- 1First Affiliated Hospital of USTC, Hefei National Laboratory for Physical Sciences at Microscale, School of Basic Medical Sciences, Division of Life Sciences and Medicine, University of Science and Technology of China, Hefei, China
- 2Jiangsu Key Laboratory for Biodiversity and Biotechnology, College of Life Sciences, Nanjing Normal University, Nanjing, China
- 3State Key Laboratory of Membrane Biology, National Engineering Laboratory for Anti-tumor Therapeutics, School of Medicine, Tsinghua University, Beijing, China
X-derived retrogenes contribute to genetic diversity in evolution and are usually specifically expressed in testis and perform important functions during spermatogenesis. Ubl4b is an autosomal retrogene with testis-specific expression derived from Ubl4a, an X-linked housekeeping gene. In the current study, we performed phylogenetic analysis and revealed that Ubl4a and Ubl4b are subject to purifying selection and may have conserved functions in evolution. Ubl4b was knocked out in mice using CRISPR/Cas9 genome editing technology and interestingly, we found no alterations in reproductive parameters of Ubl4b–/– male mice. To get insights into whether Ubl4a could compensate the absence of Ubl4b in vivo, we further obtained Ubl4a–/Y; Ubl4b–/– mice that lack both Ubl4a and Ubl4b, and the double knockout (dKO) mice also displayed normal spermatogenesis, showing that Ubl4a and Ubl4b are both dispensable for spermatogenesis. Thus, through the in vivo study of UBL4A and UBL4B, we provided a direct evidence for the first time that some X chromosome-derived autosomal retrogenes can be unfunctional in spermatogenesis, which represents an additional evolutionary type of X-derived retrogenes.
Introduction
Biological diversity depends upon the possible emergence of duplicate genes in the genome of organisms, and is also related with the changes in gene regulatory pathways (Gu et al., 2004; Johnson et al., 2007; ENCODE Project Consortium, 2012; Stunnenberg et al., 2016; Roundtree et al., 2017). Among the several mechanisms of procreating new genes, retroposition has been considered as one of the important mechanisms in diverse species (Long et al., 2003; Wang et al., 2006; Bai et al., 2007). Retroposition refers to the process where reverse transcriptase converts mRNAs of progenitor genes into DNAs followed by integration into the genome to generate retrogenes (Schrader and Schmitz, 2019). Retrogenes show diverse movement patterns because of their different genomic positions compared with progenitor genes, which contributes to genetic diversity in evolution (Dai et al., 2006; Schrader and Schmitz, 2019).
Emerging data reveal that 14 out of 20 retrogenes derived from 16 X-linked progenitor genes are specifically expressed in testis (Wang, 2004). During meiosis, most X-linked genes become transcriptionally silenced because of meiotic sex chromosome inactivation (MSCI), and their functions can be compensated by corresponding retrogenes (Wang, 2004; Turner, 2015). We previously demonstrated that the X-linked gene RPL10, encoding a ribosomal protein, and its autosomal paralog RPL10L can functionally compensate for each other in vitro and in vivo (Jiang et al., 2017). The majority of the retrotransposed genes are inactivated and become pseudogenes, and some of the retrogenes perform important functions during male meiosis (Schrader and Schmitz, 2019). For example, the knockout of the well-known testis-specific retrogenes, such as Cetn1, Cstf2t, Pgk2 and Rpl10l, can result in spermatogenic failure (Dass et al., 2007; Danshina et al., 2010; Tardif et al., 2010; Avasthi et al., 2013; Jiang et al., 2017).
Generally, ubiquitin-like (UBL) proteins modify their target substrates and may have important functions in spermatogenesis (Schwartz and Hochstrasser, 2003). As a member of the UBL proteins, Ubl4b is also an autosomal retrogene with testis-specific expression derived from Ubl4a. Ubl4a (NM_145405) maps to X chromosome and is ubiquitously expressed in different tissues (Wang et al., 2012), hence regarded as a housekeeping gene. It exerts a significant effect in protein metabolism and the maintenance of cellular homeostasis. For example, UBL4A can maintain the innate immune response through positively regulating NF-κB signaling in dendritic cells and macrophages (Liu et al., 2019). It also plays an antitumor role on autophagy-related proliferation and metastasis in pancreatic ductal adenocarcinoma by directly targeting LAMP1 (Chen et al., 2019a). Intriguingly, Ubl4a knockout (KO) mice were viable and during 6 months after the birth, no obvious abnormality was observed in the development and growth (Wang et al., 2012). What’s more, single knockout of Ubl4a does not affect spermatogenesis and fertility in mice, suggesting a possible compensatory role by the Ubl4b retrogene (Wang et al., 2012). However, the role of Ubl4b and its relationship with Ubl4a are still needed to be elucidated.
Thus, the current study was focused to investigate in vivo function of evolutionarily conserved Ubl4b gene by generation of knockout mice. We also obtained double mutant mice that lack both Ubl4a and Ubl4b to get insights into the association of Ubl4a and Ubl4b in vivo. Interestingly, we found no alterations in reproductive parameters of Ubl4b–/– male mice, and Ubl4a–/Y; Ubl4b–/– mice also displayed normal spermatogenesis. Thus, Ubl4a and Ubl4b are both dispensable for spermatogenesis through the in vivo study, which provided a direct evidence for the first time that some X chromosome-derived autosomal retrogenes can be unfunctional in spermatogenesis.
Materials and Methods
Phylogenetic Analysis and Selection Analysis of UBL4A and UBL4B
Entire coding nucleotide and amino acid sequences of UBL4A and UBL4B in different vertebrates were downloaded from National Center for Biotechnology Information (NCBI). UBL domain-based protein sequences were extracted from UniProt database. The alignments were performed using the online software MultAlin1. The phylogenetic construction was performed on MEGA 6.06 software with the following workflow: Muscle was used for the alignment of entire coding nucleotide sequences of UBL4A and UBL4B in different eukaryotic species with default settings (Edgar, 2004). Phylogenetic trees were constructed by the Neighbor-joining (NJ) method. The parameters used for the tree construction include phylogeny test (bootstrap method with 1,000 replicates), substitutions type (nucleotide), model/method (maximum composite likelihood), substitutions (transitions and transversions), rates among sites (uniform rates), pattern among lineages (same/homogeneous), gaps/missing data treatment (complete deletion), and codon positions (1st, 2nd, 3rd and non-coding sites). To investigate whether UBL4A and UBL4B genes have undergone statistically significant differences in selection pressures, we employed the branch model in the CodeML program using the phylogenetic analysis by maximum-likelihood (PAML) software version 4.4 with default settings.
