- 1School of Ecology and Nature Conservation, Beijing Forestry University, Beijing, China
- 2Beijing Engineering Research Center for Landscape Plant, Beijing Forestry University Forest Science Co. Ltd., Beijing, China
- 3College of Biological Sciences and Technology, Beijing Forestry University, Beijing, China
- 4Beijing Institute of Landscape Architecture, Beijing, China
The evening primrose family, Onagraceae, is a well defined family of the order Myrtales, comprising 22 genera widely distributed from boreal to tropical areas. In this study, we report and characterize the complete chloroplast genome sequences of 13 species in Circaea, Chamaenerion, and Epilobium using a next-generation sequencing method. We also retrieved chloroplast sequences from two other Onagraceae genera to characterize the chloroplast genome of the family. The complete chloroplast genomes of Onagraceae encoded an identical set of 112 genes (with exclusion of duplication), including 78 protein-coding genes, 30 transfer RNAs, and four ribosomal RNAs. The chloroplast genomes are basically conserved in gene arrangement across the family. However, a large segment of inversion was detected in the large single copy region of all the samples of Oenothera subsect. Oenothera. Two kinds of inverted repeat (IR) region expansion were found in Oenothera, Chamaenerion, and Epilobium samples. We also compared chloroplast genomes across the Onagraceae samples in some features, including nucleotide content, codon usage, RNA editing sites, and simple sequence repeats (SSRs). Phylogeny was inferred by the chloroplast genome data using maximum-likelihood (ML) and Bayesian inference methods. The generic relationship of Onagraceae was well resolved by the complete chloroplast genome sequences, showing potential value in inferring phylogeny within the family. Phylogenetic relationship in Oenothera was better resolved than other densely sampled genera, such as Circaea and Epilobium. Chloroplast genomes of Oenothera subsect. Oenothera, which are biparental inheritated, share a syndrome of characteristics that deviate from primitive pattern of the family, including slightly expanded inverted repeat region, intron loss in clpP, and presence of the inversion.
Introduction
Chloroplast is one of the most important organelles in plant cells and play vital metabolic roles in photosynthesis as well as amino acid and lipid synthesis (Daniell et al., 2016). It has its own genetic material that does not obey the Mendelian laws of heredity. The chloroplast genome of angiosperms often shows a stable quadripartite ring structure containing one large single copy (LSC) region and one small single copy (SSC) region separated by two copies of an inverted repeat (IR) region. It usually shows uniparental inheritance (Ravi et al., 2008), and its sequence, gene number, and gene order have been considered to be very conserved (Wolfe et al., 1987).
However, many types of mutation occur in the chloroplast genome, including single nucleotide polymorphisms (SNPs), indels, IR contraction and expansion, inversion, and translocation (Ahmed et al., 2012; Daniell et al., 2016; Liu et al., 2018; He et al., 2019; Mehmood et al., 2020), which provide potential molecular markers for phylogenetic inference, DNA barcoding, and population genetics. Studies have shown that environmental factors, such as hot, desiccation, and metal ion stress, may have an important influence on molecular evolution (such as change GC content, promote nucleotide substitution, and decrease the abundance of small RNAs) and diversification of the plant chloroplast genomes (Fitzgerald et al., 2011; Wang et al., 2011; Ivanova et al., 2017; Gao et al., 2018; Li et al., 2020). In recent years, the use of complete chloroplast genome data for phylogenetic inference has greatly deepened our insight into the evolution of plants at a wide range of taxonomic levels (Park et al., 2017; Wen et al., 2018; Li et al., 2019; Valcárcel and Wen, 2019; Wang L. et al., 2020; Brandrud et al., 2020).
The inheritance of chloroplast genomes is predominantly maternal in angiosperms. However, biparental transmission of chloroplast genome has arisen in multiple lineages of angiosperms (Hu et al., 2008). It has been estimated that approximately 20% of angiosperm species potentially have biparentally inherited chloroplast genomes (Corriveau and Coleman, 1988; Zhang et al., 2003; Zhang and Sodmergen, 2010). Biparental inheritance of chloroplast may have important impact on evolution, such as producing genetic incompatibility to arise in speciation (Greiner et al., 2011). It has also been hypothesized that the nature of chloroplast inheritance may affect its genome stability (Wicke et al., 2011). Although the underlying mechanisms are unknown, structural rearrangements in chloroplast genome in correlation with biparental inheritance had been recognized in various kinds of plant taxa (Jansen and Ruhlman, 2012; Choi et al., 2020).
The evening primrose family, Onagraceae, is composed of about 650 species of herbs, shrubs, and rarely trees distributed worldwide and species-rich in the New World (Raven, 1988). Onagraceae is characterized by flowers with four (or rarely two or five) petals, an inferior ovary, an often dehiscent capsule, and pollen grains held together by viscin threads. The family was sharply defined (Raven, 1964), but with disputed interpretation of subfamily, tribal, and some generic delimitation in its long taxonomic history (Kurabayashi et al., 1962; Raven, 1964; Munz, 1965; Wagner et al., 2007). Several molecular phylogenetic analyses using Sanger’s sequencing method have been conducted to resolve the phylogenetic relationships within Onagraceae (Martin and Dowd, 1986; Crisci et al., 1990; Bult and Zimmer, 1993; Conti et al., 1993; Levin et al., 2003, 2004; Berry et al., 2004; Hoggard et al., 2004; Evans et al., 2005; Ford and Gottlieb, 2007; Xie et al., 2009; Liu et al., 2017). Based on molecular and morphological data, a recent taxonomic monograph by Wagner et al. (2007) included 22 genera in Onagraceae. These genera were further grouped into two subfamilies: subfam. Ludwigioideae W. L. Wagner and Hoch (with only one genus, Ludwigia L.) and subfam. Onagroideae W. L. Wagner and Hoch (with six tribes and 21 genera). Onagraceae contains many popular garden plants including evening primrose (Oenothera L.) and fuchsia (Fuchsia L.). Some species of the family also have medicinal value and are widely used to make oil, spices, and nectar (Chen et al., 2007).
Inheritance of the chloroplast genome in Onagraceae has attracted great attention of botanists (Cleland, 1972; Chiu et al., 1988; Chiu and Sears, 1992; Chiu and Sears, 1993; Sears et al., 1996; Massouh et al., 2016; Sobanski et al., 2019). Both maternal and biparental inheritance of chloroplast genomes has been reported in the family (Wagner et al., 2007). Oenothera subsect. Oenothera are known to have biparentally transmitted chloroplast (Corriveau and Coleman, 1988; Wagner et al., 2007). Whereas, chloroplast genomes from Circaea L. and Fuchsia have been shown to be maternally transmitted (Corriveau and Coleman, 1988; Zhang et al., 2003). Chloroplasts of Epilobium L. were also reported to be mainly maternally transmitted, but very low proportions of paternally transmitted chloroplast were also found (Schmitz and Kowallik, 1986). As mentioned above, biparentally inherited chloroplast genomes of many plant taxa have shown extensive rearrangement of genome structure. Thus, Onagraceae provides an opportunity to better understand differences in the chloroplast genome structure and sequence diversification between the two inheritance types. However, there are still no comparative studies concerning this issue and only a limited number of complete chloroplast genomes have been published to date.
In the present study, we report the complete chloroplast genomes from three genera (Circaea, Chamaenerion Ség.1, and Epilobium) of Onagraceae, among which those of the Circaea are reported for the first time. We hypothesized that the structure and sequence variation of chloroplast genomes in Onagraceae show different structures between biparentally and maternally inherited chloroplast genomes. Thus, we compared the synteny and chloroplast genome structure across the family and investigated their chloroplast genome structure and sequence variation. We also conducted a phylogenetic study to explore the evolutionary trends of chloroplast genome variation and the potential application value of the chloroplast markers across Onagraceae.
Materials and Methods
Taxon Sampling and Next-Generation Sequencing
We sampled 16 accessions representing three genera (Circaea, Chamaenerion, and Epilobium) and 13 species of Onagraceae (Supplementary Table S1). We also retrieved all the (15 samples representing 14 species) published complete chloroplast genome sequences of Onagraceae to date, as well as two samples from Lythraceae (sister family of Onagraceae) from GenBank for phylogenetic analysis. In total, five genera (Circaea, Chamaenerion, Epilobium, Ludwigia, and Oenothera) and 27 species (31 samples) of Onagraceae were included in this study. The taxonomy of Onagraceae at generic and infrageneric level followed Wagner et al. (2007). Our sampling covered both subfamilies (subfam. Ludwigioideae and subfam. Onagroideae) and three of the total six tribes in subfam. Onagroideae. Biparentally inherited chloroplast genomes were known to have occurred in species of Oenothera subsect. Oenothera (Wagner et al., 2007). So, we used chloroplast genome of O. biennis L. as a representative of biparentally inherited chloroplast genome (also reported by Corriveau and Coleman, 1988) to compare with the maternally inherited one from Circaea and Epilobium.
Approximately 50 mg dried leaf tissue was ground for each sample. Total genomic DNA was extracted using the cetyl-trimethylammonium bromide (CTAB; Doyle and Doyle, 1987) method. The quality of DNA was assessed by 0.8% agarose gel electrophoresis, and extracted DNA was sent to Novogene (http://www.novogene.com, China) for short-insert (350 bp) library construction and next-generation sequencing. Paired-end reads of 2 × 150 bp were generated on the Illumina Hiseq 4,000 Genome Analyzer platform. We used the FASTX Toolkit (http://hannonlab.cshl.edu/fastx_toolkit) to filter the raw reads and remove the adaptors and low-quality reads to obtain high-quality data. The BLAT analysis, as implemented in a Python script (Weitemier et al., 2014), was applied to exclude nuclear and mitochondrial reads using a published complete chloroplast genome sequence of Epilobium ulleungensis as the reference (GenBank accession no. MH198310). Subsequently, the putative chloroplast reads were de novo assembled using Geneious v. Prime (Kearse et al., 2012) with a low sensitivity setting. Gaps between contigs were filled by re-mapping the entire reads to both contigs using the FineTuning program in Geneious v. Prime (iterating up to 100 times), as described by He et al. (2019). Contigs were connected into larger contigs by overlapping their terminal sequences using the RepeatFinder option in Geneious v. Prime (Kearse et al., 2012). After building an approximate 130-kb contig (including a complete SSC, a complete IRa, a complete LSC, and a partial IRb region) for each sample, the boundaries of the IR region were determined using the RepeatFinder. The IR region was manually inverted and duplicated to construct the complete chloroplast genome sequence using Geneious v. Prime (Kearse et al., 2012). The correction of the gaps and junctions between IRs and LSC/SSC regions were confirmed by PCR amplifications. The complete chloroplast genome sequences were annotated using the Plastid Genome Annotator (Qu et al., 2019) and checked manually in Geneious v. Prime (Kearse et al., 2012). Illustrations of the newly sequenced chloroplast genome sequences were drawn using the Organellar Genome DRAW tool v. 1.3.1 (Lohse et al., 2013).
