
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Genet. , 07 September 2021
Sec. Genomics of Plants and the Phytoecosystem
Volume 12 - 2021 | https://doi.org/10.3389/fgene.2021.713224
 Ahmed H. El-Sappah1,2†
Ahmed H. El-Sappah1,2† Rania G. Elbaiomy3†
Rania G. Elbaiomy3† Ahmed S. Elrys4
Ahmed S. Elrys4 Yu Wang1
Yu Wang1 Yumin Zhu1
Yumin Zhu1 Qiulan Huang5
Qiulan Huang5 Kuan Yan1
Kuan Yan1 Zhao Xianming1
Zhao Xianming1 Manzar Abbas1*
Manzar Abbas1* Khaled A. El-Tarabily6,7*
Khaled A. El-Tarabily6,7* Jia Li1*
Jia Li1*Metal tolerance proteins (MTPs) encompass plant membrane divalent cation transporters to specifically participate in heavy metal stress resistance and mineral acquisition. However, the molecular behaviors and biological functions of this family in Medicago truncatula are scarcely known. A total of 12 potential MTP candidate genes in the M. truncatula genome were successfully identified and analyzed for a phylogenetic relationship, chromosomal distributions, gene structures, docking analysis, gene ontology, and previous gene expression. M. truncatula MTPs (MtMTPs) were further classified into three major cation diffusion facilitator (CDFs) groups: Mn-CDFs, Zn-CDFs, and Fe/Zn-CDFs. The structural analysis of MtMTPs displayed high gene similarity within the same group where all of them have cation_efflux domain or ZT_dimer. Cis-acting element analysis suggested that various abiotic stresses and phytohormones could induce the most MtMTP gene transcripts. Among all MTPs, PF16916 is the specific domain, whereas GLY, ILE, LEU, MET, ALA, SER, THR, VAL, ASN, and PHE amino acids were predicted to be the binding residues in the ligand-binding site of all these proteins. RNA-seq and gene ontology analysis revealed the significant role of MTP genes in the growth and development of M. truncatula. MtMTP genes displayed differential responses in plant leaves, stems, and roots under five divalent heavy metals (Cd2+, Co2+, Mn2+, Zn2+, and Fe2+). Ten, seven, and nine MtMTPs responded to at least one metal ion treatment in the leaves, stems, and roots, respectively. Additionally, MtMTP1.1, MtMTP1.2, and MtMTP4 exhibited the highest expression responses in most heavy metal treatments. Our results presented a standpoint on the evolution of MTPs in M. truncatula. Overall, our study provides a novel insight into the evolution of the MTP gene family in M. truncatula and paves the way for additional functional characterization of this gene family.
Metals act as a cofactor, which has essential implications in inactivating enzymes in plant cells to perform specific biological reactions (Thomine and Vert, 2013). Essential metals including zinc (Zn), manganese (Mn), iron (Fe), cobalt (Co), and copper (Cu), play an essential role in plants at a low level, but excessive amounts of these ions lead to toxic effects (Kolaj-Robin et al., 2015). Moreover, a deficient concentration of non-essential metals, including mercury (Hg), silver (S), cadmium (Cd), and lead (Pb), can also cause plant cell toxicity (Clemens, 2001). Interestingly, plants are natural bio-accumulators for various heavy metals from the water and soil for appropriate plant growth and development activities (Ali et al., 2013; Elrys et al., 2018).
Plants overcome heavy metal stress by various physiological and molecular mechanisms, including genomic-level and complex biochemical processes (Liu et al., 2019). Some of these mechanisms are part of the homeostatic process and are constitutive (Rai et al., 2019). Other mechanisms are exclusively related to counter-specific metal toxicity (Gupta et al., 2019). All responses can be widely classified as being tolerant or avoidance types (Krzesłowska, 2011). Metal uptake, trafficking, storage, chelation, and efflux are plant mechanisms to maintain metal homeostasis (Montanini et al., 2007). Numerous studies have indicated the essential roles of various protein families with their specific transporters in these regulatory processes (Gao et al., 2020). The cation diffusion facilitator (CDF) family genes are integral membrane divalent cation transporters involved in divalent metal ion efflux from the cytoplasm either into subcellular compartments or outside the cell (Gustin et al., 2011). CDF transporters have been widely identified in many organisms since their first identification in the bacterial cell (Nies and Silver, 1995), which were further classified into three major groups: Mn-CDF, Zn/Fe-CDF, and Zn-CDF, based on either confirmed or hypothesized transported substrate specificities (Montanini et al., 2007).
Cation diffusion facilitator transporters are considered as metal tolerance proteins (MTPs) in plants, which were classified into seven distinct groups (1, 5, 6, 7, 8, 9, and 12) based on the annotation and phylogenetic analysis in Arabidopsis (Gustin et al., 2011). Many MTP proteins were identified in several plant species, including Arabidopsis thaliana (van der Zaal et al., 1999), Vitis vinifera (Shirazi et al., 2019), Populus trichocarpa (Gao et al., 2020), Triticum aestivum (Vatansever et al., 2017), Nicotiana tabacum, Nicotiana sylvestris, and Nicotiana tomentosiformis (Liu et al., 2019). Many Zn-CDF proteins have been studied from the first identified AtMTP1 in Arabidopsis (van der Zaal et al., 1999). Zn-CDF genes play an essential role in plant Zn tolerance. For instance, AtMTP1 and AtMTP3 of the tonoplast are involved in Zn and Co tolerance through the excess transport of Zn2+ and Co2+ ions to the vacuole (Dräger et al., 2004; Kobae et al., 2004; Arrivault et al., 2006; Kawachi et al., 2008). Moreover, two more genes of Zn-CDF family, including AtMTP5 and AtMTP12, were identified to form a functional complex during Zn transportation into the Golgi apparatus (Fujiwara et al., 2015).
Mn-CDF family members, including AtMTP8, play an essential role in the transportation of Mn besides its role in Fe and Mn localization in seeds (Delhaize et al., 2007; Eroglu et al., 2016; Chu et al., 2017). In Oryza sativa, the OsMTPs (OsMTPs8.1 and 8.2) of the tonoplast participate in Mn transport within the plant (Takemoto et al., 2017; Tsunemitsu et al., 2018). Moreover, the ShMTP gene in Stylosanthes hamata was the first group 8 member of MTPs to be identified that enhances the tolerance against Mn when overexpressed in Arabidopsis (Delhaize et al., 2007). Furthermore, the CsMTP8 gene of cucumber confers Mn tolerance when overexpressed in yeast and Arabidopsis (Migocka et al., 2014).
Medicago truncatula is a diploid plant species (2n = 2x = 16) which is comparatively small, and its genome has been sequenced which is being recursively used as a model plant for legume genetic research (Benedito et al., 2008; Young et al., 2011; Tang et al., 2014). It is one of the best essential forage crops widely cultivated across the world, the same as alfalfa (Young and Udvardi, 2009). M. truncatula is the best species to explore functional genomics of metal tolerance, although it does not accumulate metals by itself (Zhou et al., 2012).
However, only a few M. truncatula MTPs (MtMTPs) have been studied and characterized. In recent years, genome sequencing of model plants and commercially important plants were performed and provided opportunities to screen candidate genes (Edwards et al., 2017). Complete high-quality draft genomes provided a chance to systematically analyze the M. truncatula MTP gene family at the genome-wide level.
In the present study, 12 MTPs in M. truncatula were successfully identified and characterized based on their structural, functional, and evolutionary relationship. Furthermore, change in their expression level was evaluated under the stress of the following five heavy metals: Cd2+, Co2+, Mn2+, Zn2+, and Fe2+. Our findings will provide a deep insight into the MTP gene family involved in heavy metal stress response in a plant cell, founders, and biological functions of MtMTP proteins that will open new avenues of research in the area of the molecular mechanism of homeostasis and heavy metal transport and finally will help to precisely engineer M. truncatula plants for heavy metal stress.
The M. truncatula genome sequence was 2 from an online available M. truncatula genome database1, and a local database was constructed using BioEdit 7.0 software. Candidate MtMTP genes were analyzed for HMM profiling of the following two MTP domains PF16916 and PF01545 on the Pfam website2. The blast was investigated with our putative MTP protein sequences against the M. truncatula whole-genome sequence on NCBI3 and Phytozome4. All retrieved MTP protein sequences were analyzed at an E-value of <10–5 to identify the MTP domain using SMART5 tools (Letunic et al., 2004). All detailed genetic information of the putative MTP gene family, including chromosomal location and CDS, was obtained from the Phytozome database (see text footnote 4). Furthermore, MTP family proteins were analyzed for their molecular weight, number of atoms, amino acids, isoelectric point (PI), molecular weight, and instability index using ExPASy ProtoParam6 (Gasteiger et al., 2003). Finally, the subcellular localization data were predicted via the MTP amino acid sequences using protein subcellular localization prediction tools7.
In addition to M. truncatula, MTP gene family members of A. thaliana7, Cucumis sativus8, P. trichocarpa9, O. sativa10, and T. aestivum11 were also retrieved and analyzed for the phylogenetic tree of evolutionary MTP relationships. CLUSTALX version 2.0 with default parameters were used for MTP protein multiple sequence alignments. All alignments were uploaded in the MEGA 6.0.6 software with a neighbor-joining method to construct a phylogenetic tree. Finally, bootstrap analysis was performed for 1,000 iterations using a pair-wise gap deletion mode (Tamura et al., 2011).
The M. truncatula genetic database (see text footnote 4) has been analyzed to retrieve information about chromosomal localization of MTP genes, which were subsequently used to construct a genetic map using MapChart software12. Two genes of the same species located in the same clade were defined as co-paralogs to identify tandem or segmental duplication events. Simultaneously, the Phytozome database (see text footnote 4) has been analyzed to identify segmental duplication in M. truncatula genes. The paralogs were identified, which were considered the results of tandem duplication due to the splicing of two genes into five or more genes within a 100 kb region (Tang et al., 2008). Similarly, co-paralogs located within duplicated chromosomal regions were considered segmental duplications (Wei et al., 2007). The Smith–Waterman algorithm13 was used to calculate local alignments of two protein sequences. The synteny analysis of the MtMTP gene family was performed using Circos14 to localize different alleles distributed on different chromosomes (Krzywinski et al., 2009).
A structural analysis of all MTP gene family members was performed to explore the organization of intron/exon using both gDNA and CDS sequences by deploying an online tool, namely, Genes Structure Display Server program15 (Hu et al., 2015). The conserved gene family motifs were detected using Multiple EM for Motif Elicitation16 by adjusting the following parameters: a maximum of 20 motifs and 6–200 amino acids per motif (Bailey et al., 2006). The cis-acting elements (CREs) of the MtMTP promoter were identified using the PLACE17 and PlantCARE programs18 (Higo et al., 1999; Lescot et al., 2002).
The Phyre219 website was used for protein modeling, prediction, and analysis of MtMTP proteins (Kelley et al., 2015). The predicted protein model validation was assessed using Ramachandran plot analysis. Docking analysis of the ligand-binding regions in the predicted protein models was also performed using the deep site (Jiménez et al., 2017) and Proteins Plus20 online analysis software DoGSiteScorer (Volkamer et al., 2012) tools, and was finally visualized in PyMOL21.
Blast2GO v3.0.1122 and OmicsBox software were applied on all identified MTP protein sequences for GO annotation analysis (Conesa and Götz, 2008).
RNA-seq data obtained from different organs of M. truncatula was downloaded from the gene Expression Atlas Project (MtGEA)23 (He et al., 2009). Furthermore, the retrieved data were analyzed for the actual expression level of MTP genes obtained from the leaves, roots, seed coats, and flowers of M. truncatula under normal conditions. Subsequently, MTP gene expression was also analyzed by deploying cufflinks (version: 2.2.1). Finally, absolute FPKM values were divided by their mean and were transformed into a ratio of log2, and MeV 4.5 was used to cluster expression data into a heat map24 (Saeed et al., 2006; Babicki et al., 2016).
