- 1College of Biological and Pharmaceutical Engineering, West Anhui University, Lu’an, China
- 2Anhui Engineering Laboratory for Conservation and Sustainable Utilization of Traditional Chinese Medicine Resources, West Anhui University, Lu’an, China
- 3College of Life Science, Anhui Agricultural University, Hefei, China
Dendrobium is a semi-shade epiphytic Orchidaceae herb with important ornamental and medicinal value. Parts of the cultivation of Dendrobium germplasm resources, as well as the identification of medicinal components, are more studied, but the functional characterization of the flowering regulation in Dendrobium plants is less reported. Here, six PEBP family genes (DhFT3, DhFT1, DhMFT, DhTFL1b, DhFT2, and DhTFL1a) were identified from the Dendrobium huoshanense genome. The chromosome-level mapping showed that these genes were sequentially distributed on chromosomes 6, 9, 15, and 17. The paralogous gene DhTFL1b corresponded to DhTFL1a, which was determined through tandem duplication. The gene structure and conserved motif of DhPEBP indicated five PEBP genes apart from DhMFT contained four exons and three introns entirely. The phylogeny analysis showed that the PEBP gene family in A. thaliana, O. sativa, Z. mays, S. lycopersicum, and P. equestris were classified into three subclades, FT, TFL, and MFT, which maintained a high homology with D. huoshanense. The conserved domain of the amino acid demonstrated that two highly conserved short motifs (DPDXP and GXHR) embed in DhPEBPs, which may contribute to the conformation of the ligand binding bag. The 86th position of DhFTs was tyrosine (Y), while the 83th and 87th of DhTFL1s belonged to histidine (H), suggesting they should have distinct functions in flowering regulation. The promoter of six DhPEBPs contained several cis-elements related to hormone induction, light response, and abiotic stress, which indicated they could be regulated by the environmental stress and endogenous signaling pathways. The qRT-PCR analysis of DhPEBPs in short-term days induced by GA indicated the gene expressions of all DhFTs were gradually increased, whereas the expression of DhTFL1 was decreased. The results implied that DhPEBPs have various regulatory functions in modulating flowering, which will provide a scientific reference for the flowering regulation of Dendrobium plants.
Introduction
The traditional wild Dendrobium huoshanense experiences issues such as difficulty in natural pollination, fewer capsules, and low seed germination. Although asexual reproduction has been widely developed in the tissue culture of D. huoshanense, the degradation of germplasm is much more serious than sexual reproduction. Flower organs are the sexual reproduction organs of angiosperms, and the entire process of flower formation is essentially the basis for the procreation of offspring (Chen and Penfield, 2018). The flower-forming transition of plants from vegetative growth to reproductive growth is a key step to adapt to the external environment and ensure the reproduction of offspring (Liu et al., 2019). The timing of these transitions is precisely regulated by endogenous signals and the external environment, among which photoperiod signals can prompt plants to bloom at the most appropriate time (Plackett et al., 2018).
The protein encoded by phosphatidylethanolamine-binding protein (PEBP) is widely present in dicotyledonous monocots, and its family consists of three subfamilies, the FT-like, MFT-like, and TFL1-like (Auge et al., 2018). FLOWERING LOCUS T (FT), as a florigen-inducing gene, has been identified in model plants such as Arabidopsis, rice, and maize (Ahluwalia and Hatsukami, 2015; Jin et al., 2019). FT is an important integration factor in the flowering regulation pathway of plants and has been one of the key genes regulating flowering (Adeyemo et al., 2019; Bi et al., 2019). It transmits signals to the downstream flower development-related CONSTANS (CO) by sensing the vernalization pathway, gibberellin pathway, photoperiod pathway, and autonomous regulation pathways (Melzer, 2017; Xiao et al., 2018). Under long-day conditions, CO induces the expression of FT, and the FT protein binds to FLOWERING LOCUS D (FD) protein in the stem end meristem (SAM) to promote the expression of APETALA1 (AP1) (Li et al., 2015). TWIN SISTER OF FT (TSF), the homologous gene of FT, regulates the early flowering of A. thaliana and exhibits a redundant function similar to that of overexpression of FT (Wang et al., 2019). Another branch of the PEBP family is the TERMINAL FLOWER 1 (TFL1) subfamily, whose main function is to maintain vegetative growth and infinite growth of inflorescences. In Arabidopsis, TFL1 controls the morphological structure of the plant by regulating the meristem genes, LEAFY (LFY) and AP1, present in SAM (Wang et al., 2017). In addition, TFL1 plays a role in inhibiting flowering during flowering, and it exhibits completely different characteristics from FT (Jin et al., 2021). ARABIDOPSIS THALIANA CENTRORADIALIS HOMOLOGU (ATC) is classified into the same subclass as TFL1 and is homologous to the snapdragon CEN gene. ATC overexpression can complement the late flowering phenotype of tfl1, but the atc mutant has no obvious early flowering phenomenon (González-Suárez et al., 2020). Brother of FT and TFL1 (BFT) is another member of the TFL1 subfamily. Overexpression of BFT in A. thaliana shows delayed flowering, but the bft mutant does not show a similar phenotype to the tfl1 mutant (Zhang et al., 2016). MOTHER OF FT AND TFL1 (MFT) is the ancestral gene of FT and TFL1. Overexpression of AtMFT can lead to early flowering, but it is not as significant as FT. In addition, MFT is specifically expressed in seeds, mainly through ABA and GA signaling pathways to participate in seed germination regulation (Yu et al., 2019). In dicotyledonous plants like A. thaliana, tomato, grape, and poplar, 6–9 genes are contained in this PEBP family (Luo et al., 2018). However, the number of PEBP family members in monocotyledonous plants is about three times that of dicotyledonous plants. O. sativa and T. aestivum have 17 and 30 PEBP genes, respectively. Through large fragments and genome-wide replication, plants have produced a large number of repetitive genes in the process of evolution. Some functions are redundant, some genes are silent as non-function, and some neofunctionalization genes have novel functions due to mutations (Schiessl et al., 2017). Six conserved ligand-binding sites in PEBP proteins of different species form a pocket-like structure, in which two amino acids are the key sites that determine the function of FT/TFL1. TFL1 not only affects flowering time but also affects inflorescence morphology. In some plants, TFL1 homologous genes may have different regulatory functions. For example, the DET and LF genes in pea control the flowering time and the developmental state of the apical meristem, respectively (Johnson et al., 2019).
