
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Genet., 15 November 2021
Sec. RNA
Volume 12 - 2021 | https://doi.org/10.3389/fgene.2021.762678
This article is part of the Research TopicFunctional analysis of species-specific noncoding RNAs in plantsView all 14 articles
Poplar is an important bioenergy tree species. lncRNAs play important roles in various biological regulatory processes, and their expression pattern is more tissue-specific than mRNAs. In this study, P. deltoides “Danhong” (Pd) and P. simonii “Tongliao1” (Ps) with different growth rates and wood quality were used as experimental materials, and the transcriptomes of their shoot apical meristem, xylem, and phloem were sequenced. Furthermore, high-throughput RNA sequencing analysis revealed that the expression patterns of genes and lncRNAs are different between the two genotypes. 6,355 lncRNAs were identified. Based on target prediction, lncRNAs and target genes were involved in ADP binding, oxidoreductase activity, phenylpropanoid biosynthesis, and cyanoamino acid metabolism. The DElncRNAs in two poplars were co-expressed with transcription factors and structural genes of lignin and flavonoid pathways. In addition, we found the potential target lncRNAs of miRNA. This result provides basic evidence for a better understanding of the regulatory role of lncRNAs in regulating phenylalanine molecular pathways and wood formation.
Plants are unique in their ability to continuously produce new organs throughout their life cycles. The process of continuous organogenesis depends on the activity of pluripotent cells (Xue et al., 2020). In trees, this mainly refers to the shoot apical meristem (SAM) affecting high growth and the vascular cambium affecting radial growth (Elo et al., 2009). The SAM generates leaves, stems, and floral organs throughout the lifespan of higher plants (Ha et al., 2010; Xue et al., 2020). The cambium differentiates into xylem and phloem, determined cell types, and cell layers in the secondary xylem (Ye and Zhong, 2015). These complex processes are easily regulated by plant hormones, transcription factors (TFs), miRNAs, and lncRNAs (J. Zhang G. et al., 2018; Xue et al., 2020).
lncRNAs are non-coding transcripts longer than 200 nucleotides (nts), including intergenic, intronic, sense, and antisense types (Ma et al., 2013). Compared with protein-coding genes (PCgenes), most lncRNAs are less conserved between species, lower expression levels, and stronger tissue-specific expression patterns (Liu et al., 2012; Zhou et al., 2017; Xu et al., 2018). lncRNAs can regulate genes expression at transcriptional, posttranscriptional, and epigenetic levels and play an important role in genomic imprinting, chromatin remodeling, transcriptional activation, transcriptional interference, and cell cycle (Wang and Chekanova, 2017; Sun et al., 2018). With the continuous development of resequencing technology, lncRNAs of more and more species have been identified. They are widely involved in embryo development, seed formation, flower development, secondary growth of wood, and abiotic stress response (Zhou et al., 2017; Severing et al., 2018; Xu et al., 2018; Jiang et al., 2019; Wu et al., 2019). For example, lncRNAs play a potential regulatory role in endosperm and embryo development of castor bean (Xu et al., 2018). COOLAIR and COLDAIR play an important role in regulating vernalization in Arabidopsis (Heo and Sung, 2011). FLINC lncRNA participates in ambient temperature–mediated flowering time of Arabidopsis (Severing et al., 2018). lncRNAs influence the formation of tension wood by regulating ARFs in Catalpa bungei (Xiao et al., 2020). lncRNAs are widely involved in the secondary growth, GA response, heat tolerance, low nitrogen stress, salt stress, and other life processes of poplar (Chen et al., 2016; Tian et al., 2016; Ci et al., 2019; Ma et al., 2019; Song et al., 2020).
Populus is often used as a short rotation coppice (SRC) and bioenergy tree species all over the world because of its fast growth and reduced inhibitory extract from wood fermentation during bioenergy conversion (Guerra et al., 2013; Zhang et al., 2020b). So, the growth rate determines the economic benefit and the output of biomass energy. There are significant differences in the growth rate of P. deltoides “Danhong” and P. simonii “Tongliao1.” P. deltoides “Danhong” is a southern poplar characterized by fast growth and insect resistance (Zhang et al., 2008). P. simonii is a native tree species in northern China; although the growth rate is slow, it is resistant to cold and drought (Wei et al., 2013).
In order to identify the regulation mechanism of growth and wood property differences and provide theoretical basis for breeding new germplasm with fast growing ability, we selected P. deltoides “Danhong” and P. simonii “Tongliao1” as experimental materials and identified the important lncRNAs that may be involved in growth regulation by sequencing. In this study, the sequencing of lncRNA libraries was constructed from the SAM, phloem, and developing xylem of P. deltoides “Danhong” and P. simonii “Tongliao1.” P. tricorcarpa was used as the reference genome for the identification of lncRNA. We identified a total of 6,355 lncRNAs, of which 2,454 were sense_overlapping lncRNAs, 2,004 were lincRNAs, and 1,897 were antisense lncRNAs. The functional prediction of lncRNAs and their expressions as involved in wood development were examined. We investigated putative functional lncRNA candidates by differential expression analysis and co-expression network construction during SAM and xylem development. The important miRNA–lncRNA pairs in phenylalanine biosynthesis and hormone transduction were identified.
One-year-old P. deltoides “Danhong” (Pd) and P. simonii “Tongliao1” (Ps) were cultivated in the experimental field of the Chinese Academy of Forestry, Beijing, China (116.256°E, 40.007°N). We collected shoot apical meristem (SAM, Pd_S and Ps_S) and scraped phloem (inside of the bark, Pd_P and Ps_P) and developing xylem (newly formed xylem cells about 2–3 mm, Pd_X and Ps_X) from Pd and Ps, respectively, at diameter breast height (DBH) during the fast-growing period (July 20, 2019). Each tissue had three biological replicates. The samples (2 genotypes × 3 tissues × 3 biological replicates) used for RNA extraction were frozen immediately in liquid nitrogen and stored at −80°C. Shoot tips and cuneiform blocks (phloem, cambium, and xylem) at DBH for histologic analysis were fixed in a mixture of formalin, glacial acetic acid, and 70% ethanol in the ratio 5:5:90 vol.; FAA under vacuum for at least 24 h.
Stem pieces were embedded with Spurr resin as described by Zhang et al. (2020d). A cross section of 4 µm thick was obtained from the stem by Leica M205FA, while the SAM sections of 40 μm were obtained using a rotary microtome (Leica VT1200S, Wetzlar, Germany). The sections were stained using 0.05% toluidine blue O (TBO) and were examined with a microscope (Zeiss). The number and diameter of vessel cells in the same area (1,260 × 980 µm) were counted by ImageJ (version 1.8.0).
In order to understand the difference of wood properties between Pd and Ps, we measured the plant height and ground diameter and collected the stems to measure the wood properties including basic density, fiber length, fiber width, microfibril angle, cellulose, holocellulose, and lignin content of 1-year-old trees in December 2019. The basic density was determined by using the drainage method. A 10-cm-high wood segment was cut from the base of the trunk without bark and pith. It was softened by heating in 30% nitric acid and a small amount of potassium chlorate and converted into wood pulp by forced oscillation. The length and width of the fiber were measured 50 times using Shyygx Measure 2.0. Wood flour (40–60 mesh) from a 5-cm basal stem segment was used to determine the chemical composition. The content of holocellulose and lignin was calculated according to Chinese standards GB/T 2677.10-995 and GB/T 2677.8-1994, respectively. To evaluate the content of cellulose, the specimens were extracted with a mixed solvent of nitric acid and ethanol (v/v = 1/1) (Zhan et al., 2015). Three replicates were performed for each variety.