Comparative Evaluation of the Chloroplast Genome
The newly sequenced chloroplast genomes were compared with those of the other published Onagraceae species. Amino acid frequency and codon usage were calculated using the Geneious v. Prime (Kearse et al., 2012) and CodonW v. 1.4 (Peden, 1999) software, and the putative RNA editing sites in protein-coding genes were determined by the predictive RNA editor for plant chloroplasts (PREP-cp) suite (Mower, 2009). For the synteny analysis of the Onagraceae chloroplast genome, mVISTA (Frazer et al., 2004) was used in LAGAN and Shuffle-LAGAN mode, with default parameters using Epilobium sikkimense Hausskn. as reference. The contraction and expansion of the IR boundaries between the four main parts of the genome (LSC/IRb/SSC/IRa) were visualized using IRscope (Amiryousefi et al., 2018). We also conducted a sliding window analysis to identify the nucleotide variability (Pi) of the complete chloroplast genomes of the three newly sequenced genera and Oenothera using DnaSP v. 5 (Librado and Rozas, 2009).
The microsatellites were determined by MIcroSAtellite (MISA) (Varshney et al., 2005), with a minimum threshold of seven nucleotides for mononucleotide repeats, four for di-, and 3 each for tri-, tetra-, penta-, and hexanucleotide repeats. The REPuter program (Kurtz et al., 2001) was used to analyze forward (F), reverse (R), complement (C), and palindromic (P) oligonucleotide repeats with a minimum repeat size of 30 bp and similarities of 90%. Furthermore, tandem repeats were evaluated by the Tandem Repeats Finder (Benson, 1999) using default parameters.
Phylogenetic Analysis
The phylogenetic analysis was performed among 31 species of Onagraceae using two Lythraceae samples as outgroups. For phylogenetic tree reconstruction, we removed IRa from the analysis and manually reverted the inverted regions in samples of Oenothera subsect. Oenothera. We also divided the complete chloroplast genome sequences into coding regions (CDs, including protein-coding genes, tRNA genes, and rRNA genes), intergenic spacer regions (IGS), and introns. Each dataset was further divided into LSC, SSC, and IR regions. All the 13 separated and combined datasets (the complete CDs sequence, the complete IGS, the complete intron, the LSC-CDs, the LSC-IGS, the LSC intron, the SSC-CDs, the SSC-IGS, the SSC-intron, the IR-CDs, the IR-IGS, the IR-intron, and the complete chloroplast genome datasets) were then aligned using MAFFT v. 6.833 (Katoh et al., 2005) and manually adjusted by Geneious v. Prime (Kearse et al., 2012). The ambiguous alignments were removed from the datasets using a Python script (He et al., 2019).
We used both the maximum likelihood (ML) and Bayesian inference (BI) methods for phylogenetic reconstruction for each dataset. The ML tree for each dataset was generated by RAxML v.8.1.17 (Stamatakis, 2014) using the GTR + G model as suggested in the user manual. The bootstrap percentages were calculated after 500 replicates.
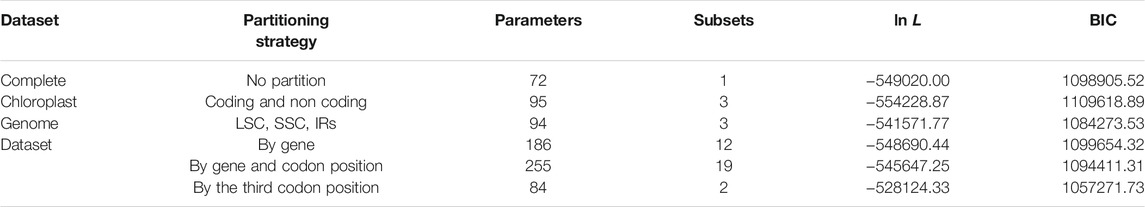
Bayesian inference for each dataset was conducted using MrBayes v3.2.3 (Ronquist and Huelsenbeck, 2003). Substitution models and data partitions of the complete chloroplast genome dataset for the Bayesian analysis were determined by PartitionFinder v2.1.1 (Lanfear et al., 2017). Six partitioning schemes were used for the complete chloroplast genome dataset: 1) no partitions, 2) partitioned by coding and non-coding regions (with the four rRNA genes as the third partition), 3) by LSC, SSC, and IRs, 4) coding region by genes (non-coding region as one partition), 5) coding region by genes and codon positions (non-coding region as one partition), 6) coding region by the third codon position (the first and second codon positions as on partition and the third position as the other partition, non-coding region as another one partition). The best scheme was selected according to the Bayesian information criterion (BIC). Partitioning of other datasets was on the basis of the result of the complete chloroplast genome dataset.
For the Bayesian inference, the default priors in MrBayes were applied for tree search. Two independent Markov chain Monte Carlo (MCMC) chains were created, each with three heated and one cold chain for 2,000,000 generations and sampling trees every 100 generations. The first 25% of the trees were discarded as burn-in, and the remaining trees were used to generate the consensus tree. All the alignments used in this study are available on Zenodo, with the identifier https://doi.org/10.5281/zenodo.5545914.
Results
Chloroplast Genome Assembly, Organization, and Nucleotide Composition Features
For each newly sampled Onagraceae species, approximately 6 Gb clean NGS data were obtained, which means that the whole genomic coverage of our NGS data ranged from ca. 6–30 × (https://cvalues.science.kew.org/). We filtered out 130,748–410,678 chloroplast reads from the samples for de novo assembly. The coverage of the chloroplast genome was from 79 to 271 ×. One to seven large contigs were retained. All the gaps between the de novo contigs were successfully bridged by re-mapping the cleaned reads to both contigs using the FineTuning program in Geneious v. Prime (Kearse et al., 2012) with 100 iterations. The correction of the gaps and junctions between IRs and LSC/SSC regions were confirmed by PCR amplifications. All the newly assembled sequences were deposited in the GenBank under accession numbers of MZ326160 and from MZ353628 to MZ353642 (Supplementary Table S1).
Chloroplast genome sequences of Circaea ranged from 155,817 bp (C. alpina subsp. micrantha (A. K. Skvortsov) Boufford) to 156,024 bp (Circaea alpina subsp. caulescens (Kom.) Tatew.) in size, and the overall GC content varied from 37.7 to 37.8%. For Chamaenerion samples, the complete chloroplast genome sequences ranged from 159,496 bp (C. conspersum (Hausskn.) Kitam.) to 160,416 bp (C. angustifolium subsp. circumvagum (Mosquin) Moldenke), and the overall GC content varied from 38.1 to 38.2%. For Epilobium chloroplast genome, the sizes ranged from 160,748 bp (Epilobium amurense subsp. amurense Hausskn.) to 161,144 bp (E. sikkimense Hausskn.), and the overall GC content varied from 38.1 to 38.2% (Supplementary Table S2).
All the newly assembled chloroplast genome sequences contained a pair of IRs (24,996–27,519 bp) separated by a LSC region (87,569–89,163 bp) and a SSC region (17,157–18,283 bp). The complete chloroplast genomes encoded an identical set of 112 genes, including 78 protein-coding genes, 30 transfer RNAs, and four ribosomal RNAs. Among these, 17 (in Circaea samples) and 18 (in Chamaenerion and Epilobium samples) genes were duplicated in IR, and 18 genes had introns (Figure 1; Table 1, and Supplementary Table S2). Among the 18 intron-containing genes, 16 (10 protein-coding genes and 6 tRNA genes) had one intron and two (ycf3 and clpP) had two introns. However, the two introns in clpP gene are absent in Oenothera sect. Oenothera samples. The longest intron (2,487 bp) was in the trnK gene of Epilobium williamsii P. H. Raven.
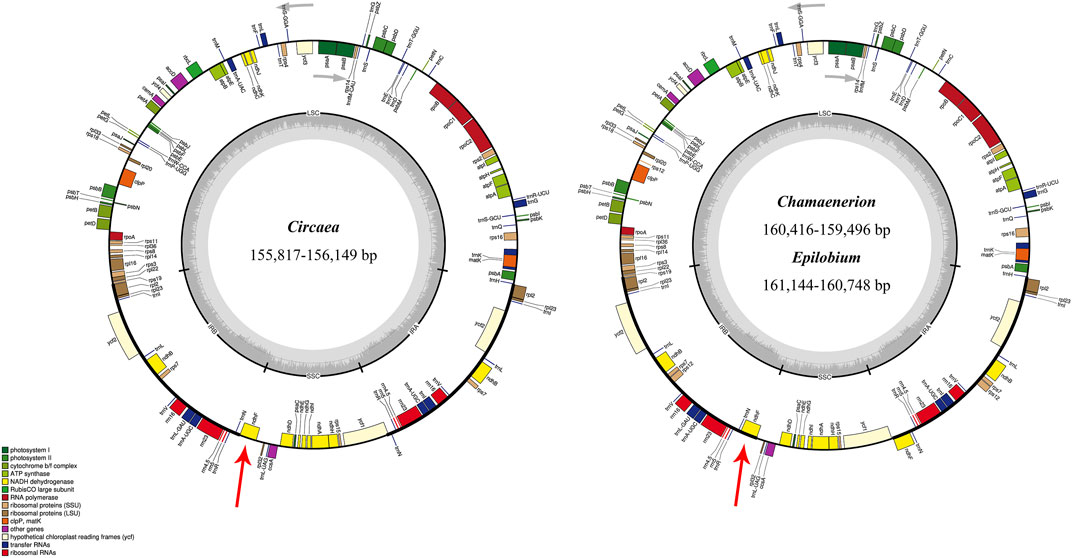
FIGURE 1. Chloroplast genome maps of Chamaenerion, Circaea, and Epilobium sampled in the present study. Thick lines on the outer circle identify inverted repeat regions (IRa and IRb). The innermost track indicates the G + C content. Genes on the outside of the map are transcribed in a clockwise direction, and genes on the inside of the map are transcribed in a counterclockwise direction. IR, inverted repeat; LSC, large single copy; SSC, small single copy. Red arrows showed the different IR-SC boundaries between the two chloroplast genome structures.
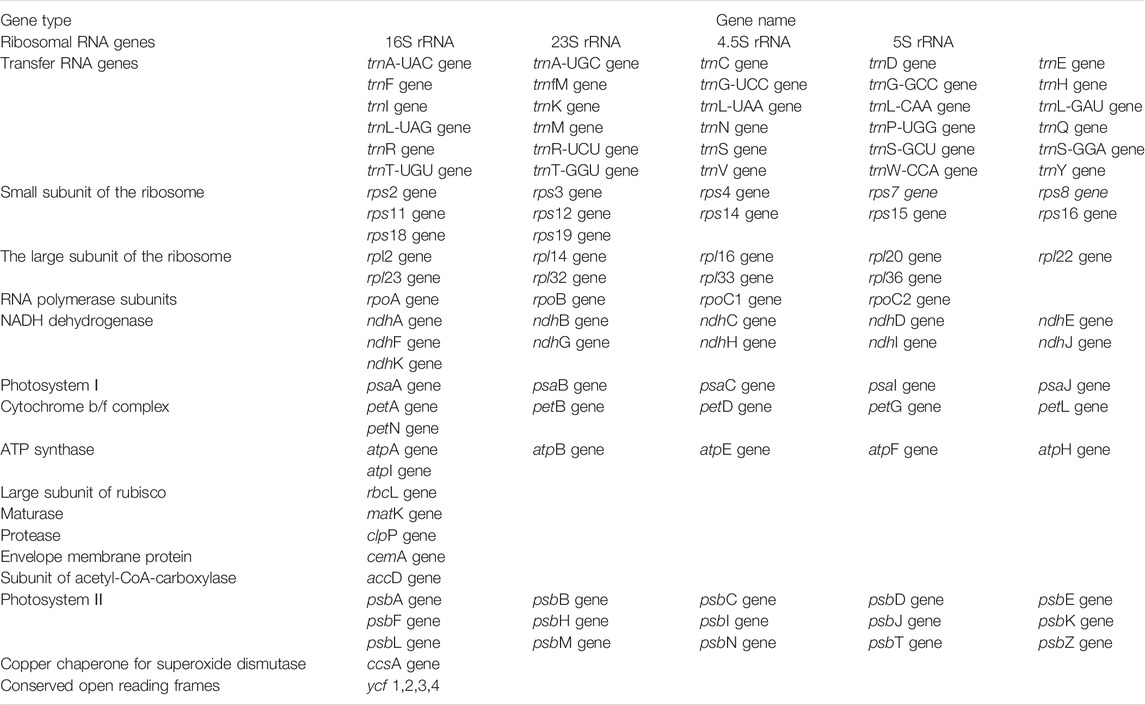
TABLE 1. Genes present in the chloroplast genome of the 16 newly sequenced Epilobium, Circaea, and Chamaenerion samples.