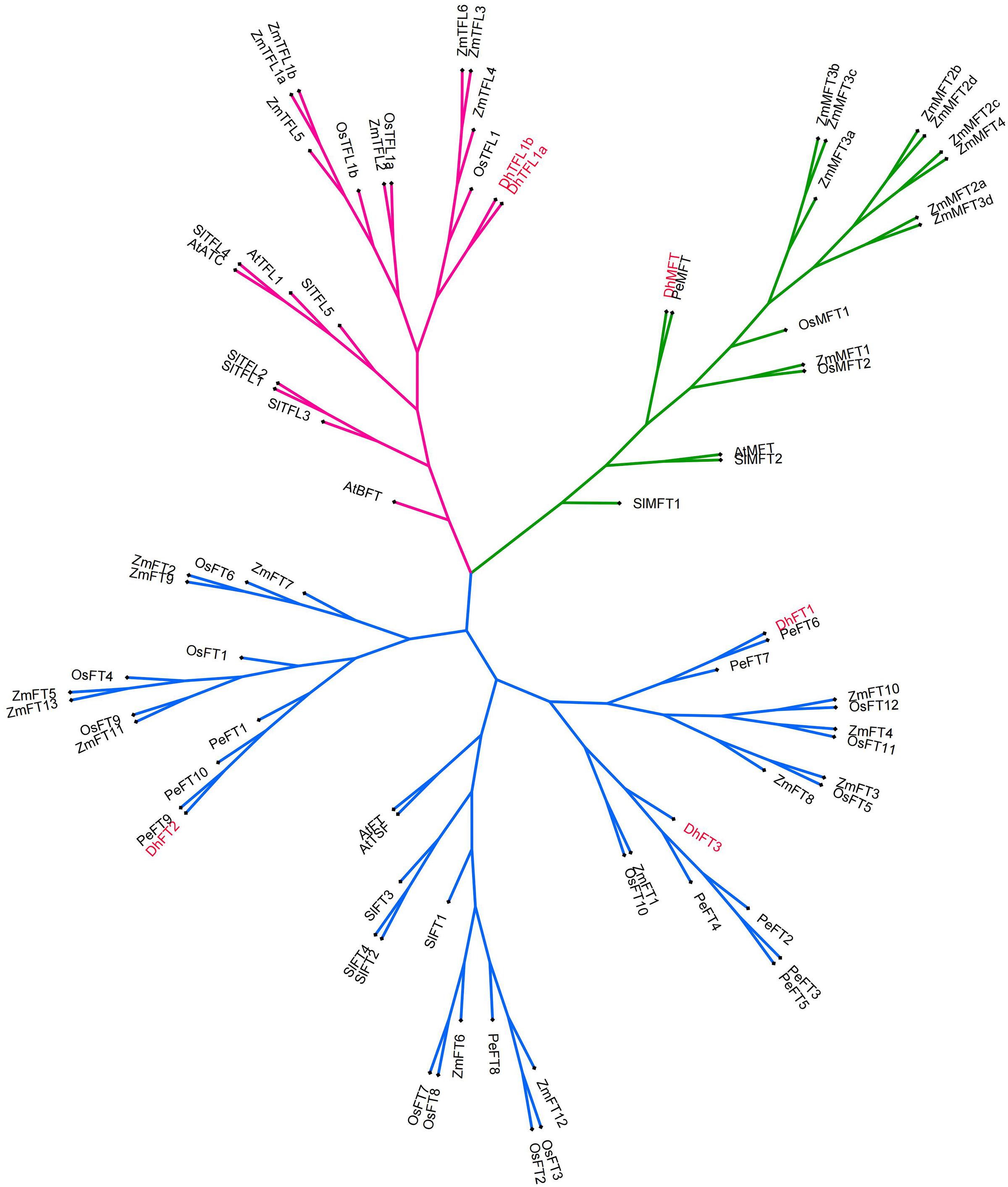
To solve the bottleneck behind the sexual reproduction of D. huoshanense, the genetic background of D. huoshanense, the regulation mechanism of key genes related to flowering need to be clarified. Based on the previous genome sequencing, six PEBP family genes were screened from the whole genome of D. huoshanense. Among them, the two subfamilies FT and TFL1 have unique regulatory effects in the flowering initiation process. Phylogeny analysis showed that MFT, FT, and TFL1 of D. huoshanense could be clustered with PEBP family of other species. By calculating the ka/ks of DhPEBP orthologous genes, all genes had suffered purified selections. Collinearity analysis revealed that the FT/TFL1 of D. huoshanense and some PEBP family genes in O. sativa, Z. mays, and G. max have collinearity. Amino acid alignment analysis showed that the 86th amino acid of DhFT2 and 87th of DhTFL1a were different. The promoter analysis indicated that signal elements related to stress and gibberellin binding possessed in DhFT and DhTFL1. Quantitative fluorescence analysis verified the flower-promoting effect of DhFT1 and DhFT3 as well as the flower-inhibiting effect of DhTFL1a. These studies would provide scientific reference for elucidating the different roles of PEBP family members in the flowering regulation of D. huoshanense.
