
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Genet., 21 September 2021
Sec. Evolutionary and Population Genetics
Volume 12 - 2021 | https://doi.org/10.3389/fgene.2021.728664
This article is part of the Research TopicCytogenomics: Structural Organization and Evolution of GenomesView all 14 articles
Diverse families of satellite DNA (satDNA) were detected in heterochromatin regions of Deschampsia. This kind of repetitive DNA consists of tandem repeat sequences forming big arrays in genomes, and can contribute to lineages differentiation. The differentiation between types of satDNA is related to their sequence identity, the size and number of monomers forming the array, and their chromosomal location. In this work, four families of satDNA (D2, D3, D12, D13), previously isolated by genomic analysis, were studied on chromosomal preparations of 12 species of Deschampsia (D. airiformis, D. antarctica, D. cespitosa, D. cordillerarum, D. elongata, D. kingii, D. laxa, D. mendocina, D. parvula, D. patula, D. venustula, and Deschampsia sp) and one of Deyeuxia (D. eminens). Despite the number of satDNA loci showing interspecific variation, the general distribution pattern of each satDNA family is maintained. The four satDNA families are AT-rich and associated with DAPI + heterochromatin regions. D2, D3, and D12 have mainly subterminal distribution, while D13 is distributed in intercalary regions. Such conservation of satDNA patterns suggests a not random distribution in genomes, where the variation between species is mainly associated with the array size and the loci number. The presence of satDNA in all species studied suggests a low genetic differentiation of sequences. On the other hand, the variation of the distribution pattern of satDNA has no clear association with phylogeny. This may be related to high differential amplification and contraction of sequences between lineages, as explained by the library model.
Repetitive DNA is one of the main components of plant genomes, reaching about 50–90% of genomic abundance (Bennett et al., 1982; Kubis et al., 1998; Heslop-Harrison, 2000), and is composed by dispersed (e.g., transposable elements) and tandem (e.g., satellite DNA) sequences, with variable abundance. Since satellite DNA (satDNA) usually form large arrays on chromosomes (frequently related to heterochromatin regions), it is detectable by fluorescence in situ hybridization (FISH) (Schwarzacher and Heslop-Harrison, 2000), allowing their study at the cytogenetical level. The main characteristics defining satDNA are monomer size over 100–150 bp and arrays up to 100 Mb with tandem disposition (Sharma and Raina, 2005; Hemleben et al., 2007; Mehrotra and Goyal, 2014). Although they are considered non-coding sequences, their monomeric size frequently varies between 150–180 and 320–360 bp, which corresponds with the structural motifs of mono- and dinucleosomes (Kubis et al., 1998; Macas et al., 2002). The current genomic sequencing methods (e.g., NGS), together with the advance of bioinformatics, constantly provide new data about the structural diversity of satDNA (Ruiz-Ruano et al., 2016; Garrido-Ramos, 2017; Lower et al., 2018), however many aspects remain to be understood.
The evolution of satDNA in genomes implies that both sequences diverge as changes in copy numbers. The abundance of satDNA in eukaryote genomes can vary widely and rapidly between generations, leading to high polymorphism in the satellite arrays’ length (Plohl et al., 2012). The sequence identity inside an array evolves according to the process called concerted evolution, which causes more similar monomers than expected due to random changes (Dover, 1986; Plohl, 2010). The monomers homogenization is much faster in species with sexual reproduction, given meiotic recombination (Mantovani et al., 1997). The chromosome organization has a fundamental influence on processes such as chromosome pairing, segregation, gene organization, and expression, and repetitive DNA such as satDNA have an important role in DNA packaging and chromatin condensation (Heslop-Harrison, 2000). Since satDNA distribution may facilitate the recognition of homologous chromosome pairs, changes between lineages have been precursors of speciation (Hemleben et al., 2007). The similarities and differences in genomic satDNA between species can be explained by the “library model”, which suggests differential amplification of satDNA between independent lineages (Garrido-Ramos, 2015). However, the patchy distribution of some satDNA types across eukaryotes (animal and plants) suggests that a scenario of multiple horizontal transfers during evolution may be considered (Yang et al., 2020).
The grass genus Deschampsia P. Beauv. is a cosmopolitan genus which includes about 30 species, 15 of them growing in South America, including D. antarctica which also occurs in Antarctica (Parodi, 1949; Chiapella and Zuloaga, 2010). Some species have difficult circumscription, forming species complexes (Chiapella et al., 2011; Tzvelev et al., 2015). Regarding Deschampsia cytogenetics, the species show a basic chromosome number of x = 13, with a few exceptions reported in the northern hemisphere. The most common chromosome number is 2n = 26, followed by 2n = 52, and the chromosomal complement has metacentric, submetacentric, and acrocentric chromosomes, in similar proportions between species (Albers, 1980; Winterfeld and Röser, 2007b; Cardone et al., 2009; González et al., 2021). In contrast with the conserved chromosome morphology, the 18-5.8-26S and 5S rDNA patterns have high intra and inter-specific variability of loci number and position, which allow the determination of chromosome markers specific to some phylogenetically related species groups (Chiapella, 2007; Saarela et al., 2017; González et al., 2021).
Several satDNA families were isolated from tribe Poeae using restriction enzymes (Grebenstein et al., 1995, 1996), and some of them are present in various genera, including Deschampsia, suggesting an ancient origin (Röser et al., 2014). Likewise, the occurrence of several specific satDNA types were reported in different groups of grasses (Anamthawat-Jónsson and Heslop-Harrison, 1993; Vershinin et al., 1995; Nakajima et al., 1996). The analysis of repetitive DNA from WGS in D. antarctica and D. cespitosa allowed researchers to recognize 34 satDNA families (González et al., 2020). This is a high satDNA diversity compared to other grasses, such as Eragrostis tef (one satDNA family), Agropyron cristatum (fourteen satDNA families), Festuca pratensis Huds. (eight satDNA families), and Poa (four satDNA families) (Gebre et al., 2016; Křivánková et al., 2017; Said et al., 2018; Wei et al., 2020). Deschampsia antarctica and D. cespitosa showed a slightly differentiated satDNA genomic pattern when considering abundance and diversity. Two types of chromosome distribution patterns were observed in the analyzed satDNA families, a C-type pattern (clustered) and M-type pattern (mixed combination of both clustered and dispersed) (González et al., 2020).
The use of C-type satDNA in Deschampsia would allow us to reveal if there exists interspecific variation of the distribution of such sequences in chromosomes, which may be related to the speciation processes. In this way, D2 (359 pb), D3 (377 pb), D12 (366 pb), and D13 (563 pb) families previously analyzed in D. antarctica and D. cespitosa (González et al., 2020), could be good markers to analyze the interspecific variation. The phylogenetic relationships between the South American species were inferred with molecular data and suggest a recent common ancestor between Deschampsia and Deyeuxia sec. Stylagrostis, which could have had chromosomal characteristics currently shared by both taxa (Saarela et al., 2017; González et al., 2021). Due to phylogenetic results, Saarela et al. (2017) suggested transferring seven South American species of Deyeuxia sect. Stylagrostis to Deschampsia, hence more studies of new sources of variation will be useful. With the aim of evaluating karyotypic variations related to phylogenetic hypotheses we analyzed chromosomal distribution of some satDNA families in twelve different Deschampsia species and one of Deyeuxia eminens.
We collected twelve species of Deschampsia (one species, vouchers MLG 79 and MLG 81, does not match any described species), Deyeuxia eminens (which belongs to sect. Stylagrostis), and Avenella flexuosa from Antarctica and Argentinian Patagonia (Table 1). Living plants were transported to the laboratory and kept in pots in culture chambers at 14°C, to obtain root tips for cytogenetic techniques. Leaves were kept in silica gel for DNA extractions. The vouchers were included in the collection of the herbarium of the Botanical Museum of Córdoba (CORD).
