- 1Laboratory of Clinical Applied Anatomy, Department of Human Anatomy, School of Basic Medical Sciences, Fujian Medical University, Fuzhou, China
- 2Department of Developmental Dentistry, School of Dentistry, The University of Texas Health Science Center at San Antonio, San Antonio, TX, United States
- 3Greehey Children’s Cancer Research Institute, The University of Texas Health Science Center at San Antonio, San Antonio, TX, United States
Bmp2 is essential for dentin development and formation. Bmp2 conditional knock-out (KO) mice display a similar tooth phenotype of dentinogenesis imperfecta (DGI). To elucidate a foundation for subsequent functional studies of cross talk between mRNAs and lncRNAs in Bmp2-mediated dentinogenesis, we investigated the profiling of lncRNAs and mRNAs using immortalized mouse dental Bmp2 flox/flox (iBmp2fx/fx) and Bmp2 knock-out (iBmp2ko/ko) papilla cells. RNA sequencing was implemented to study the expression of the lncRNAs and mRNAs. Quantitative real-time PCR (RT-qPCR) was used to validate expressions of lncRNAs and mRNAs. The Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) databases were used to predict functions of differentially expressed genes (DEGs). Protein–protein interaction (PPI) and lncRNA–mRNA co-expression network were analyzed by using bioinformatics methods. As a result, a total of 22 differentially expressed lncRNAs (16 downregulated vs 6 upregulated) and 227 differentially expressed mRNAs (133 downregulated vs. 94 upregulated) were identified in the iBmp2ko/ko cells compared with those of the iBmp2fx/fx cells. RT-qPCR results showed significantly differential expressions of several lncRNAs and mRNAs which were consistent with the RNA-seq data. GO and KEGG analyses showed differentially expressed genes were closely related to cell differentiation, transcriptional regulation, and developmentally relevant signaling pathways. Moreover, network-based bioinformatics analysis depicted the co-expression network between lncRNAs and mRNAs regulated by Bmp2 in mouse dental papilla cells and symmetrically analyzed the effect of Bmp2 during dentinogenesis via coding and non-coding RNA signaling.
Introduction
Bone morphogenetic protein 2 (Bmp2) is a multiple-functional growth factor and is involved in many organ developments (Macias et al., 1997; Schlange et al., 2000; Ou et al., 2014). The bone morphogenetic proteins (BMPs) are structurally related to the transforming growth factor β (TGF-β) superfamily. The members of the BMP family play various biological functions during embryonic development (Hogan, 1996; Shi and Massagué, 2003; Wu et al., 2003; Chen et al., 2004; Miyazono et al., 2005), including a vital role in tooth development and formation (Meguro et al., 2019). Among the BMP family members, Bmp2 has been widely investigated for its diverse biological functions, particularly during dental cell differentiation (Casagrande et al., 2010; Wang et al., 2012; Yang et al., 2012; Guo et al., 2015; Yang et al., 2017; Malik et al., 2018). Bmp2 is expressed in mesenchymal cells and promotes mesenchymal progenitor/stem cell commitment to the odontoblast lineage by regulating a series of transcription factors and others (Yamashiro et al., 2003; Chen et al., 2005; Chen et al., 2008; Cho et al., 2010; Agas et al., 2013; Yang et al., 2017).
During tooth development, at the initiation stage (E10-12), the dental lamina is formed as an epithelial clustering, and the dental epithelium and mesenchyme are distinguished. The Bmp2 gene transcript is seen in those areas of the dental lamina where it started to form a bud (Aberg et al., 1997; Heikinheimo et al., 1998; Nadiri et al., 2004; Chen et al., 2008). At the bud stage (E12-13), Bmp2 expression is detectable in the dental epithelium and mesenchyme throughout the bud period. At the cap stage, Bmp2 expression is prominent at E14 and mainly localized at the epithelial enamel knot during the late cap stage, and Bmp2 expression expands to the neighboring inner dental epithelium where the secondary enamel knots will be formed and its signal is seen in dental mesenchyme. At the bell stage, Bmp2 expression is detected in the dental mesenchymal cells (Dong et al., 2014; Dong et al., 2016; Gao et al., 2018). Later, Bmp2 expression spreads to the dental papilla and is intense in the pre-odontoblasts. At the postnatal days (PN), Bmp2 is continually expressed in odontoblasts and ameloblasts and detected in the dental papilla as well as adjacent tissues including dental follicle, periodontal ligamental cells, cementoenamel junction, and Hertwig’s epithelial root sheath (HERS) and osteoblasts in alveolar bones (Yamashiro et al., 2003; Kémoun et al., 2007).
The Bmp2 binds to two distinct types, type I serine/threonine kinase and type II receptors, which are necessary for signal transduction (Shi and Massagué, 2003; Chen et al., 2004; Miyazono et al., 2005). The Bmp signal regulates downstream gene expression through either the canonical Smad or non-canonical Smad pathway. Following heterodimerization, type I receptors are phosphorylated by type II receptors and subsequently activate the receptor-regulated R-Smad-1/-5/-8 through phosphorylation (Bhatt et al., 2013; Graf et al., 2016). The phosphorylated Smad-1/-5/-8 heterodimers form a complex with the common mediator Smad-4 (Co-Smad-4). Following nuclear translocation of the R-Smad-1/-5/-8/Co-Smad-4 complex, Bmp target gene expression is induced (Bhatt et al., 2013). On the other hand, Bmp2 also activates non-canonical Smad signaling pathways such as mitogen-activated protein kinases (MAPKs), c-Jun amino-terminal kinase (JNK), phosphoinositol-3 kinase (PI3K), Akt, and small GTPases (Derynck and Zhang, 2003). Thus, these canonical Smad pathways cooperate with non-canonical Smad pathways to regulate various cellular responses. Also, Bmp2 can crosstalk with other factors, such as Wnt, Fgf, and other factors, regulating cell proliferation, differentiation, and tissue development (Guo and Wang, 2009; O'Connell et al., 2012; Agas et al., 2013; Saito et al., 2016; Wu et al., 2016; Zhang et al., 2016; Chakka et al., 2020).
Global Bmp2 knockout (KO) mice are nonviable. Homozygous Bmp2 mutant embryos die at embryonic day 9.5 (E9.5) and exhibited defects in cardiac development, manifested by the abnormal development of the heart in the exocoelomic cavity (Zhang and Bradley, 1996). However, Bmp2 conditional KO mice were generated and viable. Teeth with Bmp2 cKO mice display the similar phenotype of dentin defects to that of dentinogenesis imperfecta (DGI) in humans and mice, the most common dentin genetic diseases, showing retardation of tooth growth, abnormal dentin structure with wide predentin, and thin dentin (Zhang et al., 2001; Lu et al., 2007; Lee et al., 2009). Accumulating evidence indicated that Bmp2 is capable of regulating a lot of bone/dentin-related gene expressions and transcriptional factors as well as dental cell proliferation and differentiation in tooth development (Chen et al., 2008; Cho et al., 2010; Oh et al., 2012; Yang et al., 2017). Additionally, cross talk has been described between Bmp signaling–associated mRNAs and non-coding RNAs during dental cell proliferation and differentiation (Wang F. et al., 2017; Wang J. et al., 2017; Liu et al., 2018; Liao et al., 2020; He et al., 2021).
High-throughput sequencing techniques have revolutionized our understanding of the human genome (Kapranov et al., 2007). The whole transcriptome study has demonstrated that although more than 90% of the genome can be transcribed into RNAs, only approximately 2% of the human genome contains protein-coding regions (Kapranov et al., 2007; Jalali et al., 2016). The remaining non-coding regions are transcribed into large amounts of non-coding RNAs (Jalali et al., 2016). lncRNAs are the largest class of non-coding RNAs composed of more than 200 nucleotides to over 100 kb in length, which lack protein-coding potential (Ponting et al., 2009). The lncRNAs comprise thousands of transcripts with distinct biogenesis, subcellular localization, and molecular functions (Chen, 2016). Studies have suggested that lncRNA genome complexity plays biological roles in regulating gene expression at transcriptional, post-transcriptional, and epigenetic levels in a lot of cellular and biological processes, such as signal transduction, cell proliferation, differentiation, and organ development (Batista and Chang, 2013; Fatica and Bozzoni, 2014; Quinn and Chang, 2016; Bunch, 2018; Statello et al., 2021). Therefore, alteration of lncRNA expression is related to numerous diseases (Fatica and Bozzoni, 2014; Wu and Du, 2017). lncRNAs can regulate the expression of growth factors, transcriptional and epigenetic factors, and vice versa (Yang et al., 2014). The pattern of cell- and tissue-specific lncRNA expression provides new insights into tissue development, diagnosis, and treatment of several diseases (Chen et al., 2021). Up to date, over 100,000 lncRNAs have been identified in the human genome and new lncRNAs are being discovered and characterized rapidly (Zhao et al., 2021).
However, the interplay of Bmp2-mediated mRNAs and lncRNAs in the regulation of odontoblastic differentiation and function has not completely been understood. Previously, our group generated immortalized mouse dental Bmp2 flox/flox (iBmp2fx/fx) and Bmp2 knock-out (iBmp2ko/ko) papilla cells. In this study, we detected the differential lncRNA and mRNA expression profiles in these two types of dental papilla cell lines by RNA sequencing. A network-based bioinformatics analysis was performed to investigate the cross talk between lncRNAs and mRNAs via integrating lncRNA–mRNA interactions, gene co-expression, and protein–protein interactions. Our results demonstrated that the synergistic or competitive lncRNA–mRNA cross talk may play an important role in Bmp2-mediated odontoblastic differentiation.
