
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Genet., 21 July 2021
Sec. Livestock Genomics
Volume 12 - 2021 | https://doi.org/10.3389/fgene.2021.697883
This article is part of the Research TopicGene Regulation Explored by Systems Biology in Livestock ScienceView all 13 articles
Deoxynivalenol (DON) caused serious cytotoxicity for animal cells. However, genes involved in regulating DON toxicity and the underlying molecular mechanisms remain largely unknown. This study explored the role of SLC4A11 and MFSD3 in alleviating DON toxicity and analyzed the DNA methylation changes of these two genes. Viability and cell cycle analysis showed that DON exposure decreased the IPEC-J2 viability (P < 0.01), blocked the cell cycle in the G2/M phase (P < 0.01), and increased the rate of apoptosis (P < 0.05). Expression of the SLC4A11 and MFSD3 genes was significantly downregulated upon DON exposure (P < 0.01). Overexpression of SLC4A11 and MFSD3 can enhance the cell viability (P < 0.01). DNA methylation assays indicated that promoter methylation of SLC4A11 (mC-1 and mC-23) and MFSD3 (mC-1 and mC-12) were significantly higher compared with those in the controls and correlated negatively with mRNA expression (P < 0.05). Further analysis showed that mC-1 of SLC4A11 and MFSD3 was located in transcription factor binding sites for NF-1 and Sp1. Our findings revealed the novel biological functions of porcine SLC4A11 and MFSD3 genes in regulating the cytotoxic effects induced by DON, and may contribute to the detection of biomarkers and drug targets for predicting and eliminating the potential toxicity of DON.
Deoxynivalenol (DON), belonging to the family of trichothecenes, is one of the metabolites of Fusarium, induces emesis in swine, leading to it being known as “vomitoxin” (Pestka and Smolinski, 2005; Pestka, 2010; Borutova et al., 2012). DON is extremely harmful to pigs, and its main toxic effects are antifeeding, vomiting, decreased immunity, necrosis of the digestive tract, decreased reproductive performance, and finally, death (Eriksen and Pettersson, 2004). Early research suggested that DON induced the apoptosis of piglet mesenteric lymph node cells, stimulated immune cells to secrete cytokines, and resulted in intestinal inflammation by destroying the intestinal barrier and inducing inflammation (Pestka, 2010; Gerez et al., 2015). Once the intestinal barrier is damaged, in addition to affecting the digestion and absorption of pigs, it is likely to cause other intestinal diseases, which will seriously affect the growth and development of pigs, resulting in huge economic losses to the pig industry. Therefore, it is necessary to determine the toxic effects and mechanism of DON, which will contribute to discovering new methods of prevention and treatment of diseases induced by DON.
Epigenetic modification is a type of gene regulation that is ubiquitous in all organisms, and is essential to maintain the normal life activities of mammals (Shi and Wu, 2009). Epigenetic modification includes DNA methylation, histone modification, and non-coding RNA regulation, among which DNA methylation has attracted much attention from epigenetic modification researchers (Dahl and Guldberg, 2003). DNA methylation regulates gene expression by affecting chromatin structure, DNA conformation, chromosome stability, and DNA-protein interaction. In eukaryotes, it plays an important role in cell differentiation, embryonic development, environmental adaptation, and the development of diseases (Deng, 2014; Dai et al., 2019). After DON exposure, the levels of methyltransferase H3K4me2 increased significantly on EZH2, encoding enhancer of Zeste 2 polycomb repressive complex 2 Subunit, a key regulator in the early development of oocytes. Thus, DON can cause the interruption of oocyte maturation by changing epigenetic modifications (Han et al., 2016). Studies have also reported increased DNA methylation levels and the expression of H3K9me3 and H4K20me3 in the oocytes of mice fed a diet containing DON, and that the expression levels of H3K27me3 and H4K20me2 were reduced, revealing that epigenetic modifications might be one of the reasons for the decrease of the level of oocyte development (Zhu et al., 2014). These studies showed that epigenetic modification plays an important regulatory role in the process of DON induction.
Our previous transcriptome analysis in IPEC-J2 cells found significant decreases in the expression levels of the SLC4A11 (encoding solute carrier family 4 member 11) and MFSD3 (encoding major facilitator superfamily domain containing 3) genes in response to DON (Wang et al., 2019). SLC4A11 and MFSD3 are both membrane-bound solute carriers (SLCs), which maintain nutrient uptake, ion transport, and waste removal associated with physiological functions (Perland et al., 2017). SLC4A11 is an electrogenic Na/borate cotransporter that stimulates cell growth and proliferation by increasing intracellular borate levels and activating the mitogen activated protein kinase (MAPK) pathway (Jiao et al., 2007; Lopez et al., 2009). MFSD3 is a kind of membrane-bound solute carrier that belongs to the major facilitator superfamily (MFS), which is the largest phylogenetic group of SLCs in humans (Nicoletti et al., 2019). Studies reported that the expression of MFSD3 was associated with nutrient intake and adipose tissue homeostasis (Hoglund et al., 2011). Therefore, SLC4A11 and MFSD3 may play an important role in DON-induced cell damage, we further explored the expression regulation mechanism of SLC4A11 and MFSD3 genes associated with the activity of IPEC-J2 cells induced by DON. We examined the effects of DON on the viability, cell cycle, and apoptosis of IPEC-J2 cells, as well as the regulation of SLC4A11 and MFSD3 expression levels in IPEC-J2 cells induced by DON, including a comprehensive analysis of the degree of methylation and expression changes of these two genes. The present study explored the regulatory role of SLC4A11 and MFSD3 in resisting DON-induced cytotoxicity. Better understanding of DON pathogenesis and identification of the responsive genes provided the theoretical basis for further study of the molecular regulation mechanism of DNA methylation modification in DON-induced cytotoxicity, and may contribute to the identification of biomarkers and drug targets for DON contamination.
The animal study proposal was approved by the Institutional Animal Care and Use Committee (IACUC) of the Yangzhou University Animal Experiments Ethics Committee [permit number: SYXK (Su) IACUC 2012-0029]. All experimental methods were conducted in accordance with the related guidelines and regulations.
The IPEC-J2 cells were preserved in our laboratory, and cultured in Dulbecco’s modified Eagle’s medium (DMEM) containing 10% fetal bovine serum (FBS) and 1% penicillin streptomycin (1 mg/mL) at 37°C with 5% CO2.
IPEC-J2 cells were cultivated in 96-well plates at a density of 2 × 103 cells/well and cultured for 24 h. Based on a previous study, (Wang et al., 2019) we could see that treatment with a DON (Sigma, Germany) concentration of 1 μg/ml for 48 h induces cytotoxicity in IPEC-J2 cells. When the cells reached 70–80% confluence, they were incubated with DON (1 μg/mL) for 24, 48, and 72 h. Cell viability was assessed using a Cell Counting Kit-8 (MedChemExpress, Monmouth Junction, NJ, United States) according to the manufacturer’s protocol. The absorbance was measured on a Tecan Infinite Pro (Sunrise, Tecan, Switzerland) at 450 nm.
IPEC-J2 cells were seeded into six-well plates at a density of 2 × 105 cells/well and randomly assigned into a control group and a DON treated group. When the cells reached 70–80% confluence, they were incubated with DON (1 μg/mL) for 48 h in the DON treated group. Subsequently, cells were collected, and stained with Annexin V-FITC according to the instructions of the Apoptosis Detection kit (Solarbio, Beijing, China). Finally, apoptosis was analyzed using a Flow Cytometer (FAC Scan, Becton Dickinson, Franklin Lakes, NJ, United States) within 1 h.
First, IPEC-J2 cells were cultured in a six-well plates and incubated at 37°C with 5% CO2 overnight and divided into a control group and a DON treated group. Then, digestion was performed with trypsin without EDTA followed by washing twice in pre-cooled phosphate-buffered saline. Cell cycle analysis was performed according to a Cell Cycle and Apoptosis Analysis Kit (Beyotime Institute of Biotechnology, Jiangsu, China) and the percentage of the cell population at a particular phase was estimated using ModFit LT for Windows V3.1 (Verity Software House, Topsham, ME, United States).
