- State Key Laboratory of Ecological Pest Control for Fujian and Taiwan Crops, Fujian Provincial Key Laboratory of Haixia Applied Plant Systems Biology, College of Life Sciences, College of Plant Protection, College of Horticulture, Fujian Agriculture and Forestry University, Fuzhou, China
SBT (Subtilisin-like serine protease), a clan of serine proteolytic enzymes, plays a versatile role in plant growth and defense. Although SBT family genes have been obtained from studies of dicots such as Arabidopsis, little is known about the potential functions of SBT in the monocots. In this study, 54 pineapple SBT genes (AcoSBTs) were divided into six subfamilies and then identified to be experienced strong purifying selective pressure and distributed on 25 chromosomes unevenly. Cis-acting element analysis indicated that almost all AcoSBTs promoters contain light-responsive elements. Further, the expression pattern via RNA-seq data showed that different AcoSBTs were preferentially expressed in different above-ground tissues. Transient expression in tobacco showed that AcoSBT1.12 was located in the plasma membrane. Moreover, Transgenic Arabidopsis ectopically overexpressing AcoSBT1.12 exhibited delayed flowering time. In addition, under the guidance of bioinformatic prediction, we found that AcoSBT1.12 could interact with AcoCWF19L, AcoPUF2, AcoCwfJL, Aco012905, and AcoSZF1 by yeast-two hybrid (Y2H). In summary, this study provided valuable information on pineapple SBT genes and illuminated the biological function of AcoSBT1.12 in floral transition.
Introduction
The normal function of plant cells is guaranteed by the precise regulation of protein level, which depends on the balance between protein synthesis and degradation (Schaller, 2004). Protein hydrolysis mediates protein degradation, not only enables amino acid recycling but also assists with post-translational modification (Have et al., 2018). Extensive proteolysis and site-specific limited proteolysis are the major mechanisms in protein hydrolysis. And the limited proteolysis, taking concerted action with other selective mechanisms, is essential for many plant biological processes such as subcellular trafficking, peptide hormone-regulated response, and immune response (Paulus et al., 2020).
Thus, evolutionarily, in order to ensure precise protein levels, a considerable number of limited proteases were widely distributed in plant cells. Among them, SBT (Subtilisin-like serine protease), a clan of serine proteolytic enzymes, were reported in numerous plants for their participation in the diverse cellular process such as protein activation. Structurally, SBTs own a highly conserved domain, named Peptidase_S8 domain (PF00082), which consists of a catalytic triad with the specific arrangement of three amino acid residues (Asp, His, and Ser) (Reichardt et al., 2018). This unique domain is served as the site of specific substrate binding (Schaller et al., 2018). Moreover, the protease-associated (PA) domain (PF02225) and the Inhibitor_I9 domain (PF05922) were also found in SBTs in plants. The conserved domains of SBTs are closely related to their versatile function evolved in plants.
Studies in model species have revealed that SBT genes exist throughout the plant kingdom and play versatile roles in plant growth and defense. SBT family has been well studied in Arabidopsis (Arabidopsis thaliana), and 56 SBTs were characterized. Among them, AtSBT1.2 was found to be specifically expressed in stomatal precursor cells and function in stomata density regulation (Zhao et al., 2020). In AtSBT2.4 deletion mutant ale1, the disrupted expression of AtSBT2.4 leads to defects in leaf shape formation and embryo development (Creff et al., 2019). Moreover, it’s also reported that AIR3 (AtSBT5.3) facilitates lateral root formation (Neuteboom et al., 1999), ARA12 (AtSBT1.7) involves in seed coat development (Rautengarten et al., 2008), and XSP1(AtSBT4.14) regulates xylem differentiation (Zhao et al., 2000). Not only their functions in plant-specific developmental processes but also their involvement in Arabidopsis responses to environmental stress were reported. AtSBT6.1, encoding the protein necessary for bZIP17 activity in the endoplasmic reticulum stress signaling pathway, was observed to initiate salt stress response in plants (Liu et al., 2007). Additionally, loss-of-function of AtSBT3.3 in deletion mutants compromises innate immune, while overexpression of AtSBT3.3 enhances the plant pathogen-resistant (Ramírez et al., 2013). In soybean, C1SBT encoded protein exhibits stringent substrate specificity and induces the decomposition of soybean seed storage protein, whereas SCS1 (Seed Coat Subtilisin 1) is preferentially expressed in soybean seed and involved in the remodeling of cell wall structure during seed coat development (Schaller et al., 2012). Also, in tomato, P69 identified as an SBT, induced by citrus exocytic viroid infection in tomato leaves, was found to behave as a pathogenesis-related protein which is important for plant-pathogen interactions (Schaller et al., 2018). More recently, the basal resistance protease tomato RCR3 is found to be activated by P69B (Paulus et al., 2020). Besides, 3 SBT genes (SlPhyts3, SlPhyts4, and SlPhyts5) in tomato were confirmed to trigger non-autolytic cell death under oxidative stress (Reichardt et al., 2018).
Pineapple (Ananas comosus L.) belongs to the family of Bromeliaceae, and it is one of the most important economic crops distributed across tropical and subtropical regions worldwide (Ming et al., 2015). In agriculture production, the flowering time of pineapples, which determines both the maximum reproductive success and the productivity of farmers’ labor, is very random and inconsistent (Bao et al., 2020; Jin et al., 2020). Although elucidation of mechanisms underlying pineapple flowering regulation will help maximize the food yield in the pineapple industry, few components regulating flowering time have been identified in pineapple. Our previous work adopted the systematic bioinformatics method WGCNA to analyze the spatio-temporal transcriptome data of pineapple floral organs and identified a series of pineapple SBT genes (AcoSBTs) located in flowering specific clusters (Wang et al., 2020). Moreover, this gene family is also reported in numerous cash crops for its versatile functions (Schaller et al., 2018; Paulus et al., 2020). Thus, it’ll be meaningful to carry out the fundamental study of the SBT gene family in pineapple. Recently, the assembled pineapple genome also provides an opportunity to genome-widely reveal the organization, evolution, and function of AcoSBTs (Ming et al., 2016).
In this study, a total of 54 AcoSBT genes were identified and classified into 6 subgroups based on their phylogenetic relationships. The results showed that pineapple AcoSBTs are located on 25 different chromosomes unevenly. We further investigated the exon-intron organization, motif structure, gene duplications, and expression profiles of AcoSBTs. Based on those bioinformatic analyses, we identified and functionally characterized the AcoSBT1.12 in pineapple. Briefly, AcoSBT1.12 interacts with five candidate substrate proteins and negatively regulates floral transition. In summary, our study provided fundamental information on AcoSBTs and further determined the crucial role of AcoSBT1.12 in controlling flowering time.
Materials and Methods
Identification and Characterization of SBTs in Pineapple
The Hidden Markov Model (HMM) of the Peptidase_S8 domain (PF00082) was obtained from the Pfam database1 as a query to search against pineapple genome database downloaded from Phytozome2 (Arango Argoty et al., 2012). After that, we further examined each of the selected candidates for having this conserved structure domain using NCBI-CDD3 (Marchler-Bauer et al., 2011). ExPAsy4 was then applied to calculate the molecular weights (MW) and isoelectric points (PI) of the AcoSBTs (Wilkins et al., 1999). Lastly, TargetP5 and SignalP6 were used for subcellular localization and signal peptide prediction, respectively (Emanuelsson et al., 2007).
Sequence Alignment and Phylogenetic Analysis
All the identified pineapple SBT protein sequences were multiply aligned with Arabidopsis SBT sequences collected from TAIR7 using Clustal Omega with default parameters. Phylogenetic analysis was conducted by MEGAX with Neighbor-joining statistical method setting default parameter except for the bootstrap replications n = 1,000. A total of 54 AcoSBTs were classified into 6 different groups according to the AtSBTs’ group scheme. The evolutionary tree was visualized by the online tool iTOL8.
Gene Structure, Motif Analysis, and Cis-Acting Elements Identification
The gene structure characteristics and exon-intron organizations of the AcoSBTs were exhibited using the TBtools program based on the comparison among the full-length genome sequences and the protein-coding sequences of the given genes (Chen et al., 2020). The web-based motif identification sever MEME (Mutiple Em for Motif Elicitation9) was used to detect potential motifs with following parameters: motif width < 50, motifs < 20, and e-value < e−5 (Granziol et al., 2019). Promoter regions, the 2,000 bp upstream regions of AcoSBT genes, were used for cis-element identification on web-based Cis-acting elements identification server PlantCARE10 (Lescot et al., 2002). The program Adobe Illustrator was adopted for result visualization.
Chromosome Distribution and Evolutionary Analyses
The location information of all identified pineapple SBTs, including start positions, located chromosomes, and chromosome lengths, were collected from Phytozome for further visualization. BLASTP with the E-value < e−5 was used to search the potential anchors between pineapple and Arabidopsis, and the top 5 matches were identified. The syntenic blocks of SBTs from both pineapple and Arabidopsis were visualized by Circos (Krzywinski et al., 2009). Both Ks and Ka values were calculated with synonymous and non-synonymous substitutions options using Dnasp (Librado and Rozas, 2009).
Plant Materials and Growth Conditions
The typical cultivated Pineapple (Ananas comosus) variety, MD2, was planted in plastic pots with long-day conditions: 30°C, 70% humidity, and 16 h light/8 h dark photocycle. MD2 pineapple organs (stamen, ovule, sepal, petal, and gynoecium) at different developmental stages were collected separately.
The Col-0 Arabidopsis was used to generate AcoSBT1.12 transgenic lines. The seeds of wildtype and transgenetic lines were germinated on 1/2 Murashige and Skoog medium after surface-sterilized with 75% ethanol, followed by vernalization in a 4°C incubator for 48 h. Plates were then transferred in the greenhouse with controlled conditions as following: 28°C, 60% humidity, and 16 h light/8 h dark photoperiod. After 2 weeks, the healthy seedlings grown on the plates were selected and planted in plastic pots with soil under 16 h light/8 h dark photocycle for further experiments.
