
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Genet. , 17 September 2021
Sec. Evolutionary and Population Genetics
Volume 12 - 2021 | https://doi.org/10.3389/fgene.2021.709388
This article is part of the Research Topic Association Between Individuals’ Genomic Ancestry and Variation in Disease Susceptibility View all 13 articles
The rapid rise and global consequences of the novel coronavirus disease 19 (COVID-19) have again brought the focus of the scientific community on the possible host factors involved in patient response and outcome to exposure to the virus. The disease severity remains highly unpredictable, and individuals with none of the aforementioned risk factors may still develop severe COVID-19. It was shown that genotype-related factors like an ABO Blood Group affect COVID-19 severity, and the risk of infection with SARS-CoV-2 was higher for patients with blood type A and lower for patients with blood type O. Currently it is not clear which specific genes are associated with COVID-19 severity. The comparative analysis of COVID-19 and other viral infections allows us to predict that the variants within the interferon pathway genes may serve as markers of the magnitude of immune response to specific pathogens. In particular, various members of Class III interferons (lambda) are reviewed in detail.
Coronavirus disease 2019 (COVID-19) is a rapidly emerging infectious disease caused by SARS-CoV-2 virus, a member of the Coronaviridae family. Since the discovery of first cases in December 2019 in Wuhan, China (World Health Organization, 2020a), the number of infected patients worldwide has been increasing logarithmically, and by December 2020 had surpassed 70 million reported cases and over 1.5 million deaths globally since the start of the pandemic (World Health Organization, 2020b).
According to WHO, mild to moderate respiratory symptoms such as fever, dry cough, upper airway congestion and sore throat are among the most common symptoms of COVID-19 (World Health Organization, 2020c; World Health Organization, 2021) which may develop over the course of 2 weeks after the exposure. However, approximately 20% of the patients develop severe or critical COVID-19 (Wu and McGoogan, 2020), characterized by pneumonia and acute respiratory distress syndrome which require hospitalization. While the overall mortality of severe COVID-19 is estimated between 1 and 4% (Ruan, 2020), in-hospital mortality in severe cases is substantially higher, reaching 28–62%, and even surpassing that in patients requiring mechanical ventilation (Weiss and Murdoch, 2020).
Multiple studies have been performed to establish the factors influencing the susceptibility to, severity and mortality of COVID-19 (Du et al., 2020; Gong et al., 2020; Vardavas and Nikitara, 2020; Verity et al., 2020). It has been shown that the severe or critical course of the disease is more likely in older adults, especially those with underlying health conditions (Centers for Disease Control and Prevention, 2020a; Garg et al., 2020), as 80% of deaths associated with COVID-19 were among adults aged 65 years or older, or those with severe comorbidities (Centers for Disease Control and Prevention, 2020b). Other proposed risk factors include smoking (Vardavas and Nikitara, 2020) and blood type (Zhao et al., 2020).
However, disease severity remains highly unpredictable, and individuals with none of the aforementioned risk factors may still develop severe COVID-19. This is particularly evident in the demographics of the disease in the United States, where even in the early stages of the outbreak 20% of the hospital admissions and 12% of the ICU admissions were attributed to people aged 20–44 years (Centers for Disease Control and Prevention, 2020b). Coupled with the first reported case of identical twins both dying of severe COVID-19 (BBC, 2020) within several days of each other, as well as data obtained in pilot studies on the heritability of COVID-19 symptoms (Williams et al., 2020), it strongly suggests that inherited DNA variants play a significant role in the severity of the disease.
The rapid rise and global consequences of the novel coronavirus disease 19 (COVID-19) has brought the focus of the research on the possible contribution of the host factors to patient response and outcome of exposure to the virus. SARS-coronavirus 2 (SARS-CoV-2), the pathogenic cause of the disease, relies on similar mechanisms of cellular entry as SARS-CoV; namely, the SARS-CoV-2 receptor angiotensin-converting enzyme II (ACE2) and the serine protease TMPRSS2, which facilitates the priming of spike (S) protein for viral entry (Hoffmann et al., 2020; Zhou et al., 2020).
Early on during the rise of the pandemic, there was a hope that variations in the ACE2 gene may account for resistance or susceptibility to COVID-19 in different populations. It was shown that populations in East Asia had higher allele frequencies in the expression quantitative trait loci (eQTL) in ACE2, which might have led to increased expression of the enzyme (Cao et al., 2020). Likewise, given that ACE2 is located on chromosome X, a hope was expressed for explaining the gender differences in response to the disease, namely the fact that men were disproportionately more susceptible to the SARS-CoV-2 virus (Majdic, 2020).
Early studies attempting to connect variation in ACE2 and TMPRSS2 loci on the risks of contracting COVID-19 in any form, so far, have produced inconclusive results, ranging from single SNP associations uncovered in small cohorts (Latini et al., 2020) to the lack thereof. The latter, possibly, is due to the dual role of ACE2, which serves both as an entry into the wells and a lung-protective molecule (Dalan et al., 2020; Nagy et al., 2021).
A large study by Lopera Maya et al. (2020) used the Lifelines cohort data to analyze the association between the variants within ACE2 or TMPRSS2 loci and cardiac, pulmonary, renal and other quantitative phenotypes, which are also pertinent to COVID-19. Despite the large sample size and wide variety of variants and quantitative phenotypes examined, no statistically significant association was detected. The study found, however, an intriguing association between the use of angiotensin receptor blockers (ARBs) and non-steroidal anti-inflammatory drugs (NSAIDs) and variants at the ACE2 and loci. As the diseases associated with the use of these medications are commonly comorbid with COVID-19 (Lopera Maya et al., 2020), these findings may eventually prove their relevance to COVID-19 severity. Drugs based on the inhibition or blockage of TMPRSS2 protease are undergoing clinical trials as a therapeutic option for COVID-19 treatment (Abbasi et al., 2021). Thus, there remains continued interest in studying the ACE2 and TMPRSS2 genes as determinants of susceptibility to SARS-CoV-2.
After the SARS-CoV outbreak in Hong Kong in 2003, researchers showed a relationship between the blood type of the participants that had been exposed to the virus and the chance of contracting infection (Cheng et al., 2005). It appeared that exposed individuals with blood group O phenotype were less susceptible to SARS-CoV infection, even while previous studies had shown that they had increased susceptibility to infection with either Norwalk virus or H. pylori (Cheng et al., 2005). The association of ABO blood groups with the risk to contract coronavirus disease has also been noted during the current pandemic. Retrospective study conducted in China showed that patients with blood group O had a significantly lower risk of infection and hospitalization with SARS-CoV-2, while patients with blood group A had a higher risk of infection and hospitalization (Li et al., 2020).
Further research has both confirmed this association and shed more light on it. Retrospective studies conducted in various regions of China, New York, Italy, Spain, and Turkey have shown a higher odds ratio for being infected with SARS-CoV-2 for patients with blood type A phenotype as well as a lower one for blood type O patients when (Focosi, 2020). A genome wide association study conducted in Italy and Spain regarding the genetic associations between individuals infected with COVID-19 and respiratory failure, confirmed that patients with blood group A had a higher risk of COVID-19-induced respiratory failure while blood group O granted patients a protective effect (The Severe Covid-19 GWAS Group, 2020). Two loci with a genome-wide significance were found, namely, the rs11385942 insertion-deletion GA at locus 3p21.31 and the rs657152 A at locus 9q34.2. The association signal at 9q34.2 coincided with the ABO locus, further implicating the connection between patient’s ABO blood group and the course and danger of the disease (The Severe Covid-19 GWAS Group, 2020). Later, a multicenter study performed in Canada showed that COVID-19 patients with blood group A or AB are at increased risk for requiring mechanical ventilation and prolonged ICU admission compared with patients with blood group O or B (Hoiland et al., 2020), thus, supporting in silico GWAS results by patient’s ward observations.
Interestingly, the viral infectivity features due to the ACE2 receptor binding, and due to contribution of the blood antigens may be related to each other. In case of SARS-CoV, the presence of anti-A antibodies, which is a characteristic of groups O and B, inhibits the adhesion of the virus to the ACE2 receptor (Guillon et al., 2008). It is tempting to speculate that this finding may be directly relevant to SARS-CoV-2 as well, given that these findings are consistent with the host response to other viruses such as measles and HIV (Arendrup et al., 1991; Preece et al., 2002) and a trend in increased efficiency of the transfusion of the convalescent plasma from O or B group donors (Hacibekiroğlu et al., 2021).