Materials and Methods
The tissue culture material of D. huoshanense used in the experiment came from the Anhui Engineering Technology Research Center of Plant Cell Engineering of West Anhui University (Lu’an City, Anhui Province). In total, 0.5 mM GA3 solution was sprayed once a day on the leaves of D. huoshanense. The samples were taken every 5 days, and each sample was biologically repeated three times. The control group was sprayed with clean water in the same way. After removing the leaves with scissors, we put them in liquid nitrogen and immediately froze them before transferring them to a 5mL screw tube to store them in a refrigerator at −80 degrees for later use.
Identification of PEBP Family Genes in D. huoshanense
Firstly, the consensus conserved seed file (PF01161) of the hidden Markov model (HMM) was downloaded from the Pfam website1. Then, the HMM profile was performed as a query to identify all PBP-containing domain in D. huoshanense by retrieving against the genome with a threshold of e-value of < e–3. All candidate DhPEBPs were verified by Pfam2, SMART3, and InterPro4 for further confirmation. The same method was used to screen out PEBP genes in A. thaliana, O. sativa subsp. japonica, Z. mays, S. lycopersicum, and P. equestris. The genome sequence and CDS files of A. thaliana, O. sativa subsp. japonica, Z. mays, and S. lycopersicum were obtained from Ensembl Plants database5. The genome sequence and CDS file of P. equestris were got from the NCBI database6. Based on the sequence alignments generated by the Muscle method in MEGA software (v.6.06), all putative redundant PEBP sequences were discarded.
The Chromosome Location and Gene Duplication Events
By using TBtools (v.1.089), the chromosome location information of the PEBP gene was obtained (Chen et al., 2020). Firstly, the gene density information from the GFF3 file was extracted, and the gene location visualization from the GTF/GFF tool was used to obtain the chromosome location map. The Genome gene dot plot tool was used to draw a dot plot of the gene duplication events. Using MEME suite7 for conservative motif analysis, the conservative motif of PEBP gene was obtained by searching the protein sequence. The number of motifs was set at 20, and the width of motifs was in 6 to 200, and select the default parameters to get the meme.xml file. TBtools was used to visualize the motif pattern and the gene structure analysis.
The Phylogeny Analysis of PEBP Genes
Neighbor-joining (NJ) and maximum-likelihood (ML) methods were used to construct the phylogenetic tree of PEBP genes in A. thaliana, O. sativa subsp. japonica, Z. mays, S. lycopersicum, and P. equestris. MEGA 6 was used to align the target protein sequence (Tamura et al., 2013). The aligned file was used to construct the phylogenetic tree and bootstrap consensus tree by using the NJ method. Bootstrap replications value is set to 1,000, and the substitution model was the Poisson model. Gaps and missing dates were treated in pairwise deletion. Using IQ-TREE (v.1.6.6), a phylogenetic tree was construct based on the ML method.
Identification of Orthologous and Paralogous Genes
Using Orthovenn28, the orthologous and paralogous genes in A. thaliana, O. sativa subsp. japonica, Z. mays, S. lycopersicum, and G. max were identified and compared to obtain the phylogenetic trees to identify the PEBP homologous genes of D. huoshanense. Through Orthovenn2 analysis, the common and unique clusters information of the five species is obtained. Using six DhPEBPs family genes as templates, the clusters associated with them are retrieved.
