- 1College of Agriculture, Nanjing Agricultural University, Nanjing, China
- 2Henan Institute of Crop Molecular Breeding, Henan Academy of Agricultural Science/Key Laboratory of Oil Crops in Huang-Huai-Hai Plains, Ministry of Agriculture/Henan Provincial Key Laboratory for Oil Crops Improvement, Zhengzhou, China
- 3Department of Horticulture, Ghazi University, Dera Ghazi Khan, Pakistan
APETALA2/ethylene response element-binding factor (AP2/ERF) transcription factors (TFs) have been found to regulate plant growth and development and response to various abiotic stresses. However, detailed information of AP2/ERF genes in peanut against drought has not yet been performed. Herein, 185 AP2/ERF TF members were identified from the cultivated peanut (A. hypogaea cv. Tifrunner) genome, clustered into five subfamilies: AP2 (APETALA2), ERF (ethylene-responsive-element-binding), DREB (dehydration-responsive-element-binding), RAV (related to ABI3/VP), and Soloist (few unclassified factors)). Subsequently, the phylogenetic relationship, intron–exon structure, and chromosomal location of AhAP2/ERF were further characterized. All of these AhAP2/ERF genes were distributed unevenly across the 20 chromosomes, and 14 tandem and 85 segmental duplicated gene pairs were identified which originated from ancient duplication events. Gene evolution analysis showed that A. hypogaea cv. Tifrunner were separated 64.07 and 66.44 Mya from Medicago truncatula L. and Glycine max L., respectively. Promoter analysis discovered many cis-acting elements related to light, hormones, tissues, and stress responsiveness process. The protein interaction network predicted the exitance of functional interaction among families or subgroups. Expression profiles showed that genes from AP2, ERF, and dehydration-responsive-element-binding subfamilies were significantly upregulated under drought stress conditions. Our study laid a foundation and provided a panel of candidate AP2/ERF TFs for further functional validation to uplift breeding programs of drought-resistant peanut cultivars.
Introduction
Transcription factors (TFs) (or trans-acting factors) are the main class of regulatory proteins that can specifically combine with DNA-binding domains and perform a key role by regulating the expression of downstream genes (Singh et al., 2002; Licausi et al., 2013). Nearly 60 different TF families have been found in higher plants, such as AP2/ERF (Gutterson and Reuber, 2004; Xu et al., 2011; Li M.-Y. et al., 2015), ARF (Finet et al., 2010; Rademacher et al., 2011), bHLH (Li et al., 2006), bZIP (Ulm et al., 2004; Liu et al., 2012), C2H2 (Tsutsui et al., 2011), MADS (Trevaskis et al., 2003; Terol et al., 2019), MYB (Dubos et al., 2010; Feller et al., 2011), NAC (Mao et al., 2012; Nakashima et al., 2012), SBP (Kandori et al., 2006; Ferreira et al., 2014), and WRKY (Rushton et al., 2010). Among these TFs, the APETALA2/ethylene-responsive element-binding factor (AP2/ERF) superfamily contains the largest group of TFs in plant, which are reportedly involved in plant growth progress and abiotic stress responsiveness according to relevant reports (Licausi et al., 2013; Feng et al., 2020). The first AP2/ERF TF was found to regulate flower development in Arabidopsis (Jofuku et al., 1994). Subsequently, AP2/ERF genes were widely found in leaf, root, seed, fruit, and other tissues (Chuck et al., 2002; Hirota et al., 2007; El-Sharkawy et al., 2009; Pietsch et al., 2009; Kitomi et al., 2011; Soares et al., 2011). Not only in plants, related AP2/ERF superfamily proteins are also found in ciliates and protists that may be associated with the His- and Asn-rich HNH class of homing endonucleases (Magnani et al., 2004; Wuitschick et al., 2004).
AP2/ERF TFs usually contain one or two AP2-conserved domains (60–70 amino acid residues) which combine with the cis-acting elements in the promoter regions of targeted genes (Okamuro et al., 1997; Allen et al., 1998; Riechmann and Meyerowitz, 1998). The AP2/ERF superfamily genes are mainly divided into AP2 (APETALA2), DREB (dehydration-responsive-element-binding), ERF (ethylene-responsive-element-binding), RAV (related to ABI3/VP), and Soloist (few unclassified factors) subfamilies based on the sequence characteristics and the number of AP2-conserved domains (Nakano et al., 2006; Tamura et al., 2011). In most cases, the AP2 subfamily contains proteins with two AP2 domains involved in regulating plant developmental processes (El et al., 2010). ERF, DREB, and RAV subfamily members contain only one single AP2 domain, while RAV members are often associated with an additional B3 DNA-binding domain (Licausi et al., 2010). Discrepancy of 14th and 19th amino acid sequences is the main differences between ERF and DREB subfamilies; the ERF subfamily consists of alanine (Ala) and aspartate (Asp) whereas the DREB subfamily consists of valine (Val) and glutamic acid (Elu) of 14th and 19th amino acid sequences, respectively (Sakuma et al., 2002). Additionally, other members with special gene structure and AP2-like domain are known as Soloist ones (Li H. et al., 2017).
With more draft genomic information of plants released, AP2/ERF superfamily members have been identified and characterized in eudicots, i.e., Arabidopsis (Sakuma et al., 2002; Nakano et al., 2006), grapevine (Licausi et al., 2010), cucumber (Hu and Liu, 2011), Chinese plum (Du et al., 2013), apple (Girardi et al., 2013), sweet orange (Ito et al., 2014), pineapple (Huang et al., 2020), canola (Ghorbani et al., 2020), Chinese cherry (Zhu et al., 2021), and dark jute (Kabir et al., 2021), and in monocots, i.e., rice (Sharoni et al., 2011), common wheat (Zhuang et al., 2011), sugarcane (Li et al., 2020), maize (Liu et al., 2013), barley (Guo et al., 2016), and foxtail millet (Lata et al., 2014). In general, AP2 TFs have been found to regulate various developmental processes, such as the development of floral organs (Irish and Sussex, 1990; Jofuku et al., 1994; Chuck et al., 1998 and, 2008; Maes et al., 2001; Aukerman and Sakai, 2003) and embryo and seed growth (Boutilier et al., 2002; Jofuku et al., 2005; Krizek and Beth, 2009). ERF and DREB subfamily proteins mainly function in the resistance to diverse biological and environmental stresses, such as biotic stresses (microbial pathogens and herbivorous insects) and abiotic stresses (drought, heat, cold, and salinity) (Feng et al., 2020). Additionally, RAV subfamily proteins play a crucial role against biotic and abiotic stress responses (Sohn et al., 2006; Li et al., 2011; Fu et al., 2014) by responding to the signal of plant hormones (ethylene and brassinosteroid) (Alonso et al., 2003; Hu et al., 2004).
Peanut, an important oil and economic crop worldwide, is used to provide oil and proteins for humans (Zhuang et al., 2019). In particular, with the characteristics of underground fruit, its yield is being devastatingly affected by drought stress (Vahdati and Lotfi, 2013). Additionally, drought will also result in the increase of aflatoxin contamination and the frequency of diseases and pests (Boyer, 1982; Nimitr et al., 2003). To date, drought has been the most serious abiotic stress which negatively affects the quality and distribution of peanut (Reddy et al., 2003; Farombi, 2006; Cardwell and Henry, 2008; Sun et al., 2013; Jayaprakash et al., 2019; Soni et al., 2020). Previous studies have demonstrated the critical role of AP2/ERF genes in mediating drought stress resistance. For example, AhERF3 and AhERF5 in root were upregulated by PEG treatment (Chen et al., 2012), indicating its association with drought stress. Notably, overexpression of AhERF019 could enhance tolerance to drought in transgenic Arabidopsis (Wan et al., 2014). Thus, there may be more important AP2/ERF members that act in enhancing the resistance to drought stress of peanut. In recent years, with the release of genome information with cultivated peanut (A. hypogaea cv. Tifrunner), an allotetraploid (AABB 2n = 4x = 40), and related wild-type ones (diploids: A. duranensis and A. ipaensis) (Bertioli et al., 2016; Zhuang et al., 2019). Several gene families, including monosaccharide transporter MST genes (Wan et al., 2020), GRF (Zhao et al., 2019), bHLH (Chao et al., 2017), and WRKY (Song et al., 2016), have been characterized at a genome scale. However, very limited information of AP2/ERF genes is available in cultivated peanut.
In the present study, 185 AP2/ERF superfamily members of A. hypogaea cv. Tifrunner were investigated by phylogenetic relationship, sequence structures, chromosomal distributions, duplication events, and promoter region analysis. The expression patterns of AhAP2/ERF under drought stress were quantified by using quantitative real-time polymerase chain reaction (qRT-PCR). Our investigation will be beneficial to identify the drought-responsive candidate genes for further functional characterization to breed drought-resistant peanut cultivars.
Materials and Methods
Screening and Identification of the AP2/ERF Superfamily Genes
To accurately collect all members of AhAP2/ERF genes and avoid nonspecific amplification, multiple-database searches were performed. The A. hypogaea cv. Tifrunner genome sequences were downloaded from peanutbase (https://www.peanutbase.org/). The AP2 domain (PF00847) profile was obtained from the pfam database (http://pfam.janelia.org), which was used to match each member of AP2/ERF protein in genomes using HMMER 3.1 software (E-value<1e−5) (Finn et al., 2011). To avoid the omission of AP2/ERF members, we also performed searches in the Transcription Factor database (http://planttfdb.cbi.pku.edu.cn/) (Jin et al., 2014). All protein sequences acquired were then verified for the AP2 domain by using the SMART (Simple Modular Architecture Research Tool: http://smart.embl-heidelberg.de/) (Letunic et al., 2012) and Pfam (http://pfam.xfam.org) databases (Finn et al., 2008) and NCBI (https://www.ncbi.nlm.nih.gov/cdd). Proteins lacking compete AP2 domains were identified by manual examination. Physichochemical profiling of AP2/ERF genes was performed by using online ExPASy (Gasteiger, 2005; Panu et al., 2012). The subcellular localization analysis of curated AP2/ERF superfamily genes was conducted on the Plant-Ploc server (http://www.csbio.sjtu.edu.cn/bioinf/plant/) (Chou and Shen, 2008).
Phylogenetic Analysis of AP2/ERF Proteins
The AP2 domains were extracted based on results of SMART (Simple Modular Architecture Research Tool: http://smart.embl-heidelberg.de/) (Letunic et al., 2012). Multiple-sequence alignment was executed by DNAMAN and CLUSTAL program (Thompson et al., 1994; Burner and Legendre, 2000). To construct phylogenetic trees, MEGA 7.0 software was used with the neighbor-joining model (1,000 replicates) (Tamura et al., 2013). AP2/ERF family gene names in A. hypogaea cv. Tifrunner were given according to the ascending order of location on chromosomes.
Gene Structure and Conserved Motif Analysis
Information of the intron–exon structure was obtained from the reference peanut genome (A. hypogaea cv. Tifrunner, https://www.peanutbase.org/). The Multiple Expectation Maximization for Motif Elicitation program (MEME, http://meme-suite.org/tools/meme) was used to identify potential conserved motifs shared by 185 AhAP2/ERF genes (Bailey et al., 2009). Basic information extraction and preliminary drawing of sequence structure were conducted using TBtools (South China Agricultural University, Guangdong, China) (Chen et al., 2020).
Chromosome Localization, Duplications, and Evolutionary Analysis of AhAP2/ERF
MapChart 2.3 software developed by Wageningen University and Research in Wageningen, Netherlands, is used to locate genes on chromosomes (Voorrips, 2002). Tandem and segmental genes, Ka/Ks values, and circos figures for chromosome locations with AP2/ERF duplication links were completed by TBtools software (South China Agricultural University, Guangdong, China) (Chen et al., 2020). Duplication and divergence time were calculated by the following formula as described by Bertioli et al. (2016):
Promoter Analysis of AhAP2/ERF Genes
Approximately 1,500-bp upstream sequences of the AhAP2/ERF genes were used to get a better knowledge of the potential function of promoter. PlantCARE (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/) (Lescot et al., 2002) was used to identify the cis-regulatory elements exited in the gene promoters related to stress responses and hormone effects, and these results were visualized by TBtools.