Codon Usage and Amino Acid Frequencies
Relative synonymous codon usage (RSCU) of the chloroplast genome sequences of the newly assembled samples was calculated using all protein-coding genes. Results of amino acid frequency, RSCU, and putative RNA editing sites are shown in Supplementary Figure S1 and Supplementary Tables S3, S4. There were 50 putative RNA editing sites detected in the 18 protein-coding genes of Epilobium, 43 sites detected in 16 protein-coding genes of Circaea, and 48 sites detected in 17 protein-coding genes of Chamaenerion. Among the three genera, the gene with the most RNA editing sites was ndhB (12 sites), and the second was ndhD (5 sites). The most common type of substitution in Epilobium was serine to leucine (26%), followed by proline to leucine (18%). This phenomenon also existed in the other two genera: the Chamaenerion chloroplast genome displayed 31.3% of editing sites substituted from serine to leucine, and 14.6% from proline to leucine; and the Circaea chloroplast genome showed 32.6% of editing sites substituted from serine to leucine, and 18.6% from proline to leucine. Among the 50 recognized RNA editing sites in Epilobium, 35 substitutions occurred at the second nucleotide position and 15 substitutions occurred at the first nucleotide position. Similar results were also detected in the other two genera.
Chloroplast Genome Comparison
To investigate the synteny and structural variation of the chloroplast genomes of Onagraceae, we performed multiple alignments of all the tested samples using mVISTA (Supplementary Figure S2). LAGAN and Shuffle-LAGAN programs were applied for this analysis. Results of generic representatives are shown in Figure 2. When using the LAGAN method, Oenothera subsect. Oenothera samples showed a large area of mismatch in their LSC region due to gene inversion. This inversion occurred between rbcL and trnQ-UUG and was approximately 56 kb in length. Typically, clpP gene has two introns in many angiosperm species. However, these two introns are absent in Oenothera sect. Oenothera samples, but still present in O. curtiflora W. L. Wagner and Hoch (sect. Gaura (L.) W. L. Wagner and Hoch). In addition, compared with other genera, some mismatch regions were found in the IR region of Oenothera, Epilobium, and Chamaenerion, which was caused by expansion of their IR zones by inclusion of the ndhF gene, and rarely other genes (described below).
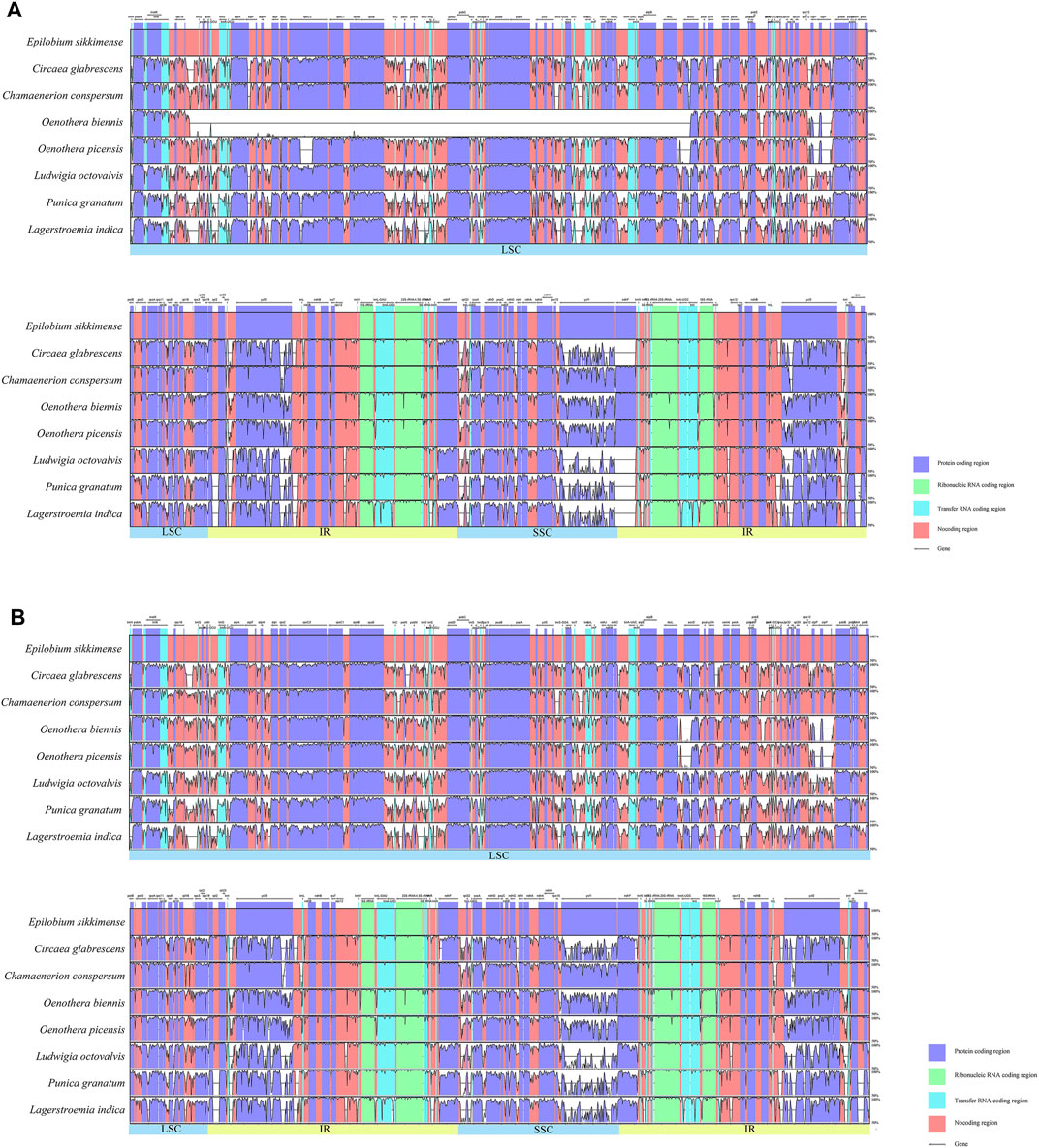
FIGURE 2. Sequence alignment of representative samples from five genera of Onagraceae and two outgroups using the mVISTA program (alignment of all 33 samples are shown in Supplementary Figure S2). A cut-off of 70% similarity was used for the plot, and the Y-scale represents the percentage similarity ranging from 50 to 100%. Blue represents coding regions, and pink represents non-coding regions. (A): LAGAN method, the large empty part of the Oenothera biennis graph is the inverted region; (B): Shuffle LAGAN method.
Subsequently, we compared the IR/SC boundary regions of 31 species of Onagraceae and two species of Lythraceae (Supplementary Figure S3). The early diverged genera of Onagraceae, Ludwigia and Circaea, have 17 genes in the IR region, which is the same with most other angiosperm genera such as Amborella Baill., Caltha L., and Arabidopsis Heynh. (Sato et al., 1999; He et al., 2019). So, the IR region of Ludwigia and Circaea can be considered as the primitive type of the family. Other tested Onagraceae genera showed more or less IR expansion. Almost all the tested samples from Chamaenerion, Epilobium, and Oenothera have 18-gene IR region (with inclusion of ndhF) (Figure 3). Two samples from Oenothera subsect. Munzia (W. Dietr.), O. picensis Phil. and O. villaricae W. Dietr., have 21-gene IR, with additional ccsA, trnL-UAG, rpl32 and ndhF genes.
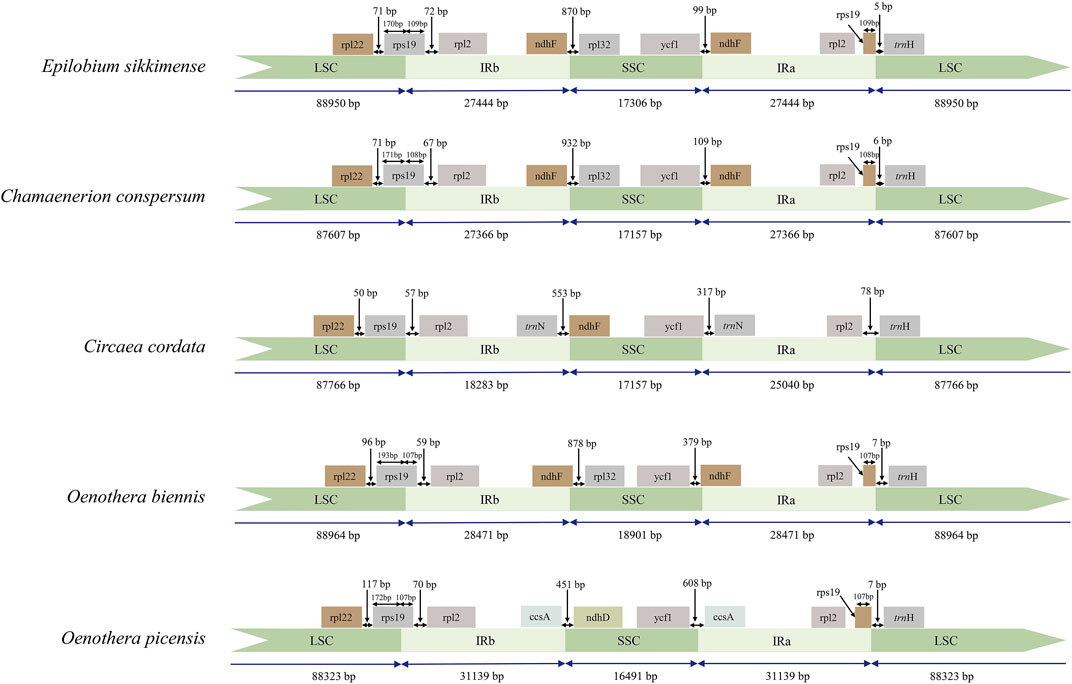
FIGURE 3. Comparison of the LSC, IR, and SSC boundary regions of representative samples the three newly sequenced genera of Onagraceae and Oenothera samples. IR: inverted repeats; LSC: large single copy; SSC: small single copy.
Sliding window analysis (Figure 4) showed that the nuclear variability of the IR region was relatively low in the three newly sequenced Onagraceae genera as well as in published Oenothera samples. Among the tested genera, Oenothera had the highest nucleotide variation. In addition, extremely high variations were discovered at both ends of the inversion in the LSC of the Oenothera chloroplast genome, which may be the main cause of the structural rearrangement of chloroplast genomes in Oenothera subsect. Oenothera.
