
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Genet. , 12 January 2022
Sec. Cancer Genetics and Oncogenomics
Volume 12 - 2021 | https://doi.org/10.3389/fgene.2021.768130
This article is part of the Research Topic Epigenetic and Transcriptional Dysregulations in Cancer and Therapeutic Opportunities View all 19 articles
 Ritu Raina1
Ritu Raina1 Abdulmajeed G. Almutary2*
Abdulmajeed G. Almutary2* Sali Abubaker Bagabir3
Sali Abubaker Bagabir3 Nazia Afroze1
Nazia Afroze1 Sharmila Fagoonee4
Sharmila Fagoonee4 Shafiul Haque5,6
Shafiul Haque5,6 Arif Hussain1*
Arif Hussain1*Purpose: Plant-derived phytochemicals have shown epigenetic modulatory effect in different types of cancer by reversing the pattern of DNA methylation and chromatin modulation, thereby restoring the function of silenced tumor-suppressor genes. In the present study, attempts have been made to explore chrysin-mediated epigenetic alterations in HeLa cells.
Methods: Colony formation and migration assays followed by methylation-specific PCR for examining the methylation status of CpG promoters of various tumor-suppressor genes (TSGs) and the expression of these TSGs at the transcript and protein levels were performed. Furthermore, global DNA methylation; biochemical activities of DNA methyltransferases (DNMTs), histone methyl transferases (HMTs), histone deacetylases (HDACs), and histone acetyl transferases (HATs) along with the expression analysis of chromatin-modifying enzymes; and H3 and H4 histone modification marks analyses were performed after chrysin treatment.
Results: The experimental analyses revealed that chrysin treatment encourages cytostatic behavior as well as inhibits the migration capacity of HeLa cells in a time- and dose-dependent manner. Chrysin reduces the methylation of various tumor-suppressor genes, leading to their reactivation at mRNA and protein levels. The expression levels of various chromatin-modifying enzymes viz DNMTs, HMTs, HDACs, and HATS were found to be decreased, and H3 and H4 histone modification marks were modulated too. Also, reduced global DNA methylation was observed following the treatment of chrysin.
Conclusion: This study concludes that chrysin can be used as a potential epigenetic modifier for cancer treatment and warrants for further experimental validation.
Regular cell functions are modifiable by different epigenetic modifications, and these alterations play a crucial role during cellular growth and development (Rahman et al., 2016; Shankar et al., 2016). The epigenetic modifications occur by the way of regulatory mechanisms involving histone modifications, DNA methylation, microRNAs, and chromatin remodeling that modulates gene expression and disturbs cellular machinery and homeostasis in cancer cells (Huang et al., 2011; Ong et al., 2011; You and Jones, 2012; Ho et al., 2013; Aggarwal et al., 2015; Busch et al., 2015; Shankar et al., 2016). DNA methylation at CpG residues of the promoters of tumor suppressor genes (TSGs) causes repression of tumor-suppressor genes (Hatzimichael and Crook, 2013; Shankar et al., 2016), which is considered as a key regulatory mechanism of gene silencing and is correlated with the overexpression of DNA methyltransferases (DNMTs) (Kogan et al., 2017; Li and Wang, 2017; Piyathilake et al., 2017). Similarly, the modification of histone proteins by epigenetic enzymes, that is, HDAC (histone deacetylase), HMT (histone methyltransferase), HATs (histone acetyltransferase), and phosphorylases, leads to the repression or activation of gene activity. The equilibrium between the erasers and the writers of histone modifications is crucial for normal expression of genes, and their dysregulation may lead to cancer development (Sharma et al., 2009).
Methylation of histones at H3 and H4 lysine residues is catalyzed by histone methyl transferases (HMTs) (Ho et al., 2013; Shankar et al., 2016). Methylation marks at H3K79, H3K36, and H3K4 are supposed to be active marks, whereas H4K20, H3K27, and H3K9 methylation marks are related with transcriptional suppression (Sharma et al., 2009; Ho et al., 2013; Shankar et al., 2016). In the past, aberrant expression of both HMTs and HDMs has been reported in cancer (McLaughlin–Drubin et al., 2011; Busch et al., 2015). Histone acetylation is another important histone modification state, and hyper-acetylation leads to the stimulation of suppressed genes. Aberrant expression of HDAC has been found in cancer and is linked with gene repression and tumorigenesis (Sharma et al., 2009; Kogan et al., 2017; Andrijauskaite et al., 2019). Likewise, aberrant expressions of HATs and HDACs as well as HMTs and HDMs have been reported in various types of cancer in the past (Lin et al., 2009; Chen et al., 2017).
In the recent times, epigenetic-based cancer treatment is gaining more interest due to its reversible nature. Several FDA-approved drugs, for example, azacytidine and decitabine (DNMT inhibitors), and vorinostat and romidepsin (HDAC inhibitors), have shown promising results in solid malignancies and myelodysplastic syndrome (Herranz and Esteller, 2007a; Ong et al., 2011; Hatzimichael and Crook, 2013; Ho et al., 2013; Guo et al., 2015; Shankar et al., 2016). The combinational cancer treatment strategy in which both HDAC inhibitors and DNMT inhibitors are being used together has proven to be more effective (Herranz and Esteller, 2007a; Herranz and Esteller, 2007b; Ho et al., 2013; Shankar et al., 2016). However, low specificity and high systemic toxicity have limited their use (Paredes-Gonzalez et al., 2014). Hence, plant-derived chemopreventive agents are the main focus of scientific scope. Earlier studies have reported that dietary agents like EGCG, quercetin, genistein, curcumin, resveratrol, luteolin, and apigenin modulate the activity of DNMT and HDAC, and can lead to re-expression of silenced TSGs (Kai et al., 2010; Kim and Kim, 2013; Aggarwal et al., 2015; Guo et al., 2015; Mocanu et al., 2015; Chang and Yu, 2016a; Kanwal et al., 2016; Loh et al., 2019; Yan et al., 2020; Ganai et al., 2021a).
Chrysin (5-dihydroxyflavone), a flavone found in honey, bee propolis, and blue passion flower (Passiflora caerulea) extract, has gained importance as an antioxidant, antiviral, and anticancer compound (Pal-Bhadra et al., 2012; Yang et al., 2014; Kanwal et al., 2016). It induces cell cycle arrest, inhibits cell adhesion and tumor cell–induced angiogenesis, and induces apoptosis in various types of cancer, and also downregulates pathways including AKT (Khoo et al., 2010; Kasala et al., 2015; Ryu et al., 2017; Lim et al., 2018). Earlier, antiproliferative and apoptosis-inducing effects of chrysin on HeLa cells (Raina et al., 2021) have been observed. However, the role and mechanistic action of chrysin in the modulation of epigenome is not fully explored, except scanty reports wherein the role of chrysin in the modulation of epigenetic enzymes has been studied. Chrysin was found to decrease the expression of HDAC two and HDAC eight, and increase the expression of H4acK16, H3acK14, and H4acK12. It decreases H3me2K9 in (melanoma cell) A375 cells and restores the transcriptional activity of the tumor-suppressor gene p21WAFI. Chrysin is capable enough to modify DNMT and HMT expressions in prostate cancer cells, and behaves as an epigenetic modifier (Pal-Bhadra et al., 2012; Kanwal et al., 2016; Ganai et al., 2021b). The precise mechanism of modulation of epigenome is not well explored and documented. Keeping the abovesaid facts in view, this study was performed to evaluate the significance of chrysin treatment on cell migration, DNA methylation, and histone modifications in human cervical cancer (HeLa) cells.
Human cervical cancer (HeLa) cells were used as an in vitro cancer model during this study. HeLa cells were maintained in complete Dulbecco’s modified Eagle medium (DMEM; Sigma-Aldrich; Merck, KGaA) containing 10% FBS (Sigma-Aldrich; Merck KGaA) and penicillin (100U/mL) (Sigma-Aldrich; Merck KGaA), and incubated at 37 °C with 5% CO2.
Chrysin (powdered, mol wt. 254.241 g/mol) was procured from Sigma-Aldrich (Merck, KGaA), and stock solution was prepared with DMSO (78.67 mM) using DMSO (stock solution). Furthermore, sub-stock (1 mM) and concentrations (5, 10, and 15 µM) of chrysin were prepared using the complete media as a diluent.
The colony-forming assay was performed following the protocol used by Crowley et al. (2016) and Sundaram et al. (2019), with minor modifications (Crowley et al., 2016; Kedhari Sundaram et al., 2019a). Briefly, ∼2.5 × 105 (Ho et al., 2013) cells were dispensed in six-well plates and incubated overnight followed by the treatment with increasing doses of chrysin (5, 10, and 15 µM) for 48 h. DMSO control (i.e., solvent control) and treated cells were collected after 48 h and plated at approximately 500 cells/well, and allowed to grow. The medium was changed time to time as per the requirement. After 14 days, the colonies formed were fixed by using absolute CH3OH and stained with crystal violet. Olympus inverted microscope (Labomed, United States) was used to obtain the images of the colonies formed. ImageJ software program was used to count the colonies in treated and DMSO control wells.
The scratch-aound assay was performed to examine the inhibitory effect of chrysin on cell migration (Yang et al., 2014; Kedhari Sundaram et al., 2019a). Approximately 2 × 105 (Ho et al., 2013) cells were plated in a six-well plate and incubated at 37°C overnight. On the next day, a “wound” or a “cell-free” line was created on a confluent monolayer of the cells by scratching the monolayer with a pipette tip. The cells were incubated in the presence of different chrysin dilutions (10 and 15 µM) and “healing” of wounds, which ensues through cell migration, and growth toward the cell-free zone was monitored on a regular basis. An inverted microscope was used for capturing the wound images in each well prior and after 24–48 h of the treatment.
The invasion assay was performed to evaluate the migratory and invasive capability of chrysin-treated HeLa cells and DMSO control using the Boyden chamber (Yang et al., 2014). Briefly, 5.0 × 103 (Aggarwal et al., 2015) cells/well were seeded on the upper side of the insert in separate inserts, and below it in the well of a 24-well plate, the medium with FBS was kept. After 48 h, absolute methanol was used to fix the cells, and 0.1% crystal violet was used for staining purposes. The cells present inside of the chamber were cleared using an ear bud. The assessment of complete migration was performed under the microscope, and any of the five fields were scanned (five fields per filter). The images were captured for each treatment and control using ×200 magnification with an inverted microscope (Olympus Corporation). ImageJ program was used for counting the colonies. The experiment was repeated three times, and mean ± SD was used to plot the graphs considering p-value ≤0.05.
Nuclear extracts from the untreated HeLa cells were prepared using the EpiQuikTM Nuclear Extraction Kit (Catalog No. OP-0002, Epigentek, United States) following the manufacturer’s protocol. The EpiQuik DNMT Activity Assay Kit (Catalog No. #P-3009, Epigentek, USA) was used to check the effect of chrysin on the DNMT activity. Briefly, chrysin (@ conc. 5, 10 and 15 µM) was added to the untreated nuclear extract, buffer, and Adomet (methyl group donor) to the cytosine-rich DNA substrate–coated assay plate and incubated for 1.5 h at 37 °C. It was followed by incubation with capture and detection antibody. After signal development, the absorbance was read on an ELISA reader at 450 nm. DNMT activity inhibition was calculated by comparing with DMSO controls. The experiment was repeated three times, and the mean ± SD was used to plot the graph. The statistical significance level was calculated using one-way ANOVA, and the p-value was maintained at ≤ 0.05.