In addition to findings reported from Italy/Spain (The Severe Covid-19 GWAS Group, 2020), a separate study comprising 3,199 hospitalized patients with COVID-19 and control individuals was released by the COVID-19 Host Genetics Initiative in which the region on chromosome 3 was the only major genetic risk factor for severe symptoms after SARS-CoV-2 infection and hospitalization at the genome-wide level (The COVID-19 Host Genetics Initiative, 2020). It is not clear which specific gene within the region identified on chromosome 3 is associated with COVID-19 severity. In particular, this region harbors CXCR6 and CCR1 genes, encoding important chemokines, which control the movement of immune cells and are critical for the innate immune system to function properly (Sokol and Luster, 2015). Another gene of this region, SLC6A20, encodes a protein that functions as a proline transporter expressed in alveolar cells, kidney and small intestine (SIT1), which is known to bind to ACE2 (Camargo et al., 2009; Vuille-dit-Bille et al., 2015; Wang et al., 2020). Notably, the entire fragment may have been inherited from Neanderthals, entering the human genome during the period of interbreeding between the two groups (Zeberg and Pääbo, 2020), and is differentially represented in human population samples.
Interferons (IFNs) are central to antiviral immunity. Previously it was shown that type I IFN deficiency in the blood could be a hallmark of severe COVID-19 and provide a rationale for combined therapeutic approaches (Hadjadj et al., 2020).
Additional studies have shown the importance of other loci in determining the genetic susceptibility of hosts to COVID-19, particularly in determining which patients are susceptible to severe manifestations of the illness. A recent study of patients with life-threatening COVID-19 pneumonia looked at thirteen loci involved in either the TLR3 or IRF7 dependent pathways for the amplification of type I IFN, and found that 3.5% of patients had deleterious variants (pLOF) in eight of the tested loci, underlining how impairment of the production of type I IFNs can lead to critical SARS-CoV-2 infection (Zhang et al., 2020). Similarly, a recent study of critically-ill COVID-19 patients in the United Kingdom used Mendelian randomization to show the potential for a causal relationship between the IFNAR2 gene which codes for a receptor subunit in interferon signalling and disease severity, and concluded likewise that the administration of interferons may aid in patient recovery, while acknowledging that it is as yet unclear when during the course of the illness they may provide therapeutic benefit (Pairo-Castineira et al., 2021). Moreover, the study was able to replicate the results of a previous study on the 3p21.31 locus, and a transcriptome-wide association study that they performed on the patient pool showed that the variant in oligoadenylate synthetase (OAS), rs10735079 affected expression of OAS3, which codes mediator involved in the degradation pathway of double-stranded RNA, which is itself involved in the replication pathway of coronaviruses (Pairo-Castineira et al., 2021).
Notably, in humans, the ACE2 belongs to a family of interferon-stimulated genes (ISGs), which typically serve to promote a complex and uniform response to an infection-related spike in interferon levels (Schneider et al., 2014). Moreover, in human lung epithelial cells, the levels of ACE2 mRNA are co-correlated with that of TMPRSS4, and many immune response pathways, including proinflammatory interleukins and IFI16 (Wruck and Adjaye, 2020). Specifically in human nasal epithelial cells, ACE2 expression is upregulated by type I (IFN-α and IFN-β) and type II (IFN-γ) interferons (Ziegler et al., 2020). The efficacy of this process may be affected by genetic variations in any part of this cascade. However, in this review, we would like to bring attention to a particular component of the interferon response, which has been massively implicated in the natural and therapeutic outcomes for other viral diseases, namely, the IFNL4 gene. This gene encodes a type III interferon IFN-λ4, capable of blocking some of the interferon signaling, resulting in poor response to HCV treatment with IFN (Sung et al., 2017). Notably, type III IFNs have been proposed as more viable therapeutic option for prevention and treatment of COVID-19 than type I IFNs, particularly because they cause fewer and milder systemic side effects (Muir et al., 2014; Prokunina-Olsson et al., 2020). It has also been shown that Type III IFNs are highly effective at preventing the viral spread from the nasal epithelium to the upper respiratory tract (Klinkhammer et al., 2018). Additional studies may be warranted to explore the mechanisms of interaction between SARS-CoV-2 and type III interferons, and to estimate how they are affected by the status of the IFNL4 gene.
It is anticipated that influence of ACE2 in COVID-19 сan potentially be exploited for the rational design of effective SARS-CoV-2 therapeutics (Ni et al., 2020; Barros et al., 2021).
Interferons are a class of cytokines that mediate the host immune response to infection by viral and non-viral pathogens (Crosse et al., 2017; Bogdan et al., 2004; Seliger et al., 2008). They are categorized into three types based on their protein sequence (O’Brien et al., 2014) (Table 1). Type I interferons are rapidly produced when viral envelope glycoproteins, CpG DNA, or dsRNA interact with host cell receptors such as mannose receptors, toll-like receptors, and cytosolic receptors (Malmgaard, 2004). Type 1 interferons can directly activate natural killers (NKs), antigen-presenting dendritic cells as well as CD4 and CD8 T cells (Hervas-Stubbs et al., 2011). All type I interferons signal through a common receptor interferon alpha receptor (IFNAR). The IFNAR induces the Janus activated kinase-signal transducer and activation of transcription (JAK-STAT) pathway that control a large collection of genes through regulated expression of various signaling intermediaries (Guan et al., 2014; Messina et al., 2016; Olex et al., 2016).
Type I interferons are rapidly produced when viral factors, such as envelope glycoproteins, CpG DNA, or dsRNA, interact with cellular pattern-recognition receptors (PRRs), such as mannose receptors, toll-like receptors (TLRs), and cytosolic receptors (Malmgaard, 2004). These interferons directly activate natural killers (NKs), antigen-presenting dendritic cells (DC) as well as CD4 and CD8 T cells (Hervas-Stubbs et al., 2011). In T cells, the signaling through the IFNAR is critical for the acquisition of effector functions (Kole et al., 2013).
Type II interferons are represented by pleiotropic Th1-type cytokine interferon-γ. The IFN-γ is induced in response to a variety of cytokines, including interleukin-2 (IL-2), IL-18, Type I IFNs alpha/beta, or by stimulation through T cell receptors (TCRs) or NK cell receptors (Malmgaard, 2004). Similar to Type I interferons, IFN-γ stimulates the JAK/STAT pathway. In addition, a number of other pathways, including MAP kinase, PI3-K, CaMKII, and NF-kappaB cross-talk with JAK-STAT signaling to fine-tune the multifaceted effects of IFNγ, which are exerted in a gene- and cell type-specific manner (Gough et al., 2008).
The type III family of interferons are comprised of IFN-λ1, IFN-λ2, and IFN-λ3 or IL-29, IL-28A, and IL-28B, respectively (Kotenko et al., 2003; Gad et al., 2009; Lin and Young., 2014; O’Brien et al., 2014). These interferons signal through a receptor complex composed of the IFN-λR1 chain (also known as IL-28RA) and the IL-10R2 chain, which is also a part of the receptor complexes for IL-10, IL-22, and IL-26. (Sheppard et al., 2003; Gad et al., 2009; Donnelly and Kotenko, 2010; Lopušná et al., 2013).
In 2013, a new member of the interferon λ (lambda) family, IFN-λ4, was described which signals through the IFNλR1 and IL-10R2 receptor chains (Hamming et al., 2013). The IFN-λ4 is encoded by the gene IFNL4, whose expression has been shown to be upregulated in response to HCV infection, but not to HBV infection (Estep et al., 2014).
Recent studies point that IFN dysregulation may be the key to determining COVID-19 pathogenesis (Andreakos and Tsiodras, 2020; Lopez et al., 2020; Meffre and Iwasaki, 2020). There is evidence that the response to class I interferons in COVID-19 is impaired. In the blood of patients with severe COVID-19, amounts of class I IFNs are much lower when compared to that of patients infected with highly pathogenic influenza viruses. Nevertheless, in the lungs, in bronchoalveolar lavage in some seriously ill COVID-19 patients, local induction of IFN genes becomes noticeable. A dysregulated interferon response is considered part of the immunomodulatory strategies used by some coronaviruses, including SARS-CoV-2 (Acharya et al., 2020). Nevertheless, a recent pan-ancestry exome-wide association study of rare genetic protein-coding variants and various t COVID-19 outcomes didn’t find any significant associations in any of the 13 interferon pathway genes (Kosmicki et al., 2021).
