- 1Department of Breast Surgery, First Hospital of Shanxi Medical University, Taiyuan, China
- 2Department of Microbiology and Immunology, Shanxi Medical University, Taiyuan, China
Background: Invasive ductal carcinoma (IDC) is the most common type of metastatic breast cancer. Due to the lack of valuable molecular biomarkers, the diagnosis and prognosis of IDC remain a challenge. A large number of studies have confirmed that coagulation is positively correlated with angiogenesis-related factors in metastatic breast cancer. Therefore, the purpose of this study was to construct a COAGULATION-related genes signature for IDC using the bioinformatics approaches.
Methods: The 50 hallmark gene sets were obtained from the molecular signature database (MsigDB) to conduct Gene Set Variation Analysis (GSVA). Gene Set Enrichment Analysis (GSEA) was applied to analyze the enrichment of HALLMARK_COAGULATION. The COAGULATION-related genes were extracted from the gene set. Then, Limma Package was used to identify the differentially expressed COAGULATION-related genes (DECGs) between ductal carcinoma in situ (DCIS) and invasive ductal carcinoma (IDC) samples in GSE26340 data set. A total of 740 IDC samples from The Cancer Genome Atlas (TCGA) database were divided into a training set and a validation set (7:3). The univariate and multivariate Cox regression analyses were performed to construct a risk signature, which divided the IDC samples into the high- and low-risk groups. The overall survival (OS) curve and receiver operating characteristic (ROC) curve were drawn in both training set and validation set. Finally, a nomogram was constructed to predict the 1-, 2-, 3-, 4-, and 5-year survival rates of IDC patients. Quantitative real-time fluorescence PCR (qRT-PCR) was performed to verify the expression levels of the prognostic genes.
Results: The “HALLMARK_COAGULATION” was significantly activated in IDC. There was a significant difference in the clinicopathological parameters between the DCIS and IDC patients. Twenty-four DECGs were identified, of which five genes (SERPINA1, CAPN2, HMGCS2, MMP7, and PLAT) were screened to construct the prognostic model. The high-risk group showed a significantly lower survival rate than the low-risk group both in the training set and validation set (p=3.5943e-06 and p=0.014243). The risk score was demonstrated to be an independent predictor of IDC prognosis. A nomogram including risk score, pathological_stage, and pathological_N provided a quantitative method to predict the survival probability of 1-, 2-, 3-, 4-, and 5-year in IDC patients. The results of decision curve analysis (DCA) further demonstrated that the nomogram had a high potential for clinical utility.
Conclusion: This study established a COAGULATION-related gene signature and showed its prognostic value in IDC through a comprehensive bioinformatics analysis, which may provide a potential new prognostic mean for patients with IDC.
Introduction
According to the GLOBOCAN 2020 cancer statistics, breast cancer (BRCA) has become one of the most common malignancies worldwide with an incidence and mortality rate of 47.8 per 100,000 and 13.6 per 100,000, respectively, and patients with breast cancer account for the highest proportion of cancers in women (Sung et al., 2021). Breast cancer, as a highly heterogeneous tumor, has different phenotypes and biological behaviors due to differences in genomics, transcriptomics, and tumor microenvironmental, thus showing different morphological manifestations, treatment response, and histological grading (Rakha et al., 2010). Among breast cancer, invasive ductal carcinoma (IDC) is the most common tissue type that accounts for 75–80% of the disease (Low et al., 2018). Compared to ductal carcinoma in situ (DCIS), IDC cells can break through the basement membrane and can metastasize to distant sites, and the high metastatic and invasive capacity of IDC is the main cause of death in breast cancer patients. The invasion and metastasis of cancer is a complex process controlled by various molecular determinants and may involve activation of oncogenes, inactivation of tumor suppressor genes, and aberration of the related signaling pathways (Fan et al., 2014; Deng et al., 2019). Therefore, uncovering the key molecular biomarkers or targets involved in breast cancer invasion and metastasis may help develop novel strategies for the diagnosis and treatment of IDC.
The main molecular biomarkers currently used in the diagnosis and treatment of breast cancer include estrogen receptor (ER), progesterone receptor (PR), cell proliferation antigen Ki67, and human epidermal growth factor receptor 2 (HER-2). These prognostic biomarkers can reflect tumor growth pattern, growth rate, malignant phenotype, recurrence, and metastasis and may often inform the prognosis of breast cancer and guide endocrine therapy and chemotherapy (Dunnwald et al., 2007; Kontzoglou et al., 2013; Burstein, 2020). However, the expression status of these traditional prognostic markers can only be obtained by biopsy puncture or postoperative pathology, and some triple-negative breast cancer patients have a higher rate of tumor recurrence and metastasis and a worse prognosis due to the lack of clear therapeutic targets. Therefore, it is necessary to discover new, easy, and convenient prognostic markers for patients with IDC (Yin et al., 2020).
The coagulation system, especially the tissue factor (TF) pathway, is an important innate defense mechanism (Khorana et al., 2019), and tumor cells use the function of these host-protective pathways to shape the tumor microenvironment (TME) and to metastasize. A large amount of experimental evidence shows that the organism of malignant tumor patients is in a hypercoagulable and hyperfibrinolytic state (a state that is caused by enhanced coagulation and massive fibrin production in the early stages of the disease), and the degree of coagulation disorders correlates with tumor invasion, metastasis and patient prognosis (Tas et al., 2013b). Inhibiting the coagulation cascade at the three stages of formation of prothrombin activator, thrombin, and fibrin (Ruf and Mueller, 2006) and blocking selective platelet activation pathways (Haemmerle et al., 2018) can prevent the spread of metastatic tumors. Many coagulation disorder-related biomarkers have shown significant prognostic relevance in various cancers (Tas et al., 2013a; Marfia et al., 2016; Ji et al., 2018; Li et al., 2019; Zhang et al., 2020).
Tissue factor (TF), an integral membrane protein, is the primary initiator of the extrinsic clotting pathway. TF promotes tumor growth, migration, and angiogenesis, independent of the coagulation cascade, via activation of protease-activated receptor (PAR)2 (Ruf et al., 2010). Some studies found that TF can mediate microvesicular shedding of MDA-MB-231 breast cancer cells and participate in cancer cell invasion and metastasis (Rondon et al., 2018). The activation of TF and thrombin in carcinoma in situ of the breast is an invasive phenomenon that promotes tumor cell infiltration and metastasis and can be used to predict the prognosis of breast cancer (Shaker et al., 2020). D-dimer (DD), as a coagulation indicator, is a product of fibrin degradation. DD is increased in breast cancer, with raised levels being found in 58% of patients with involved lymph nodes and only 8% of patients without lymph node disease (Blackwell et al., 2000). Circulating tumor cells (CTC) are associated with a hypercoagulable blood state in patients with metastatic breast cancer (Kirwan et al., 2020). Moreover, DD levels are closely related to the ability of tumor patients to tolerate chemotherapy (Yuan et al., 2017) and to treatment response (Louneva et al., 2020). Thrombin-antithrombin III (TAT) complex plasma concentration is used to assess the activation of the coagulation system as a surrogate marker for activated thrombin (Blanke et al., 1987). Plasma TAT and DD are higher in early breast cancer patients than in healthy controls (Topcu et al., 2015). Circulating fibrinogen, which is converted to fibrin by thrombin, has shown utility in distinguishing breast cancer from benign breast disease (Fu et al., 2017) and increases with disease progression (Edwards et al., 1987). In addition, high tissue expression levels of urokinase-type plasminogen activator (uPA) and plasminogen activator inhibitor-1 (PAI-1) in the fibrinolytic system are also prognostic factors in breast cancer and are associated with recurrence and metastasis (Grøndahl-Hansen et al., 1993). Therefore, we hypothesized that coagulation-related biomarkers play a crucial role in the assessment of breast cancer prognosis. To our knowledge, studies on coagulation-related genes and prognosis in invasive ductal carcinoma have rarely been reported.
In this study, we systematically evaluated the expression profile, prognostic significance, and role of COAGULATION-related genes by integrating data from the GEO database, TCGA database, and the GSEA database. Based on our analysis of those data, we constructed a prognostic model that may be used to predict individualized outcomes for patients with IDC.
Materials and Methods
Acquisition of Data Sets and Human Tumor Samples
Fifty hallmark gene sets were downloaded from Molecular signature database (MsigDB), and the raw chip data of GSE26340 data set, which included 31 ductal carcinoma in situ (DCIS) samples and 36 infiltrative ductal carcinoma (IDC) samples, were downloaded from the Gene Expression Omnibus (GEO) database.1 The IDC transcriptome and clinical data information were downloaded from The Cancer Genome Atlas (TCGA)2 that includes 740 IDC tissues.
All human breast cancer tissue samples used in the study were obtained from patients who were operated on in the First Affiliated Hospital of Shanxi Medical University, from 2019 to 2021. The tissues included 5 ductal carcinoma in situ (DCIS) samples and 10 infiltrative ductal carcinoma (IDC) samples. All tissue samples were evaluated by two experienced pathologists. This study was approved by the Medical Ethics Committee of the First Hospital of Shanxi Medical University and was performed under the approved guidelines. Informed consents were acquired from each breast cancer patient. The tumor tissue was immediately frozen and stored at −80°C.
Gene Set Variation Analysis and Gene Set Enrichment Analysis
We divided samples from the GSE26034 data set into the DCIS and IDC groups. Using the hallmark gene sets as the reference gene set and setting the p-value to <0.05 and the|t value|>2 as the cutoff criteria, we performed GSVA between DCIS and IDC patients by using the GSVA package 1.38 in R (Hänzelmann et al., 2013). The common activation/inhibition pathways of IDC were determined. GSEA was performed to analyze the difference between DCIS and IDC patients via javaGSEA 4.1.0 to show the HALLMARK_COAGULATION plot (Mootha et al., 2003; Subramanian et al., 2005).
Comparison Analysis of Clinical Characteristics Between DCIS and IDC Subtypes in the GSE26034 Data Set
The difference in each clinicopathological characteristic between the DCIS and IDC subtypes was analyzed by the chi-square test.
Differentially Expressed Analysis and Correlation of Analysis of COAGULATION-Related Gene
COAGULATION-related genes were extracted from the gene set. By using the Screening conditions of|log2FC|>0.5, value of p<0.05, the differentially expressed COAGULATION-related genes (DECGs) were determined between DCIS and IDC samples in the GSE26304 data set were analyzed using Limma Package, and then, the heat map showing the expression of DECGs was drawn via “heatmap 1.0.12” in R.3 In addition, Pearson correlation analysis was conducted between these DECGs. Pearson correlation coefficient (r)>0.6 and a value of p<0.05 were considered as medium strong correlations.