The EpiQuik HDAC Activity Assay Kit (Catalog Number P-4002, Epigentek, United States) was used for evaluating the effect of chrysin on the HDAC activity. The acetylated histone substrate–coated assay plate was prepared by adding 50 μL of the biotinylated HDAC substrate diluted in wash buffer to all wells. The assay plate was incubated at room temperature for 45 min and washed with wash buffer. HDAC assay buffer was dispensed to the wells in chrysin (5, 10, and 15 µM) treated and untreated nuclear extract placed. The plate was kept at 37°C for an hour followed by incubation with capture and detection antibody. Developing solution was used to develop the signal, stop solution stopped the reaction, and OD was read at 450 nm. The percentage inhibition of the HDAC activity against chrysin treatment was calculated by comparing with DMSO control. The experiment was repeated three times, and mean ± SD was used to plot a graph. One-way ANOVA was used to check the statistical significance at the p-value ≤ 0.05.
The assay was done by using the EpiQuik HMT H3K9 Activity Assay Kit (Catalog No. P-3003, Epigentek, USA) following the protocol given by the manufacturer. Briefly, to the histone three lysine substrate–coated assay plate, chrysin (@ conc. of 5, 10, and 15 µM) was added in separate wells, with untreated nuclear extract, buffer biotinylated substrate, and Adomet (methyl group donor), and incubated at 37 °C for 1.5 h. Following this step, capture antibody and detection antibody were added to the wells, and incubated at room temp for 30 min. Finally, a color was developed, and absorbance was measured using an ELISA reader at 450 nm. The percentage inhibition was calculated by comparing with DMSO control. The experiment was repeated three times, and the mean ± SD was calculated to plot a graph. One-way ANOVA was used for checking the statistical significance, and the p-value was maintained as ≤ 0.05.
The assay was done using the EpiQuik™ HAT Activity Assay Kit (Catalog No. P-4003 Epigenetek, United States) following the manufacturer’s protocol. Briefly, the nuclear extract and the substrate for HAT were incubated for 1 h followed by washing with the wash buffer. An inhibitor was added in the test samples, and signals were captured and detected by the capture and detection antibody, respectively. After color development, the plate was read at 450 nm using the ELISA plate reader. HAT activity and percentage inhibition were calculated by comparing with DMSO control samples. A graph was plotted by taking the mean of three experiments ± SD. One-way ANOVA was used to check the statistical significance at fixed p-value ≤ 0.05.
DNA isolation was done by using GenElute Mammalian Genomic DNA Miniprep Kit (Catalog No. G1N70, Sigma-Aldrich, Merck, KGaA) following the manufacturer’s protocol. About 1.5 × 106 cells were treated with chrysin (@ conc. 5, 10, and 15 µM for 48 h) and DMSO control. DNA was isolated from chrysin treated and the DMSO control samples, and its quality was checked by gel electrophoresis using 1% agarose gel (Catalog No. A9539, Sigma-Aldrich, Merck, KGaA) in 0.5XTBE buffer with ethidium bromide. The quantitation of DNA samples was completed by spectrophotometry using NanoDrop 2000 (Thermo Scientific™, USA) and stored at-80°C.
MethylFlash™ Methylated DNA Quantification Kit (Catalog No. P-1034, Epigentek, USA) was used to analyze the methylated DNA in treated cells (@ chrysin conc. 5, 10, and 15 µm for 48 h) and DMSO controls. The kit was used for the detection of methylated DNA using antibodies against 5-mC (cytosine) that can be analyzed calorimetrically. Optical density was measured using the ELISA reader at 450 nm wavelength. As established, the extent of methylation on the gene is directly related to the optical density. The levels of methylation were calculated in comparison with the DMSO control. The experiment was performed three times at a significance level of p-value ≤0.05.
CpG island DNA methylation quantification was conducted for estimating the percentage of methylated DNA in the total DNA content of HeLa cells by using the EpiTect Methyl II PCR System (Catalog No. 335452, Qiagen, United States). The method employed calculates the methylation of promoter regions in the input DNA after cleavage with methylation-dependent restriction enzymes and methylation-sensitive restriction enzymes that digest methylated and unmethylated DNA, respectively. Following restriction digestion, the cleaved DNA from each reaction was computed by using it as a template for Human Tumor Suppressor Genes EpiTect Methyl II Signature PCR Array (Qiagen, United States) real-time PCR in an assay plate with primers that border the promoter region of the anticipated genes. The relative amounts of unmethylated and methylated DNA were calculated by comparing the amounts of each reaction with that of a control (no enzymes added) reaction using the ∆∆CT method. The gene panel (with predesigned primers) consisted of tumor-suppressor genes comprising TP73, MGMT, APC, CDKN2A, BRCA1, PTEN, CDH1, DAPK1, CDH13, SOC51, RARB, ESR1, FHIT, RASSF1, WIF1, GSTP1, RUNX3, MLH1, NEUROG1, VHL, PDLIM4, and TIMP3.
The amount of DNA left after the restriction digestion was calculated by using qPCR array results. This was used for constructing the methylation profile of each gene with the ∆ΔCT method. The methylation and unmethylation fraction of the promoter of tested tumor-suppressor genes in chrysin-treated and untreated HeLa cells was estimated as per the protocol available with the kit. The levels of methylation were presented in the form of a graph. Statistical significance was calculated by taking the mean of three experiments by one-way ANOVA using the SPSS program with p-value ≤ 0.05.
The RNA from chrysin-treated (conc. 10 and 15 µM for 48 h) and DMSO control HeLa cells were extracted by using the GenElute Mammalian Genomic Total RNA Kit (Catalog No. RTN70 Sigma-Aldrich, Merck KGaA) and further quantified with the help of NanoDrop. The RNA (2 μg was used as a template) was then subjected to cDNA synthesis by using Applied Biosystems™ High-Capacity cDNA Reverse Transcription Kit (Catalog No. 4368814, ABI-Thermo Fisher, United States). This kit supports random primers’ scheme for initiating the synthesis of cDNA. The expression of genes related to various pathways of migration, inflammation, and TSGs was analyzed with the help of TaqMan-based custom array (4391524 and 4369514 master mix). The PCR array was run on QuantStudio3 and analyzed with the ΔΔCT method using the DataAssist™ program (Thermo Fisher, United States). GAPDH (housekeeping gene) was used for normalizing the data. Relative expression was calculated in comparison with the DMSO control. The statistical significance was calculated by maintaining p-value <0.05.
The expression analysis of TSGs, migration, and inflammation-related proteins was performed by the Proteome Profiler Array (Catalog No. ARY026, R&D, USA). The relative expression levels of 84 oncogenes were investigated with the help of this array. Briefly, 1.5×106 HeLa cells were plated in 25 cm2 flasks, and four such flasks were treated with 10 and 15 µM of chrysin for 48 h. The treated and DMSO control cells were collected and suspended in lysis buffer 17 (1 ml per 107 cells) containing 10 μg/ml each of aprotinin (Catalog No. A6279; Sigma, USA), Leupeptin (Catalog No. 1167/25, Tocris, USA), and pepstatin (Catalog No. 1190/10, Tocris, United States) and shaken gently at 2–8°C for 30 min. The lysate produced was quantitated by the Pierce BCA Assay Kit (Catalog No. 23225; Thermo-Fisher Scientific, United States) following the manufacturer’s protocol. For this assay, 400 µg of a protein in 250 µL volume of the diluted cell lysate treated with chrysin (10 and 15 µM for 48 h) and the DMSO control was used for each membrane. The signal produced was then quantified by the chemiluminescent detector gel-doc system (Bio-Rad Laboratories, United States). The expression of proteins was analyzed by the intensity of proteins in the blot using Image Lab software (version 6.1). The fold change after normalization with the reference spot was calculated by comparing the treated (chrysin) values with the DMSO control values (mean of three experiments ±SD at p-value ≤ 0.05).
Chromatin-modifying enzymes like writers—DNA and histone methyl transferases, histone acetyl transferases, and erasers like histone deacetylases and histone demethylases—help in dynamically sustaining cell metabolism and controlling processes such as cell growth propagation and gene expression by recognition of specific “marks” on histone proteins and DNA (Kouzarides, 2007). RNA was extracted from the DMSO control and chrysin-treated HeLa cells (10 and 15 µM for 48 h). RT2 Profiler™ PCR Array Human Epigenetic Chromatin Modification Enzymes (Catalog No. PAHS-085Z, Qiagen, United States) were used to check the expression of genes responsible for the modulation of DNA and histones including DNA methyl transferases, histone methyl transferases, histone acetyl transferases, and demethylases. RNA at a concentration of 1 µg was used to synthesize cDNA, and it was diluted to 1,350 µL with nuclease-free water and an equal amount of RT2 SYBR® Green qPCR Master mix (Catalog No. 330504; Qiagen, United States) was added to this. From this mixture, 25 µL was poured into each well of the array plate having predefined primers, and the plate was run on ABI Quant Studio 3. The normalization was performed using GAPDH housekeeping gene and the fold change was calculated by comparing the chrysin-treated samples with the DMSO control. The statistical significance was calculated at p-value ≤ 0.5.
In order to understand the role of chrysin as an epigenetic modifier, the Histone Extraction kit (Catalog No. ab113476, Abcam, Cambridge, UK), and Histone H3 and H4 Modification Multiplex Assay kits (Catalog Nos. ab185910 and ab185914) were procured from Abcam, Cambridge, UK. Following the extraction of histone using the Histone Extraction Kit, ∼100 ng of histone protein was used per well, and the protocol given by the manufacturer was followed. OD was measured at 405 nm, and graphs were plotted for reflecting the effect of chrysin compared to the DMSO control. The experiments were performed in triplicates, one-way ANOVA was used to determine the significance of the experiments, and p-value was maintained at ≤0.05.
Statistical analysis was performed by SPSS program (version 21). The data were examined by using one-way ANOVA followed by Tukey’s HSD post hoc analysis. All experiments were performed in triplicate. The results are expressed as the mean ± SD of three distinct experimentations. The statistical significance was set at p-value ≤0.05.
The colony-forming assay was performed to understand the long-term effect of chrysin on the growth and division of HeLa cells. After calculating the survival factor (SF), it was observed that the DMSO control had plating efficiency (PE) of 95%, whereas the survival factor for 5 and 10 µM of chrysin showed only 120 and 30 colonies, respectively. At 15 µM chrysin conc., very few colonies were formed. Hence, it can be inferred that chrysin restrained the capability of cells to form colonies. These results suggest that chrysin is not only capable of causing cell death but also leads to cytostatic state (Figure 1A and Figure 1B).
