
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Genet. , 01 October 2021
Sec. Epigenomics and Epigenetics
Volume 12 - 2021 | https://doi.org/10.3389/fgene.2021.740794
This article is part of the Research Topic Epigenetic and Transcriptional Dysregulations in Cancer and Therapeutic Opportunities View all 19 articles
Chromobox protein homolog 7 (CBX7) is a member of the Chromobox protein family and participates in the formation of the polycomb repressive complex 1(PRC1). In cells, CBX7 often acts as an epigenetic regulator to regulate gene expression. However, pathologically, abnormal expression of CBX7 can lead to an imbalance of gene expression, which is closely related to the occurrence and progression of cancers. In cancers, CBX7 plays a dual role; On the one hand, it contributes to cancer progression in some cancers by inhibiting oncosuppressor genes. On the other hand, it suppresses cancer progression by interacting with different molecules to regulate the synthesis of cell cycle-related proteins. In addition, CBX7 protein may interact with different RNAs (microRNAs, long noncoding RNAs, circular RNAs) in different cancer environments to participate in a variety of pathways, affecting the development of cancers. Furthermore, CBX7 is involved in cancer-related immune response and DNA repair. In conclusion, CBX7 expression is a key factor in the occurrence and progression of cancers.
CBX7 belongs to the Chromobox proteins family and participates in the formation of the polycomb repressive complex 1 (PRC1). The polycomb group (PcG) proteins are transcriptional inhibitors that regulate several important developmental and physiological processes in cells (Levine et al., 2002). PcG proteins are originally identified in Drosophila as epigenetic transcriptional repressors (Histones are modified to cause transcription inactivation) expressed by homologous genes (Hox). Nowadays, they have been found in various metazoans and are highly conserved in evolution (Wang et al., 2015). Two subunit complexes in PcG proteins play an important role in epigenetic control: one is the initial inhibitory complex (PRC2), with enhancer of zeste homolog 2 (EZH2), EED, and SUZ12 as the main components and the other is the maintenance inhibitory complex (PRC1), with B-lymphoma Mo-MLV insertion region 1 (BMI-1), Chromobox 7 (CBX7) and E3 ubiquitin ligase RING1A/B as the main components (Schuettengruber et al., 2007; Scott et al., 2007; Wu et al., 2009). Functionally, in the PRC2 complex, EZH2 performs methyltransferase activity on lysine 27 on histone H3(H3K27) and is involved in the mono-, di-, and tri-methylation of lysine 27 on histone H3 (H3K27me1/2/3), which is essential for inducing transcriptional inhibition and stable gene silencing (Satijn et al., 2001; Geng and Gao, 2020). Conversely, in the PRC1 complex, CBX7 can bind to the H3K27me3 with its special domain (a highly conserved N-terminal chromodomain), thereby controlling the expression of multiple genes. In addition, the E3 ubiquitin ligase RING1A/B, a member of the PRC1 complex, is also recruited unto genes to promote histone H2A lysine 119 monoubiquitination (H2AK119ub1). These histone modifications induce chromatin compaction and aggregation, thereby inhibiting the transcriptional activity of target genes (Satijn et al., 2001; Ogawa et al., 2002; Gil et al., 2005; Wu et al., 2009; Kaito and Iwama, 2020). CBX7 inhibits genes expression at the transcriptional level in the nucleus.
Recent studies have shown that CBX7 not only regulates gene expression in the nucleus but also interacts with proteins involved in cell cycle regulation in the cytoplasm. The CBX7 protein can be divided into several subtypes, the classic ones being 36 KD protein CBX7 in the nucleus and 22KD protein CBX7 in the cytoplasm. Emerging evidence has shown that p22CBX7 rather than p36CBX7, inhibits cell proliferation, further demonstrating the complex mechanism of CBX7 regulating cell development (Cho et al., 2020). P22CBX7 in the cytoplasm may interact with some other proteins in the cytoplasm that play an important role in cell cycle progression to regulate cell proliferation, or that there is a reciprocal conversion that regulates cellular homeostasis in a complementary manner between CBX7 subtypes, thus forming negative feedback in CBX7 (Cho et al., 2020).
Bioinformatic analysis of Chromobox proteins family has shown that the most significant difference between breast cancer and normal mammary tissue is the low mRNA expression of CBX7 (Li X. et al., 2020). One study of breast cancer implied that patients with low CBX7 had lower survival and were more likely to develop lymph node metastasis, P53 mutations, and the cancer further deteriorated, metastasized over time (Li X. et al., 2020). The mechanism for its carcinogenicity may be that CBX7 acts as a novel epigenetic regulator of the Wnt/β-catenin pathway in breast cancer to determine cancer progression (Li X. et al., 2020). In breast cancer, CBX7 increases Dickkopf-1 (DKK-1, a Wnt antagonist) gene transcription, thus indirectly affecting the Wnt signaling pathway (Bafico et al., 2001; Kim et al., 2015). For transcriptional activation of the DKK-1 gene, CBX7 can directly interact with p300 acetyltransferase and recruit p300/CREB binding protein (CBP) to the DKK-1 promoter, thereby inhibiting histone deacetylases (HDAC)-mediated histone deacetylation (Kim et al., 2015). In breast cancer, due to the lack of CBX7, the transcriptional activation complex p300/CBP dissociates from the promoter region of DKK-1 and restarts HDAC-mediated DKK-1 gene silencing. The decreased expression of Wnt antagonist DKK-1 can reactivate the Wnt/β-catenin/T-cell factor (TCF) pathway, leading to nuclear translocation of β-catenin and up-regulation of the expression of TCF target genes including C-MYC (Wang et al., 2008; Kim et al., 2015). Therefore, CBX7 plays a cancer-inhibiting role by guiding the synthesis of DKK-1 to attenuate the Wnt pathway in breast cancer cells. In addition, there is evidence that CBX7 also plays an important role in controlling glucose metabolism in breast cancer cells (Iqbal et al., 2021). mTORC1 signaling is a known determinant of cancer metabolism and frequently deregulated in breast cancer (Creighton, 2007; Mossmann et al., 2018). The increase of glycolysis induced by silencing CBX7 may imply that CBX7 regulates the mTORC1 pathway to control aerobic glycolysis in breast cancer (Iqbal et al., 2021). Both Chromobox 2 (CBX2) and CBX7 can be used as subunits of PRC1 to act as a reader to recognize H3K27m3, thus hindering the transcription of INK4a/ARF tumor suppressor genes (Jangal et al., 2019). Interestingly, CBX2, which is homologous to CBX7, has a high content in breast cancer (Liang et al., 2017). However, CBX7 content in breast cancer is often very low, indicating that CBX7 plays its other tumor suppressor role independently of PRC1. The reason why CBX7 is lost in breast cancer cells may be ascribed to lncRNA NEAT1 targeting CBX7 (Yan et al., 2020). It has been determined that the protein level of CBX7 is positively correlated with NEAT1 in breast cancer cells, proving that CBX7 is indeed a target gene regulated by NEAT1 (Yan et al., 2020). Moreover, miR-181b induced by high mobility group AT-hook 1 (HMGA1) interferes with CBX7 mRNA at the translation level to inhibit its expression, resulting in a lack of CBX7 in breast cancer (Mansueto et al., 2010). In these ways, CBX7 acts as an intermediate in breast cancer, creating a series of chain reactions.
The mRNA expression of CBX7 increases with liver development after birth and is maintained at a normal level to regulate the epigenome, transcriptome, and liver function (Lu et al., 2012). Accumulative evidence has shown that CBX7 content is significantly reduced in liver cancer tissues (Lu et al., 2012). In liver cancer, the expression of CBX7 mRNA is the highest in grade Ⅰ tumors among grade classification of tumor. With the increase of tumor grade, the expression of CBX7 mRNA shows a downward trend (Ning et al., 2018). In addition, the decreased expression of CBX7 is significantly correlated with liver cirrhosis by the chi-square test (Zhu et al., 2019). In patients with hepatocellular carcinoma, the high expression of CBX7 is related to a better survival rate of patients (Ning et al., 2018). One of the proposed mechanisms for cancer suppression is that CBX7 exerts a cancer suppressor effect by inhibiting the expression of cyclin E (Forzati et al., 2012). CBX7 and HDAC2 combine to form a complex and are fixed on the promoter of CCNE1, thereby repressing its transcriptional activity. The study suggested that overexpression of CBX7 would interfere with the composition of PRC1. In addition, it was proven that CBX7 dose-dependently reduced the transcriptional activity of the CCNE1 promoter (Forzati et al., 2012). We postulate that CBX7 exerts special effects independently of PRC1. On the contrary, the HMGA1b protein is a competitor of this action. The antagonistic effect of CBX7 versus HMGA1b protein on the CCNE1 promoter could inhibit cell proliferation and migration (Fusco and Fedele, 2007; Pallante et al., 2010; Forzati et al., 2012). For CBX7 itself, its expression may be regulated by non-coding RNA, such as miR-181 positively regulated by HMGA1. it may combine with the 3′-UTR of CBX7 mRNA to limit its translation (Forzati et al., 2014). Similarly, the mode of action of CBX7 in liver cancer is a continuous one-way pathway. CBX7 not only controls the synthesis of related proteins but also is restricted by other molecules.
