- 1Henan Collaborative Innovation Center of Modern Biological Breeding, Henan Institute of Sciences and Technology, Xinxiang, China
- 2State Key Laboratory of Cotton Biology, Institute of Cotton Research, Chinese Academy of Agricultural Sciences, Anyang, China
- 3Beijing River and Lake Management Office, Beijing, China
- 4Key Laboratory of Tobacco Improvement and Biotechnology, Tobacco Research Institute of Chinese Academy of Agricultural Sciences, Qingdao, China
- 5College of Medical Technology, Beihua University, Jilin City, China
RNA editing is a posttranscriptional phenomenon that includes gene processing and modification at specific nucleotide sites. RNA editing mainly occurs in the genomes of mitochondria and chloroplasts in higher plants. In recent years, pentatricopeptide repeat (PPR) proteins, which may act as trans-acting factors of RNA editing have been identified, and the study of PPR proteins has become a research focus in molecular biology. The molecular functions of these proteins and their physiological roles throughout plant growth and development are widely studied. In this minireview, we summarize the current knowledge of the PPR family, hoping to provide some theoretical reference for future research and applications.
Introduction
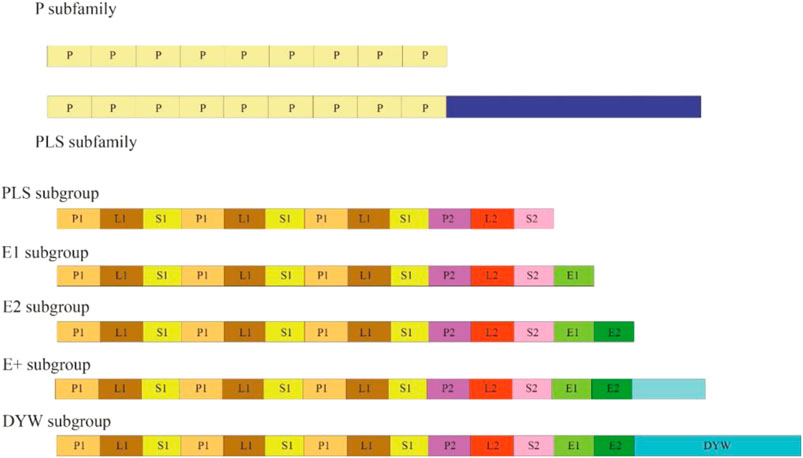
PPR family is one of the largest gene families in higher plants. PPR proteins contain an array of 2–30 tandem repetitions of a degraded unit containing 30–40 amino acid (aa) motifs (Lurin et al., 2004). PPR proteins are classified into two subfamilies based on their domain architecture: P and PPR-like (PLS), which are distinguished by motifs with no space and motifs with interspaced PPR-like motifs respectively. The PLS subfamily can be subdivided into five subgroups based on domain assembly at the C-terminus of a PPR protein: PLS, E1, E2, E+, and DYW (Figure 1) (Cheng et al., 2016; Xing et al., 2018). PPR proteins have been found in a variety of terrestrial plants since their discovery in yeast (Saccharomyces cerevisiae L.) (Manthey and McEwen, 1995). To date, PPR proteins have been found in many different plants, including Arabidopsis (Lurin et al., 2004), foxtail millet (Xing et al., 2018), poplar (Liu et al., 2016), maize (Chen L. et al., 2018), and rice (Chen G. et al., 2018), containing 441, 486, 626, 491, and 477 members of the PPR family, respectively. Additionally, PPR proteins have been discovered to have RNA-binding characteristics, allowing them to mediate gene expression via posttranscriptional mechanisms involving transcripts in the mitochondria, chloroplast, and nucleus. PPR proteins play vital roles in the plant organelle RNA editing machinery. PPRs could not only act as site recognition factors but also bind to cis-elements specifically. The resultant PPR–RNA complex and other editing factors, such as ORRM proteins and MORF proteins, can form a higher ordered editosome. As a result of their participation in different posttranscriptional processes, such as RNA editing (Hayes et al., 2015), RNA splicing (Ichinose et al., 2012), and RNA processing (Hao et al., 2019), PPR proteins are considered to have a substantial influence on organelle stability, including biogenesis and function. Furthermore, plant growth and development have been linked to the activities of PPR proteins. Previously, we briefly reviewed RNA editing in plant organelles, including the factors and mechanism of RNA editing, the editing events identified through deep sequencing data, and the roles of RNA editing (Ichinose et al., 2012). In this review, we emphasize on the recent discoveries of PPR proteins, including their roles in manipulating CMS-related genes, chloroplast biogenesis, embryogenesis, and stress responses. Furthermore, we also discuss the roles of PPR proteins in fruit growth, ripening, plant flesh colour, and fibre development.
The number of motifs in each protein can vary from 2 to 35, and the first motif can be any of P, P1, L1, S1 or SS. The E+ subgroup consists of proteins with a degenerate or truncated DYW domain.