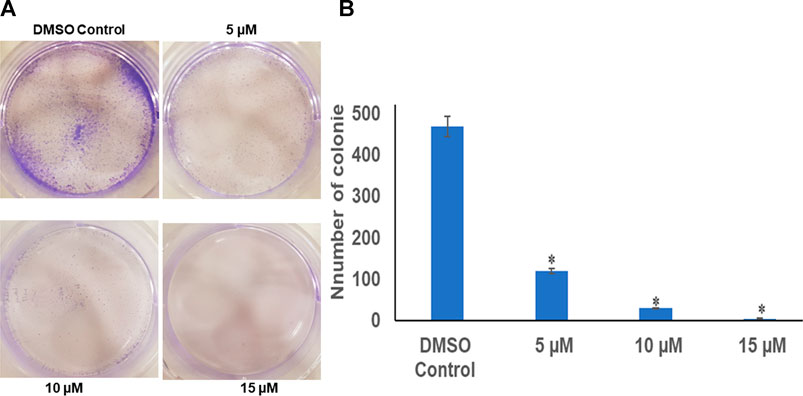
FIGURE 1. (A) Chrysin inhibits colony formation in a dose-dependent manner with almost no colonies at 15 µM chrysin treatment for 48 h. (B) Graphical representation of inhibition of colony formation.
Likewise, chrysin decreased the migration capacity of HeLa cells as demonstrated by scratch-wound and invasion assay using trans-well. Chrysin increased the wound width by 8 and 14% at 10 µM conc. treatment for 24 and 48 h and 17 and 25% at 15 µM conc. treatment for 24 and 48 h, respectively, whereas in DMSO control cells, there was almost complete wound closure after 48 h (Figure 2A and Figure 2B). This was further corroborated by significant decrease in the number of migrating cells after chrysin treatment using the trans-well assay. Only 15 and 2.5% migration at 10 and 15 µM chrysin treatment for 48 h was observed, respectively, in comparison with the DMSO control cells (Figure 2C and Figure 2D).
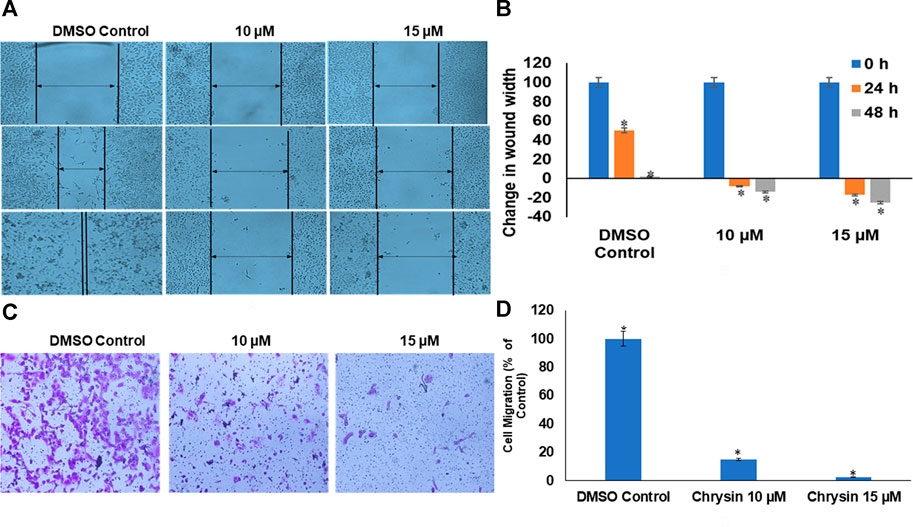
FIGURE 2. (A) Chrysin treatment prevented migration of HeLa cells, as compared to the DMSO control wells, the chrysin treatment showed increase in wound width after 10–15 µM treatment. (B) Graphical representation of increase in wound width after chrysin treatment at 10–15 µM concentrations for 24–48 h. (C) The chrysin-treated HeLa cells depicted significant decrease in cell migration using trans-well inserts. (D) Graphical representation of inhibition of cell migration by chrysin.
qPCR was done to understand the effect of decreased methylation of various tumor-suppressor genes following the treatment of chrysin. It was observed that chrysin treatment increased the expression of various TSGs (such as TIMP3, TIMP4, RARB, RASIF1, TP53, PTEN, CDH1, and SOCS1) and reduced the expression of genes responsible for metastasis (viz. MMP 2, MMP 9, MMP 14, SNAIL1, SMAD3, SMAD4, and MTA1, 2) and the genes involved in the inflammatory process (viz. IL2, IL1A, IL6, and CxCL8). Chrysin treatment also decreased the expression of oncogenes like FOS, JUN, MYC, ESR1, and TWIST1 (Figure 3A and Table 1). Relative quantification (RQ) derived from the 2–∆∆Ct method specifies the fold change in gene expression against the DMSO control after normalization with the selected endogenous gene (GAPDH). The upregulation is documented at RQ ≥ 1.5 and downregulation at RQ ≤ 0.5.
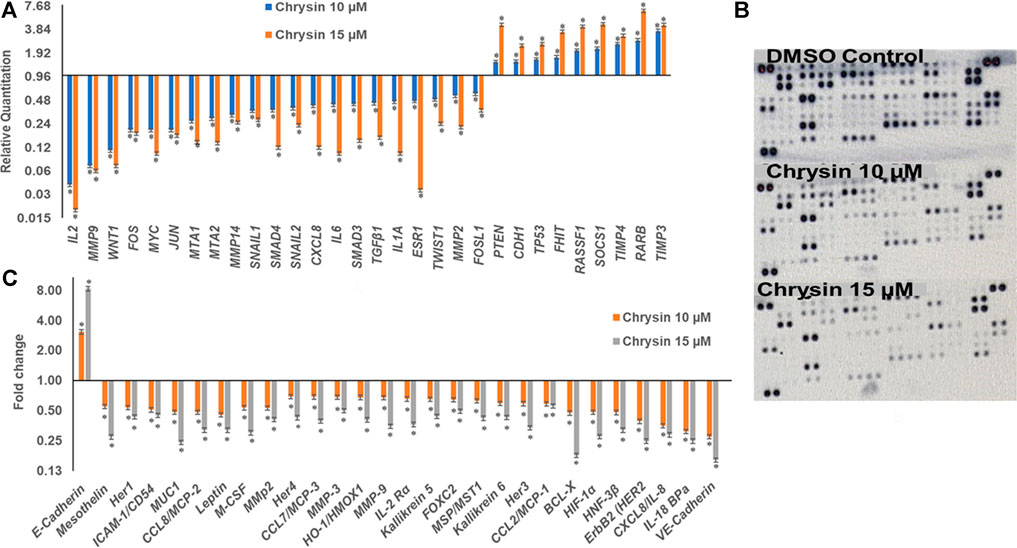
FIGURE 3. (A) Chrysin modulated the expression of various TSGs and migration related genes in a dose-dependent manner. The TSGs were reactivated, whereas inflammatory- and migration-related genes were downregulated. (B) The nitrocellulose membranes depicting the expression of different proteins. (C) Chrysin treatment modulates the proteins related to migration and inflammation in a dose-dependent manner.
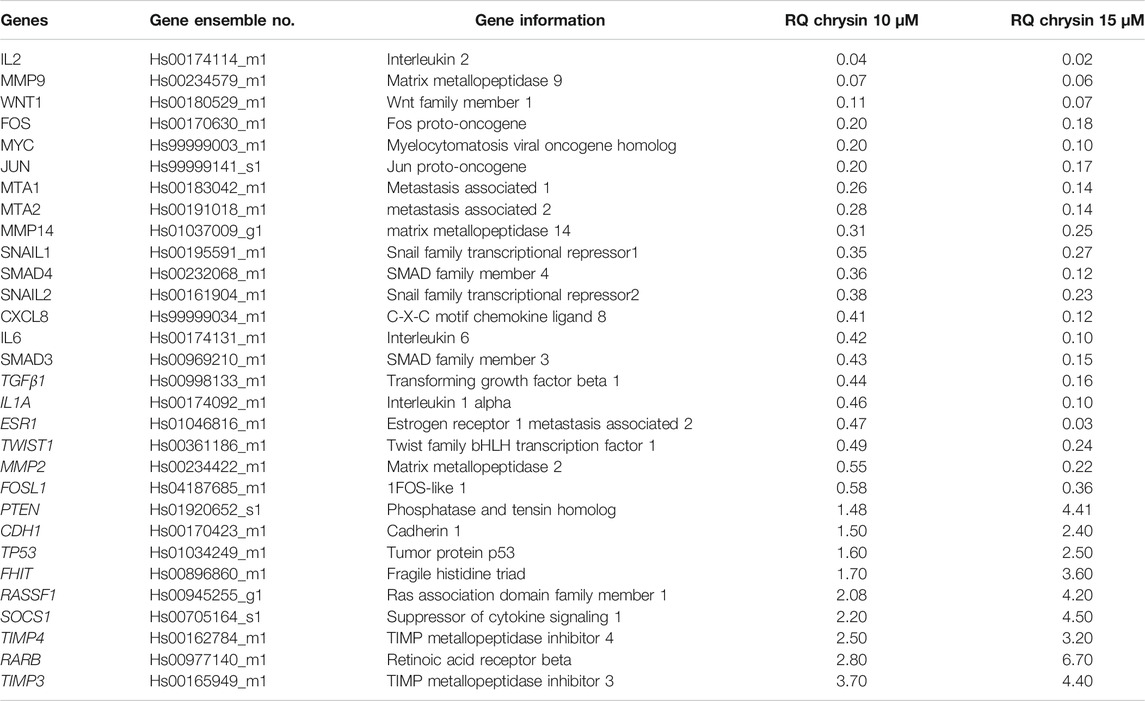
TABLE 1. Relative expression of TSGs and genes related to migration and metastasis. The values are taken as mean of three experiments ±SD (p ≤ 0.05).
Proteome profiler–based quantitation of proteins that are involved in proliferation and migration and other cellular events revealed chrysin-supported modulation was consistent with mRNA expression. The treatment of HeLa cells with 10 and 15 µM of chrysin resulted in the downregulation of the expression of various proteins related to migration viz MMP2, MMP9, and MMP3, mesothelin, MUC1, leptin, and M_CSF; inflammatory proteins like CCL8/MCP-2, CCL7/MCP-3, IL-18 BPa, CXCL8/IL-8, and IL-2 Rα; and oncogenes like HER1, 2, 3, and 4, and ICAM-1/CD54. Proteins related to cell proliferation, growth, and apoptosis like BCL-X, HIF-1α, HNF-3β, and HO-1/HMOX1 were also downregulated, that is, related to tumor progression, whereas upregulation of E-cadherin (CDH1) was observed after chrysin treatment (Figure 3B, Figure 3C and Table 2). Fold changes in protein expression were calculated by comparing the treated cells with those of the DMSO control. The upregulation was fixed at ≥1.5 fold and downregulation at ≤0.5 fold.