Construction of a Prognostic COAGULATION-Related Gene Signature Using Univariate and Stepwise Multivariate Cox Regression Analyses
Combined with the clinical information of IDC patients in the TCGA database, the genes related to the overall survival (OS) of IDC patients were screened by univariate Cox regression analysis via “survival 3.2-7” package in R, and p<0.05 was used as the criteria of selection. Then, the genes with value of p<0.05 were used for stepwise multivariate Cox regression analysis to construct a risk model. “Forestplot” package 1.10 in R was used to generate a forest map to show the HR (hazard ratio), lower/upper 95% CI, and p-value.4
Estimation of the COAGULATION-Related Gene Signature for IDC Prognosis
According to the “survival” package in R and linear model, the predict. Coxph function was used to calculate the risk score of each IDC patient in the training set and validation set. The formula is as follows:
where β is the coefficient and X is the normalized expression of each signature gene.
Then, we drew the corresponding heat map and risk curve. To determine whether the risk model has prognostic value, the Kaplan–Meier survival curves of high- and low-risk groups were drawn via the “survminer 0.4.8” package in R.5 In addition, we drew the survival curves of each gene in training set and validation set, respectively. ROC curves were drawn using the R package “survival ROC 1.0.3” for assessing and validating the effectiveness of the risk model. The area under curve (AUC) values of 1-, 2-, 3-, 4-, and 5-year survival were calculated.
Independence of the Prognostic Model From Other Clinicopathological Characteristics
To determine whether the prognostic model is independent in other clinical characteristics, all clinicopathological characteristics in the TCGA-IDC data set, such as age, gender, pathological tumor stage, pathological N stage, pathological T stage, pathological M stage, molecular subtypes, and treatment, were included for analysis. To determine which factors have the independent prognostic value, we performed univariate and multivariate Cox regression analyses on these clinical characteristics and risk scores. The factors with p<0.05 were considered to have the independent prognostic value.
Construction and Verification of the Survival Prediction Nomogram
Based on the independent prognostic analysis, we found that the prognostic model risk score, pathological_stage, Pharmaceutical_Therapy, and Both_Treatment were the independent prognostic factors that could be utilized to predict the survival rate in the TCGA-IDC data set. Therefore, the four independent prognostic factors and the molecular subtypes of IDC were used to build the nomogram via R package “rms 6.0–1.6” Besides, the survival ROC curve of these factors was used to validate the nomogram. Furthermore, the decision curve analysis (DCA), which can assess and compare prediction models that incorporate clinical consequences, was used to determine whether the nomogram was suitable for the clinical application. The x-axis indicates the percentage of threshold probability, and the y-axis represents the net benefit.
qRT-PCR Analysis
Total RNA was extracted from the tissues of infiltrative ductal carcinoma and ductal carcinoma in situ tissues using Nuclezol LS RNA Isolation Reagent Kit (ABP Biosciences Inc) following the manufacturer’s instructions.
Subsequently, cDNA was synthesized from total RNA using SureScript-First-strand-cDNA-synthesis-kit (GeneCopoeia, China). Then, RT-qPCR was performed using BlazeTaq™ SYBR ® Green qPCR Mix 2.0 (GeneCopoeia, China) on a CFX96 real-time quantitative fluorescence PCR instrument (Bio-Rad, United States) according to the following protocol: pre-denaturation at 95°C for 30s, denaturation at 95°C for 10s, annealing at 60°C for 20s, and extension at 72°C for 30s, and the whole reaction cycle 40 times.
The primers used were as follows: PLAT: fw: 5'-GCCGTGAATTTAAGGGACGC, rev: 3'-GCTCGCTGCAACCTTGGTAA, SERPINA1: fw: 5'-CCTTCACTGTCAACTTCGGG, rev: 3'-TGTGTCTCTGTCAAGCTCCT, HMGCS2: fw: 5'-TCTCTTATGGCTCTGGTTTAG, rev: 3'-GCTCTCTTTGGTTCATTATTT, MMP7: fw: 5'-TGACTCAGAAACAAAAAATGCC, rev: 3'-TACGATCCTGTAGGTGACCACT, and Gapdh: fw: 5'-CGCTGAGTACGTCGTGGAGTC, rev: 3'-GCTGATGATCTTGAGGCTGTTGTC. For each sample, qPCR assays were conducted in triplicate in a 10ml reaction volume. Using the 2−ΔΔCt method, we analyzed the RT-qPCR data by normalizing gene expression to GAPDH expression as an endogenous control.
Statistical Analysis
All the statistical analyses were performed using R software 4.0.3. Statistical significance was set as probability value p<0.05. The chi-square test was used to analyze the differences between the DCIS/IDC subtypes and clinicopathological parameters. The Kaplan–Meier survival curves were built to analyze the survival differences between the high-risk and low-risk groups. Univariate and multivariate Cox proportional hazard models were generated to estimate the hazard ratios of prognostic factors and to select independent prognostic factors. The ROC curves and DCA were compared to determine the predictive accuracy of the independent prognostic factors.
Results
“HALLMARK_COAGULATION” Pathway Is Hyperactivated in IDC Patients
We conducted GSVA of hallmark gene sets from the GSE26304 data set. The results demonstrated that the “HALLMARK_EPITHELIAL_MESENCHYMAL_TRANSITION,” “HALLMARK_ANGIOGENESIS,” “HALLMARK_UV_RESPONSE_DN,” “HALLMARK_TGF_BETA_SIGNALING,” “HALLMARK_HEDGEHOG_SIGNALING”_and “HALLMARK_COAGULATION” pathways with t value >2 were significantly activated in IDC (Figure 1A). Previous studies have revealed the correlations between these signaling pathways. Epithelial-to-mesenchymal transition (EMT) and hedgehog signaling pathways can lead to angiogenesis, loss of normal organ function, or cancer by modulating TGF-β signaling pathway in cancer progression (Wang et al., 2013; Bausch et al., 2020; Sha et al., 2020). In skin diseases, the promoted angiogenesis and lymphatic vessel enlargement were a response to ultraviolet (UV; Okazaki et al., 2012). Moreover, angiogenesis was decreased by inhibiting coagulation (Nadir, 2019). EMT also promotes basal mammary stem cell and tumor-initiating cell stemness by inducing hedgehog signaling (Guen et al., 2017). The inhibition of hedgehog signaling attenuates UVB-induced skin photoaging, reducing blood coagulation (Kim et al., 2015; Shahramian et al., 2019). CTCs expressing EMT traits could harbor enhanced procoagulant activity that could facilitate early metastasis (Bourcy et al., 2016). Hao et al. (2019) have revealed that TGF-β induces EMT to promote tumorigenesis. The inhibition of TGF-β by UV disrupted immune homeostasis in systemic lupus erythematosus (SLE; Rekik et al., 2018). In addition, several situations reported the cross talks between the TGF-β and hedgehog signaling pathways (Pelullo et al., 2019). Then, we performed GSEA of the “HALLMARK_COAGULATION” gene sets and found that it was strikingly positively enriched in IDC samples (Figure 1B). These results suggest that COAGULATION-related genes might be involved in the pathogenesis of IDC.
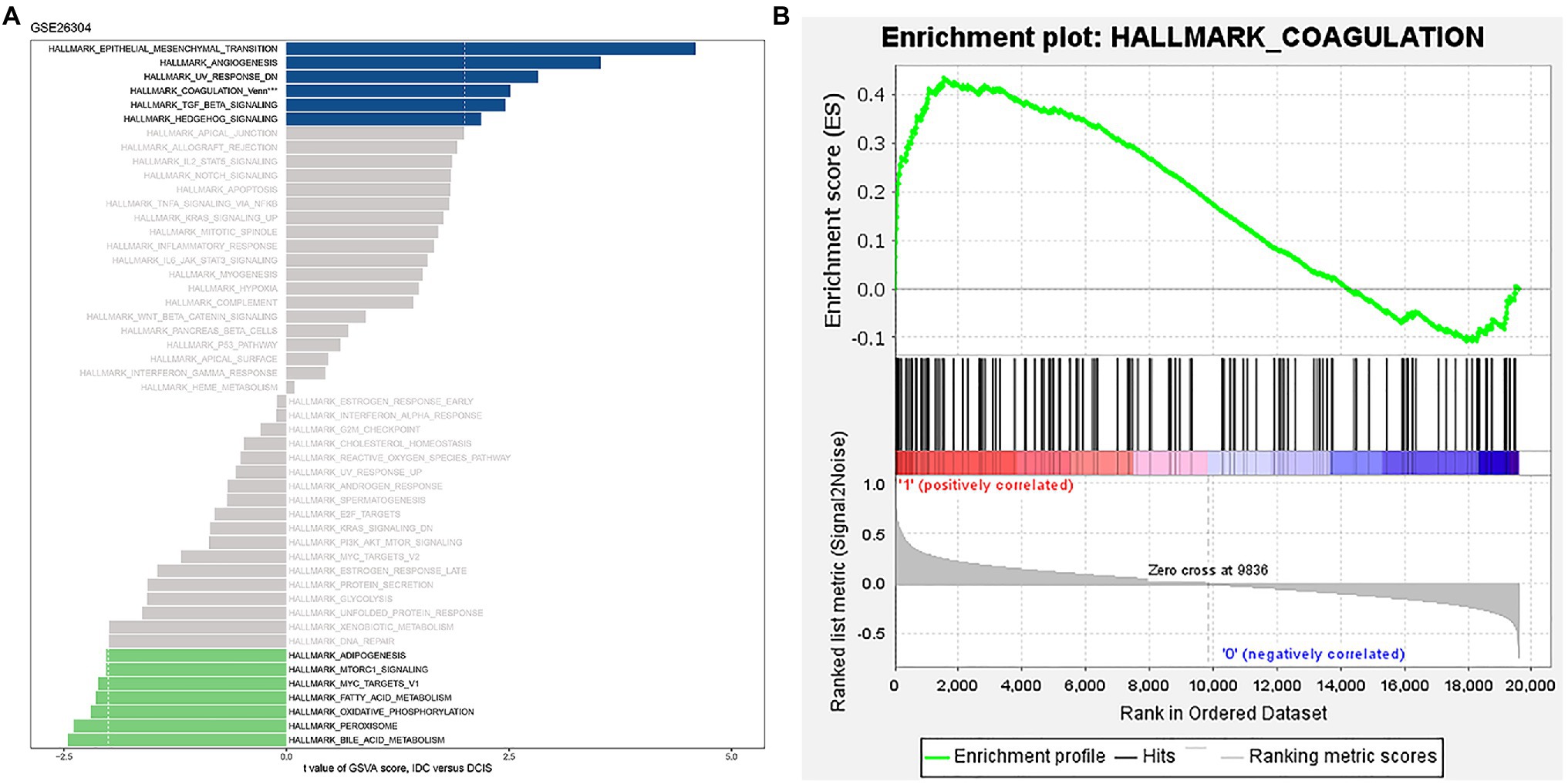
Figure 1. “HALLMARK_COAGULATION” pathway was significantly activated in IDC. (A) GSVA of the 50 hallmark gene sets in the GSE26034 data set. (B) Enrichment plot of “HALLMARK_COAGULATION” pathway.