In this study, an M. truncatula (cv. JemalongA17) line was cultivated during the autumn of 2020 at the experimental greenhouse of Yibin University (China). First, the seeds were washed with 10% hypochlorous acid and distilled water. The seeds were germinated using water-saturated filter paper and then transferred to fertilized pittmoss soil with germination conditions of 16-h light (27°C) and 8-h dark (18°C) with relative humidity of 70%. Four seeds were planted in each plastic pot. After emergence, thinning was performed to maintain two uniform seedlings per pot. Thirty-day-old seedlings of M. truncatula were placed in 1/2 Hoagland solutions (pH 6.0) with different heavy metal concentrations of 0.1 mM of CdCl2, 0.1 mM of CoCl2, 0.5 mM of FeSO4, 1 mM of MnSO4, and 0.5 mM of ZnSO4, respectively, and in normal 1/2 Hoagland solutions as the control (CK) (Desoky et al., 2020; Gao et al., 2020). The experimental pots were positioned in a completely randomized block design. The experiment comprised six treatments, as shown above, and each treatment was repeated with three pots. Then, 24 h later, the leaves and roots of tube plantlets were collected and were used as RNA extraction materials. Three biological replicates of the expression analyses were performed for each treatment.
TRIzol reagent (Invitrogen, United States) was used for RNA extraction from all plant samples (leaf, stem, and root) and subsequently reverse-transcribed to cDNA using a SuperMix Kit (Transgen, Beijing). The Primer 5.0 tool was used to design specific primers of all selected MtMTP genes, including β-actin as a housekeeping gene (Supplementary Table 1). In total, 20 μL of reaction mixture was used to perform real-time polymerase chain reaction (PCR) containing the following reagents: 10 μL of 2 × SYBR premix Taq, 1 μL of cDNA, 0.5 μL of each primer, and 8 μL of ddH2O (El-Sappah et al., 2017). Real-time PCR reaction conditions were adjusted as follows: 95°C for 10 min, 95°C for 15 s, 60°C for 60 s, and 40 cycles in total. The relative expression level was calculated by applying the Livak equation 2–ΔΔCT with three replications for each sample (Livak and Schmittgen, 2001).
Three biological replicates of expression analyses were performed using ± standard deviation at p < 0.05. The significant variations between means were compared at p < 0.05 using Student’s t-test.
In total, 27 genes have been identified using blast analysis; subsequently, genes with an incomplete functional domain were excluded for the next study, and finally, 12 candidate genes were selected for further analysis. Every gene was assigned with a specific name, i.e., MtMTP1.1, MtMTP1.2, MtMTP2, MtMTP4, MtMTP5, MtMTP7, MtMTP8.1, MtMTP8.2, MtMTP9, MtMTP10.1, MtMTP10.2, and MtMTP11. The characteristics of the MtMTP genes were analyzed in detail (Table 1). The length of the CDS sequence of MtMTP genes ranged from 1,158 bp (MtMTP1.2) to 1,476 bp (MtMTP2), whereas the length of their encoded protein ranged from 347 to 491 amino acids, and the relative MW ranged from 39,491.59 to 53,259.81 Da. Most of the MtMTP proteins have relatively low isoelectric points (pI < 7), except for MtMTP10.2 and MtMTP5, which have a pI of 7.16 and 7.76, respectively. The GRAVY of the MtMTPs ranged from −0.109 (MtMTP10.2) to 0.201 (MtMTP1.2). The total number of inter- and intra-protein ionic residues were variable, i.e., the highest anionic residues were in MtMTP1.1 and lowest in MtMTP5. Similarly, the highest cationic residues were in MtMTP10.2 and the lowest in MtMTP4 and MtMTP11. All MtMTP members harbored a variable number of introns, but MtMTP1.1, MtMTP1.2, and MtMTP4 did not have any introns. Subcellular localization prediction results showed that all MtMTP proteins were localized to the vacuole membrane (tonoplast), except for MtMTP2 in the chloroplast, MtMTP4 in the endoplasmic reticulum, and MtMTP10.2 in cytoplasm.
All MTP gene families were divided into seven groups, i.e., groups 1, 5, 6, 7, 8, 10, and 12 (Figure 1).
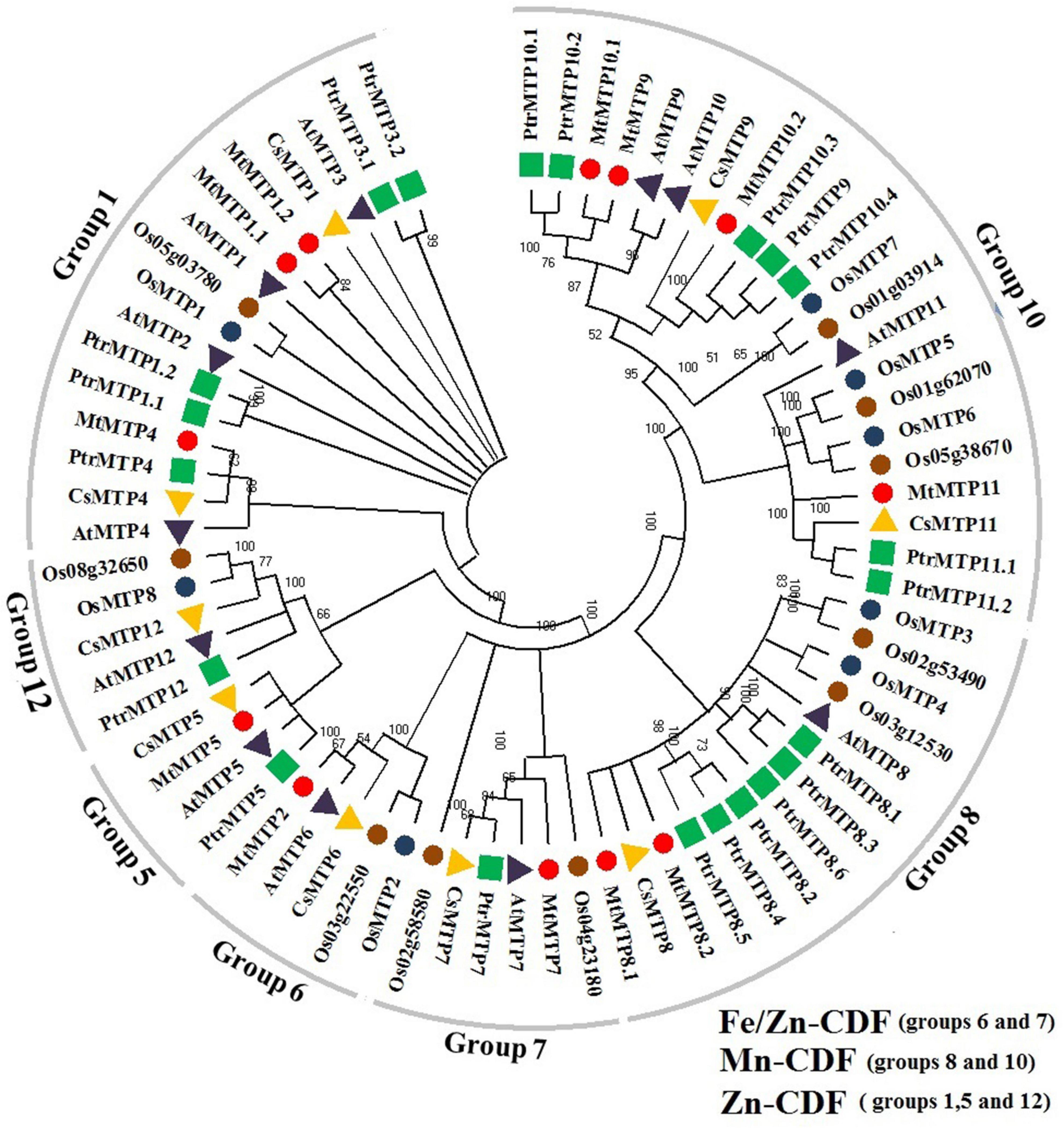
Figure 1. Phylogenetic tree of 72 MTP proteins: 12 M. truncatula (marked by red circle), 12 Arabidopsis (purple triangle), 8 wheat (blue circle), 10 rice (brown circle), 9 cucumber (yellow triangle), and 21 black poplar (blue square). ClustalX1.83 was used for protein alignments and the phylogenetic tree’s construction at the neighbor-joining (NJ) level using MEGA6.0.6 software at 1,000 replications bootstrap.
The highest number of MTPs were pooled in group 10 including MtMTP9, MtMTP10.1, MtMTP10.2, and MtMTP11 along with AtMTP9, AtMTP10, and AtMTP11; then in group 1 including MtMTP1.1, MtMTP1.2, and MtMTP4 along with AtMTP1, AtMTP2, AtMTP3, and AtMTP4; then in group 8 including MtMTP8.1 and MtMTP8.2 along with AtMTP8; then in group 7 including MtMTP7, along with AtMTP7; then in group 6 including MtMTP2, along with AtMTP6; and finally in group 5 including MtMTP5, along with AtMTP5, and no MtMTPs were placed in group 12. Noticeably, ionic clustering has revealed that four, two, and six MtMTPs were clustered in the Zn-CDFs, Fe/Zn-CDFs, and Mn-CDFs groups, respectively (Figure 1).
Synteny analyses were performed to unravel the distribution of genes on different chromosomes. MtMTP genes have been observed to be distributed among all seven chromosomes. Only segmental gene pair duplication has been observed from the Plant Genome Duplication Database. Collinearity due to excision of segmental duplication was observed in many gene pairs with a 70–100% identity percentage (Supplementary Table 2). Segment duplication resulted in many homologies of MTP genes between M. truncatula chromosome pairs, including those that occur with the genes, MtMTP1.1/MtMTP1.2, MtMTP4/MtMTP5, and MtMTP8.2/MtMTP10.2 (Figure 2). Except for MtMTP7, all the rest of the MTP genes in M. truncatula displayed single and multiple genetic duplications. Noticeably, any obvious tandem duplication was not observed among all MtMTPs.
