
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Genet., 05 October 2021
Sec. Genomics of Plants and the Phytoecosystem
Volume 12 - 2021 | https://doi.org/10.3389/fgene.2021.754237
bZIP gene family is one of the largest transcription factor families. It plays an important role in plant growth, metabolic, and environmental response. However, complete genome-wide investigation of bZIP gene family in Glycyrrhiza uralensis remains unexplained. In this study, 66 putative bZIP genes in the genome of G. uralensis were identified. And their evolutionary classification, physicochemical properties, conserved domain, functional differentiation, and the expression level under different stress conditions were further analyzed. All the members were clustered into 13 subfamilies (A–K, M, and S). A total of 10 conserved motifs were found in GubZIP proteins. Members from the same subfamily shared highly similar gene structures and conserved domains. Tandem duplication events acted as a major driving force for the evolution of bZIP gene family in G. uralensis. Cis-acting elements and protein–protein interaction networks showed that GubZIPs in one subfamily are involved in multiple functions, while some GubZIPs from different subfamilies may share the same functional category. The miRNA network targeting GubZIPs showed that the regulation at the transcriptional level may affect protein–protein interaction networks. We suspected that domain-mediated interactions may categorize a protein family into subfamilies in G. uralensis. Furthermore, the tissue-specific gene expression patterns of GubZIPs were analyzed using the public RNA-seq data. Moreover, gene expression level of 66 bZIP family members under abiotic stress treatments was quantified by using qRT-PCR. The results of this study may serve as potential candidates for functional characterization in the future.
Basic leucine zipper (bZIP) is a transcription factors (TFs) family and widely distributed in eukaryotes. The structure of bZIP protein is defined by the conserved bZIP protein and often acts as a dimer (Mair et al., 2015). They are abundant in different species. A total of 78 bZIP genes have been found in Arabidopsis (Arabidopsis thaliana) (Dröge-Laser et al., 2018), 65 in potato (Solanum tuberosum L.) (Zhao et al., 2020), 132 in tobacco (Nicotiana tabacum L.) (Li et al., 2021), 57 in Medicago sativa (Liu et al., 2021), and 86 in rice (E et al., 2014). The bZIP TF has a specific sequence consisting of a fixed motif N-X7-R/K and a leucine zipper that binds to the alkaline region. In the leucine zipper region, the highly conserved heptad repeats of leucine may be replaced by phenylalanine (F), valine (V), isoleucine (I), or methionine (M) and make bZIP protein form a variety of binding ways of homodimer and heterodimer. bZIP proteins preferentially combined with ACGT as the core motif to form a palindrome structure (Dröge-Laser et al., 2018).
In plants, bZIP TFs play important regulatory roles in plant growth, development, and response to environmental stress. In previous studies, bZIPs were found to be expressed in seeds, flowers (Strathmann et al., 2001), leaves (Van Leene et al., 2016), and roots (Ma et al., 2018). Some of them have positive responses to abiotic stress (Zhu et al., 2018) and are highly associated with the abscisic acid (ABA) activation or other metabolic pathways (Lee et al., 2010; Garg et al., 2019). AtbZIP53 promoted the transcriptional activation of seed maturation genes by forming heterodimers with bZIP1, 10, or 25 (Alonso et al., 2009). AtbZIP17 was a transcriptional activator involved in salt and stress response. After salt, heat, and ABA treatment, the C-terminal of AtbZIP17 was cleaved, and then the N-terminal was transferred to the nucleus to activate the expression of salt stress related genes (Zhou et al., 2015). AtbZIP11 (GBF6) actively regulated the expression of the proline dehydrogenase (PRODH) gene, which participated in amino acid metabolism (Hanson et al., 2008). AtbZIP11 also induced TRE1, TPP5, and TPP6 expression and regulated trehalose metabolism (Ma et al., 2011). OsbZIP16 found in rice can reduce sensitivity to abiotic stress during the germination of overexpressed seedlings (Pandey et al., 2018). It was found that nuclear-localized C subfamily member TabZIP6 in wheat (Triticum aestivum L.) could not only homodimerize but also form a dimer with other two S subfamily bZIP proteins, which are involved in cold tolerance (Cai et al., 2018). In soybean, GmbZIP15 negatively regulated GmWRKY12 and GmABF1 and made it sensitive to salt and drought stress (Zhang et al., 2020). It was found that IbbZIP1 in sweet potato was related to salt and drought tolerance and was highly responsive to ABA (Kang et al., 2019).
Licorice is a widely cultivated edible and medicinal crop with high tolerance to stress in the world, which plays very important roles in desertification control, animal husbandry, and human health (Ishimi et al., 2019; Lyu et al., 2020). Unfortunately, the content of most cultivated licorice root is not matching that of wild quality and thus is usually diverted to non-medicinal, food, or confectionary uses (Josef A. Brinckmann, 2020). Furthermore, because of global warming and the decrease of land area suitable for cultivation, there is an urgent need to improve the important traits and quality in licorice breeding. Therefore, it is necessary to make an in-depth study on the development of licorice and its response to environmental factors. The Glycyrrhiza uralensis genome was sequenced recently. Although many studies on the different gene families in G. uralensis have been reported (Tong et al., 2019; Cui et al., 2020; Goyal et al., 2020; Gao et al., 2021), but the complete genomic information of G. uralensis has not been fully explored yet. The comprehensive analysis of bZIP gene family in G. uralensis has not been reported. In this study, 66 GubZIP genes were systematically analyzed, including identification members, phylogenetic relationships, protein structure, conserved domain, duplications in the genome sequence, protein–protein interaction (PPI) network, and related targeted miRNA. The effects of ultraviolet (UV), cadmium (Cd), ABA, methyl jasmonate (MeJA), drought (PEG), and salt (NaCl) on GubZIP gene family were investigated. Finally, we performed gene network analyses on the key genes. Comprehensive analysis showed that members of GubZIP gene family played an important role in various biological processes and transcriptional regulatory networks of G. uralensis. These results lay a foundation for genetic breeding and further study of their biological functions.
The G. uralensis data used in this study were derived from the Genome sequence database of G. uralensis (http://ngs-data-archive.psc.riken.jp/Gur-genome/download.pl) (Mochida et al., 2017). The hidden Markov model (HMM) profile of the bZIP domains (PF00170 and PF07716) was downloaded from the Pfam protein family database (http://pfam.xfam.org) (El-Gebali et al., 2019). The potential bZIP genes were identified from the genome of G. uralensis using HMMER (E-value < 1 × 10–5) (http://hmmer.janelia.org/), and redundant transcripts were removed (Prakash et al., 2017). The nonredundant candidate bZIP protein sequences further confirmed the existence of the bZIP domain by using the National Center for Biotechnology Information (NCBI) Conserved Domain database (https://www.ncbi.nlm.nih.gov/cdd/) and the SMART database (http://smart.embl.de/). The ProtParam tool of the ExPASy (https://web.expasy.org/protparam/) was used to analyze the molecular weight (MW), isoelectric point (pI), instability index (II), aliphatic index (AI), and grand average of hydropathicity (GRAVY) of candidate GubZIPs. The subcellular localization of all predicted GubZIP proteins was analyzed by WoLF PSORT (https://wolfpsort.hgc.jp/) Protein Subcellular Localization Prediction system.
The bZIP protein sequences of A. thaliana and Glycine max were obtained from Plant Transcription Factor database (http://planttfdb.gao-lab.org/index.php). The full-length bZIP protein sequences of G. uralensis, A. thaliana, and G. max were aligned by ClustalW in MEGAX software using the default parameters (Kumar et al., 2018). An interspecific phylogenetic tree was constructed by the maximum likelihood (ML) method via IQ-TREE software with 1,000 ultrafast bootstrap replications (Nguyen et al., 2015). The tree was further managed by the online tool EvolView v3 (https://www.evolgenius.info/evolview/) (Subramanian et al., 2019).
The conserved motifs were analyzed and identified using MEME online (http://meme-suite.org/index.html). In order to clarify the structural diversity of GubZIP genes, the intron–exon structures of GubZIP genes were visualized using the gene structure display serving GSDS (http://gsds.cbi.pku.edu.cn/) (Hu et al., 2015). The final result was displayed by using TBtools (v1.09832) (Chen et al., 2020).
The upstream 1,500-bp regions of the transcriptional start in all candidate GubZIP coding sequences (CDSs) (GubZIP64 is less than 1,500 bp in length) were submitted to PlantPAN 3.0 (http://plantpan.itps.ncku.edu.tw/) to identify the regulatory elements (Chow et al., 2019). Based on position information provided by the G. uralensis database (http://ngs-data-archive.psc.riken.jp/Gur-genome/download.pl), we visualized the chromosomal distribution of bZIP genes in licorice using BioPerl software (Stajich et al., 2002). The similarity of two genes is more than 70%, and the chromosomal distance between the two genes is less than 100 kb, which is defined as tandem replicated gene (Cannon et al., 2004; Audemard et al., 2012). The non-synonymous (ka) and synonymous substitutions (ks) rates were calculated to evaluate the selection pressure. Divergence times (T) were estimated with equation T = Ks/2r × 10–6 (Mya) (the r was taken to be 1.5 × 10–8 for G. uralensis) (Huang et al., 2016). The graph was processed by TBtools (v1.09832) (Chen et al., 2020).
The interaction of GubZIP proteins (orthologs in A. thaliana) was performed by STRING software (https://string-db.org/), and the confidence parameter was set to 0.4. In order to predict miRNAs targeting GubZIP gene, we queried the full-length CDS of GubZIP gene using the psRNATarget service (http://plantgrn.noble.org/psRNATarget/analysis?function=2). To increase stringency, the maximum expectation value was set to 3.0, and the rest used the default parameters. The interaction network of miRNA and GubZIP genes was drawn by Cytoscape 3.8.2 software.