Total RNA was isolated from the 18 samples (SAM, phloem, and developing xylem) using the RNAprep Pure Plant Plus Kit (TIANGEN, China). An index of the reference genome (P. trichocarpa v3.0) was built using HISAT2 (Kim et al., 2015). StringTie was used to calculate FPKMs of both lncRNAs and coding genes in each sample (Pertea et al., 2015). Sequencing data are available in NCBI SRA database (SRA number: SRP2343030 to SRR13961247).
We used four filtration steps to identify lncRNAs from the transcriptome assembly: 1) Transcripts with an exon number ≥2 and length ≥200 bp were selected. 2) CuffCompare software was used to screen out transcripts that overlap with the database annotation exon field. 3) Evaluation of Coding Potential Calculator (CPC) (Kong et al., 2007), Coding Potential Assessment Tool (CPAT) (Wang et al., 2013), and Coding-Non-Coding Index (CNCI) (Sun et al., 2013) was carried out to screen whether there is coding potential. 4) They were referred to the the Hugo Gene Nomenclature Committee (HGNC) to name the novel_lncRNA of this analysis (Wright, 2014).
Two methods were used to predict lncRNA target genes. Cis target genes were predicted according to the location relationship between lncRNA and mRNA, and the screening range was within 10 kb (Jia et al., 2010). Co-expression–related target genes were predicted according to the expression correlation between lncRNA and mRNA, and the screening condition is that the correlation coefficient is greater than 0.95 and p-value< 1.68E-09. The mRNA–lncRNA regulatory network was further modeled and visualized using Cytoscape 3.8 (Otasek et al., 2019).
In order to identify lncRNAs that may be used as precursors of miRNAs, we compared the published miRNAs of P. trichocarpa in miRBase (http://www.mirbase.org/search.shtml) with lncRNAs. lncRNAs as targets of miRNAs were predicted by Novomagic, a free online platform for data analysis (https://magic.novogene.com).
To identify the differential expression of lncRNAs and mRNAs between Pd and Ps, we performed the read count of pair-wise comparisons (Pd_S vs Ps_S, Pd_P vs Ps_P, Pd_X vs Ps_X, Pd_S vs Pd_P, Pd_S vs Pd_X, Pd_P vs Pd_X, Ps_S vs Ps_P, Ps_S vs Ps_X, Ps_P vs Ps_X) by DESeq R package with a q-value < 0.05 (Love et al., 2014). Finally, those putative cis- and coexpression-targets of lncRNAs were analysed using Gene Ontology (GO) analysis (Ashburner et al., 2000; Conesa et al., 2005), and KEGG (Kyoto Encyclopedia of Genes and Genomes) enrichment of DE genes was performed based on a corrected p-value < 0.05.
We selected three DELs and three DEGs from the results of the transcriptional analysis and confirmed them through qRT-PCR. PtrActin and PtrUBQ were used as internal reference genes (Wang et al., 2020a) (Supplementary Table 1). Real-time PCR was conducted on a LightCycler 480 (Roche, Basel, Switzerland) using the SYBR Premix Ex Taq™ Kit (Takara, Dalian, China). The relative expression levels of the genes were calculated using the 2−ΔΔCT method (Livak and Schmittgen, 2001), and the data are presented as mean ± SD from three independent biological replicates.
Wood is the secondary xylem of trees, mainly composed of cellulose, hemicellulose, and lignin. All xylem cell types first undergo secondary cell wall (SCW) thickening and programmed cell death (Zhong and Ye, 2015; Zhang et al., 2020d). The plant height and ground diameter of annual Pd were significantly higher than those of Ps (Figures 1A,B). The SAM is an important regulatory site of plant height growth, and phloem and developing xylem are important parts of plant radial growth. The slice results showed that the SAM was surrounded by young leaves, and the SAM of Pd was conical and convex, while the Ps was flat, which suggested that the growth point of Ps might not be obvious enough, which caused the high growth to be slower (Figure 1C). The radial section of the stem showed that the phloem of Pd had wider phloem fibers, and the cambium was more obvious (Figure 1D). The average diameter of the vessel cells in the xylem of Pd was 72.5 μm, which was significantly larger than that of Ps (Figures 1D,E). We further determined the quality of wood. The basic density of basic Pd was less than Ps (Figure 1F), and there was no significant difference in the microfibril angle (Figure 1G). Compared with Ps, the fiber of Pd is short and thick (Figures 1H,I). Further material property determination found that the content of the three major elements of Pd is higher (Figures 1J–L).
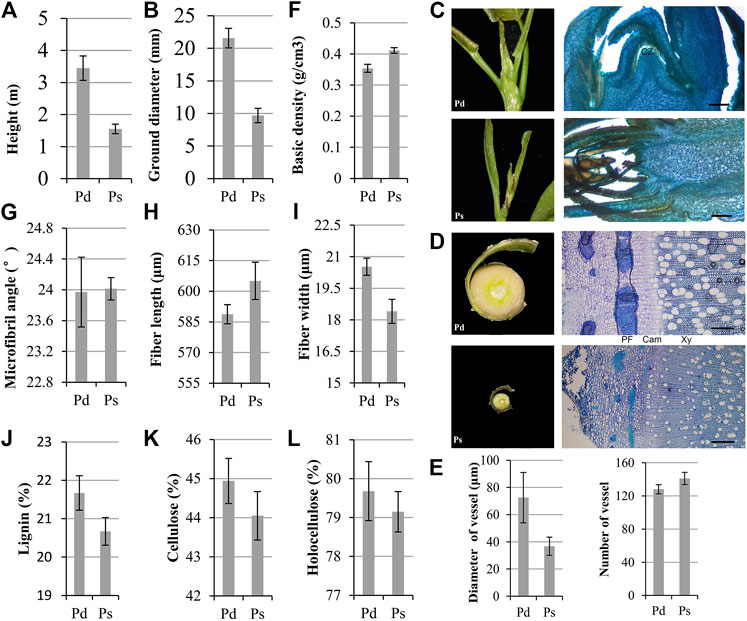
FIGURE 1. Phenotypic traits of P. deltoides “Danhong” (Pd) and P. simonii “Tongliao1” (Ps). Annual plant height (A) and ground diameter (B) of Pd and Ps. (C) Shoot apical meristem (SAM) image and section of Pd and Ps. Scale bars = 500 μm. (D) Cross sections of phloem–xylem region from 1-year-old trees. Scale bars = 200 μm. (E) Vessel size and number of vessel in the same area of Pd and Ps. CZ, central zone; PF, phloem fiber; Cam, cambium; Xy, xylem. Basic density (F), microfibril angle (G), fiber length (H), fiber width (I), lignin (J), cellulose (K), and holocellulose (L) content of Pd and Ps. Mean ± SD from three biological replicates.
As an important fast-growing tree, it is very important to understand the molecular pathways of growth and development of poplar. After trimming adapters and removing low-quality and contaminated reads, in total, 246.24 Gb clean data were obtained from 18 libraries, with an average Q30 of 93.00% (Supplementary Table 2). Finally, we identified 6,355 lncRNAs, with protections of 2,454 sense_overlapping lncRNAs, 2,004 of lincRNAs, and 1,897 of antisense lncRNAs (Table S3).
In order to analysis the characteristics of these lncRNAs, we evaluated the distribution of chromosome location, transcript length, exon number, and expression level of lncRNAs. In general, lincRNA, sense_overlapping, and antisense lncRNAs were evenly distributed on 19 chromosomes, although they had different emphases (Figure 2A). The average length of lncRNAs was 990 bp, and about 63.4% contained two exons (Figures 2B,C). Antisense lncRNAs ranged in length from 201 to 9,830 bp, and the average was 940 bp. lincRNAs ranged between 201 and 4,734 bp (average = 783 bp), and average length of sense_overlapping lncRNAs was 1,197 bp. The GC content of antisense lncRNA was 41.36%, which was significantly higher than that of lincRNAs and sense_overlapping lncRNAs (Figure 2D). For expression levels, the lncRNA expression levels were different and showed fewer average counts (FPKM = 4.44) than the coding transcripts (FPKM = 18.96) (Figures 2E,F).