Materials and Methods
In Situ Alkaline Phosphatase Assay and Alizarin Red S Staining
Mouse dental iBmp2fx/fx and iBmp2ko/ko papilla cells were generated by our group as previously described (Wu et al., 2010; Wu et al., 2015). To induce cell differentiation, the cells were cultured in α-MEM supplemented with 10% fetal bovine serum (FBS), 1% antibiotics, 10 mM sodium β-glycerophosphate, 50 μg/ml ascorbic acid, and 100 nM dexamethasone for 7 and 10 days. Then, the cells were fixed and washed in PBS. In situ alkaline phosphatase (ALP) assay was carried out in accordance with the instructions. For cell mineralization assessment, the cells were fixed in 10% formalin as well as were treated with 1% Alizarin Red S dye (pH 4.2).
RNA-Seq and Gene Expression Analysis
The iBmp2fx/fx and iBmp2ko/ko cells were cultured in 100-mm2 dishes to 80% confluence. The cells were harvested after 6, 48, or 72 h, and RNAs were extracted using TRIzol reagent (QIAGEN Inc.). The RNA quality was assessed using a bio-analyzer. The RNA-seq libraries were prepared from total RNAs in accordance with Illumina’s RNA specimen preparation protocol (Illumina Inc. San Diego, CA, United States). Paired reads to the UCSC mm9 genome build were mapped by a TopHat2 aligner. To quantify gene expression, HTSeq was used to obtain raw read counts per gene and then converted to RPKM in accordance with the gene length and total mapped read count per sample. The DEGs were calculated, as gene expression levels were measured by Log2-transformed RPKM (|log2 fold Change| >1, p value <0.05). RNA-seq data were submitted and deposited to The Gene Expression Omnibus (GEO, http://www.ncbi.nlm.nih.gov/geo/). The GEO accession number is GSE174429.
Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) Analysis
GO analysis was used to investigate the roles of all differentially expressed mRNAs (https://david.ncifcrf.gov/). DAVID-based KEGG analysis was used to determine the significant pathways related to the differentially expressed mRNAs. Fisher’s exact test and the x2 test were used to select the significant GO categories and pathways. The threshold of significance was a p value < 0.05, and a false discovery rate (FDR) was calculated to correct the p value.
Generation of the Protein–Protein Interaction (PPI) Network
The differentially expressed genes (DEGs) were imported into a STRING database, and the species was limited to “Mus musculus” to obtain PPI information. The network relationships with high confidence (≥0.75) were screened and imported into Cytoscape 3.6.2 to draw a PPI network diagram. According to the calculation method of the MCC (maximum clique centrality) algorithm, the top 20 hub genes were screened by using the “cytoHubba” plug-in. Additionally, the interactive density region was extracted using the “MCODE” plug-in in Cytoscape.
Construction of the lncRNA–mRNA Co-Expression Network
The mRNA–lncRNA co-expression network, which was used to identify interactions between the differentially expressed lncRNAs and mRNAs, was constructed based on Pearson’s correlation analysis. For each pair of genes, correlation coefficients of 0.95 or greater were selected to construct the network using the OmicStudio tools at https://www.omicstudio.cn/tool. In the network, each mRNA or lncRNA corresponds to a node, and the nodes are connected by edges.
Quantitative Reverse Transcription PCR (RT-qPCR)
Total RNA samples were isolated with TRIzol (Invitrogen, California, United States) and reverse transcribed into cDNA with the Prime Script RT reagent kit with gDNA Eraser (RR047A) following the manufacturers’ guidelines (Takara, Japan). The RNA was reverse transcribed into cDNA at 37°C for 15 min, and the reaction was stopped at 85°C for 5 s. RT-qPCR was performed using the SYBR™ Select Master Mix (Applied Biosystems, California, United States) on a CFX96 system (Bio-Rad, California, United States). The relative gene expression was calculated with the 2−ΔΔCt method using glyceraldehyde-3-phosphate dehydrogenase (GAPDH) as the reference housekeeping gene. The RT-qPCR primers are listed in Table 1.
Statistical Analysis
Data were presented as the mean ± SEM. RT-qPCR validation data presented were analyzed with Student’s t test. p < 0.05 was considered statistically significant. Significant differences are noted by asterisks, with single asterisks representing p < 0.05, two asterisks representing p < 0.01, three asterisks representing p < 0.001, and four asterisks representing p < 0.0001.
Results
Deletion of Bmp2 Causes Delay of Dental Papilla Mesenchymal Cell Differentiation and Mineralization
To determine the effect of Bmp2 on dental papilla cell differentiation and mineralization activities, we measured the alkaline phosphatase (ALP) activity by in situ ALP histochemistry since ALP is a marker of dental cell differentiation. Both cells were cultured in the calcifying medium in given time periods. This result showed delayed dental papilla cell differentiation in the iBmp2ko/ko cells compared to that in the Bmp2fx/fx cells. Additionally, deletion of the Bmp2 gene led to low activity of the dental papilla cell mineralization as observed through Alizarin Red S staining (Figure 1).
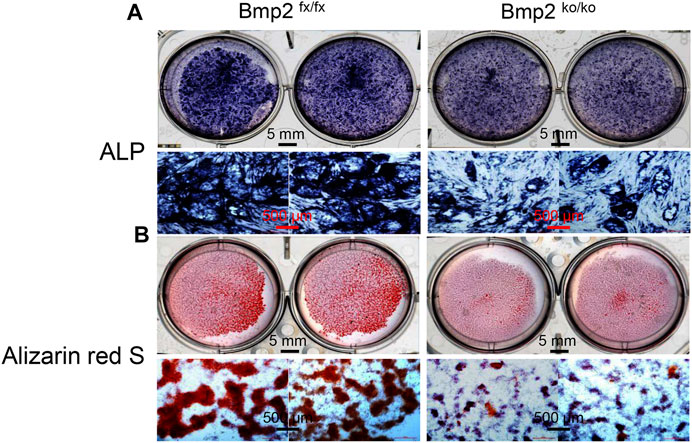
FIGURE 1. Deletion of Bmp2 delays mouse dental papilla cell differentiation and mineralization. (A) iBmp2fx/fx and iBmp2ko/ko cells were cultured in the calcifying medium for 7 days. ALP activity was analyzed using in situ ALP staining. (B) For cell mineralization assay, both the iBmp2fx/fx and iBmp2ko/ko cells were treated with the calcifying medium for 10 days. Mineralized nodules were visualized with Alizarin Red S staining. fx, floxed; ko, knock-out.
Overview of Differential lncRNA/mRNA Expression Profiling
To identify DEGs between the iBmp2fx/fx and iBmp2ko/ko cells, we performed and analyzed the RNA-seq data by using the limma package. The cutoff criteria were as follows: |log fold change| (the absolute value of log2 in the fold change of gene expression) > 1 and p value <0.05. Our data demonstrated that 94 mRNAs were upregulated, while 133 mRNAs were downregulated in the iBmp2ko/ko cells compared with those in the iBmp2fx/fx cells (Figure 2A). Likewise, 6 lncRNAs were upregulated whereas 16 lncRNAs were downregulated in the iBmp2ko/ko cells compared to those in the iBmp2fx/fx cells (Figure 2B). The volcano maps of mRNA and lncRNA DEGs were drawn by the ggplot2 package. The heatmaps of mRNA and lncRNA DEGs were shown by the heatmap package (Figure 3).
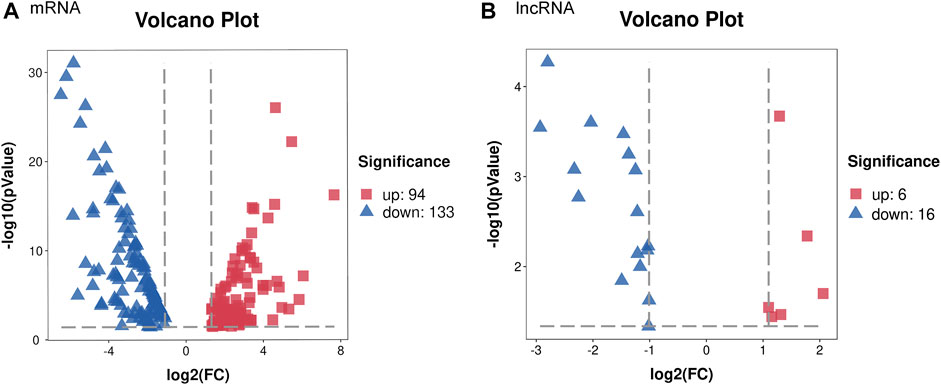
FIGURE 2. Differentially expressed genes between the iBmp2fx/fx and iBmp2ko/ko groups. Our data showed that 94 mRNAs upregulated and 133 mRNAs downregulated in the iBmp2ko/ko cells compared to those in the iBmp2fx/fx cells (A). Meanwhile, 6 lncRNAs upregulated and 16 lncRNAs downregulated (|log2FC| ≥1; Qvalue <=0.05) in the iBmp2ko/ko cells (B). Red represents upregulated DEG, blue represents downregulated DEG.
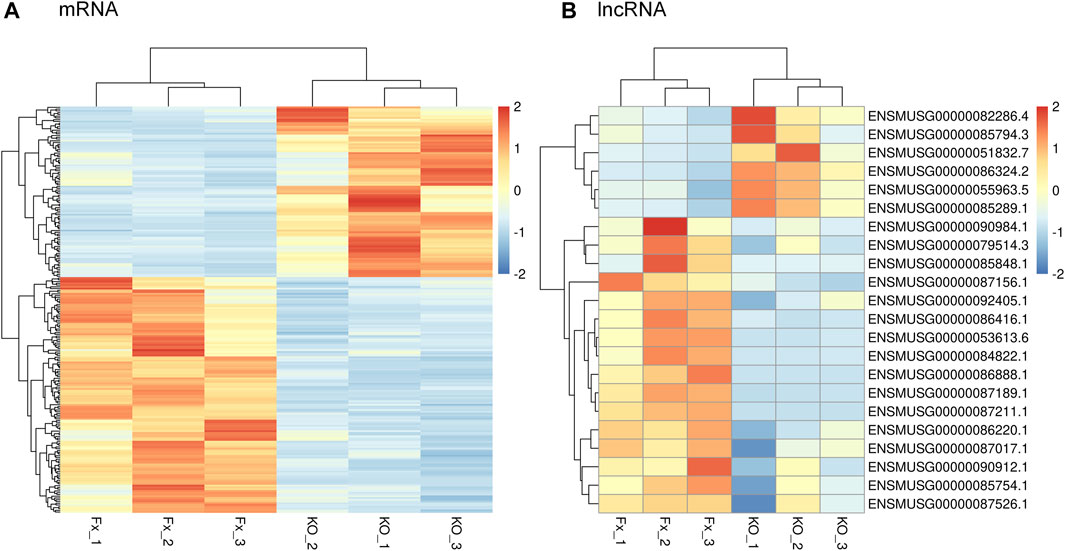
FIGURE 3. Genetic clustering of differentially expression genes (|log2FC| ≥1; Q value <=0.001). (A) mRNAs; (B) and lncRNAs. The red color represents high fold-change, and the blue color represents low fold-change.
