- 1Laboratory of Neuroepigenetics, Department of Cellular, Computational and Integrative Biology (CIBIO), University of Trento, Trento, Italy
- 2Laboratory of Neurogenomic Biomarkers, Department of Cellular, Computational and Integrative Biology (CIBIO), University of Trento, Trento, Italy
- 3Section of Hematology, Yale Cancer Center and Department of Internal Medicine, Yale University School of Medicine, New Haven, United States
- 4Laboratory of RNA and Disease Data Science, Department of Cellular, Computational and Integrative Biology (CIBIO), University of Trento, Trento, Italy
- 5Laboratory of Computational Genomics, Area of Neuroscience, International School of Advanced Studies (SISSA), Trieste, Italy
- 6Central RNA Laboratory, Italian Institute of Technology (IIT), Genova, Italy
- 7Fondazione The Microsoft Research - University of Trento Centre for Computational and Systems Biology (COSBI), Rovereto, Italy
CHD8 represents one of the highest confidence genetic risk factors implied in Autism Spectrum Disorders, with most mutations leading to CHD8 haploinsufficiency and the insurgence of specific phenotypes, such as macrocephaly, facial dysmorphisms, intellectual disability, and gastrointestinal complaints. While extensive studies have been conducted on the possible consequences of CHD8 suppression and protein coding RNAs dysregulation during neuronal development, the effects of transcriptional changes of long non-coding RNAs (lncRNAs) remain unclear. In this study, we focused on a peculiar class of natural antisense lncRNAs, SINEUPs, that enhance translation of a target mRNA through the activity of two RNA domains, an embedded transposable element sequence and an antisense region. By looking at dysregulated transcripts following CHD8 knock down (KD), we first identified RAB11B-AS1 as a potential SINEUP RNA for its domain configuration. Then we demonstrated that such lncRNA is able to increase endogenous RAB11B protein amounts without affecting its transcriptional levels. RAB11B has a pivotal role in vesicular trafficking, and mutations on this gene correlate with intellectual disability and microcephaly. Thus, our study discloses an additional layer of molecular regulation which is altered by CHD8 suppression. This represents the first experimental confirmation that naturally occurring SINEUP could be involved in ASD pathogenesis and underscores the importance of dysregulation of functional lncRNAs in neurodevelopment.
Introduction
Autism Spectrum Disorders (ASD) are a heterogeneous group of complex neurodevelopmental conditions characterized by social-communicative deficits as well as repetitive sensory-motor behaviors, appearing during early childhood (American Psychiatric Association, 2013). ASD prevalence is steadily increasing, such that the estimated global prevalence is currently 1 in 68 (Elsabbagh et al., 2012). Adding to complexity, prevalence in males is 4 to 5-fold higher than in females (Baio et al., 2018). Albeit affecting such a significant portion of the world population, the underlying mechanisms of the disease have not yet been elucidated. However, several etiological hypotheses have been proposed, with risk factors ranging from environmental, to epigenetic (Shulha et al., 2012; Ladd-Acosta et al., 2014), to genetic. Consistently with clinical heterogeneity, the genetic architecture of ASD includes variable inheritance patterns, including rare de novo variants, chromosomal alterations, and common inherited variation (De Rubeis and Buxbaum, 2015; de la Torre-Ubieta et al., 2016). To date, more than 1,000 genes have been ranked as potential risk factors for ASD [SFARI Gene (Abrahams et al., 2013)], and it is challenging to determine whether they converge on shared molecular mechanisms.
Major efforts in transcriptomics profiling are proposing a unifying model (Gandal et al., 2018), advancing the hypothesis that convergent molecular abnormalities are identifiable in autistic brains (Voineagu et al., 2011). Aberrant transcription remains a prevalent feature in ASD (Voineagu et al., 2011). Consistently with the strong impact on the transcriptome, many ASD-linked genes are chromatin modifiers and transcriptional regulators. Among them, Chromodomain Helicase DNA-Binding protein 8 (CHD8) is currently one of the highest confidence risk factors (Satterstrom et al., 2020), with de novo haploinsufficiency leading to a genetically defined ASD subtype, characterized by distinctive facial dysmorphisms, macrocephaly (Bernier et al., 2014), mild intellectual disability and postnatal overgrowth (Ostrowski et al., 2019). From a molecular standpoint, CHD8 suppression leads to changes in epigenetic marks, splicing aberrations and broad transcriptional dysregulation, impacting the coding, and non-coding transcriptome, as confirmed in independent RNA-seq studies on human neuronal models (Sugathan et al., 2014; Cotney et al., 2015; Wang et al., 2015; Wilkinson et al., 2015), cerebral organoids (Wang et al., 2017) and mouse models (Durak et al., 2016; Katayama et al., 2016; Gompers et al., 2017; Platt et al., 2017; Jung et al., 2018; Suetterlin et al., 2018). Typically, changes in coding genes expression could be directly linked to cellular pathways and biological functions, such as cell cycle, Wnt signalling, RNA transcriptional regulation and chromatin remodelling, as well as cancer-related genes, other ASD risk genes, and neural development-relevant genes (Sugathan et al., 2014; Cotney et al., 2015; Wang et al., 2015; Wilkinson et al., 2015; Durak et al., 2016; Katayama et al., 2016; Gompers et al., 2017; Platt et al., 2017; Wang et al., 2017; Jung et al., 2018; Suetterlin et al., 2018). Conversely, lncRNAs play a major role in neural functioning, development and brain disorders (van de Vondervoort et al., 2013; Chen and Zhao, 2014; Hosseini et al., 2019) and they were found broadly dysregulated in post-mortem brain samples from ASD patients (Ziats and Rennert, 2013; Parikshak et al., 2016). However, the functional implication of lncRNAs in ASD biology remains vastly unclear. These lines of evidence call for further studies about the roles of lncRNAs in nervous system pathology.
Therefore, we decided to investigate the potential presence of functional lncRNAs among the dysregulated genes in CHD8 suppression Human induced Neural Progenitor Cells (hiNPCs) model, hypothesizing that they may constitute a further layer of molecular regulation in ASD. However, in silico prediction of lncRNAs functionality is intrinsically challenging, as non-coding transcripts only rarely have a modular structure, therefore structure-to-function relationships are not always straightforward (Mattick, 2018). SINEUP is a novel class of functional antisense lncRNA, which can up-regulate protein translation of their target sense mRNAs, without altering their transcription (Zucchelli et al., 2015a; Zucchelli et al., 2015b). First discovered in mouse (Carrieri et al., 2012), where Uchl1-AS was found to up-regulate protein translation of Uchl1 mRNA, SINEUP translational increase is mediated by two functional domains, namely 1) a region overlapping the Translational Initiation Site (TIS), head-to-head antisense to the 5′ end of the target sense mRNA, which confers specificity to the protein coding transcript (binding domain, BD) and 2) a SINEB2 repeat, Alu, MIR transposable element (TE) (Carrieri et al., 2012; Patrucco et al., 2015; Zucchelli et al., 2015b; Schein et al., 2016) on the 3′ end, that mediates the effect on the target mRNA translation (effector domain, ED). Several transcripts with this structure were computationally identified, and their function as SINEUPs successfully confirmed (Carrieri et al., 2012; Schein et al., 2016). Thus, it is possible to confidently hypothesize the function of such lncRNAs based merely on their structure.
In this work, we sought to identify SINEUP-like molecules among the dysregulated transcripts in an ASD-relevant cellular model system, human neural progenitors where CHD8 expression was suppressed by approximately 50% using short hairpin RNAs (shRNAs), mimicking the haploinsufficiency condition (Sugathan et al., 2014). Among the identified candidates, we prioritized the RAB11B-AS1 lncRNA, and provided experimental evidence of its regulatory role on its sense counterpart RAB11B mRNA by means of its SINEUP-specific domains. Our results suggest that ASD transcriptional dysregulation might affect previously unrecognized lncRNAs-mediated networks and underline SINEUP molecules as unacknowledged players in ASD molecular phenotypes.
Materials and Methods
Human Induced Neural Progenitor Cells Culture
hiNPCs from fibroblasts of a control individual, GM8330-8 (Sheridan et al., 2011), were used to generate stable KD lines, where shRNAs targeting CHD8 (sh4, sh2, sh1) or GFP as control (shGFP) were delivered (Sugathan et al., 2014). Cells were cultured on poly-L-ornithine hydrobromide (20 μg/ml, #P3655 Sigma)/laminin (3 μg/ml, #23017015 Life Technologies)–coated plates in hiNPC medium [70% v/v DMEM (Life Technologies) completed with 30% v/v HAM F12 (#ECB7502L, Euroclone), 2% v/v B27 (#17504001, Life Technologies), 1% v/v Penicillin-Streptomycin solution (#15140122, Life Technologies), 1% v/v L-Glutamine (#25-005-CI, Corning), supplemented with EGF (20 ng/ml, #E9644, Sigma), bFGF (20 ng/ml, #233-FB R and D), Heparin (5 μg/ml, #H9267 Sigma)]. Semi-confluent monolayers were maintained in 5% CO2, 37°C humidified incubator.
