- 1Institute of Grassland Research, Chinese Academy of Agricultural Sciences, Hohhot, China
- 2School of Ecology and Environment, Inner Mongolia University, Hohhot, China
- 3Key Laboratory of Grassland Resources and Utilization of Ministry of Agriculture, Hohhot, China
Astragalus is the largest genus in Leguminosae. Several molecular studies have investigated the potential adulterants of the species within this genus; nonetheless, the evolutionary relationships among these species remain unclear. Herein, we sequenced and annotated the complete chloroplast genomes of three Astragalus species—Astragalus adsurgens, Astragalus mongholicus var. dahuricus, and Astragalus melilotoides using next-generation sequencing technology and plastid genome annotator (PGA) tool. All species belonged to the inverted repeat lacking clade (IRLC) and had similar sequences concerning gene contents and characteristics. Abundant simple sequence repeat (SSR) loci were detected, with single-nucleotide repeats accounting for the highest proportion of SSRs, most of which were A/T homopolymers. Using Astragalus membranaceus var. membranaceus as reference, the divergence was evident in most non-coding regions of the complete chloroplast genomes of these species. Seven genes (atpB, psbD, rpoB, rpoC1, trnV, rrn16, and rrn23) showed high nucleotide variability (Pi), and could be used as DNA barcodes for Astragalus sp. cemA and rpl33 were found undergoing positive selection by the section patterns in the coded protein. Phylogenetic analysis showed that Astragalus is a monophyletic group closely related to the genus Oxytropis within the tribe Galegeae. The newly sequenced chloroplast genomes provide insight into the unresolved evolutionary relationships within Astragalus spp. and are expected to contribute to species identification.
Introduction
Astragalus is the largest genus in Leguminosae (Li et al., 2014; Su et al., 2021) and is widely distributed in the Northern Hemisphere (Podlech, 1986; Osaloo et al., 2003), South America (Cook et al., 2017a), and Africa (Alami et al., 2019). This genus includes 11 subgenera and some 2000–3000 species (Li et al., 2014), which have been used in various fields. Most Astragalus spp. can be used as fresh herbs, forage, or silage (Li et al., 2014), and some have important medicinal values, such as Astragalus membranaceus var. mongholicus (Lei et al., 2016), whereas some can be toxic and even deadly to humans and livestock, such as Astragalus miser var. oblongitotices and Astragalus hamiensis (Martinez et al., 2019). Astragalus belongs to the tribe Galegeae in Papilionoideae; however, it has been a controversial genus concerning its inception, including at the subgenus and species levels. Astragalus spp. usually show small, patchy distribution, a pattern that may promote genetic isolation and character differentiation (Massatti et al., 2018). Extensive classical taxonomic studies have explored Astragalus spp., based on plant morphology and geography, with many focusing on the discrimination of adulterants (Cui et al., 2012; Zheng et al., 2014; Hou et al., 2016); nevertheless, the systematic evolutionary relationships among Astragalus spp. remain unclear.
The chloroplast (cp) is a significant semiautonomous organelle that can absorb carbon dioxide and release oxygen while converting light energy into chemical energy in green plants (Yin et al., 2017), phototrophic bacteria (Trüper, 1987; Mauriello, 2019), and algae (Menke et al., 1965). Chloroplasts can also be used to elucidate the genetic relationships among species and explore plant phylogeny and nuclear evolution (Daniell et al., 2016; Xiong et al., 2020; Zhao and Zhu, 2020), because of its feature of replication initiation, genome stabilization, and maternally-inherited gene conservation (Daniell et al., 2016).
Most complete cp genomes show a typical quadripartite structure with two inverted repeats (IRs) separated by two single-copy regions: a large single-copy region (LSC) and a small single-copy region (SSC). The cp genome usually encodes 120–130 genes with a size of 107–218 kb (Shinozaki et al., 1986; Osaloo et al., 2003; Chumley et al., 2006; Lin et al., 2010; Zha et al., 2020). Although the structure and gene content are relatively stable, divergence has been observed; for example, one copy of the IR was lost in some species, especially in Papilionoideae of Leguminosae, which formed a new clade, named IR lacking clade (IRLC) (Martin et al., 2014; Xiong et al., 2020). Other changes include loss of genes (Palmer and Thompson, 1981; Millen et al., 2001) and inversions (Bruneau and Palmer, 1990).
Since the complete cp genome of tobacco (Nicotiana tabacum) was first sequenced and annotated (Shinozaki et al., 1986), an increasing number of cp genomes have been reported. To date, about 26,573 vascular plant cp genomes have been deposited in the National Center for Biotechnology Information (NCBI), including 155 legumes. Within Astragalus, complete cp genomes for Astragalus laxmannii (Liu et al., 2020), A. membranaceus (Lei et al., 2016), A. mongholicus var. nakaianus (Choi et al., 2016), A. membranaceus var. membranaceus (Wang et al., 2016), Astragalus strictus, and Astragalus gummifer have been sequenced and annotated; however, the latter two can only be found in NCBI. It should be noted that, except A. laxmannii, all the other five species only have one copy of the IR region, but they all belong to the IRLC. Moreover, they have a different phylogenetic relationship with other species concerning morphological taxonomy (Choi et al., 2016; Lei et al., 2016; Wang et al., 2016; Liu et al., 2020), which further proves the controversy regarding Astragalus taxonomy.
Astragalus adsurgens, A. mongholicus var. dahuricus, and A. melilotoides belong to three different subgenera (subg. Cercidothrix, subg. Trimeniaeus, and subg. Phaca, respectively) of Astragalus; however, many of the subgenera of Astragalus are not monophyletic and their phylogenetic relationships within the genus are still poorly known (Su et al., 2021). Recent studies have shown that the taxonomic classifications within the genera based on morphology do not correspond to the phylogenetically recovered clades (Tunckol et al., 2020). Moreover, it is unclear why Astragalus and its clades have such a high number of species (Bagheri et al., 2017). Therefore, we sequenced and annotated the complete chloroplast genome of A. adsurgens, A. mongholicus var. dahuricus, and A. melilotoides to explore the relationships among Astragalus species. Then, repetitive sequences, simple sequence repeats (SSRs), nucleotide diversity (Pi), and evolution were investigated. In addition, a phylogenetic tree was constructed using the information from 37 species to examine their evolutionary relationships.
Materials and Methods
Plant Materials
Young leaves of A. adsurgens, A. mongholicus var. dahuricus, and A. melilotoides were collected at Hohhot, Inner Mongolia, China (40.57°N, 111.93°E) and deposited at the National Germplasm Perennial Herbage Nursery, Institute of Grassland Research, Chinese Academy of Agricultural Sciences.
DNA Extraction and Sequencing, Genome Assembly, and Annotation
Genomic DNA was extracted from fresh leaves using a Plant DNA Isolation Kit (Tiangen, Beijing, China) and sequenced using the MiSeq PE150 platform (Illumina, San Diego, CA, United States), yielding 150 bp paired-end reads, at Novogene Co. (Tianjing, China). The cp genome was de novo assembled using NOVOPlasty (Dierckxsens et al., 2019) with default parameters. Genomes were annotated using the plastid genome annotator (PGA) tool (Qu et al., 2019), coupled with manually edited start and stop codons using Geneious (Kearse et al., 2012). A. mongholicus cp genome sequence (NCBI accession number: NC029828) was used as a reference. The annotation results were checked using the Dual Organellar GenoMe Annotator (DOGMA) (Wyman et al., 2004) and CpGAVAS2 (Shi et al., 2019). OGDRAW1 (version 1.3.1) (Greiner et al., 2019) was used to draw the gene map of the cp genomes.
Identification of Repeat Sequences and Simple Sequence Repeats
REPuter software (Kurtz and Schleiermacher, 1999) was used to identify repeat sequences, including forward repeat (F), reverse repeat (R), complementary repeat (C), and palindromic repeat (P) in cp genomes. Detection parameter settings were as follows: minimum repeat size 30 bp and an edit distance of 3. The MIcroSAtellite identification tool (MISA2) was used for SSR identification on the cp genome sequences with the following parameter settings: unit size (nucleotide) _min-repeats: 1_8, 2_5, 3_4, 4_3, 5_3, and 6_3. The minimum distance between two SSRs was set to 100 bp.
Polymorphism Analysis and Genome Structure Comparison
Pi values and sequence polymorphisms of eight Astragalus species were analyzed using DNAsp v. 6.10 (Rozas et al., 2017). mVISTA (Frazer et al., 2014) software was used to compare the complete cp genomes of A. adsurgens, A. mongholicus var. dahuricus, and A. melilotoides that we sequenced, with four additional published cp genomes of congeneric species (A. gummifer, A. mongholicus, A. nakaianus, and A. strictus) with the shuffle-LAGAN mode and A. membranaceus var. membranaceus annotation (Wang et al., 2016) as reference.