Pathway and GO Analyses of Differentially Expressed mRNAs
The KEGG analysis showed that the DEGs were largely enriched in the TGF-β/BMP signaling pathway, Hippo signaling pathway, Wnt signaling pathway, and signaling pathways regulating pluripotency of stem cells (Figure 4A). These signal pathways are cross talks and are involved in dental and other cell proliferation and differentiation via regulating transcriptional and growth factors including Id2, Id3, Fgf2, Klf10, Lhx1, Smad7, Tcf7, Wnt1, and Wnt11 (Ho et al., 2011; Koizumi et al., 2013; Sternberg et al., 2013; Bakopoulou et al., 2015; Chen et al., 2016; Zhang et al., 2016; Wang and Martin, 2017; Neves and Sharpe, 2018; Chen et al., 2019; Liu et al., 2019; Chakka et al., 2020; Niki et al., 2021).
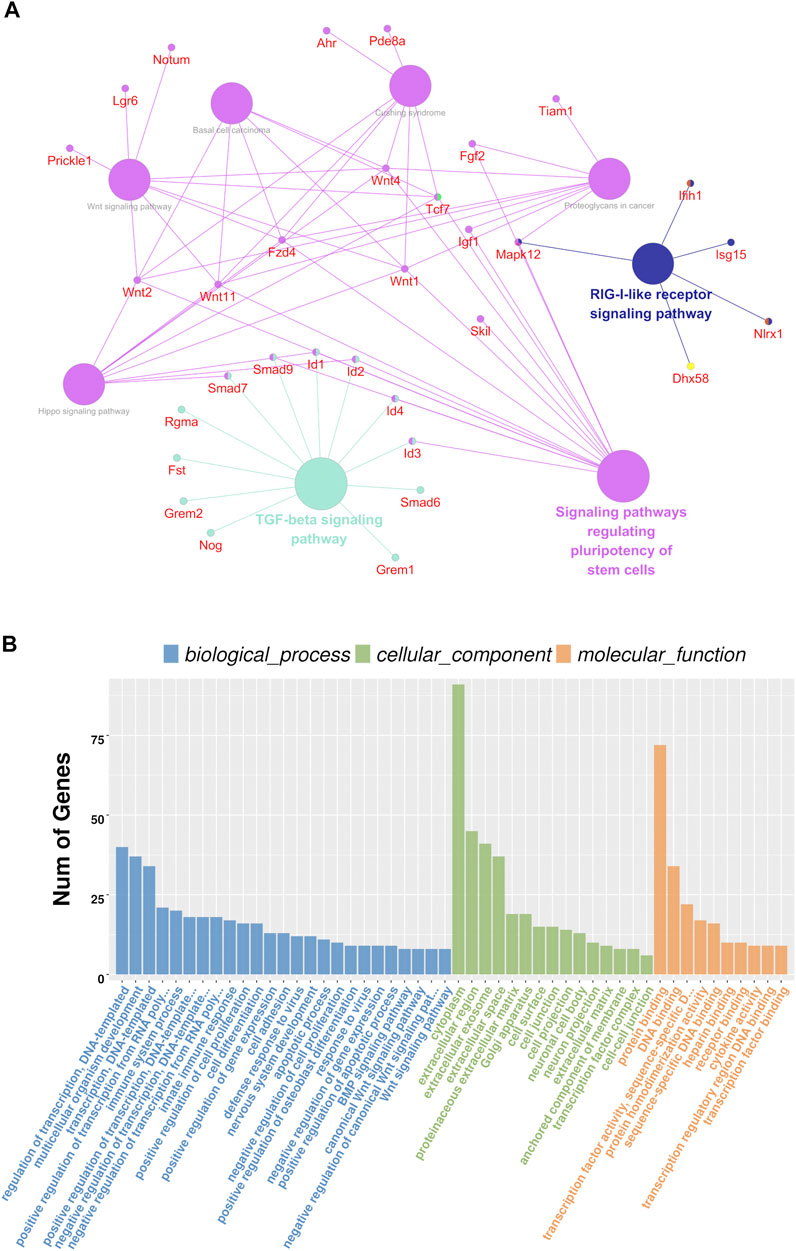
FIGURE 4. Analysis results of KEGG and GO analyses. (A) Possible signaling pathways are represented by circles, and the involved genes are represented by red characters. The possible signaling pathways showed the larger diameters of the circle; the larger the diameters are the more significant the signal pathway is. (B) GO analysis in a cellular component, biological process, and molecular function.
The GO analysis showed that the following biological pathways (BPs) were notably enriched among the DEGs: regulation of gene transcription, regulation of transcription from RNA polymerase II promoter, multicellular organism development, positive regulation of cell proliferation, cell differentiation, Wnt signaling, and BMP signaling pathways, etc. (Figure 4B). Besides, the following cellular components (CCs) were found to be largely enriched in the extracellular exosome, proteinaceous extracellular matrix, anchored component of the membrane and transcription factor complex, cell–cell junction, and cell protection (Figure 4B). Additionally, the following molecular functions (MFs) were largely enriched in protein binding, transcription factor activity, sequence-specific DNA binding, receptor binding, and transcription regulatory region DNA binding (Figure 4B). This result suggests that Bmp2 and other signaling pathways regulate dental cell proliferation and cell differentiation as well as tissue development through those aforementioned pathways (Graf et al., 2016; Salazar et al., 2016; Wu et al., 2016; Migliorini et al., 2020).
Analysis Results of the Protein–Protein Interaction (PPI) Network
To gain more insights into the role of these 227 genes in the Bmp2 network and find the hub genes, which were significantly implicated in, a PPI network based on these 227 genes (94 upregulated genes and 133 downregulated genes) was established on the STRING database for functional association analysis, and the sequential visualization was performed on Cytoscape (Figure 5A). The plug-in “cytoHubba” determined the top 20 hub genes by MCC algorithm (Figure 5B) and related the differential genes according to the association grade difference between different genes (Figure 5C). The interactive density region in the PPI network by “MCODE’’ plug-in was also discovered. The figure below showed the top dense regions (Figure 5D). The PPI analysis predicted that the 9 Bmp2-induced proteins tested can interact with each other. For instance, Fgf2 binds to Id3 and to Wnt11, Lhx1 to Wnt11, Smad7 to Klf10 and to Id2, Tcf7 to Wnt 1 and Id2, etc. On the other hand, one protein interacts with another via a third protein. For example, Id2 binds to Klf10 via Smad7; Lhx1 binds to Wnt1 via Wnt11 and Tcf7 via Wnt11; and Fgf2 binds to Id2 via Tcf7 and Lhx1 via Wnt11 as well as others. This study suggests that these proteins play synergistic roles in the regulation of dental cell proliferation and differentiation.
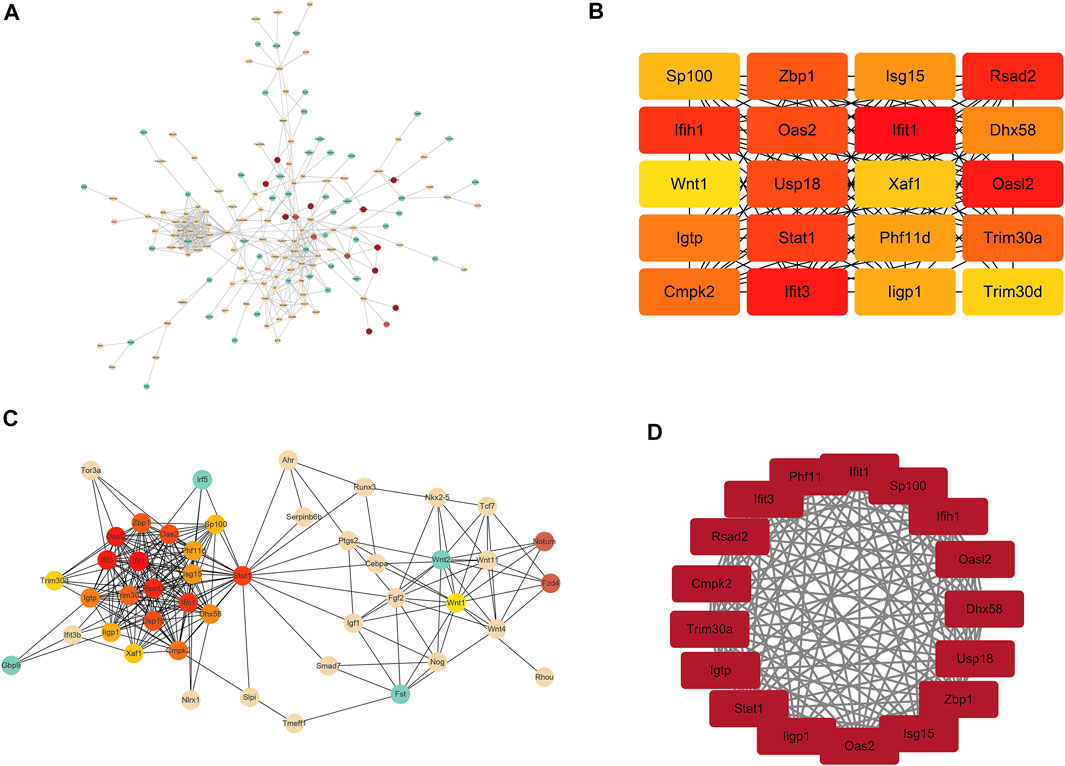
FIGURE 5. Analysis results of PPI. (A) Total network interaction of all DEGs. The redder the color, the more important it is in the network relationship. (B) Top 20 hub genes in the interaction network. (C) Total network interaction of the top 20 hub genes and the differential genes that are directly related to them. (D) Top dense regions of the interactive density region in the PPI network.
