- 1College of Animal Science and Technology, China Agricultural University, Beijing, China
- 2Guangdong Laboratory of Lingnan Modern Agriculture, Genome Analysis Laboratory of the Ministry of Agriculture and Rural Affairs, Shenzhen Branch, Agricultural Genomics Institute at Shenzhen, Chinese Academy of Agricultural Sciences, Shenzhen, China
- 3State Key Laboratory of Sheep Genetic Improvement and Healthy Breeding, Xinjiang Academy of Agricultural and Reclamation Sciences, Shihezi, China
Sheep (Ovis aries) is one of the important livestock with diverse phenotypic traits. However, little is known about the molecular mechanism of diverse phenotypic traits in domestic sheep. Using the genome-wide high-density SNP data (600K) in 253 samples from 13 populations, we conducted the tests of selective sweeps (i.e., pairwise FST and XP-CLR) associated with several important phenotypic traits (e.g., tail types, horn morphology, prolificacy, coat pigmentation, ear size, milk production, meat production, body size and wool fineness). We identified strong selective signatures in previously reported (e.g., T, RXFP2, BMPR1B, TYRP1, MSRB3, TF, CEBPA, GPR21 and HOXC8) and novel genes associated with the traits, such as CERS6, BTG1, RYR3, SLC6A4, NNAT and OGT for fat deposition in the tails, FOXO4 for fertility, PTCH1 and EMX2 for ear size, and RMI1 and SCD5 for body size. Further gene annotation analysis showed that these genes were identified to be the most probable genes accounting for the diverse phenotypic traits. Our results provide novel insights into the genetic mechanisms underlying the traits and also new genetic markers for genetic improvement in sheep and other livestock.
Introduction
Sheep (Ovis aries) is an excellent model species for investigating the genetic basis of diverse phenotypic traits under the effects of genetic drift, natural and artificial selection factors (Cao et al., 2020). Following domestication, as many as 1,400 breeds have been developed in sheep (Scherf, 2000). In particular, human-imposed selection has affected the species greatly over the past hundreds of years, and, thus, diverse phenotypic traits have been formed in different breeds, such as fat-rumped sheep (Kazakh Edilbai), thin-tailed sheep (Celle Black), Polled (Merino), high prolificacy sheep (Hu) and dairy sheep (Lacaune) (Wei et al., 2015; see Table 1).
The recent availability of genome-wide SNPs gave a new momentum to identify the genetic variants underlying phenotypic traits (Kijas et al., 2012; Xu and Li 2017; Gui et al., 2020; Abousoliman et al., 2021; Zhou et al., 2021). Previous studies have identified a number of candidate genes or variants associated with meat, growth, milk, wool, reproduction, horns and tails in sheep, most of which have employed the low-density SNPs (50K) (see the review in Xu and Li, 2017) or whole-genome sequences (Li et al., 2020). However, to date, little is known regarding the molecular mechanism of diverse phenotypic traits in sheep, such as dairy and horn traits within breeds. Here, we applied genome-wide selective scans to detect critical genes associated with the phenotypic traits based on the Ovine Infinium HD BeadChip.
Materials and Methods
Genotypic and Phenotypic Data
We collected 253 individuals from 13 domestic sheep populations with typical phenotypic traits to investigate the genetic variants under long-term artificial selection (Figure 1 and Table 1; Kijas et al., 2014; Xu et al., 2017; Zhao et al., 2017; Gao et al., 2018; Xu et al., 2018; Rochus et al., 2018; Cao et al., 2020). Whole genome SNP datasets (Ovine Infinium HD SNP BeadChip) of these individuals were obtained from previous studies (Xu et al., 2017; Zhao et al., 2017; Gao et al., 2018; Xu et al., 2018) and divided into 9 pairs populations (Table 2). We performed two different selection methods, the pairwise FST (Weir and Cockerham, 1984) and the cross-population composite likelihood ratio (XP-CLR) test (Chen et al., 2010). The identification of common signatures by the different algorithms and assumptions might be seen as good reliability of the results while reducing the likelihood of false positives.

FIGURE 1. Images of domestic sheep (A) Kazakh Edilbai sheep (B) Celle Black sheep (C) Polled Merino (D) Merino (E) Hu sheep (F) Tan sheep (G) Caucasian sheep (H) Altamurana sheep (I) Jill Wagner sheep (J) Meat Lacaune (K) Dairy Lacaune (L) Shetland (M) East Friesian sheep. Photo credits are showed in supplementary table 2.