Identification of Candidate SINEUP
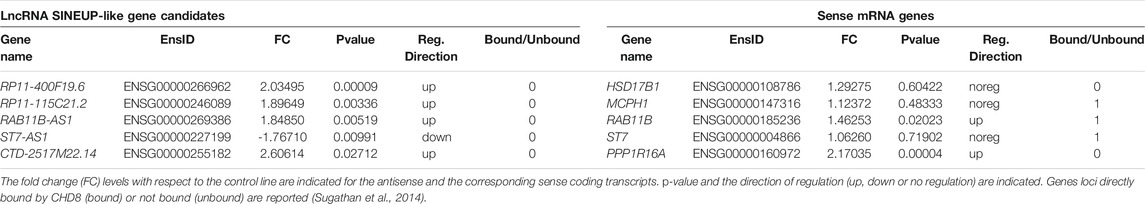
SINEUP-like transcripts were identified among the Differentially Expressed Genes (DEGs) comparing WT and CHD8 KD hiNPCs (GSE61491, GEO, NCBI) (Sugathan et al., 2014) according to a series of filtering steps (Figure 1A): 1) LncRNAs were selected relying on GENCODE v37 lncRNA gene annotation (Frankish et al., 2019); 2) Screening for SINE/Alu or SINE/MIR TEs was performed using Dfam Tool Repeat Masker v3.0 (Smit et al., 1996); 3) Antisense overlapping transcripts to sense coding mRNAs were identified using the BioConductor package GenomicRanges (Lawrence et al., 2013) (minoverlap = 1 L); 4) Transcripts overlapping the respective sense protein coding gene on the first ATG were chosen by comparing the “start codon” position of the sense mRNA to exons “start” and “end”positions of the lncRNAs from GENCODE v37 comprehensive annotation, and confirmed by using Ensembl (Yates et al., 2020) and UCSC Genome Browser (Kent et al., 2002) annotations.
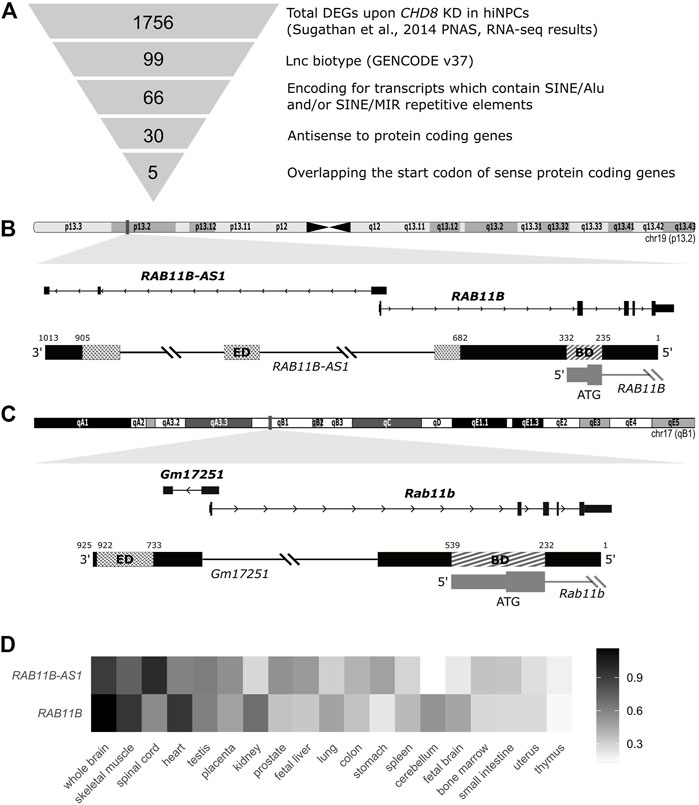
FIGURE 1. Identification of SINEUP-like molecules among deregulated transcripts following CHD8 suppression. (A) Schematic representation of the employed pipeline, with sequential filtering steps for the selection of candidate SINEUP-like molecules. (B) Integrative Genome Viewer (IGV) view of the candidate antisense lncRNA RAB11B-AS1 and the respective sense counterpart coding mRNA RAB11B (Human RefSeq annotation). Putative SINEUP functional domains are highlighted (Binding Domain, BD) (Effector Domain, ED). (C) IGV track for M. musculus RAB11B-AS1/RAB11B transcripts orthologues, respectively, Gm17251 and Rab11b. Putative functional domains are conserved and highlighted (BD) (ED). (D) Heatmap depicts normalized expression levels (2−∆∆Cq) of RAB11B and RAB11B-AS1, illustrating concordant expression across human tissues, with highest levels reported in black and lowest in white.
RNA Extraction and Retrotranscription
Total RNA was extracted using TRIzol (#15596018 Ambion, Life Technologies) following the manufacturer’s instructions. Genomic DNA was removed using DNase I (#AM2222 Ambion, Life Technologies) incubation (0.2–1 µg RNA with 2U of DNaseI) in DNase Buffer with 1U of RNase inhibitor (#AM2684, Ambion, Life Technologies), for 30′, at 37°C. Treated RNA samples were purified with RNeasy Mini Kit (#74104, QIAGEN). Reverse transcription was performed using SensiFAST™ cDNA synthesis kit (#BIO65053, Bioline) with Oligo-dT/random hexamers primers according to manufacturer’s instructions. cDNA diluted 1:10 was used for qPCR. Transcripts relative expression levels in human tissues were determined using Human Total RNA Master Panel (#636643, LOT1409502A, ClonTech). 1 µg was retro-transcribed for each tissue/cell type as previously described, and cDNA was diluted 1:20 for application in qPCR.
Quantitative PCR
Primers for qPCR were designed spanning an exon-exon junction by using the Universal Probe Library Assay Design Center (Roche Life Science, 2019). iTaq™ Universal SYBR® Green Supermix (#1725121, Biorad) was used following manufacturer’s instructions. NONO reference gene (Eisenberg and Levanon, 2013) was used for normalization, and relative expression values were calculated using the 2−∆∆Cq method (Segundo-Val and Sanz-Lozano, 2016). The co-expression pattern of S/AS pairs across Human Total RNA Master Panel was evaluated by plotting the normalized and relativized expression values (2−∆∆Cq) matrix into a heatmap. In this case, for each gene, the ∆∆Cq ratio was calculated with respect to the highest expression value across tissues. Amplicons size and specificity were verified through gel electrophoresis and Sanger sequencing.
Cloning
To clone RAB11B-AS1 we performed gene-specific (GS) retrotranscription from GM8330-8 total RNA, using RevertAid First Strand cDNA Synthesis Kit (#K1622, Thermo Fisher) according to the manufacturer’s instructions. 1 µg of total RNA was retrotranscribed with the GS primer hRAB11B-AS1-GS (5′-TCTTTAGTTCACAGATCTAGTA-3′). Primers to clone RAB11B-AS1 and RAB11B were designed on the transcript 5′ and 3′ ends, and restriction sites were added for ligation. PCR to amplify RAB11B and RAB11B-AS1 was performed using Phusion Green Hot Start II High-Fidelity PCR Master Mix (#F566S, Thermo Fisher). pcDNA™ 3.1 (-) vector (#V795-20, Invitrogen) was the backbone to clone full-length RAB11B-AS1 sequence between EcoRI, HindIII restriction sites [pcDNA 3.1 (-)-hRAB11B-AS1-WT]. Domain-targeted deletion mutants were created by using the Q5 R Site-Directed Mutagenesis kit (#E0554, NEB) according to manufacturer’s instructions. Primers for RAB11B-AS1 mutagenesis were generated using NEBase Changer™ v1.2.9 web tool (NEBaseChanger, 2019). After transformation, positive clones were Sanger sequenced to verify the proper insertion of the sequence and/or effective deletion of target domains.
hiNPCs Electroporation
Roughly 5·106 GM8330-8 hiNPCs were electroporated in 100 µl of electroporation solution (5 mM KCl, 15 mM MgCl2, 10 mM C6H12O6, and 120 mM K2HPO4/KH2PO4 1 M pH 7.2) with 5 µg of plasmid using program A-033 of the Amaxa Nucleofector™ 2b Device (#AAB1001, Lonza). After electroporation, cells were directly resuspended in hiNPCs complete medium and plated on poly-ornithine/laminin coated dishes. Cells were eventually harvested for subsequent analysis after 48 h.
