- 1Clinical College of Neurology, Neurosurgery and Neurorehabilitation, Tianjin Medical University, Tianjin, China
- 2Department of Neurosurgery, Shijiazhuang Third Hospital, Hebei, China
- 3Tianjin Key Laboratory of Cerebral Vascular and Neurodegenerative Diseases, Tianjin Neurosurgical Institute, Tianjin Huanhu Hospital, Tianjin, China
- 4Clinical Laboratory, Tianjin Huanhu Hospital, Tianjin, China
- 5Department of Neurosurgery, Tianjin Huanhu Hospital, Tianjin, China
Background: Gliomas are the most common intracranial malignant neoplasms and have high recurrence and mortality rates. Recent literatures have reported that centromere protein N (CENPN) participates in tumor development. However, the clinicopathologic significance and biological functions of CENPN in glioma are still unclear.
Methods: Clinicopathologic data and gene expression profiles of glioma cases downloaded from The Cancer Genome Atlas (TCGA) and Chinese Glioma Genome Atlas (CGGA) databases were utilized to determine the associations between the expression of CENPN and clinical features of glioma. Kaplan-Meier and ROC curves were plotted for prognostic analysis. Gene set enrichment analysis (GSEA) and single sample gene set enrichment analysis (ssGSEA) were applied to identify immune-related functions and pathways associated with CENPN’ differential expression. In vitro experiments were conducted to investigate the impacts of CENPN on human glioma cells.
Results: Elevated CENPN expression was associated with unfavorable clinical variables of glioma patients, which was validated in clinical specimens obtained from our institution by immunohistochemical staining (IHC). The GSEA and ssGSEA results revealed that CENPN expression was strongly correlated with inflammatory activities, immune-related signaling pathways and the infiltration of immune cells. Cell experiments showed that CENPN deficiency impaired cell proliferation, migration and invasion ability and increased glioma apoptosis.
Conclusion: CENPN could be a promising therapeutic target for glioma.
Introduction
Gliomas are the most common intracranial malignant neoplasms, and they present high recurrence and mortality rates (Ostrom et al., 2018). Although multimodal treatments have been developed, glioma patient’s overall survival (OS) is still limited (5 years survival of approximately 5.5%) (OmuroDeAngelis 2013). Molecular markers, such as 1p/19q codeletion status and isocitrate dehydrogenase (IDH) mutations, play valuable roles in tumor formation and progression (Kristensen et al., 2019; Wang et al., 2020). Individually tailored strategies targeting these biomarkers have been adopted in multiple clinical trials, although few have made breakthroughs. Thus, new targets for glioma therapy are urgently needed.
During mitosis, the assembly of kinetochore proteins at the centromere contributes to the accurate segregation of chromosomes (Kops et al., 2005; McKinleyCheeseman 2016). Centromeric Proteins (CENP), containing 18 subtypes, dynamically associate and dissociate onto centromeric chromatin during mitosis with microtubule regulation (CheesemanDesai 2008; WesthorpeStraight 2014; Hoischen et al., 2018). Among the CENPs, CENPN aggregates toward centromeres in the S phase and eventually dissociates in the G2 phase (Foltz et al., 2006; Carroll et al., 2009; Fang et al., 2015). Recognition of the CENPA nucleosome core region by CENPN is essential for proper chromosome division and kinetochore assembly (Carroll et al., 2009). Recently, the relevance of CENPN to the occurrence and progression of different cancers has been proposed. In oral squamous cell carcinoma, CENPN knockdown arrests the cell cycle in the G1 phase, which leads to the suppression of cellular proliferation (Oka et al., 2019). Based on bioinformatics analyses, Rahman et al. reported that CENPN was a prognostic marker for colorectal cancer (Rahman et al., 2019). Moreover, Sarah An et al. revealed that CENPN expression was involved in the malignant progression of breast cancer patients with a history of smoking (Andres et al., 2015). Most recently, CENPN was shown to promote hepatocellular carcinoma cell proliferation and CENPN deficiency was shown to increase radiotherapy-induced DNA damage (Wang et al., 2021). However, the roles of CENPN in glioma have not been clarified.
In the present study, CENPN served as a prognostic biomarker for glioma based on data obtained from public cancer databases and clinical specimens. Moreover, we investigated the potential functions of CENPN in the immune infiltration and immunoregulation of the glioma microenvironment. In vitro cell experiments were conducted to verify the reliability of bioinformatics data and provided a reference for future research.
Materials and Methods
Dataset Selection
Next-generation sequencing data (HTSeq-FPKM) and clinical variables were obtained from TCGA (Wang et al., 2016). In addition, data for 1,018 glioma cases generated with the Illumina HiSeq platform were obtained from the CGGA (Zhao et al., 2021). Apart from that, a total of 112 glioma cases enrolled from the Department of Neurosurgery (Huanhu Hospital, Tianjin, China) were used for validation.
Bioinformatics Analysis
In TCGA, the differential expression of CENPN was analyzed using GEPIA, a web database containing abundant normal specimens from GTEx database (Tang et al., 2019). Additionally, GSE16011 (Gravendeel et al., 2009), a dataset from the Gene Expression Omnibus database (GEO), was used for validation. The relationships between CENPN and clinical variables were explored using the beeswarm R package. The survival, survminer and ROC packages were used to plot receiver operating characteristic (ROC) curves and Kaplan–Meier curves.
GSEA was conducted based on the well-established gene sets of c2.cp.kegg.v7.2.symbols.gmt and c5.all.v7.2.symbols.gmt in GSEA 4.0 software. ssGSEA was used to transform the 29 immune signatures into scores for each glioma sample (Subramanian et al., 2005; Bindea et al., 2013; Zhou et al., 2020). Additionally, the CIBERSORT deconvolution algorithm was performed to evaluate the immune infiltration of 22 tumor-infiltrating immune cells (TILs) (Newman et al., 2015). The TIL signatures were downloaded from CIBERSORT (https://cibersortx.stanford.edu/). Relevant R packages were obtained from Bioconductor (http://bioconductor.org/) and CRAN (http://cran.r-project.org/). The bioinformatics analyses were conducted using R software.
Cell Culture and Transfection
The human glioblastoma cell lines LN229 and U251 were purchased from Beijing Beina Chuanglian Biotechnology Institute and cultured in DMEM containing 10% fetal bovine serum (FBS, Gibco, Invitrogen, CA, United States). The siRNAs targeting CENPN mRNA were produced by GenePharma. The sense sequences of these siRNAs were as follows: siCENPN-i, GCGUGCAAGUAUCAGUGAUTT; siCENPN-ii, GACCCUUUGUUACUCAAAUTT; and scramble (siScr), UUCUCCGAACGUGUCACGUTT. siRNA transfections were performed using Lipofectamine 2000 according to the manufacturer’s instructions (Thermo Fisher Scientific).
RNA Extraction and Real-Time PCR
TRIzol reagent was used for RNA extraction, and the SureFireRT kit (06-104; Abgen) was applied for RNA reverse transcription. Then, real-time PCR was conducted on a LightCycler 480 II (Roche). The sequences of designed PCR primers were as follows: GAPDH forward, 5′-CAATGACCCCTTCATTGACC-3′ and reverse, 5′-GACAAGCTTCCCGTTCTCAG-3′; CENPN forward, 5′- TGGATGAGACTGTTGCTGA -3′ and reverse, 5′- ACTTGCACGCTTTTCCTC -3′. The PCR conditions were 95°C for 5 min, followed by 40 cycles at 95°C for 10 s and 60°C for 1 min. Relative quantification was calculated by the 2−ΔΔCt method.