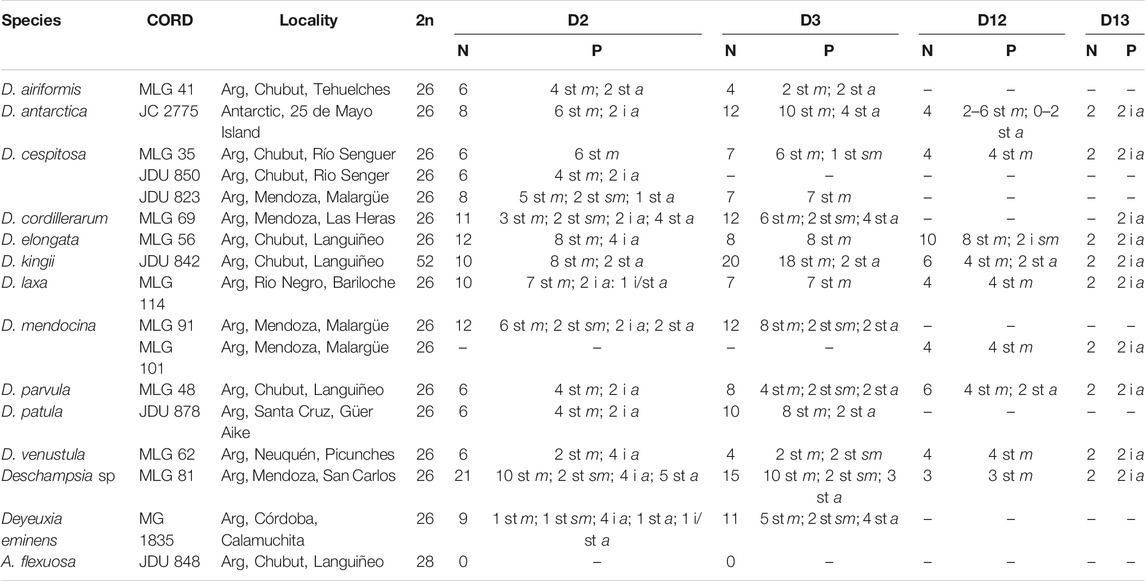
TABLE 1. Studied species and results from FISH with satDNA. CORD: herbarium number (MLG: Maria Laura González; JC: Jorge Chiapella; JDU: Juan Domingo Urdampilleta; MG: Melisa Giorgis). 2n: somatic chromosome number. N: number of hybridization signals. P: hybridization pattern (st: subterminal, i: intercalary, m: metacentric, sm: submetacentric, a: acrocentric). Arg: Argentina.
The mitotic chromosomes preparations were obtained from root meristems pretreated with 2 mM 8-hydroxyquinoline for 4–6 h at 14°C and fixed in ethanol/acetic acid (3:1, v:v). The tissues were digested with Pectinex enzyme solution (Novozimes) and squashed in 45% acetic acid. Preparations were frozen in liquid nitrogen to remove the coverslip.
Fluorescence in situ hybridization (FISH) was carried out to detect satellite DNA patterns, following protocols by Schwarzacher and Heslop-Harrison (2000). To reach stringency above 76%, the hybridization mix had 2x SSC, 50% v/v Formamide, 20% v/v dextran sulfate, 0.1% v/v SDS, and 4–6 ng/μL probes. Post-hybridization washes consisted of 2x SSC, 0.1x SSC, 2× SSC, for 10 min at 42 °C each. We selected four families with a conspicuous hybridization pattern among the previously isolated and mapped satDNA families in D. antarctica and D. cespitosa to analyze chromosomal distribution in other Deschampsia species and related taxa: D2, D3, D12, and D13. Probes were obtained by PCR as described in González et al. (2020) and labelled with biotin (Bionick, Invitrogen) (D2 and D13) or digoxigenin (D3 and D12) (DIG Nick translation mix, Roche). Double-target FISH was performed by hybridizing D2 with D3, and D12 with D13. The satDNA D2 and D3 were mapped in 14 species: D. cfr. airiformis, D. antarctica, D. cespitosa (three localities were used), D. cordillerarum, D. elongata, D. kingii, D. cfr. laxa, D. cfr. mendocina, D. parvula, D. patula, D. venustula, Deschampsia sp, A. flexuosa, and Deyeuxia eminens. The satDNA D12 and D13 were mapped in nine species: D. antarctica, D. cespitosa, D. elongata, D. kingii, D. laxa, D. mendocina, D. parvula, D. venustula, and Deschampsia sp. The detection was made with Avidin-FITC (Sigma-Aldrich) and anti-DIG antibodies conjugated with rhodamine (Roche). Chromosome metaphases were photographed using an Olympus BX61 microscope coupled with a monochromatic camera and Cytovision software (Leica Biosystems), and the images were pseudo-colored. Idiograms were constructed for Deschampsia species and Deyeuxia eminens. The karyotype conformation of each species was taken from González et al. (2021).
For the reconstruction of ancestral states of satDNA chromosomal traits, the packages ape (Paradis et al., 2004), geiger (Harmon et al., 2008), and phytools (Revell, 2012) were used in R. The phylogenetic hypothesis used was the maximum likelihood tree reconstructed with ITS, ETS, and trnL-F markers by González et al. (2021), which includes the same plant vouchers used for the cytogenetic analysis of this study. The ER (equal transition rates) and ARD (all transition rates are different) models were tested for discrete traits. For continuous traits, the models BM (Brownian motion), OU (Ornstein-Uhlenbeck), and EB (Early-burst) were tested. The models were fitted using the package geiger, and the best models were selected according to AIC criterion for each trait. The reconstruction of ancestral states of discrete traits was carried out using maximum likelihood, with the Markov k-state (Mk1) model according to the better-adjusted transition model using the package ape. The reconstruction of ancestral states of continuous traits was carried out using the package phytools. The phylogenetic signal was tested using the package geiger.
All studied species showed hybridization to all studied probes, except for A. flexuosa which did not show hybridization for D2 and D3 satDNA probes (Table 1). The four satDNA families studied here are AT-rich and were frequently observed in association with DAPI + bands observed after FISH (Supplementary Figure S1, S2, S3, and S4). The species showed variation of the number and position of loci. The satDNA D2 showed variation of the loci number, ranging from 6 to 21 hybridization signals between species (which corresponded from 2.5 to 10.5 signals per basic complement x), noting that most species showed six hybridization signals (Figure 1; Table 1). The satDNA D3 showed from 4 to 20 hybridization signals (from 2 to 7.5 per basic complement x) (Figure 2; Table 1). The satDNA D12 showed from 3 to 10 hybridization signals between species (from 1.5 to five per basic complement x), noting that most species showed four hybridization signals (Figure 3; Table 1). The satDNA D13 was constant in all studied species, showing 2 hybridization signals (corresponding with 0.5–1 locus per basic complement x, since there was one tetraploid species) (Figure 3; Table 1). Only considering D2 and D3, since they were hybridized in all species, the species with more satDNA loci per basic complement was Deschampsia sp, followed by D. mendocina and D. cordillerarum (Table 1). Likewise, D. elongata was the species with the highest number of signals observed for D12.