Protein Extraction and Western Blot
Total proteins were extracted in RIPA Buffer (#R0278, Sigma-Aldrich), with Protease Inhibitor (#88266, Thermo Fisher). After sonication (Q700, Qsonica) and centrifugation at 12.000 rpm for 20′ at 4°C, the supernatant was quantified using bicinchoninic acid (BCA) protein quantification (#23225, Thermo Fisher) following the manufacturer’s instructions. 8–15 µg of proteins were run on NuPAGE™ 4–12% Bis-Tris Protein Gel (#NP322, Invitrogen) in MOPS SDS Running Buffer (#NP0001, Novex, Life Technologies) for 2 h, 120 V. Transfer was carried out on Polyvinylidene Fluoride (PVDF) membrane in Tris-Glycine Buffer (#28363, Thermo Fisher) with 5% methanol, at 70 V for 30′ at 4°C. Membranes were blocked with 5% non-fat milk in PBS-Tween (0.1%) at room temperature (RT) and incubated with rabbit anti-RAB11B 1:1000 (#orb30974, Biorbyt or #HPA054396, Atlas Antibodies) or mouse anti-β-tubulin 1:5000 (#sc-53140, Santa Cruz Biotechnology) primary antibodies overnight at 4°C. After three washes with PBS-Tween (0.1%), membranes were incubated with HRP conjugated goat anti-rabbit (#074-1506, KPL) or goat anti-mouse (#5220-0341, KPL) secondary antibodies (1:5000), for 1 h at RT. The membrane was developed with ECL solution (#RPN2235, GE Healthcare or #EMP011005, EuroClone) using BioRad Chemidoc XRS + System. Bands analysis was performed using ImageJ-1.53a.
Statistical Analysis
Statistical analysis tests were performed using R as described in figure captions. Significance level was set to 0.05. Data were plotted using R (ggplot2) and represented as Mean ± Standard Error of the Mean (SEM), as specified in figure legends with sample sizes. The significance level was reported as NS p > 0.05, *p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.001.
Results
Identification of RAB11B-AS1 as a SINEUP-like Transcript Dysregulated Upon CHD8 Suppression
To select functional lncRNAs altered following CHD8 suppression, we applied the selection pipeline schematized in Figure 1A and detailed in Methods. We first resorted to the complete list of dysregulated genes from Sugathan et al., 2014, where 1756 DEGs were reported following CHD8 KD compared to control hiNPCs (shGFP). Selection of natural SINEUP molecules was performed based on their specific structural criteria 1) annotation as lncRNAs, 2) presence of a SINE/Alu and/or SINE/MIR TE, 3) antisense to protein coding genes, 4) overlapping the start codon of coding gene). These sequential filtering steps (Figure 1A and Methods) led to the isolation of five candidate lncRNA genes (Table 1) containing at least one inverted SINE/Alu and/or SINE/MIR repeats and overlapping in antisense orientation to the TIS of their respective sense protein coding mRNA. RAB11B-AS1 (ENSG00000269386) was identified as the most promising candidate, since the structure of the transcript precisely mirrored the one of a canonical SINEUP molecule (Figure 1B). RAB11B-AS1 transcript is the antisense counterpart of a sense, protein coding gene, RAB11B (ENSG00000185236), a small GTPase belonging to the Ras superfamily responsible for vesicle formation, transport, and fusion (Stenmark and Olkkonen, 2001). RAB11B is enriched in the brain (Lai et al., 1994), and it is involved in membrane and vesicle trafficking and apical proteins recycling (Kelly et al., 2012), processes of relevance for brain development and synaptic plasticity (Villarroel‐Campos et al., 2014). The RAB11B S/AS pairs were both significantly up-regulated upon CHD8 suppression in hiNPCs (Table 1). Notably, another previously generated and independently characterized sh-CHD8 suppression model (Cotney et al., 2015) displayed similar RAB11B-AS1 upregulation, although not nominally statistically significant after multiple test correction.
RAB11B-AS1 overlaps in opposite orientation with RAB11B, specifically with 96 nucleotides encompassing the TIS, representing the putative BD. As for the ED, RAB11B-AS1 contains two classes of partially overlapping inverted embedded TE, a FRAM repeat (free right arm monomer) and a SINE/Alu repeat. Because SINE/Alu might arise from dimerization of two different REs (Mighell et al., 1997), the 2 TEs were jointly considered as the potential RAB11B-AS1 ED, a 222 nucleotides long region near the 3′ end of the transcript (Figure 1B). Significantly, an ortholog transcript in M. musculus, Gm17251 (ENSMUSG00000090952), was identified, displaying a high sequence similarity [83% identity score, BLAST (Nucleotide BLAST, 2019)] to the human counterpart (Figure 1C). Equivalently to the human transcript, it possesses the SINEUP-like putative BD, overlapping with the sense Rab11b (ENSMUSG00000077450) on the TIS, and a putative ED consisting of a SINEB2 repeat in inverted configuration (Figure 1C). Because co-expression of the transcripts pair is essential to SINEUP protein translation function, the spatio-temporal co-expression of RAB11B and RAB11B-AS1 S/AS pair was examined. RAB11B and RAB11B-AS1 transcripts levels were quantified across an RNA panel from various human body districts. RAB11B-AS1 showed a fairly ubiquitous distribution, with detectable levels in skeletal muscle, testis and heart and highest expression in the central nervous system (spinal cord, whole brain) (Figure 1D). Importantly, RAB11B-AS1 and RAB11B displayed a concordant expression pattern primarily in whole brain, heart, and skeletal muscle (Figure 1D), thus supporting a possible S/AS functional regulatory mechanism.
Importantly, linear regression analysis performed on publicly available CAGE (Cap Analysis of Gene Expression) data from the FANTOM project (Severin et al., 2014; Lizio et al., 2015; Abugessaisa et al., 2021), derived from 1,886 human samples including primary cultures, tissues, and transformed cells, confirmed a positive significant correlation between RAB11B and RAB11B-AS1 (R = 0.25; p = 3.55E-28), consistent with our results.
RAB11B-AS1 and CHD8 Display Inversely Correlated Expression
In order to validate the transcriptional upregulation initially observed following CHD8 suppression (Sugathan et al., 2014) (Table 1), RAB11B/RAB11B-AS1 S/AS pair was quantified by qPCR in independent biological replicates of CHD8 KD hiNPCs. Conforming to initial RNA-seq results (Table 1), RAB11B exhibited a mild upregulation (p-value = 0.06), while RAB11B-AS1 dysregulation was more robust and significant in CHD8-suppressed lines (Figure 2A). We further calculated a linear regression analysis to appreciate possible correlation between the levels of CHD8 KD and the expression of the S/AS pair. By resorting to the initial logCPM from the hiNPCs models with CHD8 suppression (Sugathan et al., 2014), we uncovered a significant anti-correlation between RAB11B-AS1 and CHD8, while RAB11B correspondence was milder (Figure 2B).
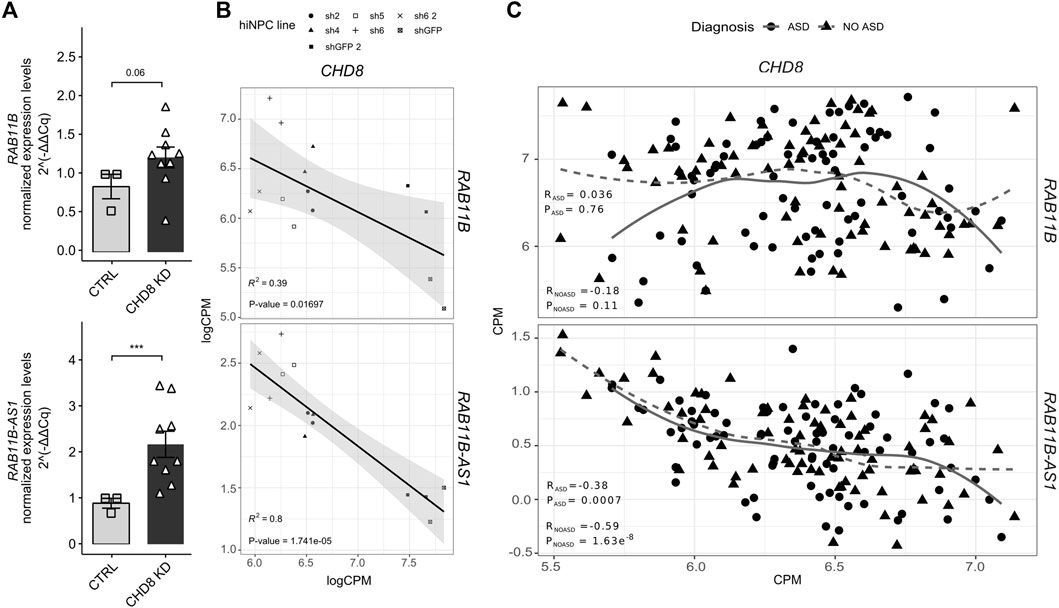
FIGURE 2. RAB11B-AS1 and CHD8 display inversely correlated expression. (A) The barplots report RAB11B (top) and RAB11B-AS1 (bottom) transcripts quantification (2−∆∆Cq), increasing upon CHD8 KD (average of sh4, sh2, sh1) with respect to control (shGFP line). Data are plotted as Mean ± SEM. For statistical analysis, un-paired one-tail Student’s t-test was performed between control and CHD8 KD lines. N = 3 for each line. ***p ≤ 0.001. (B) Dot plots with linear regression line show RAB11B (top) and RAB11B-AS1 (bottom) transcripts abundance (logCPM) with respect to CHD8 levels in hiNPCs with suppressed levels of CHD8 (Sugathan et al., 2014). Linear regression analyses unveiled a significant inverse correlation for RAB11B (R2 = 0.39, p = 0.01697) and particularly for RAB11B-AS1 (R2 = 0.8, p = 1.741e-05) compared to CHD8. (C) Dot plots display the abundance (CPM) of RAB11B (top) and RAB11B-AS1 (bottom) compared to CHD8 from RNAseq analyses of peripheral blood samples from ASD patients (ASD diagnosis) and healthy siblings (NO ASD) in the ITAN family cohort. RAB11B-AS1 transcript abundance results in significative inverse correlation relationship with CHD8 both in the ASD patients group (R = −0.38, p = 0.0007) and in the healthy family members, i.e., the control group (R = –0.59, p = 1.63e-08).