Western Blotting and Immunohistochemistry
Cells were lysed using RIPA buffer following the manufacturer’s protocols (Solarbio Co., Beijing, China). Protein (50 μg/lane) was added to the 12% SDS-PAGE gel and then transferred to a membrane (PVDF, Merck Millipore). The protein blots on the membrane blocked by BSA were incubated with antibody against CENPN (1:500 dilution; Affinity Biosciences, Jiangsu, China; DF2315) or β-tubulin (1:500 dilution; CST, United States) overnight. After incubation with HRP-linked goat anti-rabbit IgG (CST, United States) for 60 min, the membrane was exposed to enhanced chemiluminescence reagent.
Immunohistochemistry assays were conducted by the pathology department of Huanhu Hospital. Immunostaining of CENPN was performed using a rabbit polyclonal anti-CENPN antibody (1:500 dilution; Affinity Biosciences, Jiangsu, China; DF2315). The positive slides were recognized by integrated scoring, which was performed independently by two experienced pathologists. The glioma tissues were divided into high and low expression groups based on the expression density of CENPN, as previously reported (Xiao et al., 2015).
CCK-8, Colony Formation, Wound Healing, Invasion, Apoptosis and Cell Cycle Analysis
5 × 103 cells per well were seeded in 96-well plates and then transfected with siCENPN. Cells were incubated with CCK-8 reagent (K009-500, ZETA) for 30–60 min at room temperature at 0, 24, 48 and 72 h time points. Eventually, the absorbance changes were observed at 450 nm on a molecular devices microplate reader.
For the wound-healing assay, the bottom of plate was scraped by a pipette tip when 100% cell confluence. Then, wound closure (%) was observed at 0 and 24 h by ImageJ. For the Transwell assay, 5 × 104 cells with or without CENPN knockdown were seeded in the upper chamber (8 µm pore size, Corning, Cambridge, United States) coated with Matrigel (BD Biosciences) while 500 µl of medium containing 10% FBS was placed in the lower chamber. Twenty-four hours later, the upper chamber was immersed in 4% paraformaldehyde for 20 min and then subjected to 20 min crystal violet staining. Eventually, the number of penetrating cells was calculated randomly by three fields of view (FOVs). A colony formation assay was conducted to examine the glioma proliferation ability, with 500 cells per well seeded in six-well plates. Fourteen days later, the cells were fixed with 4% formaldehyde and subjected to crystal violet staining.
Cell apoptosis assay (FITC Annexin V Apoptosis Detection Kit, BD Biosciences, United States) was conducted according to the manufacturer’s protocol, as previously described (Zhou et al., 2021). For the cell cycle assay, glioma cells were harvested 48 h following transfection and then fixed with precooled 75% alcohol for 3 h. Subsequently, the cells were incubated with 0.5 ml of PI/RNase reagent (BD Biosciences, United States) at room temperature for 20 min and then analyzed by flow cytometry.
Statistical Analysis
Cases with incomplete information were removed from the corresponding analysis. Student’s t-test and one‐way analysis of variance (ANOVA) were used to test for significant differences between two groups or multiple groups, respectively. For CENPN protein expression, the chi-square test was employed. Data are presented as the mean ± standard deviation (SD) of at least three independent experiments. p < 0.05 was viewed statistically significant. All statistical tests were two-sided. R software v3.6.2. and GraphPad Prism 8 were used for the statistical analyses.
Results
Information of Datasets
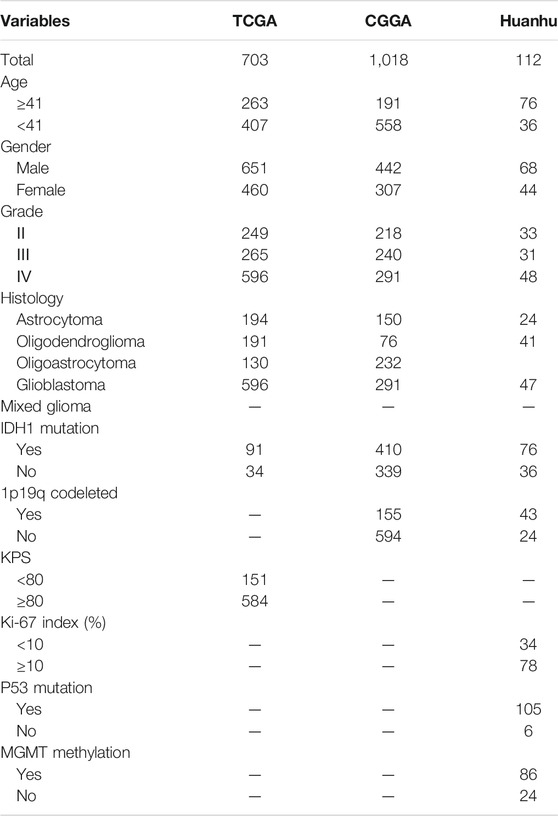
In the present study, 1833 glioma cases from the TCGA, CGGA and Huanhu datasets were included. The clinical features of these datasets are summarized in Table 1. Pathology reports containing the WHO grade, IDH1 mutation status and other information on the patients in the Huanhu dataset were provided by the Department of Pathology of Huanhu Hospital. The patients in the Huanhu cohort consisted of 44 females and 68 males, and their age ranged from 24 to 82 years old. Detailed information on the patients in the Huanhu cohort is shown in Supplementary Table S1.
mRNA Levels of CENPN in the Databases
The mRNA expression levels of CENPN in glioma were assessed based on GEPIA and GSE16011, a glioma dataset obtained from the Gene Expression Omnibus database. Compared with the values in normal samples, the CENPN mRNA level was markedly higher in the glioma samples (Figures 1A,B).
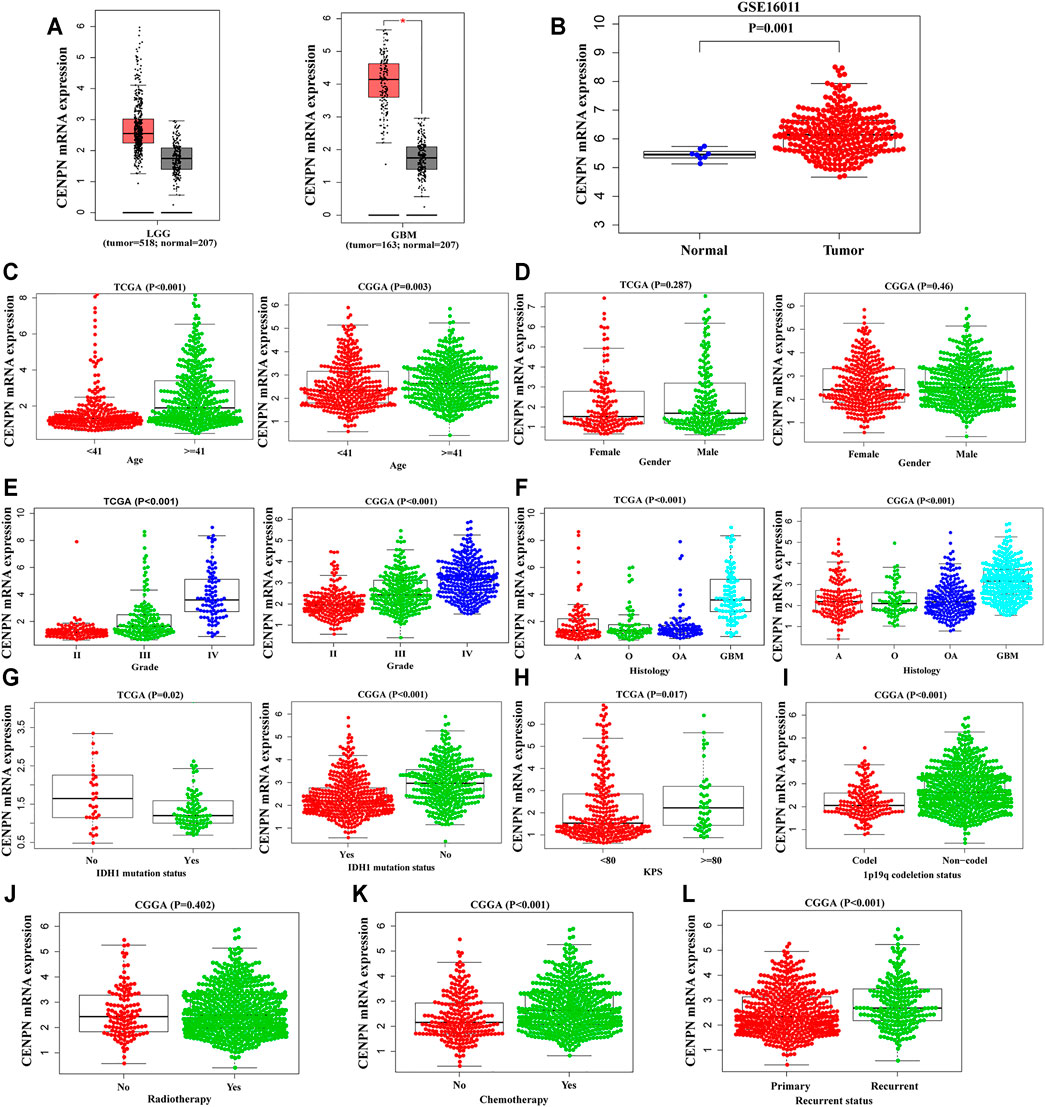
FIGURE 1. The mRNA expression profiles of CENPN in public databases. (A,B) CENPN expression levels in the GEPIA and GSE16011 datasets between tumor and normal tissues. (C–L) Associations between CENPN and clinical variables in TCGA and CGGA databases: (C) age (year); (D) gender; (E) WHO grade; (F) histological type (A, astrocytoma; O, oligodendroglioma; OA, oligoastrocytoma; GBM, glioblastoma); (G) IDH1 mutation status; (H) KPS; (I) 1p19q codeletion status; (J) radiotherapy; (K) chemotherapy; and (L) recurrence status.