Differences in Clinical Characteristics Between DCIS and IDC Subtypes
We analyzed the differences in clinical characteristics between IDC and DCIS patients. And, there were significant differences in the clinicopathological parameters between DCIS and IDC patients, especially grade (p=0.00209), tumor size (p<0.001), X-ray treatment (p=0.024), and hormone treatment (p=0.00454; Table 1). In consideration of the significant differences in clinical characteristics between IDC/DCIS and the significant enrichment of the “HALLMARK_COAGULATION” pathway in IDC, we further performed the prognostic analysis of COAGULATION-related genes in IDC.
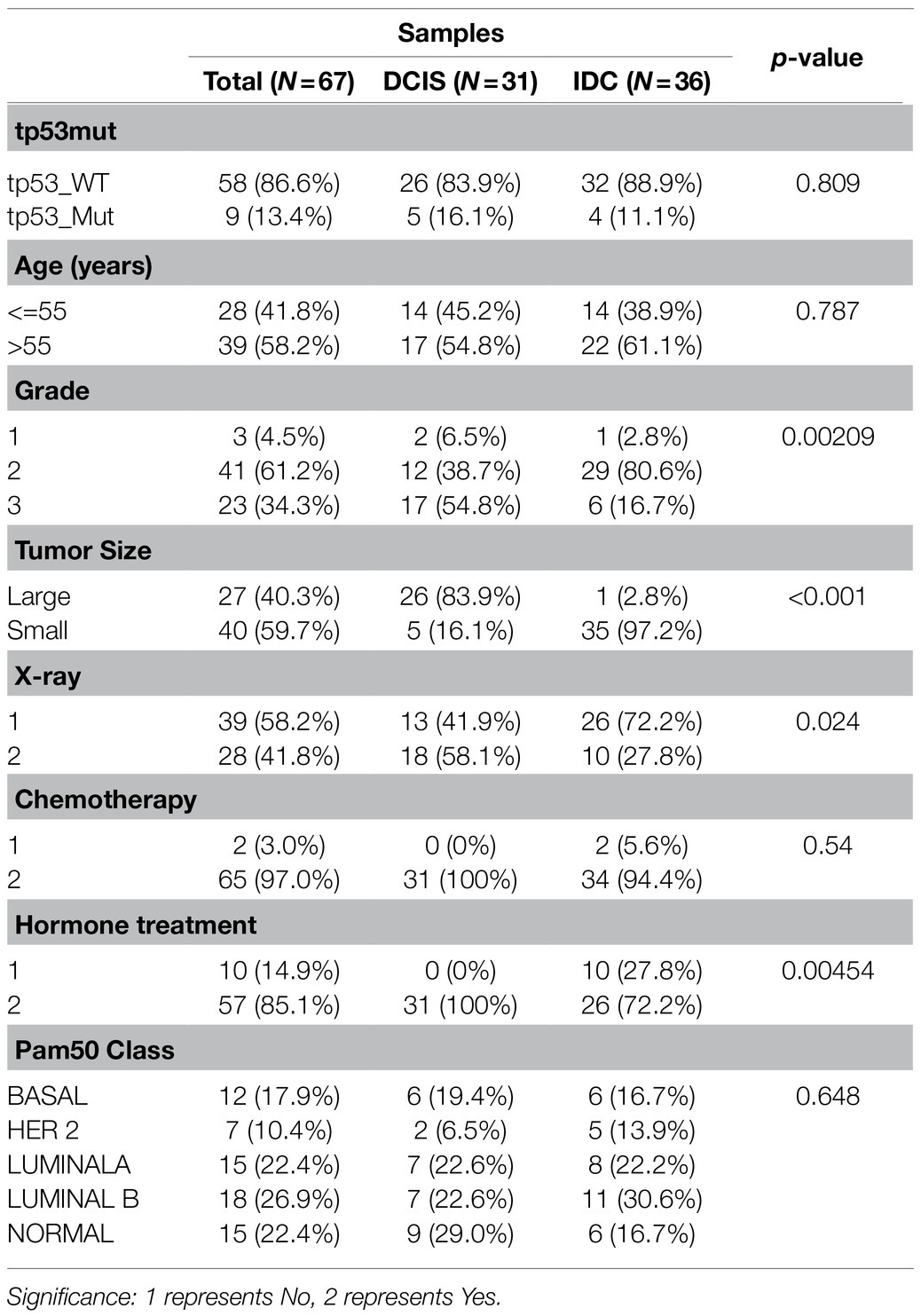
Table 1. Statistical analysis of clinical characteristics of IDC/DCIS patients in the GSE26304 data set.
COAGULATION-Related Genes Expression and Correlation Analysis
COAGULATION-related genes were extracted from the gene set. The expression levels of differentially expressed COAGULATION-related genes (DECGs) were compared between DCIS and IDC samples from the GSE26304 data set. As shown in Figures 2A,B, a total of 24 DECGs were identified. Pearson correlation analysis showed that there was a significant correlation between the genes, such as SPARC was positively correlated with HTRA1, FBN1, and FN1, and SERPINA1 was negatively correlated with CTSK, C1S, and C1R (Figure 2C).
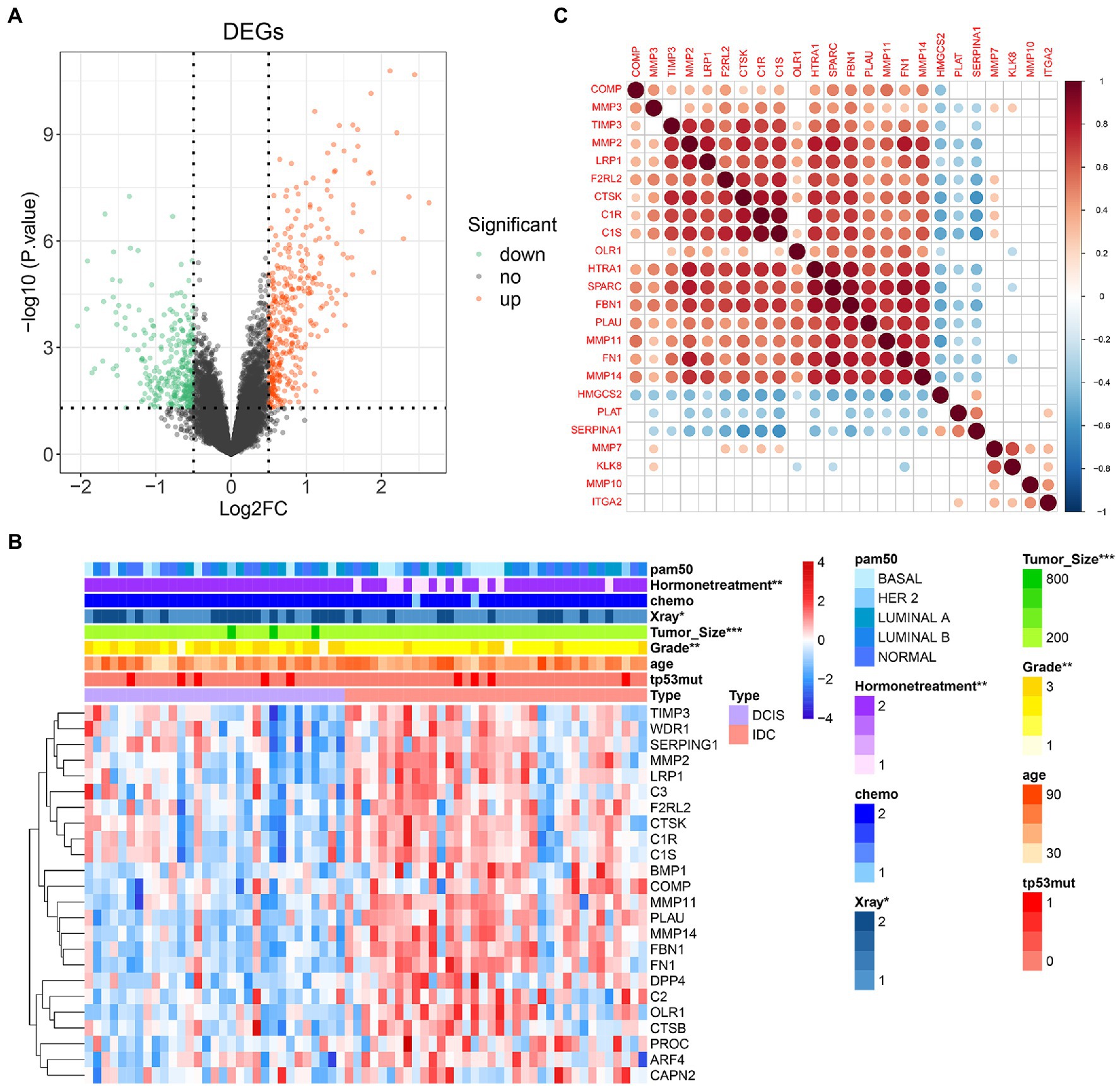
Figure 2. COAGULATION-related genes expression and correlation analysis. (A) Volcano map reveals the distribution of downregulated and upregulated DECGs. X-axis: fold change; Y-axis: −log10 p-value. (B) Heat map shows the expression of differentially expressed COAGULATION-related genes between DCIS and IDC samples. (C) Pearson correlation of these differentially expressed COAGULATION-related genes. * p < 0.05, ** p < 0.01, and *** p <0.001.