Analysis Results of Co-Expression Networks Between mRNAs and lncRNAs
Based on correlations between the differential expression levels of lncRNAs and mRNAs, a coding–noncoding gene co-expression network was established. We calculated the Pearson correlation coefficient (PCC) and used the R-value to calculate the correlation coefficient of the PCC between lncRNAs and mRNAs (lncRNA–mRNA PCC, not including lncRNA–lncRNA or mRNA–mRNA PCC). Those lncRNAs and mRNAs that had Pearson correlation coefficients (PCCs) ≥ 0.95 were selected to construct the co-expression network. The figure below showed the top 100 pairs of co-expression relationships (Figure 6A). Furthermore, visualization of the co-expression network between lncRNA and mRNAs was performed using the OmicStudio tools at https://www.omicstudio.cn/tool (Figure 6B). For instance, lncRNA ENSMUSG00000087211.1 (87,211.1) co-expressed with Id2, Klf10, Smad7, Wnt1, Wnt11, and Lhx1, while lncRNA ENSMUSG00000087189.1 (87,189.1) co-expressed with Lhx1 and Klf10, lncRNA ENSMUSG00000086888.1 (86,888.1) co-expressed with Fgf2 and Wnt1, and lncRNA ENSMUSG00000053613.6 (53,613.6) co-expressed with Fgf2 and Lhx1. Although the co-expression of lncRNAs and mRNAs was identified in the dental papilla cells, the effect of cross talk between the mRNAs and lncRNA during dentinogenesis is required for further study.
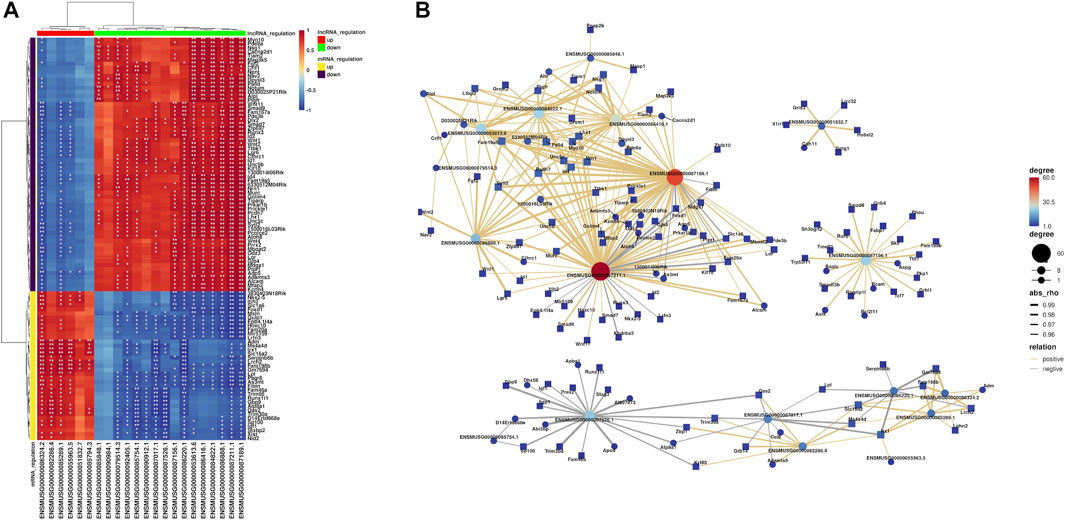
FIGURE 6. Results of co-expression networks between mRNAs and lncRNAs. (A) Heatmaps are used to describe the relationship between lncRNAs and mRNAs (different colors of lncRNA or mRNA regulation). The differences that are statistically significant are denoted by asterisks, with single asterisks representing p < 0.05 and two asterisks representing p < 0.01. (B) Hub co-expression network between lncRNAs and mRNAs. The yellow line represents the positive regulation, the gray line represents the negative regulation, and the thickness of the line represents the relative strength. In addition, the size and color depth of the points represent the number of related genes.
Validation of Differentially Expressed lncRNAs and mRNAs
In order to verify the transcriptome sequencing results between the iBmp2fx/fx and iBmp2ko/ko cells and further analyze the possible signaling network mediated by Bmp2, the 9 representative mRNAs and 3 lncRNAs were selected for validation by using RT-qPCR since the 9 Bmp signal-associated mRNAs are involved in the dental cell proliferation, differentiation, and tooth development (Ho et al., 2011; Koizumi et al., 2013; Sternberg et al., 2013; Bakopoulou et al., 2015; Chen et al., 2016; Zhang et al., 2016; Chen et al., 2019; Liu et al., 2019; Chakka et al., 2020), and three novel lncRNAs (87,211.1, 87,189.1, and 86,888.1) co-expressed with these mRNAs (Figure 6B). The RT-qPCR results showed that the expression of lncRNA87211.1 and lncRNA87189.1 decreased in the iBmp2ko/ko cells, as same as the results of RNA-seq, but lncRNA86888.1 expression was increased in the iBmp2ko/ko cells which was opposite to the results of RNA-seq (Figure 7A). The mRNA expression of Fgf2, Id2, Id3, Lhx1, Smad7, Tcf7, Wnt1, and Wnt11 by RT-qPCR analysis was consistent with the result of the RNA-seq (Figure 7B). Furthermore, the 6 validated mRNAs (Id2, Klf10, Lhx1, Smad7, Wnt1, and Wnt11) were positively correlated with the expression of lncRNA 87211.1 and lncRNA 87189.1, which was in accordance with the predicted lncRNA–mRNA co-expression network (Figure 7C). Accordingly, the PCC can be used to predict the relation network between mRNAs and lncRNAs.
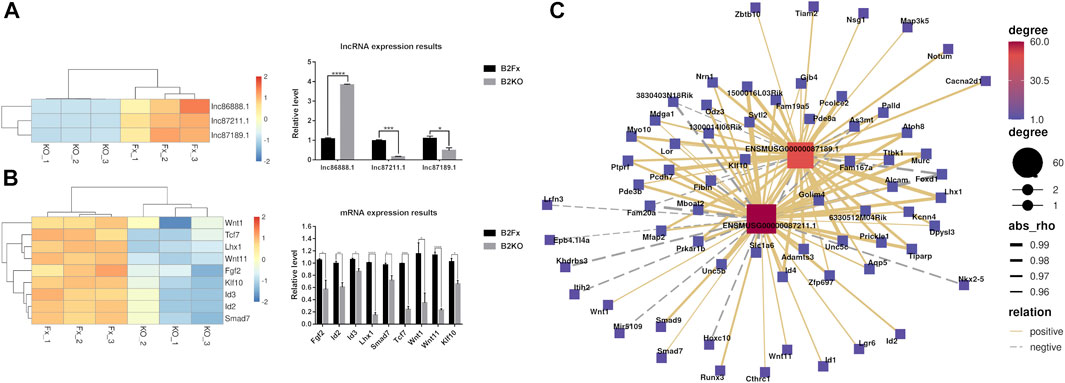
FIGURE 7. RT-qPCR relative expression results. There are 9 mRNAs and 3 hub lncRNAs validated by RT-qPCR. (A) Results of the RNA-seq and RT-qPCR from the lncRNAs of lncRNA87211.1 (ENSMUSG00000087211.1), lncRNA87189.1 (ENSMUSG00000087189.1), and lncRNA86888.1 (ENSMUSG00000086888.1). (B) Results of the RNA-seq and RT-qPCR from 9 mRNAs including Fgf2, Id2, Id3, Klf10, Lhx1, Smad7, Tcf7, Wnt1, and Wnt11. The red color represents high fold-change, and the blue color represents low fold-change. The abscissa is the gene name, and the ordinate is the relative expression of the genes. (C) Hub co-expression network between two lncRNAs lncRNA87211.1 and lncRNA87189.1 and 6 mRNAs. The solid line shows positive regulation while the dashed line represents negative regulation.
Discussion
Bone morphogenetic proteins (BMPs), members of the TGF-β superfamily, are multiple regulators for organogenesis and homeostasis. Among them, Bmp2 has been proven to be able to induce osteogenesis or dentinogenesis, independently (Bais et al., 2009; Yang et al., 2012). Several canonical and non-canonical Bmp2 signaling pathways have been reported involved in odontogenesis (Cho et al., 2010; Qin et al., 2012a; Qin et al., 2012b; Washio et al., 2012; Qin et al., 2014). Recently accumulated evidence has demonstrated that lncRNAs play biological roles in dental cell proliferation, cell proliferation, and tooth development through the regulation of growth and transcriptional factors (Zheng and Jia, 2016; Liu et al., 2018; Gil and Ulitsky, 2020; Wang et al., 2020; Li et al., 2021; Mirzadeh Azad et al., 2021). The cross talk between the Bmp2 signaling pathway and lncRNAs in dental cell proliferation, cell differentiation, and odontogenesis remains largely unclear. In the present study, we, for the first time, sequenced transcriptome in the iBmp2fx/fx and iBmp2ko/ko cells and performed bioinformatics analysis to study Bmp2-mediated signaling pathways and downstream molecules as well as to predict the co-expression network between lncRNAs and mRNAs regulated by Bmp2 in vitro.