The seeds of G. uralensis were treated with 98% sulfuric acid for 30 min to break the seed dormancy, rinsed with sterilized ultra-pure water five times, and then sowed in a seedling plate. Plants grew at 25 C under 60–70% relative humidity in a 16 h day/8 h night cycle in an artificial climate chamber. Four-leaf seedlings were transferred to the hydroponic culture system with 1/2 Murashige–Skoog (MS) liquid medium for different stress treatments. To investigate expression patterns of the genes in response to abiotic stress, PEG6000 (control, 0; drought 1, 10%; drought 2, 20%), NaCl (control, 0; salt 1, 150 mM; salt 2, 300 mM), Cd (control, 0; Cd 1, 0.02 g kg−1; Cd 2, 0.04 g kg−1), UV (control, 0; UV, 2 days, 16-h light/4-h dark/4-h UV (30 W)), MeJA (control, 0; MeJA, 100 μM), and ABA (control, 0; ABA, 100 μM) were used for abiotic stress treatment. The leaves and roots of G. uralensis seedlings were collected after 48-h treatment, then quickly frozen in liquid nitrogen, and stored at −80 C. Each treatment consisted of three biological replicates.
In order to analyze the spatial characteristics and differential expression patterns of target genes, the RNA-seq data of leaves and roots (SRP215420) (Li et al., 2020), drought, and salt stress (SRP065514) were downloaded from NCBI (https://www.ncbi.nlm.nih.gov/) and used to determine their expression patterns. The transcript levels of G. uralensis bZIP genes were normalized by transcripts per million (TPM).
Total RNA was extracted by RNAprep Pure Plant Kit (TIANGEN, Beijing, China). The concentration and quality of RNA were determined by NanoDrop 2000 spectrophotometer. cDNA was synthesized from total RNA isolated from various tissues by using TaKaRa PrimeScript™ RT Master Mix and gDNA Eraser reverse transcription system, according to the manufacturer’s protocols. Quantitative real-time PCR (qRT-PCR) was performed using QuantStudio six Flex real-time PCR system (Thermo Fisher, Waltham, MA, USA) and TB Green™ Premix Ex Taq™ II (Tli RNaseH Plus) (TaKaRa, Maebashi, Japan). The most stable housekeeping reference gene GuActin (accession number GQ404511) was selected as the internal control (Tong et al., 2019). All primers were designed using NCBI Primer Blast website (https://www.ncbi.nlm.nih.gov/tools/primer-blast/). Three independent biological and technical replicates were performed in the qRT-PCR experiments. The relative expression level of GubZIPs was measured by 2−ΔΔCt method, and the final result was visualized by TBtools. In addition, when the relative expression fold changes (treatment/control) ≥2 or ≤0.5 respectively were considered to be differentially upregulated or downregulated. Significance was confirmed by least significant difference (LSD) test (p < 0.05). The significant regulatory genes responding to different stresses were analyzed by histogram.
A total of 66 members of GubZIP gene family were obtained from the genome of G. uralensis using a series of bioinformatics methods based on HMM. The candidate GubZIPs were identified and named GubZIPX, in which X is an integer, representing the ascending order of genes on the corresponding scaffold. GubZIP34 gene has the shortest protein length with 132 amino acids, whereas GubZIP5 possesses the longest one (788 amino acids). The MW of the proteins ranged from 15.68 kDa (GubZIP34) to 85.39 kDa (GubZIP5) with an average MW of 37.90 kDa. The isoelectric points of the GubZIPs ranged from 4.97 (GubZIP25) to 10.18 (GubZIP44) with an average pI of 7.31. The proteins with an isoelectric point greater than seven accounted for 43.94% of the total number, which means that 29 of the GubZIP proteins were neutral or alkaline. The GRAVY of the 66 sequences had a maximum value of −0.329 (GubZIP43), a minimum value of −1.335 (GubZIP32), and a mean value of −0.75. All negative GRAVY values indicate their hydrophilic nature. The AI of GubZIPs ranged from 46.20 (GubZIP25) to 92.93 (GubZIP60) with an average of 68.20. Their detailed characteristics and subcellular localization are summarized in Table 1.
To elucidate the evolutionary relationship and classification of the bZIP family in different species, we constructed a phylogenetic tree using the entire amino acid of each member from G. uralensis (66 bZIP genes), model plant A. thaliana (74 bZIP genes), and cash crop G. max (149 bZIP genes) (Figure 1) (Dröge-Laser et al., 2018). As shown in Figure 1, the members of the GubZIP family were divided into 13 subfamilies (A–K, M, and S) on the basis of the classification of Arabidopsis. The S subfamily contains 13 GubZIP genes, which is the largest group in the GubZIP family, followed by the A and D subfamilies (12 members). The B, J, K, and M subfamilies contain only one GubZIP gene, which are the smallest size groups in the GubZIP family.
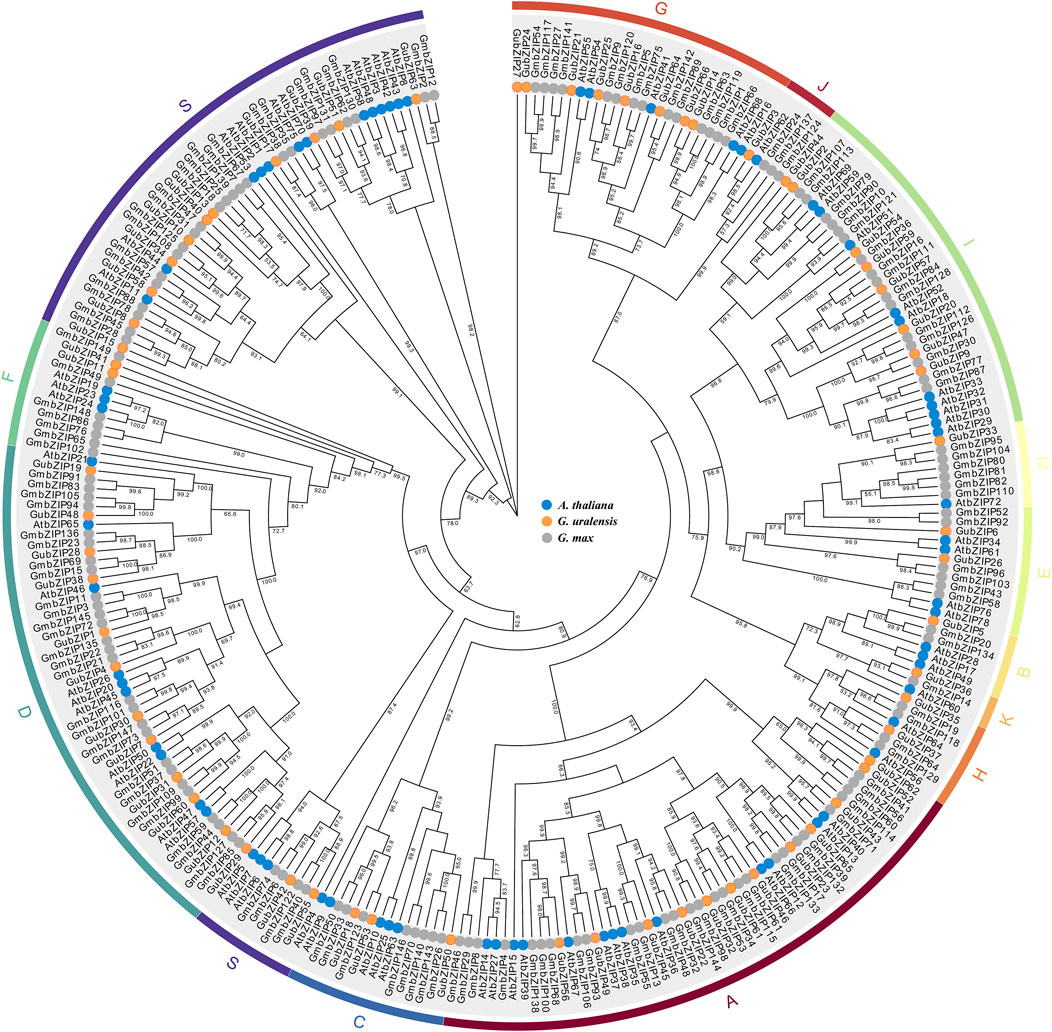
FIGURE 1. Phylogenetic analysis of bZIP proteins in Arabidopsis thaliana, Glycine max, and Glycyrrhiza uralensis. Protein sequence alignment was carried out in MEGA X by ClustalW, and the evolutionary tree was constructed by maximum likelihood method through IQ-TREE. The pilot value is based on 1,000 duplicates. Only pilot values greater than 50% are displayed. The ends of branches from different species are represented by circles of different colors. The bZIP protein is divided into 13 different evolutionary branches (A–K, M, and S), which are marked by curves with different colors.