Gene Selective Pressure Analysis
To detect whether cp genes were under selection pressure, synonymous (dS) and non-synonymous (dN) substitution rates, and the ω value (ω = dN/dS) for shared protein-coding gene in eight Astragalus cp genomes were analyzed using Phylogenetic Analysis by Maximum Likelihood 4.0 with the YN algorithm (Yang, 2007).
Phylogenetic Analysis
The three sequenced cp genomes of Astragalus, along with the genomes of 34 species (using Lotus japonicus and Glycine max as outgroups) retrieved from NCBI, were used to construct a phylogenetic tree. Multiple alignments were performed using complete cp genomes based on the conserved structure and gene order, and all nucleotide sequences were aligned using the multiple sequence alignment MAFFT software (Katoh and Standley, 2013) with default parameters. Two methods, maximum likelihood (ML) and Bayesian inference (BI), were employed to construct the phylogenetic trees. ML analyses were conducted using RAxML 8.2.11 (Stamatakis, 2014) with the GTR + Gamma nucleotide substitution model; node support was conducted by a bootstrap analysis with 1000 replicates. BI analyses were conducted using MrBayes v. 3.2.6 (Ronquist and Huelsenbeck, 2003).
Results and Discussion
Characteristics of A. adsurgens, A. mongholicus var. dahuricus, and A. melilotoides Complete Chloroplast Genomes
In the present study, we sequenced and annotated the complete cp genomes of three Astragalus species—A. adsurgens, A. mongholicus var. dahuricus, and A. melilotoides. The general gene structure and locations in the cp genomes are presented in Figure 1. All genomes were found to have lost one copy of the IR region, thereby being affiliated to IRLC in Papilionoideae, and showed the same GC content of 34% (Figure 1). The cp genomes were 122,796, 122,789, and 123,663 bp for A. adsurgens, A. mongholicus var. dahuricus, and A. melilotoides, respectively. A. melilotoides and A. adsurgens consisted of 106 genes including 76 protein-coding genes, four rRNAs, and 26 tRNAs; A. adsurgens had one tRNA (trnE-UUC) gene copy, whereas A. melilotoides and A. mongholicus var. dahuricus had two. Only A. mongholicus var. dahuricus had trnG-UCC and trnK-UCC in its genome. The species lacked trnfM-CAU and trnS-GGA, found in the cp genomes of A. adsurgens and A. melilotoides, which were replaced by trnM-CAU and trnS-GCU in the A. mongholicus var. dahuricus chloroplast genome. Thus, A. mongholicus var. dahuricus cp genome consisted of 108 genes. The numbers of tRNAs in the three species differ from those in other Astragalus spp. (Choi et al., 2016; Lei et al., 2016; Wang et al., 2016).
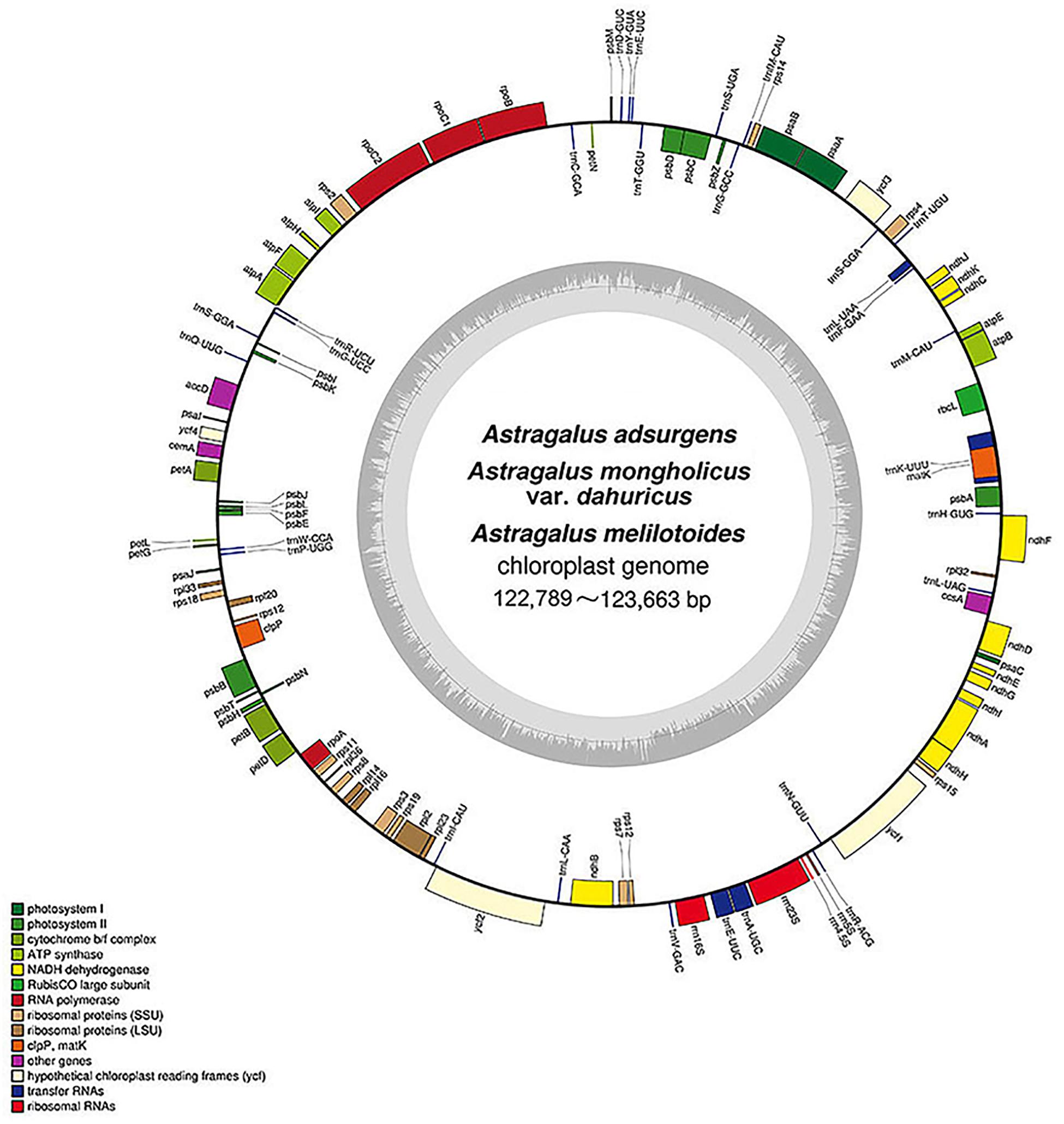
Figure 1. Structure and characteristics of the complete chloroplast genomes of three Astragalus species. Genes inside of the circle are transcribed in the clockwise direction and those outside the circle are transcribed in the counter-clockwise direction. Different colors indicate genes belonging to different functional groups. Dark gray in the inner circle indicates the GC content, and light gray indicates AT.
Among the genes in the cp genome, 45 were related to photosynthesis, including five subunits of photosystem I, 16 subunits of photosystem II, six subunits of ATP synthase, 11 subunits of NADH-dehydrogenase, and six subunits of cytochrome b/f complex as well as rbcL (a subunit of Rubisco). Genes related to self-replication included eight large subunits of ribosome, 11 small subunits of ribosome, and four DNA-dependent RNA polymerases. Genes related to self-replication were also detected, including four ribosomal RNAs, rrn5S, rrn4.5S, rrn16S, and rrn23S. In particular, there were five other genes and three genes, ycf1, ycf2, and ycf4, whose functions are unknown (Table 1). The structures and locations of the genes are shown in Figure 1. In comparison with other angiosperm plastid genomes, all three species lost rps16, rpl22, and infA, consistent with the A. membranaceus cp genome (Cook et al., 2017b). However, rps16 and rpl22 could be found in most angiosperm cp genomes (Shen et al., 2018; Biju et al., 2019; Liu et al., 2019). Their absence in the three species may be explained by genome rearrangement during the evolution process or elimination by natural selection (Daniell et al., 2016). In some species, infA has been transferred from the chloroplast to the nuclear genome (Millen et al., 2001); thus, it is reasonable to infer that lack of infA in the cp genome of the three species may be explained by a similar process. However, further studies are needed to evaluate this hypothesis. Overall, 12, 11, and 11 genes in the cp genomes of A. adsurgens, A. mongholicus var. dahuricus, and A. melilotoides, respectively, contained one intron. In addition, ycf3 had two introns in the A. adsurgens and A. mongholicus var. dahuricus cp genomes. In A. melilotoides, trnL-UAA had two introns (Table 1 and Supplementary Table 1).