Total RNA was extracted from IPEC-J2 cells using Trizol. cDNA was reverse transcribed from the mRNA was reverse transcribed using a HiScript II Q RT SuperMix kit (Vazyme Biotech Co., Ltd., Nanjing, China). The cDNA was used as the template for the quantitative real-time PCR (qPCR) step using a fast real-time PCR system (ABI Step One Plus; Applied Biosystems, Foster City, CA, United States). GAPDH (encoding glyceraldehyde-3-phosphate dehydrogenase) was used as the control gene. The relative expression levels were determined using the 2–ΔΔCT method (Livak and Schmittgen, 2001) and overexpression efficiency was also calculated using the 2–ΔΔCT method. The primers used are shown in Table 1. All primers were synthesized by Sangon (Shanghai, China).
Primers with SacI and PmeI restriction enzyme sites were designed according to the coding sequences (CDS) of porcine SLC4A11 (XM_021077564.1) and MFSD3 (XM_021090521.1) gene published in the GenBank database1 (Table 1). The primers were used to amplify the SLC4A11 and MFSD3 genes, which were ligated into the florescence expression vector, pEGFP–C1, after double digestion. The sequences of the SLC4A11 and MFSD3 overexpression vectors were confirmed by Sangon sequencing. Cells were incubated with 20 nM SLC4A11 and MFSD3 overexpression vector or negative control using Jet PRIME (Polyplus, Illkirch, France). After transfection, stable cells lines were established by incubation with geneticin (800 μg/mL) (Invivogen, Toulouse, France) for 15 days.
Analysis and identification of the promoter region, CpG islands and transcription factor binding sites in the 5′ flanking regions of SLC4A11 and MFSD3 were performed using the online tool MethPrimer and Alibaba (Li and Dahiya, 2002). Based on this analysis and the location of the predicted CpG islands, bisulfite-sequencing PCR (BSP) primers were designed using MethPrimer (Table 1).
Genomic DNA was extracted from IPEC-J2 cells using a Universal DNA purification kit (Tiangen, Beijing, China), following the manufacturer’s instructions. The genomic DNA was treated with bisulfite and used as the template for PCR (ZymoTaq PreMix; Zymo Research, Irvine, CA, United States). Purified PCR products were ligated into vector PMD-19T overnight (Bao Biological Engineering Co., Dalian, China). The recombinant clones were transformed into competent Escherichia coli DH5α cells (Tiangen, Beijing, China). 200 μL of the bacteria solution were applied to Luria-Bertani (LB) agar plates containing 100 ng/mL ampicillin, and maintained at 37°C overnight. The positive recombinant clones were selected on LB-ampicillin and confirmed by sequencing (Sangon Shanghai, China). The raw SLC4A11 and MFSD3 mRNA sequences were aligned using QUMA online software to analyze the degree of methylation at each CpG site. The methylation rate for each sample was calculated as the number of CpG methylated loci/the number of CpG loci.
One-way analysis of variance and Student’s t-test were used for comparisons between the control and DON treated groups. The association between an individual CG methylation and the expression levels of genes after DON-induced cytotoxicity in IPEC-J2 cells was analyzed using Pearson’s correlation analysis. All statistical analyses were performed using the SPSS 23.0 (IBM Corp., Armonk, NY, United States) or GraphPad Prism version 8.0 for Windows (GraphPad Software Inc., San Diego, CA, United States). P < 0.05 was considered statistically significant and all experimental samples had three replicates.
To investigate the effect of DON on the viability of IPEC-J2 cells, the cells were incubated with DON (1 μg/mL) for 24, 48, and 72 h. Cell viability was significantly lower compared with that of the control group at 24, 48, and 72 h (P < 0.01), indicating that DON induced increased cell death with time (Figure 1A). Flow Cytometry analysis also showed that DON increased the percentage of early, late, and total apoptotic cells significantly compared with that in the control group following 48 h of incubation (Figures 1B,C). In subsequent experiments, the cell samples were treated with DON for 48 h. Flow cytometry analysis of the cell cycle showed that after 48 h of DON treatment the cell cycle was arrested at the G2/M phase compared with the controls (P < 0.01), and the percentage of cells in the S phase decreased (P < 0.01) (Figure 1D).
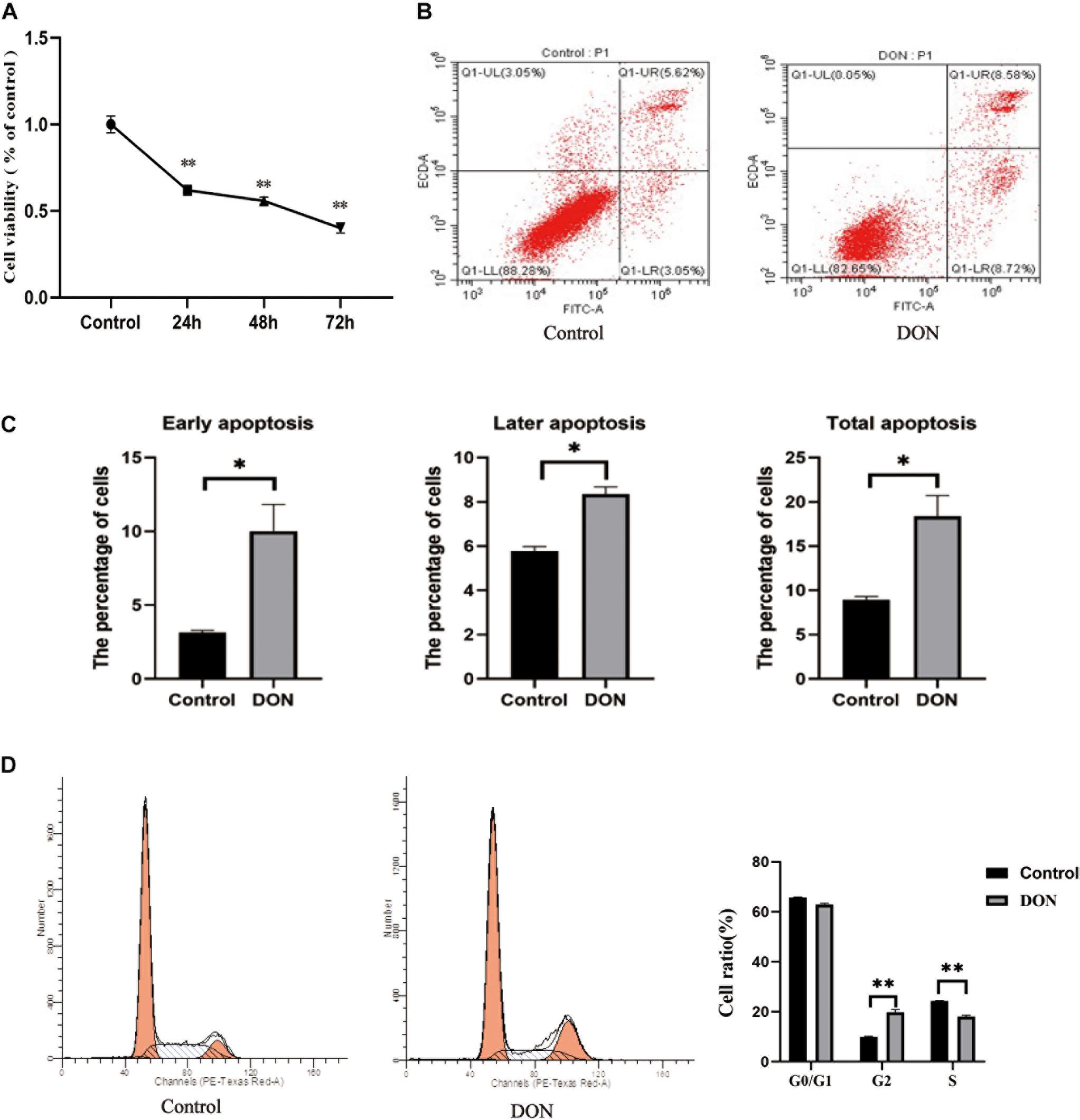
Figure 1. The viability, cell cycle, and apoptosis of IPEC-J2 cells in the control group and the DON treated group. (A) The viability of IPEC-J2 cells in the control group and the DON treated group. (B) Apoptosis of IPEC-J2 cells in the control group and the Deoxynivalenol (DON) treatment group was evaluated by measurement of Annexin V by flow cytometry. Apoptotic cells were Annexin V-positive and PI-negative. Q1-UL stands for nuclear debris, Q1-LL stands for living cells, Q1-LR stands for early apoptotic cells, and Q1-UR stands for later apoptotic cells (C) Quantification of panel (B). *P < 0.05. (D) The cell cycle distribution in the control group and the DON treated group. **P < 0.01.