Our data indicated that 94 mRNAs were upregulated, and 133 mRNAs were downregulated. Moreover, 6 lncRNAs were upregulated and 16 lncRNAs were downregulated by RNA-seq from the iBmp2 ko/ko and iBmp2 fx/fx papilla cells. GO analysis indicated that the DEGs were closely related to cell proliferation, cell differentiation, transcriptional regulation, protein–protein interactions, BMP- and Wnt-signal pathways, and so forth. Through KEGG analysis, these DEGs were found to be largely enriched in developmentally signaling pathways including TGF-β/BMP signaling pathway, Hippo signaling pathway, Wnt signaling pathway, and signaling pathways regulating pluripotency of stem cells, suggesting that Bmp2 might mediate cross-talk between these signaling pathways and regulate dental papilla mesenchymal cell proliferation, differentiation, and dentinogenesis (Graf et al., 2016; Wu et al., 2016; Neves and Sharpe, 2018; da Silva Madaleno et al., 2020). YAP (Yes-associated protein) and transcriptional co-activator with PDZ-binding motif (TAZ) function as transcriptional cofactors of the Hippo pathway, which only indirectly bind to DNA and regulate target genes through interaction with other transcription factors such as Smads, Runx1/2, and their effectors of the transcriptional factor TEA domain (TEAD) family members (Piccolo et al., 2014). The cross talk between YAP/TAZ and BMP signaling has been documented. YAP was shown to support Smad1-dependent transcription and to be required for BMP-mediated suppression of neural differentiation in mouse embryonic stem cells, providing an example of long-term modulation of BMP signaling (Alarcón et al., 2009). Moreover, following stimulation with Bmp2, YAP modulated Smad1/5 phosphorylation and target gene expression in multiple cell types, while its early interaction with nuclear Smad1/5/8 was documented to lead to a stabilization of Smad1/5/8 signaling in astrocytes (Huang et al., 2016). Likewise, YAP overexpression activated Smad-dependent BMP signaling and upregulated the mRNA and protein expression of several cementogenesis markers including ALP, Runx2, osteocalcin (Ocn), and dentin matrix acidic phosphoprotein 1 (Dmp1). Treatment with a specific BMP antagonist (LDN193189) prevented the upregulation of the mRNA levels of ALP, Runx2, Dmp1, and Ocn as well as the intensity of ALP-stained and mineralized nodules in cementoblasts (Yang et al., 2018). The PPI study predicted that Bmp-induced proteins related to tooth/bone development interact with each other, and one protein binds to another via a third protein (Figure 5). It suggests that these proteins synergically regulate tooth-related gene expression mediated by Bmp2 signaling. Furthermore, the lncRNA–mRNA co-expression network analysis revealed that lncRNAs87211.1, lnc87189.1, and lnc86888.1 co-expressed with Id2, Klf10, Lhx1, Smad7, Wnt1, and Wnt11 (Figure 6).
Based on the results of the KEGG, the PPI, and lncRNA–mRNA co-expression, the 9 mRNAs (Fgf2, Id2, Id3, Lhx1, Smad7, Tcf7, Wnt1, Wnt11, and Klf10) and 3 lncRNAs (87,211.1, 87,189.1, and 86,888.1) were selected as representative genes for RT-qPCR validation as these genes are involved in the dental cell proliferation and differentiation as well as tooth development and formation induced by Bmp2 signaling (Tsuboi et al., 2003; Koizumi et al., 2013; Sternberg et al., 2013; Chen et al., 2016; Graf et al., 2016; Wu et al., 2016; Li et al., 2019; Liu et al., 2019; Qin et al., 2019; Nottmeier et al., 2021) and three novel lncRNAs co-expressed with those mRNAs in the dental papilla cells. Our study demonstrated that the validated 9 mRNAs and 2 lncRNAs by RT-qPCR were consistent with the results of the RNA-seq, and the PCC method can be used to predict co-expression networks between mRNAs and lncRNAs.
The selected 9 mRNAs are involved in the dental cell proliferation, differentiation, and homeostasis (Ho et al., 2011; Koizumi et al., 2013; Sternberg et al., 2013; Bakopoulou et al., 2015; Chen et al., 2016; Zhang et al., 2016; Wang and Martin, 2017; Neves and Sharpe, 2018; Chen et al., 2019; Liu et al., 2019; Chakka et al., 2020; Niki et al., 2021). The loss of Bmp2 resulted in a decrease of these gene expressions detected by RNA-seq and RT-qPCR (Figures 2, 7). It suggested that Bmp2 regulates dental differentiation and dentinogenesis via these genes. It has been documented that the interplay of Bmp2 signal–associated mRNAs and lncRNAs regulates tooth development and formation (Liu et al., 2018; Zhong et al., 2020). This study showed that several novel lncRNAs such as lncRNA87211.1, lncRNA87189.1, and lncRNA86888.1 co-expressed with those mRNAs (Figure 6). However, whether the loss of Bmp2 downregulated expression of the selected 9 mRNAs through the lncRNA regulation needs to be further investigated although Liu’s and Zhong’s groups reported that lncRNAs inhibited microRNAs and indirectly upregulated Bmp2 gene expression, which enhanced the expression of Runx2, Dmp1, and Dspp genes as well as cell differentiation (Liu et al., 2018; Zhong et al., 2020).
Based on this study by the experimental and bioinformatics analyses, we suggest that Bmp2 signaling stimulates these mRNA expressions in the regulation of dental proliferation, differentiation, and homeostasis via direct or indirect lncRNA pathways. However, whether Bmp2 stimulates these 9 gene expressions via these lncRNAs during dentinogenesis needs to be further investigated.
Taken together, in the present study, we validated Bmp2 signal pathways and the possible signal molecules downstream of Bmp2 in the dental papilla cells using RNA-seq, RT-qPCR, and bioinformatics. Moreover, by performing bioinformatics analyses, we predicted several hub genes in the lncRNA–mRNA co-expression network regulated by Bmp2, which provides the direction and basis for further elucidating the regulation mechanism of Bmp2-mediated odontogenesis via the mRNA–lncRNA network.
Data Availability Statement
The authors acknowledge that the data presented in this study must be deposited and made publicly available in an acceptable repository, prior to publication. Frontiers cannot accept a article that does not adhere to our open data policies.
Ethics Statement
Animal use was approved by the Institutional Animal Care and Use Committee (IACUC) of the University of Texas Health Science Center at San Antonio, Texas, United States.
Author Contributions
FW and SC conceived and designed the experiments and revised the manuscript. FW and RT performed the experiments. YZ carried out RNA-seq. FW, ML, GG, RT, LZ, X-HH, QL, DL, and SC analyzed the data. FW and RT wrote the manuscript. All authors have read and approved the final manuscript.
Funding
This work was supported by the National Institutes of Health (NIH) grants, RO1DE019802, the Pilot Grant of School of Dentistry, UTHSCSA, the Natural Science Foundation of Fujian Province (No. 2021J01676), and the Joint Fund for Scientific and Technological Innovation of Fujian Province award (No. 2016Y9034).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors, and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We thank Aries Okungbowa-Ikponmwosa and GG for editing the manuscript.