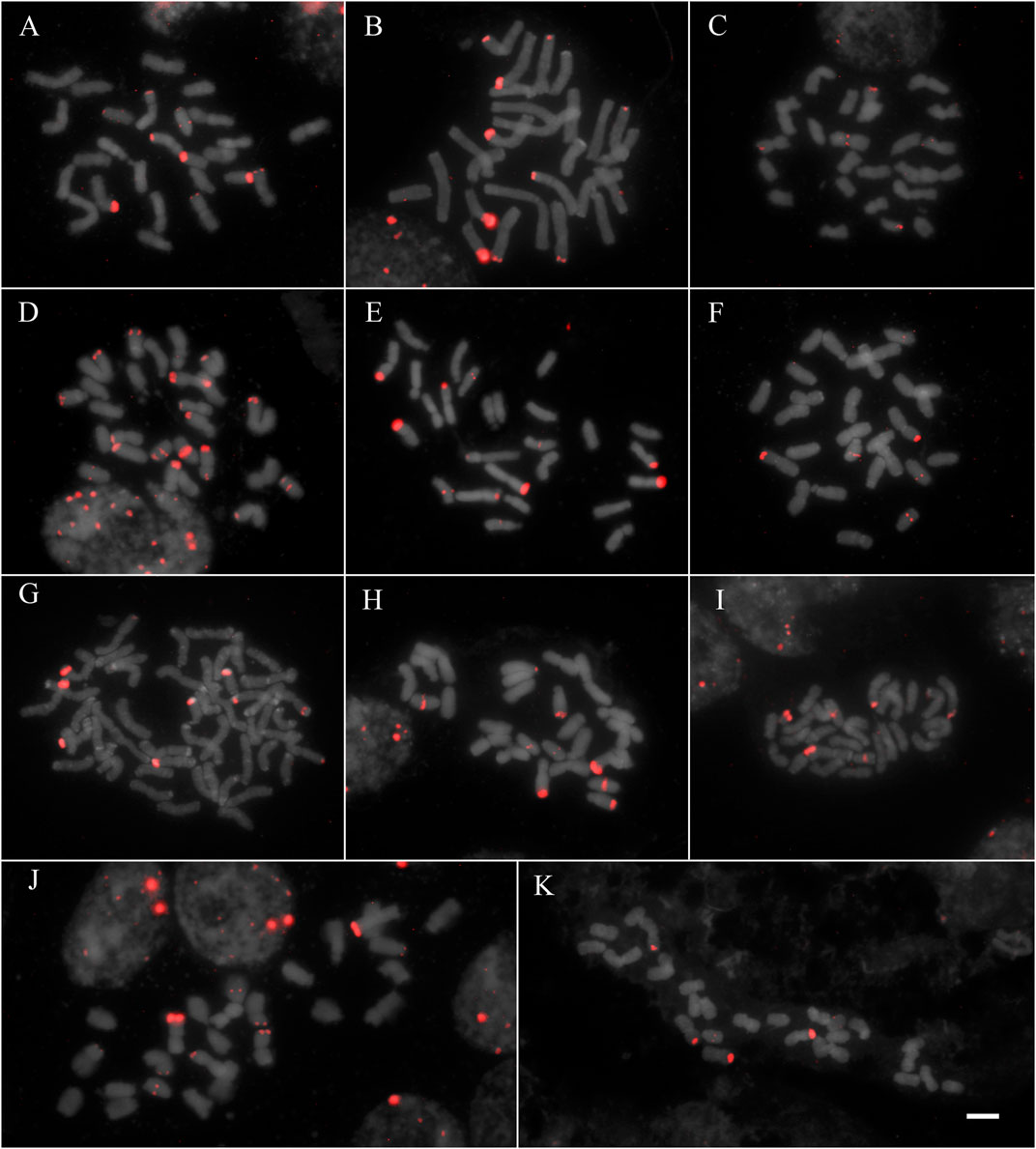
FIGURE 1. FISH of satDNA D2 (red) of studied species (A) D. cordillerarum MLG 69 (B) D. mendocina MLG 91 (C) D. airiformis MLG 41 (D) Deschampsia sp MLG 81 (E) D. elongata MLG 56 (F) D. venustula MLG 62 (G) D. kingii JDU 842 (H) Deyeuxia eminens MG 1835 (I) D. patula 878 (J) D. laxa MLG 114 (K) D. parvula MLG 48. Scale: 5 µm.
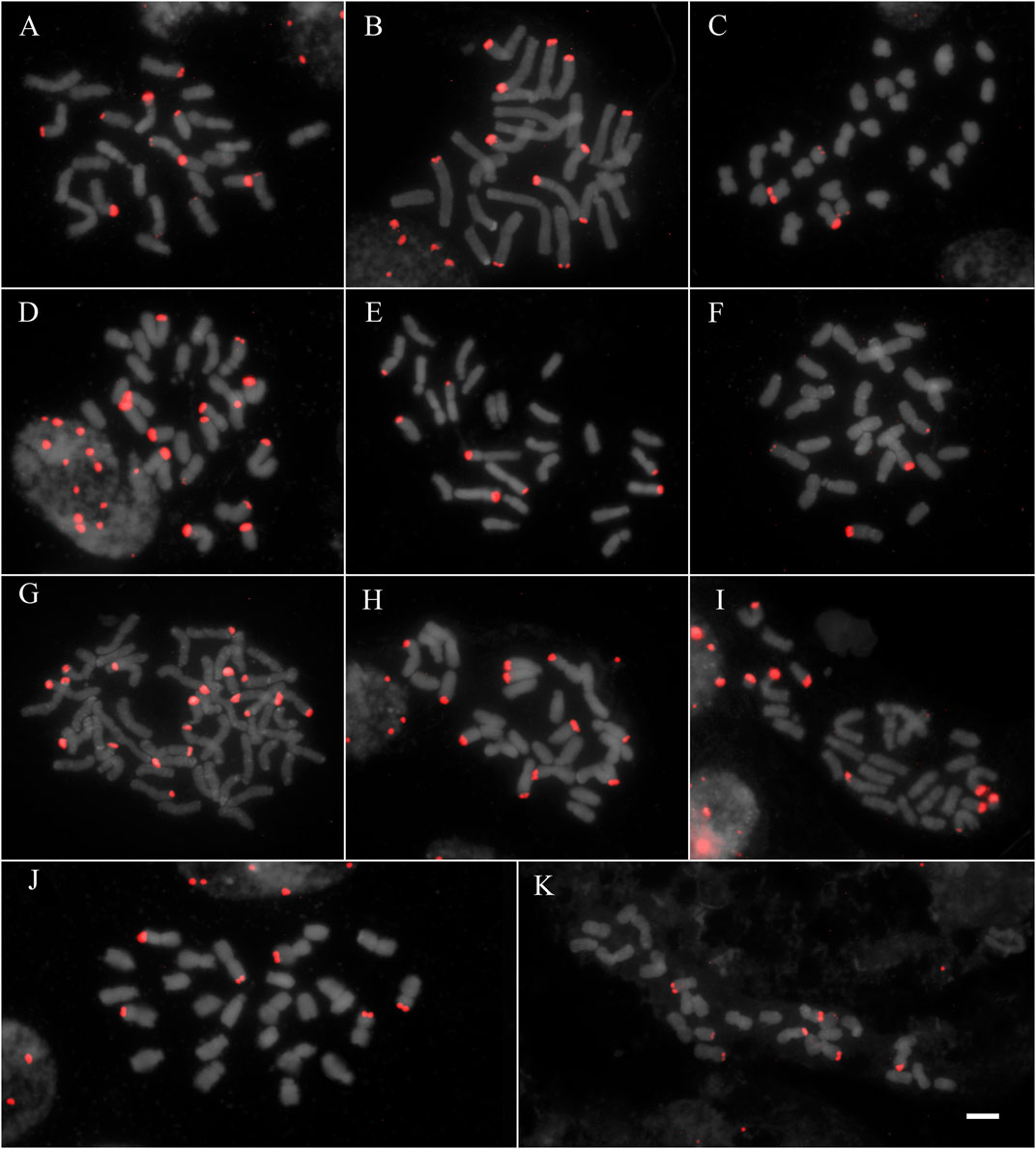
FIGURE 2. FISH of satDNA D3 (red) of studied species (A) D. cordillerarum MLG 69 (B) D. mendocina MLG 91 (C) D. airiformis MLG 41 (D) Deschampsia sp MLG 81 (E) D. elongata MLG 56 (F) D. venustula MLG 62 (G) D. kingii JDU 842 (H) Deyeuxia eminens MG 1835 (I) D. patula 878 (J) D. laxa MLG 114 (K) D. parvula MLG 48. Scale: 5 µm.