Next, to further dissect the expression crosstalk between CHD8 and RAB11B/RAB11B-AS1 S/AS pair we resorted to blood transcriptomic data of the Italian Autism Network (ITAN) (Muglia et al., 2018). RNA-seq data derived from peripheral blood samples of ASD and unaffected siblings (Filosi et al., 2020) were tested. While a modest (RASD = 0.036; RNO ASD = −0.18) anti-correlation between RAB11B and CHD8 was observed (Figure 2C, top), a significant inverse correlation between RAB11B-AS1 and CHD8 expression levels was found in both ASD and control siblings (Figure 2C, bottom). Altogether, these results suggest a possible functional suppression mechanism by CHD8 on the RAB11B/RAB11B-AS1 locus, which might be impaired in CHD8 haploinsufficiency conditions.
RAB11B-AS1 Over-Expression is Able to Enhance RAB11B Translation With No Transcriptional Alteration
Because a measurable effect of a functional SINEUP molecule is the increase in translation of its sense counterpart and considering the over-expression of RAB11B-AS in the CHD8 suppression lines, we predicted increased levels for RAB11B protein. After Western Blot quantification, densitometric analysis of the 24 kDa bands corresponding to RAB11B highlighted a significant increase in protein levels upon CHD8 suppression with respect to the control condition (Figures 3A,B). While RAB11B upregulation was solid and reproducible in CHD8-Sh2 and CHD8-Sh1, displaying roughly 50% of CHD8 KD, the data on the third KD line (CHD8-Sh4) seem to be more variable.
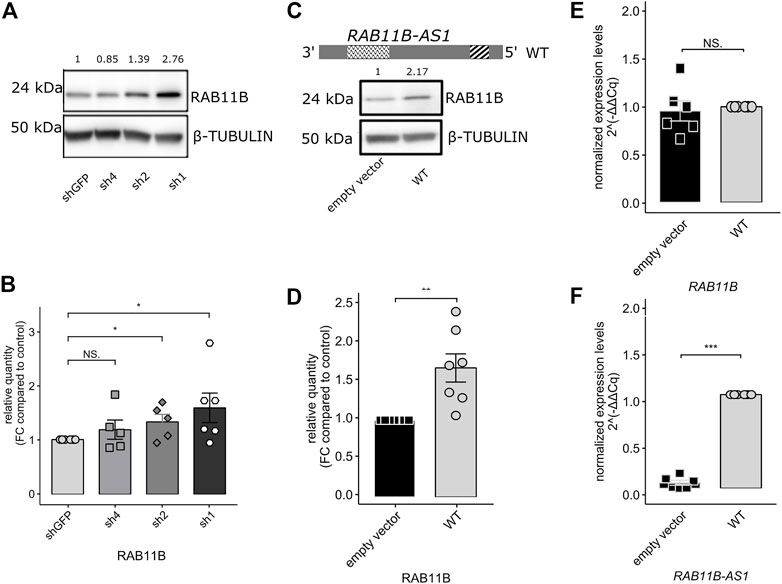
FIGURE 3. RAB11B-AS1 (WT) over-expression leads to RAB11B protein stabilization without affecting its mRNA levels. (A) Representative Western Blot shows RAB11B levels increase in CHD8 KD lines (sh4, sh2, sh1). β-TUBULIN was used as loading control. (B) The barplot reports the densitometric analysis of WB experiments. RAB11B protein levels increase in CHD8 KD lines (significance in sh2 and sh1 lines). Data are plotted as Mean ± SEM. For statistical analysis, un-paired one-tailed Student’s t-test was performed. N = 5–6. NS p > 0.05, *p ≤ 0.05. (C) The RAB11B-AS1 (WT) sequence schematic reports the location of the putative SINEUP domains (up). Representative Western Blot image (down) depicts RAB11B protein increase upon RAB11B-AS1 (WT) over-expression in GM8330-8 hiNPC line. β-TUBULIN was used as loading control. (D) Barplot of RAB11B densitometric analysis, confirming that protein increase was statistically significant upon antisense lncRNA over-expression. (E) The barplot reports RAB11B transcriptional levels, tested by qPCR analysis and expressed as 2−∆∆Cq. mRNA levels were stable upon RAB11B-AS1 over-expression (black) with respect to the control empty vector (white). (F) RAB11B-AS1 was abundantly and significantly overexpressed upon plasmid construct delivery (black) with respect to the control (white). Data are plotted as Mean ± SEM of N = 6 biological replicates. For statistical analysis, un-paired one-tailed Student’s t-test was performed. Sample size N = 6. NS p > 0.05, **p ≤ 0.01, ***p ≤ 0.001.
Finally, to functionally characterize RAB11B-AS1 as a SINEUP molecule, the full-length (WT) sequence of the transcript (Figure 3C, top) was cloned and over-expressed in GM8330-8 hiNPC parental line. As a result of RAB11B-AS1 over-expression, RAB11B protein level was increased by approximately two times compared to control (Figures 3C,D). RAB11B protein up-regulation was significant and reproducible, as confirmed by statistical analysis of replicate experiments (Figure 3D). Furthermore, qPCR experiments confirmed that, despite the RAB11B protein increase, RAB11B transcriptional levels were substantially stable (Figure 3E) while RAB11B-AS1 was abundantly over-expressed (Figure 3F). These results strongly support our initial hypothesis, as they are coherent with the functional mechanism of a SINEUP molecule.
RAB11B Translational Increase is Dependent on the Presence of RAB11B-AS1 SINEUP Functional Domains
In order to fully prove the SINEUP nature of RAB11B-AS1 lncRNA, we wanted to test whether the absence of one of the putative functional domains might impair the SINEUP-like mechanism. To this purpose, two domain-specific deletion mutants were generated by site-specific mutagenesis (∆BD or ∆ED RAB11B-AS1). RAB11B-AS1 WT, ∆BD or ∆ED were then delivered in parental GM8330-8 hiNPCs, and subsequently Western Blot experiments were performed to quantify RAB11B protein level. While the over-expression of WT, full-length RAB11B-AS1 elicited the expected increase in RAB11B protein, ∆BD, and ∆ED mutants failed to evoke RAB11B protein upregulation, in line with the anticipated SINEUP activity (Figure 4A). Such observation was confirmed by densitometric analysis on replicated experiments (n = 4–6) (Figure 4B). Importantly, qPCR revealed that RAB11B transcriptional levels were stable (Figure 4C) while RAB11B-AS1 WT and deletion mutants were significantly and strongly over-expressed (Figure 4D). These results suggest that RAB11B protein translation increase is mediated by its antisense transcript RAB11B-AS1 functional domains. Taken together, these results are reinforcing the hypothesis that RAB11B-AS1 lncRNA is a CHD8-suppression-sensitive SINEUP molecule, able to up-regulate protein translation of its target mRNA RAB11B and potentially relevant for CHD8 haploinsufficiency defined ASD.
