
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Genet. , 01 October 2021
Sec. Genomics of Plants and the Phytoecosystem
Volume 12 - 2021 | https://doi.org/10.3389/fgene.2021.762135
This article is part of the Research Topic Transcriptional/Post-Transcriptional Regulations in Agricultural Species After Stresses View all 18 articles
Prunus mume is an illustrious ornamental woody plant with colorful flowers, delicate fragrances, and graceful tree forms. Low temperature limits its geographical distribution. The basic helix-loop-helix (bHLH) proteins exist in most eukaryotes as a transcription factor superfamily, which play a crucial role in metabolism, physiology, development, and response to various stresses of higher organisms. However, the characteristics of the bHLH gene family and low-temperature response remain unknown in P. mume. In the present study, we distinguished 95 PmbHLH genes in the P. mume whole-genome and analyzed their features. PmbHLHs were divided into 23 subfamilies and one orphan by phylogenetic analysis. Similar gene structures and conserved motifs appeared in the same subfamily. These genes were situated in eight chromosomes and scaffolds. Gene duplication events performed a close relationship to P. mume, P. persica, and P. avium. Tandem duplications probably promoted the expansion of PmbHLHs. According to predicted binding activities, the PmbHLHs were defined as the Non-DNA-binding proteins and DNA-binding proteins. Furthermore, PmbHLHs exhibited tissue-specific and low-temperature induced expression patterns. By analyzing transcriptome data, 10 PmbHLHs which are responsive to low-temperature stress were selected. The qRT-PCR results showed that the ten PmbHLH genes could respond to low-temperature stress at different degrees. There were differences in multiple variations among different varieties. This study provides a basis to research the evolution and low-temperature tolerance of PmbHLHs, and might enhance breeding programs of P. mume by improving low-temperature tolerance.
Plants are subject to various unsuitable environmental stresses when they grow in a natural environment. Low-temperature stress is a severe natural disaster, which divides into chilling stress and freezing stress. Chilling stress mainly affects the process of photosynthesis and respiratory metabolism of plants, resulting in the disorder of plant cell function to make them grow abnormally and even causing growth stagnation (Pearce, 1988). Freezing stress freezes plant cells, then causes mechanical damage to the plant cell membrane and eventually might cause plant death (Mccully et al., 2004; Knight et al., 2009). To adapt and resist low-temperature stress, plants have evolved a set of complex and fine regulation mechanisms. Transcription factors are vital in plant signal regulatory networks. When plants suffer from low-temperature stress, transcription factors can activate low-temperature responsive genes by binding cis-acting elements on gene promoters. Thus, they regulate signal transduction pathways to improve low-temperature tolerance in plants.
The basic helix–loop–helix (bHLH) proteins, which belong to superfamily transcription factors are widely spread in plants, animals, and fungi (Ledent and Vervoort, 2001). The bHLH superfamily contains two highly conserved domains: the basic region and helix-loop-helix (HLH) region (Atchley et al., 1999). The basic region is composed of 15–20 amino acids and is located at the N-terminal of the bHLH domain, which can recognize and bind DNA (Atchley and Fitch, 1997). In the plant bHLH domain, 50% of the basic region contains a highly conserved His5-Glu9-Arg13 sequence, which can bind to E-box (5′-CANNTG-3′) element. This is necessary for bHLH to bind to DNA (Pires and Dolan, 2010). In the C-terminus of the bHLH domain, the HLH region is composed of about 40 amino acids. This region is characterized by two α-helices connected by a loop with variable length (Murre et al., 1989). The HLH domain can promote interaction between proteins to form homodimers or heterodimers and interact with E-box elements in the genes promoter region (Massari and Murre, 2000; Huq and Quail, 2002). Therefore, the biological functions of most bHLH transcription factors involve forming dimers.
The bHLH transcription factors are related to plant growth and development (Groszmann et al., 2008; Ding et al., 2009), floral organ formation (Buti et al., 2020), secondary metabolism (Nemesio-Gorriz et al., 2017; Wang et al., 2019), and stress resistance (Chinnusamy, 2003; Zhou et al., 2009; Seo et al., 2011; Gao et al., 2020). Under low-temperature stress, many studies have proved that bHLHs are involved in regulation. For example, ICE1 (INDUCER OF CBF EXPRESSION 1), which belongs to the bHLH transcription factor family, could activate CBF3 and COR genes in response to low temperature in Arabidopsis thaliana. Meanwhile, other bHLH transcription factors could also regulate cold tolerance in plants. In rice seedlings, cold stress specifically induced OsbHLH1 gene expression (Wang et al., 2003). Apple MdCIbHLH1 played a role in cold tolerance in a CBF dependent manner (Feng et al., 2012).
Prunus mume Sieb. et Zucc, a crucial woody plant with excellent ornamental characteristics for various colors, delicate fragrances, ample flower shapes, and abundant tree forms has been widely used for plant landscaping. P. mume originated in the Yangtze River Basin and Southwest China. The cultivar was distributed in Northern China and East Asia, with domestication taking place over a long time (Zhang et al., 2018). However, low temperature is still a limiting factor for the northward distribution of P. mume. Therefore, it is essential to enhance cold resistance to expand distribution. The bHLH transcription factors have been proved to be involved in resisting low-temperature stress. Nevertheless, the identification of bHLH genes has still not been conducted in P. mume. In the present study, we identified 95 PmbHLH genes and performed a comprehensive bioinformatics analysis based on P. mume genome-wide. We analyzed gene identification, phylogenetic tree, DNA binding activity, gene structure, conserved motifs, protein interaction, chromosomal distribution, synteny analysis, and tissue-specific expression. Combining RNA sequencing data and qRT-PCR analysis, the expression patterns of PmbHLHs were estimated under low-temperature stress. Overall, our results could form the foundation for researching the biological function of PmbHLHs and enrich the low-temperature resistance gene bank in woody plants.
We applied the genome project (http://prunusmumegenome.bjfu.edu.cn) to download the whole genome data of P. mume (Zhang et al., 2012). The sequences of A. thaliana bHLHs (AtbHLHs) were obtained from Pires and Dolan (Pires and Dolan, 2010). The Pfam database (http://pfam.xfam.org, PF00010) was used to obtain the Hidden Markov Model (HMM) profile of the HLH domain and was searched bHLH proteins of P. mume with HMMER3 software (http://hmmer.janelia.org) (Sun et al., 2015). To ensure credibility, the E-value cut-off was set at 10–5. SMART online software (http://smart.embl-heidelberg.de/) (Schultz et al., 1998) and NCBI CD-search (http://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi) (Marchler-Bauer et al., 2015) were used to confirm bHLH domains in presumptive PmbHLH proteins of P. mume.
The WoLF PSORT program (https://wolfpsort.hgc.jp/) was applied to predict the subcellular localization of PmbHLHs (Horton et al., 2007). The CDS length, molecular weights (MWs), theoretical isoelectric points (pI), amino-acid sequences (aa), the total number of positively charged residues (Arg + Lys), total number of negatively charged residues (Asp + Glu), grand averages of hydropathicity (GRAVYs), and instability index and aliphatic index of all predicted PmbHLHs were calculated using ExPASy (https://web.expasy.org/protparam/) (Artimo et al., 2012).
RAxML version eight software with maximum likelihood (ML) method was used to construct phylogenetic trees (Stamatakis, 2014). The optimal JTT (Jones-Taylor-Thornton) model of amino acid substitution was applied to construct ML phylogenetic trees. The calculated relationships of the phylogenetic tree were supported by performing 1,000 iterations of bootstrap test and visualized by iTOL (https://itol.embl.de/) (Letunic and Bork, 2006). Alignments of PmbHLHs domains were conducted by Muscle (https://www.ebi.ac.uk/Tools/msa/muscle/) (Edgar, 2004).
TBtools (Chen et al., 2020) and NCBI Batch CD-Search (https://www.ncbi.nlm.nih.gov/) (Marchler-Bauer et al., 2015) were applied to analyze and visualize gene structure and conserved domains. We applied the online MEME program to analyze motif structures of PmbHLH proteins (30 motifs were set as the maximum number), others using default parameters (Bailey et al., 2009). Jalview software (Clamp et al., 2004) and Weblogo3 (http://weblogo.berkeley.edu/logo.cgi) were used to visualize and analyze conserved domains.
We used the GDR database (https://www.rosaceae.org/) to retrieve genomes of Prunus avium and Prunus persica (Verde et al., 2013; Shirasawa et al., 2017). The P. mume genome database provided chromosomal distribution information of bHLH genes. Chromosomal location map, as well as duplication events of bHLH genes and mutation rates of Ka (nonsynonymous) versus Ks (synonymous) were predicted through TBtools (Chen et al., 2020). Syntenic relationship of bHLH genes was analyzed by MCScanX (Multiple Collinearity Scan toolkit) in P. mume, P. avium, and P. persica (Wang et al., 2012). We used the R circlize package to visualize the relationship between three varieties (Zhuo et al., 2018). The PmbHLH genes divergence time (T) was calculated through the equation: T = dS/(2λ × 106) Mya, where λ = 1.5 × 10−8 s for dicots (Lynch and Conery, 2000).
The transcriptome data of PmbHLHs in five tissues (bud, fruit, leaf, stem, and root) were downloaded from the NCBI Sequence Read Archive (accession number: SRP014885). Heat map of five tissues and low-temperature stress were drawn by TBtools (Chen et al., 2020) and iTOL (https://itol.embl.de/) based on FPKM values (Letunic and Bork, 2006).
Annual grafted seedlings of true mume ‘Beijing Yudie’ and apricot mei ‘Danfenghou’ were used for low-temperature treatment. These plant materials were exposed to the same soil humidity (60–70%), air humidity (65–70%), illumination time (12 h/12 h), and light intensity (150 μmol•m−2•s−1). Stems of P. mume were treated for chilling treatment (4°C for 8 h and 5 days) and freezing treatment (−5°C for 1 h).
To further research the regulation process of PmbHLHs under low-temperature, we carried out detailed multi-stage chilling treatment and freezing treatment.
1) Chilling treatment: in September, we took annual branches of ‘Beijing Yudie” and put branches in a 4°C low-temperature incubator and undertook sampling at 0/2/4/8/16/24/48/72 h. Stem segments without bud points were used as sampling materials.