References
Åberg, T., Wozney, J., and Thesleff, I. (1997). Expression Patterns of Bone Morphogenetic Proteins (Bmps) in the Developing Mouse Tooth Suggest Roles in Morphogenesis and Cell Differentiation. Dev. Dyn. 210, 383–396. doi:10.1002/(sici)1097-0177(199712)210:4<383:aid-aja3>3.0.co;2-c
Agas, D., Sabbieti, M. G., Marchetti, L., Xiao, L., and Hurley, M. M. (2013). FGF-2 Enhances Runx-2/Smads Nuclear Localization in BMP-2 Canonical Signaling in Osteoblasts. J. Cel. Physiol. 228, 2149–2158. doi:10.1002/jcp.24382
Alarcón, C., Zaromytidou, A.-I., Xi, Q., Gao, S., Yu, J., Fujisawa, S., et al. (2009). Nuclear CDKs Drive Smad Transcriptional Activation and Turnover in BMP and TGF-β Pathways. Cell 139, 757–769. doi:10.1016/j.cell.2009.09.035
Bais, M. V., Wigner, N., Young, M., Toholka, R., Graves, D. T., Morgan, E. F., et al. (2009). BMP2 Is Essential for post Natal Osteogenesis but Not for Recruitment of Osteogenic Stem Cells. Bone 45, 254–266. doi:10.1016/j.bone.2009.04.239
Bakopoulou, A., Leyhausen, G., Volk, J., Papachristou, E., Koidis, P., and Geurtsen, W. (2015). Wnt/β-catenin Signaling Regulates Dental Pulp Stem Cells' Responses to Pulp Injury by Resinous Monomers. Dental Mater. 31, 542–555. official publication of the Academy of Dental Materials. doi:10.1016/j.dental.2015.02.004
Batista, P. J., and Chang, H. Y. (2013). Long Noncoding RNAs: Cellular Address Codes in Development and Disease. Cell 152, 1298–1307. doi:10.1016/j.cell.2013.02.012
Bhatt, S., Diaz, R., and Trainor, P. A. (2013). Signals and Switches in Mammalian Neural Crest Cell Differentiation. Cold Spring Harbor Perspect. Biol. 5, a008326. doi:10.1101/cshperspect.a008326
Birjandi, A. A., Suzano, F. R., and Sharpe, P. T. (2020). Drug Repurposing in Dentistry: Towards Application of Small Molecules in Dentin Repair. Ijms 21, 6394. doi:10.3390/ijms21176394
Bunch, H. (2018). Gene Regulation of Mammalian Long Non-coding RNA. Mol. Genet. Genomics 293, 1–15. doi:10.1007/s00438-017-1370-9
Casagrande, L., Demarco, F. F., Zhang, Z., Araujo, F. B., Shi, S., and Nör, J. E. (2010). Dentin-derived BMP-2 and Odontoblast Differentiation. J. Dent Res. 89, 603–608. doi:10.1177/0022034510364487
Chakka, L. R. J., Vislisel, J., Vidal, C. d. M. P., Biz, M. T., Aliasger, K. S., and Salem, B. N. (2020). Application of BMP-2/FGF-2 Gene-Activated Scaffolds for Dental Pulp Capping. Clin. Oral Invest. 24, 4427–4437. doi:10.1007/s00784-020-03308-2
Chen, D., Zhao, M., and Mundy, G. R. (2004). Bone Morphogenetic Proteins. Growth Factors 22, 233–241. doi:10.1080/08977190412331279890
Chen, L.-L. (2016). Linking Long Noncoding RNA Localization and Function. Trends Biochemical Sciences 41, 761–772. doi:10.1016/j.tibs.2016.07.003
Chen, S., Gluhak-Heinrich, J., Martinez, M., Li, T., Wu, Y., Chuang, H.-H., et al. (2008). Bone Morphogenetic Protein 2 Mediates Dentin Sialophosphoprotein Expression and Odontoblast Differentiation via NF-Y Signaling. J. Biol. Chem. 283, 19359–19370. doi:10.1074/jbc.M709492200
Chen, S., Rani, S., Wu, Y., Unterbrink, A., Gu, T. T., Gluhak-Heinrich, J., et al. (2005). Differential Regulation of Dentin Sialophosphoprotein Expression by Runx2 during Odontoblast Cytodifferentiation. J. Biol. Chem. 280, 29717–29727. doi:10.1074/jbc.M502929200
Chen, X.-J., Shen, Y.-S., He, M.-C., Yang, F., Yang, P., Pang, F.-X., et al. (2019). Polydatin Promotes the Osteogenic Differentiation of Human Bone Mesenchymal Stem Cells by Activating the BMP2-Wnt/β-Catenin Signaling Pathway. Biomed. Pharmacother. 112, 108746. doi:10.1016/j.biopha.2019.108746
Chen, Y., Li, Z., Chen, X., and Zhang, S. (2021). Long Non-coding RNAs: From Disease Code to Drug Role. Acta pharmaceutica Sinica B 11, 340–354. doi:10.1016/j.apsb.2020.10.001
Chen, Z., Li, W., Wang, H., Wan, C., Luo, D., Deng, S., et al. (2016). Klf10 Regulates Odontoblast Differentiation and Mineralization via Promoting Expression of Dentin Matrix Protein 1 and Dentin Sialophosphoprotein Genes. Cell Tissue Res 363, 385–398. doi:10.1007/s00441-015-2260-2
Cho, Y.-D., Yoon, W.-J., Woo, K.-M., Baek, J.-H., Park, J.-C., and Ryoo, H.-M. (2010). The Canonical BMP Signaling Pathway Plays a Crucial Part in Stimulation of Dentin Sialophosphoprotein Expression by BMP-2. J. Biol. Chem. 285, 36369–36376. doi:10.1074/jbc.M110.103093
da Silva Madaleno, C., Jatzlau, J., and Knaus, P. (2020). BMP Signalling in a Mechanical Context - Implications for Bone Biology. Bone 137, 115416. doi:10.1016/j.bone.2020.115416
Derynck, R., and Zhang, Y. E. (2003). Smad-dependent and Smad-independent Pathways in TGF-β Family Signalling. Nature 425, 577–584. doi:10.1038/nature02006
Dinger, M. E., Pang, K. C., Mercer, T. R., and Mattick, J. S. (2008). Differentiating Protein-Coding and Noncoding RNA: Challenges and Ambiguities. Plos Comput. Biol. 4, e1000176. doi:10.1371/journal.pcbi.1000176
Dong, J., Lei, X., Wang, Y., Wang, Y., Song, H., Li, M., et al. (2016). Different Degrees of Iodine Deficiency Inhibit Differentiation of Cerebellar Granular Cells in Rat Offspring, via BMP-Smad1/5/8 Signaling. Mol. Neurobiol. 53, 4606–4617. doi:10.1007/s12035-015-9382-0
Dong, X., Shen, B., Ruan, N., Guan, Z., Zhang, Y., Chen, Y., et al. (2014). Expression Patterns of Genes Critical for BMP Signaling Pathway in Developing Human Primary Tooth Germs. Histochem. Cel Biol 142, 657–665. doi:10.1007/s00418-014-1241-y
Duan, J., Gherghe, C., Liu, D., Hamlett, E., Srikantha, L., Rodgers, L., et al. (2014). Wnt1/βcatenin Injury Response Activates the Epicardium and Cardiac Fibroblasts to Promote Cardiac Repair. EMBO J. 31, 429–442. doi:10.1038/emboj.2011.418
Fatica, A., and Bozzoni, I. (2014). Long Non-coding RNAs: New Players in Cell Differentiation and Development. Nat. Rev. Genet. 15, 7–21. doi:10.1038/nrg3606
Gao, Z., Wang, L., Wang, F., Zhang, C., Wang, J., He, J., et al. (2018). Expression of BMP2/4/7 during the Odontogenesis of Deciduous Molars in Miniature Pig Embryos. J. Mol. Hist. 49, 545–553. doi:10.1007/s10735-018-9792-1
Gil, N., and Ulitsky, I. (2020). Regulation of Gene Expression by Cis-Acting Long Non-coding RNAs. Nat. Rev. Genet. 21, 102–117. doi:10.1038/s41576-019-0184-5
Graf, D., Malik, Z., Hayano, S., and Mishina, Y. (2016). Common Mechanisms in Development and Disease: BMP Signaling in Craniofacial Development. Cytokine Growth Factor. Rev. 27, 129–139. doi:10.1016/j.cytogfr.2015.11.004
Guo, F., Feng, J., Wang, F., Li, W., Gao, Q., Chen, Z., et al. (2015). Bmp2 Deletion Causes an Amelogenesis Imperfecta Phenotype via Regulating Enamel Gene Expression. J. Cel. Physiol. 230, 1871–1882. doi:10.1002/jcp.24915
Guo, X., and Wang, X.-F. (2009). Signaling Cross-Talk between TGF-β/BMP and Other Pathways. Cell Res 19, 71–88. doi:10.1038/cr.2008.302
He, H., Yu, J., Liu, Y., Lu, S., Liu, H., Shi, J., et al. (2008). Effects of FGF2 and TGFβ1 on the Differentiation of Human Dental Pulp Stem Cells In Vitro. Cel Biol. Int. 32, 827–834. doi:10.1016/j.cellbi.2008.03.013
He, Q., Li, R., Hu, B., Li, X., Wu, Y., Sun, P., et al. (2021). Stromal Cell‐derived Factor‐1 Promotes Osteoblastic Differentiation of Human Bone Marrow Mesenchymal Stem Cells via the lncRNA‐H19/miR‐214‐5p/BMP2 axis. J. Gene Med. 23, e3366. doi:10.1002/jgm.3366
Heikinheimo, K., Bègue‐Kirn, C., Ritvos, O., Tuuri, T., and Ruch, J. V. (1998). Activin and Bone Morphogenetic Protein (BMP) Signalling during Tooth Development. Eur. J. Oral Sci. 106 (Suppl. 1), 167–173. doi:10.1111/j.1600-0722.1998.tb02171.x
Ho, C. C., and Bernard, D. J. (2010). Bone Morphogenetic Protein 2 Acts via Inhibitor of DNA Binding Proteins to Synergistically Regulate Follicle-Stimulating Hormone β Transcription with Activin A. Endocrinology 151, 3445–3453. doi:10.1210/en.2010-0071
Ho, C. C., Zhou, X., Mishina, Y., and Bernard, D. J. (2011). Mechanisms of Bone Morphogenetic Protein 2 (BMP2) Stimulated Inhibitor of DNA Binding 3 (Id3) Transcription. Mol. Cell. Endocrinol. 332, 242–252. doi:10.1016/j.mce.2010.10.019
Hogan, B. L. (1996). Bone Morphogenetic Proteins: Multifunctional Regulators of Vertebrate Development. Genes Dev. 10, 1580–1594. doi:10.1101/gad.10.13.1580