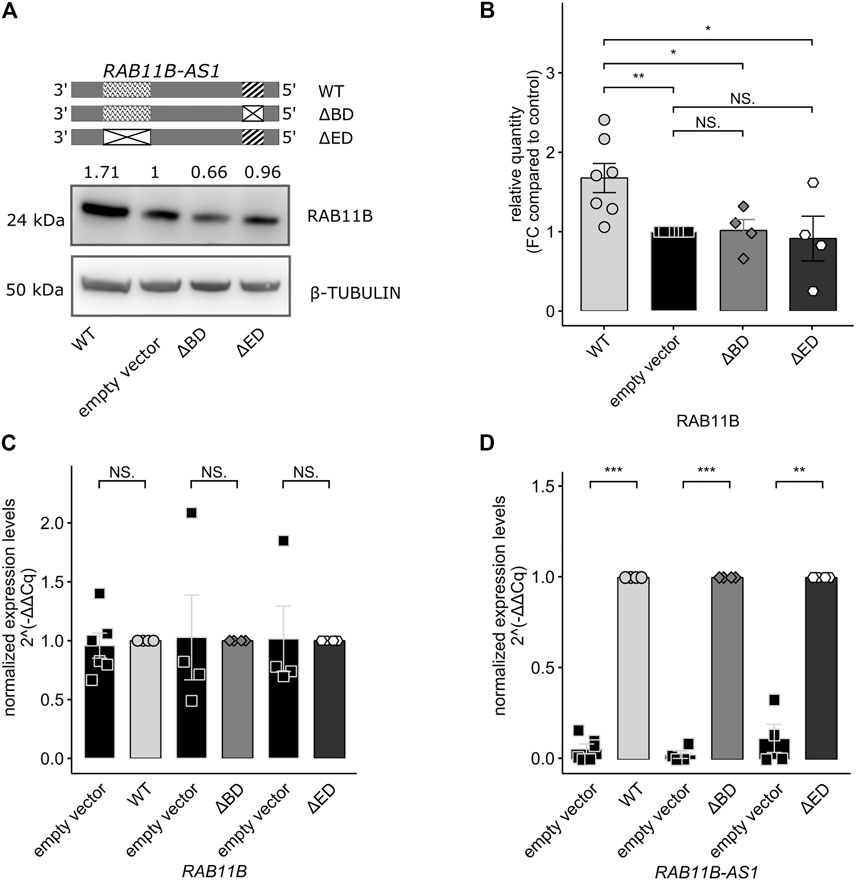
FIGURE 4. Deletion of functional SINEUP domains (BD, ED) impairs RAB11B-AS1 effect on RAB11B protein translation. (A) The RAB11B-AS1 (WT, ∆BD, and ∆ED) sequence schematics report the location of the putative SINEUP domains (up). Representative image describes a Western Blot of RAB11B after RAB11B-AS1 WT, ∆BD, and ∆ED over-expression. β-TUBULIN was used as loading control. (B) Barplot of RAB11B densitometric analysis, confirming that protein increase was statistically significant upon over-expression of RAB11B-AS1 (WT), while deletion mutants over-expression caused no protein expression difference from the control empty vector. (C) The barplot shows RAB11B transcriptional levels, tested by qPCR and expressed as 2−∆∆Cq. mRNA levels were stable upon plasmid constructs over-expression (WT = black, ∆BD = light grey, ∆ED = dark grey) with respect to the control empty vector (white). (D) RAB11B-AS1 was abundantly and significantly overexpressed upon plasmid delivery (WT = black, ∆BD = light grey, ∆ED = dark grey) with respect to the control (white). Data are plotted as Mean ± SEM, N = 4 for deletion mutants, N = 6 for WT over-expression. For statistical analysis, un-paired one-tailed Student’s t-test was performed. NS p > 0.05, *p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.001.
Discussion
NcRNAs constitute the major product of mammalian transcription (The FANTOM Consortium, 2005), however their functions are still largely unexplored. Hinting at their possible role in higher cognition, lncRNAs are primarily expressed in the brain (Mercer et al., 2009), with definite patterns across cerebral areas, and several of them exclusively described in primates (Mattick, 2018). Increasing evidence underscores their role in neuronal physiology and pathology. In fact, lncRNAs have been implicated in neural development and functioning (Andersen and Lim, 2018), brain aging and neurodegeneration (Wan et al., 2017), but also neurodevelopmental disorders, such as ASD.
In this study, we sought to characterize the effects of transcriptional dysregulation of lncRNAs in a model system of neuronal development and relevant to ASD. Specifically, we resorted to hiNPCs, where CHD8 expression was reduced by short-hairpins administration to roughly 50%. CHD8 protein haploinsufficiency represents one of the highest confidence risk factors for ASD with profound consequences for the whole transcriptome. In this work, we identified SINEUP-like antisense lncRNAs among the pool of dysregulated genes following CHD8 suppression (Sugathan et al., 2014). SINEUP ncRNAs are a class of regulatory, antisense modular transcripts, which increase protein translation of their sense mRNA by means of their characteristic functional domains (Zucchelli et al., 2015a; Zucchelli et al., 2015b). Thus, we filtered, relying merely on structural features, the 1756 DEGs upon CHD8 suppression (Sugathan et al., 2014). Only an exiguous list of candidates met our stringent criteria. However, we cannot exclude that the number of dysregulated SINEUP molecules might be underestimated in our study. In fact, due to the poly-A mRNA enrichment protocol used for library preparation (Sugathan et al., 2014), a large portion of poly-A-minus lncRNAs (Mattick, 2018), and possibly also non-polyadenylated SINEUP, might have been missed. A total of 5 SINEUP-like molecules have been identified (Table 1), however, we prioritized for further functional validation RAB11B-AS1, which displayed the structural organization more typically associated with natural SINEUP. Importantly, a murine ortholog of RAB11B-AS1 was identified, with an inverted SINEB2 TE. LncRNAs containing embedded TEs are more conserved across species with respect to non-TE-derived sequences, and display significantly less variance (Kapusta et al., 2013), sustaining the hypothesis that TEs in lncRNAs are subject to functional and/or structural constraints during evolution. Previous reports in osteosarcoma, lung, and breast cancer development described different, discrepant modes of RAB11B-AS1 regulation on RAB11B mRNA and protein levels: downregulation (Chen et al., 2018), upregulation (Li et al., 2020) or no effect (Niu et al., 2020) of the sense transcript was observed, generating an inconclusive scenario. Notably, RAB11B–the head-to-head protein coding transcript, overlapping RAB11B-AS1–has critical roles in apical recycling of cargo proteins (Delisle et al., 2009; Silvis et al., 2009; Best et al., 2011; Butterworth et al., 2012). Moreover, it was reported to inhibit Ca2+-triggered exocytosis in neuronal and neuroendocrine cells, and to be enriched in purified synaptic vesicles (Khvotchev et al., 2003). Importantly, RAB11B de novo mutations were correlated with Intellectual Disability and microcephaly (Lamers et al., 2017). Thus, these observations globally support a role for RAB11B–and possibly its overlapping lncRNA–in vesicular trafficking and synaptic activity, of relevance for ASD and other neurological conditions. Firstly, we validated the upregulation of RAB11B/RAB11B-AS1 transcripts pair following CHD8 suppression by qPCR. Secondly, we confirmed a comparable expression pattern between RAB11B-AS1/RAB11B across human body districts and CAGE data, coherently with previous observations reporting a similar spatio-temporal distribution of S/AS pairs (Chen et al., 2005). Furthermore, we uncovered an anti-correlation between CHD8 and RAB11B-AS1 in ASD-affected and healthy siblings of the ITAN cohort. Thus, aberrantly reduced expression of CHD8 seems to correlate with RAB11B-AS1 upregulation. However, in our hiNPCs model transcriptomic data, both RAB11B-AS1 and RAB11B appear to be upregulated, although with different strength and significance. While further studies will be needed to fully dissect this interplay, the observed upregulation of the sense RAB11B transcript might be directly mediated by CHD8 protein, since CHD8 binding sites were identified on RAB11B, but not on RAB11B-AS1 promoter (Sugathan et al., 2014).
Finally, we moved to the functional characterization of RAB11B-AS1 as a potential new SINEUP molecule. To this task, we cloned and overexpressed the full-length human lncRNA transcript. Over-expression of RAB11B-AS1 did not affect RAB11B transcriptional levels but led to a reproducible increase in the production of RAB11B protein. This post-transcriptional effect is consistent with a SINEUP role, as translation is typically expected to increase in the range of 1.5–3 fold (Zucchelli et al., 2015a). To further strengthen our results, we created deletion mutants of RAB11B-AS1, removing the SINEUP functional domains (BD and ED). Consistently with our hypothesis, the mutant forms of the transcript failed to exert a regulatory effect on RAB11B mRNA translation. Thus, here we propose that RAB11B-AS1 SINEUP molecule potentially represents a further indirect layer of protein translation regulation, independent of RAB11B transcriptional control. This finding seems to be discordant with previous studies (Chen et al., 2018; Li et al., 2020; Niu et al., 2020), however, AS-lncRNAs have been previously reported to have dual functions, and this could depend on the cellular context and availability of specific co-factors. To this point, Uxt-AS1, initially found to act as a SINEUP by upregulating protein translation of its sense counterpart Uxt (Carrieri et al., 2012), in a later study was, instead, found to regulate alternative splicing of UXT in human colonic carcinoma cell lines (Yin et al., 2017). Thus, alternative roles for some lncRNAs could be described when using different cell lines or other cellular contexts or tissues. This could suggest that expression of different mediators could drive different functional effects of specific AS-lncRNA on their sense counterparts.