2) Freezing treatment: in November, when the lowest ambient temperature reaches 5°C, we took branches of 'Beijing Yudie', then put them in a refrigerator at 4°C. The second day, they were put in a low-temperature incubator with step-by-step cooling (1 h/°C, keeping it from 0°C to −10°C). Stem segments were sampled at 0/2/4/6/8/10/24/48 h. The above samples were stored at −80°C for RNA extraction. There were three biological replicates.
Total RNA of low-temperature treatments was isolated by RNA Extraction Kit (Takara, Beijing, China) based on references. First-strand cDNA synthesis was reversed with DNase-treated RNA (1 µg) through PrimeScriptTMRT Reagent Kit with gDNA Eraser (Takara, Dalian, China). The template used cDNA (2 µL) in a 10 µL qRT-PCR by TB Green Ⅱ Premix Ex Taq (Takara, Dalian, China). We applied the 2ΔΔCt’ method to calculate relative expression levels and internal control used protein phosphatase 2A (PP2A)gene of P. mume (Wang et al., 2014). Supplementary Table S10 showed ten selected genes and specific primers. Each qRT-PCR was repeated at least three times.
Promoter cis-acting regulatory elements were analyzed by PlantCARE (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/) (Lescot et al., 2002). The AraNet V2 tool (Lee et al., 2015) was used to construct the protein interaction network based on homologous proteins of PmbHLHs in Arabidopsis. The protein interaction network was visualized by Cytoscape (Shannon, 2003) and STRING software (http://string-db.org/).
A total of 95 non-redundant PmbHLHs were discovered based on HMMER software from P. mume genome. According to their location position, these genes were named PmbHLH01 to PmbHLH95. The length of PmbHLH proteins ranged between 91 (PmbHLH36) and 700 (PmbHLH45) amino acids and most of these genes (74%) had lengths of 200–400 aa. The presumptive isoelectric points (pI) ranged from 4.57 (PmbHLH14) to 10.1 (PmbHLH43). About molecular weight values, the smallest was 10.27 kDa (PmbHLH67) and the largest was 78.26 kDa (PmbHLH45). The instability index varied from 36.78 to 92.34, while only one (PmbHLH66) was considered a stable protein. Predicted GRAVY values were all negative, representing all genes that possessed hydrophilic characteristics. The aliphatic index, (Asp + Glu) value and (Arg + Lys) value of PmbHLHs showed diversity features. Analysis of the gene structure of 95 PmbHLH proteins showed that coding genes of most PmbHLH proteins (92.63%) have introns. Subcellular localization of PmbHLH proteins were mostly located in the nucleus (Supplementary Table S1).
To research the evolutionary relationship of PmbHLHs, a phylogenetic tree with ML method was established by full-length amino acid sequences of 95 PmbHLHs (Figure 1). To clearly describe the classification and potential functions of PmbHLHs, we performed another ML phylogenetic tree with 95 PmbHLH proteins and 162 AtbHLHs (Supplementary Figure S1 and Supplementary Table S2). Based on verified AtbHLHs (Pires and Dolan, 2010), 95 PmbHLH proteins were divided into 23 subfamilies and one orphan (Figure 1). XII was the largest subfamily, consisting of 11 members. Five subfamilies [IVc, Va, VIIIa, VIIIc (1), XIV] contained only one bHLH protein. This indicated that PmbHLHs were distributed unevenly in different subfamilies. To date, we know very little about the biological functions of PmbHLHs except for PmICE1 (Cao et al., 2014). However, plenty of bHLH proteins have been confirmed in Arabidopsis. Through Arabidopsis ortholog analysis, PmbHLHs might play an important role in abiotic stress, hormonal regulation, development, and so on (Supplementary Table S2).
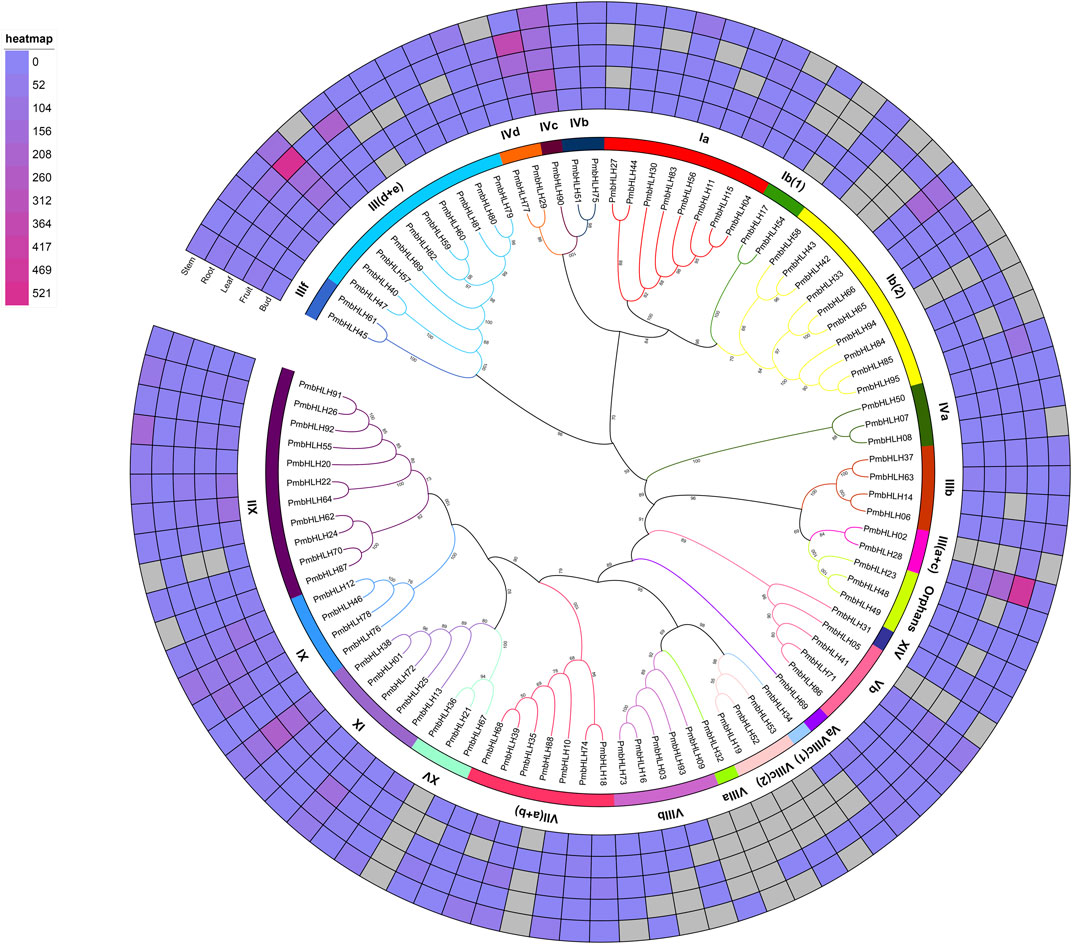
FIGURE 1. Phylogenetic analysis of PmbHLH proteins in P. mume. The RAxML version eight software was applied to draw the Maximum likelihood (ML) tree with 1,000 bootstrap replicates. Lines with roman numerals represent different PmbHLH subfamilies. A heat map of PmbHLH genes in five tissues (bud, fruit, leaf, stem, and root) was drew based on a phylogenetic tree. Color scores show the expression of PmbHLH genes in five tissues.
The bHLH domain analysis showed that PmbHLH domains consisted of four conserved regions, including the basic region, two helix regions, and a loop region (Figure 2 and Supplementary Figure S2). Additionally, 23 amino acid residues were conserved (>50% consensus ratio) in their bHLH domains. Among residues, the Arg-15, Arg-16, Leu-26, Pro-31, Leu-61 were highly conserved with a greater than 90% consensus ratio. Among the 23 conserved amino acid residues, the basic region found six conserved residues (His-8, Ala-11, Glu-12, Arg-13, Arg-15, Arg-16), first helix region found seven conserved residues (Ile-19, Asn-20, Arg-22, Leu-26, Leu-29, Val-30, Pro-31), loop region found two conserved residues (Lys-44, Asp-45), second helix region found eight conserved residues (Ala-48, Ser-49, Leu-51, Ala-54, Ile-55, Tyr-57, Lys-59, Leu-61).
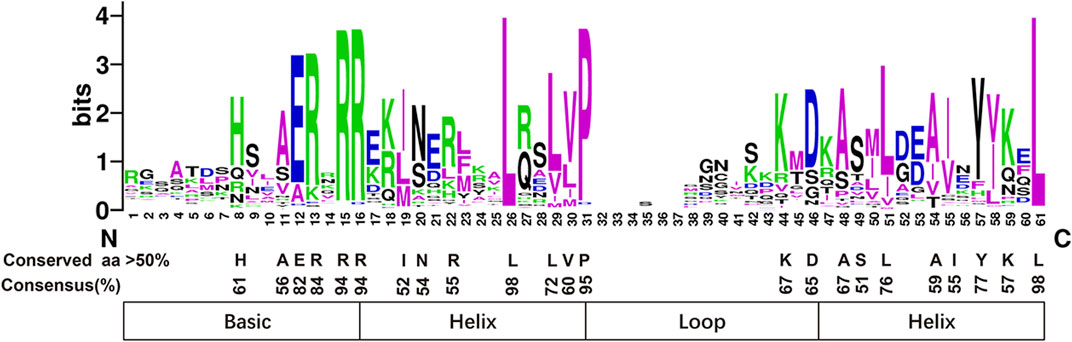
FIGURE 2. All PmbHLH proteins shown highly conserved in the domain. The overall height of every stack means sequence conservation in the corresponding position. Capital letters represent the conservation of amino acids exceed 50% among 95 PmbHLH domains.