SNP Data Quality Control
We implemented strict quality control of the SNP dataset using the PLINK v.1.09 software (Purcell et al., 2007). We removed individuals and SNPs that met any of the following criteria: 1) SNPs without chromosomal or physical locations; 2) SNPs with >0.02 missing data; 3) individuals with a genotyping rate <0.95; 4) minor allele frequency (MAF) < 0.05; and 5) the p-value of Fisher’s exact test for Hardy-Weinberg equilibrium (HWE) < 0.00001. Consequently, the final data after filtering contained various sets of SNPs and individuals in the comparison tests, such as 506,350 SNPs and 24 individuals (9 Kazakh Edilbai sheep vs 15 Celle Black Sheep) for the trait of tail shape, 485,747 SNPs and 55 individuals (36 Merino sheep vs 19 Polled Merino sheep) for horn morphology, 514,795 SNPs and 30 individuals (15 Hu sheep vs 15 Tan sheep) for fertility, 509,580 SNPs and 30 individuals (15 Celle Black Sheep vs 15 Tan sheep) for coat-color pigmentation, 529,338 SNPs and 30 individuals (15 Caucasian sheep vs 15 Altamurana sheep) for wool fineness, 519,650 SNPs and 20 individuals (11 Jill Wagner sheep vs 9 Kazakh Edilbai sheep) for ear size, 483,150 SNPs and 70 individuals (34 Meat Lacaune sheep vs 36 Dairy Lacaune sheep) for meat production, 528,576 SNPs and 20 individuals (9 Kazakh Edilbai sheep vs 11 Shetland sheep) for body size, 471,257 SNPs and 31 individuals (16 East Friesian sheep vs 15 Caucasian sheep) for milk production (Supplementary Table S1).
Genomic Selection Signals Analysis
To identify the genomic signatures of selection between pairwise populations of contrasting these phenotypes in domestic sheep, we calculated the FST values (Weir and Cockerham, 1984) for each SNP using the program Genepop v4.2 (Rousset, 2008). We took the top 0.02% of the empirical distribution of FST as the putative selective signals. Further, we calculated the XP-CLR scores for the 200 bp intervals along the chromosomes using the parameters (“-w1 0.005200 2000-p 0 0.95”). For each chromosome, we averaged the XP-CLR scores per window across non-overlapping 10 kb windows. We selected the top 0.05% of these windows as the putative selective regions.
Results and Discussion
We implemented selection screening in 9 pairs of populations: KAZ and CLS for tail types, PME and MER for presence or absence of horn, HUS and TAN for fertility, CLS and TAN for coat colors, ALT and CAU for wool fineness, WGJ and KAZ for ear size, LAC and LAM for meat types, KAZ and SHL for body size and EFR and CAU for milk production. We detected significant common signals located within or neighboring both novel and previously reported functional genes. A total of 36 genes were shared between the two selection scan metrics (Table 2). For example, seven genes (CERS6, BTG1, RYR3, T, SLC6A4, NNAT and OGT) (Figure 2 and Supplementary Tables 3, 4) were identified to be associated with different tail shapes (i.e., fat-tailed vs thin-tailed) (Joo and Yun, 2011; Tsai et al., 2013; Ruan et al., 2014; Xiao et al., 2016; Dias et al., 2016; Turner et al., 2018; Zhi et al., 2018). In the horned vs polled sheep, the well-known horn morphology-associated gene RXFP2 has been implicated as a strong candidate gene that explains the presence or absence of horn in sheep (Figure 3 and Supplementary Tables S5, 6; Hu et al., 2019). In the high prolificacy vs low prolificacy breeds, the gene BMPR1B could be involved in the variation in litter size of females (Supplementary Figure S1 and Supplementary Tables S7, 8; Rossetti et al., 2017). In addition, the two genes TYRP1 and KIT had been directly implicated in the mechanism of coat-colour pigmentation in the white vs non-white coat-colour breeds (Supplementary Figure S2 and Supplementary Tables S9, 10; Vage et al., 2003). The five genes MSI2, DSG1, HOXC8, HOXC12 and HOXC13 were crucial regulators for wool fineness in the fine-wool vs semi-fine wool sheep (Supplementary Figure S3 and Supplementary Tables S11, 12; Awgulewitsch, 2003). The genes MSRB3, PTCH1 and EMX2 were functionally associated with ear size in the large and floppy vs normal ears sheep (Supplementary Figure S4 and Supplementary Tables S13, 14; Rhodes et al., 2003; Wei et al., 2015; Shin et al., 2017). The eleven genes (CEBPA, CEBPG, DLX3, DLX4, GBAS, NSMAF, PDE3A, PEPD, SDCBP, TNRC6A and UTRN) had been reported to be involved in regulating meat production such as intramuscular fat, drip loss, marbling score, meat traceability and longissimus muscle in the meat-type vs non-meat-type sheep (Supplementary Figure S5 and Supplementary Tables S15, 16; Lobbert et al., 1996; Nonneman et al., 2013; Ayuso et al., 2015). The genes RMI1, GPR21, SCD5 and CADM1 might play important roles in regulating embryo development, body weight, lipid metabolism and energy homeostasis involved in differences in body size (Supplementary Figure S6 and Supplementary Tables S17, 18; Guo et al., 2013). The gene TF was associated with milk production in the dairy-type vs non-dairy-type sheep (Supplementary Figure S7 and Supplementary Tables S19, 20; Ju et al., 2011).