The mRNA expression levels of SLC4A11 and MFSD3 were assessed using qRT-PCR in DON-treated IPEC-J2 cells. In the DON treated group, the mRNA expression levels of SLC4A11 (P < 0.01) (Figure 2A) and MFSD3 (P < 0.01) were significantly lower compared with those in the controls (Figure 2B).
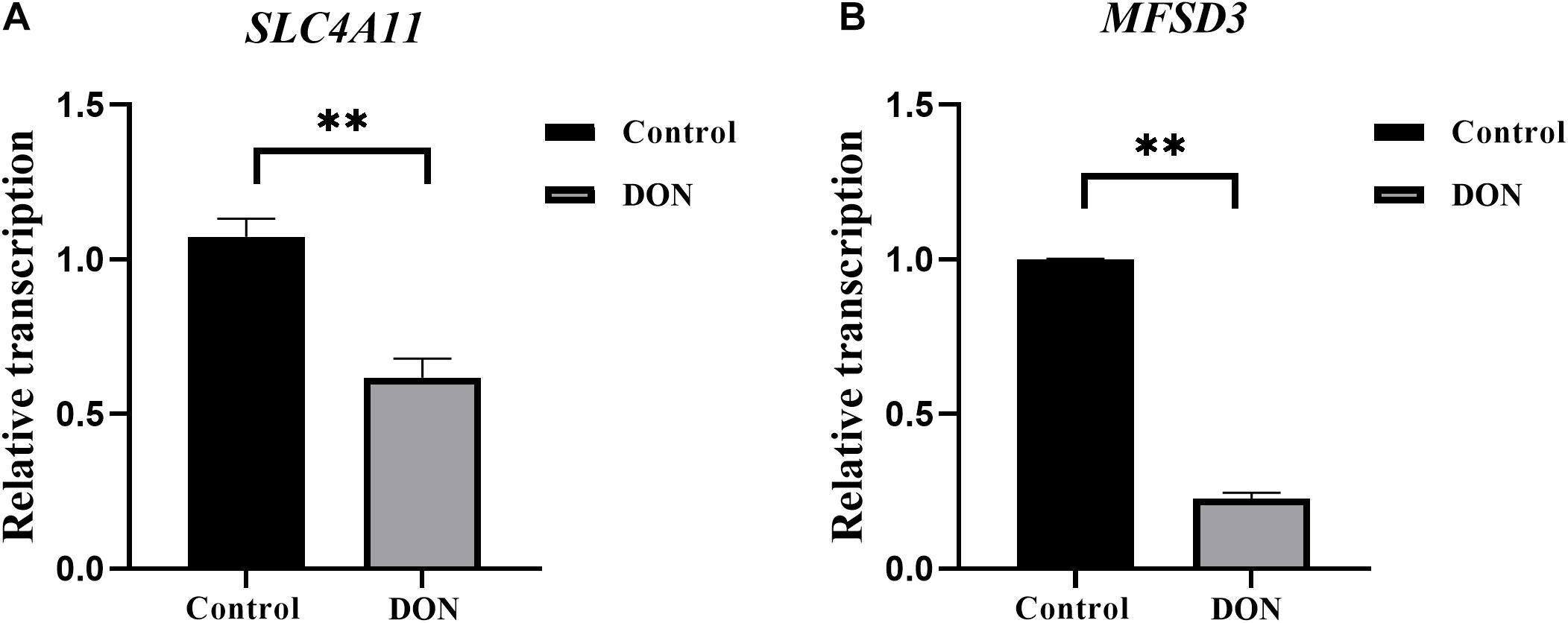
Figure 2. SLC4A11 and MFSD3 mRNA expression levels in DON-induced IPEC-J2 cells. (A) SLC4A11 mRNA expression levels induced by DON. *P < 0.05. (B) MFSD3 mRNA expression levels induced by DON. **P < 0.01.
The recombinant plasmids pEGFP-C1-SLC4A11 and pEGFP-C1-MFSD3 were verified by DNA sequencing (Supplementary Figure 1). The gene sequences of the overexpression vectors were consistent with those of the pEGFP vector, and the SLC4A11 and MFSD3 CDS, which indicated that plasmids were successfully constructed. After 15 days of screening with geneticin, green fluorescent cells could still be observed under the fluorescence microscope, which indicated that the recombinant fusion proteins of pEGFP-C1-SLC4A11 and pEGFP-C1-MFSD3 were stably expressed in IPEC-J2 cells (Supplementary Figure 2). The relative mRNA expression levels of SLC4A11 in the overexpression group represent a 585.09-fold increase compared with that in the control group (P < 0.01) (Figure 3A). The relative expression levels of the MFSD3 mRNA in the overexpression group representing a 6.68-fold increase compared with that in the control group (P < 0.01) (Figure 3B).
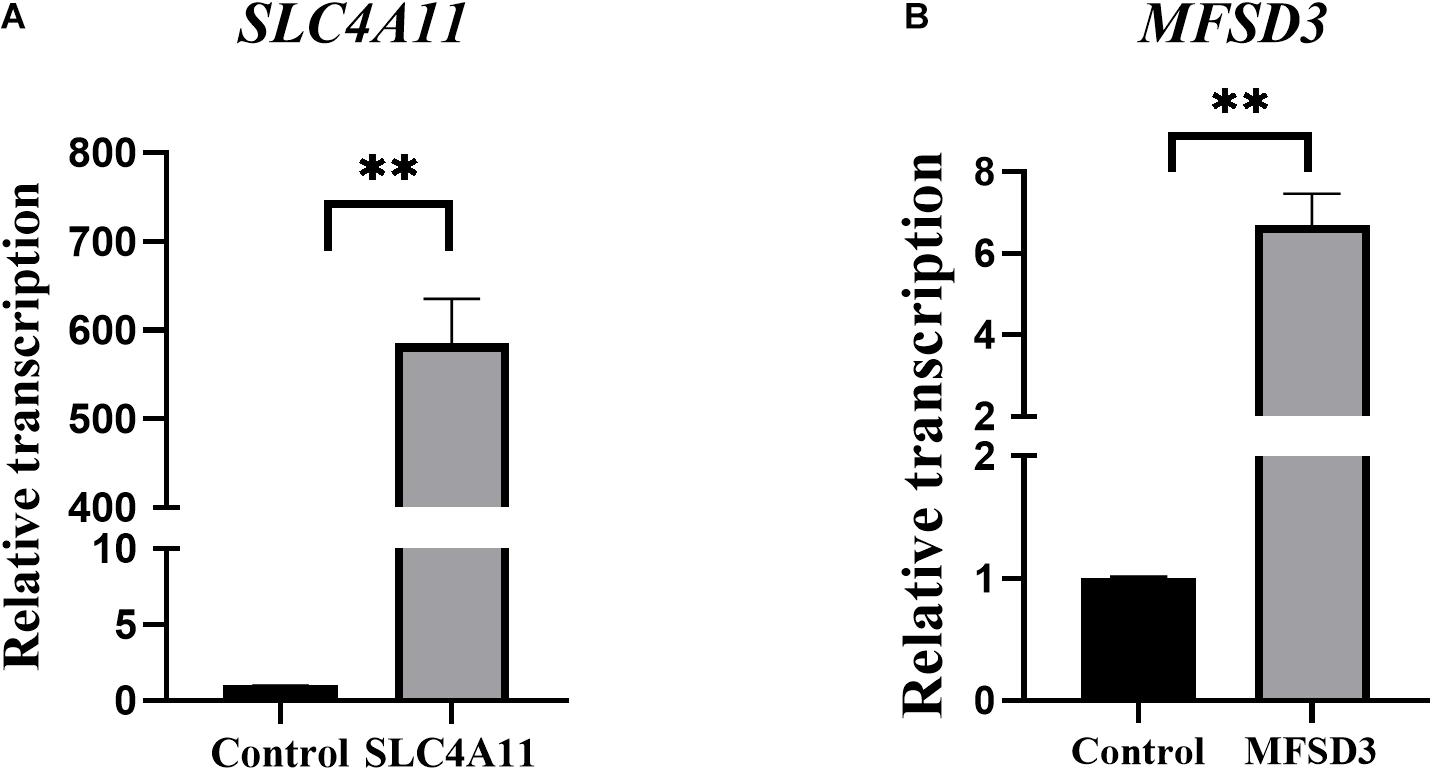
Figure 3. Construction of recombinant plasmids and cell lines overexpressing pEGFP-C1-SLC4A11, pEGFP-C1-MFSD3. (A) pEGFP-C1-SLC4A11 plasmid expression efficiency. **P < 0.01. (B) pEGFP-C1-MFSD3 plasmid expression efficiency. **P < 0.01.