The DNA binding activity of target genes was decided by the bHLH domain in the basic region. According to the classification standard in A. thaliana (Toledo-Ortiz et al., 2003), PmbHLHs were defined as Non-DNA-binding proteins and DNA-binding proteins (Figure 2 and Table 1). In addition, DNA-binding proteins were divided into E-box-binding proteins (including G-box-binding proteins) and non-E-box binding proteins according to the existence of Glu-12 and Arg-15 (positions were corresponding to positions 13 and 16 in Arabidopsis). His/Lys-8, Glu-12. Arg-16 (positions were corresponding to positions 9, 13, and 17 in Arabidopsis) are responsible for the binding of the G-box. Based on the conservation of these residues, three PmbHLHs were classified to non-E-box-binding proteins for missing Glu-12/Arg-15 residues and 37 PmbHLHs were classed to E-box-binding proteins. Among 37 E-box-binding proteins, 24 as G-box-binding proteins, while 13 proteins missed the G-box-binding site. Furthermore, 55 of the 95 PmbHLHs were classed in non-DNA-binding proteins due to less than six amino acid residues in the basic region (Table 1 and Supplementary Table S3).
Further to the analysis features of PmbHLHs, we investigated intron/exon patterns according to the phylogenetic tree with PmbHLHs sequences (Figure 3A). Analysis of genomic DNA sequences showed that number of introns changing from zero to ten (Figure 3B). Most of them usually had one to eight introns, except PmbHLH69 and PmbHLH92. Different subfamilies had different intron/exon patterns, while the same classes were similar. Seven genes of 95 PmbHLHs were intron-less, five of these were in subfamily VIIIb. Seven subfamilies [Ib (1), IIIf, IVb, Vb, VIIIb, IX, XV] had a concentrated number of exons/introns.

FIGURE 3. Phylogenetic relationship, motifs analysis, and gene structure in PmbHLH. (A) The RAxML version eight software was applied to construct a phylogenetic tree based on PmbHLH proteins. (B) Motif composition analysis of PmbHLHs by MEME. Different colorful rectangles with numbers 1–30 represent different motifs. (C) Exon-intron structure of PmbHLHs. Grey lines represent introns and green boxes represent exons. Yellow rectangles indicate the position of PmbHLHs conserved domain.
We used MEME online tool to predict thirty conserved motifs of 95 PmbHLH proteins (Figure 3C and Supplementary Table S4). The number of PmbHLHs motifs was distinctive, ranging from 1 to 16. Most of PmbHLHs shared three to six motifs. Each PmbHLH protein contained motif one and motif two except for PmbHLH31, PmbHLH58, and PmbHLH69. Different subfamilies had unique motif combinations. For instance, subfamily IVb contained motifs 1, 2, and 26, and subfamily Ib (1) contained motifs 1, 2, 5, and 8. Some motifs were unique and existed in only one subfamily. Subfamily Ⅲ (d + e) contained the most motifs and motif 9, 20, and 25 were specific to it. Motif 11, 23, and 30 were respectively observed in subfamily XI, Ⅷ (2), and XI. These conserved motifs may play special functions. The same subfamily had similar motifs, implying these PmbHLHs might have similar functions. In contrast, the kinds of motifs showed little difference in the same subfamily. For example, PmbHLH07 had motif 21 except common motifs 1, 2, 5, and eight in subfamily IVa.
According to genome annotation information, 78 PmbHLH genes were mapped on eight chromosomes, while 17 PmbHLH genes were localized on unassembled genomic scaffolds (Figure 4A and Supplementary Figure S3). The distribution of PmbHLH genes on each chromosome was irregular. 18 PmbHLH genes (18.95%) were present on chromosome 2, which was the maximum number, whereas only two PmbHLH genes (2.11%) were located on chromosome 3. The proportion of PmbHLH genes on half of the chromosomes accounted for more than 10% but less than 20%.
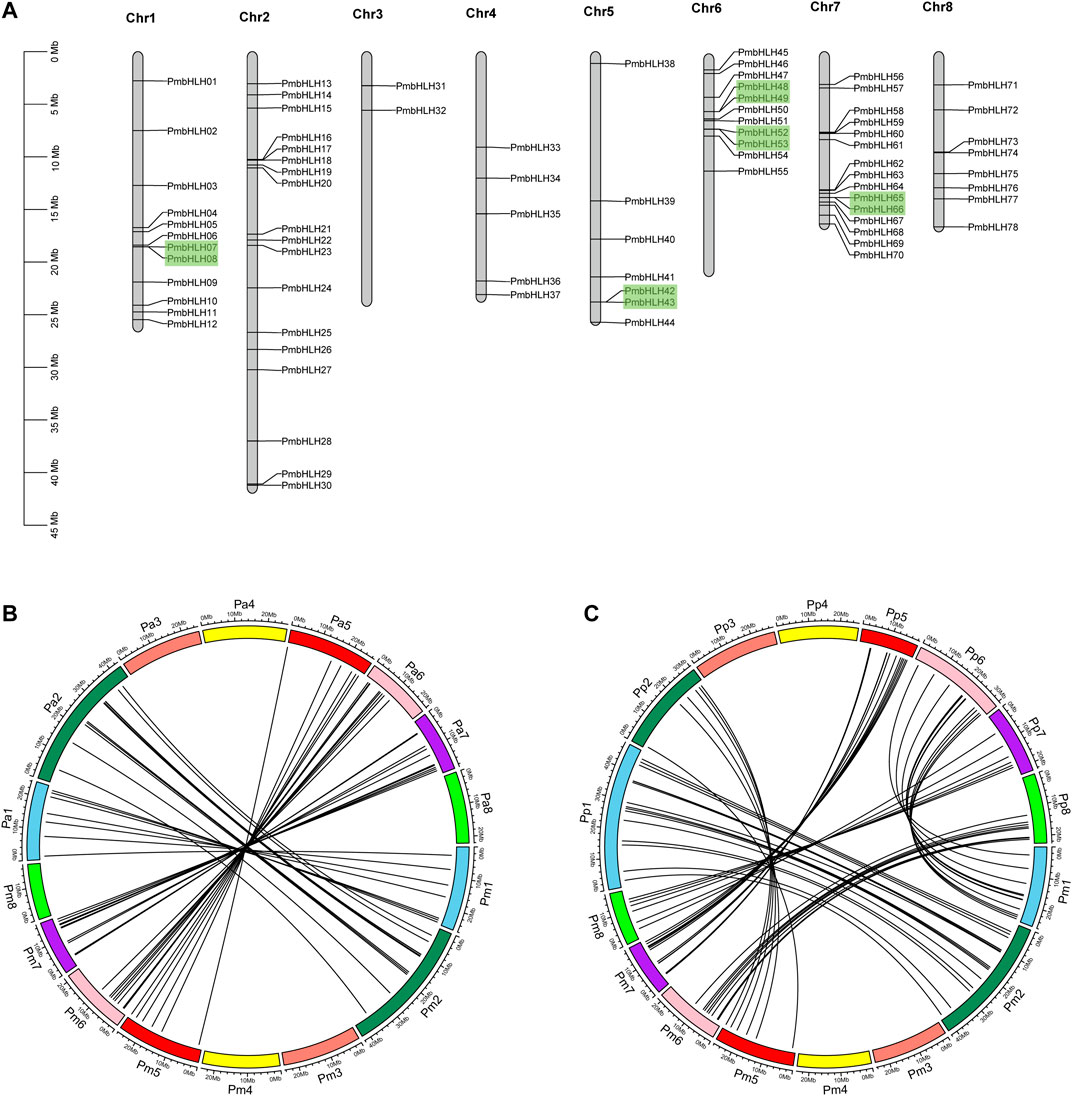
FIGURE 4. Chromosomal mapping of PmbHLHs and synteny analysis in P. mume (Pm),P. avium (Pa) and P. persica (Pp). (A) Chromosomal distribution of PmbHLH genes. 78 PmbHLH genes were unevenly mapped on eight chromosomes. Tandem duplicated gene pairs are displayed with green blocks. (B) Synteny analysis in P. mume and P. avium. The same color blocks represent the same chromosome label in P. mume and P. avium. Black lines mean orthologous bHLH in P. mume and P. avium. (C) Synteny analysis in P. persica and P. mume. The same color blocks represent the same chromosome label in P. persica and P. mume. Black lines mean orthologous bHLH in P. persica and P. mume.
Duplication events contained genome duplication, tandem duplication, segmental duplication, and transposon duplication, leading to plant evolution (Qiao et al., 2019). Duplication played an important role in PmbHLH gene expansion. Among 95 PmbHLHs, nine pairs of PmbHLH genes were described as tandem duplication, mapping on chromosome 1, 5, 6, 7, and scaffolds (Figure 4A and Supplementary Figure S3). The largest number of tandem duplications were distributed on Chromosome 6. By contrast, segmental duplication was absent in the PmbHLH gene family, which signified segmental duplication was not involved in gene expansion. The selection pressure of gene duplications was estimated by mutation rates of Ka (nonsynonymous) versus Ks (synonymous). We calculated the Ka/Ks of the PmbHLH gene family (Supplementary Table S5). Results showed that most of the Ka/Ks values of tandem duplication were <1 and changed from 0.270 to 0.949, implying a purifying selection during PmbHLH genes expansion. However, PmbHLH82-PmbHLH83 and PmbHLH85-PmbHLH86 only existed Ka value, which means that two gene pairs evolved by natural selection. The divergence time of PmbHLHs tandem duplication ranged from 3.67 to 64.06 Mya.
To further explore the evolutionary mechanism of the PmbHLH gene family, we constructed a syntenic map of P. mume associated with P. avium and P. persica. 41 syntenic orthologous gene pairs were distinguished between P. avium and P. mume, 59 pairs between P. persica and P. mume (Figure 4B and Figure 4C). This indicates that the P. avium, P. persica, and P. mume had a close relationship. Interestingly, we found that one PmbHLH gene only corresponds to one gene in these syntenic orthologous gene pairs. For further evolutionary studies, the divergence time of bHLH gene pairs was calculated in three varieties (Supplementary Table S6). Divergence time started 86.11 Mya to 0.67 Mya between P. persica and P. mume, and 1-2 Mya occurred in most duplicated events. In P. avium and P. mume, it began 60.81 Mya to 0.14 Mya and 0.5-2 Mya occurred in most of the duplicated events, which may indicate that the speciation time of these orthologous pairs was shorter in the two varieties. Additionally, higher syntenic genes appeared on chromosomes 2, 6, and 7.