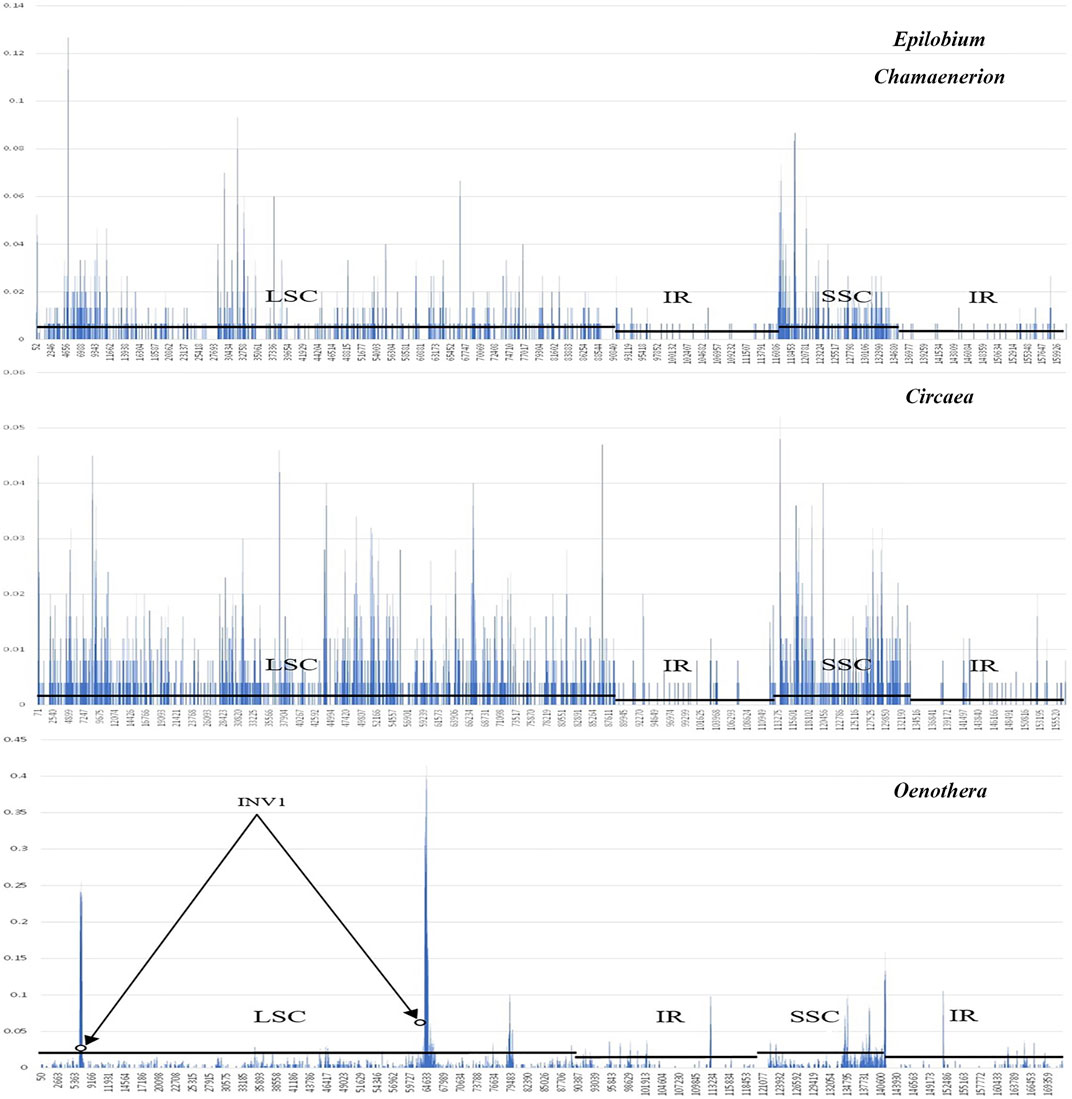
FIGURE 4. A sliding window analysis of the complete chloroplast genomes of Epilobium, Chamaenerion, Circaea, and Oenothera samples showing nulceotide variability (Pi) within each genus. Circles identify the regions bordering inversion sites of Oenothera, and lines parallel to the X-axis identify the positions of LSC, SSC, and IR regions.
SSR and Repeats Analyses
The chloroplast genome sequences are known to be highly conserved. However, chloroplast SSRs have been applied as important phylogenetic markers for unraveling polymorphisms across species and populations in plant molecular studies (Cato and Richardson, 1996; Xu et al., 2002; Wills et al., 2005; Bi et al., 2018). Furthermore, the primers for the chloroplast SSRs are conserved, which may facilitate primer design across species and genera. In this study, we detected 47–90 SSRs from chloroplast genome sequences of the three newly sequenced genera. Those in Circaea had 47–55 SSRs, which was the lowest among the three genera (Table 2). Epilobium and Chamaenerion had a higher number of SSRs, ranging from 76 to 90. The mononucleotide repeat unit (A/T) was the most common type, accounting for 85.6–97.9% of all 16 samples. The dinucleotide repeat unit (AT/TA) was the second most abundant, accounting for 1.7–9.2%. The mononucleotide repeat unit C/G existed in Chamaenerion and Circaea samples and in the chloroplast genome sequence of Epilobium sikkimense. Chamaenerion and Epilobium contained only two types of repeats: mononucleotide and dinucleotide. A trinucleotide repeat unit (AAT/ATT) was found in all samples of Circaea. Tetranucleotide repeats were found in C. alpina subsp. caulescens (AATAT/ATATA) and C. glabrescens (Pamp.) Hand.-Mazz. (AAAGG/AAGGA). Only one hexanucleotide repeat (AAATAT/ATAAAT) was present in C. alpina subsp. caulescens. These SSRs were mainly located in the IGS region and sometimes also occurred in introns and CDs. As expected, most SSRs were detected in the LSC region, followed by the SSC and IR regions.
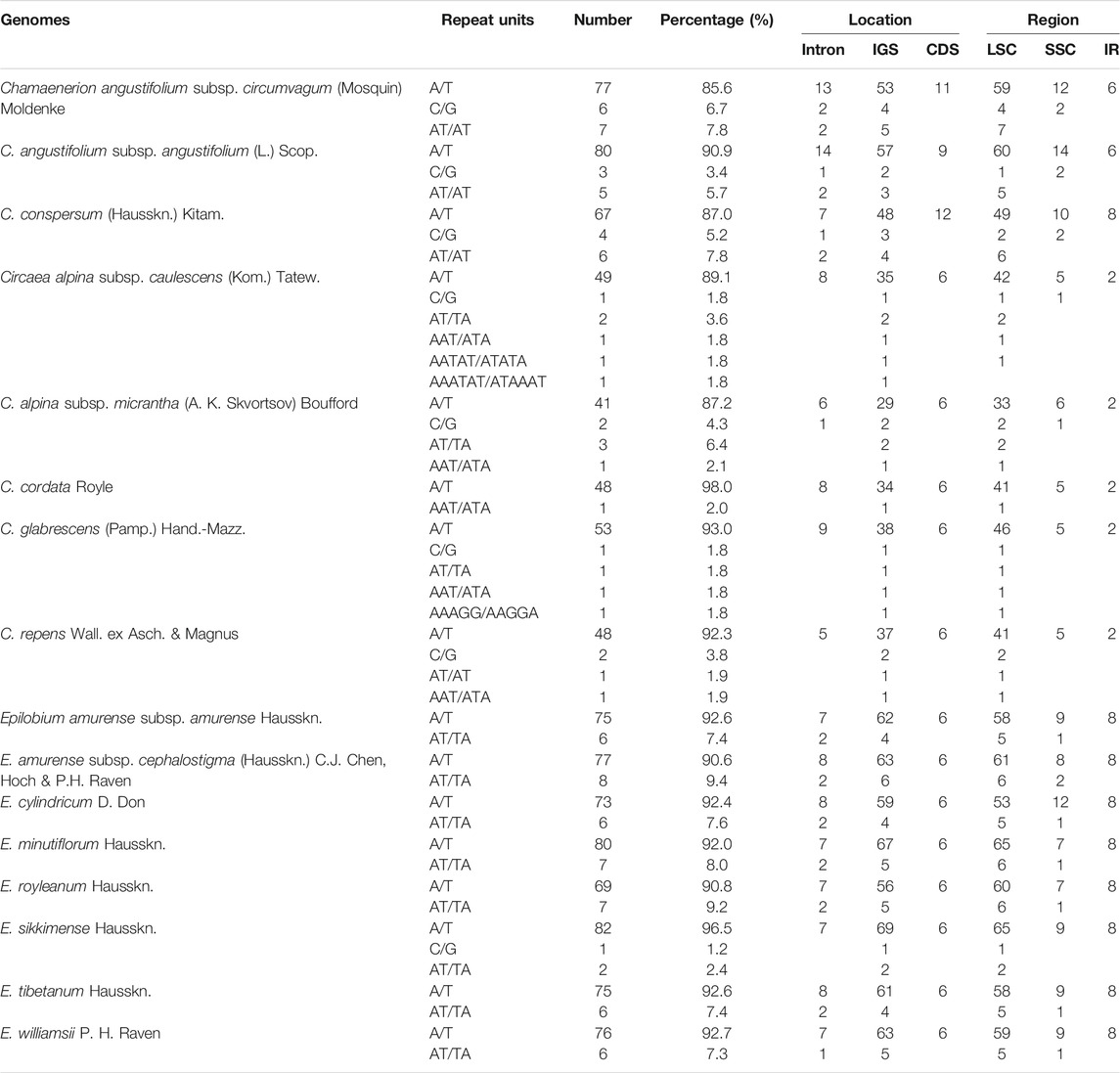
TABLE 2. Simple sequence repeats (SSRs) for the 16 newly sequenced Epilobium, Circaea, and Chamaenerion samples.
In addition to the SSRs, we also explored the role of repeats identified by REPuter (Kurtz et al., 2001). We found a total of 640 repeats in the 16 samples (Figure 5). Only palindromic and forward repeats were detected in Epilobium and Chamaenerion. In addition to these two types of repeat, a complement repeat was detected in Circaea cordata Royle, and a reverse repeat was found in C. alpina subsp. caulescens. The Circaea chloroplast genome sequence had 10–19 forward repeats, whereas Chamaenerion and Epilobium had 30–48 forward repeats. Among all the detected repeats, palindromic repeats accounted for 12.18% and forward repeats accounted for 87.5% of total repeats, whereas complement and reverse repeats only accounted for 0.032%. The repeat length of Circaea and Chamaenerion was shorter, and most repeats were between 30 and 44 bp. The repeat length of Epilobium was longer, at 30–59 bp in most samples. Much longer repeats (over 100 bp) were found in Epilobium and Chamaenerion. The longest repeat was 203 bp in length and was located in the ycf2 gene. The region containing the majority of the repeats was CDs (70%), followed by the IGS (25%), and the intron region (5%). A large number of repeats were found in the ycf2 gene (in CDs), especially in Epilobium and Chamaenerion samples. The presence of those SSRs and repeats demonstrated that the loci were potentially mutation hotspots in the chloroplast genome, and they may play an important role in developing genetic markers for future phylogenetic or population genetic studies.