Association Between CENPN and Glioma Clinical Features
To explore the CENPN expression profiles in glioma, the clinical features of glioma patients obtained from TCGA and CGGA were analyzed. As demonstrated in Figures 1C,E–G, CENPN was highly expressed in patients older than 41 years and patients with advanced grades, histopathologies and wild-type IDH1 in both TCGA and CGGA, although no association was found between CENPN expression and gender (Figure 1D). In TCGA, CENPN expression increased with higher Karnofsky Performance Status scores (KPS ≥ 80) (Figure 1H). In CGGA, some other clinical variables (radiotherapy, 1p19q codeletion, chemotherapy and recurrent status) were also analyzed (Figure 1I-L). These results revealed that CENPN upregulation could predict unfavorable glioma.
CENPN Predicts Worse Survival in Glioma
Kaplan-Meier plots were graphed to estimate the prognostic value of CENPN expression. As depicted in Figure 2A, CENPN high expression was associated with worse survival of patients in TCGA. ROC analyses demonstrated that CENPN could be a fine predictor for patient survival: 1 year (AUC = 0.800), 3 years (AUC = 0.807) and 5 years OS (AUC = 0.786) (Figure 2B). These results were verified in CGGA dataset (Figures 2C,D).
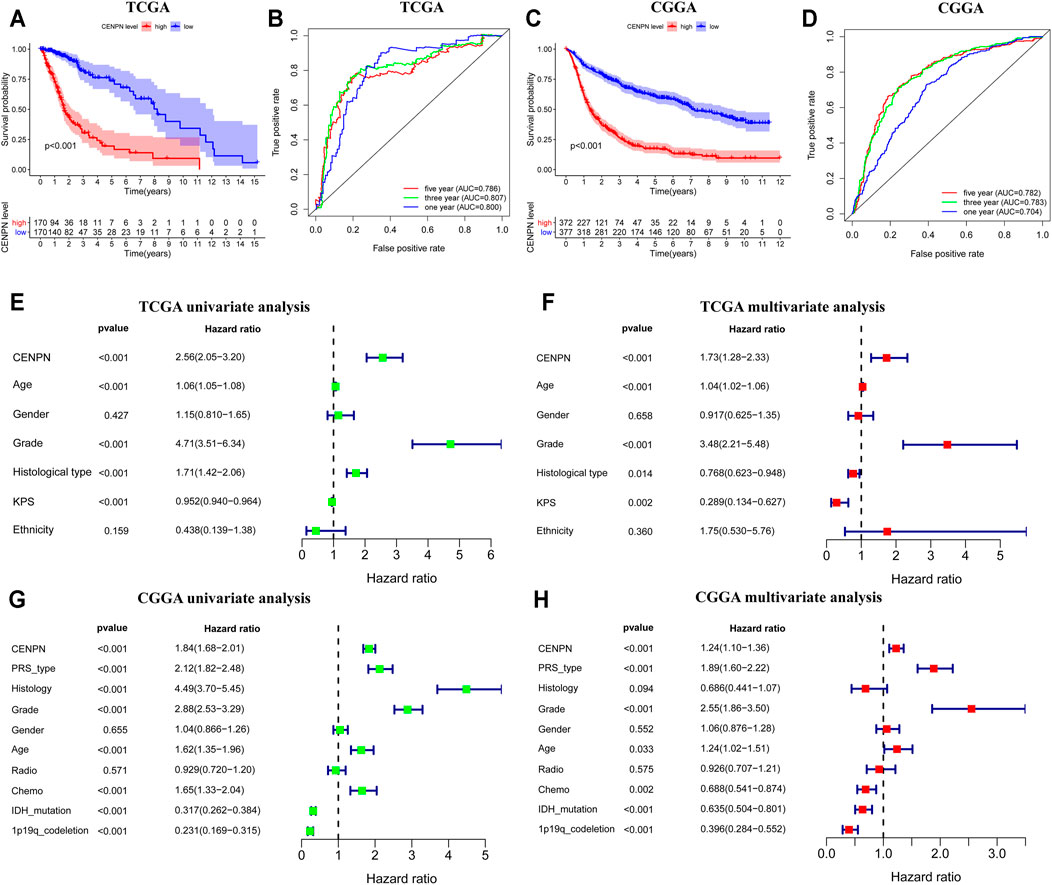
FIGURE 2. The prognostic values of CENPN in TCGA and CGGA databases. (A,C) Kaplan–Meier analysis of OS between groups with different CENPN expression; and (B,D) 1, 3- and 5 year ROC curves of OS. Univariate and multivariate Cox regression in TCGA (E,F) and CGGA (G,H). Relevant forest plots were plotted.
Furthermore, univariate and multivariate Cox regression analyses were employed to assess the independent prognostic roles of CENPN. In TCGA, univariate regression showed that patients with CENPN-high had a shorter overall survival time (hazard ratio (HR): 2.56, 95% CI [2.05–3.20], p < 0.001) (Figure 2E). The remaining clinical features associated with OS, such as age, grade and histology, are shown in Figure 2E. The multivariate regression model demonstrated that CENPN was independently correlated with patient OS (Figure 2F, HR = 1.73, 95% CI [1.28–2.33], p < 0.001). Moreover, these results were also confirmed in CGGA dataset (Figures 2G,H), indicating that CENPN could be an independent risk factor for glioma.
CENPN-Related Biological Processes and Immune Activities
The KEGG analysis based on the GSEA of genes in the high CENPN expression group in TCGA showed that some biological pathways were enriched, including the cell cycle, P53 signaling pathway and apoptosis. Moreover, some pathways related to immunity were also enriched, such as leukocyte transendothelial migration, Fc gamma r-mediated phagocytosis and antigen processing and presentation. The GO analysis indicated that immune-related biological processes, such as immunoglobulin production, leukocyte homeostasis and negative phagocytosis regulation, were enriched in the cohort with CENPN-high (Figure 3A). Similar results were also verified in the CGGA database (Figure 3B). GSEA results suggested that CENPN was associated with multiple molecular mechanisms of glioma development.
