
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Genet. , 28 September 2021
Sec. RNA
Volume 12 - 2021 | https://doi.org/10.3389/fgene.2021.744113
This article is part of the Research Topic Decoding Non-Coding RNA Implicated in Cancer Cell Survival & Growth Modulation View all 18 articles
Hepatocellular carcinoma (HCC) is one of the most general malignant tumors. Hypoxia is a critical clinical characteristic and acts as a significant part in the development and cancers’ prognosis. The prognostic value and biological functions of hypoxia-related lncRNAs in hepatocellular carcinoma is little known. Thus, we aim to establish a hypoxia-related lncRNA signature to predict the HCC patients’ survival. First, we extracted the hypoxia-related genes and expression of lncRNAs from the MSigDB and TCGA database, respectively. The co-expression analysis among hypoxia-related mRNAs and lncRNAs was employed to identify hypoxia-related lncRNAs. Then, comprehensive analyses of lncRNAs expression level and survival data were applied to establish the signature. We built a prognostic signature on the foundation of the three differently expressed hypoxia-related lncRNAs. Kaplan-Meier curves indicated the low-risk group is associated with better survival. The 1−, 3−, and 5 years AUC values of the signature were 0.805, 0.672 and 0.63 respectively. The test set performed consistent outcomes. A nomogram was built grounded on the risk score and clinicopathological features. GSEA showed the immune-related pathways in high-risk group, while metabolism-related pathways in low-risk group. Besides, we found this model was correlated with the clinical features, tumor immune cell infiltration, immune checkpoints, and m6A-related genes. Finally, a novel signature based on hypoxia-related lncRNAs was established and validated for predicting HCC patients’ survival and may offer some useful information for immunotherapies.
Hepatocellular carcinoma (HCC) one of the most general subtype of liver cancer, ranks the fifth in terms of cancer-related deaths in the world (Bray et al., 2018). The surgical treatment, radiofrequency ablation, chemotherapy and as well liver transplant are the traditional treatments for hepatocellular carcinoma patients and the therapeutic effects are limited. The majority of patients are diagnosed with advanced stage and there are distant metastasis, tumor thrombosis of portal vein, and so on, leading to extremely poor prognosis (Galle et al., 2017; Vilgrain et al., 2017). Nowadays, in China, the survival rate of 5 years of HCC patients is only 14.1% (Allemani et al., 2018). Thus, novel predictive biomarkers and therapeutic options are critically required.
Hypoxia belongs to a public microenvironmental property of most solid tumors, because of the imbalance between the tumor proliferation and oxygen supply (Hill et al., 2015; Muz et al., 2015). Previous studies have proved that hypoxia plays critical roles in angiogenesis, differentiation, proliferation and apoptosis of tumor cells (Nishida and Kudo, 2013; Wu et al., 2007). As one of the organs most prone to hypoxia, hypoxia is correlated with the metastasis, prognosis, and resistance of HCC (Erler and Giaccia, 2006; Graham and Unger, 2018). Whereas lncRNA is a subset of RNA transcripts over 200 nt, which not only regulates the mRNA’s expression but also mediates the development, occurrence, metastasis and prognosis of various cancers (Kang et al., 2019; Gao et al., 2021; Xiao et al., 2021). At present, multiple studies found that hypoxia-related lncRNAs are associated with overall survival in various tumors (Chen et al., 2020; Zhang et al., 2021). Nonetheless, whether the hypoxia-related lncRNAs are related to the prognosis of hepatocellular carcinoma is still largely not clear.
Herein, we mined the expression data of lncRNAs and the relevant clinical information of hepatocellular carcinoma samples, and hypoxia-related genes from the opening databases. Next, the prognostic signature of hypoxia-related lncRNA was built and then evaluated it based on the TCGA cohort. Lastly, we validated the predictive capability of the risk model and explored the role in diagnostic value, infiltrating immune cells, N6-methyladenosine (m6A) mRNA, and immune checkpoints.
The RNA-sequencing (FPKM) of 374 hepatocellular carcinoma samples was derived from the TCGA data portal. The corresponding data of clinical, for example, age, gender, clinical grade, clinical stage and survival information, was also downloaded from the TCGA database. The clinical information of these patients is displayed in Table 1. The Ensembl was applied to obtain the gene transfer annotation files to differentiate the messenger RNAs (mRNAs) from long non-coding lncRNAs (lncRNAs) for subsequent analyses. Besides, the merge language script Perl was used to merge the data files of RNA-sequencing into a matrix file. Then, the genes’ Ensembl IDs were translated into an array of gene symbols by utilizing the script of the Perl language.
Additionally, the Molecular Signatures database (MSigDB) (M10508, M26925) was applied to download the hypoxia-related genes. The co-expression analysis was employed to identify hypoxia-related lncRNAs. The hypoxia-related genes with correlation coefficients > 0.5 and p-value < 0.001 were regarded as hypoxia-related lncRNAs. The lncRNAs with the average expression less than 0.5 were delated. The expression profiles of hypoxia-related genes/lncRNAs of the 50 adjacent normal tissue samples and 374 samples of hepatocellular carcinoma were subjected to DE-HRGs/DE-HRlncRNAs using the “limma” package in R. The GO and KEGG were applied to explore the potential function of the differentially expressed hypoxia-related genes (DE-HRGs) (FDR < 0.05 and |log2FC| ≥ 1) using the “clusterProfiler”, “enrichplot” and “ggplot2” R packages. The samples of hepatocellular carcinoma were distributed into the train and test sets applying the “caret” package randomly. We built the prognostic signature based on the hypoxia-related lncRNAs’ expression level in the train set and validated its prognostic value in the test set.
To identify the hypoxia-related lncRNAs that related overall survival, we merged the hypoxia-related lncRNAs and survival information utilizing the package “limma”. The survival time of hepatocellular carcinoma samples ranks from 0 to 3,675 days. To improve the quality analyses, samples that lack complete survival information and those whose overall survival was not over 30 days were excluded from this analysis. Then, the univariate Cox regression analysis was applied to get survival-related hypoxia-related lncRNAs from the hypoxia-related lncRNAs (p < 0.05). These survival-related hypoxia-related lncRNAs were used for further analysis.
The LASSO Cox analysis was employed to identify the prognostic hypoxia-related lncRNAs. To prevent overfitting, we applied 1000-round cross-validation for tuning parameter selection to fulfill the minimum criteria of the partial likelihood deviance. Finally, the multivariate Cox regression analysis was fulfilled on these prognostic hypoxia-related lncRNAs to build a prognostic hypoxia-related lncRNAs signature and compute the coefficients. The risk score for each sample was counted utilizing the following formula: risk score = ∑icoefficient (hypoxia-related lncRNA) × expression of (hypoxia-related lncRNAi).
The patients of train and test sets were divided into two subgroups (low-risk vs high-risk) on the foundation of the median value of the risk score of train cohort, respectively. We applied Kaplan-Meier survival analysis to explore the difference of survival between the two groups in the train and test sets. The time-dependent receiver operating characteristic (ROC) curves of 1-, 3- and 5 years were plotted using the “survivalROC” package in two sets. Besides, to explore whether the risk score can regard as an independent biomarker for the hepatocellular carcinoma patients’ survival, the univariate and multivariate Cox regression analysis were used in the train and test sets. To prove the clinical application value of this signature, we applied the chi-square test to explore the relationship between the model and clinical variables. We compared the differences in the risk score among subgroups with different clinical features with a Wilcoxon signed-rank test.