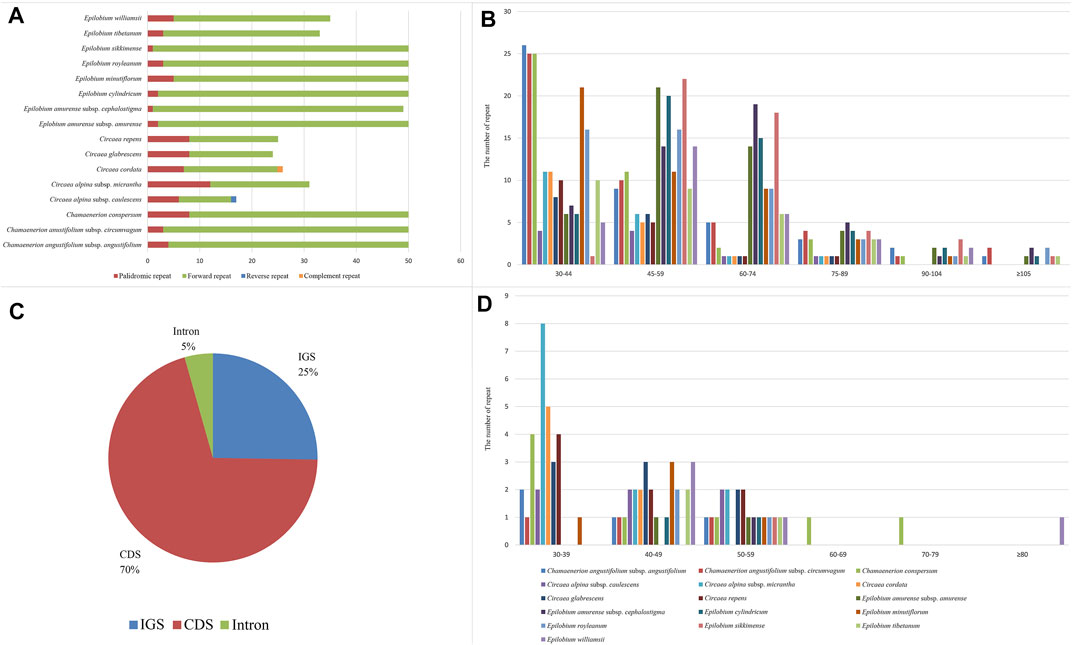
FIGURE 5. Analyses of repeated sequences in 16 newly sequenced chloroplast genomes of Onagraceae. (A): number of four repeat types; (B): frequency of direct repeats by length; (C): location of repeats; (D): frequency of palindromic repeats by length.
Biparentally vs. Maternally Inheritated Chloroplast Genome
Comparing chloroplast genome sequences of Oenothera biennis (biparental transmitted) and Circaea (maternally transmitted), several structural differences are depicted. A large inversion occurs in the LSC regions of O. biennis, whereas chloroplast genome of Circaea lacks it. Two intons in clpP genes (present in most genera of Onagraceae) are absent in the O. biennis. The IR region of the O. biennis chloroplast genome was slightly expanded with the inclusion of ndhF gene (Figures 2, 3, 6). Chloroplasts of Epilobium were also reported to be mainly (but not entirely) maternally inherited (Schmitz and Kowallik, 1986). Chloroplast genomes of Epilobium samples also lack the inversion, and their clpP have both introns, although their IR regions were also slightly expanded.
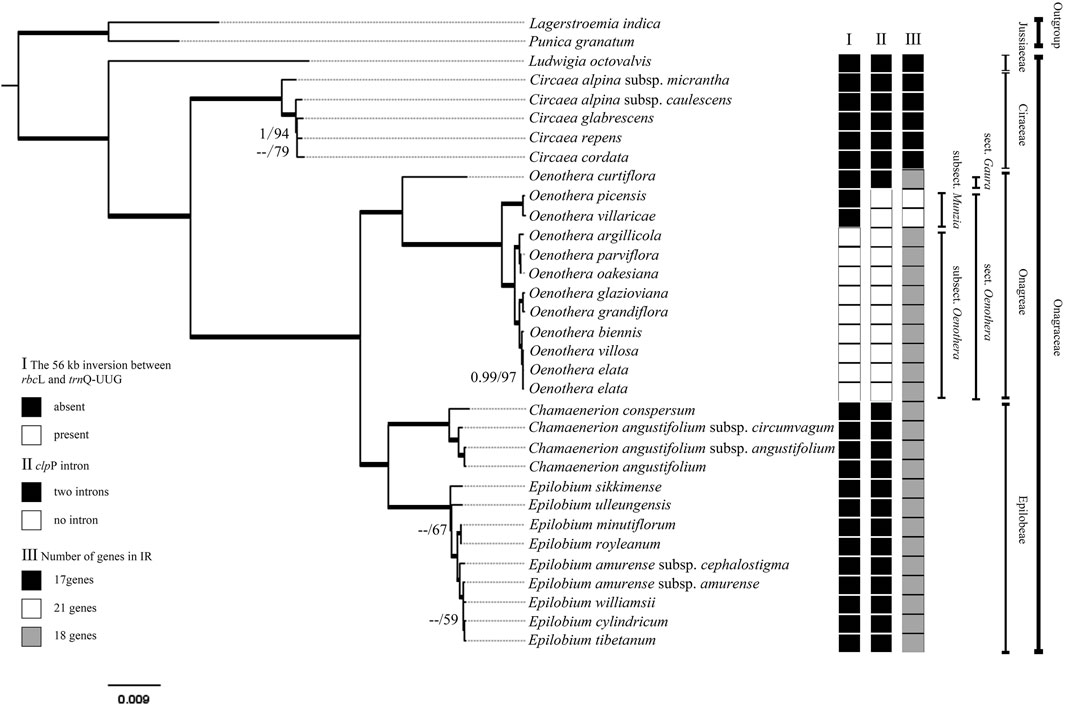
FIGURE 6. Bayesian consensus tree of Onagraceae species inferred from complete chloroplast genome sequences. Maximum likelihood (ML) bootstrap values/Posterior Probability (PP) values are shown at each node. Internal branches that are fully supported by both analyses (with 100 ML bootstrap values and 1 PP values) were thickened. ML bootstrap values < 50 and PP values < 0.95 are shown as --.
Although we cannot concluded that all the members of Oenothera subsect. Oenothera have the biparentally transmitted chloroplast, the chloroplast genome structure, gene content, and the gene arrangement of all the samples from this subsection are quite stable (Figure 6). In contrast, the chloroplast genome of Oenothera sect. Gaura are the similar to those in Chamaenerion and Epilobium rather than to other Oenothera samples. Oenothera subsect. Munzia have clpP without introns, which is similar to subsect. Oenothera, but have much more expanded IR regions (with 21 genes) and no inversion in their chloroplast genomes.
Phylogenetic Analysis
To better accommodate the heterogeneity of the data in the processes of Bayesian analysis, the complete chloroplast dataset was tested by six partitioning treatments. All the partitioning strategies showed similar results and no obvious improvement was observed among them. It seemed that partitioning the coding region by the third codon position obtained a little better result (Table 3). For this reason, we used this partition strategy for the datasets which have coding regions (the complete CDs sequence, the LSC-CDs, the SSC-CDs, the IR-CDs, and the complete chloroplast genome datasets), and GTR + I + G for each partition tested by PartitionFinder were applied for Bayesian analysis. We used GTR + G model tested by PartitionFinder (and no partitioning strategy) for the non-coding datasets (the complete IGS, the complete intron, the LSC-IGS, the LSC intron, the SSC-IGS, the SSC-intron, the IR-IGS, and the IR-intron) for both ML and Bayesian analyses. The complete chloroplast genome dataset (including LSC, SSC, and IR with an aligned length of 126,290 bp) generated a phylogeny (Figure 6), which is consistent with all the 12 separated phylogenies (Supplementary Figure S4).
Phylogenies created from all the datasets using both methods were basically the same, especially for strongly supported clades. Thus in this study, our discussion was on the basis of the phylogeny inferred by the complete chloroplast genome dataset. All the Onagraceae tribes and genera were strongly supported. The genus Ludwigia was shown to be the first diverged genus in the Onagraceae. The phylogenetic relationship within the genus Circaea was not well resolved, whereas the genus Oenothera showed a clear phylogenetic structure. Oenothera curtiflora (sect. Gaura) was revealed to be the first diverged species in the genus. Other Oenothera species formed a strongly supported clade with a long branch. Within this clade, two subsections (subsect. Munzia and subsect. Oenothera) were clearly resolved with high supporting values. The genus Epilobium was also relatively densely sampled; however, this genus was not well resolved in the basal part of the phylogeny.
Discussion
The present study reports the first chloroplast genomes of Circaea. Chloroplast genomes of some species in Chanaenerion and Epilobium were also reported for the first time. We compared the genetic diversity within each genus to obtain insight into the molecular evolution of chloroplast genomes in Onagraceae. Gene content and organization of the Onagraceae chloroplast genome were analyzed to reveal phylogenetic information pertaining to gene rearrangement. Analysis of codon usage by the chloroplast genome can aid to understand the selection pressure on genes and genome structure (Yang et al., 2014).
In the present study, the preference of codons ending with A/T in Onagraceae chloroplast genomes was confirmed (Supplementary Table S4). The same results were also observed in other angiosperm species, such as in Fabaceae, Solanaceae, Asteraceae, and many others (Nie et al., 2014; Mehmood et al., 2020; Somaratne et al., 2020). Our results also show the highest similarities of codon usage among the three newly sequenced genera (Supplementary Figure S1), indicating that these genera may have experienced similar environmental stresses in their evolutionary history. Most SSRs in the newly sequenced Onagraceae chloroplast genomes were found to be mononucleotides (A/T) (Table 2), which is similar to reports in other families of angiosperms. The genus Circaea contained more types of SSRs and repeats than the other two genera (Figure 5; Table 2). This SSR and repeat information may be helpful for the development of molecular markers for population genetics analysis and developing DNA barcodes.
The angiosperm chloroplast genomes are conserved in gene content and organization among different lineages (Palmer, 1985). However, structural variation and gene rearrangements in chloroplast genomes have been discovered in many angiosperm families, such as Anacardiaceae (Wang Y. B. et al., 2020), Apiaceae (Lee et al., 2019), Asteraceae (Walker et al., 2014), Campanulaceae (Haberle et al., 2008), Euporbiaceae (Tangphatsornruang et al., 2011), Geraniaceae (Weng et al., 2014), Fabaceae (Cai et al., 2008), Lentibulariaceae (Silva et al., 2019), Podostemaceae (Bedoya et al., 2019), and Ranunculaceae (Liu et al., 2018; He et al., 2019; Zhai et al., 2019). In Onagraceae, our results show the similarities in gene content and organization in chloroplast genome among almost all the sampled genera and species, with the exception of Oenothera subsect. Oenothera species that contain a large inversion (ca. 56 kb) in the LSC region (Supplementary Figure S2). This large gene inversion had been reported previously by Greiner et al. (2008) and is clearly a derived character (synapomorphy) for subsect. Oenothera (Figure 6). Extremely high nucleotide variations occur at both ends of this inversion, which may be the direct cause of this inversion.
