- 1Department of Pediatrics, Union Hospital, Tongji Medical College, Huazhong University of Science and Technology, Wuhan, China
- 2Department of Clinical Laboratory, Union Hospital, Tongji Medical College, Huazhong University of Science and Technology, Wuhan, China
The existing knowledge about the association between NLRP3 rs35829419/rs10754558 polymorphisms and susceptibility to autoimmune diseases (AIDs) remains controversial. Herein, a meta-analysis was performed to evaluate such association. We searched databases for relevant studies published in English up to February 2021. Stata14 was used to assess the odds ratio (OR). As for NLRP3 rs35829419, no significant association to overall AIDs was found in three genetic models [A vs. C: OR (95%CI) = 0.89 (0.69–1.14); AC vs. CC: 1.00 (0.77–1.30); AA/AC vs. CC: 0.93 (0.71–1.20)]. However, subgroup analysis by disease type showed that NLRP3 rs35829419 A allele may have a significant protective effect on rheumatoid arthritis (RA) susceptibility [A vs. C: 0.74 (0.57–0.96)]. NLRP3 rs10754558 polymorphism contributes to significantly reduce the risk of AIDs in the allelic model [G vs. C: 0.78 (0.71–0.87)], homozygote co-dominant model [GG vs. CC: 0.63 (0.51–0.77)], heterozygote co-dominant model [GC vs. CC: 0.78 (0.66–0.91)], dominant model [GG/GC vs. CC: 0.73 (0.63–0.84)], and recessive model [GG vs. GC/CC: 0.73 (0.62–0.88)]. In the subgroup analysis by ethnicity, association was observed between the NLRP3 rs10754558 G allele and AIDs in Latin Americans, but not in European, Arabian, or Asian populations. Stratification by disease type showed a significant association of the NLRP3 rs10754558 G allele with type 1 diabetes (T1D), RA, and systemic lupus erythematosus (SLE), but not with celiac disease (CD), multiple sclerosis (MS), or myasthenia gravis (MG). This meta-analysis suggests that the NLRP3 rs10754558, but not rs35829419, polymorphism is associated with susceptibility to AIDs, especially in Latin American individuals.
Introduction
Autoimmune diseases (AIDs) are characterized by a host immune response against self-antigens that damages host tissues. Autoimmune diseases are caused by a combination of genetic factors and environmental factors (Luppi et al., 1995). In particular, the role of genetic factors has been confirmed by a number of studies (Gabrielsen et al., 2016; Zhong et al., 2018; Zhang et al., 2019). Hundreds of risk susceptibility genes related to AIDs have been identified through genome-wide scans and large-scale single-nucleotide polymorphism (SNP) association studies (Lessard et al., 2012; Kochi, 2016). Many studies have confirmed the genetic background of AIDs.
Inflammasomes are highly conserved pattern recognition receptors in the cytoplasm of cells and are an important part of the innate immune system (Evavold and Kagan, 2019). The most characteristic inflammasome is the nucleotide oligomerization domain-like receptor (NLR) family pyrin domain-containing 3 (NLRP3) inflammasome. NLRP3, also known as CIAS1/NALP3/cryopyrin, is a well-known member of NLRs (de Torre-Minguela et al., 2017). NLRP3 is mainly expressed in macrophages and dendritic cells, and its main function is inflammasome assembly. The NLRP3 inflammasome is a multi-protein complex composed of NLRP3, apoptosis-associated speck-like protein (ASC), and proinflammatory caspase-1 (de Torre-Minguela et al., 2017; Wang et al., 2020). The NLRP3 inflammasome can recognize pathogen- or danger-associated molecular patterns. Once NLRP3 is activated, the adaptor protein ASC can be recruited, resulting in the cleavage of pro-caspase-1 into caspase-1. Activated caspase-1 processes pro-IL-1β and pro-IL-18 into the mature forms and secretes them outside the cell. At the same time, the activated caspase-1 fragment can induce cell membrane perforation, cell rupture, and cell pyroptosis (Fusco et al., 2020). Growing evidences indicate that NLRP3 inflammasome plays a key role in various AIDs (Shen et al., 2018; Zhao et al., 2018; Alves da Cruz et al., 2020).
The human NLRP3 gene is about 32.9-kb long. It is located on chromosome 1q44 and consists of nine exons (Villani et al., 2009). Single-nucleotide polymorphisms in the NLRP3 gene may lead to changes in its function, increasing the activation of inflammasomes and the levels of IL-1β. So far, about 60 SNPs in the NLRP3 gene have been identified. Among them, the most common polymorphisms are rs35829419 (Q705K), rs10754558, rs4612666, rs4925648, and rs10925019 (Zhang et al., 2011). This study focuses on the two most-studied polymorphic sites of NLRP3, including rs35829419, a gain-of-function polymorphism associated with pro-inflammatory phenotype (Verma et al., 2012), and rs10754558, which is located in the 3′-untranslated region (3′-UTR) of the NLRP3 gene and has a certain impact on the stability of NLRP3 mRNA (Hitomi et al., 2009). Many studies have explored the association between NLRP3 rs35829419/rs10754558 polymorphisms and AIDs, including multiple sclerosis (MS) (Imani et al., 2018), rheumatoid arthritis (RA) (Kastbom et al., 2010; Ben et al., 2012; Jenko et al., 2016; Addobbati et al., 2018), psoriatic arthritis (PsA) (Juneblad et al., 2020), celiac disease (CD) (Pontillo et al., 2011), type 1 diabetes (T1D) (Pontillo et al., 2010; Smigoc et al., 2019), myasthenia gravis (MG) (Agah et al., 2021), and systemic lupus erythematosus (SLE) (Pontillo et al., 2012; Su et al., 2020). Some studies show that these NLRP3 polymorphism sites are related to the risk of certain AIDs, but other studies suggest that NLRP3 gene polymorphism at these sites exhibit a disease protection effect. Some studies also indicate that NLRP3 rs35829419/rs10754558 polymorphisms have nothing to do with the risk of AIDs. Therefore, the views about the association between NLRP3 gene polymorphisms at these two loci and AIDs remain contradictory and inconclusive, partially because of the relatively small sample size, low statistical power, and/or disease heterogeneity in these studies. Hence, to resolve this issue, we conducted a comprehensive meta-analysis to evaluate whether NLRP3 rs35829419 C/A and rs10754558 C/G polymorphisms are associated with AIDs.
Materials and Methods
Publication Search
To explore the association of the NLRP3 rs35829419 C/A and rs10754558 C/G polymorphisms with AIDs, we searched the literature in PUBMED and Web of Science on February 1, 2021. The following keywords and medical subject heading terms were used for the search: (“NLRP3” or “CIAS1” or “NALP3” or “Cryopyrin” or “rs35829419” or “rs10754558”) and (“polymorphism” or “SNP” or “variation” or “mutation”). All eligible studies were retrieved, and their references were checked and manually searched for finding other relevant articles. The related literature published in previous meta-analyses was also searched to identify other special publications. Only articles published in English and with human subjects were selected for this meta-analysis.
Inclusion and Exclusion Criteria
Studies included in this meta-analysis had to meet all four criteria: (a) evaluation of association of rs35829419 or rs10754558 with AIDs risk; (b) original study and case-control design; (c) provision of sufficient genotype or allele data in cases and controls to calculate odds ratio (OR) and 95% confidence intervals (CIs); (d) full-text available. The definitions of patients with different types of AIDs in the selected literature followed internationally recognized guidelines such as the European League Against Rheumatism (EULAR)/American College of Rheumatology (ACR) (Wayant et al., 2020; Fanouriakis et al., 2021; Kostine et al., 2021), the American Diabetes Association (American Diabetes Association, 2020), the American College of Gastroenterology (Lacy et al., 2021), the Myasthenia Gravis Foundation of America (Narayanaswami et al., 2021), and the International Panel on Diagnosis of Multiple Sclerosis (Thompson et al., 2018). Accordingly, the exclusion criteria were as follows: (a) diseases not clearly regarded as AIDs; (b) not a case-control study; (c) no available genotype frequency; and (d) review, meta-analysis, case report, and comments. To ensure the accuracy of the included studies, all evaluations were independently performed by two separate researchers.