To provide greater insight into the gene structure of 66 bZIP family genes of G. uralensis, a rootless phylogenetic tree of GubZIPs was generated (Figure 2A). The conserved motif distribution of GubZIP proteins (Figure 2B) and the exon–intron structure (Figure 2C) were analyzed. The proteins in each subfamily contain the same conserved motifs, which further support the above result of a phylogenetic tree. However, they also have different conserved motifs among various subfamilies. Ten conserved motifs were identified using MEME software (Figure 2D). The length of the motif ranged from 15 to 100 amino acids, and the letter height of the amino acid residue represents its conservation degree. Motif1 has been recognized as a bZIP conserved domain and could be found in most subfamilies except S, K, J, and F, while some subfamilies had unique motif compositions. For example, subfamily A possesses unique motif5 and motif9, whereas motif2, 3, 6, and eight were unique to the subfamily. MEME results showed that GubZIP13, GubZIP24, GubZIP36, and GubZIP40 had only one motif, while the D subfamily contained five motifs except for GubZIP19 and GubZIP60. It was also found that the motif distribution of members in the same subfamily was often highly conservative. For example, most members of the A subfamily had motifs 1, 5, 9, and 10; while most members of the D subfamily had 1, 2, 3, 6, and 8. In addition, the exon–intron structure of the same subfamily members had a similar gene structure. For example, there were almost no introns in the F subfamily and S subfamily, whereas the G subfamily and D subfamily contained a large number of introns and exons. There were significant differences in exon–intron structure among different subfamilies, which further supported the results of the GubZIP phylogenetic tree and classification.
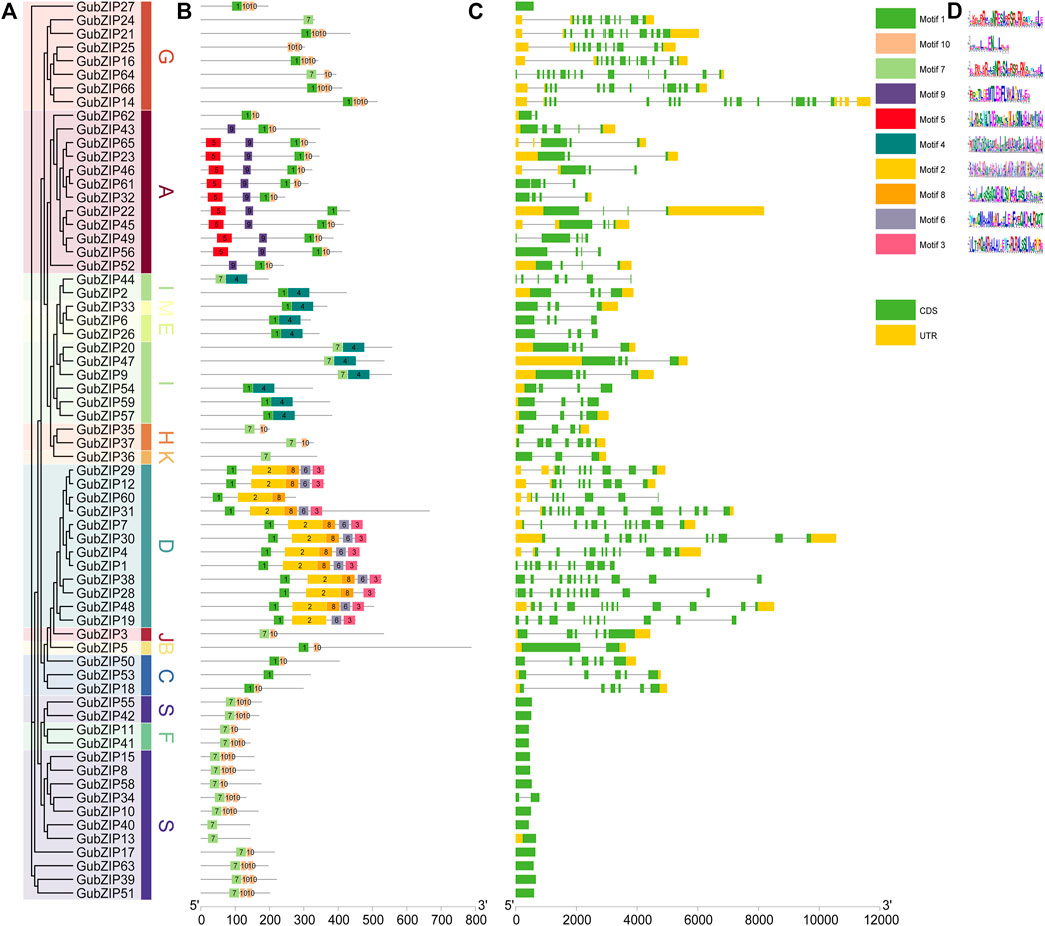
FIGURE 2. Phylogenetic relationships, gene structure, and architecture of conserved protein motifs in GubZIPs. (A) Phylogenetic tree of GubZIP genes. (B) Distribution of conserved structures in all 66 GubZIP proteins. The colorful boxes delineate different motifs (numbers 1–10). Gray lines represent non-conservative sequences. The protein length can be estimated using the scale at the bottom. (C) The exon–intron structure of GubZIP genes. The yellow and green boxes represent the untranslated region (UTR) and the coding sequence (CDS), respectively. The gray line indicates the intron. (D) Protein motifs in the bZIP members. The colorful boxes delineate different motifs.
As shown in Figure 3, bZIP genes of G. uralensis were distributed in 63 separate chromosomal scaffolds. Only scaffold13, 17, and 69 contained two bZIP genes, while other scaffolds contained only one GubZIP. New cellular functions of genes and their encoded protein products evolve through the mechanism of duplication. Due to the high similarity of retained duplicate genes, a gene is not only regulated by TFs from different families but also bound by multiple members of the same family (Panchy et al., 2016). GubZIPs were not evenly distributed across the chromosomal scaffolds. Since the genome of G. uralensis was only assembled to the chromosomal scaffold level, we used the manual blast method for duplicate gene finding, only tandem repeat genes were analyzed, and segmental duplication or transposable genes were unable to determine on this level. Then using BioPerl (Stajich et al., 2002), we analyzed the tandem duplication events among the genes. Four tandem repeat gene pairs formed by eight bZIP genes were found in G. uralensis, of which six members belonged to the D subfamily and the remaining two members belonged to the G subfamily. These lines of evidence suggested that tandem duplication events are the main driving force for the diversity of the GubZIPs.
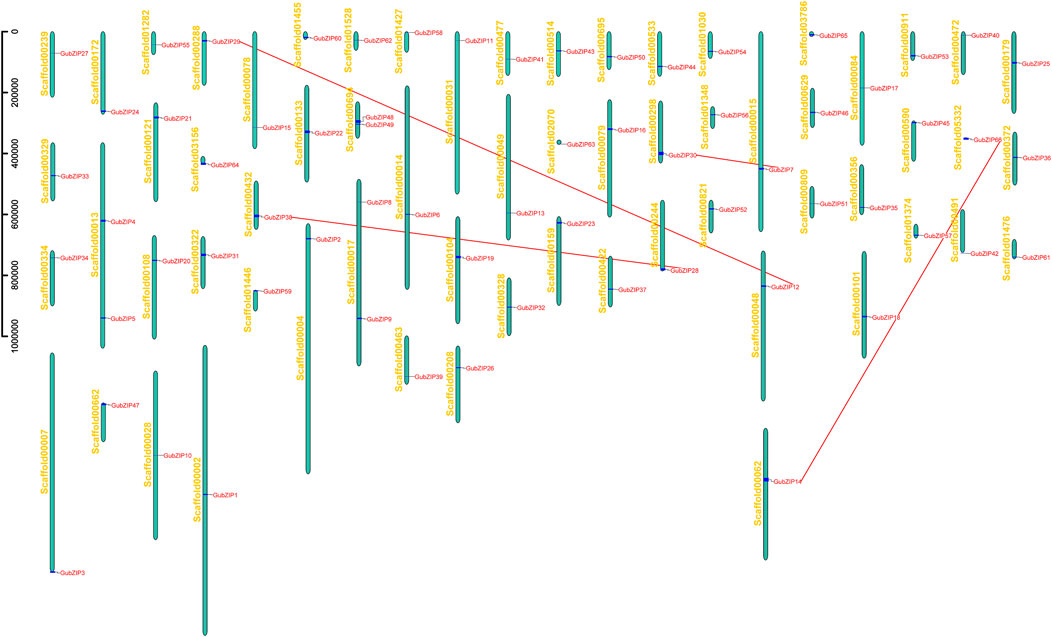
FIGURE 3. Chromosome scaffold distribution of GubZIPs. GubZIP genes were located on 63 independent scaffolds. The vertical column represents scaffold, with the number on the left. Pairs of duplicate genes are represented by red dotted lines.
To explore the evolutionary constraints of GubZIP genes, the selection pressure of replication gene pairs was analyzed. The average ratios of Ka, Ks, and Ka/Ks of all tandem repeat gene pairs were 0.0991, 0.4303, and 0.2150, respectively (Supplementary Table S3). The Ka/Ks of the four gene pairs (GubZIP7 and GubZIP30, GubZIP12 and GubZIP29, GubZIP14 and GubZIP66, and GubZIP28 and GubZIP38) were all less than 0.5. It was indicated that these gene pairs experienced strong purifying selective pressure. We further used Ks to estimate the time of GubZIP genes duplication events during the evolutionary time of the G. uralensis genome. The Ks of tandem duplications of GubZIP genes occurred from 0.43 (Ks = 0.13) mya to 2.0 (Ks = 0.60) mya, with an average of 1.43 mya.