The Pressure Analysis of Evolutionary Selection
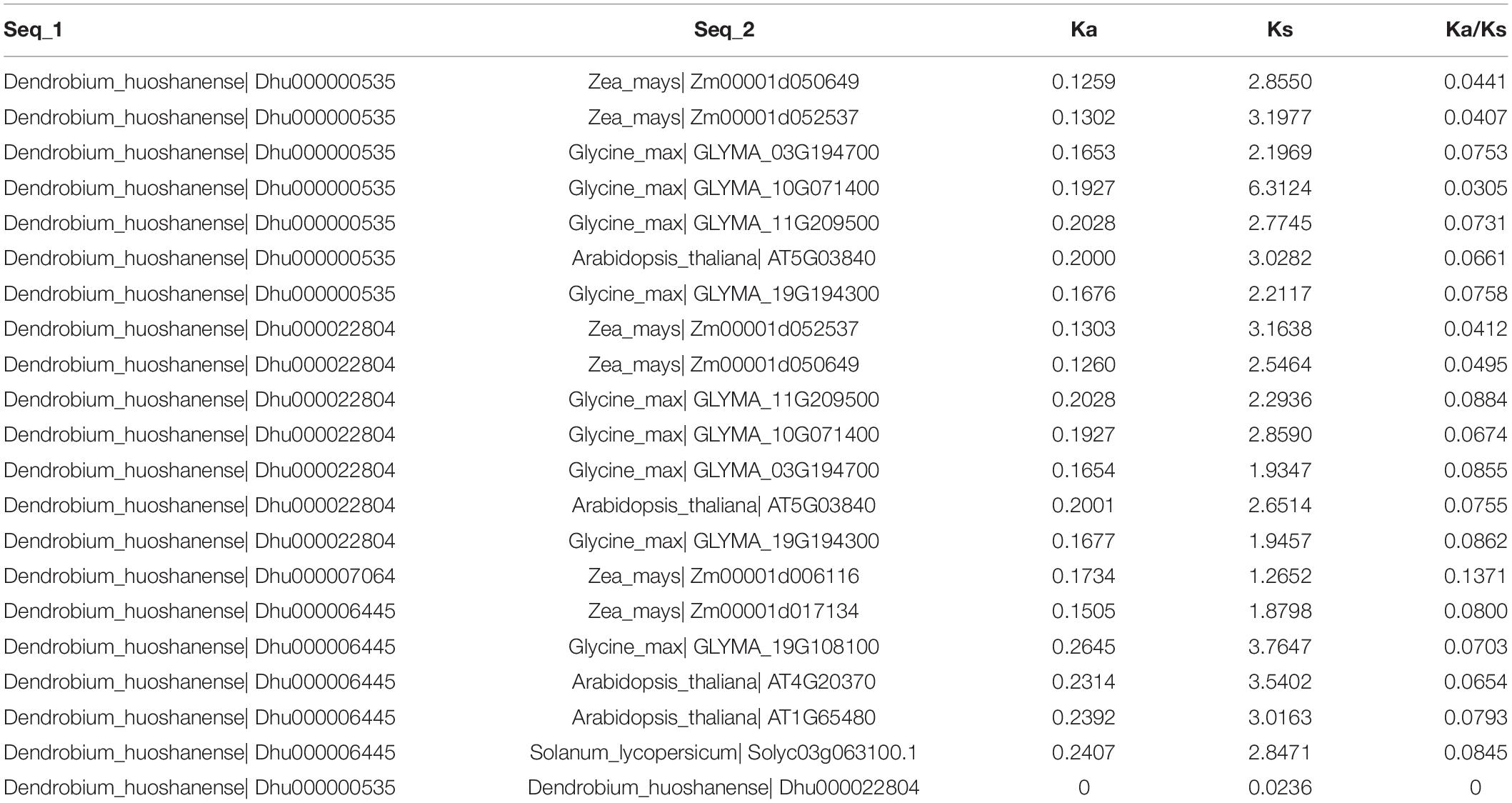
The Ka and Ks values of the obtained orthologous gene pairs were used to calculate the ka/ks ratio of all the homologous gene pairs. Meanwhile, DnaSP (v.5.10.01) was used to recalculate each gene pair to remove the gene pair with high disproportionation value, and, finally, we obtained the homologous gene pairs with normal ka/ks ratio (Ge et al., 2017).
The Collinearity Analysis of PEBP Genes
Based on the genome sequence and gene annotation files, we used the one-step MCScanX plug-in in TBtools to obtain the collinearity files between every two species, including collinearity, gene linkage, and basic gene replication.
Amino Acid Alignment and the Conserved Domain Analysis
MEGA 6 was used to compare the PEBP protein sequences of A. thaliana, O. sativa subsp. japonica, Z. mays, S. lycopersicum, and P. equestris. According to the level of amino acid homology, Genedoc9 was used for amino acid coloring.
The cis-Acting Element Analysis of DhPEBPs
To find out the responsive elements in the promoter, TBtools was used to extract the upstream 2,000 bp of gene sequence from the genome sequence of D. huoshanense (Carvalho et al., 2015). First, the GFF/GTF sequence extractor tool in the TBtools software was used to obtain the promoter regions of all genes. Then, the quick fasta extractor or filter tool was used to extract the promoter sequence of all PEBP family genes. PlantCARE10 was used to analyze the cis-regulatory elements in the sequences. The BioSequence Viewer tool was used to visualize the promoter elements of DhPEBPs.
The qRT-PCR Analysis of DhPEBPs
RNAprep Pure Plant Kit (Takara, Japan) was used to extract total RNA from Dendrobium leaves of CK group and GA-treated groups for 5 and 10 days. An ultra-micro spectrophotometer was used to detect RNA concentration and quality. The 7,500 series real-time fluorescent quantitative PCR (Bio-RAD, America) was used for the quantitative analysis. The CDS sequences of the six genes DhFT3, DhFT1, DhMFT, DhTFL1b, DhFT2, and DhTFL1a were obtained from the genome of D. huoshanense, and the primers for the fluorescent qPCR were designed respectively (Supplementary Table 1). The total RNA of the above samples was reverse transcribed into cDNA with PrimeScript RT reagent Kit (Takara, Japan). A total of 20 μl reaction system was used for qPCR: 10 μl SYBR Premix Ex Taq II (2×), 2 μl cDNA, 0.8 μl DhFT-RT-F, and DhFT-RT-R. The PCR reaction program was as follows: 50°C 2 min, 95°C 30 s, 95°C 5 s, 60°C 34 s, 40 cycles; and 72°C 10 min. With β-actin as the internal reference gene, the 2-ΔΔCt method was used to calculate the relative gene expression, and the experiment was repeated three times.
Results and Discussion
Identification of PEBP Family in D. huoshanense
Many studies had confirmed the PEBP genes were involved in flowering regulation (Ahluwalia and Hatsukami, 2015). We used the established hidden Markov model of PEBP protein (PF01161) to conduct preliminary screening of PEBP genes in the whole genome of D. huoshanense (Han et al., 2020). A total of six PEBP family genes were identified, and we further verified the specific PBP-containing domain through pfam, SMART, and the InterPro database, which confirmed these six genes were PEBP genes.