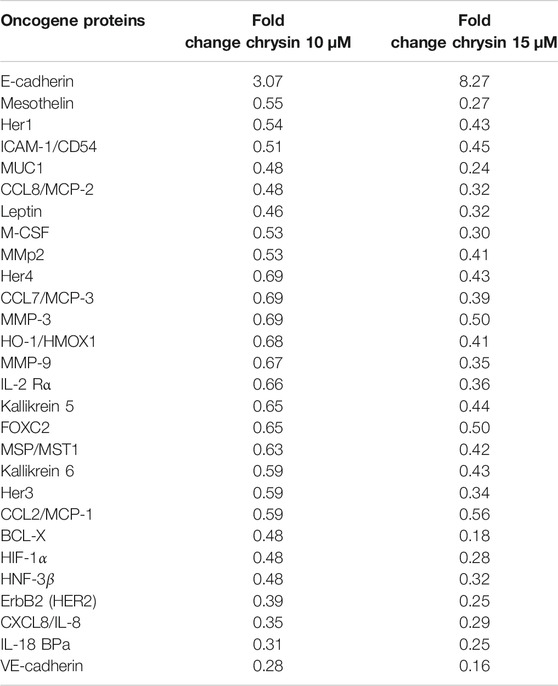
TABLE 2. Expression of proteins involved in migration and inflammation. The values are taken as mean of three experiments ±SD (p ≤ 0.05).
Chrysin inhibited DNMT activity in HeLa cells in a dose-dependent manner. The incubation of the nuclear extract with 5, 10, and 15 µM of chrysin resulted in the inhibition of DNMT activity by 35, 54, and 61%, respectively, compared to the DMSO control (Figure 4A).
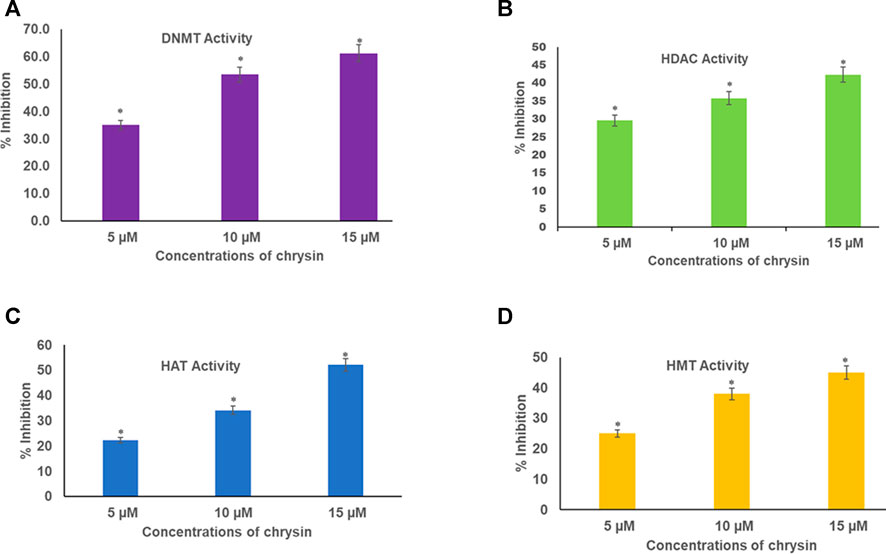
FIGURE 4. (A) Chrysin decreased DNMT activity in HeLa cells irrespective of their location on the genome in a concentration-dependent manner. (B) Chrysin inhibited HDAC activity in HeLa cells in a concentration-dependent manner. The activity decreased with increase in the dose of chrysin (C) Chrysin decreased the activity of HAT in a dose-dependent manner. As the concentration of chrysin increased, the inhibition percentage increased. (D) Chrysin decreased HMT H3K9 enzyme activity in HeLa cells, irrespective of their location on the genome in a concentration-dependent way.
Nuclear extracts were incubated with increasing concentrations (5, 10, and 15 µM) of chrysin; it was found that it inhibited the activity of HDACs by 30, 36, and 42% in a dose-dependent response compared with the DMSO control (Figure 4B).
Histone acetyl transferases cause acetylation at N-terminal tails of histone proteins. The incubation of nuclear extract with varying concentrations of chrysin (5, 10, and 15 µM) showed decline in HAT activity in the treated cells compared to the DMSO control. A decrease of 22, 34, and 52% was observed at 5, 10, and 15 µM conc. of chrysin treatment, respectively (Figure 4C).
HMT H3K9 can add methyl groups at histone three and lysine 9. All the methylation marks—mono, di, and trimethylation—are repressive marks. The incubation of HeLa cell nuclear extracts with 5, 10, and 15 µM conc. of chrysin reduced the activity of the enzyme by 25, 38, and 45%, respectively (Figure 4D).
RT (Rahman et al., 2016) Profiler™ PCR Array Human Epigenetic Chromatin Modification Enzymes (Catalog No. PAHS-085Z; Qiagen, USA) were used to check the expression of various chromatin-modifying enzymes following the treatment of chrysin (@ conc. 10 and 15 µM) for 48 h compared to the DMSO control. Chrysin treatment down-regulated the expression of DNA methyltransferases like DNMT1, 3A, and 3B significantly. HDAC1, 2, 3, 4, and 11 also showed a steep decline after the above-stated chrysin treatment. Remarkably, downregulation of WHSC1, AURKA, AURKB, and AURKX. EHM2, PRMT8, and HAT1 were also observed after 10 and 15 µM of chrysin treatment. However, enhanced expression of SETD2, ESC O 2, and CIITA was found after the same chrysin treatment (RQ in Figure 5A and Table 3).
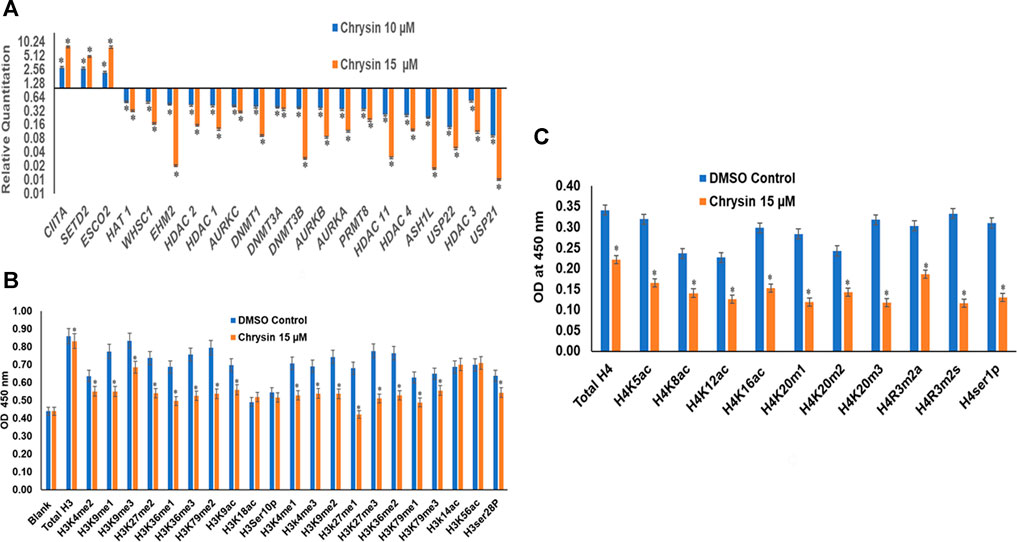
FIGURE 5. (A) Treatment of HeLa cells with chrysin at 10 and 15 µM for 48 h modulated the expression of epigenetic enzymes (HDACs, DNMTs, HATs, HMTs etc.) in a dose-dependent manner. (B) Chrysin modulates the H3 acetylation and methylation histone marks at 15–48 h. (C) H4 histone marks modulated by chrysin as compared to the DMSO controls.
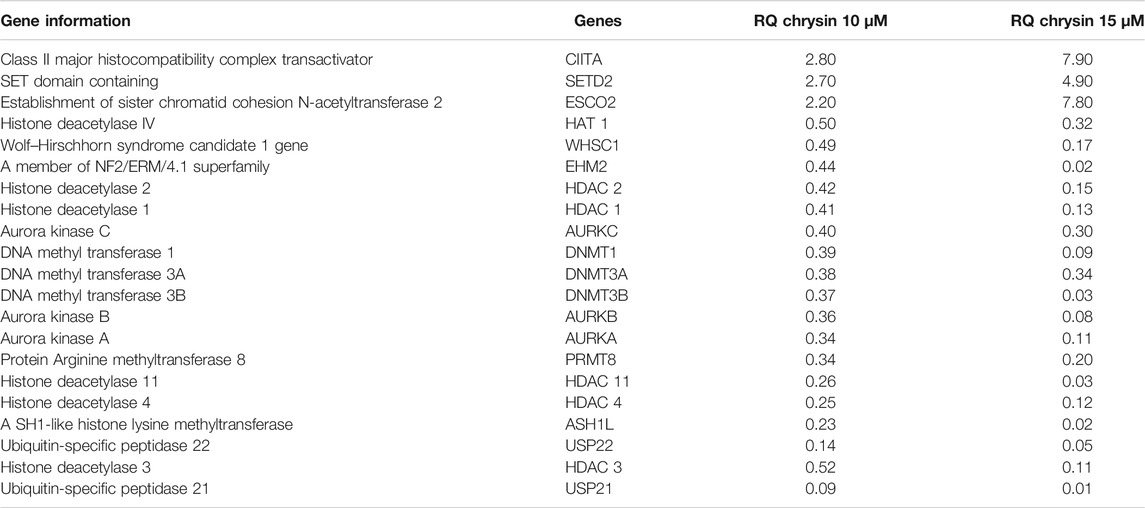
TABLE 3. RQ values of chromatin-modifying enzymes after chrysin treatment. The values are taken as mean of three experiments ±SD (p ≤ 0.05).
Chrysin modulated the expression of methylation, acetylation, and phosphorylation H3 and H4 marks. H3K9me1, H3K9me2, H3K9me3, H3K27me1, H3K27me2, H3K27me3, H3K36me1, H3K36me3, H3K79me1, H3K79me2, and H3K79me3 marks were reduced after the treatment of HeLa cells with 15 µM of chrysin for 48 h; similarly, H3 acetylation marks were diminished after treatment with 15 µM of chrysin (Figure 5B). The expression of H3K9ac, H3K18ac, H3K14ac, and H3K56ac was reduced after chrysin treatment. Likewise, the acetylation marks at H4 were also modulated against chrysin treatment including H4K5ac, H4K8ac, H4K12ac, and H4K16ac. H4 methylation marks, like H4K20me1, H4K20me2, and H4K20me3, showed decreased expression after chrysin treatment (Figure 5C). Phosphorylation marks of H3ser28p, H4ser10 p, H4R3m2a, and H4Rm2s were also decreasingly expressed after 15 µM chrysin treatment of HeLa cells for 48 h (Figure 5B and 5C).
An obvious decrease in global methylation was observed after 48 h treatment of 5, 10, and 15 µM of chrysin against HeLa cells. Global DNA methylation was studied by comparing with the DMSO control. This was reduced to 61, 44, and 30% against 5, 10, and 15 µM chrysin treatment of HeLa cells, respectively (Figure 6A).