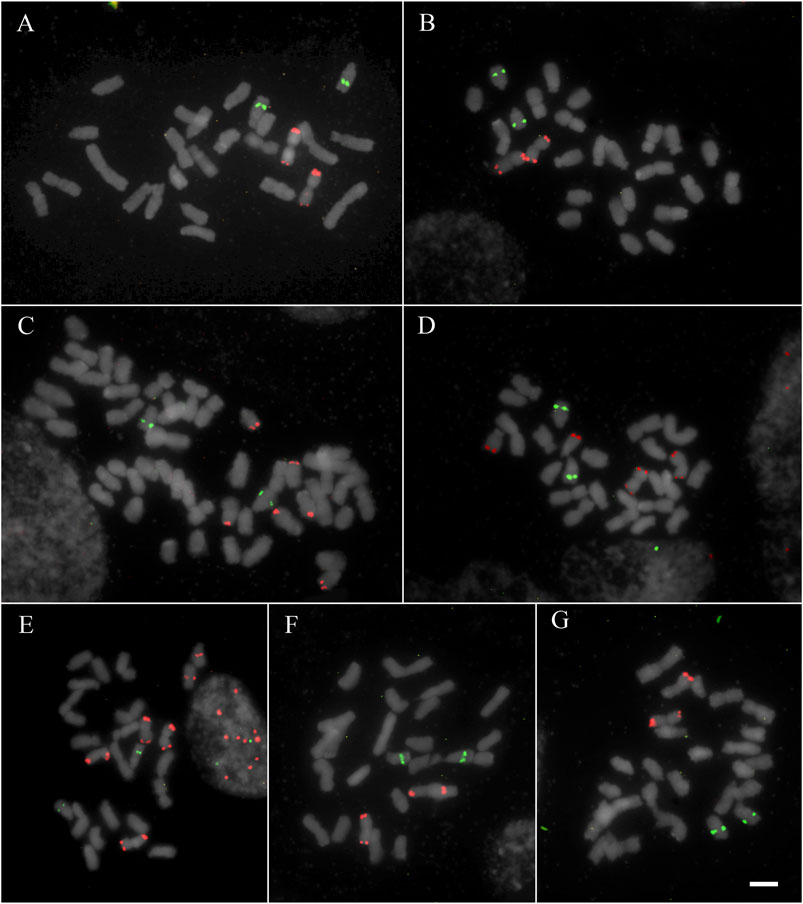
FIGURE 3. FISH of satDNAs D12 (red) and D13 (green) of studied species (A) D. venustula MLG 62 (B) D. laxa MLG 114 (C) D. kingii JDU 842 (D) D. parvula MLG 48 (E) D. elongata MLG 56 (F) D. mendocina MLG 101 (G) Deschampsia sp MLG 81. Scale: 5 µm.
The satDNA family D2 was mostly located at subterminal regions of different chromosome types, forming blocks of variable size. All species showed subterminal loci of D2 on metacentric chromosomes, and the species group formed by D. cordillerarum, D. mendocina, Deyeuxia eminens, and Deschampsia sp showed subterminal loci on submetacentric chromosomes. Also, some loci were found in acrocentric chromosomes, at an intercalary position in most species (except for D. kingii and D. airiformis) and at a subterminal position for D. cordillerarum, D. mendocina, Deyeuxia eminens, Deschampsia sp, D. airiformis, and D. kingii (Figure 1 and Supplementary Figure S4).
The satDNA family D3 was exclusively observed at subterminal regions. All species showed D3 loci on metacentric chromosomes, and most species also showed on the long arm of submetacentric chromosomes except for D. laxa, D. airiformis, D. elongata, D. antarctica, and D. patula. The satDNA D3 was also observed on acrocentric chromosomes at a subterminal position in most species, except for D. cespitosa, D. laxa, D. venustula, and D. elongata. This satDNA was frequently co-localized with D2 (Figures 1, 2 and Supplementary Figure S1, S2, S3, and S4).
The satDNA family D12 was detected at a subterminal position on metacentric chromosomes in all studied species. Particularly for D12, most of the studied species showed metacentric chromosome pairs with hybridization signals in both arms (symmetric chromosome). In general, the species showed only one chromosome pair with satDNA D12 (in one or two chromosomal arms), except for D. kingii, D. antarctica, and D. elongata, which showed three or four pairs. This satDNA was also detected in acrocentric chromosomes at a subterminal position in D. antarctica, D. parvula, and D. kingii. Deschampsia elongata was the only species showing one locus of D12 on submetacentric chromosomes (Figure 3 and Supplementary Figure S4).
The chromosomal distribution of satDNA family D13 was conserved in the analyzed species (Figure 3 and Supplementary Figure S4). We observed only one locus at an intercalary position on an acrocentric chromosome pair for all the species, inclusive for the tetraploid D. kingii.
The hybridization pattern of D2 and D3 satDNA was studied in three and two localities of D. cespitosa, respectively, revealing intraspecific variation. The satDNA D2 showed six loci in two localities separated by approximately 26 km (JDU 850 and MLG 35), and eight loci for the most distant locality (JDU 823). The satDNA D3 showed three loci for both studied localities (JDU 823 and MLG 35). In addition to the numerical variation, the disposition of loci showed variation for both probes in all localities (Table 1; Figure 4).
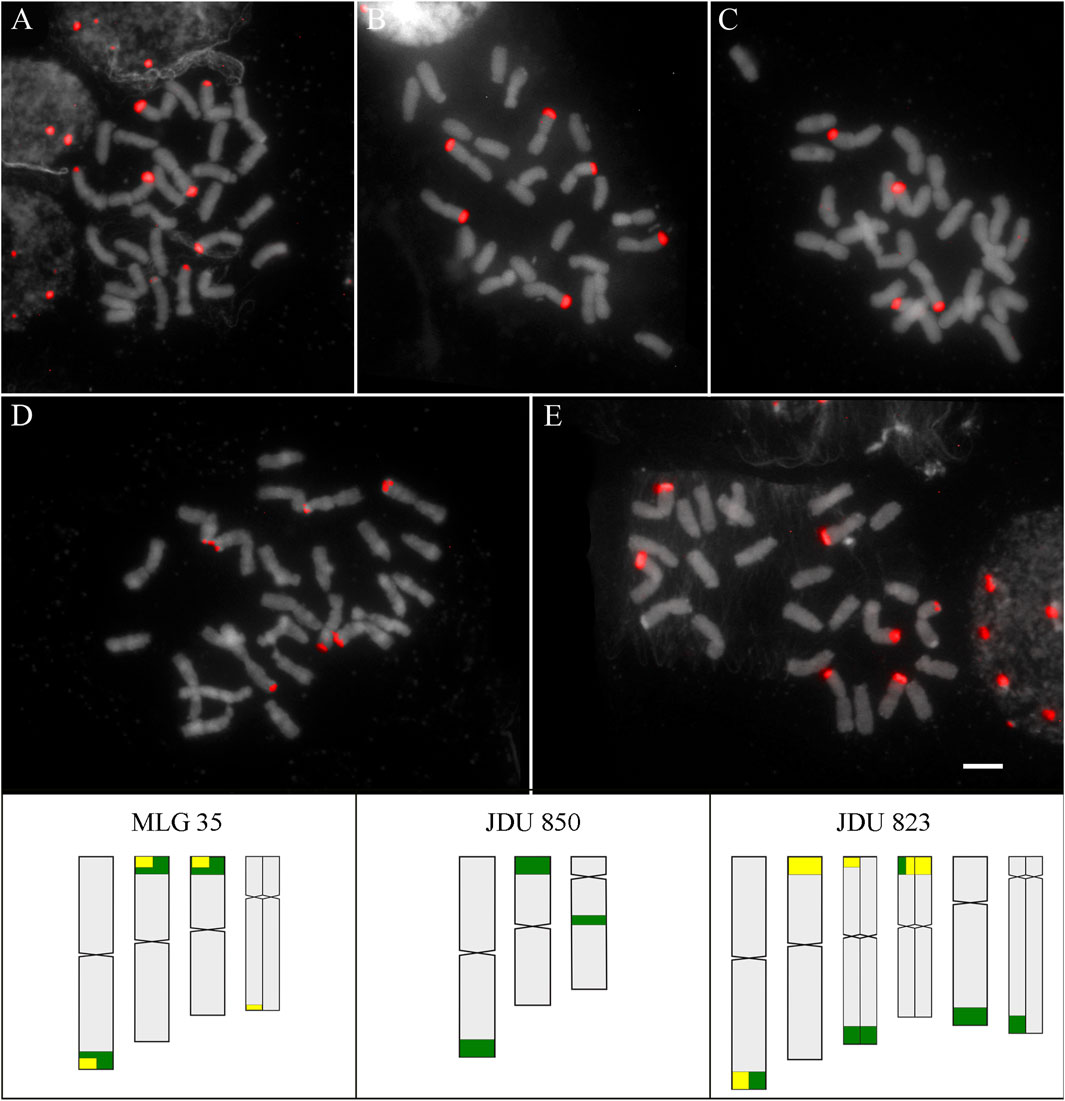
FIGURE 4. SatDNA D2 and D3 in D. cespitosa. Above: FISH of satDNA D2 and D3 of D. cespitosa (A) D2 of JDU 823 (B) D2 of MLG 35 (C) D2 of JDU 850 (D) D3 of MLG 35 (E) D3 of JDU 823. Scale: 5 µm. Below: Idiograms of the three localities, showing the position of satDNA D2 (green) and D3 (yellow). Heteromorphic chromosome pairs are shown divided into two.