PPR Proteins Manipulate Cytoplasmic Male Sterility-Related Genes
Plant cytoplasmic male sterility (CMS) is a maternally inherited trait that maintains female fertility but results in abortive pollen. At present, CMS is considered to be jointly controlled by mitochondrial genes and their corresponding nuclear restorer (RF) genes, which can change the expression of CMS-related genes in mitochondria. Most Rf genes belong to the PPR gene family with several exceptions. Radish Rfo is a PPR protein containing 16 motifs. Inhibition of the Rfo translation process by binding to an orf125-transcribed mRNA inhibits the accumulation of the CMS-inducible protein ORF125 and restores CMS fertility (Koizuka et al., 2003). In addition, it is worth noting that sorghum Rf1 encodes PPR13, which belongs to the PLS-E class and can restore male fertility by recruiting RNA editing enzymes (Klein et al., 2005). OsRF1A encodes pentatricopeptide repeat proteins, plays an additional role in promoting the editing of atp6 mRNAs, and restores cytoplasmic male sterility in rice (Wang et al., 2006). Moreover, the PPR protein OsRF5 forms a complex with GRP162 to process atp6-orf79 and restore fertility in the Honglian CMS line (Hu et al., 2012). Similarly, the interaction between the PPR proteins OsRF6 and Oshxk6 for coprocessing atp6-orf79 rescues the male fertility of Honglian CMS lines (Huang et al., 2015). PPR756, a member of the PLS-E subclass, participates in RNA editing events of atp6, ccmC, and nad7. The loss of PPR756 could cause abortive pollen development in rice (Zhang Q. et al., 2020). The P-subfamily PPR protein OsPPR939, which can be phosphorylated by OsS6K1, regulates plant growth and pollen development by splicing mitochondrial nad5 introns 1, 2, and 3 (Zheng et al., 2021). PPS1 is a mitochondria-localized PPR protein, while OsPGLl is a PPR protein that is localized to both mitochondria and chloroplasts. PPS1 is involved in 5 consecutive editing sites of the nad3 transcript in mitochondria, while OsPGL1 is involved in RNA editing at a single site in both chloroplast and mitochondrial transcripts. The function of these two PPR proteins results in a decrease in the RNA editing efficiency at the specific site and ultimately in a defective phenotype concerning reproductive growth in rice.
PPRs Are Required for Chloroplast Biogenesis
Previous studies have reported that 21 PPR and PPR-related proteins are required for plastid RNA editing in thale cress (Lu, 2018). Two PPR proteins (OsPPR4 and OsPPR6) have been shown to participate in the editing of a single plastid RNA editing site in rice; they are also involved in the editing of chloroplast RNA and are required for chloroplast biogenesis (Asano et al., 2013; Tang et al., 2017). The PPR protein AtPDM2 is located in plastids and regulates the expression of plastid genes related to chloroplast development by interacting with the organelle RNA editing factors morf2 and morf9. T-DNA insertion of the AtPDM2 leads to the loss of pigment in Arabidopsis cotyledons and cotyledon albinism (Du et al., 2017). Another PPR protein in Arabidopsis, Hcf152 is located in chloroplasts and regulates the metabolism of chloroplast RNA by combining with the petb operon. This gene mutation will affect the accumulation of the cytochrome b6f complex (Meierhoff et al., 2003); A PPR gene in Poaceae, Clb19 is required for editing the chloroplast transcripts rpoA and rpoB. Mutation of this gene will lead to impaired chloroplast development, a yellowing phenotype of seedlings and even death (Hein and Knoop, 2018). OsPPR16, a PLS-DYW subfamily PPR protein, is responsible for RNA editing of the RNA polymerase subunit RpoB and affects Chl synthesis and efficient chloroplast development in rice (Huang et al., 2020). The PPR protein DUA1 interacts with sigma factor 1 to form the PPR-SIG complex, and the module regulates chloroplast gene expression and chloroplast development in response to light and temperature (Du et al., 2021). The PPR protein OsPGL1 disrupts chloroplast RNA editing of ndhD-878, which is involved in the conversion of serine to leucine. Loss of OsPGL1 leads to the dysfunction of chloroplasts and the photosynthetic complex (Xiao et al., 2018). The PPR-SMR protein ATP4 participates in C-to-U editing of rps8 RNA in rice and maize, which is required for the formation of photosynthetic complexes (Zhang J. et al., 2020).