TFs regulate the target genes both spatially and temporally through the specific binding of cis-regulatory elements (CREs) present in their promoters (Qiu, 2003). In order to explore the CREs of GubZIP gene family, the 1.5-kb genomic sequence upstream of each gene was extracted and matched to the PlantPAN 3.0 database. The CREs of the GubZIPs are listed in Figure 4. The CREs of GubZIPs were involved in transcriptional initiation (transcriptional start, promoter, and enhancer regions), phytohormone responses (ABA, MeJA, gibberellin, ethylene, and auxin response elements), and stress responses (drought, light response, and low temperature). Several CREs were identified to be involved in the hormonal response, such as MeJA (TGACG-motif), auxin (TGA) response elements, and gibberellin response element (P-box). At the same time, stress-response elements related to ABA (ABRE), low-temperature reactivity (LTR), ethylene (ERE) responses, and the MYB binding site (MBS) involved in drought induction were also identified. ABA response elements were detected in GubZIP29, 3, 4, 42, 44, and 48, which belonged to subfamilies J, D, S, and I. The MYB binding site (MBS) involved in drought induction was detected in 20 members. Moreover, a total of 18 CREs with light-responsive components were identified. MYB binding sites (MRE) involved in light response, light-responsive elements (GT1-motif), cis-acting regulatory elements about light responsiveness (G-Box), part of a conserved DNA module involved in light responsiveness (Box4), and light-responsive elements (Box I) were detected in five, nine, 13, 59, and 25 members, respectively. The results showed that GubZIP genes may play important roles in plant growth and stress responses, and especially in the response to light.

FIGURE 4. Cis-regulatory element analysis of GubZIP gene promoter. The promoter sequence (−1,500 bp) of GubZIP genes (−1,000 bp for GubZIP64) was inferred on PlantPAN 3.0. The upstream length of each cis-regulatory element to the translation start point can be inferred from the scale at the bottom.
Based on the studied AtbZIPs and the orthologs in Arabidopsis, we could speculate about the functions of most GubZIP genes. To analyze them, we constructed an interaction network analysis by using the STRING database based on search for protein families (Figure 5A) and multiple sequences (Figure 5B). Through the GubZIP protein family interaction network, it was found that there were 14 members involved in the ABA activation signal pathway (NOG243340), of which 10 members belonged to the A subfamily (83.3% of the A subfamily), three belonged to the S subfamily (23.1% of the S subfamily), and one belonged to the G subfamily (12.5% of the G subfamily). The members involved in primary cell wall formation (COG1215), UDP-glycosyltransferase (KOG1192), flower development regulation (NOG259341), and positive regulation of seed maturation (NOG10040) were from H, G, D, and C subfamilies. The members involved in seed germination (NOG257560) were GubZIP49 and GubZIP56, both belonging to the A subfamily. The results suggested that the interaction network of the GubZIP family was spread around the ABA signal pathway, and their regulation network plays an important role in the development of G. uralensis. Wang et al. showed a similar phenomenon in GHbZIPs (Wang et al., 2020). As shown in Figure 5B, bZIP53 (GubZIP13 and 40) interacted directly with BZIP17 (GubZIP15), GBF3 (GubZIP21 and 24), ABI5 (GubZIP56), GBF6 (GubZIP10), HYH (GubZIP35), bZIP16 (GubZIP66), bZIP44 (GubZIP 8, 15, and 58), and bZIP68 (GubZIP14). It was suggested that bZIP53 is involved in cellular response to abiotic stress response and positive regulation of seed maturation (Alonso et al., 2009). The direct interactions of bZIP68 (GubZIP14) and HYH (GubZIP35) with GBF1 (GubZIP16 and 25), GBF4 (GubZIP43, 52, and 62) with GBF3 (GubZIP21 and 24), and AREB3 (GubZIP23, 32, 46, 61, and 65) with GBF6 (GubZIP10) were found in PPI network. The interaction between GBF4 (homolog of GubZIP43, 52, and 62) and bZIP68 (homolog of GubZIP14) was regulated by light or other hormones (Terzaghi et al., 1997).
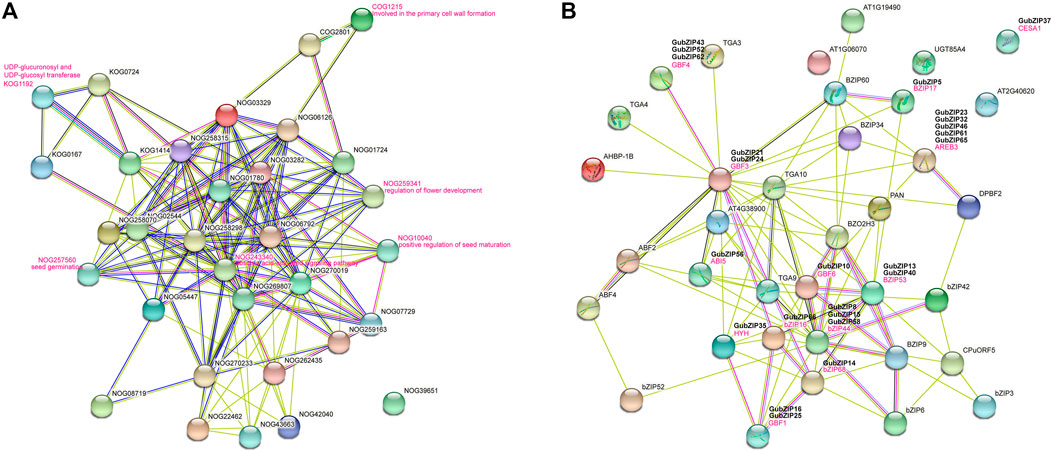
FIGURE 5. Interaction network of GubZIP proteins. (A) Search for the orthologs in Arabidopsis matching GubZIPs. (B) The GubZIP protein–protein interaction network. This network was predicted by online software STRING. The GubZIP proteins related to abiotic stresses are shown in the bold black font above the Arabidopsis orthologs.
With the use of the psRNATarget server, the miRNAs associated with GubZIP genes were predicted based on annotated data of A. thaliana. The results showed that six miRNA families were identified. The targeted GubZIPs (11 members) belonged to K, B, D, H, and A subfamilies (Figure 6), which were involved in the development, abiotic stress, lateral root formation, and endoplasmic reticulum process. The results showed that not only one miRNA family targeted unique GubZIP, but also one miRNA family could target multiple GubZIPs with different functions. It is suggested that miRNA plays a potential role in the PPI relationship of GubZIPs. The complicated regulatory relationship of miRNAs and multi-functional GubZIPs provided the opportunity for the collaboration of genes with different functions.
To verify the expression patterns of GubZIP family members, the RNA-seq data of leaves and roots under drought and salt stress were accessed in NCBI. GubZIP genes showed different expression patterns under different spatial and abiotic stresses (Supplementary Figure S1). GubZIP members were clustered into three categories in different tissues (Supplementary Figure S1A). One group was highly expressed in roots containing 31 members (46.97%) in total, while 23 members (34.85%) were specifically expressed in leaves. The rest of the members were expressed in both leaves and roots. In the abiotic stress expression (Supplementary Figure S1), GubZIP members showed various expression patterns under different stress conditions. For example, GubZIP40 was downregulated under salt stress but upregulated under drought stress; GubZIP6, 9, 33, 54, and 57 were upregulated under salt stress but downregulated under drought stress. Different expression patterns reflected the different roles of GubZIP genes in the corresponding pathways, which provide a reference for the identification of functional genes. In order to further study the biological function of GubZIP genes, qRT-PCR was used to analyze the expression patterns of GubZIP genes under various abiotic stresses condition. As shown in Figure 7, the expression pattern of GubZIPs under salt and drought stress was similar to that of RNA-seq data.
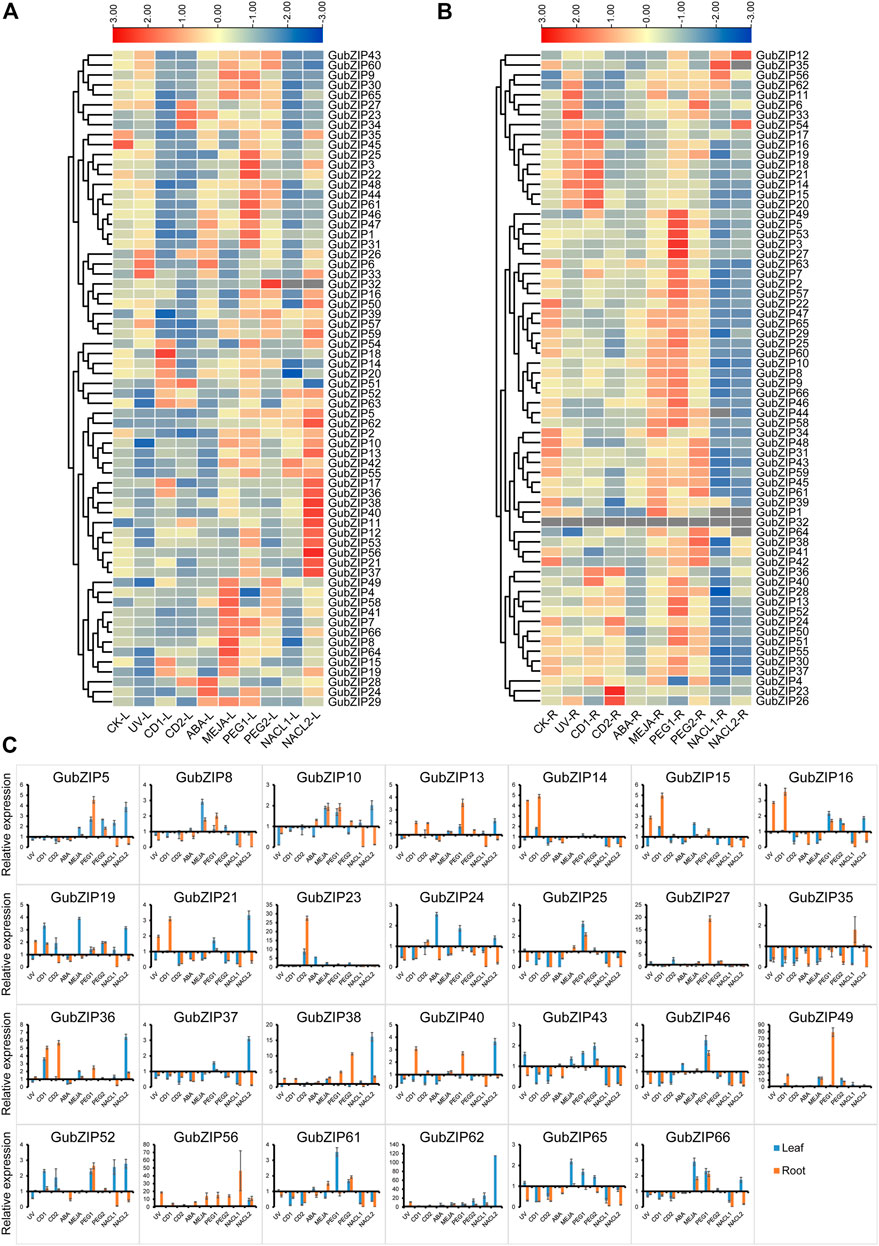
FIGURE 7. Expression pattern of bZIP genes in Glycyrrhiza uralensis under abiotic stress. (A) Heat maps of the relative expression of bZIP genes in leaf tissue of G. uralensis under abiotic stress. (B) Heat maps of the relative expression of bZIP genes in root tissue of G. uralensis under abiotic stress. The level of expression is indicated by the graded color code. (C) The qRT-PCR expression profile of GubZIP genes under abiotic stress. The expression level of GubZIPs in the control group was standardized as “1.” The vertical bar represents the standard error of the average. Significance is indicated on the bar graph by a–i for the difference between gene expression; significance was confirmed by least significant difference (LSD) test (p < 0.05).