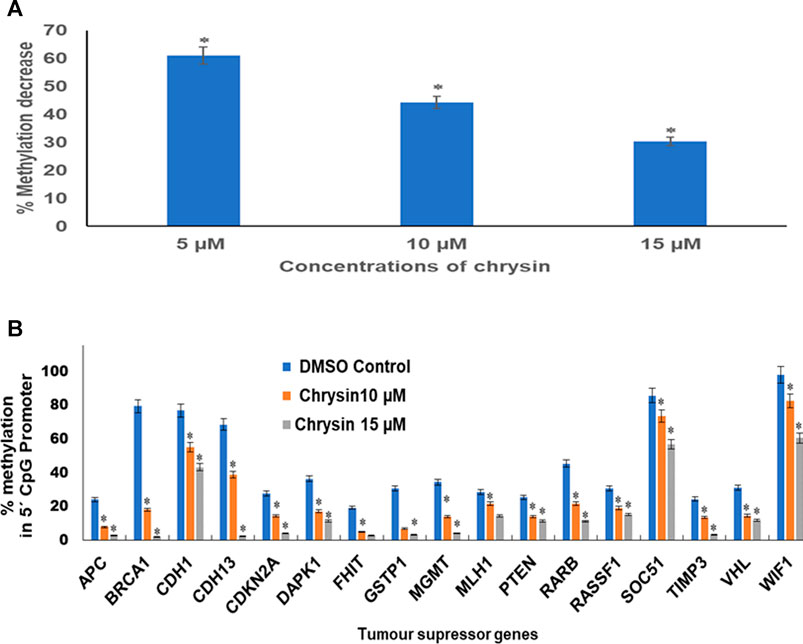
FIGURE 6. (A) Chrysin treatment at 5, 10, and 15 µM–48 h decreased the global DNA methylation in a dose-dependent manner. (B) Chrysin treatment of HeLa cells at 10 and 15 µM for 48 h demonstrated profound decrease in percent methylation in 5′ CpG promoter regions of TSGs as compared to the DMSO controls in a dose-dependent manner.
Methylation-sensitive restriction enzyme PCR revealed that chrysin decreased the promoter methylation of crucial tumor-suppressor genes of HeLa cells. These TSGs included APC, BRCA1, CDH1, PTEN, GSTP1, FHIT, DAPK1, CDH13, CDKN2A, MGMT, MLH1, RARB, RASSF1, SOCS1, VHL, WIFI, and TIMP3. The methylation percentage of the abovesaid genes decreased significantly as APC (8%, 3%), BRCA1 (18%, 2%), CDH1 (55%, 43%), CDH13 (39%, 3%), CDKN2A (14%, 4%), DAPK1 (17%, 11%), FHIT (5%, 3%), GSTP1 (7%, 3%), MGMT (14%, 4%) MLH1 (22%, 15%), PTEN (14%, 11%), RARB (22%, 11%), RASSF1 (19%, 15%), SOCS1 (73%, 56%), TIMP3 (13%, 3%), VHL (15%, 12%), and WIFI (82%, 61%) at 10 and 15 µM chrysin, respectively, compared to the DMSO control, wherein the methylation percentage was much higher (Figure 6B and Table 4).
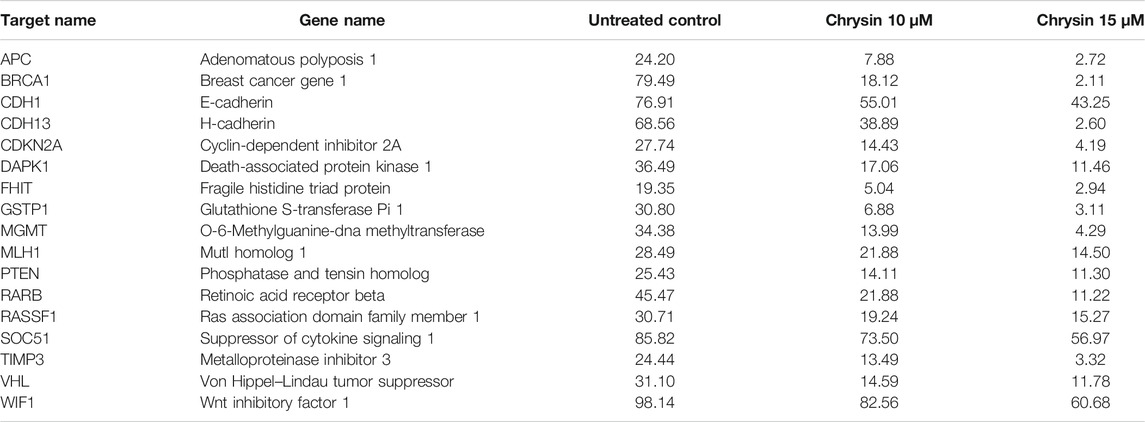
TABLE 4. Percentage of CpG promoter methylation after chrysin treatment as compared to untreated control. The values are taken as mean of three experiments ±SD (p ≤ 0.05).
Epigenetic alterations are commonly associated with carcinogenesis and metastasis (Wang et al., 2018a). Cancer metastasis is the major cause of treatment failure and mortality in women detected with cervical cancer. This suggests that the inhibition of metastasis serves a pivotal role in survival improvement, and hence can be exploited as a potential target for cancer treatment and prevention (Chatterjee et al., 2018). Additionally, epigenetic alteration in key metastatic genes is one of the reasons of metastasis (Chatterjee et al., 2018). The modifiable nature of epigenetics makes the epigenetic regulation an attractive target for cancer prevention and treatment (Busch et al., 2015; Kanwal et al., 2015). Currently available synthetic drugs direct at crucial epigenetic signature enzymes, for example, HDACs and DNMTs. Nevertheless, these synthetic drugs have shown many adverse side effects (Ho et al., 2013; Heerboth et al., 2014); therefore, it is crucial to probe the natural agents which are derived from plants that can regulate all cell processes including epigenetic mechanisms and can potentially reverse malignancy-associated epigenetic patterns (Shankar et al., 2016).
Earlier, studies on flavonoids targeting various types of cancer have demonstrated their anticancer effect by modulation of various molecular pathways involved in migration and epigenetics (Pal-Bhadra et al., 2012; Kanwal et al., 2016; Liskova et al., 2020). Considering the anticancer potential of plant flavonoids in view, the present study was carried out to explore the antiproliferative, antimigratory, and modulatory effects of chrysin on DNA methylation and histone modification on human cervical cancer (HeLa) cells. Previously, it was reported from the lab that chrysin inhibits the proliferation of HeLa cells in a dose- and time-dependent manner, induces apoptosis, and modulates various signaling pathways (Raina et al., 2021).
In the present study, 5 and 10 µM chrysin-treated HeLa cells showed reduced colony formation as 120 and 30 colonies were formed, respectively, after 48 h, whereas at 15 µM chrysin, there was no noticeable colony formation (Figure 1A and 1B). To understand chrysin-mediated antimigratory effects, the scratch-wound assay that depicted significant inhibition of cell migration was carried out. Chrysin treatment of HeLa cells at a conc. of 10 and 15 µM showed wound width increment by 8 and 17%, respectively, after 24 h incubation, whereas the wound width was increased by 14 and 25% after 48 h compared to the DMSO control, where complete wound closure was found after 48 h (Figure 2A and 2B). The trans-well assay also depicted the inhibition of migration at varying concentrations of chrysin (Figure 2C and Figure 2D). Hence, it can be inferred that chrysin at 10 and 15 μM at 48 h is a strong inhibitor of migration. Earlier studies from various research groups have also reported that chrysin bears antiproliferative and cytostatic effects and inhibits migration and invasion in various cancer cell lines (Kasala et al., 2015; Wang et al., 2018b).
Matrix metalloproteinases are important proteolytic enzymes involved in the cancer cell invasion process. Epithelial mesenchymal transition (EMT) increases cell migration, and the transforming growth factor pathway has an important role to play in epithelial mesenchymal transition; it induces EMT either by transforming growth factor-β (TGF-β)/SMAD pathway or via the non-SMAD pathway by activating the AKT/PI3K pathway, thus triggering migration (Wu et al., 2021; Xue et al., 2012). The downregulation of TGF-β/SMAD mediates reduction in the expression of MMPS and TWIST1 (Wu et al., 2021; Xue et al., 2012; Baruah et al., 2016). Cadherins are a class of type-1 transmembrane proteins that maintain the adhesion between cells, their loss lead to invasion and metastasis, and snails are their inhibitors. MTA1 and MTA2 are metastasis promoters, and their reduction leads to the inhibition of metastasis (Wu et al., 2021; Xue et al., 2012; Baruah et al., 2016). In the study, chrysin-mediated inhibition of migration was found to be well correlated with the downregulation of metalloproteases MMP 9, MMP 2, and MMP 14 and their co-operators like SMAD3, SMAD4, SNAIL1, SNAIL2, MTA1, and MTA2, and upregulation of their inhibitors like TIMP3, TIMP4, and CDH1, hence endorsing the inhibitory effect of chrysin on migration. In addition, the downregulation of genes related to inflammation like IL2, IL1A, and IL6 and oncogenes like Fos, Jun, Myc, WNT1, and FOSL1 (Figure 3A and Table 1) was observed. Inflammation subsequent to viral infection is a power tool that accelerates cancer progression; hence, downregulation of inflammatory proteins aids in cancer prevention and treatment (Deivendran et al., 2014).
In the study, gene expression studies at mRNA levels were found to be consistent with their protein level expression, and various genes involved in inflammation and migration such as HER1, HER2, Her3, and Her4, and MMPs like MMP 9, MMP 2, MMP 3, FOXC2, IL-2 Rα, IL-18 BPa, CXCL8/IL-8, CCL2/MCP-1, CL8/MCP-2, CCL7/MCP-3, Mesothelin, ICAM-1/CD54, MUC1, Leptin, M-CSF, H O-1/HMOX1, Kallikrein 5, MSP/MST1, Kallikrein 6, HIF-1α, and HNF-3β were significantly downregulated against chrysin treatment, while upregulation of E-cadherins was observed at different chrysin concentrations (Figure 3B, 3C and Table 2). Our current findings are in line with the available reports wherein in vitro/in vivo models have shown that chrysin inhibits tumor metastasis by decreasing the expression of MMP9 and MMP2 as well as COX-2 and i-NOS, and modulates the PI3K/AKT signaling pathway (Yang et al., 2014; Xia et al., 2015; Koosha et al., 2016; Zam and Khadour, 2017).
Furthermore, epigenetic modulations induced by chrysin were also analyzed in this study. Epigenetic modifications including aberrant DNA methylation and histone modifications interactions are crucial for controlling the operational activities of the genome by changing the chromatin structure, thereby leading to silencing of various tumor-suppressor genes (Guo et al., 2015; Chistiakov et al., 2017). DNMTs catalyze the transfer of the acetyl group onto 5′cytosine at promoter CpG island of TSGs. Hypermethylation of CpG islands in the promoter region of TSGs leads to silencing of these genes (Ali Khan et al., 2015). DNMTs are found to be upregulated in cervical cancer cells, and their expression levels are correlated to disease progression (Piyathilake et al., 2017; Charostad et al., 2019). The analysis of biochemical activity of DNMTs after chrysin treatment was performed, and it was observed that chrysin decreased the biochemical activity of DNMTs in a dose-dependent manner, and it was reduced by 35, 53.5, and 61.2% at 5, 10, and 15 µM chrysin treatment, respectively (Figure 4A).