SLC4A11 and MFSD3 overexpressing cells were incubated with DON (1 μg/mL) for 24, 48, and 72 h to detect cell viability. The results showed that the viability of the SLC4A11 overexpression group under DON treatment was significantly increased compared to the control group at 24, 48, and 72 h (P < 0.01) (Figure 4A). For the MFSD3 overexpression group under DON treatment, the viability was significantly lower that of the control group at 24 and 48 h (P < 0.05). There was an increasing trend at 72 h, but the changes were not significant (P > 0.05) (Figure 4B).
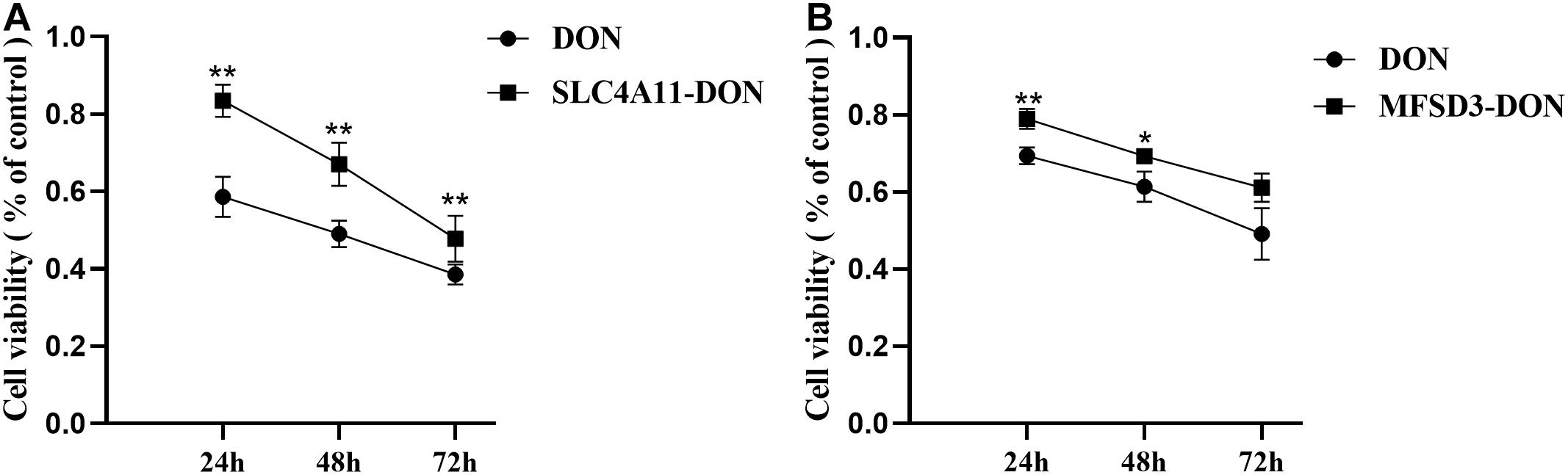
Figure 4. Effects of DON induction on the viability of SLC4A11 and MFSD3 overexpressing IPEC-J2 cell lines after 24, 48, and 72 h. (A) The cell viability of the SLC4A11 cell line and DON treated group induced by DON. **P < 0.01. (B) The cell viability of the MFSD3 cell line DON treated group. *P < 0.05. **P < 0.01.
The CpG islands in the porcine SLC4A11 and MFSD3 5′ flanking region were predicted by MethPrimer, respectively. The analysis indicated that the SLC4A11 5′ flanking region contained two CpG islands, and primers were designed to amplify a 345 bp fragment (Figure 5A). The MFSD3 5′ flanking region also contained two CpG islands, and primers were designed to amplify a 260 bp fragment (Figure 5B). After determining the CpG island regions of the porcine SLC4A11 and MFSD3 genes, we analyzed the rest of the gene sequences and found that the SLC4A11 gene contained 28 CpG sites (Figure 6A) and the MFSD3 gene contained 19 CpG sites (Figure 6D). Methylation levels of both genes in the DON group (48 h of treatment) and the control group were very high. In addition, there was no significant difference in the overall methylation degree of SLC4A11 and MFSD3 between the DON treated group and the control group (Figures 6B,E). Interestingly, the methylation levels of the mC-1 and mC-23 sites of SLC4A11 and the mC-1 and mC-12 sites of MFSD3 were significantly higher compared with those in the controls (P < 0.05) (Figures 6C,F).
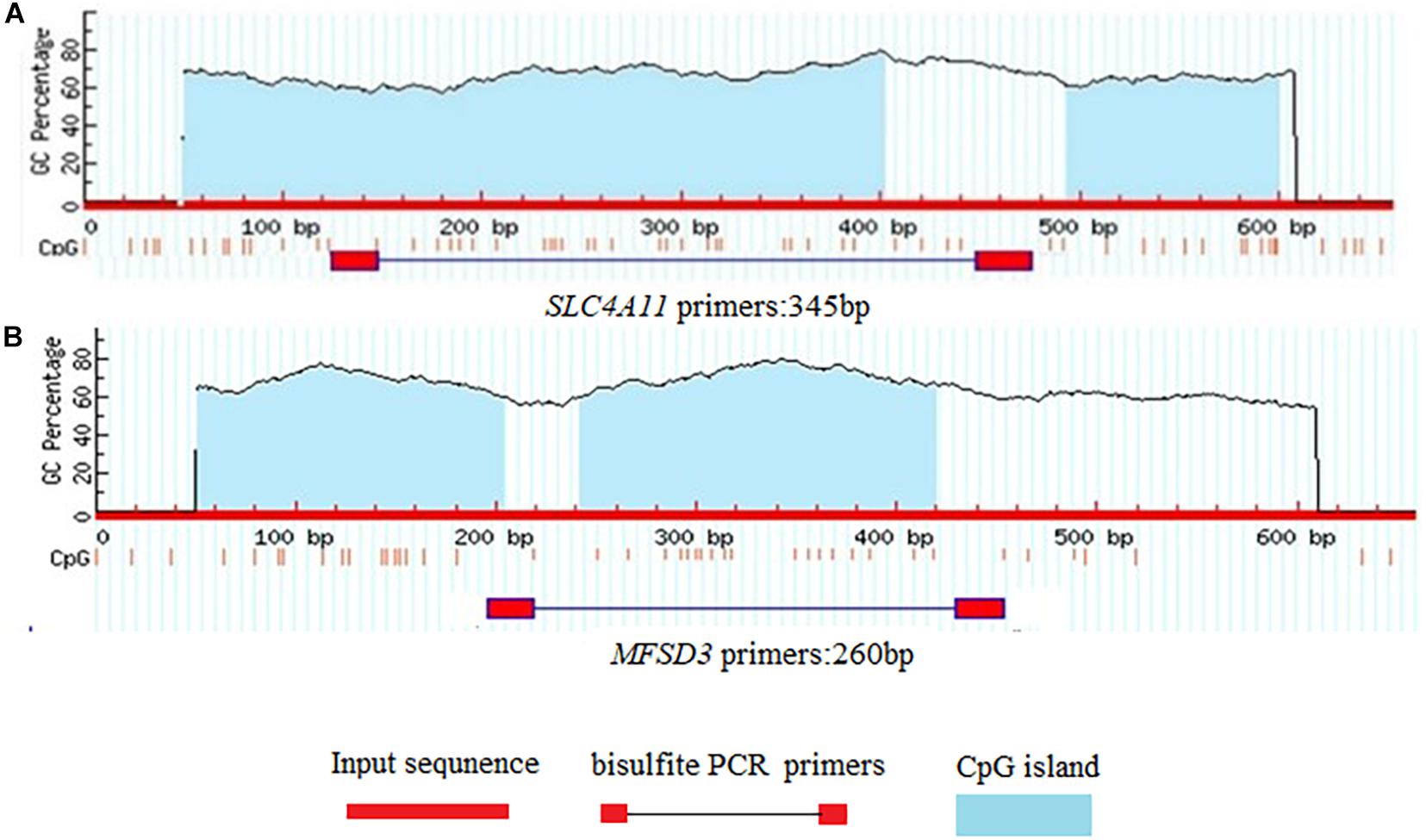
Figure 5. Prediction of CpG islands in gene promoter regions. (A) CpG islands in the SLC4A11 gene promoter region. (B) CpG islands in the MFSD3 gene promoter region.