Huang, Z., Sun, D., Hu, J.-X., Tang, F.-L., Lee, D.-H., Wang, Y., et al. (2016). Neogenin Promotes BMP2 Activation of YAP and Smad1 and Enhances Astrocytic Differentiation in Developing Mouse Neocortex. J. Neurosci. 36, 5833–5849. doi:10.1523/jneurosci.4487-15.2016
Jalali, S., Gandhi, S., and Scaria, V. (2016). Navigating the Dynamic Landscape of Long Noncoding RNA and Protein-Coding Gene Annotations in GENCODE. Hum. Genomics 10, 35. doi:10.1186/s40246-016-0090-2
Kapranov, P., Cheng, J., Dike, S., Nix, D. A., Duttagupta, R., Willingham, A. T., et al. (2007). RNA Maps Reveal New RNA Classes and a Possible Function for Pervasive Transcription. Science 316, 1484–1488. doi:10.1126/science.1138341
Kémoun, P., Laurencin-Dalicieux, S., Rue, J., Vaysse, F., Roméas, A., Arzate, H., et al. (2007). Localization of STRO-1, BMP-2/-3/-7, BMP Receptors and Phosphorylated Smad-1 during the Formation of Mouse Periodontium. Tissue and Cell 39, 257–266. doi:10.1016/j.tice.2007.06.001
Koizumi, Y., Kawashima, N., Yamamoto, M., Takimoto, K., Zhou, M., Suzuki, N., et al. (2013). Wnt11 Expression in Rat Dental Pulp and Promotional Effects of Wnt Signaling on Odontoblast Differentiation. Congenit. Anom. 53, 101–108. doi:10.1111/cga.12011
Lee, S.-K., Lee, K.-E., Jeon, D., Lee, G., Lee, H., Shin, C.-U., et al. (2009). A Novel Mutation in the DSPP Gene Associated with Dentinogenesis Imperfecta Type II. J. Dent Res. 88, 51–55. doi:10.1177/0022034508328168
Li, J., Jin, F., Cai, M., Lin, T., Wang, X., and Sun, Y. (2021). LncRNA Nron Inhibits Bone Resorption in Periodontitis. J. Dent Res. 220345211019689, 002203452110196. doi:10.1177/00220345211019689
Li, J., Yang, Y., Fan, J., Xu, H., Fan, L., Li, H., et al. (2019). Long Noncoding RNA ANCR Inhibits the Differentiation of Mesenchymal Stem Cells toward Definitive Endoderm by Facilitating the Association of PTBP1 with ID2. Cell Death Dis 10, 492. doi:10.1038/s41419-019-1738-3
Li, Q., Hua, Y., Yang, Y., He, X., Zhu, W., Wang, J., et al. (2018). T Cell Factor 7 (TCF7)/TCF1 Feedback Controls Osteocalcin Signaling in Brown Adipocytes Independent of the Wnt/β-Catenin Pathway. Mol. Cel Biol 38. doi:10.1128/mcb.00562-17
Liang, Z., Hore, Z., Harley, P., Uchenna Stanley, F., Michrowska, A., Dahiya, M., et al. (2020). A Transcriptional Toolbox for Exploring Peripheral Neuroimmune Interactions. Pain 161, 2089–2106. doi:10.1097/j.pain.0000000000001914
Liao, C., Zhou, Y., Li, M., Xia, Y., and Peng, W. (2020). LINC00968 Promotes Osteogenic Differentiation In Vitro and Bone Formation In Vivo via Regulation of miR-3658/RUNX2. Differentiation 116, 1–8. doi:10.1016/j.diff.2020.09.005
Liu, J., Zhang, Z. Y., Yu, H., Yang, A. P., Hu, P. F., Liu, Z., et al. (2018). Long Noncoding RNA C21orf121/bone Morphogenetic Protein 2/microRNA‐140‐5p Gene Network Promotes Directed Differentiation of Stem Cells from Human Exfoliated Deciduous Teeth to Neuronal Cells. J. Cel Biochem 120, 1464–1476. doi:10.1002/jcb.27313
Liu, Z., Chen, T., Bai, D., Tian, W., and Chen, Y. (2019). Smad7 Regulates Dental Epithelial Proliferation during Tooth Development. J. Dent Res. 98, 1376–1385. doi:10.1177/0022034519872487
Lu, Y., Ye, L., Yu, S., Zhang, S., Xie, Y., Mckee, M. D., et al. (2007). Rescue of Odontogenesis in Dmp1-Deficient Mice by Targeted Re-expression of DMP1 Reveals Roles for DMP1 in Early Odontogenesis and Dentin Apposition In Vivo. Dev. Biol. 303, 191–201. doi:10.1016/j.ydbio.2006.11.001
Macias, D., Ganan, Y., Sampath, T. K., Piedra, M. E., Ros, M. A., and Hurle, J. M. (1997). Role of BMP-2 and OP-1 (BMP-7) in Programmed Cell Death and Skeletogenesis during Chick Limb Development. Development 124, 1109–1117. doi:10.1242/dev.124.6.1109
Malik, Z., Alexiou, M., Hallgrimsson, B., Economides, A. N., Luder, H. U., and Graf, D. (2018). Bone Morphogenetic Protein 2 Coordinates Early Tooth Mineralization. J. Dent Res. 97, 835–843. doi:10.1177/0022034518758044
Meguro, F., Porntaveetus, T., Kawasaki, M., Kawasaki, K., Yamada, A., Kakihara, Y., et al. (2019). Bmp Signaling in Molar Cusp Formation. Gene Expr. Patterns 32, 67–71. doi:10.1016/j.gep.2019.04.002
Migliorini, E., Guevara-Garcia, A., Albiges-Rizo, C., and Picart, C. (2020). Learning from BMPs and Their Biophysical Extracellular Matrix Microenvironment for Biomaterial Design. Bone 141, 115540. doi:10.1016/j.bone.2020.115540
Mirzadeh Azad, F., Polignano, I. L., Proserpio, V., and Oliviero, S. (2021). Long Noncoding RNAs in Human Stemness and Differentiation. Trends Cell Biology 31, 542–555. doi:10.1016/j.tcb.2021.02.002
Miyazono, K., Maeda, S., and Imamura, T. (2005). BMP Receptor Signaling: Transcriptional Targets, Regulation of Signals, and Signaling Cross-Talk. Cytokine Growth Factor. Rev. 16, 251–263. doi:10.1016/j.cytogfr.2005.01.009
Murayama, K., Kato-Murayama, M., Itoh, Y., Miyazono, K., Miyazawa, K., and Shirouzu, M. (2020). Structural Basis for Inhibitory Effects of Smad7 on TGF-β Family Signaling. J. Struct. Biol. 212, 107661. doi:10.1016/j.jsb.2020.107661
Nadiri, A., Kuchler-Bopp, S., Haikel, Y., and Lesot, H. (2004). Immunolocalization of BMP-2/-4, FGF-4, and WNT10b in the Developing Mouse First Lower Molar. J. Histochem. Cytochem. 52, 103–112. doi:10.1177/002215540405200110
Neves, V. C. M., and Sharpe, P. T. (2018). Regulation of Reactionary Dentine Formation. J. Dent Res. 97, 416–422. doi:10.1177/0022034517743431
Niki, Y., Kobayashi, Y., and Moriyama, K. (2021). Expression Pattern of Transcriptional Enhanced Associate Domain Family Member 1 (Tead1) in Developing Mouse Molar Tooth. Gene Expr. Patterns 40, 119182. doi:10.1016/j.gep.2021.119182
Nottmeier, C., Liao, N., Simon, A., Decker, M. G., Luther, J., Schweizer, M., et al. (2021). Wnt1 Promotes Cementum and Alveolar Bone Growth in a Time-dependent Manner. J. Dent Res. 100, 1501–1509. doi:10.1177/00220345211012386
O’Connell, D. J., Ho, J. W. K., Mammoto, T., Turbe-Doan, A., O’Connell, J. T., Haseley, P. S., et al. (2012). A Wnt-Bmp Feedback Circuit Controls Intertissue Signaling Dynamics in Tooth Organogenesis. Sci. Signal. 5, ra4. doi:10.1126/scisignal.2002414
Oh, S. H., Hwang, Y. C., Yang, H., Kang, J. H., Hur, S. W., Jung, N. R., et al. (2012). SHP Is Involved in BMP2-Induced Odontoblast Differentiation. J. Dent Res. 91, 1124–1129. doi:10.1177/0022034512461916
Ou, M., Zhao, Y., Zhang, F., and Huang, X. (2014). Bmp2 and Bmp4 Accelerate Alveolar Bone Development. Connect. Tissue Res. 56, 204–211. doi:10.3109/03008207.2014.996701
Pandur, P., Läsche, M., Eisenberg, L. M., and Kühl, M. (2002). Wnt-11 Activation of a Non-canonical Wnt Signalling Pathway Is Required for Cardiogenesis. Nature 418, 636–641. doi:10.1038/nature00921
Piccolo, S., Dupont, S., and Cordenonsi, M. (2014). The Biology of YAP/TAZ: Hippo Signaling and beyond. Physiol. Rev. 94, 1287–1312. doi:10.1152/physrev.00005.2014
Ponting, C. P., Oliver, P. L., and Reik, W. (2009). Evolution and Functions of Long Noncoding RNAs. Cell 136, 629–641. doi:10.1016/j.cell.2009.02.006
Qin, W., Lin, Z. M., Deng, R., Li, D. D., Song, Z., Tian, Y. G., et al. (2012a). p38a MAPK Is Involved in BMP-2-Induced Odontoblastic Differentiation of Human Dental Pulp Cells. Int. Endod. J. 45, 224–233. doi:10.1111/j.1365-2591.2011.01965.x
Qin, W., Liu, P., Zhang, R., Huang, S., Gao, X., Song, Z., et al. (2014). JNK MAPK Is Involved in BMP-2-Induced Odontoblastic Differentiation of Human Dental Pulp Cells. Connect. Tissue Res. 55, 217–224. doi:10.3109/03008207.2014.882331
Qin, W., Yang, F., Deng, R., Li, D., Song, Z., Tian, Y., et al. (2012b). Smad 1/5 Is Involved in Bone Morphogenetic Protein-2-Induced Odontoblastic Differentiation in Human Dental Pulp Cells. J. endodontics 38, 66–71. doi:10.1016/j.joen.2011.09.025
Qin, X., Jiang, Q., Miyazaki, T., and Komori, T. (2019). Runx2 Regulates Cranial Suture Closure by Inducing Hedgehog, Fgf, Wnt and Pthlh Signaling Pathway Gene Expressions in Suture Mesenchymal Cells. Hum. Mol. Genet. 28, 896–911. doi:10.1093/hmg/ddy386
Quinn, J. J., and Chang, H. Y. (2016). Unique Features of Long Non-coding RNA Biogenesis and Function. Nat. Rev. Genet. 17, 47–62. doi:10.1038/nrg.2015.10