CBX7 is associated with multiple clinicopathologic parameters in colon cancer. Compared with normal colonic mucosa, CBX7 expression is reduced or absent in a large number of colon cancer specimens, and the absence of CBX7 expression is remarkably correlated with the poor prognosis of colon cancer patients (Pallante et al., 2010). The mRNA level of CBX7 in colon cancer samples is lower than that in normal colonic tissues, suggesting that the CBX7 expression is inhibited at the transcriptional level (Pallante et al., 2010). However, the mechanism by which it is suppressed at the transcriptional level remains unclear. Furthermore, CBX7 negatively regulates cyclin E which is involved in a G1-S phase transition. CBX7 is positively correlated with E-cadherin expression (Federico et al., 2009; Pallante et al., 2010). However, two components of the PRC1 which includes CBX7 and BMI-1 regulate the expression of E-cadherin differently. Studies had shown that BMI-1 significantly down-regulates the content of E-cadherin in colon cancer and promotes the epithelial-mesenchymal transition (EMT) process (Zhang et al., 2016). The different regulatory effects on the same protein between the two reveal that CBX7 may be independent of PRC1 composed of BMI-1 to achieve EMT inhibition. Not surprisingly, like CBX7 in breast cancer, abnormal activation of the Wnt signaling pathway is found in 90% of colon cancer, and one of the reasons may be the absence of secreted frizzled-related protein 4(sFRP4) (Liu et al., 2020). The sFRP4 could compete with Wnt proteins via binding their receptor (Frizzled) to act as an antagonist of the Wnt signaling pathway in colonic cancer (Liu et al., 2006; Liu et al., 2020). Interestingly, both DKK-1 and sFRP4 are antagonists of the Wnt signaling pathway, but the effect of CBX7 in regulating sFRP4 in colon cancer is quite different from the effect of regulating DKK-1 in breast cancer. CBX7 and other PcG proteins, such as EZH2 and Jumonji, AT rich interactive domain 2 (JARID2) are found to be enriched in the SFRP4 gene promoter region to regulate gene expression without DNA methylation (Liu et al., 2020). Perhaps it is the synergistic effect of CBX7, EZH2, and JARID2 on histone methylation modification that leads to epigenetic gene silencing (Walters et al., 2014; Pallante et al., 2015). Therefore, silencing the sFRP4 gene indirectly enhances the Wnt pathway, thereby promoting the development of cancer. The contradiction between the two roles of CBX7 in colon cancer further suggests that multiple CBX7 regulatory pathways may exist. In cancer progression, different cancer-promoting or anti-cancer pathways have gained corresponding advantages due to their expression intensity. It would be beneficial to further actively explore the pathways of CBX7 involvement.
It has been reported that the loss of CBX7 is associated with a highly malignant phenotype in thyroid cancer (Pallante et al., 2008). CBX7mRNA is high in most of follicular thyroid adenomas and rarely develops into cancer (Pallante et al., 2008; Monaco et al., 2014). However, a low level of CBX7 is shown in thyroid cancer. For example, Hurtle adenoma is a rare differentiated thyroid tumor with the highest incidence of metastasis, and as the tumor worsens, CBX7 gradually decreases at the transcriptional level and the tumor develops into Hurtle carcinoma (Cheung et al., 2000; Monaco et al., 2014). The decrease of CBX7 in thyroid cancer may be due to the negative regulation of tumor protein HMGA1 (Chiappetta et al., 1995). A loss of epithelial features and an acquisition of mesenchymal phenotypes are signs of tumor aggressiveness, which are often related to a lack of E-cadherin (Mansueto et al., 2010). It has been confirmed that CBX7 can actively regulate the expression of E-cadherin by interacting with HDAC2 and inhibiting its effect on the E-cadherin promoter (Federico et al., 2009). In addition, HDAC2 catalyzes the acetyl transfer of core histones and is generally considered to be a transcriptional repressor due to its ability to induce gene silencing (Federico et al., 2009; Mansueto et al., 2010). In thyroid cancer, H3k27me3 catalyzed by EZH2 is overexpressed (Tsai et al., 2019). However, CBX7, which has the recognition function of H3k27me3, did not follow its increasing trend to show high expression. This indicated that CBX7 did not act as a member of PRC1 to link up EZH2-mediated H3k27me3 to assist gene silencing. Instead, CBX7 can up-regulate the expression of FOS, FOSB, and EGR1, which are mostly involved in the physiological process of inhibiting cancer (Pallante et al., 2014). The exact molecular mechanism of promoting the expression of these genes is unclear. There is clear evidence that CBX7 is associated with osteopontin protein, which is encoded by the SPP1 gene (Sepe et al., 2015). Osteopontin is a well-known protein involved in cancer progression by promoting cell invasion and migration, leading to tumor metastasis (Rohde et al., 2007). On the one hand, CBX7 regulates cell migration by blocking HMGA1b and inhibiting SPP1 gene expression; on the other hand, CBX7 and HMGA1b limit SPP1 gene expression by regulating the activity of nuclear factor kB (NF-κB) (Sepe et al., 2015).
In gliomas, unsurprisingly, CBX7 and HDAC2 jointly address the CCNE1 gene promoter induced G1/S phase arrest, which is similar to the effect of CBX7 in liver cancer, and CBX7 also enhances the expression of DKK-1 by binding to the DKK-1 promoter, which is similar to the effect of CBX7 in breast cancer (Mansueto et al., 2010; Kim et al., 2015; Bao et al., 2017; Yu et al., 2017). Not only that, there is a strong connection between CBX7 and the Hippo signaling pathway in gliomas (Nawaz et al., 2016). CBX7 inhibits YAP/TAZ, down-regulates CTGF (adverse prognostic product of tumor) with PRC2 as a member of PRC1, and reduces the phosphorylation level of c-Jun NH2-terminal kinase (JNK) (Nawaz et al., 2016). Among these, connective tissue growth factor (CTGF) plays an auxiliary role in gliomas by activating the ITGB1-TrkA-NF-kB pathway, which can inhibit E-cadherin at the transcription level and enhance the invasion and migration of glioma cells (Edwards et al., 2011). Thus, CBX7 indirectly inhibits the decrease of E-cadherin, plays an important role in its quantity homeostasis. For CBX7 expression, the orphan nuclear receptor TLX (NR2E1, a transcription repressor) is related to the regulation of CBX7 expression (OʼLoghlen et al., 2015b). The ectopic expression of NR2E1 binds to the CBX7 locus, activates CBX7 expression. In contrast, CBX7 binds to the NR2E1 locus as a member of the PRC1 and inhibits it as part of a regulatory feedback loop (OʼLoghlen et al., 2015b). These also imply that different pathways can also influence each other to regulate cancer progression.
In pancreatic cancer, the expression of CBX7 is lower than that of normal pancreatic tissues (Ni et al., 2017). Microarray and GO-pathway analysis predict that CBX7 promotes the activation of Phosphatase and tensin homolog (PTEN) and then inhibits the downstream phosphatidylinositol 3-phosphate kinase (PI3K)/AKT pathway (Ni et al., 2017). Interestingly, the mechanism of increasing PTEN transcription may be the same as the enhanced DKK-1 expression in breast cancer, where CBX7 recruits p300 independently of PRC1 to the promoter region to participate in epigenetic changes (Kim et al., 2015; Ni et al., 2017). It is well known that PTEN is a classic tumor suppressor (Ying et al., 2011). Once PTEN is abnormally expressed, it will lead to the activation of the PI3K/AKT signaling pathway, then target NF-kB and C-MYC transcription factors, which are conducive to the survival and development of cancers (Asano et al., 2004; Ying et al., 2011).
In gastric cancer, CBX7 is highly expressed, contrary to its low expression in all of the above-mentioned cancers. It has been proven that CBX7 acts as a carcinogen mainly by inducing the down-regulation of p16INK4a/ARF, which is blocked by the BMI-1 protein (Guo et al., 2007; Zhang et al., 2010). Besides, accumulated evidence shows that CBX7 has a similar function to BMI-1 in gastric cancer, suggesting that CBX7 may participate and function in a way related to BMI-1 regulation (Guo et al., 2007; Ni et al., 2018). Although multivariate Cox proportional risk model analysis shows that CBX7 is not an independent prognostic factor for gastric cancer deterioration (Zhang et al., 2010), the carcinogenesis role of CBX7 should not be underestimated. In gastric cancer, CBX7 actively regulates the stem cell characteristics of gastric cancer cells through the AKT-NF-kB-miR-21 pathway (Ni et al., 2018). Therefore, CBX7 utilizes at least the two pathways mentioned above to play a cancer-promoting role in gastric cancer (Zhang et al., 2010; Ni et al., 2018). It is worth considering that miR-21 in the microRNA family is a downstream target of CBX7. However, in the presence of miR-421 inhibitors, the growth of gastric cancer cells is inhibited. Therefore, miR-421 may act as an upstream molecule to regulate the target CBX7, suggesting a deeper link between CBX7 and the microRNA family (Jiang et al., 2010; Ni et al., 2018).