Repeat Sequences and SSRs Analysis
Repetitive sequences are the primary source of repeat, deletion, and rearrangement events in the chloroplast genome (Li and Zheng, 2018). Furthermore, nuclear and genome rearrangements contribute to the majority of repetitive sequences. Herein, 50 scattered repetitive sequences with lengths of no more than 30 bp, including forward, reverse, complementary, and palindromic repeats, were detected in the three species of Astragalus. The proportions of each type of repetitive sequence differed slightly among species. In the A. adsurgens cp genome, palindromic repeats were the most common (44%), followed by forward (42%), complementary (8%), and reverse (2%). Equal numbers of forward and palindromic (42%) as well as of complementary and reverse repeats (8%) were detected in A. mongholicus var. dahuricus genomes. Forward (48%) was the most common type of repeat in the A. melilotoides cp genome, followed by palindromic (36%), reverse (12%), and complimentary (4%) repeats (Figure 2). Those with lengths of 30–40 bp accounted for the majority of repetitive sequences (Supplementary Table 2). Compared with A. membranaceus (Lei et al., 2016), all three species in this study lacked tandem repeat sequences, suggesting that the mutation frequencies and rate of evolution are high in A. membranaceus (Saltonstall and Lambertini, 2012).
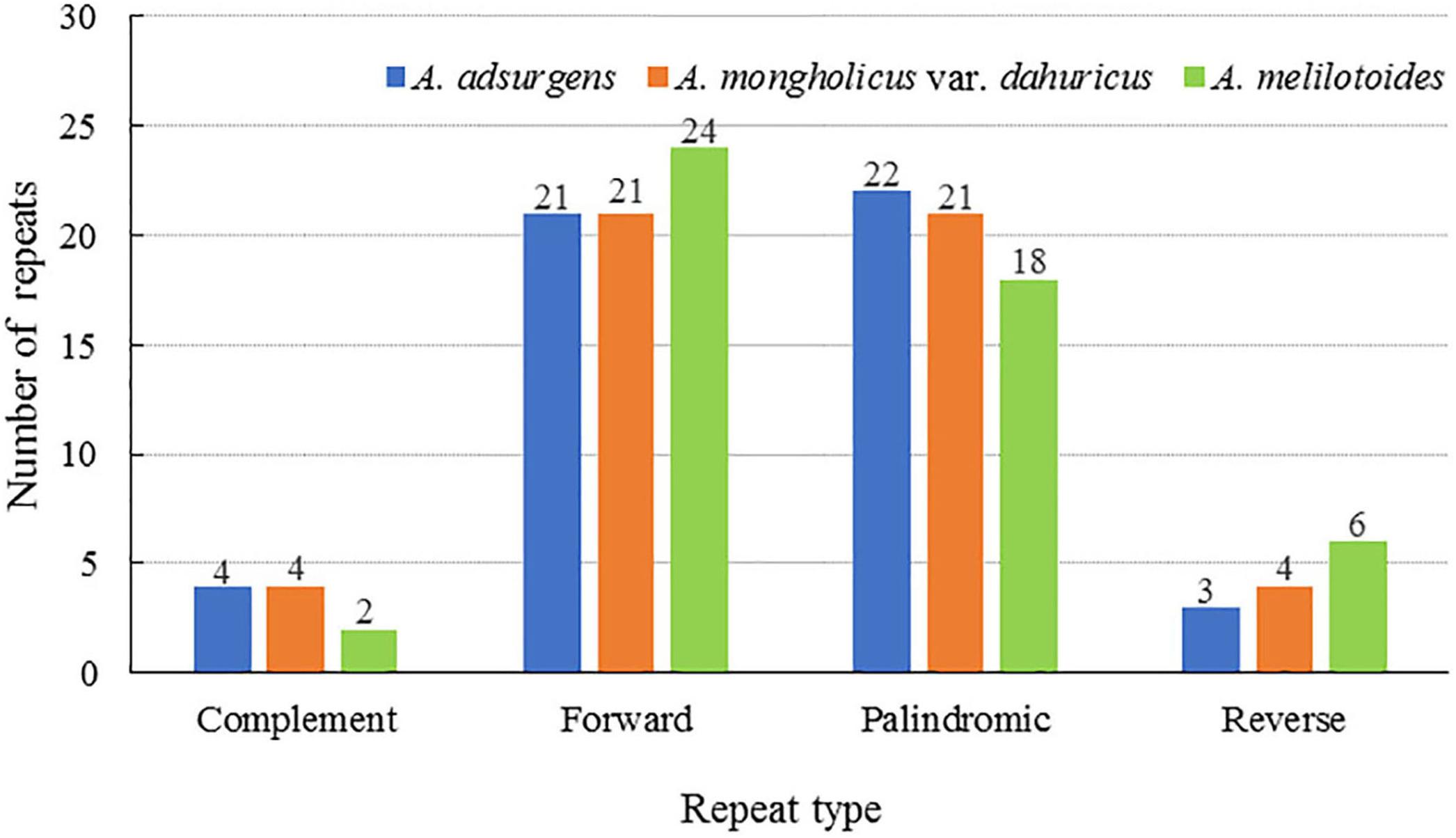
Figure 2. Number and proportion of repetitive sequences in the cp complete genomes of three Astragalus species.
Molecular markers can be used for genome mapping, identification of genetic relationships, and systematic classification of species (Kapoor et al., 2020). Among different types of DNA molecular markers, SSRs are highly polymorphic, codominant, and widely distributed across genomes and therefore are useful for studies of genetic diversity and relationships among plant populations (Saha et al., 2019; Li et al., 2020). The chloroplast SSRs (cp SSRs) are maternally inherited, thus they are considered to be highly efficient tools in the studies of population structure, genetic variation, species identification, and phylogenetic relationships analyses (Saski et al., 2005). In particular, 146 SSRs (8–298 bp) were detected in the cp genome of A. melilotoides, and 129 SSRs (8–335 bp) were detected in the A. adsurgens and in the A. mongholicus var. dahuricus cp genomes. The same number of SSRs can also be found in Lupinus albus and Lupinus luteus (Zha et al., 2020). In addition, the numbers of mononucleotide, dinucleotide, trinucleotide, tetranucleotide, and pentanucleotide repeats were the same in the A. adsurgens and A. mongholicus var. dahuricus cp genomes, which had no hexanucleotides; however, the types were slightly different (Figure 3A and Supplementary Table 3). Among the three species, mononucleotides were the most frequent repeat type, and most of them were A/T homopolymers, accounting for 59.59% of all SSRs in A. melilotoides and 51.94% in A. adsurgens and A. mongholicus var. dahuricus cp genomes. There were 12 dinucleotides in three species, which were AT/TA or TA/AT, accounting for 8.22–9.30% of the SSRs, and no more than four trinucleotides and seven tetranucleotides in the three complete cp genomes. All the species had one pentanucleotide, and only A. melilotoides had one hexanucleotide. The cp SSRs identified in the species, mainly poly-A/T and C/G, are rare, even for multiple base repeats. These results are consistent with those for most species sequenced in IRL clade in Papilionoideae (Lei et al., 2016; Liu et al., 2016; Somaratne et al., 2019; Wei et al., 2020). Furthermore, compound SSRs accounted for 23.56–32.56% of the three cp genomes. Although the richness of SSRs was similar within Astragalus, the differences in SSR count may be a useful molecular marker for species identification (Figure 3B and Supplementary Table 3). However, using SSRs to elucidate ecological and evolutionary processes has yet to be fully achieved (Ebert and Peakal, 2009). The herein described SSRs in the cp genomes of Astragalus may pave the way for exploring evolutionary processes at the population level.