PPRs Regulate Embryogenesis
Previous studies have demonstrated that PPR proteins are essential for Arabidopsis and maize kernel formation, with the loss of function of specific PPR proteins resulting in empty pericarps and tiny, malformed kernels in various genetic backgrounds. AtEmb175 is located in chloroplasts and is the first PPR gene found in Arabidopsis that is related to early embryo death. Its mutation will lead to continuous cell division, embryo expansion, and abnormal tissue formation (Cushing et al., 2005). ZmSmk1 encodes a protein containing a PPR structural domain that regulates seed embryo and endosperm development by regulating RNA editing of the mitochondrial gene nad7 in maize (Li et al., 2014). The maize PPR-like protein EMP9 (EMPTY PERICARP9) regulates seed development by regulating RNA editing of the mitochondrial genes ccmB and rps4 (Yang et al., 2017). ZmEMP21, a PPR-DYW protein that is needed for the editing of 81 mitochondrial target sites, is required for mitochondrial complex assembly as well as embryo and endosperm development (Wang et al., 2019). ZmPPR-SMR1 interacts with ZmCSF1 and is essential for the splicing of numerous group II introns, mitochondrial functions, embryogenesis, and endosperm development (Chen et al., 2019). The E-subgroup PPR protein DEK55 affects mitochondrial RNA processing, which is important for maize kernel formation (Ren et al., 2020). ZmPPR27 interacts with ZmMORF1 (MULTIPLE ORGANELLAR RNA EDITING FACTOR 1), regulates the RNA editing rate of mitochondrial genes such as ccmFN and affects the formation of the key mitochondrial protein complex, thus hindering seed embryo and endosperm development (Liu et al., 2020). The maize DEK46 (DEFECTIVE KERNEL 46) gene encodes a protein containing a PPR domain that edits a specific site in the intron of the mitochondrial nad7 gene. When DEK46 is functionally absent, the percentage of selective splicing of the intron of the mitochondrial nad7 gene is reduced, which in turn affects normal seed development in maize (Xu et al., 2020). A DYW domain–containing PPR protein, PPR2263, was found to be involved in RNA editing in the mitochondrial NADH dehydrogenase 5 (nad5) and cytochrome b (cob) in maize. The ppr2263 mutant showed reduced embryo and endosperm growth, resulting in growth defects in kernels and seedlings (Sosso et al., 2012). A PPR protein, defective kernel 2 (Dek2), is required for nad1 mRNA splicing in maize. The dek2 mutant displayed small kernel and tardy development. (Qi et al., 2017). Another E+ subgroup PPR protein, DEK40, involved in the processing of cox3, nad2, and nad5 was identified to be essential for mitochondrial function and kernel development in maize (Ren et al., 2019).
PPRs Participate in Stress Responses
RNA editing may have contributed to the adaptation of land plants to extreme temperature, UV, and oxidative stress during the early stages of land plant formation (Fujii and Small, 2011). Increasing molecular evidence has revealed that many PPRs are involved in the response to a variety of biotic and abiotic stresses. In Arabidopsis, salt, oxidative, and ABA stressors all increased the expression of the PPR96 gene (Oren et al., 2001). The PPR protein GUN1 is associated with plastid-to-nucleus retrograde communication, control of ABI4 expression, and photooxidative stress responses in Arabidopsis (Koussevitzky et al., 2007). In Arabidopsis, PPR40 is known to offer a signaling connection between mitochondrial electron transport elements. PPR40 knockout resulted in increased reactive oxygen species (ROS) accumulation, lipid peroxidation, and superoxide dismutase activity (Zsigmond et al., 2008). The PPR protein ABA overly sensitive 5 (ABO5/At1g51965) is required for NADH dehydrogenase subunit 2 (NAD2) intron 3 splicing in mitochondria. Compared to the wild type, the abo5 mutant accumulated more H2O2 in roots (Liu et al., 2010). MITOCHONDRIAL RNA EDITING FACTOR 11 (MEF11)/LOVASTATIN INSENSITIVE 1 (LOI1) controls isoprenoid production, which is known to influence defense gene expression in response to wounding and pathogen infection (Kobayashi et al., 2007; Tang et al., 2010).
The Arabidopsis PPR-like protein AHG11 (ABA HYPERSENSITIVE GERMINATION 11) can edit the mRNA of the mitochondrial gene nad4 to maintain normal intracellular levels of reactive oxygen species (Murayama et al., 2012). PGN (PENTATRICOPEPTIDE REPEAT PROTEIN FOR GERMINATION ON NACl) has been shown to be involved in biotic and abiotic stress responses (Laluk et al., 2011). Functional disruption of the PPR protein SLG1 impacts mitochondrial RNA editing, plant growth, and abiotic stress responses in Arabidopsis (Yuan and Liu, 2012).
Plant development is also regulated by another PPR protein, SLO2. Stress-sensitive genes have higher transcript levels in the slo2 mutant. Furthermore, the slo2 mutant is hypersensitive to osmotic and ABA stressors at various phases of seed germination, although their mature plants have a high resistance to salt and drought stresses (Zhu et al., 2012; Zhu et al., 2014). SVR7 (SUPPRESSOR OF VARIATION 7) is needed for chloroplast ATP synthase subunit translation in Arabidopsis. SVR7 knockout led to increased ROS production, increased sensitivity to H2O2, and decreased photosynthetic activity (Lv et al., 2014). SOAR1 (suppressor of ABAR-overexpressor 1), which encodes a nucleocytoplasmic localized PPR protein, has recently been discovered to be a positive regulator of the responses to different stresses, including those to drought, salt, and cold (Jiang et al., 2015).
