- 1School of Clinical Medicine, Weifang Medical University, Weifang, China
- 2School of Basic Medicine Sciences, Weifang Medical University, Weifang, China
- 3Institute of Stem Cell and Regenerative Medicine, Department of Basic Medicine, Qingdao University Medical College, Qingdao, China
- 4The 80th Group Army Hospital of Chinese People’ Liberation Army, Weifang, China
The N6-methyladenosine (m6A) modification is the most abundant epitranscriptomic modification in eukaryotic messenger RNA (mRNA). The m6A modification process is jointly regulated by various enzymes and proteins, such as methyltransferases, demethylases and related m6A-binding proteins. The process is dynamic and reversible, and it plays an essential role in mRNA metabolism and various biological activities. Recently, an increasing number of researchers have confirmed that the onset and development of many diseases are closely associated with the molecular biological mechanism of m6A RNA methylation. This study focuses on the relationship between m6A RNA modification and atherosclerosis (AS). It thoroughly summarizes the mechanisms and processes of m6A RNA modification in AS-related cells and the relationships between m6A RNA modification and AS risk factors, and it provides a reference for exploring new targets for the early diagnosis and treatment of AS.
Introduction
The morbidity and mortality associated with cardiovascular diseases are increasing every year. Atherosclerosis (AS) is the underlying cause of cardiovascular diseases such as myocardial infarction, stroke, and coronary heart disease (Virani et al., 2020). AS refers to the continuous buildup of plaques composed of lipids and fibrous components in the vascular wall and eventually the formation of atherosclerotic necrotic lesions. Additionally, AS is considered a systemic chronic inflammatory disease caused by the activation of inflammatory cells or inflammatory cytokines after cell injury (Fu et al., 2017; Zhu et al., 2018). Unfortunately, however, the mechanism of AS is still unclear.
Recently, epitranscriptomics has become a prominent and emerging research field in explaining AS pathogenesis. An increasing number of studies have shown that epitranscriptomic modification is closely associated with the metabolic processes of tissues and cells in AS (Herrington et al., 2016; Quiles-Jiménez et al., 2020). This article reviews the occurrence of the N6-methyladenosine (m6A) modification in tissues and cells during the development of AS, as well as the relationships between the m6A modification and AS risk factors, such as oxidative stress, obesity, and smoking, to provide new ideas for the study of molecules targeting AS.
The m6A Ribonucleic Acid Modification
The use of m (Dominissini et al., 2012)A-specific methylated RNA immunoprecipitation and next-generation sequencing (MeRIP-Seq) has significantly increased the efficiency of identifying m6A-modified target mRNAs. Transcriptome-wide sequencing methods are vital for determining methyl modification sites, quantitatively analyzing modifications, and identifying regulatory enzymes or proteins associated with m6A (Dominissini et al., 2012). Three types of enzymes are involved in m6A RNA modification: methyltransferases (MTs) (writers), demethylases (erasers), and m6A-related binding proteins (readers) (Shi et al., 2019; Frye et al., 2018). In various cell lines under different processing environments, the expression levels and localization of m6A-related proteins in cells vary. In addition, their diversity, richness, and varied intracellular localizations make RNA molecules intrinsically heterogeneous, making it quite challenging to study the mechanisms of m6A RNA modification (Marinov et al., 2014; Adivarahan et al., 2018). According to the epitranscriptomic modification database (http://mods.rna.albany.edu/) and relevant studies, there are more than 100 epitranscriptomic modifications, among which the m6A modification is the most abundant in eukaryotic cells (Cantara et al., 2011; Jia et al., 2013; Yue et al., 2015). The m6A modification was first identified in 1974 (Desrosiers et al., 1974) and refers to the methylation of the 6th nitrogen atom of adenine, a nucleotide concentrated in stop codons, the 3′ untranslated region (3′-UTR), and the consensus sequence RRACH (R = A/G, H = A/C/U) in exons. However, subsequently, species-specific distributions of m6A signatures have been found in 5′-UTRs (Linder et al., 2015), around stop codons, and even around start codons in Arabidopsis thaliana (Dominissini et al., 2012; Meyer et al., 2012; Luo et al., 2014; Chen et al., 2015; Cao et al., 2016).
The m6A RNA modification process is dynamic and reversible, and it relies on the regulation of MTs, demethylases, and related specific binding proteins. Studies have shown that the m6A modification is associated with RNA metabolism, including RNA cleavage, degradation, and translation (Camper et al., 1984; Klungland and Dahl, 2014; Zhao et al., 2014). Here, we present a summary of m6A-modification-related enzymes and proteins.
m6A Writers: Methyltransferases
Methyltransferase complexes, also known as encoders or writers, play essential roles in catalyzing methyl groups (writers). These MTs are multicomponent N6-adenosine MT complexes composed of multiple subunits, including methyltransferase-like 3 (METTL3), METTL14, and other critical regulatory factors, such as Wilms’ tumor 1-associated protein (WTAP) and METTL16 (Bokar et al., 1997; Liu et al., 2014; Ping et al., 2014; Koh et al., 2019). METTL3 is a catalytically active subunit that was first identified based on S-adenosylmethionine (SAM) playing a critical role by providing a methyl group (Liu et al., 2014; Wang et al., 2016; Gao et al., 2021). Studies have shown that METTL14 plays an important role structurally supporting RNA binding by providing an RNA-binding scaffold, which synergistically promotes the binding of the transferase to RNA substrates and enhances the complex’s stability (Koh et al., 2019; Gao et al., 2021; Zheng et al., 2013). According to the latest research, METTL14, an important part of the heterodimeric N6-methyltransferase complex, is a noncatalytic subunit (Wang et al., 2016; Liu et al., 2021). In addition to the METTL3-METTL14 complex, a regulatory subunit, WTAP, is noteworthy. Although WTAP does not have methyl catalytic activity, it can bind to the METTL3-METTL14 complex, and plays an essential role in regulating the localization of the METTL3-METTL14 complex to nuclear foci, thereby affecting the efficiency of methyl modification (Ping et al., 2014; Liu et al., 2014). Moreover, the regulation of the process may strongly depend on METTL14 (Liu et al., 2014).
m6A Erasers: Demethylases
Demethylases are also known as erasers. Fat mass and obesity-associated protein (FTO) and alkylation repair homolog protein 5 (ALKBH5) are both demethylases that belong to the AlkB family (Zheng et al., 2013; Jia et al., 2011) and can remove m6A methylation in an FeII/α-ketoglutarate (α-KG)- and 2-oxoglutarate (2OG)-dependent manner (Gerken et al., 2007).
The FTO oxidation reaction produces two intermediate products, N6-hydroxymethyladenosine (hm6A) and N6-formyladenosine (f6A), and the final decomposition products are adenine and formaldehyde, which function in an FeII/α-KG- and 2OG-dependent manner (Gerken et al., 2007). Notably, hm6a and f6A are not detected in samples treated with only ALKBH5, and ALKBH5 can directly and efficiently demethylate m6A sites with its DBSH domain combined with the ATP domain of DDX3 (Zheng et al., 2013; Shah et al., 2017; Wei et al., 2018). Therefore, the mechanisms by which FTO and ALKBH5 demethylate are different (Zheng et al., 2013; Gerken et al., 2007; Fu et al., 2013). The deletion of ALKBH5 leads to decreased RNA metabolism (Zheng et al., 2013). Several studies have shown that demethylase dysregulation is closely associated with the occurrence or progression of various diseases, such as AS, metabolic syndrome, brain malformation, growth retardation and even cancer (Dina et al., 2007; Scuteri et al., 2007; Fu et al., 2013; Wang et al., 2020a). These topics may be important research hotspots in the future.
m6A Readers: m6A-binding Proteins
m6A-binding proteins, also known as readers, are proteins that specifically bind to m6A, and they include YT521-B homology (YTH) domain-containing family (YTHDF) proteins and RNA-binding proteins (RBPs) localized in the nucleus. YT521-B proteins contain a YTH domain and can be divided into 2 subfamilies, YTHDF (YTHDF1-3) and YTHDC (YTHDC1-2) (Wang et al., 2014; Zhou et al., 2021). YTHDF1 is an important m6A-binding protein, and Wang et al. (2015) showed that YTHDF1 is positively correlated with the translation efficiency of its target transcripts and the degree of ribosome occupancy. Zhou et al. (2015) found that under certain stressors, such as heat shock, YTHDF2 can inhibit FTO to facilitate the demethylation of m6A at the 5′-UTR, thus stabilizing m6A methylation at the site; in addition, YTHDF2 can reduce the stability of target transcripts (Wang et al., 2015). YTHDF3 is often synergistic with YTHDF1, thereby enhancing translation efficiency, and it is synergistic with YTHDF2, thereby reducing the stability of m6A-modified transcripts (Wang et al., 2015; Shi et al., 2017). In addition, Lan et al. (Roundtree et al., 2017) found that the YTHDC1 level negatively correlated with the level of m6A methylated mRNA in the nucleus and that YTHDC1 directly or indirectly promoted the export of m6A-modified mRNAs from the nucleus to the cytoplasm through SRSF3 and NXF1, thus promoting translation.