Prediction of the Protein Interaction Network
Prediction of the protein interaction network was conducted on the basis of the STRING database (https://string-db.org/. accessed on January 28, 2021) (Szklarczyk et al., 2019). Arabidopsis thaliana L., the well-characterized model plant, was the subject organism (combined score≥ 0.4). PPI networks were constructed by Cytoscape software v 3.8.0 (Shannon et al., 2003).
Plant Materials and Drought Stress Treatment
Seeds of Arachis hypogaea L., ‘YUANZA 9102’, laboratory homozygous material, were sown in 392 cm3 (7 cm long, 7 cm wide, and 8 cm high) pots which were filled with a mixture of vermiculite and perlite (3:1 v/v). Plants were put in a fully controlled growth room (relative humidity: 70%, 16 h/8 h light/dark; 30°C/28°C day/night; light intensity 17,000 lx). Watering was stopped in one part of pots (drought treatment) when seedlings were 5 weeks old, whereas the watering regime remained unchanged in the control plants (every 4 days). Roots were collected from the control and treatment groups every 4 days from seedlings aged 5–9 weeks. All samples were frozen in liquid nitrogen immediately and then stored at -80°C for RNA isolation.
RNA Isolation and Quantitative RT-PCR Assays
RNAprep Pure Plant Plus Kit (Tiangen Biotech, Co., Beijing, China) was utilized to extract RNA from control and treated peanut samples. The cDNA was prepared by following the user manual of the PrimeScrit™ RT Kit with gDNA eraser (perfect real-time, Takara Biomedical Technology, Ltd., Beijing, China). Thirty-five genes from each family were designed by Primer Premier 5.0 and are shown in Supplementary Table S1. The alcohol dehydrogenase class III (AhADH3, Arahy. VYWU26.2) (forward primer: 5′-GACGCTTGGCGAGATCAACA-3′, reverse primer: 5′-AACCGGACAACCACCACATG-3′) was selected as the internal reference control (Brand and Hovav, 2010). Subsequently, quantitative RT-PCR was performed using the ABI QS5 qRT-PCR detection system (ABI, United States) and SYBR Green Kit (Tiangen, Beijing, China). An ABI QS5 real-time PCR system was used under the following procedure: 95°C for 15 min, followed by 40 cycles of 95°C for 10 s, and 60°C for 32 s in a 20 µl volume. Each PCR assay was carried out in three biological replicates, of which each replicate corresponded to three technical repeats. Relative expression levels of the genes were calculated using the 2-△△Ct method (Livak and Schmittgen, 2001).
Results
Characterization and Phylogenetic Analysis of AhAP2/ERF Family Proteins in Cultivated Peanut
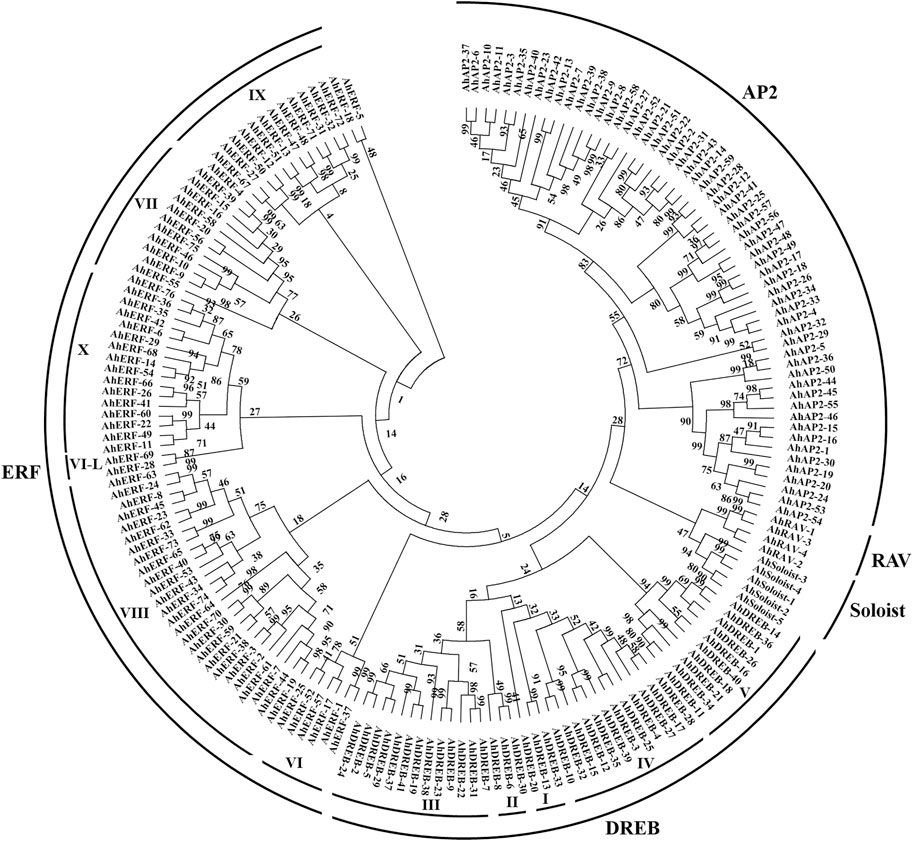
A total of 185 unigenes with the AP2 domain were characterized as AP2/ERF TFs in A. hypogaea cv. Tifrunner (Supplementary Table S2). Depending on the sequence characteristics, the AP2 domains, phylogenetic tree analysis, and the classification system established by the group of Yamaguchi-Shinozaki (Tamura et al., 2011) and Shinshi (Nakano et al., 2006), the AP2/ERF superfamily genes are mainly classified into AP2 (APETALA2), ERF (VI-X, ethylene-responsive-element-binding), DREB (I-V, dehydration-responsive-element-binding), RAV (related to ABI3/VP), and Soloist (few unclassified factors) subfamilies (Figure 1 and Supplementary Figure S1). Among these, the 27 to encode two AP2 domains and the 4 to one AP2 domain together with one B3 domain were thus assigned to the AP2 and RAV families, respectively. Based on the similarity of amino acid sequences with the AP2 domain, the 117 genes were further assigned to the ERF 76) and DREB 41) subfamilies, respectively. Thirty-two members with a single AP2 domain but were distinct from the ERF or DREB subfamily were classified into the AP2 subfamily (Supplementary Table S2). The remaining five genes with independent clades from others were identified as Soloist genes. Subsequently, all superfamily members were named according to the order on the chromosomes of each family member to distinguish from each other for the study (Supplementary Table S3).
To evaluate the phylogenic relationship and classification of the ERF and DREB subfamily, multiple-alignment analyses was performed on the protein sequences with the AP2 domain acquired from peanut (117), Arabidopsis (139), and rice (139), as suggested by Nakano et al. (2006) (Supplementary Figures S2, S3). The NJ phylogenetic tree divided the ERF and DREB subfamilies of peanut and Arabidopsis into 10 subgroups (DREB-I-V and ERF-VI to X) following the classification as described by Nakano et al. (2006) (Supplementary Figure S4). The phylogenetic tree of ERF and DREB subfamily proteins of peanut and rice also exhibited similar results (Supplementary Figure S5). Overall, current findings of the phylogenetic tree demonstrated that classification of the peanut ERF and DREB subfamily proteins is similar to the Arabidopsis and rice ERF family (Supplementary Table S4).
Molecular property analysis showed that MW of AP2/ERF superfamily members varied from 14.46 to 82.54 kDa. Most of AP2/ERF superfamily genes showed their localization in the nucleus. Moreover, the negative GRAVITY values suggested the globular hydrophilic nature of the AP2/ERF proteins. Interestingly, the members of the same families or clades shared similar physical properties, indicating the functions conservatively in the same clades and differentially among subfamilies.
Structure Analysis of AhAP2/ERF
Structural analysis of AP2/ERF genes is helpful for us to fully understand the conservative characteristics of peanut AP2/ERF protein and analyze its evolutionary differences. Numbers of introns varied among AP2/ERF subfamilies (Figure 2). All the AP2 family genes contain 3–10 introns, whereas most members of ERF, RAV, and Soloist subfamilies have only 1–2 introns or do not possess introns. A similar phenomenon was discovered in Arabidopsis thaliana L. and Cucumis sativus L., where most Arabidopsis genes of the ERF family do not possess introns (Sakuma et al., 2002), and 83% of CsERF genes do not have intron (Ito et al., 2014). What is more, members of the ERF family clustered into one branch have a similar gene structure (Figure 2). There was some subgroup specificity: members of groups DREB-III, DREB-II, DREB-I, and ERF-VI did not possess introns, and the genes owning two introns were in the groups ERF-VII and ERF-X, possibly attributed to number changes of introns during evolution, whereas the number and position of the introns were relatively conserved in the same group of plant species.
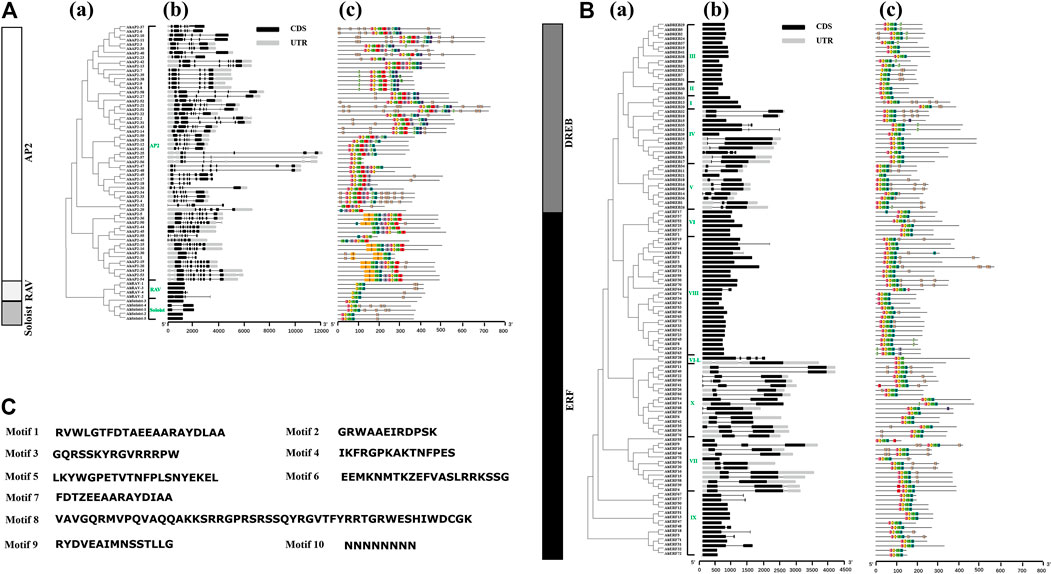
FIGURE 2. Intron–exon structures and conserved motif analysis of AhAP2/ERF genes according to the phylogenetic relationship. (A) Related information of AP2, RAV, and Soloist subfamily. (B) Related information of ERF and DREB subfamily. (C) The amino acid composition of each motif. (a) The phylogenetic tree. (b) The exon–intron structure of AP2/ERF genes. (c) The distribution of conserved motifs in AP2/ERF proteins. Each conserved motif is represented by various-colored rectangles. Box length corresponds to motif length. Color blocks of different colors represent different family and group members.
To provide evidence for the classification and the functional conversation of AP2/ERF superfamily genes among different groups, 10 conserved motifs (motifs 1–10) were analyzed by using MEME software (Figure 2). Among these, motifs 1 and 2 were dominantly present in the AP2 domain regions of all family members. The proteins of the same group showed identical numbers and arrangements of motifs, which are different among the various clades. For example, nine motifs were detected in AP2 and ERF families, and four motifs in RAV and Soloist families. Furthermore, the number and arrangement of motifs in the RAV (motifs 2, 1, 4, 10), ERF, and Soloist (motifs 3, 2, 1, 4) families showed high similarity. Motif 8 was only detected in few AP2 family members, signifying the meticulousness of the motif in the AP2 family. Remarkably, motifs in the same group showed great similarity, indicating the functional conservation in different groups. Comparing the intron–exon structure and conserved motif analysis, it is clear that the members of the same group showed great similarity of characteristics, indicating that most of AhAP2/ERF genes were highly conserved among groups.