PPRs Regulate Fruit Growth, Ripening, Plant Flesh Colour, and Fibre Development
With the discovery of growing molecular evidence, researchers have found that PPR genes are involved in a variety of fruit functions, including as growth, ripening, colouration, and fiber formation. GUN1, which encodes a plastid-localized PPR protein, has been involved in the plastid-to-nucleus retrograde signalling route during tomato fruit development and ripening (Pesaresi et al., 2014). A number of tomato mutants, such as Cnr, were found to have dramatically reduced the expression of ripening-related PPR genes, resulting in mature fruits with colorless pericarp tissue, indicating that PPR proteins play a role in the development of fruits (Eriksson et al., 2004). CmPPR1, which encodes a plastid-targeted P-type PPR protein in melon (Cucumis melo L.), has been identified as a potential main quantitative trait locus (QTL) that determines flesh colour intensity (Galpaz et al., 2018). It was shown that genotyping 70 lines utilizing four SNPs from four ClaPPRs resulted in match rates above 0.87 for each validated SNP in association with the distinct phenotypes of skin colour. These findings contribute to a better knowledge of PPR genes and their functions in watermelon fruit growth and ripening, which may be useful in watermelon cultivar improvement (Subburaj et al., 2020). GhImA, which encodes a PPR protein, is involved in mitochondrial nad7 splicing, respiratory metabolism, and cotton fibre formation via ATP supply and ROS balancing (Zhang et al., 2021).
Conclusion and Perspectives
In general, PPR proteins participate in plant growth and development by RNA editing or RNA stabilization and splicing. Recently, many attempts have been made to engineer special proteins for efficient RNA editing in plant organelles. Shen et al. observed the interactions between different PPR codes and RNA bases at the atomic level, revealing the molecular basis for the modular and specific recognition patterns of the RNA bases U, C, A, and G (Zhang et al., 2021). PPR family proteins have the ability to enter organelles and bind to single-stranded RNA, and more recognition codes are being studied and verified. On this basis, according to the single-stranded sequence characteristics of target RNA, the corresponding PPR proteins have been artificially designed. Developing RNA-targeting tools will likely accelerate the functional study of RNA editing and the biological study of chloroplasts and mitochondria. By fusing the corresponding functional domains, the artificial design of fused PPR proteins is expected to become the next generation of biotechnology that can regulate the expression of organelle genes.
Author Contributions
WH and GL contributed equally to the design and coordination of the study; TQ, PZ, and JS collected the data; With the help from the YPZ, QY, YXZ, WW, ZC, TM, and YYZ wrote the manuscript. All of the authors reviewed and edited the manuscript.
Funding
This research was supported by Science and Technology Program of Sichuan Province (2020YJ0406), National Natural Science Foundation of China (31900256), Natural Science Foundation of Shandong Province (ZR2020QC026), Modern Agricultural Industry Technical Economic Evaluation System Green Development Position of Henan Province, Soft Science Project of Henan Province (202400410185) and State Key Laboratory of Cotton Biology Open Fund (CB2021A02).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Asano, T., Miyao, A., Hirochika, H., Kikuchi, S., and Kadowaki, K.-i. (2013). A Pentatricopeptide Repeat Gene of rice Is Required for Splicing of Chloroplast Transcripts and RNA Editing of ndhA. Plant Biotechnol. 12, 1217. doi:10.5511/plantbiotechnology.12.1217a
Chen, G., Zou, Y., Hu, J., and Ding, Y. (2018a). Genome-wide Analysis of the rice PPR Gene Family and Their Expression Profiles under Different Stress Treatments. BMC genomics 19 (1), 1–14. doi:10.1186/s12864-018-5088-9