Since the beginning of the pandemics, interferons were repeatedly seen as a viable option for boosting the host’s defences against SARS-CoV-2. Indeed, early evidence suggests that SARS-CoV-2 may be more susceptible to pretreatment with type I IFNs, even more so than SARS-CoV (Lokugamage et al., 2020; Sallard et al., 2020). Later, in human intestinal cells, the treatment with interferon-lambda and respective responses showed efficiency at controlling SARS-CoV-2 replication (Stanifer et al., 2020). In this light, a renewed attention was paid to type III interferons, which have being tested as therapeutics in COVID-19 outpatients, either with no success (Jagannathan et al., 2021) or with limited virological response detected (Feld et al., 2021). The difference in outcomes of the interferon-lambda based therapeutics may be explained by the varied presence of neutralizing IFNL3 autoantibodies pre-existing in patients that later develop severe COVID-19 (Credle et al., 2021).
The IFNL4 gene is located on chromosome 19q13, just over 1 kb upstream of, and in the same orientation as, the gene encoding IFN-λ3 (Figure 1). It is extremely conserved in all mammals, indicating its functional importance (Key et al., 2014). The ancestral allele of IFNL4, contains a guanine residue at position 342 of the coding sequence (referred to as “ΔG”). It encodes a functional IFN-λ4 peptide.
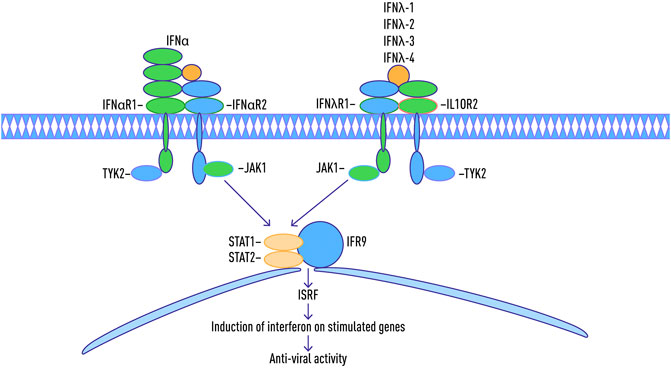
FIGURE 1. Location of common SNPs in IFNL4 Locus on Chromosome 19, and the map of IFNL4 exons. Adapted from: Stephen M. Laidlaw and Lynn B. Dustin, 2014, with changes.
The IFNL4 locus is known to contain a number of medically relevant single nucleotide polymorphisms (SNPs). One of these, rs368234815 or ss469415590 (TT) is characterized by the substitution of the G nucleotide with two thymine residues (TT) resulting in a nonsense mutation. As a result, IFN-λ4 can be generated only by individuals, who carry the ΔG allele, To date, the majority of the studies of IFNL4 locus have been performed in the context of hepatitis C virus (HCV). In contrast to other IFNs, expression of IFNλ4 is associated with decreased clearance of HCV in the human population; by contrast, a natural frameshift mutation that abrogates IFNλ4 production improves HCV clearance. The ΔG allele is associated with adverse outcomes of infection and interferon-based treatments (Amanzada et al., 2013; Franco et al., 2014 AIDS; Aka et al., 2014; Nozawa et al., 2014; Jouvin-Marche et al., 2014; Stättermayer et al., 2014) while the TT allele is associated with the spontaneous clearance of HCV and interferon responsiveness. It is presently the strongest known host factor for predicting clearance of HCV (O’Brien et al., 2014). Another SNP, known as rs12979860, located within the intron of IFNL4 gene, is closely linked to the rs368234815 allele and is significantly associated with sustained viral response (SVR) in HCV patients (Younossi et al., 2012).
In a genome-wide association study published in 2009, the presence of a rs12979860 with a “C” allele was strongly associated with spontaneous viral clearance and treatment response (Ge et al., 2009). Patients who were homozygous for the presence of “C” allele had a greater than 2-fold increase in rates of SVR as compared to patients with heterozygosity of this locus (C/T allele combination) and homozygous state T/T (Younossi et al., 2012; Meissner et al., 2014; Stättermayer et al., 2014). In addition to increased SVR rates, patients homozygous for C allele (C/C) were more likely to demonstrate spontaneous clearance of HCV (Thomas et al., 2009). Additionally, the presence of SVR-promoting rs12979860 allele of IL28B locus was associated with lower baseline inflammation and possible suppression of apoptosis in peripheral blood mononuclear cell (PBMCs) evaluated during early phase of the treatment as compared to the presence of deleterious allele (Younossi et al., 2012). These findings were confirmed in the 2013 GWAS performed in 13 international multicenter study sites (Duggal et al., 2013).
Interpretation of these findings relies on the proximity linkage of rs12979860 (IL28B) to rs368234815 (IFNL4) that is functionally responsible for effects of both variants. Due to shorter average size of haplotype blocks in individuals of African ancestry, rs368234815 is more strongly associated with HCV clearance in these ethnicities, whereas in Europeans and Asians it performs similarly to rs12979860 (Prokunina-Olsson et al., 2013).
Non-functional rs368234815-TT allele is specific for humans and is common in all human populations. In HapMap collection, it is detected in 93% of Asians genomes, 68% of European genomes, and 23% of Africans genomes (Prokunina-Olsson et al., 2013).
Similar frequencies of distribution were observed in the 1000 Genomes Project samples: the TT allele is present in 89.8–95.2% of Chinese genomes, in 68.9% of European genomes and in 29.3% of African genomes (The 1000 Genomes Project Consortium, 2015).
Linkage disequilibrium between rs368234815-TT allele and rs12979860-C allele results in the same frequencies distribution pattern for the latter genetic variant: C allele is present in 89.8–95.2% Chinese genomes, in 69.1% of European genomes and in 33.1% of African genomes (The 1000 Genomes Project Consortium, 2015). Frequency of the rs12979860 C/C genotype in IFNL4 gene was significantly lower in COVID-19 patients (p < 0.001) (Saponi-Cortes et al., 2021).
Linkage between these two genetic variations rises from Africans (R2 = 0.8318) through Europeans (R2 = 0.9815) and is absolute in Chinese populations (R2 = 1.0) (Machiela and Chanock, 2015).
It is theorized that it emerged right before the onset of the “out-of-Africa” migration and was immediately supported in its spread by positive selection in European and Asian populations resulting in the high frequency observed today (Key et al., 2014).
It is most likely that the selective force that was driving elimination of IFN-λ4 in a majority of human populations was an exposure to a certain pathogen (or pathogens), most likely a virus. However, this pathogen is unlikely to be HCV, which is known for its relatively slow progress toward symptomatic phase (Fan et al., 2016). There is also no association between IFNL4 polymorphisms and HBV susceptibility or natural clearance (Fan et al., 2016) and the advantages or disadvantages of IFN-λ4 expression in case of infection with a majority of non-HCV non-HBV viruses remain unknown.
A number of recent studies showed that IFN-λ4 possesses strong antiviral activity toward HCV and coronaviruses (Hamming et al., 2013; Prokunina-Olsson et al., 2013). When over-expressed in a hepatoma cells, IFNL4 induces STAT1/STAT2 phosphorylation and expression of interferon-stimulated genes (Prokunina-Olsson et al., 2013; O’Brien et al., 2014; Randall and Goodburn, 2008; Ank et al., 2006) (Figure 2). Interestingly, when studied against either HCV or coronavirus (HCoV-229E and MERS-CoV) challenges tested in either human ciliated airway epithelial cell (HAE) or hepatocyte cultures, the antiviral activities of recombinant IFNλ3 and IFNλ4 were similar (Hamming et al., 2013). Another recent comparative study of Type III interferons, this time performed using transcriptome sequencing, also failed to reveal any crucial differences between particular members of this family, with the majority of the identified genes being similarly regulated in hepatocytes as well as airway epithelial cells (Lauber et al., 2015). Hence, it looks like the differences in mode of action for various IFN-λ may be due to their direct binding to some cellular or viral targets rather than to the transcription programs they stimulate.