In conclusion, we provided evidence that naturally occurring SINEUP could be involved in ASD pathogenesis, highlighting the importance of dysregulation of functional lncRNAs during brain development.
Data Availability Statement
Publicly available datasets were analyzed in this study. This data can be found here: http://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1405266111/-/DCSupplemental/pnas.1405266111.sd01.xlsx, “Sugathan A, Biagioli M, Golzio C, Erdin S et al. CHD8 regulates neurodevelopmental pathways associated with autism spectrum disorder in neural progenitors. Proc Natl Acad Sci U S A 2014 Oct 21;111 (42):E4468-77. PMID: 25294932”, GSE61491, GEO, NCBI; CAGE data were retrieved from ZENBU Severin J, Lizio M, Harshbarger J, Kawaji H, Daub CO, Hayashizaki Y; FANTOM Consortium, Bertin N, Forrest AR. Interactive visualization and analysis of large-scale sequencing datasets using ZENBU. Nat Biotechnol. 2014 Mar;32 (3):217-9. doi: 10.1038/nbt.2840. PMID: 24727769.
Ethics Statement
The studies involving human participants were reviewed and approved by the Verona Hospital Ethical Review board (study protocol AUT-SFK001, CE1419) which approved the study protocol in first instance, followed by the Ethical Review Committees of each recruiting site for the ITAN collection. Written informed consent to participate in this study was provided by the participants’ legal guardian/next of kin.
Author Contributions
MB, FD, GZ conceptualized and designed the study. GZ identified the candidate lncRNAs under TT advice and supervision, and cloned the candidate lncRNA. GZ, FD performed lncRNA characterization experiments. AB performed protein quantification experiments. GZ, MA performed the overexpression experiments, transcript, and protein quantification experiments. MB, FD directed experimental data collection. GZ, MA performed statistical analysis of experimental results. MF performed ITAN transcriptomic data analyses and linear regression, and provided critical advice on statistical analyses. ED supervised ITAN data analyses. GS performed linear regression analyses of CAGE data. RS, SG supervised CAGE data analyses. GZ, MA, FD, MB wrote the article. MB, ED, SG supervised the project. All authors revised, read and approved the submitted version.
Funding
This work was supported by Department CIBIO Institutional funding to MB.
Conflict of Interest
SG declares competing financial interests as cofounder and member of TranSINE Therapeutics.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
The authors wish to thank Nythia Arumugam, former Master student in the lab for initial contribution to the identification of RAB11B-AS1. We are grateful to the members of the NeuroEpigenetics team for stimulating discussion. We thank Dr. Haggarthy and Dr. Talkowski (Massachusetts General Hospital and Harvard Medical School, Boston, United States ) for providing the stable hiNPCs cell lines for CHD8 haploinsufficiency modelling (GM8330-8, shGFP, sh1, sh2, sh4). We thank the ITAN–Italian Autism Network Foundation for RNAseq data availability.
References
Abrahams, B. S., Arking, D. E., Campbell, D. B., Mefford, H. C., Morrow, E. M., Weiss, L. A., et al. (2013). SFARI Gene 2.0: a Community-Driven Knowledgebase for the Autism Spectrum Disorders (ASDs). Mol. Autism 4, 36. doi:10.1186/2040-2392-4-36
Abugessaisa, I., Ramilowski, J. A., Lizio, M., Severin, J., Hasegawa, A., Harshbarger, J., et al. (2021). FANTOM Enters 20th Year: Expansion of Transcriptomic Atlases and Functional Annotation of Non-coding RNAs. Nucleic Acids Res. 49, D892–D898. doi:10.1093/nar/gkaa1054
American Psychiatric Association (2013). Diagnostic and Statistical Manual of Mental Disorders. Fifth Edition. Washington,U.S: American Psychiatric Association. doi:10.1176/appi.books.9780890425596
Andersen, R. E., and Lim, D. A. (2018). Forging Our Understanding of lncRNAs in the Brain. Cell Tissue Res. 371, 55–71. doi:10.1007/s00441-017-2711-z
Baio, J., Wiggins, L., Christensen, D. L., Maenner, M. J., Daniels, J., Warren, Z., et al. (2018). Prevalence of Autism Spectrum Disorder Among Children Aged 8 Years - Autism and Developmental Disabilities Monitoring Network, 11 Sites, United States, 2014. MMWR Surveill. Summ. 67, 1–23. doi:10.15585/mmwr.ss6706a1
Bernier, R., Golzio, C., Xiong, B., Stessman, H. A., Coe, B. P., Penn, O., et al. (2014). Disruptive CHD8 Mutations Define a Subtype of Autism Early in Development. Cell 158, 263–276. doi:10.1016/j.cell.2014.06.017
Best, J. M., Foell, J. D., Buss, C. R., Delisle, B. P., Balijepalli, R. C., January, C. T., et al. (2011). Small GTPase Rab11b Regulates Degradation of Surface Membrane L-type Cav1.2 Channels. Am. J. Physiol. Cell Physiol. 300, C1023–C1033. doi:10.1152/ajpcell.00288.2010
Butterworth, M. B., Edinger, R. S., Silvis, M. R., Gallo, L. I., Liang, X., Apodaca, G., et al. (2012). Rab11b Regulates the Trafficking and Recycling of the Epithelial Sodium Channel (ENaC). Am. J. Physiol. Renal Physiol. 302, F581–F590. doi:10.1152/ajprenal.00304.2011
Carrieri, C., Cimatti, L., Biagioli, M., Beugnet, A., Zucchelli, S., Fedele, S., et al. (2012). Long Non-coding Antisense RNA Controls Uchl1 Translation through an Embedded SINEB2 Repeat. Nature 491, 454–457. doi:10.1038/nature11508
Chen, L.-L., and Zhao, J. C. (2014). Functional Analysis of Long Noncoding RNAs in Development and Disease. Adv. Exp. Med. Biol. 825, 129–158. doi:10.1007/978-1-4939-1221-6_4
Chen, J., Sun, M., Hurst, L., Carmichael, G., and Rowley, J. (2005). Genome-wide Analysis of Coordinate Expression and Evolution of Human Encoded Sense-Antisense Transcripts. Trends Genet. 21, 326–329. doi:10.1016/j.tig.2005.04.006
Chen, Z., Liu, Z., Yang, Y., Zhu, Z., Liang, R., Huang, B., et al. (2018). Long Non-coding RNA RAB11B-AS1 Prevents Osteosarcoma Development and Progression via its Natural Antisense Transcript RAB11B. Oncotarget 9, 26770–26786. doi:10.18632/oncotarget.24247
Cotney, J., Muhle, R. A., Sanders, S. J., Liu, L., Willsey, A. J., Niu, W., et al. (2015). The Autism-Associated Chromatin Modifier CHD8 Regulates Other Autism Risk Genes during Human Neurodevelopment. Nat. Commun. 6, 1–11. doi:10.1038/ncomms7404
de la Torre-Ubieta, L., Won, H., Stein, J. L., and Geschwind, D. H. (2016). Advancing the Understanding of Autism Disease Mechanisms through Genetics. Nat. Med. 22, 345–361. doi:10.1038/nm.4071
De Rubeis, S., and Buxbaum, J. D. (2015). Genetics and Genomics of Autism Spectrum Disorder: Embracing Complexity. Hum. Mol. Genet. 24, R24–R31. doi:10.1093/hmg/ddv273
Delisle, B. P., Underkofler, H. A. S., Moungey, B. M., Slind, J. K., Kilby, J. A., Best, J. M., et al. (2009). Small GTPase Determinants for the Golgi Processing and Plasmalemmal Expression of Human Ether-A-Go-Go Related (hERG) K+ Channels. J. Biol. Chem. 284, 2844–2853. doi:10.1074/jbc.M807289200