Chen, L., Li, Y. X., Li, C., Shi, Y., Song, Y., Zhang, D., et al. (2018b). Genome-wide Analysis of the Pentatricopeptide Repeat Gene Family in Different maize Genomes and its Important Role in Kernel Development. BMC Plant Biol. 18 (1), 366–414. doi:10.1186/s12870-018-1572-2
Chen, Z., Wang, H.-C., Shen, J., Sun, F., Wang, M., Xu, C., et al. (2019). PPR-SMR1 Is Required for the Splicing of Multiple Mitochondrial Introns, Interacts with Zm-mCSF1, and Is Essential for Seed Development in maize. J. Exp. Bot. 70 (19), 5245–5258. doi:10.1093/jxb/erz305
Cheng, S., Gutmann, B., Zhong, X., Ye, Y., Fisher, M. F., Bai, F., et al. (2016). Redefining the Structural Motifs that Determine RNA Binding and RNA Editing by Pentatricopeptide Repeat Proteins in Land Plants. Plant J. 85 (4), 532–547. doi:10.1111/tpj.13121
Cushing, D. A., Forsthoefel, N. R., Gestaut, D. R., and Vernon, D. M. (2005). Arabidopsis Emb175 and Other Ppr Knockout Mutants Reveal Essential Roles for Pentatricopeptide Repeat (PPR) Proteins in Plant Embryogenesis. Planta 221 (3), 424–436. doi:10.1007/s00425-004-1452-x
Du, L., Zhang, J., Qu, S., Zhao, Y., Su, B., Lv, X., et al. (2017). The Pentratricopeptide Repeat Protein Pigment-Defective Mutant2 Is Involved in the Regulation of Chloroplast Development and Chloroplast Gene Expression in Arabidopsis. Plant Cel Physiol. 58 (4), 747–759. doi:10.1093/pcp/pcx004
Du, Y., Mo, W., Ma, T., Tang, W., Tian, L., and Lin, R. (2021). A Pentatricopeptide Repeat Protein DUA1 Interacts with Sigma Factor 1 to Regulate Chloroplast Gene Expression in Rice. Photosynth Res. 147 (2), 131–143. doi:10.1007/s11120-020-00793-0
Eriksson, E. M., Bovy, A., Manning, K., Harrison, L., Andrews, J., De Silva, J., et al. (2004). Effect of the Colorless Non-ripening Mutation on Cell wall Biochemistry and Gene Expression during Tomato Fruit Development and Ripening. Plant Physiol. 136 (4), 4184–4197. doi:10.1104/pp.104.045765
Fujii, S., and Small, I. (2011). The Evolution of RNA Editing and Pentatricopeptide Repeat Genes. New Phytol. 191 (1), 37–47. doi:10.1111/j.1469-8137.2011.03746.x
Galpaz, N., Gonda, I., Shem‐Tov, D., Barad, O., Tzuri, G., Lev, S., et al. (2018). Deciphering Genetic Factors that Determine Melon Fruit‐quality Traits Using RNA ‐Seq‐based High‐resolution QTL and eQTL Mapping. Plant J. 94 (1), 169–191. doi:10.1111/tpj.13838
Hao, Y., Wang, Y., Wu, M., Zhu, X., Teng, X., Sun, Y., et al. (2019). The Nuclear-Localized PPR Protein OsNPPR1 Is Important for Mitochondrial Function and Endosperm Development in rice. J. Exp. Bot. 70 (18), 4705–4720. doi:10.1093/jxb/erz226
Hayes, M. L., Dang, K. N., Diaz, M. F., and Mulligan, R. M. (2015). A Conserved Glutamate Residue in the C-Terminal Deaminase Domain of Pentatricopeptide Repeat Proteins Is Required for RNA Editing Activity. J. Biol. Chem. 290 (16), 10136–10142. doi:10.1074/jbc.m114.631630
Hein, A., and Knoop, V. (2018). Expected and Unexpected Evolution of Plant RNA Editing Factors CLB19, CRR28 and RARE1: Retention of CLB19 Despite a Phylogenetically Deep Loss of its Two Known Editing Targets in Poaceae. BMC Evol. Biol. 18 (1), 85–15. doi:10.1186/s12862-018-1203-4
Hu, J., Wang, K., Huang, W., Liu, G., Gao, Y., Wang, J., et al. (2012). The Rice Pentatricopeptide Repeat Protein RF5 Restores Fertility in Hong-Lian Cytoplasmic Male-Sterile Lines via a Complex with the Glycine-Rich Protein GRP162. The Plant Cell 24 (1), 109–122. doi:10.1105/tpc.111.093211
Huang, W., Yu, C., Hu, J., Wang, L., Dan, Z., Zhou, W., et al. (2015). Pentatricopeptide-repeat Family Protein RF6 Functions with Hexokinase 6 to rescue rice Cytoplasmic Male Sterility. Proc. Natl. Acad. Sci. USA 112 (48), 14984–14989. doi:10.1073/pnas.1511748112
Huang, W., Zhang, Y., Shen, L., Fang, Q., Liu, Q., Gong, C., et al. (2020). Accumulation of the RNA Polymerase Subunit RpoB Depends on RNA Editing by OsPPR16 and Affects Chloroplast Development during Early Leaf Development in rice. New Phytol. 228 (4), 1401–1416. doi:10.1111/nph.16769