Mouse Models
CRISPR/Cas9 technology was utilized to make Ubl4b mutant mice as we described previously (Wang et al., 2013). The following sequence 5′TCAGCACCTACAGGTGCCCG3′ was designed to target the exon 1 of Ubl4b. Sanger sequencing of toe biopsies was carried out and heterozygous founder mice were bred to obtain homozygous mice. All mice were nourished with proper food and ddH2O, and kept in definite photoperiod (lights on 08:00–20:00). All animal experiments were approved by the Institutional Animal Care Committee of the University of Science and Technology of China. Primers that were used for genotyping are listed in Supplementary Table 1.
Western Blot
Testes from 10-week-old mice were homogenized in lysis buffer (50 mM Tris, pH 7.5, 150 mM NaCl, 0.5% Triton X–100, 5 mM EDTA and 1 mM Na3VO4) containing protease inhibitors (Roche). Western blot was carried out as described previously (Jiang et al., 2014). The primary antibodies are rabbit anti-β-actin (Abcam, ab8227; 1:3,000), rabbit anti-UBL4A (Yang et al., 2007) and rabbit anti-UBL4B (Yang et al., 2007). The secondary antibody is HRP-conjugated donkey anti-rabbit IgG (BioLegend, 406401; 1:10,000).
Fertility Test
To check the fertility status, each 10-week-old male mouse was mated with two 8-week-old wild-type (WT) females (C57BL/6J) for 3 months. All the females were monitored for pregnancy. Dates of birth, numbers of pups and offspring sex ratios were recorded for all the litters.
Hematoxylin and Eosin (H&E) Staining
Adult mice at 10 weeks old were sacrificed, then their testes and epididymides were detached immediately and fixed overnight in Bouin’s solution. Further H&E staining was carried out as explained previously (Jiang et al., 2015). To reduce the experimental variations, all these procedures were performed simultaneously in WT and KO mice.
Sperm Analysis
For sperm counting, epididymides were removed and chopped into small pieces. After incubation at 37°C for 30 min, a hemocytometer was used for cell counting. For sperm morphology, smear slides were analyzed by H&E staining. The percentages of morphologically normal sperm were quantified with at least 400 sperm examined for each mouse. For sperm motility, cauda epididymides from 10-week-old mice were collected and sperm were incubated in human tubal fluid (HTF) media (Millipore) supplemented with 10% FBS at 37°C for 5 min. Sperm samples were diluted and analyzed using Hamilton Thorne’s Ceros II system.
Statistical Analysis
Student’s t-test was performed to statistically compare the teste/body weight ratios, litter sizes, and sperm parameters. Results were presented as mean ± SEM from at least three mice for each group and p < 0.05 was considered significant.
Results
Conserved Evolution of UBL4A and Its Retrogene UBL4B
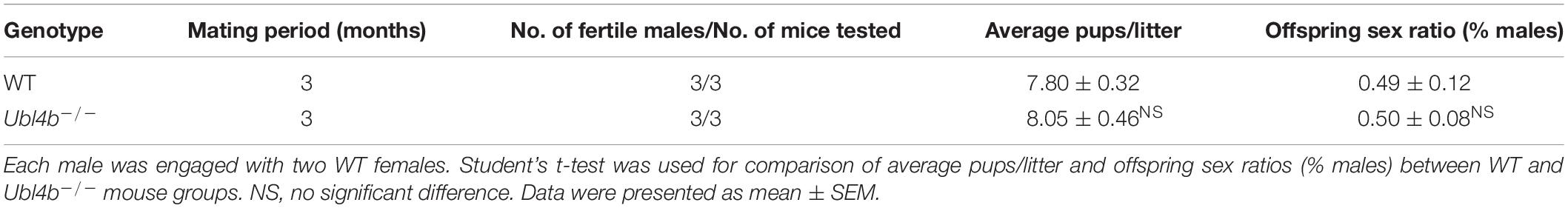
To explore the origin of UBL4A and UBL4B individually, the genetic relationships among various organisms were included. Since it has been previously reported that retroposition of Ubl4b probably occurred at least 170 million years ago, prior to the radiation of therian mammals (Yang et al., 2007), we collected detailed information (Taxonomy ID, Organism name, Gene ID, Chromosome and Exon count) of UBL4A and UBL4B among vertebrates from NCBI database (Supplementary Table 2). We found that UBL4A genes are detected in vertebrates such as fishes, amphibians, reptiles, birds and mammals, while UBL4B members are only present in reptiles and mammals (Supplementary Table 2). Almost the half of the UBL4A genes are located on X chromosomes with four exons, while most of the UBL4B genes exist on autosomes with only one exon (Supplementary Table 2). The entire coding nucleotide and amino acid sequences of UBL4A and UBL4B among vertebrates were downloaded from NCBI database. Multiple sequence alignments of the entire coding nucleotide and amino acid sequences revealed a high similarity in UBL4A and UBL4B (Supplementary Figures 1–4). In particular, amino acid sequence alignments from UBL domains of UBL4A and UBL4B in various organisms showed a higher similarity (Figures 1A,B). Phylogenetic trees were then constructed based on entire coding nucleotide sequences of UBL4A and UBL4B using MEGA 6.06 software and NJ method, and further revealed that UBL4A and UBL4B among vertebrates may derive from the corresponding ancestral genes, respectively (Figures 1C,D). Thus, these results based on sequence and phylogenetic analysis indicated that UBL4A and UBL4B are conserved in vertebrates.