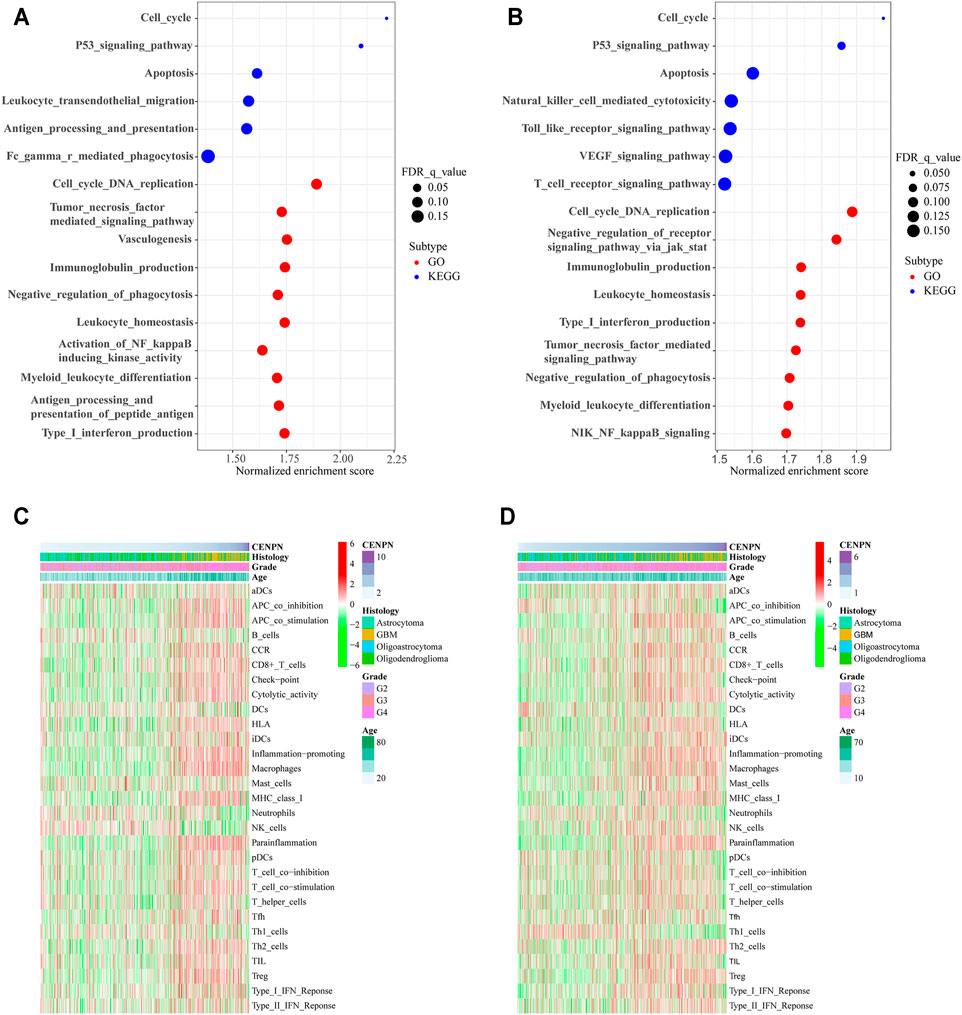
FIGURE 3. CENPN-related biological processes and immune activities. (A,B) GO and KEGG pathway analyses in TCGA and CGGA. A false discovery rate (FDR) < 0.25 was viewed significantly enriched. (C,D) Heatmaps based on ssGSEA exhibiting CENPN expression, clinical variables, and 29 immune-related gene sets.
To further explore the role of CENPN in the immune microenvironment, ssGSEA was applied to assess the enrichment scores of immune-related pathways, immune cells and immune-related functions grounded on the 29 gene sets derived from previous studies (Subramanian et al., 2005; Bindea et al., 2013; Zhou et al., 2020). In TCGA, the heatmap (Figure 3C) shows that some immune-related functions were positively correlated with CENPN expression. APC coinhibition, APC costimulation, chemokine receptor (CCR), cytolytic activity, immune checkpoint, inflammation promoting, human leukocyte antigen (HLA), parainflammation, major histocompatibility complex (MHC) class I, type I interferon (IFN) responses, T cell coinhibition and T cell costimulation were enriched in CENPN-high patients. Additionally, several immune cell types, such as CD8+ T cells, macrophages and regulatory T cells (Tregs), were infiltrated in glioma cases with CENPN-high, while neutrophils, dendritic cells (DCs) and natural killer cells (NK cells) showed opposite trends. Meanwhile, similar results were also obtained in CGGA database (Figure 3D). These results indicated that CENPN has a close relationship with immunomodulation of glioma.
Evaluation of Immune Infiltration
To systematically estimate the relationship between CENPN and tumor-infiltrating lymphocytes (TILs), the CIBERSORT algorithm method was employed to infer the fractions of 22 immune cell types in tumor samples based on gene expression profiles. Consistent with the ssGSEA results, CD8+ T cells, follicular helper T cells, Tregs, gamma delta T cells, M0, M1 and M2 macrophages in TCGA and follicular helper T cells and M0 macrophages in CGGA were enriched in the CENPN-high group. However, memory resting CD4+ T cells, NK cells activated, dendritic cells and mast cells, monocytes and eosinophils in TCGA and CD4 naïve T cells, monocytes, mast cells and NK cells activated in CGGA were enriched in the CENPN-low group (Figure 4). These findings indicated an important role of CENPN in the glioma immune microenvironment.
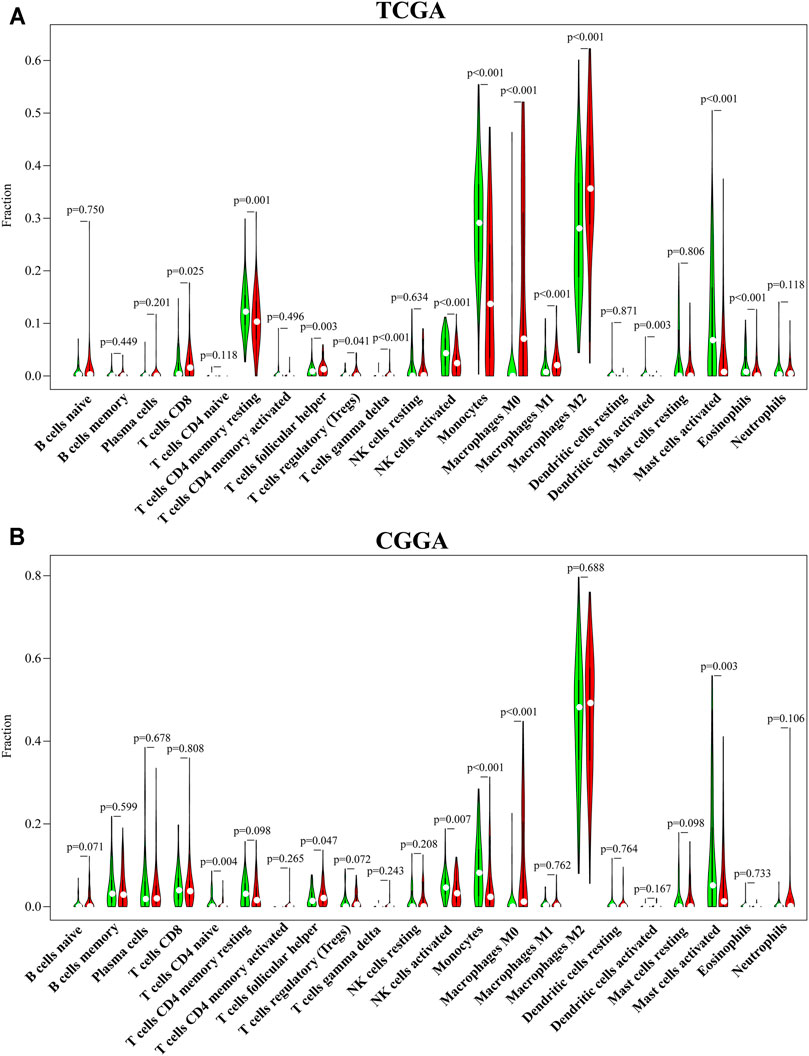
FIGURE 4. CENPN-related immune infiltration. (A,B) The fractions of 22 TILs infiltrating the glioma microenvironment in TCGA and CGGA based on the CIBERSORT algorithm.