In prostate cancer, CBX7 is also positively correlated to cancer progression (Bernard et al., 2005). Overexpression of CBX7 contributes to the down-regulation of the expression of the tumor suppressor gene INK4a/ARF (Figure 1) (Bernard et al., 2005). Generally, androgens are favorable factors for prostatic cancer development, but CBX7 plays an important role in maintaining prostate cancer cells growth by cooperating with C-MYC with androgen-independent transformation (Bernard et al., 2005). Recently, it has been reported that GOLPH3 can bind to the CBX7 protein in prostatic cancer to promote cell proliferation and inhibit cell apoptosis (Gong et al., 2020). More importantly, circGOLPH3, like most of circRNAs, has abundant microRNA binding sites (Gong et al., 2020). CircGOLPH3 may have a “sponge effect”, absorbing microRNAs and reducing the number of microRNA bound to target mRNA, thus increasing the expression level of target protein such as CBX7 (Gong et al., 2020). While this clearly shows the interaction in RNAs to regulate CBX7, for the binding between GOLPH3 and CBX7, it is questionable whether circGOLPH3 can also directly regulate the activity of CBX7 protein in post-translational modification.
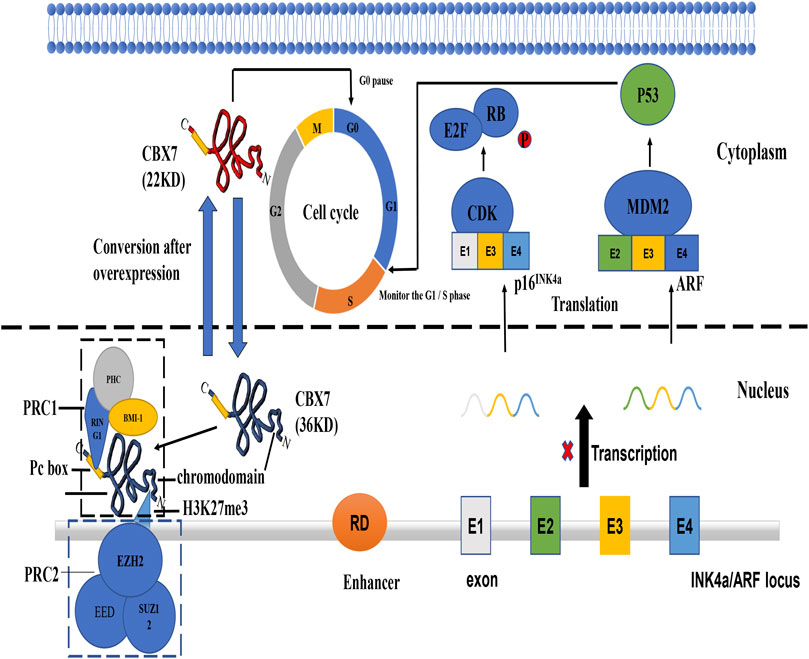
FIGURE 1. The mechanism of tumor suppressor gene INK4a/ARF and functions of different CBX7 isoforms. (Kim and Sharpless, 2006) INK4a/ARF can express two co-exon products p16 and ARF. P16 INK4a can inhibit the phosphate of RB protein by interacting with cycle-dependent kinase. Inactivation of CDK4/6 promotes Rb/E2F1 association triggering G1/S transition. ARF can bind to Mouse Double Minute 2 protein (MMD2) protein to inactivate it, thereby releasing its inhibitory effect on p53 protein. P36CBX7 inhibits the expression of ARF in the nucleus to cause p53 protein abnormalities and make the cell cycle out of control at the G1/S regulatory point. P22CBX7 interacts with related proteins in the cytoplasm to keep the cell cycle in the G0 phase. Both have opposite effects on cell proliferation, showing the duality of CBX7.
Finally, CBX7 is also highly expressed in lymphomas. CBX7 promotes T-cell lymphoma, and when sensitized oncogenes are directed to B-cell loci, CBX7 also promotes aggressive B-cell lymphoma, with a presence and latency similar to that of BMI-1 (Scott et al., 2007). CBX7 may cause the occurrence of lymphoma by enhancing stem cell self-renewal or increasing the replication potential of cancer stem cells (Scott et al., 2007). Although CBX7 may affect the transcription of multiple genes, inhibiting INK4a/ARF seems to be more important for its carcinogenic potential (Scott et al., 2007). However, CBX7 does not regulate cancer progression alone, the long latency and incomplete penetrance observed after overexpression of CBX7 suggest that aberrant CBX7 expression is not sufficient for causing lymphoma. CBX7 usually cooperates with C-MYC in the formation of aggressive lymphomas during lymphocyte genesis (Scott et al., 2007). More commonly, CBX7 is co-expressed with ZBTB7 in human follicular lymphoma, increasing the possibility that these proteins collectively control INK4a/ARF expression (Maeda et al., 2005). It also should be noted that CBX7 often works with other proteins, such as B-cell lymphoma 2(BCL-2), to accelerate the progress of lymphoma (Ramaswamy et al., 2001; Scott et al., 2007). Therefore, CBX7 could work with different partner molecules to produce different effects (pro-cancer or anti-cancer). Determining the specific molecular mechanism of CBX7 is very interesting and deserves further study.
MicroRNAs have obvious tissue specificity and can interact with CBX7 to affect cancer progression (Correia de Sousa et al., 2019). Many members of the microRNA family are involved in interference with CBX7 expression. For example, luciferase reporter assay has shown that miR-19 promotes the proliferation, migration of lung cancer cells by binding to the 3′-UTR of CBX7 mRNA and inhibits CBX7 expression at the translation level (Peng et al., 2018). Similarly, in lung adenocarcinomas, miR-181 overexpression promotes epithelial-mesenchymal transition (EMT) by directly targeting CBX7 (Pei et al., 2020). In addition, miR-375 plays a cancer-promoting role in prostate cancer by influencing the epigenetic regulation of transcriptional programs through its ability to directly target the polycomb complex member CBX7 (Pickl et al., 2016). Moreover, compared with CBX7+/+MEFs(mouse embryonic fibroblasts), CBX7−/−MEFs show a higher level of miR-181 in breast cancer while CBX7 ± MEFs express a moderate level, suggesting that CBX7 negatively regulates the expression of miR-181 (Pallante et al., 2015). Therefore, on the one hand, CBX7 can negatively regulate the expression of miR-181. On the other hand, since miR-181 binds to the mRNA 3′-UTR of CBX7, the expression of CBX7 is restricted, thus forming a synergistic cycle that promotes cancers (Pallante et al., 2015; Peng et al., 2018; Yan et al., 2020). MiR-155 is positively regulated by CBX7 in MEFs and colon carcinomas. Moreover, it has been determined that K-ras is a target of miR-155, the overexpression of miR-155 causes a sharp drop in K-ras mRNA and protein levels (Forzati et al., 2017). In glioma, the in vitro and in vivo functional assays results indicate that overexpression of miR-18a induced by miR-18a mimics may promote cell proliferation and migration of liver cancer cells by acting on CBX7 (Wu et al., 2017). On the contrary, miR-18a shows a cancer suppressor effect in primary ovarian tumors, due to the dual role of CBX7 as a bridge molecule in different cancers progression (Zhao et al., 2020). In addition, it has been reported that miR-9 can induce cell senescence without affecting the expression of CBX7, but the expression of CBX7 weakens its ability to induce senescence, which may be related to the regulation of senescence involved in p16INK4a (OʼLoghlen et al., 2015a).
There are few studies on the regulatory relationship between long noncoding RNA lncRNA and CBX7. Some lncRNAs can directly or indirectly influence the role of CBX7 in cancer progression. In breast cancer, lncRNA NEAT1 is highly expressed in breast cancer tissues and closely related to clinical stage, lymph node metastasis (Li et al., 2017; Yan et al., 2020). It has been reported that with the down-regulation of NEAT1, the expression of CBX7 protein in breast cancer cells is also significantly down-regulated (p < 0.01) (Yan et al., 2020). lncRNA NEAT1 is mainly located in the cytoplasm but also exists in the nucleus. It is speculated that lncRNA NEAT1 may play a role in regulating CBX7 expression in terms of chromatin status, DNA binding, or the fate of newly transcribed mRNA in the nucleus (Yan et al., 2020). lncRNA ANRIL can coordinate or reverse the “reader” function of CBX7 at the transcriptional level (Yap et al., 2010). The p16 gene and the gene transcribed into ANRIL are located on the same chromosome, and CBX7 chromodomain uses distinct regions and residues for binding H3K27me or lncRNA (Pasmant et al., 2007; Yap et al., 2010). Both PRC1 and PRC2 are retained at a repression site of a target gene through their association with the nascent ANRIL transcripts of Pol II, ANRIL/CBX7 binding could result in dissociation of PRC1 from H3K27me, leading to a reversal of transcriptional repression of the target gene (Yap et al., 2010). In addition, lncRNA SNHG7 can also indirectly influence the expression of CBX7 by regulating miR-181 in lung adenocarcinoma (Pei et al., 2020). It can be seen that the interaction between CBX7 and lncRNA family is not one-to-one, but a complex regulatory network between lncRNA and microRNA families.