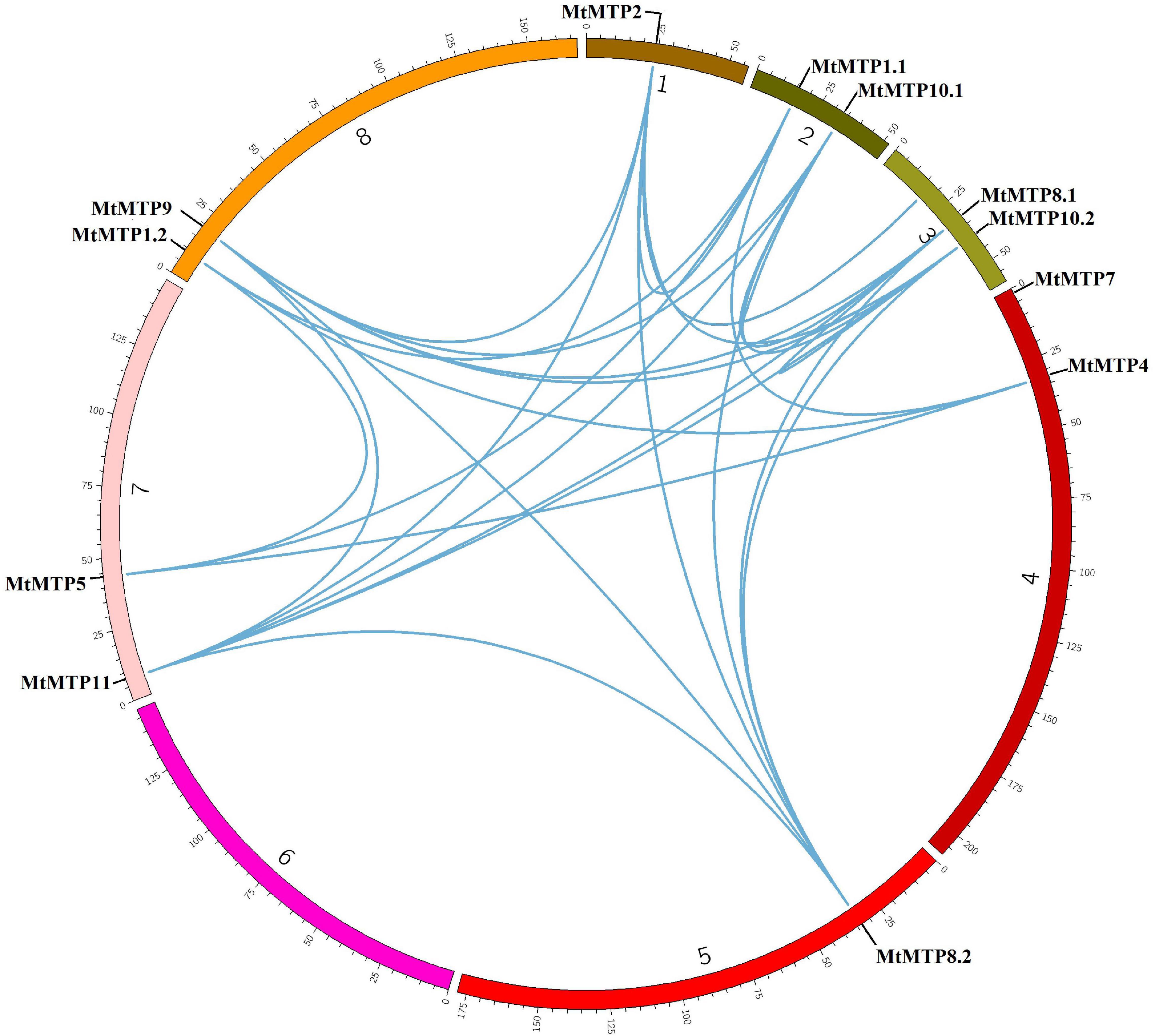
Figure 2. Genome-wide synteny analysis of MTP gene family among eight M. truncatula chromosomes. The blue lines represented the syntenic orthologs and paralogs and displayed segmental duplication.
All M. truncatula MTP family genes were further divided into six subfamilies (A, B, C, D, E, and F) (Figure 3A). Subfamilies A and F were the largest among all subfamilies with six members each, followed by subfamily B with two members, whereas subfamilies C, D, and E contained only one gene each (Figure 3A). Intron and exon analysis of all MTP genes revealed that each retrieved sequence of the MtMTP gene family is a correct and true member of the six subfamilies (Figure 3C). Although the size and location of the intron and exon of all MtMTP genes varied, the similarity index was higher among all subfamilies, which proved a close evolutionary relationship among MtMTP gene family members. All MTP family genes contain a variable number of introns, but all members of subfamily F did not have any introns. Amino acid sequence-based conserved motifs of MTP were analyzed using Multiple EM for Motif Elicitation (Figure 3B and Supplementary Table 3). All conserved motifs comprised 50 amino acids, except for motif 10 which comprised only 41 amino acids. The largest motifs were 3 and 6, which were observed in all subfamilies, followed by motif 10, which was 83.3% of the aforementioned motif. Noticeably, the number, type, and order of motifs were more similar in the intrasubfamily than in the intersubfamily.
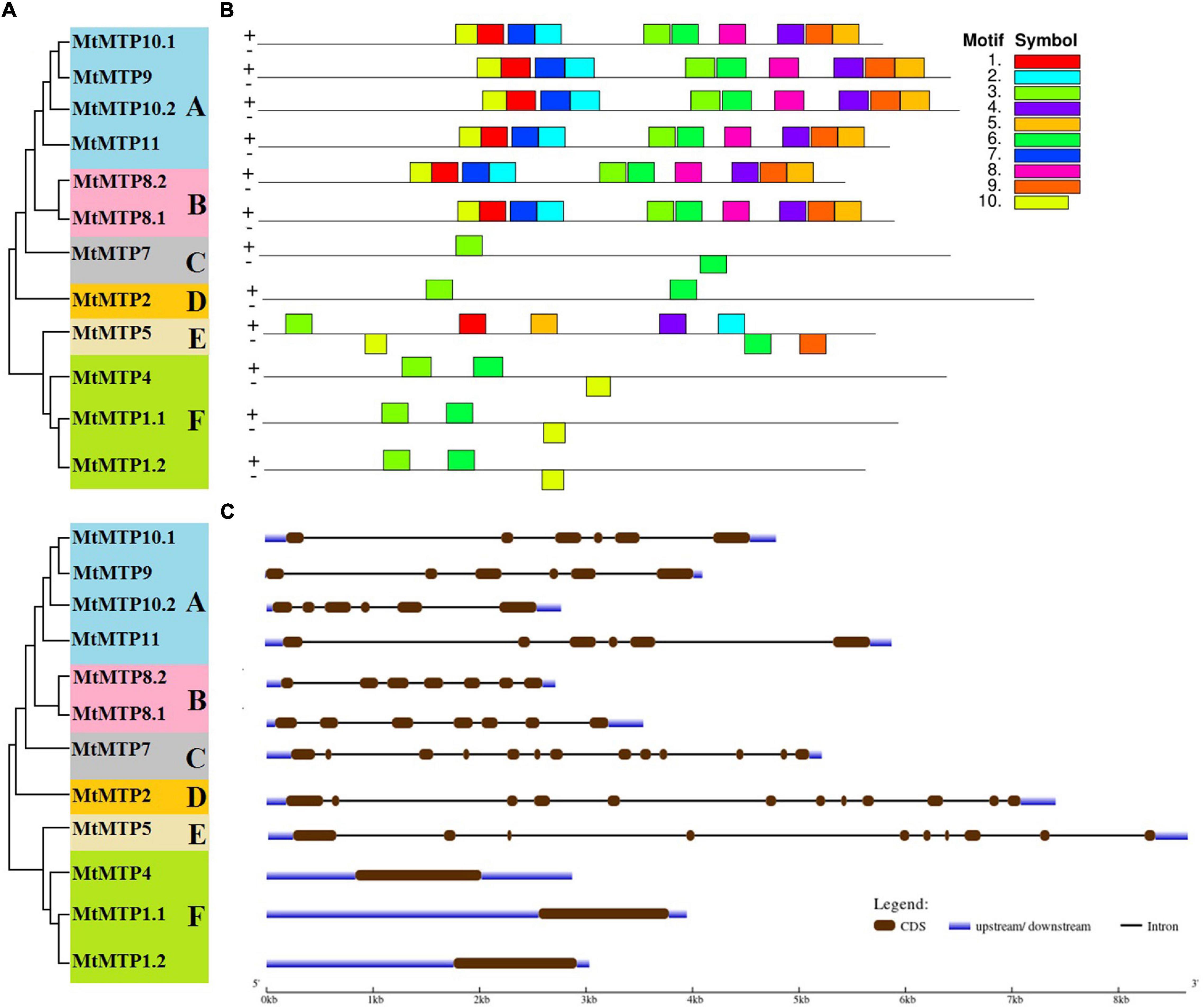
Figure 3. Phylogenetic relationship, gene structure, and conserved motif analysis of MtMTP genes. (A) The neighbor-joining phylogenetic tree was constructed using MEGA6.0.6 using MtMTP amino acid sequences with 1,000 times replication. (B) The motif composition of MtMTP proteins using 10 conserved motifs is represented by the unique color mentioned in the box on the top left. (C) The exon-intron structure of M. truncatula MTP proteins, where dark green boxes represent the exons, and the black lines represent the introns. The blue boxes represent the untranslated regions (UTRs), with size scales detailed at the bottom.
Cis-elements related to different stress responses (ARE, WUN-motif, LTR, and MBS) were also found in the promoters of most MtMTPs (Figure 4). Furthermore, cis-elements involved in plant development (MBS1, CAT-box, ERE, O2– site, and EBRE) were found in the promoter region of almost all MtMTPs. Meanwhile, the cis-elements of the whole MtMTP family were divided into different categories based on function prediction. The result showed that the largest category of cis-elements was hormone-related, followed by stress-related and developmental-related cis-elements. However, many motifs have not yet been functionally characterized, and whether these motifs confer unique functional roles to MtMTPs remains to be further investigated.
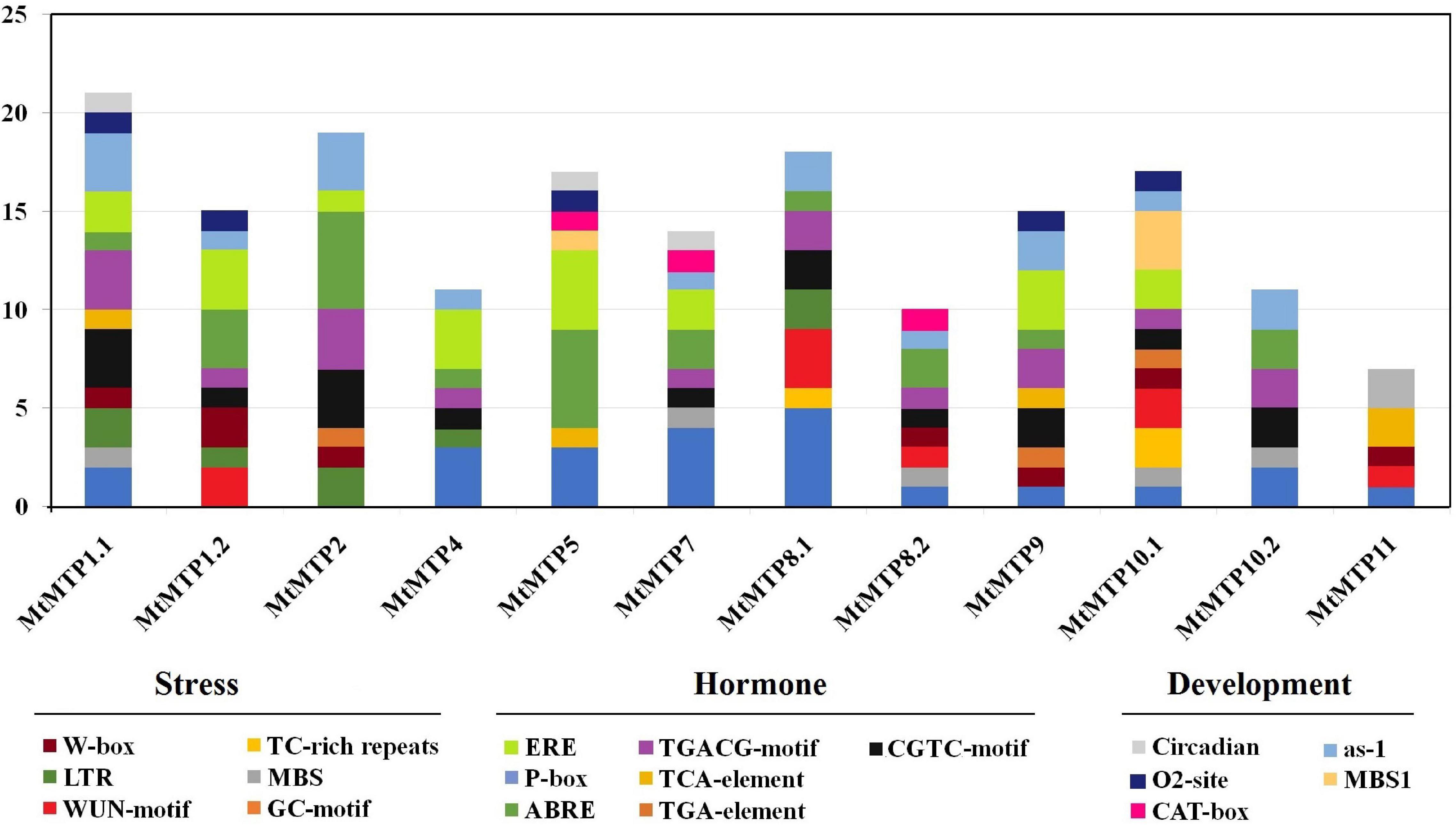
Figure 4. Cis-regulatory elements in the promoter region of MtMTP genes. Cis-regulatory elements were identified in 2,000 bp promoter sequences upstream of the start codon of MtMTP genes using the online tools PlantCARE and PLACE. These elements are related to different functional diversity, represented by different colors.