Protein Expression Level of CENPN in Glioma Tissues
Our study further evaluated the protein level of CENPN in 112 glioma specimens by IHC. We found that CENPN was upregulated in 72.3% (81/112) of glioma tissues (Supplementary Table S1). Representative IHC images of samples with grades II, III and IV are displayed in Figure 5A. CENPN expression was positively correlated with some adverse outcome clinicopathological variables, such as age, WHO grade and histology (Figure 5B). However, no correlation was found between CENPN expression and 1p19q codeletion or P53 mutation.
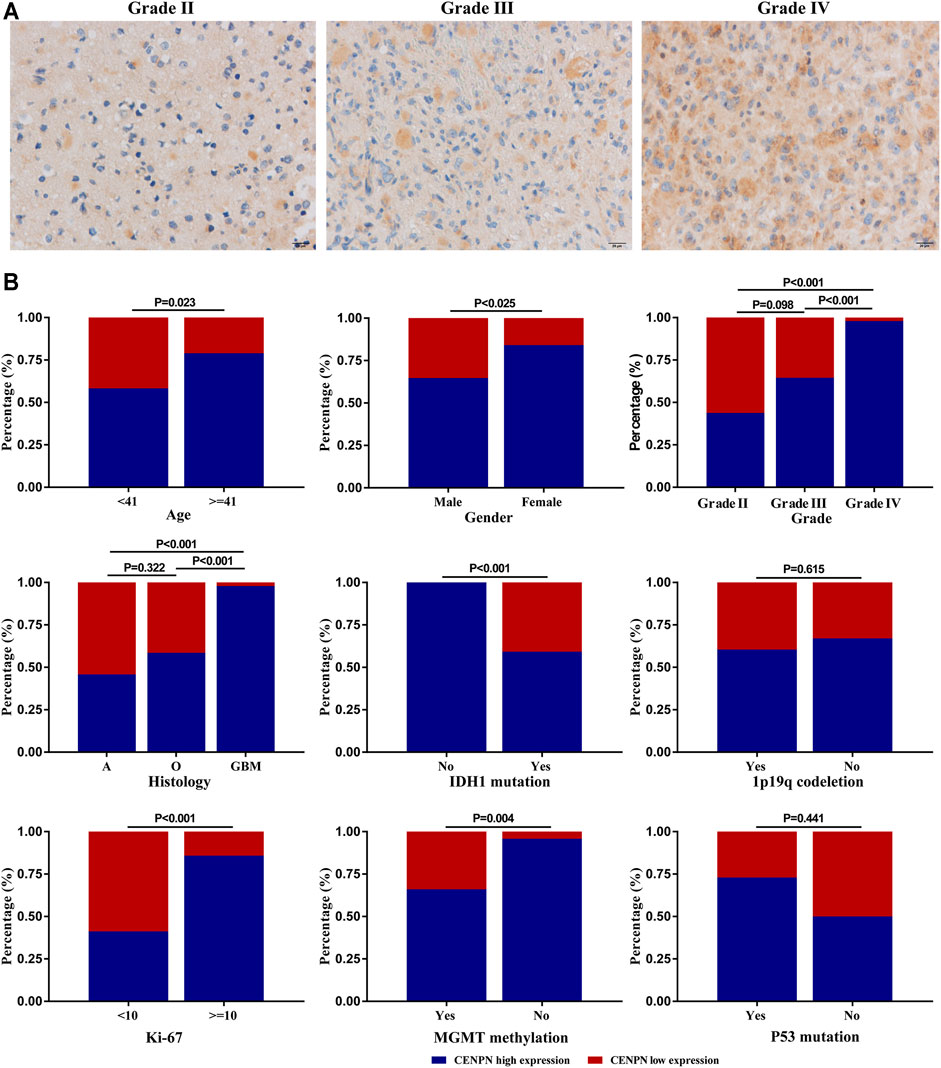
FIGURE 5. CENPN protein expression in glioma tissues from the Huanhu cohort. (A) Representative IHC images of grade Ⅱ, Ⅲ and IV samples. (B) Associations between CENPN expression and clinical characteristics in the Huanhu cohort.
Biological Functions of CENPN in Human Glioma Cells
To explore the functions of CENPN in glioma, LN229 and U251 cells were transfected with two siRNAs (siCENPN-i and siCENPN-ii) to silence CENPN transcription. The CENPN siRNAs resulted in significant declines in both mRNA and protein levels (Figures 6A,B). The CCK-8 assay showed that transfection with siCENPN significantly reduced the proliferation of the cells (Figure 6C). The colony formation assay displayed a similar result (Figure 6D). Furthermore, wound healing and Transwell assays showed that downregulation of CENPN suppressed the migration and invasion ability of glioma cells compared with that in the siScr group (Figures 6E,F). In addition, we found that higher apoptotic percentages were detected in the siCENPN groups than in the siScr groups (Figure 6G). However, the cell cycle assay showed that there were no differences in the fractions of cells in the G0/G1, G2/M, and S phases after CENPN suppression compared with those subjected to siScr (Supplementary Figure S1). These results suggest that CENPN could promote malignant phenotypes of glioma cells in vitro.