RNA Extraction, RNA-Seq, and qRT-PCR
The developing samples of pineapple MD2 organs (sepal, petal, stamen, and ovule) were collected according to a previous paper (Wang et al., 2020). After sampling, three independent samples were collected and treated with liquid nitrogen rapidly, and then RNA extraction was performed immediately using the Trizol extraction kit (Omega Bio Tek, China) following the manufacturer’s guidance. The quality of RNA was confirmed by both agarose gel electrophoresis and spectrophotometer (NanoDrop, United States). The transcriptome data were downloaded from the NCBI database11 and the European Nucleotide Archive (accession number: PRJEB38680). The collected FPKM values (Fragments per kilobase of exon model per million mapped reads) were log2 transformed and visualized using the R package and genesis, respectively. qRT-PCR assays were carried out on the Bio-Rad Real-time PCR system (Foster, United States) using SYBR Premix Ex TaqII (Takara, Dalian, China), and the program was: 95°C for 30 s, 40 cycles of 95°C for 5 s, and 60°C for 30 s, followed by the last 95°C for 15 s. In each case, three technical replicates and three biological repeats were carried out. AcoPP2A (F: TTGTCATCGCTTCCTCCAAG; R: GTGTTGTCCACCACAGTATGA) and AtACTIN (F: TGCCAA TCTACGAGGGTTTC; R: TCTCTTACAATTTCCCGCTCTG) were selected as reference genes for pineapple samples and Arabidopsis samples, respectively.
Vector Construction and Subcellular Localization
The 35S: AcoSBT1.12 vector was constructed as the following procedure: amplifying the full-length of AcoSBT1.12 CDS sequence without termination codon and then cloning the amplified fragment into the Penter/D-TOPO vector, followed by recombining the positive clone into the destination vector pGWB605 harboring 35s promoter by LR reaction. The resulting vector was transformed into the Agrobacterium tumefaciens (GV3103) by electroporation and further transformed into Arabidopsis (Col-0) through the floral dip method. The multiple independent T1 plants confirmed by qRT-PCR were analyzed in further experiments. Above mentioned vector was introduced into Nicotiana bethamiana leaf epidermal cells by agroinfiltration. After 3 days growing at 28°C under 16 h light/8 h dark conditions, the leaves were subjected to the observation of the subcellular location of AcoSBT1.12-GFP under a Zeiss LSM750 confocal laser-scanning microscope.
Yeast Two-Hybrid Assay
The coding sequence of AcoSBT1.12 was cloned into pGBDT7 vector while the coding sequences of AcoCWF19L, AcoPUF2, AcoCwfJL, Aco012905, AcoPM1P, Aco009239, AcoSZF1, and AcoCZF1 were cloned into pGADT7 vector, respectively. Then the AD and BD constructs were co-transformed into AH109 yeast strain. Firstly, the co-transformed yeasts were seeded on the synthetic dropout medium without Leu and Trp (SD/- Leu/- Trp), and then transferred to the synthetic dropout medium without Leu, Trp and adenine (SD/- Leu/- Trp/- His) for 4 days before observation.
Statistical Analysis
Experiments were carried out with at least three biological repeats and three technical repeats, and Y2H was carried out with three technical repeats. The difference significances were statistically analyzed using the Student’s t-test. Asterisks denote significant differences between two groups of data (∗P < 0.05; ∗∗P < 0.01; ∗∗∗P < 0.001).
Results
Identification and Characterization of SBTs in Pineapple
To genome-widely identify SBTs in pineapple, the Hidden Markov Model (HMM) tool was used to search all the possible SBTs against the pineapple genome database with Peptidase_S8 domain (PF00082) as the query. After that, NCBI-CDD was employed to ensure that all the identified sequences contain the target domain (Marchler-Bauer et al., 2011). 54 AcoSBTs were identified in the pineapple genome and named according to their phylogenetic relationships with Arabidopsis SBT proteins. The length of 54 pineapple SBT proteins varied from 228 (AcoSBT5.9) to 2,759 aa (AcoSBT2.5), with the corresponding molecular weight ranged from 23.66 to 297.43 Kd. The isoelectric points (PI) of the AcoSBT varied from 5.29 (AcoSBT1.11) to 9.77 (AcoSBT1.15). Among them, 32 SBTs contain the plant-unique domain, PA domain, which is reported to determine the specificities of protein-protein interactions. In comparison, 40 SBTs possesses an additional Peptidase inhibitor I9 domain, which is characterized to function in the activation of the pro-enzyme, indicating the functional diversification of AcoSBTs (Arango Argoty et al., 2012). The detailed information about AcoSBTs, including transcript ID, protein size, chromosome location, and protein isoelectric point, were listed in Supplementary Table 1.
Phylogenetic Analysis of Pineapple SBTs
To investigate the evolutionary relationship of AcoSBTs, we constructed the phylogenetic tree of AcoSBT genes along with Arabidopsis SBTs using the Neighbor-joining statistical method. Full-length protein sequences of 56 Arabidopsis SBTs (AtSBTs) and 54 pineapple SBTs (AcoSBTs) were aligned to generate a neighbor-joining phylogenetic tree (Figure 1A). The SBT genes were divided into 6 subfamilies as previously reported, namely, Group I, Group II, Group III, Group IV, Group V, and Group VI. Among them, Group I was the largest subfamily with 9 AtSBTs and 30 AcoSBTs. Compared to Arabidopsis, the much greater number of SBTs in pineapple indicates that group I SBT members in pineapple might have a broader function and that Group I genes might have undergone the evolutionary divergence between dicotyledons and monocotyledons. The smallest group, Group VI, possesses only two genes in pineapple and Arabidopsis evenly, suggesting that this subfamily may not have undergone evolutionary divergence between dicotyledonous and monocotyledonous plants.
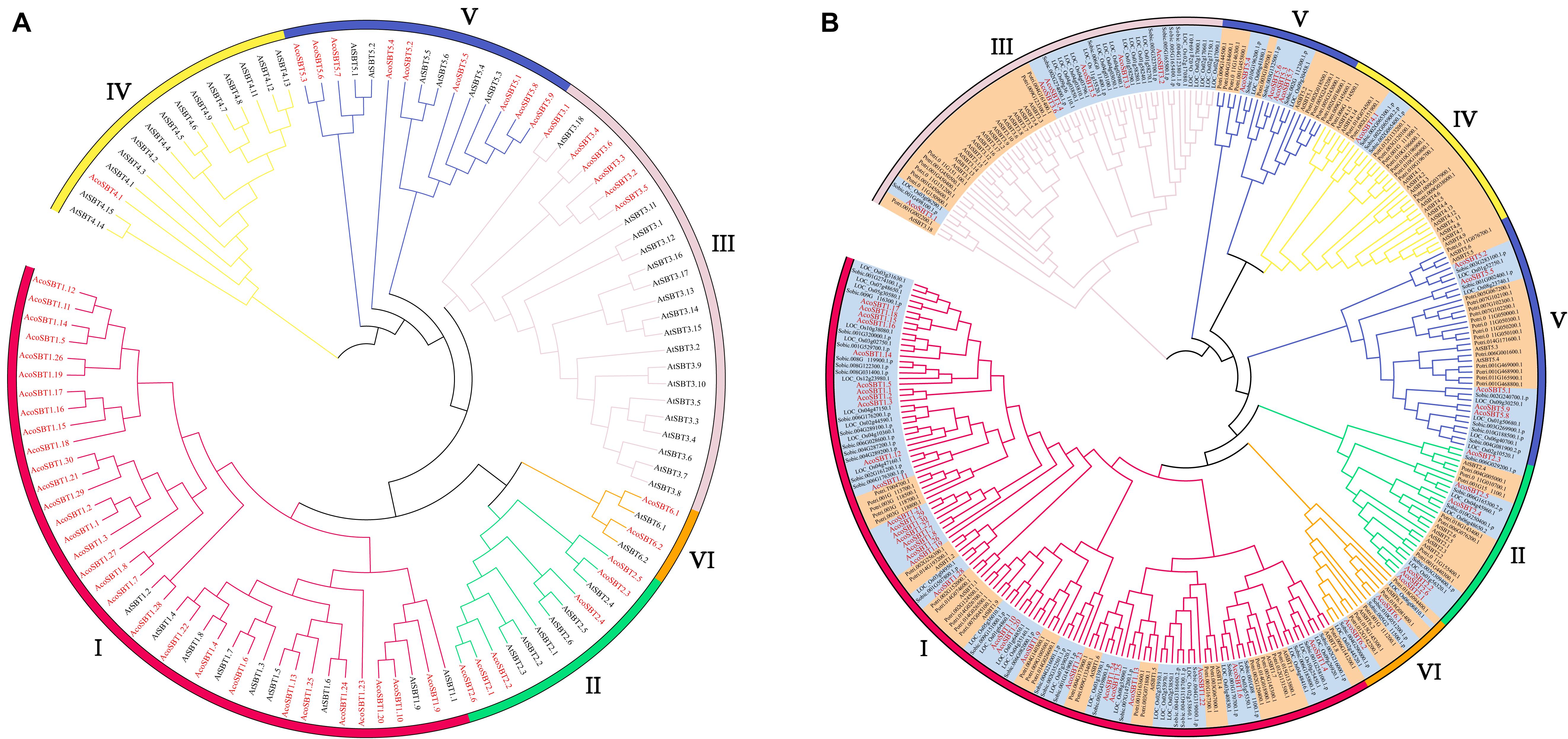
Figure 1. The evolutionary relationship of SBTs. (A) Phylogenetic relationship of the SBT proteins of Aco (Pineapple), At (Arabidopsis thaliana). (B) Phylogenetic relationship of the SBT proteins of Aco, At, Potri (Populus trichocarpa), Os (Oryza sativa), and Sbicolor (Sorghum bicolor). The phylogenetic tree was generated using neighbor-joining with 1,000. The phylo-genetic tree divides SBTs into six subfamilies, and the subfamilies are represented with different colors. Members from monocotyledonous plants are highlighted in orange while genes from dicotyledonous plants are highlighted in bright blue. SBT members from pineapple are marked in red font.
The evolutionary of SBTs between monocotyledons and dicotyledons were further explored based on the phylogenetic tree with monocotyledonous plants (Ananas comosus, Oryza sativa, and Sorghum bicolor) and dicotyledons (Arabidopsis thaliana and Populus trichocarpa) SBT genes (Figure 1B). In general, members from the same class tend to gather in the given subfamily. Among them, Group I is the largest subfamily, containing 126 members. Of the 126 members, 87 are monocots members accounting for 69%, and only 39 are dicots members accounting for 31%. Consistent with the speculation above, this disparity indicates Group I members probably experienced the differentiation between monocotyledons and dicotyledons. Moreover, Group VI is the smallest subfamily, with only seven genes in monocots and dicots evenly, further confirming the speculation that this subfamily is conserved in evolutionary divergence.
Gene Structure Characterization and Protein Motif Identification
To further understand their gene structure diversity, we analyzed the exon-intron organization of AcoSBT genes. As shown in Figure 2A, the exon numbers of the AcoSBTs range from 1 to 46. AcoSBT2.5 contains the most exons, whereas 12 genes (AcoSBT1.5, AcoSBT1.6, AcoSBT1.7, AcoSBT1.11, AcoSBT1.13, AcoSBT1.12, AcoSBT1.14, AcoSBT1.15, AcoSBT1.22, AcoSBT1.24, AcoSBT1.26, and AcoSBT1.30) harbor only one exon. Genes from the same subgroup generally show similar gene structures, especially in the number and length of exons. All the genes with only one exon are from Group I, and most other Group I genes also contain fewer exons. All genes of Group II harbor more than 8 exons. This conserved gene structure of each subgroup suggests that SBT genes with high homologous sequence similarity tend to possess the same number of exons (Figure 2A).