Based on the FPKM values from RNA sequencing data, we investigated the expression pattern of PmbHLHs among different tissues and different low-temperature treatments. The heatmap of Figure 1 and Supplementary Figure S5 show that the PmbHLH gene family presented clear tissue-specific expression. Among 95 PmbHLHs, 53 genes were expressed in five tissues (bud, fruit, leaf, stem, and root), implying that these PmbHLHs may participate in the development and growth process of tissues. While, PmbHLH21 and PmbHLH32 (they belonged to subfamily XV and VIIa, respectively) lacked expression in all detected tissues. We discovered that nine PmbHLHs were expressed in only one tissue. They included two genes (PmbHLH54 and PmbHLH88) only in fruit, seven genes (PmbHLH02, PmbHLH05, PmbHLH19, PmbHLH42, PmbHLH43, PmbHLH53 and PmbHLH94) only in root. This probably indicated that the nine genes have special functions in fruit or root. Moreover, most of remaining PmbHLHs were expressed in three or four detected tissues (Supplementary Table S7). According to the phylogenetic tree analysis, PmbHLHs from the same subfamily had a similar expression pattern (Figure 1). For instance, IX subfamily (included PmbHLH01, PmbHLH13, PmbHLH25, PmbHLH38 and PmbHLH72) exhibited expression in all detected tissues. Furthermore, Ib (2) subfamily genes (included PmbHLH42, PmbHLH43, PmbHLH65, PmbHLH66, PmbHLH84, PmbHLH85, PmbHLH94, and PmbHLH95) were highly expressed in root, while PmbHLH33 and PmbHLH58 had a high expression level in leaf and stem respectively, implying that the functions of genes in the same family gradually differ in the evolution process.
Previous studies have proved that the cold resistance of apricot mei is stronger than that of true mume. Therefore, annual plants of apricot mei ‘Danfenghou’ and true mume ‘Beijing Yudie’ were treated with low temperatures. Based on transcriptome data, this study analyzed differences in the expression level of the PmbHLHs gene family during these three periods. FPKM value greater than one is an effective expression. 62 genes that were effectively expressed after low-temperature stress were detected. The FPKM value of these genes was plotted as a heat map.
As shown in Figure 5, according to different expression patterns in different periods, PmbHLHs genes in the two varieties were clustered into four groups (Supplementary Table S8). PmbHLH genes of the ‘Danfenghou’ group Ⅰ were highly expressed during freezing treatments and had a similar expression pattern to the ‘Beijing Yudie’ group Ⅲ. There are six genes (PmbHLH64, PmbHLH25, PmbHLH10, PmbHLH78, PmbHLH31, and PmbHLH47) in the two varieties with similar expression patterns, which can respond to freezing stress. The expression level of PmbHLHs in the ‘Danfenghou’ group Ⅱ and ‘Beijing Yudie’ group Ⅰ gradually decreased with the extension of treatment time. The expression profiles of PmbHLHs in the ‘Danfenghou’ group Ⅲ and ‘Beijing Yudie’ group Ⅱ were similar. 14 genes increased expression levels when two varieties were treated at 4°C for 8 h. They might be involved in the perception and transport of cold signals. The expression levels of ‘Danfenghou’ group Ⅱ and ‘Beijing Yudie’ group Ⅰ both increased at 4°C for 5 days, but there were no overlapping genes in the two cultivars. This indicated that the function of PmbHLH genes in the two cultivars after prolonged low-temperature acclimation has a difference.
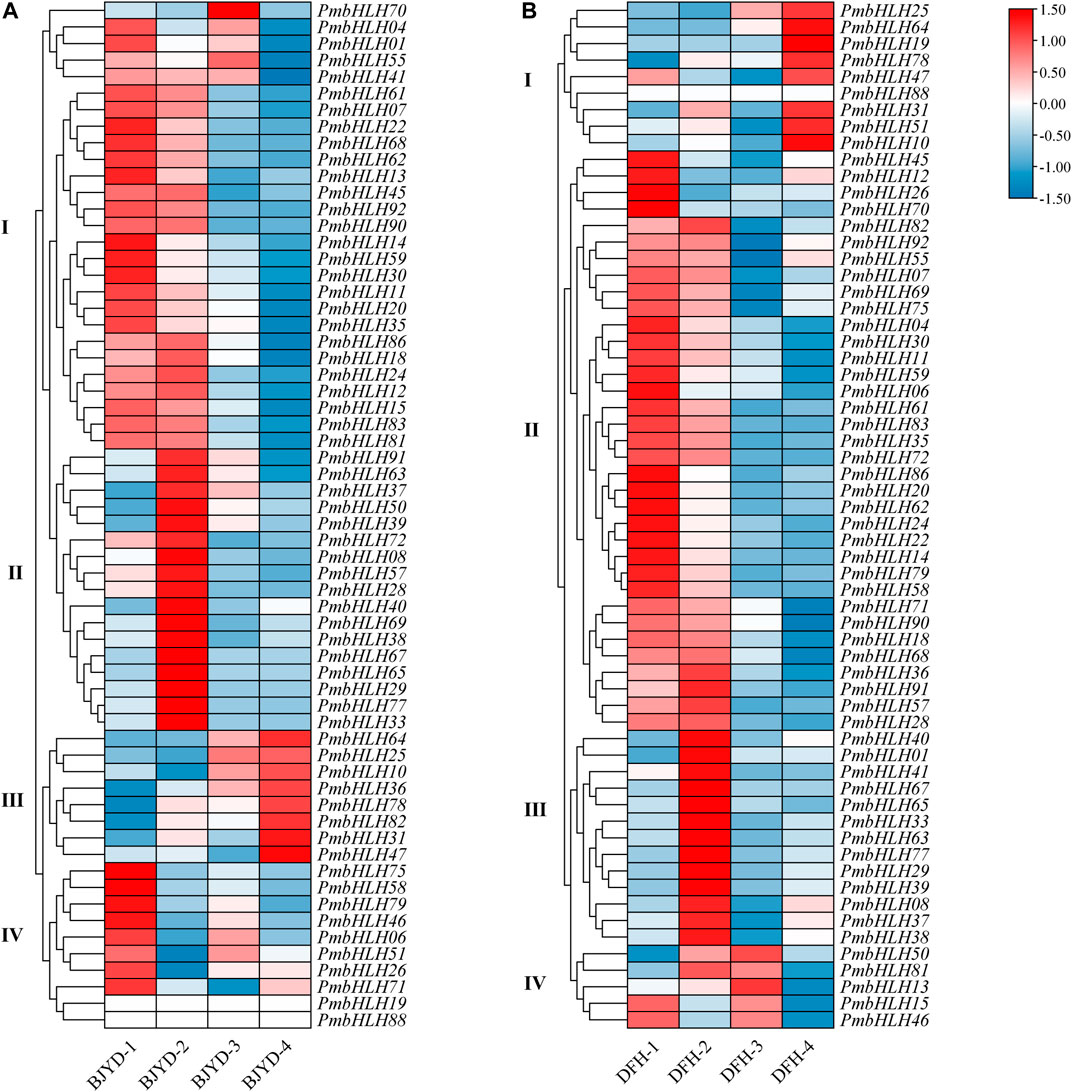
FIGURE 5. Hierarchical clustering of the expression profile of PmbHLHs in ‘Beijing Yudie’ (A) and ‘Danfenghou’ (B) under low-temperature treatment. “1” represents stems that were treated at 25°C, “2” represents stems that were treated at 4°C for 8 h, “3” represents stems that were treated at 4°C for 5 days, “4” represents stems that were treated at −5°C for 1 h. Heat maps were generated with FPKM values. Colorful scale means relative expression level and is shown at the top. Red represents high expression and blue represents low expression. Hierarchical clustering groups are displayed by roman numerals.
According to FPKM value and fold log2 change value, 10 PmbHLH genes were screened out. Compared with control material that has not been treated with low temperature, the transcriptional expression levels of six PmbHLHs genes (PmbHLH25, PmbHLH28, PmbHLH38, PmbHLH40, PmbHLH57, and PmbHLH78) increased by 2–5 times at chilling treatment (4°C for 8 h and 5 days), which may be involved in the response of P. mume to chilling treatment. The expression levels of PmbHLH4, PmbHLH6, PmbHLH26, and PmbHLH46 gradually decreased with the extension of 4°C treatment time. Compared with the control, the highest reduction factor reached 5 times. They might negatively regulate downstream low-temperature response genes or proteins to participate in the chilling stress response of P. mume. We used transcriptome to further analyze the expression patterns of PmbHLHs genes in different varieties. The trend of expression levels with these 10 differential genes showed the same in strong cold-resistant ‘Danfenghou’ and weaker cold-resistant ‘Beijing Yudie’. However, there were differences in multiple variations among different varieties. For example, the expression of the PmbHLH28 gene in ‘Beijing Yudie’ was 4.8 times higher than that in the control, but only 1.1 times higher in the ‘Danfenghou’ group.
To further investigate PmbHLHs function in low temperature comprehensively, the 10 genes were detected by qRT-PCR experiments in which the stems of P. mume were treated for chilling treatment (4°C for 8 h and 5 days) and freezing treatment (−5°C for 1 h). The expression of PmbHLH genes was distinct in two varieties of P. mume (Figure 6A). In true mume ‘Beijing Yudie’, six genes were up-regulated in varying degrees under low-temperature stress. Remaining genes were down-regulated. Among up-regulated genes, PmbHLH25, PmbHLH40, PmbHLH46, and PmbHLH57 were highly expressed under 4°C and −5°C treatment. The greatest expression of PmbHLH25 and PmbHLH40 was found in 4°C treatments only. In addition, low temperature induced most of the genes to change significantly in apricot mei ‘Danfenghou’. PmbHLH46 and PmbHLH25 were up-regulated at 4°C treatment and at −5°C treatment, respectively. PmbHLH40 and PmbHLH57 were highly expressed under 4°C and −5°C treatment. In two varieties, four genes (PmbHLH25, PmbHLH38, PmbHLH40, and PmbHLH78) could be induced to high expression in true mume ‘Beijing Yudie’ and apricot mei ‘Danfenghou’. In addition, the expression of four PmbHLHs (PmbHLH38, PmbHLH40, PmbHLH57, and PmbHLH78) in apricot mei ‘Danfenghou’ higher than in true mume ‘Beijing Yudie’, further showing that apricot mei ‘Danfenghou’ was more resistant than true mume ‘Beijing Yudie’.