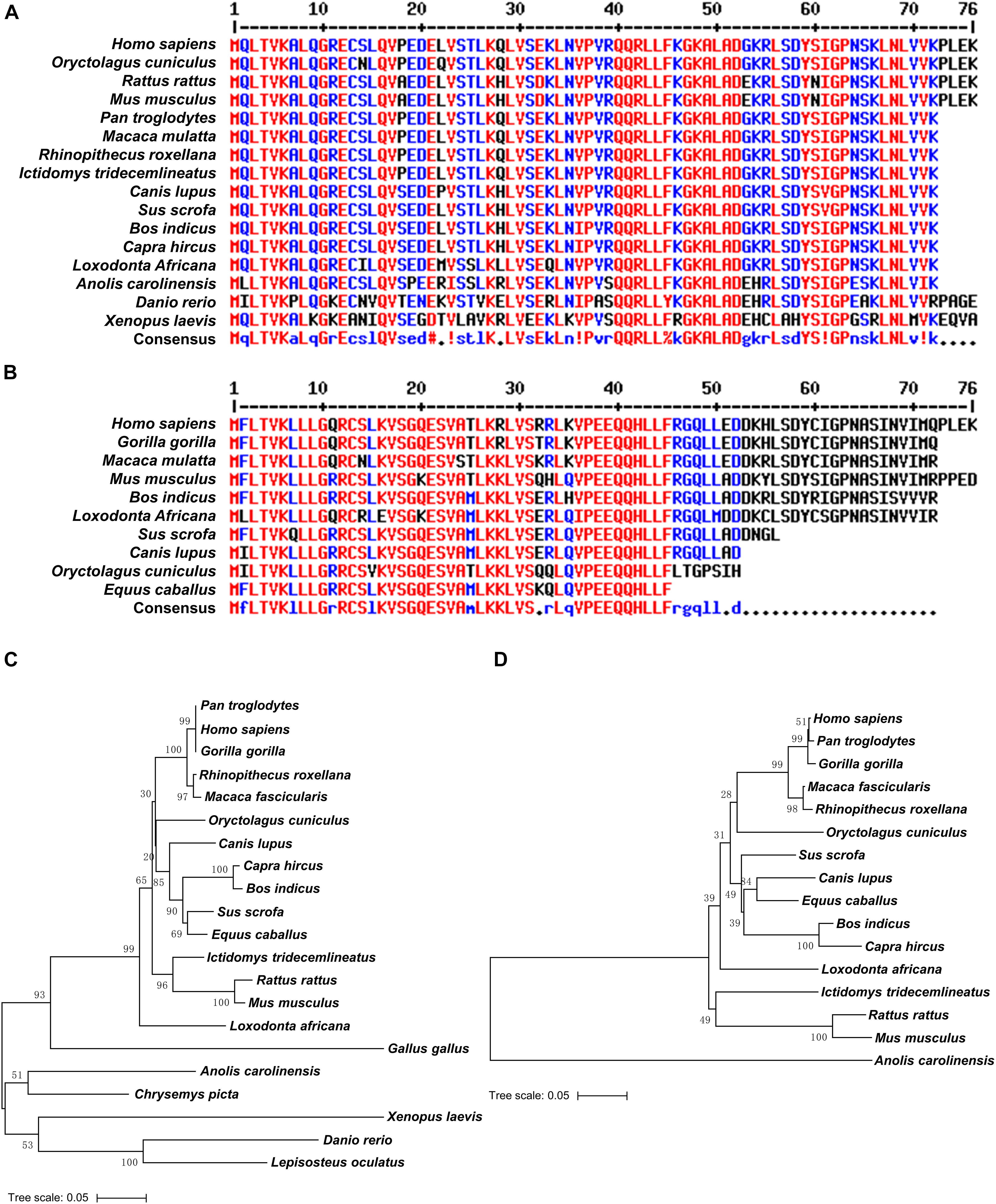
Figure 1. UBL4A and its retrogene UBL4B are evolutionarily conserved. (A,B) Amino acid sequence alignments of UBL domains in UBL4A (A) and UBL4B (B) from various organisms. Residues that are identical appear in red and as uppercase letters in the consensus line. Residues highly similar are indicated by red symbols (!, any one of I and V;%, any one of F and Y; #, any one of N, D, Q, E, B, and Z). Unconserved residues are written in blue or as asterisks in the consensus line. (C,D) The phylogenetic trees based on entire coding nucleotide sequences of UBL4A (C) and UBL4B (D) in various organisms. The bootstrap confidence values of nodes are displayed.
To trace the evolution of UBL4A and UBL4B, we performed phylogenetic analysis on UBL4A and UBL4B among vertebrates simultaneously and found that UBL4A and UBL4B may originate from a common ancestral gene (Figure 2). Furthermore, UBL4A and UBL4B in mammalian vertebrates notably gather together, respectively, in the phylogenetic tree (Figure 2). To better understand which selection UBL4A and UBL4B underwent in the course of evolution, we utilized the branch model in the CodeML program for the phylogenetic analysis with PAML software 4.4. The non-synonymous substitution per non-synonymous site (dN)/synonymous substitution per synonymous site (dS), termed as the ω value, indicates the nature of selective forces. We defined the phylogeny of UBL4A in mammalian vertebrates as a foreground branch, the other phylogenies as a background branch, and calculated ω and p-values (Figure 2). The ω and p-values for above group were 0.07033 and 0.00067, showing that the branch of UBL4A in mammalian vertebrates evolves under purifying selection. Similarly, we defined the phylogeny of UBL4B in mammalian vertebrates as a foreground branch, the other phylogenies as a background branch, and figured ω and p-values (Figure 2). The ω and p-values for this group were 0.00303 and 0.00204, revealing that the branch of UBL4B in mammalian vertebrates is under strong purifying selection process in evolution. Thus, these results suggested that UBL4A and UBL4B may have conserved functions in the course of evolution.
Figure 2. Purifying selection of UBL4A and UBL4B in various organisms. The phylogenetic tree based on entire coding nucleotide sequences of UBL4A and UBL4B in various organisms. The number on the branch shows the ω value. The ω values refer to the non-synonymous substitution per non-synonymous site (dN)/synonymous substitution per synonymous site (dS). The bootstrap confidence values of nodes are also displayed.
Ubl4b Is Dispensable for Male Fertility and Spermatogenesis
In order to investigate the function of Ubl4b in vivo, we generated Ubl4b knockout mice using CRISPR/Cas9 technology (Supplementary Figure 5A). The knockout of Ubl4b was firstly validated by a large deletion at the genomic DNA level (Supplementary Figure 5B). Sanger sequencing confirmed an 80 base pairs (bp) deletion in Ubl4b, which introduced a frameshift mutation at amino acid 13 (Supplementary Figure 5C). Then, Western blot confirmed a complete absence of UBL4B protein in Ubl4b–/– testis, which further confirmed the successful deletion of Ubl4b in our gene modified mice (Figure 3A).