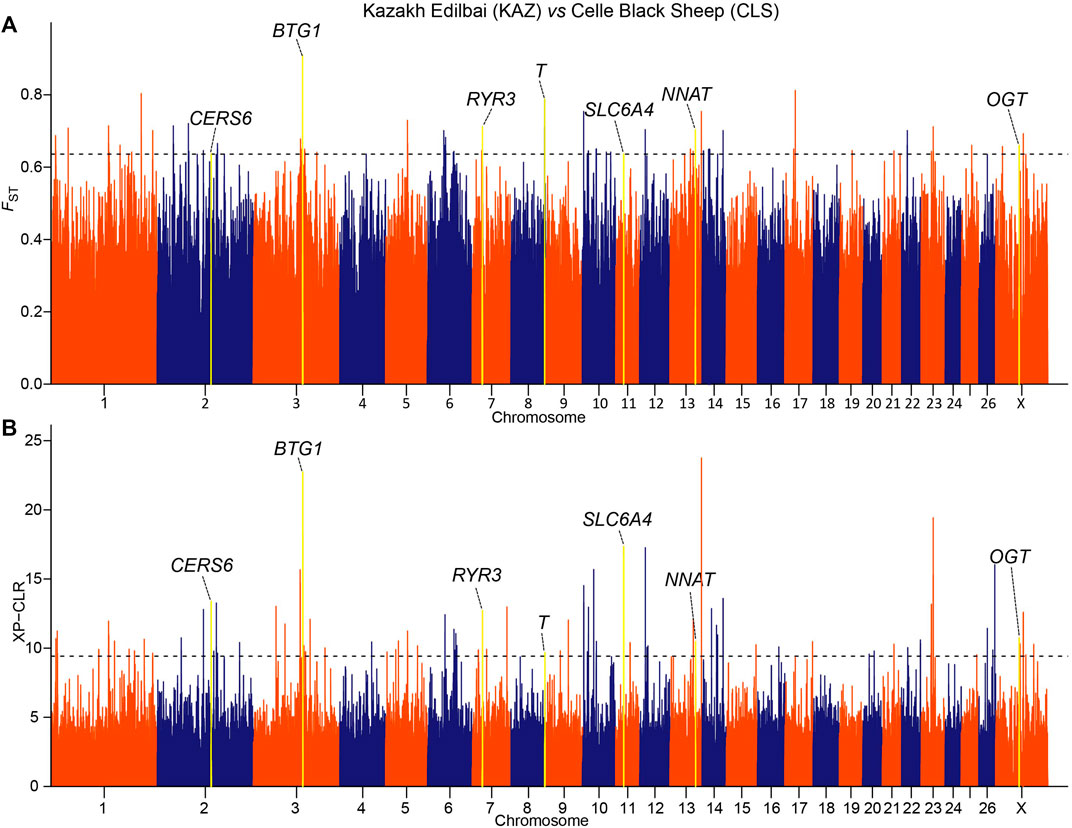
FIGURE 2. Manhattan plots of (A) pairwise FST and (B) XP-CLR selection tests with tail shapes in the comparison of Kazakh Edilbai (KAZ) and Celle Black Sheep (CLS) populations. The top 0.02% of the empirical distribution of FST and 0.05% of the XP-CLR scores are indicated by dotted lines, respectively.