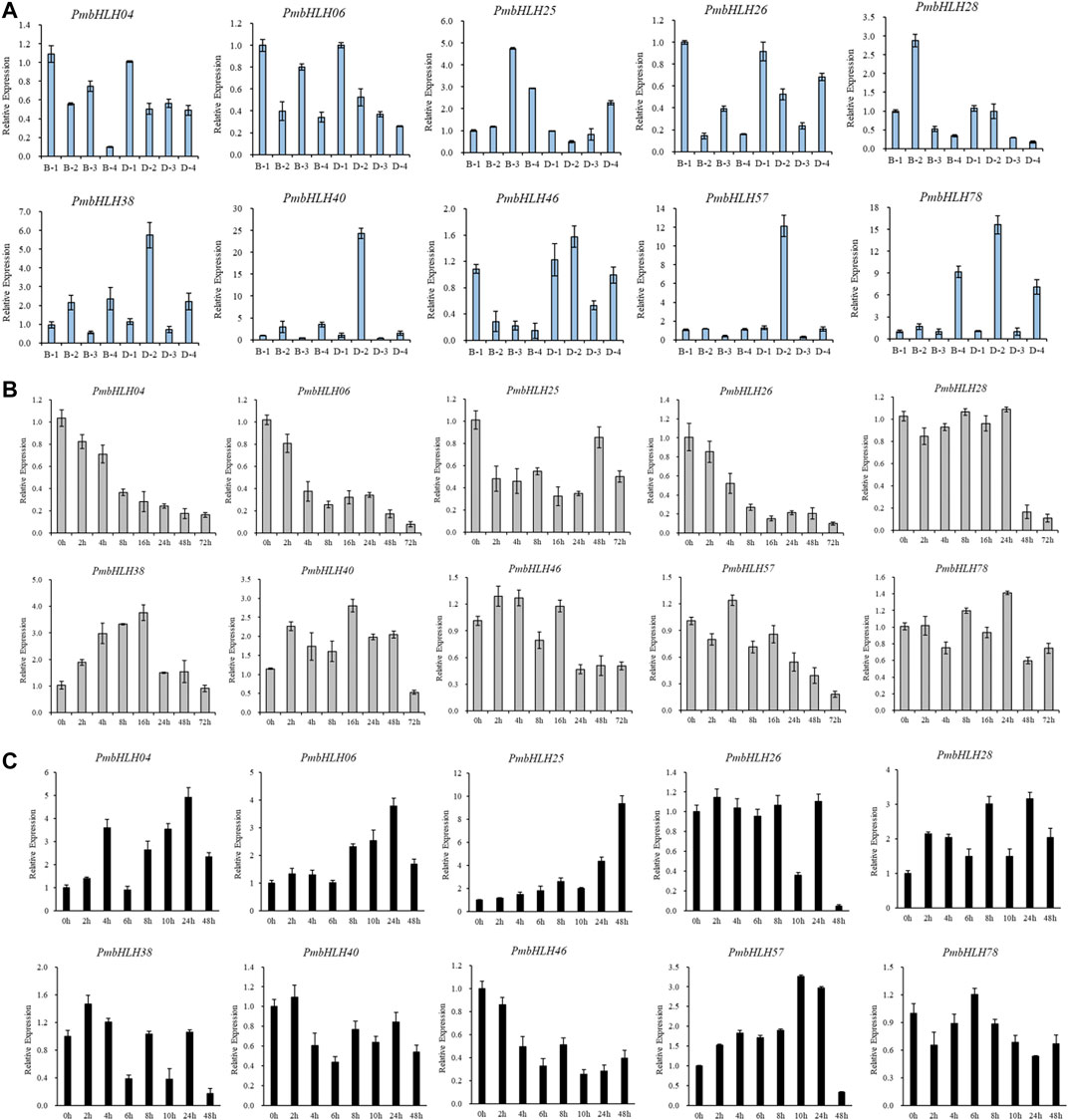
FIGURE 6. Expression patterns of 10 candidate PmbHLHs in ‘Beijing Yudie’ and ‘Danfenghou’ under low-temperature treatments by qRT-PCR. (A) Expression patterns under cold treatment (4°C for 8 h and 5 days) and freezing treatment (−5°C for 1 h). “1” represented stems that were treated at 25°C, “2” represented stems that were treated at 4°C for 8 h, “3” represented stems that were treated at 4°C for 5 days, “4” represented stems that were treated at −5°C for 1 h. (B) Expression patterns exposed to cold treatment for different times (0/2/4/8/16/24/48/72 h). (C) Expression patterns exposed to freezing treatment for different times (0/2/4/6/8/10/24/48 h). The standard deviation of three independent replicates was represented by Error bars.
To explore the regulation process of PmbHLHs, we tested the expression pattern of 10 genes by qRT-PCR at chilling treatment (0/2/4/8/16/24/48/72 h) and freezing treatment (0/2/4/6/8/10/24/48 h) for different periods using true mume ‘Beijing Yudie’. As shown in Figure 6B, the expression level of PmbHLH04, PmbHLH25, PmbHLH26, PmbHLH46, and PmbHLH57 were reduced with chilling treatment. However, PmbHLH38 and PmbHLH40 were induced to high expression and up-regulation expression peaked on treating with 16 h. Under freezing treatment, the expression level of PmbHLH26, PmbHLH40, and PmbHLH78 changed slightly. PmbHLH38 and PmbHLH40 presented a trend of decreasing. After prolonging the freezing treatment time, PmbHLH04, PmbHLH06, PmbHLH25, PmbHLH28, and PmbHLH57 were induced to high expression. Especially, PmbHLH25 had the largest expression level increase (approximately 10-fold) at 48 h after being exposed to freezing conditions (Figure 6C). These results imply that PmbHLH genes may play a role in resisting low-temperature stress.
The promoter cis-elements (1500 bp) were analyzed through PlantCARE software and the predicted regulation mechanisms of PmbHLHs. The result showed that PmbHLH genes were abundant in abiotic and biotic elements (LTR, MBS, WRE3, WUN-motif, ARE), light-responsive elements (AAAC-motif, ATCT-motif, LAMP-element, Box 4, I-box, and G-box), plant growth and development-related elements (MSA-like, GCN4_motif, and RY-element). In addition, hormone-responsive elements (ABRE, TGACG-motif, G-box, MYC, P-box, and TATC-box) exhibited a wide range of positions in the promoter. This means that PmbHLHs may extensively participate in various physiological biochemistry pathways of P. mume (Supplementary Figure S4).
To further predict the functions of PmbHLHs, we constructed the interaction network using AraNet V2 based on homologous proteins of Arabidopsis (Lee et al., 2015). A total of 56 PmbHLH proteins had orthologs in Arabidopsis and predicted about 640 interaction protein pairs (Figure 7 and Supplementary Table S9). The interaction network showed that PmbHLHs might interact with MYB, bHLH, NAC, bZIP, HB, WRKY, ERF, and so on, implying PmbHLHs might exert functions by interacting with other genes. The above results predicted that PmbHLHs might be involved in the response of P. mume to low temperature. To further research ten PmbHLHs, we constructed and analyzed the interaction network of candidate ten genes. Among the ten PmbHLHs, PmbHLH04, PmbHLH26, and PmbHLH38 were absent in homologous proteins of Arabidopsis. The three candidate genes might be novel and potential proteins in response to low temperature. The other seven candidate PmbHLHs formed an interactive network centered on PmbHLH40, which showed high homology to JAM2. Moreover, they might strongly interact with MYB124, TIFY, NAC, WRKY, and IAA to drive function when suffered from low-temperature stress in P. mume. Supplementary Figure S6 shows that PmbHLH25 (AKS2) could directly interact with PmbHLH28 (AT5G57150). PmbHLH06 (bHLH93) could directly interact with PmbHLH38 (FBH4). This indicated that these PmbHLH might work by forming dimers under low-temperature stress. Overall, interaction networks could provide a crucial reference for investigating the regulation mechanism of PmbHLHs.
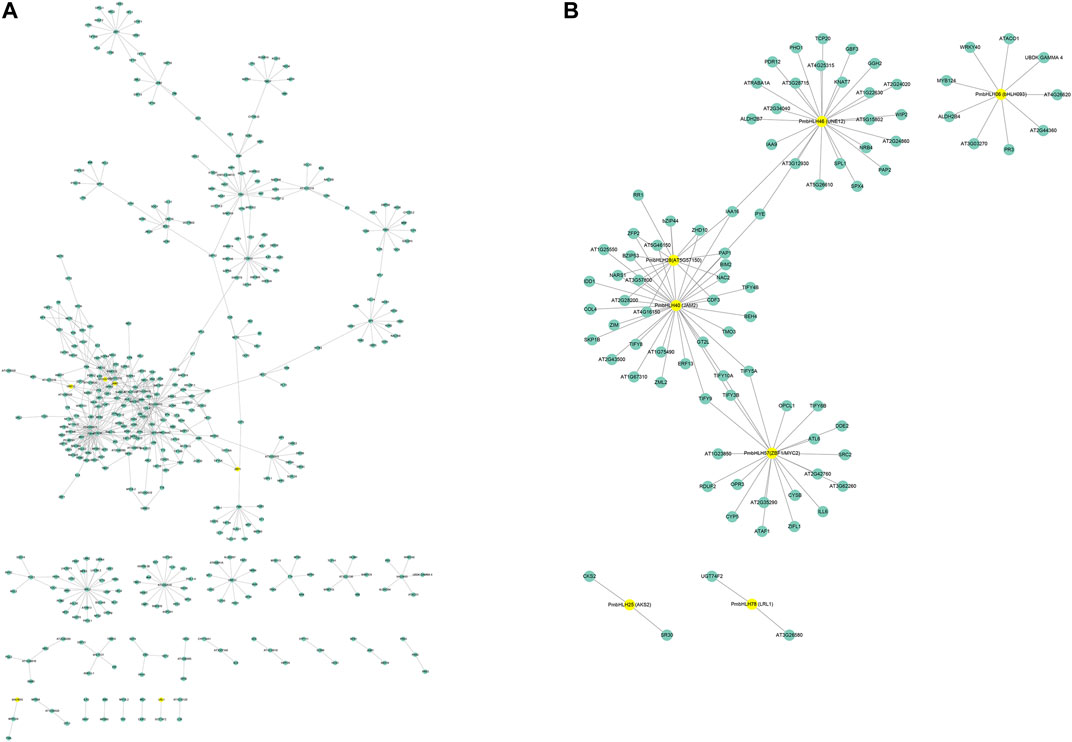
FIGURE 7. The interaction network for bHLHs in P. mume based on orthologs in Arabidopsis. (A) The whole interaction network for PmbHLHs. (B) Protein interaction network of seven candidate PmbHLHs. Yellow circles represent PmbHLHs and blue circles represent other genes.