Gene Duplication of D. huoshanense and the Chromosomal Location of the PEBP Genes
Large-scale gene duplication and recombination are one of the important driving forces of species evolution (Jiang et al., 2020). However, few reports have focused on the gene duplication event in Dendrobium plants. In this study, we analyzed the gene duplication events in the evolution of D. huoshanense. The dot plot shows that D. huoshanense has experienced at least two large-scale WGD events since ancient times. There are a large number of WGD events that occurred in chromosome 1, chromosome 4, chromosome 16, chromosome 17, and chromosome 19. A large number of WGT events in chromosome 5, chromosome 6, chromosome 7, chromosome 14, and chromosome 18 were observed. In addition, the ancient γ-WGD events also appeared in great numbers, including chromosome 5, chromosome 10, chromosome 12, chromosome 19, and chromosome 17. The tandem duplication were scattered on the diagonal (Figure 1). These results indicated that D. huoshanense has undergone such ancient polyploidization events to adapt to changes in the external environment, which are consistent with previous results (Ospina-Zapata et al., 2020). The phylogeny tree indicated that two ancient PEBP duplication events in the lineage leading to the common ancestor of angiosperms after its split with gymnosperms. The first duplication gave rise to the MFT-like subfamily and the ancient lineage of the TFL1-like and FT-like subfamilies, which experienced a second duplication to create the two subfamilies. The TFL1 ancestor underwent two separate duplication events in the common ancestor of angiosperms, which created three daughter lineages corresponding to BFT, TFL1, and ATC in A. thaliana (Wang et al., 2015). The chromosome mapping showed that the PEBP family genes were distributed on chromosomes 6, 9, 17, and 15, among which DhFT1, DhFT2, and DhFT3 were located on chromosomes 9, 15, and 17, respectively. The two tandem duplicated genes of the DhTFL1 family are located on chromosome 6 (Figure 2).
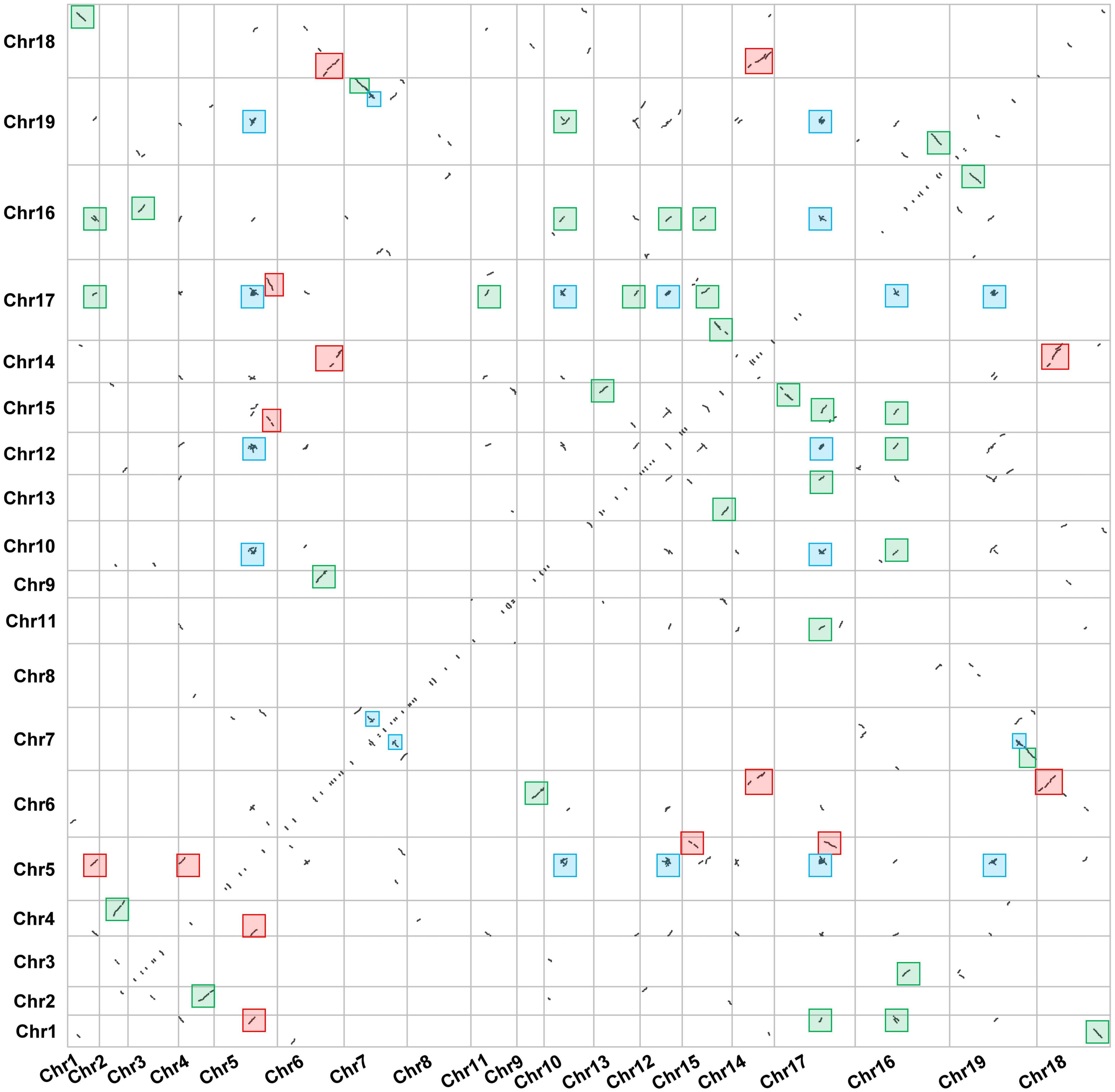
Figure 1. The genome dotplot of D. huoshanense. The dot lines denote the duplication events that occur between two chromosomes. The blue boxes represented the ancient WGD events. The red boxes represented the large-scale WGT events. The green boxes represented the recent WGD events.