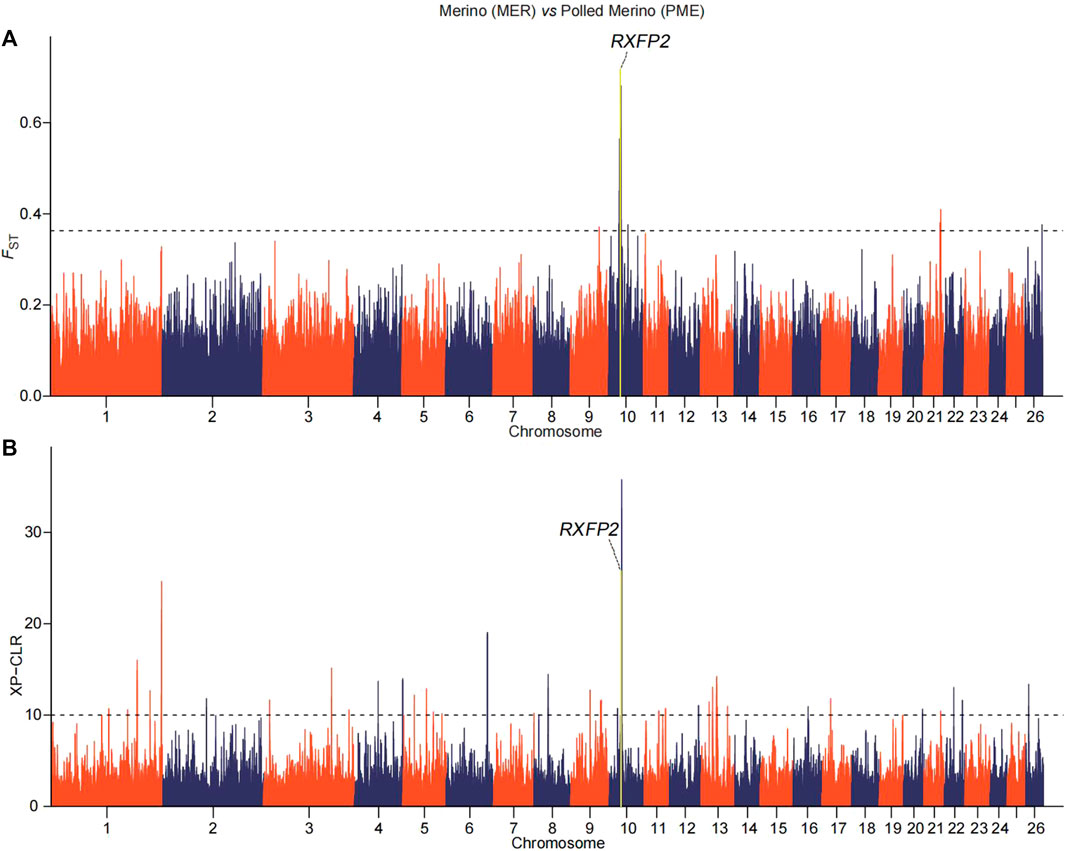
FIGURE 3. Manhattan plots of (A) pairwise FST and (B) XP-CLR with horn morphology in the comparison of Polled Merino (PME) and Merino (MER) breeds. The top 0.02% of the empirical distribution of FST and 0.05% of the XP-CLR scores are indicated by dotted lines, respectively.
In particular, we detected novel genes with functions associated with specific traits in sheep, such as CERS6, BTG1, NNAT and OGT for fat deposition in the tail of sheep, FOXO4 for fertility, PTCH1 and EMX2 for ear size, and SCD5 for body size. As a negative regulator of β-oxidation, the expression of CERS6 was significantly increased in subcutaneous fat of obese subjects with type 2 diabetes (Raichur et al., 2019). The BTG1 gene plays a key role in intramuscular fat deposition by regulating adipose-derived stem cell differentiation to osteocytes and myocytes (Moisa et al., 2015). The gene NNAT, as an endoplasmic reticulum proteolipid implicated in the intracellular signalling, is associated with severe obesity (Scott et al., 2013). The gene OGT is an important determinant of fatty acid synthesis in the mouse liver, which plays a critical role in fat deposition (Guinez et al., 2011; Kos et al., 2009). The FOXO4 gene has an important role in the activity of corpus luteum that is linked to folliculogenesis (Pisarska et al., 2009). The PTCH1 gene plays a critical role in the microcephaly, developmental delay, short stature, and facial dysmorphism by stimulating sonic hedgehog homolog (SHH) pathway (Derwinska et al., 2009). The EMX2 gene is highly expressed in mouse inner ear, with the role of activating early hair cell development (Holley et al., 2010). The SCD5 gene is linked to the regulator of sterol regulatory element-binding proteins involved in the development of body size (Baeza et al., 2013). Taken together, the apparent differences in the phenotypic traits among the breeds might be explained by diverse regulation mechanisms.