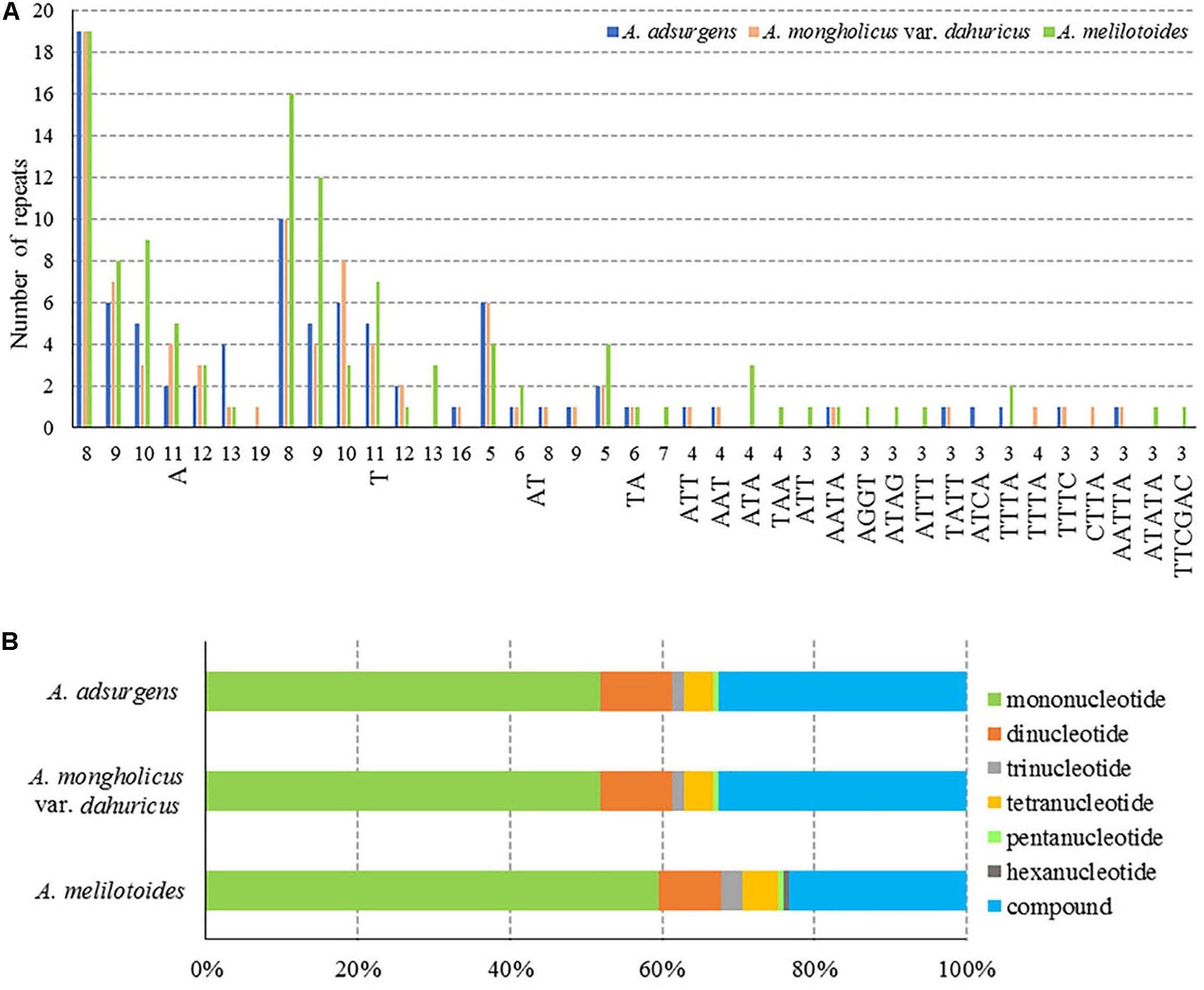
Figure 3. Numbers and proportions of repetitive sequences in the cp complete genomes of three Astragalus species. (A) Number of SSR motifs of different repeat types. (B) Number of repeat sequences.
Comparative Genome Analysis and Sequence Variation
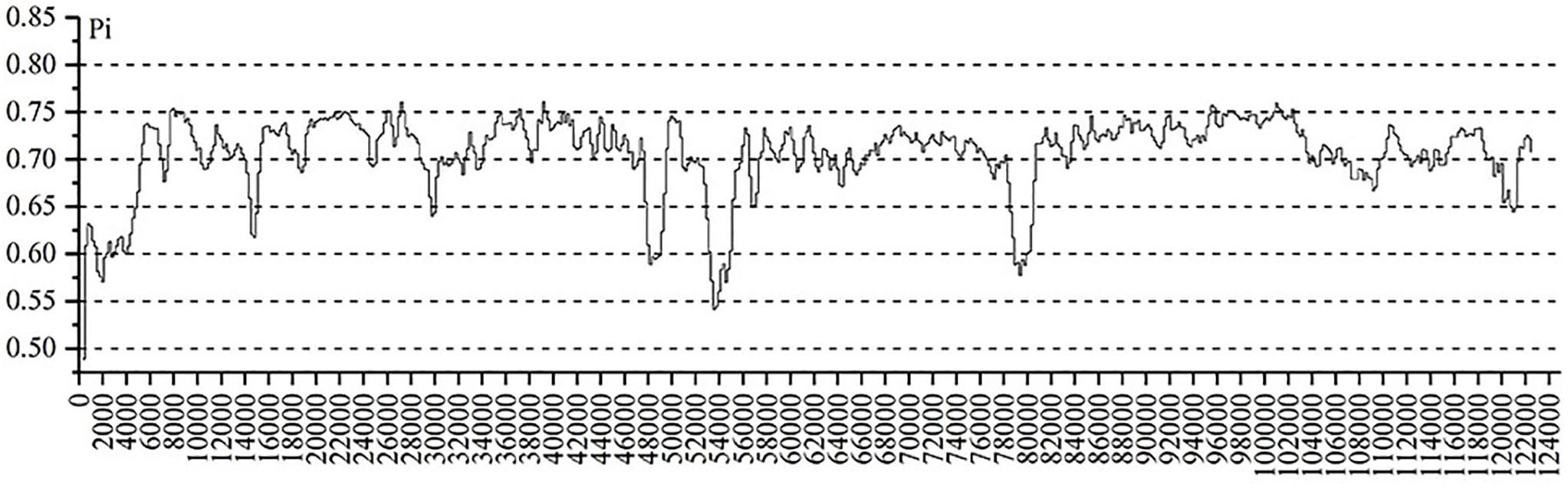
The highly variable regions of the cp genome can be used to identify closely related species and provide abundant information for further phylogenetic studies (Cui et al., 2020). Setting A. membranaceus var. membranaceus as reference, we used mVISTA to compare the cp genomes of seven species of Astragalus species, including the newly sequenced genomes and data deposited in the NCBI database, to explore sequence variation (Figure 4). The cp genome length varied among species, being A. mongholicus var. dahuricus genome (122,789 bp) the shortest and that of A. nakaianus (123,633 bp) the longest. In general, there was high sequence similarity among the cp genomes of the seven species, with high conservation of size and gene order. However, sequence variation was higher in conserved non-coding sequences (CNS) regions than in other regions. In addition to start–trnH–GTG, atpE–trnM–CAT, trnT–TGT–rps4, rps14–trnfM–GCC, psbJ–psbL, trnW–CCA–petG, psbN–psbH, and ndhG–ndhE, almost all other regions had variation. Previous studies have shown that trnH–psbA, rps16–trnQ (Dong et al., 2012), atpH–atpI, and psaA–ycf3 (Cui et al., 2020) can be used as DNA barcodes in other plant taxa. Further studies are needed to confirm whether these CNS regions can be used to identify closely related species in Astragalus. These highly variable regions may also resolve the interspecific relationships of Astragalus in the legume phylogeny. A. adsurgens, A. mongholicus var. dahuricus, and A. melilotoides had lower levels of divergence concerning non-coding regions. However, there was less variation in the coding than in the non-coding regions. To further clarify the variation in the coding regions, Pi was also calculated (Figure 5). atpB, psbD, rpoB, rpoC1, trnV, rrn16, and rrn23 all had high Pi values, exceeding 0.75. atpB and psbD encode proteins involved in photosynthesis, in which transcription is affected by light conditions; accordingly, high Pi values may reflect adaptation to different environmental light conditions (Christopher and Mullet, 1994). These highly variable regions may also resolve the interspecific relationships of Astragalus in the legume phylogeny.