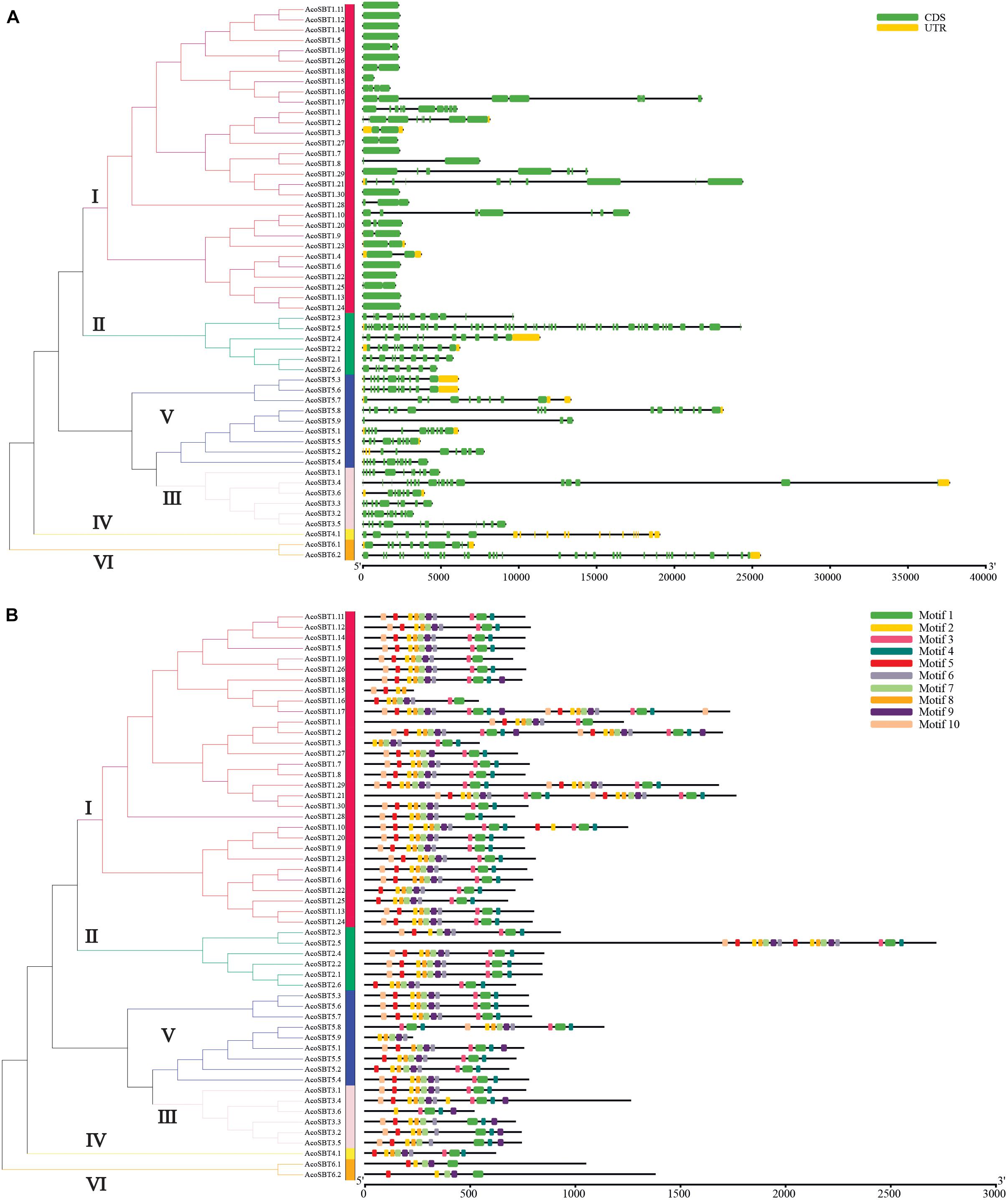
Figure 2. Gene structure and architecture of conserved protein motifs. (A) The Exon-intron structure of AcoSBTs. Green boxes represent CDS regions, yellow boxes represent UTR regions, and black lines represent untranslated introns. (B) The conserved motif composition of AcoSBTs. Different motifs are displayed by boxes with different colors.
Protein motifs are basic units of protein structure, which directly determined the function of given proteins. To elucidate the diversification of AcoSBT proteins, the conserved and diverged motifs were further identified by MEME with setting 10 motifs (Granziol et al., 2019). The detailed logo of each letter represents the level of conservation of amino acids (Supplementary Figure 1 and Supplementary Table 2). As displayed in Figure 2B, the number of motifs in pineapple SBT proteins ranged from 4 to 10. Motif 1 was founded in almost all AcoSBTs, except AcoSBT1.15, and AcoSBT5.9. For all AcoSBTs from Group II, their last motif on the C-terminal was motif 4, while almost all other group members ended with diverse motifs. 5 proteins (AcoSBT3.2, AcoSBT3.3, AcoSBT3.4, AcoSBT3.5, and AcoSBT3.6) from Group III, contained similar motif arrangement. Almost all the Group IV members, except AcoSBT3.1, ended with the conserved fragment of peptidases S8 domain Motif 9. The Group VI members showed the same protein structures in terms of motif number and motif type. For instance, members of this group did not preserve motif 10, which is acting as the “temporary inhibitors and assisting in the peptidase folding.” In conclusion, although a slight difference in motif arrangement was shown in some clusters, AcoSBTs from the same group generally possessed a similar protein structure.
Chromosome Location and Evolutionary Analyses of AcoSBTs
All of 54 AcoSBT genes were distributed on 25 different chromosomes unevenly, as 7 genes were mapped onto linkage group (LG) 3, followed by 6 genes located in LG8, and 5 genes were distributed in LG7. While, 8 LGs, namely LG5, LG14, LG16, LG21, LG22, LG23, LG24, and LG25, were found to harbor only one AcoSBT gene.
To obtain the possible mechanism of the expansion of AcoSBTs, we investigated the gene duplication events in pineapple (Figure 3). A total of 5 duplicated pineapple SBT pairs indicating duplication events were identified: AcoSBT1.10/AcoSBT1.9, AcoSBT5.1/AcoSBT5.5, AcoSBT2.1/AcoSBT2.2, AcoSBT1.22/Aco SBT1.4, and AcoSBT1.11/AcoSBT1.12. Except for the gene pair AcoSBT1.11/AcoSBT1.12, which is a tandem duplication, all the other duplication events founded in the pineapple genome are segmental duplications. The result suggested that segmental duplication plays a more crucial role in the amplification process of SBTs in the pineapple genome. Comprehensive syntenic analysis of SBTs between pineapple and Arabidopsis was further conducted to illustrate the possible evolutionary mechanism of AcoSBTs. As a result, a total of 3 one-to-one syntenic orthologous gene pairs (AtSBT2.1/AcoSBT2.2, AtSBT1.3/AcoSBT1.12, and AtSBT5.5/AcoSBT5.2) were identified, suggesting that these genes derived from the same ancestor before the divergence of monocotyledonous plants and dicotyledonous plants. We also found other kinds of syntenic orthologous gene pairs, such as one AcoSBT corresponds to two AtSBTs (AcoSBT1.6-AtSBT1.7/AtSBT1.9), or one AtSBTs corresponds to two AcoSBTs (AtSBT1.4-AcoSBT1.22/AtSBT1.4). These pairs might form after the divergence of monocotyledon and dicotyledon. We further calculated the Ka/Ks values of the gene pairs shown on the comparative synteny map (Supplementary Table 3). All the Ka/Ks values of SBT gene pairs are less than 1, with the highest value shown in the AcoSBT1.22/AcoSBT1.4 pair (Ka/Ks = 0.506). These results suggested that the SBT gene family in pineapple experienced strong purifying selective pressure, as Ka/Ks > 1 means positive selection, Ka/Ks = 1 indicates neutral selection, and Ka/Ks < 1 represents negative selection (Zhang et al., 2020).

Figure 3. Synteny analysis of SBT genes between pineapple and Arabidopsis. Schematic representation for both chromosomal distribution and interchromosomal relationships of SBT genes.
Given that comparative syntenic maps help to the study of evolutionary trait, we also have set up two comparative syntenic maps of pineapple associated with the above-mentioned four species, namely, Arabidopsis, Populus, rice, and Sorghum (Figure 4). According to the syntenic analyze results, we found 17 and 28 orthologous gene pairs in Arabidopsis and P. trichocarapa, while 28 gene pairs in both O. sativa and S. bicolor (Supplementary Table 3 and Figure 4). Some AcoSBTs had associated with multiple orthologous genes. Actually, many AcoSBTs from Group I are often corresponding to many SBTs from monocotyledons. For example, AcoSBT1.11 is associated with Sobic.002G161200/Sobic.006G176300 and LOC_Os04g47160, but we couldn’t find its orthologous gene in Arabidopsis and Populus. Additionally, AcoSBT1.22 is associated with 3 monocots genes (AcoSBT1.4, LOC_Os02g53860 and Sobic.004G319000) and 2 dicots genes (Potri.001G167300 and Potri.003G067000), AcoSBT1.4 is associated with 4 monocots genes (LOC_Os09g26920, LOC_Os10g25450, Sobic.001G258100, and Sobic.002G216000) but not with dicots gene. The results support our speculation in the evolutionary tree analysis that the Group I members may have undergone evolutionary divergence between dicotyledonous and monocotyledonous species. However, similar numbers of orthologous genes could not be found in Group VI. For example, AcoSBT6.1 existed one gene pair in P. trichocarapa (Potri.018G081400) and S. bicolor (Sobic.010G051200), indicating these group genes may appear before the divergence of dicotyledonous and monocotyledonous. We calculated the Ka, Ks, and Ka/Ks of the gene pairs (Supplementary Table 3) and found that all the Ka/Ks values of SBT gene pairs are less than 1, further confirming that the SBT gene family experienced strong negative selective pressure.