For the reconstruction of ancestral states of satDNA, we used a total of six chromosomal traits, two discrete and four continuous (Supplementary Table 1). None of these showed a significant phylogenetic signal according to parameter λ (Pagel, 1999), despite some of them showing values of 1. For discrete traits, the analysis with ER (the better fitted model) did not reflect high probabilities on the ancestral state (presence/absence) in most of the nodes, except in common ancestors of small clades. The continuous traits fitted with a BM model, and does not suggested any trend in the number of satDNA loci in Deschampsia. However, increases or decreases of loci can be seen for small groups or species, as an increase of satDNA D3 in the Deschampsia sp/D. cordillerarum/D. mendocina clade and the D. antarctica/D. patula clade. Particularly, Deschampsia sp shows an increase in the number of signals for both satDNA D2 and D3, while D. elongata shows an increase in D12. The satDNA D13 was constant in terms of loci number per basic complement, with the exception of a decrease in D. kingii, a polyploid species (Supplementary Figure S5).
Constitutive heterochromatin is an important genomic component, essential in nuclear architecture, DNA repair, and genome stability (Strom et al., 2017). The first analyses of heterochromatin in Deschampsia were made on D. cespitosa from Europe, by the use of the C-band technique, which detected big heterochromatin blocks at subterminal regions of most chromosomes and at intercalary regions of a few chromosomes (Garcia-Suarez et al., 1997), a frequent feature of several genera of tribe Poeae. Heterochromatin is commonly composed of several kinds of tandem repetitive sequences (satDNA), usually with unknown origin and function, considered as fast-changing genomic components (Fuchs et al., 1994; Kuipers et al., 2002; Winterfeld and Röser, 2007a). The variation of such repetitive DNA between related species could provide information about its evolution, and clarify the phylogenetic relationships between lineages (Beckmann and Soller, 1986; King et al., 1995). Highly repetitive sequences were detected for the first time in D. cespitosa by membrane hybridization of four satDNA previously isolated from Helictotrichon (which belong to the same tribe, Poeae) and were reported in several other grasses (CON1, CON2, COM1, and COM2) (Grebenstein et al., 1995, 1996). Further studies of the satellitome corroborated the presence of these satDNAs in South American D. cespitosa and D. antarctica (except for COM2), as well as described another 31 satDNA families, with two kinds of hybridization patterns: “mainly clustered”, and mixed “clustered and dispersed” (González et al., 2018, 2020). The satDNA here studied correspond with the pool of clustered satDNA, of which D2 and D3 are the most abundant satDNA of D. antarctica and D. cespitosa genomes, while D12 and D13 (homologous with CON1 and CON2, respectively) are commonly detected in the genome of several Poaceae (Alix et al., 1998; Röser et al., 2014; Zhao et al., 2017).
As originally described, the species of Deschampsia are characterized by their appearance in subterminal regions of AT-rich heterochromatin (as observed with DAPI staining post-FISH), and these regions also show several kinds of clustered satDNA as we found here for D2, D3, and D12 (Garcia-Suarez et al., 1997; Winterfeld and Röser, 2007a; Amosova et al., 2017; González et al., 2020, 2021). The presence of AT-rich repetitive sequences has been frequently detected at the terminal region of other grass genomes (Flavell, 1986; Kubis et al., 1998; Winterfeld and Röser, 2007a; Hemleben et al., 2007), and such repetitive DNA accumulation may be caused by the low recombination rate in the telomeric chromosome regions (Charlesworth et al., 1994). On the other hand, intercalary heterochromatic bands are less common in Deschampsia chromosomes, but they may also coincide with satDNA. Here we found intercalary distribution for D13 and D2 in acrocentric chromosomes only, however, previous studies showed that chromosome intercalary regions in Deschampsia are also rich in dispersed satDNA (González et al., 2020).
The variability of loci number and chromosome distribution of repetitive DNA between and within species is common for Deschampsia. Studies of rDNA distribution (18-5.8-26S and 5S) have shown chromosomal variation related with phylogeny of species and geographic distances (González et al., 2016, 2021). Here we found high variation of the distribution pattern of satDNA D2, D3, and D12 between Deschampsia species. Particularly, D13 was the only satDNA which did not show any variation in its distribution between the studied species. Even D. kingii, which is tetraploid with 52 chromosomes, showed only one locus of D13 in the same position and chromosome type as the rest of the species. Previous observations between D. antarctica and D. cespitosa showed the same hybridization patterns only for two of the thirteen studied satDNA families, D13 and D14 (homologous to satDNA pSc200, widely spread in tribes Poeae and Triticeae), which indicates that this conservation of the distribution pattern is not the most frequently observed in satDNA families (Bedbrook et al., 1980; González et al., 2020). Despite the fact that satDNA could enhance the phylogenetic information of species (Plohl et al., 2012), we found a lack of association between phylogenetic relationships of Deschampsia and the chromosomal distribution and the loci number of satDNA. The genomic abundance of satDNA changes widely and rapidly between generations, leading to polymorphism of arrays (Plohl et al., 2012), and therefore some variation between and within species may be expected.
Here we report different hybridization patterns of D2 and D3 between localities of D. cespitosa from South America. The intraspecific variation of chromosomal distribution of repetitive DNA has been previously found for Deschampsia, by studying the rDNA and satDNA in several localities of D. antarctica, and rDNA in localities of D. cespitosa (Winterfeld and Röser, 2007b; González et al., 2016, 2018, 2021; Navrotska et al., 2018). In both cases, this variability seems to be related with geographic distances since the nearest localities share more loci with each other, and satDNA showed more variation than rDNA. On the other hand, the Patagonian localities of other species, such as D. elongata and D. parvula also showed different hybridization patterns of rDNA than localities from other world regions (United States and Falkland Islands, respectively) (Amosova et al., 2017, 2021).
The intraspecific chromosomal variability of satDNA could explain the lack of association between chromosomal changes and phylogenetic relationships observed in Deschampsia species (Figure 5). Since the chromosome morphologies and karyotype composition remain highly conserved in Deschampsia (Kawano, 1963; Albers, 1980; Winterfeld and Röser, 2007b; Cardone et al., 2009; González et al., 2016, 2021), the high variability of sequences distribution is likely a consequence of differential amplification and loss of repetitive DNA loci between lineages (Fry and Salser, 1977; Ruiz-Ruano et al., 2016). Also small rearrangements can be involved, without notable changes in chromosome numbers and morphologies (Raskina et al., 2008; Nani et al., 2016). The divergent evolution of satDNA may be involved in micro and macroevolutionary processes, facilitating the reproductive isolation of groups by the emergence of chromosomal barriers, eventually giving rise to speciation (Noor et al., 2001; Widmer et al., 2009; Plohl et al., 2012). At the same time, the eventual secondary contacts of lineages with a differentiated pattern of repetitive DNA could give rise to allopolyploids (Mallet, 2007; Soltis and Soltis, 2009; González et al., 2016).