Gene Structure Analysis and Conserved Motifs of DhPEBPs
The gene structure of DhPEBP showed that, except for DhMFT, the other five genes contained four exons and three introns (Figure 3A). The conserved motifs in the six DhPEBP proteins were identified using the MEME suite (Figure 3B). In total, 20 motifs were identified in DhPEBP proteins, named motifs 1 to 20, and the motifs identified were 6 to 121 amino acids in length. Motif 1, motif 2, and motif 3 are the three main conserved domains in the PEBP gene (Figure 3C). The number of mainly conserved motifs in each PEBP varied between three and four, indicating that the same subgroup of PEBP protein members shared one or more identical motifs. All of the PEBP genes contained motif 2. Except for the DhMFT, the other five PEBP genes had similar motif composition, which suggested their similar functions. However, some motifs were only presented in DhMFT, indicating that they may perform its particular functions (Zhao et al., 2020).
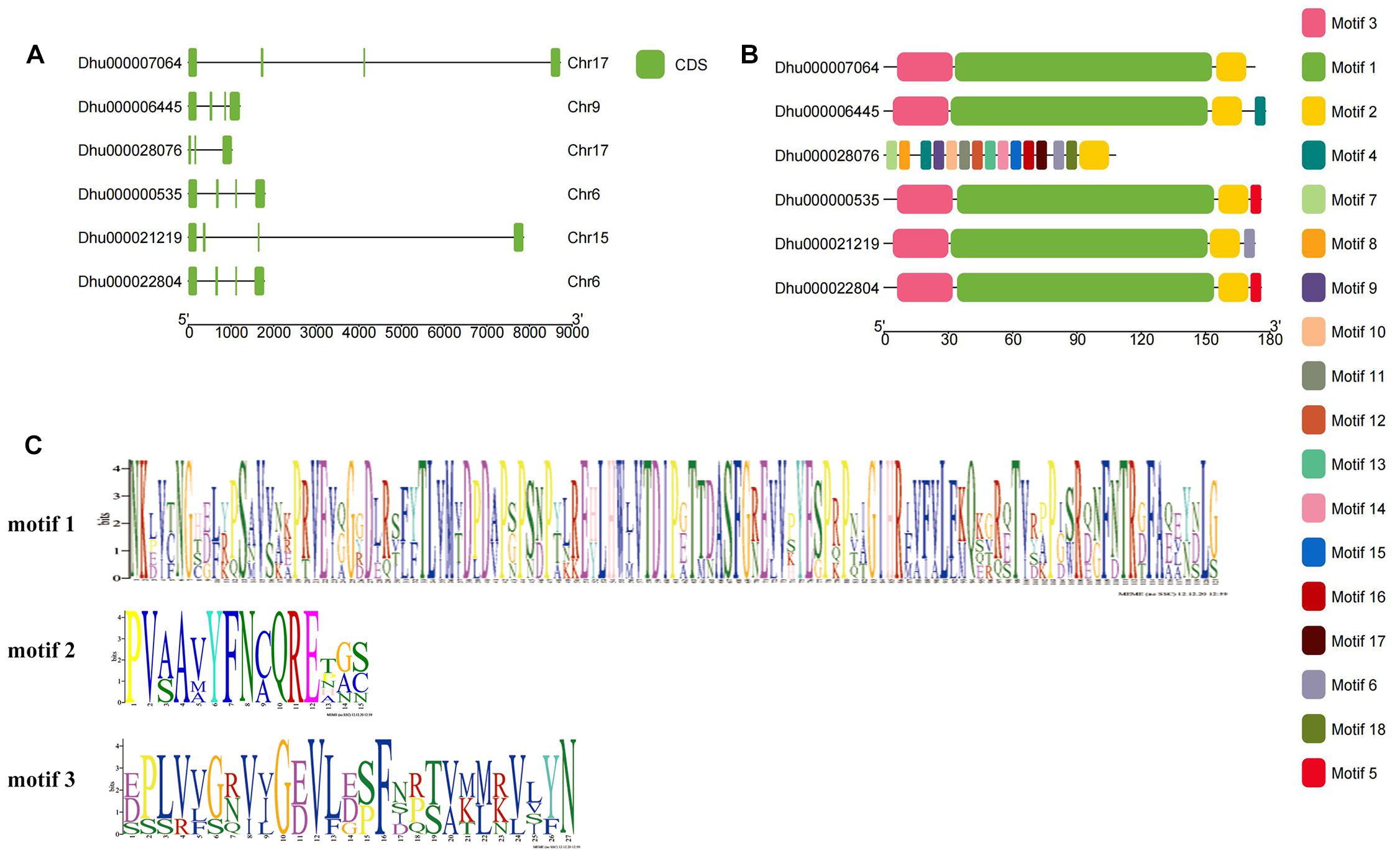
Figure 3. The gene structure and the conserved motif of PEBP genes in D. huoshanense. (A) The gene structure of exon and intron. (B) The conserved motif distributed in PEBP genes. (C) The composition of three main motifs.
The Phylogenetic Analysis of PEBP Genes
In this experiment, MEGA 6 was used to perform homology alignment and phylogeny analysis of 81 PEBP-like protein sequences from six species. The evolutionary tree constructed by the NJ method showed that these genes were clearly divided into three clades. According to the classification of the PEBP family in A. thaliana, we divided these genes into FT subclade, TFL1 subclade and MFT subclade (Supplementary Figure 1). D. huoshanense PEBP protein maintained high homology with genes in the same clade of other species. To improve the reliability and accuracy of the established phylogenetic tree, we used the IQ-TREE tools to build an ML tree based on the optimal model (Nguyen et al., 2015). The optimal model is determined by calculation to be JTT + G4. Using this mode, further phylogeny analysis showed that DhTFL1 is the closest to OsTFL1, DhMFT is the closest to PeMFT, and DhFT1, DhFT2, and DhFT3 are the closest to PeFT6, PeFT9, and PeFT4, respectively. These results are also consistent with the adjacent evolutionary relationship between Dendrobium and Phalaenopsis genus (Figure 4).