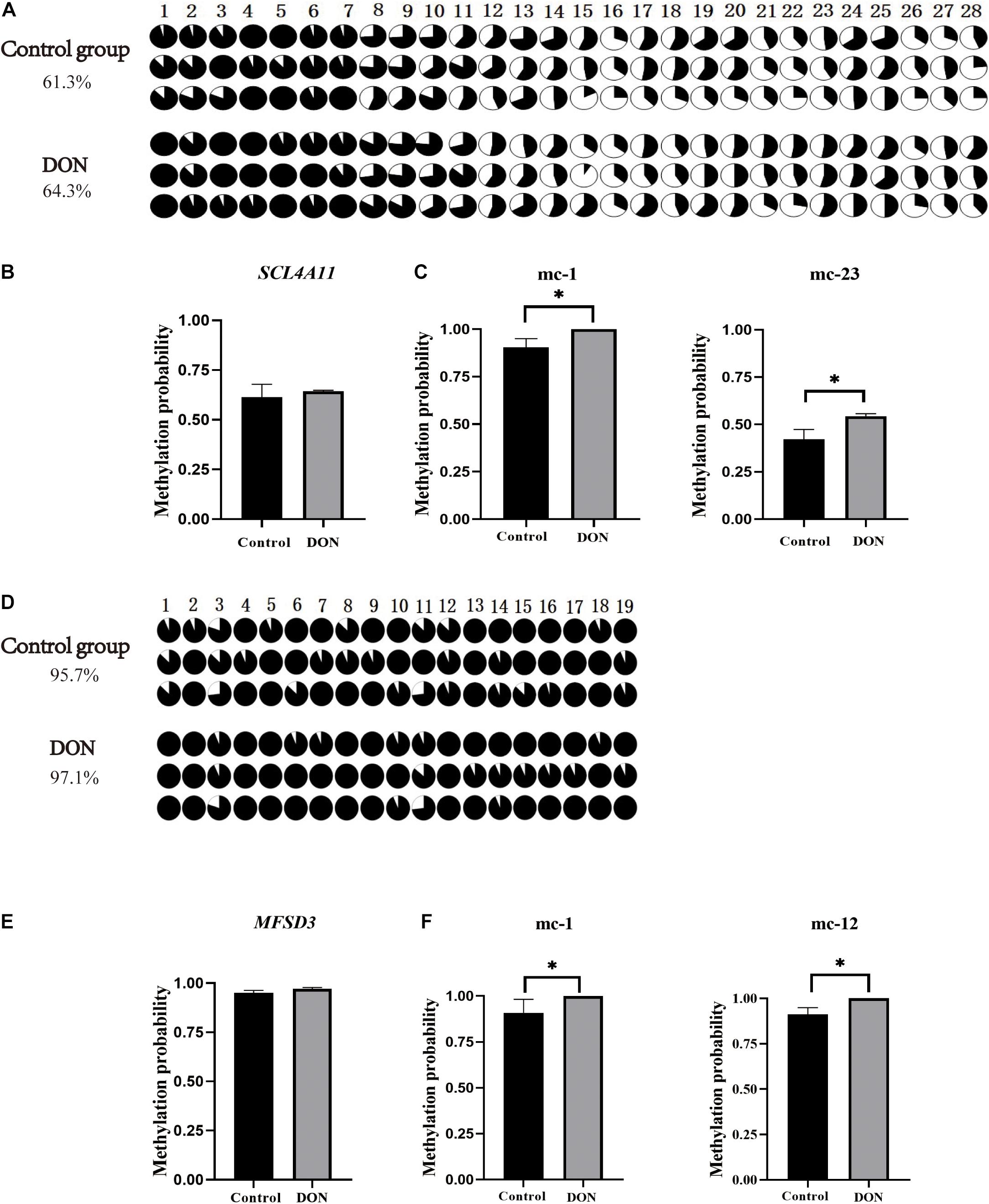
Figure 6. CpG island methylation analysis of the SLC4A11 and MFSD3 gene promoter regions. (A) The methylation level of CpG site of the SLC4A11 gene. (B) Degree of methylation of the SLC4A11 gene between the DON treated and control groups. (C) the methylation levels of the mC-1 and mC-23 sites of SLC4A11 between the DON treated and control groups. *P < 0.05. (D) The methylation level of CpG site of the MFSD3 gene. (E) Degree of methylation of the MFSD3 gene between the two groups. (F) the methylation levels of the mC-1 and mC-12 sites of MFSD3 between the DON treated and control group. *P < 0.05.
Pearson analysis was performed to examine the correlation between the methylation degree of CpG islands in the promoter regions of SLC4A11 and MFSD3 genes and the mRNA expression level. The methylation level of 19 (out of 28) CpG sites in SLC4A11 CpG islands correlated negatively with the mRNA expression level, in which the methylation levels of mC-1 and mC-23 correlated significantly and negatively with the mRNA expression level (P < 0.05) (Figure 7A). The methylation level of 11 (out of 19) CpG sites in MFSD3 correlated negatively with the mRNA expression level (Figure 7B), in which the methylation level of mC-1 and mC-12 correlated significantly and negatively with the mRNA expression level (P < 0.05).
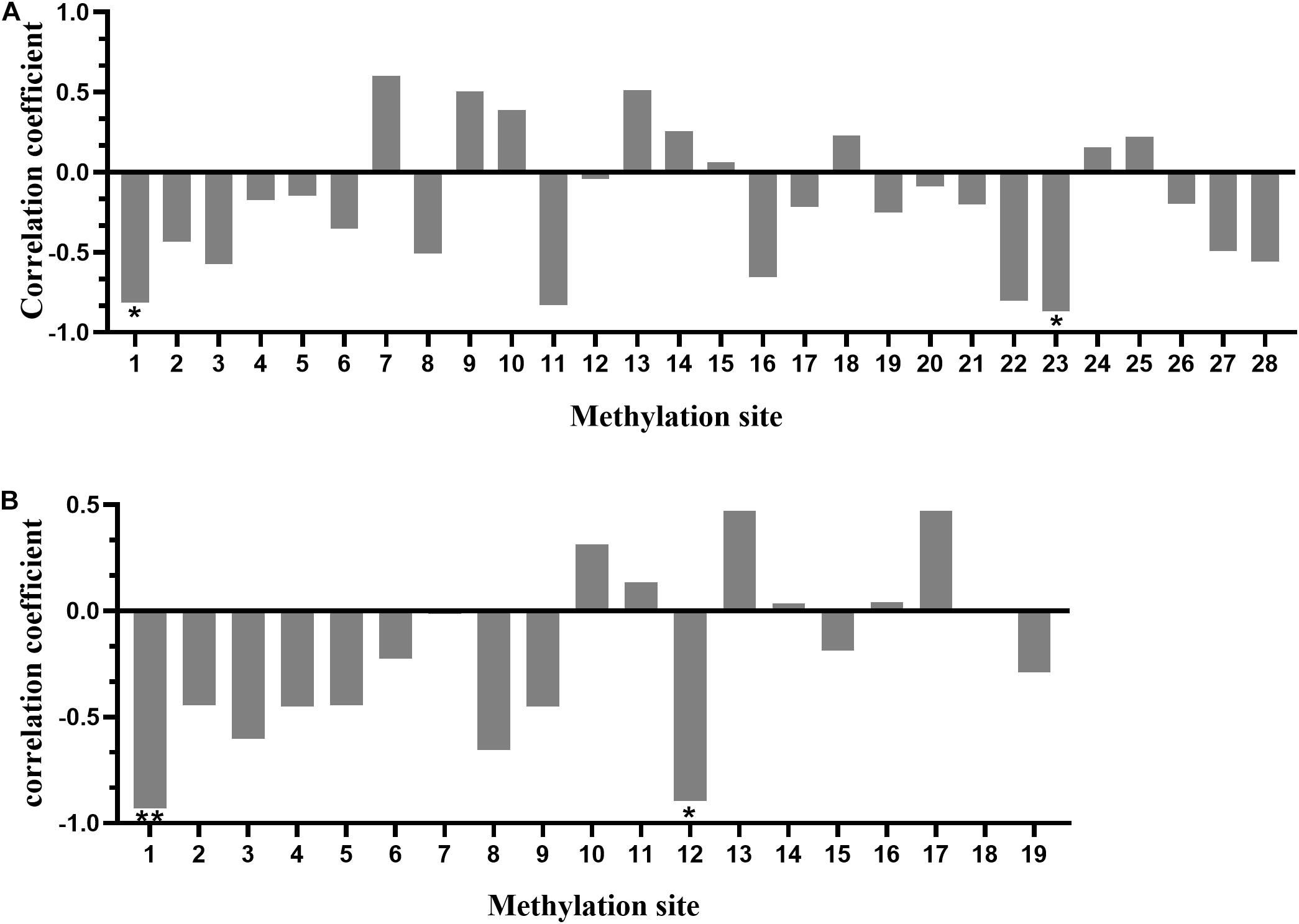
Figure 7. Correlation between the degree of methylation of CpG islands in the gene promoter region and mRNA expression. (A) Correlation between the CpG island methylation degree in the promoter region of SLC4A11 gene and mRNA expression. *P < 0.05. (B) Correlation between CpG island methylation degree in the promoter region of the MFSD3 gene and mRNA expression. *P < 0.05; **P < 0.01.