Data Extraction
All the data from all eligible studies were independently extracted by two investigators using the selection criteria above and checked by other authors. Any disagreement was resolved by discussion. We collected the following information from each study: ages and genders of research subjects, name of first author, year of publication, country, patient ethnicity, disease type, genotyping method, and total numbers and genotype/allele frequency distributions of both cases and controls.
Quality Assessment
We used the Newcastle–Ottawa Quality Assessment Scale to evaluate the quality score of the included studies. High-quality research was defined with a score ≥7.
Statistical Analysis
Hardy–Weinberg equilibrium (HWE) was tested by the Chi-square test. The crude OR and its 95% CIs in each case-control study were used to evaluate the strength of association between NLRP3 rs35829419 or rs10754558 polymorphisms and the risk of AIDs. The statistical significance of the pooled OR was determined by the Z-test, and a p-value of <0.05 was considered statistically significant. The results were summarized in the following models: an allele model (A vs. C for rs35829419; G vs. C for rs10754558), a dominant model (AA/AC vs. CC for rs35829419; GG/GC vs. CC for rs10754558), a recessive model (AA vs. AC/CC for rs35829419; GG vs. GC/CC for rs10754558), a homozygote co-dominant model (AA vs. CC for rs35829419; GG vs. CC for rs10754558), and a heterozygote co-dominant model (AC vs. CC for rs35829419; GC vs. CC for rs10754558), respectively. Subgroup analyses were conducted by ethnicity and disease type. Heterogeneity analysis was examined by the Chi-square based on Q-test. If the result of this heterogeneity test was p < 0.05, then the pooled OR was analyzed using a random-effects model (the DerSimonian and Laird method) to evaluate the association of NLRP3 Single-nucleotide polymorphisms with AIDs. Otherwise, a fixed-effects model (the Mantel–Haenszel method) was selected. Besides, in order to quantitatively estimate heterogeneity, we used another measure, I2 = 100% × (Q – df)/Q. The I2 statistic estimated the degree of inconsistency in the studies (I2 < 25% no heterogeneity; I2 = 25–50% moderate heterogeneity; I2 > 50% extreme heterogeneity). Publication bias was investigated using Begg's funnel plot and Eegg's test. If the funnel plot was asymmetric, and the Eegg's result was p < 0.05, it was considered that there might be a publication bias. Meanwhile, we performed a sensitivity analysis in which one study was removed, and the rest were conducted to evaluate whether the results were affected statistically significantly. In this study, the meta-analysis was performed by using the software STATA version 14.0 (Stata Corporation, College Station, TX, USA).
Results
Baseline Characteristics of Included Studies
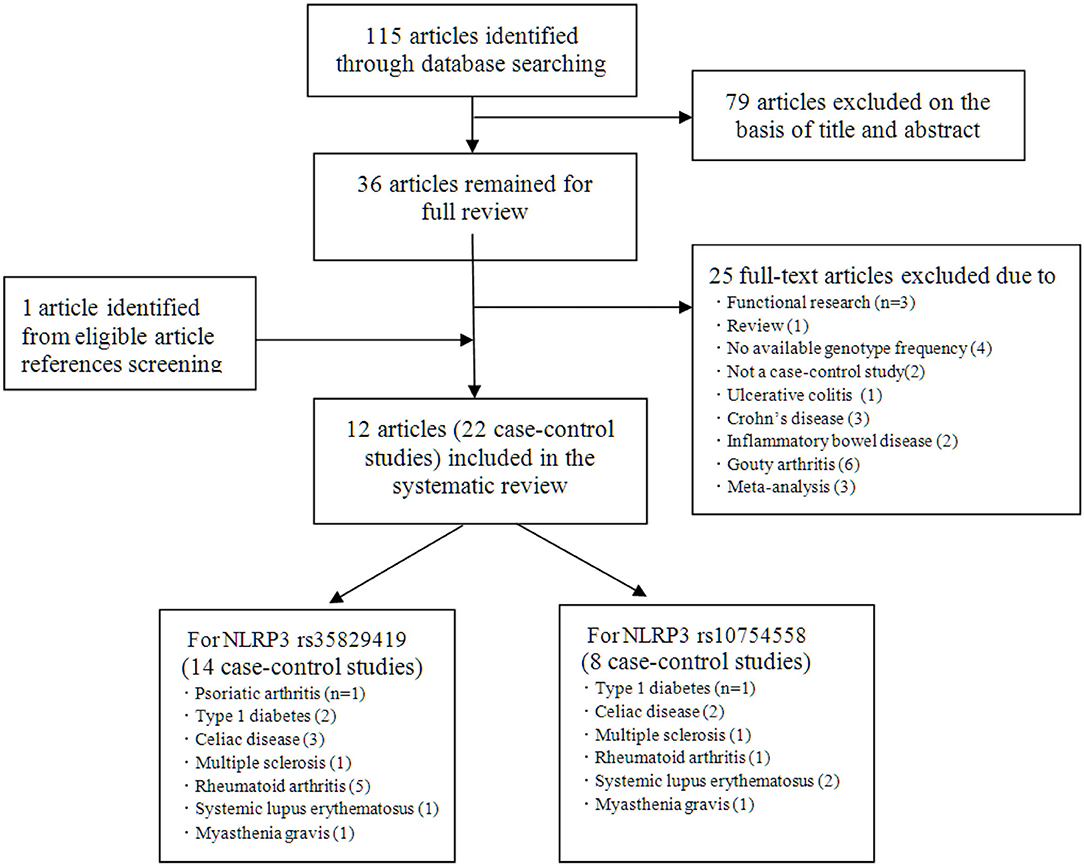
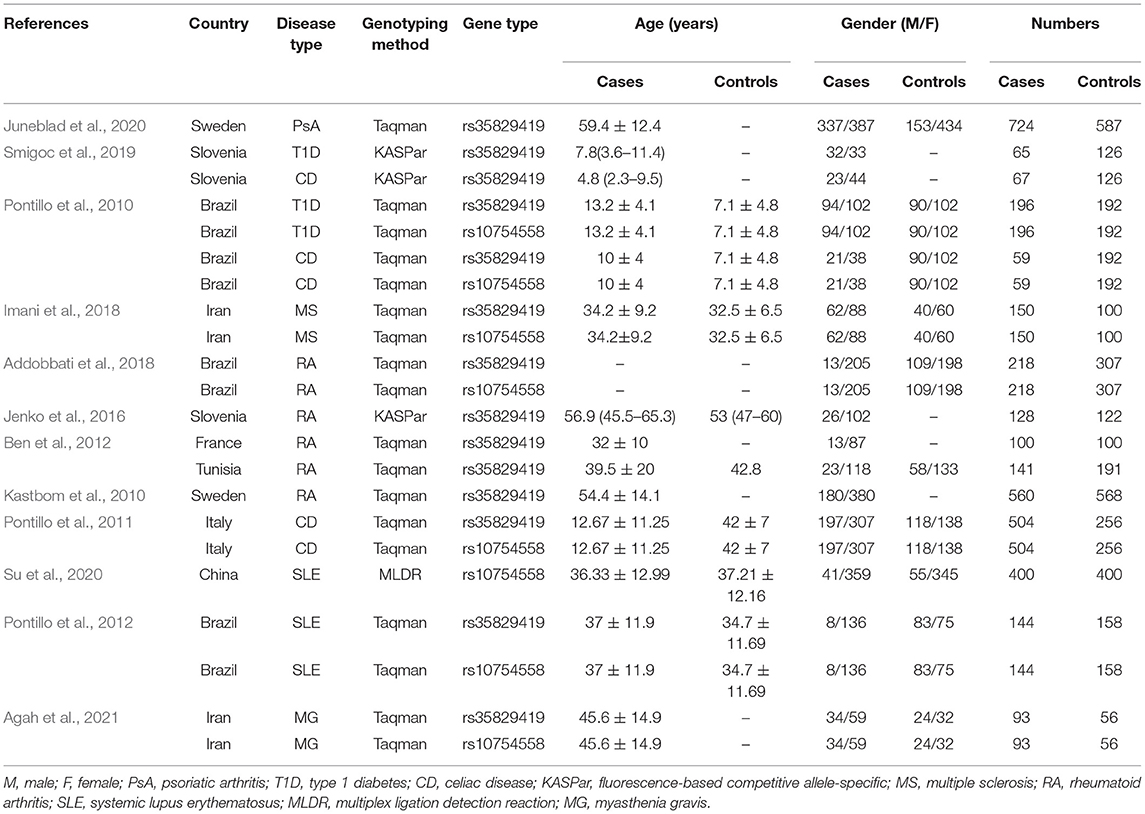
Initially, 115 studies were identified by electronic searches. Among them, according to the title and abstract, a total of 79 non-inflammatory susceptibility studies were excluded. Finally, 36 full-text articles were included in our detailed analysis. After careful evaluation of the published literature based on the inclusion and exclusion criteria, we finally identified 12 articles to be included in this meta-analysis (Figure 1). For the NLRP3 rs35829419 polymorphic site, there were 11 articles in total. In these articles, sometimes one article involved several AIDs, and we regarded one of the AIDs as a case-control study. In the end, it included 14 case-control studies with 3,149 cases and 3,081 controls to assess the relationship between NLRP3 rs35829419 and AIDs. For the NLRP3 rs10754558 polymorphic site, there were a total of six articles. Eight case-control studies with 1,764 cases and 1,661 controls were included to evaluate the relationship between NLRP3 rs10754558 and AIDs. All studies were of case-control design and targeted at RA (n = 4), T1D (2), CD (3), SLE (2), psoriatic arthritis (PsA, 1), MS (1), and MG (1). All cases were patients with AIDs, and the controls were free of AIDs. In the included articles, although three articles were published by the same author, the research populations did not overlap with each other. The genotyping methods included fluorescence-based competitive allele-specific (KASPar), TaqMan, and multiplex ligation detection reaction (MLDR). The baseline characteristics of the eligible studies are summarized in Table 1.