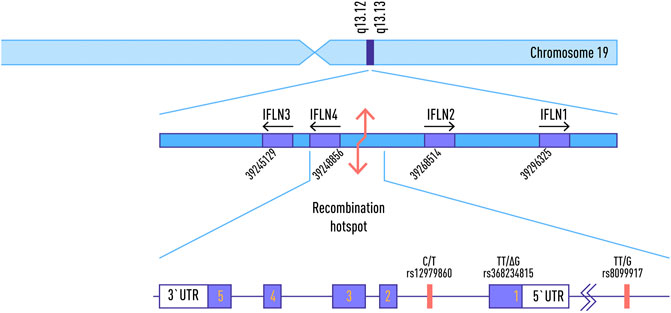
FIGURE 2. Schematic map of Jak-STAT pathway during an immune response with type 1-3 interferon antiviral activity.
IFNLR is expressed at relatively high levels in respiratory epithelial cells, and mice treated with IFN-λ prior to infection with human metapneumovirus (HMPV) develop lower viral titers and reduced inflammatory responses. On the other hand, Ifnlr1 −/− mice exhibit increased susceptibility to respiratory viral infections, including influenza virus, HMPV, respiratory syncytial virus, and SARS coronavirus (Lazear et al., 2015).
In contrast, Prokunina-Olsson proffered the hypothesis that functional IFN-λ4 protein may compete with the IL28B/IFN-λ3 receptors and apparently cause a pre-activation of the interferon-dependent genes, thus, reducing overall responsiveness to Type I and III interferon (Prokunina-Olsson et al., 2013). This hypothesis is a good agreement with previous findings that SVR-promoting alleles are associated with lower baseline inflammation (Younossi et al., 2012). Notably, in a small study of rs12979860 allele distribution in COVID-19 patients and controls, the “C” allele, previously associated with favourable HCV outcomes and lower baseline inflammation, showed association both with higher susceptibility to coronavirus and with poorer outcomes of SARS-CoV-2 disease (Agwa et al., 2021).
There is some evidence that the polymorphisms in IFNλ4 may influence outcomes of non-HCV non-coronavirus types of acute and chronic infections. In particular, solid-organ transplant recipients homozygous for the active, ancestral rs368234815 allele (ΔG) are more susceptible to CMV replication, especially in absence of antiviral prophylaxis (Egli et al., 2014; Manuel et al., 2015). Another study showed that the same allele is associated with increased susceptibility to AIDS-related CMV retinitis (Bibert et al., 2014).
Findings related to IFN-lambda gene variants in patients with HIV infection remain controversial. One study showed that, in Caucasian populations, the CC genotype of rs12979860, which is associated with favourable HCV outcomes, is also associated with spontaneous control of human immunodeficiency virus (HIV) viremia (Machmach et al., 2013). In cohorts of African Americans these findings, however, were not replicated (Sajadi et al., 2011; Salgado et al., 2011). In a study of Real and co-authors, pseudogenized allele rs368234815-TT that protects against infection with HCV was also associated with decreased likelihood of HIV-1 infection in male intravenous drug users [odds ratio (OR): 0.3; p = 0.006], and this association was not modified by the genotype of CCR5 (Real et al., 2015). Another recent study of rs368234815 variant showed that carriers of its active, ancestral variant ΔG have a higher occurrence of AIDS-defining illnesses and lower CD4 T-cell counts (Machmach et al., 2015). These results suggest that genetic susceptibility to HCV and HIV-1 infection may share a common molecular pathway (Real et al., 2015).
It should be noted that the relationships between pre-existing HIV infection and COVID-19 are still unclear (Centers for Disease Control and Prevention, 2020b), most likely due to limited cross-testing (Jones et al., 2020). While the use of protease inhibitors such as lopinavir and ritonavir had a positive effect on patients with MERS-CoV, recent research suggests that in patients with SARS-CoV-2 these compounds do not work (Jones et al., 2020; Jothimani et al., 2020). Attempts to utilize known anti-HCV treatments in COVID-19 wards had failed in a similar way (Huang et al., 2020).
Nevertheless, recent events have unequivocally shown that the coronaviruses, in general, and the SARS-CoV-2, in particular, should be regarded as yet another evolutionary driver for the fine-tuning of human interferon response to existing and emerging pathogens. Population frequencies of IFNL4 and other interferon-encoding gene variants may reflect a sum of past exposures to the various pathogens, and the subsequent bottlenecks which may or may not be related to epidemic events.
Genetic resistance to severe viral diseases shares common molecular pathways for various viral infections. Interferon expression pathways in host cells make a crucial contribution to the proinflammatory response to infectious agent appearance. The presence of some variants in the loci of interferon gene sequences reduces the natural immunity, and stimulates a susceptibility to severe viral disease.
LG and AB substantially contributed to the conception of the review. LG and KP drafted the outline of the review. DN and AS contributed to the genetic and clinical aspects of the review. AB, LG, and IK contributed to the immunological aspects of the review. AB, DN, AS, and IK reviewed and edited the draft with input from all authors. DN prepared and created figures. AB supervised and managed the review planning and execution. All have read and approved the submitted version.
Authors DN, AS, and AB were employed by the company Atlas Biomed Group Limited.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Abbasi, A. Z., Kiyani, D. A., Hamid, S. M., Saalim, M., Fahim, A., and Jalal, N. (2021). Spiking Dependence of SARS‐CoV‐2 Pathogenicity on TMPRSS2. J. Med. Virol. 93, 4205–4218. doi:10.1002/jmv.26911
Acharya, D., Liu, G., and Gack, M. U. (2020). Dysregulation of Type I Interferon Responses in COVID-19. Nat. Rev. Immunol. 20 (7), 397–398. doi:10.1038/s41577-020-0346-x
Agwa, S. H. A., Kamel, M. M., Elghazaly, H., Abd Elsamee, A. M., Hafez, H., Girgis, S. A., et al. (2021). Association between Interferon-Lambda-3 Rs12979860, TLL1 Rs17047200 and DDR1 Rs4618569 Variant Polymorphisms with the Course and Outcome of SARS-CoV-2 Patients. Genes 12 (6), 830. doi:10.3390/genes12060830
Aka, P. V., Kuniholm, M. H., Pfeiffer, R. M., Wang, A. S., Tang, W., Chen, S., et al. (2014). Association of the IFNL4-Δg Allele with Impaired Spontaneous Clearance of Hepatitis C Virus. J. Infect. Dis. 209 (3), 350–354. doi:10.1093/infdis/jit433
Amanzada, A., Kopp, W., Spengler, U., Ramadori, G., and Mihm, S. (2013). Interferon-λ4 (IFNL4) Transcript Expression in Human Liver Tissue Samples. PloS one 8 (12), e84026. doi:10.1371/journal.pone.0084026
Andreakos, E., and Tsiodras, S. (2020). COVID ‐19: Lambda Interferon against Viral Load and Hyperinflammation. EMBO Mol. Med. 12 (6), e12465. doi:10.15252/emmm.202012465
Ank, N., West, H., and Paludan, S. R. (2006). IFN-λ: Novel Antiviral Cytokines. J. Interferon Cytokine Res. 26 (6), 373–379. doi:10.1089/jir.2006.26.373
Arendrup, M., Hansen, J.-E. S., Clausen, H., Nielsen, C., Mathiesen, L. R., and Nielsen, J. O. (1991). Antibody to Histo-Blood Group A Antigen Neutralizes HIV Produced by Lymphocytes from Blood Group A Donors but Not from Blood Group B or O Donors. Aids 5 (4), 441–444. doi:10.1097/00002030-199104000-00014
Barros, E. P., Casalino, L., Gaieb, Z., Dommer, A. C., Wang, Y., Fallon, L., et al. (2021). The Flexibility of ACE2 in the Context of SARS-CoV-2 Infection. Biophysical J. 120 (6), 1072–1084. doi:10.1016/j.bpj.2020.10.036
BBC (2020). Coronavirus: Twin sisters Katy and Emma Davis die with Covid-19. Southampton: BBC News. Available at: https://www.bbc.com/news/uk-england-hampshire-52409765.
Bibert, S., Wojtowicz, A., Taffé, P., Manuel, O., Bernasconi, E., Furrer, H., et al. (2014). The IFNL3/4 ΔG Variant Increases Susceptibility to Cytomegalovirus Retinitis Among HIV-Infected Patients. AIDS 28 (13), 1885–1889. doi:10.1097/qad.0000000000000379
Bogdan, C., Mattner, J., and Schleicher, U. (2004). The Role of Type I Interferons in Non-viral Infections. Immunol. Rev. 202, 33–48. doi:10.1111/j.0105-2896.2004.00207.x
Camargo, S. M. R., Singer, D., Makrides, V., Huggel, K., Pos, K. M., Wagner, C. A., et al. (2009). Tissue-Specific Amino Acid Transporter Partners ACE2 and Collectrin Differentially Interact with Hartnup Mutations. Gastroenterology 136 (3), 872–882. doi:10.1053/j.gastro.2008.10.055
Cao, Y., Li, L., Feng, Z., Wan, S., Huang, P., Sun, X., et al. (2020). Comparative Genetic Analysis of the Novel Coronavirus (2019-nCoV/SARS-CoV-2) Receptor ACE2 in Different Populations. Cell Discov. 6, 11. doi:10.1038/s41421-020-0147-1
Centers for Disease Control and Prevention (2020a). COVID-19 and HIV. HIV. Available at: https://www.cdc.gov/hiv/basics/covid-19.html (Accessed November 11, 2020).