A nomogram was built on the foundation of the risk score and other clinical biomarkers for predicting the 1-, 3-, 5 years overall survival of hepatocellular carcinoma patients. We applied the calibration curves to assess the nomogram’ accuracy. The Gene set enrichment analyses (GSEA) was performed to investigate the KEGG pathways between two risk (low vs high) groups in whole cohort of TCGA-LIHC using GSEA 4.0.1 software. p < 0.05 and FDR < 5% were considered as statistical significance.
To explore the association between the risk score and features of immune cell infiltration, we used the various acknowledged methods to count the tumor infiltrating immune cells levels among the patients from the whole cohort, including CIBERSORT-ABS, XCELL, EPIC, MCPcounter, TIMER, QUANTISEQ, and CIBERSORT. We used the Wilcoxon signed-rank test to explore the difference in immune cell infiltration level between two risk groups. To explore the association between two risk groups and immune status, we quantified the tumor-infiltrating cells related pathways with ssGSEA (Xiao et al., 2020), namely, APC_co_inhibition, APC_co_stimulation, CCR, check-point, cytolytic activity, HLA, inflammation promoting, MHC_class_I, parainflammation T cell co-inhibition, T cell co-stimulation, type I_IFN response, and type II_IFN response. Previous studies have proved that m6a-related genes were associated with prognosis in hepatocellular carcinoma (Zhao et al., 2018; Wu et al., 2020). What is more, m6a-related genes can mediate the expression of lncRNAs, which dyregulate the tumorigenesis, development, metastasis, as well as prognosis (Lan et al., 2019; Wu et al., 2021a). Thus, we evaluated the association between the risk score and N6-methyladenosine (m6A) mRNAs as well as immune checkpoints in the whole cohort. The m6A mRNAs and immune checkpoints were obtained from previous literature.
We obtained 76 DE-HRGs including 58 upregulated and 18 downregulated genes (Supplementary Table S1). The most enriched GO terms were CC (cellular component) such as membrane raft, endoplasmic reticulum lumen, secretory granule lumen, membrane microdomain and cytoplasmic vesicle lumen; BP (biological process), for example, glucose catabolic process to pyruvate, NADH regeneration, carbohydrate biosynthetic process, glucose metabolic process and canonical glycolysis; and MF (molecular function) such as glycosaminoglycan binding, monosaccharide binding, carbohydrate binding, growth factor binding, and cell adhesion molecule binding (Figure 1A, Supplementary Table S2). The KEGG pathways of 76 DE-HRGs were predominantly related to glycolysis/gluconeogenesis, starch and sucrose metabolism, fructose and mannose metabolism, as well as pentose phosphate pathway (Figure 1B, Supplementary Table S2).
We obtained 200 hypoxia-related genes from the MSigDB database (Supplementary Table S3). Then, a total of 171 hypoxia-related lncRNAs were identified by applying co-expression analysis between hypoxia-related genes and lncRNAs. Among these hypoxia-related lncRNAs, 111 lncRNAs were differentially expressed hypoxia-related lncRNAs (Supplementary Figures S1A,B). Then, we added the hypoxia-related lncRNAs with survival data by merging complete survival material of samples and hypoxia-related lncRNAs. Next, 343 hepatocellular carcinoma samples were distributed into the train (243) and test (100) sets applying the “caret“ package randomly. The samples in the train set were employed for constructing the prognostic model.
First, we identified 21 prognostic hypoxia-related lncRNAs utilizing univariate Cox regression analysis, as shown in Supplementary Table S4. Then, utilizing LASSO regression analysis, 16 hypoxia-related lncRNAs were removed (Figures 2A,B). Finally, three hypoxia-related lncRNAs (AC099850.4, MIR210HG, and NRAV) were obtained through using multivariate Cox regression analysis (Figure 2C). The hypoxia-related lncRNAs signature was constructed based on these three lncRNAs and corresponding coefficients: risk score = (0.08283797* expression of AC099850.4) + (0.10722211* expression of MIR210HG) + (0.25250738* expression of NRAV). Besides, the association between three hypoxia-related lncRNAs in the signature and hypoxia-related genes is displayed in Figure 2D.
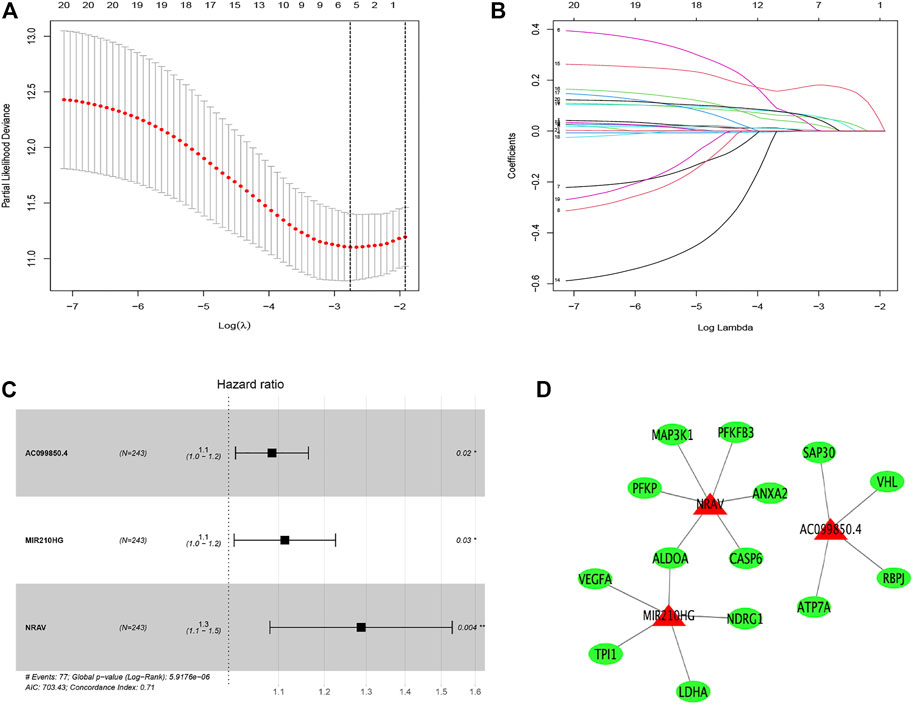
FIGURE 2. Distinction of hypoxia-related lncRNAs signature: LASSO (A,B) and multivariate (C) Cox regression analysis. The relationship between the prognostic lncRNAs (red) and genes (green) (D).
To explore the underlying prognostic capability of the hypoxia-related lncRNA signature, we assigned the patients in train and test sets into two groups (low-risk vs high-risk) on the foundation of the median of risk score. The Kaplan-Meier analysis demonstrated the high-risk group was correlated with worse survival (p < 0.001, Figure 3A) in the train set and the K-M analysis in the test set revealed the similar result (p = 0.002, Figure 3B). Meanwhile, the 1-, 3-, and 5 years AUC values of ROC curve were 0.805, 0.672 and 0.63, respectively, in the train set (Figure 3C). The corresponding values in the test set were 0.739, 0.762 and 0.667, respectively (Figure 3D). Utilizing the risk survival status plots of patients, we revealed that the risk scores of patients were negatively correlated with the hepatocellular carcinoma patients’ survival in train and test sets. Besides, the heatmaps showed that these three lncRNAs were positively associated with the risk signature in two sets (Figures 3E,F).