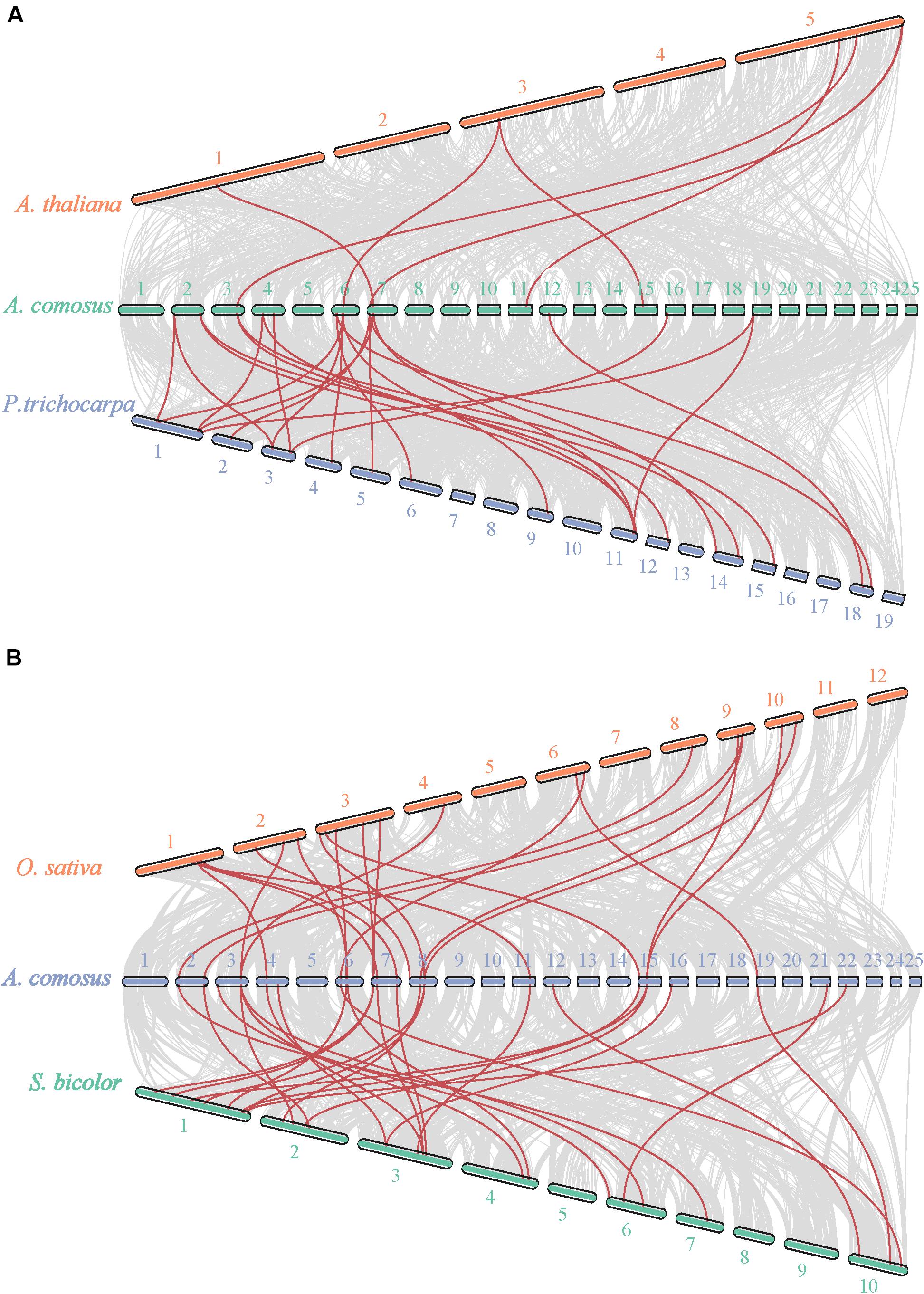
Figure 4. Synteny analysis of SBT genes between pineapple and Arabidopsis, populus, rice, and Sorghum. (A) Synteny analysis of SBT genes between pineapple and two dicotyledonous plants, Arabidopsis and populus. (B) Synteny analysis of SBT genes between pineapple and two monocotyledonous plants, rice and Sorghum. Gray lines in the background indicate the collinear blocks within pineapple and other plant genomes, while the red lines highlight the syntenic SBT gene pairs.
Cis-Acting Elements Identification in Promoter Region of AcoSBTs
To detect the potential biological functions of AcoSBT genes in pineapple, we collected the 2,000 bp upstream regions of all SBT genes and identified their Cis-acting elements using the PlantCARE program (Lescot et al., 2002). Within these 2,000 bp regions, 8 genes (AcoSBT1.1, AcoSBT1.2, AcoSBT1.9, AcoSBT1.10, AcoSBT1.16, AcoSBT1.28, AcoSBT1.29, and AcoSBT5.1) have overlapped regions with their upstream genes; thus, those overlapped regions were deleted, and the remaining promoter sequences were used for cis-elements analysis. A total 1,246 Cis-acting elements of 21 types were identified in the promoter region of 54 genes, classified into three categories: plant growth and development response elements, stress-related response elements, and phytohormone-related response elements (Figure 5, Supplementary Table 4, and Supplementary Figure 2). The plant growth and development response elements category include 7 types of Cis-acting elements (AE-box, ACE, ATCT-motif, I-box, Box-4, G-box, GATA-motif). And the top three Cis-acting components in this category are Box 4 (42.16%), G-box (32.39%), and GATA-motif (9.76%) (Figure 5 and Supplementary Table 4). Interestingly, all the three elements with the highest proportion belong to the parts of the module involved in light responsiveness (Jeong and Shih, 2003; Mande et al., 2018; Zheng et al., 2021). Moreover, only 20.92% of identified elements were classified into the stress-related response elements category. In this category, elements essential for the anaerobic induction (ARE, 37.7%), wound-responsive elements (WUN-motif, 15.58%), MYB binding site involving in drought-inducibility (MBS, 15.21%), elements involving in low-temperature reaction (LTR, 11.59%), elements responsive to pathogens (W-box, 11.23%), and elements related to defense responsiveness (TC-rich repeats, 8.70%) were identified. 32.20% of elements in the third category are phytohormone-related response elements category. Most of the elements in this category were responsive to abscisic acid (ABRE, 41.59%), ethylene (ERE, 23.00%), methyl jasmone (CGTCA-motif, 20.79%), auxin (TGA-element, 3.27%; AuxRR-core, 2.33%), and gibberellin (GARE-motif, 3.27%), etc.
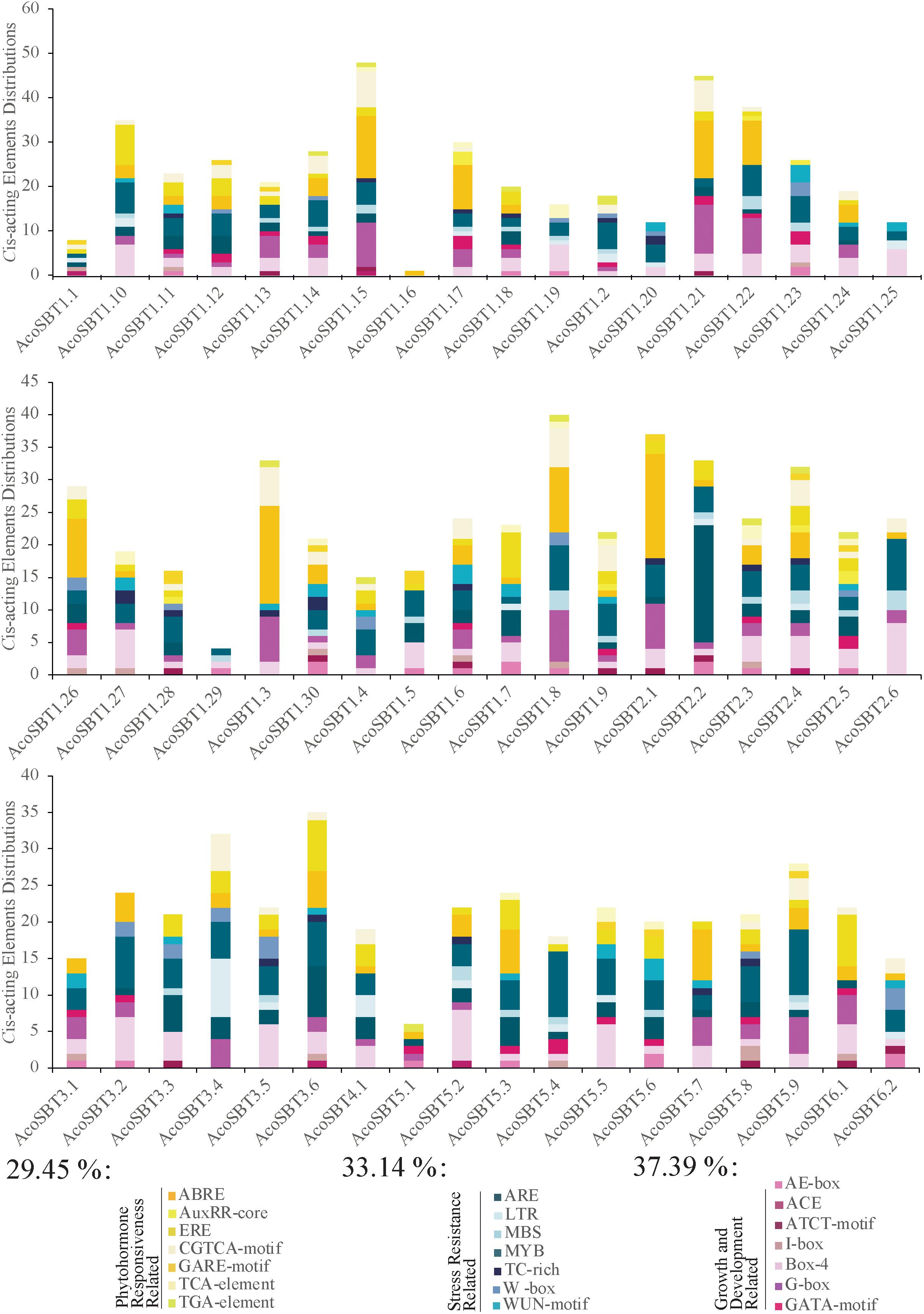
Figure 5. Information of Cis-acting elements in promoter region AcoSBT genes. Distribution, classification and proportion of Cis-acting elements in the promoter region of each gene.