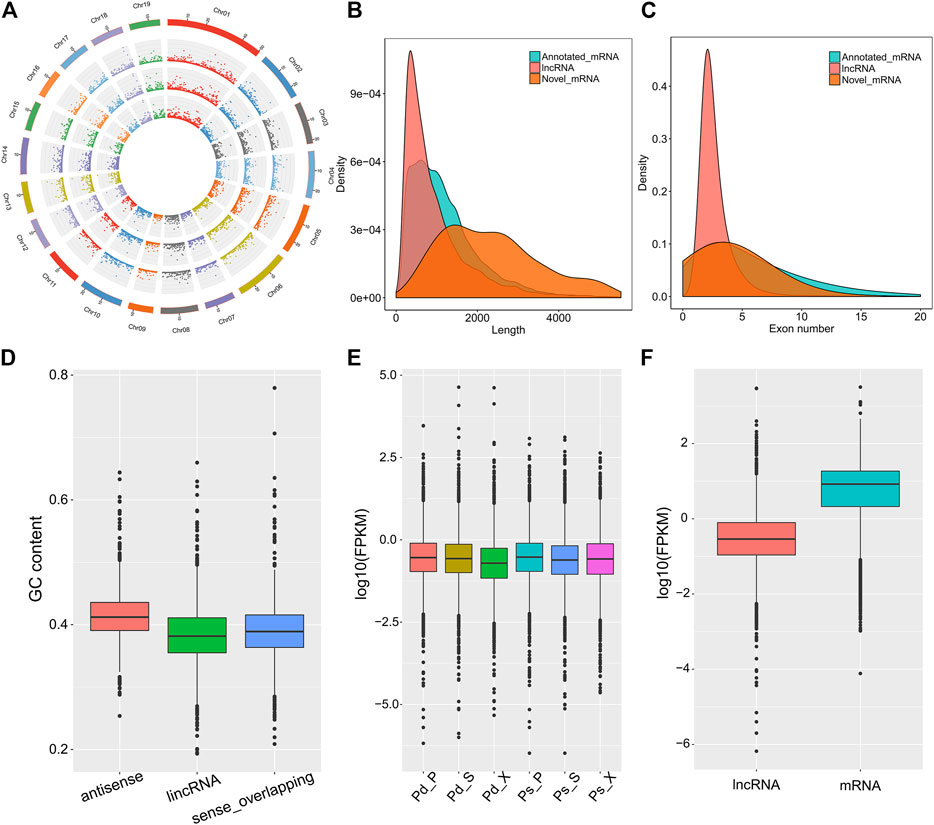
FIGURE 2. Characterization of cassava lncRNAs in Pd and Ps. (A) Distribution of lncRNAs along each chromosome for the SAM, phloem, and xylem in Pd from inside to outside. Distributions of length density (B) and exon numbers (C) in mRNAs, novel mRNAs, and novel lncRNAs. (D) GC content of lncRNAs. (E) Fragments per kilobase per million read (FPKM) values of lncRNAs in different samples. (F) Comparison of the expression levels of lncRNA and mRNA.
A principal component analysis (PCA) plot of the whole dataset revealed a sequential order of the different samples. The results showed that the SAM, xylem, and phloem of the two species were clustered into three groups, and the similarity of lncRNAs in tissues was greater than that between genotypes (Figure 3A).
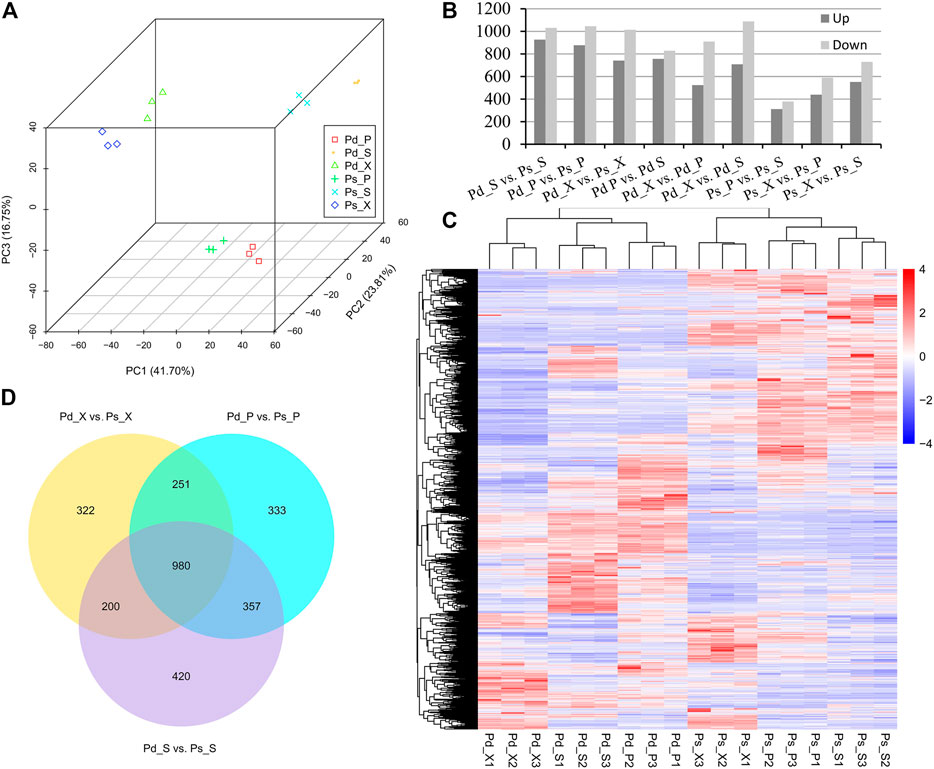
FIGURE 3. Differentially expressed (DE) lncRNAs identified in pair-wise comparison of 18 samples. (A) Principal component analysis (PCA) 3D of the expressed genes showing sample separation. (B) Number of DElncRNAs in different comparisons. (C) Heat map of DElncRNAs in the two genotypes. (D) Venn diagram showing DElncRNAs of Pd_X_vs_Ps_X, Pd_P_vs_Ps_P, and Pd_S_vs_Ps_S.
In order to further analyze whether these genes were differentially expressed between the two genotypes and different tissues, nine comparative combinations were carried out. Finally, 3,572 differentially expressed (DE) lncRNAs and 27,582 DEmRNAs were obtained. Among them, the DElncRNAs of Pd_S vs Ps_S were the highest in number, including 1957 lncRNAs (Figures 3B,C; Supplementary Figure 1). DEmRNAs participated in molecular functions such as “ADP binding” and “catalytic activity” (Supplementary Figure 1C). We also compared the DElncRNAs in three tissue difference genes between the two genotypes, and there were 980 DElncRNAs in the three comparison combinations (Pd_ X vs Ps_ X, Pd_ P vs Ps_ P and Pd_ S vs Ps_ S) and 322, 333, and 420 specifically expressed lncRNAs in the SAM, xylem, and phloem, respectively (Figure 3D).
Since lncRNAs play important roles in regulating gene expression, identification and analysis of their target genes may help us explore their potential functions. We calculated and predicted 13,932 co-localization pairs consisting of 3,413 lncRNAs and 10,627 RNAs and identified 72,038 co-expression pairs consisting of 1,975 lncRNAs and 11,709 RNAs (Tables S4, S5). To further analyze the function of these lncRNAs, we performed GO and KEGG analyses on their target genes. The colocation target genes of DElncRNAs were mainly enriched in 60 GO terms such as “ADP binding” and “nucleoside binding” (Supplementary Figure 2). Some target genes were enriched in the photosynthesis pathway, including 39 lncRNAs and 68 mRNAs. For example, TCONS_00135489 showed the same trend as its target, and the expression level of related genes was high in the SAM (Supplementary Figure 3). And 3,489 genes were found by co-location and co-expression of lncRNAs. They participated in immune response, cell death, purine nucleotide binding, and ATP binding progress, and the KEGG enrichment analysis shows that they were enriched in cytochrome P450, chalcone synthase, and phenylalanine ammonia-lyase which were important parts of phenylpropanoid biosynthesis (Figure S4).