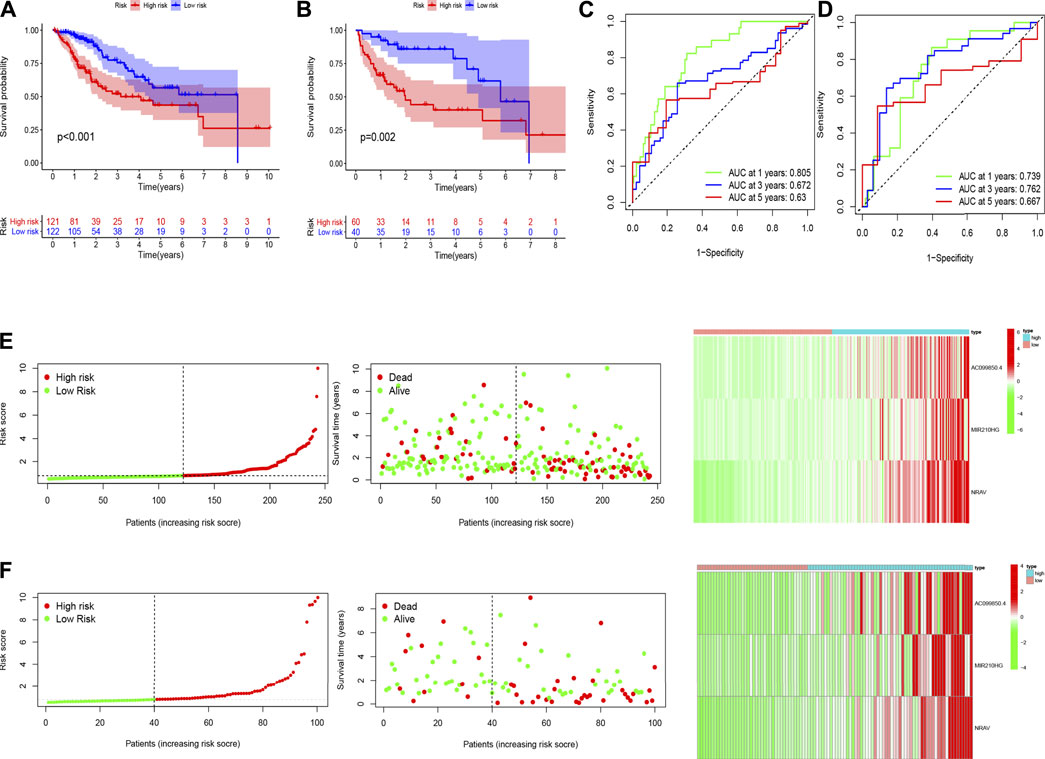
FIGURE 3. The results of Kaplan-Meier analysis, ROC curves, and risk survival status plots in train (A,C,E) and test (B,D,F) sets.
The univariate Cox regression analysis revealed that risk score (HR = 1.163, 95% CI: 1.100–1.231, p < 0.001) and stage (HR = 1.558, 95% CI: 1.313–1.848, p < 0.001) were remarkably associated with overall survival in the train set (Figure 4A), and risk score (HR = 1.218, 95% CI: 1.065–1.394, p = 0.004) and stage (HR = 1.314, 95% CI: 1.040–1.660, p = 0.022) were also associated with overall survival in the test set (Figure 4C). Furthermore, the multivariate Cox regression analysis also demonstrated that risk score (HR = 1.153, 95% CI: 1.318–1.953, p < 0.001) and stage (HR = 1.604, 95% CI: 1.313–1.848, p < 0.001) were the independent factors to predict the hepatocellular carcinoma patients’ prognosis in the train set (Figure 4B), and risk score (HR = 1.160, 95% CI: 0.993–1.355, p = 0.061) and stage (HR = 1.319, 95% CI: 1.023–1.701, p = 0.032) were also the independent factors in the test set (Figure 4D). Subsequently, we applied the ROC curve to appraise the accurateness of clinical variables in predicting the HCC patients’ overall survival. As shown in Supplementary Figure S1C, the AUC value of clinical variables was lower than that of risk score. Besides, to further explore the signature’s prognostic value in hepatocellular carcinoma patients arranged by clinical variables, the patients were divided into distinct subgroups according to clinical variables (age, sex, stage and clinical grade). Figure 5 shows that the survival of patients in high-risk group was worse than those in the low-risk group in most of subgroups.
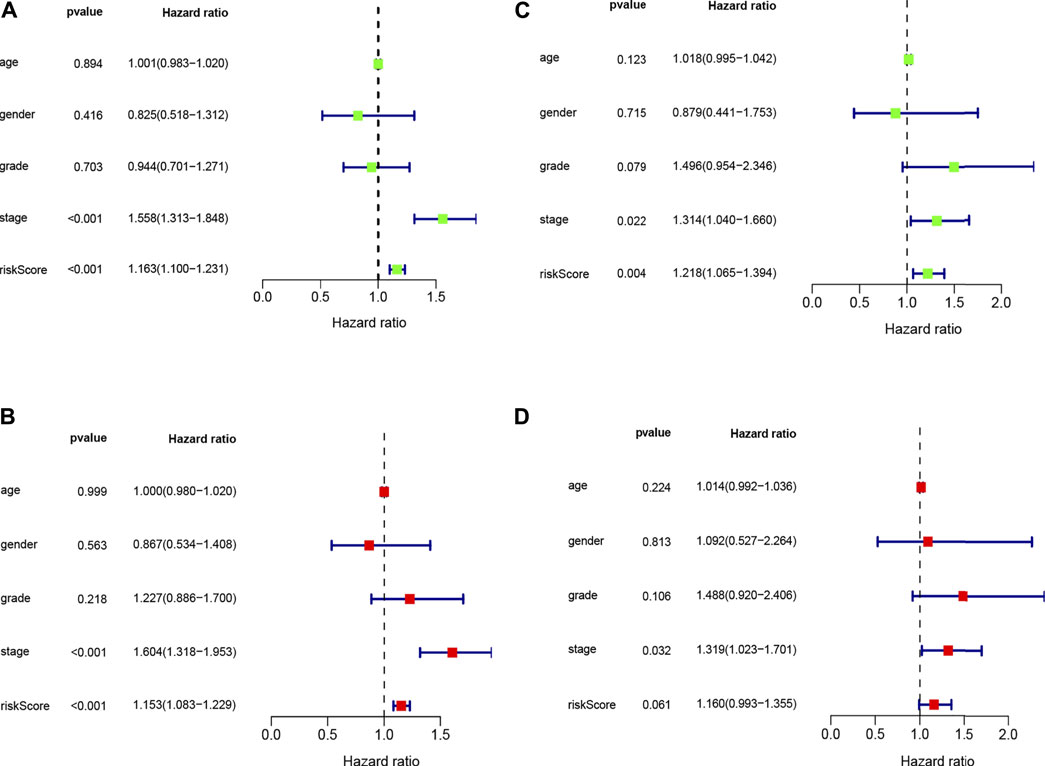
FIGURE 4. Univariate (A,C) and multivariate (B,D) COX analysis for risk score and clinical variables in train and test sets.