Identification of Homologous Genes With Selection Pressure Analysis
To further clarify the homologous duplication of DhPEBPs in the evolutionary process, the plausible functions of homologous genes were speculated. The Orthovenn 2 was used to analyze the homology of five species of A. thaliana, D. huoshanense, Z. mays, S. lycopersicum, and G. max. The results showed that these species formed 23,615 clusters, including 23,049 orthologous clusters (at least contains two species) and 566 single-copy gene clusters. Among them, the genes from D. huoshanense were distributed in 8,714 clusters (Supplementary Figure 2). We further analyzed the clusters related to DhPEBP and found that DhPEBPs scattered in cluster 734, cluster 2,452, and cluster 19,662, indicating these genes are orthologous genes (Supplementary Figure 3). In order to further reveal the evolutionary selection pressure of these homologous genes, we used DnaSP 5.0 to calculate the ka and ks values of each gene pair and found that the ka/ks ratio of all gene pairs was far less than 1, indicating that they were all subjected to a strongly purified selection (Table 1).
Colinearity and Microsynteny Analysis
Collinearity analysis is often used to reveal the genetic relationship between the same species or several different species. In this experiment, we performed microsynteny analysis on the genomes of A. thaliana, D. huoshanense, Z. mays, S. lycopersicum, and G. max. It was indicated that DhTFL1/TFL2 and GmTFL1 (KRG96189) have collinearity, DhFT2 and OsFT8, DhFT1, and OsFT2, and DhTFL1/TFL2 and OsTFL1b have collinearity. DhFT1 and ZmFT3, DhFT2, and ZmFT13 have collinearity (Figure 5). In addition, a pair of tandem duplicated genes DhTFL1 and DhTFL2 are paralogs. These results indicated there was a homologous evolutionary relationship on PEBP genes between D. huoshanense and other species.
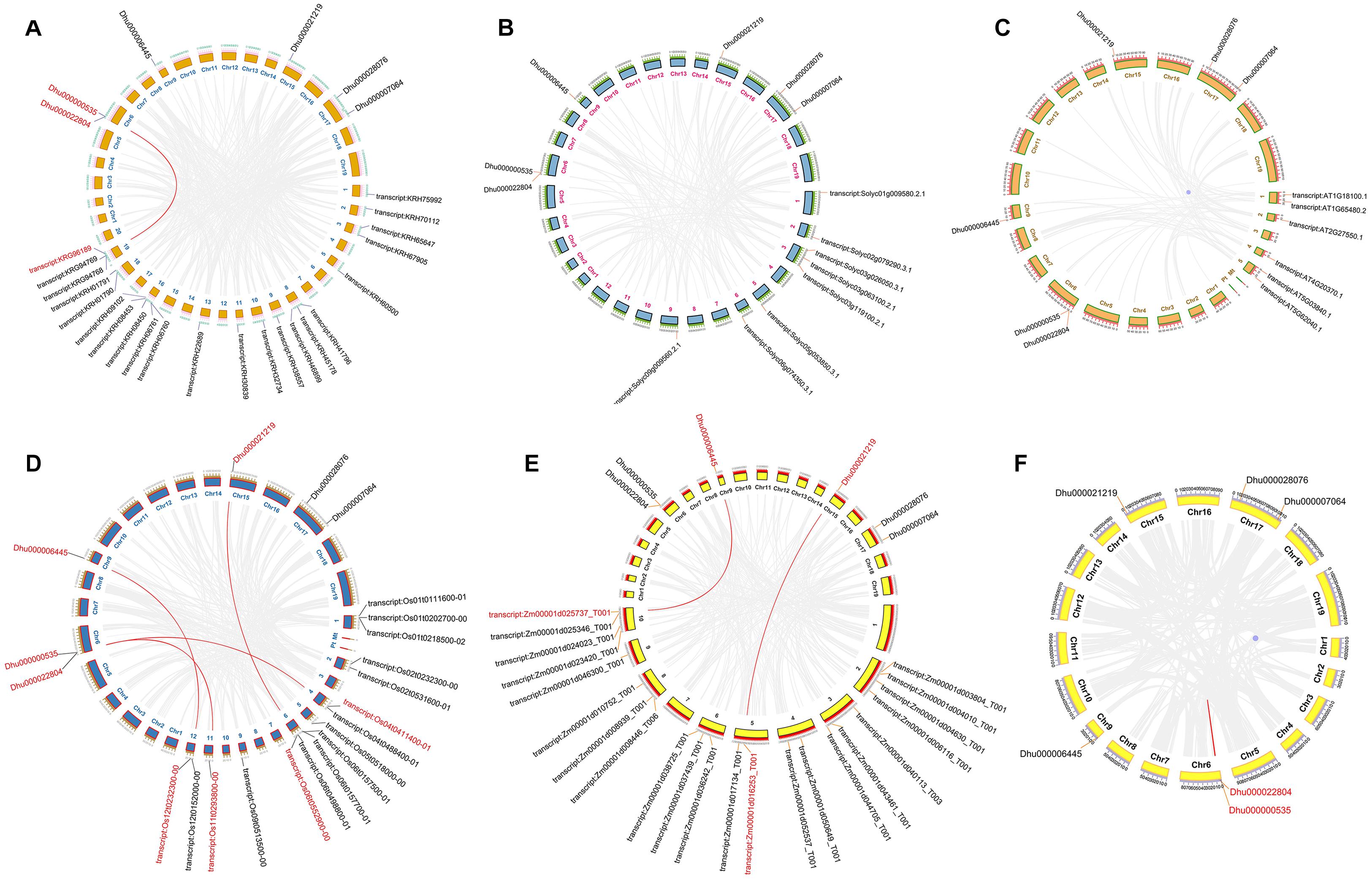
Figure 5. The collinearity and mcirosynteny of D. huoshanense and other species. (A) D. huoshanense and G. max. (B) D. huoshanense and A. thaliana. (C) D. huoshanense and Z.mays. (D) D. huoshanense and S. lycopersicum. (E) D. huoshanense and O. sativa. (F) D. huoshanense and D. huoshanense.