Ichinose, M., Tasaki, E., Sugita, C., and Sugita, M. (2012). A PPR-DYW Protein Is Required for Splicing of a Group II Intron of Cox1 Pre-mRNA in Physcomitrella Patens. Plant J. 70 (2), 271–278. doi:10.1111/j.1365-313x.2011.04869.x
Jiang, S. C., Mei, C., Liang, S., Yu, Y. T., Lu, K., Wu, Z., et al. (2015). Crucial Roles of the Pentatricopeptide Repeat Protein SOAR1 in Arabidopsis Response to Drought, Salt and Cold Stresses. Plant Mol. Biol. 88 (4), 369–385. doi:10.1007/s11103-015-0327-9
Klein, R. R., Klein, P. E., Mullet, J. E., Minx, P., Rooney, W. L., and Schertz, K. F. (2005). Fertility Restorer Locus Rf1 of Sorghum (Sorghum Bicolor L.) Encodes a Pentatricopeptide Repeat Protein Not Present in the Colinear Region of rice Chromosome 12. Theor. Appl. Genet. 111 (6), 994–1012. doi:10.1007/s00122-005-2011-y
Kobayashi, K., Suzuki, M., Tang, J., Nagata, N., Ohyama, K., Seki, H., et al. (2007). Lovastatin Insensitive 1, a Novel Pentatricopeptide Repeat Protein, Is a Potential Regulatory Factor of Isoprenoid Biosynthesis in Arabidopsis. Plant Cel. Physiol. 48 (2), 322–331. doi:10.1093/pcp/pcm005
Koizuka, N., Imai, R., Fujimoto, H., Hayakawa, T., Kimura, Y., Kohno-Murase, J., et al. (2003). Genetic Characterization of a Pentatricopeptide Repeat Protein Gene, Orf687, that Restores Fertility in the Cytoplasmic Male-Sterile Kosena Radish. Plant J. 34 (4), 407–415. doi:10.1046/j.1365-313x.2003.01735.x
Koussevitzky, S., Nott, A., Mockler, T. C., Hong, F., Sachetto-Martins, G., Surpin, M., et al. (2007). Signals from Chloroplasts Converge to Regulate Nuclear Gene Expression. Science 316 (5825), 715–719. doi:10.1126/science.1140516
Laluk, K., AbuQamar, S., and Mengiste, T. (2011). The Arabidopsis Mitochondria-Localized Pentatricopeptide Repeat Protein PGN Functions in Defense against Necrotrophic Fungi and Abiotic Stress Tolerance. Plant Physiol. 156 (4), 2053–2068. doi:10.1104/pp.111.177501
Li, X.-J., Zhang, Y.-F., Hou, M., Sun, F., Shen, Y., Xiu, Z.-H., et al. (2014). Small Kernel 1encodes a Pentatricopeptide Repeat Protein Required for Mitochondrialnad7transcript Editing and Seed Development in maize(Zea mays)and rice(Oryza Sativa). Plant J. 79 (5), 797–809. doi:10.1111/tpj.12584
Liu, J. M., Xu, Z. S., Lu, P. P., Li, W. W., Chen, M., Guo, C. H., et al. (2016). Genome-wide Investigation and Expression Analyses of the Pentatricopeptide Repeat Protein Gene Family in Foxtail Millet. BMC genomics 17 (1), 1–16. doi:10.1186/s12864-016-3184-2
Liu, R., Cao, S.-K., Sayyed, A., Yang, H.-H., Zhao, J., Wang, X., et al. (2020). The DYW-Subgroup Pentatricopeptide Repeat Protein PPR27 Interacts with ZmMORF1 to Facilitate Mitochondrial RNA Editing and Seed Development in maize. J. Exp. Bot. 71 (18), 5495–5505. doi:10.1093/jxb/eraa273
Liu, Y., He, J., Chen, Z., Ren, X., Hong, X., and Gong, Z. (2010). ABA Overly-Sensitive 5 (ABO5), Encoding a Pentatricopeptide Repeat Protein Required for Cis-Splicing of Mitochondrial Nad2 Intron 3, Is Involved in the Abscisic Acid Response in Arabidopsis. Plant J. 63 (5), 749–765. doi:10.1111/j.1365-313x.2010.04280.x
Lu, Y. (2018). RNA Editing of Plastid-Encoded Genes. Photosynt. 56 (1), 48–61. doi:10.1007/s11099-017-0761-9
Lurin, C., Andreés, C., Aubourg, S., Bellaoui, M., Bitton, F., Bruyère, C., et al. (2004). Genome-Wide Analysis of Arabidopsis Pentatricopeptide Repeat Proteins Reveals Their Essential Role in Organelle Biogenesis[W]. The Plant Cell 16 (8), 2089–2103. doi:10.1105/tpc.104.022236
Lv, H.-X., Huang, C., Guo, G.-Q., and Yang, Z.-N. (2014). Roles of the Nuclear-Encoded Chloroplast SMR Domain-Containing PPR Protein SVR7 in Photosynthesis and Oxidative Stress Tolerance in Arabidopsis. J. Plant Biol. 57 (5), 291–301. doi:10.1007/s12374-014-0041-1