Furthermore, the downregulation of DNMT activity against chrysin treatment was verified by downregulation of DNMT1, DNMT3A, and DNMT3B in a dose-dependent manner at transcript levels (Figure 5A and Table 3). Various flavonoids have attracted attention because of their chemopreventive and antitumor effects including chrysin, luteolin, and apigenin, and are known to inhibit DNMTs and histone methyl transferases (Busch et al., 2015; Kanwal et al., 2016). The decrease in DNMT expression after chrysin treatment was well correlated with the decrease in global DNA methylation (Figure 6A) and modulation pattern of CpG promoter methylation of TSGs (APC, BRCA1, FHIT, CDH1, CDH13, MGMT, MLH1, GSTP1, TIMP3, RARB, RASSIF1, SOCS1, PTEN, VHL, and WIFI) (Figure 6B) after chrysin treatment of 10 and 15 µM for 48 h. This study revealed that chrysin treatment downregulated global DNA methylation levels by decreasing to 61, 44, and 30% at 5, 10, and 15 µM chrysin treatment, respectively, for 48 h compared to DMSO controls (Figure 6A). Hypermethylation of TSGs leads to silencing of these genes and has been found to be well correlated with the overexpression of various DNMTs in cervical cancer (Guo et al., 2015; Jiménez–wences et al., 2014). Interestingly, this is the very first time it has been reported that chrysin treatment significantly decreases the methylation levels at the promoter region of several TSGs viz APC, CDH1, CDH13, BRCA1, CDKN2A, DAPK1, FHIT, GSTP1, MGMT, MLH1, PTEN, RARB, RASSF1, SOCS1, TIMP3, and WIFI (Figure 6B and Table 4), which are found to be hypermethylated in many cancers and have critical roles to play in various cellular processes (Mukherjee et al., 2015). Hypermethylation of PTEN and RASSIF1 is a common feature in cervical cancers. PTEN has an important role in cell migration and proliferation, and inhibits migration by being the antagonist of MMPs (Salimi Sartakhti et al., 2017), whereas reduced RASSIF1 quenches cell death by the receptor mode. VHL is important for stabilization of HIF1 and HIF2, and methylation of other TSGs like RARB and FHIT leads to uncontrolled proliferation; GSTP1 is involved in detoxification of harmful compounds, and MGMT is important for DNA repair (Mukherjee et al., 2015). A reduction of CpG methylation at the abovementioned gene loci can be correlated to reactivation of these genes at the transcription level; chrysin treatment upregulated the expression of PTEN, CDH1, TP53, FHIT, RASSIFI, SOCS1, RARB, TIMP3, and TIMP4 (Figure 3A) in our study. Several polyphenols including chrysin and luteolin have shown modulation of methylation, and thus reactivation of the silenced TSGs (Aggarwal et al., 2015; Ali Khan et al., 2015; Busch et al., 2015; Kanwal et al., 2016; Carlos-Reyes et al., 2019).
Apart from DNA modification, histone modifications like histone acetylation and histone methylation influence the expression of various genes that have an important role in cancer cell proliferation and migration (Chakravarthy et al., 2005; Dueñas–González et al., 2005; Soto et al., 2017). HDACs deacetylate histone and non-histone proteins such as TP53, rendering them non-functional (Chakrabarti et al., 2015). Overexpression of HDAC1, HDAC2, HDAC3, and HDAC6 has been reported in different cancers (Kogan et al., 2017; Lin et al., 2009; Huang et al., 2005; Zhang et al., 2016; Ahn and Yoon, 2017). HDAC overexpression together with DNA methylation and other histone modifications silences tumor-suppressor genes (Rose and Klose, 2014; Jenuwein and Allis, 2001). In our study, chrysin was found to decrease the expression of various HDACs (1, 2, 3, 11, and 4), HAT1, EHM2, AURKA, AURKB, PRMT 8, ASH1l, USP21, and USP22 at the transcript level, and increased the expression of ESCO2 and CIITA in a dose-dependent manner (RQs are given in Table 3 and Figure 5A). Chrysin treatment decreased the HDAC activity in a dose-dependent manner; HDAC activity decreased by 30, 36, and 42% after 5, 10, and 15 µM chrysin treatment, respectively (Figure 4B). This was further endorsed by decrease in the expression at the transcript level of HDACs 1, 2, 3, 4, and 11 in a significant manner at 10 and 15 µM chrysin treatment for 48 h (Figure 5A). HAT1 is upregulated in cervical cancer and is responsible for the induction of colony formation (Kedhari Sundaram et al., 2019b). Chrysin downregulated the activity of HAT by 22, 34, and 52% at 5, 10, and 15 µM chrysin, respectively (Figure 4C). It also reduced the expression of HAT in a dose-dependent manner at the transcript level with RQ of 0.31 at 15 µM (Figure 5A) and thus correlated with complete inhibition of colony formation. Similar results were reported by other researchers who observed inhibition of HDAC two and eight and upregulation of H3acK14, H4acK12, and H4acK16, and decrease in H3me2K9 methylation in different cell lines (Pal-Bhadra et al., 2012; Sun et al., 2012).
CIITA and ESCO2 were up-regulated after chrysin treatment (Figure 5A). CIITA positively regulates the expression of class II major histocompatibility complex and is often found to be methylated in cancer cells (Ramia et al., 2019). ESCO2 histone acetyltransferase curbs MMP2 and also encourages apoptosis in cancer cells (Guo et al., 2018). Trimethylation of lysine 9 and 27 of histone 3 (H3K9 and H3K27) at the promoter region is related to reduced TSG expression (Trievel, 2004; Daniel et al., 2005; Lachner et al., 2001). These marks are found to be overactive in cervical cancer (Chen et al., 2017). Remarkably, in the current study, chrysin downregulated all mono, di, and trimethylation marks at H3K9 (Figure 5B), and this was further verified by the assessment of H3K9 methyltransferase activity after incubation of HeLa cells with chrysin. Also, it was found that H3K9 HMT activity was significantly reduced by 25, 38, and 45% against 5, 10, and 15 µM chrysin treatment (Figure 4D). Moreover, chrysin also downregulated EHM2 expression (Figure 5A), which is responsible for methylation of H3K9. All the methylation marks at H3K27, H3K36, H3K79, and H3K4 were downregulated after chrysin treatment (Figure 5B). H3K4 mono and demethylation are related to transcription activation (Chang and Yu, 2016b). H4 acetylation marks such as H4K5, H4K8, H4K12, and H4k16 were also reduced by chrysin (Figure 5C). This is in line with the previously published report on a flavone luteolin, wherein it blocks the acetylation of histone H4 and controls the activity of c-FOS, p21, and other genes related to cell cycle control (Izzo et al., 2020). Thus, it can be suggested that chrysin is a potent inhibitor of DNA methyl transferases and histone methyltransferases, and thus modulates the methylation of TSGs.
The overexpression of any one of the EMT inducers such as Twist TGF-β1 or Snail upregulates FOXC2 expression and can lead to the initiation of EMT (Mani et al., 2007). In fact, a significant cadherin switch from E-cadherin to N-cadherin is expressed in cancer progression (Kouzarides, 2007). VE-cadherin, another cadherin, mediates cell-to-cell bonding by holding the catenin in between, which in turn connects the actin cytoskeleton of the cells. Both E cadherin and VE-cadherin are down-regulated in cancer progression (Ramis-Conde et al., 2009). It was observed that chrysin (10 and 15 µM for 48 h) decreased the expression of SNAIL, TWIST (Figure 3A), and FOXC2 (Figure 3B and 3C), and increased the expression of E cadherin (Figure 3B). The suppressor of cytokine signaling 1 (SOCS1) is a tumor-suppressor gene and suppresses cytokine signaling and destroys the HPV E7 protein. SOCS1 is hypermethylated in cervical cancer and renewal of its expression upsurges Rb protein thereby inhibits cell proliferation (Kamio et al., 2004; Sobti et al., 2011). A decrease in E cadherin can be linked to WNT signaling which prevents phosphorylation of SNAIL, allowing it to accumulate and repress cadherin (Loh et al., 2019). Based upon the findings from the present study, it can be proposed that the restoration of transcription in the tumor-suppressor genes plays a crucial role in the anticancer potential of chrysin against HeLa cells, as it can directly influence cell proliferation and cell migration. Our current results are based on the chrysin efficacy on HeLa cells, but can further be extrapolated on other cell lines and animal models.
Chrysin appears to be a promising natural chemopreventive agent which is cytotoxic to cancer cells and inhibits migration, diminishes CpG promoter methylation of TSG modulates, and causes re-expression of TSG, and downregulation of genes related to migration and inflammation. Hence, chrysin can be exploited for further use at a clinical setup after experimental validation and checking its pharmacokinetic properties involving human subjects.
The original contributions presented in the study are included in the article, further inquiries can be directed to the corresponding authors.
RR, NA, and AH contributed to conception, design of study, experimental work, and manuscript preparation; AA and SB contributed to data validation and review; SH and SF contributed to data validation and statistical analysis.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors, and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The authors are thankful to MAHE DUBAI Internal research grant (R and DP/MAHE DUBAI/RL-02/19) for the financial support. The author (AM) is thankful to the Deanship of Scientific Research, Qassim University, for funding the publication of this project.