The co-expression genes of Pd_ X vs Ps_ X, Pd_ P vs Ps_ P, and Pd_ S vs Ps_ S were mainly related to “ADP binding”, “heme binding”, and other biological functions (Figure 4A). And they were significantly enriched in the “phenylpropanoid biosynthesis” and “cyanoamino acid metabolism” pathway (Figure 4B). These possible target genes provide new insights into the role of lncRNAs in poplar development.
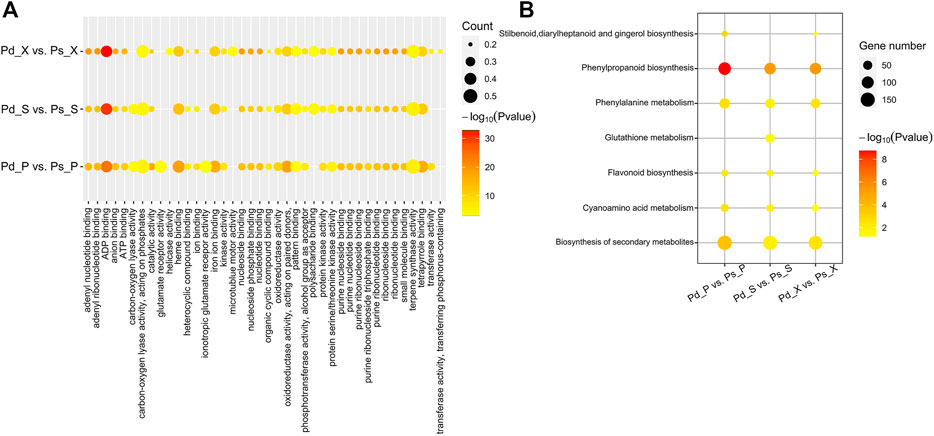
FIGURE 4. Co-expression mRNA of DElncRANs, GO, and KEGG enrichment. (A) GO enrichment analysis of differentially expressed lncRNA-target genes. (B) KEGG enrichment analysis of differentially expressed lncRNA-target genes. Node color represents −log10 corrected p-value.
Based on the predicted GO conditions of growth differential lncRNAs and the pathways associated with target genes, we speculated that lncRNAs might play an important role in phenylalanine biosynthesis in poplar. Phenylpropanoids are a group of plant secondary metabolites derived from phenylalanine, which has a variety of structural and signal molecular functions (Costa et al., 2003). It is the starting compound for biosynthesis of lignin, flavonoids, anthocyanins, etc., and a core mediator of crosstalk between development- and defense-related pathways (Alessandra et al., 2010). Further analysis co-expression network of these lncRNAs, and structure genes of lignins, and flavonoid biosynthesis found that TCONS_00128372, lincRNA, was located in Chr12 and interacted with MYB46, secondary wall–associated NAC domain2 (SND2), cinnamate-4-hydroxylase (C4H), caffeoyl-CoA 3-O-methyltransferase (CcoAMT), and laccase (LAC). And sense_overlapping lncRNA TCONS_00079190 co-expressed with MYB83, MYB46, NAC secondary wall thickening promoting factor1 (NST1), and LAC (Figure 5A). Similarly, the results of the study by Quan et al. (2019) and Zhou et al. (2017) showed that lncRNAs and miRNAs regulated lignin biosynthesis by regulating TFs such as bHLHs and bZIPs or directly coexpressing with structural genes in the process of lignin biosynthesis. And TCONS_00174042, TCONS_00101258, and TCONS_00136338 regulated the structural genes of flavonoid biosynthesis. TCONS_00174042 and TCONS_00101258 co-expressed with MYB3, chalcone and stilbene synthase (CHS), leucoanthocyanidin reductase (LAR), leucoanthocyanidin dioxygenase (LDOX), and naringenin 3- dioxygenase (F3H). In addition, TCONS_00136338 can coexpress with LDOX, CHS, UFGT, LAR, and dihydroflavonol 4-reductase (DFR) (Figure 5B). Lignin-related lncRNAs and structural genes were highly expressed in xylem of Pd and Ps, while flavonoid-related genes were highly expressed in the SAM (Figure S5).
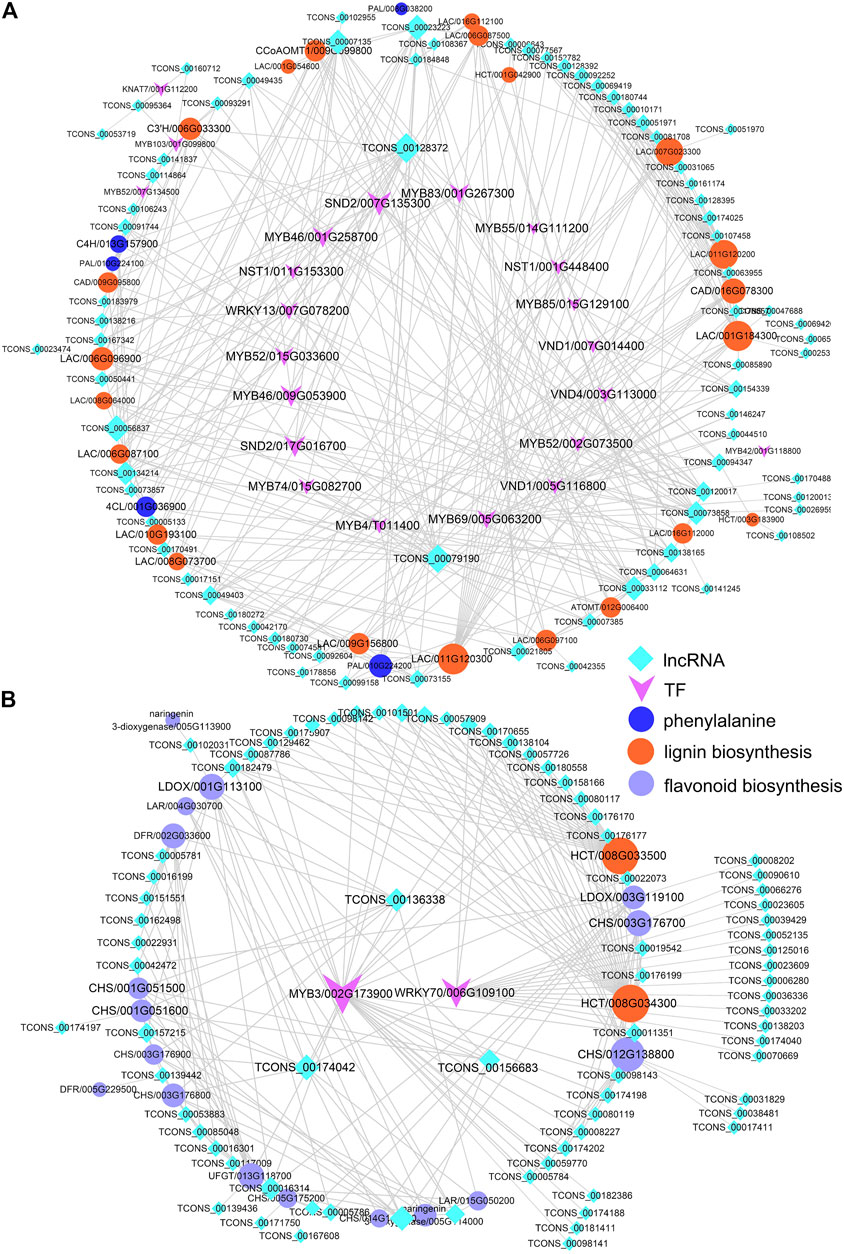
FIGURE 5. Interaction network diagrams. Interaction networks of differentially expressed lncRNAs, DEmRNAs, and transcription factors (TFs) of phenylalanine biosynthesis, including lignin (A) and flavonoid (B) biosynthesis.