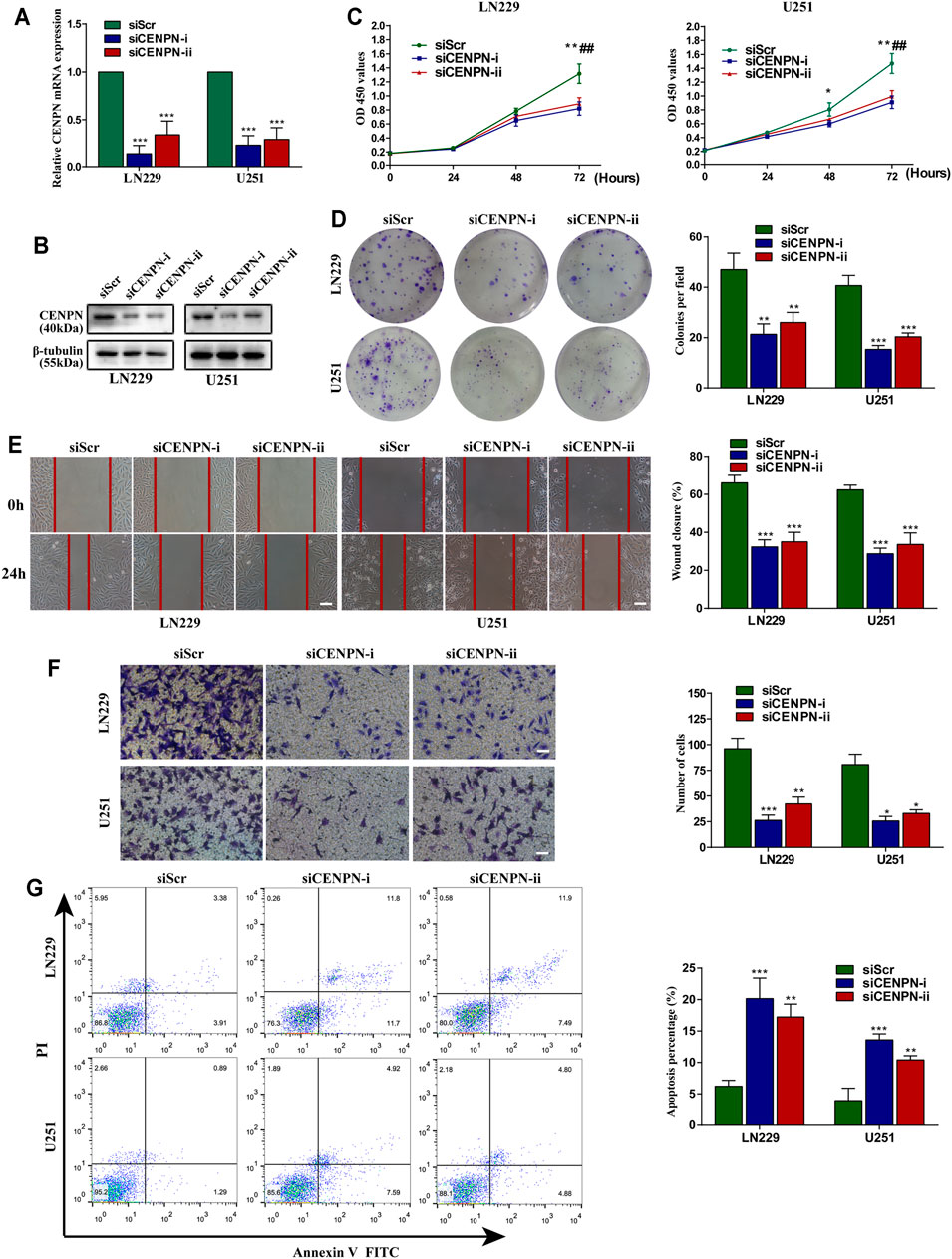
FIGURE 6. Effects of CENPN knockdown on glioma cells in vitro. (A,B) Expression of CENPN detected after transfection with siRNA by PCR and western blot. *p < 0.05, **p < 0.01, ***p < 0.001. (C) CCK-8 assay was used to estimate the effect of CENPN knockdown on cell proliferation. *p < 0.05, **p < 0.01, ***p < 0.001, siCENPN-i vs siScr group; #p < 0.05, ##p < 0.01, ###p < 0.001, siCENPN-ii vs siScr group. (D) Cell proliferation was estimated by colony formation assay. (E) Wound-healing assay was used to analyze the migration ability of LN229 and U251 cells. Scale bar, 400 μm. (F,G) Invasive ability and the degree of apoptosis were estimated by Transwell assay (F) and FITC annexin V (G), respectively. Data are the mean ± SD. *p < 0.05, **p < 0.01, ***p < 0.001, one-way ANOVA. Scale bar, 200 μm.
Discussion
The process of tumorigenesis is complicated and involves abnormalities in various genes and signaling pathways (Losurdo et al., 2018; Ronchi et al., 2019; Wang et al., 2019). The lack of timely and effective treatments may lead to the malignant transformation of low-grade glioma, aggravating the prognosis of patients (HanahanWeinberg 2000; FernaldKurokawa 2013; Li et al., 2017; Cai et al., 2018). Therefore, it is essential to clarify the specific mechanisms of glioma occurrence and progression for tumor early diagnosis and treatment.
Taking advantage of the substantial data gained from public databases, we performed comprehensive bioinformatic analyses for CENPN expression in glioma. The results showed that upregulated CENPN expression was correlated with advanced clinicopathological parameters (age, grade, histological type and KPS, etc.) and represented an independent prognostic factor of glioma. These results were validated in clinical samples obtained from our hospital. GSEA and ssGSEA revealed that some immune-related pathways and functions were differentially enriched in the group with CENPN-high. The CIBERSORT algorithm was further used to investigate the influence of CENPN on the infiltration of 22 TILs in the tumor microenvironment. In addition, in vitro cell experiments were conducted. Knockdown of CENPN impaired the proliferation, migration and invasion and increased the apoptosis of glioma cell lines. These results demonstrated that CENPN could be a promising target in glioma therapies.
The centromere is a chromosome site that contains chromatin and protein complexes that bind microtubules during mitosis that allows chromosomes to be equally separated (WesthorpeStraight 2014). During the mitosis process, many CENPs associate with and dissociate dynamically from centromeres (CheesemanDesai 2008). In humans, CENPA, also called the histone H3 variant, is always present in the functional centromere. Disruption or mutation of CENPA leads to a complete failure in centromere formation (Palmer et al., 1991; CheesemanDesai 2008). Nevertheless, the biological functions of CENPN are not well clarified. CENP-N interacts directly with the centromere-targeting domain of CENPA to facilitate centromere assembly (Tian et al., 2018), and the depletion of CENPN can induce the loss of many other CENPs (Foltz et al., 2006; Carroll et al., 2009). Therefore, CENPN seems to be essential in centromere assembly, similar to CENPA. However, studies have reported that CENPN is not indispensable in sustaining existing centromeres and recruiting kinetochore proteins because CENPN protein levels decrease before mitosis (McClelland et al., 2007; Hellwig et al., 2011). In cancers, Wang et al. (2021) and Oka et al. (2019) reported that CENPN knockdown could arrest cell cycle at the G1 phase in hepatocellular carcinoma and oral squamous cell carcinoma, respectively. However, in this study, significant changes were not observed in the distribution of cell cycle phases in glioma cells subjected to CENPN knockdown. CENPN may not promote glioma cell progression by influencing the cell cycle; therefore, additional cell experiments are needed to clarify its specific functions in glioma.
In the past decade, immunotherapies have gained great success in multiple tumor treatments (Newick et al., 2017; Ronchi et al., 2020). However, the therapeutic effects of immunotherapies in glioma are less than satisfactory (Lim et al., 2018). As previously reported, the immune infiltration profiles of the tumor microenvironment can directly reflect the immune status and predict the outcomes of cancers (Bindea et al., 2013; Cheng et al., 2016). In our study, patients with different CENPN expression levels had differences in immune infiltration. Based on the CIBERSORT algorithm, five immune cells had the same infiltration trends in both the TCGA and CGGA cohorts. Among these immune cells, the proportions of activated NK cells and monocytes were lower in the CENPN-high group than in the CENPN-low group (Figure 4). Activated NK cells are powerful innate immune cells that can help enhance antitumor immunity by releasing perforin and granzyme and secreting various cytokines (Andre et al., 2018). Monocytes have the property of transforming into macrophages and can function as antigen-presenting cells to strengthen adaptive immunity (Jakubzick et al., 2017). Given these findings, CENPN upregulation might promote glioma progression by suppressing the innate immune system. Glioma cells can release multiple immunosuppressive cytokines, such as TGF-β and IL-10, to generate a “cold” tumor microenvironment (Gieryng et al., 2017). Thus, strategies that can reverse the protumor infiltration of immune cells are urgently needed.
Our work presented certain limitations that should be noted. First, the sample size of controls cases in both public databases and clinical cohorts from our institution was small; therefore, more studies with sufficient normal samples must be conducted in the future. Second, CENPN knockdown did not affect the cell cycle distribution, which was inconsistent with previous reports (Oka et al., 2019; Wang et al., 2021). Thus, more glioma cell lines should be adopted for cell biological function assays. At last, functional enrichment analyses revealed that some immune-related functions, immune cells and immune-related pathways were correlated with CENPN expression. The mechanisms by which CENPN may immunomodulate the microenvironment of glioma remain to be understood.