The phylogenetic analysis in this study also clearly demonstrated the evolutionary trends of the other two structural variations (IR expansion and intron loss in clpP) of the chloroplast genome in Onagraceae. Previous studies have shown that expansion/contraction of the IR region is common in angiosperm chloroplast genomes and is the major cause of length variation in chloroplast genomes (Goulding et al., 1996; Kim and Lee, 2004). IR expansion that results in the duplication of genes has been reported in various plant taxa (Chumley et al., 2006; Lee et al., 2007; Park et al., 2017; Liu et al., 2018). Typically, there are 17 genes in the IR region in a wide range of angiosperm taxa (He et al., 2019). In Onagraceae, the early diverged genera, Ludwigia and Circaea, have 17 genes in their IR region, which may represent primitive state of this character in the family. For the other samples, two kinds of IR expansion were discovered. Chloroplast genomes of Chamaenerion, Epilobium, and Oenothera sect. Gaura have 18-gene IR regions, whereas, Oenothera subsect. Munzia have 21-gene IR regions. From our phylogenetic analysis, the 18-gene IR region can be seen as a derived state from the 17-gene IR regions, and the 21-gene IR region maybe further derived from the 18-gene IR region (Figure 6). The IR regions in Onagraceae seem to evolve toward gradual expansion, and no IR contraction was detected in the family by our analysis. In addition to inversion and IR expansion, another derived character, i.e., introns loss in clpP (Figure 6), is present in Oenothera sect. Oenothera, but not in sect. Gaura and other genera. In addition, the occurrence order of the three structural variations can also be inferred by our phylogenetic analysis. The IR expansion (from 17 genes to 18 genes) in Oenothera, Chamaenerion, and Epilobium, happened before the loss of clpP introns in Oenothera sect. Oenothera, and then followed by the acquisition of the large inversion in subsect. Oenothera.
Oenothera subsect. Oenothera seemed to be a very distinctive group carrying almost all specialized chloroplast genome variation in Onagraceae. Species of this subsection are not only known to have biparentally inherited chloroplast genomes but also known to have permanent translocation heterozygosity (PTH), a specialized system in which all seven pairs of chromosomes exchange their arms during meiosis (Cleland, 1972; Raven, 1979; Harte, 1994; Dietrich, 1997; Wagner et al., 2007). In our chloroplast genome analysis, three derived characters, presence of a large inversion, intron loss in clpP, and 18-gene IR, are concentrated in Oenothera subsect. Oenothera. Among them, presence of inversion is only found in this subsection. Although the presence of inversion and biparental transmission of the chloroplast genome are only possessed by Oenothera subsect. Oenothera, we still cannot tell whether biparental transmission has triggered the large inversion or vice versa, because there are many chloroplast genomes with inversions in other plant taxa (such as in Ranunculaceae, He et al., 2019) that do not have biparental plastid transmission.
The phylogenetic relationship resolved in this study is basically consistent with that reported in previous studies (Levin et al., 2003; Levin et al., 2004). However, phylogeny inferred from the complete chloroplast genome sequences was better resolved and better supported statistically than from previous studies using Sanger’s sequencing method (Bult and Zimmer, 1993; Conti et al., 1993; Levin et al., 2003; Levin et al., 2004; Sytsma et al., 2004), demonstrating that chloroplast genome sequences may be a good molecular marker for resolving phylogeny of Onagraceae at generic level. Within each genus, species phylogeny was better resolved in Oenothera than in Circaea and Epilobium, which is due to the higher level of variation in Oenothera chloroplast genome (Figure 6). This result indicates that the chloroplast genome sequences can be applied for inferring phylogenetic relationship of Oenothera at sectional or even species level. However, Oenothera has 18 sections (Wagner et al., 2007) and the chloroplast genome from only two sections have been reported. Further studies are needed to be done in the future because it is possible that the other unsampled sections might have their own distinguishing characteristics in the chloroplast genome.
Conclusion
The complete chloroplast genome sequences of 16 samples representing 13 species in Circaea, Chamaenerion, and Epilobium (Onagraceae) were assembled in this study. We compared chloroplast genomes across the Onagraceae samples and obtained comprehensive molecular information including nucleotide content, codon usage, RNA editing sites, structural variation, and simple sequence repeats (SSRs) through bioinformatic analyses. Phylogeny of Onagraceae was inferred using maximum-likelihood (ML) and Bayesian inference (BI) methods to understand generic and specific relationships. The results of the present study showed potential values of the complete chloroplast genome sequences in inferring phylogeny of the family and may provide powerful genetic resources for future studies.
Data Availability Statement
The data presented in the study are deposited in the GenBank repository, accession number MZ326160, and from MZ353628 to MZ353642.
Author Contributions
Investigation and writing: YL; formal analysis: JH; data curation: RL and JX; investigation and resources: WL, MY, and LP; conceptualization, supervision, and funding acquisition: JC and JL; writing review and editing, project administration, and supervision: LX. All authors read and agreed to the published version of the article.
Funding
This study was supported by the National Natural Science Foundation of China (grant number 31670207).
Conflict of Interests
Author LP was employed by the company Beijing Forestry University Forest Science Co. Ltd.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2021.730495/full#supplementary-material
Footnotes
1The genus name Chamaenerion is accepted in this study according to Sennikov (2011).
References
Ahmed, I., Biggs, P. J., Matthews, P. J., Collins, L. J., Hendy, M. D., and Lockhart, P. J. (2012). Mutational Dynamics of Aroid Chloroplast Genomes. Genome Biol. Evol. 4, 1316–1323. doi:10.1093/gbe/evs110
Amiryousefi, A., Hyvönen, J., and Poczai, P. (2018). IRscope: an Online Program to Visualize the Junction Sites of Chloroplast Genomes. Bioinformatics. 34, 3030–3031. doi:10.1093/bioinformatics/bty220
Bedoya, A. M., Ruhfel, B. R., Philbrick, C. T., Madriñán, S., Bove, C. P., Mesterházy, A., et al. (2019). Plastid Genomes of Five Species of Riverweeds (Podostemaceae): Structural Organization and Comparative Analysis in Malpighiales. Front. Plant Sci. 10, 1035. doi:10.3389/fpls.2019.01035
Benson, G. (1999). Tandem Repeats Finder: a Program to Analyze DNA Sequences. Nucleic Acids Res. 27, 573–580. doi:10.1093/nar/27.2.573
Berry, P. E., Hahn, W. J., Sytsma, K. J., Hall, J. C., and Mast, A. (2004). Phylogenetic Relationships and Biogeography of Fuchsia (Onagraceae) Based on Noncoding Nuclear and Chloroplast DNA Data. Am. J. Bot. 91 (4), 601–614. doi:10.3732/ajb.91.4.601
Bi, Y., Zhang, M. F., Xue, J., Dong, R., Du, Y. P., and Zhang, X. H. (2018). Chloroplast Genomic Resources for Phylogeny and DNA Barcoding: a Case Study on Fritillaria. Sci. Repsci. Rep. 8 (1), 1184–1212. doi:10.1038/s41598-018-19591-9
Brandrud, M. K., Baar, J., Lorenzo, M. T., Athanasiadis, A., Bateman, R. M., Chase, M. W., et al. (2020). Phylogenomic Relationships of Diploids and the Origins of Allotetraploids in Dactylorhiza (Orchidaceae). Syst. Biol. 69, 91–109. doi:10.1093/sysbio/syz035
Bult, C. J., and Zimmer, E. A. (1993). Nuclear Ribosomal RNA Sequences for Inferring Tribal Relationships Within Onagraceae. Syst. Bot. 18, 48–63. doi:10.2307/2419787
Cai, Z., Guisinger, M., Kim, H.-G., Ruck, E., Blazier, J. C., McMurtry, V., et al. (2008). Extensive Reorganization of the Plastid Genome of Trifolium Subterraneum (Fabaceae) Is Associated With Numerous Repeated Sequences and Novel DNA Insertions. J. Mol. Evol. 67, 696–704. doi:10.1007/s00239-008-9180-7
Cato, S. A., and Richardson, T. E. (1996). Inter- and Intraspecific Polymorphism at Chloroplast SSR Loci and the Inheritance of Plastids in Pinus Radiata D. Don. Theoret. Appl. Genet. 93 (4), 587–592. doi:10.1007/bf00417952
Chen, J., Hoch, P. C., Raven, P. H., Boufford, D. E., and Wagner, W. L. (2007). ““Onagraceae,” in Flora of China,”. Editors Z. Y. Wu, and P. Raven (Beijing: Missouri Botanical Garden Press), 13, 400–427.
Chiu, W.-L., and Sears, B. B. (1992). Electron Microscopic Localization of Replication Origins in Oenothera Chloroplast DNA. Mol. Gen. Genet. 232, 33–39. doi:10.1007/BF00299134
Chiu, W.-L., Stubbe, W., and Sears, B. B. (1988). Plastid Inheritance in Oenothera: Organelle Genome Modifies the Extent of Biparental Plastid Transmission. Curr. Genet. 13, 181–189. doi:10.1007/BF00365653
Chiu, W. L., and Sears, B. B. (1993). Plastome-Genome Interactions Affect Plastid Transmission in Oenothera. Genetics. 133, 989–997. doi:10.1093/genetics/133.4.989
Choi, J. W., Graf, L., Peters, A. F., Cock, J. M., Nishitsuji, K., Arimoto, A., et al. (2020). Organelle Inheritance and Genome Architecture Variation in Isogamous Brown Algae. Sci. Rep. 10 (1), 1–12. doi:10.1038/s41598-020-58817-7
Chumley, T. W., Palmer, J. D., Mower, J. P., Fourcade, H. M., Calie, P. J., Boore, J. L., et al. (2006). The Complete Chloroplast Genome Sequence of Pelargonium × Hortorum: Organization and Evolution of the Largest and Most Highly Rearranged Chloroplast Genome of Land Plants. Mol. Biol. Evol. 23, 2175–2190. doi:10.1093/molbev/msl089
Conti, E., Fischbach, A., and Sytsma, K. J. (1993). Tribal Relationships in Onagraceae: Implications From rbcL Sequence Data. Ann. Mo. Bot. Garden. 80, 672–685. doi:10.2307/2399853
Corriveau, J. L., and Coleman, A. W. (1988). Rapid Screening Method to Detect Potential Biparental Inheritance of Plastid Dna and Results for over 200 Angiosperm Species. Am. J. Bot. 75, 1443–1458. doi:10.1002/j.1537-2197.1988.tb11219.x
Crisci, J. V., Zimmer, E. A., Hoch, P. C., Johnson, G. B., Mudd, C., and Pan, N. S. (1990). Phylogenetic Implications of Ribosomal DNA Restriction Site Variation in the Plant Family Onagraceae. Ann. Mo. Bot. Garden. 77, 523–538. doi:10.2307/2399516
Daniell, H., Lin, C. S., Yu, M., and Chang, W. J. (2016). Chloroplast Genomes: Diversity, Evolution, and Applications in Genetic Engineering. Genome Biol. 17, 134–229. doi:10.1186/s13059-016-1004-2
Dietrich, W. (1977). The South American Species of Oenothera Sect. Oenothera (Raimannia, Renneria; Onagraceae). Ann. Mo. Bot. Garden. 64, 425–626. doi:10.2307/2395257
Doyle, J. J., and Doyle, J. L. (1987). A Rapid DNA Isolation Procedure for Small Quantities of Fresh Leaf Tissue. Phytochem. Bull. 19, 11–15.