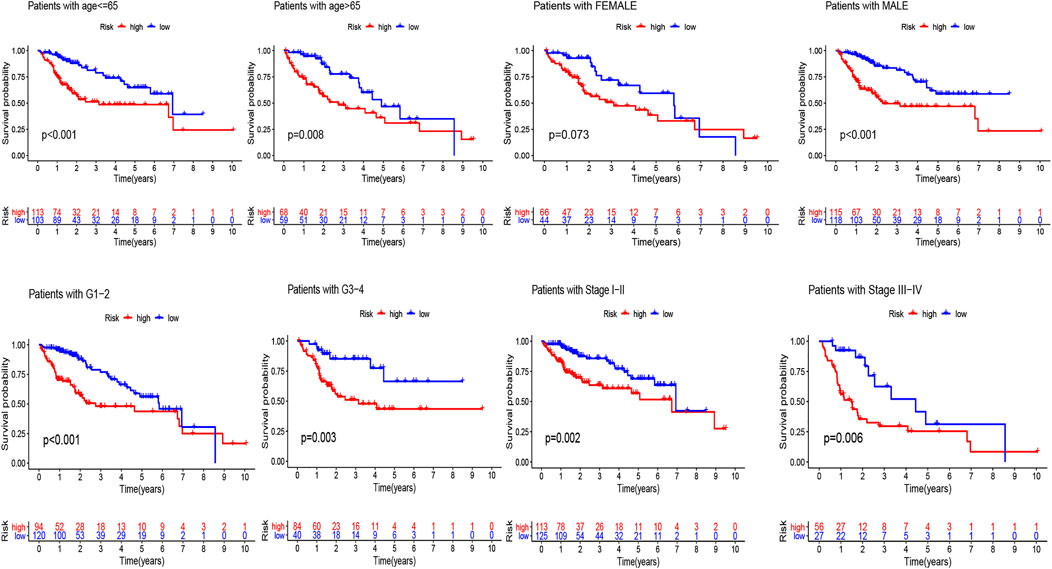
FIGURE 5. Kaplan-Meier analyses for the two subgroups (high-risk vs low-risk) categorized by clinical variables, comprising age, gender, grade, and stage.
The nomogram based on the risk score and clinical features was constructed for predicting the 1-, 3- and 5 years overall survival of the hepatocellular carcinoma patients (Figure 6A). The 1-, 3-, and 5 years calibration curves showed stable and accurate performance (Figures 6B–D); therefore, they may be used for hepatocellular carcinoma patients’ clinical management.
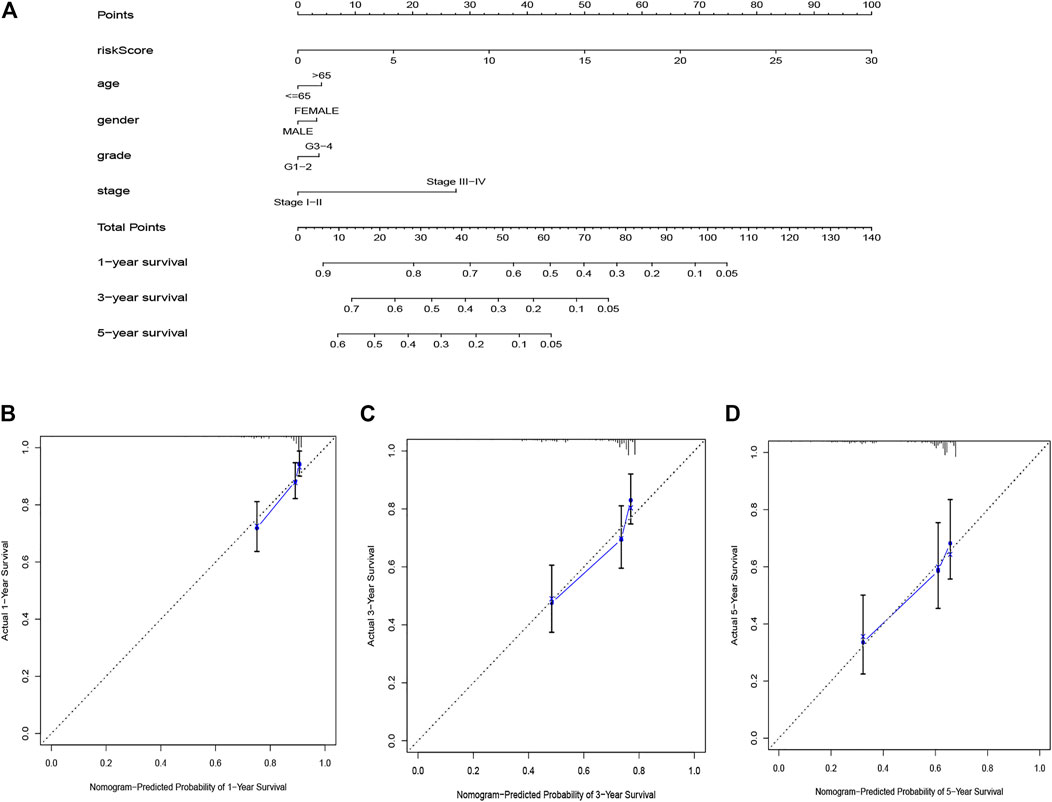
FIGURE 6. The nomogram for predicting HCC patients’ survival (A). Calibration plots applied for predicting the 1-year (B), 3-year (C) and 5-year (D) survival.
GSEA demonstrated the prognostic hypoxia-related lncRNAs signature was enriched in immune-, metabolism- and tumor-related pathways. The high-risk group was enriched in B cell receptor signaling pathway, T cell receptor signaling pathway, P53 signaling pathway, TGF_BETA signaling pathway and WNT signaling pathway, while the low-risk group was enriched in butanoate metabolism, fatty acid metabolism, retinol metabolism, tryptophan metabolism and glycine serine and threonine metabolism (Figure 7).
The heatmap of tumor immune cell infiltration based on TIMER, EPIC, CIBERSORT−ABS, QUANTISEQ, XCELL, MCP counter and CIBERSORT algorithms is displayed in Figure 8 and Supplementary Table S5. The difference of immune cell related functions indicated that cytolytic activity, type II IFN response and MHC class I were remarkably different between the two risk groups applying the single-sample gene set enrichment analysis (ssGSEA) (Figure 9A). Considering the significance of immune checkpoints in immunotherapies, we next investigated the difference of immune checkpoints’ expression between two risk groups. The boxplot showed most of these genes, for example, PDCD-1 (PD-1), CD86, LAIR1, CTLA4 and CD70, existed significantly different between the two groups (Figure 9B). Besides, most of m6A-related genes’ expression also had significant difference between two risk groups except for FMR1 and ZC3H13 (Figure 9C).