Conclusion
This study showed that upregulated CENPN expression was associated with adverse clinical outcomes of glioma patients. CENPN is also associated with multiple immune processes and immune cell infiltration. In addition, in vitro experiments showed that CENPN deficiency impaired the proliferation, migration and invasion and increased the apoptosis of glioma cell lines. All in all, these findings indicate that CENPN might be a valid therapeutic target for glioma.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Ethics Statement
The studies involving human participants were reviewed and approved by Ethics Committee of Huanhu Hospital of Tianjin Medical University. The patients/participants provided their written informed consent to participate in this study.
Author Contributions
Study design and data collection: HailW, YZ, and HaiyW. Data analysis and interpretation: HaiyW, LX, YY. Writing and review of the manuscript: YZ, HY, HaiyW, and XT
Funding
This study was funded by National Natural Science Foundation of China (No. 81972349) and Tianjin Municipal Science and Technology Commission (No. 20JCQNJC00410).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We gratefully acknowledge contributions from Departments of Neurosurgery and Pathology of Huanhu hospital, and TCGA and CGGA databases. We thank American Journal Experts (AJE) for language editing.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2021.732376/full#supplementary-material
References
André, P., Denis, C., Soulas, C., Bourbon-Caillet, C., Lopez, J., Arnoux, T., et al. (2018). Anti-NKG2A mAb Is a Checkpoint Inhibitor that Promotes Anti-tumor Immunity by Unleashing Both T and NK Cells. Cell 175 (7), 1731–1743. e13. doi:10.1016/j.cell.2018.10.014
Andres, S. A., Bickett, K. E., Alatoum, M. A., Kalbfleisch, T. S., Brock, G. N., and Wittliff, J. L. (2015). Interaction between Smoking History and Gene Expression Levels Impacts Survival of Breast Cancer Patients. Breast Cancer Res. Treat. 152 (3), 545–556. doi:10.1007/s10549-015-3507-z
Bindea, G., Mlecnik, B., Tosolini, M., Kirilovsky, A., Waldner, M., Obenauf, A. C., et al. (2013). Spatiotemporal Dynamics of Intratumoral Immune Cells Reveal the Immune Landscape in Human Cancer. Immunity 39 (4), 782–795. doi:10.1016/j.immuni.2013.10.003
Cai, J., Chen, Q., Cui, Y., Dong, J., Chen, M., Wu, P., et al. (2018). Immune Heterogeneity and Clinicopathologic Characterization of IGFBP2 in 2447 Glioma Samples. Oncoimmunology 7 (5), e1426516. doi:10.1080/2162402X.2018.1426516
Carroll, C. W., Silva, M. C. C., Godek, K. M., Jansen, L. E. T., and Straight, A. F. (2009). Centromere Assembly Requires the Direct Recognition of CENP-A Nucleosomes by CENP-N. Nat. Cel Biol 11 (7), 896–902. doi:10.1038/ncb1899
Cheeseman, I. M., and Desai, A. (2008). Molecular Architecture of the Kinetochore-Microtubule Interface. Nat. Rev. Mol. Cel Biol 9 (1), 33–46. doi:10.1038/nrm2310
Cheng, W., Ren, X., Zhang, C., Cai, J., Liu, Y., Han, S., et al. (2016). Bioinformatic Profiling Identifies an Immune-Related Risk Signature for Glioblastoma. Neurology 86 (24), 2226–2234. doi:10.1212/WNL.0000000000002770
Fang, J., Liu, Y., Wei, Y., Deng, W., Yu, Z., Huang, L., et al. (2015). Structural Transitions of Centromeric Chromatin Regulate the Cell Cycle-dependent Recruitment of CENP-N. Genes Dev. 29 (10), 1058–1073. doi:10.1101/gad.259432.115
Fernald, K., and Kurokawa, M. (2013). Evading Apoptosis in Cancer. Trends Cel Biol. 23 (12), 620–633. doi:10.1016/j.tcb.2013.07.006
Foltz, D. R., Jansen, L. E. T., Black, B. E., Bailey, A. O., Cleveland, D. W., and Cleveland, J. R. (2006). The Human CENP-A Centromeric Nucleosome-Associated Complex. Nat. Cel Biol 8 (5), 458–469. doi:10.1038/ncb1397
Gieryng, A., Pszczolkowska, D., Walentynowicz, K. A., Rajan, W. D., and Kaminska, B. (2017). Immune Microenvironment of Gliomas. Lab. Invest. 97 (5), 498–518. doi:10.1038/labinvest.2017.19
Gravendeel, L. A. M., Kouwenhoven, M. C. M., Gevaert, O., de Rooi, J. J., Stubbs, A. P., Duijm, J. E., et al. (2009). Intrinsic Gene Expression Profiles of Gliomas Are a Better Predictor of Survival Than Histology. Cancer Res. 69 (23), 9065–9072. doi:10.1158/0008-5472.CAN-09-2307
Hanahan, D., and Weinberg, R. A. (2000). The Hallmarks of Cancer. Cell 100 (1), 57–70. doi:10.1016/s0092-8674(00)81683-9
Hellwig, D., Emmerth, S., Ulbricht, T., Döring, V., Hoischen, C., Martin, R., et al. (2011). Dynamics of CENP-N Kinetochore Binding during the Cell Cycle. J. Cel Sci 124 (Pt 22), 3871–3883. doi:10.1242/jcs.088625
Hoischen, C., Yavas, S., Wohland, T., and Diekmann, S. (2018). CENP-C/H/I/K/M/T/W/N/L and hMis12 but Not CENP-S/X Participate in Complex Formation in the Nucleoplasm of Living Human Interphase Cells outside centromeresL and hMis12 but Not CENP-S/X Participate in Complex Formation in the Nucleoplasm of Living Human Interphase Cells outside Centromeres. PLoS One 13 (3), e0192572. doi:10.1371/journal.pone.0192572
Jakubzick, C. V., Randolph, G. J., and Henson, P. M. (2017). Monocyte Differentiation and Antigen-Presenting Functions. Nat. Rev. Immunol. 17 (6), 349–362. doi:10.1038/nri.2017.28
Kops, G. J. P. L., Weaver, B. A. A., and Cleveland, D. W. (2005). On the Road to Cancer: Aneuploidy and the Mitotic Checkpoint. Nat. Rev. Cancer 5 (10), 773–785. doi:10.1038/nrc1714
Kristensen, B. W., Priesterbach-Ackley, L. P., Petersen, J. K., and Wesseling, P. (2019). Molecular Pathology of Tumors of the central Nervous System. Ann. Oncol. 30 (8), 1265–1278. doi:10.1093/annonc/mdz164
Li, G., Wang, Z., Zhang, C., Liu, X., Cai, J., Wang, Z., et al. (2017). Molecular and Clinical Characterization of TIM-3 in Glioma through 1,024 Samples. Oncoimmunology 6 (8), e1328339. doi:10.1080/2162402X.2017.1328339
Lim, M., Xia, Y., Bettegowda, C., and Weller, M. (2018). Current State of Immunotherapy for Glioblastoma. Nat. Rev. Clin. Oncol. 15 (7), 422–442. doi:10.1038/s41571-018-0003-5
Losurdo, G., Principi, M., Girardi, B., Pricci, M., Barone, M., Ierardi, E., et al. (2018). Histamine and Histaminergic Receptors in Colorectal Cancer: From Basic Science to Evidence-Based Medicine. Acamc 18 (1), 15–20. doi:10.2174/1871520616666160321115349