Genome Distribution of AP2/ERF Genes
AP2/ERF genes showed random distribution on the 20 chromosomes of the peanut genome (Figure 3 and Supplementary Table S3). Maximum numbers of AhAP2/ERF genes (16 genes) were located on Chromosomes 6 and 15, while Chr17 had the least number of genes (4 genes). Other chromosomes had a random number of allocated AhAP2/ERF genes (5–13 genes). Interestingly, most members are distributed at both ends of the chromosome. A similar study on Arabidopsis and other species showed consistent findings (Nakano et al., 2006; Guo et al., 2016). This location similarity of genes on chromosomes indicates functional consistency.
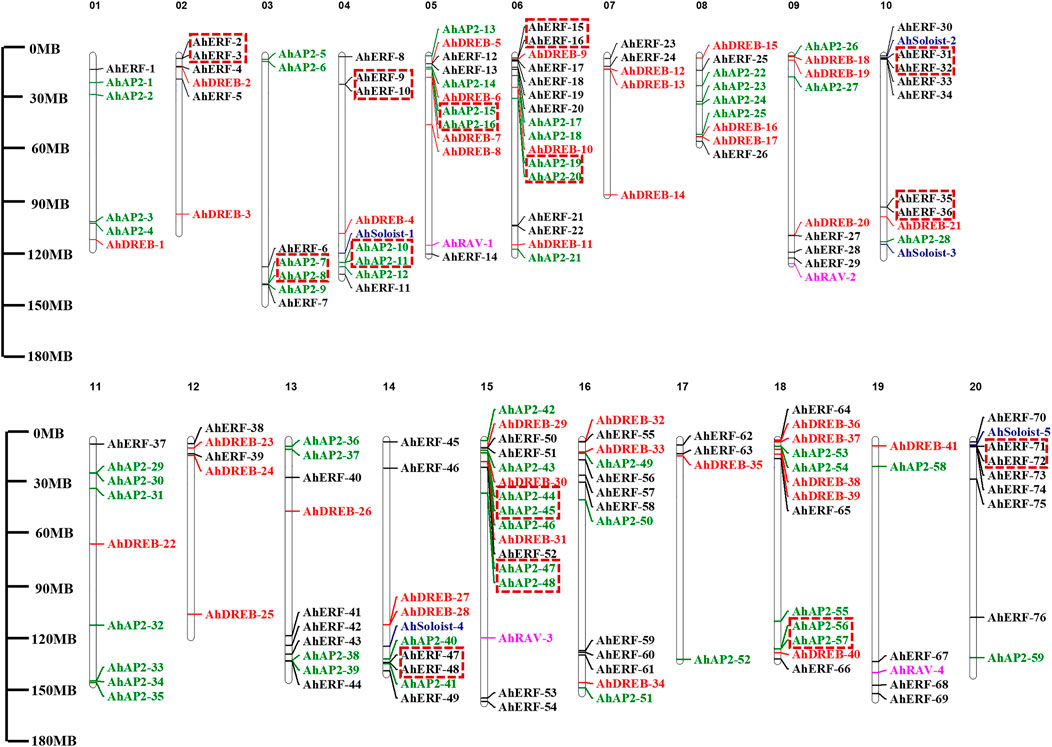
FIGURE 3. Chromosome mapping and duplication of AhAP2/ERF genes. On the right of the chromosome is the gene name. Scale represents a 30-Mb chromosomal distance. Genes in tandem repeats are shown in the red box.
Duplication Events of AP2/ERF Genes and Synteny Analysis
Gene duplication events (segmental or tandem) play a significant role in the expansion and evolution of gene families in plant species (Baloglu, 2014). In total, 99 duplicated gene pairs, which were also named as homoeologous genes, were identified: 14 tandem and 85 segmental duplications (Figure 4). Segmental gene duplication mainly occurred in the A. hypogaea cv. Tifrunner genome rather than tandem duplication event (Figure 4 and Supplementary Table S5). No duplication events occurred in group I of the ERF family, whereas more segmental duplication events occurred in other groups of the ERF family, implying that the biggest members of the families might arise from a higher frequency of segmental duplication, when adapting to various environmental shifts. In contrast, tandem duplication has a confined benefaction to the gene family expansion as compared to the segmental duplication. Similar studies on Arabidopsis, rice, sorghum (Wang et al., 2011), common bean (Kavas et al., 2015; 2016), and cucumber (Baloglu, 2014) showed compatible findings. In general, segmental duplication might be the main driving force for the AP2/ERF gene family expansion in peanut genome.
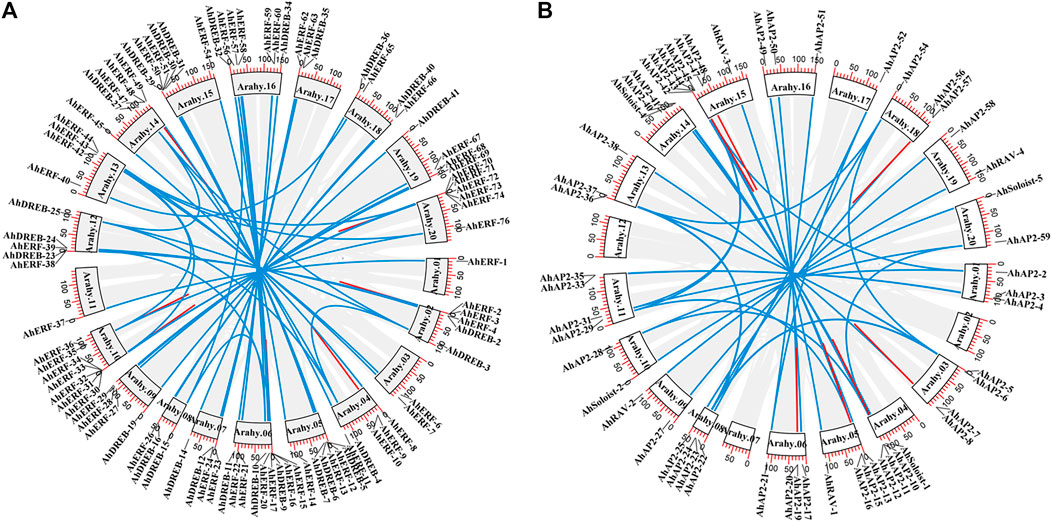
FIGURE 4. Circos figures for chromosome locations with AP2/ERF duplication links. (A) ERF and DREB subfamily duplication links. (B) AP2, RAV, and Soloist subfamily duplication links. Blue and red lines indicated segmented and tandem duplicated gene pairs, respectively.
Moreover, comparative orthologous analysis was conducted among A. hypogaea cv. Tifrunner, A. duranensis, A. ipaensis, Medicago truncatula L., and Glycine max L. to characterize the evolutionary patterns of AhAP2/ERF genes with Leguminosae species (Figure 5). In total, 140, 133, 314, and 145 orthologous gene pairs were found with A. duranensis, A. ipaensis, Medicago truncatula L., and Glycine max L., respectively (Supplementary Tables S6–S9). The Ka/Ks for segmental duplication was 0.02–0.92 with an average of 0.28, while the ratio of tandem duplication ranged from 0.29 to 0.68 with an average of 0.42 (Supplementary Table S5). These segmental and tandem duplications may occur in ∼3.82–43.68 Mya, respectively. In addition, the Ka/Ks ratio of ortholog gene pairs between A. hypogaea cv. Tifrunner and Medicago truncatula L. (0.25) and A. hypogaea cv. Tifrunner and Glycine max L. (0.24) were strongly subjected to pure selection (Lynch and Conery, 2000). The divergence times were 64.07 and 66.44 Mya for Medicago truncatula L. and Glycine max L., respectively.
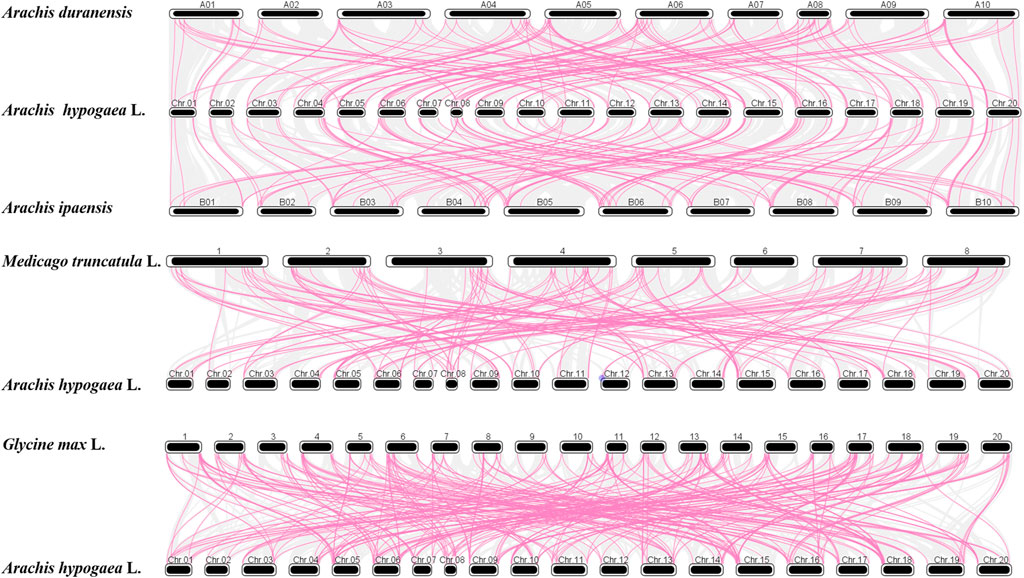
FIGURE 5. Synteny of AP2/ERF genes in the different genomes of A. duranensis, A. ipaensis, Medicago truncatula L., Glycine max L., and A. hypogaea cv. Tifrunner.
Prediction Analysis of cis-Acting Elements With AhAP2/ERF
Specific cis-element motifs can be recognized by TFs and participate in gene expression regulation. In order to further study the potential regulatory mechanism of the AP2/ERF gene in diversified biological processes, especially in plant drought stress response, the 1.5 kb upstream sequence of the AP2/ERF gene translation start site was submitted to the PLANTCARE database to detect cis elements (Cui et al., 2018; Meng et al., 2020). A total of 56 known cis-elements (30 light-related elements, 11 hormone-related elements, 8 tissue-specific elements, and 7 stress-related elements) were detected (Figure 6 and Supplementary Table S10). ABRE, AuxRR core, CGTCA motifs, GARE motifs, O2 site, P-box, TATC-box, TCA element, TGA element, and TGACG motifs involved in hormonal responses are found in 63.8, 5.9, 49.2, 9.2%,18.9, 18.4, 10.8, 32.4, 25.9, and 49.2% of AhAP2/ERF promoters, respectively. Meanwhile, there are a large number of stress-related elements, including MBS (drought inducibility), TC-rich repeats (defense and stress responsiveness), WUN motif (wound responsiveness), LTR (low-temperature responsiveness), ARE (essential for the anaerobic induction), and GC motif (anoxic specific inducibility) (Supplementary Table S10). Moreover, there was a divergence in the percentage of cis-acting elements in promoter regions of various families (Figure 6D). For example, all the RAV family members contained ARE and O2-site elements, whereas 74.6, 80.0, and 60.7% family members of AP2, Soloist, and ERF families possessed ARE elements, and 10.2, 20, and 20.5% of those families exhibited O2-site elements in the promoter region, respectively. Notably, MBS, an important cis-element related to the plant drought-inducibility process, was detected in the promoters of 25.4% of AP2, 50% of RAV, 60% of Soloist, and 22.2% of ERF family members. As a major hormone in plant response to drought stress, ABRE possessed 57.6% of AP2, 75% of RAV, 40% of Soloist, and 67.5% of ERF family members. TC-rich repeats, a cis-acting element involved in defense and stress responsiveness, were discovered only in the promoters of AP2 and ERF family members. WUN motif, a wound-responsive element, was only detected in the ERF family. The variants in the characterization of cis-acting elements implied the functional discrepancy in different families.