The protein structures of all the candidate MTP proteins were modeled at >90% confidence using the Phyre 2 web portal19 (Supplementary Figure 1 and Supplementary Table 4), and their potential active ligand-binding sites were also identified. According to the protein structure results, different active ligand-binding sites were predicted to be MTP proteins (Supplementary Figure 2). Some diversity in the protein structure may reflect their various roles in the transmembrane transport process in response to multiple environments. Additionally, the binding region/active sites of MTP proteins were predicted.
Based on the results, different pockets were observed, and the key amino acids involved in the function of MTP proteins were predicted (Figure 5). The GLY, ILE, LEU, and MET amino acids were predicted to be the binding residues in the ligand-binding site of all candidate MTP proteins (Figure 6). Followed by ALA, SER, THR, and VAL, the frequency of amino acid residues present in each pocket site was 96% (Figure 6). Overall, GLY, ILE, LEU, and MET are recognized as the key residues in the predicted pocket sites in MTP proteins. These results suggest the importance of these residues in these positions on the DNA molecule and finally, the cellular functional performance.
The subcellular localization, molecular function, and biological process were predicted using GO enrichment analysis (Figure 7 and Supplementary Table 5). In subcellular localization analysis, the predicted distribution scores of MTP proteins were as follows: 12/60% in all membranes, 3/15% in the plasma membrane and vacuole, and 1/5% in the Golgi apparatus and root hair. Noticeably, the MtMTP1.2 gene was localized in 12 subcellular compartments out of all 14, which underlined the significant role of MtMTP1.2 in metal stress resistance. The collective scores of MTP protein molecules during biological processes were as follows: transmembrane transport of Zn+ and Mn+ ions was 3/43%, whereas transmembrane transport of cations was 1/14%. More precisely, MtMTP1.1, MtMTP1.2, and MtMTP4 play a key role in transmembrane transport of Zn2+, whereas MtMTP8.1, MtMTP8.2, and MtMTP11 play a crucial role in transmembrane transport of Mn2+. Molecular function analysis revealed the significant roles of MtMTP8.2 and MtMTP11 in heavy metal processes.
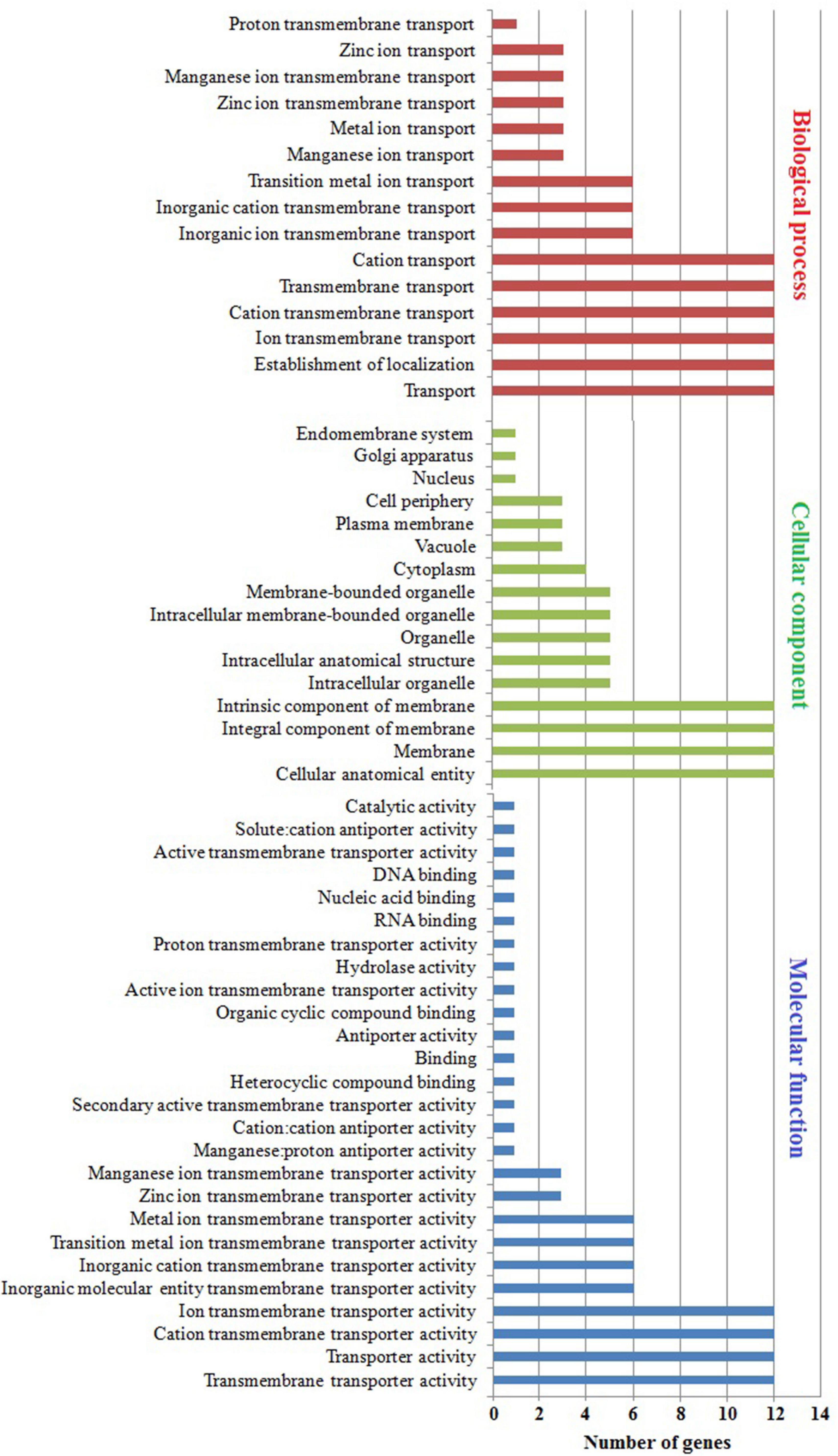
Figure 7. Gene ontology analysis of M. truncatula MTP genes. Gene ontology showed the distribution of every MtMTP gene in the plant, where a red-colored column shows the cellular component. Conversely, the biological processes in which the MTP family participate are shown by a blue-colored column, and the molecular function is shown by a mauve color.
A heatmap diagram was constructed to show the differential expression level of each MtMTP gene in all tissues (Figure 8 and Supplementary Table 6). Comparatively, MtMTP1.1 displayed the highest expression level in a pod, whereas MtMTP1.2 displayed the highest expression level in the root tip. Similarly, the highest expression level of MtMTP2 was observed in plant shoots, whereas the expression level of MtMTP5 was mild in the hypocotyl, root, and root tip. The expression level of MtMTP7 was also mild in the buds and higher in the shoot. MtMTP11 displayed the highest expression level in approximately all tissues, MtMTP10.1 only in the hypocotyl and seed coat, MtMTP8.1 only in the root, and MtMTP9 only in the flower tissue. Noticeably, MtMTP8.2 and MtMTP10.2 displayed the lowest expression level in all tissues and the highest expression in the roots.
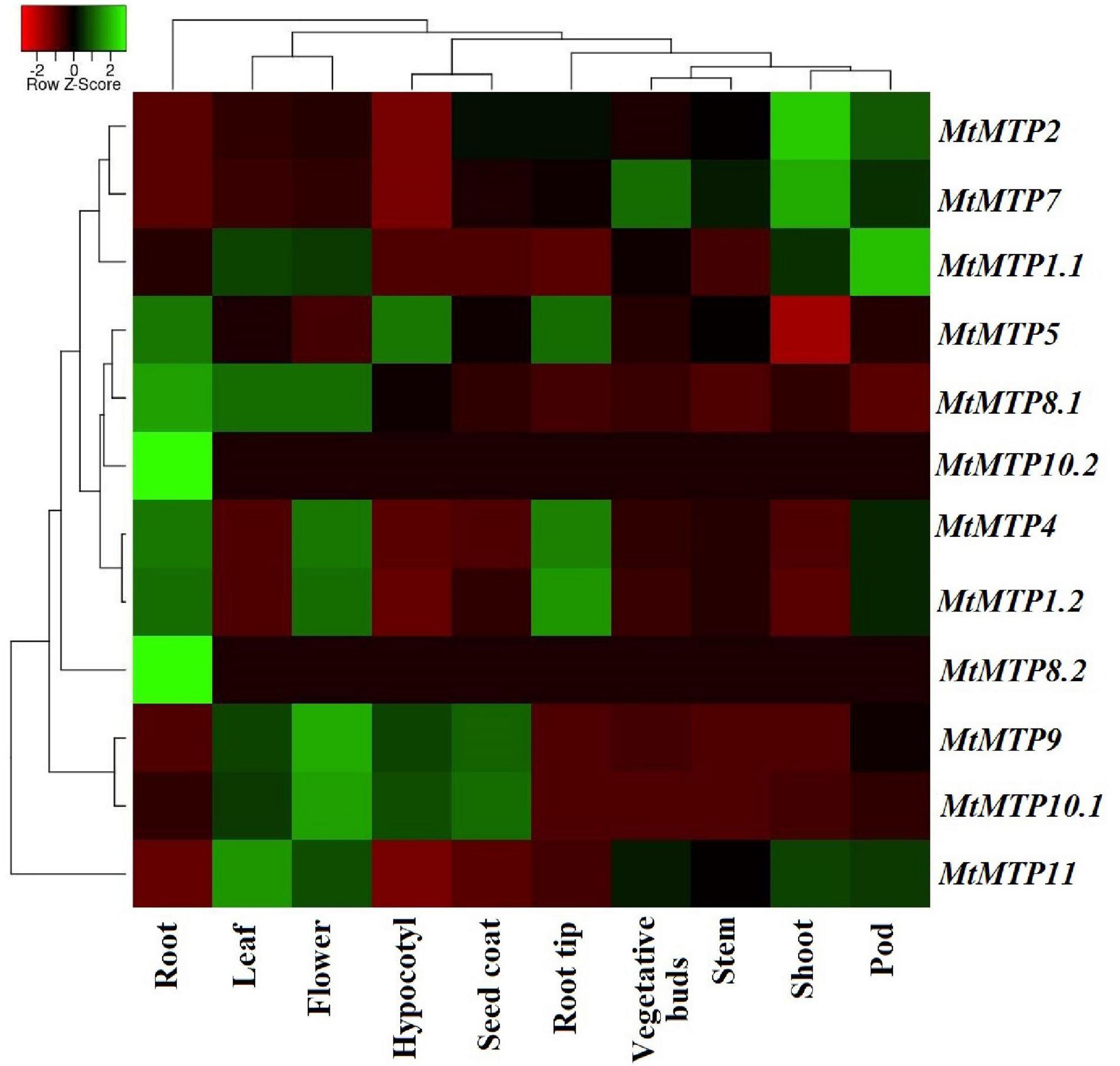
Figure 8. The heat map of 12 M. truncatula MTP gene expression profiles based on RNA-seq data. The previous expression has been shown in the root, leaf, flower, hypocotyl, seed coat, root tip, vegetative bud, stem, shoot, and pod tissues.