The bHLH transcription factor family plays a positive or negative role in the physiological and biochemical processes of the development of plant trichome, root hair, photomorphogenesis, light signal transmission, and development of plant tissues and organs. In addition, the transcription factor family has contributed to resisting adverse environmental factors in plants, such as drought resistance, salt tolerance, cold tolerance, plant iron deficiency stress, and so on (Toledo-Ortiz et al., 2003). A great many bHLH genes have been distinguished in the plant kingdom, including A. thaliana (162) (Pires and Dolan, 2010), Hibiscus hamabo (162) (Ni et al., 2021), Helianthus annuus (183) (Li et al., 2021), Capsicum annuum (107) (Liu et al., 2021), Sorghum bicolor (174) (Fan et al., 2021), Osmanthus fragrans (206) (Li et al., 2020), Camellia sinensis (134) (Liu et al., 2021), Juglans regia (102) (Zhao et al., 2021), Pyrus bretschneideri (197) (Dong et al., 2021). The number of bHLH transcription factors changed widely among various plants. However, the characteristic of bHLH genes remains unknown in P. mume. In our research, we used P. mume genome to distinguish 95 PmbHLH genes. The number of PmbHLHs was the same as bHLH in P. persica, which further proved that the genetic relationship of P. persica and P. mume was closer.
PmbHLHs were inhomogeneous on eight chromosomes and scaffolds. These results showed that the bHLH gene family existed specific evolution patterns in different varieties. In a previous study, it was suggested that the expansion of bHLH might derive from gene duplication during evolution with a high proportion of segmental duplications and tandem duplications (Cannon et al., 2004; Kavas et al., 2016). In P. mume, nine pairs of PmbHLH genes were described as tandem duplication (18.95%) and absent in segmental duplications. Tandem duplication possibly promoted PmbHLHs expansion. The rate of gene duplication in this study was lower than in other plants, suggesting gene duplication was the secondary formation in PmbHLH gene expansion or PmbHLHs, and that loss-functions or redundancies may be lost during evolution. This conclusion was similar to the PmWRKY genes family (Bao et al., 2019).
The bHLH domain consists of the basic region, two helix regions, and a loop region (Massari and Murre, 2000). DNA binding activity was determined by the basic region of the bHLH domain and the HLH region was essential in homodimer or heterodimer formation (Carretero-Paulet et al., 2010). In the present study, 23 amino acid residues in the PmbHLHs domains were conserved (>50% consensus ratio). Among them, residues Arg-15, Arg-16, Leu-26, Pro-31, Leu-61 (corresponding to Arg-16, Arg-17, Leu-27, Pro-32, Leu-61 in AtbHLHs) showed highly conserved with greater than 90% consensus ratio. Particularly, residues Leu-26 and Leu-61 were more conservative (Figure 2). DNA binding activity in the basic region was decided by greater than five basic amino acid residues (Toledo-Ortiz et al., 2003). According to the classification standard, PmbHLHs were defined as the Non-DNA-binding proteins and DNA-binding proteins (Figure 2). Furthermore, Glu-13 could specifically identify E-box and the position of Glu-13 was stabilized through Arg-16. And the existence of His/Lys-9, Glu-13, and Arg-17 could discern G-box binding motif. Therefore, DNA-binding proteins were divided into non-G-box binding proteins, G-box-binding proteins, and non-E-box binding proteins based on these factors. In addition, Leu-27 and Leu-61 played an important role in protein interaction in the helix region (Carretero-Paulet et al., 2010). 98% PmbHLHs had Leu-26 and Leu-61 (positions were equivalent to positions 27 and 61 in Arabidopsis), implying that PmbHLHs possessed dimerization capacity.
Expression profiles of genes could present their functions. Therefore, expression patterns of PmbHLHs in five tissues were analyzed. The expression pattern of PmbHLHs represented clear tissue-specific expression based on transcriptome data (Figure 1). Among them, PmbHLH17 and PmbHLH54 had a high expression level in fruit, suggesting that they might associate with embryonic development. RGE1 had PmbHLH17 and PmbHLH54 homologous proteins in Arabidopsis, which could result in the retarded growth of embryos (Kondou et al., 2008). Meanwhile, PmbHLH27, PmbHLH30, and PmbHLH44 were expressed in leaf and stem. The homolog FAMA, SPCH, and MUTE could control meristem differentiation during stomatal development (Ohashi-Ito and Bergmann, 2006; Pillitteri et al., 2007; Marcos et al., 2017). In subfamily IIIb, PmbHLH37 was expressed highly in stem, homolog, and AtICE was implicated in cold acclimation response and freezing tolerance (Chinnusamy, 2003). Additionally, PmbHLH02 were expressed in the root and the expression of AtFIT was up-regulated when it suffered from iron deficiency stress in Arabidopsis roots (Ling et al., 2002). These results contributed to predicting the functional regions and further understanding the functions of PmbHLHs.
Low temperature is an important environmental factor affecting plant yield and distribution. When plants suffer from adversity stress at low temperatures, they could feel low-temperature signals to produce a series of physiological and biochemical reactions and regulate gene expression (Zhu, 2016). Previous reports have demonstrated that the bHLH transcription factor had a crucial influence in resisting low-temperature stress (Nakamura et al., 2011). In the present study, we distinguished 10 PmbHLH genes that might possess low-temperature resistance based on transcriptome data. By analyzing qRT-PCR, the expression of PmbHLH genes was distinct in two varieties of P. mume. Among up-regulated PmbHLHs, four genes could be induced to express in true mume ‘Beijing Yudie’ and apricot mei ‘Danfenghou’. Simultaneously, the expression of four PmbHLHs in apricot mei ‘Danfenghou’ was higher than in true mume ‘Beijing Yudie’, further showing that apricot mei ‘Danfenghou’ was more resistant than true mume ‘Beijing Yudie’. The remaining four genes were down-regulated when they suffer low-temperature stress. To further research the regulation mode of the 10 PmbHLHs, fine regulation detection was carried out. The result showed that PmbHLHs were more strongly expressed in −5°C treatment than 4°C treatment. In the meantime, the expression of PmbHLHs increased with the prolongation of −5°C treatment time, suggesting PmbHLHs could respond to deep freezing. Especially PmbHLH25, which possibly has great potential in breeding frost resistance. It was noteworthy that AtAKS2 (ABA-responsive kinase substrates 2) was PmbHLH25 homologous proteins in Arabidopsis. ABA could induce phosphorylation of AKS to change stomatal opening or close, then enable plants to adapt to changing environmental conditions (Takahashi et al., 2013). We speculated that PmbHLH25 might play an essential role in the process of low-temperature signal perception and transduction.
In our study, we identified 95 PmbHLHs from P. mume genome and comprehensively analyzed their characterization, including gene identification, gene structure, conserved motifs, DNA binding activity, and chromosomal distribution, protein interaction, synteny analysis, and expression profiling. According to the phylogenetic tree, 95 PmbHLHs were classified into 23 subfamilies. The protein interaction and synteny analysis further expounded the potential functions and evolutionary mechanisms of PmbHLHs. Moreover, tissue-specific expression revealed that PmbHLHs might widely participate in the development of tissues. Combining with the qRT-PCR date, PmbHLH04, PmbHLH06, PmbHLH25, PmbHLH28, PmbHLH38, PmbHLH40, and PmbHLH57 may play a major role in resisting low-temperature stress. These results provide a molecular basis and valuable insights for further studying the functions of PmbHLHs in regulating low temperature in P. mume.
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
QZ conceived and designed the experiments. FB revised the manuscript. AD and AD performed data analysis and experiments. AD drafted the manuscript. PL, JW, and TC contributed reagents, materials, and analysis tools. All authors have read and approved the final manuscript.