Allele and Genotype Frequencies of Included Studies
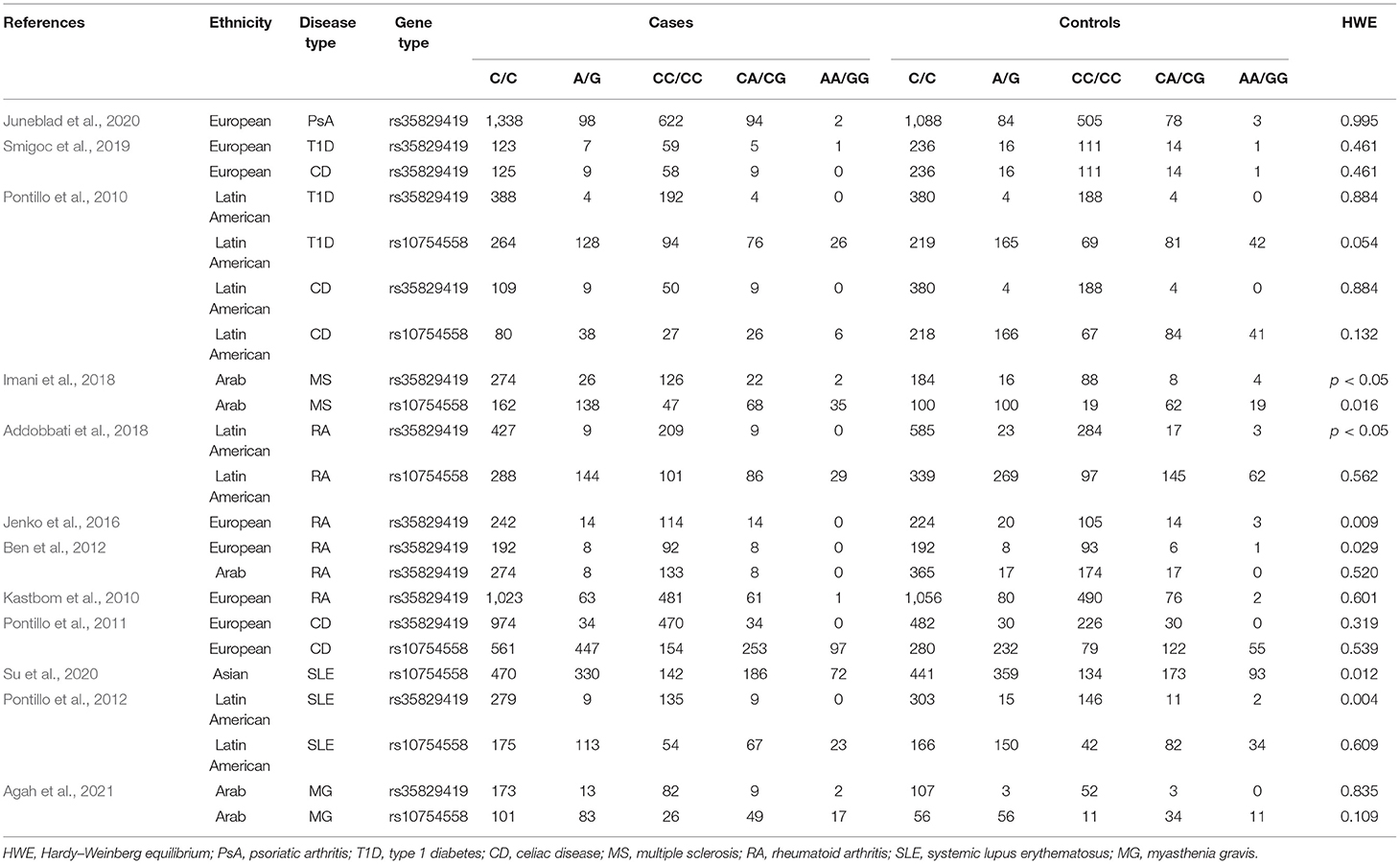
Details of allele and genotype frequencies are shown in Table 2. In several articles, the data in the control group deviated from the HWE (p < 0.05). Among these articles, three articles came from the Latin American populations, six articles came from the European populations, three articles came from the Arab populations, and one article came from the Asian populations. Ethnicity-subgroup meta-analysis was performed to determine the association of the NLRP3 rs35829419 and rs10754558 polymorphisms with AIDs in different races. Furthermore, disease-subgroup meta-analysis was conducted to explore the association of the NLRP3 rs35829419 and rs10754558 polymorphisms with PsA, T1D, CD, MS, RA, SLE, and MG.
Meta-Analysis for Association Between Nucleotide Oligomerization Domain-Like Receptor Family Pyrin Domain-Containing 3 rs35829419 Polymorphisms and Risks of Autoimmune Diseases
In the included studies, we observed significant heterogeneity in some models, so the random effects model was applied, and the M–H fixed effects model was used in the other models. In all, when all eligible studies were pooled into the meta-analyses, we found that no significant AIDs risk was associated with the NLRP3 rs35829419 polymorphism in the allelic model (A vs. C: OR = 0.89, 95% CI = 0.69–1.14), the heterozygote co-dominant model (AC vs. CC: OR = 1.00, 95% CI = 0.77–1.30), or the dominant model (AA/AC vs. CC: OR = 0.93, 95% CI = 0.71–1.20). However, a protected effect was found in the recessive model (AA vs. AC/CC: OR = 0.45, 95% CI = 0.22–0.92, p = 0.029) and the homozygote co-dominant model (AA vs. CC: OR = 0.45, 95% CI = 0.22–0.93, p = 0.030). In the subgroup analysis by ethnicity, statistically slightly decreased AIDs risks were found among European populations in the allelic model (A vs. C: OR = 0.83, 95% CI = 0.69–0.99, p = 0.043). However, in other genetic models, no association between NLRP3 rs35829419 polymorphism and AIDs susceptibility was observed. More importantly, among the Latin American and Arabian populations, we also found no association between NLRP3 rs35829419 polymorphism and AIDs risk in any genetic model. In the subgroup of disease type, there were no significantly decreased or increased risks of PsA, T1D, CD, MS, SLE, and MG in any genetic model. However, in the RA group, remarkably decreased risks were found in the allelic model (A vs. C: OR = 0.74, 95% CI = 0.57–0.96, p = 0.024) and the homozygote co-dominant model (AA vs. CC: OR = 0.27, 95% CI = 0.08–0.97, p = 0.045). No significant associations were found in any other genetic model. Details of the meta-analysis are shown in Table 3 and Supplementary Table 1.