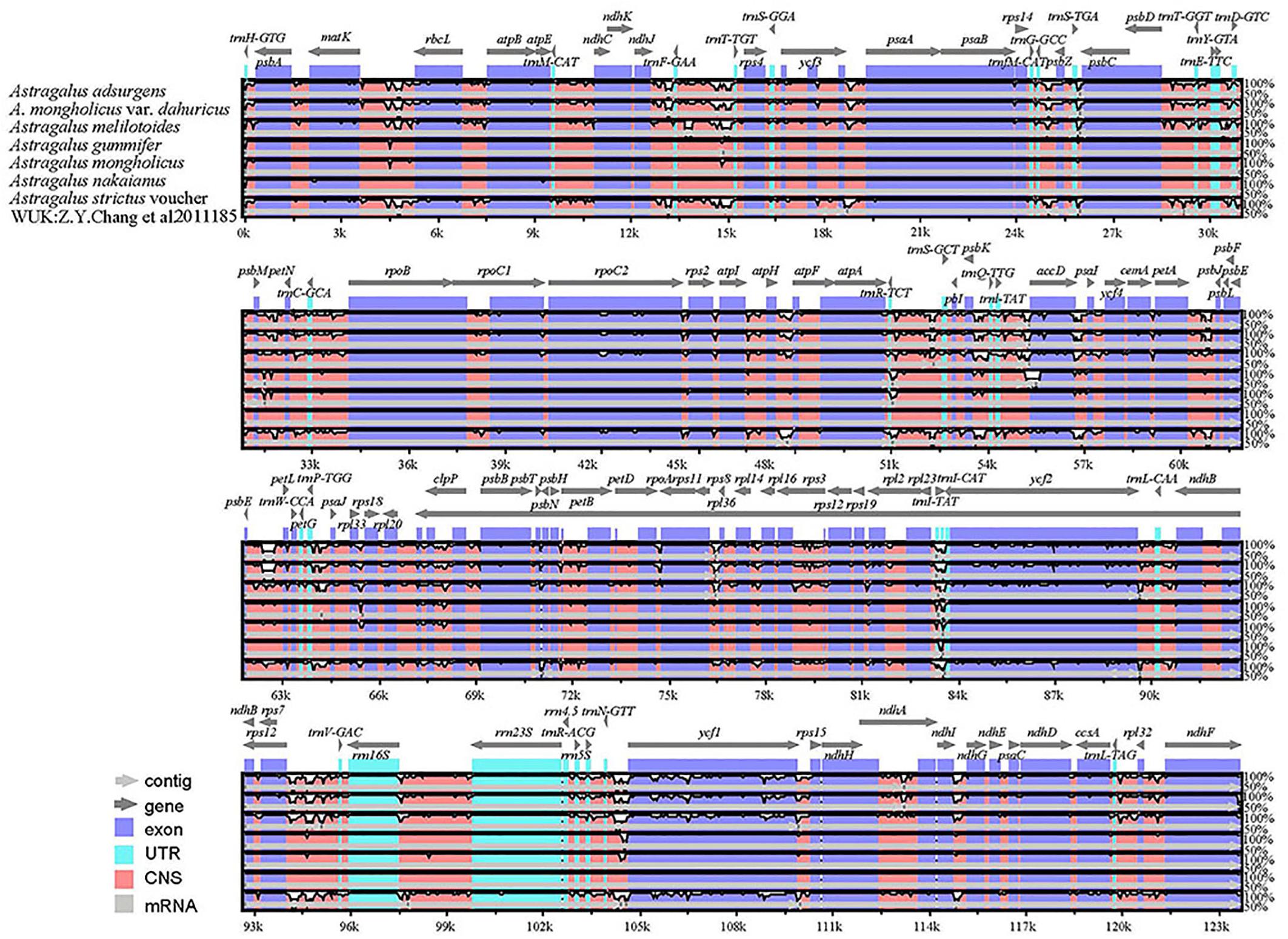
Figure 4. Global alignment of seven cp genomes of Astragalus generated using mVISTA. The y-axis represents the range of identity (50–100%). The x-axis indicates the coordinate in the cp genome. Annotated genes are shown along the top. The alignment was generated using A. membranaceus var. membranaceus as reference. Genomic regions are color-coded to indicate protein-coding regions, exons, UTRs, and CNS.
Selection on Functional Genes
The synonymous substitution rates (dS) of the four species in Astragalus ranged from 0.0000 to 0.0280 (ycf2), and the non-synonymous substitution rates (dN) ranged from 0.0000 to 0.0752 (psbZ). The ω value for 74 shared protein-coding genes within the species showed that cemA (encoding an envelope membrane protein) and rpl33 (encoding the ribosomal protein L33) underwent positive selection (ω > 1), with the highest ω values (1.6545) being identified for cemA between A. melilotoides–A. adsurgens and A. melilotoides–A. mongholicus var. dahuricus (Figure 6 and Supplementary Table 4). The dN/dS ratio (ω) in the chloroplast genome provides important insights into adaptive molecular evolution (Dos Reis, 2015). The substitution rates in the cp genome are affected by both lineage-specific and locus-specific events; additionally, rate heterogeneity is mainly related to non-synonymous substitutions (Muse and Gaut, 1994). Synonymous variation is low in the cp genome; however, rates of non-synonymous changes are lower than those of synonymous changes (Volff et al., 2008), and most protein-coding genes related to photosynthesis undergo purifying selection (Jin et al., 2016). Positive selection based on high dN/dS substitution ratio is rare (Endo et al., 1996). Our results are consistent with these previous findings. Genes undergoing positive selection are mainly self-replication genes and those with unknown functions (Hong et al., 2020). In addition, rearrangements in the chloroplast genome may be subjected to positive selection (Sanderson and Doyle, 1993).
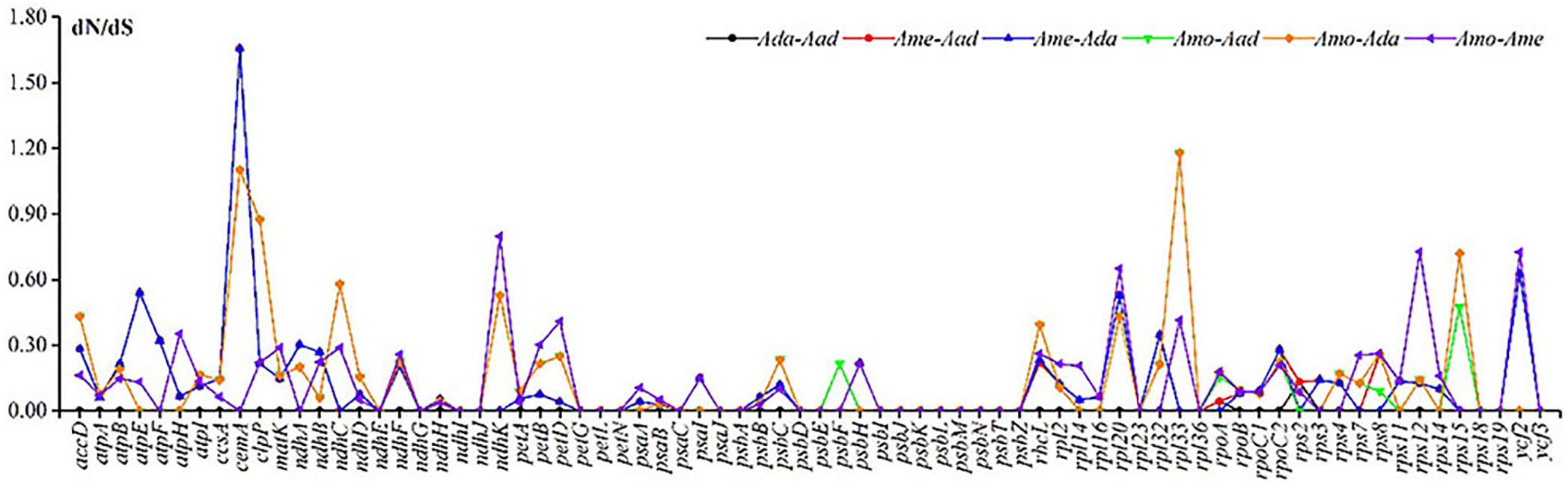
Figure 6. The non-synonymous/synonymous substitution rates (dN/dS) calculated using 74 shared genes in four Astragalus species.
Comparative Genome Analysis and Sequence Variation
The topological structure of the phylogenetic tree of 35 species belonging to 18 genera in Papilionoideae as well as L. japonicus and G. max, which were used as outgroups, was consistent with the classification of Papilionoideae with strong bootstrap support (Figure 7). Six species of Astragalus formed a well-supported clade that included two major groups. A. adsurgens and A. mongholicus var. dahuricus showed the closest relationship among all Astragalus spp. Additionally, the genus Astragalus was monophyletic (Sanderson and Doyle, 1993; Wojciechowski et al., 1993) and was closely related to the clade that comprises the Oxytropis genus (Zimmers et al., 2017) and Sphaerophysa salsula within the Galegeae tribe. Previous studies have shown that there are 10 clades within Astragalus, including a new one, Pseudosesbanella, recovered in a recent phylogenetic analysis of coding sequences (Azani et al., 2019; Su et al., 2021). Our results confirm that A. mongholicus and A. nakaianus are in the Cenentrum section of Phaca, and A. melilotoides with A. mongholicus var. dahuricus belong to different sections (Su et al., 2021). The results of our phylogenetic analysis add to knowledge of previous studies and indicate that the cp genome can be used to construct relationships among species in this genus.