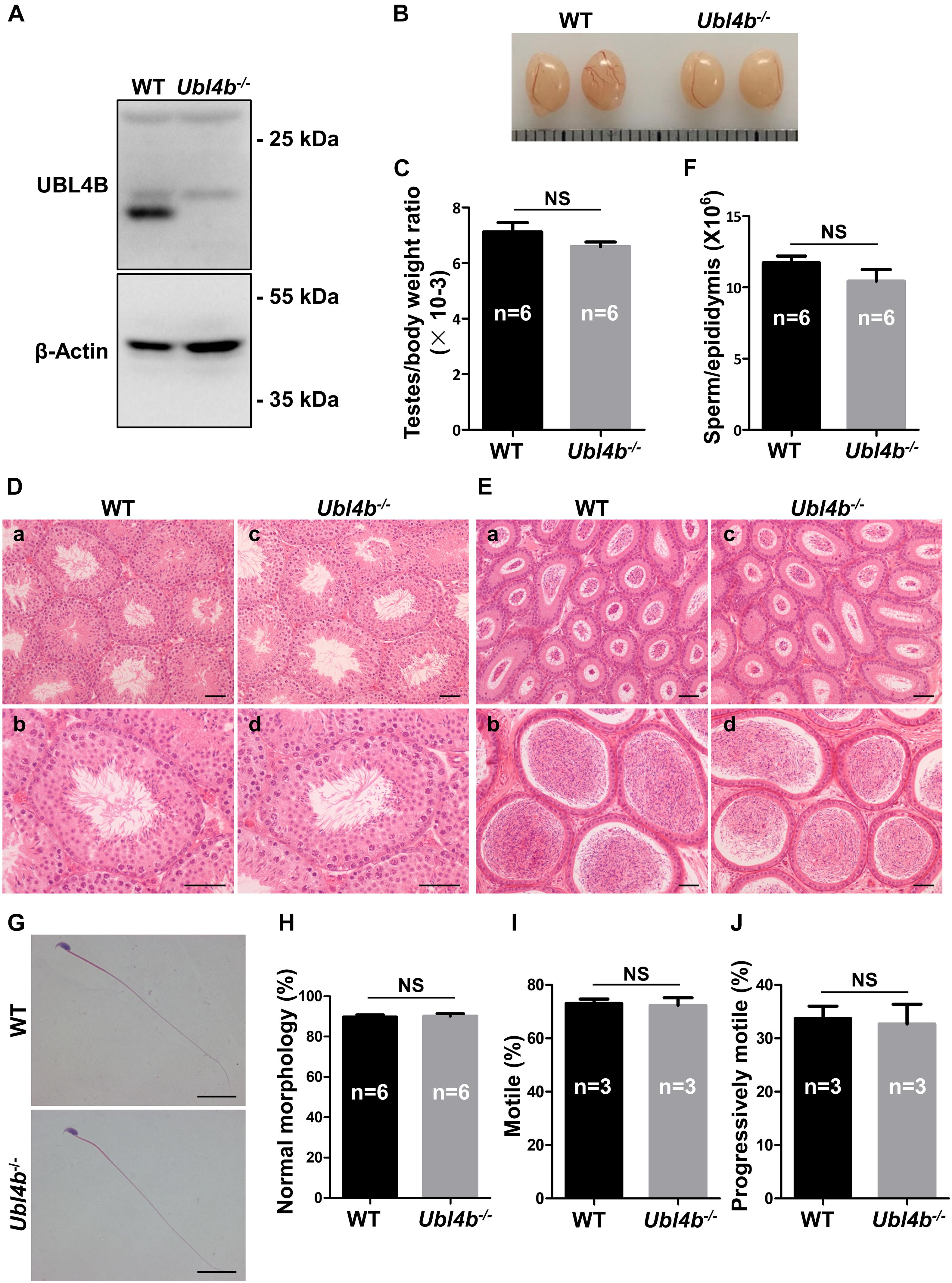
Figure 3. Spermatogenesis in Ubl4b knockout mice. (A) Western blot analysis of UBL4B expression in testes from 10-week-old WT and Ubl4b–/– mice. β-Actin was used as the loading control. (B) Representative image of testes from 10-week-old WT and Ubl4b–/– mice. (C) Ratios of testes to body weight were determined from 10-week-old WT and Ubl4b–/– mice. (D) H&E staining of testes from 10-week-old WT (a,b) and Ubl4b–/– (c, d) mice. Scale bars, 50 μm. (E) H&E staining of caput and cauda epididymides from 10-week-old WT (a, b) and Ubl4b–/– (c,d) mice. Scale bars, 50 μm. (F) Average sperm count in unilateral epididymis from 10-week-old WT and Ubl4b–/– mice. (G) H&E staining of sperm in cauda epididymides from 10-week-old WT and Ubl4b–/– mice. Scale bars, 20 μm. (H) Percentages of sperm with normal morphology were shown. (I,J) Percentages of motile sperm (I) and progressively motile sperm (J) from 10-week-old WT and Ubl4b–/– mice. n, the number of animals. The data shown were represented as the mean ± SEM. Student’s t-test was performed between WT and Ubl4b–/– mice. NS, no significant difference.
Ubl4b–/– mice were outward normal, and showed normal growth and development similar to their WT littermates. Subsequently, the fertility status was analyzed by keeping WT and Ubl4b–/– male mice with WT females, respectively. The statistical analysis showed that the Ubl4b–/– male mice had comparable number of pups per litter and offspring sex ratios to the WT group, which suggested that the deficiency of Ubl4b does not affect male fertility (Table 1).
To determine the possible role of Ubl4b in spermatogenesis, we firstly analyzed the testes from adult WT and Ubl4b–/– mice and found no any noteworthy between-group difference in the testis morphology and testes to body weight ratios (Figures 3B,C). Moreover, the histology of testes by H&E staining revealed intact seminiferous tubule architecture along with the presence of all types of germ cells from spermatogonia to spermatozoa in Ubl4b–/– mice (Figure 3D). Additionally, the cauda epididymides of adult Ubl4b–/– mice were filled with abundant mature spermatozoa (Figure 3E). Unilateral epididymal sperm from adult WT and Ubl4b–/– mice were counted and statistical analysis showed that there is no significant difference (Figure 3F). Then we further compared the sperm parameters of WT and Ubl4b–/– mice. Epididymal sperm from adult Ubl4b–/– mice displayed normal morphology, similar to that of WT mice (Figures 3G,H). Furthermore, we did not observe any remarkable difference in sperm motility between adult WT and Ubl4b–/– mice (Figures 3I,J). Taken altogether, these findings suggested that Ubl4b is not required for normal fertility and spermatogenesis in mice.
Double Knockout of Ubl4a and Ubl4b Has No Effect on Fertility and Spermatogenesis
As Ubl4b–/– mice showed normal fertility and spermatogenesis, we hypothesized that Ubl4a and Ubl4b may be functionally redundant. In order to investigate the functions of these two genes in vivo, we crossed Ubl4a–/Y mice with Ubl4b–/– female mice (Wang et al., 2012). After two generations of breeding, we obtained desired Ubl4a–/Y; Ubl4b–/– mice. The genotypes of Ubl4a–/Y; Ubl4b–/– mice were firstly verified on genomic level (Supplementary Figure 6). Then, the observation of complete absence of both UBL4A and UBL4B proteins in Ubl4a–/Y; Ubl4b–/– testes confirmed the successful deletion of Ubl4a and Ubl4b simultaneously (Figure 4A).