Centers for Disease Control and Prevention (2020b). Severe Outcomes Among Patients with Coronavirus Disease 2019 (COVID-19) — United States, February 12–March 16, 2020. MMWR Morb Mortal Wkly Rep. 69, 343–346. Available at: https://www.cdc.gov/mmwr/volumes/69/wr/mm6912e2.htm. doi:10.15585/mmwr.mm6912e2
Cheng, Y., Cheng, G., Chui, C. H., Lau, F. Y., Chan, P. K. S., Ng, M. H. L., et al. (2005). ABO Blood Group and Susceptibility to Severe Acute Respiratory Syndrome. JAMA 293 (12), 1447–1451. doi:10.1001/jama.293.12.1450-c
Credle, J. J., Gunn, J., Sangkhapreecha, P., Monaco, D. R., Zheng, X. A., Tsai, H. J., et al. (2021). Neutralizing IFNL3 Autoantibodies in Severe COVID-19 Identified Using Molecular Indexing of Proteins by Self-Assembly. bioRxiv, 432977. [Preprint]. doi:10.1101/2021.03.02.432977
Crosse, K. M., Monson, E. A., Beard, M. R., and Helbig, K. J. (2017). Interferon-Stimulated Genes as Enhancers of Antiviral Innate Immune Signaling. J. Innate Immun. 10 (2), 85–93. doi:10.1159/000484258
Dalan, R., Bornstein, S. R., El-Armouche, A., Rodionov, R. N., Markov, A., Wielockx, B., et al. (2020). The ACE-2 in COVID-19: Foe or Friend?. Horm. Metab. Res. 52 (5), 257–263. doi:10.1055/a-1155-0501
Donnelly, R. P., and Kotenko, S. V. (2010). Interferon-lambda: a New Addition to an Old Family. J. Interferon Cytokine Res. 30 (8), 555–564. doi:10.1089/jir.2010.0078
Du, R.-H., Liang, L.-R., Yang, C.-Q., Wang, W., Cao, T.-Z., Li, M., et al. (2020). Predictors of Mortality for Patients with COVID-19 Pneumonia Caused by SARS-CoV-2: a Prospective Cohort Study. Eur. Respir. J. 55 (5), 2000524. doi:10.1183/13993003.00524-2020
Duggal, P., Thio, C. L., Wojcik, G. L., Goedert, J. J., Mangia, A., Latanich, R., et al. (2013). Genome-Wide Association Study of Spontaneous Resolution of Hepatitis C Virus Infection: Data from Multiple Cohorts. Ann. Intern. Med. 158 (4), 235. doi:10.7326/0003-4819-158-4-201302190-00003
Egli, A., Levin, A., Santer, D. M., Joyce, M., O’Shea, D., Thomas, B. S., et al. (2014). Immunomodulatory Function of Interleukin 28B during Primary Infection with Cytomegalovirus. J. Infect. Dis. 210 (5), 717–727. doi:10.1093/infdis/jiu144
Estep, M., Perry, K., Tavakolian, K., Younoszai, Z., Stepanova, M., Noorzad, A., and Younossi, Z. (2014). Interferon Lambda-4 (IFNL4) TT Allele Is Associated with Lower Expression of Genes Associated with Early Inflammation after Initiation of Treatment: 1835. 65th Annual Meeting of the American Association for the Study of Liver Diseases: The Liver Meeting 2014, 182A. doi:10.1002/hep.27415
Fan, J. H., Hou, S. H., Qing-Ling, L., Hu, J., Peng, H., and Guo, J. J. (2016). Association of HLA-DQ and IFNL4 Polymorphisms with Susceptibility to Hepatitis B Virus Infection and Clearance. Ann. Hepatol. 15 (4), 532–539. doi:10.5604/16652681.1202946
Feld, J. J., Kandel, C., Biondi, M. J., Kozak, R. A., Zahoor, M. A., Lemieux, C., et al. (2021). Peginterferon Lambda for the Treatment of Outpatients with COVID-19: a Phase 2, Placebo-Controlled Randomised Trial. Lancet Respir. Med. 9 (5), 498–510. doi:10.1016/S2213-2600(20)30566-X
Focosi, D. (2020). Anti‐A Isohaemagglutinin Titres and SARS‐CoV‐2 Neutralization: Implications for Children and Convalescent Plasma Selection. Br. J. Haematol. 190 (3), 148–150. doi:10.1111/bjh.16932
Franco, S., Aparicio, E., Parera, M., Clotet, B., Tural, C., and Martinez, M. A. (2014). IFNL4 Ss469415590 Variant Is a Better Predictor Than Rs12979860 of Pegylated Interferon-Alpha/ribavirin Therapy Failure in Hepatitis C virus/HIV-1 Coinfected Patients. AIDS 28 (1), 133–136. doi:10.1097/QAD.0000000000000052
Gad, H. H., Dellgren, C., Hamming, O. J., Vends, S., Paludan, S. R., and Hartmann, R. (2009). Interferon-λ Is Functionally an Interferon but Structurally Related to the Interleukin-10 Family. J. Biol. Chem. 284 (31), 20869–20875. doi:10.1074/jbc.m109.002923
Garg, S., Kim, L., Whitaker, M., O'Halloran, A., Cummings, C., Holstein, R., et al. (2020). Hospitalization Rates and Characteristics of Patients Hospitalized with Laboratory-Confirmed Coronavirus Disease 2019 — COVID-NET, 14 States, March 1–30, 2020. MMWR Morb Mortal Wkly Rep. 69, 458–464. doi:10.15585/mmwr.mm6915e3
Ge, D., Fellay, J., Thompson, A. J., Simon, J. S., Shianna, K. V., Urban, T. J., et al. (2009). Genetic Variation in IL28B Predicts Hepatitis C Treatment-Induced Viral Clearance. Nature 461, 399–401. doi:10.1038/nature08309
Gong, J., Ou, J., Qiu, X., Jie, Y., Chen, Y., Yuan, L., et al. (2020). A Tool to Early Predict Severe Corona Virus Disease 2019 (COVID-19) : A Multicenter Study Using the Risk Nomogram in Wuhan and Guangdong, China. Clin. Infect. Dis 71, 833–840. Cold Spring Harbor Laboratory. doi:10.1093/cid/ciaa443
Gough, D. J., Levy, D. E., Johnstone, R. W., and Clarke, C. J. (2008). IFNgamma Signaling-Does it Mean JAK-STAT?. Cytokine Growth Factor. Rev. 19 (5-6), 383–394. doi:10.1016/j.cytogfr.2008.08.004
Guan, J., Miah, S. M. S., Wilson, Z. S., Erick, T. K., Banh, C., and Brossay, L. (2014). Role of Type I Interferon Receptor Signaling on NK Cell Development and Functions. PLoS ONE 9 (10), e111302. doi:10.1371/journal.pone.0111302
Guillon, P., Clément, M., Sébille, V., Rivain, J.-G., Chou, C.-F., Ruvoën-Clouet, N., et al. (2008). Inhibition of the Interaction between the SARS-CoV Spike Protein and its Cellular Receptor by Anti-histo-blood Group Antibodies. Glycobiology 18 (12), 1085–1093. doi:10.1093/glycob/cwn093
Hacibekiroğlu, T., Kalpakçı, Y., Genç, A. C., Hacibekiroğlu, İ., Sunu, C., Saricaoğlu, A., et al. (2021). Efficacy of Convalescent Plasma According to Blood Groups in COVID-19 Patients. Turk J. Med. Sci. 51 (1), 45–48. doi:10.3906/sag-2007-59
Hadjadj, J., Yatim, N., Barnabei, L., Corneau, A., Boussier, J., Smith, N., et al. (2020). Impaired Type I Interferon Activity and Inflammatory Responses in Severe COVID-19 Patients. Science 369 (6504), 718–724. doi:10.1126/science.abc6027
Hamming, O. J., Terczyńska-Dyla, E., Vieyres, G., Dijkman, R., Jørgensen, S. E., Akhtar, H., et al. (2013). Interferon Lambda 4 Signals via the IFNλ Receptor to Regulate Antiviral Activity against HCV and Coronaviruses. EMBO J. 32 (23), 3055–3065. doi:10.1038/emboj.2013.232
Hervas-Stubbs, S., Perez-Gracia, J. L., Rouzaut, A., Sanmamed, M. F., Le Bon, A., and Melero, I. (2011). Direct Effects of Type I Interferons on Cells of the Immune System. Clin. Cancer Res. 17 (9), 2619–2627. doi:10.1158/1078-0432.CCR-10-1114