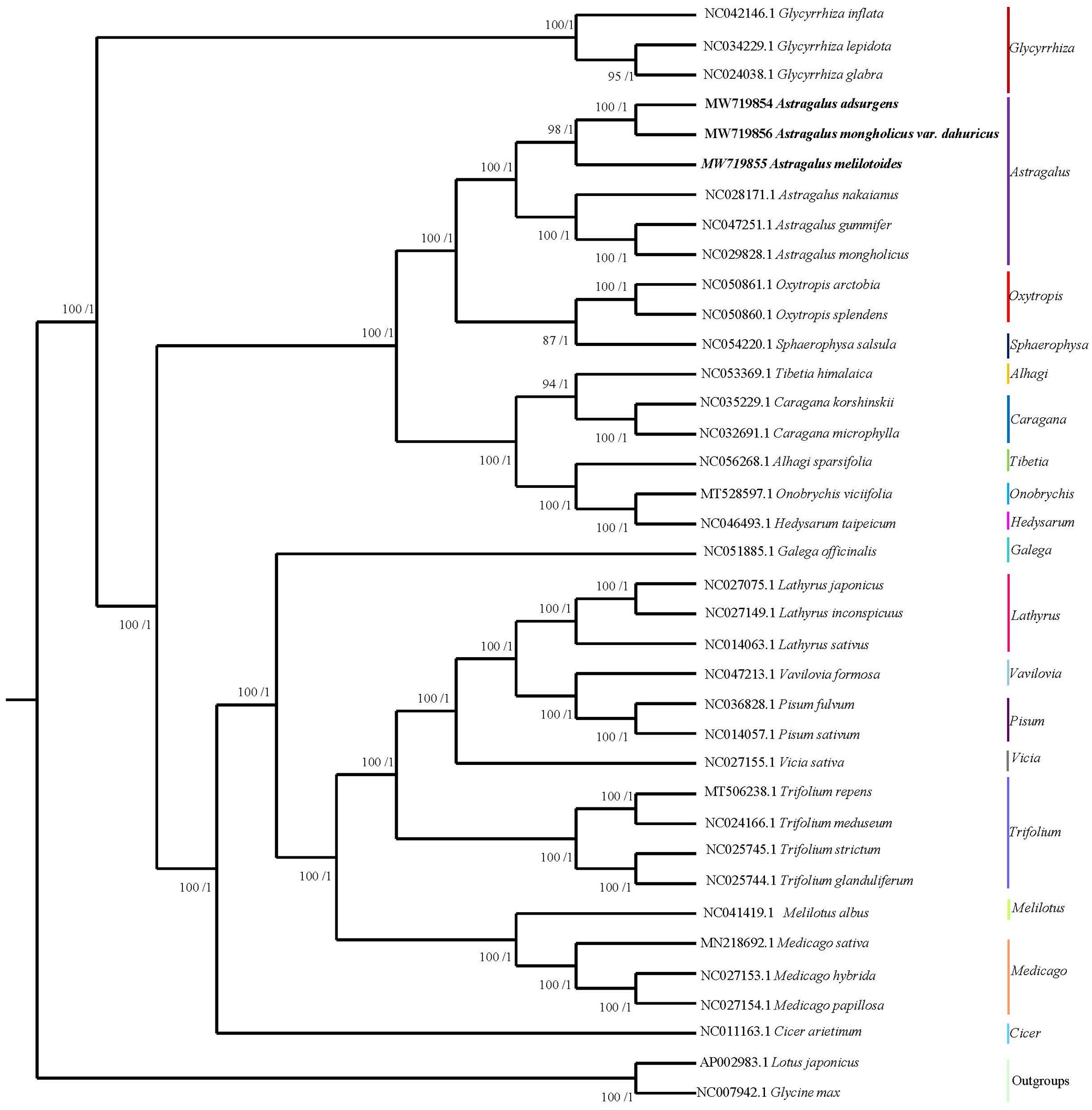
Figure 7. Phylogenetic tree of 35 species in Papilionoideae generated using maximum likelihood and Bayesian inferences (Lotus japonicus and Glycine max were used as outgroups) based on their complete cp genomes. Numbers associated with branches are ML values/BI values (posterior probability).
Conclusion
In the present study, we sequenced and annotated the cp genomes of A. adsurgens, A. mongholicus var. dahuricus, and A. melilotoides in Papilionoideae (Leguminosae). All these species belong to the IRLC, and their genomes include repeat sequence and abundant SSRs. Using A. membranaceus var. membranaceus as reference, the divergence was evident in most coding regions of cp genomes of Astragalus, and seven genes can be used as candidate DNA barcodes. Most protein-coding genes undergo purifying selection, and only cemA and rpl33 are under positive selection. Astragalus is a monophyletic group and is closely related to Oxytropis. Our analysis provides useful information for the identification and phylogenetic analyses of the IR lacking species.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/, SRR13870432, SRR13870430, and SRR13870431.
Author Contributions
CT collected the plant materials, did the analysis, and wrote the first manuscript. ZW designed the experiment and performed data analysis. XL, ZL, XH, and FL contributed to the result interpretation and manuscript revision. All authors read and agreed to the published version of the manuscript.
Funding
The research was funded by the National Natural Sciences Foundation of China (No. 31502008), the Central Public-interest Scientific Institution Fundamental Research Fund (1610332020002), and the Natural Science Foundation of Inner Mongolia (No. 2018MS03001).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2021.705482/full#supplementary-material
Footnotes
References
Alami, S., Lamin, H., Bouhnik, O., El Faik, S., Filali-Maltouf, A., Abdelmoumen, H., et al. (2019). Astragalus algarbiensis is nodulated by the genistearum symbiovar of Bradyrhizobium spp. in Morocco. Syst. Appl. Microbiol. 42, 440–447. doi: 10.1016/j.syapm.2019.03.004
Azani, N., Bruneau, A., Wojciechowski, M. F., and Zarre, S. (2019). Miocene climate change as a driving force for multiple origins of annual species in Astragalus (Fabaceae, Papilionoideae). Mol. Phylogenet Evol. 137, 210–221. doi: 10.1016/j.ympev.2019.05.008
Bagheri, A., Maassoumi, A. A., Rahiminejad, M. R., Brassac, J., and Blattner, F. R. (2017). Molecular phylogeny and divergence times of Astragalus section Hymenostegis: An analysis of a rapidly diversifying species group in Fabaceae. Sci. Rep. 7, 14033. doi: 10.1038/s41598-017-14614-3
Biju, V. C., Shidhi, P. R., Vijayan, S., Rajan, V. S., Sasi, A., Janardhanan, A., et al. (2019). The complete chloroplast genome of Trichopus zeylanicus, and phylogenetic analysis with Dioscoreales. Plant Genome 12, 1–11. doi: 10.3835/plantgenome2019.04.0032
Bruneau, A., and Palmer, D. J. D. (1990). A chloroplast DNA inversion as a subtribal character in the Phaseoleae (Leguminosae). Systematic Botany 15, 378–386. doi: 10.2307/2419351
Choi, I. S., Kim, J. H., and Choi, B. H. (2016). Complete plastid genome of Astragalus mongholicus var. nakaianus (Fabaceae). Mitochondrial DNA Part A 27, 2838–2839. doi: 10.3109/19401736.2015.1053118
Christopher, D. A., and Mullet, J. E. (1994). Separate photosensory pathways coregulate blue light/ultraviolet-A-activated psbD-psbC transcription and light-induced D2 and CP43 degradation in barley (Hordeum vulgare) chloroplasts. Plant Physiol. 104, 1119–1129. doi: 10.1104/pp.104.4.1119
Chumley, T. W., Palmer, J. D., Mower, J. P., Fourcade, H. M., Calie, P. J., Boore, J. L., et al. (2006). The complete chloroplast genome sequence of Pelargonium ×hortorum: organization and evolution of the largest and most highly rearranged chloroplast genome of land plants. Mol. Biol. Evol. 23, 2175–2190. doi: 10.1093/molbev/msl089
Cook, D., Gardner, D. R., Martinez, A., Robles, C. A., and Pfister, J. A. (2017a). Screening for swainsonine among South American Astragalus species. Toxicon 139, 54–57. doi: 10.1016/j.toxicon.2017.09.014
Cook, D., Gardner, D. R., Pfister, J. A., Lee, S. T., Welch, K. D., and Welsh, S. L. (2017b). A Screen for Swainsonine in Select North American Astragalus Species. Chem. Biodivers. 14, e1600364. doi: 10.1002/cbdv.201600364
Cui, N., Liao, B., Liang, C., Li, S., Zhang, H., Xu, J., et al. (2020). Complete chloroplast genome of Salvia plebeia: organization, specific barcode and phylogenetic analysis. Chin. J. Nat. Med. 18, 563–572. doi: 10.1016/S1875-5364(20)30068-6
Cui, Z., Li, Y., Yuan, Q., Zhou, L., and Li, M. (2012). Molecular identification of astragali radix and its adulterants by ITS sequences. Zhongguo Zhong Yao Za Zhi 37, 3773–3776.