This research was funded by the National Key R and D Program of China (2019YFD1001500), the open funds of the State Key Laboratory of Plant Physiology and Biochemistry (Grant/Award Number: SKLPPBKF 2005) and Special Fund for Beijing Common Construction Project.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
We greatly appreciate the Frontiers editors and reviewers for handling our manuscript and providing critical suggestions.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2021.762135/full#supplementary-material
Artimo, P., Jonnalagedda, M., Arnold, K., Baratin, D., Csardi, G., de Castro, E., et al. (2012). ExPASy: SIB Bioinformatics Resource portal. Nucleic Acids Res. 40, W597–W603. doi:10.1093/nar/gks400
Atchley, W. R., and Fitch, W. M. (1997). A Natural Classification of the Basic helix-loop-helix Class of Transcription Factors. Proc. Natl. Acad. Sci. 94, 5172–5176. doi:10.1073/pnas.94.10.5172
Atchley, W. R., Terhalle, W., and Dress, A. (1999). Positional Dependence, Cliques, and Predictive Motifs in the bHLH Protein Domain. J. Mol. Evol. 48, 501–516. doi:10.1007/pl00006494
Bailey, T. L., Boden, M., Buske, F. A., Frith, M., Grant, C. E., Clementi, L., et al. (2009). MEME SUITE: Tools for Motif Discovery and Searching. Nucleic Acids Res. 37, W202–W208. doi:10.1093/nar/gkp335
Bao, F., Ding, A., Cheng, T., Wang, J., and Zhang, Q. (2019). Genome-Wide Analysis of Members of the WRKY Gene Family and Their Cold Stress Response in Prunus Mume. Genes 10, 911. doi:10.3390/genes10110911
Buti, S., Hayes, S., and Pierik, R. (2020). The bHLH Network Underlying Plant Shade‐avoidance. Physiol. Plantarum 169, 312–324. doi:10.1111/ppl.13074
Cannon, S. B., Mitra, A., Baumgarten, A., Young, N. D., and May, G. (2004). The Roles of Segmental and Tandem Gene Duplication in the Evolution of Large Gene Families in Arabidopsis thaliana. Bmc Plant Biol. 4, 10. doi:10.1186/1471-2229-4-10
Cao, N., Zhang, Q. X., Hao, R. J., Xu, Z. D., Wang, T., and Yang, W. R. (2014). Molecular Cloning and Expression Analysis of Cold-Resistant Transcription Factor PmICE1 from Prunus Mume. J. Northeast. For. Univ 42, 21–25. doi:10.13759/j.cnki.dlxb.2014.04.005
Carretero-Paulet, L., Galstyan, A., Roig-Villanova, I., Martínez-García, J. F., Bilbao-Castro, J. R., and Robertson, D. L. (2010). Genome-Wide Classification and Evolutionary Analysis of the bHLH Family of Transcription Factors in Arabidopsis, Poplar, Rice, Moss, and Algae. Plant Physiol. 153, 1398–1412. doi:10.1104/pp.110.153593
Chen, C., Chen, H., Zhang, Y., Thomas, H. R., Frank, M. H., He, Y., et al. (2020). TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 13, 1194–1202. doi:10.1016/j.molp.2020.06.009
Chinnusamy, V. (2003). ICE1: A Regulator of Cold-Induced Transcriptome and Freezing Tolerance in Arabidopsis. Gene Dev. 17, 1043–1054. doi:10.1101/gad.1077503
Clamp, M., Cuff, J., Searle, S. M., and Barton, G. J. (2004). The Jalview Java Alignment Editor. Bioinformatics 20, 426–427. doi:10.1093/bioinformatics/btg430
de Marcos, A., Houbaert, A., Triviño, M., Delgado, D., Martín-Trillo, M., Russinova, E., et al. (2017). A Mutation in the bHLH Domain of the SPCH Transcription Factor Uncovers a BR-dependent Mechanism for Stomatal Development. Plant Physiol. 174, 823–842. doi:10.1104/pp.17.00615
Ding, W., Yu, Z., Tong, Y., Huang, W., Chen, H., and Wu, P. (2009). A Transcription Factor with a bHLH Domain Regulates Root Hair Development in rice. Cell Res 19, 1309–1311. doi:10.1038/cr.2009.109
Dong, H., Chen, Q., Dai, Y., Hu, W., Zhang, S., and Huang, X. (2021). Genome-wide Identification of PbrbHLH Family Genes, and Expression Analysis in Response to Drought and Cold Stresses in Pear (Pyrus Bretschneideri). BMC Plant Biol. 21, 86. doi:10.1186/s12870-021-02862-5
Edgar, R. C. (2004). MUSCLE: Multiple Sequence Alignment with High Accuracy and High Throughput. Nucleic Acids Res. 32, 1792–1797. doi:10.1093/nar/gkh340
Fan, Y., Yang, H., Lai, D., He, A., Xue, G., Feng, L., et al. (2021). Genome-wide Identification and Expression Analysis of the bHLH Transcription Factor Family and its Response to Abiotic Stress in Sorghum [Sorghum Bicolor (L.) Moench]. BMC Genomics 22, 415. doi:10.1186/s12864-021-07652-9
Feng, X.-M., Zhao, Q., Zhao, L.-L., Qiao, Y., Xie, X.-B., Li, H.-F., et al. (2012). The Cold-Induced Basic helix-loop-helix Transcription Factor Gene MdCIbHLH1encodes an ICE-like Protein in Apple. Bmc Plant Biol. 12, 22. doi:10.1186/1471-2229-12-22
Gao, F., Robe, K., Bettembourg, M., Navarro, N., Rofidal, V., Santoni, V., et al. (2020). The Transcription Factor bHLH121 Interacts with bHLH105 (ILR3) and its Closest Homologs to Regulate Iron Homeostasis in Arabidopsis. Plant Cell 32, 508–524. doi:10.1105/tpc.19.00541
Groszmann, M., Paicu, T., and Smyth, D. R. (2008). Functional Domains of SPATULA, a bHLH Transcription Factor Involved in Carpel and Fruit Development in Arabidopsis. Plant J. 55, 40–52. doi:10.1111/j.1365-313X.2008.03469.x
Horton, P., Park, K.-J., Obayashi, T., Fujita, N., Harada, H., Adams-Collier, C. J., et al. (2007). WoLF PSORT: Protein Localization Predictor. Nucleic Acids Res. 35, W585–W587. doi:10.1093/nar/gkm259
Huq, E., and Quail, P. H. (2002). PIF4, a Phytochrome-Interacting bHLH Factor, Functions as a Negative Regulator of Phytochrome B Signaling in Arabidopsis. Embo J. 21, 2441–2450. doi:10.1093/emboj/21.10.2441
Kavas, M., Baloğlu, M. C., Atabay, E. S., Ziplar, U. T., Daşgan, H. Y., and Ünver, T. (2016). Genome-wide Characterization and Expression Analysis of Common Bean bHLH Transcription Factors in Response to Excess Salt Concentration. Mol. Genet. Genomics 291, 129–143. doi:10.1007/s00438-015-1095-6
Kim, Y. S., Lee, M., Lee, J.-H., Lee, H.-J., and Park, C.-M. (2015). The Unified ICE-CBF Pathway Provides a Transcriptional Feedback Control of Freezing Tolerance during Cold Acclimation in Arabidopsis. Plant Mol. Biol. 89, 187–201. doi:10.1007/s11103-015-0365-3
Knight, H., Mugford, S. G., Ülker, B., Gao, D., Thorlby, G., and Knight, M. R. (2009). Identification of SFR6, a Key Component in Cold Acclimation Acting post-translationally on CBF Function. Plant J. 58, 97–108. doi:10.1111/j.1365-313X.2008.03763.x
Kondou, Y., Nakazawa, M., Kawashima, M., Ichikawa, T., Yoshizumi, T., Suzuki, K., et al. (2008). RETARDED GROWTH of EMBRYO1, a New Basic Helix-Loop-Helix Protein, Expresses in Endosperm to Control Embryo Growth. Plant Physiol. 147, 1924–1935. doi:10.1104/pp.108.118364
Kurbidaeva, A., Ezhova, T., and Novokreshchenova, M. (2014). Arabidopsis thaliana ICE 2 Gene: Phylogeny, Structural Evolution and Functional Diversification from ICE1. Plant Sci. 229, 10–22. doi:10.1016/j.plantsci.2014.08.011
Ledent, V., and Vervoort, M. (2001). The Basic helix-loop-helix Protein Family: Comparative Genomics and Phylogenetic Analysis. Genome Res. 11, 754–770. doi:10.1101/gr.177001
Lee, T., Yang, S., Kim, E., Ko, Y., Hwang, S., Shin, J., et al. (2015). AraNet V2: an Improved Database of Co-functional Gene Networks for the Study of Arabidopsis thaliana and 27 Other Nonmodel Plant Species. Nucleic Acids Res. 43, D996–D1002. doi:10.1093/nar/gku1053
Lescot, M., Déhais, P., Thijs, G., Marchal, K., Moreau, Y., Van de Peer, Y., et al. (2002). PlantCARE, a Database of Plant Cis-Acting Regulatory Elements and a portal to Tools for In Silico Analysis of Promoter Sequences. Nucleic Acids Res. 30, 325–327. doi:10.1093/nar/30.1.325
Letunic, I., and Bork, P. (2006). Interactive Tree of Life (iTOL): an Online Tool for Phylogenetic Tree Display and Annotation. Bioinformatics 23, 127–128. doi:10.1093/bioinformatics/btl529
Li, J., Li, X., Han, P., Liu, H., Gong, J., Zhou, W., et al. (2021). Genome-wide Investigation of bHLH Genes and Expression Analysis under Different Biotic and Abiotic Stresses in Helianthus Annuus L. Int. J. Biol. Macromolecules 189, 72–83. doi:10.1016/j.ijbiomac.2021.08.072
Li, Y., Li, L., Ding, W., Li, H., Shi, T., Yang, X., et al. (2020). Genome-wide Identification of Osmanthus Fragrans bHLH Transcription Factors and Their Expression Analysis in Response to Abiotic Stress. Environ. Exp. Bot. 172, 103990. doi:10.1016/j.envexpbot.2020.103990
Ling, H.-Q., Bauer, P., Bereczky, Z., Keller, B., and Ganal, M. (2002). The Tomato Fer Gene Encoding a bHLH Protein Controls Iron-Uptake Responses in Roots. Proc. Natl. Acad. Sci. 99, 13938–13943. doi:10.1073/pnas.212448699
Liu, R., Song, J., Liu, S., Chen, C., Zhang, S., Wang, J., et al. (2021). Genome-wide Identification of the Capsicum bHLH Transcription Factor Family: Discovery of a Candidate Regulator Involved in the Regulation of Species-specific Bioactive Metabolites. BMC Plant Biol. 21, 262. doi:10.1186/s12870-021-03004-7