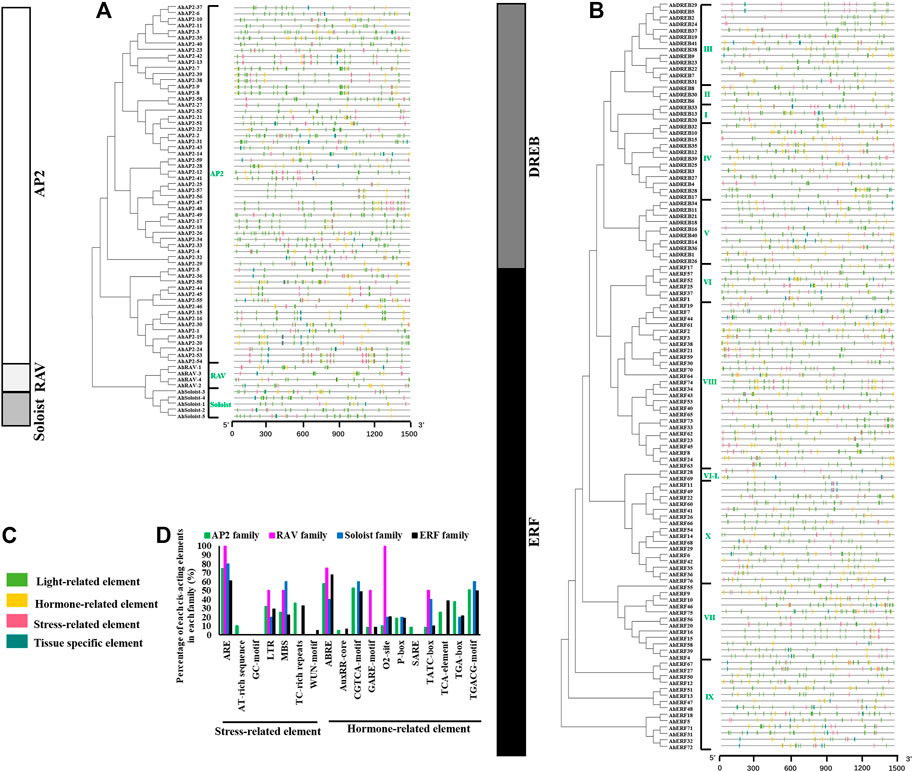
FIGURE 6. Identification of the cis-acting element in the 1.5-kb promoter region of AhAP2/ERF genes. (A) cis-Acting elements of AhAP2, AhRAV, and AhSoloist family genes. (B) cis-Acting elements of AhERF and AhDREB subfamily genes. (C) The classification and annotation of cis-acting elements. Each type of element is represented by a number of colored rectangles. Box length corresponds to element length. Color blocks of different colors represent different family and group members. (D) Percentage of each cis-acting element in promoter of the AhAP2/ERF superfamily.
What is more, certain ERF members in groups III (AhDREB5/29/41), IV (AhDREB15/27/39), V (AhDREB21/26), VI (AhERF25/52), VII (AhERF56), VIII (AhERF3/19/30/34/43/53/62/64/65/70/74), and IX (AhERF5/31/71/72) possess a relatively large number of MBS elements in the promoter regions, implying the members’ main role in the regulation of plant drought responsiveness and the function variations among groups (d). In other words, cis-acting elements in the same group showed great similarity, indicating the functional conservation in the same groups or clades (Figure 6D).
Interaction Network Analysis of AhAP2/ERF Proteins
To understand the synergy among peanut AP2/ERF TFs during their regulatory process, an interaction network was drawn using Arabidopsis ortholog genes (Figure 7). A sum of 31 gene pairs with a combined score over zero value was deliberated to have the interaction with others (Supplementary Table S11). AhAP2-29, AhAP2-37, AhERF-47, and AhRAV-1 had more than three nodes and protein pairs and were involved in more powerful crossing networks, suggesting their core role in peanut. However, other members were only regulated by a few numbers of genes, indicating its less important role in transcriptional level regulation (Figure 7). Interestingly, proteins from different families showed complex interaction. For example, core members AP2-39 or RAV-3 interact with genes from DREB and ERF subfamilies, implying the exitance of functional interaction among subfamilies. These interrelationships will provide a reference for studying the regulatory functions of AhAP2/ERF genes in peanut.
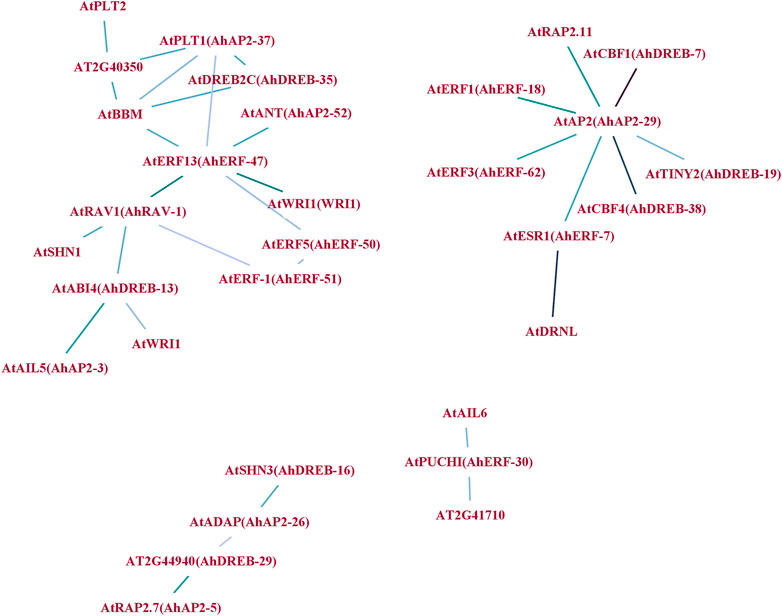
FIGURE 7. Protein–protein interaction (PPI) network analysis of AhAP2/ERF TF proteins. Specific protein interactions between AP2/ERF transcription factors in peanut were determined using String (Szklarczyk et al., 2019). Strong associations are represented by thicker lines.
Expression Profiles of AhAP2/ERF Genes Under Drought Stress Using qRT-PCR
For better knowing the possible regulatory roles of AP2/ERF family genes in peanut response to drought stress, 35 representative genes from AP2 (5), RAV (3), and ERF (27, members from each group) were selected to verify whether their expression levels would be induced under drought stress conditions by qRT-PCR (Figure 8), especially for the ones that possess MBS elements (drought-inducibility), ABRE, TC-rich elements, ARE, and WUN-motif elements in the promoter regions.
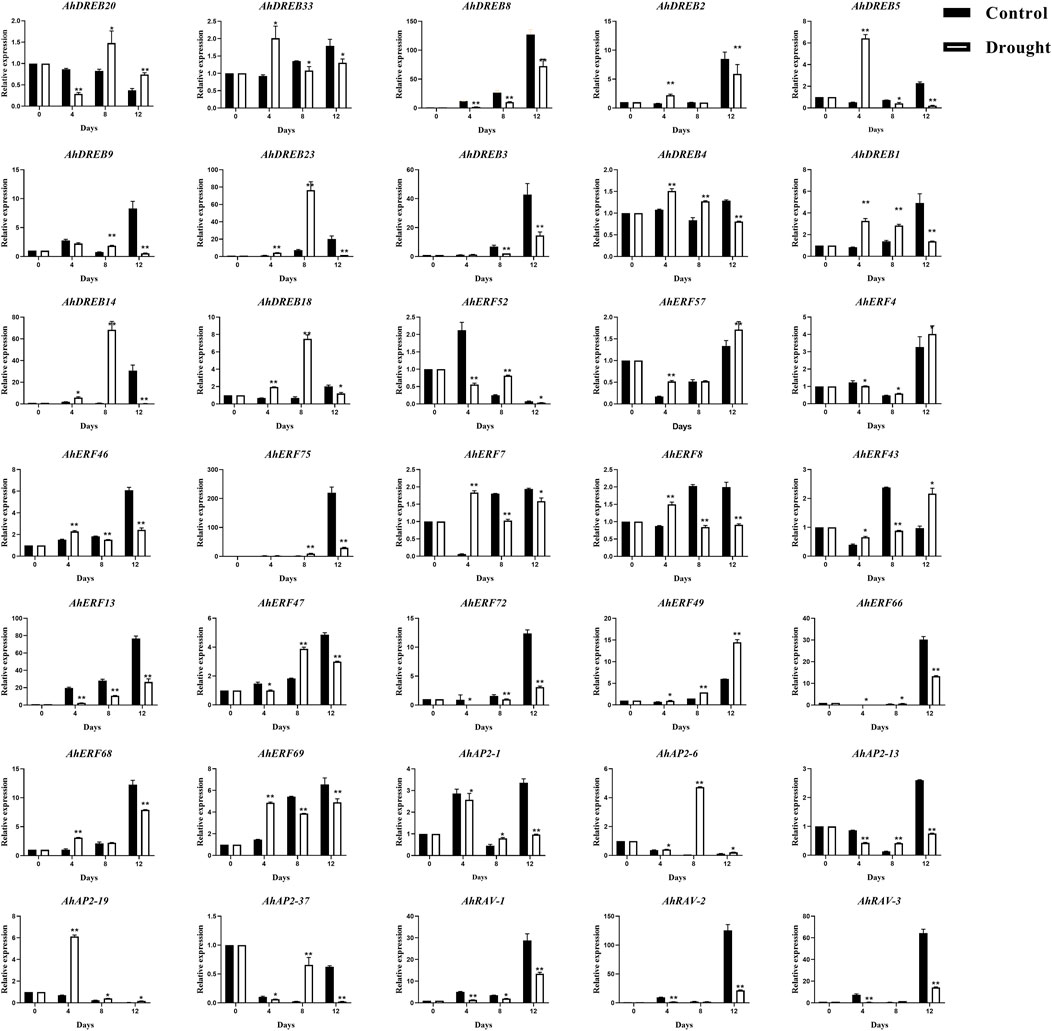
FIGURE 8. qRT-PCR expression analysis of 35 selected AhAP2/ERF genes under drought stress conditions. The expression levels of the untreated (0 h) group were normalized to 1 as a control. Error bars were obtained from three biological replicates. Values are means ± standard errors (SEs) of three independent biological replicates (n = 3). Asterisks indicate a significant difference between the control group and treatment group at each time point as determined by Student’s t-test (*p < 0.05; **p < 0.01).
The treatment group with no watering was used to stimulate the expression of plant defense genes. The expression analysis of AhAP2/ERF genes responsive to drought stress could present useful information to identify their implied role as candidate genes to mitigate drought stress severity. As shown in Figure 8, under the no watering condition, except AhDREB-3, AhDREB8, AhERF-4, AhERF-13, AhERF-66, AhERF-72, AhERF-75, AhAP2-1, AhAP2-6, AhAP2-13, AhRAV-1, AhRAV-2, and AhRAV-3 exhibited downregulation, whereas the remaining genes were significantly upregulated and subsequently downregulated, at all time points compared with those at 0 h. Furthermore, the peak expression of AhDREB-1, AhDREB-4, AhDREB-5, AhDREB-33, AhERF-7, AhERF-8, AhERF-69, and AhAP2-19 was discovered on the 4th day, but that of AhDREB-14, AhDREB-18, AhDREB-20, AhDREB-23, AhERF-47, AhAP2-6, and AhAP2-37 was detected at the 8th day. Interestingly, the members in the same group showed semblable expression trends, which indicates the function consistency. Notably, collinear genes AhDREB-9 and AhDREB-23, AhAP-26 and AhAP2-37, AhRAV-1, and AhRAV-3 showed highly similar expression patterns under drought stress treatment, indicating that their biological functions also have a certain similarity.