In this study, expression patterns of MtMTPs were investigated using qPCR under different metals treatment. All MtMTP genes displayed different gene expression levels under treatment with different types of heavy metals investigated in the following tissues: root, stem, and leaf (Figure 9). In the roots, MtMTP1.2 and MtMTP4 displayed the highest expression level, whereas MtMTP5, MtMTP7, and MtMTP9 displayed the lowest expression level under Cd2+ treatment. Similarly, MtMTP1.1 and MtMTP11 displayed the highest expression level, whereas MtMTP7 displayed the lowest expression level under Co2+ treatment. MtMTP1.1, MtMTP4, MtMTP5, MtMTP8.1, MtMTP8.2, and MtMTP11 displayed the highest expression level, whereas MtMTP10.2 displayed the lowest expression level under Fe2+ treatment. MtMTP1.1 and MtMTP4 displayed the highest expression level, whereas MtMTP5 displayed the lowest expression level under the Mn2+ treatment. MtMTP1.1, MtMTP1.2, and MtMTP4 displayed the highest expression level, whereas MtMTP2, MtMTP5, MtMTP7, and MtMTP11 displayed the lowest expression level under Zn2+ treatment.
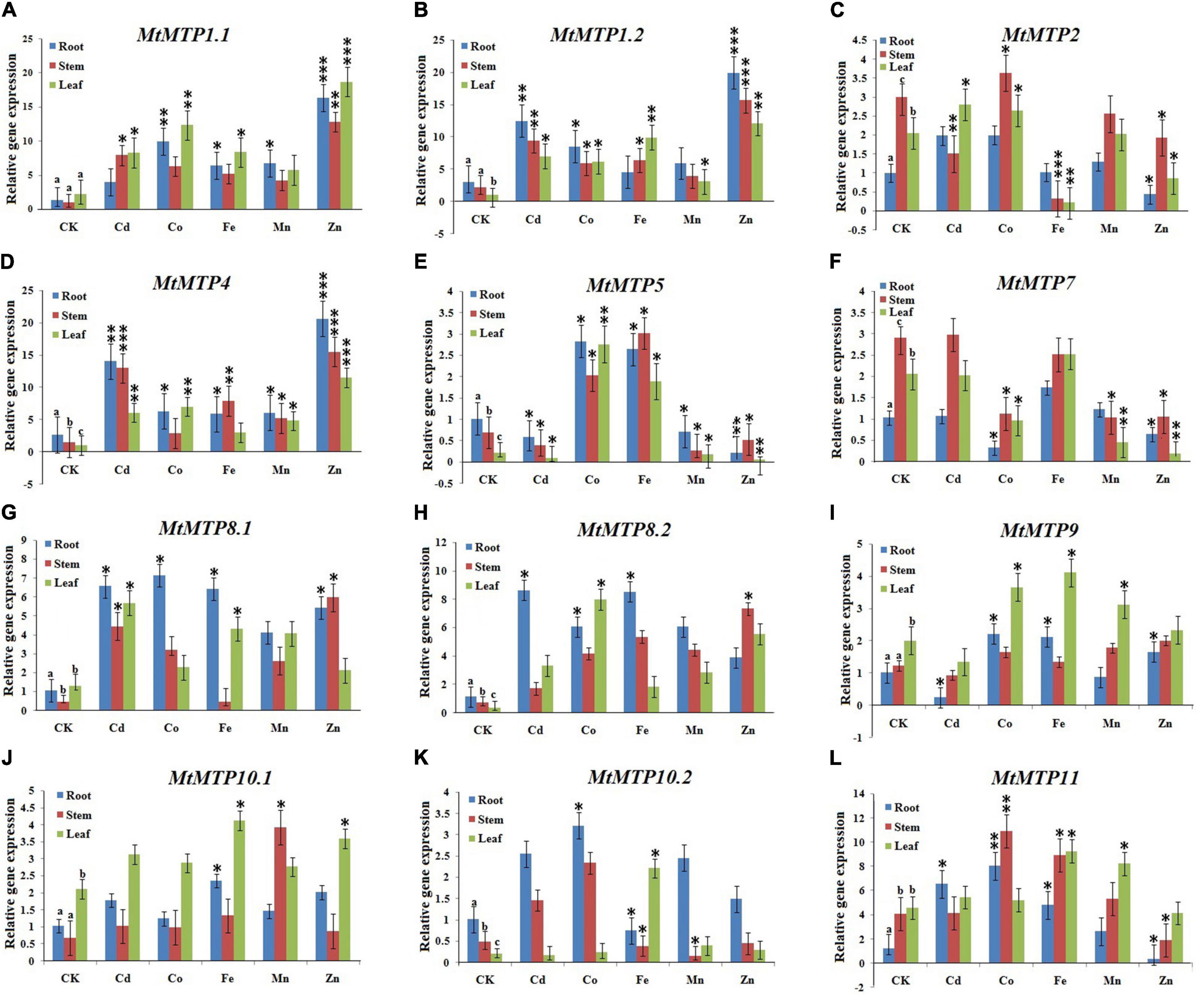
Figure 9. The qRT-PCR expression of the M. truncatula MTP genes from root, stem and leaf samples where the genes where showed alphabetically from (A–L). The reactions were normalized using the β-actin reference gene. The standard deviations have been represented by the error bars from three independent technical replicates. The mean expression levels of the three replicates were analyzed with the five heavy metal treatments (Cd2+, Co2+, Fe2+, Mn2+, and Zn2+) using t-tests (p < 0.05), where as the CK represents the control samples. Different letters (a, b and c) indicate significant differences between the roots, stems, and leaves under normal conditions. Asterisks indicate significant differences between the treatment samples and the corresponding control samples in the roots, stems, and leaves (n = 9, p < 0.05, Student’s t-test).
In the stem, Cd2+ treatment resulted in MtMTP1.2 and MtMTP4 upregulation, but downregulated MtMTP2 and MtMTP5. Similarly, Co2+ treatment significantly upregulated MtMTP11 but downregulated MtMTP7. Fe2+ treatment upregulated MtMTP4, MtMTP5, and MtMTP11 but downregulated MtMTP2 and MtMTP10.2. Mn2+ treatment upregulated MtMTP4 and MtMTP10.1 but downregulated MtMTP5, MtMTP7, and MtMTP10.2. Finally, Zn2+ upregulated MtMTP1.1, MtMTP1.2, and MtMTP but downregulated MtMTP5, MtMTP7, and MtMTP11.
In the leaf, the expression level of MtMTP4 was increased in response to Cd2+ treatment while MtMTP5 was downregulated. Similarly, Co2+ treatment upregulated MtMTP1.1, MtMTP4, and MtMTP5 but downregulated MtMTP7. Fe2+ treatment upregulated MtMTP1.2 but downregulated MtMTP2. Furthermore, expression levels of MtMTP1.1 and MtMTP4 were increased in response to Mn2+ treatment, while MtMTP1.2, MtMTP5, and MtMTP7 were downregulated. Finally, Zn2+ treatment upregulated MtMTP1.1, MtMTP1.2, and MtMTP4 but downregulated MtMTP2, MtMTP5, and MtMTP7.
Heavy metals have the biggest impact on the ecosystem and make it unfit for human consumption (El-Sappah et al., 2012; Chidambaram, 2015). Once released into the environment, they accumulate in plants and then in other living tissues via the food chain and cause toxicity even at lower concentrations (Samuel et al., 2020). MTP genes (membrane divalent cation transporters) are essential for transporting various heavy metals and enhancing plant tolerance against heavy metal stress (Ricachenevsky et al., 2013). They also have an expected role in plant mineral nutrition maintenance (Liu et al., 2019). Moreover, these metal-binding proteins are now being used as bioenvironmental markers for predicting heavy metal contamination based on their expression levels (Samuel et al., 2021). The MTP family has previously been studied in several plants, including Arabidopsis (van der Zaal et al., 1999), tobacco (Liu et al., 2019), wheat (Vatansever et al., 2017), and black poplar (Gao et al., 2020), whereas this is the first genomic identification study of the MTP family in M. truncatula. A total of 12 putative MTP genes in M. truncatula have been identified, and they were named based on the sequence similarities and orthologous relationships between them and AtMTPs.
The broad range of basic physicochemical properties of the MtMTP gene including CDS length, protein size, MW, pI, GRAVY, and subcellular localization, was analyzed and was found to be consistent with previous studies indicating huge probabilities of amino acid in metal tolerance (Vatansever et al., 2017; Büyükköroǧlu et al., 2018). MtMTP proteins showed diversity in terms of physicochemical properties, indicating that these gene family members are involved in various pathways and biochemical networks (Ahmadizadeh et al., 2020; Musavizadeh et al., 2021). Consistent with previous studies (Vatansever et al., 2017), our genes are localized to the tonoplast (the vacuole membrane), whereas some might also be localized in the cytoplasm or chloroplast, suggesting that MtMTPs might function as vacuole-localized cation transporters. The phylogeny of MTP proteins between M. truncatula and the other five studies was compared (Figure 1). The phylogeny results were aligned to that of the previous studies conducted in various plant species. Multiple homologous pairs were observed in M. truncatula, whereas no such pairs were observed in Arabidopsis, showing that the MtMTP gene family might have undergone gene expansion and/or gene loss in its evolutionary history, probably due to polyploidization events.
About four, two, and six MtMTP genes were grouped into Zn-CDFs, Zn/Fe-CDFs, and Mn-CDFs, respectively (Figure 2), considering the implications of phylogenetic distributions in inferring structure and functional roles across species (Eroglu et al., 2017). The various groups are involved in specific mechanisms, and this information could provide clues to predict their function in different species.
In our study, to obtain more knowledge about the gene annotation and expansion mechanism of the MTP gene family in M. truncatula, gene synteny and duplication analysis were investigated (Figure 2 and Supplementary Table 2). Two or more genes on the same chromosome are often related to tandem duplication, whereas segmental duplication often occurs in different chromosomes (Schlueter et al., 2007). Our study did not show any of the tandem duplication pairs, whereas 30 segmental duplications occurred, which included MtMTP2/MtMTP8.1, MtMTP2/MtMTP10.2, and MtMTP8.2/MtMTP9. During the evolution process of a plant gene, family duplication events occur, followed by divergence considering standard features that are more related to secondary plant metabolism genes (Ober, 2005).
Almost all subfamilies contained the same numbers of introns and motif sequences that are consistent with the previous studies wherein a similar gene structure was found within the same subfamilies (Liu et al., 2018). For example, all of the gene members of subfamily A contained five introns. However, subfamily F members did not contain any introns. These outcomes indicated that during the M. truncatula evolution events of MtMTPs, some intron gain and loss occurred. Some genes have no introns but have one exon causing the lower ability of exons in the gain/loss rate due to higher selection pressure in the exon sequences (Harrow et al., 2006). Thus, with all these observations, it is probable that the placement divergences in intron number consider shared events related to the gene family evolution (Jeffares et al., 2006; Rogozin et al., 2012; Babicki et al., 2016).