In addition to the YTHDF proteins, the heterogeneous nuclear ribonucleoprotein C (HNRNPC) family and insulin-like growth factor 2 (IGF2) mRNA binding protein 2 (IGF2BP2/IMP2) can also act as m6A methylation readers (Huang et al., 2018). HNRNP plays an important role in promoting the maturation of pre-microRNAs (miRNAs) and regulating the stability of target RNAs by binding to m6A-methylated transcripts (Liu et al., 2015). IGF2BP2 is a type 2 diabetes (T2D)-related gene. An increasing number of studies have shown that IGF2BP2 can bind to m6A modifications. In contrast to the role YTHDF2 plays in m6A modifications, IGF2BP2 can promote the structural stability of m6A-modified mRNAs and thus participate in the occurrence of various diseases (Dai et al., 2011).
m6A modification can induce the exposure of RNA-protein binding sites and affect the biological regulation of RNA-protein interactions, referred to as the m6A switch mechanism (Liu et al., 2015). An abundant RNA-binding protein, HNRNPC readily binds to uracil (U) on single-stranded RNA and participates in precursor RNA processing. The m6A modification of single-stranded RNA can induce the exposure of RNA-protein binding sites, thereby promoting HNRNPC binding, affecting substrate mRNA content, and influencing mRNA alternative splicing. These findings provide new directions for epitranscriptomic research (Figure 1).
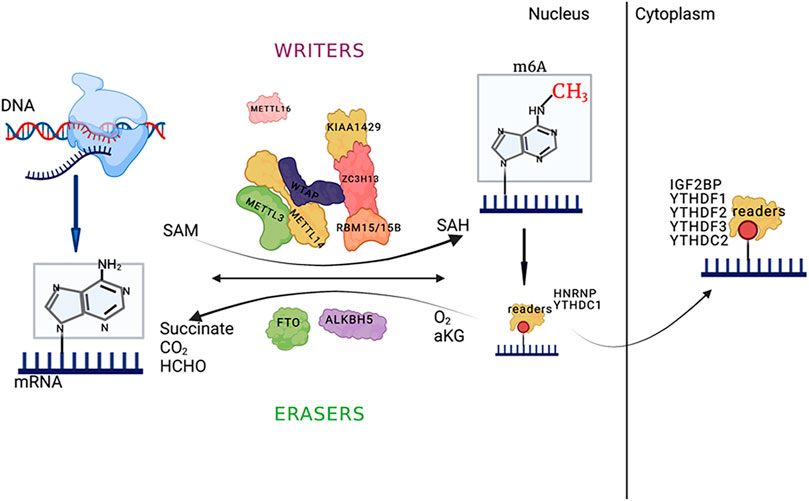
FIGURE 1. The main processes of m6A RNA modification and the major enzymes involved; and the picture was created with BioRender. The key enzymes that catalyze m6A methylation, such as methyltransferase (MT) complexes (METTL3/METTL14/Wilms tumor 1-associated protein (WTAP) and other regulatory cofactors that form the MT complex), or via methylation by METTL16 alone, are called “writers.” The demethylases that catalyze m6A demethylation (FTO/ALKBH5) are called “erasers.” YTHDF1-3/YTHDC1-2 and IGF2BP1/2/3 are called “readers.” The reaction uses s-adenosylmethionine (SAM) as a methyl donor, and the methyl group is removed to form SAH. The demethylation process for m6A RNA depends on Fe (II)/α-ketoglutarate (α-KG) and 2-oxoglutarate (2OG). The main functions of the binding proteins are to affect splicing, export and translation efficiency. The schematic diagram was created with BioRender.
Atherosclerosis and the m6A Ribonucleic Acid Modification
AS could cause vascular endothelial cell function or structure change, causes the lining the lower blood lipid and inflammatory cells abnormal deposition, migration of smooth muscle cells lining to the proliferation of macrophage origin and smooth muscle cells source sex foam cells, resulting in the formation of atherosclerotic plaque (Falk, 2006). Chemical modifications provide additional novel functional features for RNA, and one of the most common and characteristic mRNA modifications is m6A. This modification has been shown to be important in many aspects of the mRNA life cycle, from splicing to nuclear transport and stabilization to translational regulation. The relationship between atherosclerosis and m6A modification is highlighted below.
An increasing number of studies have shown that m6A RNA modification can affect the development and progression of AS (Jian et al., 2020; Quiles-Jiménez et al., 2020; Chen et al., 2021). An AS study found that the expression levels of zinc finger NFX1-type containing 1 (ZNFX1) antisense RNA1 (ZFAS1) and the downstream ADAM10/RAB22A were important in the development of vascular inflammation and cholesterol metabolism (Tang et al., 2020). Chen et al. (2016) showed that the m6A modification of ZFAS1 in AS patients was significantly higher than that in a control group and that m6A modification of ZFAS1 was regulated by METTl14. METTl14 affects the expression of downstream ADAM10/RAB22A by affecting the m6A modification of ZFAS1, thus participating in cholesterol metabolism and vascular inflammation and ultimately regulating the occurrence and development of AS (Quiles-Jiménez et al., 2020; Gong et al., 2021). These results provide an important reference for the study of m6A RNA modification and AS.
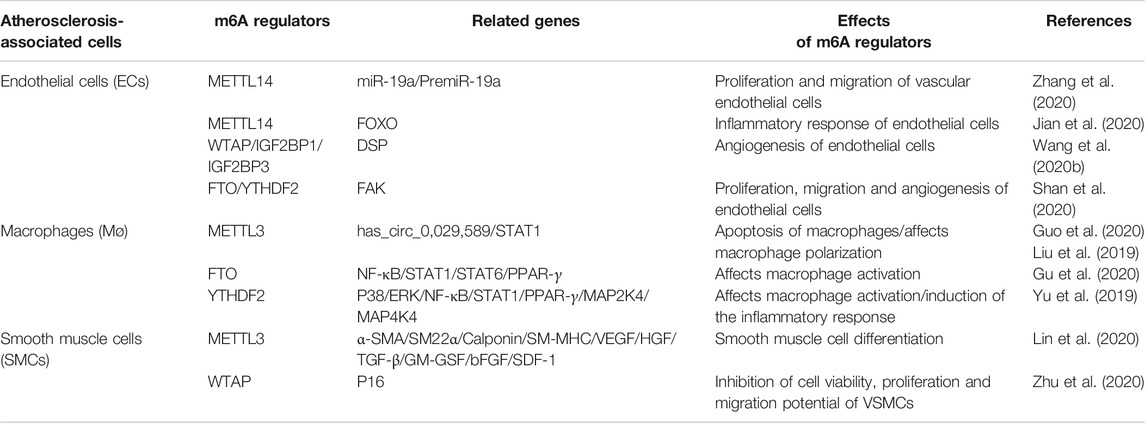
The occurrence of AS is closely related to the cells associated with the occurrence of plaques, such as vascular endothelial cells, macrophages and SMCs. m6A RNA modification may be involved in AS progression by affecting the functions of these cells. Thus, we have summarized the relationship between the regulation of m6A RNA modification-related enzymes and the cells involved in AS, as shown in Table 1.
m6A Ribonucleic Acid Modification and Vascular Endothelial Cells
AS is often accompanied by structural or functional damage to endothelial cells, so the involvement of vascular endothelial cells in angiogenesis is critical in atherosclerotic vascular repair. Jian et al. (2020) screened and identified m6A-modified mRNA via methylated RNA immunoprecipitation (RIP) sequencing and selected the forkhead box, class O (FOXO) gene as a potential target. Subsequently, the m6A modification of FOXO was found to increase the inflammatory response. However, when METTL14 was knocked out, the expression of inflammation-induced FOXO was significantly reduced. A study by Jian et al. (2020) demonstrated that m6A modification was involved in the induction of inflammatory responses in endothelial cells and played an essential role in the formation of plaques in AS. In addition, WTAP acts as a crucial regulatory subunit in the MT complex, and several studies have shown that WTAP expression affects m6A modification at the mRNA level in endothelial cells. For instance, Wang et al. (2020b) used RNA transcriptome sequencing (RNA-Seq) and found that in brain arteriovenous malformations (AMVs), the expression level of WTAP was significantly decreased but could inhibit endothelial cell angiogenesis. Interestingly, there were no significant differences in the expression of METTL3 or METTL14. MeRIP-Seq was further used to conclude that WTAP affects desmoplakin (DSP) methylation to influence angiogenesis, and the m6A readers IGF2BP1 and IGF2BP3 can stabilize methylated DSP mRNAs and prevent degradation (Wang et al., 2020b). Other studies have shown that the demethylase FTO is significantly increased, and in neovascularized corneas and ECs, the expression of FTO is increased (Shan et al., 2020). The m6A modification of FAK increased under pathological conditions. Interestingly, YTHDF2, an m6A RNA reader, can reduce the stability and increase the decay of m6A-modified RNA (Shan et al., 2020). The above results show that the m6A modification can affect biological functions, thereby causing corresponding diseases.