Discussion
In plants, AP2/ERF TFs play diverse roles in multiple growth processes and work against environmental factors through transcriptional regulation (Sakuma et al., 2002; Nakano et al., 2006; Woo et al., 2010; Li A. et al., 2017; Yao et al., 2017). Therefore, studying the biological functions and molecular mechanisms of these proteins will facilitate obtaining a deeper understanding of the pathways adapting to environmental pressures during plant growth.
In this study, 185 AP2/ERF genes with at least one AP2 conserved domain were identified in the A. hypogaea cv. Tifrunner genome through genome-wide analysis. Similar to other plants, all putative AP2/ERF superfamily genes were identified as five subfamilies: AP2, ERF, DREB, RAV, and Soloist (Supplementary Table S2). Each subfamily has 59, 76, 41, 4, and 5 members, respectively. In different plants, the numbers of AP2/ERF proteins vary significantly depending on the genome size (eudicot or monocot) (Supplementary Table S12), which may result in gene evolution and duplication. On the other hand, the number of each subfamily follows the regular pattern: the number of the ERF subfamily is the largest, followed by DREB, AP2, and RAV or Soloist (Supplementary Table S12), suggesting that the composition of the AP2/ERF superfamily TFs is highly conserved in plants and may share a common ancestor before separation. Moreover, the largest number of ERF and DREB subfamilies strongly implies its main role in plant growth and development process. The differences in the values of molecular weight (14.46–82.54 kDa) and pI (4.55–10.58) of AhAP2/ERF suggest the putative differences in AhAP2/ERF (Lata et al., 2014). Subcellular localization predicted that AhAP2/ERF TFs are mainly localized to the nucleus and thus validates the posttranscriptional regulatory mechanism of the proteins (Karniely and Pines, 2005). Further analysis showed that the AP2/ERFs exhibit certain subfamily characteristics in intron/exon patterns, motif structures, and phylogenetic relationships (Figures 1, 2). This high evolutionary conservation can be used as an important basis for subfamily classification.
Chromosomal mapping showed an uneven distribution of AhAP2/ERF genes on 20 chromosomes (Figure 3). There were hot regions or gene clusters on chr06 and chr15. Generally, tandem, large-scale chromosome segmental duplication and transposition were identified as the main evolutionary mechanisms that cause the expansion of the gene family (Lynch and Conery, 2000; Cannon et al., 2004). In total, 14 gene pairs showed tandem duplication and 85 gene pairs revealed segmental duplication, which sustain the overall 8.6% tandem duplication of AP2/ERF in A. hypogaea cv. Tifrunner (Zhuang et al., 2019) (Figure 4). The numbers of duplicated gene pairs vary between crops, such as the number of duplication pairs which is 90 in sunflower, 76 in grape, 51 in Arabidopsis, 41 in rice, and 11 in dark jute, all of which were lower than in peanut. Hence, this variation in AP2/ERF gene numbers in plants might be due to the different duplication events. The microsyntenic analysis of these AP2/ERF gene families across the Leguminosae family could provide valuable information about their evolution. Our findings demonstrated that a strong association between AhAP2/ERF genes of cultivated peanut and wild species was observed (Figure 5). Among them, there were an equal number of pairs of syntenic relationships in the genome of A. hypogaea cv. Tifrunner with A. duranensis and A. ipaensis (Supplementary Tables S6, S7). Notably, most AhAP2/ERF genes of A. duranensis and A. ipaensis might have more than one ortholog in cultivated peanut. These results suggested that cultivated peanut, an allotetraploid plant, likely contained twice the number of AP2/ERF observed for wild-type peanut. The mean Ka/Ks value of peanut with Medicago truncatula L. and Glycine max L. suggests a purifying selection of AhAP2/ERF genes that have undergone great selective constraint and substitution elimination by natural selection (Supplementary Tables S8, S9).
Abiotic stresses do a great harm to the regular growth in peanut at early stages; thus, seedlings at 5 weeks of age were used for drought tolerance (Passioura, 1983; Pierret et al., 2007; Songsri et al., 2008). As an important organ for plants to absorb water and mineral elements, roots directly experience soil drought, and thus, the expression pattern of AhAP2/ERF genes under drought stress in roots is essential to clarifying functional divergence (Passioura, 1983; Songsri et al., 2008). The prediction of peanut AP2/ERF protein function by constructing a protein interaction network (Letovsky and Kasif, 2003) proposed the interaction among AP2, ERF, and RAV families, thus implying its interactive function in response to various stresses. For example, AhAP2-29 showed a strong interaction with AtRAP2.11, AtCBF1, AtTINY2, and AtESR1, which are members of the AP2/ERF family that participate in the stress tolerance in Arabidopsis (Banno et al., 2001; Novillo et al., 2004; Wei et al., 2005; Min et al., 2012), implying the potential function of AhAP2-29 in peanut response to drought stress. The same phenomenon is discovered in the AP2/ERF family members, including AhAP2-37, AhERF47, and AhRAV-1 which are known to function against stress in Arabidopsis, thus indicating that there are strong and complex interactions of AhAP2, AhERF, and AhRAV members in peanut response to drought stress. However, under no-watering condition, the expression of RAVs was extremely downregulated, perhaps suggesting the indirect role in peanut against drought stress, which is in-line with the previous studies in other crops (Hu et al., 2004; Sohn et al., 2006; Je et al., 2010; Li X.-J. et al., 2015; Wei et al., 2018).
The promoter sequence possesses vital information about gene functional components (i.e., cis-acting elements) and reflects potential function of the gene (Kabir et al., 2021). In this study, four distinct types of cis-acting elements were found, of which hormone-related and stress-related elements are ones having close relationships with plant stress conditions. Moreover, ABA, as a stress signal, is essential during plant growth and development. It integrates various stress signals and controls downstream stress responses to make plants adapt to various stress environments through uninterrupted adjustment (Tuteja, 2007). Promoter analysis showed that almost all AhAP2/ERF members have ABA response elements, especially ones from AP2, ERF, and DREB subfamilies. Other considerable elements which are related to their function in peanut were TC-rich elements, MBS, and other hormone-related and stress-related elements, which interact in a way. For ERF, DREB, and AP2 subfamily genes, it appears that, except AhDREB-3, AhERF-13, AhERF-66, and AhERF-72, one owns any cis-acting elements of ABRE, TC-rich, MBS, or ARE which may be upregulated by the no-watering treatment, implying the main role of cis-acting elements of the promoter in peanut response to drought stress. However, AhRAV genes were all downregulated during drought regardless if they are in the promoter region. For the complex interaction of ERF, DREB, and AP2 (Figure 7), AhRAV may be strongly regulated by other subfamilies. These results suggest that AhAP2, AhERF, and AhDREB genes may play pivotal roles in response to drought stress.
Conclusion
All in all, a total of 185 AP2/ERF genes were identified in the A. hypogaea cv. Tifrunner genome and divided into AP2 (59), ERF (76), DREB (41), RAV (4), and Soloist 5) subfamilies. Members in the same family or group shared great similarity of exon–intron structure and conserved motifs. Segmental duplication contributed to the expansion of AhAP2/ERF genes, and these duplication pair genes had evolved under strong purifying selection. cis-Element analysis suggested that the expression of AhAP2/ERF can be regulated by hormones and various environmental factors. Protein interaction predicted complex interaction relationships among or within groups of ERF, DREB, and AP2 members. The expression profile under drought stress by qRT-PCR showed that some AhAP2/ERF were significantly upregulated, indicating their potential roles in response to drought stress.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author Contributions
MC, SH, and MH conceived the study and carried out the bioinformatic analysis. MC, PC, JG, and HL collected the plant materials and performed the experiments. PD, WD, and BH participated in handling figures and tables. MC and ZZ drafted the manuscript. MH, LS, and XZ revised the manuscript. All authors read and approved the final manuscript.
Funding
This work was supported by the National Natural Science Foundation of China (No. 31871663), the China Agriculture Research System (CARS-13), the Major Technology Research and Development of Henan Province, China (201300111000) and the Henan Provincial Agriculture Research System, China (S2012-5).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2021.750761/full#supplementary-material
References
Allen, M. D., Yamasaki, K., Ohme-Takagi, M., Tateno, M., and Suzuki, M. (1998). A Novel Mode of DNA Recognition by a β-Sheet Revealed by the Solution Structure of the GCC-Box Binding Domain in Complex with DNA. EMBO J. 17, 5484–5496. doi:10.1093/emboj/17.18.5484
Alonso, J. M., Stepanova, A. N., Solano, R., Wisman, E., Ferrari, S., Ausubel, F. M., et al. (2003). Five Components of the Ethylene-Response Pathway Identified in a Screen for Weak Ethylene-Insensitive Mutants in Arabidopsis. Proc. Natl. Acad. Sci. 100, 2992–2997. doi:10.1073/pnas.0438070100
Artimo, P., Jonnalagedda, M., Arnold, K., Baratin, D., Csardi, G., de Castro, E., et al. (2012). ExPASy: SIB Bioinformatics Resource portal. Nucleic Acids Res. 40, W597–W603. doi:10.1093/nar/gks400
Aukerman, M. J., and Sakai, H. (2003). Regulation of Flowering Time and floral Organ Identity by a microRNA and its APETALA2-Like Target Genes. Plant Cell 15, 2730–2741. doi:10.1105/tpc.016238