Manthey, G. M., and McEwen, J. E. (1995). The Product of the Nuclear Gene PET309 Is Required for Translation of Mature mRNA and Stability or Production of Intron-Containing RNAs Derived from the Mitochondrial COX1 Locus of Saccharomyces cerevisiae. EMBO J. 14 (16), 4031–4043. doi:10.1002/j.1460-2075.1995.tb00074.x
Meierhoff, K., Felder, S., Nakamura, T., Bechtold, N., and Schuster, G. (2003). HCF152, an Arabidopsis RNA Binding Pentatricopeptide Repeat Protein Involved in the Processing of Chloroplast psbB-psbT-psbH-petB-petD RNAs. Plant Cell 15 (6), 1480–1495. doi:10.1105/tpc.010397
Murayama, M., Hayashi, S., Nishimura, N., Ishide, M., Kobayashi, K., Yagi, Y., et al. (2012). Isolation of Arabidopsis Ahg11, a Weak ABA Hypersensitive Mutant Defective in Nad4 RNA Editing. J. Exp. Bot. 63 (14), 5301–5310. doi:10.1093/jxb/ers188
Oren, R., Ellsworth, D. S., Johnsen, K. H., Phillips, N., Ewers, B. E., Maier, C., et al. (2001). Soil Fertility Limits Carbon Sequestration by forest Ecosystems in a CO2-enriched Atmosphere. Nature 411 (6836), 469–472. doi:10.1038/35078064
Pesaresi, P., Mizzotti, C., Colombo, M., and Masiero, S. (2014). Genetic Regulation and Structural Changes during Tomato Fruit Development and Ripening. Front. Plant Sci. 5, 124. doi:10.3389/fpls.2014.00124
Qi, W., Yang, Y., Feng, X., Zhang, M., and Song, R. (2017). Mitochondrial Function and maize Kernel Development Requires Dek2, a Pentatricopeptide Repeat Protein Involved in Nad1 mRNA Splicing. Genetics 205 (1), 239–249. doi:10.1534/genetics.116.196105
Ren, R. C., Yan, X. W., Zhao, Y. J., Wei, Y. M., Lu, X., Zang, J., et al. (2020). The Novel E-Subgroup Pentatricopeptide Repeat Protein DEK55 Is Responsible for RNA Editing at Multiple Sites and for the Splicing of Nad1 and Nad4 in maize. BMC Plant Biol. 20 (1), 1–15. doi:10.1186/s12870-020-02765-x
Ren, R. C., Lu, X., Zhao, Y. J., Wei, Y. M., Wang, L. L., Zhang, L., et al. (2019). Pentatricopeptide Repeat Protein DEK40 Is Required for Mitochondrial Function and Kernel Development in maize. J. Exp. Bot. 70 (21), 6163–6179. doi:10.1093/jxb/erz391
Sosso, D., Mbelo, S., Vernoud, V., Gendrot, G., Dedieu, A., Chambrier, P., et al. (2012). PPR2263, a DYW-Subgroup Pentatricopeptide Repeat Protein, Is Required for Mitochondrial Nad5 and Cob Transcript Editing, Mitochondrion Biogenesis, and maize Growth. Plant Cell 24 (2), 676–691. doi:10.1105/tpc.111.091074
Subburaj, S., Tu, L., Lee, K., Park, G.-S., Lee, H., Chun, J.-P., et al. (2020). A Genome-Wide Analysis of the Pentatricopeptide Repeat (PPR) Gene Family and PPR-Derived Markers for Flesh Color in Watermelon (Citrullus lanatus). Genes 11 (10), 1125. doi:10.3390/genes11101125
Tang, J., Zhang, W., Wen, K., Chen, G., Sun, J., Tian, Y., et al. (2017). OsPPR6, a Pentatricopeptide Repeat Protein Involved in Editing and Splicing Chloroplast RNA, Is Required for Chloroplast Biogenesis in rice. Plant Mol. Biol. 95 (4), 345–357. doi:10.1007/s11103-017-0654-0
Tang, J., Kobayashi, K., Suzuki, M., Matsumoto, S., and Muranaka, T. (2010). The Mitochondrial PPR Protein LOVASTATIN INSENSITIVE 1 Plays Regulatory Roles in Cytosolic and Plastidial Isoprenoid Biosynthesis through RNA Editing. Plant J. 61 (3), 456–466. doi:10.1111/j.1365-313x.2009.04082.x
Wang, Y., Liu, X.-Y., Yang, Y.-Z., Huang, J., Sun, F., Lin, J., et al. (2019). Empty Pericarp21 Encodes a Novel PPR-DYW Protein that Is Required for Mitochondrial RNA Editing at Multiple Sites, Complexes I and V Biogenesis, and Seed Development in maize. Plos Genet. 15 (8), e1008305. doi:10.1371/journal.pgen.1008305
Wang, Z., Zou, Y., Li, X., Zhang, Q., Chen, L., Wu, H., et al. (2006). Cytoplasmic Male Sterility of rice with Boro II Cytoplasm Is Caused by a Cytotoxic Peptide and Is Restored by Two Related PPR Motif Genes via Distinct Modes of mRNA Silencing. The Plant Cell 18 (3), 676–687. doi:10.1105/tpc.105.038240