Daniell, H., Lin, C. S., Yu, M., and Chang, W. J. (2016). Chloroplast genomes: diversity, evolution, and applications in genetic engineering. Genome Biol. 17, 134. doi: 10.1186/s13059-016-1004-2
Dierckxsens, N., Mardulyn, P., and Smits, G. (2019). Unraveling heteroplasmy patterns with NOVOPlasty. NAR Genom. Bioinform. 2, lqz011. doi: 10.1093/nargab/lqz011
Dong, W., Liu, J., Yu, J., Wang, L., and Zhou, S. (2012). Highly variable chloroplast markers for evaluating plant phylogeny at low taxonomic levels and for DNA barcoding. PLoS ONE 7:e35071. doi: 10.1371/journal.pone.0035071
Dos Reis, M. (2015). How to calculate the non-synonymous to synonymous rate ratio of protein-coding genes under the Fisher-Wright mutation-selection framework. Biol. Lett. 11, 20141031. doi: 10.1098/rsbl.2014.1031
Ebert, D., and Peakal, R. (2009). Chloroplast simple sequence repeats (cpSSRs): technical resources and recommendations for expanding cpSSR discovery and applications to a wide array of plant species. Mol. Ecol. Resour. 9, 673–690. doi: 10.1111/j.1755-0998.2008.02319.x
Endo, T., Ikeo, K., and Gojobori, T. (1996). Large-scale search for genes on which positive selection may operate. Mol. Biol. Evol. 13, 685–690. doi: 10.1093/oxfordjournals.molbev.a025629
Frazer, K. A., Pachter, L., Poliakov, A., Rubin, E. M., and Dubchak, I. (2014). VISTA: Computational tools for comparative genomics. Nucleic Acids Res. 32, W273–W279. doi: 10.1093/nar/gkh458
Greiner, S., Lehwark, P., and Bock, R. (2019). OrganellarGenomeDRAW (OGDRAW) version 1.3.1: expanded toolkit for the graphical visualization of organellar genomes. Nucleic Acids Res. 47, W59–W64. doi: 10.1093/nar/gkz238
Hong, Z., Wu, Z., Zhao, K., Yang, Z., Zhang, N., Guo, J., et al. (2020). Comparative Analyses of Five Complete Chloroplast Genomes from the Genus Pterocarpus (Fabacaeae). Int. J. Mol. Sci. 21, 3758. doi: 10.3390/ijms21113758
Hou, Y., Shi, Y., Zhang, Y., Liu, X., and Chen, Y. (2016). Astragali Radix and Hedysari Radix molecular identification of SSR primers screening and fingerprints code. Zhongguo Zhong Yao Za Zhi. 41, 1819–1822. doi: 10.4268/cjcmm20161010
Jin, J., Kong, J., Qiu, J., Zhu, H., Peng, Y., and Jiang, H. (2016). High level of microsynteny and purifying selection affect the evolution of WRKY family in Gramineae. Dev. Genes Evol. 226, 15–25. doi: 10.1007/s00427-015-0523-2
Kapoor, M., Mawal, P., Sharma, V., and Gupta, R. C. (2020). Analysis of genetic diversity and population structure in Asparagus species using SSR markers. J. Genet. Eng. Biotechnol. 18, 50. doi: 10.1186/s43141-020-00065-3
Katoh, K., and Standley, D. M. (2013). MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 30, 772–780. doi: 10.1093/molbev/mst010
Kearse, M., Moir, R., Wilson, A., Stones-Havas, S., Cheung, M., Sturrock, S., et al. (2012). Geneious Basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28, 1647–1649. doi: 10.1093/bioinformatics/bts199
Kurtz, S., and Schleiermacher, C. (1999). REPuter: Fast computation of maximal repeats in complete genomes. Bioinformatics 15, 426–427. doi: 10.1093/bioinformatics/15.5.426
Lei, W., Ni, D., Wang, Y., Shao, J., Wang, X., Yang, D., et al. (2016). Intraspecific and heteroplasmic variations, gene losses and inversions in the chloroplast genome of Astragalus membranaceus. Sci. Rep. 6, 21669. doi: 10.1038/srep21669
Li, B., Lin, F., Huang, P., Guo, W., and Zheng, Y. (2020). Development of nuclear SSR and chloroplast genome markers in diverse Liriodendron chinense germplasm based on low-coverage whole genome sequencing. Biol. Res. 53, 21. doi: 10.1186/s40659-020-00289-0
Li, B., and Zheng, Y. (2018). Dynamic evolution and phylogenomic analysis of the chloroplast genome in Schisandraceae. Sci. Rep. 8, 9285. doi: 10.1038/s41598-018-27453-7
Li, X., Qu, L., Dong, Y., Han, L., Liu, E., Fang, S., et al. (2014). A review of recent research progress on the Astragalus genus. Molecules 19, 18850–18880. doi: 10.3390/molecules191118850
Lin, C., Huang, J., Wu, C., Hsu, C., and Chaw, S. (2010). Comparative chloroplast genomics reveals the evolution of Pinaceae genera and subfamilies. Genome Biol. Evol. 2, 504–517. doi: 10.1093/gbe/evq036
Liu, B., Duan, N., Zhang, H., Liu, S., Shi, J., and Chai, B. (2016). Characterization of the whole chloroplast genome of Caragana microphylla Lam (Fabaceae). Conserv. Genet. Resour. 8, 371–373. doi: 10.1007/s12686-016-0561-8
Liu, E., Yang, C., Liu, J., Jin, S., Harijati, N., Hu, Z., et al. (2019). Comparative analysis of complete chloroplast genome sequences of four major Amorphophallus species. Sci. Rep. 9, 809. doi: 10.1038/s41598-018-37456-z
Liu, Y., Chen, Y., and Fu, X. (2020). The complete chloroplast genome sequence of medicinal plant: Astragalus laxmannii (Fabaceae). Mitochondrial DNA B Resour. 5, 3661–3662. doi: 10.1080/23802359.2020.1829122
Martin, G. E., Rousseau-Gueutin, M., Cordonnier, S., Lima, O., Michon-Coudouel, S., Naquin, D., et al. (2014). The first complete chloroplast genome of the Genistoid legume Lupinus luteus: evidence for a novel major lineage-specific rearrangement and new insights regarding plastome evolution in the legume family. Ann. Bot. 113, 1197–1210. doi: 10.1093/aob/mcu050
Martinez, A., Robles, C. A., Roper, J. M., Gardner, D. R., Neyaz, M. S., Joelson, N. Z., et al. (2019). Detection of swainsonine-producing endophytes in Patagonian Astragalus species. Toxicon 171, 1–6. doi: 10.1016/j.toxicon.2019.09.020
Massatti, R., Belus, M. T., Dowlatshahi, S., and Allan, G. J. (2018). Genetic analyses of Astragalus sect. Humillimi (Fabaceae) resolve taxonomy and enable effective conservation. Am. J. Bot. 105, 1703–1711. doi: 10.1002/ajb2.1157
Mauriello, E. (2019). How bacteria arrange their organelles. Elife 8, e43777. doi: 10.7554/eLife.43777
Menke, W., French, C. S., and Butler, W. L. (1965). On absorption changes in chloroplast and algen caused by drying. Z. Naturforsch. B 20, 482–487.
