Artavanis-Tsakonas, S. (1990). Deltex, a Locus Interacting with the Neurogenic Genes, Notch, Delta and Mastermind in Drosophila melanogaster. Genetics 126, 665–677. doi:10.1093/genetics/126.3.665
Yamada, K., Fuwa, T. J., Ayukawa, T., Tanaka, T., Nakamura, A., Wilkin, M. B., et al. (2011). Roles of Drosophila Deltex in Notch Receptor Endocytic Trafficking and Activation. Genes Cells 16, 261–272. doi:10.1111/j.1365-2443.2011.01488.x
Ye, Y., and Rape, M. (2009). Building Ubiquitin Chains: E2 Enzymes at Work. Nat. Rev. Mol. Cel Biol 10, 755–764. doi:10.1038/nrm2780
Yeh, E., Dermer, M., Commisso, C., Zhou, L., McGlade, C. J., and Boulianne, G. L. (2001). Neuralized Functions as an E3 Ubiquitin Ligase during Drosophila Development. Curr. Biol. 11, 1675–1679. doi:10.1016/s0960-9822(01)00527-9
Yeh, E., Zhou, L., Rudzik, N., and Boulianne, G. L. (2000). Neuralized Functions Cell Autonomously to Regulate Drosophila Sense Organ Development. EMBO J. 19, 4827–4837. doi:10.1093/emboj/19.17.4827
Yeh, T.-C., and Bratton, S. B. (2013). Caspase-dependent Regulation of the Ubiquitin-Proteasome System through Direct Substrate Targeting. Proc. Natl. Acad. Sci. 110, 14284–14289. doi:10.1073/pnas.1306179110
Yousefian, J., Troost, T., Grawe, F., Sasamura, T., Fortini, M., and Klein, T. (2013). Dmon1 Controls Recruitment of Rab7 to Maturing Endosomes in Drosophila. J. Cel Sci. 126, 1583–1594. doi:10.1242/jcs.114934
Keywords: ubiquitin conjugating enzyme, UbcD1, Notch, Drosophila, wing
Citation: Zhang F, Chen Y, Shen J and Zhang J (2021) The Ubiquitin Conjugating Enzyme UbcD1 is Required for Notch Signaling Activation During Drosophila Wing Development. Front. Genet. 12:770853. doi: 10.3389/fgene.2021.770853
Received: 05 September 2021; Accepted: 30 September 2021;
Published: 12 October 2021.
Edited by:
Wei Zhang, Peking University, ChinaCopyright © 2021 Zhang, Chen, Shen and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Junzheng Zhang, emhhbmdqekBjYXUuZWR1LmNu
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.