Bailey, T. L., Bodén, M., Buske, F. A., Frith, M., Grant, C. E., Clementi, L., et al. (2009). MEME SUITE: Tools for Motif Discovery and Searching. Nucleic Acids Res. 37, w202–w208. doi:10.1093/nar/gkp335
Baloglu, M. C. (2014). Genome-Wide In Silico Identification and Comparison of Growth Regulating Factor (GRF) Genes in Cucurbitaceae Family. Plant Omics 7, 260–270. doi:10.1097/01.tp.0000332556.64365.e7
Banno, H., Ikeda, Y., Niu, Q.-W., and Chua, N.-H. (2001). Overexpression of Arabidopsis ESR1 Induces Initiation of Shoot Regeneration. Plant Cell 13, 2609–2618. doi:10.2307/387152210.1105/tpc.010234
Bertioli, D. J., Cannon, S. B., Froenicke, L., Huang, G., Farmer, A. D., Cannon, E. K. S., et al. (2016). The Genome Sequences of Arachis Duranensis and Arachis Ipaensis, the Diploid Ancestors of Cultivated Peanut. Nat. Genet. 48, 438–446. doi:10.1038/ng.3517
Boutilier, K., Offringa, R., Sharma, V. K., Kieft, H., Ouellet, T., Zhang, L., et al. (2002). Ectopic Expression of BABY BOOM Triggers a Conversion from Vegetative to Embryonic Growth. Plant Cell 14, 1737–1749. doi:10.1105/tpc.001941
Boyer, J. S. (1982). Plant Productivity and Environment. Science 218, 443–448. doi:10.1126/science.218.4571.443
Brand, Y., and Hovav, R. (2010). Identification of Suitable Internal Control Genes for Quantitative Real-Time PCR Expression Analyses in Peanut (Arachis hypogaea). Peanut Sci. 37, 12–19. doi:10.3146/PS09-014.1
Cannon, S. B., Mitra, A., Baumgarten, A., Young, N. D., and May, G. (2004). The Roles of Segmental and Tandem Gene Duplication in the Evolution of Large Gene Families in Arabidopsis thaliana. Bmc Plant Biol. 4, 10. doi:10.1186/1471-2229-4-10
Cardwell, K. F., and Henry, S. H. (2008). Risk of Exposure to and Mitigation of Effect of Aflatoxin on Human Health: A West African Example. J. Toxicol. Toxin Rev. 23, 217–247. doi:10.1081/TXR-200027817
Chen, C., Chen, H., Zhang, Y., Thomas, H. R., Frank, M. H., He, Y., et al. (2020). TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 13, 1194–1202. doi:10.1016/j.molp.2020.06.009
Chen, N., Yang, Q., Su, M., Pan, L., Chi, X., Chen, M., et al. (2012). Cloning of Six ERF Family Transcription Factor Genes from Peanut and Analysis of Their Expression during Abiotic Stress. Plant Mol. Biol. Rep. 30, 1415–1425. doi:10.1007/s11105-012-0456-0
Chou, K.-C., and Shen, H.-B. (2008). Cell-PLoc: A Package of Web Servers for Predicting Subcellular Localization of Proteins in Various Organisms. Nat. Protoc. 3, 153–162. doi:10.1038/nprot.2007.494
Chuck, G., Meeley, R. B., and Hake, S. (1998). The Control of Maize Spikelet Meristem Fate by the APETALA2-Like Gene Indeterminate Spikelet1. Genes Develop. 12, 1145–1154. doi:10.1101/gad.12.8.1145
Chuck, G., Meeley, R., and Hake, S. (2008). Floral Meristem Initiation and Meristem Cell Fate Are Regulated by the maize AP2 Genes Ids1 and Sid1. Development 135, 3013–3019. doi:10.1242/dev.024273
Chuck, G., Muszynski, M., Kellogg, E., Hake, S., and Schmidt, R. J. (2002). The Control of Spikelet Meristem Identity by the Branched Silkless1 Gene in Maize. Science 298, 1238–1241. doi:10.1126/science.1076920
Cui, M., Wang, C., Zhang, W., Pervaiz, T., Haider, M. S., Tang, W., et al. (2018). Characterization of Vv-miR156: Vv-SPL Pairs Involved in the Modulation of Grape Berry Development and Ripening. Mol. Genet. Genomics 293, 1333–1354. doi:10.1007/s00438-018-1462-1
Du, D., Hao, R., Cheng, T., Pan, H., Yang, W., Wang, J., et al. (2013). Genome-Wide Analysis of the AP2/ERF Gene Family in Prunus Mume. Plant Mol. Biol. Rep. 31, 741–750. doi:10.1007/s11105-012-0531-6
Dubos, C., Stracke, R., Grotewold, E., Weisshaar, B., Martin, C., Lepiniec, L., et al. (2010). MYB Transcription Factors in Arabidopsis. Trends Plant Sci. 15, 573–581. doi:10.1016/j.tplants.2010.06.005
El Ouakfaoui, S., Schnell, J., Abdeen, A., Colville, A., Labbé, H., Han, S., et al. (2010). Control of Somatic Embryogenesis and Embryo Development by AP2 Transcription Factors. Plant Mol. Biol. 74, 313–326. doi:10.1007/s11103-010-9674-8
El-Sharkawy, I., Sherif, S., Mila, I., Bouzayen, M., and Jayasankar, S. (2009). Molecular Characterization of Seven Genes Encoding Ethylene-Responsive Transcriptional Factors during Plum Fruit Development and Ripening. J. Exp. Bot. 60, 907–922. doi:10.1093/jxb/ern354
Farombi, E. O. (2006). Aflatoxin Contamination of Foods in Developing Countries: Implications for Hepatocellular Carcinoma and Chemopreventive Strategies. Afr. J. Biotech. 5, 1–14. doi:10.1016/j.enzmictec.2005.08.024
Feller, A., Machemer, K., Braun, E. L., and Grotewold, E. (2011). Evolutionary and Comparative Analysis of MYB and bHLH Plant Transcription Factors. Plant J. 66, 94–116. doi:10.1111/j.1365-313X.2010.04459.x
Feng, K., Hou, X.-L., Xing, G.-M., Liu, J.-X., Duan, A.-Q., Xu, Z.-S., et al. (2020). Advances in AP2/ERF Super-Family Transcription Factors in Plant. Crit. Rev. Biotechnol. 40, 750–776. doi:10.1080/07388551.2020.1768509
Finet, C., Fourquin, C., Vinauger, M., Berne-Dedieu, A., Chambrier, P., Paindavoine, S., et al. (2010). Parallel Structural Evolution of Auxin Response Factors in the Angiosperms. Plant J. 63, 952–959. doi:10.1111/j.1365-313X.2010.04292.x
Finn, R. D., Tate, J., Mistry, J., Coggill, P. C., Sammut, S. J., Hotz, H. R., et al. (2008). The Pfam Protein Families Database. Nucleic Acids Res. 36, D281–D288. doi:10.1093/nar/gkm960
Finn, R. D., Clements, J., and Eddy, S. R. (2011). HMMER Web Server: Interactive Sequence Similarity Searching. Nucleic Acids Res. 39, W29–W37. doi:10.1093/nar/gkr367
Fu, M., Kang, H. K., Son, S.-H., Kim, S.-K., and Nam, K. H. (2014). A Subset of Arabidopsis RAV Transcription Factors Modulates Drought and Salt Stress Responses Independent of ABA. Plant Cel Physiol 55, 1892–1904. doi:10.1093/pcp/pcu118
Gao, C., Sun, J., Wang, C., Dong, Y., Xiao, S., Wang, X., et al. (2017). Genome-Wide Analysis of Basic/Helix-Loop-Helix Gene Family in Peanut and Assessment of its Roles in Pod Development. PLoS ONE 12, e0181843. doi:10.1371/journal.pone.0181843
Gasteiger, E., Hoogland, C., Gattiker, A., Duvaud, S. e., Wilkins, M. R., Appel, R. D., et al. (2005). Protein Identification and Analysis Tools on the ExPASy Server. The Proteomics Protoc. handbook, 571–607. doi:10.1385/1-59259-890-0:571
Ghorbani, R., Zakipour, Z., Alemzadeh, A., and Razi, H. (2020). Genome-Wide Analysis of AP2/ERF Transcription Factors Family in Brassica Napus. Physiol. Mol. Biol. Plants 26, 1463–1476. doi:10.1007/s12298-020-00832-z
Girardi, C. L., Rombaldi, C. V., Dal Cero, J., Nobile, P. M., Laurens, F., Bouzayen, M., et al. (2013). Genome-Wide Analysis of the AP2/ERF Superfamily in Apple and Transcriptional Evidence of ERF Involvement in Scab Pathogenesis. Scientia Horticulturae 151, 112–121. doi:10.1016/j.scienta.2012.12.017
Guo, B., Wei, Y., Xu, R., Lin, S., Luan, H., Lv, C., et al. (2016). Genome-Wide Analysis of APETALA2/Ethylene-Responsive Factor (AP2/ERF) Gene Family in Barley (Hordeum Vulgare L.). PLoS ONE 11, e0161322. doi:10.1371/journal.pone.0161322
Gutterson, N., and Reuber, T. L. (2004). Regulation of Disease Resistance Pathways by AP2/ERF Transcription Factors. Curr. Opin. Plant Biol. 7, 465–471. doi:10.1016/j.pbi.2004.04.007
Hirota, A., Kato, T., Fukaki, H., Aida, M., and Tasaka, M. (2007). The Auxin-Regulated AP2/EREBP GenePUCHIIs Required for Morphogenesis in the Early Lateral Root Primordium ofArabidopsis. Plant Cell 19, 2156–2168. doi:10.1105/tpc.107.050674
Hu, L., and Liu, S. (2011). Genome-Wide Identification and Phylogenetic Analysis of the ERF Gene Family in Cucumbers. Genet. Mol. Biol. 34, 624–634. doi:10.1590/S1415-47572011005000054
Hu, Y. X., Wang, Y. H., Liu, X. F., and Li, J. Y. (2004). Arabidopsis RAV1 Is Down-Regulated by Brassinosteroid and May Act as a Negative Regulator during Plant Development. Cell Res 14, 8–15. doi:10.1038/sj.cr.7290197
Huang, Y., Liu, Y., Zhang, M., Chai, M., He, Q., Jakada, B. H., et al. (2020). Genome-Wide Identification and Expression Analysis of the ERF Transcription Factor Family in Pineapple (Ananas Comosus (L.) Merr.). Peer J. 8, e10014. doi:10.7717/peerj.10014
Irish, V. F., and Sussex, I. M. (1990). Function of the Apetala-1 Gene during Arabidopsis floral Development. Plant Cell 2, 741–753. doi:10.1105/tpc.2.8.741
Ito, T. M., Polido, P. B., Rampim, M. C., Kaschuk, G., and Souza, S. G. H. (2014). Genome-Wide Identification and Phylogenetic Analysis of the AP2/ERF Gene Superfamily in Sweet orange (Citrus Sinensis). Genet. Mol. Res. 13, 7839–7851. doi:10.4238/201410.4238/2014.september.26.22
Jayaprakash, A., Thanmalagan, R. R., Roy, A., Arunachalam, A., and Lakshmi, P. (2019). Strategies to Understand Aspergillus flavus Resistance Mechanism in Arachis hypogaea L. Curr. Plant Biol. 20, 100123. doi:10.1016/j.cpb.2019.100123
Je, B. I., Piao, H. L., Park, S. J., Park, S. H., Kim, C. M., Xuan, Y. H., et al. (2010). RAV-Like1 Maintains Brassinosteroid Homeostasis via the Coordinated Activation of BRI1 and Biosynthetic Genes in Rice. Plant Cell 22, 1777–1791. doi:10.4161/psb.5.11.1335710.1105/tpc.109.069575
Jin, J., Zhang, H., Kong, L., Gao, G., and Luo, J. (2014). PlantTFDB 3.0: A portal for the Functional and Evolutionary Study of Plant Transcription Factors. Nucl. Acids Res. 42, D1182–D1187. doi:10.1093/nar/gkt1016
Jofuku, K. D., den Boer, B. G., Van Montagu, M., and Okamuro, J. K. (1994). Control of Arabidopsis Flower and Seed Development by the Homeotic Gene APETALA2. Plant Cell 6, 1211–1225. doi:10.1105/tpc.6.9.1211
Jofuku, K. D., Omidyar, P. K., Gee, Z., and Okamuro, J. K. (2005). Control of Seed Mass and Seed Yield by the floral Homeotic Gene APETALA2. Proc. Natl. Acad. Sci. 102, 3117–3122. doi:10.1073/pnas.0409893102
Kabir, S. M. T., Hossain, M. S., Bashar, K. K., Honi, U., Ahmed, B., Emdad, E. M., et al. (2021). Genome-Wide Identification and Expression Profiling of AP2/ERF Superfamily Genes under Stress Conditions in Dark Jute (Corchorus Olitorius L.). Ind. Crops Prod. 166, 113469. doi:10.1016/j.indcrop.2021.113469