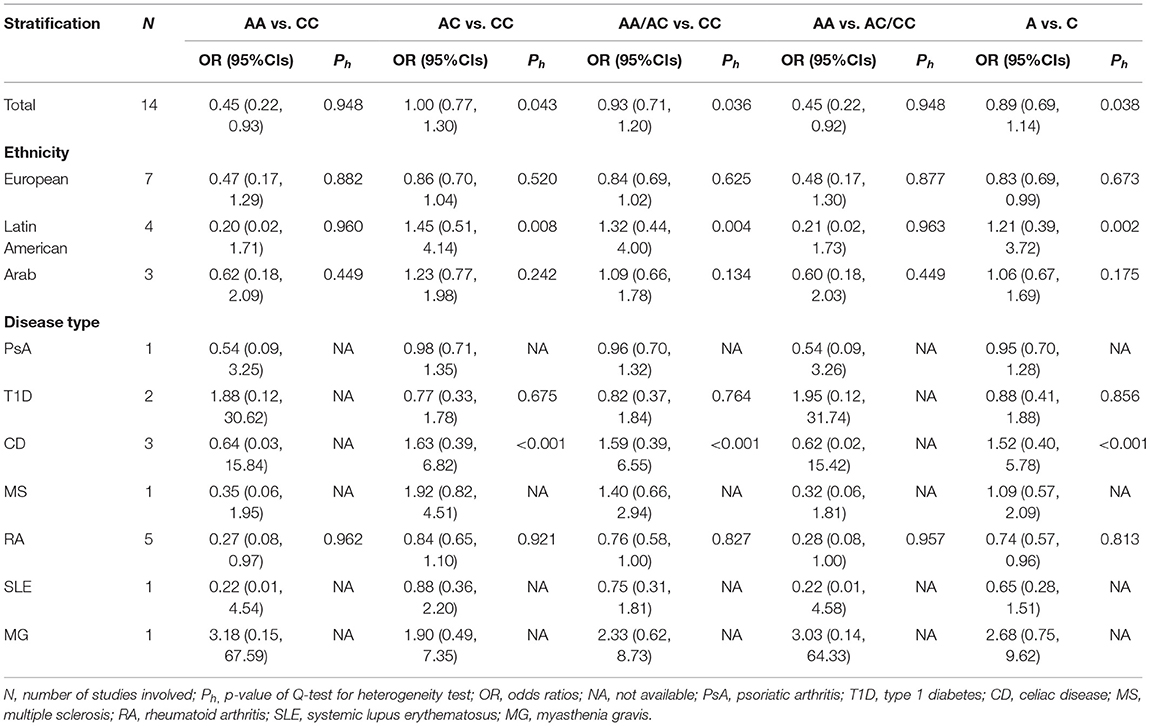
Table 3. Summary of odd ratios (OR) and 95% CIs of NLRP3 rs35829419 polymorphism and autoimmune diseases (AIDs) susceptibility for various comparisons.
Meta-Analysis for Association Between Nucleotide Oligomerization Domain-Like Receptor Family Pyrin Domain-Containing 3 rs10754558 Polymorphisms and Autoimmune Diseases Risks
Since there was no significant heterogeneity in the following eight articles, the M–H fixed effects model was used for meta-analysis. On the whole, NLRP3 rs10754558 polymorphism contributed to reducing the risk of AIDs in all genetic models, including the allelic model (G vs. C: OR = 0.78, 95% CI = 0.71–0.87, p < 0.001), the homozygote co-dominant model (GG vs. CC: OR = 0.63, 95% CI = 0.51–0.77, p < 0.001), the heterozygote co-dominant model (GC vs. CC: OR = 0.78, 95% CI = 0.66–0.91, p=0.002), the dominant model (GG/GC vs. CC: OR = 0.73, 95% CI = 0.63–0.84, p < 0.001), and the recessive model (GG vs. GC/CC: OR = 0.73, 95% CI = 0.62-0.88, p = 0.001). The mutation of allele from C to G can play a protective role against AIDs. Besides, in the subgroup analysis by ethnicity, significantly decreased AIDs risks were also observed among the Latin American populations in all genetic models. Among the Arabian populations, we only observed a significant reduction in the risk of AIDs in the dominant model and the heterozygote co-dominant model. In other genetic models, no association between NLRP3 rs10754558 polymorphism and AIDs susceptibility was observed. However, among European and Asian populations, we found no relationship between NLRP3 rs10754558 polymorphism and AIDs risk in any genetic model. In the subgroup of disease type, no significantly decreased or increased risks of CD and MG were found in any genetic model. However, in the RA group, remarkably decreased risks were found in all genetic models. In the T1D group, other genetic models in addition to the heterozygote co-dominant model showed that NLRP3 rs10754558 polymorphism was associated with a decreased risk of T1D. In the SLE group, the results of the allelic model, the homozygote co-dominant model, and the recessive model indicate that NLRP3 rs10754558 polymorphism can help reduce the risk of SLE. In the MS group, the results of the heterozygote co-dominant model and the dominant model also show the protective effect of NLRP3 rs10754558 polymorphism. No significant associations were found in any other genetic models. Details of the meta-analysis are shown in Table 4 and Supplementary Table 2.
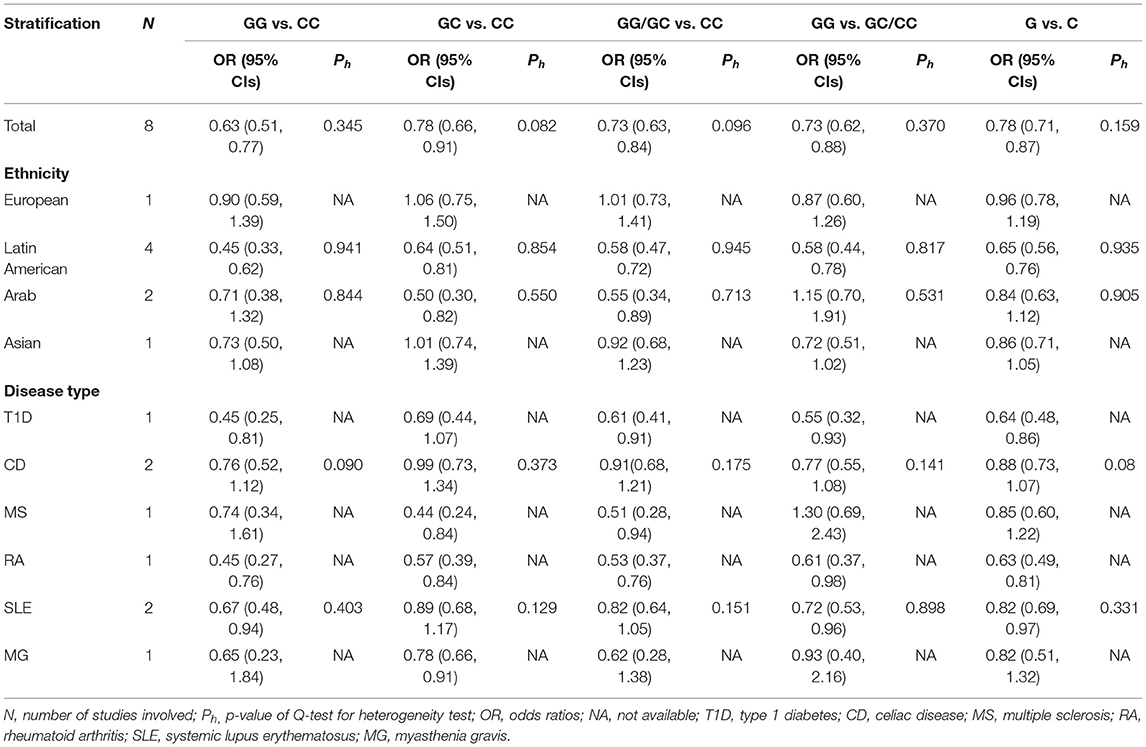
Table 4. Summary of OR and 95% CIs of NLRP3 rs10754558 polymorphism and AIDs susceptibility for various comparisons.