CircRNA has been proved to be an important molecule that can be used as a therapeutic target for tumors. However, studies on the relationship between circRNA and CBX7 are few and currently limited to prostate cancer (Chen et al., 2019; Gong et al., 2020). In the study of prostate cancer, it is found that CBX7 can specifically bind to circGOLPH3 based on the results of mass spectrometry, but the detailed mechanism of circRNA-CBX7 binding remains unclear (Gong et al., 2020). The answer to the direct and specific binding of GOLPH3 to CBX7 may be that the two directly bind to prevent CBX7 degradation. Moreover, in prostate cancer, circCSNK1G3 promotes cell growth by interacting with miR-181 involved in the regulation of CBX7 (Chen et al., 2019; Pei et al., 2020). Of note, circRNAs can bind, store, classify and isolate proteins to specific subcellular locations (Huang et al., 2020). Given that the above discussion in various cancers, CBX7 mainly functions as an epigenetic regulator in the nucleus. Therefore, it is possible to speculate whether CBX7 can be dispersed into different subcellular locations under the action of circular RNAs to show a confusing result, which is mistaken for the duality of CBX7. For example, CBX7 is highly expressed in cells, however, the amount of CBX7 in the nucleus is small and its regulatory role is not significant. The relationship between circRNA family and CBX7 may further uncover the complex mechanism of CBX7.
It has been reported that CBX7-deficient CD4+T cells express more FasL and display the FasL gene promoter demethylation after antigen-specific TCR activation (Li et al., 2014). It is suggested that CBX7 can inhibit the expression of FasL in CD4+ T cells, resulting in significantly reduced demethylation of the FasL gene promoter. It is well known that the Fas-FasL association is a typical pathway that induces T cell apoptosis (Fisher et al., 1995). Therefore, CBX7 can inhibit activation-induced T cell apoptosis, indicating that it plays a role in the adaptive immune response. Interestingly, a small proportion of people infected with HIV remain asymptomatic and maintain a high CD4 + T cell count after years of seroconversion (Imami et al., 2013). A meta-analysis showed that the CBX7 gene is highly correlated with viral control (Ding et al., 2019). In addition, changes in somatic cell copy numbers of the CBX family inhibit the infiltration of immune cells in melanoma (Li D. et al., 2020). The analysis of immune infiltration for the CBX family showed that the expression of CBX7 is related to the abundance of these six immune cells (B cells, CD8+ T cells, CD4+ T cells, macrophages, neutrophils, and dendritic cells) (Li D. et al., 2020). Moreover, in prostate cancer, C-C motif chemokine ligand 2 (CCL2) is a main target of PRC1, and the H3K27me3 inhibitory marker that is bound with CBX7 is relatively low in the promoter region of the CCL2 gene, which leads to massive production of CCL2 (Su et al., 2019). The cytokine CCL2 not only promotes self-renewal by binding to C-C motif chemokine receptor 4 (CCR4) on prostate cancer cells but also recruits tumor-associated macrophages and Tregs in cancer metastasis sites by paracrine, thus creating a profound immunosuppression and angiogenesis microenvironment, confirming once again the importance of CBX7 in tumor immunity (Josefowicz et al., 2012; Su et al., 2019).
Immune cells infiltrating in the tumor microenvironment can regulate cancer progression, which is the current trend of cancer therapy (Quail and Joyce, 2013). CBX7 can play its duality in different cancer tissues, but it could be beneficial to the immune system of the human body. It would be interesting to assess whether CBX7 can be used as an intermediate molecule between cancers and immunity.
It is well known that the source of cancers can be naturalized with proto-oncogene mutations and cancer suppressor gene deletions. The fundamental reason for cancer lies within the problems within DNA damage and repair (OʼConnor, 2015). Recently, the contact between CBX7 and DNA damage repair has emerged in cancers. In melanoma, the downstream kinase targets of CBX7 are Polo-like kinase 1(PLK1) and cyclin-dependent kinase 1(CDK1), which are related to genome stability. Therefore, CBX7 is indirectly involved in DNA damage repair (Li D. et al., 2020). Moreover, in melanoma studies, it has been shown that in the presence of CBX7 inhibitors, DNA damage repair is inhibited (Connelly et al., 2016). In addition, RING1, a member of the PRC1, has been shown to possess ubiquitin ligase activity toward histone H2A, suggesting that the recruitment of polycomb proteins may be responsible for the ubiquitination of histone H2A at DNA lesions. CBX7 is also a member of the PRC1. In other words, CBX7 may locate at DNA damage sites with the recruitment of PRC1 containing RING1 (Chou et al., 2010; Pei et al., 2020). All these suggest the importance of CBX7 in DNA damage repair.
Clinically, the expression of CBX7 in different cancers is inconsistent for cancer progression, showing double-sidedness. CBX7 is lowly expressed in cancers of the breast, pancreas, liver, thyroid, colon, and glioma, but highly expressed in cancers of the stomach, prostate, and lymph. We outline above that CBX7 may regulate cell proliferation, apoptosis, and metastasis through different targets in different tissues. Based on the circumstances regarding CBX7 in cancers, it may be bold to speculate that the double-sidedness of CBX7 in cancer progression is closely related to its regulatory pathways (Table 1). Limited by our understanding of various cancer pathways, we describe some of the regulatory pathways in which CBX7 participates in cancers.
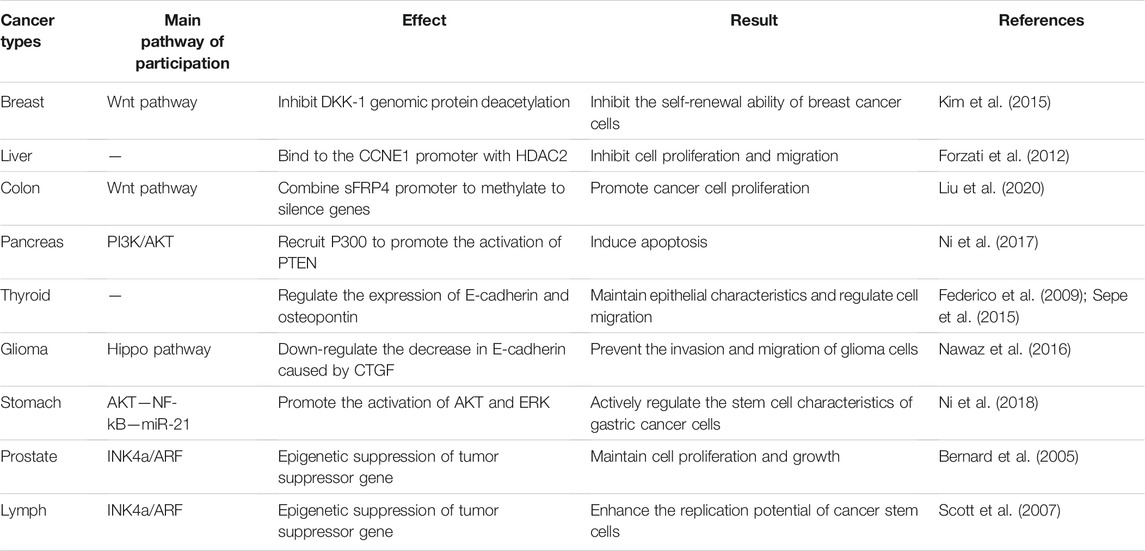
TABLE 1. CBX7 participates in various pathways and plays different physiological effects in different cancers.
In general, the current consensus on the carcinogenicity of CBX7 is that CBX7 inhibits the oncosuppressor gene INK4a/ARF (Figure 1). For its anti-cancer effect, CBX7 participates in different pathways according to the different tissue environments (Table 1). It mainly interacts with other molecules to epigenetically control the expression of related proteins, thereby regulating some cell signaling pathways to combat cancer manifestations (Such as proliferation, migration, invasion, etc. Figure 2). It needs to be pointed out that different RNA molecules have a complex relationship with CBX7, forming an intricate network between them, making the effect of CBX7 on cancer is not absolute (Figure 3).