The potential transcription factor binding sites in the regions containing the CpG islands in the SLC4A11 and MFSD3 genes were predicted. The results showed that mC-1 of SLC4A11 was located in the NF-1 and Antp binding region, while mC-23 was not located in a transcription binding site (Figure 8A). The mC-1 site of MFSD3 was located in the Sp1 binding region, while mC-12 was not located in a transcription binding site (Figure 8B).
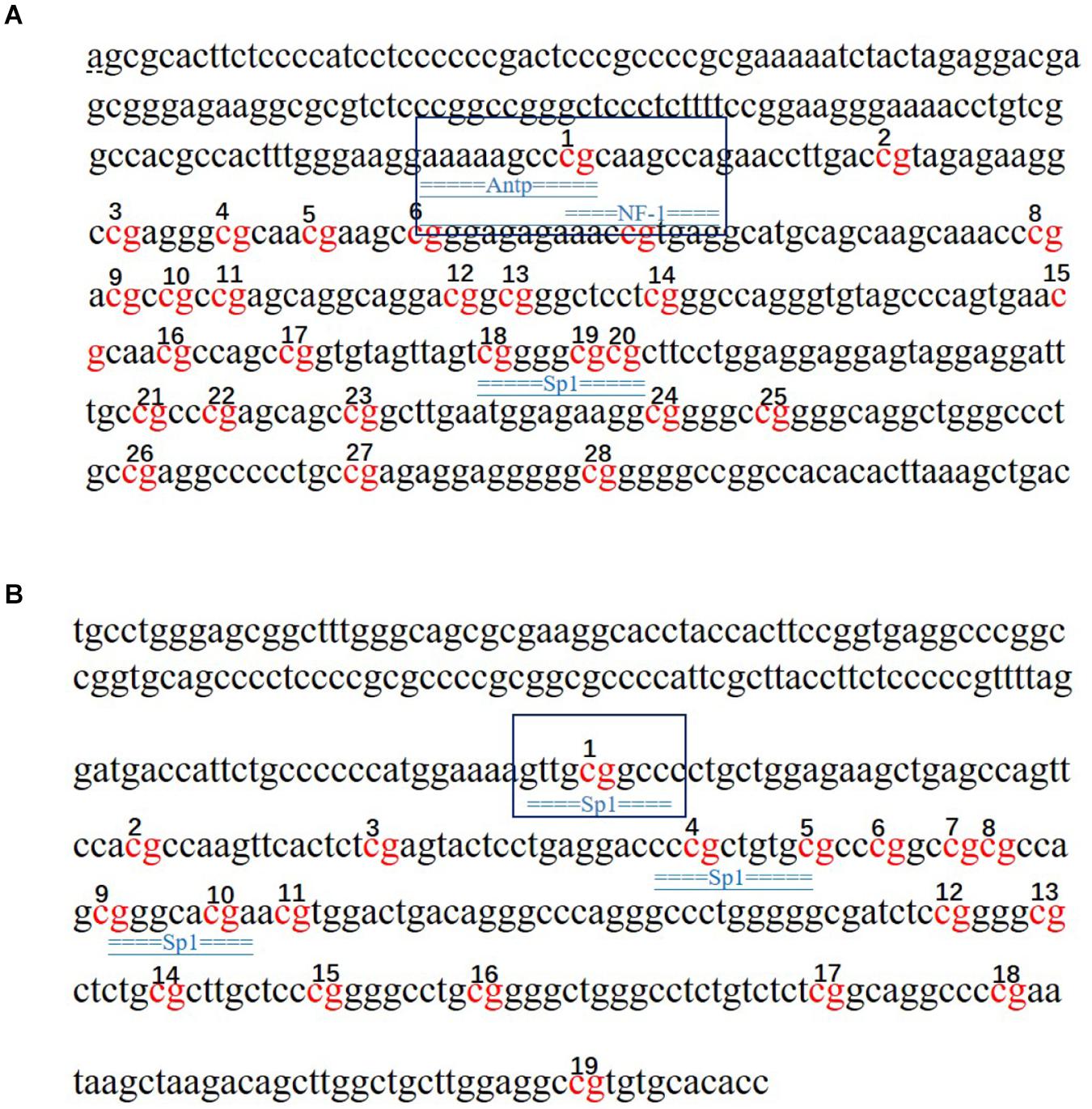
Figure 8. Prediction of transcription factors that bind to the SLC4A11 and MFSD3 genes. (A) Prediction of transcription factor binding sites in the SLC4A11 gene promoter region. (B) Prediction of transcription factor binding sites in the MFSD3 gene promoter region. The underlined regions represent the transcription factor binding sites, and the red bases are the CpG sites.
The growth and function of intestinal epithelial cells might be affected by DON through various pathological mechanisms, including the activation of cell signaling and ribosomal stress (Diesing et al., 2012). In pigs, the intestines are the main organ that absorbs DON, and most of it is absorbed in the jejunum (Diesing et al., 2011). The porcine intestinal epithelial IPEC-J2 cell line provides an ideal in vitro model system to study porcine-specific pathogenesis, which can show basic features similar to those in vivo (Goyarts and Dänicke, 2006). In this study, we examined the effects of DON on the viability, cell cycle, and apoptosis of IPEC-J2 cells. We found that DON significantly decreased cell viability, induced cell cycle arrest in the G2/M phase, and increased the apoptosis rate. Previously, it was shown that DON affects cell growth significantly by inducing IPEC-J2 cell apoptosis and arresting the cell cycle in the G2/M phase (Schierack et al., 2006), which is consistent with our results. Wang et al. (2019) showed that DON significantly increased the expression of the BCL10 and AEN genes, whose protein products function in the induction and enhancement of apoptosis. Previous research also showed that DON decreases the activity of porcine endometrial cells, resulting in apoptotic phenotypes, such as mitochondrial swelling, membrane rupture, and cytoplasmic vacuolization, which hinders the synthesis of DNA, arrest the cell cycle at the G0 and G1 phases, and reduces the expression of PCNA, a key protein in the cell cycle, thus inhibiting cell proliferation (Tiemann et al., 2003; Diesing et al., 2011). Other studies have indicated the toxicological effects of DON on cell growth of different cell types via activation of the apoptosis and cell cycle arrest pathway. Our previous transcriptome analysis found that the expression levels of SLC4A11 and MFSD3 decreased significantly in the DON treated group. DON activates MAPKs related to differentiation and apoptosis, thereby disrupting normal cell functions. Previous research showed DON induces apoptosis and disrupts cellular homeostasis through MAPK signaling pathways in bovine mammary epithelial cells (Lee et al., 2019) and SLC4A11 activated the MAPK pathway to stimulate cell growth and proliferation (Lopez et al., 2009). As MAPKs are important molecules in regulation of cell cycle and growth (Vithana et al., 2006), we speculate that SLC4A11 and MFSD3 may play a central role in DON-induced cell damage. In the present study, the expression levels of SLC4A11 and MFSD3 in the DON treated group were significantly lower than those in the control group. The cell viability of SLC4A11 and MFSD3 IPEC-J2 overexpressing cell lines under DON induction enhanced compared with the control group. SLC4A11 and MFSD3 can maintain nutrient physiological functions, and overexpression of SLC4A11 and MFSD3 may help resist the toxicity of DON. A previous study showed that relative to SLC4A11 wild-type cells, SLC4A11 knockout caused obvious oxidative damage to the corneal endothelial cells and depressed glutamine (Gln) catabolism, suggesting that SLC4A11 can protect cells from Gln-induced toxicity (Bonanno and Ogando, 2018). A recent study found that SLC4A11 plays a key role in the oxidative stress response in human corneal endothelial cells (HCEnC) mediated by the NRF2 gene, and overexpression of SLC4A11 in HEK 293 cells resulted in a significant increase in cell viability and reduced reactive oxygen species (ROS) in these cells compared with those in the control cells (Guha et al., 2017). In addition, DON exerts a direct toxic effect on the IPEC-J2 cells by enhancing ROS accumulation, activating NF-kB and apoptotic signaling pathways (Ruifen et al., 2019). Therefore, we hypothesized that the downregulation of SLC4A11 and MFSD3 expression is closely related to growth inhibition induced by DON.