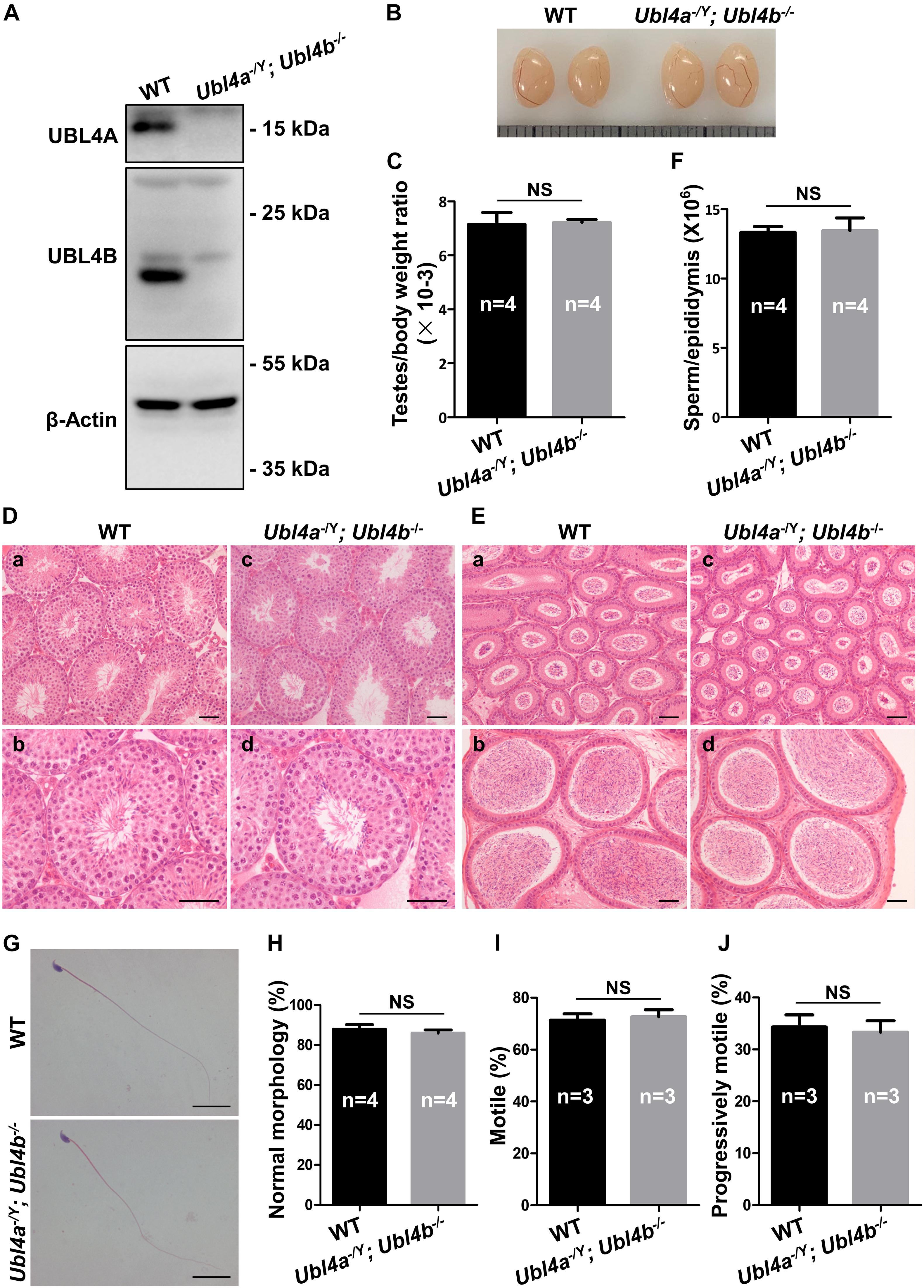
Figure 4. Spermatogenesis in Ubl4a and Ubl4b double knockout mice. (A) Western blot analysis of UBL4A and UBL4B expression in testes from 10-week-old WT and Ubl4a–/Y; Ubl4b–/– mice. β-Actin was used as the loading control. (B) Representative image of testes from 10-week-old WT and Ubl4a–/Y; Ubl4b–/– mice. (C) Ratios of testes to body weight were determined from 10-week-old WT and Ubl4a–/Y; Ubl4b–/– mice. (D) H&E staining of testes from 10-week-old WT (a,b) and Ubl4a–/Y; Ubl4b–/– (c,d) mice. Scale bars, 50 μm. (E) H&E staining of caput and cauda epididymides from 10-week-old WT (a,b) and Ubl4a–/Y; Ubl4b–/– (c,d) mice. Scale bars, 50 μm. (F) Average sperm count in unilateral epididymis from 10-week-old WT and Ubl4a–/Y; Ubl4b–/– mice. (G) H&E staining of sperm in cauda epididymides from 10-week-old WT and Ubl4a–/Y; Ubl4b–/– mice. Scale bars, 20 μm. (H) Percentages of sperm with normal morphology were shown. (I,J) Percentages of motile sperm (I) and progressively motile sperm (J) from 10-week-old WT and Ubl4a–/Y; Ubl4b–/– mice. n, the number of animals. The data shown were represented as the mean ± SEM. Student’s t-test was performed between WT and Ubl4a–/Y; Ubl4b–/– mice. NS, no significant difference.
After the confirmation of successful generation of Ubl4a–/Y; Ubl4b–/– mice, we assessed the fertility of WT and dKO males. Mice lacking both Ubl4a and Ubl4b showed normal growth and development. The average pups per litter and offspring sex ratios of adult Ubl4a–/Y; Ubl4b–/– mice were not significantly different from those in WT group (Table 2).
The testis morphology and testes to body weight ratios of adult Ubl4a–/Y; Ubl4b–/– mice did not exhibit notable difference from those of WT group (Figures 4B,C). Subsequently, histological analysis showed normal spermatogenesis in seminiferous tubules and abundant sperm in epididymides of Ubl4a–/Y; Ubl4b–/– mice (Figures 4D,E). Epididymal sperm count did not significantly differ between adult WT and Ubl4a–/Y; Ubl4b–/– mice (Figure 4F). Moreover, sperm morphology was analyzed by H&E staining and we found no obvious difference between adult WT and Ubl4a–/Y; Ubl4b–/– mice (Figures 4G,H). Additionally, sperm motility of adult WT and Ubl4a–/Y; Ubl4b–/– mice was indistinguishable from each other (Figures 4I,J). Altogether, these data strongly demonstrated that the simultaneously deletion of Ubl4a and its retrogene, Ubl4b, did not affect male fertility and spermatogenesis in mice.
Discussion
The mammalian genome contains various retrotransposed genes and most of them are transcriptionally silent (Venter et al., 2001; Brosius, 2019). Here we showed that Ubl4a and Ubl4b are subject to purifying selection in phylogenetic analysis and may have conserved functions in evolution. In order to determine the functional role of testis-specific X-derived retrogene Ubl4b in mice, we generated a knockout mouse line by CRISPR/Cas9 technology and found no alternations in reproductive parameters. Subsequently, to explore the possibility of compensation by the ubiquitously expressed Ubl4a gene, we further produced double mutant mice lacking both Ubl4a and Ubl4b genes. Interestingly, adult Ubl4a–/Y; Ubl4b–/– mice also displayed normal fertility and spermatogenesis, showing dispensable roles of both genes in mouse reproduction.