Protein structures are precisely associated with the functions of genes and can reflect the phylogenetic relationships among them (Zhang et al., 2016; Faraji et al., 2020). The protein structures of all the candidate MTP proteins were modeled at >90% confidence, and their potential active ligand-binding sites were also identified. Some diversity in these protein structures may reflect their different roles in the transmembrane transport process in response to multiple environments. Based on the three-dimensional structure analysis, it can be mentioned that these proteins from multiple clades belong to functionally diverse groups but share a common catalytic mechanism in the stabilization of membrane potential and cellular metal ion homeostasis under stresses, supporting the fact that MTP proteins can play an essential role in intracellular signaling pathways in response to unfavorable conditions.
The GLY, ILE, LEU, MET, ALA, SER, THR, VAL, ASN, and PHE amino acids were predicted as the binding residues in the ligand-binding site of nearly all candidate MTP proteins, which may manifest the importance of these residues in those positions on the DNA molecule and their cellular functional performance. For instance, SER, VAL, and LEU have been identified as the key amino acids involved in regulating responses to stress (Galili and Höfgen, 2002; Beauregard and Hefford, 2006). CREs are non-coding DNA binding by transcription factors and/or other regulatory molecules for regulating the transcription of neighboring genes (Wittkopp and Kalay, 2012). Furthermore, CREs play an essential role in plant response to environmental cues (Yamaguchi-Shinozaki and Shinozaki, 2005), and many of those have been found in the MtMTP genes in this study (Supplementary Figure 1). The CRE analysis in the promoter regions of the MtMTP genes showed the potential regulatory mechanisms controlling their expression.
The ABRE element is important in ABA signaling and plant response to drought and salt stress in Arabidopsis (Narusaka et al., 2003), and it is the most abundant element present in MtMTPs. The SA signaling pathway is mediated by the TCA element that is found in four of the MtMTPs and is responsible for different stress reduction (Mou et al., 2013; Ali et al., 2015). The ARE element was found to be both necessary and sufficient for anaerobic induction of genes (Ali et al., 2021) and as part of the 10 MtMTP genes. MBS was found in five of the studied MtMTPs in M. truncatula and was reported to bind to MYB transcriptional factors involved in drought stress signaling (Shukla et al., 2015). Five of MtMTP promoters harbor LTR and are important to induce cold-regulated genes (Brown et al., 2001). Furthermore, only two genes MtMTP8.1 and MtMTP10.1, harbor a TC-rich repeat where they are involved in defense and stress responsiveness (Sazegari et al., 2015). The CGTCA motif is responsible for methyl jasmonate-responsive elements and is important in stress induction (Ali et al., 2018). The detection of these stress-responsive cis-elements indicates that the proteins encoding the MtMTP genes may have a functional role in heavy metal stress tolerance in M. truncatula.
In a detailed evaluation of the MTP proteins, their 3D configuration was predicted, which was considered to be a supportive tool for inspecting their function (Büyükköroǧlu et al., 2018). The four temples in M. truncatula MTP proteins indicated that these proteins with heavy metal, where these transport proteins are in plants, are classified into metal-uptake proteins that transport essential and toxic heavy metals to the cytoplasm and metal-uptake proteins. Simultaneously, the other is metal-efflux proteins that help the cell remove any excess heavy metals (Mani and Sankaranarayanan, 2018). Conversely, protein–protein interaction analysis provided us with more knowledge about the plant developmental processes with their interactions with the environment (Struk et al., 2019).
In contrast, gene ontology is a fundamental analysis to predict putative functional contributions across living organisms (Consortium, 2010). Moreover, gene ontology classes and concepts have been used to define the relationships and gene functions existing between these concepts (Purwantini et al., 2014). Our gene ontology analysis has revealed the significant role of the M. truncatula MTP genes with heavy metals (Figure 5 and Supplementary Table 5). Furthermore, GO showed the molecular functions of the MtMTPs, where most of them participate in metal-related processes, including transmembrane transporter activity, cation transmembrane transporter activity, transporter activity, and ion transmembrane transporter activity.
Transcriptomic analysis is a proper tool for detecting the existence, structure, and quantity of the RNA in any abiological sample under certain conditions (Zambounis et al., 2020). Thus, the expression profile of all members of the MTP gene family from previously published RNA-sequencing data has been investigated, which showed the expression of all gene members in all selected M. truncatula tissues (Figure 6 and Supplementary Table 6). Digital data analysis showed that the significant roles of the MTP gene could contribute significantly to growth and development. Noteworthy evidence has been obtained about the essential roles of M. truncatula MTPs after tissue expression evaluation. For instance, the exclusive expression of the two genes MtMTP2 and MtMTP7 was in the root tip more than in any other root part, whereas MtMTP11 was most abundant in the vegetative buds, indicating that they might be involved in the early root and bud development. Besides the expected vital roles of MtMTP11 in pod maturation and development, its expression has increased. However, only MtMTP8.2 and MtMTP10.2 were weakly expressed in all examined tissues from all MtMTPs.
The documented downregulation in some gene expressions is essential for maintaining gene duplicates and ancestral functions (Qian et al., 2010). Hence in our study, the downregulation of MtMTP8.2 and MtMTP10.2 expression is expected to be vital to keep their biological functions and to maintain them from losing them during cell evaluation. The reliability of the transcriptome data was further validated using quantitative reverse transcription-polymerase chain reaction (qRT-PCR) (Figure 7); however, the minor asymmetry between both analyses may be due to different growth conditions and M. truncatula varieties, which finally affected the spatial expression. The expression behavior of MTP genes was examined under five divalent metals (Mn2+, Cd2+, Co2+, Fe2+, and Zn2+). Numerous studies in other plants indicated the significant roles of the MTP gene family to enhance plant tolerance against these metals (Montanini et al., 2007; Gao et al., 2020) as it was found that metal-efflux transporters from the cytoplasm mainly transport Zn2+ but also transport Ni+2, Co2+, Cd2+, Fe2+, and Mn2+ (Ricachenevsky et al., 2013).
The transcript accumulation transcription of MTPs in response to various heavy metals is varied and complicated, although the gene expression response to different stresses is usually reflected in corresponding gene roles. In Arabidopsis, the tonoplast-localized Zn transporter AtMTP1 showed slight changes in expression with excess Zn exposure at both transcription and translation levels (Dräger et al., 2004; Kobae et al., 2004). Moreover, despite the high expression of the CsMTP1 encoded protein, the gene expression was steady under the high concentration of Zn2+ in cucumber (Migocka et al., 2015).
As mentioned before, the AtMTP12 upregulation was not due to Zn+ treatment but due to the formation of a heterodimeric complex with AtMTP5 for Zn+ transport (Fujiwara et al., 2015), same in tobacco (Liu et al., 2019). Moreover, variable concentrations of Mn2+ slightly affect the expression of Mn-CDFs (AtMTP8, AtMTP9, AtMTP10, and AtMTP11) (Delhaize et al., 2007), same with tobacco (Liu et al., 2019). Contrarily, all Zn-CDF members displayed significant variation in their expression under Zn2+ treatment, such that MtMTP2 and MtMTP7 of Zn/Fe-CDFs were downregulated under treatment of a higher concentration of Zn2+. Furthermore, MtMTP10.1 of the Mn-CDF class was highly affected by Mn2+ accumulation only in the stem. Our findings provide deep insights in the molecular function of the MTP genes in M. truncatula under various heavy metal stresses, which will invite researchers to precisely identify the function of the desired MTP genes in M. truncatula via wet lab experiments.
Generally, these results would provide essential clues in clarifying the roles of MtMTPs in heavy metal tolerance and the mechanism of heavy metal transport mediated by MtMTP proteins. Altogether, these results would lay a theoretical and practical foundation for the functional characterization of MtMTP genes in future studies. Furthermore, the highest expressed MTPs (MtMTP1.1, MtMTP1.2, and MtMTP4) can be used as bioenvironmental markers for predicting heavy metal contamination based on their expression levels.
The first genome-wide study of the MTP gene family in M. truncatula has been provided, providing important comparative data for evolutionary relationships. Twelve identified MTP genes were phylogenetically divided into three major substrate-specific clusters (Zn/Fe-CDFs, Mn-CDFs, and Zn-CDFs), and seven groups seemed to have undergone expansion and gene loss after polyploidization through segmental duplication. The MtMTP transcripts under plant hormones and heavy metal stresses showed that they might participate in the plant response signaling pathway, which was related to the wide distribution of the CREs in their upstream regions. GLY, ILE, LEU, MET, ALA, SER, THR, VAL, ASN, and PHE amino acids were predicted to be the binding residues in the ligand-binding site of all these proteins. The expression patterns of MtMTP members in response to various heavy metals at different tissues indicated the significant role of these genes in the growth and development of M. truncatula. Furthermore, our gene expression analysis of various heavy metals has revealed the significant roles of the MTPs, particularly those of MtMTP1.1, MtMTP1.2, and MtMTP4, in plant tolerance to heavy metal stresses.
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author/s.
AE-S, JL, ZX, and KE-T: conceptualization. AE-S and RE: writing original draft. AE-S, RE, KE-T, and JL: bioinformatics and formal analysis. AE-S: drawing figures. AE-S, JL, and RE: experiment design and perform. AE-S, AE, KE-T, YW, QH, KE, YZ, and MA: editing. AE-S, YZ, JL, QY, and MA: review. AE-S, KE-T, and RE: writing final manuscript. All authors critically revised the manuscript and approved the final version.
This work was supported by the Innovation Research Team of Yibin University (Nos. 2017TD01 and 2018TD04) to JL and Abu Dhabi Department of Education and Knowledge (Grant No. 21S105) to KE-T.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Many thanks to Wang Ling, the president of Yibin University, for her support. At the same time, thanks to Mrs. Zhou Lei, Jiang Qianwen, and Li Shengnan, the international office team, for their continuous help to achieve a suitable environment for research. KE-T would like to thank the library at Murdoch University, Australia, for the valuable online resources and comprehensive databases.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2021.713224/full#supplementary-material
Ahmadizadeh, M., Chen, J.-T., Hasanzadeh, S., Ahmar, S., and Heidari, P. (2020). Insights into the genes involved in the ethylene biosynthesis pathway in Arabidopsis thaliana and Oryza sativa. J. Genet. Eng. Biotechnol. 18, 62–62.
Ali, E., Hussain, N., Shamsi, I. H., Jabeen, Z., Siddiqui, M. H., and Jiang, L.-X. (2018). Role of jasmonic acid in improving tolerance of rapeseed (Brassica napus L.) to Cd toxicity. J. Zhejiang Univ. Sci. B 19, 130–146. doi: 10.1631/jzus.b1700191
Ali, E., Maodzeka, A., Hussain, N., Shamsi, I. H., and Jiang, L. (2015). The alleviation of cadmium toxicity in oilseed rape (i) by the application of salicylic acid. Plant Growth Regul. 75, 641–655. doi: 10.1007/s10725-014-9966-0
Ali, E., Saand, M. A., Khan, A. R., Shah, J. M., Feng, S., Ming, C., et al. (2021). Genome-wide identification and expression analysis of detoxification efflux carriers (DTX) genes family under abiotic stresses in flax. Physiol. Plant. 171, 483–501. doi: 10.1111/ppl.13105
Ali, I., Asim, M., and Khan, T. A. (2013). Arsenite removal from water by electro-coagulation on zinc–zinc and copper–copper electrodes. Int. J. Environ. Sci. Technol. 10, 377–384. doi: 10.1007/s13762-012-0113-z
Arrivault, S., Senger, T., and Krämer, U. (2006). The Arabidopsis metal tolerance protein AtMTP3 maintains metal homeostasis by mediating Zn exclusion from the shoot under Fe deficiency and Zn oversupply. Plant J. 46, 861–879. doi: 10.1111/j.1365-313x.2006.02746.x
Babicki, S., Arndt, D., Marcu, A., Liang, Y., Grant, J. R., Maciejewski, A., et al. (2016). Heatmapper: web-enabled heat mapping for all. Nucleic Acids Res. 44, W147–W153.