DNA methylation is a significant epigenetic modification, which can regulate gene expression by promoting or inhibiting the ability of transcription factors to bind to DNA (Keshet et al., 1985; Lister et al., 2009). Liu et al. (2020) showed that DON affected the expression of growth related-genes in liver through DNA methylation. This suggested that epigenetic modifications play an important regulatory role in the mechanism of DON’s effects. In view of the role of SLC4A11 and MFSD3 gene expression on DON-induced injury, the present study explored the regulatory mechanism of promoter methylation on DON-induced toxicity. The results showed that the methylation levels of SLC4A11 gene methylation sites (mC-1 and mC-23) and MFSD3 methylation sites (mC-1 and mC-12) correlated significantly and negatively with the mRNA expression level. Previous research showed that promoter region methylation was associated with inhibition of gene expression, which occurs mainly through interference, in which the binding of transcription factors to the target gene or histone deacetylase (HDACs) is hindered (Keshet et al., 1985; Nan et al., 1998). Our study found that the mC-1 site of SLC4A11 is located in an NF1 transcription factor binding domain and the mC-1 site of MFSD3 is located in an Sp1 transcription factor binding domain. Sp1-like transcription regulators participate in the regulation of cell function, including cell proliferation, apoptosis, differentiation, and tumor transformation, by regulating the expression of many genes with CG-rich promoters (Black et al., 2001). NF-1 is considered as a common transcription factor (Inoue et al., 1990). However, the interaction with specific NF-1 subtypes in different cell types might contribute to the selective transcriptional activation or silencing of target genes (Chaudhry et al., 1997). ANTP belongs to Transcription factors of the homeodomain family (Bernd et al., 2008) NF-1, ANTP and Sp1 transcription factors fulfill important roles in cell proliferation and growth. Various components in different CpG islands play their respective roles in gene regulation. Some specific CpG sites have a crucial impact on the function of CpG islands and are decisive factors for the methylation of CpG islands (Mikeska et al., 2007; Barrera and Peinado, 2012). A single methylation site in the CCR3 (C–C motif chemokine receptor 3) region is a candidate region for causing narcolepsy (Mihoko et al., 2018). Wang et al. (2020) found that changes in the methylation degree of mC-6 site would significantly change the expression of gonadotropin releasing hormone 1 (GNRH), indicating that the methylation level of specific sites would affect the expression level of the gene. Therefore, it can be speculated that in the regulatory regions of SLC4A11 and MFSD3, hypermethylation of mC-1 in SLC4A11 and MFSD3 induced by DON in IPEC-J2 cells might hinder the binding of transcription factors to their target sequences, which would inhibit gene expression.
In the present study, we found that overexpression of SLC4A11 and MFSD3 can enhance the cell viability and alleviate the cytotoxicity of DON. The methylation levels of the mC-1 and mC-23 sites of SLC4A11, and the mC-1 and mC-12 sites of MFSD3 showed significantly negative correlations with mRNA expression. These findings indicated that the SLC4A11 and MFSD3 genes may play important roles in regulating the growth of IPEC-J2 cells and the DON-induced cell damages. This study provided novel insights into the biological functions of SLC4A11 and MFSD3 genes in regulating the cytotoxic effects induced by DON, which lays important foundations for future studies on the identification of functional gene and toxic mechanisms associated with DON. Further studies are needed to use chip-assay to confirm transcription factor binding, to reveal the detailed relationship between SLC4A11 and MFSD3 promoter methylation and the specific mechanism of DON-induced cytotoxicity.
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
WB and SW designed the study. YX, LY, and YW acquired and interpreted the data. YX analyzed the data and was a major contributor in writing the manuscript. YX and XC prepared figures and tables. ZW, HW, and WB prepared the manuscript and supervised the study. All authors read and approved the final manuscript.
This research was funded by Key Research and Development Project (Modern Agriculture) of Jiangsu Province (BE2019341) and College Students’ Innovation and Entrepreneurship Training Program of Yangzhou University (X20200649) and the Priority Academic Program Development of Jiangsu Higher Education Institutions.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2021.697883/full#supplementary-material
Barrera, V., and Peinado, M. A. (2012). Evaluation of single CpG sites as proxies of CpG island methylation states at the genome scale. Nucleic Acids Res. 40, 11490–11498. doi: 10.1093/nar/gks928
Bernd, S., Kai, K., Mansi, S., Daniel, R., Rosengarten, R. D., Dellaporta, S. L., et al. (2008). The early antp gene repertoire: insights from the placozoan genome. PLoS One 3:e2457. doi: 10.1371/journal.pone.0002457
Black, A. R., Black, J. D., and Cliffod, J. A. (2001). Sp1 and krüppel-like factor family of transcription factors in cell growth regulation and cancer. J. Cell. Physiol. 188, 143–160. doi: 10.1002/jcp.1111
Bonanno, J., and Ogando, D. (2018). SLC4A11 provides NH3 sensitive mitocho ndri- al uncoupling and ROS prevention that facilitates glutamine catabolism. Free Radic. Biol. Med. 128:S80.
Borutova, R., Aragon, Y. A., NäHrer, K., and Berthiller, F. (2012). Co-occurrence and statistical correlations between mycotoxins in feedstuffs collected in the Asia–Oceania in 2010. Anim. Feed Sci. Tech. 178, 190–197. doi: 10.1016/j.anifeedsci.2012.09.015
Chaudhry, A. Z., Lyons, G. E., and Gronostajski, R. M. (1997). Expression patterns of the four nuclear factor I genes during mouse embryogenesis indicate a potential role in development. Dev. Dyn. 208, 313–325.
Dahl, C., and Guldberg, P. (2003). DNA methylation analysis techniques. Biogerontology. 4, 233–250. doi: 10.1023/A:1025103319328
Dai, C., Yang, L., Jin, J., Wang, H., and Bao, W. (2019). Regulation and molecular mechanism of TLR5 on resistance to Escherichia coli F18 in Weaned Piglets. Animals 9:735. doi: 10.3390/ani9100735
Deng, D. J. (2014). DNA methylation and demethylation: current status and future perspective. Heredity 36, 403–410. doi: 10.3724/SP.J.1005.2014.0403
Diesing, A. K., Nossol, C., Panther, P., Walk, N., Post, A., Kluess, J., et al. (2011). Mycotoxin deoxynivalenol (DON) mediates biphasic cellular response in intestinal porcine epithelial cell lines IPEC-1 and IPEC-J2. Toxicol. Lett. 200, 8–18. doi: 10.1016/j.toxlet.2010.10.006
Diesing, A. K., Nossol, S., Ponsuksili, S., Wimmers, K., Kluess, J., Walk, N., et al. (2012). Gene regulation of intestinal porcine epithelial cells IPEC-J2 is dependent on the site of deoxynivalenol toxicological action. PLoS One 7:e34136. doi: 10.1371/journal.pone.0034136
Eriksen, G. S., and Pettersson, H. (2004). Toxicological evaluation of trichothecenes in animal feed. Anim. Feed Sci. Tech. 114, 205–239.