Durak, O., Gao, F., Kaeser-Woo, Y. J., Rueda, R., Martorell, A. J., Nott, A., et al. (2016). Chd8 Mediates Cortical Neurogenesis via Transcriptional Regulation of Cell Cycle and Wnt Signaling. Nat. Neurosci. 19, 1477–1488. doi:10.1038/nn.4400
Eisenberg, E., and Levanon, E. Y. (2013). Human Housekeeping Genes, Revisited. Trends Genet. 29, 569–574. doi:10.1016/j.tig.2013.05.010
Elsabbagh, M., Divan, G., Koh, Y.-J., Kim, Y. S., Kauchali, S., Marcín, C., et al. (2012). Global Prevalence of Autism and Other Pervasive Developmental Disorders. Autism Res. 5, 160–179. doi:10.1002/aur.239
Filosi, M., Kam-Thong, T., Kam-Thong, T., Essioux, L., Muglia, P., Trabetti, E., et al. (2020). Transcriptome Signatures from Discordant Sibling Pairs Reveal Changes in Peripheral Blood Immune Cell Composition in Autism Spectrum Disorder. Transl. Psychiatry 10, 106. doi:10.1038/s41398-020-0778-x
Frankish, A., Diekhans, M., Ferreira, A.-M., Johnson, R., Jungreis, I., Loveland, J., et al. (2019). GENCODE Reference Annotation for the Human and Mouse Genomes. Nucleic Acids Res. 47, D766–D773. doi:10.1093/nar/gky955
Gandal, M. J., Zhang, P., Hadjimichael, E., Walker, R. L., Chen, C., Liu, S., et al. (2018). Transcriptome-wide Isoform-Level Dysregulation in ASD, Schizophrenia, and Bipolar Disorder. Science 362, eaat8127. doi:10.1126/science.aat8127
Gompers, A. L., Su-Feher, L., Ellegood, J., Copping, N. A., Riyadh, M. A., Stradleigh, T. W., et al. (2017). Germline Chd8 Haploinsufficiency Alters Brain Development in Mouse. Nat. Neurosci. 20, 1062–1073. doi:10.1038/nn.4592
Hosseini, E., Bagheri-Hosseinabadi, Z., De Toma, I., Jafarisani, M., and Sadeghi, I. (2019). The Importance of Long Non-coding RNAs in Neuropsychiatric Disorders. Mol. Aspects Med. 70, 127–140. doi:10.1016/j.mam.2019.07.004
Jung, H., Park, H., Choi, Y., Kang, H., Lee, E., Kweon, H., et al. (2018). Sexually Dimorphic Behavior, Neuronal Activity, and Gene Expression in Chd8-Mutant Mice. Nat. Neurosci. 21, 1218–1228. doi:10.1038/s41593-018-0208-z
Kapusta, A., Kronenberg, Z., Lynch, V. J., Zhuo, X., Ramsay, L., Bourque, G., et al. (2013). Transposable Elements Are Major Contributors to the Origin, Diversification, and Regulation of Vertebrate Long Noncoding RNAs. Plos Genet. 9, e1003470. doi:10.1371/journal.pgen.1003470
Katayama, Y., Nishiyama, M., Shoji, H., Ohkawa, Y., Kawamura, A., Sato, T., et al. (2016). CHD8 Haploinsufficiency Results in Autistic-like Phenotypes in Mice. Nature 537, 675–679. doi:10.1038/nature19357
Kelly, E. E., Horgan, C. P., and McCaffrey, M. W. (2012). Rab11 Proteins in Health and Disease. Biochem. Soc. Trans. 40, 1360–1367. doi:10.1042/BST20120157
Kent, W. J., Sugnet, C. W., Furey, T. S., Roskin, K. M., Pringle, T. H., Zahler, A. M., et al. (2002). The Human Genome Browser at UCSC. Genome Res. 12, 996–1006. doi:10.1101/gr.229102
Khvotchev, M. V., Ren, M., Takamori, S., Jahn, R., and Südhof, T. C. (2003). Divergent Functions of Neuronal Rab11b in Ca2+-Regulated versus Constitutive Exocytosis. J. Neurosci. 23, 10531–10539. doi:10.1523/jneurosci.23-33-10531.2003
Ladd-Acosta, C., Hansen, K. D., Briem, E., Fallin, M. D., Kaufmann, W. E., and Feinberg, A. P. (2014). Common DNA Methylation Alterations in Multiple Brain Regions in Autism. Mol. Psychiatry 19, 862–871. doi:10.1038/mp.2013.114
Lai, F., Stubbs, L., and Artzt, K. (1994). Molecular Analysis of Mouse Rab11b: a New Type of Mammalian YPT/Rab Protein. Genomics 22, 610–616. doi:10.1006/geno.1994.1434
Lamers, I. J. C., Reijnders, M. R. F., Venselaar, H., Kraus, A., Jansen, S., de Vries, B. B. A., et al. (2017). Recurrent De Novo Mutations Disturbing the GTP/GDP Binding Pocket of RAB11B Cause Intellectual Disability and a Distinctive Brain Phenotype. Am. J. Hum. Genet. 101, 824–832. doi:10.1016/j.ajhg.2017.09.015
Lawrence, M., Huber, W., Pagès, H., Aboyoun, P., Carlson, M., Gentleman, R., et al. (2013). Software for Computing and Annotating Genomic Ranges. PLOS Comput. Biol. 9, e1003118. doi:10.1371/journal.pcbi.1003118
Li, T., Wu, D., Liu, Q., Wang, D., Chen, J., Zhao, H., et al. (2020). Upregulation of Long Noncoding RNA RAB11B-AS1 Promotes Tumor Metastasis and Predicts Poor Prognosis in Lung Cancer. Ann. Transl. Med. 8, 582. doi:10.21037/atm.2020.04.52
Lizio, M., Harshbarger, J., Harshbarger, J., Shimoji, H., Severin, J., Kasukawa, T., et al. (2015). Gateways to the FANTOM5 Promoter Level Mammalian Expression Atlas. Genome Biol. 16, 22. doi:10.1186/s13059-014-0560-6
Mattick, J. S. (2018). The State of Long Non-coding RNA Biology. ncRNA 4, 17. doi:10.3390/ncrna4030017
Mercer, T. R., Dinger, M. E., and Mattick, J. S. (2009). Long Non-coding RNAs: Insights into Functions. Nat. Rev. Genet. 10, 155–159. doi:10.1038/nrg2521
Mighell, A. J., Markham, A. F., and Robinson, P. A. (1997). Alusequences. FEBS Lett. 417, 1–5. doi:10.1016/S0014-5793(97)01259-3
Muglia, P., Filosi, M., Filosi, M., Da Ros, L., Kam-Thong, T., Nardocci, F., et al. (2018). The Italian Autism Network (ITAN): a Resource for Molecular Genetics and Biomarker Investigations. BMC Psychiatry 18, 369. doi:10.1186/s12888-018-1937-y
NEBaseChanger (2019). NEBaseChanger. Available at: https://nebasechanger.neb.com/(Accessed November 19, 2019).
Niu, Y., Bao, L., Chen, Y., Wang, C., Luo, M., Zhang, B., et al. (2020). HIF2-Induced Long Noncoding RNA RAB11B-AS1 Promotes Hypoxia-Mediated Angiogenesis and Breast Cancer Metastasis. Cancer Res. 80, 964–975. doi:10.1158/0008-5472.CAN-19-1532
Nucleotide BLAST (2019). Nucleotide BLAST. Available at: https://blast.ncbi.nlm.nih.gov/Blast.cgi?PROGRAM=blastn&PAGE_TYPE=BlastSearch&LINK_LOC=blasthome (Accessed November 19, 2019).
Ostrowski, P. J., Zachariou, A., Loveday, C., Beleza‐Meireles, A., Bertoli, M., Dean, J., et al. (2019). The CHD8 Overgrowth Syndrome: A Detailed Evaluation of an Emerging Overgrowth Phenotype in 27 Patients. Am. J. Med. Genet. 181, 557–564. doi:10.1002/ajmg.c.31749
Parikshak, N. N., Swarup, V., Belgard, T. G., Irimia, M., Ramaswami, G., Gandal, M. J., et al. (2016). Genome-wide Changes in lncRNA, Splicing, and Regional Gene Expression Patterns in Autism. Nature 540, 423–427. doi:10.1038/nature20612
Patrucco, L., Chiesa, A., Soluri, M. F., Fasolo, F., Takahashi, H., Carninci, P., et al. (2015). Engineering Mammalian Cell Factories with SINEUP Noncoding RNAs to Improve Translation of Secreted Proteins. Gene 569, 287–293. doi:10.1016/j.gene.2015.05.070
Platt, R. J., Zhou, Y., Slaymaker, I. M., Shetty, A. S., Weisbach, N. R., Kim, J.-A., et al. (2017). Chd8 Mutation Leads to Autistic-like Behaviors and Impaired Striatal Circuits. Cel Rep. 19, 335–350.‘ doi:10.1016/j.celrep.2017.03.052
Roche Life Science (2019). Probes and The Universal ProbeLibrary. Available at: https://lifescience.roche.com/en_it/brands/universal-probe-library.html#assay-design-center (Accessed November 19, 2019).