Aggarwal, R., Jha, M., Shrivastava, A., and Jha, A. K. (2015). Natural Compounds: Role in Reversal of Epigenetic Changes. Biochem. Mosc. 80, 972–989. doi:10.1134/S0006297915080027
Ahn, M.-Y., and Yoon, J.-H. (2017). Histone Deacetylase 7 Silencing Induces Apoptosis and Autophagy in Salivary Mucoepidermoid Carcinoma Cells. J. Oral Pathol. Med. 46, 276–283. doi:10.1111/jop.12560
Ali Khan, M., Kedhari Sundaram, M., Hamza, A., Quraishi, U., Gunasekera, D., Ramesh, L., et al. (2015). Sulforaphane Reverses the Expression of Various Tumor Suppressor Genes by Targeting DNMT3B and HDAC1 in Human Cervical Cancer Cells. Evidence-Based Complement. Altern. Med. 2015, 1–12. doi:10.1155/2015/412149
Andrijauskaite, K., Morris, J., and Wargovich, M. J. (2019). “Natural Anticancer Agents,” in Epigenetics of Cancer Prevention (Elsevier), 49–73. doi:10.1016/b978-0-12-812494-9.00003-2
Baruah, M. M., Khandwekar, A. P., and Sharma, N. (2016). Quercetin Modulates Wnt Signaling Components in Prostate Cancer Cell Line by Inhibiting Cell Viability, Migration, and Metastases. Tumor Biol. 37, 14025–14034. doi:10.1007/s13277-016-5277-6
Busch, C., Burkard, M., Leischner, C., Lauer, U. M., Frank, J., and Venturelli, S. (2015). Epigenetic Activities of Flavonoids in the Prevention and Treatment of Cancer. Clin. Epigenet. 7, 64. doi:10.1186/s13148-015-0095-z
Carlos-Reyes, Á., López-González, J. S., Meneses-Flores, M., Gallardo-Rincón, D., Ruíz-García, E., Marchat, L. A., et al. (2019). Dietary Compounds as Epigenetic Modulating Agents in Cancer. Front. Genet. 10, 1–14. doi:10.3389/fgene.2019.00079
Chakrabarti, A., Oehme, I., Witt, O., Oliveira, G., Sippl, W., Romier, C., et al. (2015). HDAC8: a Multifaceted Target for Therapeutic Interventions. Trends Pharmacol. Sci. 36, 481–492. doi:10.1016/j.tips.2015.04.013
Chakravarthy, S., Park, Y.-J., Chodaparambil, J., Edayathumangalam, R. S., and Luger, K. (2005). Structure and Dynamic Properties of Nucleosome Core Particles. FEBS Lett. 579, 895–898. doi:10.1016/j.febslet.2004.11.030
Chang, L.-C., and Yu, Y.-L. (2016). Dietary Components as Epigenetic-Regulating Agents against Cancer. BioMed 6, 9–16. doi:10.7603/s40681-016-0002-8
Chang, L. C., and Yu, Y. L. (2016). Dietary Components as Epigenetic-Regulating Agents against Cancer. Biomedicine (Taipei) 6, 2–8. doi:10.7603/s40681-016-0002-8
Charostad, J., Astani, A., Goudarzi, H., and Faghihloo, E. (2019). DNA Methyltransferases in Virus-Associated Cancers. Rev. Med. Virol. 29, e2022. doi:10.1002/rmv.2022
Chatterjee, A., Rodger, E. J., and Eccles, M. R. (2018). Epigenetic Drivers of Tumourigenesis and Cancer Metastasis. Semin. Cancer Biol. 51, 149–159. doi:10.1016/j.semcancer.2017.08.004
Chen, R.-J., Shun, C.-T., Yen, M.-L., Chou, C.-H., and Lin, M.-C. (2017). Methyltransferase G9a Promotes Cervical Cancer Angiogenesis and Decreases Patient Survival. Oncotarget 8, 62081–62098. doi:10.18632/oncotarget.19060
Chistiakov, D. A. D., Myasoedova, V. A. V., Orekhov, A. N., and Bobryshev, Y. V. (2017). Epigenetically Active Drugs Inhibiting DNA Methylation and Histone Deacetylation. Cpd 23, 1167–1174. doi:10.2174/1381612822666161021110827
Crowley, L. C., Marfell, B. J., Scott, A. P., Boughaba, J. A., Chojnowski, G., Christensen, M. E., et al. (2016). Dead Cert: Measuring Cell Death. Cold Spring Harb. Protoc. 2016, pdb.top070318–1072. doi:10.1101/pdb.top070318
Daniel, J. A., Pray-Grant, M. G., and Grant, P. A. (2005). Effector Proteins for Methylated Histones: an Expanding Family. Cell Cycle 4, 919. doi:10.4161/cc.4.7.1824
Deivendran, S., Marzook, K. H., and Radhakrishna Pillai, M. (2014). The Role of Inflammation in Cervical Cancer. Adv. Exp. Med. Biol. 816, 377–399. doi:10.1007/978-3-0348-0837-8_15
Dueñas-González, A., Lizano, M., Candelaria, M., Cetina, L., Arce, C., and Cervera, E. (2005). Epigenetics of Cervical Cancer. An Overview and Therapeutic Perspectives. Mol. Cancer 4, 1–24. doi:10.1186/1476-4598-4-38
Ganai, S. A., Sheikh, F. A., Baba, Z. A., Mir, M. A., Mantoo, M. A., and Yatoo, M. A. (2021). Anticancer Activity of the Plant Flavonoid Luteolin against Preclinical Models of Various Cancers and Insights on Different Signalling Mechanisms Modulated. Phytotherapy Res. 35, 3509–3532. doi:10.1002/ptr.7044
Ganai, S. A., Sheikh, F. A., and Baba, Z. A. (2021). Plant Flavone Chrysin as an Emerging Histone Deacetylase Inhibitor for Prosperous Epigenetic‐based Anticancer Therapy. Phytotherapy Res. 35, 823–834. doi:10.1002/ptr.6869
Guo, X.-B., Huang, B., Pan, Y.-H., Su, S.-G., and Li, Y. (2018). ESCO2 Inhibits Tumor Metastasis via Transcriptionally Repressing MMP2 in Colorectal Cancer. Cmar 10, 6157–6166. doi:10.2147/cmar.s181265
Guo, Y., Su, Z.-Y., and Kong, A.-N. T. (2015). Current Perspectives on Epigenetic Modifications by Dietary Chemopreventive and Herbal Phytochemicals. Curr. Pharmacol. Rep. 1, 245–257. doi:10.1007/s40495-015-0023-0
Hatzimichael, E., and Crook, T. (2013). Cancer Epigenetics: New Therapies and New Challenges. J. Drug Deliv. 2013, 529312. doi:10.1155/2013/529312
Heerboth, S., Lapinska, K., Snyder, N., Leary, M., Rollinson, S., and Sarkar, S. (2014). Use of Epigenetic Drugs in Disease: An Overview. Genet. Epigenet. 6, GEG.S12270–19. doi:10.4137/GeG.s12270
Herranz, M., and Esteller, M. (2007). “DNA Methylation and Histone Modifications in Patients with Cancer,” in Target Discovery and Validation Reviews and Protocols (Springer), 25–62.
Herranz, M., and Esteller, M. (2007). DNA Methylation and Histone Modifications in Patients with Cancer: Potential Prognostic and Therapeutic Targets. Methods Mol. Biol. 361, 25–62. doi:10.1385/1-59745-208-4:25
Ho, A. S., Turcan, S., and Chan, T. A. (2013). Epigenetic Therapy: Use of Agents Targeting Deacetylation and Methylation in Cancer Management. Onco Targets Ther. 6, 223–232. doi:10.2147/OTT.S34680
Huang, B. H., Laban, M., Leung, C. H.-W., Lee, L., Lee, C. K., Salto-Tellez, M., et al. (2005). Inhibition of Histone Deacetylase 2 Increases Apoptosis and p21Cip1/WAF1 Expression, Independent of Histone Deacetylase 1. Cell Death Differ. 12, 395–404. doi:10.1038/sj.cdd.4401567
Huang, J., Plass, C., and Gerhauser, C. (2011). Cancer Chemoprevention by Targeting the Epigenome. Cdt 12, 1925–1956. doi:10.2174/138945011798184155
Izzo, S., Naponelli, V., and Bettuzzi, S. (2020). Flavonoids as Epigenetic Modulators for Prostate Cancer Prevention. Nutrients 12, 1010–1024. doi:10.3390/nu12041010
Jenuwein, T., and Allis, C. D. (2001). Translating the Histone Code. Science 293, 1074–1080. doi:10.1126/science.1063127
Jiménez-wences, H., Peralta-Zaragoza, O., and Fernández-tilapa, G. (2014). Human Papilloma Virus, DNA Methylation and microRNA Expression in Cervical Cancer (Review). Oncol. Rep. 31, 2467–2476. doi:10.3892/or.2014.3142
Kai, L., Samuel, S. K., and Levenson, A. S. (2010). Resveratrol Enhances P53 Acetylation and Apoptosis in Prostate Cancer by Inhibiting MTA1/NuRD Complex. Int. J. Cancer 126, 1538–1548. doi:10.1002/ijc.24928
Kamio, M., Yoshida, T., Ogata, H., Douchi, T., Nagata, Y., Inoue, M., et al. (2004). SOC1 Inhibits HPV-E7-Mediated Transformation by Inducing Degradation of E7 Protein. Oncogene 23, 3107–3115. doi:10.1038/sj.onc.1207453
Kanwal, R., Gupta, K., and Gupta, S. (2015). “Aberrant DNA Methylation Is One of the Most Important Epigenetic Modifications in Cancer Cells and it Is Also Associated with Histone Modifications and Their Interaction of Is Crucial to Regulate the Functioning of the Genome by Changing Chromatin Archite,” in Cancer Epigenetics (Springer), 3–25.
Kanwal, R., Datt, M., Liu, X., and Gupta, S. (2016). Dietary Flavones as Dual Inhibitors of DNA Methyltransferases and Histone Methyltransferases. PLoS One 11, e0162956. doi:10.1371/journal.pone.0162956
Kasala, E. R., Bodduluru, L. N., Madana, R. M., V, A. K., Gogoi, R., Barua, C. C., et al. (2015). Chemopreventive and Therapeutic Potential of Chrysin in Cancer: Mechanistic Perspectives. Toxicol. Lett. 233, 214–225. doi:10.1016/j.toxlet.2015.01.008
Kedhari Sundaram, M., Raina, R., Afroze, N., Bajbouj, K., Hamad, M., Haque, S., et al. (2019). Quercetin Modulates Signaling Pathways and Induces Apoptosis in Cervical Cancer Cells. Biosci. Rep. 39. doi:10.1042/BSR20190720
Kedhari Sundaram, M., Hussain, A., Haque, S., Raina, R., and Afroze, N. (2019). Quercetin Modifies 5′CpG Promoter Methylation and Reactivates Various Tumor Suppressor Genes by Modulating Epigenetic marks in Human Cervical Cancer Cells. J. Cel. Biochem. 120, 18357–18369. doi:10.1002/jcb.29147
Khoo, B. Y., Chua, S. L., and Balaram, P. (2010). Apoptotic Effects of Chrysin in Human Cancer Cell Lines. Ijms 11, 2188–2199. doi:10.3390/ijms11052188
Kim, S. O., and Kim, M. R. (2013). (-)-Epigallocatechin 3-gallate Inhibits Invasion by Inducing the Expression of Raf Kinase Inhibitor Protein in AsPC-1 Human Pancreatic Adenocarcinoma Cells through the Modulation of Histone Deacetylase Activity. Int. J. Oncol. 42, 349–358. doi:10.3892/ijo.2012.1686
Kogan, E. A., Unanyan, A. L., Kadyrova, A. E., Demura, T. A., Sidorova, I. S., Faizullin, R. I., et al. (2017). Immunohistochemical Analysis of Epigenetic Markers in Cervical Pathologies Associated with Human Papillomavirus Infection. BioNanoSci. 7, 284–287. doi:10.1007/s12668-016-0339-1
Koosha, S., Alshawsh, M. A., Looi, C. Y., Seyedan, A., and Mohamed, Z. (2016). An Association Map on the Effect of Flavonoids on the Signaling Pathways in Colorectal Cancer. Int. J. Med. Sci. 13, 374–385. doi:10.7150/ijms.14485
Kouzarides, T. (2007). Chromatin Modifications and Their Function. Cell 128, 693–705. doi:10.1016/j.cell.2007.02.005
Lachner, M., O'Carroll, D., Rea, S., Mechtler, K., and Jenuwein, T. (2001). Methylation of Histone H3 Lysine 9 Creates a Binding Site for HP1 Proteins. Nature 410, 116–120. doi:10.1038/35065132
Li, L-L., and Wang, S-S. (2017). DNA Methyltransferase (DNMTs) Expression in Cervical Cancer Tissues and its Relationship with HPV Infection and Tumor Malignancy. J. Hainan Med. Univ. 23, 136–139.