Evans, M. E. K., Hearn, D. J., Hahn, W. J., Spangle, J. M., and Venable, D. L. (2005). Climate and Life-History Evolution in Evening Primroses (Oenothera, Onagraceae): a Phylogenetic Comparative Analysis. Evol. 59, 1914–1927. doi:10.1111/j.0014-3820.2005.tb01061.x10.1554/04-708.1
Fitzgerald, T. L., Shapter, F. M., McDonald, S., Waters, D. L. E., Chivers, I. H., Drenth, A., et al. (2011). Genome Diversity in Wild Grasses Under Environmental Stress. Proc. Natl. Acad. Sci. 108 (52), 21140–21145. doi:10.1073/pnas.1115203108
Ford, V. S., and Gottlieb, L. D. (2007). Tribal Relationships within Onagraceae Inferred From PgiC Sequences. Syst. Bot. 32 (2), 348–356. doi:10.1600/036364407781179725
Frazer, K. A., Pachter, L., Poliakov, A., Rubin, E. M., and Dubchak, I. (2004). VISTA: Computational Tools for Comparative Genomics. Nucleic Acids Res. 32, W273–W279. doi:10.1093/nar/gkh458
Gao, R., Wang, W., Huang, Q., Fan, R., Wang, X., Feng, P., et al. (2018). Complete Chloroplast Genome Sequence of Dryopteris Fragrans (L.) Schott and the Repeat Structures Against the Thermal Environment. Sci. Rep. 8 (1), 1–11. doi:10.1038/s41598-018-35061-8
Goulding, S. E., Wolfe, K. H., Olmstead, R. G., and Morden, C. W. (1996). Ebb and Flow of the Chloroplast Inverted Repeat. Mol. Gen. Genet. 252, 195–206. doi:10.1007/BF02173220
Greiner, S., Rauwolf, U., Meurer, J., and Herrmann, R. G. (2011). The Role of Plastids in Plant Speciation. Mol. Ecol. 20 (4), 671–691. doi:10.1111/j.1365-294X.2010.04984.x
Greiner, S., Wang, X., Rauwolf, U., Silber, M. V., Mayer, K., Meurer, J., et al. (2008). The Complete Nucleotide Sequences of the Five Genetically Distinct Plastid Genomes of Oenothera , Subsection Oenothera : I. Sequence Evaluation and Plastome Evolution †. Nucleic Acids Res. 36 (7), 2366–2378. doi:10.1093/nar/gkn081
Haberle, R. C., Fourcade, H. M., Boore, J. L., and Jansen, R. K. (2008). Extensive Rearrangements in the Chloroplast Genome of Trachelium Caeruleum Are Associated With Repeats and tRNA Genes. J. Mol. Evol. 66, 350–361. doi:10.1007/s00239-008-9086-4
He, J., Yao, M., Lyu, R.-D., Lin, L.-L., Liu, H.-J., Pei, L.-Y., et al. (2019). Structural Variation of the Complete Chloroplast Genome and Plastid Phylogenomics of the Genus Asteropyrum (Ranunculaceae). Sci. Rep. 9, 1–13. doi:10.1038/s41598-019-51601-2
Hoggard, G. D., Kores, P. J., Molvray, M., and Hoggard, R. K. (2004). The Phylogeny of Gaura (Onagraceae) Based on ITS, ETS, Andtrn L- F Sequence Data. Am. J. Bot. 91 (1), 139–148. doi:10.3732/ajb.91.1.139
Hu, Y., Zhang, Q., Rao, G., and Sodmergen, (2008). Occurrence of Plastids in the Sperm Cells of Caprifoliaceae: Biparental Plastid Inheritance in Angiosperms Is Unilaterally Derived From Maternal Inheritance. Plant Cell Physiol. 49, 958–968. doi:10.1093/pcp/pcn069
Ivanova, Z., Sablok, G., Daskalova, E., Zahmanova, G., Apostolova, E., Yahubyan, G., et al. (2017). Chloroplast Genome Analysis of Resurrection Tertiary Relict Haberlea Rhodopensis Highlights Genes Important for Desiccation Stress Response. Front. Plant Sci. 8, 204. doi:10.3389/fpls.2017.00204
Jansen, R. K., and Ruhlman, T. A. (2012). “Plastid Genomes of Seed Plants,” in Genomics of Chloroplasts and Mitochondria (Dordrecht: Springer), 103–126. doi:10.1007/978-94-007-2920-9_5
Katoh, K., Kuma, K. I., Toh, H., and Miyata, T. (2005). MAFFT Version 5: Improvement in Accuracy of Multiple Sequence Alignment. Nucleic Acids Res. 33 (2), 511–518. doi:10.1093/nar/gki198
Kearse, M., Moir, R., Wilson, A., Stones-Havas, S., Cheung, M., Sturrock, S., et al. (2012). Geneious Basic: an Integrated and Extendable Desktop Software Platform for the Organization and Analysis of Sequence Data. Bioinformatics. 28, 1647–1649. doi:10.1093/bioinformatics/bts199
Kim, K.-J., and Lee, H. L. (2004). Complete Chloroplast Genome Sequences From Korean Ginseng (Panax Schinseng Nees) and Comparative Analysis of Sequence Evolution Among 17 Vascular Plants. DNA Res. 11, 247–261. doi:10.1093/dnares/11.4.247
Kurabayashi, M., Lewis, H., and Raven, P. H. (1962). A Comparative Study of Mitosis in the Onagraceae. Am. J. Bot. 49, 1003–1026. doi:10.1002/j.1537-2197.1962.tb15040.x
Kurtz, S., Choudhuri, J. V., Ohlebusch, E., Schleiermacher, C., Stoye, J., and Giegerich, R. (2001). REPuter: the Manifold Applications of Repeat Analysis on a Genomic Scale. Nucleic Acids Res. 29, 463–4642. doi:10.1093/nar/29.22.4633
Lanfear, R., Frandsen, P. B., Wright, A. M., Senfeld, T., and Calcott, B. (2017). PartitionFinder 2: New Methods for Selecting Partitioned Models of Evolution for Molecular and Morphological Phylogenetic Analyses. Mol. Biol. Evol. 34, msw260–773. doi:10.1093/molbev/msw260
Lee, H.-L., Jansen, R. K., Chumley, T. W., and Kim, K.-J. (2007). Gene Relocations within Chloroplast Genomes of Jasminum and Menodora (Oleaceae) Are Due to Multiple, Overlapping Inversions. Mol. Biol. Evol. 24, 1161–1180. doi:10.1093/molbev/msm036
Lee, H. O., Joh, H. J., Kim, K., Lee, S.-C., Kim, N.-H., Park, J. Y., et al. (2019). Dynamic Chloroplast Genome Rearrangement and DNA Barcoding for Three Apiaceae Species Known as the Medicinal Herb "Bang-Poong". Int. J. Mol. Sci. 20, 2196. doi:10.3390/ijms20092196
Levin, R. A., Wagner, W. L., Hoch, P. C., Hahn, W. J., Rodriguez, A., Baum, D. A., et al. (2004). Paraphyly in Tribe Onagreae: Insights Into Phylogenetic Relationships of Onagraceae Based on Nuclear and Chloroplast Sequence Data. Syst. Bot. 29, 147–164. doi:10.1600/036364404772974293
Levin, R. A., Wagner, W. L., Hoch, P. C., Nepokroeff, M., Pires, J. C., Zimmer, E. A., et al. (2003). Family‐Level Relationships of Onagraceae Based on Chloroplast Rbc L and Ndh F Data. Am. J. Bot. 90, 107–115. doi:10.3732/ajb.90.1.107
Li, H.-T., Yi, T.-S., Gao, L.-M., Ma, P.-F., Zhang, T., Yang, J.-B., et al. (2019). Origin of Angiosperms and the Puzzle of the Jurassic gap. Nat. Plants. 5, 461–470. doi:10.1038/s41477-019-0421-0
Li, L. L., Li, A. R., Guo, Y. F., Li, G. L., Xiao, W. J., and Zeng, Y. L. (2020). Effects of Titanium Ion Implantation on Chloroplast DNA, Chlorophyll Conten and Photosynthetic Parameters of Camellia Oleifera. J. South. Agricult. 51, 2738–2746. (in Chinese with English abstract). doi:10.3969/j.issn.2095-1191.2020.11.017
Librado, P., and Rozas, J. (2009). DnaSP V5: a Software for Comprehensive Analysis of DNA Polymorphism Data. Bioinformatics. 25, 1451–1452. doi:10.1093/bioinformatics/btp187
Liu, H., He, J., Ding, C., Lyu, R., Pei, L., Cheng, J., et al. (2018). Comparative Analysis of Complete Chloroplast Genomes of Anemoclema, Anemone, Pulsatilla, and Hepatica Revealing Structural Variations Among Genera in Tribe Anemoneae (Ranunculaceae). Front. Plant Sci. 9, 1097. doi:10.3389/fpls.2018.01097
Liu, S.-H., Hoch, P. C., Diazgranados, M., Raven, P. H., and Barber, J. C. (2017). Multi-locus Phylogeny of Ludwigia (Onagraceae): Insights on Infra-Generic Relationships and the Current Classification of the Genus. Taxon. 66, 1112–1127. doi:10.12705/665.7
Lohse, M., Drechsel, O., Kahlau, S., and Bock, R. (2013). OrganellarGenomeDRAW-a Suite of Tools for Generating Physical Maps of Plastid and Mitochondrial Genomes and Visualizing Expression Data Sets. Nucleic Acids Res. 41, W575–W581. doi:10.1093/nar/gkt289
Martin, P. G., and Dowd, J. M. (1986). Phylogenetic Studies Using Protein Sequences Within the Order Myrtales. Ann. Mo. Bot. Garden. 73, 442–448. doi:10.2307/2399122
Massouh, A., Schubert, J., Yaneva-Roder, L., Ulbricht-Jones, E. S., Zupok, A., Johnson, M. T. J., et al. (2016). Spontaneous Chloroplast Mutants Mostly Occur by Replication Slippage and Show a Biased Pattern in the Plastome of Oenothera. Plant Cell. 28, 911–929. doi:10.1105/tpc.15.00879
Mehmood, F., Abdullah, I., Shahzadi, I., Ahmed, I., Waheed, M. T., and Mirza, B. (2020). Characterization of Withania Somnifera Chloroplast Genome and its Comparison With Other Selected Species of Solanaceae. Genomics. 112, 1522–1530. doi:10.1016/j.ygeno.2019.08.024
Mower, J. P. (2009). The PREP Suite: Predictive RNA Editors for Plant Mitochondrial Genes, Chloroplast Genes and User-Defined Alignments. Nucleic Acids Res. 37, W253–W259. doi:10.1093/nar/gkp337
Munz, P. A. (1965). ““Onagraceae,” in North American Flora Series Ⅱ, Part 5,”. Editor P. A. Munz (New York: The New York Botanical Garden Press), 1–278.