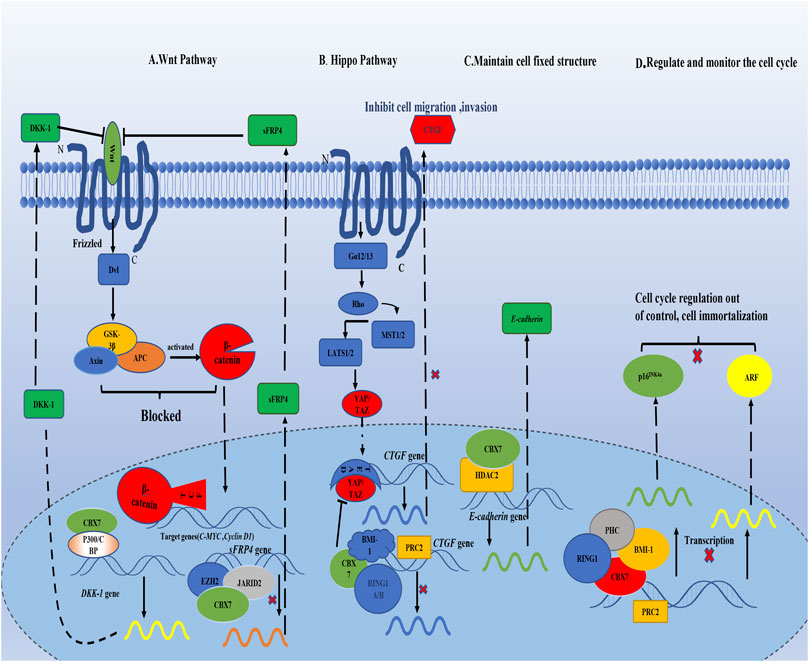
FIGURE 2. CBX7 interacts with proteins in the nucleus, transcriptionally controls the synthesis of related proteins, and regulates different signaling pathways. (A) CBX7 enhances the expression of Wnt antagonist DKK-1 by interacting with other molecules in epigenetics, leading to the blockage of downstream signal transduction of Wnt pathway, and then the expression of C-MYC and Cyclin D are restricted to maintain normal cell proliferation. But CBX7 inhibits the expression of sFRP4, which is also a Wnt antagonist.(B) On the one hand, CBX7 directly inhibits CTGF expression epigenetically; on the other hand, CBX7 antagonizes YAP/TAZ to indirectly restrict CTGF expression, which inhibits cell invasion and migration. (C) CBX7 cooperates with HDAC2 to promote the synthesis of E-cadherin in transcription to maintain cell fixed structure. (D) CBX7 relies on PRC1, and cooperates with PRC2 to epigenetically inhibit the expression of INK4a/ARF, making cell proliferation out of control.
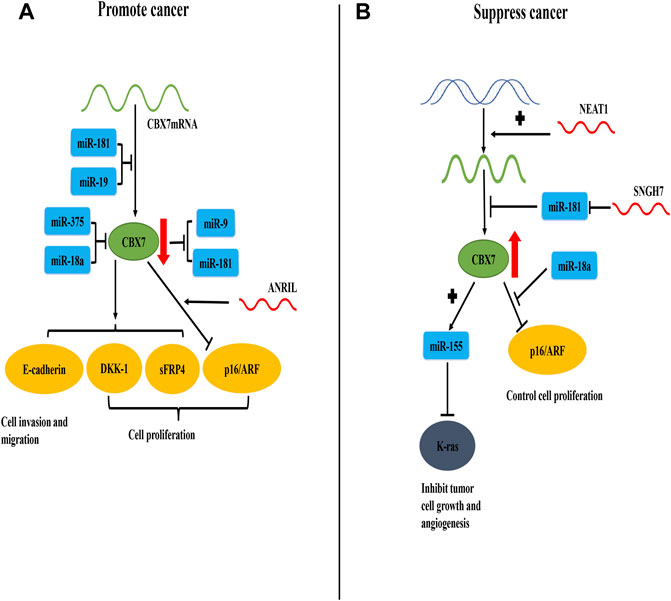
FIGURE 3. CBX7 expression is regulated by RNA molecules and the interaction between CBX7 and RNA molecules promotes or inhibits cancer progression. (A) In terms of quantity, miR-181 and miR-19 combine with the 3′-UTR of CBX7 mRNA to inhibit its translation and reduce its content. In terms of function, miR-375 and miR-18a act on CBX7 protein to decrease its activity. Therefore, the expression of DKK-1, sFRP4 and E-cadherin mediated by CBX7 decreases, resulting in uncontrolled cell proliferation, cell migration and invasion. CBX7 can also strengthen the inhibition of INK4a/ARF expression under the action of ANRIL, leading to uncontrolled cell cycle regulation and cell proliferation. Meanwhile, CBX7 can also inhibit miR-9 and miR-181. (B) In terms of quantity, NEAT1 promotes the synthesis of CBX7 at the transcription level, and SNGH7 indirectly facilitates the synthesis of CBX7 protein by blocking miR-181 at the translation level. Therefore, high content of CBX7 protein can inhibit abnormal K-ras protein via miR-155, thereby controlling tumor cell growth and angiogenesis. Meanwhile, miR-18a can relieve the transcriptional inhibition of INK4a/ARF by CBX7, thereby restoring normal cell proliferation.
CBX7 is involved in the reading recognition of H3K27me3 with its special structure and causes gene silencing, therefore CBX7 may be used as a clinical cancer treatment target. Due to the high degree of homology in the CBX family, antagonists that specifically target the CBX7 reader protein binding to histone methylated lysine have not yet been discovered, but some antagonists have a higher selectivity for CBX7 than CBX2/6/8 (Simhadri et al., 2016). Small molecules such as suramin and Ms37452 can inhibit the binding of lysine methylated polypeptides to the CBX7 chromodomain, which can be used as a feasible cancer suppressor in some cancers with high CBX7 expression, such as prostate cancer (Ren et al., 2015). Furthermore, CBX7 can also increase the sensitivity of cancer cells to chemical drugs (Cacciola et al., 2015; Iqbal et al., 2021). For example, the sensitivity of lung carcinoma cells to irinotecan and the sensitivity of breast cancer cells to FDA-approved drugs. It is worth noting that when defining the function of inhibitory polycomb complexes and evaluating the anti-cancer drugs for CBX7, the overall expression pattern should be given priority, rather than the expression of a single complex member. Of course, the complete mechanism of CBX7 has not been clarified so far. As a subunit of the PcG proteins, CBX7 not only functions independently of PRC1 but also has the function of inhibiting gene expression together with PRC1 as a whole and PCR2 (Scott et al., 2007; Pallante et al., 2015). These phenomena deserve further investigation to elucidate their mechanisms and may provide evidence for the clinical prognostic value and therapeutic targets of cancer.
More importantly, in this review, we briefly illustrated the duality of the interaction between CBX7 and various RNAs in cancers (Figure 3). Similarly, other molecules that use CBX7 as an intermediate for physiological effects can also play a dual role in different cancer progression, such as miR-18a (Wu et al., 2017; Zhao et al., 2020). It is indisputable that the specificity of the environment in which the tissue cells are located affects the duality of CBX7. Additionally, the physical and chemical environment of the tissue may also greatly influence the expression of CBX7 (Chiu et al., 2020). It has been reported that under hypoxic conditions activated hypoxia-inducible factor-1α (HIF-1α) directly binds to the CBX7 gene promoter and activates the expression of CBX7, and hypoxia-induced overexpression of CBX7 stimulates the proliferation of nasopharyngeal carcinoma cells in the ischemic brain (Chiu et al., 2020).
The most important function of the CBX7 protein is to regulate gene expression by epigenetics. In some cases, it can control cell proliferation, cell apoptosis, and self-renewal. Besides, the cellular effects mediated by CBX7 are diverse in different cancer environments. Therefore, actively guiding the expression of CBX7 in vivo according to cancer types may be helpful in guiding the clinical progress of cancer treatment.
TO proposed ideas for this article and participated in revising the article. JL wrote, revised the paper, and made diagrams. ML, TH, AM, WM, and NZ reviewed and made significant revisions to the manuscript. All authors have read and approved the final manuscript.
The National Natural Science Foundation of China, grant no. 81760447 (TO); The National Natural Science Foundation of China, grant no. 81960247 (NZ); Project of Science and Technology Department of Jiangxi Province, grant no. S2019QNJJB1056 (NZ); Project of Science and Technology Department of Jiangxi Province, grant no.20202BABL206099 (WM); Jiangxi Provincial Education Department Project, grant no. GJJ180054 (TO); Jiangxi Provincial Education Department Project, grant no. GJJ180116 (NZ).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
We thank Emmy Graham for the language polish of this review and put forward suggestions.