There were eight and 56 GubZIP genes that were differentially expressed in roots and leaves under salt stress, respectively. GubZIP12, 17, 35, 56, and 62 were upregulated in roots; GubZIP5, 11, 28, and 62 were upregulated in leaves in low salt concentration levels; GubZIP6, 12, 38, 54, and 56 were upregulated in roots; and 26 genes were upregulated in leaves under low salt concentration stress condition. In roots, the expression level of GubZIP12 increased with the increasing NaCl concentration, whereas the expression of GubZIP56 was upregulated compared with control but decreased with the increasing NaCl concentration. The expression of GubZIP17, 35, and 62 was upregulated from low salt concentration but decreased at high salt concentration. On the contrary, the expression of GubZIP6, 38, and 54 was upregulated in high salt concentration. In leaves, the expression levels of GubZIP3, 21, 26, 37, and 56 were downregulated at low salt concentration but upregulated at high salt concentration. The expression of GubZIP62 was upregulated in both roots and leaves at low salt concentration.
There were 35 and 41 GubZIP genes that were differentially expressed in roots and leaves, respectively, under drought stress. A total of 27 GubZIP genes were upregulated, four GubZIP genes (GubZIP4, 11, 26, and 34) were downregulated in roots under low concentration drought stress, 25 GubZIP genes were upregulated, and only GubZIP4 was downregulated in leaves. Ten GubZIP genes were upregulated in roots, three GubZIP genes (GubZIP21, 35, and 63) were downregulated in roots under high concentration drought stress, and 14 genes were upregulated and 10 GubZIP genes were downregulated in leaves. In roots, the expression level of GubZIP6 and 38 increased with the increasing drought stress concentration, while the expression of GubZIP27 and 49 increased compared with that of the control but decreased with the increasing PEG concentration. The expression level of GubZIP11 changed from downregulation under low drought stress to upregulation under high drought stress. In leaves, the expression level of GubZIP3 changed from upregulated under low drought stress to downregulated under high drought stress. And the expression of GubZIP62 was upregulated in roots and leaves under low concentration drought stress.
GubZIP genes were differentially expressed in roots (16 GubZIPs) and leaves (34 GubZIPs) under the MeJA treatment condition. There were 10 and 19 GubZIP genes upregulated in roots and leaves, while six and 14 genes were downregulated, respectively. The expression of GubZIP6, 33, and 56 was upregulated in roots but downregulated in leaves, whereas GubZIP42 showed the opposite expression pattern. The expression of GubZIP11, 38, 49, 58, 62, and 64 was upregulated in roots and leaves, whereas the expression of GubZIP16 and 35 was downregulated in roots and leaves.
There were 30 and 25 GubZIP genes differentially expressed in roots and leaves, respectively, under ABA treatment conditions. Furthermore, the expression levels of GubZIP18, 22, 36, 37, and 42 were downregulated in both roots and leaves. GubZIP24 and 31 showed a downregulated pattern in roots and upregulated levels in leaves, whereas GubZIP56 showed opposite expression tendency in roots and leaves. The expression level of GubZIP62 was upregulated in both roots and leaves.
There were 45 and 51 differentially expressed genes in roots and leaves, respectively, under cadmium stress conditions. Fourteen GubZIP genes were upregulated and 24 GubZIP genes were downregulated in roots under low cadmium concentration, while nine GubZIP genes were upregulated and 29 GubZIP genes were downregulated in leaves. GubZIP23, 26, 36, and 56 were upregulated in roots, while 30 of GubZIP genes were downregulated in high cadmium concentration. In roots, the expression of GubZIP56 was upregulated when compared with that of the control, but it decreased with the increase of cadmium concentration. The expression levels of GubZIP23 and 26 were downregulated under low cadmium stress condition but upregulated under high-stress conditions, while GubZIP12, 17, and 18 showed opposite expression patterns. In leaves, the expression levels of GubZIP11 and 51 increased with the increase in cadmium concentration, while the expression levels of GubZIP17 and 18 were upregulated under low cadmium stress conditions but downregulated under high concentration. GubZIP23, 26, 27, and 34 showed a downregulated expression pattern under low cadmium stress but upregulated under high cadmium stress. The expression levels of GubZIP1, 3, 6, 22, 31, 35, 41, 48, 59, 60, and 65 were downregulated in roots and leaves. Interestingly, the expression of GubZIP26 was downregulated at low cadmium concentration but upregulated at high cadmium concentration in both roots and leaves.
There were 36 and 23 differentially expressed genes in roots and leaves, respectively, under UV stress conditions. The expression levels of GubZIP22, 24, 35, 42, and 63 were downregulated in both roots and leaves; while GubZIP6, 11, 26, 33, and 54 showed opposite expression patterns in roots and leaves. The expression level of GubZIP39 was downregulated in roots and upregulated in leaves. The expression levels of GubZIP12, 15, 18, 21, 38, and 56 were upregulated in roots but downregulated in leaves.
The results revealed the different response mechanisms of GubZIPs under abiotic stress. The function of GubZIP genes can be more effectively estimated in the future.
The bZIP TFs exist widely in the plant kingdom and play an important role in plant growth, development, and the response to corresponding environmental changes (Herath and Verchot, 2021). bZIP proteins have been identified in many plant species, including A. thaliana (Dröge-Laser et al., 2018), soybean (Zhang et al., 2018), poplar (Zhao et al., 2021), and Vitis vinifera (Liu et al., 2014). However, up to now, no systematic study on bZIP gene family of G. uralensis has been reported. The genome-wide analysis of GubZIPs would aid in their further functional analyses as well as licorice breeding research. In this study, a total of 66 bZIP genes of G. uralensis were identified in the genome. The genes belonging to the same subfamily in the phylogenetic tree had a relatively conservative relationship, and GubZIPs were divided into 13 subfamilies, which is consistent with A. thaliana. The stability of GubZIP protein was estimated by II (Table 1). A protein whose II is smaller than 40 was predicted as stable. The II values for 66 GubZIP proteins were all above 40, predicted as unstable. The AI of the protein was used to evaluate the thermal stability of GubZIP protein (Gasteiger et al., 2005). In this study, the AI of D and S subfamily proteins was generally higher than that of G subfamily proteins, indicating that D and S subfamily GubZIP proteins have higher thermal stability than G subfamily proteins. The prediction results of subcellular localization of GubZIP showed that subfamilies with multiple members would not be distributed on the same subcellular structure. In eukaryote cells, members of the multigene family were localized to specific subcellular compartments, suggesting phylogenetic divergence and distinct functional roles in vivo (Vierling, 1991).
Tandem replication is an evolutionary process whereby a segment of DNA was replicated and proximally inserted. It was considered the main driving force that expands gene families, which is a key driver of adaptive evolution in species that are currently facing widespread environmental challenges (Song et al., 2019). Comparative genomic analysis between closely related species has revealed that tandem duplication is one of the major mechanisms creating new genes, particularly genes clustered into a gene family, which have been documented in many organisms (Anderson and Roth, 1977; Hazkani-Covo and Graur, 2007; Fan et al., 2008). bZIP gene family, as one of the largest known TF families in plants, has a large number of subfamilies and members (Liu and Chu, 2015). However, the Ka/Ks ratios of GubZIP tandem repeat gene pairs were all lower than 0.5, which indicated that these repetitive GubZIP genes may be affected by strong purification selection and maintain a relatively conservative function in different species. For example, as tandem repeat pairs, GubZIP14 and 66 were upregulated in the root under UV and hormone treatment conditions, respectively, which showed a highly similar expression profile to the regulation of homologous in A. thaliana (bZIP68 and bZIP16) by light-induced or hormone-controlled stimuli (Shen et al., 2008). Repetitive genes also provide templates for new genes, which have new functions (Fan et al., 2008).