Construction of the Prognostic Gene Signature in IDC
We then performed a prognostic analysis of 24 DECGs in the TCGA database. In the present study, 740 IDC patients were randomly divided into training set and validation set according to the 7:3 ratio, and 6 of 24 DEGCs with p<0.05 were screened by univariate Cox hazard ratio regression analysis (Figure 3A), which was used to perform a stepwise multivariate Cox proportional hazard analysis. Four genes (SERPINA1, HMGCS2, MMP7, and PLAT) were selected as prognostic gene signatures in IDC patients (Figure 3B). The expression levels of the four prognostic genes were significantly downregulated in the IDC patient (Figure 3C). Notably, the expression of SERPINA1 and PLAT significantly correlated with the prognosis of IDC patients, respectively (p=0.00249 and 0.0312). However, HMGCS2 and MMP7 showed no significant prognostic significance in the IDC patients in the training set (Figure 3D). In addition, while SERPINA1 and HMGCS2 demonstrated significant prognostic significance in IDC patients (p=0.004735 and 0.018575, respectively), PLAT and MMP7 showed no significant prognostic significance in validation set (Figure 3E).
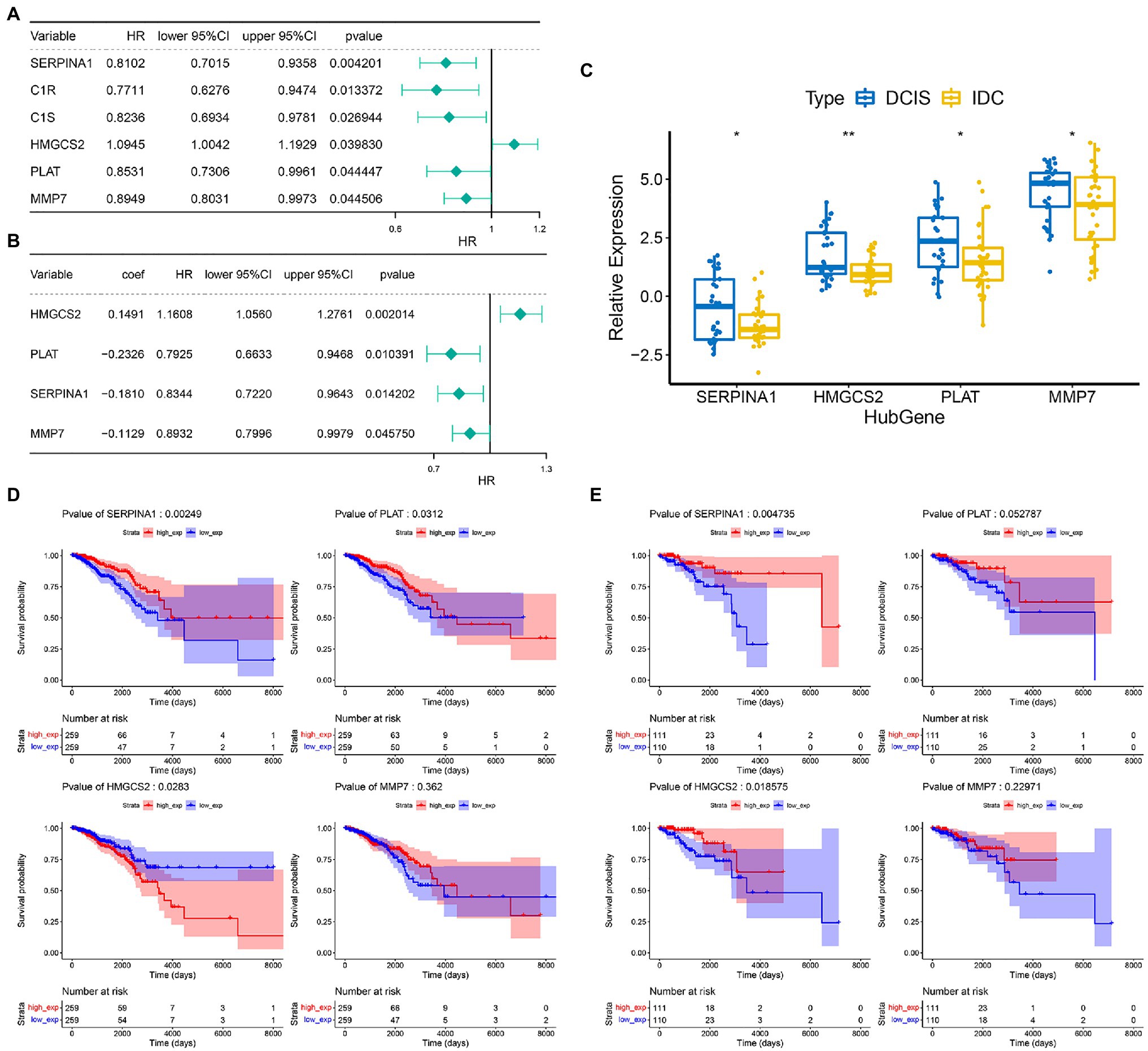
Figure 3. Construction of prognostic gene signature of 4 COAGULATION-related genes in IDC. (A) Univariate Cox proportional risk regression forest plot including p-values, lower/upper 95%CI, and HR. (B) Forest plot for multivariate Cox regression analysis. (C) The expression levels of the four prognostic genes between the DCIS and IDC samples in the GSE26304 data set. (D) Kaplan–Meier survival curves of model genes in the training set. (E) Kaplan–Meier survival curves of model genes in the validation set. * p < 0.05 and ** p < 0.01.
Evaluation of the Prognostic COAGULATION-Related Gene Signature
Based on the 4 genes signature, we calculated the risk score for each IDC sample, and then, patients were equally divided into low-risk and high-risk groups in the training set. The risk curve and survival state distribution and risk score heat map in low-risk and high-risk groups were shown in Figures 4A,B. The results of Kaplan–Meier analysis indicated that the survival rate of patients with low-risk score was higher than that of patients with a high-risk score (p<0.0001; Figure 4C). Time-dependent ROC analysis showed that the AUC of the COAGULATION-related gene signature was 0.693 for 1year, 0.638 for 2years, 0.628 for 3years, 0.633 for 4years, and 0.664 for 5years, respectively (Figure 4D).

Figure 4. Evaluation of the prognostic gene signature of 4 COAGULATION-related genes. (A) Risk curve and its corresponding scatter plots in the training set. (B) Heat map of risk model genes in the training set. (C) Kaplan–Meier survival curves of high- and low-risk groups in the training set. Red represents high risk, and blue represents low risk. (D) Survival ROC curves for 1-, 2-, 3-, 4-, and 5-year survival.
Validation of the Prognostic COAGULATION-Related Gene Signature
To further assess the prediction value of this COAGULATION-related gene signature, we constructed risk curve, survival state distribution, overall survival, and ROC curve between low-risk and high-risk groups in the validation set. The risk curve and survival state distribution and risk score heat map were similar to the training set (Figures 5A,B). We observed significantly higher survival rates in the low-risk group than that in the high-risk group in the validation set (Figure 5C). The 1-, 2-, 3-, 4-, and 5-year prognostic accuracies were 0.710, 0.681, 0.513, 0.592, and 0.621, respectively (Figure 5D).
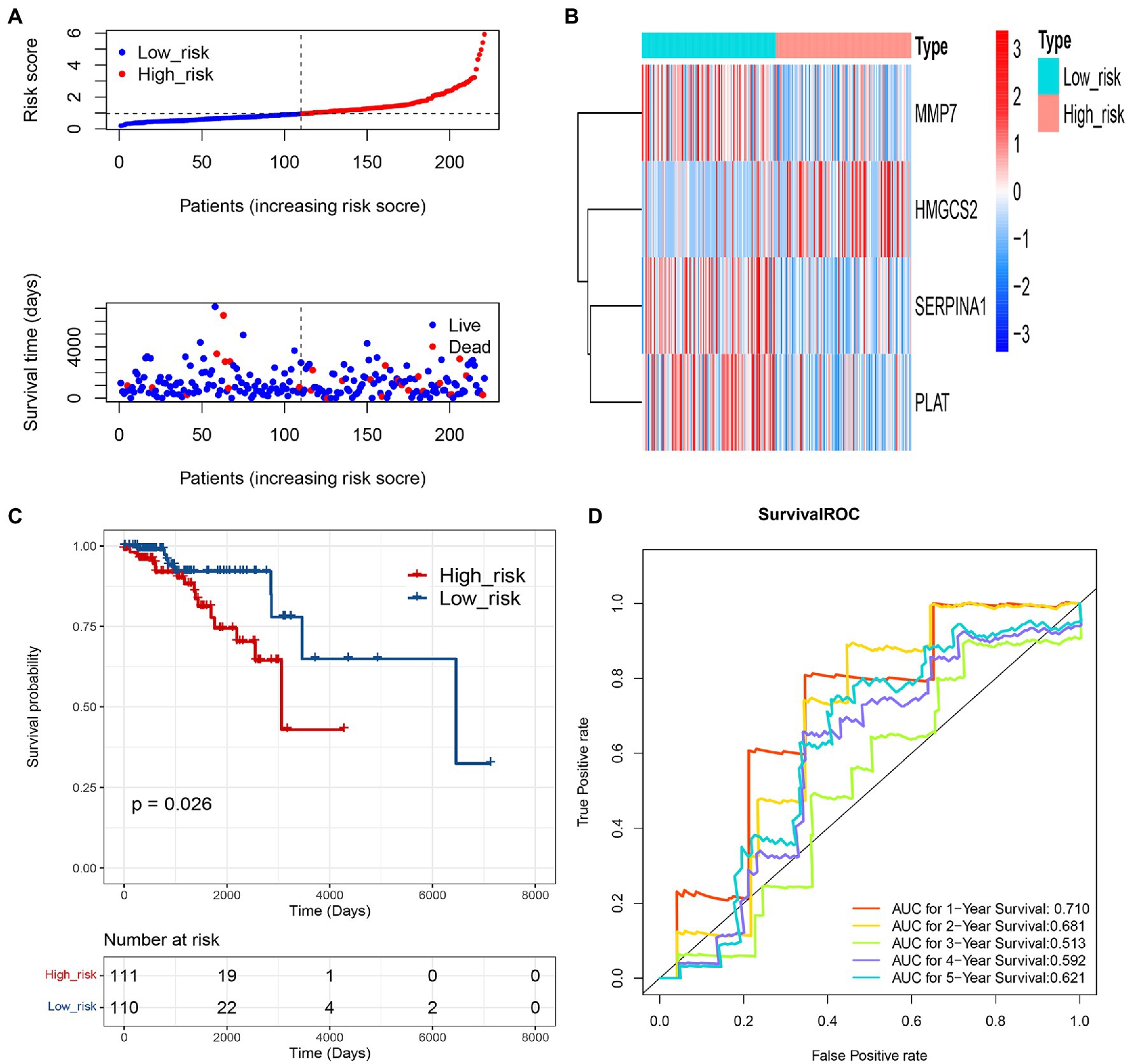
Figure 5. Validation of the prognostic gene signature of 4 COAGULATION-related genes. (A) Risk curve and its corresponding scatter plots in the validation set. (B) Heat map of risk model genes in the validation set. (C) Kaplan–Meier survival curves of high- and low-risk groups in the validation set. Red represents high risk, and blue represents low risk. (D) Survival ROC curves for 1-, 2-, 3-, 4-, and 5-year survival.