Asano, T., Yao, Y., Zhu, J., Li, D., Abbruzzese, J. L., and Reddy, S. A. G. (2004). The PI 3-kinase/Akt Signaling Pathway Is Activated Due to Aberrant Pten Expression and Targets Transcription Factors NF-Κb and C-Myc in Pancreatic Cancer Cells. Oncogene 23, 8571–8580. doi:10.1038/sj.onc.1207902
Bafico, A., Liu, G., Yaniv, A., Gazit, A., and Aaronson, S. A. (2001). Novel Mechanism of Wnt Signalling Inhibition Mediated by Dickkopf-1 Interaction with LRP6/Arrow. Nat. Ce.l Biol. 3, 683–686. doi:10.1038/35083081
Bao, Z., Xu, X., Liu, Y., Chao, H., Lin, C., Li, Z., et al. (2017). CBX7 Negatively Regulates Migration and Invasion in Glioma via Wnt/β-Catenin Pathway Inactivation. Oncotarget 8, 39048–39063. doi:10.18632/oncotarget.16587
Bernard, D., Martinez-Leal, J. F., Rizzo, S., Martinez, D., Hudson, D., Visakorpi, T., et al. (2005). CBX7 Controls the Growth of normal and Tumor-Derived Prostate Cells by Repressing the Ink4a/Arf Locus. Oncogene 24, 5543–5551. doi:10.1038/sj.onc.1208735
Cacciola, N. A., Sepe, R., Forzati, F., Federico, A., Pellecchia, S., Malapelle, U., et al. (2015). Restoration of CBX7 Expression Increases the Susceptibility of Human Lung Carcinoma Cells to Irinotecan Treatment. Naunyn-Schmiedeberg's Arch. Pharmacol. 388, 1179–1186. doi:10.1007/s00210-015-1153-y
Chen, S., Huang, V., Xu, X., Livingstone, J., Soares, F., Jeon, J., et al. (2019). Widespread and Functional RNA Circularization in Localized Prostate Cancer. Cell 176, 831–843.e822. doi:10.1016/j.cell.2019.01.025
Cheung, C. C., Ezzat, S., Ramyar, L., Freeman, J. L., and Asa, S. L. (2000). Molecular Basis of Hurthle Cell Papillary Thyroid Carcinoma1. J. Clin. Endocrinol. Metab. 85, 878–882. doi:10.1210/jcem.85.2.6404
Chiappetta, G., Bandiera, A., Berlingieri, M. T., Visconti, R., Manfioletti, G., Battista, S., et al. (1995). The Expression of the High Mobility Group HMGI (Y) Proteins Correlates with the Malignant Phenotype of Human Thyroid Neoplasias. Oncogene 10, 1307–1314.
Chiu, H. Y., Lee, H. T., Lee, K. H., Zhao, Y., Hsu, C. Y., and Shyu, W. C. (2020). Mechanisms of Ischaemic Neural Progenitor Proliferation: a Regulatory Role of the HIF‐1α‐CBX7 Pathway. Neuropathol. Appl. Neurobiol. 46, 391–405. doi:10.1111/nan.12585
Cho, K.-W., Andrade, M., Zhang, Y., and Yoon, Y.-S. (2020). Mammalian CBX7 Isoforms P36 and P22 Exhibit Differential Responses to Serum, Varying Functions for Proliferation, and Distinct Subcellular Localization. Sci. Rep. 10, 8061. doi:10.1038/s41598-020-64908-2
Chou, D. M., Adamson, B., Dephoure, N. E., Tan, X., Nottke, A. C., Hurov, K. E., et al. (2010). A Chromatin Localization Screen Reveals Poly (ADP Ribose)-Regulated Recruitment of the Repressive Polycomb and NuRD Complexes to Sites of DNA Damage. Proc. Natl. Acad. Sci. 107, 18475–18480. doi:10.1073/pnas.1012946107
Connelly, K. E., Martin, E. C., and Dykhuizen, E. C. (2016). CBX Chromodomain Inhibition Enhances Chemotherapy Response in Glioblastoma Multiforme. Yale J. Biol. Med. 89, 431–440.
Correia de Sousa, M., Gjorgjieva, M., Dolicka, D., Sobolewski, C., and Foti, M. (2019). Deciphering miRNAs' Action through miRNA Editing. Ijms 20, 6249. doi:10.3390/ijms20246249
Creighton, C. J. (2007). A Gene Transcription Signature of the Akt/mTOR Pathway in Clinical Breast Tumors. Oncogene 26, 4648–4655. doi:10.1038/sj.onc.1210245
Ding, J., Ma, L., Zhao, J., Xie, Y., Zhou, J., Li, X., et al. (2019). An Integrative Genomic Analysis of Transcriptional Profiles Identifies Characteristic Genes and Patterns in HIV-Infected Long-Term Non-Progressors and Elite Controllers. J. Transl Med. 17, 35. doi:10.1186/s12967-019-1777-7
Edwards, L. A., Woolard, K., Son, M. J., Li, A., Lee, J., Ene, C., et al. (2011). Effect of Brain- and Tumor-Derived Connective Tissue Growth Factor on Glioma Invasion. J. Natl. Cancer Inst. 103, 1162–1178. doi:10.1093/jnci/djr224
Federico, A., Pallante, P., Bianco, M., Ferraro, A., Esposito, F., Monti, M., et al. (2009). Chromobox Protein Homologue 7 Protein, with Decreased Expression in Human Carcinomas, Positively Regulates E-Cadherin Expression by Interacting with the Histone Deacetylase 2 Protein. Cancer Res. 69, 7079–7087. doi:10.1158/0008-5472.can-09-1542
Fisher, G. H., Rosenberg, F. J., Straus, S. E., Dale, J. K., Middelton, L. A., Lin, A. Y., et al. (1995). Dominant Interfering Fas Gene Mutations Impair Apoptosis in a Human Autoimmune Lymphoproliferative Syndrome. Cell 81, 935–946. doi:10.1016/0092-8674(95)90013-6
Forzati, F., De Martino, M., Esposito, F., Sepe, R., Pellecchia, S., Malapelle, U., et al. (2017). miR-155 Is Positively Regulated by CBX7 in Mouse Embryonic Fibroblasts and colon Carcinomas, and Targets the KRAS Oncogene. BMC Cancer 17, 170. doi:10.1186/s12885-017-3158-z
Forzati, F., Federico, A., Pallante, P., Abbate, A., Esposito, F., Malapelle, U., et al. (2012). CBX7 Is a Tumor Suppressor in Mice and Humans. J. Clin. Invest. 122, 612–623. doi:10.1172/jci58620
Forzati, F., Federico, A., Pallante, P., Colamaio, M., Esposito, F., Sepe, R., et al. (2014). CBX7 Gene Expression Plays a Negative Role in Adipocyte Cell Growth and Differentiation. Biol. open 3, 871–879. doi:10.1242/bio.20147872
Fusco, A., and Fedele, M. (2007). Roles of HMGA Proteins in Cancer. Nat. Rev. Cancer 7, 899–910. doi:10.1038/nrc2271
Geng, Z., and Gao, Z. (2020). Mammalian PRC1 Complexes: Compositional Complexity and Diverse Molecular Mechanisms. Ijms 21, 8594. doi:10.3390/ijms21228594
Gil, J., Bernard, D., and Peters, G. (2005). Role of Polycomb Group Proteins in Stem Cell Self-Renewal and Cancer. DNA Cel. Biol. 24, 117–125. doi:10.1089/dna.2005.24.117
Gong, L., Tang, Y., Jiang, L., Tang, W., and Luo, S. (2020). Regulation of circGOLPH3 and its Binding Protein CBX7 on the Proliferation and Apoptosis of Prostate Cancer Cells. Biosci. Rep. 40, BSR20200936. doi:10.1042/bsr20200936
Guo, W.-J., Zeng, M.-S., Yadav, A., Song, L.-B., Guo, B.-H., Band, V., et al. (2007). Mel-18 Acts as a Tumor Suppressor by Repressing Bmi-1 Expression and Down-Regulating Akt Activity in Breast Cancer Cells. Cancer Res. 67, 5083–5089. doi:10.1158/0008-5472.can-06-4368
Huang, A., Zheng, H., Wu, Z., Chen, M., and Huang, Y. (2020). Circular RNA-Protein Interactions: Functions, Mechanisms, and Identification. Theranostics 10, 3503–3517. doi:10.7150/thno.42174
Imami, N., Westrop, S. J., Grageda, N., and Herasimtschuk, A. A. (2013). Long-Term Non-Progression and Broad HIV-1-Specific Proliferative T-Cell Responses. Front. Immunol. 4, 58. doi:10.3389/fimmu.2013.00058