To further investigate whether the Cis-acting elements were correlated with phylogenetic groups, we analyzed the Cis-acting elements for different subgroups. The results showed that the distribution of the Cis-acting elements in the promoter region did not show a strong correlation with phylogenetic groups. AcoSBT1.15, which possesses 48 Cis-acting elements, is the gene with the most Cis-acting, including 14 ABRE, 10 G-box, 8 CGTCA-motif. ABRE, CGTCA-motif, and TGAGC-motif are all components of phytohormone responsiveness, suggesting that AcoSBT1.15 may have potential functions in the plant hormone pathway. With 45 Cis-acting elements, AcoSBT1.21 harbored the second large number of Cis-acting elements, mainly composed of 13 ABRE, 11 G-box, and 7 CGTCA-motifs, indicating that AcoSBT1.21 may function in both the phytohormone responsiveness and light-responsive development. Moreover, as shown in Figure 5, Box 4, a conserved DNA module involved in light responsiveness was identified in almost all of the AcoSBT genes. Take together, it is suggested that the above study implies that the AcoSBT gene family may be closely associated with light-responsive growth and development in pineapple, which will provide clues for further studies on SBTs in pineapple.
Spatio-Temporal Expression Profiles of AcoSBTs in Pineapple
To explore the possible role of AcoSBT genes in pineapple growth and development, we analyzed the expression profiles of all SBT genes in different tissues and developmental stages using publicly available transcriptome databases. The 54 AcoSBTs exhibit diverse organ-specific expression patterns, which may help to illustrate the functional divergence of SBT gene family genes in pineapple growth and development (Supplementary Table 5). As shown in Figures 3, 6 Group I genes, AcoSBT1.23, AcoSBT1.10, and AcoSBT1.20, were specifically expressed in 5 developmental stages of pineapple female sexual organ ovule. In contrast, 2 Group V genes, AcoSBT5.1 and AcoSBT5.4, had analogical transcription profiles that were specifically expressed in pineapple stamen. Those results implicate that Group I and Group V genes might play essential roles in the female and male gametophyte development, respectively. In Group III, almost all genes except AcoSBT3.4 were poorly expressed in all tested samples, illustrating that the genes in this group might not be involved in the developmental processes in pineapple. Meanwhile, 6 genes (AcoSBT2.4, AcoSBT1.24, AcoSBT6.2, AcoSBT1.6, AcoSBT1.13, and AcoSBT1.22) were ubiquitously expressed in all tested tissues (7 ovule developmental tissues, 4 sepal developmental tissues, 6 stamen developmental tissues, 3 petal developmental tissues, 6 fruit developmental tissues, as well as root, flower and leaf samples), suggesting that these 6 genes might exert necessary functions for both vegetative and reproductive growth in pineapple.
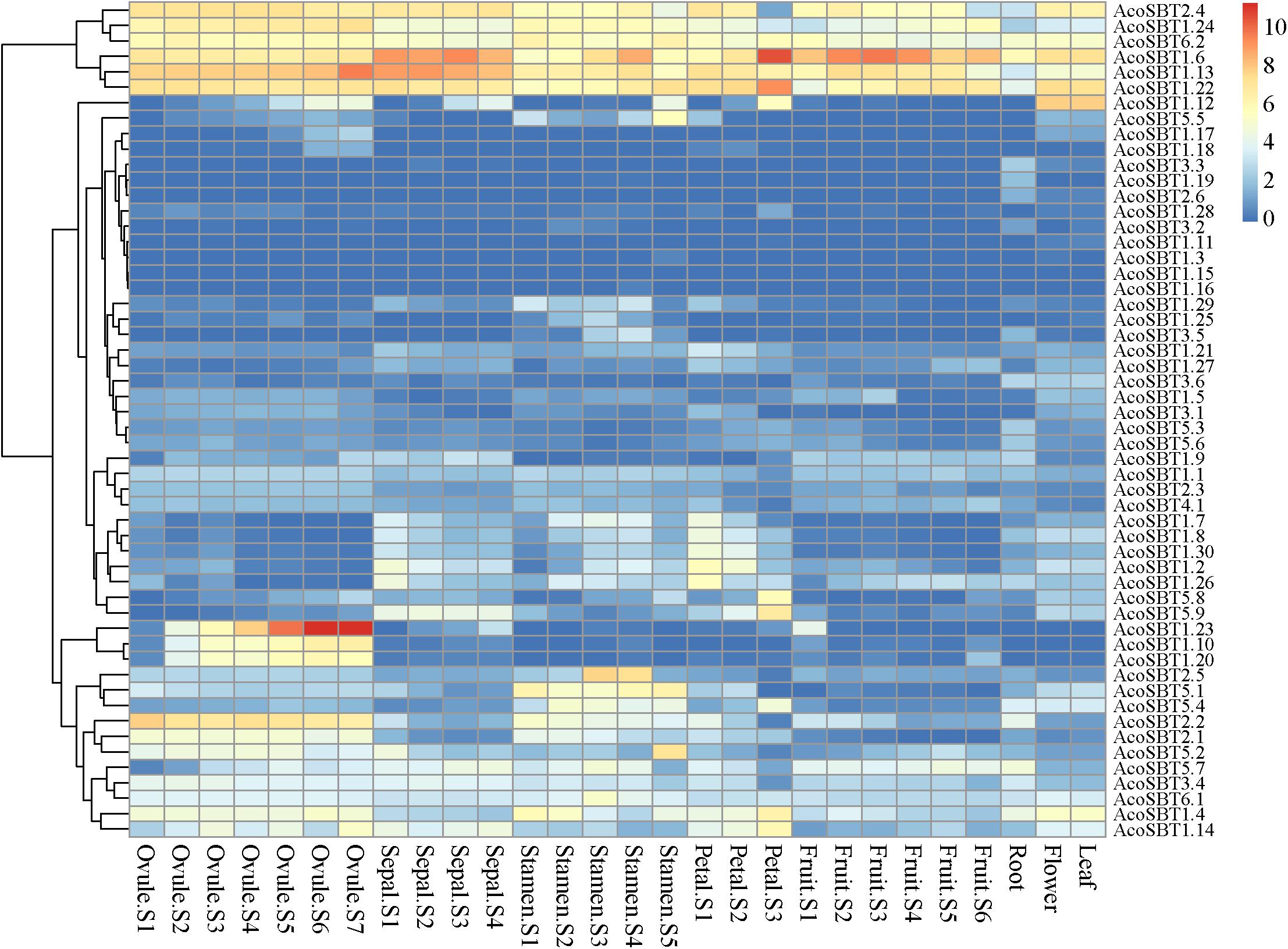
Figure 6. Tissue-specific expression profiles of AcoSBTs in pineapple. Heat-map of tissue-specific expression profiles of AcoSBTs in pineapple. Samples are mentioned at the top of each lane: ovule S1–S7, sepal S1–S4, stamen S1–S5, petal S1–S3, root, leaf, flower, fruit S1–S7. “S” is the abbreviation of the word “stage.”
We also verified the reliability of transcriptome data using qRT-PCR technology. In this analysis, the mixed stage samples of 5 tissues (leaf, sepal, petal, stamen, and ovule) were used to investigate the transcription of 12 abundantly or specifically expressed genes, namely, AcoSBT1.12, AcoSBT1.13, AcoSBT1.22, AcoSBT1.23, AcoSBT1.24, AcoSBT1.4, AcoSBT1.6, AcoSBT2.2, AcoSBT2.4, AcoSBT2.5, AcoSBT5.2, and AcoSBT6.2. The results showed that the expression patterns of qRT-PCR were generally consistent with that given by RNA-seq analysis (Supplementary Figure 3).
Subcellular Localization of AcoSBT1.12 Proteins
The Cis-acting elements profile showed that AcoSBT1.12 harbored various Cis-acting elements in its promoter region, and the expression pattern revealed that AcoSBT1.12 specifically expresses in flower and leaf, suggesting that AcoSBT1.12 might play an essential role in the development of above-ground organs in pineapple. As previously reported, SBTI genes play essential roles in plant growth and development (Schaller et al., 2012). According to the phylogenetic tree constructed in this study, AcoSBT1.12 was clustered into the subclade with widely reported SBTI genes.
To further investigate the subcellular localization of AcoSBT1.12 in plant cells, We fused the coding sequence of AcoSBT1.12 to the area between the CaMV 35S promoter and the GFP encoding region. The transient expression leaves in tobacco (Nicotiana benthamiana) showed that the AcoSBT1.12-GFP protein was mainly distributed in the plasma membrane, whereas the control protein produced by the empty vector (35S:GFP) was equally distributed in the plasma membrane and the nucleus (Figure 7).
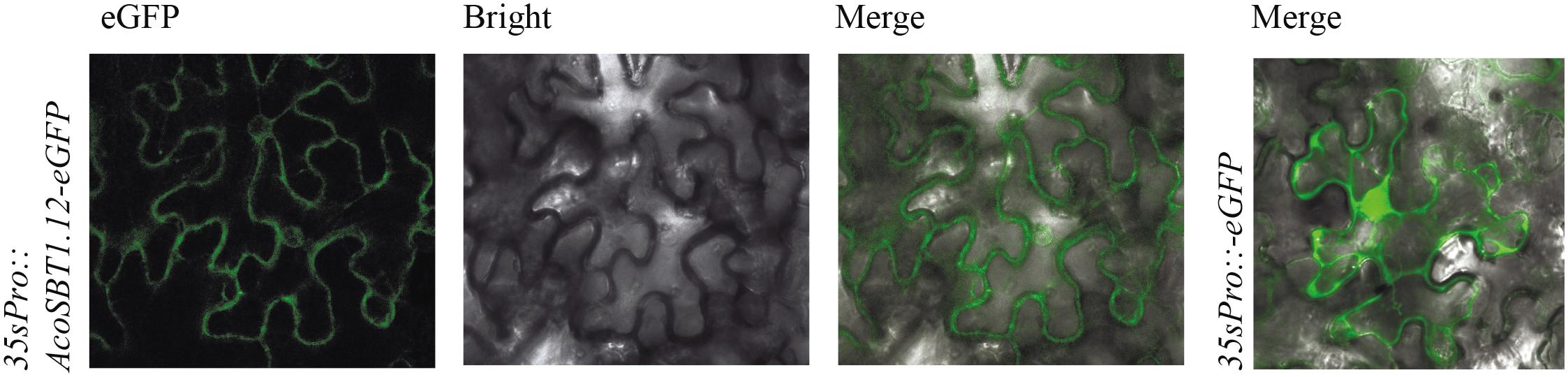
Figure 7. Subcellular location of AcoSBT1.12. The subcellular location of AcoSBT1.12 and blank vector were detected on the surface cells of Nicotiana Benthamiana’s leave.
AcoSBT1.12 Delays Floral Transition in Arabidopsis
To further confirm the hypothesis that AcoSBT1.12 is involved in plant growth, we overexpressed AcoSBT1.12 in Arabidopsis driving by 35S promoter (Supplementary Table 6). Two AcoSBT1.12 T3 transgenic lines (OE-24# and OE-25#) with positive PCR bands and high AcoSBT1.12 transcription abundance were selected for phenotyping (Figure 8).