Millen, R. S., Olmstead, R. G., Adams, K. L., Palmer, J. D., Lao, N. T., Heggie, L., et al. (2001). Many parallel losses of infA from chloroplast DNA during angiosperm evolution with multiple independent transfers to the nucleus. Plant Cell 13, 645–658. doi: 10.1105/tpc.13.3.645
Muse, S. V., and Gaut, B. S. (1994). A likelihood approach for comparing synonymous and nonsynonymous nucleotide substitution rates, with application to the chloroplast genome. Mol. Biol. Evol. 11, 715–724. doi: 10.1093/oxfordjournals.molbev.a040152
Osaloo, S. K., Maassoumi, A. A., and Murakami, N. (2003). Molecular systematics of the genus Astragalus L. (Fabaceae): Phylogenetic analyses of nuclear ribosomal DNA internal transcribed spacers and chloroplast gene ndhF sequences. Plant System Evol. 242, 1–32. doi: 10.1007/s00606-003-0014-1
Palmer, J. D., and Thompson, W. F. (1981). Rearrangements in the chloroplast genomes of mung bean and pea. Proc. Natl. Acad. Sci. USA. 78, 5533–5537. doi: 10.1073/pnas.78.9.5533
Podlech, D. (1986). Taxonomic and phytogeographical problems in Astragalus of the Old World and South-West Asia. Proceedings of the Royal Society of Edinburgh. 89, 37–43. doi: 10.1017/S0269727000008885
Qu, X., Moore, M. J., Li, D., and Yi, T. (2019). PGA: a software package for rapid, accurate, and flexible batch annotation of plastomes. Plant Methods 15, 1–12. doi: 10.1186/s13007-019-0435-7
Ronquist, F., and Huelsenbeck, J. P. (2003). MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19, 1572–1574. doi: 10.1093/bioinformatics/btg180
Rozas, J., Ferrer, M. A., Sánchez-DelBarrio, J. C., Guirao, R. S., Librado, P., Ramos-Onsins, S. E., et al. (2017). DnaSP v6: DNA sequence polymorphism analysis of large datasets. Mol. Biol. Evol. 34, 3299–3302. doi: 10.1093/molbev/msx248
Saha, D., Rana, R. S., Das, S., Datta, S., Jiban, M., Sylvie, J. C., et al. (2019). Genome-wide regulatory gene-derived SSRs reveal genetic differentiation and population structure in fiber flax genotypes. J. Appl. Genetics 60, 13–25. doi: 10.1007/s13353-018-0476-z
Saltonstall, K., and Lambertini, C. (2012). The value of repetitive sequences in chloroplast DNA for phylogeographic inference: A comment on Vachon & Freeland 2011. Mol. Ecol. Resour. 12, 581–585. doi: 10.1111/j.1755-0998.2012.03146.x
Sanderson, M. J., and Doyle, J. J. (1993). Phylogenetic relationships in North American Astragalus (Fabaceae) based on chloroplast DNA restriction site variation. Systematic Botany 18, 395–408. doi: 10.2307/2419416
Saski, C., Lee, S. B., Daniell, H., Wood, T. C., Tomkins, J., Kim, H. G., et al. (2005). Complete chloroplast genome sequence of Gycine max and comparative analyses with other legume genomes. Plant Mol. Biol. 59, 309–322. doi: 10.1007/s11103-005-8882-0
Shen, X., Guo, S., Yin, Y., Zhang, J., Yin, X., Liang, C., et al. (2018). Complete chloroplast genome sequence and phylogenetic analysis of Aster tataricus. Molecules 23, 2426. doi: 10.3390/molecules23102426
Shi, L., Chen, H., Jiang, M., Wang, L., Wu, X., Huang, L., et al. (2019). CPGAVAS2, an integrated plastome sequence annotator and analyzer. Nucleic Acids Res. 47, W65–W73. doi: 10.1093/nar/gkz345
Shinozaki, K., Ohme, M., Tanaka, M., Wakasugi, T., Hayashida, N., Matsubayashi, T., et al. (1986). The complete nucleotide sequence of the tobacco chloroplast genome: its gene organization and expression. EMBO J. 5, 2043–2049. doi: 10.1002/j.1460-2075.1986.tb04464.x
Somaratne, Y., Guan, D. L., Wang, W. Q., Zhao, L., and Xu, S. Q. (2019). The complete chloroplast genomes of two Lespedeza species: insights into codon usage bias, RNA editing sites, and phylogenetic relationships in Desmodieae (Fabaceae: Papilionoideae). Plants (Basel) 9, 56. doi: 10.3390/plants9010051
Stamatakis, A. (2014). RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30, 1312–1313. doi: 10.1093/bioinformatics/btu033
Su, C., Duan, L., Liu, P., Liu, J., Chang, Z., and Wen, J. (2021). Chloroplast phylogenomics and character evolution of eastern Asian Astragalus (Leguminosae): Tackling the phylogenetic structure of the largest genus of flowering plants in Asia. Mol. Phylogenet Evol. 156, 107025. doi: 10.1016/j.ympev.2020.107025
Trüper, H. G. (1987). Phototrophic bacteria (an incoherent group of prokaryotes). A taxonomic versus phylogenetic survey. Microbiologia 3, 71–89.
Tunckol, B., Ayta, Z., Aksoy, N., and Fisne, A. (2020). Astragalus bartinense (Fabaceae), a new species from Turkey. Acta botanica Croatica 79, 131–136. doi: 10.37427/botcro-2020-023
Volff, J. N., Erixon, P., and Oxelman, B. (2008). Whole-gene positive selection, elevated synonymous substitution rates, duplication, and indel evolution of the chloroplast clpP1 Gene. PLoS ONE 3:e1386. doi: 10.1371/journal.pone.0001386
Wang, B., Chen, H., Ma, H., Zhang, H., Lei, W., Wu, W., et al. (2016). Complete plastid genome of Astragalus membranaceus (Fisch.) Bunge var. membranaceus. Mitochondrial DNA B Resour. 1, 517–519. doi: 10.1080/23802359.2016.1197057
Wei, F., Tang, D., Wei, K., Qin, F., Li, L., Lin, Y., et al. (2020). The complete chloroplast genome sequence of the medicinal plant Sophora tonkinensis. Sci. Rep. 10, 12473. doi: 10.1038/s41598-020-69549-z
Wojciechowski, M. F., Sanderson, M. J., Baldwin, B. G., and Donoghue, M. J. (1993). Monophyly of aneuploid Astragalus (Fabaceae): evidence from nuclear ribosomal DNA internal transcribed spacer sequences. American Journal of Botany 80, 711–722. doi: 10.2307/2419416
Wyman, S. K., Jansen, R. K., and Boore, J. L. (2004). Automatic annotation of organellar genomes with DOGMA. Bioinformatics 20, 3252–3255. doi: 10.1093/bioinformatics/bth352
Xiong, Y., Xiong, Y., He, J., Yu, Q., Zhao, J., Lei, X., et al. (2020). The Complete chloroplast genome of two important annual clover species, Trifolium alexandrinum and T. resupinatum: genome structure, comparative analyses and phylogenetic relationships with relatives in Leguminosae. Plants (Basel) 9, 478. doi: 10.3390/plants9040478
Yang, Z. (2007). PAML 4: Phylogenetic Analysis by Maximum Likelihood. Mol. Biol. Evol. 24, 1586–1591. doi: 10.1093/molbev/msm088
Yin, D., Wang, Y., Zhang, X., Ma, X., He, X., and Zhang, J. (2017). Development of chloroplast genome resources for peanut (Arachis hypogaea L.) and other species of Arachis. Sci. Rep. 7, 11649. doi: 10.1038/s41598-017-12026-x
Zha, X., Wang, X., Li, J., Gao, F., and Zhou, Y. (2020). Complete chloroplast genome of Sophora alopecuroides (Papilionoideae): molecular structures, comparative genome analysis and phylogenetic analysis. J. Genet. 99, 13. doi: 10.1007/s12041-019-1173-3
Zhao, X., and Zhu, Z. (2020). Comparative genomics and phylogenetic analyses of Christia vespertilionis and Urariopsis brevissima in the Tribe Desmodieae (Fabaceae: Papilionoideae) based on complete chloroplast genomes. Plants (Basel) 9, 1116. doi: 10.3390/plants9091116
Zheng, S., Liu, D., Ren, W., Fu, J., Huang, L., and Chen, S. (2014). Integrated analysis for identifying astragali radix and its adulterants based on DNA barcoding. Evid. Based Complement Alternat. Med. 2014, 843923. doi: 10.1155/2014/843923
Keywords: Astragalus, complete chloroplast genome, IR lacking, genetic diversity, phylogenetic analysis
Citation: Tian C, Li X, Wu Z, Li Z, Hou X and Li FY (2021) Characterization and Comparative Analysis of Complete Chloroplast Genomes of Three Species From the Genus Astragalus (Leguminosae). Front. Genet. 12:705482. doi: 10.3389/fgene.2021.705482
Received: 05 May 2021; Accepted: 16 July 2021;
Published: 05 August 2021.
Edited by:
Frank M. You, Agriculture and Agri-Food Canada (AAFC), CanadaReviewed by:
Sachin Kumar, Chaudhary Charan Singh University, IndiaLei Xie, Beijing Forestry University, China
Copyright © 2021 Tian, Li, Wu, Li, Hou and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zinian Wu, d3V6aW5pYW5AY2Fhcy5jbg==; Zhiyong Li, emhpeW9uZ2xpMTIxNkAxMjYuY29t
 Chunyu Tian1,2
Chunyu Tian1,2 Zinian Wu
Zinian Wu