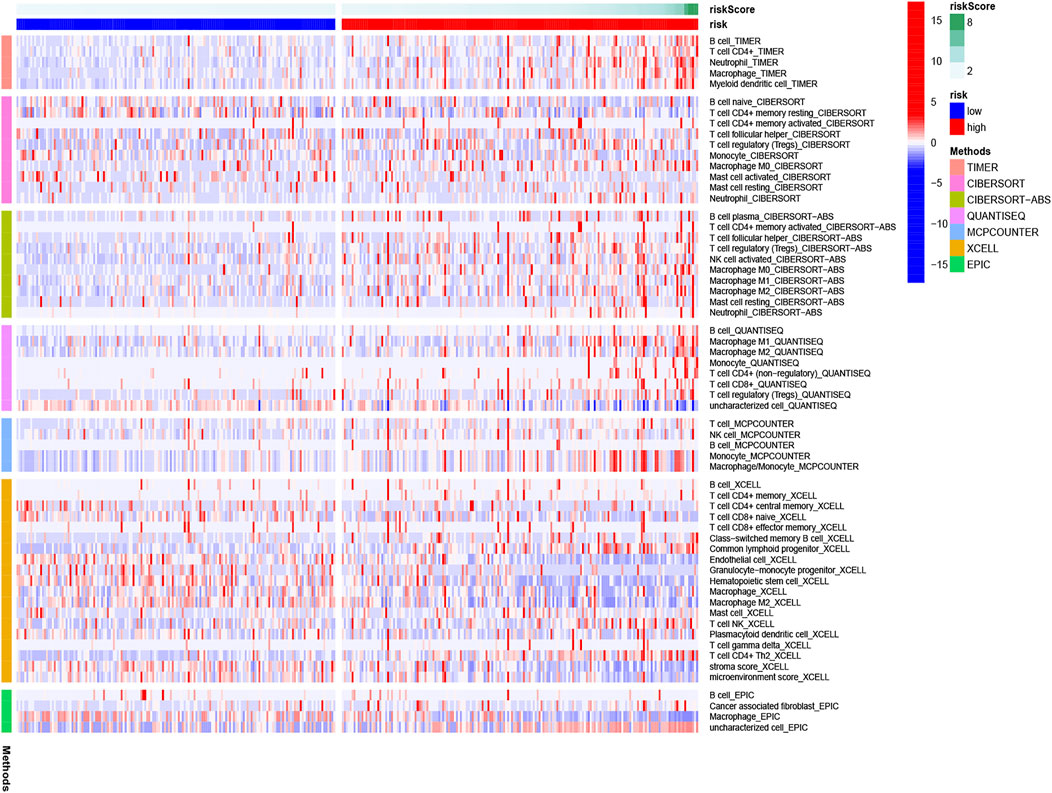
FIGURE 8. Heatmap for immune cells based on EPIC, CIBERSORT−ABS, CIBERSORT, QUANTISEQ, XCELL, MCP counter, and TIMER algorithms between two risk groups.
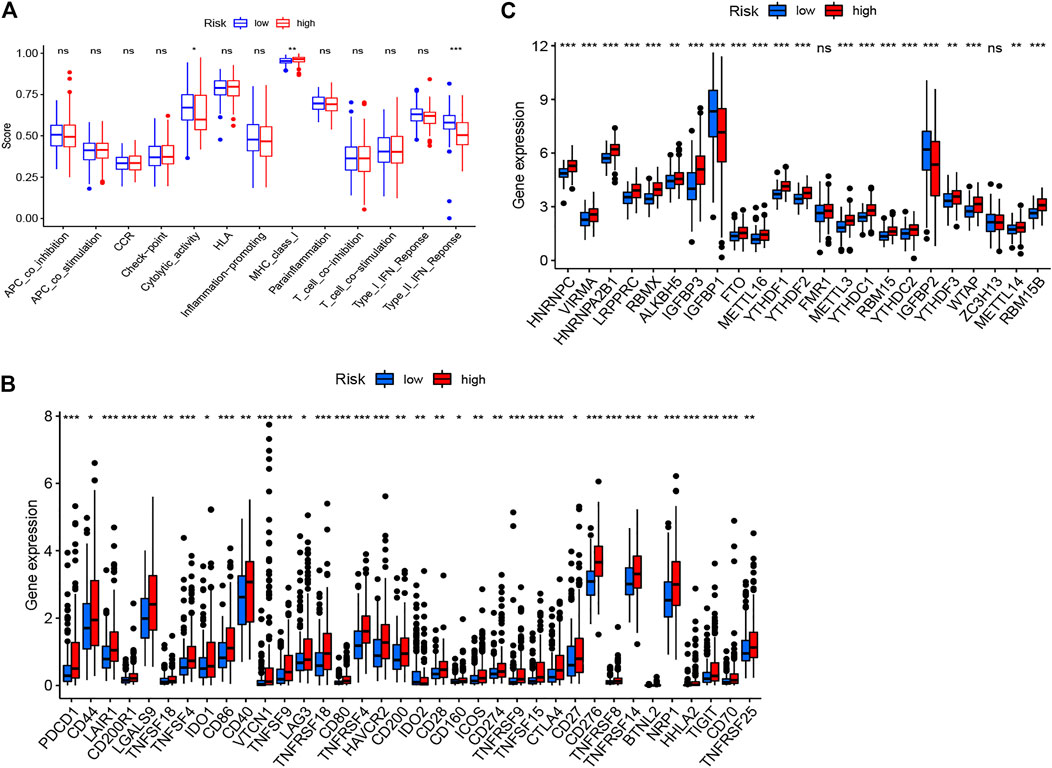
FIGURE 9. The immune cells related functions (A), immune checkpoints (B) and m6A-related genes (C) between two risk groups.
Despite the development of surgical resection, liver transplantation, chemotherapy, immunotherapy, and targeted therapy in the therapy of hepatocellular carcinoma, the hepatocellular carcinoma patients’ prognosis is still poor (Maluccio and Covey, 2012; Xu et al., 2020a). The studies showed that the treatment response and prognosis are influenced by tumor molecular characteristics, although patients own homogeneous clinical risk variables. Thus, it is urgent to identify novel molecular prognostic markers that distinguish from common clinical risk features. Previous studies have proved that hypoxia is associated with tumor angiogenesis, metabolic alteration, metastasis, and prognosis (Krock et al., 2011; Eales et al., 2016; Sun et al., 2020). Meanwhile, lncRNAs play a crucial role in regulating the hypoxia-related genes’ expression. Therefore, we built a signature based on the hypoxia-related lncRNAs and explored tumor immune cell infiltration, immune checkpoints and m6A-related genes in the prognosis of hepatocellular carcinoma, which may offer underlying biomarkers and therapeutic targets for HCC.