Evaluation of COAGULATION-Related Gene Signature in the Four Molecular Subtypes of IDC
Having shown that the signature based on COAGULATION-related genes can serve as a potent prognostic signature for IDC, we further explored the potential effect of prognosis prediction for the molecular subtypes of IDC (Figure 6). The K-M analysis showed that there was a significant difference in OS between the high-risk groups and low-risk groups of LuA patients (p=0.013). The ROC curve analyses at 1-, 2-, 3-, 4-, and 5-year for this prognostic gene signature were conducted to evaluate the predictive efficiency. The results showed that the risk score had high accuracy in predicting the 1- and 2-year survival of LuA patients. However, this signature based on COAGULATION-related genes could not predict the other molecular subtypes, such as LuB, HER2, and basal-like.
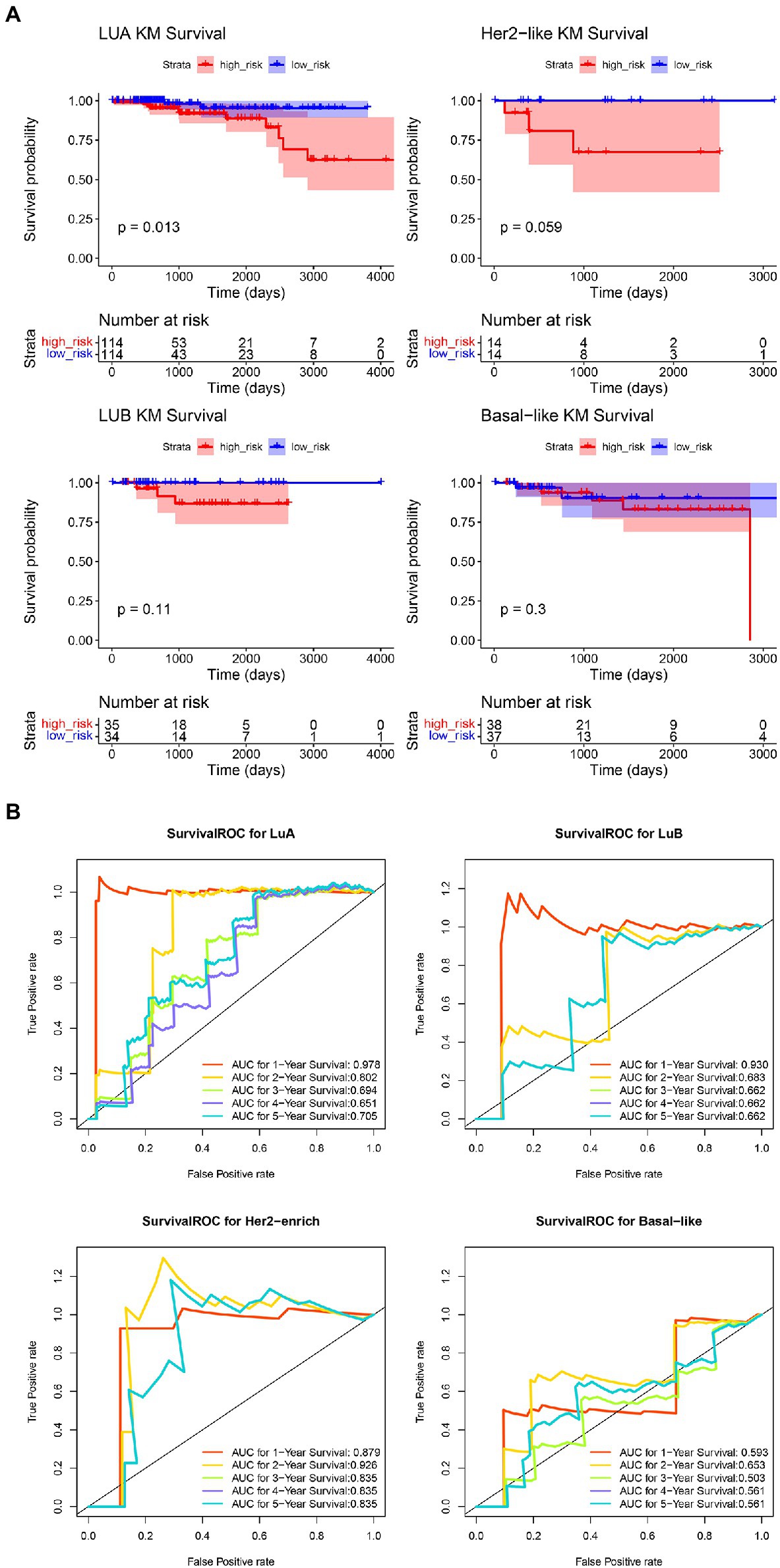
Figure 6. Evaluation of COAGULATION-related gene signature in the four molecular subtypes of IDC. (A) The K-M curves of high-risk groups and low-risk groups in the four molecular subtypes of IDC. (B) The AUC of ROC curves in predicting 1-, 2-, 3-, 4-, and 5-year OS.
Univariate and Multivariate Cox Regression Analyses of the Independent Prognostic Ability of 4-Gene Signature
Next, we performed univariate and multivariate Cox regression analyses on risk score and other clinical characteristics. The results revealed that the COAGULATION-related gene signature constructed by SERPINA1, HMGCS2, MMP7, and PLAT, pathological_stage, Pharmaceutical_Therapy, and Both_Treatment were the independent prognostic factors that could be used to predict the survival rate of IDC patients (Figures 7A,B).
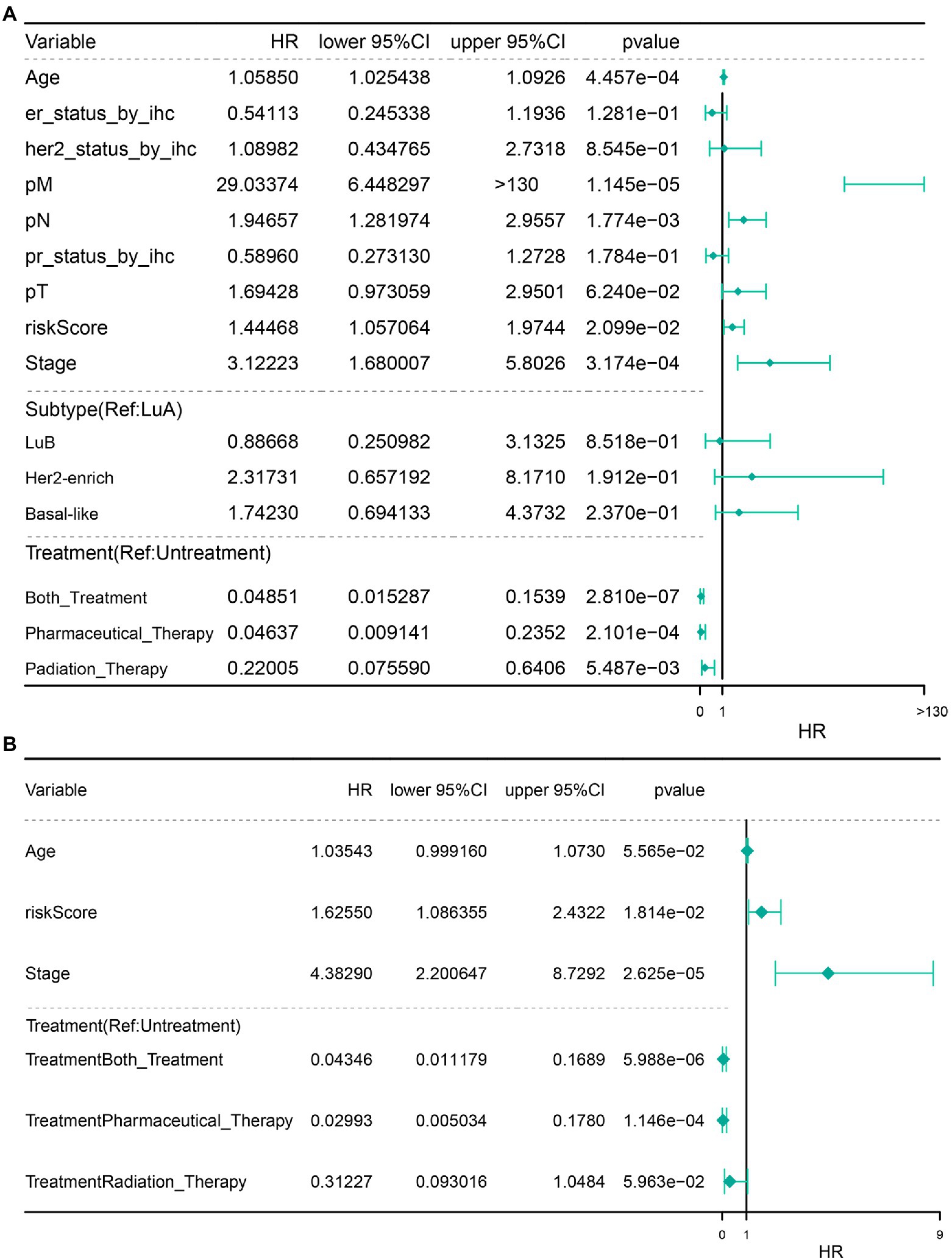
Figure 7. Independent prognostic analysis. (A) Independent prognostic univariate Cox regression analysis. (B) Independent prognostic multivariate analysis.