Iqbal, M. A., Siddiqui, S., Ur Rehman, A., Siddiqui, F. A., Singh, P., Kumar, B., et al. (2021). Multiomics Integrative Analysis Reveals Antagonistic Roles of CBX2 and CBX7 in Metabolic Reprogramming of Breast Cancer. Mol. Oncol. 15, 1450–1465. doi:10.1002/1878-0261.12894
Jangal, M., Lebeau, B., and Witcher, M. (2019). Beyond EZH2: Is the Polycomb Protein CBX2 an Emerging Target for Anti-cancer Therapy. Expert Opin. Ther. Targets 23, 565–578. doi:10.1080/14728222.2019.1627329
Jiang, Z., Guo, J., Xiao, B., Miao, Y., Huang, R., Li, D., et al. (2010). Increased Expression of miR-421 in Human Gastric Carcinoma and its Clinical Association. J. Gastroenterol. 45, 17–23. doi:10.1007/s00535-009-0135-6
Josefowicz, S. Z., Lu, L.-F., and Rudensky, A. Y. (2012). Regulatory T Cells: Mechanisms of Differentiation and Function. Annu. Rev. Immunol. 30, 531–564. doi:10.1146/annurev.immunol.25.022106.141623
Kaito, S., and Iwama, A. (2020). Pathogenic Impacts of Dysregulated Polycomb Repressive Complex Function in Hematological Malignancies. Int. J. Mol. Sci. 22, 74. doi:10.3390/ijms22010074
Kim, H. Y., Park, J. H., Won, H. Y., Lee, J. Y., and Kong, G. (2015). CBX7 Inhibits Breast Tumorigenicity through DKK‐1‐mediated Suppression of the Wnt/β‐Catenin Pathway. FASEB j. 29, 300–313. doi:10.1096/fj.14-253997
Kim, W. Y., and Sharpless, N. E. (2006). The Regulation of INK4/ARF in Cancer and Aging. Cell 127, 265–275. doi:10.1016/j.cell.2006.10.003
Levine, S. S., Weiss, A., Erdjument-Bromage, H., Shao, Z., Tempst, P., and Kingston, R. E. (2002). The Core of the Polycomb Repressive Complex Is Compositionally and Functionally Conserved in Flies and Humans. Mol. Cel. Biol. 22, 6070–6078. doi:10.1128/mcb.22.17.6070-6078.2002
Li, D., Liu, Y., Hao, S., Chen, B., and Li, A. (2020a). Mining Database for the Clinical Significance and Prognostic Value of CBX Family in Skin Cutaneous Melanoma. J. Clin. Lab. Anal. 34, e23537. doi:10.1002/jcla.23537
Li, J., Li, Y., Cao, Y., Yuan, M., Gao, Z., Guo, X., et al. (2014). Polycomb Chromobox (Cbx) 7 Modulates Activation-Induced CD4+ T Cell Apoptosis. Arch. Biochem. Biophys. 564, 184–188. doi:10.1016/j.abb.2014.10.004
Li, X., Gou, J., Li, H., and Yang, X. (2020b). Bioinformatic Analysis of the Expression and Prognostic Value of Chromobox Family Proteins in Human Breast Cancer. Sci. Rep. 10, 17739. doi:10.1038/s41598-020-74792-5
Li, X., Wang, S., Li, Z., Long, X., Guo, Z., Zhang, G., et al. (2017). The lncRNA NEAT1 Facilitates Cell Growth and Invasion via the miR-211/HMGA2 axis in Breast Cancer. Int. J. Biol. macromolecules 105, 346–353. doi:10.1016/j.ijbiomac.2017.07.053
Liang, Y.-K., Lin, H.-Y., Chen, C.-F., and Zeng, D. (2017). Prognostic Values of Distinct CBX Family Members in Breast Cancer. Oncotarget 8, 92375–92387. doi:10.18632/oncotarget.21325
Liu, T.-H., Raval, A., Chen, S.-S., Matkovic, J. J., Byrd, J. C., and Plass, C. (2006). CpG Island Methylation and Expression of the Secreted Frizzled-Related Protein Gene Family in Chronic Lymphocytic Leukemia. Cancer Res. 66, 653–658. doi:10.1158/0008-5472.can-05-3712
Liu, Y., Yu, J., Xie, Y., Li, M., Wang, F., Zhang, J., et al. (2020). EZH2 Regulates sFRP4 Expression without Affecting the Methylation of sFRP4 Promoter DNA in Colorectal Cancer Cell Lines. Exp. Ther. Med. 20, 33. doi:10.3892/etm.2020.9160
Lu, H., Cui, J., Gunewardena, S., Yoo, B., Zhong, X.-B., and Klaassen, C. (2012). Hepatic Ontogeny and Tissue Distribution of mRNAs of Epigenetic Modifiers in Mice Using RNA-Sequencing. Epigenetics 7, 914–929. doi:10.4161/epi.21113
Maeda, T., Hobbs, R. M., Merghoub, T., Guernah, I., Zelent, A., Cordon-Cardo, C., et al. (2005). Role of the Proto-Oncogene Pokemon in Cellular Transformation and ARF Repression. Nature 433, 278–285. doi:10.1038/nature03203
Mansueto, G., Forzati, F., Ferraro, A., Pallante, P., Bianco, M., Esposito, F., et al. (2010). Identification of a New Pathway for Tumor Progression: MicroRNA-181b Up-Regulation and CBX7 Down-Regulation by HMGA1 Protein. Genes & Cancer 1, 210–224. doi:10.1177/1947601910366860
Monaco, M., Chiappetta, G., Aiello, C., Federico, A., Sepe, R., Russo, D., et al. (2014). CBX7 Expression in Oncocytic Thyroid Neoplastic Lesions (Hürthle Cell Adenomas and Carcinomas). Eur. Thyroid J. 3, 211–216. doi:10.1159/000367989
Mossmann, D., Park, S., and Hall, M. N. (2018). mTOR Signalling and Cellular Metabolism Are Mutual Determinants in Cancer. Nat. Rev. Cancer 18, 744–757. doi:10.1038/s41568-018-0074-8
Nawaz, Z., Patil, V., Arora, A., Hegde, A. S., Arivazhagan, A., Santosh, V., et al. (2016). Cbx7 Is Epigenetically Silenced in Glioblastoma and Inhibits Cell Migration by Targeting YAP/TAZ-Dependent Transcription. Sci. Rep. 6, 27753. doi:10.1038/srep27753
Ni, S.-J., Zhao, L.-Q., Wang, X.-F., Wu, Z.-H., Hua, R.-X., Wan, C.-H., et al. (2018). CBX7 Regulates Stem Cell-Like Properties of Gastric Cancer Cells via P16 and AKT-NF-Κb-miR-21 Pathways. J. Hematol. Oncol. 11, 17. doi:10.1186/s13045-018-0562-z
Ni, S., Wang, H., Zhu, X., Wan, C., Xu, J., Lu, C., et al. (2017). CBX7 Suppresses Cell Proliferation, Migration, and Invasion through the Inhibition of PTEN/Akt Signaling in Pancreatic Cancer. Oncotarget 8, 8010–8021. doi:10.18632/oncotarget.14037
Ning, G., Huang, Y.-L., Zhen, L.-M., Xu, W.-X., Jiao, Q., Yang, F.-J., et al. (2018). Transcriptional Expressions of Chromobox 1/2/3/6/8 as Independent Indicators for Survivals in Hepatocellular Carcinoma Patients. Aging 10, 3450–3473. doi:10.18632/aging.101658
O'Loghlen, A., Brookes, S., Martin, N., Rapisarda, V., Peters, G., and Gil, J. (2015a). CBX7 and miR-9 Are Part of an Autoregulatory Loop Controlling p16INK4a. Aging cell 14, 1113–1121. doi:10.1111/acel.12404
O'Loghlen, A., Martin, N., Krusche, B., Pemberton, H., Alonso, M. M., Chandler, H., et al. (2015b). The Nuclear Receptor NR2E1/TLX Controls Senescence. Oncogene 34, 4069–4077. doi:10.1038/onc.2014.335
O’Connor, M. J. (2015). Targeting the DNA Damage Response in Cancer. Mol. Cel 60, 547–560. doi:10.1016/j.molcel.2015.10.040
Ogawa, H., Ishiguro, K., Gaubatz, S., Livingston, D. M., and Nakatani, Y. (2002). A Complex with Chromatin Modifiers that Occupies E2F- and Myc-Responsive Genes in G0 Cells. Science (New York, NY) 296, 1132–1136. doi:10.1126/science.1069861
Pallante, P., Forzati, F., Federico, A., Arra, C., and Fusco, A. (2015). Polycomb Protein Family Member CBX7 Plays a Critical Role in Cancer Progression. Am. J. Cancer Res. 5, 1594–1601.