m6A Ribonucleic Acid Modification and Macrophages
AS is a chronic inflammatory disease. Macrophages, vital effector cells of the immune system, are inseparable from the vascular inflammatory process in AS (Koelwyn et al., 2018). Circular RNAs (circRNAs) are a new group of noncoding RNAs that always form circular structures with 5′-3′-phosphodiester bonds and do not contain free 5′ or 3′ ends. They are highly stable in cells (Li et al., 2018). A growing body of research suggests that circRNA is closely associated with cardiovascular disease (Kishore et al., 2020; Cao et al., 2020). Studies have shown that the expression of macrophage hsa_circ_0,029,589 in acute coronary syndrome is significantly reduced and that the expression and methylation of METTL3 (an m6A MT) are significantly increased; when METTL3 expression is inhibited, the methylation of hsa_circ_0,029,589 decreases, and the expression of hsa_circ_0,029,589 increases. Interestingly, interferon regulatory factor-1 (IRF-1) overexpression promotes METTL3 expression in macrophages and the m6A methylation of hsa_circ_0,029,589. Therefore, IRF-1 may inhibit the expression of hsa_circ_0,029,589 through hsa_circ_0,029,589 methylation, thereby regulating macrophage apoptosis and controlling the occurrence of inflammation (Guo et al., 2020). In addition, Liu et al. (2019) found that the expression of the m6A MT METTL3 was closely associated with the M1 polarization of mouse macrophages. METTL3 can increase m6A modifications in the coding sequence (CDS) and 3′-UTR of signal transducers and activators of transcription 1 (STAT1), increase the stability of STAT1, and participate in the inflammatory response.
An association between the demethylase FTO and macrophage polarization has also been reported. Gu et al. (2020) found that FTO can affect the nuclear factor kappa B (NF-κB) signaling pathway and that YTHDF2 can affect the stability of STAT1 and peroxisome proliferator-activated receptor-γ (PPAR-γ). When the m6A-binding protein YTHDF2 was knocked out, the mRNA stability and mRNA expression levels of STAT1 and PPAR-γ increased, and both STAT1 and PPAR-γ participated in macrophage activation. In addition, when YTHDF2 was knocked out in lipopolysaccharide (LPS)-stimulated RAW 264.7 cells, the stability of m6A-modified mitogen-activated protein 2 kinase 4 (MAP2K4) and mitogen-activated mitogen-activated protein 4 kinase 4 (MAP4K4) was enhanced. The expression levels of related inflammatory pathways, such as the p38, extracellular signal-regulated kinase (ERK) and NF-κB pathways. And the downstream related inflammatory molecules, such as interleukin (IL)-6, tumor necrosis factor (TNF)-α, IL-1β and IL-12, were increased, suggesting that YTHDF2 regulates the LPS-induced inflammatory process in macrophages (Yu et al., 2019). Similarly, studies have reported that exposure to macrophage mono-(2-ethylhexyl) phthalate (MEHP) can affect the m6A modification of the scavenger receptor B type 1 (SR-B1) gene associated with cholesterol efflux and the m6A modification of related miRNAs and can participate in macrophage-mediated cholesterol efflux (Park et al., 2020), providing strong evidence supporting the involvement of the RNA m6A modification in macrophages in the occurrence of AS.
The m6A Ribonucleic Acid Modification and Vascular Smooth Muscle Cells
Mature VSMCs have a high degree of plasticity and can transform from a quiescent contractile phenotype to a migratory proliferative and synthetic secretory phenotype. Under the continuous action of a specific environment or stimulating factors, the ability of SMCs to mechanically contract significantly decreases, and the ability to secrete extracellular matrix is enhanced, eventually leading to a thickening of the vascular wall and a narrowing of the lumen (Miano et al., 2021). Studies have shown that hypoxia can affect METTL3 expression and further affect the m6A modification of related factors, such as vascular endothelial growth factor (VEGF) and transforming growth factor (TGF)-beta, thereby inducing the differentiation of adipose-derived stem cells (ADSCs) into VSMCs (Lin et al., 2020). The occurrence of AS and restenosis after angioplasty are closely related to this process (Frismantiene et al., 2018). In a rat model of carotid balloon injury, Zhu et al. (2020) measured m6A modification in SMCs. The results showed that m6A modification in rats with carotid balloon injury was 0.59 times that in rats in the control group and that m6A methylation was reduced. Additionally, quantitative polymerase chain reaction (qPCR) analysis showed that WTAP expression was significantly decreased, and after treatment with total Panax notoginseng saponin (TPNS), WTAP expression increased and m6A modification of the downstream target gene P16 increased, indicating that the WTAP-p16 signaling axis plays a key role in the regulation of endometrial hyperplasia through m6A modification. The study by Zhu et al. (2020) not only examined the effect of TPNS in the treatment of endometrial hyperplasia but also provided potential biomarkers and new research ideas for the treatment of AS.
Relationship Between m6A Ribonucleic Acid Modification and Atherosclerosis Risk Factors
In recent years, substantial progress has been made in the treatment of cardiovascular diseases, but cardiovascular diseases are still the leading cause of death (Iyen et al., 2019; Libby et al., 2013). An increasing number of studies indicate that dynamic m6A modification plays an important regulatory role in cardiovascular diseases, especially AS (Quiles-Jiménez et al., 2020). Strong risk factors for AS, such as oxidative stress, obesity, and smoking, are closely associated with the dynamic regulation of m6A RNA methylation. Research progress on the relationship between m6A RNA modification and AS risk factors is summarized (Figure 2).
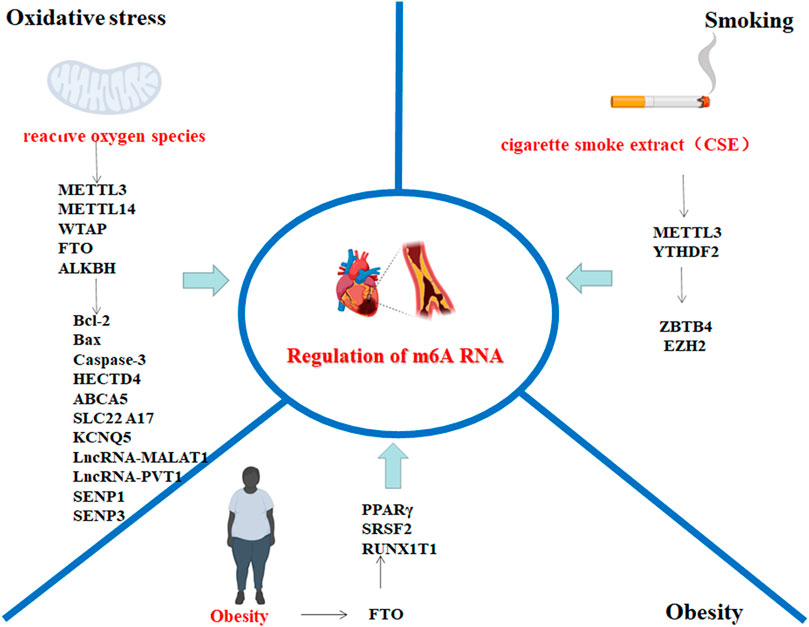
FIGURE 2. AS risk factors involved in the regulation of m6A methylation-related target genes; the picture was created with BioRender. The genes associated with oxidative stress and m6A RNA modification include HECTD4, ABCA5, SLC22A17, KCNQ5, lncRNA-MALAT1, and lncRNA-PVT1; genes associated with obesity and m6A RNA modification include SRSF2 and PPAR-γ; and genes associated with smoking and m6A modification include ZBTB4, EZH2 and H3K27me3. The above genes may be involved in m6A RNA modification, which may be involved in the AS disease process. The schematic diagram was created with BioRender.