Xiao, H., Zhang, Q., Qin, X., Xu, Y., Ni, C., Huang, J., et al. (2018). Rice PPS1 Encodes a DYW Motif-Containing Pentatricopeptide Repeat Protein Required for Five Consecutive RNA-Editing Sites of Nad3 in Mitochondria. New Phytol. 220 (3), 878–892. doi:10.1111/nph.15347
Xing, H., Fu, X., Yang, C., Tang, X., Guo, L., Li, C., et al. (2018). Genome-wide Investigation of Pentatricopeptide Repeat Gene Family in poplar and Their Expression Analysis in Response to Biotic and Abiotic Stresses. Sci. Rep. 8 (1), 2817–2819. doi:10.1038/s41598-018-21269-1
Xu, C., Song, S., Yang, Y. Z., Lu, F., Zhang, M. D., Sun, F., et al. (2020). DEK46 Performs C‐to‐U Editing of a Specific Site in Mitochondrial Nad7 Introns that Is Critical for Intron Splicing and Seed Development in maize. Plant J. 103 (5), 1767–1782. doi:10.1111/tpj.14862
Yang, Y. Z., Ding, S., Wang, H. C., Sun, F., Huang, W. L., Song, S., et al. (2017). The Pentatricopeptide Repeat Protein EMP 9 Is Required for Mitochondrial ccmB and Rps4 Transcript Editing, Mitochondrial Complex Biogenesis and Seed Development in maize. New Phytol. 214 (2), 782–795. doi:10.1111/nph.14424
Yuan, H., and Liu, D. (2012). Functional Disruption of the Pentatricopeptide Protein SLG1 Affects Mitochondrial RNA Editing, Plant Development, and Responses to Abiotic Stresses in Arabidopsis. Plant J. 70 (3), 432–444. doi:10.1111/j.1365-313x.2011.04883.x
Zhang, D., Chen, C., Wang, H., Niu, E., Zhao, P., Fang, S., et al. (2021). Cotton Fiber Development Requires the Pentatricopeptide Repeat Protein GhIm for Splicing of Mitochondrial Nad7 mRNA. Genetics 217 (1), 1–17. doi:10.1093/genetics/iyaa017
Zhang, J., Guo, Y., Fang, Q., Zhu, Y., Zhang, Y., Liu, X., et al. (2020a). The PPR-SMR Protein ATP4 Is Required for Editing the Chloroplast Rps8 mRNA in rice and maize. Plant Physiol. 184 (4), 2011–2021. doi:10.1104/pp.20.00849
Zhang, Q., Xu, Y., Huang, J., Zhang, K., Xiao, H., Qin, X., et al. (2020b). The rice Pentatricopeptide Repeat Protein PPR756 Is Involved in Pollen Development by Affecting Multiple RNA Editing in Mitochondria. Front. Plant Sci. 11, 749. doi:10.3389/fpls.2020.00749
Zheng, P., Liu, Y., Liu, X., Huang, Y., Sun, F., Wang, W., et al. (2021). OsPPR939, a Nad5 Splicing Factor, Is Essential for Plant Growth and Pollen Development in rice. Theor. Appl. Genet. 134 (3), 923–940. doi:10.1007/s00122-020-03742-6
Zhu, Q., Dugardeyn, J., Zhang, C., Mühlenbock, P., Eastmond, P. J., Valcke, R., et al. (2014). The Arabidopsis thaliana RNA Editing Factor SLO2, Which Affects the Mitochondrial Electron Transport Chain, Participates in Multiple Stress and Hormone Responses. Mol. Plant 7 (2), 290–310. doi:10.1093/mp/sst102
Zhu, Q., Dugardeyn, J., Zhang, C., Takenaka, M., Kühn, K., Craddock, C., et al. (2012). SLO2, a Mitochondrial Pentatricopeptide Repeat Protein Affecting Several RNA Editing Sites, Is Required for Energy Metabolism. Plant J. 71 (5), 836–849. doi:10.1111/j.1365-313x.2012.05036.x
Keywords: pentatricopeptide repeat, RNA editing, biogenesis, development, mechanism
Citation: Qin T, Zhao P, Sun J, Zhao Y, Zhang Y, Yang Q, Wang W, Chen Z, Mai T, Zou Y, Liu G and Hao W (2021) Research Progress of PPR Proteins in RNA Editing, Stress Response, Plant Growth and Development. Front. Genet. 12:765580. doi: 10.3389/fgene.2021.765580
Received: 27 August 2021; Accepted: 04 October 2021;
Published: 18 October 2021.
Edited by:
Yanqiang Li, Boston Children’s Hospital and Harvard Medical School, United StatesReviewed by:
Mingkun Huang, The Chinese University of Hong Kong, ChinaZhengrui Qin, Zhejiang University, China
Copyright © 2021 Qin, Zhao, Sun, Zhao, Zhang, Yang, Wang, Chen, Mai, Zou, Liu and Hao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Guoxiang Liu, bGl1Z3VveGlhbmdAY2Fhcy5jbg==; Wei Hao, aGFvd2VpOTExMUBnbWFpbC5jb20=
†These authors have contributed equally to this work
 Tengfei Qin
Tengfei Qin Pei Zhao2†
Pei Zhao2† Jialiang Sun
Jialiang Sun Yaxin Zhang
Yaxin Zhang Qiuyue Yang
Qiuyue Yang Weipeng Wang
Weipeng Wang Zhuanqing Chen
Zhuanqing Chen Wei Hao
Wei Hao