Construction of Survival Prediction Nomogram in IDC
Based on the independent prognostic factors and molecular subtypes, we constructed a nomogram for predicting 1-, 2-, 3-, 4-, and 5-year survival rates. The nomogram containing pathological_stage, Pharmaceutical_Therapy, Both_Treatment, and molecular subtypes may provide a quantitative method for predicting the 1-, 2-, 3-, 4-, and 5-year survival rate of IDC patients (Figure 8A, c-index=0.82). Every patient got a point for each prognostic parameter, and a higher score indicated a worse prognosis. ROC analyses indicated that the prognostic accuracy of the COAGULATION-related gene signature-based prognostic nomogram was compared with those of pathological_stage treatment and molecular subtypes of IDC (Figure 8B). The results of 3- and 5-year DCA demonstrated that the independent prognostic factors had a high potential for clinical application (Figures 8C,D).

Figure 8. Construction and validation of survival prediction nomogram in IDC. (A) A nomogram for predicting the survival rates of 1-, 2-, 3-, 4-, and 5-year. (B) ROC of prognostic signature of 4 COAGULATION-related genes, pathological_stage treatment, and molecular subtypes of IDC. (C) DCA decision curve for 3-year survival prediction. (D) DCA decision curve for 5-year survival predicting.
Validation of Prognostic Signature Genes Expression in IDC/DCIS Tissues
To assess the genes that are significant for the construction of the risk score signature, we performed the qRT-PCR assay. The results of the qRT-PCR analysis demonstrated that PLAT, SERPINA1, HMGCS2, and MMP7 were significantly downregulated in IDC tissues, compared with the DCIS tissues (all p<0.05, Figures 9A–D), which were consistent with the results obtained from the GSE26304 data set.
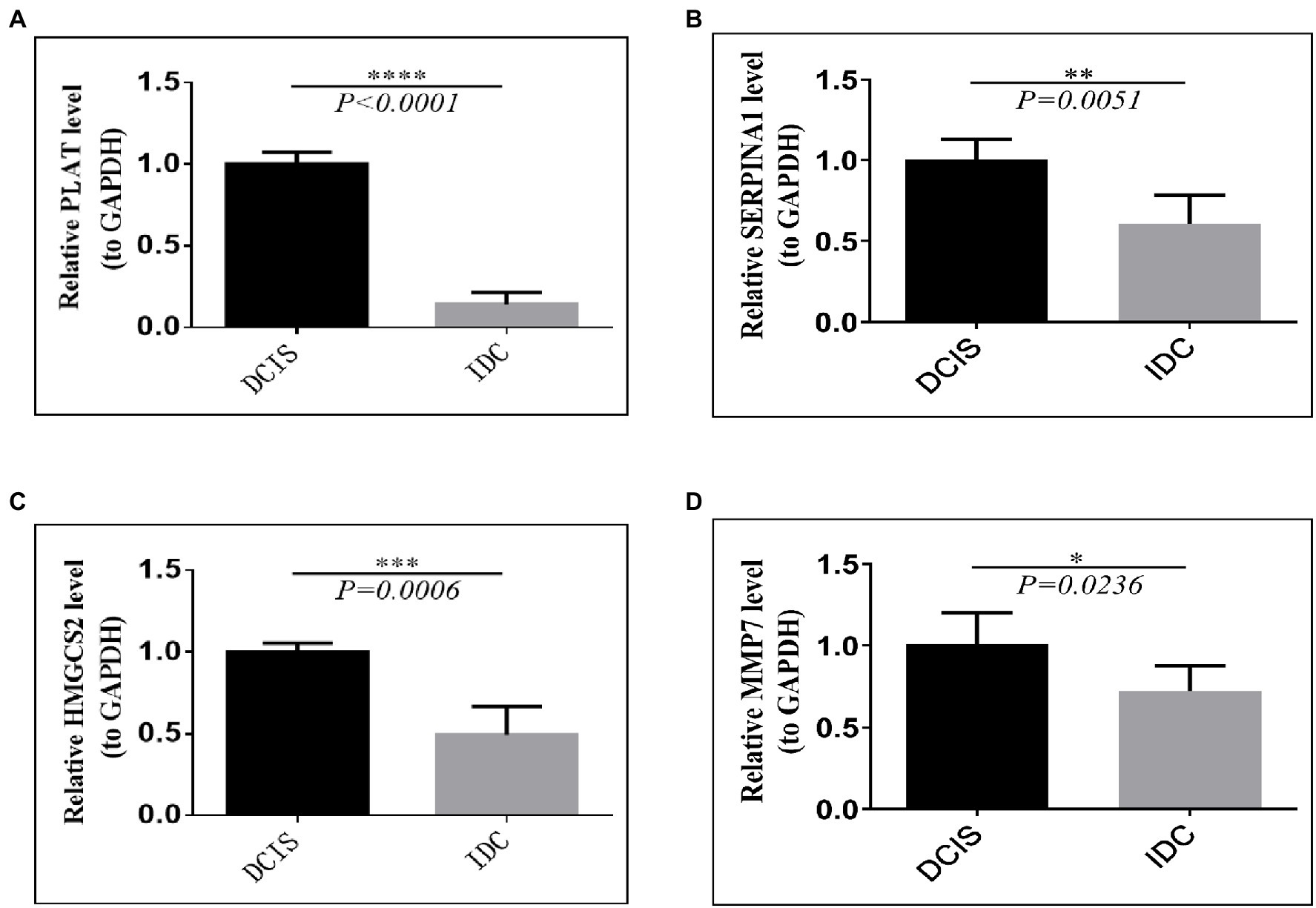
Figure 9. Validation of prognostic signature gene expression in IDC/DCIS tissues. The mRNA expression patterns of four prognostic between DCIS and IDC tissues. (A) PLAT, (B) SERPINA1, (C) HMGCS2, and (D) MMP7. All experimental data were expressed in means ± SD (IDC: n=10, DCIS: n=5). * p<0.05, ** p<0.01, *** p<0.001, **** vs. the DCIS group.
Discussion
The incidence of breast cancer is increasing year by year, and despite the improvement of treatments, it is very difficult to treat in the event of recurrence or metastasis. Therefore, identifying effective prognostic biomarkers is an indispensable step in predicting breast cancer patients’ disease and controlling the progress of treatment. With the rapid development of molecular genomics, highly specific diagnostic and prognostic biomarkers are providing molecular evidence to improve conventional diagnostic and therapeutic options.
In this study, we first found that the “HALLMARK_COAGULATION” pathway was activated in IDC based on the results of GSVA analysis. The enrichment of the most COAGULATION genes in IDC indicates a potential role of these genes in the tumorigenesis and development of IDC. There were studies reporting found that patients with IDC had higher DD levels, hypercoagulation, secondary fibrinolysis, and increased fibrin degradation (Bazzi et al., 2016; Dai et al., 2018; Shaker et al., 2020). There is evidence that fibrin formed by fibrinogen decomposition not only provides scaffolding for tumor cells but also acts as a ligand to promote adhesion binding between platelets and tumor cells, thus promoting the infiltration and metastasis of tumor cells (Mego et al., 2015). Secondly, patients with IDC have high PLT aggregation activity and low erythrocyte count, which may increase erythrocyte aggregation and plasma viscosity, resulting in impaired oxygen transport efficiency and microcirculatory hypoxia, which is conducive to tumor sedimentation, metastasis, and thrombosis (Tikhomirova et al., 2016). In a hypercoagulable state, tumor cells can interact with PLT immediately (within 2min) after leaving the primary site and entering the bloodstream, and PLT aggregates and secretes VEGF, which increases the local vascular density of the tumor (Krenn-Pilko et al., 2015), immediately surrounding the tumor cells and protecting them from blood impact and NK cell-based immune responses (Placke et al., 2011). In addition, PLT induces EMT in tumor cells (Labelle et al., 2011), giving the cells the characteristics of tumor stem cells with increased malignancy (Scheel et al., 2011; Ye and Weinberg, 2015). It also contributes to the exudation of tumor cells from the blood by secreting chemokines CXCL5 and CXCL7 that activate the granulocyte expression receptor CXCR2. Therefore, in the infiltration and angiogenesis of breast malignancies, tumor development forms a vicious circle with the activation of coagulation-related factors and cells.
We used transcriptomic data and clinical data of IDC patients in the TCGA database to construct a prognostic model of four genes (SERPINA1, HMGCS2, MMP7, and PLAT) related to the coagulation pathway. We further verified the actual expression of these four genes in the GSE26304 clinical samples using PCR technique. Among them, SERPINA1 and PLAT showed significant correlations. SERPINA1, a plasma protein, is synthesized mainly in hepatocytes, which is mainly involved in angiogenesis, intravascular fibrinolysis, wound healing, tumor invasion, and metastasis. It has been reported that the expression of SERPINA1 was correlated with the prognosis of metastasis in a variety of cancers, including lung, colon, and bladder cancers (Zhang et al., 2014; Kwon et al., 2015; Ercetin et al., 2019). Our study showed that SERPINA1 is a potential protective gene expressed in breast cancer, which is consistent with previous studies (Chan et al., 2015). PLAT is a gene encoding tissue-type plasminogen activator (tPA), which is synthesized and secreted mainly by vascular endothelial cells. PLAT has a role in promoting fibrinolysis and antagonizing thrombosis and is likely involved in tumor oncogenesis and progression (Jia et al., 2019). HMGCS2 is localized at 1p12, which encodes a mitochondrial hydroxymethylglutaryl coenzyme A synthase (HMGCS) found mainly in the liver and testicular tissues (Hu et al., 2017). It is the first rate-limiting enzyme in ketone body synthesis and can drive malignancy progression and metastasis by promoting ketone body production and reuse (Koutnik et al., 2020; Wang et al., 2020). MMP-7 is a stromal lysin protein, and many studies have shown that MMP-7 plays a significant regulatory role in apoptosis, immunity, cell migration, and angiogenesis pathways. The function of MMP7 usually complements the classical tumor properties, leading to invasion, immune immunity, and metastasis of tumor cells and plays an important role in the carcinogenesis process (Jobin et al., 2017). Many malignant tumors including gastrointestinal tumors, breast cancer, and head and neck tumors are found to have a high expression of MMP-7 (Gobin et al., 2019; Dirks et al., 2020).
This study, combining with clinical features, demonstrated that pathological_stage, Pharmaceutical_Therapy, Both_Treatment, and molecular subtypes have independent prognostic value, and based on this finding, we constructed a nomogram to predict IDC survival from 1 to 5years, which may provide a platform for clinical assessment of breast cancer. However, further studies with more clinical samples and information are needed to test the benefits and value of this prognostic model.