Risk of Bias Across Studies and Within Studies
Begger's funnel plot and Egger's test were performed to assess the publication bias of the literature. In all comparative models, the shape of the funnel plots was asymmetrical, which shows some evidence of publication bias (Figure 2). However, Egger's test can provide enough statistical evidence about funnel plot asymmetry. Results do not show any evidence of publication bias (A vs. C: t = 1.36, p = 0.199; AA/AC vs. CC: t = 1.36, p = 0.199; G vs. C: t = −1.25, p = 0.259; GG/GC vs. CC: t = −1.81, p = 0.120; Figure 2). In addition, individual studies were assessed for bias using the Newcastle–Ottawa Scale for case-control studies (Supplementary Table 3). Two studies scored 9 points, 13 studies 8 points, and the rest scored 7 points out of a total 10 points.
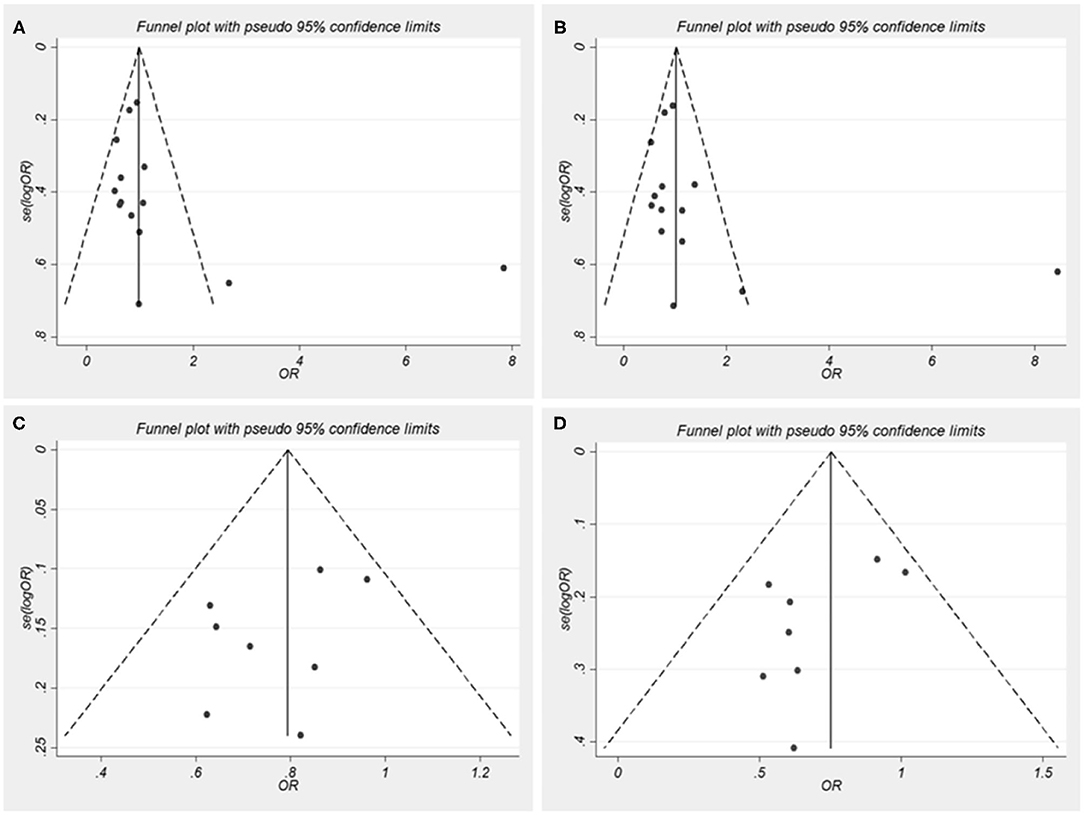
Figure 2. Funnel plots for nucleotide oligomerization domain-like receptor family pyrin domain-containing 3 (NLRP3) single nucleotide polymorphisms (SNPs) and autoimmune diseases (AIDs) risk. OR, odds ratios. (A) A vs. C for rs35829419; (B) AA/AC vs. CC for rs35829419; (C) G vs. C for rs10754558; (D) GG/GC vs. CC for rs10754558.
Forest Plot and Sensitivity Analysis
Pooling the eligible studies, we found no association of NLRP3 rs35829419 with AIDs risk in the allele/dominant model (p = 0.339/p = 0.565, Figures 3A,B). However, we found a significant association between NLRP3 rs10754558 and AIDs risk in the allele/dominance model, and the G allele showed a protective effect (p < 0.001/p < 0.001, Figures 3C,D). In addition, sensitivity analysis was conducted to assess whether each eligible study affected the final results. After sequential removal of each study, we found the pooled OR was not influenced by any single study (Figure 4).
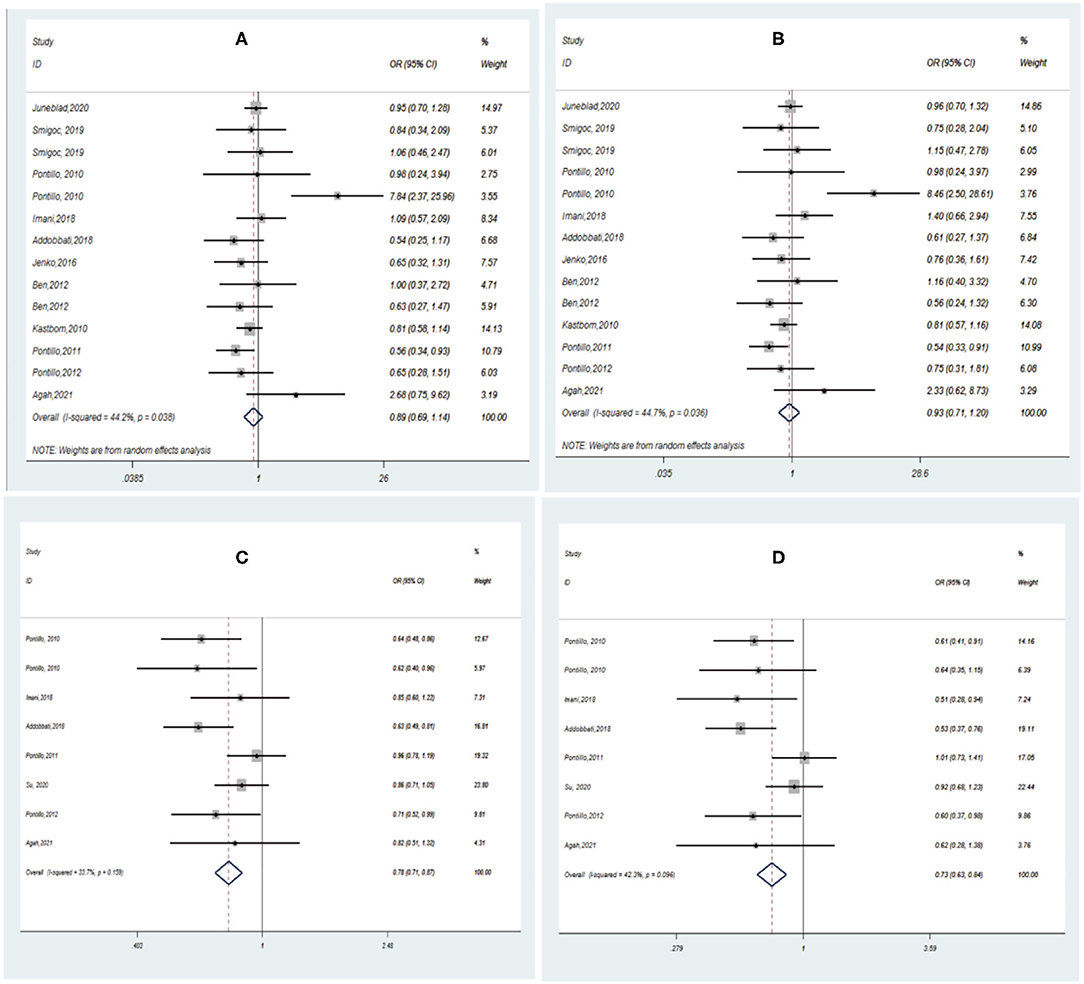
Figure 3. Forest plot of AIDs risk associated with NLRP3 SNPs under the allele/dominant model contrast. (A) A vs. C for rs35829419; (B) AA/AC vs. CC for rs35829419; (C) G vs. C for rs10754558; (D) GG/GC vs. CC for rs10754558.