During male meiosis, most of the X-linked genes are transcriptionally silent due to formation of heterochromatin XY body and the functions of these genes can be compensated by their corresponding autosomal retrogenes so that spermatocytes can complete meiosis (Richler et al., 1994; Turner et al., 2002). The expression profile indicates that Ubl4a transcription is transiently silenced during MSCI, while Ubl4b mRNA could be detected from pachytene stage, indicating that Ubl4b may compensate the silence of Ubl4a in MSCI (Yang et al., 2007). Previously knockout of such types of retrogenes, including Cetn1, Cstf2t, Pgk2 and Rpl10l, always resulted in spermatogenic abnormalities and male infertility (Dass et al., 2007; Danshina et al., 2010; Tardif et al., 2010; Avasthi et al., 2013; Jiang et al., 2017). However, the deletion of either Ubl4a or Ubl4b in mice showed no overt abnormalities in spermatogenesis and fertility (Wang et al., 2012). What is more, the mice with dKO of Ubl4a and Ubl4b also displayed normal spermatogenesis with no obvious alternation in fertility status. Thus, our unexpected results revealed that both genes have no roles in fertility and spermatogenesis.
Several hypotheses have been described about the evolution and function of retrogenes in mammals. One of them is the compensation hypothesis of X-derived retrogenes, for example, we previously demonstrated that the X-linked gene RPL10, encoding a ribosomal protein, and its autosomal paralog RPL10L can functionally compensate for each other (Jiang et al., 2017). Another hypothesis about retrogene evolution is the specialization hypothesis in which many X-originated retrogenes have evolved novel functions to meet the special needs of germ cells. This is supported by different mechanisms including alternative splicing, polyadenylation and apoptosis in somatic and germ cells, and the obvious examples of genes are hnRNPG-T, CSTF2T, and BIRC8 (Elliott et al., 2000; Richter et al., 2001; Dass et al., 2002; Grozdanov et al., 2018; Bousoik et al., 2019; Zhou et al., 2019; Yan et al., 2021). RBMX protein has been implicated in splice site selection, and RBMX-derived retrogene product, hnRNPG-T, could be specialized in alternative splicing in germ cells (Elliott et al., 2000; Zhou et al., 2019; Yan et al., 2021). CSTF2T could function in the non-canonical polyadenylation of mRNAs in germ cells (Dass et al., 2002; Grozdanov et al., 2018). Another well-known hypothesis regarding X-derived retrogenes is the haploid syncytium hypothesis based on non-equality of sex chromosomes in haploid germ cells and this disequilibrium is fulfilled by the presence of “back up” autosomal retrogenes (Dahl et al., 1990; Casola and Betran, 2017). An explanatory example of this hypothesis is that G6pd2, a spermatid specific expressed gene, is considered as a “back up” gene for equal amount of its product for both X and Y bearing spermatids (Hendriksen et al., 1997; Homolka et al., 2011). In addition, the possession of novel adjacent sequences in the form of untranslated regions (UTRs) to attain a special regulatory mechanism was put forward to explain the emergence of retrogenes from X chromosomes (Shiao et al., 2007). The function and evolution of these autosomal retrogenes derived from X-linked genes might be unrelated with MSCI during male meiosis and an explanatory example is that CHML, an ubiquitous expressed gene, has been reported to have function in cancer but not in reproduction (Chen et al., 2019b). However, none of these hypotheses can explain the observation that both X-linked Ubl4a and its autosomal retrogene, Ubl4b are dispensable in spermatogenesis.
In the current study, we found Ubl4a and Ubl4b double knockout mice evinced normal fertility and spermatogenesis similar to Ubl4a and Ubl4b single knockout mice, which does not support the existing hypotheses for X-derived retrogenes. We deduce that the evolutionarily conserved Ubl4a and Ubl4b genes may initially have essential functions in spermatogenesis, but are no longer required during evolution. On the other hand, it is also plausible that Ubl4b may be derived from insertion of Ubl4a mRNA into autosomal genomes by accident, which did not lead to any deleteriousness and thus was kept during evolution or has not yet begun its degeneration to oblivion. What is more, Ubl4b and Ubl4b may be hitchhiked by other genes under purifying selection in the genome. For example, Ubl4a is close to mouse Slc10a3, while Ubl4b is near Slc6a17, in which both Slc10a3 and Slc6a17 are regarded as housekeeping genes that play important functions in many biological processes. Alternatively, a few substitutions are sufficient to acquire a new function, and UBL4A and UBL4B may have obtained some important functions that is not related to male fertility. Thus, we propose a new “junk” hypothesis that some autosomal retrogenes derived from X-linked progenitor genes may not have functions in spermatogenesis, which is supported by the in vivo study of Ubl4a and Ubl4b. In future, more intensive and sophisticated studies are needed to reinforce this hypothesis by other groups of X-linked genes and autosomal counterparts. Additionally, all the examinations were performed under normal laboratory mating conditions and challenging the UBL system in mice, such as a toxicology study, deserves further consideration.
Conclusion
We generated Ubl4b–/– and Ubl4a–/Y; Ubl4b–/– mice which displayed normal fertility and spermatogenesis, proposing that both X-linked Ubl4a and its autosomal counterpart Ubl4b are not necessary for male fertility and providing the first direct evidence for the “junk” hypothesis regarding X-derived retrogenes.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author/s.
Ethics Statement
The animal study was reviewed and approved by the Institutional Animal Care Committee of the University of Science and Technology of China.
Author Contributions
QS, XJ, and CY: conceptualization. XJ and CY: formal analysis. CY and RD: investigation. QS, ZC, and CD: resources. RK and CY: writing—original draft preparation. XJ, HM, and CY: writing—review and editing. QS: project administration and funding acquisition. All authors contributed to the article and approved the submitted version.
Funding
This research was funded by the National Key Research and Developmental Program of China (2018YFC1003400 and 2018YFC1004700), the Strategic Priority Research Program of the Chinese Academy of Sciences (XDB19000000), the National Natural Science Foundation of China (31890780, 31630050, 32061143006, 31871514, and 82071709), the Fundamental Research Funds for the Central Universities (YD2070002006), and the Natural Science Foundation of Qinghai (2019-HZ-823).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Dr. P. Jeremy Wang for anti-UBL4A and anti-UBL4B antibodies. We thank Haidi Chen for evolutionary analysis.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2021.689902/full#supplementary-material
Supplementary Figure 1 | CDS sequence alignment of UBL4A in various organisms.
Supplementary Figure 2 | CDS sequence alignment of UBL4B in various organisms.
Supplementary Figure 3 | Amino acid sequence alignment of UBL4A in various organisms. Residues that are identical appear in red and as uppercase letters in the consensus line. Residues highly similar are indicated by red symbols (!, any one of I and V; $, any one of L and M;%, any one of F and Y; #, any one of N, D, Q, E, B, and Z). Unconserved residues are written in blue or as asterisks in the consensus line.
