
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
EDITORIAL article
Front. Genet. , 23 June 2021
Sec. Evolutionary and Population Genetics
Volume 12 - 2021 | https://doi.org/10.3389/fgene.2021.708222
This article is part of the Research Topic Current and Emerging Trends in Human Identification and Molecular Anthropology View all 11 articles
Editorial on the Research Topic
Current and Emerging Trends in Human Identification and Molecular Anthropology
Recent developments in the DNA analysis technologies such as Massively Parallel Sequencing (MPS) have found prolific use in forensic applications. MPS not only allows the identification of a given individual using traditional forensic genetic markers but also the prediction of his/her age, appearance, and ancestry (Børsting and Morling, 2015). This can be very useful when conventional identification attempts using the comparative DNA profiling systems [e.g., short tandem repeats (STRs)] reach a dead end due to the absence of requisite DNA reference samples and/or potential matches in the respective databases. DNA-based estimation of ancestry and physical characteristics, now collectively described as forensic DNA phenotyping (FDP), has also led to the emergence of the concept of “biological witness” that can potentially provide investigative leads in such cases (Kayser, 2015). For instance, the prediction of the eye and hair color of King Richard III of England (1452–1485) and World Word II victims were all made possible with FDP (King et al., 2014; Chaitanya et al., 2017). A growing number of allele frequency data for diverse reference populations has facilitated increasingly better estimates of individual biogeographic ancestry (Pakstis et al., 2015, 2019; Bulbul et al., 2018). In other words, FDP may help us better understand the recent or distant past via the “reconstruction” of otherwise unidentifiable human remains. Many facets of human identification in a forensic context overlap with various aspects of human population genetics and molecular anthropology. Accordingly, in this Research Topic, we sought manuscripts that would provide a snapshot of the current and emerging trends in human identification or molecular anthropology, especially at the interface of the two fields (Figure 1).
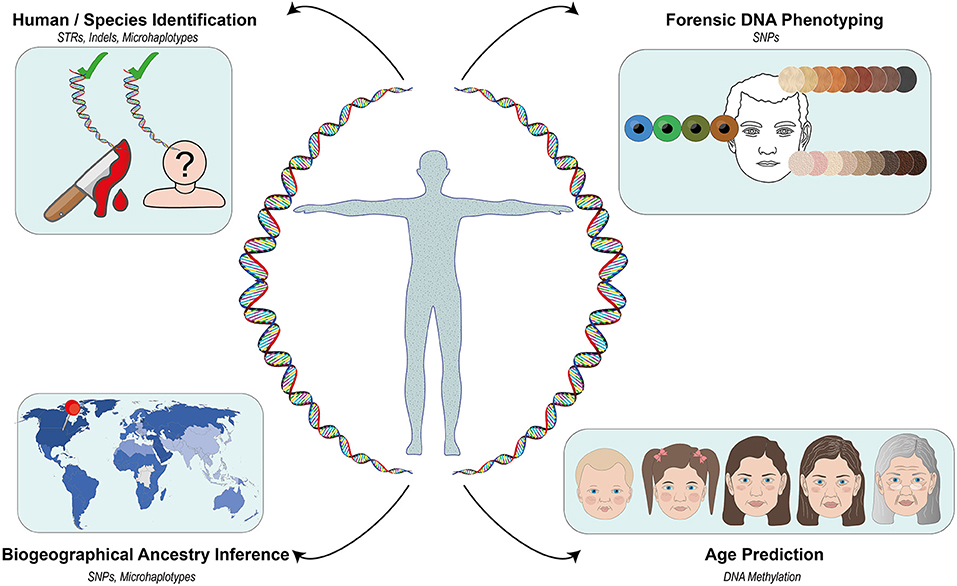
Figure 1. An overview of the genetic markers covered in the Current and Emerging Trends in Human Identification and Molecular Anthropology Research Topic.
A significant new trend in human identification and molecular anthropology is the emergence of a new genetic marker called microhaplotypes. Since the introduction of the concept less than a decade ago (Kidd et al., 2013), these very short segments (<300 bp) of the genome with multiple variants such that the loci are multiallelic have been getting increasing attention (Oldoni et al., 2019). Use of microhaplotypes is exemplified by de la Puente et al. who present a validation of a panel of 113 loci showing their power for biogeographic ancestry inference as well as familial relationship detection. Furthermore, Qu et al. have broadened the concept of the microhaplotype to include multiple small insertions/deletions allowing the markers to be studied using a conventional method like capillary electrophoresis (CE). Accordingly, these new microhaplotype loci may provide a useful supplement to standard CE-based STR typing using the same equipment. The growing number of published microhaplotype loci and the accumulating population data on them has been summarized in a new database called MicroHapDB by Standage and Mitchell. This extensible database will certainly be an important resource as the trend for more microhaplotype studies continues.
Another emerging trend is the gradual introduction of FDP in more routine casework investigations, and to this end, Atwood et al., report the selection process for the provision of FDP services to be used by the Australian law enforcement agencies. Here, a comparison of FDP services offered by six different providers was carried out that led to the successful selection of a provider for the prediction of biogeographical ancestry, hair, and eye color. A parallel trend in forensic research, which can also be regarded complementary to FDP, is the use of molecular techniques to determine the age of the person who left a DNA sample (Naue et al., 2018). Freire-Aradas et al. present a methodological comparison of the current DNA-methylation based forensic age prediction models using four different technologies available, and report largely comparable age predictions results. In fact, DNA methylation-based investigations are part of the growing field of forensic epigenomics whereby it has already been shown to distinguish homo-zygotic twins and the source tissue of a given sample, but also in the near future, even hold the potential for the prediction of lifestyle and environmental exposures of unknown perpetrators (Vidaki and Kayser, 2017).