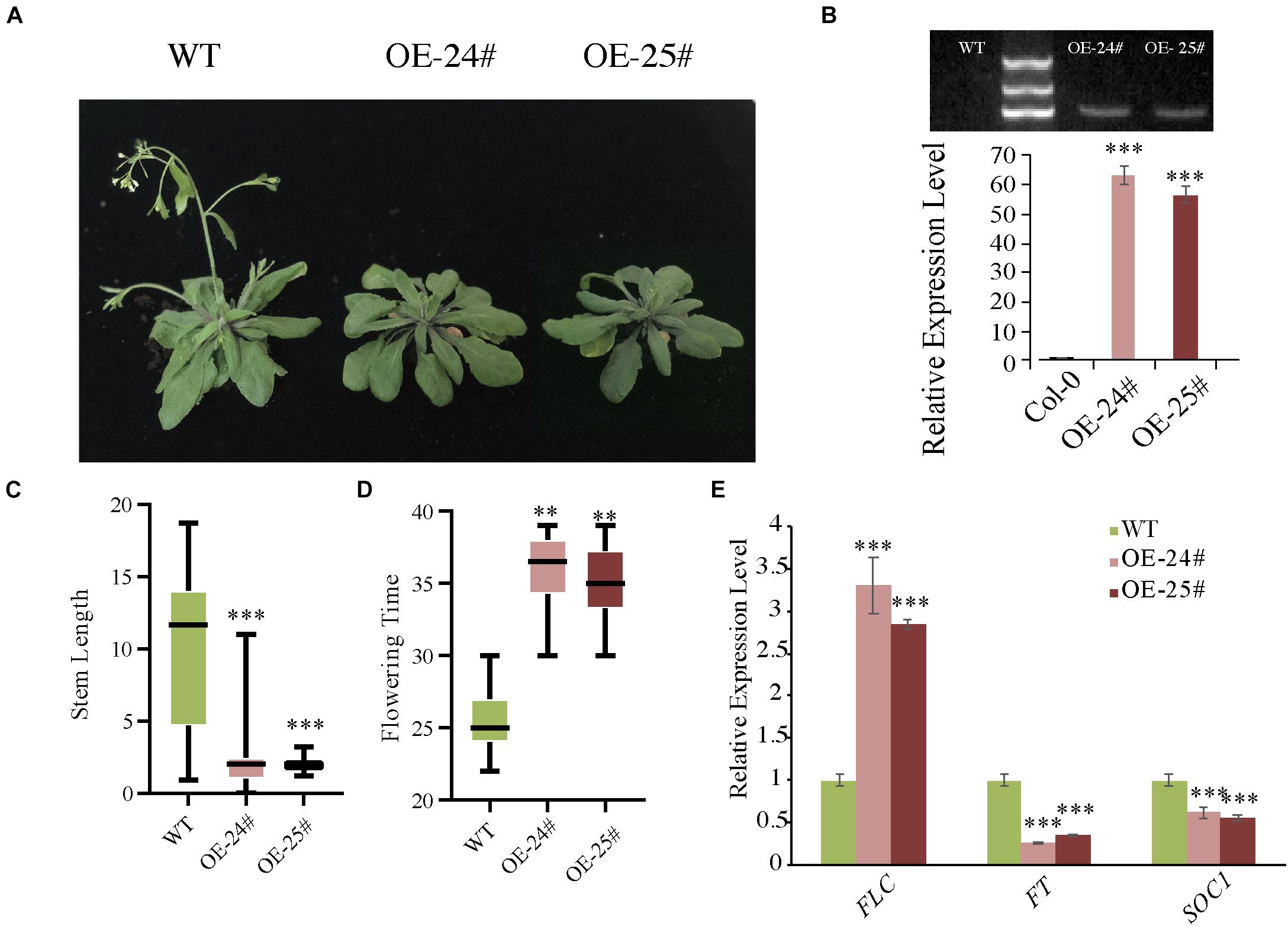
Figure 8. Overexpression of AcoSBT1.12 in Arabidopsis delayed floral transition with the expression level of FLC increased while FT and SOC1 decreased. (A) Characterization of the flowering phenotype of AcoSBT1.12 OE lines. (B) The gel electrophoresis results and qPT-PCR results confirmed the reliability of OE lines. (C) Stem length on the 40th day, the error bars indicate + SD. (D) Flowering time of WT and OE lines, the error bars indicate ± SD. (E) Relative expression level of flowering regulator genes. Asterisks indicate significant differences for the indicated comparisons based on Student’s t-test (∗∗∗p < 0.001; ∗∗p < 0.01).
Under LD (Long-day) conditions, transgenic lines showed delayed floral transition with better-developed rosette leaves than WT (Figures 8A,C). Normally, WT plants bolt in about 25 days, while the overexpressing lines are delayed by about 10 days (Figure 8D). Meanwhile, the expression levels of three critical floral transition regulating genes were changed significantly. The MADS-box gene FLC (FLOWERING LOCUS C), the inhibitor of flowering event, was significantly up-regualated in all three OE lines (Figure 8E). While FT (FLOWERING LOCUS T) and SOC1 (SUPPRESSOR OF OVEREXPRESSION OF CONSTANS1), function in flowering induction in LD, were down-regulated in the three OE lines (Figure 8E). However, under SD (Short day) conditions, the transgenic lines didn’t show any difference from WT plants. Collectively, these results indicate that AcoSBT1.12 might participate in floral transition under LD conditions.
Since the SBT family genes encode proteases, identifying the potential substrates of AcoSBTs will help to understand their function further. Using STRING database, 10 pineapple proteins that might interact with AcoSBT1.12 were predicted. Subsequently, the coding sequences of eight of these genes (AcoCWF19L, AcoPUF2, AcoCwfJL, Aco012905, AcoPM1P, Aco009239, AcoSZF1, and AcoCZF1) were fused into the pGADT7 as AD constructs, while the CDS of AcoSBT1.12 was cloned into the pGBDT7 as BD construct. Yeast two-hybrid assay showed that AcoSBT1.12 could interact with AcoCWF19L, AcoPUF2, AcoCwfJL, Aco012905, and AcoSZF1 (Figure 9 and Supplementary Table 6). Although litter is known about these proteins, these conformed protein-protein interactions will provide reference information for studying the regulatory functions of AcoSBT genes in pineapple.
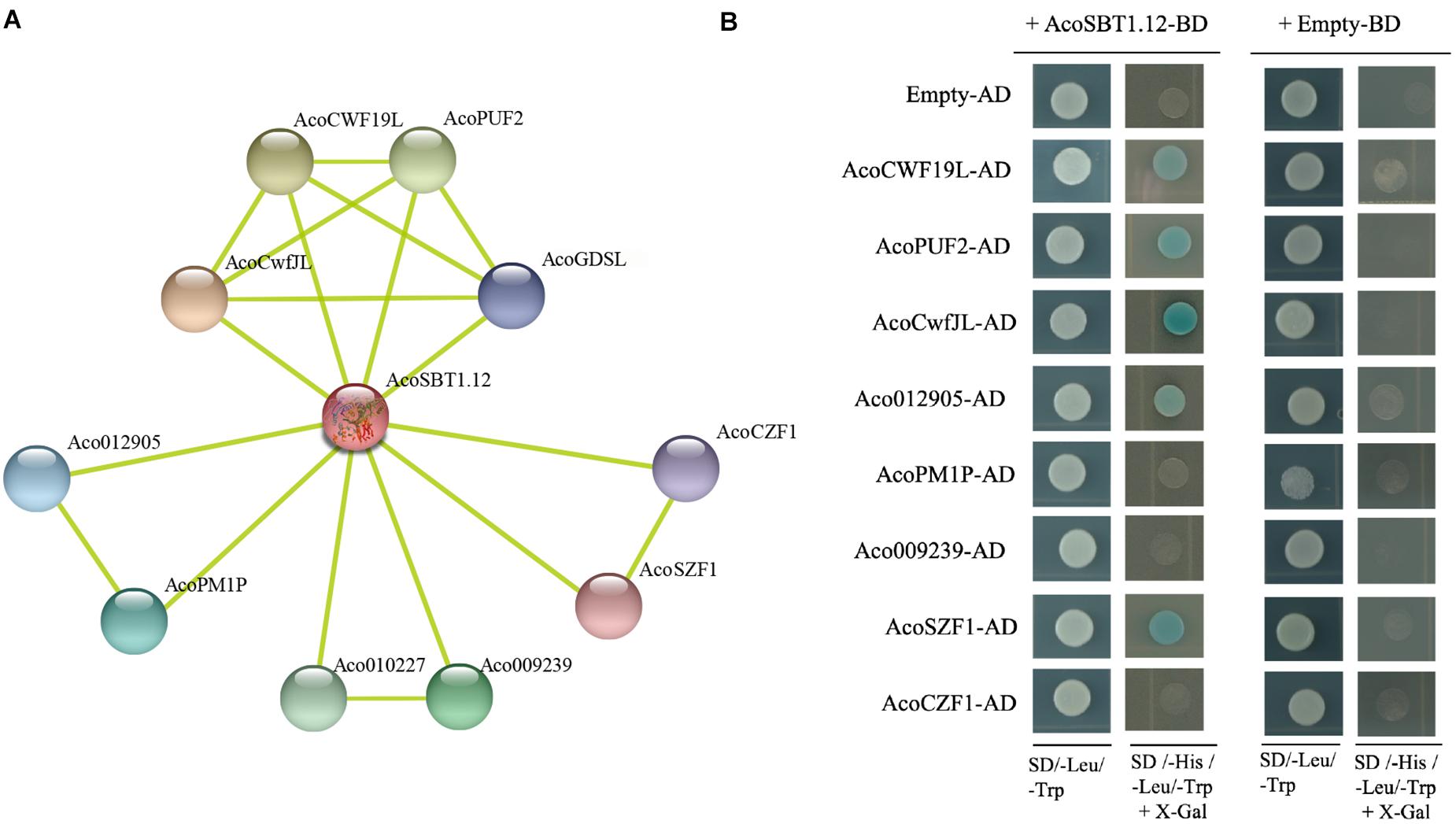
Figure 9. AcoSBT1.12 influences flowering time. (A) The network prediction of AcoSBT1.12 and its interaction protein based on STRING database. (B) Protein interactions between AcoSBT1.12 and predicted protein by yeast two-hybrid assay.