Conclusion
Overall, through analyzing the “HALLMARK_COAGULATION” pathway in IDC and its potential prognostic value in IDC, we constructed a COAGULATION-related gene-based prognostic model and developed a nomogram for predicting the survival of IDC patients within 1–5years. In this model, the expression of SERPINA1 and PLAT was significantly correlated with the survival of IDC. Thus, we propose that the expressions of SERPINA1 and PLAT warrant further investigation for their potential as new predictive biomarkers for IDC.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/supplementary material.
Author Contributions
All authors participated in the design of this study and read and approved the final version of the manuscript. JD was in charge of data obtainment and analysis. JL was in charge of statistical analysis and drafted the manuscript. YW and HJ supervised this study and revised the manuscript.
Funding
The research fund was provided by General Project of Natural Science Foundation of Shanxi Province (201901D111347).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We thank all authors of the studies included in this study. We also thank all database of the studies for providing the free data and the free R software which was used for analysis.
Footnotes
1. ^https://www.ncbi.nlm.nih.gov/geo/
2. ^https://portal.gdc.cancer.gov/
3. ^https://CRAN.R-project.org/package=pheatmap
4. ^https://CRAN.R-project.org/package=forestplot
References
Bausch, D., Fritz, S., Bolm, L., Wellner, U. F., Fernandez-Del-Castillo, C., Warshaw, A. L., et al. (2020). Hedgehog signaling promotes angiogenesis directly and indirectly in pancreatic cancer. Angiogenesis 23, 479–492. doi: 10.1007/s10456-020-09725-x
Bazzi, Z. A., Lanoue, D., El-Youssef, M., Romagnuolo, R., Tubman, J., Cavallo-Medved, D., et al. (2016). Activated thrombin-activatable fibrinolysis inhibitor (TAFIa) attenuates breast cancer cell metastatic behaviors through inhibition of plasminogen activation and extracellular proteolysis. BMC Cancer 16:328. doi: 10.1186/s12885-016-2359-1
Blackwell, K., Haroon, Z., Broadwater, G., Berry, D., Harris, L., Iglehart, J. D., et al. (2000). Plasma D-dimer levels in operable breast cancer patients correlate with clinical stage and axillary lymph node status. J. Clin. Oncol. 18, 600–608. doi: 10.1200/JCO.2000.18.3.600
Blanke, H., Praetorius, G., Leschke, M., Seitz, R., Egbring, R., and Strauer, B. E. (1987). Significance of the thrombin-antithrombin III complex in the diagnosis of pulmonary embolism and deep venous thrombosis--comparison with fibrinopeptide A, platelet factor 4 and beta-thromboglobulin. Klin. Wochenschr. 65, 757–763. doi: 10.1007/BF01743250
Bourcy, M., Suarez-Carmona, M., Lambert, J., Francart, M. E., Schroeder, H., Delierneux, C., et al. (2016). Tissue factor induced by epithelial-mesenchymal transition triggers a procoagulant state that drives metastasis of circulating tumor cells. Cancer Res. 76, 4270–4282. doi: 10.1158/0008-5472.CAN-15-2263
Burstein, H. J. (2020). Systemic therapy for estrogen receptor-positive, HER2-negative breast cancer. N. Engl. J. Med. 383, 2557–2570. doi: 10.1056/NEJMra1307118
Chan, H. J., Li, H., Liu, Z., Yuan, Y. C., Mortimer, J., and Chen, S. (2015). SERPINA1 is a direct estrogen receptor target gene and a predictor of survival in breast cancer patients. Oncotarget 6, 25815–25827. doi: 10.18632/oncotarget.4441
Dai, H., Zhou, H., Sun, Y., Xu, Z., Wang, S., Feng, T., et al. (2018). D-dimer as a potential clinical marker for predicting metastasis and progression in cancer. Biomed. Rep. 9, 453–457. doi: 10.3892/br.2018.1151
Deng, J. L., Xu, Y. H., and Wang, G. (2019). Identification of potential crucial genes and key pathways in breast cancer using bioinformatic analysis. Front. Genet. 10:695. doi: 10.3389/fgene.2019.00695
Dirks, R. C., Burney, H. N., Anjanappa, M., Sandusky, G. E., Hao, Y., Liu, Y., et al. (2020). Breast heterogeneity: obstacles to developing universal biomarkers of breast cancer initiation and progression. J. Am. Coll. Surg. 231, 85–96. doi: 10.1016/j.jamcollsurg.2020.03.035
Dunnwald, L. K., Rossing, M. A., and Li, C. I. (2007). Hormone receptor status, tumor characteristics, and prognosis: a prospective cohort of breast cancer patients. Breast Cancer Res. 9:R6. doi: 10.1186/bcr1639
Edwards, R. L., Rickles, F. R., Moritz, T. E., Henderson, W. G., Zacharski, L. R., Forman, W. B., et al. (1987). Abnormalities of blood coagulation tests in patients with cancer. Am. J. Clin. Pathol. 88, 596–602. doi: 10.1093/ajcp/88.5.596
Ercetin, E., Richtmann, S., Delgado, B. M., Gomez-Mariano, G., Wrenger, S., Korenbaum, E., et al. (2019). Clinical significance of SERPINA1 gene and its encoded alpha1-antitrypsin protein in NSCLC. Cancers 11:1306. doi: 10.3390/cancers11091306
Fan, L., Strasser-Weippl, K., Li, J. J., St Louis, J., Finkelstein, D. M., Yu, K. D., et al. (2014). Breast cancer in China. Lancet Oncol. 15, e279–e289. doi: 10.1016/S1470-2045(13)70567-9
Fu, S., Yun, Z. Y., Cui, M. M., Meng, H., Qian, C., Liu, T., et al. (2017). Cancer antigen 15-3, platelet distribution width, and fibrinogen in combination to distinguish breast cancer from benign breast disease in non-conclusive mammography patients. Oncotarget 8, 67829–67836. doi: 10.18632/oncotarget.18870
Gobin, E., Bagwell, K., Wagner, J., Mysona, D., Sandirasegarane, S., Smith, N., et al. (2019). A pan-cancer perspective of matrix metalloproteases (MMP) gene expression profile and their diagnostic/prognostic potential. BMC Cancer 19:581. doi: 10.1186/s12885-019-5768-0
Grøndahl-Hansen, J., Christensen, I. J., Rosenquist, C., Brünner, N., Mouridsen, H. T., Danø, K., et al. (1993). High levels of urokinase-type plasminogen activator and its inhibitor PAI-1 in cytosolic extracts of breast carcinomas are associated with poor prognosis. Cancer Res. 53, 2513–2521.
Guen, V. J., Chavarria, T. E., Kröger, C., Ye, X., Weinberg, R. A., and Lees, J. A. (2017). EMT programs promote basal mammary stem cell and tumor-initiating cell stemness by inducing primary ciliogenesis and hedgehog signaling. Proc. Natl. Acad. Sci. U. S. A. 114, E10532–e10539. doi: 10.1073/pnas.1711534114
Haemmerle, M., Stone, R. L., Menter, D. G., Afshar-Kharghan, V., and Sood, A. K. (2018). The platelet lifeline to cancer: challenges and opportunities. Cancer Cell 33, 965–983. doi: 10.1016/j.ccell.2018.03.002
Hänzelmann, S., Castelo, R., and Guinney, J. (2013). GSVA: gene set variation analysis for microarray and RNA-seq data. BMC Bioinform. 14:7. doi: 10.1186/1471-2105-14-7
Hao, Y., Baker, D., and Ten Dijke, P. (2019). TGF-β-mediated epithelial-mesenchymal transition and cancer metastasis. Int. J. Mol. Sci. 20:2767. doi: 10.3390/ijms20112767
Hu, L. T., Zhu, B. L., Lai, Y. J., Long, Y., Zha, J. S., Hu, X. T., et al. (2017). HMGCS2 promotes autophagic degradation of the amyloid-β precursor protein through ketone body-mediated mechanisms. Biochem. Biophys. Res. Commun. 486, 492–498. doi: 10.1016/j.bbrc.2017.03.069
Ji, R., Ren, Q., Bai, S., Wang, Y., and Zhou, Y. (2018). Prognostic significance of pretreatment plasma fibrinogen in patients with hepatocellular and pancreatic carcinomas: a meta-analysis. Medicine (Baltimore) 97:e10824. doi: 10.1097/MD.0000000000010824
Jia, N., Tong, H., Zhang, Y., Katayama, H., Wang, Y., Lu, W., et al. (2019). CeRNA expression profiling identifies KIT-related circRNA-miRNA-mRNA networks in gastrointestinal stromal tumour. Front. Genet. 10:825. doi: 10.3389/fgene.2019.00825
Jobin, P. G., Butler, G. S., and Overall, C. M. (2017). New intracellular activities of matrix metalloproteinases shine in the moonlight. Biochim. Biophys. Acta, Mol. Cell Res. 1864, 2043–2055. doi: 10.1016/j.bbamcr.2017.05.013
Khorana, A. A., Soff, G. A., Kakkar, A. K., Vadhan-Raj, S., Riess, H., Wun, T., et al. (2019). Rivaroxaban for thromboprophylaxis in high-risk ambulatory patients with cancer. N. Engl. J. Med. 380, 720–728. doi: 10.1056/NEJMoa1814630
Kim, W., Kim, E., Yang, H. J., Kwon, T., Han, S., Lee, S., et al. (2015). Inhibition of hedgehog signalling attenuates UVB-induced skin photoageing. Exp. Dermatol. 24, 611–617. doi: 10.1111/exd.12735
Kirwan, C. C., Descamps, T., and Castle, J. (2020). Circulating tumour cells and hypercoagulability: a lethal relationship in metastatic breast cancer. Clin. Transl. Oncol. 22, 870–877. doi: 10.1007/s12094-019-02197-6
Kontzoglou, K., Palla, V., Karaolanis, G., Karaiskos, I., Alexiou, I., Pateras, I., et al. (2013). Correlation between Ki67 and breast cancer prognosis. Oncology 84, 219–225. doi: 10.1159/000346475