First, we explored the function of 76 hypoxia related DEGs. The KEGG analysis indicated these genes mainly enriched in glycolysis/gluconeogenesis, fructose and mannose metabolism, pentose phosphate pathway, as well as starch and sucrose metabolism. Under hypoxia, cancer cells could retain survival and stimulate tumor proliferation, migration, and invasion via glycolysis (Huang and Zong, 2017). Wu et al. reported that OMA, an ATP-dependent zinc metalloprotease, promotes cancer development by reprogramming metabolic colorectal cancer under hypoxia (Wu et al., 2021b). A recent study revealed that hypoxia downregulates the enzymes of pentose phosphate pathway and upregulates the enzymes of glycolysis in tumor and then stimulate tumor migration while inhibiting tumor proliferation (Kathagen-Buhmann et al., 2016). Besides, found that hypoxia and the hypoxia inducible factors (HIFs) step in the metabolic crosstalk between tumor cells and their microenvironment (Mucaj et al., 2012). Then, we established a prognostic signature based on three hypoxia-related lncRNAs (AC099850.4, MIR210HG, and NRAV). Wang et al. revealed that high expression of MIR210HG in hepatocellular carcinoma samples and cells not only associated with clinicopathological characteristics, for example, stage, tumor size, vascular invasion, and histological differentiation but also with poor prognosis (Wang et al., 2019). Du et al. revealed MIR210HG is a hypoxia-related lncRNA in triple-negative breast cancer and promotes the Warburg effect by directly regulating HIF-1α (Du et al., 2020). Besides, overexpression of MIR210HG promotes the progression of colorectal adenocarcinoma via modulating hypoxia (He et al., 2019) and was associated with worse overall survival in colon cancer patients (Ruan et al., 2019). AC099850.4 is one of the top lncRNA in miRNA–lncRNA network and may be associated with tumor division, tumor proliferation, cell cycle and so on in ovarian cancer (Zhao et al., 2020). Higher expression of NRAV was associated with advanced clinical stage, poor prognosis and immunologic characteristics in hepatocellular carcinoma patients (Feng et al., 2021; Xu et al., 2021). However, to date, there are few studies on these lncRNAs in hepatocellular carcinoma under hypoxia. These findings may offer crucial insights into the cancers’ control in the future.
Herein, we divided the samples into the two risk subgroups on the foundation of the hypoxia-related lncRNA signature to explore the underlying roles in HCC. Hypoxia acts as an important role in anti-tumor immune responses via mediating the expression levels of immune checkpoints (Multhoff and Vaupel, 2020). There are very little studies exploring the connection between immune checkpoint inhibitors and hypoxia at present. We compared the difference of immune checkpoints between two risk group and discovered that most of them exist remarkably difference, which may offer underlying therapeutic targets for hepatocellular carcinoma. More and more studies show that miRNA and lncRNA act as an important role in hypoxia. Hypoxia-inducible factor (HIF) is a critical to cellular responses to hypoxic stress. LncRNA SNHG11 promotes invasion and metastasis of colorectal cancer cells by regulating HIF-1α (Xu et al., 2020b). Hypoxia activated lncRNA HABON enhances the proliferation and growth of hepatocarcinoma cells by mediating HIF-1α (Ma et al., 2021). Hypoxia-inducible miR-182 accelerates tumor angiogenesis and growth by regulating the expression level of HIF1α in prostate cancer cells (Li et al., 2015).
Besides, hypoxia can change the tumor cells’ interplay and crosstalk with the tumor micro-environment, resulting to immune suppression and resistance, which contribute tumor cells to escape immune surveillance (Jing et al., 2019; Noman et al., 2019). To explore the relationship between our signature and tumor micro-environment, we fulfilled GSEA. The outcomes revealed that various immune-related pathways participated in the high-risk group, while the metabolism-related pathways participated in the low-risk group. We next plotted the heatmap of immune cell infiltration of HCC patients to explore the difference of immune microenvironment between two risk groups by using multiple algorithms. We found that T cell follicular helper, NK cell, macrophage M0, macrophages M2, neutrophil, and mast cells resting were highly infiltrated in HCC, associating with tumorigenesis, progression, and metastasis (Zhang et al., 2017; Aponte-López and Muñoz-Cruz, 2020; Ngabire et al., 2020). These results showed that this hypoxia-related lncRNA signature can partially reflect tumor immune cell infiltration and may offer some useful information for immunotherapies. Moreover, majority of m6a-related genes were highly expressed in high risk group, which is similar to previous studies. Chen et al. revealed that WTAP was extremely overexpressed and regarded as an independent prognostic biomarker for HCC patients (Chen et al., 2019). It was reported that FTO was over-expressed in HCC tissue and associated with poor prognosis of HCC patients as well as promoted cell proliferation by mediating the demethylation of PKM2 (Li et al., 2019). YTHDF1 was remarkably up-regulated and positively associated with pathology stage in HCC. K-M curve revealed that higher expression of YTHDF1 is correlated with worse survival (Zhao et al., 2018). What is more, most of immune checkpoints were over-expressed in high risk group, which may provide some underlying targets for immunotherapy. Some of them have proved correlated with poor prognosis in HCC, such as LAG3 (Yarchoan et al., 2017), NRP1 (Lin et al., 2018) and LAIR1 (Wu et al., 2019).
The data of this analysis was downloaded from TCGA database, including complete survival and clinicopathological information of most patients of HCC. Besides, the HCC samples were sufficient enough to divide into train set and test set. Thus, the prognostic signature was constructed and validated using the patients from the single database. However, there were some limitations. First, we did not validate the prognostic model in external database and lack of the validation in clinical practice without enough HCC samples. Second, we did not perform basic experiment to verify the lncRNAs in this study.
Specific hypoxia associated lncRNAs provide a novel prognostic signature for hepatocellular carcinoma patients and may improve the individualized treatment strategies.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
CZ designed the project and wrote the manuscript; HZ support data analysis and discussion. LL guided the project and critically reviewed the manuscript. All authors have contributed to the preparation of the manuscript and reviewed/approved it in its final form.
CZ designed the project and wrote the manuscript; HZ support data analysis and discussion. LL guided the project and critically reviewed the manuscript. All authors have contributed to the preparation of the manuscript and reviewed/approved it in its final form. CZ was financially supported by CSC scholarship (The China Scholarship Council).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, orclaim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2021.744113/full#supplementary-material.