Saito, E., Saito, A., Kato, H., Shibukawa, Y., Inoue, S., Yuge, F., et al. (2016). A Novel Regenerative Technique Combining Bone Morphogenetic Protein-2 with Fibroblast Growth Factor-2 for Circumferential Defects in Dog Incisors. J. Periodontol. 87, 1067–1074. doi:10.1902/jop.2016.150746
Salazar, V. S., Gamer, L. W., and Rosen, V. (2016). BMP Signalling in Skeletal Development, Disease and Repair. Nat. Rev. Endocrinol. 12, 203–221. doi:10.1038/nrendo.2016.12
Schlange, T., Andrée, B., Arnold, H.-H., and Brand, T. (2000). BMP2 Is Required for Early Heart Development during a Distinct Time Period. Mech. Dev. 91, 259–270. doi:10.1016/s0925-4773(99)00311-1
Sepponen, K., Lundin, K., Knuus, K., Väyrynen, P., Raivio, T., Tapanainen, J. S., et al. (2017). The Role of Sequential BMP Signaling in Directing Human Embryonic Stem Cells to Bipotential Gonadal Cells. J. Clin. Endocrinol. Metab. 102, 4303–4314. doi:10.1210/jc.2017-01469
Shi, Y., and Massagué, J. (2003). Mechanisms of TGF-β Signaling from Cell Membrane to the Nucleus. Cell 113, 685–700. doi:10.1016/s0092-8674(03)00432-x
Statello, L., Guo, C.-J., Chen, L.-L., and Huarte, M. (2021). Gene Regulation by Long Non-coding RNAs and its Biological Functions. Nat. Rev. Mol. Cel Biol 22, 96–118. doi:10.1038/s41580-020-00315-9
Sternberg, H., Kidd, J., Murai, J. T., Jiang, J., Rinon, A., Erickson, I. E., et al. (2013). Seven Diverse Human Embryonic Stem Cell-Derived Chondrogenic Clonal Embryonic Progenitor Cell Lines Display Site-specific Cell Fates. Regenerative Med. 8, 125–144. doi:10.2217/rme.12.117
Touat‐Todeschini, L., Shichino, Y., Dangin, M., Thierry‐Mieg, N., Gilquin, B., Hiriart, E., et al. (2017). Selective Termination of Lnc RNA Transcription Promotes Heterochromatin Silencing and Cell Differentiation. Embo J. 36, 2626–2641. doi:10.15252/embj.201796571
Tsuboi, T., Mizutani, S., Nakano, M., Hirukawa, K., and Togari, A. (2003). Fgf-2 Regulates Enamel and Dentine Formation in Mouse Tooth Germ. Calcified Tissue Int. 73, 496–501. doi:10.1007/s00223-002-4070-2
Vijaykumar, A., Root, S. H., and Mina, M. (2020). Wnt/β-Catenin Signaling Promotes the Formation of Preodontoblasts In Vitro. J. Dent Res. 100, 387–396. doi:10.1177/0022034520967353
Wang, F., Li, Y., Wu, X., Yang, M., Cong, W., Fan, Z., et al. (2017a). Transcriptome Analysis of Coding and Long Non-coding RNAs Highlights the Regulatory Network of cascade Initiation of Permanent Molars in Miniature Pigs. BMC Genomics 18, 148. doi:10.1186/s12864-017-3546-4
Wang, J., and Martin, J. F. (2017). Hippo Pathway: An Emerging Regulator of Craniofacial and Dental Development. J. Dent Res. 96, 1229–1237. doi:10.1177/0022034517719886
Wang, J., Samuels, D., Zhao, S., Xiang, Y., Zhao, Y.-Y., and Guo, Y. (2017b). Current Research on Non-coding Ribonucleic Acid (RNA). Genes 8, 366. doi:10.3390/genes8120366
Wang, Y., Li, L., Zheng, Y., Yuan, G., Yang, G., He, F., et al. (2012). BMP Activity Is Required for Tooth Development from the Lamina to Bud Stage. J. Dent Res. 91, 690–695. doi:10.1177/0022034512448660
Wang, Z., Huang, Y., and Tan, L. (2020). Downregulation of lncRNA DANCR Promotes Osteogenic Differentiation of Periodontal Ligament Stem Cells. BMC Dev. Biol. 20, 2. doi:10.1186/s12861-019-0206-8
Washio, A., Kitamura, C., Morotomi, T., Terashita, M., and Nishihara, T. (20122012). Possible Involvement of Smad Signaling Pathways in Induction of Odontoblastic Properties in KN-3 Cells by Bone Morphogenetic Protein-2: A Growth Factor to Induce Dentin Regeneration. Int. J. dentistry 2012, 1–6. doi:10.1155/2012/258469
Wu, L. A., Feng, J., Wang, L., Mu, Y.-d., Baker, A., Donly, K. J., et al. (2010). Immortalized Mouse Floxed Bmp2 Dental Papilla Mesenchymal Cell Lines Preserve Odontoblastic Phenotype and Respond to BMP2. J. Cel. Physiol. 225, 132–139. doi:10.1002/jcp.22204
Wu, L., Wang, F., Donly, K. J., Wan, C., Luo, D., Harris, S. E., et al. (2015). Establishment of Immortalized Mouse Bmp2 Knock-Out Dental Papilla Mesenchymal Cells Necessary for Study of Odontoblastic Differentiation and Odontogenesis. J. Cel. Physiol. 230, 2588–2595. doi:10.1002/jcp.25061
Wu, M., Chen, G., and Li, Y.-P. (2016). TGF-β and BMP Signaling in Osteoblast, Skeletal Development, and Bone Formation, Homeostasis and Disease. Bone Res. 4, 16009. doi:10.1038/boneres.2016.9
Wu, T., and Du, Y. (2017). LncRNAs: From Basic Research to Medical Application. Int. J. Biol. Sci. 13, 295–307. doi:10.7150/ijbs.16968
Wu, X.-B., Li, Y., Schneider, A., Yu, W., Rajendren, G., Iqbal, J., et al. (2003). Impaired Osteoblastic Differentiation, Reduced Bone Formation, and Severe Osteoporosis in Noggin-Overexpressing Mice. J. Clin. Invest. 112, 924–934. doi:10.1172/jci15543
Yamashiro, T., Tummers, M., and Thesleff, I. (2003). Expression of Bone Morphogenetic Proteins and Msx Genes during Root Formation. J. Dent Res. 82, 172–176. doi:10.1177/154405910308200305
Yang, B., Sun, H., Song, F., Wu, Y., and Wang, J. (2018). Yes-associated Protein 1 Promotes the Differentiation and Mineralization of Cementoblast. J. Cel Physiol 233, 2213–2224. doi:10.1002/jcp.26089
Yang, G., Yuan, G., MacDougall, M., Zhi, C., and Chen, S. (2017). BMP-2 Induced Dspp Transcription Is Mediated by Dlx3/Osx Signaling Pathway in Odontoblasts. Sci. Rep. 7, 10775. doi:10.1038/s41598-017-10908-8
Yang, G., Lu, X., and Yuan, L. (2014). LncRNA: a Link between RNA and Cancer. Biochim. Biophys. Acta (Bba) - Gene Regul. Mech. 1839, 1097–1109. doi:10.1016/j.bbagrm.2014.08.012
Yang, W., Harris, M. A., Cui, Y., Mishina, Y., Harris, S. E., and Gluhak-Heinrich, J. (2012). Bmp2 Is Required for Odontoblast Differentiation and Pulp Vasculogenesis. J. Dent Res. 91, 58–64. doi:10.1177/0022034511424409
Yin, Y., Yan, P., Lu, J., Song, G., Zhu, Y., Li, Z., et al. (2015). Opposing Roles for the lncRNA Haunt and its Genomic Locus in Regulating HOXA Gene Activation during Embryonic Stem Cell Differentiation. Cell stem cell 16, 504–516. doi:10.1016/j.stem.2015.03.007
Yuan, X., Liu, M., Cao, X., and Yang, S. (2019). Ciliary IFT80 Regulates Dental Pulp Stem Cells Differentiation by FGF/FGFR1 and Hh/BMP2 Signaling. Int. J. Biol. Sci. 15, 2087–2099. doi:10.7150/ijbs.27231
Zhang, F., Song, J., Zhang, H., Huang, E., Song, D., Tollemar, V., et al. (2016). Wnt and BMP Signaling Crosstalk in Regulating Dental Stem Cells: Implications in Dental Tissue Engineering. Genes Dis. 3, 263–276. doi:10.1016/j.gendis.2016.09.004
Zhang, H., and Bradley, A. (1996). Mice Deficient for BMP2 Are Nonviable and Have Defects in Amnion/chorion and Cardiac Development. Development 122, 2977–2986. doi:10.1242/dev.122.10.2977
Zhang, X., Zhao, J., Li, C., Gao, S., Qiu, C., Liu, P., et al. (2001). DSPP Mutation in Dentinogenesis Imperfecta Shields Type II. Nat. Genet. 27 (2), 151–152. doi:10.1038/84765
Zhao, L., Wang, J., Li, Y., Song, T., Wu, Y., Fang, S., et al. (2021). NONCODEV6: an Updated Database Dedicated to Long Non-coding RNA Annotation in Both Animals and Plants. Nucleic Acids Res. 49, D165–d171. doi:10.1093/nar/gkaa1046
Zheng, Y., and Jia, L. (2016). Long Noncoding RNAs Related to the Odontogenic Potential of Dental Mesenchymal Cells in Mice. Arch. Oral Biol. 67, 1–8. doi:10.1016/j.archoralbio.2016.03.001
Keywords: bone morphogenetic protein 2, dental mesenchymal papilla cells, coding and non-coding RNAs, bioinformatics, signal pathways
Citation: Wang F, Tao R, Zhao L, Hao X-H, Zou Y, Lin Q, Liu MM, Goldman G, Luo D and Chen S (2021) Differential lncRNA/mRNA Expression Profiling and Functional Network Analyses in Bmp2 Deletion of Mouse Dental Papilla Cells. Front. Genet. 12:702540. doi: 10.3389/fgene.2021.702540
Received: 29 April 2021; Accepted: 29 November 2021;
Published: 22 December 2021.
Edited by:
Liborio Stuppia, University of Studies G. d’Annunzio Chieti and Pescara, ItalyCopyright © 2021 Wang, Tao, Zhao, Hao, Zou, Lin, Liu, Goldman, Luo and Chen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shuo Chen, Y2hlbnMwQHV0aHNjc2EuZWR1; Daoshu Luo, bHVvZHMyMDA0QDE2My5jb20=
†These authors have contributed equally to this work and share first authorship
 Feng Wang
Feng Wang Ran Tao
Ran Tao Li Zhao1
Li Zhao1 Daoshu Luo
Daoshu Luo Shuo Chen
Shuo Chen