Supplementary Figure 4 | Amino acid sequence alignment of UBL4B in various organisms. Residues that are identical appear in red and as uppercase letters in the consensus line. Residues highly similar are indicated by red symbols (!, any one of I and V; $, any one of L and M; #, any one of N, D, Q, E, B, and Z). Unconserved residues are written in blue or as asterisks in the consensus line.
Supplementary Figure 5 | Generation of Ubl4b knockout mice. (A) Schematic strategy of sgRNA designed for Ubl4b knockout. Diagram of the mouse Ubl4b locus is based on Ensembl data (transcript ENSMUST00000052853.7), with blue and white bars representing coding and non-coding regions, respectively. PAM, protospacer adjacent motif. (B) PCR using genomic DNA (gDNA) from WT and Ubl4b–/– mice was performed to verify the deletion of Ubl4b in the knockout mice. M, marker. (C) Representative Sanger sequencing chromatograms confirmed the genotypes of WT and Ubl4b–/– mice. Red arrow above the chromatograms in aligned sequences indicates the deletion.
Supplementary Figure 6 | Generation of Ubl4a and Ubl4b double knockout mice. PCR using gDNA from WT and Ubl4a–/Y; Ubl4b–/– mice was performed to verify the deletions of Ubl4a and Ubl4b in the double knockout mice. M, marker.
Footnotes
References
Avasthi, P., Scheel, J. F., Ying, G., Frederick, J. M., Baehr, W., and Wolfrum, U. (2013). Germline deletion of Cetn1 causes infertility in male mice. J. Cell Sci. 126, 3204–3213.
Bai, Y., Casola, C., Feschotte, C., and Betran, E. (2007). Comparative genomics reveals a constant rate of origination and convergent acquisition of functional retrogenes in Drosophila. Genome Biol. 8:R11.
Bousoik, E., Nabiee, R., Amirrad, F., Nichols, A., Witt, R., Mahdipoor, P., et al. (2019). Heterogeneity and plasticity of human breast cancer cells in response to molecularly-targeted drugs. Front. Oncol. 9:1070.
Brosius, J. (2019). Exaptation at the molecular genetic level. Sci. China Life Sci. 62, 437–452. doi: 10.1007/s11427-018-9447-8
Casola, C., and Betran, E. (2017). The genomic impact of gene retrocopies: what have we learned from comparative genomics, population genomics, and transcriptomic analyses? Genome Biol. Evol. 9, 1351–1373. doi: 10.1093/gbe/evx081
Chen, H., Li, L., Hu, J., Zhao, Z., Ji, L., Cheng, C., et al. (2019a). UBL4A inhibits autophagy-mediated proliferation and metastasis of pancreatic ductal adenocarcinoma via targeting LAMP1. J. Exp. Clin. Cancer Res. 38:297.
Chen, T. W., Yin, F. F., Yuan, Y. M., Guan, D. X., Zhang, E., Zhang, F. K., et al. (2019b). CHML promotes liver cancer metastasis by facilitating Rab14 recycle. Nat. Commun. 10:2510.
ENCODE Project Consortium (2012). An integrated encyclopedia of DNA elements in the human genome. Nature 489, 57–74. doi: 10.1038/nature11247
Dahl, H.-H., Brown, R., Hutchison, W., Maragos, C., and Brown, G. (1990). A testis-specific form of the human pyruvate dehydrogenase E1α subunit is coded for by an intronless gene on chromosome 4. Genomics 8, 225–232. doi: 10.1016/0888-7543(90)90275-y
Dai, H., Yoshimatsu, T. F., and Long, M. (2006). Retrogene movement within- and between-chromosomes in the evolution of Drosophila genomes. Gene 385, 96–102. doi: 10.1016/j.gene.2006.04.033
Danshina, P. V., Geyer, C. B., Dai, Q., Goulding, E. H., Willis, W. D., Kitto, G. B., et al. (2010). Phosphoglycerate kinase 2 (PGK2) is essential for sperm function and male fertility in mice. Biol. Reprod. 82, 136–145. doi: 10.1095/biolreprod.109.079699
Dass, B., Mcdaniel, L., Schultz, R. A., Attaya, E., and Macdonald, C. C. (2002). The gene CSTF2T, encoding the human variant CstF-64 polyadenylation protein tauCstF-64, lacks introns and may be associated with male sterility. Genomics 80, 509–514. doi: 10.1006/geno.2002.6862
Dass, B., Tardif, S., Park, J. Y., Tian, B., Weitlauf, H. M., Hess, R. A., et al. (2007). Loss of polyadenylation protein tauCstF-64 causes spermatogenic defects and male infertility. Proc. Natl. Acad. Sci. U.S.A. 104, 20374–20379. doi: 10.1073/pnas.0707589104
Edgar, R. C. (2004). MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797. doi: 10.1093/nar/gkh340
Elliott, D. J., Venables, J. P., Newton, C. S., Lawson, D., Boyle, S., Eperon, I. C., et al. (2000). An evolutionarily conserved germ cell-specific hnRNP is encoded by a retrotransposed gene. Hum. Mol. Genet. 9, 2117–2124. doi: 10.1093/hmg/9.14.2117
Grozdanov, P. N., Li, J., Yu, P., Yan, W., and Macdonald, C. C. (2018). Cstf2t Regulates expression of histones and histone-like proteins in male germ cells. Andrology 6, 605–615. doi: 10.1111/andr.12488
Gu, Z., Rifkin, S. A., White, K. P., and Li, W. H. (2004). Duplicate genes increase gene expression diversity within and between species. Nat. Genet. 36, 577–579. doi: 10.1038/ng1355
Hendriksen, P. J., Hoogerbrugge, J. W., Baarends, W. M., De Boer, P., Vreeburg, J. T., Vos, E. A., et al. (1997). Testis-specific expression of a functional retroposon encoding glucose-6-phosphate dehydrogenase in the mouse. Genomics 41, 350–359. doi: 10.1006/geno.1997.4673
Homolka, D., Ivanek, R., Forejt, J., and Jansa, P. (2011). Differential expression of non-coding RNAs and continuous evolution of the X chromosome in testicular transcriptome of two mouse species. PLoS One 6:e17198. doi: 10.1371/journal.pone.0017198
Jiang, L., Li, T., Zhang, X., Zhang, B., Yu, C., Li, Y., et al. (2017). RPL10L is required for male meiotic division by compensating for RPL10 during meiotic sex chromosome inactivation in mice. Curr. Biol. 27, 1498–1505.e1496.
Jiang, X., Ma, T., Zhang, Y., Zhang, H., Yin, S., Zheng, W., et al. (2015). Specific deletion of Cdh2 in Sertoli cells leads to altered meiotic progression and subfertility of mice. Biol. Reprod. 92:79.