Of course, studies on more traditional markers such STRs is still ongoing with several new applications. Cui et al. have taken STRs beyond humans to identify loci that can be used to identify different species of interest in a given sample. Their panel can identify DNA samples from 10 different species in addition to human using pairs of species-specific STR loci, and as such it may not only find use in forensic species identification, but also in the detection of meat fraud and adulteration. Gomes et al. focus on the unique inheritance and population genetic characteristics of markers located on the X chromosome. While noting an unexpected decrease in the number of new forensic investigations in the literature over the last two decades using X chromosome markers, nevertheless, the authors show the utility of these markers, primarily STRs, that so far have been identified. On a different note, Wyner et al. question the validity of the common assumption that the standard forensic STR loci are not associated with any phenotypes. In the light of increasing amount of compelling evidence toward the associations between forensic STR loci and certain phenotypes, authors point out to the presence of numerous legal and ethical implications associated with the already accumulated and expanding data using these markers, and suggest follow-ups and appropriate counter measures to minimize any misuse of such additional information.
SNPs continue to be investigated for their ability to infer ancestry of individuals and genetic affinities between populations. Pereira et al. have studied one admixed population in Brazil and shown dependencies in assessing admixture in ancestry of individuals on the numbers of markers used and the criteria used to identify the markers. Liu et al. have studied many populations in East Asia to assess the ancestry of Tibetans and their relationships to other Asian populations.
CG, OB, and KK co-edited the Research Topic and co-wrote, co-edited, and co-approved the final version of the Editorial for publication. All authors contributed to the article and approved the submitted version.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The guest editors wish to thank all the authors and peer-reviewers for their valuable contributions to this Research Topic, Huseyin Sevay for the rendering of double helix DNA structure in Figure 1 and Sumeyye Zulal Simsek for general help in putting together Figure 1.
Børsting, C., and Morling, N. (2015). Next generation sequencing and its applications in forensic genetics. Forensic Sci. Int. Genet. 18, 78–89. doi: 10.1016/j.fsigen.2015.02.002
Bulbul, O., Speed, W. C., Gurkan, C., Soundararajan, U., Rajeevan, H., Pakstis, A. J., et al. (2018). Improving ancestry distinctions among Southwest Asian populations. Forensic Sci. Int. Genet. 35, 14–20. doi: 10.1016/j.fsigen.2018.03.010
Chaitanya, L., Pajnič, I. Z., Walsh, S., BalaŽic, J., Zupanc, T., and Kayser, M. (2017). Bringing colour back after 70 years: predicting eye and hair colour from skeletal remains of World War II victims using the HIrisPlex system. Forensic Sci. Int. Genet. 26, 48–57. doi: 10.1016/j.fsigen.2016.10.004
Kayser, M. (2015). Forensic DNA Phenotyping: Predicting human appearance from crime scene material for investigative purposes. Forensic Sci. Int. Genet. 18. doi: 10.1016/j.fsigen.2015.02.003
Kidd, K. K., Pakstis, A. J., Speed, W. C., Lagace, R., Chang, J., Wootton, S., et al. (2013). Microhaplotype loci are a powerful new type of forensic marker. Forensic Sci. Int. Genet. Suppl. Ser. 4, e123–e124. doi: 10.1016/j.fsigss.2013.10.063
King, T. E., Fortes, G. G., Balaresque, P., Thomas, M. G., Balding, D., Delser, P. M., et al. (2014). Identification of the remains of King Richard III. Nat. Commun. 5, 1–8. doi: 10.1038/ncomms6631
Naue, J., Sänger, T., Hoefsloot, H. C. J., Lutz-Bonengel, S., Kloosterman, A. D., and Verschure, P. J. (2018). Proof of concept study of age-dependent DNA methylation markers across different tissues by massive parallel sequencing. Forensic Sci. Int. Genet. 36, 152–159. doi: 10.1016/j.fsigen.2018.07.007
Oldoni, F., Kidd, K. K., and Podini, D. (2019). Microhaplotypes in forensic genetics. Forensic Sci. Int. Genet. 38, 54–69. doi: 10.1016/j.fsigen.2018.09.009
Pakstis, A. J., Gurkan, C., Dogan, M., Balkaya, H. E., Dogan, S., Neophytou, P. I., et al. (2019). Genetic relationships of European, Mediterranean, and SW Asian populations using a panel of 55 AISNPs. Eur. J. Hum. Genet. 27, 1885–1893. doi: 10.1038/s41431-019-0466-6
Pakstis, A. J., Haigh, E., Cherni, L., Elgaaied, A. B. A., Barton, A., Evsanaa, B., et al. (2015). 52 additional reference population samples for the 55 AISNP panel. Forensic Sci. Int. Genet. 19, 269–271. doi: 10.1016/j.fsigen.2015.08.003
Keywords: ancestry informative markers, forensic DNA phenotyping, age prediction, microhaplotypes, genotyping
Citation: Gurkan C, Bulbul O and Kidd KK (2021) Editorial: Current and Emerging Trends in Human Identification and Molecular Anthropology. Front. Genet. 12:708222. doi: 10.3389/fgene.2021.708222
Received: 11 May 2021; Accepted: 24 May 2021;
Published: 23 June 2021.
Edited and reviewed by: Denis Baurain, University of Liège, Belgium
Copyright © 2021 Gurkan, Bulbul and Kidd. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Cemal Gurkan, Y2VtYWwuZ3Vya2FuQGVtdS5lZHUudHI=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.