Discussion
It is extensively reported that SBT proteins exist in all three domains of life and are relatively conserved across different plant species. Beginning from the first description of 15 SBT genes in tomato (Lycopersicon esculentum), the gene members from this family were further identified and well-characterized in many dicotyledons, including Arabidopsis (56 gene members), poplar (Populus trichocarpa, 90), potato (Solanum tuberosum, 82), and melon (Cucumis melo L.) (Yamagata et al., 1994; Meichtry et al., 1999; Rautengarten et al., 2005; Tian et al., 2005; Schaller et al., 2012; Norero et al., 2016). In this study, 54 AcoSBTs were identified via the HMM-based method in the pineapple genome. Compared with polar and potato, the pineapple SBT family is relatively small, suggesting that some SBT members may have undergone an evolutionary selection and get lost in pineapple during evolution. To reveal the phylogenetic relationship of pineapple SBT genes, a phylogenetic tree was constructed, and the pineapple SBT members were classified into 6 groups according to their phylogenetic relationship (Figure 1A).
In the subsequent studies on gene structure and motif composition, we further proved the relative conservation among the members of the same subfamily (Figure 2). The investigation of gene duplication events illustrated the potential expansion mechanism of AcoSBTs and showed that the duplication gene pairs tend to be from the same subfamily (Figures 3, 4). Subsequent Cis-acting element analysis showed that AcoSBTs probably function in light-responsiveness events. The expression pattern analysis showed that the expression patterns of AcoSBT genes showed an organ-specific tendency (Figures 5, 6), which is consistent with the previous findings in Arabidopsis and rice. These results indicated that the AcoSBT genes might be regulated by light and might participate in many aspects of photomorphogenesis in pineapple (Jonesb et al., 2004). It has been previously reported that several Group I members are involved in many aspects of plant growth and development, including embryo development, stomatal density regulation, and reproductive development. For example, SBT1.12, also named as SDD1 represent for Stomatal Density and Distribution 1, highly expressed in stomata initials but undetectable in mature guard cells, functions in stomata development suppression via TMM (Too Many Mouths)—dependent pathway in both Arabidopsis and Solanum lycopersicum (Groll et al., 2002; Morales et al., 2018). In Arabidopsis, SBT1.4 (SASP), expressed in all above-ground organs, down-regulates silique production and branched inflorescences during reproductive development (Wang et al., 2018). In legumes, including Medicago truncatula and Pisum sativum, SBT1.1 proteins locate in the endosperm, controlling seed size variation through regulating embryo cell division during reproductive development (D’Erfurth et al., 2012). The apparent relevance of SBT I subfamily with plant development, together with the significant expansion and uneven distribution of SBT I members in pineapple genome indicates the unique role of this gene subfamily in pineapple development.
Among all AcoSBT I members, the transcription level of AcoSBT1.12 was relatively high in leave and flower. Interestingly, we also found AcoSBT1.12 shared conserved domains with SDD and SASP in many species, both of which are involved in above-ground organ development (Groll et al., 2002; Morales et al., 2018). These results suggested that AcoSBT1.12 genes may have functions in pineapple growth and development. Due to the limitations of pineapple transformation technology, we constructed AcoSBT1.12 OE lines in Arabidopsis. Our results showed that overexpression of pineapple AcoSBT1.12 delayed the flowering time of Arabidopsis under LD conditions. Moreover, The core flowering regulating genes, FT, FLC, and SOC1, were up-regulated in the transgenic Arabidopsis OE lines (Figure 8). In angiosperms, the flowering transition located at the converter node from vegetative growth to reproductive growth is the most vital phase transition in plant development (Domagalska et al., 2010). Plant flowering is regulated by six major flowering genetic pathways to shape maximum reproductive success, including photoperiod, thermosensory, age, autonomous, vernalization, and GA pathways (Coupland, 1995; Koornneef et al., 1998; Fornara et al., 2010). The MADS-box gene FLC (FLOWERING LOCUS C), the first reported gene regulating flowering transition, locating in the central position of the flowering network, acts as a canary in the coal mine (Sheldon et al., 2000; Mentzer et al., 2010). And FT (FLOWERING LOCUS T), another widely studied florigen in the flowering regulatory network, is also acknowledged as the basis of numerous signaling pathway directly affect floral transition (Liu et al., 2007; Eckardt, 2010). Ectopic expression of AcoSBT1.12 affected the expression abundance of FT and FLC in Arabidopsis, which confirmed that SBT1.12 is involved in floral transition controlling in plants. Base on bioinformatics analysis, we conducted a yeast two-hybrid experiment and confirmed the interaction relationships between AcoSBT1.12 and PLANT-UNIQUE RAB5 EFFECTOR 2 (AcoPUF2). Due to the lack of molecular biological and genetic studies done in pineapple, further research needs to be done to elusive the molecular mechanism underlying AcoSBT1.12-mediated flowering transition. However, the valuable information in this study will shed light on the understanding of SBT-involved development in pineapple.
Conclusion
In this study, 54 AcoSBTs were firstly characterized base on genome-wide identification of the SBT gene family in pineapple. The results of the evolution scenario analysis of AcoSBTs suggested that AcoSBTs are highly conserved compared with their homologs from other monocotyledon plants. In addition, the expression profiling accompany with Cis-elements analysis showed that AcoSBT genes have essential roles in controlling plant growth and development. Overexpression of AcoSBT1.12 in Arabidopsis delayed flowering time and altered the expression level of FLC, FT, and SOC1. In conclusion, these results provide valuable information for further studying the roles of AcoSBTs in plant development and growth.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ebi.ac.uk/ena, PRJEB38680.
Author Contributions
XJ performed vector construction and phenotype analysis. XJ and YL performed RNA-seq and transformation. YZ, ZH, and YF calculated all the data. ZH and HC performed qRT-PCR analysis. XJ and YQ wrote the manuscript. YC, YQ, and HC revised the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the Science and Technology Program of Fujian Province (2019N5008) and the National Natural Science Foundation of China (31970333) to YQ.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2021.730821/full#supplementary-material
Supplementary Figure 1 | The detailed logo of each letter represents the level of conservation of amino acid of each Motifs.
Supplementary Figure 2 | Distribution of motifs in promoter region of each gene.
Supplementary Figure 3 | Validation of AcoSBT genes expression profile by qRT-PCR at five different tissues.
Supplementary Table 1 | Detailed information of pineapple SBTs, including sequenced ID, chromosome locations, isoelectric point (pI), molecular weight (KD), protein length and function domains.
Supplementary Table 2 | The multilevel consensus sequence and description of 10 motifs.
Supplementary Table 3 | The original data of Ka and Ks value.
Supplementary Table 4 | The distribution of Cis-acting elements.
Supplementary Table 5 | The original data of FPKM value.
Supplementary Table 6 | Information about AcoSBT1.12 and genes encoding interacting proteins.
Footnotes
- ^ http://pfam.xfam.org/
- ^ https://phytozome-next.jgi.doe.gov
- ^ https://www.ncbi.nlm.nih.gov/cdd
- ^ http://web.expasy.org
- ^ http://www.cbs.dtu.dk.services/TargetP/
- ^ http://www.cbs.dtu.dk.services/SignalP/
- ^ http://www.arabidopsis.org
- ^ https://itol.embl.de
- ^ http://meme-suite.org/tools/meme
- ^ http://bioinformatics.psb.ugent.be/webtools/plantcare/html
- ^ http://www.ncbi.nlm.nih.gov/Traces/study/?acc=SRP067011
References
Arango Argoty, G. A., Ruiz-Munoz, J. F., Jaramillo-Garzon, J. A., and Castellanos-Dominguez, C. G. (2012). An adaptation of Pfam profiles to predict protein sub-cellular localization in Gram positive bacteria. Annu. Int. Conf. IEEE Eng. Med. Biol. Soc. 2012, 5554–5557.
Bao, S., Hua, C., Shen, L., and Yu, H. (2020). New insights into gibberellin signaling in regulating flowering in Arabidopsis. J. Integr. Plant Biol. 62, 118–131. doi: 10.1111/jipb.12892
Chen, C., Chen, H., Zhang, Y., Thomas, H. R., Frank, M. H., and He, Y. (2020). TBtools: an integrative toolkit developed for interactive analyses of big biological data. Mol. Plant. 13, 1194–1202. doi: 10.1016/j.molp.2020.06.009
Coupland, G. (1995). Genetic and environmental control of flowering time in Arabidopsis. Trends Genet. 11, 393–397. doi: 10.1016/s0168-9525(00)89122-2
Creff, A., Brocard, L., Joubes, J., Taconnat, L., Doll, N. M., Marsollier, A. C., et al. (2019). A stress-response-related inter-compartmental signalling pathway regulates embryonic cuticle integrity in Arabidopsis. PLoS Genet. 15:e1007847. doi: 10.1371/journal.pgen.1007847
D’Erfurth, I., Le Signor, C., Aubert, G., Sanchez, M., Vernoud, V., Darchy, B., et al. (2012). A role for an endosperm-localized subtilase in the control of seed size in legumes. New Phytol. 196, 738–751. doi: 10.1111/j.1469-8137.2012.04296.x
Domagalska, M. A., Sarnowska, E., Nagy, F., and Davis, S. J. (2010). Genetic analyses of interactions among gibberellin, abscisic acid, and brassinosteroids in the control of flowering time in Arabidopsis thaliana. PLoS One 5:e14012. doi: 10.1371/journal.pone.0014012
Eckardt, N. A. (2010). Dissecting cis-regulation of FLOWERING LOCUS T. Plant Cell 22:1422. doi: 10.1105/tpc.110.220511
Emanuelsson, O., Brunak, S., Von Heijne, G., and Nielsen, H. (2007). Locating proteins in the cell using TargetP, SignalP and related tools. Nat. Protoc. 2, 953–971. doi: 10.1038/nprot.2007.131
Fornara, F., De Montaigu, A., and Coupland, G. (2010). SnapShot: control of flowering in Arabidopsis. Cell 141, 550–552. doi: 10.1016/j.cell.2010.04.024
Granziol, D., Ru, B., Zohren, S., Dong, X., Osborne, M., and Roberts, S. (2019). MEMe: an accurate maximum entropy method for efficient approximations in large-scale machine learning. Entropy 21:551. doi: 10.3390/e21060551
Groll, U. V., Berger, D., and Altmann, T. (2002). The subtilisin-like serine protease SDD1 mediates cell-to-cell signaling during Arabidopsis stomatal development. Plant Cell 14, 1527–1539. doi: 10.1105/tpc.001016
Have, M., Balliau, T., Cottyn-Boitte, B., Derond, E., Cueff, G., Soulay, F., et al. (2018). Increases in activity of proteasome and papain-like cysteine protease in Arabidopsis autophagy mutants: back-up compensatory effect or cell-death promoting effect? J. Exp. Bot. 69, 1369–1385. doi: 10.1093/jxb/erx482
Jeong, M.-J., and Shih, M.-C. (2003). Interaction of a GATA factor with cis-acting elements involved in light regulation of nuclear genes encoding chloroplast glyceraldehyde-3-phosphate dehydrogenase in Arabidopsis. Biochem. Biophys. Res. Commun. 300, 555–562. doi: 10.1016/s0006-291x(02)02892-9
Jin, X., Hou, Z., Zhao, L., Liu, L., Priyadarshani, S. V. G. N., Wang, L., et al. (2020). Genome-wide identification and evaluation of new reference genes in pineapple (Ananas comosus L.) during stamen and ovule development. Trop. Plant Biol. 13, 371–381. doi: 10.1007/s12042-020-09269-w
Jonesb, A. M., Dickermanc, A. W., and Beersa, E. P. (2004). The S8 serine, C1A cysteine and A1 aspartic protease families in Arabidopsis. Phytochemistry 65, 43–58. doi: 10.1016/j.phytochem.2003.09.005
Koornneef, M., Alonso-Blanco, C., Peeters, A. J., and Soppe, W. (1998). Genetic control of flowering time in Arabidopsis. Annu. Rev. Plant Physiol. Plant Mol. Biol. 49, 345–370.