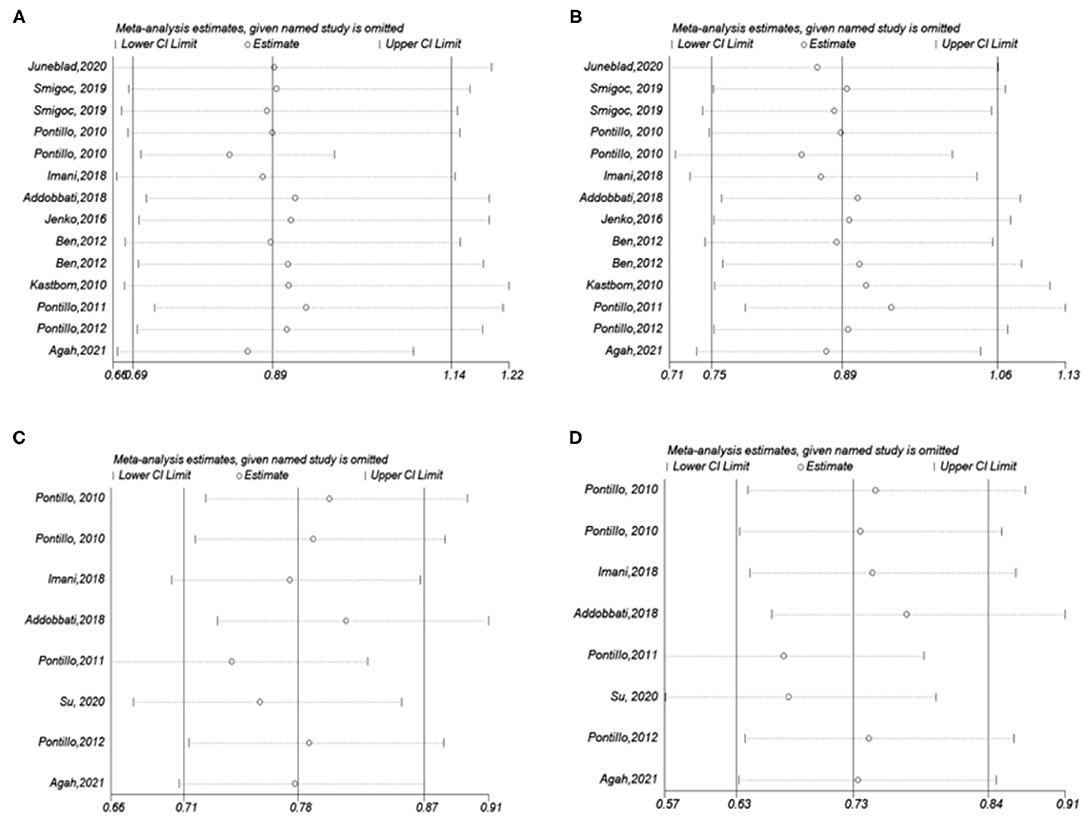
Figure 4. Influence analysis for NLRP3 SNPs in the overall meta-analysis. The middle vertical axis indicates the overall OR and the other two vertical axes indicate the 95% CI. (A) A vs. C for rs35829419; (B) AA/AC vs. CC for rs35829419; (C) G vs. C for rs10754558; (D) GG/GC vs. CC for rs10754558.
Discussion
Single-nucleotide polymorphisms of DNA sequence are caused by nucleotide variation at the chromosome genome level and are among the important causes for drug responsiveness, disease susceptibility, and ethnic difference (Shastry, 2003). Recently, more researchers are devoted to revealing the relationship between NLRP3 SNPs and the susceptibility of inflammatory diseases. We systematically analyzed the relationships of NLRP3 rs35829419 and rs10754558 polymorphisms with susceptibility to AIDs through the existing research, so as to clarify the contradictory results brought by individual research. Our meta-results show that the NLRP3 rs35829419 polymorphism is unrelated with risk of occurrence of AIDs. However, this polymorphism may reduce the risk of RA susceptibility. In addition, significant association was observed between the NLRP3 rs10754558 polymorphism and susceptibility to AIDs, which tended to have a protective effect. Among the different races, this association was more significant among the Latin Americans. Analysis by the types of diseases showed that NLRP3 rs10754558 polymorphism was significantly related to the susceptibility of T1D, SLE, and RA, but not to other types of diseases.
Compared with previous meta-analyses, the published articles chosen in our research were stricter and more comprehensive. We searched all articles concerning the relationship between the NLRP3 rs35829419/rs10754558 polymorphisms and AIDs susceptibility and published in recent years through several databases, and excluded those inflammatory diseases such as Crohn's disease, ulcerative colitis, primary gout, and atherosclerosis, which were not removed in other meta-analyses. Previous meta-analyses focused on the relationship between NLRP3 polymorphisms and all inflammatory diseases, but our research was mainly concerned with the relationship between NLRP3 polymorphisms and AIDs. Finally, 12 qualified case–control studies were chosen in our research.
The NLRP3 rs35829419 variant is a C>A polymorphism located in exon 3 of its gene (Theodoropoulou et al., 2020). Considering that this variant is a gain-of-function mutation, it may lead to excessive production of IL-1β and IL-18. Therefore, researchers are keen to explore the role of NLRP3 rs35829419 in a variety of inflammatory diseases. However, current studies show it may have a different relationship with AIDs. Reportedly, the minor A allele of NLRP3 rs35829419 polymorphism seemingly exerts a protective role against the development of CD (Pontillo et al., 2011). However, another article about CD demonstrates that the NLRP3 rs35829419 A allele and especially the CA genotype seem to confer a higher risk for developing CD (Pontillo et al., 2010). Moreover, more evidence shows that NLRP3 rs35829419 is unrelated to the occurrence of AIDs, such as RA, CD, MS, and SLE. In addition, a meta-analysis by Zhang et al. includes multiple human diseases and shows that the NLRP3 rs35829419 gene polymorphism is generally a disease susceptibility factor related to various diseases (Zhang et al., 2015). In our meta-analysis, except for the homozygote co-dominant and recessive models, we found no significant association between NLRP3 rs35829419 and the overall AIDs, suggesting that polymorphism of this site may not play a key role in AIDs development. In a subgroup analysis by ethnicity, the allelic model showed a slightly, but not significantly, reduced risk of AIDs in the European population. Our meta-analysis results are consistent with Lee, who did not observe an association between autoimmune or inflammatory diseases and NLRP3 rs35829419 (Lee and Bae, 2016). Stratification by ethnicity also showed no association in the European, Latin American, or Polynesian populations. After stratification by disease type, the allelic and homozygote co-dominant models show that NLRP3 rs35829419 is significantly correlated to the susceptibility of RA, and the carrying of A allele contributes to the reduction of risk. In contrast to our results, an earlier meta-analysis shows that NLRP3 rs35829419 is not associated with the susceptibility to RA (Yang et al., 2017). In other AIDs, we found no association between NLRP3 rs35829419 and risk of AIDs in any genetic model. Although our analysis included more research data on RA, we cannot completely deny the irrelevance of NLRP3 rs35829419 gene polymorphism with RA. In fact, more research is required to explore the relationship.