m6A Ribonucleic Acid Modification and Oxidative Stress
Oxidative DNA damage caused by reactive oxygen species (ROS) can be involved in the occurrence of many diseases, and increasing evidence shows that excessive oxidative stress plays a key role in inducing cell damage and promoting AS progression, which is an important cause of cardiovascular diseases (Incalza et al., 2018; Krylatov et al., 2018). Epitranscriptomics plays a critical role in the study of cardiovascular disease. Some studies have shown that the m6A-modified form of the 5′-UTR can affect mRNA translation in a cap-independent manner (Linder et al., 2015; Jackson et al., 2010). These results provide a reliable theoretical basis for studying the relationship between m6A RNA modifications and some pathogenic factors. An increasing number of studies have found that translation in a cap-independent manner is associated with cell stress, infection, cancer and other diseases (Stoneley and Willis, 2004). It has not been reported whether ROS are involved in the process of AS by affecting m6A modification of mRNA and then regulating RNA translation. Wu et al. (Stoneley and Willis, 2004) showed that compared with normal mice, aflatoxin-treated mice had increased ROS levels, which in turn led to the increased expression of the MT METTL3 and the decreased expression of the demethylase FTO in the liver tissue of mice treated with ROS; therefore, the m6A modification of related RNA, such as B-cell lymphoma 2 (Bcl-2), cysteine aspartyl-specific protease (caspase)-3, and Bcl-2-associated X protein (Bax), increased, thus participating in the pathological process of liver injury. Studies have shown that resveratrol may reverse ROS-induced liver injury caused by aflatoxin by reducing RNA methylation (Wu et al., 2020; Ko et al., 2017). Zhao et al. (2019) confirmed that arsenous-acid-induced oxidative stress can affect the expression levels of m6A MT and demethylases, especially the expression levels of WTAP and METTL14, thereby regulating the m6A modification of genetic polymorphisms of C12orf51 (HECTD4), ATP-binding cassette transporter A5 (ABCA5), solute carrier family 22 member 17 (SLC22 A17) and potassium voltage-gated channel subfamily Q member 5 (KCNQ5) genes and influencing the transcription and translation of the genes, thus participating in subsequent pathophysiological processes. Exposure to arsenic is closely associated with the induction of human diseases, such as various skin diseases and cancer (Martinez et al., 2011; Argos, 2015). Cui et al. (Argos, 2015) discovered that the FTO expression level is increased in keratinocytes treated with arsenic and that the m6A RNA modification level decreases. The authors also showed that arsenic can inhibit FTO degradation by inhibiting P62 transcription, consequently promoting FTO stabilization. The changes in epitranscriptomic modification caused by arsenic-induced oxidative damage have great significance in understanding disease progression (Cui et al., 2021).
The RNA demethylase ALKBH5 is also a key regulator in protecting cells from DNA damage and apoptosis during ROS-induced stress. ROS can increase the overall m6A modification of mRNA in human HEK293T and HeLa cells. In one study, ROS inhibited the activity of ALKBH5 via ERK/c-Jun NH 2-terminal kinase (JNK), which promoted the phosphorylation of ALKBH5 on serine residues S87 and S325. Furthermore, phosphorylated ALKBH5 further promotes the interaction between ALKBH5 and small ubiquitin-related modifier (SUMO)ylated E2 UBC9. At the same time, the ERK/JNK/ALKBH5 PTM/m6A axis was activated in hematopoietic stem/progenitor cells (HSPCs), which also contributed to maintaining stable genomic integrity and thereby participated in the DNA damage response (Yu et al., 2021). In addition, in other studies, ALKBH5 is closely associated with sentrin/SUMO-specific proteases 1 and 3 (SENP1 and SENP3) under oxidative stress (Xu et al., 2008; Huang et al., 2009; Wang et al., 2012). In addition, in the presence of cadmium sulfate-induced oxidative damage, FTO expression decreases, the m6A modification of the long noncoding RNAs (lncRNAs) lncRNA-MALAT1 and lncRNA-PVT1 increases, and the expression levels of lncRNA-MALAT1 and lncRNA-PVT1 decrease, which in turn induces biological activities, such as ROS accumulation, malondialdehyde (MDA) accumulation and decreased superoxide dismutase (SOD) activity (Qu et al., 2021). Chromium (Cr) (VI) is a toxic substance, and Lv et al. (2021) found that Cr(VI) can cause mitochondrial damage in spermatogonial stem cells/progenitors, resulting in increased ROS production, autophagy, and loss of cell viability. In turn, this damage may cause various reproductive diseases, such as disorders of spermatogenesis. The authors explored the mechanism and found that the m6A RNA modification level decreased in spermatogonial stem cells/progenitors treated with Cr (VI), while melatonin restored the expression of METTL3. Cr(VI)-induced damage was reversed by restoring METTL3-mediated RNA m6A modification and activating mitofusin 2 (MFN2) and optic atrophy 1 (OPA1), as well as inhibiting the Bcl-2/adenovirus E1B 19-kDa-interacting protein 3 (BNIP3)/NIX mitophagy receptor pathway (Lv et al., 2021). However, studies on the relationship between AS, epitranscriptomics, and ROS are still lacking; their relationship requires further investigation.
m6A Ribonucleic Acid Modification and Obesity
Poor dietary habits can increase the incidence of nutritional imbalance or obesity and can also seriously threaten cardiovascular and cerebrovascular health. According to statistics from the World Health Organization (WHO), by 2015, there were more than 700 million people with obesity worldwide, indicating that many people are overweight (Morabia and Abel, 2006). FTO was first identified as a demethylase in 2011 and is involved in m6A RNA methylation. Zhang et al. (2015) proposed that FTO was necessary for preadipocyte differentiation and may affect adipocyte differentiation by activating the WNT signaling pathway to inhibit the activity of PPAR-γ. Subsequent studies have shown that FTO gene mutations, such as single nucleotide polymorphisms (SNPs), are closely associated with the probability of obesity in children and adults (Dina et al., 2007; Keller et al., 2017)). In addition, Zhao et al. (2014) found that the m6A-modified region spatially overlapped with mRNA splicing regulatory serine/arginine-rich (SR) protein exonic splicing enhancer binding regions and the serine/arginine-rich splicing factor 2 (SRSF2) mRNA binding clusters, indicating that FTO plays an important role in regulating SRSF2 mRNA splicing and adipocyte differentiation. In addition, FTO has been shown to regulate the splicing of the exon region of adipogenic regulator runt-related transcription factor 1 translocation partner 1 (RUNX1T1) by regulating m6A modification, which provides new insights for the study of m6A modification in adipocyte differentiation (Zhao et al., 2014). Church et al. (2010) found that after FTO overexpression, the fat content and body weight of mice fed a normal diet and a high-fat diet showed dose-dependent increases, confirming that FTO is an important factor involved in the development of obesity. Therefore, in-depth studies of the relationship between obesity and m6A modification are playing key roles in revealing the mechanism of AS, especially intervention targets.
m6A Ribonucleic Acid Modification and Smoking
Smoking is a risk factor for the development of AS. Increasing evidence shows that cigarette smoke extract (CSE) is not only potentially associated with vascular endothelial dysfunction (Pepine et al., 1998) but can also induce endothelial cell pyrolysis (Wang et al., 2019). In addition, CSE is closely associated with a variety of biological activities, such as LDL oxidation, VSMC proliferation, and inflammatory cell aggregation (Yamaguchi et al., 2005; Guo et al., 2017; Zong et al., 2019). In recent years, there have been several studies on the role of CSE in the regulation of dynamic m6A RNA modification in disease mechanisms (Kupsco et al., 2020). Cheng et al. (2021) found that in CSE-treated human bronchial epithelial (HBE) cells, the expression of the MT METTL3 increased, resulting in increased m6A modification of zinc finger and BTB domain containing 4 (ZBTB4) and that m6A-modified ZBTB4 was recognized by YTHDF2, resulting in decreased stability and decreased ZBTB4 expression; interestingly, low ZBTB4 expression can lead to the increased expression of enhancer of zeste homolog 2 (EZH2), which is involved in the induction of the epithelial-mesenchymal transition (EMT). Increasing evidence shows that smoking plays an important role in the regulation of m6A modification, providing a reliable reference for the study and interpretation of the occurrence and development of smoking-induced diseases.
Conclusion and Prospects
AS is by far the most common vascular disease and is the main cause of coronary heart disease, cerebral infarction, and peripheral vascular disease (Falk, 2006). The development of AS involves interactions among multiple factors, such as oxidative stress and proinflammatory cytokine stimulation (Libby, 2000; Zaman et al., 2000; Singh and Bedi, 2013). Therefore, the mechanism of AS is very complex and is still not fully understood. It is undeniable that cells involved in the development of AS, such as vascular endothelial cells, macrophages and SMCs, play important roles. By regulating the levels of MTs and demethylases, m6A RNA modification can be dynamically affected, and then, through binding to specific m6A-binding proteins, the splicing, folding, transport, translocation, degradation and translation of RNA are affected, ultimately determining the progression and outcome of AS.
This study investigated the relationships between m6A RNA modification and strong risk factors for AS, such as oxidative stress, obesity, and smoking, and found that these factors can affect the m6A modification of specific genes to regulate gene expression and ultimately participate in the occurrence and development of a variety of diseases. Interestingly, the m6A RNA modification is preferentially found at the 5′-UTRs of newly transcribed mRNAs, and the m6A modification level is increased under certain stressful situations. Zhou et al. (2015) found that under heat shock, proteins modified by m6A, such as Hsp70, and the m6A modification level of Hsp70 mRNA in the 5′-UTR were increased, which led YTHDF2 to localize to the nucleus and suppressed FTO expression. Then, they showed that the 5′-UTR, which was m6A modified, could enable translation initiation, and this progress was independent of the m7G cap at the 5′ end (Zhou et al., 2015). Can atherosclerotic risk factors also be considered a stressor that regulates the modification of m6A mRNA and participates in mRNA translation in a cap-independent manner? The stress pathways that regulate m6A modification at the 5′-UTR to influence AS remain unknown. The studies in this field are still insufficient, and extensive study is urgently needed.