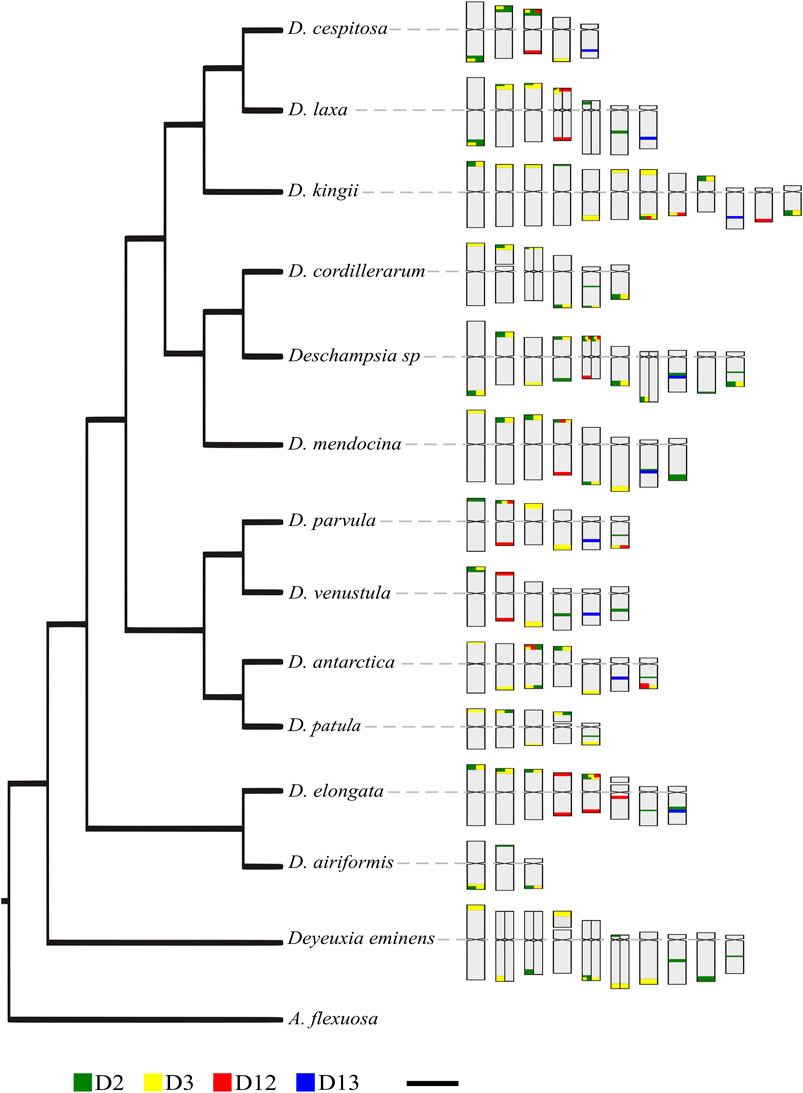
FIGURE 5. Phylogenetic relationships of studied species and idiograms showing the chromosome position of satDNA. The phylogeny is based on González et al. (2021), trimming the taxa is not studied here. Heteromorphic chromosome pairs are shown divided into two. Scale: 5 µm.
Despite the fact that most repetitive DNA are considered as fast evolutionary elements, mainly in abundance but also in sequence, some satDNA may persist over long evolutionary periods (Plohl, 2010). This may be the case of several satDNAs found in Deschampsia which show homology with sequences previously described in Poaceae (Röser et al., 2014; González et al., 2020). The homology of D12 and D13, with the previously described CON1 and CON2 was of 74 and 78% (González et al., 2018). The repetitive DNA shared between distant Poaceae tribes or genus may be related with an ancestral repetitive DNA pool in grasses which evolved differentially between lineages (Hemleben et al., 2007). This is known as the “library model”, and implies that the satDNA profile of a species is the result of differential amplification or contraction from a sequences set of an ancestral genome (Fry and Salser, 1977; Plohl et al., 2012). Some satDNAs may persist in genomes as low copy number sequences over long evolutionary periods and eventually amplify giving rise to high repetitive DNA (Ugarkovic, 2008; Röser et al., 2014). Other satDNA can be lost in some lineages as a consequence of an unequal exchange, which would eventually cause a single copy sequence and then be lost by drift (Charlesworth et al., 1986). One possible reason for low evolutionary rates is the preference of some monomers over others due to their potential function (Plohl, 2010; Plohl et al., 2012). Also, the heterochromatic environment can cause an extreme conservation of the sequence due to low recombination frequency, which together with the concerted evolution would cause a prolonged persistence of repetitive sequences throughout evolutionary time (Charlesworth et al., 1986).
The satDNA D12 (CON1) and D13 (CON2) could have had an early appearance in Poaceae; specifically, D13 seems to belong exclusively to tribe Poeae, while D12 is widespread in Poaceae including members of subfamilies Pooideae, Panicoideae, Chloridideae, and Oryzoideae (Grebenstein et al., 1996; Alix et al., 1998; Winterfeld and Röser, 2007a; Röser et al., 2014; Zhao et al., 2017). The other two satDNA here studied, D2 and D3, seem to be exclusively of Deschampsia lineage (González et al., 2020). Since we found D2 and D3 in all studied Deschampsia and Deyeuxia eminens, it is likely these satDNAs originated in a recent common ancestor between these two groups. As has been previously found, the sect. Stylagrostis from Deyeuxia (where D. eminens belong) seems to be highly related with Deschampsia (Saarela et al., 2017; González et al., 2021), and the presence of D2 and D3 in the D. eminens genome supports their monophyletic relationship.
Despite the chromosomal distribution variability of repetitive DNA (González et al., 2020, 2021), the presence of a clear hybridization for all probes in all Deschampsia species indicates likely conservation of the monomer sequence in the group. This can be a consequence of low evolutionary rates of the studied satDNA, or a recent species radiation. Either way, since satDNA are considered rapid evolution DNA, the presence of all studied satDNA in Deschampsia and Deyeuxia eminens genomes reinforce the idea of a low genetic differentiation between species (in terms of DNA sequences), as previously found in another studies of this group (Chiapella, 2007; Chiapella et al., 2011; Fasanella et al., 2017; González et al., 2020, 2021). In fact, previous studies suggest that karyotype differentiation between Deschampsia species is more associated with changes in chromosomal distribution than changes in the DNA sequence of satDNA (González et al., 2020; Amosova et al., 2021).
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
MG designed the study, made the experiments, and write the manuscript. JU designed, supervised the study, and review the manuscript. JC review the manuscript.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
We thank CONICET, ANPCyT-FONCyT, and SECyTUNC for financial support; and Dirección Nacional del Antártico for logistic support in Antarctica. This work was supported by ANPCyT-FONCyT under project PICT-2018–1691; the International Association for Plant Taxonomy (IAPT) by 2018 Research Grants; and CONICET under project PIP-11220150100392CO.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2021.728664/full#supplementary-material
Albers, F. (1980). Vergleichende Karyologie der Gräser-SubtribenAristaveninae undAirinae (Poaceae-Aveneae). Pl Syst. Evol. 136, 137–167. doi:10.1007/BF01004624
Alix, K., Baurens, F.-C., Paulet, F., Glaszmann, J.-C., and D'Hont, A. (1998). Isolation and Characterization of a Satellite DNA Family in the Saccharum Complex. Genome 41, 854–864. doi:10.1139/g98-076
Amosova, A. V., Bolsheva, N. L., Zoshchuk, S. A., Twardovska, M. O., Yurkevich, O. Y., Andreev, I. O., et al. (2017). Comparative Molecular Cytogenetic Characterization of Seven Deschampsia (Poaceae) Species. PLoS One 12, e0175760. doi:10.1371/journal.pone.0175760
Amosova, A. V., Ghukasyan, L., Yurkevich, O. Y., Bolsheva, N. L., Samatadze, T. E., Zoshchuk, S. A., et al. (2021). Cytogenomics of Deschampsia P. Beauv. (Poaceae) Species Based on Sequence Analyses and Fish Mapping of CON/COM Satellite Dna Families. Plants 10, 1105. doi:10.3390/plants10061105
Anamthawat-Jónsson, K., and Heslop-Harrison, J. S. (1993). Isolation and Characterization of Genome-specific DNA Sequences in Triticeae Species. Mol. Gen. Genet. 240, 151–158. doi:10.1007/BF00277052
Beckmann, J. S., and Soller, M. (1986). Restriction Fragment Length Polymorphisms and Genetic Improvement of Agricultural Species. Euphytica 35, 111–124. doi:10.1007/BF00028548
Bedbrook, J. R., Jones, J., O'Dell, M., Thompson, R. D., and Flavell, R. B. (1980). A Molecular Description of Telomeric Heterochromatin in Secale Species. Cell 19, 545–560. doi:10.1016/0092-8674(80)90529-2
Bennett, M. D., Smith, J. B., and Smith, R. I. L. (1982). DNA Amounts of Angiosperms from the Antarctic and South Georgia. Environ. Exp. Bot. 22, 307–318. doi:10.1016/0098-8472(82)90023-5