Koutnik, A. P., Poff, A. M., Ward, N. P., Deblasi, J. M., Soliven, M. A., Romero, M. A., et al. (2020). Ketone bodies attenuate wasting in models of atrophy. J. Cachexia. Sarcopenia Muscle 11, 973–996. doi: 10.1002/jcsm.12554
Krenn-Pilko, S., Langsenlehner, U., Stojakovic, T., Pichler, M., Gerger, A., Kapp, K. S., et al. (2015). An elevated preoperative plasma fibrinogen level is associated with poor disease-specific and overall survival in breast cancer patients. Breast 24, 667–672. doi: 10.1016/j.breast.2015.08.003
Kwon, C. H., Park, H. J., Choi, J. H., Lee, J. R., Kim, H. K., Jo, H. J., et al. (2015). Snail and serpinA1 promote tumor progression and predict prognosis in colorectal cancer. Oncotarget 6, 20312–20326. doi: 10.18632/oncotarget.3964
Labelle, M., Begum, S., and Hynes, R. O. (2011). Direct signaling between platelets and cancer cells induces an epithelial-mesenchymal-like transition and promotes metastasis. Cancer Cell 20, 576–590. doi: 10.1016/j.ccr.2011.09.009
Li, M., Wu, Y., Zhang, J., Huang, L., Wu, X., and Yuan, Y. (2019). Prognostic value of pretreatment plasma fibrinogen in patients with colorectal cancer: a systematic review and meta-analysis. Medicine (Baltimore) 98:e16974. doi: 10.1097/MD.0000000000016974
Louneva, N., Maity, A., and Kennedy, A. R. (2020). Plasma D-dimer levels are elevated in radiation oncology patients. Radiat. Res. 193, 46–53. doi: 10.1667/RR15429.1
Low, S. K., Zembutsu, H., and Nakamura, Y. (2018). Breast cancer: the translation of big genomic data to cancer precision medicine. Cancer Sci. 109, 497–506. doi: 10.1111/cas.13463
Marfia, G., Navone, S. E., Fanizzi, C., Tabano, S., Pesenti, C., Abdel Hadi, L., et al. (2016). Prognostic value of preoperative von willebrand factor plasma levels in patients with glioblastoma. Cancer Med. 5, 1783–1790. doi: 10.1002/cam4.747
Mego, M., Karaba, M., Minarik, G., Benca, J., Sedlácková, T., Tothova, L., et al. (2015). Relationship between circulating tumor cells, blood coagulation, and urokinase-plasminogen-activator system in early breast cancer patients. Breast J. 21, 155–160. doi: 10.1111/tbj.12388
Mootha, V. K., Lindgren, C. M., Eriksson, K. F., Subramanian, A., Sihag, S., Lehar, J., et al. (2003). PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat. Genet. 34, 267–273. doi: 10.1038/ng1180
Nadir, Y. (2019). Decreasing tumor growth and angiogenesis by inhibition of coagulation. Semin. Thromb. Hemost. 45, 622–628. doi: 10.1055/s-0039-1693473
Okazaki, H., Hirakawa, S., Shudou, M., Nakaoka, Y., Shirakata, Y., Miyata, K., et al. (2012). Targeted overexpression of Angptl6/angiopoietin-related growth factor in the skin promotes angiogenesis and lymphatic vessel enlargement in response to ultraviolet B. J. Dermatol. 39, 366–374. doi: 10.1111/j.1346-8138.2011.01396.x
Pelullo, M., Zema, S., Nardozza, F., Checquolo, S., Screpanti, I., and Bellavia, D. (2019). Wnt, notch, and TGF-β pathways impinge on hedgehog Signaling complexity: an open window on cancer. Front. Genet. 10:711. doi: 10.3389/fgene.2019.00711
Placke, T., Kopp, H. G., and Salih, H. R. (2011). Modulation of natural killer cell anti-tumor reactivity by platelets. J. Innate Immun. 3, 374–382. doi: 10.1159/000323936
Rakha, E. A., Reis-Filho, J. S., Baehner, F., Dabbs, D. J., Decker, T., Eusebi, V., et al. (2010). Breast cancer prognostic classification in the molecular era: the role of histological grade. Breast Cancer Res. 12:207. doi: 10.1186/bcr2607
Rekik, R., Smiti Khanfir, M., Larbi, T., Zamali, I., Beldi-Ferchiou, A., Kammoun, O., et al. (2018). Impaired TGF-β signaling in patients with active systemic lupus erythematosus is associated with an overexpression of IL-22. Cytokine 108, 182–189. doi: 10.1016/j.cyto.2018.04.011
Rondon, A. M. R., De Almeida, V. H., Gomes, T., Verçoza, B. R. F., Carvalho, R. S., König, S., et al. (2018). Tissue factor mediates microvesicles shedding from MDA-MB-231 breast cancer cells. Biochem. Biophys. Res. Commun. 502, 137–144. doi: 10.1016/j.bbrc.2018.05.136
Ruf, W., and Mueller, B. M. (2006). Thrombin generation and the pathogenesis of cancer. Semin. Thromb. Hemost. 32(Suppl. 1), 61–68. doi: 10.1055/s-2006-939555
Ruf, W., Yokota, N., and Schaffner, F. (2010). Tissue factor in cancer progression and angiogenesis. Thromb. Res. 125(Suppl. 2), S36–S38. doi: 10.1016/S0049-3848(10)70010-4
Scheel, C., Eaton, E. N., Li, S. H., Chaffer, C. L., Reinhardt, F., Kah, K. J., et al. (2011). Paracrine and autocrine signals induce and maintain mesenchymal and stem cell states in the breast. Cell 145, 926–940. doi: 10.1016/j.cell.2011.04.029
Sha, Y., Wang, S., Bocci, F., Zhou, P., and Nie, Q. (2020). Inference of intercellular communications and multilayer gene-regulations of epithelial-mesenchymal transition from single-cell transcriptomic data. Front. Genet. 11:604585. doi: 10.3389/fgene.2020.604585
Shahramian, K., Abdulmajeed, A., Kangasniemi, I., Söderling, E., and Närhi, T. (2019). TiO(2) coating and UV photofunctionalization enhance blood coagulation on zirconia surfaces. Biomed. Res. Int. 2019:8078230. doi: 10.1155/2019/8078230
Shaker, H., Bundred, N. J., Landberg, G., Pritchard, S. A., Albadry, H., Nicholson, S. L., et al. (2020). Breast cancer stromal clotting activation (tissue factor and thrombin): a pre-invasive phenomena that is prognostic in invasion. Cancer Med. 9, 1768–1778. doi: 10.1002/cam4.2748
Subramanian, A., Tamayo, P., Mootha, V. K., Mukherjee, S., Ebert, B. L., Gillette, M. A., et al. (2005). Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U. S. A. 102, 15545–15550. doi: 10.1073/pnas.0506580102
Sung, H., Ferlay, J., Siegel, R. L., Laversanne, M., Soerjomataram, I., Jemal, A., et al. (2021). Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 71, 209–249. doi: 10.3322/caac.21660
Tas, F., Kilic, L., Bilgin, E., Keskin, S., Sen, F., Ciftci, R., et al. (2013a). Clinical and prognostic significance of coagulation assays in advanced epithelial ovarian cancer. Int. J. Gynecol. Cancer 23, 276–281. doi: 10.1097/IGC.0b013e31827b8796
Tas, F., Kilic, L., Serilmez, M., Keskin, S., Sen, F., and Duranyildiz, D. (2013b). Clinical and prognostic significance of coagulation assays in lung cancer. Respir. Med. 107, 451–457. doi: 10.1016/j.rmed.2012.11.007
Tikhomirova, I., Petrochenko, E., Malysheva, Y., Ryabov, M., and Kislov, N. (2016). Interrelation of blood coagulation and hemorheology in cancer. Clin. Hemorheol. Microcirc. 64, 635–644. doi: 10.3233/CH-168037
Topcu, T. O., Kavgacı, H., Canyılmaz, E., Orem, A., Yaman, H., Us, D., et al. (2015). The effect of adjuvant chemotherapy on plasma TAT and F 1+2 levels in patients with breast cancer. Biomed. Pharmacother. 73, 19–23. doi: 10.1016/j.biopha.2015.05.003
Wang, X., Abraham, S., Mckenzie, J. A. G., Jeffs, N., Swire, M., Tripathi, V. B., et al. (2013). LRG1 promotes angiogenesis by modulating endothelial TGF-β signalling. Nature 499, 306–311. doi: 10.1038/nature12345
Wang, Y. H., Suk, F. M., and Liao, Y. J. (2020). Loss of HMGCS2 enhances lipogenesis and attenuates the protective effect of the ketogenic diet in liver cancer. Cancers 12:1797. doi: 10.3390/cancers12071797
Ye, X., and Weinberg, R. A. (2015). Epithelial-mesenchymal plasticity: a central regulator of cancer progression. Trends Cell Biol. 25, 675–686. doi: 10.1016/j.tcb.2015.07.012
Yin, L., Duan, J. J., Bian, X. W., and Yu, S. C. (2020). Triple-negative breast cancer molecular subtyping and treatment progress. Breast Cancer Res. 22:61. doi: 10.1186/s13058-020-01296-5
Yuan, Y., Vora, N., Sun, C. L., Li, D., Soto-Perez-De-Celis, E., Mortimer, J., et al. (2017). Association of pre-chemotherapy peripheral blood pro-inflammatory and coagulation factors with reduced relative dose intensity in women with breast cancer. Breast Cancer Res. 19:101. doi: 10.1186/s13058-017-0895-5
Zhang, Y., Cao, J., Deng, Y., Huang, Y., Li, R., Lin, G., et al. (2020). Pretreatment plasma fibrinogen level as a prognostic biomarker for patients with lung cancer. Clinics 75:e993. doi: 10.6061/clinics/2020/e993
Keywords: invasive ductal carcinoma, coagulation, SERPINA1, HMGCS2, MMP7, plat, prognosis
Citation: Li J, Du J, Wang Y and Jia H (2021) A Coagulation-Related Gene-Based Prognostic Model for Invasive Ductal Carcinoma. Front. Genet. 12:722992. doi: 10.3389/fgene.2021.722992
Edited by:
Hongmin Cai, South China University of Technology, ChinaReviewed by:
Tao Qin, Sun Yat-sen Memorial Hospital, ChinaShankar Suman, The Ohio State University, United States
Copyright © 2021 Li, Du, Wang and Jia. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hongyan Jia, c3dhbGxvd19qaHlAMTYzLmNvbQ==; Yanhong Wang, d2FuZ3lhbmhvbmdtYWlsQDEyNi5jb20=
 Jing Li
Jing Li Jiajia Du
Jiajia Du Yanhong Wang2*
Yanhong Wang2* Hongyan Jia
Hongyan Jia