Hormones such as auxin, cytokinin, and gibberellin play important roles in primary and secondary growth (Nieminen et al., 2008; Xu et al., 2019). In order to further analyze the relationship between lncRNAs and plant hormones, we constructed a co-expression network including the structural genes of auxin, cytokinin, and gibberellin biosynthesis and their possible regulatory lncRNAs (Figure 6A). For example, TCONS_00134627 could be coexpressed with GA2OX8 (Potri.011G134000), GASA10 (Potri.009G092600), and SAUR94 (Potri.009G127300) (Figure 6A). The expression levels of DEGs and DElncRNAs related to auxin, cytokinin, and gibberellin biosynthesis were shown in the heatmap (Figure 6B). The differential genes related to auxins and gibberellins were mainly concentrated in the high expression of Pd_S and Pd_X, while only a few genes are highly expressed in the phloem. And the genes related to cytokinins are highly expressed in Ps_P (Figure 6B).
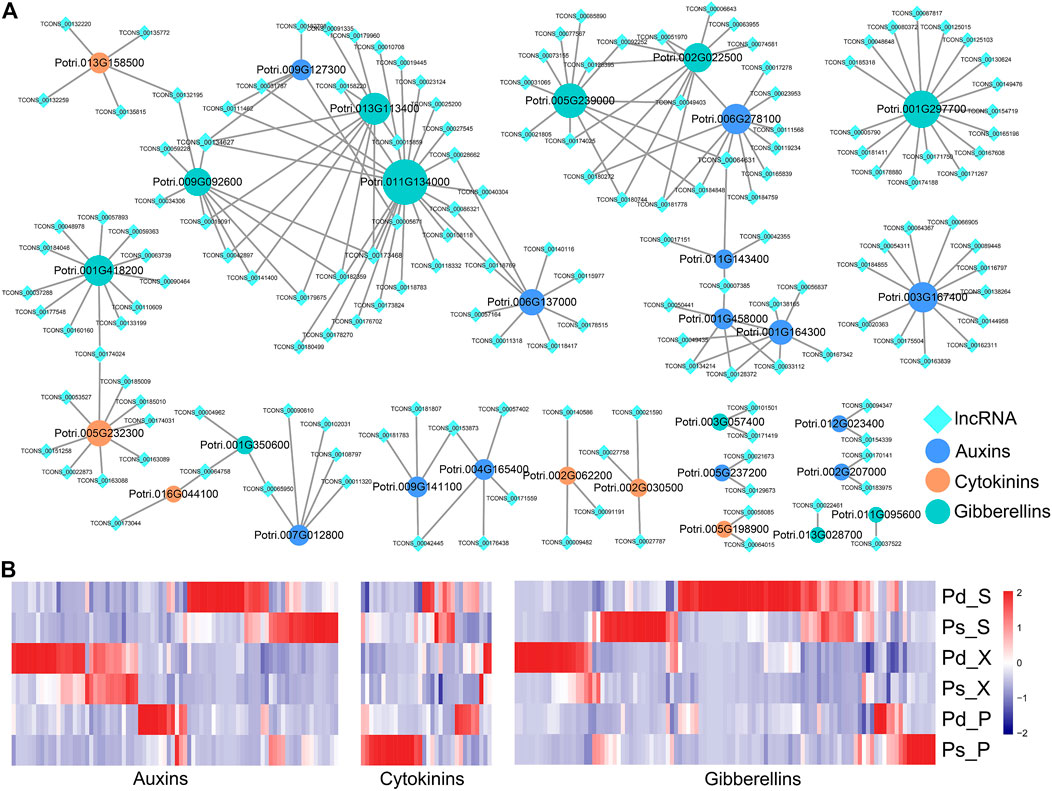
FIGURE 6. lncRNAs related to plant hormone biosynthesis and signal transduction. (A) Co-expression network of DElncRNAs and protein coding genes (PCgenes) involved in auxin, cytokinin, and gibberellin biosynthesis and signal transduction. (B) Expression of selected lncRNAs and their predicted co-expression target PCgenes involved in plant hormone biosynthesis signal transduction.
To further analyze whether lncRNAs were used as ceRNAs to absorb miRNAs, inhibited the effect of miRNAs, and promoted mRNA expression, we conducted sequential analysis. MicroRNAs, a major class of small RNAs with 20–24 nucleotides, create various aspects of plant development and stress responses through posttranscriptionally regulated gene expression (Yu et al., 2019). A total of 658 miRNA–lncRNA pairs consisting of 188 miRNAs and 200 lncRNAs were identified, including 19 plant hormone–related pairs and 28 phenylalanine-related pairs (Supplementary Table 6). TCONS_00066905, hormone-related lncRNA, was predicted to be a target mimic of miR396a and miR396b. In addition, phenylalanine-related TCONS_00023606 and TCONS_00093325 were the target mimics of flavonoid-related regulatory genes miR156h and miR828a (Figure 7A). miR396-GRF was an important regulatory module of plant growth and development. We found that 12 GRFs were differentially expressed and highly expressed in the SAM of Pd, which may be an important reason for the rapid growth of Pd (Figure 7B).
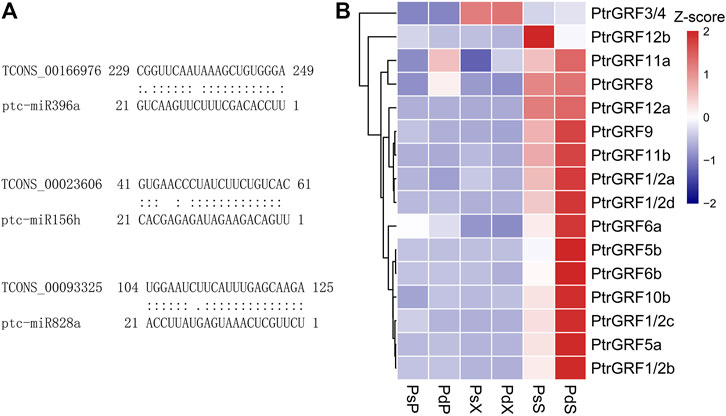
FIGURE 7. Expression analysis of related lncRNAs as potential targets or target mimics of miRNAs. (A) Predicted binding alignment of lncRNAs with ptc-miR396a, ptc-miR156h, and ptc-miR828a. (B) Expression patterns of differentially expressed GRFs and the target genes of miR396 in the two species.
In order to verify the accuracy of transcriptome data, differential genes related to xylem development and hormone signal transduction were selected for qRT-PCR verification. The results showed that the expression trends of three lncRNAs and three mRNAs in six tissues were consistent with those of qRT-PCR, which proved the reliability of transcriptome data (Figure S6).
Poplar is an important bioenergy tree in the world. Its growth rate and wood quality determine its economic value. With the development of RNA-seq technology, genome-wide mapping has been proved to be a powerful tool for studying primary and secondary growth of poplar. Pd and Ps have great differences in plant height, ground diameter, and other growth characters, as well as wood qualities, such as basic density and lignin content (Figure 1). In this study, lncRNAs of woody plants was comprehensively analyzed to study their growth and development and the regulation of wood quality. We identified 6,355 lncRNAs, including 3,572 DElncRNAs and 27, 582 DEGs. lncRNAs in Pd and Ps show similar characteristics with other species (Ci et al., 2019). They are characterized by high tissue specificity and short length. The length of lncRNAs was about 991 bp, and the cis- and trans- effects were recognized. In this study, we identified a large number of trans-regulatory networks, mainly acting on the phenylalanine pathway. These differentially expressed genes may be an important reason for the differences in growth rate and wood quality.