McClelland, S. E., Borusu, S., Amaro, A. C., Winter, J. R., Belwal, M., McAinsh, A. D., et al. (2007). The CENP-A NAC/CAD Kinetochore Complex Controls Chromosome Congression and Spindle Bipolarity. EMBO J. 26 (24), 5033–5047. doi:10.1038/sj.emboj.7601927
McKinley, K. L., and Cheeseman, I. M. (2016). The Molecular Basis for Centromere Identity and Function. Nat. Rev. Mol. Cel Biol 17 (1), 16–29. doi:10.1038/nrm.2015.5
Newick, K., O'Brien, S., Moon, E., and Albelda, S. M. (2017). CAR T Cell Therapy for Solid Tumors. Annu. Rev. Med. 68, 139–152. doi:10.1146/annurev-med-062315-120245
Newman, A. M., Liu, C. L., Green, M. R., Gentles, A. J., Feng, W., Xu, Y., et al. (2015). Robust Enumeration of Cell Subsets from Tissue Expression Profiles. Nat. Methods 12 (5), 453–457. doi:10.1038/nmeth.3337
Oka, N., Kasamatsu, A., Endo-Sakamoto, Y., Eizuka, K., Wagai, S., Koide-Ishida, N., et al. (2019). Centromere Protein N Participates in Cellular Proliferation of Human Oral Cancer by Cell-Cycle Enhancement. J. Cancer 10 (16), 3728–3734. doi:10.7150/jca.32281
Omuro, A., and DeAngelis, L. M. (2013). Glioblastoma and Other Malignant Gliomas. JAMA 310 (17), 1842–1850. doi:10.1001/jama.2013.280319
Ostrom, Q. T., Gittleman, H., Truitt, G., Boscia, A., Kruchko, C., and Barnholtz-Sloan, J. S. (2018). CBTRUS Statistical Report: Primary Brain and Other Central Nervous System Tumors Diagnosed in the United States in 2011-2015, Neuro Oncol, 20, Suppl. l_4, iv1–iv86. doi:10.1093/neuonc/noy131
Palmer, D. K., O'Day, K., Trong, H. L., Charbonneau, H., and Margolis, R. L. (1991). Purification of the Centromere-specific Protein CENP-A and Demonstration that it Is a Distinctive Histone. Proc. Natl. Acad. Sci. 88 (9), 3734–3738. doi:10.1073/pnas.88.9.3734
Rahman, M. R., Islam, T., Gov, E., Turanli, B., Gulfidan, G., Shahjaman, M., et al. (2019). Identification of Prognostic Biomarker Signatures and Candidate Drugs in Colorectal Cancer: Insights from Systems Biology Analysis. Medicina 55 (1), 20. doi:10.3390/medicina55010020
Ronchi, A., Montella, M., Cozzolino, I., Argenziano, G., Moscarella, E., Piccolo, V., et al. (2020). The Potential Diagnostic and Predictive Role of Anaplastic Lymphoma Kinase (ALK) Gene Alterations in Melanocytic Tumors. Eur. Rev. Med. Pharmacol. Sci. 24 (7), 3829–3838. doi:10.26355/eurrev_202004_20849
Ronchi, A., Cozzolino, I., Montella, M., Panarese, I., Zito Marino, F., Rossetti, S., et al. (2019). Extragonadal Germ Cell Tumors: Not Just a Matter of Location. A Review about Clinical, Molecular and Pathological Features. Cancer Med. 8 (16), 6832–6840. doi:10.1002/cam4.2195
Subramanian, A., Tamayo, P., Mootha, V. K., Mukherjee, S., Ebert, B. L., Gillette, M. A., et al. (2005). Gene Set Enrichment Analysis: a Knowledge-Based Approach for Interpreting Genome-wide Expression Profiles. Proc. Natl. Acad. Sci. 102 (43), 15545–15550. doi:10.1073/pnas.0506580102
Tang, Z., Kang, B., Li, C., Chen, T., and Zhang, Z. (2019). GEPIA2: an Enhanced Web Server for Large-Scale Expression Profiling and Interactive Analysis. Nucleic Acids Res. 47 (W1), W556–W560. doi:10.1093/nar/gkz430
Tian, T., Li, X., Liu, Y., Wang, C., Liu, X., Bi, G., et al. (2018). Molecular Basis for CENP-N Recognition of CENP-A Nucleosome on the Human Kinetochore. Cell Res 28 (3), 374–378. doi:10.1038/cr.2018.13
Wang, Q., Yu, X., Zheng, Z., Chen, F., Yang, N., and Zhou, Y. (2021). Centromere Protein N May Be a Novel Malignant Prognostic Biomarker for Hepatocellular Carcinoma. PeerJ 9, e11342. doi:10.7717/peerj.11342
Wang, Y., Tang, C., Yao, S., Lai, H., Li, R., Xu, J., et al. (2019). Discovery of a Novel Protein Kinase C Activator from Croton Tiglium for Inhibition of Non-small Cell Lung Cancer. Phytomedicine 65, 153100. doi:10.1016/j.phymed.2019.153100
Wang, Y., Tang, S., Lai, H., Jin, R., Long, X., Li, N., et al. (2020). Discovery of Novel IDH1 Inhibitor through Comparative Structure-Based Virtual Screening. Front. Pharmacol. 11, 579768. doi:10.3389/fphar.2020.579768
Wang, Z., Jensen, M. A., and Zenklusen, J. C. (2016). A Practical Guide to the Cancer Genome Atlas (TCGA). Methods Mol. Biol. 1418, 111–141. doi:10.1007/978-1-4939-3578-9_6
Westhorpe, F. G., and Straight, A. F. (2014). The Centromere: Epigenetic Control of Chromosome Segregation during Mitosis. Cold Spring Harb Perspect. Biol. 7 (1), a015818. doi:10.1101/cshperspect.a015818
Xiao, Y., Yuan, Y., Zhang, Y., Li, J., Liu, Z., Zhang, X., et al. (2015). CMTM5 Is Reduced in Prostate Cancer and Inhibits Cancer Cell Growth In Vitro and In Vivo. Clin. Transl Oncol. 17 (6), 431–437. doi:10.1007/s12094-014-1253-z
Zhao, Z., Zhang, K.-N., Wang, Q., Li, G., Zeng, F., Zhang, Y., et al. (2021). Chinese Glioma Genome Atlas (CGGA): A Comprehensive Resource with Functional Genomic Data from Chinese Glioma Patients. Genomics, Proteomics & Bioinformatics. doi:10.1016/j.gpb.2020.10.005
Zhou, X., Qiu, S., Nie, L., Jin, D., Jin, K., Zheng, X., et al. (2020). Classification of Muscle-Invasive Bladder Cancer Based on Immunogenomic Profiling. Front. Oncol. 10, 1429. doi:10.3389/fonc.2020.01429
Keywords: CENPN, biomarker, bioinformatics analysis, glioma, immune infiltration
Citation: Wu H, Zhou Y, Wu H, Xu L, Yan Y, Tong X and Yan H (2021) CENPN Acts as a Novel Biomarker that Correlates With the Malignant Phenotypes of Glioma Cells. Front. Genet. 12:732376. doi: 10.3389/fgene.2021.732376
Received: 29 June 2021; Accepted: 15 September 2021;
Published: 27 September 2021.
Edited by:
Jian-Bing Fan, Illumina, United StatesReviewed by:
Lucia Taja-Chayeb, National Institute of Cancerology (INCAN), MexicoGuojun Zhang, Capital Medical University, China
Jinquan Cai, Harbin Medical University, China
Copyright © 2021 Wu, Zhou, Wu, Xu, Yan, Tong and Yan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hua Yan, eWFuaHVhMjAwNDIwMDdAc2luYS5jb20=; Xiaoguang Tong, dG9uZ3hnQHlhaG9vLmNvbQ==
†These authors have contributed equally to this work
 Hailong Wu
Hailong Wu Yan Zhou1†
Yan Zhou1† Haiyang Wu
Haiyang Wu Lixia Xu
Lixia Xu Hua Yan
Hua Yan