Hoffmann, M., Kleine-Weber, H., Schroeder, S., Krüger, N., Herrler, T., Erichsen, S., et al. (2020). SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell 181 (2), 271–280. doi:10.1016/j.cell.2020.02.052
Hoiland, R. L., Fergusson, N. A., Mitra, A. R., Griesdale, D. E. G., Devine, D. V., Stukas, S., et al. (2020). The Association of ABO Blood Group with Indices of Disease Severity and Multiorgan Dysfunction in COVID-19. Blood Adv. 4 (20), 4981–4989. doi:10.1182/bloodadvances.2020002623
Huang, Y. Q., Tang, S. Q., Xu, X. L., Zeng, Y. M., He, X. Q., Li, Y., et al. (2020). No Statistically Apparent Difference in Antiviral Effectiveness Observed Among Ribavirin Plus Interferon-Alpha, Lopinavir/Ritonavir Plus Interferon-Alpha, and Ribavirin Plus Lopinavir/Ritonavir Plus Interferon-Alpha in Patients with Mild to Moderate Coronavirus Disease 2019: Results of a Randomized, Open-Labeled Prospective Study. Front. Pharmacol. 11, 1071. doi:10.3389/fphar.2020.01071
Jagannathan, P., Andrews, J. R., Bonilla, H., Hedlin, H., Jacobson, K. B., Balasubramanian, V., et al. (2021). Peginterferon Lambda-1a for Treatment of Outpatients with Uncomplicated COVID-19: a Randomized Placebo-Controlled Trial. Nat. Commun. 12 (1), 1967. doi:10.1038/s41467-021-22177-1
Jones, R., Nelson, M., Bracchi, M., Asboe, D., and Boffito, M. (2020). COVID-19 in Patients with HIV. The Lancet HIV. 7 (6), e383. doi:10.1016/s2352-3018(20)30139-9
Jothimani, D., Venugopal, R., Abedin, M. F., Kaliamoorthy, I., and Rela, M. (2020). COVID-19 and the Liver. J. Hepatol. 73 (5), 1231–1240. doi:10.1016/j.jhep.2020.06.006
Jouvin-Marche, E., Macek Jílková, Z., Thelu, M.-A., Marche, H., Fugier, E., Van Campenhout, N., et al. (2014). Lymphocytes Degranulation in Liver in Hepatitis C Virus Carriers Is Associated with IFNL4 Polymorphisms and ALT Levels. J. Infect. Dis. 209 (12), 1907–1915. doi:10.1093/infdis/jiu016
Key, F. M., Peter, B., Dennis, M. Y., Huerta-Sánchez, E., Tang, W., Prokunina-Olsson, L., et al. (2014). Selection on a Variant Associated with Improved Viral Clearance Drives Local, Adaptive Pseudogenization of Interferon Lambda 4 (IFNL4). Plos Genet. 10 (10), e1004681. doi:10.1371/journal.pgen.1004681
Klinkhammer, J., Schnepf, D., Ye, L., Schwaderlapp, M., Gad, H. H., Hartmann, R., et al. (2018). IFN-λ Prevents Influenza Virus Spread from the Upper Airways to the Lungs and Limits Virus Transmission. eLife 7, e33354. doi:10.7554/eLife.33354
Kole, A., He, J., Rivollier, A., Silveira, D. D., Kitamura, K., Maloy, K. J., et al. (2013). Type I IFNs Regulate Effector and Regulatory T Cell Accumulation and Anti-inflammatory Cytokine Production during T Cell-Mediated Colitis. J. Immunol. 191 (5), 2771–2779. doi:10.4049/jimmunol.1301093
Kosmicki, J. A., Horowitz, J. E., Banerjee, N., Lanche, R., Marcketta, A., Maxwell, E., et al. (2021). Pan-ancestry Exome-wide Association Analyses of COVID-19 Outcomes in 586,157 Individuals. Am. J. Hum. Genet. 108 (7), 1350–1355. doi:10.1016/j.ajhg.2021.05.017
Kotenko, S. V., Gallagher, G., Baurin, V. V., Lewis-Antes, A., Shen, M., Shah, N. K., et al. (2003). IFN-lambdas Mediate Antiviral protection through a Distinct Class II Cytokine Receptor Complex. Nat. Immunol. 4 (1), 69–77. doi:10.1038/ni875
Latini, A., Agolini, E., Novelli, A., Borgiani, P., Giannini, R., Gravina, P., et al. (2020). COVID-19 and Genetic Variants of Protein Involved in the SARS-CoV-2 Entry into the Host Cells. Genes (Basel) 11 (9), 1010. doi:10.3390/genes11091010
Lauber, C., Vieyres, G., Terczyńska-Dyla, E., Dijkman, R., Gad, H. H., Akhtar, H., et al. (2015). Transcriptome Analysis Reveals a Classical Interferon Signature Induced by IFNλ4 in Human Primary Cells. Genes Immun. 16, 414–421. doi:10.1038/gene.2015.23
Lazear, H. M., Nice, T. J., and Diamond, M. S. (2015). Interferon-λ: Immune Functions at Barrier Surfaces and beyond. Immunity 43 (1), 15–28. doi:10.1016/j.immuni.2015.07.001
Li, J., Wang, X., Chen, J., Cai, Y., Deng, A., and Yang, M. (2020). Association between ABO Blood Groups and Risk of SARS‐CoV‐2 Pneumonia. Br. J. Haematol. 190 (1), 24–27. doi:10.1111/bjh.16797
Lin, F. C., and Young, H. A. (2014). Interferons: Success in Anti-viral Immunotherapy. Cytokine Growth Factor. Rev. 25 (4), 369–376. doi:10.1016/j.cytogfr.2014.07.015
Lokugamage, K. G., Hage, A., de Vries, M., Valero-Jimenez, A. M., Schindewolf, C., Dittmann, M., et al. (2020). Type I Interferon Susceptibility Distinguishes SARS-CoV-2 from SARS-CoV. J. Virol. 94, e01410. Cold Spring Harbor Laboratory. doi:10.1128/JVI.01410-20
Lopera Maya, E. A., van der Graaf, A., Lanting, P., van der Geest, M., Fu, J., Swertz, M., et al. (2020). Lack of Association between Genetic Variants at ACE2 and TMPRSS2 Genes Involved in SARS-CoV-2 Infection and Human Quantitative Phenotypes. Front. Genet. 11, 613. doi:10.3389/fgene.2020.00613
Lopez, L., Sang, P. C., Tian, Y., and Sang, Y. (2020). Dysregulated Interferon Response Underlying Severe COVID-19. Viruses 12 (12), 1433. doi:10.3390/v12121433
Lopušná, K., Režuchová, I., Betáková, T., Skovranová, L., Tomašková, J., Lukáčiková, L., et al. (2013). Interferons Lambda, New Cytokines with Antiviral Activity. Acta Virol. 57 (2), 171–179. doi:10.4149/av_2013_02_171
Machiela, M. J., and Chanock, S. J. (2015). LDlink: a Web-Based Application for Exploring Population-specific Haplotype Structure and Linking Correlated Alleles of Possible Functional Variants: Fig. 1. Bioinformatics 31 (21), 3555–3557. doi:10.1093/bioinformatics/btv402
Machmach, K., Abad-Molina, C., Romero-Sánchez, M. C., Abad, M. A., Ferrando-Martínez, S., Genebat, M., et al. (2013). IL28B Single-Nucleotide Polymorphism Rs12979860 Is Associated with Spontaneous HIV Control in White Subjects. J. Infect. Dis. 207 (4), 651–655. doi:10.1093/infdis/jis717
Machmach, K., Abad-Molina, C., Romero-Sánchez, M. C., Dominguez-Molina, B., Moyano, M., Rodriguez, M. M., et al. (2015). IFNL4 Ss469415590 Polymorphism Is Associated with Unfavourable Clinical and Immunological Status in HIV-Infected Individuals. Clin. Microbiol. Infect. 21 (3), 289. doi:10.1016/j.cmi.2014.10.012
Majdic, G. (2020). Could Sex/Gender Differences in ACE2 Expression in the Lungs Contribute to the Large Gender Disparity in the Morbidity and Mortality of Patients Infected with the SARS-CoV-2 Virus?. Front. Cell Infect. Microbiol. 10, 327. doi:10.3389/fcimb.2020.00327