Noteworthy, we did not detect previously reported important functional genes associated with specific traits, for example, PDGFD and BMP15, which are associated with fat deposition in the tails of sheep (Li et al., 2020) and litter size (Xu et al., 2018), respectively. The main reason could be complex genetic mechanisms of phenotypic traits, for example, fertility was regulated by different major functional genes BMP15, NCOA1 and NF1 for Wadi, Icelandic and Finnsheep, respectively (Xu et al., 2018). In addition, we identified candidate functional genes different from those identified in earlier investigations, which could be due to that the power for general linear models to detect such associations will be weak when treating quantitative traits given the small sample size (Xu et al., 2018). Furthermore, these breeds could have been subjected to selection on specific traits (e.g., body weight) through environmental variables such as climate, diet and diseases. However, we did not obtain detailed information for these variables in our data analysis. Thus, these variables were not taken into account in our data analysis, which would be essential for future study.
In conclusion, we detected some novel and previously reported functional genes associated with particular phenotypic traits under strong and long-term artificial selection. Nevertheless, associations between these genes detected in two tests and the specific traits should be worthy of further exploration in future investigations. These findings contribute to understanding of the genomic consequences of artificial selection in the genomes of domestic sheep.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material further inquiries can be directed to the corresponding author.
Author Contributions
FL and S-SX conceived and designed the project. LG and MS collected the samples. S-SX and LG analyzed the data. S-SX and LG wrote the manuscript with contributions from FL.
Funding
This project work was supported by the National Natural Science Foundation of China (31972527, 31660651, 30760661), the Chinese Universities Scientific Fund 2021TC053 and the Guide Project of State Key Laboratory of Sheep Genetic Improvement and Healthy Production (SKLSGIHP 2017A01).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2021.738879/full#supplementary-material
References
Abousoliman, I., Reyer, H., Oster, M., Murani, E., Mohamed, I., and Wimmers, K. (2021). Genome-Wide SNP Analysis for Milk Performance Traits in Indigenous Sheep: A Case Study in the Egyptian Barki Sheep. Animals 11, 1671. doi:10.3390/ani11061671
Awgulewitsch, A. (2003). Hox in Hair Growth and Development. Naturwissenschaften 90, 193–211. doi:10.1007/s00114-003-0417-4
Ayuso, M., Óvilo, C., Rodríguez-Bertos, A., Rey, A. I., Daza, A., Fenández, A., et al. (2015). Dietary Vitamin A Restriction Affects Adipocyte Differentiation and Fatty Acid Composition of Intramuscular Fat in Iberian Pigs. Meat Sci. 108, 9–16. doi:10.1016/j.meatsci.2015.04.017
Baeza, M. C., Corva, P. M., Soria, L. A., Pavan, E., Rincon, G., and Medrano, J. F. (2013). Genetic Variants in a Lipid Regulatory Pathway as Potential Tools for Improving the Nutritional Quality of Grass-Fed Beef. Anim. Genet. 44, 121–129. doi:10.1111/j.1365-2052.2012.02386.x