In summary, m6A RNA modification is dynamic and reversible and has great importance in the prevention and treatment of disease progression. Further studies are needed to identify additional regulators, in addition to the currently known methylases, demethylases, and related specific m6A-binding proteins, to further explain their relationships with the mechanisms of disease occurrence and development and provide a new direction for research on and the treatment of AS.
Author Contributions
JF and XhC, Writing-original draft; XdC and ZG Writing, review and editing; XZ and XL review; MC and XC, Funding acquisition. The manuscript was reviewed and approved by all authors.
Funding
This work was supported by the Natural Science Foundation of Shandong Province (grants ZR2020MH020), the National Natural Science Foundation of China (grants 81,870,237 and 81,700,406), The National Key Research and Development Program of China (2018YFA0109800).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Adivarahan, S., Livingston, N., Nicholson, B., Rahman, S., Wu, B., Rissland, O. S., et al. (2018). Spatial Organization of Single mRNPs at Different Stages of the Gene Expression Pathway. Mol. Cel 72 (4), 727–738. doi:10.1016/j.molcel.2018.10.010
Argos, M. (2015). Arsenic Exposure and Epigenetic Alterations: Recent Findings Based on the Illumina 450K DNA Methylation Array. Curr. Envir Health Rpt 2 (2), 137–144. doi:10.1007/s40572-015-0052-1
Bokar, J. A., Shambaugh, M. E., Polayes, D., Matera, A. G., and Rottman, F. M. (1997). Purification and cDNA Cloning of the AdoMet-Binding Subunit of the Human mRNA (N6-Adenosine)-Methyltransferase. Rna 3 (11), 1233–1247.
Camper, S. A., Albers, R. J., Coward, J. K., and Rottman, F. M. (1984). Effect of Undermethylation on mRNA Cytoplasmic Appearance and Half-Life. Mol. Cel Biol 4 (3), 538–543. doi:10.1128/mcb.4.3.53810.1128/mcb.4.3.538-543.1984
Cantara, W. A., Crain, P. F., Rozenski, J., McCloskey, J. A., Harris, K. A., Zhang, X., et al. (2011). The RNA Modification Database, RNAMDB: 2011 Update. Nucleic Acids Res. 39, D195–D201. doi:10.1093/nar/gkq1028
Cao, G., Li, H.-B., Yin, Z., and Flavell, R. A. (2016). Recent Advances in Dynamic M 6 A RNA Modification. Open Biol. 6 (4), 160003. doi:10.1098/rsob.160003
Cao, Q., Guo, Z., Du, S., Ling, H., and Song, C. (2020). Circular RNAs in the Pathogenesis of Atherosclerosis. Life Sci. 255, 117837. doi:10.1016/j.lfs.2020.117837
Chen, L., Yao, H., Hui, J.-y., Ding, S.-h., Fan, Y.-l., Pan, Y.-h., et al. (2016). Global Transcriptomic Study of Atherosclerosis Development in Rats. Gene 592 (1), 43–48. doi:10.1016/j.gene.2016.07.023
Chen, T., Hao, Y.-J., Zhang, Y., Li, M.-M., Wang, M., Han, W., et al. (2015). m6A RNA Methylation Is Regulated by MicroRNAs and Promotes Reprogramming to PluripotencyA RNA Methylation Is Regulated by microRNAs and Promotes Reprogramming to Pluripotency. Cell Stem Cell 16 (3), 289–301. doi:10.1016/j.stem.2015.01.016
Chen, Y.-s., Ouyang, X.-p., Yu, X.-h., Novák, P., Zhou, L., He, P.-p., et al. (2021). N6-Adenosine Methylation (m6A) RNA Modification: an Emerging Role in Cardiovascular Diseases. J. Cardiovasc. Trans. Res. doi:10.1007/s12265-021-10108-w
Cheng, C., Wu, Y., Xiao, T., Xue, J., Sun, J., Xia, H., et al. (2021). METTL3-mediated m6A Modification of ZBTB4 mRNA Is Involved in the Smoking-Induced EMT in Cancer of the Lung. Mol. Ther. - Nucleic Acids 23, 487–500. doi:10.1016/j.omtn.2020.12.001
Church, C., Moir, L., McMurray, F., Girard, C., Banks, G. T., Teboul, L., et al. (2010). Overexpression of Fto Leads to Increased Food Intake and Results in Obesity. Nat. Genet. 42 (12), 1086–1092. doi:10.1038/ng.713
Cui, Y.-H., Yang, S., Wei, J., Shea, C. R., Zhong, W., Wang, F., et al. (2021). Autophagy of the m6A mRNA Demethylase FTO Is Impaired by Low-Level Arsenic Exposure to Promote Tumorigenesis. Nat. Commun. 12 (1), 2183. doi:10.1038/s41467-021-22469-6
Dai, N., Rapley, J., Angel, M., Yanik, M. F., Blower, M. D., and Avruch, J. (2011). mTOR Phosphorylates IMP2 to Promote IGF2 mRNA Translation by Internal Ribosomal Entry. Genes Develop. 25 (11), 1159–1172. doi:10.1101/gad.2042311
Desrosiers, R., Friderici, K., and Rottman, F. (1974). Identification of Methylated Nucleosides in Messenger RNA from Novikoff Hepatoma Cells. Proc. Natl. Acad. Sci. 71 (10), 3971–3975. doi:10.1073/pnas.71.10.3971
Dina, C., Meyre, D., Gallina, S., Durand, E., Körner, A., Jacobson, P., et al. (2007). Variation in FTO Contributes to Childhood Obesity and Severe Adult Obesity. Nat. Genet. 39 (6), 724–726. doi:10.1038/ng2048
Dominissini, D., Moshitch-Moshkovitz, S., Schwartz, S., Salmon-Divon, M., Ungar, L., Osenberg, S., et al. (2012). Topology of the Human and Mouse m6A RNA Methylomes Revealed by m6A-Seq. Nature 485 (7397), 201–206. doi:10.1038/nature11112
Falk, E. (2006). Pathogenesis of Atherosclerosis. J. Am. Coll. Cardiol. 47 (8 Suppl. l), C7–C12. doi:10.1016/j.jacc.2005.09.068
Frismantiene, A., Philippova, M., Erne, P., and Resink, T. J. (2018). Smooth Muscle Cell-Driven Vascular Diseases and Molecular Mechanisms of VSMC Plasticity. Cell Signal. 52, 48–64. doi:10.1016/j.cellsig.2018.08.019
Frye, M., Harada, B. T., Behm, M., and He, C. (2018). RNA Modifications Modulate Gene Expression during Development. Science 361 (6409), 1346–1349. doi:10.1126/science.aau1646
Fu, Y., Jia, G., Pang, X., Wang, R. N., Wang, X., Li, C. J., et al. (2013). FTO-mediated Formation of N6-Hydroxymethyladenosine and N6-Formyladenosine in Mammalian RNA. Nat. Commun. 4, 1798. doi:10.1038/ncomms2822
Fu, Z., Zhou, E., Wang, X., Tian, M., Kong, J., Li, J., et al. (2017). Oxidized Low-Density Lipoprotein-Induced Microparticles Promote Endothelial Monocyte Adhesion via Intercellular Adhesion Molecule 1. Am. J. Physiology-Cell Physiol. 313 (5), C567–C574. doi:10.1152/ajpcell.00158.2016
Gao, R., Ye, M., Liu, B., Wei, M., Ma, D., and Dong, K. (2021). m6A Modification: A Double-Edged Sword in Tumor Development. Front. Oncol. 11, 679367. doi:10.3389/fonc.2021.679367
Gerken, T., Girard, C. A., Tung, Y.-C. L., Webby, C. J., Saudek, V., Hewitson, K. S., et al. (2007). The Obesity-Associated FTO Gene Encodes a 2-oxoglutarate-dependent Nucleic Acid Demethylase. Science 318 (5855), 1469–1472. doi:10.1126/science.1151710
Gong, C., Fan, Y., and Liu, J. (2021). METTL14 Mediated m6A Modification to LncRNA ZFAS1/RAB22A: A Novel Therapeutic Target for Atherosclerosis. Int. J. Cardiol. 328, 177. doi:10.1016/j.ijcard.2020.12.002
Gu, X., Zhang, Y., Li, D., Cai, H., Cai, L., and Xu, Q. (2020). N6-methyladenosine Demethylase FTO Promotes M1 and M2 Macrophage Activation. Cell Signal. 69, 109553. doi:10.1016/j.cellsig.2020.109553