The NLRP3 rs10754558 variant is a C>G polymorphism located in the 3′-UTR. Mutations in the 3′-UTR of NLRP3 gene may alter the activity of the inflammasome pathway by affecting the stability of mRNA and ultimately regulate the production of inflammatory factors including IL-1β and IL-18 (Hitomi et al., 2009). Compared with the C allele, the G allele of NLRP3 rs10754558 contributes to improving the stability of its mRNA, and the more stable NLRP3 expression may have a protective effect on the occurrence of T1D (Pontillo et al., 2010). However, the mechanism behind this seemingly contradictory phenomenon is unknown. Whether this enhanced NLRP3 mRNA stability affects the activation of inflammasomes and subsequent IL-1β secretion remains unknown. In this meta-analysis, we determine that NLRP3 rs10754558 in all genetic models exhibits a protective effect on reducing the risks of AIDs. The subgroup analysis of ethnicity shows a significant association between the NLRP3 rs10754558 polymorphism and AIDs in the Latin Americans, but not in the European, Arabian, or Asian populations. Ethnic differences in the population may be one of the root causes of this phenomenon, as the frequency of NLRP3 polymorphism may be different in different ethnic groups. This finding is consistent with a previous meta-analysis that includes other inflammatory diseases in addition to some AIDs (Lee and Bae, 2016). Meta-analysis by disease type shows no association between the NLRP3 rs10754558 polymorphism and MS, CD, or MG. However, in a variety of genetic models, meta-analysis results indicate that NLRP3 rs10754558 polymorphism is related to the susceptibility to T1D, RA, and SLE. NLRP3 rs10754558 polymorphism shows different associations with different AIDs, which may be related to the heterogeneity of the disease. Therefore, for the NLRP3 rs10754558 variant, a further study to confirm its protective effect against AIDs and its detailed mechanisms in AIDs progress is warranted. We speculate that this may be the result of a combination of innate immunity, local inflammation, and systemic environment.
There are some limitations that we cannot avoid. First, although the results of publication bias in our study are not significant, publication bias may also be caused by some unpublished studies of negative results or some articles ignored during the literature search. Second, some included studies have relatively small sample sizes, which may lead to lower statistical power, especially in the stratified analysis. Third, during subgroup analyses based on ethnicity and disease types, the numbers of studies in some subgroups are too small, which may reduce our analytical power. Moreover, in some studies, the control population does not conform to HWE, which may influence the accuracy of results. In addition, we found that although the authors of the original literature included in this meta-analysis did not statistically analyze age factors between the case and control groups for CD and T1D, there appeared to be significant age differences between the two groups. Whether it affects the association between NLRP3 polymorphism and susceptibility to CD or T1D is unclear. Finally, the research objects of our meta-analysis mainly focus on AIDs, including systemic AIDs and organ-specific AIDs. Different types of AIDs may have different pathogenesis due to their different pathophysiology and disease severity. NLPR3 may play a different role in different types and severity of AIDs. We speculate that these factors may have some influence on the results of the meta-analysis of the association of NLPR3 gene polymorphisms in autoimmune diseases.
In conclusion, the NLRP3 rs35829419 polymorphism may not be related to the risk of AIDs. However, NLRP3 rs10754558 allele G may have a protective effect on the risk of AIDs, especially in the Latin American population. In addition, NLRP3 rs10754558 polymorphism is associated with the occurrence of various AIDs, such as SLE, RA, and T1D. Further clinical research and functional analysis are needed to explore whether NLRP3 rs10754558 affects the production of cytokines through the inflammasome pathway.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author/s.
Author Contributions
ZW conducted the statistical analysis and revised the manuscript. TL designed the study and wrote the manuscript. SW collected the data and all authors contributed to the completion of the final manuscript.
Funding
This study was supported by the Health Commission of Hubei Province Scientific Research Project (No. WJ2021M239) and the Special Fund of Union Hospital of Tongji Medical College, Huazhong University of Science and Technology (Grant No. 02.03.2018-130).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We were also grateful to the reviewers for giving valuable advice and comments in the review.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2021.690860/full#supplementary-material
References
Addobbati, C., Alves da Cruz, H. L., Adelino, J. E., Melo, T. R. A. L., Fragoso, T. S., Domingues, A., et al. (2018). Polymorphisms and expression of inflammasome genes are associated with the development and severity of rheumatoid arthritis in Brazilian patients. Inflamm. Res. 67, 255–264. doi: 10.1007/s00011-017-1119-2
Agah, E., Nafissi, S., Saleh, F., Sarraf, P., Tafakhori, A., et al. (2021). Investigating the possible association between NLRP3 gene polymorphisms and myasthenia gravis. Muscle Nerve. 63, 730–736. doi: 10.1002/mus.27193
Alves da Cruz, H. L., Cavalcanti, C. A. J., de Azêvedo Silva, J., de Lima, C. A. D., Fragoso, T. S., Barbosa, A. D., et al. (2020). Differential expression of the inflammasome complex genes in systemic lupus erythematosus. Immunogenetics 72, 217–224. doi: 10.1007/s00251-020-01158-6
American Diabetes Association (2020). 2. Classification and diagnosis of diabetes: standards of medical care in diabetes-2020. Diabetes Care 43, S14–S31. doi: 10.2337/dc20-S002
Ben, H. M., Cornelis, F., Marzouk, S., Chabchoub, G., Bahloul, Z., Rebai, A., et al. (2012). Association study of CARD8 (p.C10X) and NLRP3 (p.Q705K) variants with rheumatoid arthritis in French and Tunisian populations. Int. J. Immunogenet. 39, 131–136. doi: 10.1111/j.1744-313X.2011.01070.x
de Torre-Minguela, C., Mesa, D. C. P., and Pelegrin, P. (2017). The NLRP3 and pyrin inflammasomes: implications in the pathophysiology of autoinflammatory diseases. Front. Immunol. 8:43. doi: 10.3389/fimmu.2017.00043
Evavold, C. L., and Kagan, J. C. (2019). Inflammasomes: threat-assessment organelles of the innate immune system. Immunity 51, 609–624. doi: 10.1016/j.immuni.2019.08.005
Fanouriakis, A., Tziolos, N., Bertsias, G., and Boumpas, D. T. (2021). Update omicronn the diagnosis and management of systemic lupus erythematosus. Ann. Rheum. Dis. 80, 14–25. doi: 10.1136/annrheumdis-2020-218272
Fusco, R., Siracusa, R., Genovese, T., Cuzzocrea, S., Cuzzocrea, S., and Di Paola, R. (2020). Focus on the role of NLRP3 inflammasome in diseases. Int. J. Mol. Sci. 21:4223. doi: 10.3390/ijms21124223
Gabrielsen, I. S., Amundsen, S. S., Helgeland, H., Flam, S. T., Hatinoor, N., Holm, K., et al. (2016). Genetic risk variants for autoimmune diseases that influence gene expression in thymus. Hum. Mol. Genet. 25, 3117–3124. doi: 10.1093/hmg/ddw152
Hitomi, Y., Ebisawa, M., Tomikawa, M., Imai, T., Komata, T., Hirota, T., et al. (2009). Associations of functional NLRP3 polymorphisms with susceptibility to food-induced anaphylaxis and aspirin-induced asthma. J. Allergy Clin. Immunol. 124, 779.e6–785.e6. doi: 10.1016/j.jaci.2009.07.044
Imani, D., Azimi, A., Salehi, Z., Rezaei, N., Emamnejad, R., Sadr, M., et al. (2018). Association of nod-like receptor protein-3 single nucleotide gene polymorphisms and expression with the susceptibility to relapsing-remitting multiple sclerosis. Int. J. Immunogenet. 45, 329–336. doi: 10.1111/iji.12401