Bailey, T. L., Williams, N., Misleh, C., and Li, W. W. (2006). MEME: discovering and analyzing DNA and protein sequence motifs. Nucleic Acids Res. 34, W369–W373.
Beauregard, M., and Hefford, M. A. (2006). Enhancement of essential amino acid contents in crops by genetic engineering and protein design. Plant Biotechnol. J. 4, 561–574.
Benedito, V. A., Torres-Jerez, I., Murray, J. D., Andriankaja, A., Allen, S., Kakar, K., et al. (2008). A gene expression atlas of the model legume Medicago truncatula. Plant J. 55, 504–513.
Brown, A., Dunn, M., Goddard, N., and Hughes, M. (2001). Identification of a novel low-temperature-response element in the promoter of the barley (Hordeum vulgare L) gene blt101. 1. Planta 213, 770–780. doi: 10.1007/s004250100549
Büyükköroǧlu, G., Dora, D. D., Özdemir, F., and Hızel, C. (2018). “Chapter 15 Techniques for Protein Analysis,” in Omics Technologies and Bio-Engineering, eds D. Barh V. Azevedo (Cambridge, MA: Academic Press), 317–351.
Chidambaram, R. (2015). Isotherm modelling, kinetic study and optimization of batch parameters using response surface methodology for effective removal of Cr (VI) using fungal biomass. PLoS One 10:e0116884. doi: 10.1371/journal.pone.0116884
Chu, H.-H., Car, S., Socha, A. L., Hindt, M. N., Punshon, T., and Guerinot, M. L. (2017). The Arabidopsis MTP8 transporter determines the localization of manganese and iron in seeds. Sci. Rep. 7:11024.
Clemens, S. (2001). Molecular mechanisms of plant metal tolerance and homeostasis. Planta 212, 475–486. doi: 10.1007/s004250000458
Conesa, A., and Götz, S. (2008). Blast2GO: a comprehensive suite for functional analysis in plant genomics. Int. J. Plant Genomics 2008:619832.
Consortium, G. O. (2010). The Gene Ontology in 2010: extensions and refinements. Nucleic Acids Res. 38, D331–D335.
Delhaize, E., Gruber, B. D., Pittman, J. K., White, R. G., Leung, H., Miao, Y., et al. (2007). A role for the AtMTP11 gene of Arabidopsis in manganese transport and tolerance. Plant J. 51, 198–210. doi: 10.1111/j.1365-313x.2007.03138.x
Desoky, E.-S. M., El-Maghraby, L. M., Awad, A. E., Abdo, A. I., Rady, M. M., and Semida, W. M. (2020). Fennel and ammi seed extracts modulate antioxidant defence system and alleviate salinity stress in cowpea (Vigna unguiculata). Sci. Hortic. 272:109576. doi: 10.1016/j.scienta.2020.109576
Dräger, D. B., Desbrosses-Fonrouge, A. G., Krach, C., Chardonnens, A. N., Meyer, R. C., Saumitou-Laprade, P., et al. (2004). Two genes encoding Arabidopsis halleri MTP1 metal transport proteins co-segregate with zinc tolerance and account for high MTP1 transcript levels. Plant J. 39, 425–439. doi: 10.1111/j.1365-313x.2004.02143.x
Edwards, K., Fernandez-Pozo, N., Drake-Stowe, K., Humphry, M., Evans, A., Bombarely, A., et al. (2017). A reference genome for Nicotiana tabacum enables map-based cloning of homeologous loci implicated in nitrogen utilization efficiency. BMC Genomics 18:448. doi: 10.1186/s12864-017-3791-6
Elrys, A. S., Merwad, A.-R. M., Abdo, A. I., Abdel-Fatah, M. K., and Desoky, E.-S. M. (2018). Does the application of silicon and Moringa seed extract reduce heavy metals toxicity in potato tubers treated with phosphate fertilizers? Environ. Sci. Pollut. Res. 25, 16776–16787. doi: 10.1007/s11356-018-1823-7
El-Sappah, A., Shawky, A., Sayed-Ahmad, M., and Youssef, M. (2012). Nile tilapia as bio indicator to estimate the contamination of water using SDS-PAGE and RAPDPCR techniques. Egypt. J. Genet. Cytol. 41, 209–227.
El-Sappah, A. H., Shawky, A., Sayed-Ahmad, M. S., and Youssef, M. (2017). Estimation of heat shock protein 70 (hsp 70) gene expression in nile tilapia (Oreochromis niloticus) using quantitative Real-Time PCR. Zagazig J. Agric. Res. 44, 1003–1015. doi: 10.21608/zjar.2017.52300
Eroglu, S., Filiz, E., and Vatansever, R. (2017). Genome-wide exploration of metal tolerance protein (MTP) genes in common wheat (Triticum aestivum): insights into metal homeostasis and biofortification. Biometals 30, 217–235.
Eroglu, S., Meier, B., Von Wirén, N., and Peiter, E. (2016). The vacuolar manganese transporter MTP8 determines tolerance to iron deficiency-induced chlorosis in Arabidopsis. Plant Physiol. 170, 1030–1045. doi: 10.1104/pp.15.01194
Faraji, S., Filiz, E., Kazemitabar, S. K., Vannozzi, A., Palumbo, F., Barcaccia, G., et al. (2020). The AP2/ERF gene family in Triticum durum: genome-wide identification and expression analysis under drought and salinity stresses. Genes 11:1464. doi: 10.3390/genes11121464
Fujiwara, T., Kawachi, M., Sato, Y., Mori, H., Kutsuna, N., Hasezawa, S., et al. (2015). A high molecular mass zinc transporter MTP 12 forms a functional heteromeric complex with MTP 5 in the Golgi in Arabidopsis thaliana. FEBS J. 282, 1965–1979. doi: 10.1111/febs.13252
Galili, G., and Höfgen, R. (2002). Metabolic engineering of amino acids and storage proteins in plants. Metab. Eng. 4, 3–11. doi: 10.1006/mben.2001.0203
Gao, Y., Yang, F., Liu, J., Xie, W., Zhang, L., Chen, Z., et al. (2020). Genome-wide identification of metal tolerance protein genes in Populus trichocarpa and their roles in response to various heavy metal stresses. Int. J. Mol. Sci. 21:1680. doi: 10.3390/ijms21051680
Gasteiger, E., Gattiker, A., Hoogland, C., Ivanyi, I., Appel, R. D., and Bairoch, A. (2003). ExPASy: the proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res. 31, 3784–3788. doi: 10.1093/nar/gkg563
Gupta, N., Yadav, K. K., Kumar, V., Kumar, S., Chadd, R. P., and Kumar, A. (2019). Trace elements in soil-vegetables interface: translocation, bioaccumulation, toxicity and amelioration-A review. Sci. Total Environ.651, 2927–2942. doi: 10.1016/j.scitotenv.2018.10.047
Gustin, J. L., Zanis, M. J., and Salt, D. E. (2011). Structure and evolution of the plant cation diffusion facilitator family of ion transporters. BMC Evol. Biol. 11:76. doi: 10.1186/1471-2148-11-76
Harrow, J., Denoeud, F., Frankish, A., Reymond, A., Chen, C.-K., Chrast, J., et al. (2006). GENCODE: producing a reference annotation for ENCODE. Genome Biol. 7, 1–9.
He, J., Benedito, V. A., Wang, M., Murray, J. D., Zhao, P. X., Tang, Y., et al. (2009). The Medicago truncatula gene expression atlas web server. BMC Bioinformatics 10:441. doi: 10.1186/1471-2105-10-441
Higo, K., Ugawa, Y., Iwamoto, M., and Korenaga, T. (1999). Plant cis-acting regulatory DNA elements (PLACE) database: 1999. Nucleic Acids Res. 27, 297–300. doi: 10.1093/nar/27.1.297
Hu, B., Jin, J., Guo, A.-Y., Zhang, H., Luo, J., and Gao, G. (2015). GSDS 2.0: an upgraded gene feature visualization server. Bioinformatics 31, 1296–1297. doi: 10.1093/bioinformatics/btu817
Jiménez, J., Doerr, S., Martínez-Rosell, G., Rose, A. S., and De Fabritiis, G. (2017). DeepSite: protein-binding site predictor using 3D-convolutional neural networks. Bioinformatics 33, 3036–3042. doi: 10.1093/bioinformatics/btx350
Kawachi, M., Kobae, Y., Mimura, T., and Maeshima, M. (2008). Deletion of a histidine-rich loop of AtMTP1, a vacuolar Zn2+/H+ antiporter of Arabidopsis thaliana, stimulates the transport activity. J. Biol. Chem. 283, 8374–8383. doi: 10.1074/jbc.m707646200
Kelley, L. A., Mezulis, S., Yates, C. M., Wass, M. N., and Sternberg, M. J. E. (2015). The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 10, 845–858. doi: 10.1038/nprot.2015.053
Kobae, Y., Uemura, T., Sato, M. H., Ohnishi, M., Mimura, T., Nakagawa, T., et al. (2004). Zinc transporter of Arabidopsis thaliana AtMTP1 is localized to vacuolar membranes and implicated in zinc homeostasis. Plant Cell Physiol. 45, 1749–1758. doi: 10.1093/pcp/pci015
Kolaj-Robin, O., Russell, D., Hayes, K. A., Pembroke, J. T., and Soulimane, T. (2015). Cation diffusion facilitator family: structure and function. FEBS Lett. 589, 1283–1295. doi: 10.1016/j.febslet.2015.04.007
Krzesłowska, M. (2011). The cell wall in plant cell response to trace metals: polysaccharide remodeling and its role in defense strategy. Acta Physiol. Plant. 33, 35–51. doi: 10.1007/s11738-010-0581-z
Krzywinski, M., Schein, J., Birol, I., Connors, J., Gascoyne, R., Horsman, D., et al. (2009). Circos: an information aesthetic for comparative genomics. Genome Res. 19, 1639–1645. doi: 10.1101/gr.092759.109
Lescot, M., Déhais, P., Thijs, G., Marchal, K., Moreau, Y., Van De Peer, Y., et al. (2002). PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 30, 325–327. doi: 10.1093/nar/30.1.325
Letunic, I., Copley, R. R., Schmidt, S., Ciccarelli, F. D., Doerks, T., Schultz, J., et al. (2004). SMART 4.0: towards genomic data integration. Nucleic Acids Res. 32, D142–D144.
Liu, J., Gao, Y., Tang, Y., Wang, D., Chen, X., Yao, Y., et al. (2019). Genome-wide identification, comprehensive gene feature, evolution, and expression analysis of plant metal tolerance proteins in tobacco under heavy metal toxicity. Front. Genet. 10:345. doi: 10.3389/fgene.2019.00345
Liu, J., Pang, X., Cheng, Y., Yin, Y., Zhang, Q., Su, W., et al. (2018). The Hsp70 gene family in Solanum tuberosum: genome-wide identification, phylogeny, and expression patterns. Sci. Rep. 8:16628.