The SCW provides rigidity and strength for plants to support their body weight and ensure water and nutrient transport (Oda and Fukuda, 2012; Wang and Dixon, 2012). It is mainly composed of lignin, cellulose, and hemicellulose, and their biosynthesis was highly related to the transformation and production of biofuels and biological products (Mohnen, 2008; Hussey et al., 2019). It is regulated by microRNA, MYB, NAC, and WRKY in the SCW complex regulatory network (Zhang et al., 2018b). As an important support to the SCW, lignin determines the conversion efficiency of poplar as biomass energy. In previous studies, lncRNAs participate in the lignin biosynthesis of poplar with TFs and miRNAs (Quan et al., 2019; Zhang et al., 2020c). Also, in our research, we found that 75 lncRNAs, including TCONS_00079190, TCONS_00128372, and TCONS_00007135, can be directly coexpressed with MYBs, VNDs, and lignin structural genes (Figure 5A). Flavonoids and lignin are the metabolic pathways of phenylalanine metabolism, and there are some common structural genes. At the same time, flavonoids are important compounds for plants to respond to biological and abiotic stresses. lncRNAs are involved in regulating the anthocyanin biosynthetic pathways in strawberry, buckthorn, and apple (Zhang et al., 2018a; Lin et al., 2018; Yang et al., 2019). We identified a large number of lncRNAs coexpressed with structural genes of the flavonoid pathway, such as CHS and DFR (Figure 5B). The insertion of five cisgenes encoding gibberellin metabolism or signal proteins affects plant growth (Han et al., 2011). The auxin-mediated Aux/IAA-ARF-HB signal cascade regulates the development of the secondary xylem of poplar (Xu et al., 2019). And it was found that a large number of auxin- and gibberellin-related lncRNA–mRNA coexpression networks were identified (Figure 6), which was similar to endogenous hormone regulation in the secondary xylem and during tension wood formation in Catalpa bungei (Xiao et al., 2020). Therefore, lncRNAs were widely involved in lignin and flavonoid metabolism and plant growth of Pd and Ps, and they affect their differences.
Posttranscriptional regulation is an important process affecting gene expression and plant development (Li and Zhang, 2016; Yu et al., 2019). miR156 and miR828 are involved in the biosynthesis of flavonoids and anthocyanins by regulating MYBs in many species (Tirumalai et al., 2019; B. Zhang et al., 2020a; Wang et al., 2020b). We identified TCONS_00023606 and TCONS_00093325 as the target genes of miRNA156 and miR828, respectively, so they may indirectly participate in the transcriptional regulation of flavonoids through this pathway (Figure 7; Supplementary Table 3). miR396, GRFs, and GRF-INTERACTING FACTORS (GIFs) have been proven to control the growth of multiple tissues and organs of multiple species (Debernardi et al., 2014; Liebsch and Palatnik, 2020). GRFs are important regulators of the SAM, which are the starting sites of leaf and stem development (Wang Y. et al., 2020). The high expression of GRFs in Pd_S may be an important reason for the rapid growth of Pd. lncRNAs–miRNAs–TFs–mRNAs play an important role in regulating the growth of poplars. Therefore, the differences in the growth rate and wood quality of the two poplars may be caused by the joint regulation of these factors, which requires our follow-up for further functional verification.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
JH conceived and designed the project. LZ, XG, JD, XC, and XP participated in the experiments and data analysis. LZ drafted the manuscript. LZ and JH modified the manuscript. All authors read and approved the final manuscript.
This study was supported by the National Key Research and Development Program of China (2021YFD2200201), the National Natural Science Foundation (32071797), and the National Key Program on Transgenic Research (2018ZX08020002).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations or those of the publisher, the editors, and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2021.762678/full#supplementary-material
Supplementary Figure 1 | Differentially expressed mRNAs (DEGs) in pair-wise comparison of 18 samples. Number of different genes (A) and heat maps (B) of different combinations were compared. (C) Gene Ontology (GO) enrichment of DEGs in different comparisons. It is detailed enrichment of the biological process (BP). Node color represents −log10 corrected p-value.
Supplementary Figure 2 | GO enrichment of lncRNA colocation target genes in nine comparative combinations.
Supplementary Figure 3 | Expression profile of lncRNAs and mRNAs related to photosynthesis in different tissues.
Supplementary Figure 4 | GO (A) and KEGG (B) analyses of co-location and co-expression mRNAs. BP, biological process; MF, molecular function; CC, cellular component.
Supplementary Figure 5 | Expression patterns of lncRNAs (A) and mRNAs (B) related to lignin and flavonoid biosynthesis.
Supplementary Figure 6 | lncRNA and mRNA results from RNA-seq verified by qRT-PCR. Mean ± SD from three biological replicates.
Supplementary Table 1 | Primer pairs for qRT- PCR.
Supplementary Table 2 | RNA-seq filter data.
Supplementary Table 3 | Basic information of lncRNAs was identified in P. deltoides “Danhong” and P. simonii “Tongliao1.”
Supplementary Table 4 | Co-location target mRNAs of lncRNAs.
Supplementary Table 5 | Co-expression mRNAs of lncRNAs.
Supplementary Table 6 | Phenylpropanoid biosynthesis–related lncRNAs targeted by miRNAs.
Alessandra, L., Luca, P., and Adriano, M. (2010). Differential Gene Expression in Kernels and Silks of maize Lines with Contrasting Levels of Ear Rot Resistance after Fusarium Verticillioides Infection. J. Plant Physiol. 167, 1398–1406. doi:10.1016/j.jplph.2010.05.015
Ashburner, M., Ball, C. A., Blake, J. A., Botstein, D., Butler, H., Cherry, J. M., et al. (2000). Gene Ontology: Tool for the Unification of Biology. Nat. Genet. 25, 25–29. doi:10.1038/75556
Chen, M., Wang, C., Bao, H., Chen, H., and Wang, Y. (2016). Genome-wide Identification and Characterization of Novel lncRNAs in Populus under Nitrogen Deficiency. Mol. Genet. Genomics 291, 1663–1680. doi:10.1007/s00438-016-1210-3
Ci, D., Tian, M., Song, Y., Du, Q., Quan, M., Xuan, A., et al. (2019). Indole-3-acetic Acid Has Long-Term Effects on Long Non-coding RNA Gene Methylation and Growth in Populus Tomentosa. Mol. Genet. Genomics 294, 1511–1525. doi:10.1007/s00438-019-01593-5