Cao, Y. H., Xu, S. S., Shen, M., Chen, Z. H., Gao, L., Lv, F. H., et al. (2020). Historical Introgression from Wild Relatives Enhanced Climatic Adaptation and Resistance to Pneumonia in Sheep. Mol. Biol. Evol. 38, 838–855. doi:10.1093/molbev/msaa236
Chen, H., Patterson, N., and Reich, D. (2010). Population Differentiation as a Test for Selective Sweeps. Genome Res. 20, 393–402. doi:10.1101/gr.100545.109
Derwińska, K., Smyk, M., Cooper, M. L., Bader, P., Cheung, S. W., and Stankiewicz, P. (2009). PTCH1 Duplication in a Family with Microcephaly and Mild Developmental Delay. Eur. J. Hum. Genet. 17, 267–271. doi:10.1038/ejhg.2008.176
Dias, H., Muc, M., Padez, C., and Manco, L. (2016). Association of Polymorphisms in 5-HTT (SLC6A4) and MAOA Genes with Measures of Obesity in Young Adults of Portuguese Origin. Arch. Physiol. Biochem. 122, 8–13. doi:10.3109/13813455.2015.1111390
Gao, L., Xu, S. S., Yang, J. Q., Shen, M., and Li, H. H. (2018). Genome-Wide Association Study Reveals Novel Genes for the Ear Size in Sheep (Ovis aries). Anim. Genet. 49, 345–48. doi:10.1111/age.12670
Gui, L.-S., Raza, S. H. A., Zhou, L., Garcia, M., Abd El-Aziz, A. H., Wei, D., et al. (2020). Association between Single Nucleotide Polymorphisms in SIRT1 and SIRT2 Loci and Growth in Tibetan Sheep. Animals 10, 1362. doi:10.3390/Ani10081362
Guinez, C., Filhoulaud, G., Rayah-Benhamed, F., Marmier, S., Dubuquoy, C., Dentin, R., et al. (2011). O-GlcNAcylation Increases ChREBP Protein Content and Transcriptional Activity in the Liver. Diabetes 60, 1399–1413. doi:10.2337/db10-0452
Guo, Y., Lanktree, M. B., Taylor, K. C., Hakonarson, H., Lange, L. A., Keating, B. J., et al. (2013). Gene-Centric Meta-Analyses of 108 912 Individuals Confirm Known Body Mass Index Loci and Reveal Three Novel Signals. Hum. Mol. Genet. 22, 184–201. doi:10.1093/hmg/dds396
Holley, M., Rhodes, C., Kneebone, A., Herde, M. K., Fleming, M., and Steel, K. P. (2010). Emx2 and Early Hair Cell Development in the Mouse Inner Ear. Develop. Biol. 340, 547–556. doi:10.1016/j.ydbio.2010.02.004
Hu, X.-J., Yang, J., Xie, X.-L., Lv, F.-H., Cao, Y.-H., Li, W.-R., et al. (2019). The Genome Landscape of Tibetan Sheep Reveals Adaptive Introgression from Argali and the History of Early Human Settlements on the Qinghai-Tibetan Plateau. Mol. Biol. Evol. 36, 283–303. doi:10.1093/molbev/msy208
Joo, J. I., and Yun, J. W. (2011). Gene Expression Profiling of Adipose Tissues in Obesity Susceptible and Resistant Rats under a High Fat Diet. Cell Physiol. Biochem. 27, 327–340. doi:10.1159/000327959
Ju, Z. H., Li, Q. L., Huang, J. M., Hou, M. H., Li, R. L., Li, J. B., et al. (2011). Three Novel SNPs of the Bovine Tf Gene in Chinese Native Cattle and Their Associations with Milk Production Traits. Genet. Mol. Res. 10, 340–352. doi:10.4238/vol10-1gmr1038
Kijas, J. W., Lenstra, J. A., Hayes, B., Boitard, S., Porto Neto, L. R., San Cristobal, M., et al. (2012). Genome-Wide Analysis of the World's Sheep Breeds Reveals High Levels of Historic Mixture and strong Recent Selection. PLoS Biol. 10, e1001258. doi:10.1371/journal.pbio.1001258
Kijas, J. W., Porto-Neto, L., Dominik, S., Reverter, A., Bunch, R., McCulloch, R., et al. (2014). Linkage Disequilibrium over Short Physical Distances Measured in Sheep Using a High-Density SNP Chip. Anim. Genet. 45, 754–757. doi:10.1111/age.12197