Cardone, S., Sawatani, P., Rush, P., García, A. M., Poggio, L., and Schrauf, G. (2009). Karyological Studies in Deschampsia antarctica Desv. (Poaceae). Polar Biol. 32, 427–433. doi:10.1007/s00300-008-0535-8
Charlesworth, B., Langley, C. H., and Stephan, W. (1986). The Evolution of Restricted Recombination and the Accumulation of Repeated DNA Sequences. Genetics 112, 947–962. doi:10.1093/genetics/112.4.947
Charlesworth, B., Sniegowski, P., and Stephan, W. (1994). The Evolutionary Dynamics of Repetitive DNA in Eukaryotes. Nature 371, 215–220. doi:10.1038/371215a0
Chiapella, J., and Zuloaga, F. O. (2010). A Revision of Deschampsia, Avenella, and Vahlodea (Poaceae, Poeae, Airinae) in South America1. Ann. Mo. Bot. Garden 97, 141–162. doi:10.3417/2008115
Chiapella, J. O., DeBoer, V. L., Amico, G. C., and Kuhl, J. C. (2011). A Morphological and Molecular Study in theDeschampsia Cespitosacomplex (Poaceae; Poeae; Airinae) in Northern North America. Am. J. Bot. 98, 1366–1380. doi:10.3732/ajb.1000495
Chiapella, J. O. (2007). A Molecular Phylogenetic Study of Deschampsia (Poaceae: Aveneae) Inferred from Nuclear ITS and Plastid trnL Sequence Data: Support for the Recognition of Avenella and Vahlodea. Taxon 55, 55–64. doi:10.2307/25065735
Dover, G. A. (1986). Molecular Drive in Multigene Families: How Biological Novelties Arise, Spread and Are Assimilated. Trends Genet. 2, 159–165. doi:10.1016/0168-9525(86)90211-8
Fasanella, M., Premoli, A. C., Urdampilleta, J. D., González, M. L., and Chiapella, J. O. (2017). How Did a Grass Reach Antarctica? the Patagonian Connection of Deschampsia antarctica (Poaceae). Bot. J. Linn. Soc. 185, 511–524. doi:10.1093/botlinnean/box070
Flavell, R. B. (1986). Repetitive DNA and Chromosome Evolution in Plants. Phil. Trans. R. Soc. Lond. B 312, 227–242. doi:10.1098/rstb.1986.0004
Fry, K., and Salser, W. (1977). Nucleotide Sequences of HS-α Satellite DNA from Kangaroo Rat dipodomys Ordii and Characterization of Similar Sequences in Other Rodents. Cell 12, 1069–1084. doi:10.1016/0092-8674(77)90170-2
Fuchs, J., Pich, U., Meister, A., and Schubert, I. (1994). Differentiation of Field Bean Heterochromatin Byin Situ Hybridization with a repeatedFokI Sequence. Chromosome Res. 2, 25–28. doi:10.1007/BF01539450
Garcia-Suarez, R., Alonso-Blanco, C., Fernandez-Carvajal, M. C., Fernandez-Prieto, J. A., Roca, A., and Giraldez, R. (1997). Diversity and Systematics ofDeschampsia Sensu Lato (Poaceae), Inferred from Karyotypes, Protein Electrophoresis, Total Genomic DNA Hybridization and Chloroplast DNA Analysis. Pl Syst. Evol. 205, 99–110. doi:10.1007/BF00982800
Garrido-Ramos, M. A. (2015). Satellite DNA in Plants: More Than Just Rubbish. Cytogenet. Genome Res. 146, 153–170. doi:10.1159/000437008
Gebre, Y. G., Bertolini, E., Pè, M. E., and Zuccolo, A. (2016). Identification and Characterization of Abundant Repetitive Sequences in Eragrostis Tef Cv. Enatite Genome. BMC Plant Biol. 16, 1–13. doi:10.1186/s12870-016-0725-4
González, M. L., Urdampilleta, J. D., Fasanella, M., Premoli, A. C., and Chiapella, J. O. (2016). Distribution of rDNA and Polyploidy in Deschampsia antarctica E. Desv. In Antarctic and Patagonic Populations. Polar Biol. 39, 1663–1677. doi:10.1007/s00300-016-1890-5
González, M. L., Chiapella, J. O., and Urdampilleta, J. D. (2018). Characterization of Some Satellite DNA Families in Deschampsia antarctica (Poaceae). Polar Biol. 41, 457–468. doi:10.1007/s00300-017-2205-1
González, M. L., Chiapella, J., Topalian, J., and Urdampilleta, J. D. (2020). Genomic Differentiation of Deschampsia antarctica and D. Cespitosa (Poaceae) Based on Satellite DNA. Bot. J. Linn. Soc. 194, 326–341. doi:10.1093/botlinnean/boaa045
González, M. L., Chiapella, J. O., and Urdampilleta, J. D. (2021). The Antarctic and South American Species of Deschampsia: Phylogenetic Relationships and Cytogenetic Differentiation. Syst. Biodivers. 19, 453–470. doi:10.1080/14772000.2020.1860151
Grebenstein, B., Grebenstein, O., Sauer, W., and Hemleben, V. (1995). Characterization of a Highly Repeated DNA Component of Perennial Oats (Helictotrichon, Poaceae) with Sequence Similarity to a A-genome-specific Satellite DNA of rice (Oryza). Theoret. Appl. Genet. 90, 1101–1105. doi:10.1007/BF00222928
Grebenstein, B., Grebenstein, O., Sauer, W., and Hemleben, V. (1996). Distribution and Complex Organization of Satellite DNA Sequences in Aveneae Species. Genome 39, 1045–1050. doi:10.1139/g96-131
Harmon, L. J., Weir, J. T., Brock, C. D., Glor, R. E., and Challenger, W. (2008). GEIGER: Investigating Evolutionary Radiations. Bioinformatics 24, 129–131. doi:10.1093/bioinformatics/btm538
Hemleben, V., Kovarik, A., Torres‐Ruiz, R. A., Volkov, R. A., and Beridze, T. (2007). Plant Highly Repeated Satellite DNA: Molecular Evolution, Distribution and Use for Identification of Hybrids. Syst. Biodivers. 5, 277–289. doi:10.1017/s147720000700240x
Heslop-Harrison, J. S. (2000). Comparative Genome Organization in Plants: from Sequence and Markers to Chromatin and Chromosomes. Plant Cell 12, 617–636. doi:10.1105/tpc.12.5.61710.2307/3870990
Kawano, S. (1963). Cytogeography and Evolution of the Desghampsia Caespitosa Complex. Can. J. Bot. 41, 719–742. doi:10.1139/b63-059
King, K., Jobst, J. r., and Hemleben, V. (1995). Differential Homogenization and Amplification of Two Satellite DNAs in the Genus Cucurbita (Cucurbitaceae). J. Mol. Evol. 41, 996–1005. doi:10.1007/BF00173181
Křivánková, A., Kopecký, D., Stočes, Š., Doležel, J., and Hřibová, E. (2017). Repetitive DNA: A Versatile Tool for Karyotyping in Festuca Pratensis Huds. Cytogenet. Genome Res. 151, 96–105. doi:10.1159/000462915
Kubis, S., Schmidt, T., and Heslop-Harrison, J. S. (1998). Repetitive DNA Elements as a Major Component of Plant Genomes. Ann. Bot. 82, 45–55. doi:10.1006/anbo.1998.0779
Kuipers, A. G. J., Kamstra, S. A., De Jeu, M. J., and Visser, R. G. F. (2002). Molecular Characterization and Physical Localization of Highly Repetitive DNA Sequences from Brazilian Alstroemeria Species. Chromosom. Res. 10, 389–398. doi:10.1023/A:1016801702777
Lower, S. S., McGurk, M. P., Clark, A. G., and Barbash, D. A. (2018). Satellite DNA Evolution: Old Ideas, New Approaches. Curr. Opin. Genet. Dev. 49, 70–78. doi:10.1016/j.gde.2018.03.003
Macas, J., Mészáros, T., and Nouzová, M. (2002). PlantSat: A Specialized Database for Plant Satellite Repeats. Bioinformatics 18, 28–35. doi:10.1093/bioinformatics/18.1.28