Satterstrom, F. K., Kosmicki, J. A., Wang, J., Breen, M. S., De Rubeis, S., An, J. Y., et al. (2020). Large-Scale Exome Sequencing Study Implicates Both Developmental and Functional Changes in the Neurobiology of Autism. Cell 180, 568–e23. doi:10.1016/j.cell.2019.12.036
Schein, A., Zucchelli, S., Kauppinen, S., Gustincich, S., and Carninci, P. (2016). Identification of Antisense Long Noncoding RNAs that Function as SINEUPs in Human Cells. Sci. Rep. 6, 33605. doi:10.1038/srep33605
San Segundo-Val, I., and Sanz-Lozano, C. S. (2016). Introduction to the Gene Expression Analysis. Methods Mol. Biol. Clifton NJ 1434, 29–43. doi:10.1007/978-1-4939-3652-6_3
Severin, J., Lizio, M., Lizio, M., Harshbarger, J., Kawaji, H., Daub, C. O., et al. (2014). Interactive Visualization and Analysis of Large-Scale Sequencing Datasets Using ZENBU. Nat. Biotechnol. 32, 217–219. doi:10.1038/nbt.2840
Sheridan, S. D., Theriault, K. M., Reis, S. A., Zhou, F., Madison, J. M., Daheron, L., et al. (2011). Epigenetic Characterization of the FMR1 Gene and Aberrant Neurodevelopment in Human Induced Pluripotent Stem Cell Models of Fragile X Syndrome. PLoS ONE 6, e26203. doi:10.1371/journal.pone.0026203
Shulha, H. P., Cheung, I., Whittle, C., Wang, J., Virgil, D., Lin, C. L., et al. (2012). Epigenetic Signatures of Autism. Arch. Gen. Psychiatry 69, 314–324. doi:10.1001/archgenpsychiatry.2011.151
Silvis, M. R., Bertrand, C. A., Ameen, N., Golin-Bisello, F., Butterworth, M. B., Frizzell, R. A., et al. (2009). Rab11b Regulates the Apical Recycling of the Cystic Fibrosis Transmembrane Conductance Regulator in Polarized Intestinal Epithelial Cells. MBoC 20, 2337–2350. doi:10.1091/mbc.e08-01-0084
Smit, A. F. A., Hubley, R., and Green, P. (1996). RepeatMasker Open-3.0. Available at: http://www.repeatmasker.org (Accessed February 22, 2021).
Stenmark, H., and Olkkonen, V. M. (2001). The Rab GTPase Family. Genome Biol. 2, REVIEWS3007. doi:10.1186/gb-2001-2-5-reviews3007
Suetterlin, P., Hurley, S., Mohan, C., Riegman, K. L. H., Pagani, M., Caruso, A., et al. (2018). Altered Neocortical Gene Expression, Brain Overgrowth and Functional Over-connectivity in Chd8 Haploinsufficient Mice. Cereb. Cortex N. Y. N. 28, 2192–2206. doi:10.1093/cercor/bhy058
Sugathan, A., Biagioli, M., Golzio, C., Erdin, S., Blumenthal, I., Manavalan, P., et al. (2014). CHD8 Regulates Neurodevelopmental Pathways Associated with Autism Spectrum Disorder in Neural Progenitors. Proc. Natl. Acad. Sci. USA 111, E4468–E4477. doi:10.1073/pnas.1405266111
The FANTOM Consortium (2005). The Transcriptional Landscape of the Mammalian Genome. Science 309, 1559–1563. doi:10.1126/science.1112014
van deVondervoort, I. I. G. M., Gordebeke, P. M., Khoshab, N., Tiesinga, P. H. E., Buitelaar, J. K., Kozicz, T., et al. (2013). Long Non-coding RNAs in Neurodevelopmental Disorders. Front. Mol. Neurosci. 6, 53. doi:10.3389/fnmol.2013.00053
Villarroel-Campos, D., Gastaldi, L., Conde, C., Caceres, A., and Gonzalez-Billault, C. (2014). Rab-mediated Trafficking Role in Neurite Formation. J. Neurochem. 129, 240–248. doi:10.1111/jnc.12676
Voineagu, I., Wang, X., Johnston, P., Lowe, J. K., Tian, Y., Horvath, S., et al. (2011). Transcriptomic Analysis of Autistic Brain Reveals Convergent Molecular Pathology. Nature 474, 380–384. doi:10.1038/nature10110
Wan, P., Su, W., and Zhuo, Y. (2017). The Role of Long Noncoding RNAs in Neurodegenerative Diseases. Mol. Neurobiol. 54, 2012–2021. doi:10.1007/s12035-016-9793-6
Wang, P., Lin, M., Pedrosa, E., Hrabovsky, A., Zhang, Z., Guo, W., et al. (2015). CRISPR/Cas9-mediated Heterozygous Knockout of the Autism Gene CHD8 and Characterization of its Transcriptional Networks in Neurodevelopment. Mol. Autism 6, 55. doi:10.1186/s13229-015-0048-6
Wang, P., Mokhtari, R., Pedrosa, E., Kirschenbaum, M., Bayrak, C., Zheng, D., et al. (2017). CRISPR/Cas9-mediated Heterozygous Knockout of the Autism Gene CHD8 and Characterization of its Transcriptional Networks in Cerebral Organoids Derived from iPS Cells. Mol. Autism 8, 11. doi:10.1186/s13229-017-0124-1
Wilkinson, B., Grepo, N., Thompson, B. L., Kim, J., Wang, K., Evgrafov, O. V., et al. (2015). The Autism-Associated Gene Chromodomain Helicase DNA-Binding Protein 8 (CHD8) Regulates Noncoding RNAs and Autism-Related Genes. Transl. Psychiatry 5, e568. doi:10.1038/tp.2015.62
Yates, A. D., Achuthan, P., Akanni, W., Allen, J., Allen, J., Alvarez-Jarreta, J., et al. (2020). Ensembl 2020. Nucleic Acids Res. 48, D682–D688. doi:10.1093/nar/gkz966
Yin, J., Luo, W., Zeng, X., Zeng, L., Li, Z., Deng, X., et al. (2017). UXT-AS1-induced Alternative Splicing of UXT Is Associated with Tumor Progression in Colorectal Cancer. Am. J. Cancer Res. 7, 462–472.
Ziats, M. N., and Rennert, O. M. (2013). Aberrant Expression of Long Noncoding RNAs in Autistic Brain. J. Mol. Neurosci. 49, 589–593. doi:10.1007/s12031-012-9880-8
Zucchelli, S., Cotella, D., Takahashi, H., Carrieri, C., Cimatti, L., Fasolo, F., et al. (2015a). SINEUPs: A New Class of Natural and Synthetic Antisense Long Non-coding RNAs that Activate Translation. RNA Biol. 12, 771–779. doi:10.1080/15476286.2015.1060395
Keywords: autism spectrum disorders (ASD), CHD8, lncRNA, natural antisense transcript (NAT), SINEUP, post-transcriptional regulation, neurodeveloment, RAB11 GTPase
Citation: Zarantonello G, Arnoldi M, Filosi M, Tebaldi T, Spirito G, Barbieri A, Gustincich S, Sanges R, Domenici E, Di Leva F and Biagioli M (2021) Natural SINEUP RNAs in Autism Spectrum Disorders: RAB11B-AS1 Dysregulation in a Neuronal CHD8 Suppression Model Leads to RAB11B Protein Increase. Front. Genet. 12:745229. doi: 10.3389/fgene.2021.745229
Received: 21 July 2021; Accepted: 20 October 2021;
Published: 22 November 2021.
Edited by:
Elisabetta Morini, Massachusetts General Hospital and Harvard Medical School, United StatesReviewed by:
Alex Star Nord, University of California, Davis, United StatesRincic Martina, University of Zagreb, Croatia
Copyright © 2021 Zarantonello, Arnoldi, Filosi, Tebaldi, Spirito, Barbieri, Gustincich, Sanges, Domenici, Di Leva and Biagioli. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Marta Biagioli, bWFydGEuYmlhZ2lvbGlAdW5pdG4uaXQ=
 Giulia Zarantonello
Giulia Zarantonello Michele Arnoldi
Michele Arnoldi Michele Filosi
Michele Filosi Toma Tebaldi
Toma Tebaldi Giovanni Spirito
Giovanni Spirito Anna Barbieri
Anna Barbieri Stefano Gustincich
Stefano Gustincich Remo Sanges
Remo Sanges Enrico Domenici
Enrico Domenici Francesca Di Leva
Francesca Di Leva Marta Biagioli
Marta Biagioli