Malmgaard, L. (2004). Induction and Regulation of IFNs during Viral Infections. J. Interferon Cytokine Res. 24 (8), 439–454. doi:10.1089/1079990041689665
Manuel, O., Wójtowicz, A., Bibert, S., Mueller, N. J., van Delden, C., Hirsch, H. H., et al. (2015). Influence of IFNL3/4 Polymorphisms on the Incidence of Cytomegalovirus Infection after Solid-Organ Transplantation. J. Infect. Dis. 211 (6), 906–914. doi:10.1093/infdis/jiu557
Meffre, E., and Iwasaki, A. (2020). Interferon Deficiency Can lead to Severe COVID. Nature 587 (7834), 374–376. doi:10.1038/d41586-020-03070-1
Meissner, E. G., Bon, D., Prokunina-Olsson, L., Tang, W., Masur, H., O'Brien, T. R., et al. (2014). IFNL4-ΔG Genotype Is Associated with Slower Viral Clearance in Hepatitis C, Genotype-1 Patients Treated with Sofosbuvir and Ribavirin. J. Infect. Dis. 209 (11), 1700–1704. doi:10.1093/infdis/jit827
Messina, N. L., Clarke, C. J., and Johnstone, R. W. (2016). Constitutive IFNα/β Signaling Maintains Expression of Signaling Intermediaries for Efficient Cytokine Responses. JAK-STAT 5 (1), e1173804. doi:10.1080/21623996.2016.1173804
Muir, A. J., Arora, S., Everson, G., Flisiak, R., George, J., Ghalib, R., et al. (2014). A Randomized Phase 2b Study of Peginterferon Lambda-1a for the Treatment of Chronic HCV Infection. J. Hepatol. 61 (6), 1238–1246. doi:10.1016/j.jhep.2014.07.022
Nagy, B., Fejes, Z., Szentkereszty, Z., Sütő, R., Várkonyi, I., Ajzner, É., et al. (2021). A Dramatic Rise in Serum ACE2 Activity in a Critically Ill COVID-19 Patient. Int. J. Infect. Dis. 103, 412–414. doi:10.1016/j.ijid.2020.11.184
Ni, W., Yang, X., Yang, D., Bao, J., Li, R., Xiao, Y., et al. (2020). Role of Angiotensin-Converting Enzyme 2 (ACE2) in COVID-19. Crit. Care. 24, 422. doi:10.1186/s13054-020-03120-0
Nozawa, Y., Umemura, T., Katsuyama, Y., Shibata, S., Kimura, T., Morita, S., et al. (2014). Genetic Polymorphism in IFNL4 and Response to Pegylated Interferon-α and Ribavirin in Japanese Chronic Hepatitis C Patients. Tissue Antigens 83 (1), 45–48. doi:10.1111/tan.12264
O'Brien, T. R., Prokunina-Olsson, L., and Donnelly, R. P. (2014). IFN-λ4: the Paradoxical New Member of the Interferon Lambda Family. J.Iinterferon Cytokine Res.: official J. Int. Soc. Interferon Cytokine Res. 34 (11), 829–838. doi:10.1089/jir.2013.0136
Olex, A. L., Turkett, W. H., Brzoza-Lewis, K. L., Fetrow, J. S., and Hiltbold, E. M. (2016). Impact of the Type I Interferon Receptor on the Global Gene Expression Program during the Course of Dendritic Cell Maturation Induced by Polyinosinic Polycytidylic Acid. J. interferon Cytokine Res. : official J. Int. Soc. Interferon Cytokine Res. 36 (6), 382–400. doi:10.1089/jir.2014.0150
Pairo-Castineira, E., Clohisey, S., Klaric, L., Bretherick, A. D., Rawlik, K., Pasko, D., et al. (2021). Genetic Mechanisms of Critical Illness in COVID-19. Nature 591, 92–98. doi:10.1038/s41586-020-03065-y
Preece, A. F., Strahan, K. M., Devitt, J., Yamamoto, F., and Gustafsson, K. (2002). Expression of ABO or Related Antigenic Carbohydrates on Viral Envelopes Leads to Neutralization in the Presence of Serum Containing Specific Natural Antibodies and Complement. Blood 99 (7), 2477–2482. doi:10.1182/blood.v99.7.2477
Prokunina-Olsson, L., Muchmore, B., Tang, W., Pfeiffer, R. M., Park, H., Dickensheets, H., et al. (2013). A Variant Upstream of IFNL3 (IL28B) Creating a New Interferon Gene IFNL4 Is Associated with Impaired Clearance of Hepatitis C Virus. Nat. Genet. 45 (2), 164–171. doi:10.1038/ng.2521
Prokunina-Olsson, L., Alphonse, N., Dickenson, R. E., Durbin, J. E., Glenn, J. S., Hartmann, R., et al. (2020). COVID-19 and Emerging Viral Infections: The Case for Interferon Lambda. J. Exp. Med. 217 (5), e20200653. doi:10.1084/jem.20200653
Randall, R. E., and Goodbourn, S. (2008). Interferons and Viruses: an Interplay between Induction, Signalling, Antiviral Responses and Virus Countermeasures. J. Gen. Virol. 89 (Pt 1), 1–47. doi:10.1099/vir.0.83391-0
Real, L. M., Herrero, R., Rivero-Juárez, A., Camacho, Á., Macías, J., Vic, S., et al. (2015). IFNL4 Rs368234815 Polymorphism Is Associated with Innate Resistance to HIV-1 Infection. AIDS 29 (14), 1895–1897. doi:10.1097/qad.0000000000000773
Ruan, S. (2020). Likelihood of Survival of Coronavirus Disease 2019. Lancet Infect. Dis. 20 (6), 630–631. doi:10.1016/s1473-3099(20)30257-7
Sajadi, M. M., Shakeri, N., Talwani, R., Howell, C. D., Pakyz, R., Redfield, R. R., et al. (2011). IL28B Genotype Does Not Correlate with HIV Control in African Americans. Clin. Translational Sci. 4 (4), 282–284. doi:10.1111/j.1752-8062.2011.00307.x
Salgado, M., Kirk, G. D., Cox, A., Rutebemberwa, A., Higgins, Y., Astemborski, J., et al. (2011). Protective interleukin-28B Genotype Affects Hepatitis C Virus Clearance, but Does Not Contribute to HIV-1 Control in a Cohort of African-American Elite Controllers/suppressors. AIDS (London, England) 25 (3), 385–387. doi:10.1097/QAD.0b013e328341b86a
Sallard, E., Lescure, F.-X., Yazdanpanah, Y., Mentre, F., and Peiffer-Smadja, N. (2020). Type 1 Interferons as a Potential Treatment against COVID-19. Antiviral Res. 178, 104791. doi:10.1016/j.antiviral.2020.104791
Saponi-Cortes, J. M. R., Rivas, M. D., Calle, F., Sanchez Muñoz-Torrero, J. F., Costo, A., Martin, C., et al. (2021). IFNL4 Genetic Variant Can Predispose to COVID-19. medRxiv Cold Spring Harbor Laboratory. doi:10.1101/2021.03.01.21252696
Schneider, W. M., Chevillotte, M. D., and Rice, C. M. (2014). Interferon-stimulated Genes: a Complex Web of Host Defenses. Annu. Rev. Immunol. 32, 513–545. doi:10.1146/annurev-immunol-032713-120231
Seliger, B., Ruiz-Cabello, F., and Garrido, F. (2008). IFN Inducibility of Major Histocompatibility Antigens in Tumors. Adv. Cancer Res. 101, 249–276. doi:10.1016/S0065-230X(08)00407-7
Sheppard, P., Kindsvogel, W., Xu, W., Henderson, K., Schlutsmeyer, S., Whitmore, T. E., et al. (2003). IL-28, IL-29 and Their Class II Cytokine Receptor IL-28R. Nat. Immunol. 4 (1), 63–68. doi:10.1038/ni873
Sokol, C. L., and Luster, A. D. (2015). The Chemokine System in Innate Immunity. Cold Spring Harbor Perspect. Biol. 7 (5), a016303. doi:10.1101/cshperspect.a016303