CREs play critical roles in the regulation of plant stress responses. ABA-responsive elements (ABREs) are involved in the plant response to ABA hormone treatment, drought, and salt stress. LTR responds to low-temperature reactivity. The TCA and ERE element are correlated with the expression level of MeJA and ethylene. There were 14 members of GubZIPs involved in the ABA-activated signaling pathway (NOG243340), which cis-acting regulatory elements associated with light, low temperature, drought, ethylene, and MeJA located in their upstream sequences of promoters. Several cis-acting elements responding to light and MeJA were found in the promoter regions of GubZIP37 that were involved in primary cell wall formation (COG1215). The expression of GubZIP37 was downregulated under cadmium, ABA, MeJA, and salt stress conditions, suggesting that the cell damage caused by stress may be due to the effect of related cis-acting regulatory elements. The upstream sequences of GubZIP1 and 64 that were involved in the regulation of flower development (NOG259341) and UDP-glycosyltransferase (KOG1192) contain several cis-acting elements related to light response. The expression level of GubZIP64 was downregulated under UV stress and upregulated under MeJA and low concentration drought stress, which was consistent with previous studies that moderate drought could increase the content of secondary metabolites in G. uralensis (Li et al., 2011). A great deal of cis-elements responding to environmental stress were found in the promoter regions of the GubZIP50, such as ERE (ethylene response), MRE (metal-responsive element), and LTR (low-temperature reactivity). GubZIP50 was downregulated in leaves under a high concentration of cadmium treatment, which may indicate that GubZIP50 was involved in the response to heavy metal tress that may be related to the decrease of mitotic index caused by cytotoxicity (Chidambaram et al., 2009). The analysis of cis-acting elements and the prediction of the protein interaction network of 66 GubZIP family members indicated the enrichment of cis-acting elements, such as ABRE, ERE, Box 4, GT 1-motif, or G-Box; and the interaction network together with homologs in Arabidopsis suggested that GubZIP family members also play wide roles in the light response, hormone response, and growth and development of licorice plants.
Gene expression is regulated by cis-acting elements of the upstream promoter, which are sites involved in transcriptional initiation and regulating the specific binding of proteins and often determine the transcriptional process of gene expression (Zou et al., 2011; Hernandez-Garcia and Finer, 2014). As shown in Supplementary Figure S1, the expression trend of GubZIP TFs showed different profiles under abiotic stress, which were based on RNA-seq data (from public databases). The results of qRT-PCR verification (Figure 7) showed the expression profiles of bZIPs in leaves and roots of G. uralensis under various abiotic stresses. The expression pattern of GubZIPs under salt and drought stress treatments was similar to that of RNA-seq data. GubZIP11 belongs to the F subfamily. In root tissues, Cd, NaCl, and low drought (PEG) stress can downregulate its expression, while MeJA and UV can upregulate its expression. In leaf tissue, the expression of six kinds of abiotic stress was upregulated. He et al. proved that soybean GmbZIP19 was positively regulated by ABA, jasmonic acid (JA), and salicylic acid (SA) and negatively regulated by salt and drought, showing tolerance to plant pathogens (He et al., 2020). The phylogenetic tree showed that GubZIP11 was highly homologous GmbZIP19, which belongs to the F subfamily. This suggested that GubZIP11 may have a function similar to that of GmbZIP19 in improving the tolerance of pathogens.
Some members of the S subfamily in licorice showed specific expression patterns under salt and drought stress and contained a MeJA response element (TGACG-motif) in the promoter region. Yang et al. reported that the S subfamily GmbZIP2 may enhance drought and salt resistance by regulating reactive oxygen species (ROS) in soybean (G. max) (Yang et al., 2020). For example, GubZIP42 and 55, the homologs of GmbZIP2, were upregulated in leaves and downregulated in roots under salt stress. In addition, both of them had TGACG-motif, which suggested that they could improve their tolerance to salt stress via the process of hormone regulation. It suggested that GubZIPs of the S subfamily may be responsible for the resistance to salt and drought stress.
Li et al. reported that CabZIP25 (member of subfamily A in Capsicum annuum) not only can maintain the stability of chlorophyll in pepper (C. annuum) to enhance salt tolerance but also can increase the germination rate, fresh weight, and root length of overexpressed A. thaliana (Gai et al., 2020). Gai et al. have also proved that NtbZIP62, which belongs to subfamily A in tobacco (N. tabacum), can be induced by salt and ABA to enhance its salt tolerance (Li et al., 2021, 62). GubZIP52 and 62, which belong to the same A subfamily as CabZIP25 and NtbZIP62, were also highly expressed under low salt concentration and ABA treatment, so their high expression may have a positive effect on the salt tolerance of licorice. In the present study, almost all members of the A subfamily in G. uralensis were homologous to ABI5 and AREB3 in A. thaliana (Figure 5B), involved in the ABA signaling process. For example, GubZIP56 and 62 were upregulated in root under the treatment of ABA, MeJA, NaCl, and PEG, whereas they were downregulated in leaf under the treatment of ABA and MeJA and then were upregulated at a later time. All the results suggested that members of subfamily A in licorice may also play wide roles in ABA response.
The cis-acting elements analysis also showed that both GubZIP16 and 25 (belonged to the G subfamily) had light-regulated elements. The expressions of GubZIP16 and 25 were upregulated and downregulated respectively under UV treatment, while they showed a downregulated pattern under NaCl treatment in roots. It has been reported that GBF1, the homolog in A. thaliana, was a negative regulator of blue light-dependent hypocotyl expansion (Gangappa et al., 2013) and can trigger ROS accumulation (Giri et al., 2017, 1). ChbZIP1 that belongs to the G subfamily may enhance antioxidation by regulating genes related to oxidant detoxification in Alkaliphilic Microalgae Chlorella to adapt to abiotic stress (Qu et al., 2021). These results suggested that the G subfamily of GubZIPs may respond to pathogen invasion and environmental stress factors by regulating the accumulation of ROS.
The bZIP gene family plays an important role in plant growth, development, and response to biotic and abiotic stress. We undertook a comprehensive genome-wide characterization and expression analysis of bZIP gene family in licorice under different abiotic stresses. A total of 66 GubZIP genes were identified and classified into 13 subfamilies. Proteins within the same subfamilies contained very similar gene structures and protein motifs. We detected a large number of tandem duplication events, which suggested that tandem duplication events were the main driving force for the evolution of bZIP gene family in licorice. The expression patterns of the GubZIP family were verified by heat map and qRT-PCR. It was showed that certain genes were significantly upregulated or downregulated under abiotic stresses. Gene expression patterns can provide important clues for gene function. It was found that GubZIP11, an ortholog of GmbZIP19, showed specific response to MeJA and UV treatments in root tissue, suggesting that it might be a candidate gene to improve the tolerance to pathogens in licorice. And our findings also indicated that several genes (such as GubZIP56, GubZIP62, GubZIP64, and GubZIP42) played key roles in abiotic stress tolerance. The comprehensive understandings of GubZIP gene family provide useful information for further functional studies to elucidate their regulation mechanism and lay the foundation for cultivating high-quality cultivars of G. uralensis through molecular breeding methods in the future.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
YH conceived, designed, and performed the experiments. ZH validated the data. YH and QH performed the formal analysis. YH and QH participated in original draft preparation. XZ and KY obtained the funding. RH supervised the research. ZL administered this project. All authors have read and approved the manuscript.
This study was supported by the Key Research and Development Program of Shaanxi Province (2017TSCL-SF-11-2, 2020ZDLSF05-02, and 2019TSLSF02-02).
Authors XZ and KY were employed by the company Tasly Holding Group Co., Ltd.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
We thank the colleagues in the plant group of Zhejiang Sci-Tech University, whose comments and suggestions helped us to enhance the quality of this paper.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2021.754237/full#supplementary-material
Alonso, R., Oñate-Sánchez, L., Weltmeier, F., Ehlert, A., Diaz, I., Dietrich, K., et al. (2009). A Pivotal Role of the Basic Leucine Zipper Transcription Factor bZIP53 in the Regulation of Arabidopsis Seed Maturation Gene Expression Based on Heterodimerization and Protein Complex Formation. Plant Cell 21, 1747–1761. doi:10.1105/tpc.108.062968
Anderson, R. P., and Roth, J. R. (1977). Tandem Genetic Duplications in Phage and Bacteria. Annu. Rev. Microbiol. 31, 473–505. doi:10.1146/annurev.mi.31.100177.002353
Audemard, E., Schiex, T., and Faraut, T. (2012). Detecting Long Tandem Duplications in Genomic Sequences. BMC Bioinf. 13, 83. doi:10.1186/1471-2105-13-83
Brinckmann, J. A. (2020). The Long Road to Sustainable Licorice. United Plant Savers. Available at: https://unitedplantsavers.org/the-long-road-to-sustainable-licorice/(Accessed July 1, 2021).
Cai, W., Yang, Y., Wang, W., Guo, G., Liu, W., and Bi, C. (2018). Overexpression of a Wheat (Triticum aestivum L.) bZIP Transcription Factor Gene, TabZIP6, Decreased the Freezing Tolerance of Transgenic Arabidopsis Seedlings by Down-Regulating the Expression of CBFs. Plant Physiol. Biochem. 124, 100–111. doi:10.1016/j.plaphy.2018.01.008
Cannon, S. B., Mitra, A., Baumgarten, A., Young, N. D., and May, G. (2004). The Roles of Segmental and Tandem Gene Duplication in the Evolution of Large Gene Families in Arabidopsis thaliana. BMC Plant Biol. 4, 10. doi:10.1186/1471-2229-4-10
Chen, C., Chen, H., Zhang, Y., Thomas, H. R., Frank, M. H., He, Y., et al. (2020). TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 13, 1194–1202. doi:10.1016/j.molp.2020.06.009
Chidambaram, A., Sundaramoorthy, P., Murugan, A., Ganesh, K., and Baskaran, L. (2009). Chromium Induced Cytotoxicity in Blackgram (Vigna Mungo L.). J. Environ. Health Sci. Eng. 6, 17–22.