Kos, K., Harte, A. L., O’Hare, P. J., Kumar, S., and McTernan, P. G. (2009). Ghrelin and the Differential Regulation of Des-Acyl (DSG) and Oct-Anoyl Ghrelin (OTG) in Human Adipose Tissue (AT). Clin. Endocrinol. 70, 383–389. doi:10.1111/j.1365-2265.2008.03321.x
Li, X., Yang, J., Shen, M., Xie, X.-L., Liu, G.-J., Xu, Y.-X., et al. (2020). Whole-genome Resequencing of Wild and Domestic Sheep Identifies Genes Associated with Morphological and Agronomic Traits. Nat. Commun. 11, 2815. doi:10.1038/s41467-020-16485-1
Löbbert, R. W., Winterpacht, A., Seipel, B., and Zabel, B. U. (1996). Molecular Cloning and Chromosomal Assignment of the Human Homologue of the Rat cGMP-Inhibited Phosphodiesterase 1 (PDE3A)-A Gene Involved in Fat Metabolism Located at 11p15.1. Genomics 37, 211–218. doi:10.1006/geno.1996.0544
Moisá, S. J., Shike, D. W., Shoup, L., Rodriguez-Zas, S. L., and Loor, J. J. (2015). Maternal Plane of Nutrition during Late Gestation and Weaning Age Alter Angus × Simmental Offspring Longissimus Muscle Transcriptome and Intramuscular Fat. PLoS One 10, e0131478. doi:10.1371/journal.pone.0131478
Nonneman, D. J., Shackelford, S. D., King, D. A., Wheeler, T. L., Wiedmann, R. T., Snelling, W. M., et al. (2013). Genome-Wide Association of Meat Quality Traits and Tenderness in Swine. J. Anim. Sci. 91, 4043–4050. doi:10.2527/jas.2013-6255
Pisarska, M. D., Kuo, F.-T., Tang, D., Zarrini, P., Khan, S., and Ketefian, A. (2009). Expression of Forkhead Transcription Factors in Human Granulosa Cells. Fertil. Steril. 91, 1392–1394. doi:10.1016/j.fertnstert.2008.04.054
Purcell, S., Neale, B., Todd-Brown, K., Thomas, L., Ferreira, M. A. R., Bender, D., et al. (2007). PLINK: A Tool Set for Whole-Genome Association and Population-Based Linkage Analyses. Am. J. Hum. Genet. 81, 559–575. doi:10.1086/519795
Raichur, S., Brunner, B., Bielohuby, M., Hansen, G., Pfenninger, A., Wang, B., et al. (2019). The Role of C16:0 Ceramide in the Development of Obesity and Type 2 Diabetes: CerS6 Inhibition as a Novel Therapeutic Approach. Mol. Metab. 21, 36–50. doi:10.1016/j.molmet.2018.12.008
Rhodes, C. R., Parkinson, N., Tsai, H., Brooker, D., Mansell, S., Spurr, N., et al. (2003). The Homeobox Gene Emx2 Underlies Middle Ear and Inner Ear Defects in the Deaf Mouse Mutant Pardon. J. Neurocytol. 32, 1143–1154. doi:10.1023/b:neur.0000021908.98337.91
Rochus, C. M., Tortereau, F., Plisson-Petit, F., Restoux, G., Moreno-Romieux, C., Tosser-Klopp, G., et al. (2018). Revealing the Selection History of Adaptive Loci Using Genome-Wide Scans for Selection: An Example from Domestic Sheep. BMC Genom. 19, 71. doi:10.1186/s12864-018-4447-x
Rossetti, R., Ferrari, I., Bonomi, M., and Persani, L. (2017). Genetics of Primary Ovarian Insufficiency. Clin. Genet. 91, 183–198. doi:10.1111/cge.12921
Rousset, F. (2008). genepop'007: A Complete Re-Implementation of the Genepop Software for Windows and Linux. Mol. Ecol. Resour. 8, 103–106. doi:10.1111/j.1471-8286.2007.01931.x
Ruan, H.-B., Dietrich, M. O., Liu, Z.-W., Zimmer, M. R., Li, M.-D., Singh, J. P., et al. (2014). O-GlcNAc Transferase Enables AgRP Neurons to Suppress Browning of White Fat. Cell 159, 306–317. doi:10.1016/j.cell.2014.09.010
Scherf, B. D. (2000). World Watch List for Domestic Animal Diversity. Ed. 3. Rome: Food and Agriculture Organization of the United Nations. Available at: https://agris.fao.org/agris-search/search.do?recordID=XF9660113 (Accessed July 15, 2020).