Guo, M., Yan, R., Ji, Q., Yao, H., Sun, M., Duan, L., et al. (2020). IFN Regulatory Factor-1 Induced Macrophage Pyroptosis by Modulating m6A Modification of Circ_0029589 in Patients with Acute Coronary Syndrome. Int. Immunopharmacology 86, 106800. doi:10.1016/j.intimp.2020.106800
Guo, T., Chai, X., Liu, Q., Peng, W., Peng, Z., and Cai, Y. (2017). Downregulation of P16 Promotes Cigarette Smoke Extract-Induced Vascular Smooth Muscle Cell Proliferation via Preventing G1/S Phase Transition. Exp. Ther. Med. 14 (1), 214–220. doi:10.3892/etm.2017.4468
Herrington, W., Lacey, B., Sherliker, P., Armitage, J., and Lewington, S. (2016). Epidemiology of Atherosclerosis and the Potential to Reduce the Global Burden of Atherothrombotic Disease. Circ. Res. 118 (4), 535–546. doi:10.1161/CIRCRESAHA.115.307611
Huang, C., Han, Y., Wang, Y., Sun, X., Yan, S., Yeh, E. T. H., et al. (2009). SENP3 Is Responsible for HIF-1 Transactivation under Mild Oxidative Stress via P300 De-SUMOylation. Embo j 28 (18), 2748–2762. doi:10.1038/emboj.2009.210
Huang, H., Weng, H., Sun, W., Qin, X., Shi, H., Wu, H., et al. (2018). Recognition of RNA N6-Methyladenosine by IGF2BP Proteins Enhances mRNA Stability and Translation. Nat. Cel Biol 20 (3), 285–295. doi:10.1038/s41556-018-0045-z
Incalza, M. A., D'Oria, R., Natalicchio, A., Perrini, S., Laviola, L., and Giorgino, F. (2018). Oxidative Stress and Reactive Oxygen Species in Endothelial Dysfunction Associated with Cardiovascular and Metabolic Diseases. Vasc. Pharmacol. 100, 1–19. doi:10.1016/j.vph.2017.05.005
Iyen, B., Qureshi, N., Kai, J., Akyea, R. K., Leonardi-Bee, J., Roderick, P., et al. (2019). Risk of Cardiovascular Disease Outcomes in Primary Care Subjects with Familial Hypercholesterolaemia: A Cohort Study. Atherosclerosis 287, 8–15. doi:10.1016/j.atherosclerosis.2019.05.017
Jackson, R. J., Hellen, C. U. T., and Pestova, T. V. (2010). The Mechanism of Eukaryotic Translation Initiation and Principles of its Regulation. Nat. Rev. Mol. Cel Biol 11 (2), 113–127. doi:10.1038/nrm2838
Jia, G., Fu, Y., and He, C. (2013). Reversible RNA Adenosine Methylation in Biological Regulation. Trends Genet. 29 (2), 108–115. doi:10.1016/j.tig.2012.11.003
Jia, G., Fu, Y., Zhao, X., Dai, Q., Zheng, G., Yang, Y., et al. (2011). N6-methyladenosine in Nuclear RNA Is a Major Substrate of the Obesity-Associated FTO. Nat. Chem. Biol. 7 (12), 885–887. doi:10.1038/nchembio.687
Jian, D., Wang, Y., Jian, L., Tang, H., Rao, L., Chen, K., et al. (2020). METTL14 Aggravates Endothelial Inflammation and Atherosclerosis by Increasing FOXO1 N6-Methyladeosine Modifications. Theranostics 10 (20), 8939–8956. doi:10.7150/thno.45178
Keller, M., Hopp, L., Liu, X., Wohland, T., Rohde, K., Cancello, R., et al. (2017). Genome-wide DNA Promoter Methylation and Transcriptome Analysis in Human Adipose Tissue Unravels Novel Candidate Genes for Obesity. Mol. Metab. 6 (1), 86–100. doi:10.1016/j.molmet.2016.11.003
Kishore, R., Garikipati, V. N. S., and Gonzalez, C. (2020). Role of Circular RNAs in Cardiovascular Disease. J. Cardiovasc. Pharmacol. 76 (2), 128–137. doi:10.1097/fjc.0000000000000841
Klungland, A., and Dahl, J. A. (2014). Dynamic RNA Modifications in Disease. Curr. Opin. Genet. Develop. 26, 47–52. doi:10.1016/j.gde.2014.05.006
Ko, J.-H., Sethi, G., Um, J.-Y., Shanmugam, M. K., Arfuso, F., Kumar, A. P., et al. (2017). The Role of Resveratrol in Cancer Therapy. Int. J. Mol. Sci. 18 (12), 2589. doi:10.3390/ijms18122589
Koelwyn, G. J., Corr, E. M., Erbay, E., and Moore, K. J. (2018). Regulation of Macrophage Immunometabolism in Atherosclerosis. Nat. Immunol. 19 (6), 526–537. doi:10.1038/s41590-018-0113-3
Koh, C. W. Q., Goh, Y. T., and Goh, W. S. S. (2019). Atlas of Quantitative Single-Base-Resolution N6-Methyl-Adenine Methylomes. Nat. Commun. 10 (1), 5636. doi:10.1038/s41467-019-13561-z
Krylatov, A. V., Maslov, L. N., Voronkov, N. S., Boshchenko, A. A., Popov, S. V., Gomez, L., et al. (2018). Reactive Oxygen Species as Intracellular Signaling Molecules in the Cardiovascular System. Curr. Cardial.Rev. 14 (4), 290–300. doi:10.2174/1573403x14666180702152436
Kupsco, A., Gonzalez, G., Baker, B. H., Knox, J. M., Zheng, Y., Wang, S., et al. (2020). Associations of Smoking and Air Pollution with Peripheral Blood RNA N6-Methyladenosine in the Beijing Truck Driver Air Pollution Study. Environ. Int. 144, 106021. doi:10.1016/j.envint.2020.106021
Li, X., Yang, L., and Chen, L.-L. (2018). The Biogenesis, Functions, and Challenges of Circular RNAs. Mol. Cel 71 (3), 428–442. doi:10.1016/j.molcel.2018.06.034
Libby, P. (2000). Changing Concepts of Atherogenesis. J. Intern. Med. 247 (3), 349–358. doi:10.1046/j.1365-2796.2000.00654.x
Libby, P., Lichtman, A. H., and Hansson, G. K. (2013). Immune Effector Mechanisms Implicated in Atherosclerosis: from Mice to Humans. Immunity 38 (6), 1092–1104. doi:10.1016/j.immuni.2013.06.009
Lin, J., Zhu, Q., Huang, J., Cai, R., and Kuang, Y. (2020). Hypoxia Promotes Vascular Smooth Muscle Cell (VSMC) Differentiation of Adipose-Derived Stem Cell (ADSC) by Regulating Mettl3 and Paracrine Factors. Stem Cell Int. 2020, 1–11. doi:10.1155/2020/2830565
Linder, B., Grozhik, A. V., Olarerin-George, A. O., Meydan, C., Mason, C. E., and Jaffrey, S. R. (2015). Single-nucleotide-resolution Mapping of m6A and m6Am throughout the Transcriptome. Nat. Methods 12 (8), 767–772. doi:10.1038/nmeth.3453
Liu, J., Yue, Y., Han, D., Wang, X., Fu, Y., Zhang, L., et al. (2014). A METTL3-METTL14 Complex Mediates Mammalian Nuclear RNA N6-Adenosine Methylation. Nat. Chem. Biol. 10 (2), 93–95. doi:10.1038/nchembio.1432
Liu, N., Dai, Q., Zheng, G., He, C., Parisien, M., and Pan, T. (2015). N6-methyladenosine-dependent RNA Structural Switches Regulate RNA-Protein Interactions. Nature 518 (7540), 560–564. doi:10.1038/nature14234
Liu, X., Wang, H., Zhao, X., Luo, Q., Wang, Q., Tan, K., et al. (2021). Arginine Methylation of METTL14 Promotes RNA N6-Methyladenosine Modification and Endoderm Differentiation of Mouse Embryonic Stem Cells. Nat. Commun. 12 (1), 3780. doi:10.1038/s41467-021-24035-6
Liu, Y., Liu, Z., Tang, H., Shen, Y., Gong, Z., Xie, N., et al. (2019). The N6-Methyladenosine (m6A)-Forming Enzyme METTL3 Facilitates M1 Macrophage Polarization through the Methylation of STAT1 mRNA. Am. J. Physiology-Cell Physiol. 317 (4), C762–C775. doi:10.1152/ajpcell.00212.2019
Luo, G.-Z., MacQueen, A., Zheng, G., Duan, H., Dore, L. C., Lu, Z., et al. (2014). Unique Features of the m6A Methylome in Arabidopsis thaliana. Nat. Commun. 5, 5630. doi:10.1038/ncomms6630
Lv, Y., Li, T., Yang, M., Su, L., Zhu, Z., Zhao, S., et al. (2021). Melatonin Attenuates Chromium (VI)-Induced Spermatogonial Stem Cell/Progenitor Mitophagy by Restoration of METTL3-Mediated RNA N6-Methyladenosine Modification. Front. Cel Dev. Biol. 9, 684398. doi:10.3389/fcell.2021.684398