Chow, C.-N., Lee, T.-Y., Hung, Y.-C., Li, G.-Z., Tseng, K.-C., Liu, Y.-H., et al. (2019). PlantPAN3.0: a New and Updated Resource for Reconstructing Transcriptional Regulatory Networks from ChIP-Seq Experiments in Plants. Nucleic Acids Res. 47, D1155–D1163. doi:10.1093/nar/gky1081
Cui, Y. X., Xu, Z. C., Chen, X. L., Nie, L. P., Wu, L. W., Wang, Y., et al. (2020). Genome-wide Identification of Abscisic Acid (ABA) Receptor Pyrabactin Resistance 1-like Protein (PYL) Family Members and Expression Analysis of PYL Genes in Response to Different Concentrations of ABA Stress in Glycyrrhiza Uralensis. Chin. J. Nat. Med. 18, 606–611. doi:10.1016/S1875-5364(20)30072-8
Dröge-Laser, W., Snoek, B. L., Snel, B., and Weiste, C. (2018). The Arabidopsis bZIP Transcription Factor Family - an Update. Curr. Opin. Plant Biol. 45, 36–49. doi:10.1016/j.pbi.2018.05.001
E, Z. G., Zhang, Y. P., Zhou, J. H., and Wang, L. (2014). Mini Review Roles of the bZIP Gene Family in rice. Genet. Mol. Res. 13, 3025–3036. doi:10.4238/2014.april.16.11
El-Gebali, S., Mistry, J., Bateman, A., Eddy, S. R., Luciani, A., Potter, S. C., et al. (2019). The Pfam Protein Families Database in 2019. Nucleic Acids Res. 47, D427–D432. doi:10.1093/nar/gky995
Fan, C., Chen, Y., and Long, M. (2008). Recurrent Tandem Gene Duplication Gave Rise to Functionally Divergent Genes in Drosophila. Mol. Biol. Evol. 25, 1451–1458. doi:10.1093/molbev/msn089
Gai, W. X., Ma, X., Qiao, Y. M., Shi, B. H., ul Haq, S., Li, Q. H., et al. (2020). Characterization of the bZIP Transcription Factor Family in Pepper (Capsicum Annuum L.): CabZIP25 Positively Modulates the Salt Tolerance. Front. Plant Sci. 11, 139. doi:10.3389/fpls.2020.00139
Gangappa, S. N., Srivastava, A. K., Maurya, J. P., Ram, H., and Chattopadhyay, S. (2013). Z-box Binding Transcription Factors (ZBFs): A New Class of Transcription Factors in Arabidopsis Seedling Development. Mol. Plant 6, 1758–1768. doi:10.1093/mp/sst140
Gao, L., Wang, F., Xie, S-q., Chen, X-f., Shen, H-t., and Li, H-b. (2021). Genome-wide Identification and Expression Analysis of CBL Gene Family in Glycyrrhiza Uralensis. Biotechnol. Bull. 37, 18–27. doi:10.13560/j.cnki.biotech.bull.1985.2020-1128
Garg, A., Kirchler, T., Fillinger, S., Wanke, F., Stadelhofer, B., Stahl, M., et al. (2019). Targeted Manipulation of bZIP53 DNA-Binding Properties Influences Arabidopsis Metabolism and Growth. J. Exp. Bot. 70, 5659–5671. doi:10.1093/jxb/erz309
Gasteiger, E., Hoogland, C., Gattiker, A., Duvaud, S. e., Wilkins, M. R., Appel, R. D., et al. (2005). “Protein Identification and Analysis Tools on the ExPASy Server,” in The Proteomics Protocols Handbook Springer Protocols Handbooks. Editor J. M. Walker (Totowa, NJ: Humana Press), 571–607. doi:10.1385/1-59259-890-0:571
Giri, M. K., Singh, N., Banday, Z. Z., Singh, V., Ram, H., Singh, D., et al. (2017). GBF1 Differentially Regulates CAT2 and PAD4 Transcription to Promote Pathogen Defense in Arabidopsis thaliana. Plant J. 91, 802–815. doi:10.1111/tpj.13608
Goyal, P., Manzoor, M. M., Vishwakarma, R. A., Sharma, D., Dhar, M. K., and Gupta, S. (2020). A Comprehensive Transcriptome-wide Identification and Screening of WRKY Gene Family Engaged in Abiotic Stress in Glycyrrhiza Glabra. Sci. Rep. 10, 373. doi:10.1038/s41598-019-57232-x
Hanson, J., Hanssen, M., Wiese, A., Hendriks, M. M., and Smeekens, S. (2008). The Sucrose Regulated Transcription Factor bZIP11 Affects Amino Acid Metabolism by Regulating the Expression of ASPARAGINE SYNTHETASE1 and PROLINE DEHYDROGENASE2. Plant J. 53, 935–949. doi:10.1111/j.1365-313X.2007.03385.x
Hazkani-Covo, E., and Graur, D. (2007). A Comparative Analysis of Numt Evolution in Human and Chimpanzee. Mol. Biol. Evol. 24, 13–18. doi:10.1093/molbev/msl149
He, Q., Cai, H., Bai, M., Zhang, M., Chen, F., Huang, Y., et al. (2020). A Soybean bZIP Transcription Factor GmbZIP19 Confers Multiple Biotic and Abiotic Stress Responses in Plant. Ijms 21, 4701. doi:10.3390/ijms21134701
Herath, V., and Verchot, J. (2021). Insight into the bZIP Gene Family in Solanum tuberosum: Genome and Transcriptome Analysis to Understand the Roles of Gene Diversification in Spatiotemporal Gene Expression and Function. Int. J. Mol. Sci. 22, 253. doi:10.3390/ijms22062837
Hernandez-Garcia, C. M., and Finer, J. J. (2014). Identification and Validation of Promoters and Cis-Acting Regulatory Elements. Plant Sci. 217-218, 109–119. doi:10.1016/j.plantsci.2013.12.007
Hu, B., Jin, J., Guo, A.-Y., Zhang, H., Luo, J., and Gao, G. (2015). GSDS 2.0: an Upgraded Gene Feature Visualization Server. Bioinformatics 31, 1296–1297. doi:10.1093/bioinformatics/btu817
Huang, Z., Duan, W., Song, X., Tang, J., Wu, P., Zhang, B., et al. (2016). Retention, Molecular Evolution, and Expression Divergence of the Auxin/Indole Acetic Acid and Auxin Response Factor Gene Families inBrassica RapaShed Light on Their Evolution Patterns in Plants. Genome Biol. Evol. 8, 302–316. doi:10.1093/gbe/evv259
Ishimi, Y., Takebayashi, J., Tousen, Y., Yamauchi, J., Fuchino, H., Kawano, T., et al. (2019). Quality Evaluation of Health Foods Containing Licorice in the Japanese Market. Toxicol. Rep. 6, 904–913. doi:10.1016/j.toxrep.2019.08.013
Kang, C., Zhai, H., He, S., Zhao, N., and Liu, Q. (2019). A Novel Sweetpotato bZIP Transcription Factor Gene, IbbZIP1, Is Involved in Salt and Drought Tolerance in Transgenic Arabidopsis. Plant Cel Rep 38, 1373–1382. doi:10.1007/s00299-019-02441-x
Kumar, S., Stecher, G., Li, M., Knyaz, C., and Tamura, K. (2018). MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 35, 1547–1549. doi:10.1093/molbev/msy096
Lee, S.-j., Kang, J.-y., Park, H.-J., Kim, M. D., Bae, M. S., Choi, H.-i., et al. (2010). DREB2C Interacts with ABF2, a bZIP Protein Regulating Abscisic Acid-Responsive Gene Expression, and its Overexpression Affects Abscisic Acid Sensitivity. Plant Physiol. 153, 716–727. doi:10.1104/pp.110.154617
Li, W. D., Hou, J. L., Wang, W. Q., Tang, X. M., Liu, C. L., and Xing, D. (2011). Effect of Water Deficit on Biomass Production and Accumulation of Secondary Metabolites in Roots of Glycyrrhiza Uralensis. Russ. J. Plant Physiol. 58, 538–542. doi:10.1134/s1021443711030101
Li, Y., Chen, C., Xie, Z., Xu, J., Wu, B., and Wang, W. (2020). Integrated Analysis of mRNA and microRNA Elucidates the Regulation of Glycyrrhizic Acid Biosynthesis in Glycyrrhiza Uralensis Fisch. Ijms 21, 3101. doi:10.3390/ijms21093101
Li, Z., Chao, J., Li, X., Li, G., Song, D., Guo, Y., et al. (2021). Systematic Analysis of the bZIP Family in Tobacco and Functional Characterization of NtbZIP62 Involvement in Salt Stress. Agronomy 11, 148. doi:10.3390/agronomy11010148
Liu, X., and Chu, Z. (2015). Genome-Wide Evolutionary Characterization and Analysis of bZIP Transcription Factors and Their Expression Profiles in Response to Multiple Abiotic Stresses in Brachypodium Distachyon. BMC Genomics 16, 227. doi:10.1186/s12864-015-1457-9
Liu, J., Chen, N., Chen, F., Cai, B., Dal Santo, S., Tornielli, G. B., et al. (2014). Genome-wide Analysis and Expression Profile of the bZIP Transcription Factor Gene Family in Grapevine (Vitis vinifera). BMC Genomics 15, 281. doi:10.1186/1471-2164-15-281
Liu, S.-X., Qin, B., Fang, Q.-x., Zhang, W.-J., Zhang, Z.-Y., Liu, Y.-C., et al. (2021). Genome-wide Identification, Phylogeny and Expression Analysis of the bZIP Gene Family in Alfalfa (Medicago Sativa). Biotechnol. Biotechnol. Equip. 35, 905–916. doi:10.1080/13102818.2021.1938674