Karniely, S., and Pines, O. (2005). Single Translation-Dual Destination: Mechanisms of Dual Protein Targeting in Eukaryotes. EMBO Rep. 6, 420–425. doi:10.1038/sj.embor.7400394
Kavas, M., Kizildogan, A., Gökdemir, G., and Baloglu, M. C. (2015). Genome-Wide Investigation and Expression Analysis of AP2-ERF Gene Family in Salt Tolerant Common Bean. EXCLI J. 14, 1187–1206. doi:10.17179/excli2015-600
Kavas, M., Baloğlu, M. C., Atabay, E. S., Ziplar, U. T., Daşgan, H. Y., and Ünver, T. (2016). Genome-Wide Characterization and Expression Analysis of Common Bean bHLH Transcription Factors in Response to Excess Salt Concentration. Mol. Genet. Genomics. 291, 129–143. doi:10.1007/s00438-015-1095-6
Kim, M. J., Ruzicka, D., Shin, R., and Schachtman, D. P. (2012). The Arabidopsis AP2/ERF Transcription Factor RAP2.11 Modulates Plant Response to Low-Potassium Conditions. Mol. Plant 5, 1042–1057. doi:10.1093/mp/sss003
Kitomi, Y., Ito, H., Hobo, T., Aya, K., Kitano, H., and Inukai, Y. (2011). The Auxin Responsive AP2/ERF Transcription Factor Crown Rootless5 Is Involved in crown Root Initiation in rice through the Induction of OsRR1, a Type-A Response Regulator of Cytokinin Signaling. Plant J. 67 (3), 472–484. doi:10.1111/j.1365-313x.2011.04610.x
Krizek, B., and Beth, A. (2009). Aintegumenta and Aintegumenta-LIKE6 Act Redundantly to Regulate Arabidopsis Floral Growth and Patterning. Plant Physiol. 150, 1916–1929. doi:10.1104/pp.109.141119
Lata, C., Mishra, A. K., Muthamilarasan, M., Bonthala, V. S., Khan, Y., and Prasad, M. (2014). Genome-Wide Investigation and Expression Profiling of AP2/ERF Transcription Factor Superfamily in Foxtail Millet (Setaria Italica L.). PLoS ONE 9, e113092. doi:10.1371/journal.pone.0113092
Lescot, M., Déhais, P., Thijs, G., Marchal, K., Moreau, Y., Van de Peer, Y., et al. (2002). PlantCARE, a Database of Plant Cis-Acting Regulatory Elements and a portal to Tools for In Silico Analysis of Promoter Sequences. Nucleic Acids Res. 30, 325–327. doi:10.1093/nar/30.1.325
Letovsky, S., and Kasif, S. (2003). Predicting Protein Function from Protein/protein Interaction Data: A Probabilistic Approach. Bioinformatics 19, i197–i204. doi:10.1093/bioinformatics/btg1026
Letunic, I., Doerks, T., and Bork, P. (2012). SMART 7: Recent Updates to the Protein Domain Annotation Resource. Nucleic Acids Res. 40, D302–D305. doi:10.1093/nar/gkr931
Li, A., Yu, X., Cao, B. B., Peng, L. X., Gao, Y., Feng, T., et al. (2017a). LkAP2L2, an AP2/ERF Transcription Factor Gene of Larix Kaempferi, with Pleiotropic Roles in Plant branch and Seed Development. Russ. J. Genet. 53 (12), 1335–1342. doi:10.1134/S1022795417120079
Li, C.-W., Su, R.-C., Cheng, C.-P., Sanjaya, S. J., You, S.-J., Hsieh, T.-H., et al. (2011). Tomato RAV Transcription Factor Is a Pivotal Modulator Involved in the AP2/EREBP-Mediated Defense Pathway. Plant Physiol. 156, 213–227. doi:10.1104/pp.111.174268
Li, H., Wang, Y., Wu, M., Li, L., Li, C., Han, Z., et al. (2017b). Genome-Wide Identification of AP2/ERF Transcription Factors in Cauliflower and Expression Profiling of the ERF Family under Salt and Drought Stresses. Front. Plant Sci. 8, 946. doi:10.3389/fpls.2017.00946
Li, M.-Y., Xu, Z.-S., Huang, Y., Tian, C., Wang, F., and Xiong, A.-S. (2015b). Genome-wide Analysis of AP2/ERF Transcription Factors in Carrot (Daucus Carota L.) Reveals Evolution and Expression Profiles under Abiotic Stress. Mol. Genet. Genomics 290, 2049–2061. doi:10.1007/s00438-015-1061-3
Li, P., Chai, Z., Lin, P., Huang, C., Huang, G., Xu, L., et al. (2020). Genome-Wide Identification and Expression Analysis of AP2/ERF Transcription Factors in Sugarcane (Saccharum Spontaneum L.). BMC Genomics 21, 685. doi:10.1186/s12864-020-07076-x
Li, X.-J., Li, M., Zhou, Y., Hu, S., Hu, R., Chen, Y., et al. (2015a). Overexpression of Cotton RAV1 Gene in Arabidopsis Confers Transgenic Plants High Salinity and Drought Sensitivity. PLoS ONE 10, e0118056. doi:10.1371/journal.pone.0118056
Li, X., Duan, X., Jiang, H., Sun, Y., Tang, Y., Yuan, Z., et al. (2006). Genome-Wide Analysis of Basic/Helix-Loop-Helix Transcription Factor Family in rice and Arabidopsis. Plant Physiol. 141, 1167–1184. doi:10.1104/pp.106.080580
Licausi, F., Giorgi, F. M., Zenoni, S., Osti, F., Pezzotti, M., and Perata, P. (2010). Genomic and Transcriptomic Analysis of the AP2/ERF Superfamily in Vitis vinifera. BMC Genomics 11, 719. doi:10.1186/1471-2164-11-719
Licausi, F., Ohme-Takagi, M., and Perata, P. (2013). APETALA2/Ethylene Responsive Factor (AP2/ERF) Transcription Factors: Mediators of Stress Responses and Developmental Programs. New Phytol. 199, 639–649. doi:10.1111/nph.12291
Liu, C., Wu, Y., and Wang, X. (2012). bZIP Transcription Factor OsbZIP52/RISBZ5: A Potential Negative Regulator of Cold and Drought Stress Response in rice. Planta 235, 1157–1169. doi:10.1007/s00425-011-1564-z
Liu, S., Wang, X., Wang, H., Xin, H., Yang, X., Yan, J., et al. (2013). Genome-Wide Analysis of ZmDREB Genes and Their Association with Natural Variation in Drought Tolerance at Seedling Stage of Zea mays L. Plos Genet. 9, e1003790. doi:10.1371/journal.pgen.1003790
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2−ΔΔCT Method. Methods 25, 402–408. doi:10.1006/meth.2001.1262
Lynch, M., and Conery, J. S. (2000). The Evolutionary Fate and Consequences of Duplicate Genes. Science 290, 1151–1155. doi:10.1126/science.290.5494.1151
Maes, T., Van de Steene, N., Zethof, J., Karimi, M., D’Hauw, M., Mares, G., et al. (2001). Petunia Ap2-Like Genes and Their Role in Flower and Seed Development. Plant Cell 13, 229–244. doi:10.1105/tpc.13.2.229
Magnani, E., Sjölander, K., and Hake, S. (2004). From Endonucleases to Transcription Factors: Evolution of the AP2 DNA Binding Domain in Plants[W]. The Plant Cell 16, 2265–2277. doi:10.1105/tpc.104.023135
Manning, K., Tör, M., Poole, M., Hong, Y., Thompson, A. J., King, G. J., et al. (2006). A Naturally Occurring Epigenetic Mutation in a Gene Encoding an SBP-Box Transcription Factor Inhibits Tomato Fruit Ripening. Nat. Genet. 38, 948–952. doi:10.1038/ng1841
Mao, X., Zhang, H., Qian, X., Li, A., Zhao, G., and Jing, R. (2012). TaNAC2, a NAC-Type Wheat Transcription Factor Conferring Enhanced Multiple Abiotic Stress Tolerances in Arabidopsis. J. Exp. Bot. 63, 2933–2946. doi:10.1093/jxb/err462
Meng, J., Yang, J., Peng, M., Liu, X., and He, H. (2020). Genome-Wide Characterization, Evolution, and Expression Analysis of the Leucine-Rich Repeat Receptor-Like Protein Kinase (LRR-RLK) Gene Family in Medicago Truncatula. Life 10, 176. doi:10.3390/life10090176
Nakano, T., Suzuki, K., Fujimura, T., and Shinshi, H. (2006). Genome-Wide Analysis of the ERF Gene Family in Arabidopsis and Rice. Plant Physiol. 140, 411–432. doi:10.1104/pp.105.073783
Nakashima, K., Takasaki, H., Mizoi, J., Shinozaki, K., and Yamaguchi-Shinozaki, K. (2012). NAC Transcription Factors in Plant Abiotic Stress Responses. Biochim. Biophys. Acta (Bba) - Gene Regul. Mech. 1819, 97–103. doi:10.1016/j.bbagrm.2011.10.005
Nimitr, V., Patcharin, S., Chutipong, A., Sanun, J., and Aran, P. (2003). Effect of Water Stress on Yield and Agronomic Characters of Peanut (Arachis hypogaea L.). Songklanakar J. Sci. Tech. 25, 283–288.
Novillo, F., Alonso, J. M., Ecker, J. R., and Salinas, J. (2004). CBF2/DREB1C Is a Negative Regulator of CBF1/DREB1B and CBF3/DREB1A Expression and Plays a central Role in Stress Tolerance in Arabidopsis. Proc. Natl. Acad. Sci. 101, 3985–3990. doi:10.1073/pnas.0303029101
Okamuro, J. K., Caster, B., Villarroel, R., Van Montagu, M., and Jofuku, K. D. (1997). The AP2 Domain of APETALA2 Defines a Large New Family of DNA Binding Proteins in Arabidopsis. Proc. Natl. Acad. Sci. 94, 7076–7081. doi:10.1073/pnas.94.13.7076
Pan, Y.-B., Burner, D. M., and Legendre, B. L. (2000). An Assessment of the Phylogenetic Relationship Among Sugarcane and Related Taxa Based on the Nucleotide Sequence of 5S rRNA Intergenic Spacers. Genetica 108, 285–295. doi:10.1023/A:1004191625603
Passioura, J. B. (1983). Roots and Drought Resistance. Agric. Water Manage. 7, 265–280. doi:10.1016/0378-3774(83)90089-6
Pierret, A., Doussan, C., Capowiez, Y., Bastardie, F., and Pagès, L. (2007). Root Functional Architecture: A Framework for Modeling the Interplay between Roots and Soil. Vadose Zone J. 6, 269–281. doi:10.2136/vzj2006.0067
Pietsch, C., Sreenivasulu, N., Wobus, U., and Röder, M. S. (2009). Linkage Mapping of Putative Regulator Genes of Barley Grain Development Characterized by Expression Profiling. BMC Plant Biol. 9, 4. doi:10.1186/1471-2229-9-4
Rademacher, E. H., Möller, B., Lokerse, A. S., Llavata-Peris, C. I., van den Berg, W., and Weijers, D. (2011). A Cellular Expression Map of the Arabidopsis AUXIN Response Factor Gene Family. Plant J. 68, 597–606. doi:10.1111/j.1365-313X.2011.04710.x
Reddy, T. Y., Reddy, V. R., and Anbumozhi, V. (2003). Physiological Responses of Groundnut (Arachis Hypogea L.) to Drought Stress and its Amelioration: A Critical Review. Plant Growth Regul. 41, 75–88. doi:10.1023/A:1027353430164
Riechmann, J. L., and Meyerowitz, E. M. (1998). The AP2/EREBP Family of Plant Transcription Factors. Biol. Chem. 379, 633–646. doi:10.1515/bchm.1998.379.6.633
Rushton, P. J., Somssich, I. E., Ringler, P., and Shen, Q. J. (2010). WRKY Transcription Factors. Trends Plant Sci. 15, 247–258. doi:10.1016/j.tplants.2010.02.006
Sakuma, Y., Liu, Q., Dubouzet, J. G., Abe, H., Shinozaki, K., and Yamaguchi-Shinozaki, K. (2002). DNA-Binding Specificity of the ERF/AP2 Domain of Arabidopsis DREBs, Transcription Factors Involved in Dehydration- and Cold-Inducible Gene Expression. Biochem. Biophysical Res. Commun. 290, 998–1009. doi:10.1006/bbrc.2001.6299
Shannon, P., Markiel, A., Ozier, O., Baliga, N. S., Wang, J. T., Ramage, D., et al. (2003). Cytoscape: A Software Environment for Integrated Models of Biomolecular Interaction Networks. Genome Res. 13, 2498–2504. doi:10.1101/gr.1239303
Sharoni, A. M., Nuruzzaman, M., Satoh, K., Shimizu, T., Kondoh, H., Sasaya, T., et al. (2011). Gene Structures, Classification and Expression Models of the AP2/EREBP Transcription Factor Family in rice. Plant Cel Physiol 52, 344–360. doi:10.1093/pcp/pcq196