Jenko, B., Praprotnik, S., Tomsic, M., and Dolzan, V. (2016). NLRP3 and CARD8 polymorphisms influence higher disease activity in rheumatoid arthritis. J. Med. Biochem. 35, 319–323. doi: 10.1515/jomb-2016-0008
Juneblad, K., Kastbom, A., Johansson, L., Rantapaa-Dahlqvist, S., Soderkvist, P., and Alenius, G. M. (2020). Association between inflammasome-related polymorphisms and psoriatic arthritis. Scand. J. Rheumatol. 50, 206–212. doi: 10.1080/03009742.2020.1834611
Kastbom, A., Johansson, M., Verma, D., Soderkvist, P., and Rantapaa-Dahlqvist, S. (2010). CARD8 p.C10X polymorphism is associated with inflammatory activity in early rheumatoid arthritis. Ann. Rheum. Dis. 69, 723–726. doi: 10.1136/ard.2008.106989
Kochi, Y. (2016). Genetics of autoimmune diseases: perspectives from genome-wide association studies. Int. Immunol. 28, 155–161. doi: 10.1093/intimm/dxw002
Kostine, M., Finckh, A., Bingham, C. O., Visser, K., Leipe, J., Schulze-koops, H., et al. (2021). EULAR points to consider for the diagnosis and management of rheumatic immune-related adverse events due to cancer immunotherapy with checkpoint inhibitors. Ann. Rheum. Dis. 80, 36–48. doi: 10.1136/annrheumdis-2020-217139
Lacy, B. E., Pimentel, M., Brenner, D. M., Chey, W. D., Keefer, L. A., Long, M. D., et al. (2021). ACG clinical guideline: management of irritable bowel syndrome. Am. J. Gastroenterol. 116, 17–44. doi: 10.14309/ajg.0000000000001036
Lee, Y. H., and Bae, S. C. (2016). Association between functional NLRP3 polymorphisms and susceptibility to autoimmune and inflammatory diseases: a meta-analysis. Lupus 25, 1558–1566. doi: 10.1177/0961203316644336
Lessard, C. J., Ice, J. A., Adrianto, I., Wiley, G. B., Kelly, J. A., Gaffney, P. M., et al. (2012). The genomics of autoimmune disease in the era of genome-wide association studies and beyond. Autoimmun. Rev. 11, 267–275. doi: 10.1016/j.autrev.2011.10.003
Luppi, P., Rossiello, M. R., Faas, S., and Trucco, M. (1995). Genetic background and environment contribute synergistically to the onset of autoimmune diseases. J. Mol. Med. 73, 381–393. doi: 10.1007/BF00240137
Narayanaswami, P., Sanders, D. B., Wolfe, G., Benatar, M., Cea, G., Evoli, A., et al. (2021). International consensus guidance for management of myasthenia gravis: 2020 update. Neurology 96, 114–122. doi: 10.1212/WNL.0000000000011124
Pontillo, A., Brandao, L., Guimaraes, R., Segat, L., Araujo, J., and Crovella, S. (2010). Two SNPs in NLRP3 gene are involved in the predisposition to type-1 diabetes and celiac disease in a pediatric population from northeast Brazil. Autoimmunity 43, 583–589. doi: 10.3109/08916930903540432
Pontillo, A., Girardelli, M., Kamada, A. J., Pancotto, J. A., Donadi, E. A., Crovella, S., et al. (2012). Polimorphisms in inflammasome genes are involved in the predisposition to systemic lupus erythematosus. Autoimmunity 45, 271–278. doi: 10.3109/08916934.2011.637532
Pontillo, A., Vendramin, A., Catamo, E., Fabris, A., and Crovella, S. (2011). The missense variation Q705K in CIAS1/NALP3/NLRP3 gene and an NLRP1 haplotype are associated with celiac disease. Am. J. Gastroenterol. 106, 539–544. doi: 10.1038/ajg.2010.474
Shastry, B. S. (2003). SNPs and haplotypes: genetic markers for disease and drug response (review). Int. J. Mol. Med. 11, 379–382. doi: 10.3892/ijmm.11.3.379
Shen, H. H., Yang, Y. X., Meng, X., Luo, X. Y., Li, X. M., Shuai, Z. W., et al. (2018). NLRP3: A promising therapeutic target for autoimmune diseases. Autoimmun. Rev. 17, 694–702. doi: 10.1016/j.autrev.2018.01.020
Smigoc, S. D., Goricar, K., Hovnik, T., Mendez, A., Bratina, N., Brecelj, J., et al. (2019). Dual role of PTPN22 BUT Not NLRP3 inflammasome polymorphisms in type 1 diabetes and celiac disease in children. Front Pediatr 7:63. doi: 10.3389/fped.2019.00063
Su, Z., Niu, Q., Huang, Z., Yang, B., and Zhang, J. (2020). Association of nucleotide-binding oligomerization domain-like receptor family pyrin domain-containing protein 3 polymorphisms with systemic lupus erythematosus disease activity and biomarker levels: a case-control study in Chinese population. Medicine 99:e21888. doi: 10.1097/MD.0000000000021888
Theodoropoulou, K., Wittkowski, H., Busso, N., Von Scheven-Gete, A., Moix, I., Vanoni, F., et al. (2020). Increased prevalence of NLRP3 Q703K variant among patients with autoinflammatory diseases: an international multicentric study. Front. Immunol. 11:877. doi: 10.3389/fimmu.2020.00877
Thompson, A. J., Banwell, B. L., Barkhof, F., Carroll, W. M., Coetzee, T., Comi, G., et al. (2018). Diagnosis of multiple sclerosis: 2017 revisions of the McDonald criteria. Lancet Neurol. 17, 162–173. doi: 10.1016/S1474-4422(17)30470-2
Verma, D., Sarndahl, E., Andersson, H., Eriksson, P., Fredrikson, M., Jonsson, J. I., et al. (2012). The Q705K polymorphism in NLRP3 is a gain-of-function alteration leading to excessive interleukin-1β and IL-18 production. PLoS ONE 7:e34977. doi: 10.1371/journal.pone.0034977
Villani, A. C., Lemire, M., Fortin, G., Louis, E., Silverberg, M. S., Collette, C., et al. (2009). Common variants in the NLRP3 region contribute to Crohn's disease susceptibility. Nat. Genet. 41, 71–76. doi: 10.1038/ng.285
Wang, Z., Zhang, S., Xiao, Y., Zhang, W., Wu, S., Qin, T., et al. (2020). NLRP3 inflammasome and inflammatory diseases. Oxid. Med. Cell. Longev. 2020:4063562. doi: 10.1155/2020/4063562
Wayant, C., Walters, C., Zaaza, Z., Gilstrap, C., Combs, T., Crow, H., et al. (2020). Evaluation of financial conflicts of interest among physician-authors of American College of Rheumatology Clinical Practice Guidelines. Arthr. Rheumatol. 72, 1427–1434. doi: 10.1002/art.41224
Yang, Z., Cao, J., Yang, Q., Zhang, Y., and Han, L. (2017). NLRP3 p.Q705K and CARD8 p.C10X single nucleotide polymorphisms are not associated with susceptibility to rheumatoid arthritis: a meta-analysis. Int. J. Rheum. Dis. 20, 1481–1491. doi: 10.1111/1756-185X.13016
Zhang, A. Q., Zeng, L., Gu, W., Zhang, L. Y., Zhou, J., Jiang, D. P., et al. (2011). Clinical relevance of single nucleotide polymorphisms within the entire NLRP3 gene in patients with major blunt trauma. Crit. Care 15:R280. doi: 10.1186/cc10564
Zhang, Q., Fan, H. W., Zhang, J. Z., Wang, Y. M., and Xing, H. J. (2015). NLRP3 rs35829419 polymorphism is associated with increased susceptibility to multiple diseases in humans. Genet. Mol. Res. 14, 13968–13980. doi: 10.4238/2015.October.29.17
Zhang, R., Li, H., Bai, L., and Duan, J. (2019). Association between T-Cell Immunoglobulin and Mucin Domain 3 (TIM-3) genetic polymorphisms and susceptibility to autoimmune diseases. Immunol. Invest. 48, 563–576. doi: 10.1080/08820139.2019.1599009
Zhao, C., Gu, Y., Zeng, X., and Wang, J. (2018). NLRP3 inflammasome regulates Th17 differentiation in rheumatoid arthritis. Clin. Immunol. 197, 154–160. doi: 10.1016/j.clim.2018.09.007
Keywords: NLRP3, single nucleotide polymorphisms, autoimmune diseases, susceptibility, meta-analysis
Citation: Wu Z, Wu S and Liang T (2021) Association of NLRP3 rs35829419 and rs10754558 Polymorphisms With Risks of Autoimmune Diseases: A Systematic Review and Meta-Analysis. Front. Genet. 12:690860. doi: 10.3389/fgene.2021.690860
Received: 15 April 2021; Accepted: 22 June 2021;
Published: 22 July 2021.
Edited by:
Ramcés Falfán-Valencia, Instituto Nacional de Enfermedades Respiratorias-México (INER), MexicoReviewed by:
Gloria Pérez-Rubio, Instituto Nacional de Enfermedades Respiratorias-México (INER), MexicoPradeep Kumar, Veer Bahadur Singh Purvanchal University, India
Marco Antonio Ponce-Gallegos, Civil Hospital of Guadalajara, Mexico
Copyright © 2021 Wu, Wu and Liang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Tao Liang, bHQtbGlhbmd0YW8tbHRAMTYzLmNvbQ==
 Zubo Wu
Zubo Wu Suyuan Wu2
Suyuan Wu2 Tao Liang
Tao Liang