Liu, R., Wang, Y., Tang, S., Cai, J., Liu, S., Zheng, P., et al. (2021). Genome-wide Identification of the tea Plant bHLH Transcription Factor Family and Discovery of Candidate Regulators of Trichome Formation. Sci. Rep. 11, 10764. doi:10.1038/s41598-021-90205-7
Lynch, M., and Conery, J. S. (2000). The Evolutionary Fate and Consequences of Duplicate Genes. Science 290, 1151–1155. doi:10.1126/science.290.5494.1151
Marchler-Bauer, A., Derbyshire, M. K., Gonzales, N. R., Lu, S., Chitsaz, F., Geer, L. Y., et al. (2015). CDD: NCBI's Conserved Domain Database. Nucleic Acids Res. 43, D222–D226. doi:10.1093/nar/gku1221
Massari, M. E., and Murre, C. (2000). Helix-Loop-Helix Proteins: Regulators of Transcription in Eucaryotic Organisms. Mol. Cel Biol 20, 429–440. doi:10.1128/MCB.20.2.429-440.2000
Mccully, M. E., Canny, M. J., and Huang, C. X. (2004). The Management of Extracellular Ice by Petioles of Frost-Resistant Herbaceous Plants. Ann. Bot-london 94, 665–674. doi:10.1093/aob/mch191
Murre, C., Mccaw, P. S., and Baltimore, D. (1989). A New DNA Binding and Dimerization Motif in Immunoglobulin Enhancer Binding, Daughterless, MyoD, and Myc Proteins. Cell 56, 777–783. doi:10.1016/0092-8674(89)
Nakamura, J., Yuasa, T., Huong, T. T., Harano, K., Tanaka, S., Iwata, T., et al. (2011). Rice Homologs of Inducer of CBF Expression (OsICE) Are Involved in Cold Acclimation. Plant Biotechnol. 28, 303–309. doi:10.5511/plantbiotechnology.11.0421a
Nakata, M., Mitsuda, N., Herde, M., Koo, A. J. K., Moreno, J. E., Suzuki, K., et al. (2013). A bHLH-type Transcription Factor, ABA-INDUCIBLE BHLH-TYPE TRANSCRIPTION FACTOR/JA-ASSOCIATED MYC2-LIKE1, Acts as a Repressor to Negatively Regulate Jasmonate Signaling in Arabidopsis. The Plant Cell 25, 1641–1656. doi:10.1105/tpc.113.111112
Nemesio-Gorriz, M., Blair, P. B., Dalman, K., Hammerbacher, A., Arnerup, J., Stenlid, J., et al. (2017). Identification of norway spruce MYB-bHLH-WDR Transcription Factor Complex Members Linked to Regulation of the Flavonoid Pathway. Front. Plant Sci. 8, 305–319. doi:10.3389/fpls.2017.00305
Ni, L., Wang, Z., Fu, Z., Liu, D., Yin, Y., Li, H., et al. (2021). Genome-wide analysis of basic helix-loop-helix family genes and expression analysis in response to drought and salt stresses in Hibiscus hamabo Sieb. et Zucc. Int. J. Mol. Sci. 22, 8748. doi:10.3390/ijms22168748
Ohashi-Ito, K., and Bergmann, D. C. (2006). Arabidopsis FAMA Controls the Final Proliferation/Differentiation Switch during Stomatal Development. The Plant Cell 18, 2493–2505. doi:10.1105/tpc.106.046136
Pearce, R. S. (1988). Extracellular Ice and Cell Shape in Frost-Stressed Cereal Leaves: a Low-Temperature Scanning-Electron-Microscopy Study. Planta 175, 313–324. doi:10.1007/BF00396336
Pillitteri, L. J., Sloan, D. B., Bogenschutz, N. L., and Torii, K. U. (2007). Termination of Asymmetric Cell Division and Differentiation of Stomata. Nature 445, 501–505. doi:10.1038/nature05467
Pires, N., and Dolan, L. (2010). Origin and Diversification of Basic-Helix-Loop-Helix Proteins in Plants. Mol. Biol. Evol. 27, 862–874. doi:10.1093/molbev/msp288
Qiao, X., Li, Q., Yin, H., Qi, K., Li, L., Wang, R., et al. (2019). Gene Duplication and Evolution in Recurring Polyploidization-Diploidization Cycles in Plants. Genome Biol. 20, 38. doi:10.1186/s13059-019-1650-2
Schultz, J., Milpetz, F., Bork, P., and Ponting, C. P. (1998). SMART, a Simple Modular Architecture Research Tool: Identification of Signaling Domains. Proc. Natl. Acad. Sci. 95, 5857–5864. doi:10.1073/pnas.95.11.5857
Seo, J.-S., Joo, J., Kim, M.-J., Kim, Y.-K., Nahm, B. H., Song, S. I., et al. (2011). OsbHLH148, a Basic helix-loop-helix Protein, Interacts with OsJAZ Proteins in a Jasmonate Signaling Pathway Leading to Drought Tolerance in rice. Plant J. 65, 907–921. doi:10.1111/j.1365-313X.2010.04477.x
Shannon, P. (2003). Cytoscape: a Software Environment for Integrated Models of Biomolecular Interaction Networks. Genome Res. 13, 2498–2504. doi:10.1101/gr.1239303
Shirasawa, K., Isuzugawa, K., Ikenaga, M., Saito, Y., Yamamoto, T., Hirakawa, H., et al. (2017). The Genome Sequence of Sweet Cherry (Prunus Avium) for Use in Genomics-Assisted Breeding. Dna Res. 24, 499–508. doi:10.1093/dnares/dsx020
Stamatakis, A. (2014). RAxML Version 8: a Tool for Phylogenetic Analysis and post-analysis of Large Phylogenies. Bioinformatics 30, 1312–1313. doi:10.1093/bioinformatics/btu033
Sun, H., Fan, H.-J., and Ling, H.-Q. (2015). Genome-wide Identification and Characterization of the bHLH Gene Family in Tomato. Bmc Genomics 16, 9. doi:10.1186/s12864-014-1209-2
Takahashi, Y., Ebisu, Y., Kinoshita, T., Doi, M., Okuma, E., Murata, Y., et al. (2013). bHLH Transcription Factors that Facilitate K+ Uptake during Stomatal Opening Are Repressed by Abscisic Acid through Phosphorylation. Sci. Signaling 6, ra48. doi:10.1126/scisignal.2003760
Toledo-Ortiz, G., Huq, E., and Quail, P. H. (2003). The Arabidopsis Basic/Helix-Loop-Helix Transcription Factor Family[w]. The Plant Cell 15, 1749–1770. doi:10.1105/tpc.013839
Verde, I., Abbott, A. G., Abbott, A. G., Scalabrin, S., Jung, S., Shu, S., et al. (2013). The High-Quality Draft Genome of Peach (Prunus Persica) Identifies Unique Patterns of Genetic Diversity, Domestication and Genome Evolution. Nat. Genet. 45, 487–494. doi:10.1038/ng.2586
Wang, H., Li, Y., Pan, J., Lou, D., Hu, Y., and Yu, D. (2017). The bHLH Transcription Factors MYC2, MYC3, and MYC4 Are Required for Jasmonate-Mediated Inhibition of Flowering in Arabidopsis. Mol. Plant 10, 1461–1464. doi:10.1016/j.molp.2017.08.007
Wang, L., Tang, W., Hu, Y., Zhang, Y., Sun, J., Guo, X., et al. (2019). A MYB/bHLH Complex Regulates Tissue‐specific Anthocyanin Biosynthesis in the Inner Pericarp of Red‐centered Kiwifruit Actinidia Chinensis Cv. Hongyang. Plant J. 99, 359–378. doi:10.1111/tpj.14330
Wang, T., Lu, J., Xu, Z., Yang, W., Wang, J., Cheng, T., et al. (2014). Selection of Suitable Reference Genes for miRNA Expression Normalization by qRT-PCR during Flower Development and Different Genotypes of Prunus Mume. Scientia Horticulturae 169, 130–137. doi:10.1016/j.scienta.2014.02.006
Wang, Y.-J., Zhang, Z.-G., He, X.-J., Zhou, H.-L., Wen, Y.-X., Dai, J.-X., et al. (2003). A rice Transcription Factor OsbHLH1 Is Involved in Cold Stress Response. Theor. Appl. Genet. 107, 1402–1409. doi:10.1007/s00122-003-1378-x
Wang, Y., Tang, H., Debarry, J. D., Tan, X., Li, J., Wang, X., et al. (2012). MCScanX: A Toolkit for Detection and Evolutionary Analysis of Gene Synteny and Collinearity. Nucleic Acids Res. 40, e49. doi:10.1093/nar/gkr1293
Zhang, Q., Chen, W., Sun, L., Zhao, F., Huang, B., Yang, W., et al. (2012). The Genome of Prunus Mume. Nat. Commun. 3. doi:10.1038/ncomms2290
Zhang, Q., Zhang, H., Sun, L., Fan, G., Ye, M., Jiang, L., et al. (2018). The Genetic Architecture of floral Traits in the Woody Plant Prunus Mume. Nat. Commun. 9, 1318. doi:10.1038/s41467-018-04093-z
Zhao, W., Liu, Y., Li, L., Meng, H., Yang, Y., Dong, Z., et al. (2021). Genome-Wide Identification and Characterization of bHLH Transcription Factors Related to Anthocyanin Biosynthesis in Red walnut (Juglans Regia L.). Front. Genet. 12, 632509. doi:10.3389/fgene.2021.632509
Zhou, J., Li, F., Wang, J.-l., Ma, Y., Chong, K., and Xu, Y.-y. (2009). Basic helix-loop-helix Transcription Factor from Wild rice (OrbHLH2) Improves Tolerance to Salt- and Osmotic Stress in Arabidopsis. J. Plant Physiol. 166, 1296–1306. doi:10.1016/j.jplph.2009.02.007
Zhu, J.-K. (2016). Abiotic Stress Signaling and Responses in Plants. Cell 167, 313–324. doi:10.1016/j.cell.2016.08.029
Keywords: Prunus mume, basic helix-loop-helix gene family, genome-wide analysis, expression pattern, low temperature stress 3
Citation: Ding A, Ding A, Li P, Wang J, Cheng T, Bao F and Zhang Q (2021) Genome-Wide Identification and Low-Temperature Expression Analysis of bHLH Genes in Prunus mume. Front. Genet. 12:762135. doi: 10.3389/fgene.2021.762135
Received: 21 August 2021; Accepted: 16 September 2021;
Published: 01 October 2021.
Edited by:
Suxu Tan, Michigan State University, United StatesReviewed by:
Yong Wang, Jiangsu University, ChinaCopyright © 2021 Ding, Ding, Li, Wang, Cheng, Bao and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Fei Bao, YmFvZmVpQGJqZnUuZWR1LmNu; Qixiang Zhang, enF4YmpmdUAxMjYuY29t
†These authors share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.