Marinov, G. K., Williams, B. A., McCue, K., Schroth, G. P., Gertz, J., Myers, R. M., et al. (2014). From Single-Cell to Cell-Pool Transcriptomes: Stochasticity in Gene Expression and RNA Splicing. Genome Res. 24 (3), 496–510. doi:10.1101/gr.161034.113
Martinez, V. D., Vucic, E. A., Becker-Santos, D. D., Gil, L., and Lam, W. L. (2011). Arsenic Exposure and the Induction of Human Cancers. J. Toxicol. 2011, 1–13. doi:10.1155/2011/431287
Meyer, K. D., Saletore, Y., Zumbo, P., Elemento, O., Mason, C. E., and Jaffrey, S. R. (2012). Comprehensive Analysis of mRNA Methylation Reveals Enrichment in 3′ UTRs and Near Stop Codons. Cell 149 (7), 1635–1646. doi:10.1016/j.cell.2012.05.003
Miano, J. M., Fisher, E. A., and Majesky, M. W. (2021). Fate and State of Vascular Smooth Muscle Cells in Atherosclerosis. Circulation 143 (21), 2110–2116. doi:10.1161/circulationaha.120.049922
Morabia, A., and Abel, T. (2006). The WHO Report "Preventing Chronic Diseases: a Vital Investment" and Us. Soz.-Präventivmed. 51 (2), 74. doi:10.1007/s00038-005-0015-7
Park, M. H., Jeong, E., and Choudhury, M. (2020). Mono-(2-Ethylhexyl)phthalate Regulates Cholesterol Efflux via MicroRNAs Regulated m6A RNA Methylation. Chem. Res. Toxicol. 33 (2), 461–469. doi:10.1021/acs.chemrestox.9b00367
Pepine, C. J., Schlaifer, J. D., Mancini, G. B. J., Pitt, B., O'Neill, B. J., and Haber, H. E. (1998). Influence of Smoking Status on Progression of Endothelial Dysfunction. Clin. Cardiol. 21 (5), 331–334. doi:10.1002/clc.4960210506
Ping, X.-L., Sun, B.-F., Wang, L., Xiao, W., Yang, X., Wang, W.-J., et al. (2014). Mammalian WTAP Is a Regulatory Subunit of the RNA N6-Methyladenosine Methyltransferase. Cell Res 24 (2), 177–189. doi:10.1038/cr.2014.3
Qu, T., Mou, Y., Dai, J., Zhang, X., Li, M., Gu, S., et al. (2021). Changes and Relationship of N6-Methyladenosine Modification and Long Non-coding RNAs in Oxidative Damage Induced by Cadmium in Pancreatic β-cells. Toxicol. Lett. 343, 56–66. doi:10.1016/j.toxlet.2021.02.014
Quiles-Jiménez, A., Gregersen, I., Mittelstedt Leal de Sousa, M., Abbas, A., Kong, X. Y., Alseth, I., et al. (2020). N6-methyladenosine in RNA of Atherosclerotic Plaques: An Epitranscriptomic Signature of Human Carotid Atherosclerosis. Biochem. Biophysical Res. Commun. 533 (4), 631–637. doi:10.1016/j.bbrc.2020.09.057
Roundtree, I. A., Luo, G.-Z., Zhang, Z., Wang, X., Zhou, T., Cui, Y., et al. (2017). YTHDC1 Mediates Nuclear export of N6-Methyladenosine Methylated mRNAs. Elife 6, 6. doi:10.7554/eLife.31311
Scuteri, A., Sanna, S., Chen, W.-M., Uda, M., Albai, G., Strait, J., et al. (2007). Genome-wide Association Scan Shows Genetic Variants in the FTO Gene Are Associated with Obesity-Related Traits. Plos Genet. 3 (7), e115. doi:10.1371/journal.pgen.0030115
Shah, A., Rashid, F., Awan, H. M., Hu, S., Wang, X., Chen, L., et al. (2017). The DEAD-Box RNA Helicase DDX3 Interacts with m6A RNA Demethylase ALKBH5. Stem Cell Int. 2017, 1–11. doi:10.1155/2017/8596135
Shan, K., Zhou, R.-m., Xiang, J., Sun, Y.-n., Liu, C., Lv, M.-w., et al. (2020). FTO Regulates Ocular Angiogenesis via m6A-YTHDF2-dependent Mechanism. Exp. Eye Res. 197, 108107. doi:10.1016/j.exer.2020.108107
Shi, H., Wang, X., Lu, Z., Zhao, B. S., Ma, H., Hsu, P. J., et al. (2017). YTHDF3 Facilitates Translation and Decay of N6-Methyladenosine-Modified RNA. Cel Res 27 (3), 315–328. doi:10.1038/cr.2017.15
Shi, H., Wei, J., and He, C. (2019). Where, when, and How: Context-dependent Functions of RNA Methylation Writers, Readers, and Erasers. Mol. Cel 74 (4), 640–650. doi:10.1016/j.molcel.2019.04.025
Singh, M., and Bedi, U. S. (2013). Is Atherosclerosis Regression a Realistic Goal of Statin Therapy and what Does that Mean? Curr. Atheroscler. Rep. 15 (1), 294. doi:10.1007/s11883-012-0294-4
Stoneley, M., and Willis, A. E. (2004). Cellular Internal Ribosome Entry Segments: Structures, Trans-acting Factors and Regulation of Gene Expression. Oncogene 23 (18), 3200–3207. doi:10.1038/sj.onc.1207551
Tang, X., Yin, R., Shi, H., Wang, X., Shen, D., Wang, X., et al. (2020). LncRNA ZFAS1 Confers Inflammatory Responses and Reduces Cholesterol Efflux in Atherosclerosis through Regulating miR-654-3p-Adam10/rab22a axis. Int. J. Cardiol. 315, 72–80. doi:10.1016/j.ijcard.2020.03.056
Virani, S. S., Alonso, A., Benjamin, E. J., Bittencourt, M. S., Callaway, C. W., Carson, A. P., et al. (2020). Heart Disease and Stroke Statistics-2020 Update: A Report from the American Heart Association. Circulation 141 (9), e139–e596. doi:10.1161/CIR.0000000000000757
Wang, J., Wang, J., Gu, Q., Ma, Y., Yang, Y., Zhu, J., et al. (2020). The Biological Function of m6A Demethylase ALKBH5 and its Role in Human Disease. Cancer Cel Int 20, 347. doi:10.1186/s12935-020-01450-1
Wang, L. j., Xue, Y., Li, H., Huo, R., Yan, Z., Wang, J., et al. (2020). Wilms' Tumour 1‐associating Protein Inhibits Endothelial Cell Angiogenesis by m6A‐dependent Epigenetic Silencing of Desmoplakin in Brain Arteriovenous Malformation. J. Cel Mol Med 24 (9), 4981–4991. doi:10.1111/jcmm.15101
Wang, X., Bian, Y., Zhang, R., Liu, X., Ni, L., Ma, B., et al. (2019). Melatonin Alleviates Cigarette Smoke-Induced Endothelial Cell Pyroptosis through Inhibiting ROS/NLRP3 axis. Biochem. Biophysical Res. Commun. 519 (2), 402–408. doi:10.1016/j.bbrc.2019.09.005
Wang, X., Feng, J., Xue, Y., Guan, Z., Zhang, D., Liu, Z., et al. (2016). Structural Basis of N6-Adenosine Methylation by the METTL3-METTL14 Complex. Nature 534 (7608), 575–578. doi:10.1038/nature18298
Wang, X., Lu, Z., Gomez, A., Hon, G. C., Yue, Y., Han, D., et al. (2014). N6-methyladenosine-dependent Regulation of Messenger RNA Stability. Nature 505 (7481), 117–120. doi:10.1038/nature12730
Wang, X., Zhao, B. S., Roundtree, I. A., Lu, Z., Han, D., Ma, H., et al. (2015). N6-methyladenosine Modulates Messenger RNA Translation Efficiency. Cell 161 (6), 1388–1399. doi:10.1016/j.cell.2015.05.014
Wang, Y., Yang, J., Yang, K., Cang, H., Huang, X.-z., Li, H., et al. (2012). The Biphasic Redox Sensing of SENP3 Accounts for the HIF-1 Transcriptional Activity Shift by Oxidative Stress. Acta Pharmacol. Sin 33 (7), 953–963. doi:10.1038/aps.2012.40
Wei, J., Liu, F., Lu, Z., Fei, Q., Ai, Y., He, P. C., et al. (2018). Differential m6A, m6Am, and m1A Demethylation Mediated by FTO in the Cell Nucleus and Cytoplasm. Mol. Cel 71 (6), 973–985. doi:10.1016/j.molcel.2018.08.011
Wu, J., Gan, Z., Zhuo, R., Zhang, L., Wang, T., and Zhong, X. (2020). Resveratrol Attenuates Aflatoxin B1-Induced ROS Formation and Increase of m6A RNA Methylation. Animals 10 (4), 677. doi:10.3390/ani10040677
Xu, Z., Lam, L. S. M., Lam, L. H., Chau, S. F., Ng, T. B., and Au, S. W. N. (2008). Molecular Basis of the Redox Regulation of SUMO Proteases: a Protective Mechanism of Intermolecular Disulfide Linkage against Irreversible Sulfhydryl Oxidation. FASEB j. 22 (1), 127–137. doi:10.1096/fj.06-7871com