Lim, W., Ryu, S., Bazer, F. W., Kim, S.-M., and Song, G. (2018). Chrysin Attenuates Progression of Ovarian Cancer Cells by Regulating Signaling Cascades and Mitochondrial Dysfunction. J. Cel. Physiol. 233, 3129–3140. doi:10.1002/jcp.26150
Lin, Z., Bazzaro, M., Wang, M.-C., Chan, K. C., Peng, S., and Roden, R. B. S. (2009). Combination of Proteasome and HDAC Inhibitors for Uterine Cervical Cancer Treatment. Clin. Cancer Res. 15, 570–577. doi:10.1158/1078-0432.CCR-08-1813
Liskova, A., Koklesova, L., Samec, M., Smejkal, K., Samuel, S. M., Varghese, E., et al. (2020). Flavonoids in Cancer Metastasis. Cancers 12, 1498. doi:10.3390/cancers12061498
Loh, C.-Y., Chai, J., Tang, T., Wong, W., Sethi, G., Shanmugam, M., et al. (2019). The E-Cadherin and N-Cadherin Switch in Epithelial-To-Mesenchymal Transition: Signaling, Therapeutic Implications, and Challenges. Cells 8, 1118. doi:10.3390/cells8101118
Mani, S. A., Yang, J., Brooks, M., Schwaninger, G., Zhou, A., Miura, N., et al. (2007). Mesenchyme Forkhead 1 (FOXC2) Plays a Key Role in Metastasis and Is Associated with Aggressive Basal-like Breast Cancers. Proc. Natl. Acad. Sci. 104, 10069–10074. doi:10.1073/pnas.0703900104
McLaughlin-Drubin, M. E., Crum, C. P., and Münger, K. (2011). Human Papillomavirus E7 Oncoprotein Induces KDM6A and KDM6B Histone Demethylase Expression and Causes Epigenetic Reprogramming. Proc. Natl. Acad. Sci. USA 108, 2130–2135. doi:10.1073/pnas.1009933108
Mocanu, M.-M., Nagy, P., and Szöllősi, J. (2015). Chemoprevention of Breast Cancer by Dietary Polyphenols. Molecules 20, 22578–22620. doi:10.3390/molecules201219864
Mukherjee, N., Kumar, A. P., and Ghosh, R. (2015). DNA Methylation and Flavonoids in Genitourinary Cancers. Curr. Pharmacol. Rep. 1, 112–120. doi:10.1007/s40495-014-0004-8
Ong, T. P., Moreno, F. S., and Ross, S. A. (2011). Targeting the Epigenome with Bioactive Food Components for Cancer Prevention. J. Nutrigenet. Nutrigenomics 4, 275–292. doi:10.1159/000334585
Pal-Bhadra, M., Ramaiah, M. J., Reddy, T. L., Krishnan, A., Pushpavalli, S., Babu, K. S., et al. (2012). Plant HDAC Inhibitor Chrysin Arrest Cell Growth and Induce P21 WAF1 by Altering Chromatin of STAT Response Element in A375 Cells. BMC Cancer 12, 180. doi:10.1186/1471-2407-12-180
Paredes-Gonzalez, X., Fuentes, F., Su, Z.-Y., and Kong, A.-N. T. (2014). Apigenin Reactivates Nrf2 Anti-oxidative Stress Signaling in Mouse Skin Epidermal JB6 P + Cells through Epigenetics Modifications. AAPS J. 16, 727–735. doi:10.1208/s12248-014-9613-8
Piyathilake, C., Badiga, S., Borak, S., Weragoda, J., Bae, S., Matthews, R., et al. (2017). A Higher Degree of Expression of DNA Methyl Transferase 1 in Cervical Cancer Is Associated with Poor Survival Outcome. Ijwh 9, 413–420. doi:10.2147/IJWH.S133441
Rahman, M. S., Jamil, H. M., Akhtar, N., Islam, R., Abdul-Awal, S. M., Rana, M. M., et al. (2016). Cancer Epigenetics and Epigenetical Therapy. J. Exp. Integr. Med. 6, 1. doi:10.5455/jeim.270616.rw.016
Raina, R., Afroze, N., Kedhari Sundaram, M., Haque, S., Bajbouj, K., Hamad, M., et al. (2021). Chrysin Inhibits Propagation of HeLa Cells by Attenuating Cell Survival and Inducing Apoptotic Pathways. Eur. Rev. Med. Pharmacol. Sci. 25, 2206–2220. doi:10.26355/eurrev_202103_25253
Ramia, E., Chiaravalli, A. M., Bou Nasser Eddine, F. F., Tedeschi, A., Sessa, F., Accolla, R. S., et al. (2019). CIITA-related Block of HLA Class II Expression, Upregulation of HLA Class I, and Heterogeneous Expression of Immune Checkpoints in Hepatocarcinomas: Implications for New Therapeutic Approaches. Oncoimmunology 8, 1548243. doi:10.1080/2162402X.2018.1548243
Ramis-Conde, I., Chaplain, M. A., Anderson, A. R., and Drasdo, D. (2009). Multi-scale Modelling of Cancer Cell Intravasation: the Role of Cadherins in Metastasis. Phys. Biol. 6, 016008. doi:10.1088/1478-3975/6/1/016008
Rose, N. R., and Klose, R. J. (2014). Understanding the Relationship between DNA Methylation and Histone Lysine Methylation. Biochim. Biophys. Acta (Bba) - Gene Regul. Mech. 1839, 1362–1372. doi:10.1016/j.bbagrm.2014.02.007
Ryu, S., Lim, W., Bazer, F. W., and Song, G. (2017). Chrysin Induces Death of Prostate Cancer Cells by Inducing ROS and ER Stress. J. Cel. Physiol. 232, 3786–3797. doi:10.1002/jcp.25861
Salimi Sartakhti, J., Manshaei, M. H., and Sadeghi, M. (2017). MMP-TIMP Interactions in Cancer Invasion: An Evolutionary Game-Theoretical Framework. J. Theor. Biol. 412, 17–26. doi:10.1016/j.jtbi.2016.09.019
Shankar, E., Kanwal, R., Candamo, M., and Gupta, S. (2016). Dietary Phytochemicals as Epigenetic Modifiers in Cancer: Promise and Challenges. Semin. Cancer Biol. 40-41, 82–99. doi:10.1016/j.semcancer.2016.04.002
Sharma, S., Kelly, T. K., and Jones, P. A. (2009). Epigenetics in Cancer. Carcinogenesis 31, 27–36. doi:10.1093/carcin/bgp220
Sobti, R. C., Singh, N., Hussain, S., Suri, V., Nijhawan, R., Bharti, A. C., et al. (2011). Aberrant Promoter Methylation and Loss of Suppressor of Cytokine Signalling-1 Gene Expression in the Development of Uterine Cervical Carcinogenesis. Cell Oncol. 34, 533–543. doi:10.1007/s13402-011-0056-2
Soto, D., Song, C., and McLaughlin-Drubin, M. E. (2017). Epigenetic Alterations in Human Papillomavirus-Associated Cancers. Viruses 9, 248. doi:10.3390/v9090248
Sun, L.-P., Chen, A.-L., Hung, H.-C., Chien, Y.-H., Huang, J.-S., Huang, C.-Y., et al. (2012). Chrysin: a Histone Deacetylase 8 Inhibitor with Anticancer Activity and a Suitable Candidate for the Standardization of Chinese Propolis. J. Agric. Food Chem. 60, 11748–11758. doi:10.1021/jf303261r
Trievel, R. C. (2004). Structure and Function of Histone Methyltransferases. Crit. Rev. Eukaryot. Gene Expr. 14, 147–170. doi:10.1615/CritRevEukaryotGeneExpr.v14.i3.10
Wang, J., Wang, H., Sun, K., Wang, X., Pan, H., Zhu, J., et al. (2018). Chrysin Suppresses Proliferation, Migration, and Invasion in Glioblastoma Cell Lines via Mediating the ERK/Nrf2 Signaling Pathway. Dddt 12, 721–733. doi:10.2147/dddt.s160020
Wang, X., Jiang, Z., An, J., Mao, X., Lin, F., and Sun, P. (2018). Effect of a Synthetic Inhibitor of Urokinase Plasminogen Activator on the Migration and Invasion of Human Cervical Cancer Cells In Vitro. Mol. Med. Rep. 17, 4273–4280. doi:10.3892/mmr.2018.8414
Wu, D., Zhao, B., Song, Y., Chi, X., Fu, H., Guan, T., et al. (2021). Nogo-B Receptor Is Required for Stabilizing TGF-β Type Ireceptor and Promotes the TGF-Β1-Induced Epithelial-To-Mesenchymal Transition of Non-small Cell Lung Cancer. J. Cancer 12, 717–725. doi:10.7150/jca.50483
Xia, Y., Lian, S., Khoi, P. N., Yoon, H. J., Joo, Y. E., Chay, K. O., et al. (2015). Chrysin Inhibits Tumor Promoter-Induced MMP-9 Expression by Blocking AP-1 via Suppression of ERK and JNK Pathways in Gastric Cancer Cells. PLoS One 10, e0124007. doi:10.1371/journal.pone.0124007
Xue, G., Restuccia, D. F., Lan, Q., Hynx, D., Dirnhofer, S., Hess, D., et al. (2012). Akt/PKB-Mediated Phosphorylation of Twist1 Promotes Tumor Metastasis via Mediating Cross-Talk between PI3K/Akt and TGF-β Signaling Axes. Cancer Discov. 2, 248–259. doi:10.1158/2159-8290.cd-11-0270
Yan, W., Wu, T. H. Y., Leung, S. S. Y., and To, K. K. W. (2020). Flavonoids Potentiated Anticancer Activity of Cisplatin in Non-small Cell Lung Cancer Cells In Vitro by Inhibiting Histone Deacetylases. Life Sci. 258, 118211. doi:10.1016/j.lfs.2020.118211
Yang, B., Huang, J., Xiang, T., Yin, X., Luo, X., Huang, J., et al. (2014). Chrysin Inhibits Metastatic Potential of Human Triple-Negative Breast Cancer Cells by Modulating Matrix Metalloproteinase-10, Epithelial to Mesenchymal Transition, and PI3K/Akt Signaling Pathway. J. Appl. Toxicol. 34, 105–112. doi:10.1002/jat.2941
You, J. S., and Jones, P. A. (2012). Cancer Genetics and Epigenetics: Two Sides of the Same Coin? Cancer Cell 22, 9–20. doi:10.1016/j.ccr.2012.06.008
Zam, W., and Khadour, A. (2017). Impact of Phytochemicals and Dietary Patterns on Epigenome and Cancer. Nutr. Cancer 69, 184–200. doi:10.1080/01635581.2017.1263746
Keywords: phytochemicals, epigenetic modification, DNA methylation, epigenome, chrysin
Citation: Raina R, Almutary AG, Bagabir SA, Afroze N, Fagoonee S, Haque S and Hussain A (2022) Chrysin Modulates Aberrant Epigenetic Variations and Hampers Migratory Behavior of Human Cervical (HeLa) Cells. Front. Genet. 12:768130. doi: 10.3389/fgene.2021.768130
Received: 31 August 2021; Accepted: 06 December 2021;
Published: 12 January 2022.
Edited by:
Ata Abbas, Case Western Reserve University, United StatesReviewed by:
Lubna Wasim, All India Institute of Medical Sciences, IndiaCopyright © 2022 Raina, Almutary, Bagabir, Afroze, Fagoonee, Haque and Hussain. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Arif Hussain, ZHIuYXJpZmh1c3NhaW5AeWFob28uY28uaW4=; Abdulmajeed G. Almutary, YWJkdWxtYWplZWQuYWxtdXRhcnlAcXUuZWR1LnNh
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.