Nie, X., Deng, P., Feng, K., Liu, P., Du, X., You, F. M., et al. (2014). Comparative Analysis of Codon Usage Patterns in Chloroplast Genomes of the Asteraceae Family. Plant Mol. Biol. Rep. 32 (4), 828–840. doi:10.1007/s11105-013-0691-z
Palmer, J. D. (1985). Comparative Organization of Chloroplast Genomes. Annu. Rev. Genet. 19, 325–354. doi:10.1146/annurev.ge.19.120185.001545
Park, I., Kim, W.-j., Yang, S., Yeo, S.-M., Li, H., and Moon, B. C. (2017). The Complete Chloroplast Genome Sequence of Aconitum Coreanum and Aconitum Carmichaelii and Comparative Analysis With Other Aconitum Species. PLoS One. 12, e0184257. doi:10.1371/journal.pone.0184257
Qu, X.-J., Moore, M. J., Li, D.-Z., and Yi, T.-S. (2019). PGA: a Software Package for Rapid, Accurate, and Flexible Batch Annotation of Plastomes. Plant Methods. 15, 1–12. doi:10.1186/s13007-019-0435-7
Raven, P. H. (1979). A Survey of Reproductive Biology in Onagraceae. New Zealand J. Bot. 17, 575–593. doi:10.1080/0028825X.1979.10432572
Raven, P. H. (1988). “Onagraceae as a Model of Plant Evolution,” in ”Plant Evolutionary Biology. Editors L. D. Gottlieb, and S. K. Jain (London: Chapman & Hall), 85–107. doi:10.1007/978-94-009-1207-6_4
Raven, P. H. (1964). The Generic Subdivision of Onagraceae, Tribe Onagreae. Brittonia. 16, 276–288. doi:10.2307/2805062
Ravi, V., Khurana, J. P., Tyagi, A. K., and Khurana, P. (2008). An Update on Chloroplast Genomes. Plant Syst. Evol. 271, 101–122. doi:10.1007/s00606-007-0608-0
Ronquist, F., and Huelsenbeck, J. P. (2003). MrBayes 3: Bayesian Phylogenetic Inference Under Mixed Models. Bioinformatics. 19, 1572–1574. doi:10.1093/bioinformatics/btg180
Sato, S., Nakamura, Y., Kaneko, T., Asamizu, E., and Tabata, S. (1999). Complete Structure of the Chloroplast Genome of Arabidopsis thaliana. DNA Res. 6 (5), 283–290. doi:10.1093/dnares/6.5.283
Schmitz, U. K., and Kowallik, K.-V. (1986). Plastid Inheritance in Epilobium. Curr. Genet. 11 (1), 1–5. doi:10.1007/BF00389419
Sears, B. B., Stoike, L. L., and Chiu, W. L. (1996). Proliferation of Direct Repeats Near the Oenothera Chloroplast DNA Origin of Replication. Mol. Biol. Evol. 13, 850–863. doi:10.1093/oxfordjournals.molbev.a025645
Sennikov, A. N. (2011). Chamerion or Chamaenerion (Onagraceae )? The Old story in New Words. Taxon. 60 (5), 1485–1488. doi:10.1002/tax.605028
Silva, S. R., Pinheiro, D. G., Penha, H. A., Płachno, B. J., Michael, T. P., Meer, E. J., et al. (2019). Intraspecific Variation within the Utricularia Amethystina Species Morphotypes Based on Chloroplast Genomes. Int. J. Mol. Sci. 20, 6130. doi:10.3390/ijms20246130
Sobanski, J., Giavalisco, P., Fischer, A., Kreiner, J. M., Walther, D., Schöttler, M. A., et al. (2019). Chloroplast Competition Is Controlled by Lipid Biosynthesis in Evening Primroses. Proc. Natl. Acad. Sci. USA. 116, 5665–5674. doi:10.1073/pnas.1811661116
Somaratne, Y., Guan, D.-L., Wang, W.-Q., Zhao, L., and Xu, S.-Q. (2020). The Complete Chloroplast Genomes of Two Lespedeza Species: Insights into Codon Usage Bias, rNA Editing Sites, and Phylogenetic Relationships in Desmodieae (Fabaceae: Papilionoideae). Plants. 9 (1), 51. doi:10.3390/plants9010051
Stamatakis, A. (2014). RAxML Version 8: a Tool for Phylogenetic Analysis and post-analysis of Large Phylogenies. Bioinformatics. 30, 1312–1313. doi:10.1093/bioinformatics/btu033
Sytsma, K. J., Litt, A., Zjhra, M. L., Chris Pires, J., Nepokroeff, M., Conti, E., et al. (2004). Clades, Clocks, and Continents: Historical and Biogeographical Analysis of Myrtaceae, Vochysiaceae, and Relatives in the Southern Hemisphere. Int. J. Plant Sci. 165, S85–S105. doi:10.1086/421066
Tangphatsornruang, S., Uthaipaisanwong, P., Sangsrakru, D., Chanprasert, J., Yoocha, T., Jomchai, N., et al. (2011). Characterization of the Complete Chloroplast Genome of Hevea Brasiliensis Reveals Genome Rearrangement, RNA Editing Sites and Phylogenetic Relationships. Gene. 475, 104–112. doi:10.1016/j.gene.2011.01.002
Valcárcel, V., and Wen, J. (2019). Chloroplast Phylogenomic Data Support Eocene Amphi‐Pacific Early Radiation for the Asian Palmate Core Araliaceae. Jnl Sytematics Evol. 57, 547–560. doi:10.1111/jse.12522
Varshney, R. K., Graner, A., and Sorrells, M. E. (2005). Genic Microsatellite Markers in Plants: Features and Applications. Trends Biotechnol. 23, 48–55. doi:10.1016/j.tibtech.2004.11.005
Wagner, W. L., Hoch, P. C., and Raven, P. H. (2007). Revised Classification of the Onagraceae. Syst. Bot. Monogr. 83, 1–240.
Walker, J. F., Zanis, M. J., and Emery, N. C. (2014). Comparative Analysis of Complete Chloroplast Genome Sequence and Inversion Variation in Lasthenia Burkei (Madieae, Asteraceae). Am. J. Bot. 101, 722–729. doi:10.3732/ajb.1400049
Wang, L., He, N., Li, Y., Fang, Y., and Zhang, F. (2020a). Complete Chloroplast Genome Sequence of Chinese Lacquer Tree (Toxicodendron Vernicifluum, Anacardiaceae) and its Phylogenetic Significance. Biomed. Res. Int. 2020, 1–13. doi:10.1155/2020/9014873
Wang, Y. B., Liu, B. B., Nie, Z. L., Chen, H. F., Chen, F. J., Figlar, R. B., et al. (2020b). Major Clades and a Revised Classification of Magnolia and Magnoliaceae Based on Whole Plastid Genome Sequences via Genome Skimming. J. Syst. Evol. 58, 673–695. doi:10.1111/jse.12588
Wang, L., Yu, X., Wang, H., Lu, Y.-Z., de Ruiter, M., Prins, M., et al. (2011). A Novel Class of Heat-Responsive Small RNAs Derived From the Chloroplast Genome of Chinese Cabbage (Brassica Rapa). BMC Genomics. 12 (1), 115. doi:10.1186/1471-2164-12-289
Weitemier, K., Straub, S. C. K., Cronn, R. C., Fishbein, M., Schmickl, R., McDonnell, A., et al. (2014). Hyb-Seq: Combining Target Enrichment and Genome Skimming for Plant Phylogenomics. Appl. Plant Sci. 2, 1400042. doi:10.3732/apps.1400042
Wen, J., Harris, A., Kalburgi, Y., Zhang, N., Xu, Y., Zheng, W., et al. (2018). Chloroplast Phylogenomics of the New World Grape Species ( Vitis , Vitaceae). Jnl Sytematics Evol. 56, 297–308. doi:10.1111/jse.12447
Weng, M.-L., Blazier, J. C., Govindu, M., and Jansen, R. K. (2014). Reconstruction of the Ancestral Plastid Genome in Geraniaceae Reveals a Correlation Between Genome Rearrangements, Repeats, and Nucleotide Substitution Rates. Mol. Biol. Evol. 31, 645–659. doi:10.1093/molbev/mst257
Wicke, S., Schneeweiss, G. M., Depamphilis, C. W., Müller, K. F., and Quandt, D. (2011). The Evolution of the Plastid Chromosome in Land Plants: Gene Content, Gene Order, Gene Function. Plant Mol. Biol. 76, 273–297. doi:10.1007/s11103-011-9762-4
Wills, D. M., Hester, M. L., Liu, A., and Burke, J. M. (2005). Chloroplast SSR Polymorphisms in the Compositae and the Mode of Organellar Inheritance in Helianthus Annuus. Theor. Appl. Genet. 110 (5), 941–947. doi:10.1007/s00122-004-1914-3
Wolfe, K. H., Li, W. H., and Sharp, P. M. (1987). Rates of Nucleotide Substitution Vary Greatly Among Plant Mitochondrial, Chloroplast, and Nuclear DNAs. Proc. Natl. Acad. Sci. 84, 9054–9058. doi:10.1073/pnas.84.24.9054
Xie, L., Wagner, W. L., Ree, R. H., Berry, P. E., and Wen, J. (2009). Molecular Phylogeny, Divergence Time Estimates, and Historical Biogeography of Circaea (Onagraceae) in the Northern Hemisphere. Mol. Phylogenet. Evol. 53 (3), 995–1009. doi:10.1016/j.ympev.2009.09.009
Xu, D., Abe, J., Gai, J., and Shimamoto, Y. (2002). Diversity of Chloroplast DNA SSRs in Wild and Cultivated Soybeans: Evidence for Multiple Origins of Cultivated Soybean. Theor. Appl. Genet. 105 (5), 645–653. doi:10.1007/s00122-002-0972-7
Yang, X., Luo, X., and Cai, X. (2014). Analysis of Codon Usage Pattern in Taenia Saginata Based on a Transcriptome Dataset. Parasites Vectors. 7, 1–11. doi:10.1186/s13071-014-0527-1
Zhai, W., Duan, X., Zhang, R., Guo, C., Li, L., Xu, G., et al. (2019). Chloroplast Genomic Data Provide New and Robust Insights into the Phylogeny and Evolution of the Ranunculaceae. Mol. Phylogenet. Evol. 135, 12–21. doi:10.1016/j.ympev.2019.02.024
Zhang, Q., Liu, Y., and Sodmergen, (2003). Examination of the Cytoplasmic DNA in Male Reproductive Cells to Determine the Potential for Cytoplasmic Inheritance in 295 Angiosperm Species. Plant Cell Physiol. 44, 941–951. doi:10.1093/pcp/pcg121
Keywords: chloroplast genome, inversion, Onagraceae, phylogeny, RNA editing, biparental inheritance, IR expansion
Citation: Luo Y, He J, Lyu R, Xiao J, Li W, Yao M, Pei L, Cheng J, Li J and Xie L (2021) Comparative Analysis of Complete Chloroplast Genomes of 13 Species in Epilobium, Circaea, and Chamaenerion and Insights Into Phylogenetic Relationships of Onagraceae. Front. Genet. 12:730495. doi: 10.3389/fgene.2021.730495
Received: 25 June 2021; Accepted: 20 October 2021;
Published: 04 November 2021.
Edited by:
Deepmala Sehgal, International Maize and Wheat Improvement Center, MexicoReviewed by:
Swarup Roy Choudhury, Indian Institute of Science Education and Research, Tirupati, IndiaJoseph Charboneau, University of Arizona, United States
Copyright © 2021 Luo, He, Lyu, Xiao, Li, Yao, Pei, Cheng, Li and Xie. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Lei Xie, eGllbGVpQGJqZnUuZWR1LmNu
†These authors have contributed equally to this work
 Yike Luo
Yike Luo Jian He
Jian He Rudan Lyu
Rudan Lyu Jiamin Xiao1†
Jiamin Xiao1† Jin Cheng
Jin Cheng Jinyu Li
Jinyu Li Lei Xie
Lei Xie