Allemani, C., Matsuda, T., Di Carlo, V., Harewood, R., Matz, M., Nikšić, M., et al. (2018). Global Surveillance of Trends in Cancer Survival 2000-14 (CONCORD-3): Analysis of Individual Records for 37 513 025 Patients Diagnosed with One of 18 Cancers from 322 Population-Based Registries in 71 Countries. Lancet 391 (10125), 1023–1075. doi:10.1016/S0140-6736(17)33326-3
Aponte-López, A., and Muñoz-Cruz, S. (2020). Mast Cells in the Tumor Microenvironment. Adv. Exp. Med. Biol. 1273, 159–173. doi:10.1007/978-3-030-49270-0_9
Bray, F., Ferlay, J., Soerjomataram, I., Siegel, R. L., Torre, L. A., and Jemal, A. (2018). Global Cancer Statistics 2018: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA: A Cancer J. Clinicians 68 (6), 394–424. doi:10.3322/caac.21492
Chen, Q., Hu, L., and Chen, K. (2020). Construction of a Nomogram Based on a Hypoxia-Related lncRNA Signature to Improve the Prediction of Gastric Cancer Prognosis. Front. Genet. 11, 570325. doi:10.3389/fgene.2020.570325
Chen, Y., Peng, C., Chen, J., Chen, D., Yang, B., He, B., et al. (2019). WTAP Facilitates Progression of Hepatocellular Carcinoma via m6A-HuR-dependent Epigenetic Silencing of ETS1. Mol. Cancer 18 (1), 127. doi:10.1186/s12943-019-1053-8
Du, Y., Wei, N., Ma, R., Jiang, S.-H., and Song, D. (2020). Long Noncoding RNA MIR210HG Promotes the Warburg Effect and Tumor Growth by Enhancing HIF-1α Translation in Triple-Negative Breast Cancer. Front. Oncol. 10, 580176. doi:10.3389/fonc.2020.580176
Eales, K. L., Hollinshead, K. E. R., and Tennant, D. A. (2016). Hypoxia and Metabolic Adaptation of Cancer Cells. Oncogenesis 5–e190, e190. doi:10.1038/oncsis.2015.50
Erler, J. T., and Giaccia, A. J. (2006). Lysyl Oxidase Mediates Hypoxic Control of Metastasis: Figure 1. Cancer Res. 66 (21), 10238–10241. doi:10.1158/0008-5472.CAN-06-3197
Feng, Y., Hu, X., Ma, K., Zhang, B., and Sun, C. (2021). Genome-Wide Screening Identifies Prognostic Long Noncoding RNAs in Hepatocellular Carcinoma. Biomed. Res. Int. 2021, 1–16. doi:10.1155/2021/6640652
Galle, P. R., Tovoli, F., Foerster, F., Wörns, M. A., Cucchetti, A., and Bolondi, L. (2017). The Treatment of Intermediate Stage Tumours beyond TACE: From Surgery to Systemic Therapy. J. Hepatol. 67 (1), 173–183. doi:10.1016/j.jhep.2017.03.007
Gao, K., Lv, C.-L., Zhang, T., Zhang, K. Z.-L., and Lv, C. L. (2021). LncRNA Airsci Increases the Inflammatory Response after Spinal Cord Injury in Rats through the Nuclear Factor Kappa B Signaling Pathway. Neural Regen. Res. 16 (4), 772–777. doi:10.4103/1673-5374.295335
Graham, K., and Unger, E. (2018). Overcoming Tumor Hypoxia as a Barrier to Radiotherapy, Chemotherapy and Immunotherapy in Cancer Treatment. Ijn 13, 6049–6058. doi:10.2147/IJN.S140462
He, Z., Dang, J., Song, A., Cui, X., Ma, Z., and Zhang, Z. (2019). Identification of LINC01234 and MIR210HG as Novel Prognostic Signature for Colorectal Adenocarcinoma. J. Cel Physiol 234 (5), 6769–6777. doi:10.1002/jcp.27424
Hill, R. P., Bristow, R. G., Fyles, A., Koritzinsky, M., Milosevic, M., and Wouters, B. G. (2015). Hypoxia and Predicting Radiation Response. Semin. Radiat. Oncol. 25 (4), 260–272. doi:10.1016/j.semradonc.2015.05.004
Huang, R., and Zong, X. (2017). Aberrant Cancer Metabolism in Epithelial-Mesenchymal Transition and Cancer Metastasis: Mechanisms in Cancer Progression. Crit. Rev. Oncology/Hematology 115, 13–22. doi:10.1016/j.critrevonc.2017.04.005
Jing, X., Yang, F., Shao, C., Wei, K., Xie, M., Shen, H., et al. (2019). Role of Hypoxia in Cancer Therapy by Regulating the Tumor Microenvironment. Mol. Cancer 18 (1), 157. doi:10.1186/s12943-019-1089-9
Kang, C. L., Qi, B., Cai, Q. Q., Fu, L. S., Yang, Y., Tang, C., et al. (2019). LncRNA AY Promotes Hepatocellular Carcinoma Metastasis by Stimulating ITGAV Transcription. Theranostics 9 (15), 4421–4436. doi:10.7150/thno.32854
Kathagen-Buhmann, A., Schulte, A., Weller, J., Holz, M., Herold-Mende, C., Glass, R., et al. (2016). Glycolysis and the Pentose Phosphate Pathway Are Differentially Associated with the Dichotomous Regulation of Glioblastoma Cell Migration versus Proliferation. Neuonc 18 (9), 1219–1229. doi:10.1093/neuonc/now024
Krock, B. L., Skuli, N., and Simon, M. C. (2011). Hypoxia-induced Angiogenesis: Good and Evil. Genes & Cancer 2 (12), 1117–1133. doi:10.1177/1947601911423654
Lan, T., Li, H., Zhang, D., Xu, L., Liu, H., Hao, X., et al. (2019). KIAA1429 Contributes to Liver Cancer Progression through N6-methyladenosine-dependent post-transcriptional Modification of GATA3. Mol. Cancer 18 (1), 186. doi:10.1186/s12943-019-1106-z
Li, J., Zhu, L., Shi, Y., Liu, J., Lin, L., and Chen, X. (2019). m6A Demethylase FTO Promotes Hepatocellular Carcinoma Tumorigenesis via Mediating PKM2 Demethylation. Am. J. Transl Res. 11 (9), 6084–6092.
Li, Y., Zhang, D., Wang, X., Yao, X., Ye, C., Zhang, S., et al. (2015). Hypoxia-inducible miR-182 Enhances HIF1α Signaling via Targeting PHD2 and FIH1 in Prostate Cancer. Sci. Rep. 5, 12495. doi:10.1038/srep12495
Lin, J., Zhang, Y., Wu, J., Li, L., Chen, N., Ni, P., et al. (2018). Neuropilin 1 (NRP1) Is a Novel Tumor Marker in Hepatocellular Carcinoma. Clinica Chim. Acta 485, 158–165. doi:10.1016/j.cca.2018.06.046
Ma, C.-N., Wo, L.-L., Wang, D.-F., Zhou, C.-X., Li, J.-C., Zhang, X., et al. (2021). Hypoxia Activated Long Non-coding RNA HABON Regulates the Growth and Proliferation of Hepatocarcinoma Cells by Binding to and Antagonizing HIF-1 Alpha. RNA Biol. 21 (1), 1–16. doi:10.1080/15476286.2020.1871215
Maluccio, M., and Covey, A. (2012). Recent Progress in Understanding, Diagnosing, and Treating Hepatocellular Carcinoma. CA: A Cancer J. Clinicians 62 (6), 394–399. doi:10.3322/caac.21161
Mucaj, V., Shay, J. E. S., and Simon, M. C. (2012). Effects of Hypoxia and HIFs on Cancer Metabolism. Int. J. Hematol. 95 (5), 464–470. doi:10.1007/s12185-012-1070-5
Multhoff, G., and Vaupel, P. (2020). Hypoxia Compromises Anti-cancer Immune Responses. Adv. Exp. Med. Biol. 1232, 131–143. doi:10.1007/978-3-030-34461-0_18