Jiang, X., Zhang, H., Yin, S., Zhang, Y., Yang, W., Zheng, W., et al. (2014). Specific deficiency of Plzf paralog, Zbtb20, in Sertoli cells does not affect spermatogenesis and fertility in mice. Sci. Rep. 4:7062.
Johnson, D. S., Mortazavi, A., Myers, R. M., and Wold, B. (2007). Genome-wide mapping of in vivo protein-DNA interactions. Science 316, 1497–1502. doi: 10.1126/science.1141319
Liu, C., Zhou, Y., Li, M., Wang, Y., Yang, L., Yang, S., et al. (2019). Absence of GdX/UBL4A protects against inflammatory diseases by regulating NF-small ka, cyrillicB signaling in macrophages and dendritic cells. Theranostics 9, 1369–1384. doi: 10.7150/thno.32451
Long, M., Betran, E., Thornton, K., and Wang, W. (2003). The origin of new genes: glimpses from the young and old. Nat. Rev. Genet. 4, 865–875. doi: 10.1038/nrg1204
Richler, C., Ast, G., Goitein, R., Wahrman, J., Sperling, R., and Sperling, J. (1994). Splicing components are excluded from the transcriptionally inactive XY body in male meiotic nuclei. Mol. Biol. Cell 5, 1341–1352. doi: 10.1091/mbc.5.12.1341
Richter, B. W., Mir, S. S., Eiben, L. J., Lewis, J., Reffey, S. B., Frattini, A., et al. (2001). Molecular cloning of ILP-2, a novel member of the inhibitor of apoptosis protein family. Mol. Cell Biol. 21, 4292–4301. doi: 10.1128/mcb.21.13.4292-4301.2001
Roundtree, I. A., Evans, M. E., Pan, T., and He, C. (2017). Dynamic RNA modifications in gene expression regulation. Cell 169, 1187–1200. doi: 10.1016/j.cell.2017.05.045
Schrader, L., and Schmitz, J. (2019). The impact of transposable elements in adaptive evolution. Mol. Ecol. 28, 1537–1549. doi: 10.1111/mec.14794
Schwartz, D. C., and Hochstrasser, M. (2003). A superfamily of protein tags: ubiquitin, SUMO and related modifiers. Trends Biochem. Sci. 28, 321–328. doi: 10.1016/s0968-0004(03)00113-0
Shiao, M. S., Khil, P., Camerini-Otero, R. D., Shiroishi, T., Moriwaki, K., Yu, H. T., et al. (2007). Origins of new male germ-line functions from X-derived autosomal retrogenes in the mouse. Mol. Biol. Evol. 24, 2242–2253. doi: 10.1093/molbev/msm153
Stunnenberg, H. G. International Human Epigenome Consortium and Hirst, M. (2016). The international human epigenome consortium: a blueprint for scientific collaboration and discovery. Cell 167, 1145–1149.
Tardif, S., Akrofi, A. S., Dass, B., Hardy, D. M., and Macdonald, C. C. (2010). Infertility with impaired zona pellucida adhesion of spermatozoa from mice lacking TauCstF-64. Biol. Reprod. 83, 464–472. doi: 10.1095/biolreprod.109.083238
Turner, J. M. (2015). Meiotic silencing in mammals. Annu. Rev. Genet. 49, 395–412. doi: 10.1146/annurev-genet-112414-055145
Turner, J. M., Mahadevaiah, S. K., Elliott, D. J., Garchon, H. J., Pehrson, J. R., Jaenisch, R., et al. (2002). Meiotic sex chromosome inactivation in male mice with targeted disruptions of Xist. J. Cell Sci. 115, 4097–4105. doi: 10.1242/jcs.00111
Venter, J. C., Adams, M. D., Myers, E. W., Li, P. W., Mural, R. J., Sutton, G. G., et al. (2001). The sequence of the human genome. Science 291, 1304–1351.
Wang, H., Yang, H., Shivalila, C. S., Dawlaty, M. M., Cheng, A. W., Zhang, F., et al. (2013). One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering. Cell 153, 910–918. doi: 10.1016/j.cell.2013.04.025
Wang, P. J. (2004). X chromosomes, retrogenes and their role in male reproduction. Trends Endocrinol. Metab. 15, 79–83. doi: 10.1016/j.tem.2004.01.007
Wang, W., Zheng, H., Fan, C., Li, J., Shi, J., Cai, Z., et al. (2006). High rate of chimeric gene origination by retroposition in plant genomes. Plant Cell 18, 1791–1802. doi: 10.1105/tpc.106.041905
Wang, Y., Wang, D., Ren, F., Zhang, Y., Lin, F., Hou, N., et al. (2012). Generation of mice with conditional null allele for GdX/Ubl4A. Genesis 50, 534–542. doi: 10.1002/dvg.20832
Yan, Q., Zeng, P., Zhou, X., Zhao, X., Chen, R., Qiao, J., et al. (2021). RBMX suppresses tumorigenicity and progression of bladder cancer by interacting with the hnRNP A1 protein to regulate PKM alternative splicing. Oncogene 40, 2635–2650. doi: 10.1038/s41388-021-01666-z
Yang, F., Skaletsky, H., and Wang, P. J. (2007). Ubl4b, an X-derived retrogene, is specifically expressed in post-meiotic germ cells in mammals. Gene Expr. Patterns 7, 131–136. doi: 10.1016/j.modgep.2006.06.002
Keywords: X chromosome, retrogene, evolution, Ubl4a, Ubl4b, spermatogenesis
Citation: Yu C, Diao R, Khan R, Deng C, Ma H, Chang Z, Jiang X and Shi Q (2021) The Dispensable Roles of X-Linked Ubl4a and Its Autosomal Counterpart Ubl4b in Spermatogenesis Represent a New Evolutionary Type of X-Derived Retrogenes. Front. Genet. 12:689902. doi: 10.3389/fgene.2021.689902
Received: 01 April 2021; Accepted: 02 June 2021;
Published: 25 June 2021.
Edited by:
Tarun Jairaj Narwani, Institut National de la Santé et de la Recherche Médicale (INSERM), FranceReviewed by:
Izabela Makałowska, Adam Mickiewicz University, PolandSayaka Miura, Temple University, United States
Copyright © 2021 Yu, Diao, Khan, Deng, Ma, Chang, Jiang and Shi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Qinghua Shi, cXNoaUB1c3RjLmVkdS5jbg==; Xiaohua Jiang, YmlvanhoQHVzdGMuZWR1LmNu
 Changping Yu
Changping Yu Runjie Diao2
Runjie Diao2 Ranjha Khan
Ranjha Khan Zhijie Chang
Zhijie Chang Xiaohua Jiang
Xiaohua Jiang Qinghua Shi
Qinghua Shi