Scott, W. R., Gelegen, C., Chandarana, K., Karra, E., Yousseif, A., Amouyal, C., et al. (2013). Differential Pre-mRNA Splicing Regulates Nnat Isoforms in the Hypothalamus after Gastric Bypass Surgery in Mice. PLoS One 8, e59407. doi:10.1371/journal.pone.0059407
Shin, J.-O., Ankamreddy, H., Jakka, N. M., Lee, S., Kim, U.-K., and Bok, J. (2017). Temporal and Spatial Expression Patterns of Hedgehog Receptors in the Developing Inner and Middle Ear. Int. J. Dev. Biol. 61, 557–563. doi:10.1387/ijdb.170155jb
Tsai, S.-H., Chang, E. Y.-C., Chang, Y.-C., Hee, S.-W., Tsai, Y.-C., Chang, T.-J., et al. (2013). Knockdown of RyR3 Enhances Adiponectin Expression through an Atf3-dependent Pathway. Endocrinology 154, 1117–1129. doi:10.1210/en.2012-1515
Turner, N., Lim, X. Y., Toop, H. D., Osborne, B., Brandon, A. E., Taylor, E. N., et al. (2018). A Selective Inhibitor of Ceramide Synthase 1 Reveals a Novel Role in Fat Metabolism. Nat. Commun. 9, 3165. doi:10.1038/S41467-018-05613-7
Vage, D. I., Fleet, M. R., Ponz, R., Olsen, R. T., Monteagudo, L. V., Tejedor, M. T., et al. (2003). Mapping and Characterization of the Dominant Black Colour Locus in Sheep. Pigment Cell Res. 16, 693–697. doi:10.1046/j.1600-0749.2003.00090.x
Wei, C., Wang, H., Liu, G., Wu, M., Cao, J., Liu, Z., et al. (2015). Genome-Wide Analysis Reveals Population Structure and Selection in Chinese Indigenous Sheep Breeds. BMC Genom. 16, 194. doi:10.1186/s12864-015-1384-9
Weir, B. S., and Cockerham, C. C. (1984). Estimating F -Statistics for the Analysis of Population Structure. Evolution 38, 1358–1370. doi:10.1111/j.1558-5646.1984.tb05657.x
Xiao, F., Deng, J., Guo, Y., Niu, Y., Yuan, F., Yu, J., et al. (2016). BTG1 Ameliorates Liver Steatosis by Decreasing Stearoyl-CoA Desaturase 1 (SCD1) Abundance and Altering Hepatic Lipid Metabolism. Sci. Signal. 9, ra50. doi:10.1126/scisignal.aad8581
Xu, S.-S., Gao, L., Xie, X.-L., Ren, Y.-L., Shen, Z.-Q., Wang, F., et al. (2018). Genome-Wide Association Analyses Highlight the Potential for Different Genetic Mechanisms for Litter Size Among Sheep Breeds. Front. Genet. 9, 118. doi:10.3389/fgene.2018.00118
Xu, S.-S., and Li, M.-H. (2017). Recent Advances in Understanding Genetic Variants Associated with Economically Important Traits in Sheep (Ovis Aries) Revealed by High-Throughput Screening Technologies. Front. Agr. Sci. Eng. 4, 279–288. doi:10.15302/J-FASE-2017151
Xu, S. S., Ren, X., Yang, G. L., Xie, X. L., Zhao, Y. X., Zhang, M., et al. (2017). Genome-Wide Association Analysis Identifies the Genetic Basis of Fat Deposition in the Tails of Sheep (Ovis Aries). Anim. Genet. 48, 560–569. doi:10.1111/age.12572
Zhao, Y.-X., Yang, J., Lv, F.-H., Hu, X.-J., Xie, X.-L., Zhang, M., et al. (2017). Genomic Reconstruction of the History of Native Sheep Reveals the Peopling Patterns of Nomads and the Expansion of Early Pastoralism in East Asia. Mol. Biol. Evol. 34, 2380–2395. doi:10.1093/molbev/msx181
Zhi, D., Da, L., Liu, M., Cheng, C., Zhang, Y., Wang, X., et al. (2018). Whole Genome Sequencing of Hulunbuir Short-Tailed Sheep for Identifying Candidate Genes Related to the Short-Tail Phenotype. G3 (Bethesda) 8, 377–383. doi:10.1534/g3.117.300307
Keywords: artificial selection, genome-wide SNPs, sheep, phenotypic traits, genetic improvement
Citation: Xu S-S, Gao L, Shen M and Lyu F (2021) Whole-Genome Selective Scans Detect Genes Associated With Important Phenotypic Traits in Sheep (Ovis aries). Front. Genet. 12:738879. doi: 10.3389/fgene.2021.738879
Received: 09 July 2021; Accepted: 28 October 2021;
Published: 18 November 2021.
Edited by:
Duy Ngoc Do, Dalhousie University, CanadaReviewed by:
Sayed Haidar Abbas Raza, Northwest A and F University, ChinaLoan To Nguyen, University of Queensland, Australia
Copyright © 2021 Xu, Gao, Shen and Lyu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Fenghua Lyu, bHZmZW5naHVhQGNhdS5lZHUuY24=
†These authors have contributed equally to this work
 Song-Song Xu
Song-Song Xu Lei Gao3†
Lei Gao3† Fenghua Lyu
Fenghua Lyu