Yamaguchi, Y., Haginaka, J., Morimoto, S., Fujioka, Y., and Kunitomo, M. (2005). Facilitated Nitration and Oxidation of LDL in Cigarette Smokers. Eur. J. Clin. Invest. 35 (3), 186–193. doi:10.1111/j.1365-2362.2005.01472.x
Yu, F., Wei, J., Cui, X., Yu, C., Ni, W., Bungert, J., et al. (2021). Post-translational Modification of RNA m6A Demethylase ALKBH5 Regulates ROS-Induced DNA Damage Response. Nucleic Acids Res. 49 (10), 5779–5797. doi:10.1093/nar/gkab415
Yu, R., Li, Q., Feng, Z., Cai, L., and Xu, Q. (2019). m6A Reader YTHDF2 Regulates LPS-Induced Inflammatory Response. Ijms 20 (6), 1323. doi:10.3390/ijms20061323
Yue, Y., Liu, J., and He, C. (2015). RNAN6-methyladenosine Methylation in post-transcriptional Gene Expression Regulation. Genes Dev. 29 (13), 1343–1355. doi:10.1101/gad.262766.115
Zaman, A. G., Helft, G., Worthley, S. G., and Badimon, J. J. (2000). The Role of Plaque Rupture and Thrombosis in Coronary Artery Disease. Atherosclerosis 149 (2), 251–266. doi:10.1016/s0021-9150(99)00479-7
Zhang, B. Y., Han, L., Tang, Y. F., Zhang, G. X., Fan, X. L., Zhang, J. J., et al. (2020). METTL14 Regulates M6A Methylation-Modified Primary miR-19a to Promote Cardiovascular Endothelial Cell Proliferation and Invasion. Eur. Rev. Med. Pharmacol. Sci. 24 (12), 7015–7023. doi:10.26355/eurrev_202006_21694
Zhang, M., Zhang, Y., Ma, J., Guo, F., Cao, Q., Zhang, Y., et al. (2015). The Demethylase Activity of FTO (Fat Mass and Obesity Associated Protein) Is Required for Preadipocyte Differentiation. PLoS One 10 (7), e0133788. doi:10.1371/journal.pone.0133788
Zhao, T., Li, X., Sun, D., and Zhang, Z. (2019). Oxidative Stress: One Potential Factor for Arsenite-Induced Increase of N6-Methyladenosine in Human Keratinocytes. Environ. Toxicol. Pharmacol. 69, 95–103. doi:10.1016/j.etap.2019.04.005
Zhao, X., Yang, Y., Sun, B.-F., Shi, Y., Yang, X., Xiao, W., et al. (2014). FTO-dependent Demethylation of N6-Methyladenosine Regulates mRNA Splicing and Is Required for Adipogenesis. Cel Res 24 (12), 1403–1419. doi:10.1038/cr.2014.151
Zheng, G., Dahl, J. A., Niu, Y., Fedorcsak, P., Huang, C.-M., Li, C. J., et al. (2013). ALKBH5 Is a Mammalian RNA Demethylase that Impacts RNA Metabolism and Mouse Fertility. Mol. Cel 49 (1), 18–29. doi:10.1016/j.molcel.2012.10.015
Zhou, B., Liu, C., Xu, L., Yuan, Y., Zhao, J., Zhao, W., et al. (2021). N 6 ‐Methyladenosine Reader Protein YT521‐B Homology Domain‐Containing 2 Suppresses Liver Steatosis by Regulation of mRNA Stability of Lipogenic Genes. Hepatology 73 (1), 91–103. doi:10.1002/hep.31220
Zhou, J., Wan, J., Gao, X., Zhang, X., Jaffrey, S. R., and Qian, S.-B. (2015). Dynamic m6A mRNA Methylation Directs Translational Control of Heat Shock Response. Nature 526 (7574), 591–594. doi:10.1038/nature15377
Zhu, B., Gong, Y., Shen, L., Li, J., Han, J., Song, B., et al. (2020). Total Panax Notoginseng Saponin Inhibits Vascular Smooth Muscle Cell Proliferation and Migration and Intimal Hyperplasia by Regulating WTAP/p16 Signals via m6A Modulation. Biomed. Pharmacother. 124, 109935. doi:10.1016/j.biopha.2020.109935
Zhu, Y., Xian, X., Wang, Z., Bi, Y., Chen, Q., Han, X., et al. (2018). Research Progress on the Relationship between Atherosclerosis and Inflammation. Biomolecules 8 (3), 80. doi:10.3390/biom8030080
Zong, D., Liu, X., Li, J., Ouyang, R., and Chen, P. (2019). The Role of Cigarette Smoke-Induced Epigenetic Alterations in Inflammation. Epigenetics & Chromatin 12 (1), 65. doi:10.1186/s13072-019-0311-8
Glossary
AS atherosclerosis
m6A N6-methyladenosine
SAM S-adenosyl-l-methionine
SAH S-Adenosyl-l-homocysteine
METTL3 methyltransferase like protein 3
METTL14 methyltransferase like protein 14
FTO fat mass and obesity-associated protein
ALKBH5 alkylation repair homolog protein 5
hm6A N6-hydroxymethyladenosine
f6A N6-formyladenosine
YTH YT521-B homology
YTHDF YT521-B homology domain-containing family
HNRNPC heterogeneous nuclear ribonucleoprotein C
IGF2 insulin-like growth factor 2
T2D type 2 diabetes
U uracil
ZNFX1 zinc finger NFX1-type containing 1
ZFAS1 zinc finger antisense RNA1
SMCs smooth muscle cells
RIP RNA immunoprecipitation
FOXO fork head box class O
IRF-1 interferon regulatory factor-1
STAT1 signal transducers and activators of transcription 1
PPAR-γ peroxisome proliferator-activated receptor-γ
LPS lipopolysaccharide
MAP2K4 mitogen-activated protein kinase kinase 4
ERK extracellular signal‐regulated kinase
TNF tumor necrosis factor
IL interleukin
MEHP macrophage mono-(2-ethylhexyl) phthalate
SR-B1 scavenger receptor B type 1
VSMCs vascular smooth muscle cells
VEGF vascular endothelial growth factor
TGF transforming growth factor
ADSCs adipose-derived stem cells
qPCR quantitative polymerase chain reaction
TPNS total Panax notoinseng saponin
ROS reactive oxygen species
Bcl-2 B-cell lymphoma 2
caspase cysteine aspartyl-specific protease
bax,Bcl-2-associated X protein
ABCA5 ATP-binding cassette transporter A5
SLC22 A17 solute carrier family 22 member 17
KCNQ5 potassium voltage-gated channel subfamily Q member 5
lncRNA long noncoding RNA
MDA malondialdehyde
SOD superoxide dismutase
WHO world health organization
SNPs single nucleotide polymorphisms
SR serine/arginine-rich
SRSF2 serine/arginine-rich splicing factor 2
RUNX1T1 runt-related transcription factor 1 translocation partner 1
CSE cigarette smoke extract
LDL low-density lipoprotein
HBE human bronchial epithelial
EZH2 enhancer of zeste homolog 2
EMT epithelial-mesenchymal transformation
H3K27me3 histone 3 lysine 27 trimethylation
DSP desmoplakin
IGF2BP2 insulin-like growth factor 2 binding proteins 2
IGF2BP3 insulin-like growth factor 2 binding proteins 3
SENP1 sentrin/sumo-specific proteases1
SENP3 sentrin/sumo-specific proteases3
Keywords: N6methyladenosine (m6A) modification, atherosclerosis, methyltransferase, erasers-demethylases, vascular endothelial cells
Citation: Fu J, Cui X, Zhang X, Cheng M, Li X, Guo Z and Cui X (2021) The Role of m6A Ribonucleic Acid Modification in the Occurrence of Atherosclerosis. Front. Genet. 12:733871. doi: 10.3389/fgene.2021.733871
Received: 02 July 2021; Accepted: 02 September 2021;
Published: 16 September 2021.
Edited by:
Julian J-L. Chen, Arizona State University, United StatesReviewed by:
Kausik Chakrabarti, University of North Carolina at Charlotte, United StatesJiangbo Wei, University of Chicago, United States
Copyright © 2021 Fu, Cui, Zhang, Cheng, Li, Guo and Cui. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhiliang Guo, ZHJ6bGd1b0AxNjMuY29t; Xiaodong Cui, eGlhb2RvbmdjdWlAd2ZtYy5lZHUuY24=
†These authors have contributed equally to this work and share first authorship
 Jie Fu
Jie Fu Xinghui Cui
Xinghui Cui Xiaoyun Zhang
Xiaoyun Zhang Min Cheng
Min Cheng Xiaoxia Li
Xiaoxia Li Zhiliang Guo
Zhiliang Guo Xiaodong Cui
Xiaodong Cui