Silva, G. F. F. E., Silva, E. M., da Silva Azevedo, M., Guivin, M. A. C., Ramiro, D. A., Figueiredo, C. R., et al. (2014). MicroRNA156-Targeted SPL/SBP Box Transcription Factors Regulate Tomato Ovary and Fruit Development. Plant J. 78, 604–618. doi:10.1111/tpj.12493
Singh, K., Foley, R. C., and Luis, O. S. (2002). Transcription Factors in Plant Defense and Stress Responses. Curr. Opin. Plant Biol. 5, 430–436. doi:10.1016/s1369-5266(02)00289-3
Soares, V. L. F., Rodrigues, S. M., de Oliveira, T. M., de Queiroz, T. O., Lima, L. S., Hora-Júnior, B. T., et al. (2011). Unraveling New Genes Associated with Seed Development and Metabolism in Bixa Orellana L. By Expressed Sequence Tag (EST) Analysis. Mol. Biol. Rep. 38 (2), 1329–1340. doi:10.1007/s11033-010-0234-8
Sohn, K. H., Lee, S. C., Jung, H. W., Hong, J. K., and Hwang, B. K. (2006). Expression and Functional Roles of the Pepper Pathogen-Induced Transcription Factor RAV1 in Bacterial Disease Resistance, and Drought and Salt Stress Tolerance. Plant Mol. Biol. 61, 897–915. doi:10.1007/s11103-006-0057-0
Song, H., Wang, P., Lin, J.-Y., Zhao, C., Bi, Y., and Wang, X. (2016). Genome-Wide Identification and Characterization of WRKY Gene Family in Peanut. Front. Plant Sci. 7, 534. doi:10.3389/fpls.2016.00534
Songsri, P., Jogloy, S., Vorasoot, N., Akkasaeng, C., Patanothai, A., and Holbrook, C. C. (2008). Root Distribution of Drought-Resistant Peanut Genotypes in Response to Drought. J. Agron. Crop Sci. 194, 92–103. doi:10.1111/j.1439-037X.2008.00296.x
Soni, P., Gangurde, S. S., Ortega-Beltran, A., Kumar, R., Parmar, S., Sudini, H. K., et al. (2020). Functional Biology and Molecular Mechanisms of Host-Pathogen Interactions for Aflatoxin Contamination in Groundnut (Arachis hypogaea L.) and maize (Zea mays L.). Front. Microbiol. 11, 227. doi:10.3389/fmicb.2020.00227
Sun, L., Hu, R., Shen, G., and Zhang, H. (2013). Genetic Engineering Peanut for Higher Drought- and Salt-Tolerance. Food Nutr. Sci. 04, 1–7. doi:10.4236/fns.2013.46A001
Szklarczyk, D., Gable, A. L., Lyon, D., Junge, A., Wyder, S., Huerta-Cepas, J., et al. (2019). STRING V11: Protein-Protein Association Networks with Increased Coverage, Supporting Functional Discovery in Genome-Wide Experimental Datasets. Nucleic Acids Res. 47, D607–D613. doi:10.1093/nar/gky1131
Tamura, K., Peterson, D., Peterson, N., Stecher, G., Nei, M., and Kumar, S. (2011). MEGA5: Molecular Evolutionary Genetics Analysis Using Maximum Likelihood, Evolutionary Distance, and Maximum Parsimony Methods. Mol. Biol. Evol. 28, 2731–2739. doi:10.1093/molbev/msr121
Tamura, K., Stecher, G., Peterson, D., Filipski, A., and Kumar, S. (2013). MEGA6: Molecular Evolutionary Genetics Analysis Version 6.0. Mol. Bio. Evol. 30, 2725–2729. doi:10.1093/molbev/mst197
Terol, J., Nueda, M. J., Ventimilla, D., Tadeo, F., and Talon, M. (2019). Transcriptomic Analysis of Citrus Clementina Mandarin Fruits Maturation Reveals a MADS-Box Transcription Factor that Might Be Involved in the Regulation of Earliness. BMC Plant Biol. 19, 47. doi:10.1186/s12870-019-1651-z
Thompson, J. D., Higgins, D. G., and Gibson, T. J. (1994). CLUSTAL W: Improving the Sensitivity of Progressive Multiple Sequence Alignment through Sequence Weighting, Position-specific gap Penalties and Weight Matrix Choice. Nucl. Acids Res. 22, 4673–4680. doi:10.1093/nar/22.22.4673
Trevaskis, B., Bagnall, D. J., Ellis, M. H., Peacock, W. J., and Dennis, E. S. (2003). MADS Box Genes Control Vernalization-Induced Flowering in Cereals. Proc. Natl. Acad. Sci. 100, 13099–13104. doi:10.1073/pnas.1635053100
Tsutsui, T., Yamaji, N., and Feng Ma, J. (2011). Identification of a Cis-Acting Element of ART1, a C2H2-Type Zinc-Finger Transcription Factor for Aluminum Tolerance in rice. Plant Physiol. 156, 925–931. doi:10.1104/pp.111.175802
Tuteja, N. (2007). Abscisic Acid and Abiotic Stress Signaling. Plant Signaling Behav. 2, 135–138. doi:10.4161/psb.2.3.4156
Ulm, R., Baumann, A., Oravecz, A., Mate, Z., Adam, E., Oakeley, E. J., et al. (2004). Genome-Wide Analysis of Gene Expression Reveals Function of the bZIP Transcription Factor HY5 in the UV-B Response of Arabidopsis. Proc. Natl. Acad. Sci. 101, 1397–1402. doi:10.1073/pnas.0308044100
Vahdati, K., and Lotfi, N. (2013). “Abiotic Stress Tolerance in Plants with Emphasizing on Drought and Salinity Stresses in walnut,” in Abiotic Stress-Plant Responses and Applications in Agriculture. Editors K Vahdati, and C Leslie (Rijeka: InTech), 307–365. doi:10.5772/56078
Voorrips, R. E. (2002). MapChart: Software for the Graphical Presentation of Linkage Maps and QTLs. J. Hered. 93, 77–78. doi:10.1093/jhered/93.1.77
Wan, L., Ren, W., Miao, H., Zhang, J., and Fang, J. (2020). Genome-Wide Identification, Expression, and Association Analysis of the Monosaccharide Transporter (MST) Gene Family in Peanut (Arachis hypogaea L.). 3 Biotech. 10, 1–16. doi:10.1007/s13205-020-2123-8
Wan, L., Wu, Y., Huang, J., Dai, X., Lei, Y., Yan, L., et al. (2014). Identification of ERF Genes in Peanuts and Functional Analysis of AhERF008 and AhERF019 in Abiotic Stress Response. Funct. Integr. Genomics 14, 467–477. doi:10.1007/s10142-014-0381-4
Wang, J., Zhou, J., Zhang, B., Vanitha, J., Ramachandran, S., and Jiang, S.-Y. (2011). Genome-Wide Expansion and Expression Divergence of the Basic Leucine Zipper Transcription Factors in Higher Plants with an Emphasis on SorghumF. J. Integr. Plant Biol. 53, 212–231. doi:10.1111/j.1744-7909.2010.01017.x
Wei, G., Pan, Y., Lei, J., and Zhu, Y.-X. (2005). Molecular Cloning, Phylogenetic Analysis, Expressional Profiling and In Vitro Studies of TINY2 from Arabidopsis thaliana. BMB Rep. 38, 440–446. doi:10.5483/BMBRep.2005.38.4.440
Wei, Y., Chang, Y., Zeng, H., Liu, G., He, C., and Shi, H. (2018). RAV Transcription Factors Are Essential for Disease Resistance against Cassava Bacterial Blight via Activation of Melatonin Biosynthesis Genes. J. Pineal Res. 64, e12454. doi:10.1111/jpi.12454
Woo, H. R., Kim, J. H., Kim, J., Kim, J., Lee, U., Song, I.-J., et al. (2010). The RAV1 Transcription Factor Positively Regulates Leaf Senescence in Arabidopsis. J. Exp. Bot. 61, 3947–3957. doi:10.1093/jxb/erq206
Wuitschick, J. D., Lindstrom, P. R., Meyer, A. E., and Karrer, K. M. (2004). Homing Endonucleases Encoded by Germ Line-Limited Genes in Tetrahymena Thermophila Have APETELA2 DNA Binding Domains. Eukaryot. Cel 3, 685–694. doi:10.1128/EC.3.3.685-694.2004
Xu, Z.-S., Chen, M., Li, L.-C., and Ma, Y.-Z. (2011). Functions and Application of the AP2/ERF Transcription Factor Family in Crop ImprovementF. J. Integr. Plant Biol. 53, 570–585. doi:10.1111/j.1744-7909.2011.01062.x
Yao, Y., He, R. J., Xie, Q. L., Zhao, X. H., Deng, X. M., He, J. B., et al. (2017). Ethylene Response Factor 74 ( ERF74 ) Plays an Essential Role in Controlling a Respiratory Burst Oxidase Homolog D (RbohD)‐dependent Mechanism in Response to Different Stresses in Arabidopsis. NEW PHYTOL. 213, 1667–1681. doi:10.1111/nph.14278
Zhao, K., Li, K., Ning, L., He, J., Ma, X., Li, Z., et al. (2019). Genome-Wide Analysis of the Growth-Regulating Factor Family in Peanut (Arachis hypogaea L.). Int. J. Mol. Sci. 20, 4120. doi:10.3390/ijms20174120
Zhu, Y., Liu, X., Gao, Y., Li, K., and Guo, W. (2021). Transcriptome-Based Identification of AP2/ERF Family Genes and Their Cold-Regulated Expression during the Dormancy Phase Transition of Chinese Cherry Flower Buds. Scientia Horticulturae 275, 109666. doi:10.1016/j.scienta.2020.109666
Zhuang, J., Chen, J.-M., Yao, Q.-H., Xiong, F., Sun, C.-C., Zhou, X.-R., et al. (2011). Discovery and Expression Profile Analysis of AP2/ERF Family Genes from Triticum aestivum. Mol. Biol. Rep. 38, 745–753. doi:10.1007/s11033-010-0162-7
Zhuang, W., Chen, H., Yang, M., Wang, J., Pandey, M. K., Zhang, C., et al. (2019). The Genome of Cultivated Peanut Provides Insight into Legume Karyotypes, Polyploid Evolution and Crop Domestication. Nat. Genet. 51, 865–876. doi:10.1038/s41588-019-0402-2
Glossary
ABRE abscisic acid responsiveness element
Ala alanine
AP2/ERF APETALA2/ethylene response element-binding factor
Asp aspartate
AuxRR auxin responsiveness
ARE anaerobic response regulatory elements
ARF auxin response factors
bHLH basic helix–loop–helix
bZIP basic leucine zipper
Cs Cucumis sativus
DREB dehydration-responsive-element-binding
Elu glutamic acid
GRF growth-regulating factor
GARE gibberellin-responsive element
KDa kilodalton
LTR low-temperature response elements
MST monosaccharide transporter
MEGA Molecular Evolutionary Genetics Analysis
MEME Multiple Expectation Maximization for Motif Elicitation
Mya million years ago
NAC NAM, ATAF, and CUC
NCBI National Center for Biotechnology Information
PEG polyethylene glycol
pI isoelectric point
qRT-PCR quantitative real-time
RAV related to ABI3/VP
SBP SQUAMOSA promoter-binding protein
SMART imple Modular Architecture Research Tool
TFs transcription factors
Val valine
WUN wound.
Keywords: AP2/ERF1, peanut, phylogenetic analysis, evolution analysis, drought stress
Citation: Cui M, Haider M, Chai P, Guo J, Du P, Li H, Dong W, Huang B, Zheng Z, Shi L, Zhang X and Han S (2021) Genome-Wide Identification and Expression Analysis of AP2/ERF Transcription Factor Related to Drought Stress in Cultivated Peanut (Arachis hypogaea L.). Front. Genet. 12:750761. doi: 10.3389/fgene.2021.750761
Received: 31 July 2021; Accepted: 17 September 2021;
Published: 13 October 2021.
Edited by:
Abhishek Bohra, Indian Institute of Pulses Research (ICAR), IndiaReviewed by:
Hui Song, Qingdao Agricultural University, ChinaCheng Qin, Zunyi Vocational and Technical College, China
Copyright © 2021 Cui, Haider, Chai, Guo, Du, Li, Dong, Huang, Zheng, Shi, Zhang and Han. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xinyou Zhang, aGFhc3hpbnlvdUAxNjMuY29t; Suoyi Han, c3VveWlfaGFuQDEyNi5jb20=
 Mengjie Cui
Mengjie Cui Muhammad Salman Haider
Muhammad Salman Haider Pengpei Chai2
Pengpei Chai2 Pei Du
Pei Du Zheng Zheng
Zheng Zheng Suoyi Han
Suoyi Han