Conesa, A., Götz, S., García-Gómez, J. M., Terol, J., Talón, M., and Robles, M. (2005). Blast2GO: a Universal Tool for Annotation, Visualization and Analysis in Functional Genomics Research. Bioinformatics 21, 3674–3676. doi:10.1093/bioinformatics/bti610
Costa, M. A., Collins, R. E., Anterola, A. M., Cochrane, F. C., Davin, L. B., and Lewis, N. G. (2003). An In Silico Assessment of Gene Function and Organization of the Phenylpropanoid Pathway Metabolic Networks in Arabidopsis thaliana and Limitations Thereof. Phytochemistry 64, 1097–1112. doi:10.1016/S0031-9422(03)00517-X
Debernardi, J. M., Mecchia, M. A., Vercruyssen, L., Smaczniak, C., Kaufmann, K., Inze, D., et al. (2014). Post-transcriptional Control ofGRFtranscription Factors by microRNA miR396 and GIF Co-activator Affects Leaf Size and Longevity. Plant J. 79, 413–426. doi:10.1111/tpj.12567
Elo, A., Immanen, J., Nieminen, K., and Helariutta, Y. (2009). Stem Cell Function during Plant Vascular Development. Semin. Cel Dev. Biol. 20, 1097–1106. doi:10.1016/j.semcdb.2009.09.009
Guerra, F. P., Wegrzyn, J. L., Sykes, R., Davis, M. F., Stanton, B. J., and Neale, D. B. (2013). Association Genetics of Chemical wood Properties in Black poplar ( Populus Nigra ). New Phytol. 197, 162–176. doi:10.1111/nph.12003
Ha, C. M., Jun, J. H., and Fletcher, J. C. (2010). Shoot Apical Meristem Form and Function. Curr. Top. Dev. Biol. 91, 103–140. doi:10.1016/S0070-2153(10)91004-1
Han, K. M., Dharmawardhana, P., Arias, R. S., Ma, C., Busov, V., and Strauss, S. H. (2011). Gibberellin-associated Cisgenes Modify Growth, Stature and wood Properties in Populus. Plant Biotechnol. J. 9, 162–178. doi:10.1111/j.1467-7652.2010.00537.x
Heo, J. B., and Sung, S. (2011). Vernalization-mediated Epigenetic Silencing by a Long Intronic Noncoding RNA. Science 331, 76–79. doi:10.1126/science.1197349
Hussey, S. G., Grima-Pettenati, J., Myburg, A. A., Mizrachi, E., Brady, S. M., Yoshikuni, Y., et al. (2019). A Standardized Synthetic Eucalyptus Transcription Factor and Promoter Panel for Re-engineering Secondary Cell Wall Regulation in Biomass and Bioenergy Crops. ACS Synth. Biol. 8, 463–465. doi:10.1021/acssynbio.8b00440
Jia, H., Osak, M., Bogu, G. K., Stanton, L. W., Johnson, R., and Lipovich, L. (2010). Genome-wide Computational Identification and Manual Annotation of Human Long Noncoding RNA Genes. RNA 16, 1478–1487. doi:10.1261/rna.1951310
Jiang, H., Jia, Z., Liu, S., Zhao, B., Li, W., Jin, B., et al. (2019). Identification and Characterization of Long Non-coding RNAs Involved in Embryo Development of Ginkgo Biloba. Plant Signaling Behav. 14, 1674606. doi:10.1080/15592324.2019.1674606
Kim, D., Langmead, B., and Salzberg, S. L. (2015). HISAT: a Fast Spliced Aligner with Low Memory Requirements. Nat. Methods 12, 357–360. doi:10.1038/nmeth.3317
Kong, L., Zhang, Y., Ye, Z.-Q., Liu, X.-Q., Zhao, S.-Q., Wei, L., et al. (2007). CPC: Assess the Protein-Coding Potential of Transcripts Using Sequence Features and Support Vector Machine. Nucleic Acids Res. 35, W345–W349. doi:10.1093/nar/gkm391
Li, C., and Zhang, B. (2016). MicroRNAs in Control of Plant Development. J. Cel. Physiol 231, 303–313. doi:10.1002/jcp.25125
Liebsch, D., and Palatnik, J. F. (2020). MicroRNA miR396, GRF Transcription Factors and GIF Co-regulators: a Conserved Plant Growth Regulatory Module with Potential for Breeding and Biotechnology. Curr. Opin. Plant Biol. 53, 31–42. doi:10.1016/j.pbi.2019.09.008
Lin, Y., Jiang, L., Chen, Q., Li, Y., Zhang, Y., Luo, Y., et al. (2018). Comparative Transcriptome Profiling Analysis of Red- and White-Fleshed Strawberry (Fragaria×ananassa) Provides New Insight into the Regulation of the Anthocyanin Pathway. Plant Cel Physiol 59, 1844–1859. doi:10.1093/pcp/pcy098
Liu, J., Jung, C., Xu, J., Wang, H., Deng, S., Bernad, L., et al. (2012). Genome-wide Analysis Uncovers Regulation of Long Intergenic Noncoding RNAs in. Plant Cell 24, 4333–4345. doi:10.1105/tpc.112.102855
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2−ΔΔCT Method. Methods 25, 402–408. doi:10.1006/meth.2001.1262
Love, M. I., Huber, W., and Anders, S. (2014). Moderated Estimation of Fold Change and Dispersion for RNA-Seq Data with DESeq2. Genome Biol. 15, 550. doi:10.1186/s13059-014-0550-8
Ma, J., Bai, X., Luo, W., Feng, Y., Shao, X., Bai, Q., et al. (2019). Genome-wide Identification of Long Noncoding RNAs and Their Responses to Salt Stress in Two Closely Related Poplars. Front. Genet. 10, 777. doi:10.3389/fgene.2019.00777
Ma, L., Bajic, V. B., and Zhang, Z. (2013). On the Classification of Long Non-coding RNAs. RNA Biol. 10, 924–933. doi:10.4161/rna.24604
Mohnen, D. (2008). Pectin Structure and Biosynthesis. Curr. Opin. Plant Biol. 11, 266–277. doi:10.1016/j.pbi.2008.03.006
Nieminen, K., Immanen, J., Laxell, M., Kauppinen, L., Tarkowski, P., Dolezal, K., et al. (2008). Cytokinin Signaling Regulates Cambial Development in poplar. Proc. Natl. Acad. Sci. 105, 20032–20037. doi:10.1073/pnas.0805617106
Oda, Y., and Fukuda, H. (2012). Secondary Cell wall Patterning during Xylem Differentiation. Curr. Opin. Plant Biol. 15, 38–44. doi:10.1016/j.pbi.2011.10.005
Otasek, D., Morris, J. H., Bouças, J., Pico, A. R., and Demchak, B. (2019). Cytoscape Automation: Empowering Workflow-Based Network Analysis. Genome Biol. 20, 185. doi:10.1186/s13059-019-1758-4
Pertea, M., Pertea, G. M., Antonescu, C. M., Chang, T.-C., Mendell, J. T., and Salzberg, S. L. (2015). StringTie Enables Improved Reconstruction of a Transcriptome from RNA-Seq Reads. Nat. Biotechnol. 33, 290–295. doi:10.1038/nbt.3122
Quan, M., Du, Q., Xiao, L., Lu, W., Wang, L., Xie, J., et al. (2019). Genetic Architecture Underlying the Lignin Biosynthesis Pathway Involves Noncoding RNAs and Transcription Factors for Growth and wood Properties in Populus. Plant Biotechnol. J. 17, 302–315. doi:10.1111/pbi.12978
Severing, E., Faino, L., Jamge, S., Busscher, M., Kuijer-Zhang, Y., Bellinazzo, F., et al. (2018). Arabidopsis thaliana Ambient Temperature Responsive lncRNAs. BMC Plant Biol. 18, 145. doi:10.1186/s12870-018-1362-x
Song, Y., Chen, P., Liu, P., Bu, C., and Zhang, D. (2020). High-temperature-responsive poplar lncRNAs Modulate Target Gene Expression via RNA Interference and Act as RNA Scaffolds to Enhance Heat Tolerance. Ijms 21, 6808. doi:10.3390/ijms21186808