Mantovani, B., Tinti, F., Bachmann, L., and Scali, V. (1997). The Bag320 Satellite DNA Family in Bacillus Stick Insects (Phasmatodea): Different Rates of Molecular Evolution of Highly Repetitive DNA in Bisexual and Parthenogenic Taxa. Mol. Biol. Evol. 14, 1197–1205. doi:10.1093/oxfordjournals.molbev.a025729
Mehrotra, S., and Goyal, V. (2014). Repetitive Sequences in Plant Nuclear DNA: Types, Distribution, Evolution and Function. Genomics, Proteomics Bioinf. 12, 164–171. doi:10.1016/j.gpb.2014.07.003
Nakajima, R., Noma, K., Ohtsubo, H., and Ohtsubo, E. (1996). Identification and Characterization of Two Tandem Repeat Sequences (TrsB and TrsC) and a Retrotransposon (RIRE1) as Genome-General Sequences in rice. Genes Genet. Syst. 71, 373–382. doi:10.1266/ggs.71.373
Nani, T. F., Pereira, D. L., Souza Sobrinho, F., and Techio, V. H. (2016). Physical Map of Repetitive DNA Sites in Brachiaria spp.: Intravarietal and Interspecific Polymorphisms. Crop Sci. 56, 1769–1783. doi:10.2135/cropsci2015.12.0760
Navrotska, D., Andreev, I., Betekhtin, A., Rojek, M., Parnikoza, I., Myryuta, G., et al. (2018). Assessment of the Molecular Cytogenetic, Morphometric and Biochemical Parameters of Deschampsia antarctica from its Southern Range Limit in Maritime Antarctic. Polish Polar Res. 39, 525–548. doi:10.24425/118759
Noor, M. A. F., Grams, K. L., Bertucci, L. A., and Reiland, J. (2001). Chromosomal Inversions and the Reproductive Isolation of Species. Proc. Natl. Acad. Sci. 98, 12084–12088. doi:10.1073/pnas.221274498
Paradis, E., Claude, J., and Strimmer, K. (2004). APE: Analyses of Phylogenetics and Evolution in R Language. Bioinformatics 20, 289–290. doi:10.1093/bioinformatics/btg412
Plohl, M., Meštrovic, N., and Mravinac, B. (2012). “Satellite DNA Evolution,” in Repetitive DNA (Granada: Karger Publishers), 126–152. doi:10.1159/000337122
Raskina, O., Barber, J. C., Nevo, E., and Belyayev, A. (2008). Repetitive DNA and Chromosomal Rearrangements: Speciation-Related Events in Plant Genomes. Cytogenet. Genome Res. 120, 351–357. doi:10.1159/000121084
Revell, L. J. (2012). Phytools: An R Package for Phylogenetic Comparative Biology (And Other Things). Methods Ecol. Evol. 3, 217–223. doi:10.1111/j.2041-210X.2011.00169.x
Röser, M., Winterfeld, G., Döring, E., and Schneider, J. (2014). Chromosome Evolution in Grass Tribes Aveneae/Poeae (Poaceae): Insights from Karyotype Structure and Molecular Phylogeny. Schlechtendalia 28, 1–22.
Ruiz-Ruano, F. J., López-León, M. D., Cabrero, J., and Camacho, J. P. M. (2016). High-throughput Analysis of the Satellitome Illuminates Satellite DNA Evolution. Sci. Rep. 6, 1–14. doi:10.1038/srep28333
Saarela, J. M., Bull, R. D., Paradis, M. J., Ebata, S. N., Peterson, P. M., Soreng, R. J., et al. (2017). Molecular Phylogenetics of Cool-Season Grasses in the Subtribes Agrostidinae, Anthoxanthinae, Aveninae, Brizinae, Calothecinae, Koeleriinae and Phalaridinae (Poaceae, Pooideae, Poeae, Poeae Chloroplast Group 1). Pk 87, 1–139. doi:10.3897/phytokeys.87.12774
Said, M., Hřibová, E., Danilova, T. V., Karafiátová, M., Čížková, J., Friebe, B., et al. (2018). The Agropyron Cristatum Karyotype, Chromosome Structure and Cross-Genome Homoeology as Revealed by Fluorescence In Situ Hybridization with Tandem Repeats and Wheat Single-Gene Probes. Theor. Appl. Genet. 131, 2213–2227. doi:10.1007/s00122-018-3148-9
Schwarzacher, T., and Heslop-Harrison, P. (2000). Practical in Situ Hybridization. Norwich: BIOS Scientific Publishers Ltd.
Sharma, S., and Raina, S. N. (2005). Organization and Evolution of Highly Repeated Satellite DNA Sequences in Plant Chromosomes. Cytogenet. Genome Res. 109, 15–26. doi:10.1159/000082377
Soltis, P. S., and Soltis, D. E. (2009). The Role of Hybridization in Plant Speciation. Annu. Rev. Plant Biol. 60, 561–588. doi:10.1146/annurev.arplant.043008.092039
Strom, A. R., Emelyanov, A. V., Mir, M., Fyodorov, D. V., Darzacq, X., and Karpen, G. H. (2017). Phase Separation Drives Heterochromatin Domain Formation. Nature 547, 241–245. doi:10.1038/nature22989
Tzvelev, N. N., Probatova, N. S., Probatova, N. S., and Chiapella, J. (2015). New Taxa of Deschampsia P. Beauv. (Poaceae) from Russia. Bot. Pac. 4, 1–6. doi:10.17581/bp.2015.04105
Ugarkovic, D. (2008). Satellite DNA Libraries and Centromere Evolution. Toevolj 2, 1–6. doi:10.2174/1874404400802010001
Vershinin, A. V., Schwarzacher, T., and Heslop-Harrison, J. S. (1995). The Large-Scale Genomic Organization of Repetitive DNA Families at the Telomeres of rye Chromosomes. Plant Cell 7, 1823–1833. doi:10.1105/tpc.7.11.1823
Wei, L., Liu, B., Zhang, C., Yu, Y., Yang, X., Dou, Q., et al. (2020). Identification and Characterization of Satellite DNAs in Poa L. Mol. Cytogenet. 13, 1–8. doi:10.1186/s13039-020-00518-x
Widmer, A., Lexer, C., and Cozzolino, S. (2009). Evolution of Reproductive Isolation in Plants. Heredity 102, 31–38. doi:10.1038/hdy.2008.69
Winterfeld, G., and Röser, M. (2007a). Chromosomal Localization and Evolution of Satellite DNAs and Heterochromatin in Grasses (Poaceae), Especially Tribe Aveneae. Plant Syst. Evol. 264, 75–100. doi:10.1038/ejhg.2015.20410.1007/s00606-006-0482-1
Winterfeld, G., and Röser, M. (2007b). Disposition of Ribosomal DNAs in the Chromosomes of Perennial Oats (Poaceae: Aveneae). Bot. J. Linn. Soc. 155, 193–210. doi:10.1111/j.1095-8339.2007.00690.x
Yang, J., Yuan, B., Wu, Y., Li, M., Li, J., Xu, D., et al. (2020). The Wide Distribution and Horizontal Transfers of Beta Satellite DNA in Eukaryotes. Genomics 112, 5295–5304. doi:10.1016/j.ygeno.2020.10.006
Keywords: Deschampsia, cytogenetics, repetitive DNA, satellite DNA, FISH
Citation: González ML, Chiapella JO and Urdampilleta JD (2021) Chromosomal Differentiation of Deschampsia (Poaceae) Based on Four Satellite DNA Families. Front. Genet. 12:728664. doi: 10.3389/fgene.2021.728664
Received: 21 June 2021; Accepted: 06 September 2021;
Published: 21 September 2021.
Edited by:
Magdalena Vaio, Universidad de la República, UruguayReviewed by:
Sonia Garcia, Consejo Superior de Investigaciones Científicas (CSIC), SpainCopyright © 2021 González, Chiapella and Urdampilleta. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Juan Domingo Urdampilleta, anVyZGFtcGlsbGV0YUBpbWJpdi51bmMuZWR1LmFy
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.