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Mani, A., and Sankaranarayanan, K. (2018). “Heavy metal and mineral element-induced abiotic stress in rice plant,” in Rice Crop — Current Developments, eds S. Farooq, K. Z. Hayat, and I. Amjad (London: IntechOpen). doi: 10.5772/intechopen.76080
Migocka, M., Kosieradzka, A., Papierniak, A., Maciaszczyk-Dziubinska, E., Posyniak, E., Garbiec, A., et al. (2015). Retracted: Two metal-tolerance proteins, MTP1 and MTP4, are involved in Zn homeostasis and Cd sequestration in cucumber cells. J. Exp. Bot. 66, 1001–1015. doi: 10.1093/jxb/eru459
Migocka, M., Papierniak, A., Maciaszczyk-Dziubińska, E., Poździk, P., Posyniak, E., Garbiec, A., et al. (2014). Retracted: Cucumber metal transport protein MTP8 confers increased tolerance to manganese when expressed in yeast and Arabidopsis thaliana. J. Exp. Bot. 65, 5367–5384. doi: 10.1093/jxb/eru295
Montanini, B., Blaudez, D., Jeandroz, S., Sanders, D., and Chalot, M. (2007). Phylogenetic and functional analysis of the Cation Diffusion Facilitator (CDF) family: improved signature and prediction of substrate specificity. BMC Genomics 8:107. doi: 10.1186/1471-2164-8-107
Mou, S., Liu, Z., Guan, D., Qiu, A., Lai, Y., and He, S. (2013). Functional analysis and expressional characterization of rice ankyrin repeat-containing protein, Os PIANK1, in basal defense against Magnaporthe oryzae attack. PLoS One 8:e59699. doi: 10.1371/journal.pone.0059699
Musavizadeh, Z., Najafi-Zarrini, H., Kazemitabar, S. K., Hashemi, S. H., Faraji, S., Barcaccia, G., et al. (2021). Genome-wide analysis of potassium channel genes in rice: expression of the OsAKT and OsKAT genes under salt stress. Genes 12:784. doi: 10.3390/genes12050784
Narusaka, Y., Nakashima, K., Shinwari, Z. K., Sakuma, Y., Furihata, T., Abe, H., et al. (2003). Interaction between two cis-acting elements, ABRE and DRE, in ABA-dependent expression of Arabidopsis rd29A gene in response to dehydration and high-salinity stresses. Plant J. 34, 137–148. doi: 10.1046/j.1365-313x.2003.01708.x
Nies, D. H., and Silver, S. (1995). Ion efflux systems involved in bacterial metal resistances. J. Ind. Microbiol. 14, 186–199. doi: 10.1007/bf01569902
Ober, D. (2005). Seeing double: gene duplication and diversification in plant secondary metabolism. Trends Plant Sci. 10, 444–449. doi: 10.1016/j.tplants.2005.07.007
Purwantini, E., Torto-Alalibo, T., Lomax, J., Setubal, J. C., Tyler, B. M., and Mukhopadhyay, B. (2014). Genetic resources for methane production from biomass described with the Gene Ontology. Front. Microbiol. 5:634. doi: 10.3389/fmicb.2014.00634
Qian, W., Liao, B.-Y., Chang, A. Y.-F., and Zhang, J. (2010). Maintenance of duplicate genes and their functional redundancy by reduced expression. Trends Genet. 26, 425–430. doi: 10.1016/j.tig.2010.07.002
Rai, P. K., Lee, S. S., Zhang, M., Tsang, Y. F., and Kim, K.-H. (2019). Heavy metals in food crops: Health risks, fate, mechanisms, and management. Environ. Int. 125, 365–385. doi: 10.1016/j.envint.2019.01.067
Ricachenevsky, F. K., Menguer, P. K., Sperotto, R. A., Williams, L. E., and Fett, J. P. (2013). Roles of plant metal tolerance proteins (MTP) in metal storage and potential use in biofortification strategies. Front. Plant Sci. 4:144. doi: 10.3389/fpls.2013.00144
Rogozin, I. B., Carmel, L., Csuros, M., and Koonin, E. V. (2012). Origin and evolution of spliceosomal introns. Biol. Direct 7, 1–28.
Saeed, A. I., Bhagabati, N. K., Braisted, J. C., Liang, W., Sharov, V., Howe, E. A., et al. (2006). [9] TM4 microarray software suite. Methods Enzymol. 411, 134–193. doi: 10.1016/s0076-6879(06)11009-5
Samuel, M. S., Datta, S., Khandge, R. S., and Selvarajan, E. (2021). A state of the art review on characterization of heavy metal binding metallothioneins proteins and their widespread applications. Sci. Total Environ. 775:145829. doi: 10.1016/j.scitotenv.2021.145829
Samuel, M. S., Selvarajan, E., Subramaniam, K., Mathimani, T., Seethappan, S., and Pugazhendhi, A. (2020). Synthesized β-cyclodextrin modified graphene oxide (β-CD-GO) composite for adsorption of cadmium and their toxicity profile in cervical cancer (HeLa) cell lines. Process Biochem. 93, 28–35. doi: 10.1016/j.procbio.2020.02.014
Sazegari, S., Niazi, A., and Ahmadi, F. S. (2015). A study on the regulatory network with promoter analysis for Arabidopsis DREB-genes. Bioinformation 11, 101–106. doi: 10.6026/97320630011101
Schlueter, J. A., Lin, J.-Y., Schlueter, S. D., Vasylenko-Sanders, I. F., Deshpande, S., Yi, J., et al. (2007). Gene duplication and paleopolyploidy in soybean and the implications for whole genome sequencing. BMC Genomics 8:330. doi: 10.1186/1471-2164-8-330
Shirazi, Z., Abedi, A., Kordrostami, M., Burritt, D. J., and Hossain, M. A. (2019). Genome-wide identification and characterization of the metal tolerance protein (MTP) family in grape (Vitis vinifera L.). 3 Biotech 9:199.
Shukla, P. S., Agarwal, P., Gupta, K., and Agarwal, P. K. (2015). Molecular characterization of an MYB transcription factor from a succulent halophyte involved in stress tolerance. AoB Plants 7:plv054. doi: 10.1093/aobpla/plv054
Struk, S., Jacobs, A., Sánchez Martín-Fontecha, E., Gevaert, K., Cubas, P., and Goormachtig, S. (2019). Exploring the protein–protein interaction landscape in plants. Plant Cell Environ. 42, 387–409. doi: 10.1111/pce.13433
Takemoto, Y., Tsunemitsu, Y., Fujii-Kashino, M., Mitani-Ueno, N., Yamaji, N., Ma, J. F., et al. (2017). The tonoplast-localized transporter MTP8. 2 contributes to manganese detoxification in the shoots and roots of Oryza sativa L. Plant Cell Physiol. 58, 1573–1582. doi: 10.1093/pcp/pcx082
Tamura, K., Peterson, D., Peterson, N., Stecher, G., Nei, M., and Kumar, S. (2011). MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28, 2731–2739. doi: 10.1093/molbev/msr121
Tang, H., Bowers, J. E., Wang, X., Ming, R., Alam, M., and Paterson, A. H. (2008). Synteny and collinearity in plant genomes. Science 320, 486–488. doi: 10.1126/science.1153917
Tang, H., Krishnakumar, V., Bidwell, S., Rosen, B., Chan, A., Zhou, S., et al. (2014). An improved genome release (version Mt4.0) for the model legume Medicago truncatula. BMC Genomics 15:312. doi: 10.1186/1471-2164-15-312
Thomine, S., and Vert, G. (2013). Iron transport in plants: better be safe than sorry. Curr. Opin. Plant Biol. 16, 322–327. doi: 10.1016/j.pbi.2013.01.003
Tsunemitsu, Y., Yamaji, N., Ma, J. F., Kato, S.-I., Iwasaki, K., and Ueno, D. (2018). Rice reduces Mn uptake in response to Mn stress. Plant Signal. Behav. 13:e1422466. doi: 10.1080/15592324.2017.1422466
van der Zaal, B. J., Neuteboom, L. W., Pinas, J. E., Chardonnens, A. N., Schat, H., Verkleij, J. A., et al. (1999). Overexpression of a novel Arabidopsis gene related to putative zinc-transporter genes from animals can lead to enhanced zinc resistance and accumulation. Plant Physiol. 119, 1047–1056. doi: 10.1104/pp.119.3.1047
Vatansever, R., Filiz, E., and Eroglu, S. (2017). Genome-wide exploration of metal tolerance protein (MTP) genes in common wheat (Triticum aestivum): insights into metal homeostasis and biofortification. Biometals 30, 217–235. doi: 10.1007/s10534-017-9997-x
Volkamer, A., Kuhn, D., Grombacher, T., Rippmann, F., and Rarey, M. (2012). Combining global and local measures for structure-based druggability predictions. J. Chem. Inf. Model. 52, 360–372. doi: 10.1021/ci200454v
Wei, F., Coe, E., Nelson, W., Bharti, A. K., Engler, F., Butler, E., et al. (2007). Physical and genetic structure of the maize genome reflects its complex evolutionary history. PLoS Genet. 3:e123. doi: 10.1371/journal.pgen.0030123
Wittkopp, P. J., and Kalay, G. (2012). Cis-regulatory elements: molecular mechanisms and evolutionary processes underlying divergence. Nat. Rev. Genet. 13, 59–69. doi: 10.1038/nrg3095
Yamaguchi-Shinozaki, K., and Shinozaki, K. (2005). Organization of cis-acting regulatory elements in osmotic-and cold-stress-responsive promoters. Trends Plant Sci. 10, 88–94. doi: 10.1016/j.tplants.2004.12.012
Young, N. D., Debellé, F., Oldroyd, G. E. D., Geurts, R., Cannon, S. B., Udvardi, M. K., et al. (2011). The Medicago genome provides insight into the evolution of rhizobial symbioses. Nature 480, 520–524.
Young, N. D., and Udvardi, M. (2009). Translating Medicago truncatula genomics to crop legumes. Curr. Opin. Plant Biol. 12, 193–201. doi: 10.1016/j.pbi.2008.11.005
Zambounis, A., Ganopoulos, I., Valasiadis, D., Karapetsi, L., and Madesis, P. (2020). RNA sequencing-based transcriptome analysis of kiwifruit infected by Botrytis cinerea. Physiol. Mol. Plant Pathol. 111:101514. doi: 10.1016/j.pmpp.2020.101514
Zhang, S., Yang, W., Zhao, Q., Zhou, X., Jiang, L., Ma, S., et al. (2016). Analysis of weighted co-regulatory networks in maize provides insights into new genes and regulatory mechanisms related to inositol phosphate metabolism. BMC Genomics 17:129. doi: 10.1186/s12864-016-2476-x
Keywords: Medicago truncatula, heavy metals, metal tolerance protein (MTP), genome-wide identification, gene expression
Citation: El-Sappah AH, Elbaiomy RG, Elrys AS, Wang Y, Zhu Y, Huang Q, Yan K, Xianming Z, Abbas M, El-Tarabily KA and Li J (2021) Genome-Wide Identification and Expression Analysis of Metal Tolerance Protein Gene Family in Medicago truncatula Under a Broad Range of Heavy Metal Stress. Front. Genet. 12:713224. doi: 10.3389/fgene.2021.713224
Received: 22 May 2021; Accepted: 30 June 2021;
Published: 07 September 2021.
Edited by:
Reyazul Rouf Mir, Sher-e-Kashmir University of Agricultural Sciences and Technology of Kashmir, IndiaReviewed by:
Parviz Heidari, Shahrood University of Technology, IranCopyright © 2021 El-Sappah, Elbaiomy, Elrys, Wang, Zhu, Huang, Yan, Xianming, Abbas, El-Tarabily and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jia Li, amlhLmxpMTZAYWxpeXVuLmNvbQ==; amlhLmxpMTBAYWxpeXVuLmNvbQ==; Khaled A. El-Tarabily, a3RhcmFiaWx5QHVhZXUuYWMuYWU=; Manzar Abbas, YWJiYXMyNDcyQGhvdG1haWwuY29t
†These authors have contributed equally to this work and share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.