Sun, L., Luo, H., Bu, D., Zhao, G., Yu, K., Zhang, C., et al. (2013). Utilizing Sequence Intrinsic Composition to Classify Protein-Coding and Long Non-coding Transcripts. Nucleic Acids Res. 41, e166. doi:10.1093/nar/gkt646
Sun, X., Zheng, H., and Sui, N. (2018). Regulation Mechanism of Long Non-coding RNA in Plant Response to Stress. Biochem. Biophysical Res. Commun. 503, 402–407. doi:10.1016/j.bbrc.2018.07.072
Tian, J., Song, Y., Du, Q., Yang, X., Ci, D., Chen, J., et al. (2016). Population Genomic Analysis of Gibberellin-Responsive Long Non-coding RNAs inPopulus. Exbotj 67, 2467–2482. doi:10.1093/jxb/erw057
Tirumalai, V., Swetha, C., Nair, A., Pandit, A., and Shivaprasad, P. V. (2019). miR828 and miR858 Regulate VvMYB114 to Promote Anthocyanin and Flavonol Accumulation in Grapes. J. Exp. Bot. 70, 4775–4792. doi:10.1093/jxb/erz264
Wang, H.-L. V., and Chekanova, J. A. (2017). Long Noncoding RNAs in Plants. Adv. Exp. Med. Biol. 1008, 133–154. doi:10.1007/978-981-10-5203-3_5
Wang, H.-Z., and Dixon, R. A. (2012). On-off Switches for Secondary Cell wall Biosynthesis. Mol. Plant. 5, 297–303. doi:10.1093/mp/ssr098
Wang, J., Zhou, H., Zhao, Y., Sun, P., Tang, F., Song, X., et al. (2020a). Characterization of poplar Growth-Regulating Factors and Analysis of Their Function in Leaf Size Control. BMC Plant Biol. 20, 509. doi:10.1186/s12870-020-02699-4
Wang, L., Park, H. J., Dasari, S., Wang, S., Kocher, J.-P., and Li, W. (2013). CPAT: Coding-Potential Assessment Tool Using an Alignment-free Logistic Regression Model. Nucleic Acids Res. 41, e74. doi:10.1093/nar/gkt006
Wang, Y., Liu, W., Wang, X., Yang, R., Wu, Z., Wang, H., et al. (2020b). MiR156 Regulates Anthocyanin Biosynthesis through SPL Targets and Other microRNAs in poplar. Hortic. Res. 7, 118. doi:10.1038/s41438-020-00341-w
Wei, Z., Du, Q., Zhang, J., Li, B., and Zhang, D. (2013). Genetic Diversity and Population Structure in Chinese Indigenous poplar (Populus Simonii) Populations Using Microsatellite Markers. Plant Mol. Biol. Rep. 31, 620–632. doi:10.1007/s11105-012-0527-2
Wright, M. W. (2014). A Short Guide to Long Non-coding RNA Gene Nomenclature. Hum. Genomics 8, 7. doi:10.1186/1479-7364-8-7
Wu, X., Shi, T., Iqbal, S., Zhang, Y., Liu, L., and Gao, Z. (2019). Genome-wide Discovery and Characterization of Flower Development Related Long Non-coding RNAs in Prunus Mume. BMC Plant Biol. 19, 64. doi:10.1186/s12870-019-1672-7
Xiao, Y., Yi, F., Ling, J., Yang, G., Lu, N., Jia, Z., et al. (2020). Genome-wide Analysis of lncRNA and mRNA Expression and Endogenous Hormone Regulation during Tension wood Formation in Catalpa Bungei. BMC Genomics 21, 609. doi:10.1186/s12864-020-07044-5
Xu, C., Shen, Y., He, F., Fu, X., Yu, H., Lu, W., et al. (2019). Auxin‐mediated Aux/IAA ‐ ARF ‐ HB Signaling cascade Regulates Secondary Xylem Development in Populus. New Phytol. 222, 752–767. doi:10.1111/nph.15658
Xu, W., Yang, T., Wang, B., Han, B., Zhou, H., Wang, Y., et al. (2018). Differential Expression Networks and Inheritance Patterns of Long Non-coding RNAs in castor Bean Seeds. Plant J. 95, 324–340. doi:10.1111/tpj.13953
Xue, Z., Liu, L., and Zhang, C. (2020). Regulation of Shoot Apical Meristem and Axillary Meristem Development in Plants. Ijms 21, 2917. doi:10.3390/ijms21082917
Yang, T., Ma, H., Zhang, J., Wu, T., Song, T., Tian, J., et al. (2019). Systematic Identification of Long Noncoding RNA S Expressed during Light‐induced Anthocyanin Accumulation in Apple Fruit. Plant J. 100, 572–590. doi:10.1111/tpj.14470
Ye, Z.-H., and Zhong, R. (2015). Molecular Control of wood Formation in Trees. Exbotj 66, 4119–4131. doi:10.1093/jxb/erv081
Yu, Y., Zhang, Y., Chen, X., and Chen, Y. (2019). Plant Noncoding RNAs: Hidden Players in Development and Stress Responses. Annu. Rev. Cel Dev. Biol. 35, 407–431. doi:10.1146/annurev-cellbio-100818-125218
Zhan, T., Lu, J., Zhou, X., and Lu, X. (2015). Representative Volume Element (RVE) and the Prediction of Mechanical Properties of Diffuse Porous Hardwood. Wood Sci. Technol. 49, 147–157. doi:10.1007/s00226-014-0687-3
Zhang, B., Yang, H.-J., Yang, Y.-Z., Zhu, Z.-Z., Li, Y.-N., Qu, D., et al. (2020a). mdm-miR828 Participates in the Feedback Loop to Regulate Anthocyanin Accumulation in Apple Peel. Front. Plant Sci. 11, 608109. doi:10.3389/fpls.2020.608109
Zhang, C., Li, S., Zhao, Z., Hu, J., and Han, Y. (2008). A New poplar Variety Populus deltoides 'Danhong'. Scientia Silvae Sinicae 44, 169. doi:10.11707/j.1001-7488.20080127
Zhang, G., Chen, D., Zhang, T., Duan, A., Zhang, J., and He, C. (2018a). Transcriptomic and Functional Analyses Unveil the Role of Long Non-coding RNAs in Anthocyanin Biosynthesis during Sea Buckthorn Fruit Ripening. DNA Res. 25, 465–476. doi:10.1093/dnares/dsy017
Zhang, J., Song, X., Zhang, L., Jia, H., Peng, X., Zhao, Z., et al. (2020b). Agronomic Performance of 27 Populus Clones Evaluated after Two 3‐year Coppice Rotations in Henan, China. GCB Bioenergy 12, 168–181. doi:10.1111/gcbb.12662
Zhang, J., Tuskan, G. A., Tschaplinski, T. J., Muchero, W., and Chen, J.-G. (2020c). Transcriptional and post-transcriptional Regulation of Lignin Biosynthesis Pathway Genes in Populus. Front. Plant Sci. 11, 652. doi:10.3389/fpls.2020.00652
Zhang, J., Xie, M., Tuskan, G. A., Muchero, W., and Chen, J.-G. (2018b). Recent Advances in the Transcriptional Regulation of Secondary Cell wall Biosynthesis in the Woody Plants. Front. Plant Sci. 9, 1535. doi:10.3389/fpls.2018.01535
Zhang, L., Liu, B., Zhang, J., and Hu, J. (2020d). Insights of Molecular Mechanism of Xylem Development in Five Black poplar Cultivars. Front. Plant Sci. 11, 620. doi:10.3389/fpls.2020.00620
Zhong, R., and Ye, Z.-H. (2015). Secondary Cell walls: Biosynthesis, Patterned Deposition and Transcriptional Regulation. Plant Cel Physiol 56, 195–214. doi:10.1093/pcp/pcu140
Keywords: lncRNA–mRNA, poplar, phenylalanine biosynthesis, xylem, hormone
Citation: Zhang L, Ge X, Du J, Cheng X, Peng X and Hu J (2021) Genome-Wide Identification of Long Non-Coding RNAs and Their Potential Functions in Poplar Growth and Phenylalanine Biosynthesis. Front. Genet. 12:762678. doi: 10.3389/fgene.2021.762678
Received: 22 August 2021; Accepted: 11 October 2021;
Published: 15 November 2021.
Edited by:
Deqiang Zhang, Beijing Forestry University, ChinaCopyright © 2021 Zhang, Ge, Du, Cheng, Peng and Hu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiaopeng Peng, eHBAY2FmLmFjLmNu; Jianjun Hu, aHVqakBjYWYuYWMuY24=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.