Pallante, P., Federico, A., Berlingieri, M. T., Bianco, M., Ferraro, A., Forzati, F., et al. (2008). Loss of the CBX7 Gene Expression Correlates with a Highly Malignant Phenotype in Thyroid Cancer. Cancer Res. 68, 6770–6778. doi:10.1158/0008-5472.can-08-0695
Pallante, P., Sepe, R., Federico, A., Forzati, F., Bianco, M., and Fusco, A. (2014). CBX7 Modulates the Expression of Genes Critical for Cancer Progression. PLoS One 9, e98295. doi:10.1371/journal.pone.0098295
Pallante, P., Terracciano, L., Carafa, V., Schneider, S., Zlobec, I., Lugli, A., et al. (2010). The Loss of the CBX7 Gene Expression Represents an Adverse Prognostic Marker for Survival of colon Carcinoma Patients. Eur. J. Cancer 46, 2304–2313. doi:10.1016/j.ejca.2010.05.011
Pasmant, E., Laurendeau, I., Héron, D., Vidaud, M., Vidaud, D., and Bièche, I. (2007). Characterization of a Germ-Line Deletion, Including the Entire INK4/ARF Locus, in a Melanoma-Neural System Tumor Family: Identification of ANRIL, an Antisense Noncoding RNA Whose Expression Coclusters with ARF. Cancer Res. 67, 3963–3969. doi:10.1158/0008-5472.can-06-2004
Pei, Y.-F., He, Y., Hu, L.-Z., Zhou, B., Xu, H.-Y., and Liu, X.-Q. (2020). The Crosstalk between lncRNA-SNHG7/miRNA-181/cbx7 Modulates Malignant Character in Lung Adenocarcinoma. Am. J. Pathol. 190, 1343–1354. doi:10.1016/j.ajpath.2020.02.011
Peng, X., Guan, L., and Gao, B. (2018). miRNA-19 Promotes Non-Small-Cell Lung Cancer Cell Proliferation via Inhibiting CBX7 Expression. Onco Targets Ther. 11, 8865–8874. doi:10.2147/ott.s181433
Pickl, J. M. A., Tichy, D., Kuryshev, V. Y., Tolstov, Y., Falkenstein, M., Schüler, J., et al. (2016). Ago-RIP-Seq Identifies Polycomb Repressive Complex I Member CBX7 as a Major Target of miR-375 in Prostate Cancer Progression. Oncotarget 7, 59589–59603. doi:10.18632/oncotarget.10729
Quail, D. F., and Joyce, J. A. (2013). Microenvironmental Regulation of Tumor Progression and Metastasis. Nat. Med. 19, 1423–1437. doi:10.1038/nm.3394
Ramaswamy, S., Tamayo, P., Rifkin, R., Mukherjee, S., Yeang, C.-H., Angelo, M., et al. (2001). Multiclass Cancer Diagnosis Using Tumor Gene Expression Signatures. Proc. Natl. Acad. Sci. 98, 15149–15154. doi:10.1073/pnas.211566398
Ren, C., Morohashi, K., Plotnikov, A. N., Jakoncic, J., Smith, S. G., Li, J., et al. (2015). Small-Molecule Modulators of Methyl-Lysine Binding for the CBX7 Chromodomain. Chem. Biol. 22, 161–168. doi:10.1016/j.chembiol.2014.11.021
Rohde, F., Rimkus, C., Friederichs, J., Rosenberg, R., Marthen, C., Doll, D., et al. (2007). Expression of Osteopontin, a Target Gene of De-regulated Wnt Signaling, Predicts Survival in colon Cancer. Int. J. Cancer 121, 1717–1723. doi:10.1002/ijc.22868
Satijn, D. P. E., Hamer, K. M., den Blaauwen, J., and Otte, A. P. (2001). The Polycomb Group Protein EED Interacts with YY1, and Both Proteins Induce Neural Tissue in Xenopus Embryos. Mol. Cel. Biol. 21, 1360–1369. doi:10.1128/mcb.21.4.1360-1369.2001
Schuettengruber, B., Chourrout, D., Vervoort, M., Leblanc, B., and Cavalli, G. (2007). Genome Regulation by Polycomb and Trithorax Proteins. Cell 128, 735–745. doi:10.1016/j.cell.2007.02.009
Scott, C. L., Gil, J., Hernando, E., Teruya-Feldstein, J., Narita, M., Martínez, D., et al. (2007). Role of the Chromobox Protein CBX7 in Lymphomagenesis. Proc. Natl. Acad. Sci. 104, 5389–5394. doi:10.1073/pnas.0608721104
Sepe, R., Formisano, U., Federico, A., Forzati, F., Bastos, A. U., D’Angelo, D., et al. (2015). CBX7 and HMGA1b Proteins Act in Opposite Way on the Regulation of the SPP1 Gene Expression. Oncotarget 6, 2680–2692. doi:10.18632/oncotarget.2777
Simhadri, C., Gignac, M. C., Anderson, C. J., Milosevich, N., Dheri, A., Prashar, N., et al. (2016). Structure-Activity Relationships of Cbx7 Inhibitors, Including Selectivity Studies against Other Cbx Proteins. ACS Omega 1, 541–551. doi:10.1021/acsomega.6b00120
Su, W., Han, H. H., Wang, Y., Zhang, B., Zhou, B., Cheng, Y., et al. (2019). The Polycomb Repressor Complex 1 Drives Double-Negative Prostate Cancer Metastasis by Coordinating Stemness and Immune Suppression. Cancer Cell 36, 139–155.e110. doi:10.1016/j.ccell.2019.06.009
Tsai, C.-C., Chien, M.-N., Chang, Y.-C., Lee, J.-J., Dai, S.-H., and Cheng, S.-P. (2019). Overexpression of Histone H3 Lysine 27 Trimethylation Is Associated with Aggressiveness and Dedifferentiation of Thyroid Cancer. Endocr. Pathol. 30, 305–311. doi:10.1007/s12022-019-09586-1
Walters, Z. S., Villarejo-Balcells, B., Olmos, D., Buist, T. W. S., Missiaglia, E., Allen, R., et al. (2014). JARID2 Is a Direct Target of the PAX3-FOXO1 Fusion Protein and Inhibits Myogenic Differentiation of Rhabdomyosarcoma Cells. Oncogene 33, 1148–1157. doi:10.1038/onc.2013.46
Wang, J., Wang, H., Li, Z., Wu, Q., Lathia, J. D., McLendon, R. E., et al. (2008). c-Myc Is Required for Maintenance of Glioma Cancer Stem Cells. PLoS One 3, e3769. doi:10.1371/journal.pone.0003769
Wang, W., Qin, J.-J., Voruganti, S., Nag, S., Zhou, J., and Zhang, R. (2015). Polycomb Group (PcG) Proteins and Human Cancers: Multifaceted Functions and Therapeutic Implications. Med. Res. Rev. 35, 1220–1267. doi:10.1002/med.21358
Wu, J. I., Lessard, J., and Crabtree, G. R. (2009). Understanding the Words of Chromatin Regulation. Cell 136, 200–206. doi:10.1016/j.cell.2009.01.009
Wu, W., Zhou, X., Yu, T., Bao, Z., Zhi, T., Jiang, K., et al. (2017). The Malignancy of miR-18a in Human Glioblastoma via Directly Targeting CBX7. Am. J. Cancer Res. 7, 64–76.
Yan, L., Zhang, Z., Yin, X., and Li, Y. (2020). lncRNA NEAT1 Facilitates Cell Proliferation, Invasion and Migration by Regulating CBX7 and RTCB in Breast Cancer. Onco Targets Ther. 13, 2449–2458. doi:10.2147/ott.s240769
Yap, K. L., Li, S., Muñoz-Cabello, A. M., Raguz, S., Zeng, L., Mujtaba, S., et al. (2010). Molecular Interplay of the Noncoding RNA ANRIL and Methylated Histone H3 Lysine 27 by Polycomb CBX7 in Transcriptional Silencing of INK4a. Mol. Cel. 38, 662–674. doi:10.1016/j.molcel.2010.03.021
Ying, H., Elpek, K. G., Vinjamoori, A., Zimmerman, S. M., Chu, G. C., Yan, H., et al. (2011). PTEN Is a Major Tumor Suppressor in Pancreatic Ductal Adenocarcinoma and Regulates an NF-Κb-Cytokine Network. Cancer Discov. 1, 158–169. doi:10.1158/2159-8290.cd-11-0031
Yu, T., Wu, Y., Hu, Q., Zhang, J., Nie, E., Wu, W., et al. (2017). CBX7 Is a Glioma Prognostic Marker and Induces G1/S Arrest via the Silencing of CCNE1. Oncotarget 8, 26637–26647. doi:10.18632/oncotarget.15789
Zhang, X.-W., Zhang, L., Qin, W., Yao, X.-H., Zheng, L.-Z., Liu, X., et al. (2010). Oncogenic Role of the Chromobox Protein CBX7 in Gastric Cancer. J. Exp. Clin. Cancer Res. 29, 114. doi:10.1186/1756-9966-29-114
Zhang, Z., Bu, X., Chen, H., Wang, Q., and Sha, W. (2016). Bmi-1 Promotes the Invasion and Migration of colon Cancer Stem Cells through the Downregulation of E-Cadherin. Int. J. Mol. Med. 38, 1199–1207. doi:10.3892/ijmm.2016.2730
Zhao, Y., Liu, X. L., Huang, J. H., Yin, A. J., and Zhang, H. (2020). MicroRNA-18a Suppresses Ovarian Carcinoma Progression by Targeting CBX7 and Regulating ERK/MAPK Signaling Pathway and Epithelial-To-Mesenchymal Transition. Eur. Rev. Med. Pharmacol. Sci. 24, 5292–5302. doi:10.26355/eurrev_202005_21311
Keywords: CBX7, cancer, molecular interaction, PRC1, RNAs
Citation: Li J, Ouyang T, Li M, Hong T, Alriashy M, Meng W and Zhang N (2021) CBX7 is Dualistic in Cancer Progression Based on its Function and Molecular Interactions. Front. Genet. 12:740794. doi: 10.3389/fgene.2021.740794
Received: 22 July 2021; Accepted: 17 September 2021;
Published: 01 October 2021.
Edited by:
Rais Ahmad Ansari, Nova Southeastern University, United StatesReviewed by:
Luigi Aloia, University College London, United KingdomCopyright © 2021 Li, Ouyang, Li, Hong, Alriashy, Meng and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Wei Meng, NjAxMTkyNzk1QHFxLmNvbQ==; Na Zhang, aHVzdHpoYW5nbmFAMTYzLmNvbQ==
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.