Krzywinski, M., Schein, J., Birol, I., Connors, J., Gascoyne, R., Horsman, D., et al. (2009). Circos: an information aesthetic for comparative genomics. Genome Res. 19, 1639–1645. doi: 10.1101/gr.092759.109
Lescot, M., Dehais, P., Thijs, G., Marchal, K., Moreau, Y., Van De Peer, Y., et al. (2002). PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 30, 325–327. doi: 10.1093/nar/30.1.325
Librado, P., and Rozas, J. (2009). DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25, 1451–1452. doi: 10.1093/bioinformatics/btp187
Liu, J. X., Srivastava, R., Che, P., and Howell, S. H. (2007). Salt stress responses in Arabidopsis utilize a signal transduction pathway related to endoplasmic reticulum stress signaling. Plant J. 51, 897–909. doi: 10.1111/j.1365-313x.2007.03195.x
Mande, X., Yan, L., Zhiqiang, Z., Gege, H., Ke, H., Tianbao, Z., et al. (2018). Isolation and characterization of a green-tissue promoter from common wild rice (Oryza rufipogon Griff.). Int. J. Mol. Sci. 19:2009. doi: 10.3390/ijms19072009
Marchler-Bauer, A., Lu, S., Anderson, J. B., Chitsaz, F., Derbyshire, M. K., Deweese-Scott, C., et al. (2011). CDD: a conserved domain database for the functional annotation of proteins. Nucleic Acids Res. 39, 225–229.
Meichtry, J., Amrhein, N., and Schaller, A. (1999). Characterization of the subtilase gene family in tomato (Lycopersicon esculentum Mill.). Plant Mol. Biol. 39, 749–760.
Mentzer, L., Yee, T., Wang, T. Y., and Himelblau, E. (2010). FLOWERING LOCUS C influences the timing of shoot maturation in Arabidopsis thaliana. Genesis 48, 680–683. doi: 10.1002/dvg.20683
Ming, R., Vanburen, R., Wa, C. M., Tang, H., Schatz, M. C., Bowers, J. E., et al. (2015). The pineapple genome and the evolution of CAM photosynthesis. Nat. Genet. 47, 1435–1442.
Ming, R., Wai, C., and Guyot, R. (2016). Pineapple genome: a reference for monocots and CAM photosynthesis. Trends Genet. 32, 690–696. doi: 10.1016/j.tig.2016.08.008
Morales, S., Pérez, R., Ortega, A., Serrano, A. D. M., Mena, M., Fenoll, C., et al. (2018). Overexpression of a SDD1-Like gene from wild tomato decreases stomatal density and enhances dehydration avoidance in Arabidopsis and cultivated tomato. Front. Plant Sci. 9:940. doi: 10.3389/fpls.2018.00940
Neuteboom, L., Veth-Tello, L., Clijdesdale, O., Hooykaas, P., and Van Der Zaal, B. (1999). A novel subtilisin-like protease gene from Arabidopsis thaliana is expressed at sites of lateral root emergence. DNA Res. 6, 13–19. doi: 10.1093/dnares/6.1.13
Norero, N., Castellote, M. A., Canal, L., and Feingold, D. L. (2016). Genome-wide analyses of subtilisin-like serine proteases on Solanum tuberosum. Am. J. Potato Res. 93, 485–496. doi: 10.1007/s12230-016-9525-5
Paulus, J. K., Kourelis, J., Ramasubramanian, S., Homma, F., Godson, A., Horger, A. C., et al. (2020). Extracellular proteolytic cascade in tomato activates immune protease Rcr3. Proc. Natl. Acad. Sci. U.S.A. 117, 17409–17417. doi: 10.1073/pnas.1921101117
Ramírez, V., Lo’Pez, A., Mauch-Mani, B., Gil, M. J., and Vera, P. (2013). An extracellular subtilase switch for immune priming in Arabidopsis. PLoS Pathog. 9:e1003445. doi: 10.1371/journal.ppat.1003445
Rautengarten, C., Steinhauser, D., Bussis, D., Stintzi, A., Schaller, A., Kopka, J., et al. (2005). Inferring hypotheses on functional relationships of genes: analysis of the Arabidopsis thaliana subtilase gene family. PLoS Comput. Biol. 1:e40. doi: 10.1371/journal.pcbi.0010040
Rautengarten, C., Usadel, B., Neumetzler, L., Hartmann, J., Bussis, D., and Altmann, T. (2008). A subtilisin-like serine protease essential for mucilage release from Arabidopsis seed coats. Plant J. 54, 466–480. doi: 10.1111/j.1365-313x.2008.03437.x
Reichardt, S., Repper, D., Tuzhikov, A. I, Raisa, A. G., Planas-Marquès, M., Chichkova, N. V., et al. (2018). The tomato subtilase family includes several cell death-related proteinases with caspase specificity. Sci. Rep. 8:10531.
Schaller, A. (2004). A cut above the rest: the regulatory function of plant proteases. Planta 220, 183–197. doi: 10.1007/s00425-004-1407-2
Schaller, A., Stintzi, A., and Graff, L. (2012). Subtilases – versatile tools for protein turnover, plant development, and interactions with the environment. Physiol. Plant. 145, 52–66. doi: 10.1111/j.1399-3054.2011.01529.x
Schaller, A., Stintzi, A., Rivas, S., Serrano, I., Chichkova, N. V., Vartapetian, A. B., et al. (2018). From structure to function - a family portrait of plant subtilases. New Phytol 218, 901–915. doi: 10.1111/nph.14582
Sheldon, C. C., Rouse, D. T., Finnegan, E. J., Peacock, W. J., and Dennis, E. S. (2000). The molecular basis of vernalization: the central role of FLOWERING LOCUS C (FLC). Proc. Natl. Acad. Sci. U.S.A. 97, 3753–3758. doi: 10.1073/pnas.97.7.3753
Tian, M., Benedetti, B., and Kamoun, S. (2005). A Second Kazal-like protease inhibitor from Phytophthora infestans inhibits and interacts with the apoplastic pathogenesis-related protease P69B of tomato. Plant Physiol. 138, 1785–1793. doi: 10.1104/pp.105.061226
Wang, L., Li, Y., Jin, X., Liu, L., Dai, X., Liu, Y., et al. (2020). Floral transcriptomes reveal gene networks in pineapple floral growth and fruit development. Commun. Biol. 3:500.
Wang, Q., Guo, Q., Guo, Y., Yang, J., Wang, M., Duan, X., et al. (2018). Arabidopsis subtilase SASP is involved in the regulation of ABA signaling and drought tolerance by interacting with OPEN STOMATA 1. J. Exp. Bot. 69, 4403–4417. doi: 10.1093/jxb/ery205
Wilkins, M. R., Gasteiger, E., Bairoch, A., Sanchez, J. C., Williams, K. L., Appel, R. D., et al. (1999). Protein identification and analysis tools in the ExPASy server. Methods Mol. Biol. 112, 531–552. doi: 10.1385/1-59259-584-7:531
Yamagata, H., Masuzawa, T., Nagaoka, Y., Ohnishi, T., and Iwasaki, T. (1994). Cucumisin, a serine protease from melon fruits, shares structural homology with subtilisin and is generated from a large precursor. J. Biol. Chem. 269, 32725–32731. doi: 10.1016/s0021-9258(20)30051-x
Zhang, M., Liu, Y., He, Q., Chai, M., Huang, Y., Chen, F., et al. (2020). Genome-wide investigation of calcium-dependent protein kinase gene family in pineapple: evolution and expression profiles during development and stress. BMC Genomics 21:72. doi: 10.1186/s12864-020-6501-8
Zhao, C., Johnson, B., Kositsup, B., and Beers, E. (2000). Exploiting secondary growth in Arabidopsis. Construction of xylem and bark cDNA libraries and cloning of three xylem endopeptidases. Plant Physiol. 123, 1185–1196. doi: 10.1104/pp.123.3.1185
Zhao, P., Miao, Z., Jing, Z., Chen, S., Liu, Q., and Xiang, C. (2020). MADS-box factor AGL16 negatively regulates drought resistance via stomatal density and stomatal movement. J. Exp. Bot. 71, 6092–6106. doi: 10.1093/jxb/eraa303
Keywords: subtilisin-like serine protease, pineapple, genome-wide, expression pattern, functional analysis
Citation: Jin X, Liu Y, Hou Z, Zhang Y, Fang Y, Huang Y, Cai H, Qin Y and Cheng Y (2021) Genome-Wide Investigation of SBT Family Genes in Pineapple and Functional Analysis of AcoSBT1.12 in Floral Transition. Front. Genet. 12:730821. doi: 10.3389/fgene.2021.730821
Received: 25 June 2021; Accepted: 20 July 2021;
Published: 07 September 2021.
Edited by:
Zefeng Yang, Yangzhou University, ChinaReviewed by:
Jian Wu, Yangzhou University, ChinaXiumei Zhang, South Subtropical Crops Research Institute, Chinese Academy of Tropical Agricultural Sciences, China
Copyright © 2021 Jin, Liu, Hou, Zhang, Fang, Huang, Cai, Qin and Cheng. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yuan Qin, eXVhbnFpbkBmYWZ1LmVkdS5jbg==; Yan Cheng, Y2hlbmd5YW4xMjIwQGhvdG1haWwuY29t
†These authors have contributed equally to this work
 Xingyue Jin
Xingyue Jin Yanhui Liu
Yanhui Liu Zhimin Hou
Zhimin Hou Yunfei Zhang
Yunfei Zhang Yunying Fang
Yunying Fang Youmei Huang
Youmei Huang Hanyang Cai
Hanyang Cai Yuan Qin
Yuan Qin Yan Cheng
Yan Cheng