Gerez, J. R., Pinton, P., Callu, P., Grosjean, F., Oswald, I. P., and Bracarense, A. P. F. I. (2015). Deoxynivalenol alone or in combination with nivalenol and zear alenone induce systemic histological changes in pigs. Exp. Toxicol. Pathol. 67, 89–98. doi: 10.1016/j.etp.2014.10.001
Goyarts, T., and Dänicke, S. (2006). Bioavailability of the Fusarium toxin deoxynival- enol (DON) from naturally contaminated wheat for the pig. Toxicol. Lett. 163, 171–182. doi: 10.1016/j.toxlet.2005.10.007
Guha, S., Chaurasia, S., Ramachandran, C., and Roy, S. (2017). SLC4A11 depletion impairs NRF2 mediated antioxidant signaling and increases reactive oxygen species in human corneal endothelial cells during oxidative stress. Sci. Rep. 7:4074. doi: 10.1038/s41598-017-03654-4
Han, J., Wang, Q. C., Zhu, C. C., Liu, J., Zhang, Y., Cui, X. S., et al. (2016). Deoxynivalenol exposure induces autophagy/apoptosis and epigenetic modification changes during porcine oocyte maturation. Toxicol. Appl. Pharmacol. 300, 70–67. doi: 10.1016/j.taap.2016.03.006
Hoglund, P. J., Nordstrom, K. J., Schioth, H. B., and Fredriksson, R. (2011). The solute carrier families have a remarkably long evolutionary history with the majority of the human families present before divergence of Bilaterian species. Mol. Biol. Evol. 28, 153–141. doi: 10.1093/molbev/msq350
Inoue, T., Tamura, T., Furuichi, T., and Mikoshiba, K. (1990). Isolation of complementary DNAs encoding a cerebellum-enriched nuclear factor I family that activates transcription from the mouse myelin basic protein promoter. J. Biol. Chem. 265, 19065–19070. doi: 10.1016/0005-2728(90)90052-6
Jiao, X., Sultana, A., Garg, P., Ramamurthy, B., Vemuganti, G. K., Gangopadhyay, N., et al. (2007). Autosomal recessive corneal endothelial dystrophy (CHED2) is associated with mutations in SLC4A11. J. Med. Genet. 44:407. doi: 10.1136/jmg.2006.044644
Keshet, I., Yisraeli, J., and Cedar, H. (1985). Effect of regional DNA methylation on gene expression. Proc. Natl. Acad. Sci. U. S. A. 82, 2560–2564. doi: 10.2307/25215
Lee, J. Y., Lim, W., Park, S., Kim, J., You, S., and Song, G. (2019). Deoxynivalenol induces apoptosis and disrupts cellular homeostasis through MAPK signaling pathways in bovine mammary epi- thelial cells. Environ. Pollut. 252, 879–887.
Li, L. C., and Dahiya, R. (2002). Methprimer: designing primers for methylation PCRs. Bioinformatics 18, 1427–1431. doi: 10.1093/bioinformatics/18.11.1427
Lister, R., Pelizzola, M., Dowen, R. H., Hawkins, R. D., Hon, G., Tonti-Filippini, J., et al. (2009). Human DNA methylomes at base resolution show wide spread epigenomic differences. Nature 462, 315–322. doi: 10.1038/nature08514
Liu, A., Hu, S., Zhu, X., Ihsan, A., Wu, Q., Sun, L., et al. (2020). Dietary exposure of deoxynivalenol affected cytochrome P450 and growth related-gene expression via DNA methylation in piglet liver. Research square. Preprint. doi: 10.21203/rs.3.rs-34338/v1
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2–ΔΔCT method. Methods 25, 402–408. doi: 10.1006/meth.2001
Lopez, I. A., Rosenblatt, M. I., Kim, C., Galbraith, G. C., Jones, S. M., Kao, L., et al. (2009). SLC4A11 gene disruption in mice. J. Biol. Chem. 284, 26882–26896. doi: 10.1074/jbc.M109.008102
Mihoko, S., Taku, M., Hiromi, T., Katsushi, T., and Makoto, H. (2018). Epigeno mew- ide association study of DNA methylation in narcolepsy: an integrated genetic an- d epigenetic approach. Sleep 41, 32–42. doi: 10.1093/sleep/zsy019
Mikeska, T., Bock, C., El-Maarri, O., Hübner, A., Ehrentrau, D., Schramm, J., et al. (2007). Optimization of quantitative MGMT promoter methylation analysis using pyrosequencing and combined bisulfite restriction analysis. J. Mol. Diagn. 9, 368–381. doi: 10.2353/jmoldx.2007.060167
Nan, X., Ng, H. H., Johnson, C. A., Laherty, C. D., Turner, B. M., Eisenman, R. N., et al. (1998). Transcriptional repression by the methyl-CpG-binding protein MeCP2 involves a histone deacetylase complex. Nature 393, 386–389. doi: 10.1038/30764
Nicoletti, C. F., Pinhel, M. A. S., Noronha, N. Y., Jacomo, A., Crujeiras, A. B., and Nonino, C. B. (2019). Association of MFSD3 promoter methylation level and weight regain after gastric bypass: assessment for three years after surgery. Nutrition 70:110499. doi: 10.1016/j.nut.2019.04.010
Perland, E., Hellsten, S. V., Lekholm, E., Eriksson, M. M., Arapi, V., and Fredriksson, R. (2017). The novel membrane-bound proteins MFSD1 and MFSD3 are putative SLC transporters affected by altered nutrient intake. J. Mol. Neurosci. 61, 1–16. doi: 10.1007/s12031-016-0867-8
Pestka, J. J. (2010). Deoxynivalenol-induced proinflammatory gene expression: mechanisms and pathological sequelae. Toxins 2, 1300–1317.
Pestka, J. J., and Smolinski, A. T. (2005). Deoxynivalenol: toxicology and potential effects on humans. J. Toxicol. Environ. Health B 8, 39–69. doi: 10.1080/10937400590889458
Ruifen, K., Ruonan, L., Pengyuan, D., Zhaojian, L., Yansen, L., Chunmei, L., et al. (2019). Deoxynivalenol induced apoptosis and inflammation of IPEC-J2 cells by promoting ROS production. Environ. Pollut. 251, 689–698. doi: 10.1016/j.envpol.2019.05.026
Schierack, P., Nordhoff, M., Pollmann, M., Weyrauch, K. D., and Amasheh, S. (2006). Characterization of a porcine intestinal epithelial cell line for in vitro studies of microbial pathogenesis in swine. Histochem. Cell Biol. 125, 293–305. doi: 10.1007/s00418-005-0067-z
Shi, L., and Wu, J. (2009). Epigenetic regulation in mammalian preimplantation embryo development. Reprod. Biol. Endocrinol. 7:59. doi: 10.118-6/1477-7827-7-59
Tiemann, U., Viergutz, T., Jonas, L. F., and Schneider, F. (2003). Influence of the my- cotoxins α- and β zearalenol and deoxynivalenol on the cell cycle of cultured por- cine endometrial cells. Reprod. Toxicol. 17, 209–218.
Vithana, E. N., Morgan, P., Sundaresan, P., Ebenezer, N. D., Tan, D. T. H., Mohamed, M. D., et al. (2006). Mutations in sodium-borate cotransporter SLC4A11 cause recessive congenital hereditary endothelial dystro- phy (CHED2). Nat. Genet. 38, 755–757. doi: 10.1038/ng1824
Wang, H. F., Zong, Q. F., Wang, S. Q., Zhao, C. X., Wu, S. L., and Bao, W. B. (2019). Genome-wide DNA methylome and transcriptome analysis of porcine intestinal epithelial cells upon deoxynivalenol exposure. J. Agric. Food Chem. 67, 6423–6431. doi: 10.1021/acs.jafc.9b00613
Wang, X., Li, X. J., Chen, J., Xiao, J., Li, K., and Zhou, Y. (2020). The effect of miR-29 on the methylation of specific sites in the promoter region of GnRH gene. J. Biol. 37, 2095–1736. (In Chinese)
Keywords: pig, DON, SLC4A11, MFSD3, methylation regulation
Citation: Xu Y, Chen X, Yu L, Wang Y, Wang H, Wu Z, Wu S and Bao W (2021) SLC4A11 and MFSD3 Gene Expression Changes in Deoxynivalenol Treated IPEC-J2 Cells. Front. Genet. 12:697883. doi: 10.3389/fgene.2021.697883
Received: 20 April 2021; Accepted: 21 June 2021;
Published: 21 July 2021.
Edited by:
Aline Silva Mello Cesar, University of São Paulo, BrazilReviewed by:
Doreen Becker, Leibniz Institute for Farm Animal Biology (FBN), GermanyCopyright © 2021 Xu, Chen, Yu, Wang, Wang, Wu, Wu and Bao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Wenbin Bao, d2JiYW9AeXp1LmVkdS5jbg==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.