Muz, B., de la Puente, P., Azab, F., and Azab, A. K. (2015). The Role of Hypoxia in Cancer Progression, Angiogenesis, Metastasis, and Resistance to Therapy. Hp 3, 83–92. doi:10.2147/HP.S93413
Ngabire, D., Niyonizigiye, I., Patil, M. P., Seong, Y.-A., Seo, Y. B., and Kim, G.-D. (2020). M2 Macrophages Mediate the Resistance of Gastric Adenocarcinoma Cells to 5-Fluorouracil through the Expression of Integrin β3, Focal Adhesion Kinase, and Cofilin. J. Immunol. Res. 2020, 1–9. doi:10.1155/2020/1731457
Nishida, N., and Kudo, M. (2013). Oxidative Stress and Epigenetic Instability in Human Hepatocarcinogenesis. Dig. Dis. 31 (5-6), 447–453. doi:10.1159/000355243
Noman, M. Z., Hasmim, M., Lequeux, A., Xiao, M., Duhem, C., Chouaib, S., et al. (2019). Improving Cancer Immunotherapy by Targeting the Hypoxic Tumor Microenvironment: New Opportunities and Challenges. Cells 8 (9), 1083. doi:10.3390/cells8091083
Ruan, Z., Xu, Z., Li, Z., and Lv, Y. (2019). Integral Analyses of Survival-related L-ong N-on-coding RNA MIR210HG and its Prognostic Role in colon Cancer. Oncol. Lett. 18 (2), 1107–1116. doi:10.3892/ol.2019.10435
Sun, J., Zhao, T., Zhao, D., Qi, X., Bao, X., Shi, R., et al. (2020). Development and Validation of a Hypoxia-Related Gene Signature to Predict Overall Survival in Early-Stage Lung Adenocarcinoma Patients. Ther. Adv. Med. Oncol. 12, 175883592093790. doi:10.1177/1758835920937904
Vilgrain, V., Pereira, H., Assenat, E., Guiu, B., Ilonca, A. D., Pageaux, G.-P., et al. (2017). Efficacy and Safety of Selective Internal Radiotherapy with Yttrium-90 Resin Microspheres Compared with Sorafenib in Locally Advanced and Inoperable Hepatocellular Carcinoma (SARAH): an Open-Label Randomised Controlled Phase 3 Trial. Lancet Oncol. 18 (12), 1624–1636. doi:10.1016/S1470-2045(17)30683-6
Wang, Y., Li, W., Chen, X., Li, Y., Wen, P., and Xu, F. (2019). MIR210HG Predicts Poor Prognosis and Functions as an Oncogenic lncRNA in Hepatocellular Carcinoma. Biomed. Pharmacother. 111, 1297–1301. doi:10.1016/j.biopha.2018.12.134
Wu, J., Pang, R., Li, M., Chen, B., Huang, J., and Zhu, Y. (2021). m6A-Induced LncRNA MEG3 Suppresses the Proliferation, Migration and Invasion of Hepatocellular Carcinoma Cell through miR-544b/BTG2 Signaling. Ott 14, 3745–3755. doi:10.2147/OTT.S289198
Wu, X.-Z., Xie, G.-R., and Chen, D. (2007). Hypoxia and Hepatocellular Carcinoma: The Therapeutic Target for Hepatocellular Carcinoma. J. Gastroenterol. Hepatol. 22 (8), 1178–1182. doi:10.1111/j.1440-1746.2007.04997.x
Wu, X., Zhang, L., Zhou, J., Liu, L., Fu, Q., Fu, A., et al. (2019). Clinicopathologic Significance of LAIR-1 Expression in Hepatocellular Carcinoma. Curr. Probl. Cancer 43 (1), 18–26. doi:10.1016/j.currproblcancer.2018.04.005
Wu, X., Zhang, X., Tao, L., Dai, X., and Chen, P. (2020). Prognostic Value of an m6A RNA Methylation Regulator-Based Signature in Patients with Hepatocellular Carcinoma. Biomed. Res. Int. 2020, 1–11. doi:10.1155/2020/2053902
Wu, Z., Zuo, M., Zeng, L., Cui, K., Liu, B., Yan, C., et al. (2021). OMA1 Reprograms Metabolism under Hypoxia to Promote Colorectal Cancer Development. EMBO Rep. 22 (1), e50827. doi:10.15252/embr.202050827
Xiao, B., Liu, L., Chen, Z., Li, A., Wang, P., Xiang, C., et al. (2021). Identification of Epithelial-Mesenchymal Transition-Related Prognostic lncRNAs Biomarkers Associated with Melanoma Microenvironment. Front. Cel Dev. Biol. 9, 679133. doi:10.3389/fcell.2021.679133
Xiao, B., Liu, L., Li, A., Xiang, C., Wang, P., Li, H., et al. (2020). Identification and Verification of Immune-Related Gene Prognostic Signature Based on ssGSEA for Osteosarcoma. Front. Oncol. 10, 607622. doi:10.3389/fonc.2020.607622
Xu, L., Huan, L., Guo, T., Wu, Y., Liu, Y., Wang, Q., et al. (2020). LncRNA SNHG11 Facilitates Tumor Metastasis by Interacting with and Stabilizing HIF-1α. Oncogene 39 (46), 7005–7018. doi:10.1038/s41388-020-01512-8
Xu, Q., Wang, Y., and Huang, W. (2021). Identification of Immune-Related lncRNA Signature for Predicting Immune Checkpoint Blockade and Prognosis in Hepatocellular Carcinoma. Int. Immunopharmacology 92, 107333. doi:10.1016/j.intimp.2020.107333
Xu, Y., Sun, C., Han, B., Xi, Y., Zhang, M., Yang, J., et al. (2020). High KIAA1522 Expression Predicts a Poor Prognosis in Patients with Hepatocellular Carcinoma. Oncol. Lett. 20 (1), 509–516. doi:10.3892/ol.2020.11588
Yarchoan, M., Xing, D., Luan, L., Xu, H., Sharma, R. B., Popovic, A., et al. (2017). Characterization of the Immune Microenvironment in Hepatocellular Carcinoma. Clin. Cancer Res. 23 (23), 7333–7339. doi:10.1158/1078-0432.CCR-17-0950
Zhang, H., Qin, C., Liu, H. W., Guo, X., and Gan, H. (2021). An Effective Hypoxia-Related Long Non-coding RNAs Assessment Model for Prognosis of Clear Cell Renal Carcinoma. Front. Oncol. 11, 616722. doi:10.3389/fonc.2021.616722
Zhang, H., Yue, R., Zhao, P., Yu, X., Li, J., Ma, G., et al. (2017). Proinflammatory Follicular Helper T Cells Promote Immunoglobulin G Secretion, Suppress Regulatory B Cell Development, and Correlate with Worse Clinical Outcomes in Gastric Cancer. Tumour Biol. 39 (6), 101042831770574. doi:10.1177/1010428317705747
Zhao, J., Song, X., Xu, T., Yang, Q., Liu, J., Jiang, B., et al. (2020). Identification of Potential Prognostic Competing Triplets in High-Grade Serous Ovarian Cancer. Front. Genet. 11, 607722. doi:10.3389/fgene.2020.607722
Keywords: hepatocellular carcinoma, hypoxia, lncRNA, prognosis, immune cells infiltration, immune checkpoint, m6A
Citation: Zhou C, Zhang H and Lu L (2021) Identification and Validation of Hypoxia-Related lncRNA Signature as a Prognostic Model for Hepatocellular Carcinoma. Front. Genet. 12:744113. doi: 10.3389/fgene.2021.744113
Received: 03 August 2021; Accepted: 07 September 2021;
Published: 28 September 2021.
Edited by:
Wei Jiang, Nanjing University of Aeronautics and Astronautics, ChinaReviewed by:
Huiyu Li, University of Texas Southwestern Medical Center, United StatesCopyright © 2021 Zhou, Zhang and Lu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Liqing Lu, bGlxaW5nMTk5MkBjc3UuZWR1LmNu
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.