Lyu, Y., Shi, P., Han, G., Liu, L., Guo, L., Hu, X., et al. (2020). Desertification Control Practices in China. Sustainability 12, 3258. doi:10.3390/su12083258
Ma, J., Hanssen, M., Lundgren, K., Hernández, L., Delatte, T., Ehlert, A., et al. (2011). The Sucrose‐regulated Arabidopsis Transcription Factor bZIP11 Reprograms Metabolism and Regulates Trehalose Metabolism. New Phytol. 191, 733–745. doi:10.1111/j.1469-8137.2011.03735.x
Ma, H., Liu, C., Li, Z., Ran, Q., Xie, G., Wang, B., et al. (2018). ZmbZIP4 Contributes to Stress Resistance in Maize by Regulating ABA Synthesis and Root Development. Plant Physiol. 178, 753–770. doi:10.1104/pp.18.00436
Mair, A., Pedrotti, L., Wurzinger, B., Anrather, D., Simeunovic, A., Weiste, C., et al. (2015). SnRK1-Triggered Switch of bZIP63 Dimerization Mediates the Low-Energy Response in Plants. eLife 4, e05828. doi:10.7554/eLife.05828
Mochida, K., Sakurai, T., Seki, H., Yoshida, T., Takahagi, K., Sawai, S., et al. (2017). Draft Genome Assembly and Annotation of Glycyrrhiza Uralensis , a Medicinal Legume. Plant J. 89, 181–194. doi:10.1111/tpj.13385
Nguyen, L.-T., Schmidt, H. A., von Haeseler, A., and Minh, B. Q. (2015). IQ-TREE: A Fast and Effective Stochastic Algorithm for Estimating Maximum-Likelihood Phylogenies. Mol. Biol. Evol. 32, 268–274. doi:10.1093/molbev/msu300
Panchy, N., Lehti-Shiu, M., and Shiu, S.-H. (2016). Evolution of Gene Duplication in Plants. Plant Physiol. 171, 2294–2316. doi:10.1104/pp.16.00523
Pandey, A. S., Sharma, E., Jain, N., Singh, B., Burman, N., and Khurana, J. P. (2018). A rice bZIP Transcription Factor, OsbZIP16, Regulates Abiotic Stress Tolerance when Over-expressed in Arabidopsis. J. Plant Biochem. Biotechnol. 27, 393–400. doi:10.1007/s13562-018-0448-8
Prakash, A., Jeffryes, M., Bateman, A., and Finn, R. D. (2017). The HMMER Web Server for Protein Sequence Similarity Search. Curr. Protoc. Bioinf. 60, 3.23.1–3.15.23. doi:10.1002/cpbi.40
Qiu, P. (2003). Recent Advances in Computational Promoter Analysis in Understanding the Transcriptional Regulatory Network. Biochem. Biophys. Res. Commun. 309, 495–501. doi:10.1016/j.bbrc.2003.08.052
Qu, D., Show, P.-L., and Miao, X. (2021). Transcription Factor ChbZIP1 from Alkaliphilic Microalgae Chlorella Sp. BLD Enhancing Alkaline Tolerance in Transgenic Arabidopsis thaliana. Ijms 22, 2387. doi:10.3390/ijms22052387
Shen, H., Cao, K., and Wang, X. (2008). AtbZIP16 and AtbZIP68, Two New Members of GBFs, Can Interact with Other G Group bZIPs in Arabidopsis thaliana. BMB Rep. 41, 132–138. doi:10.5483/bmbrep.2008.41.2.132
Song, H., Sun, J., and Yang, G. (2019). Old and Young Duplicate Genes Reveal Different Responses to Environmental Changes in Arachis Duranensis. Mol. Genet. Genomics 294, 1199–1209. doi:10.1007/s00438-019-01574-8
Stajich, J. E., Block, D., Boulez, K., Brenner, S. E., Chervitz, S. A., Dagdigian, C., et al. (2002). The Bioperl Toolkit: Perl Modules for the Life Sciences. Genome Res. 12, 1611–1618. doi:10.1101/gr.361602
Strathmann, A., Kuhlmann, M., Heinekamp, T., and Dröge-Laser, W. (2001). BZI-1 Specifically Heterodimerises with the Tobacco bZIP Transcription Factors BZI-2, BZI-3/TBZF and BZI-4, and Is Functionally Involved in Flower Development. Plant J. 28, 397–408. doi:10.1046/j.1365-313x.2001.01164.x
Subramanian, B., Gao, S., Lercher, M. J., Hu, S., and Chen, W.-H. (2019). Evolview V3: a Webserver for Visualization, Annotation, and Management of Phylogenetic Trees. Nucleic Acids Res. 47, W270–W275. doi:10.1093/nar/gkz357
Terzaghi, W. B., Bertekap, R. L., and Cashmore, A. R. (1997). Intracellular Localization of GBF Proteins and Blue Light-Induced Import of GBF2 Fusion Proteins into the Nucleus of Cultured Arabidopsis and Soybean Cells. Plant J. 11, 967–982. doi:10.1046/j.1365-313x.1997.11050967.x
Tong, X., Cao, A., Wang, F., Chen, X., Xie, S., Shen, H., et al. (2019). Calcium-Dependent Protein Kinase Genes in Glycyrrhiza Uralensis Appear to Be Involved in Promoting the Biosynthesis of Glycyrrhizic Acid and Flavonoids under Salt Stress. Molecules 24, 1837. doi:10.3390/molecules24091837
Van Leene, J., Blomme, J., Kulkarni, S. R., Cannoot, B., De Winne, N., Eeckhout, D., et al. (2016). Functional Characterization of the Arabidopsis Transcription Factor bZIP29 Reveals its Role in Leaf and Root Development. Exbotj 67, 5825–5840. doi:10.1093/jxb/erw347
Vierling, E. (1991). The Roles of Heat Shock Proteins in Plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 42, 579–620. doi:10.1146/annurev.pp.42.060191.003051
Wang, X., Lu, X., Malik, W. A., Chen, X., Wang, J., Wang, D., et al. (2020). Differentially Expressed bZIP Transcription Factors Confer Multi-Tolerances in Gossypium Hirsutum L. Int. J. Biol. Macromol. 146, 569–578. doi:10.1016/j.ijbiomac.2020.01.013
Yang, Y., Yu, T.-F., Ma, J., Chen, J., Zhou, Y.-B., Chen, M., et al. (2020). The Soybean bZIP Transcription Factor Gene GmbZIP2 Confers Drought and Salt Resistances in Transgenic Plants. Ijms 21, 670. doi:10.3390/ijms21020670
Zhang, M., Liu, Y., Shi, H., Guo, M., Chai, M., He, Q., et al. (2018). Evolutionary and Expression Analyses of Soybean Basic Leucine Zipper Transcription Factor Family. BMC Genomics 19, 159. doi:10.1186/s12864-018-4511-6
Zhang, M., Liu, Y., Cai, H., Guo, M., Chai, M., She, Z., et al. (2020). The bZIP Transcription Factor GmbZIP15 Negatively Regulates Salt- and Drought-Stress Responses in Soybean. Ijms 21, 7778. doi:10.3390/ijms21207778
Zhao, P., Ye, M., Wang, R., Wang, D., and Chen, Q. (2020). Systematic Identification and Functional Analysis of Potato (Solanum tuberosum L.) bZIP Transcription Factors and Overexpression of Potato bZIP Transcription Factor StbZIP-65 Enhances Salt Tolerance. Int. J. Biol. Macromol. 161, 155–167. doi:10.1016/j.ijbiomac.2020.06.032
Zhao, K., Chen, S., Yao, W., Cheng, Z., Zhou, B., and Jiang, T. (2021). Genome-wide Analysis and Expression Profile of the bZIP Gene Family in poplar. BMC Plant Biol. 21, 122. doi:10.1186/s12870-021-02879-w
Zhou, S. F., Sun, L., Valdés, A. E., Engström, P., Song, Z. T., Lu, S. J., et al. (2015). Membrane‐associated Transcription Factor Peptidase, Site‐2 Protease, Antagonizes ABA Signaling in Arabidopsis. New Phytol. 208, 188–197. doi:10.1111/nph.13436
Zhu, M., Meng, X., Cai, J., Li, G., Dong, T., and Li, Z. (2018). Basic Leucine Zipper Transcription Factor SlbZIP1 Mediates Salt and Drought Stress Tolerance in Tomato. BMC Plant Biol. 18, 83. doi:10.1186/s12870-018-1299-0
Keywords: bZIP transcription factor, licorice, abiotic stress, interaction network, expression
Citation: Han Y, Hou Z, He Q, Zhang X, Yan K, Han R and Liang Z (2021) Genome-Wide Characterization and Expression Analysis of bZIP Gene Family Under Abiotic Stress in Glycyrrhiza uralensis. Front. Genet. 12:754237. doi: 10.3389/fgene.2021.754237
Received: 06 August 2021; Accepted: 13 September 2021;
Published: 05 October 2021.
Edited by:
Ashish Kumar Srivastava, Bhabha Atomic Research Centre (BARC), IndiaReviewed by:
Xiaojun Nie, Northwest A and F University, ChinaCopyright © 2021 Han, Hou, He, Zhang, Yan, Han and Liang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zongsuo Liang, bGlhbmd6c0B6c3R1LmVkdS5jbg==; Ruilian Han, aGFucmxAbndhZnUuZWR1LmNu
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.