Stanifer, M. L., Kee, C., Cortese, M., Zumaran, C. M., Triana, S., Mukenhirn, M., et al. (2020). Critical Role of Type III Interferon in Controlling SARS-CoV-2 Infection in Human Intestinal Epithelial Cells. Cell Rep. 32 (1), 107863. doi:10.1016/j.celrep.2020.107863
Stättermayer, A. F., Strassl, R., Maieron, A., Rutter, K., Stauber, R., Strasser, M., et al. (2014). Polymorphisms of Interferon-Λ4 and IL28B - Effects on Treatment Response to Interferon/ribavirin in Patients with Chronic Hepatitis C. Aliment. Pharmacol. Ther. 39 (1), 104–111. doi:10.1111/apt.12547
Sung, P. S., Hong, S. H., Chung, J. H., Kim, S., Park, S. H., Kim, H. M., et al. (2017). IFN-λ4 Potently Blocks IFN-α Signalling by ISG15 and USP18 in Hepatitis C Virus Infection. Sci. Rep. 7, 3821. doi:10.1038/s41598-017-04186-7
The 1000 Genomes Project Consortium (2015). Corresponding authors., Auton, A. et al. A global reference for human genetic variation. Nature 526, 68–74. doi:10.1038/nature15393
The COVID-19 Host Genetics Initiative (2020). The COVID-19 Host Genetics Initiative, a Global Initiative to Elucidate the Role of Host Genetic Factors in Susceptibility and Severity of the SARS-CoV-2 Virus Pandemic. Eur. J. Hum. Genet. 28, 715–718. doi:10.1038/s41431-020-0636-6
The Severe Covid-19 GWAS Group (2020). Genomewide Association Study of Severe Covid-19 with Respiratory Failure. New Engl. J. Med. 383 (16), 1522–1534. doi:10.1056/nejmoa2020283
Thomas, D. L., Thio, C. L., Martin, M. P., Qi, Y., Ge, D., O'Huigin, C., et al. (2009). Genetic Variation in IL28B and Spontaneous Clearance of Hepatitis C Virus. Nature 461 (7265), 798–801. doi:10.1038/nature08463
Vardavas, C. I., and Nikitara, K. (2020). COVID-19 and Smoking: A Systematic Review of the Evidence. Tob. Induc Dis. 18, 20. doi:10.18332/tid/119324
Verity, R., Okell, L. C., Dorigatti, I., Winskill, P., Whittaker, C., Imai, N., et al. (2020). Estimates of the Severity of Coronavirus Disease 2019: a Model-Based Analysis. Lancet Infect. Dis. 20 (6), 669–677. doi:10.1016/s1473-3099(20)30243-7
Vuille-dit-Bille, R. N., Camargo, S. M., Emmenegger, L., Sasse, T., Kummer, E., Jando, J., et al. (2015). Human Intestine Luminal ACE2 and Amino Acid Transporter Expression Increased by ACE-Inhibitors. Amino Acids. 47, 693–705. doi:10.1007/s00726-014-1889-6
Wang, A., Chiou, J., Poirion, O. B., Buchanan, J., Valdez, M. J., Verheyden, J. M., et al. (2020). Single-cell Multiomic Profiling of Human Lungs Reveals Cell-type-specific and Age-Dynamic Control of SARS-CoV2 Host Genes. ELife 9. doi:10.7554/elife.62522
Weiss, P., and Murdoch, D. R. (2020). Clinical Course and Mortality Risk of Severe COVID-19. The Lancet 395 (10229), 1014–1015. doi:10.1016/s0140-6736(20)30633-4
Williams, F. M., Freidin, M. B., Mangino, M., Couvreur, S., Visconti, A., Bowyer, R. C., et al. (2020). Self-reported Symptoms of Covid-19 Including Symptoms Most Predictive of SARS-CoV-2 Infection, Are Heritable. Twin. Res. Hum. Genet. 23, 316–321. Cold Spring Harbor Laboratory. doi:10.1017/thg.2020.85
World Health Organization (2021). Coronavirus. Available at: https://www.who.int/health-topics/coronavirus#tab=tab_1 (Accessed May 11, 2021).
World Health Organization (2020c). Coronavirus Disease (COVID-19). Available at: https://www.who.int/news-room/q-a-detail/q-a-coronaviruses (Accessed November 11, 2020).
World Health Organization (2020a). Novel Coronavirus - China. Available at: https://www.who.int/csr/don/12-january-2020-novel-coronavirus-china/en/ (Accessed November 11, 2020).
World Health Organization (2020b). Weekly Epidemiological and Operational Updates December 2020. Available at: https://www.who.int/emergencies/diseases/novel-coronavirus-2019/situation-reports (Accessed November 11, 2020).
Wruck, W., and Adjaye, J. (2020). SARS-CoV-2 Receptor ACE2 Is Co-expressed with Genes Related to Transmembrane Serine Proteases, Viral Entry, Immunity and Cellular Stress. Sci. Rep. 10 (1), 21415. doi:10.1038/s41598-020-78402-2
Wu, Z., and McGoogan, J. M. (2020). Characteristics of and Important Lessons from the Coronavirus Disease 2019 (COVID-19) Outbreak in China: Summary of a Report of 72 314 Cases from the Chinese Center for Disease Control and Prevention. JAMA 323 (13), 1239–1242. doi:10.1001/jama.2020.2648
Younossi, Z. M., Birerdinc, A., Estep, M., Stepanova, M., Afendy, A., Baranova, A., et al. (2012). The Impact of IL28B Genotype on the Gene Expression Profile of Patients with Chronic Hepatitis C Treated with Pegylated Interferon Alpha and Ribavirin. J. Transl Med. 10, 25. doi:10.1186/1479-5876-10-25
Zeberg, H., and Pääbo, S. (2020). The Major Genetic Risk Factor for Severe COVID-19 Is Inherited from Neandertals. Nature 587, 610–612. Cold Spring Harbor Lab.. doi:10.1038/s41586-020-2818-3
Zhang, Q., Bastard, P., Liu, Z., Le Pen, J., Moncada-Velez, M., Chen, J., et al. (2020). Inborn Errors of Type I IFN Immunity in Patients with Life-Threatening COVID-19. Science 370 (6515), eabd4570. doi:10.1126/science.abd4570
Zhao, J., Yang, Y., Huang, H., Li, D., Gu, D., Lu, X., et al. (2020). Relationship between the ABO Blood Group and the COVID-19 Susceptibility. Cold Spring Harbor Lab.. doi:10.1101/2020.03.11.20031096
Zhou, P., Yang, X. L., Wang, X. G., Hu, B., Zhang, L., Zhang, W., et al. (2020). A Pneumonia Outbreak Associated with a New Coronavirus of Probable Bat Origin. Nature 579, 270–273. doi:10.1038/s41586-020-2012-7
Ziegler, C. G. K., Allon, S. J., Nyquist, S. K., Mbano, I. M., Miao, V. N., Tzouanas, C. N., et al. (2020). SARS-CoV-2 Receptor ACE2 Is an Interferon-Stimulated Gene in Human Airway Epithelial Cells and Is Detected in Specific Cell Subsets across Tissues. Cell 181 (5), 1016–1035. doi:10.1016/j.cell.2020.04.035
Keywords: COVID-19, interferons, SARS-CoV-2, signaling, type I interferon, differential activity
Citation: Gozman L, Perry K, Nikogosov D, Klabukov I, Shevlyakov A and Baranova A (2021) A Role of Variance in Interferon Genes to Disease Severity in COVID-19 Patients. Front. Genet. 12:709388. doi: 10.3389/fgene.2021.709388
Received: 13 May 2021; Accepted: 01 September 2021;
Published: 17 September 2021.
Edited by:
Ranajit Das, Yenepoya University, IndiaReviewed by:
Eaaswarkhanth Muthukrishnan, Dasman Diabetes Institute, KuwaitCopyright © 2021 Gozman, Perry, Nikogosov, Klabukov, Shevlyakov and Baranova. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ancha Baranova, YWJhcmFub3ZAZ211LmVkdQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.