Amino Acid Alignment and the Conserved Domain Analysis
Within amino acid comparison, the conserved structural domains of some proteins could be obtained, accompanied by the conserved catalytic sites and DNA-binding sites for the subsequent research (Li et al., 2014). In this experiment, we performed the amino acid alignment on the aligned sequences of 81 proteins from six species. It was indicated that the 86th position of DhFTs was tyrosine (Y), while 83th and 87th positions of of DhTFL1s were both histidine (H), suggesting they should have distinct functions in flowering regulation (Figure 6). PEBP proteins are characterized by the presence of two highly conserved short motifs, DPDxP and GxHR, which presumably contribute to the conformation of the ligand binding pocket (Guo et al., 2015; Mackenzie et al., 2019). It was reported that substitution of the single amino acid, Tyr85 to His, in FT partially converts FT function to TFL1 function probably through discrimination of structurally related interactors (Yang et al., 2019). In addition, the amino acid sequence encoded by the fourth exon plays a critical role to determine FT/TFL1 protein functions, which are divided into four segments (A–D). Segment B and segment C containing the LYN/IYN triplet conserved motif are especially important for the determination of functional specifcity between FT and TFL1.
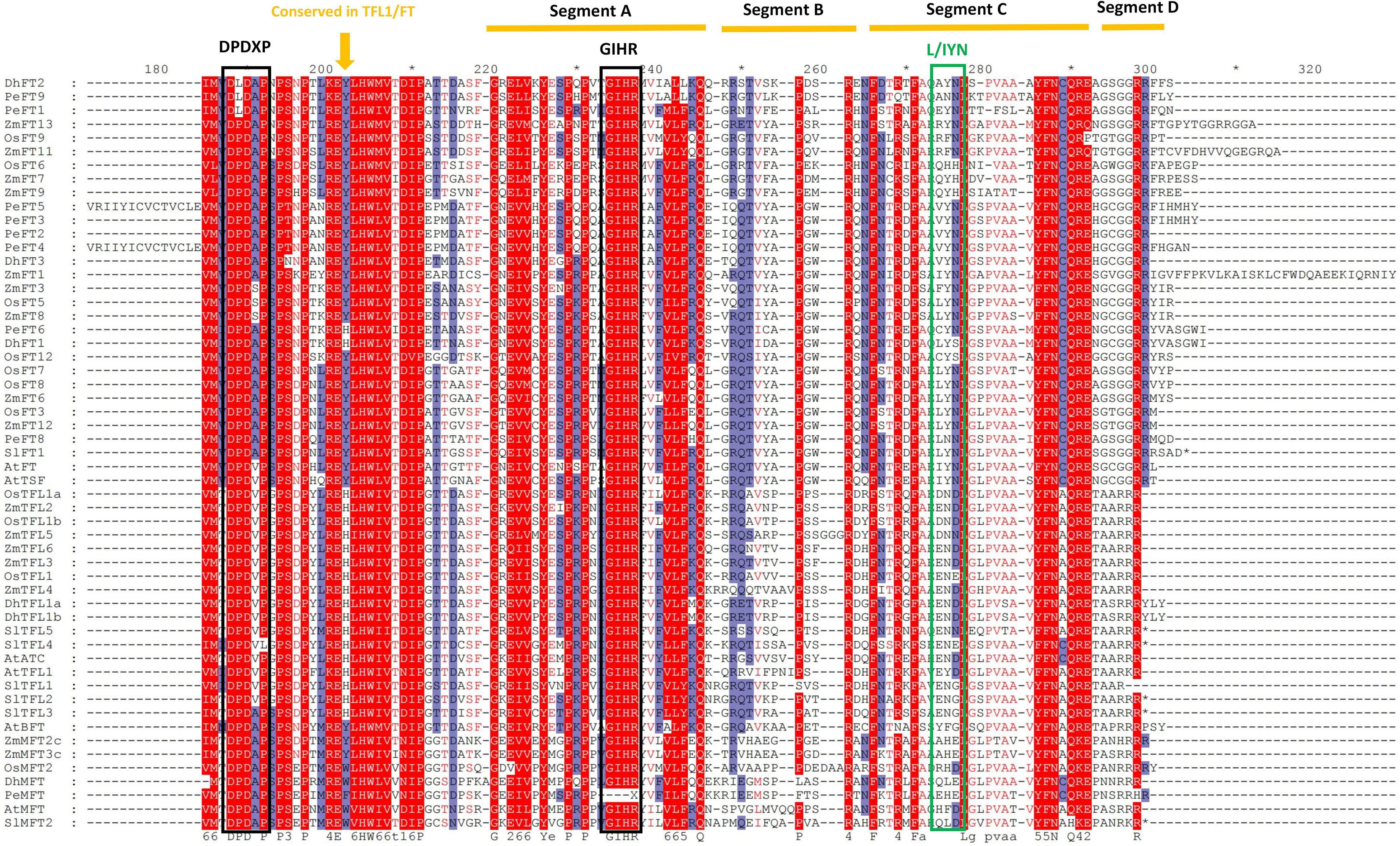
Figure 6. The amino acid comparison and conserved domains of PEBP proteins. The brown arrow indicates a key amino acid residue that determines FT-like and TFL1-like functions. Black boxes denote the conserved DPDxP, GIHR motif, and L/IYN motif. Underlines represent segments A–D.
Analysis of cis-Acting Elements
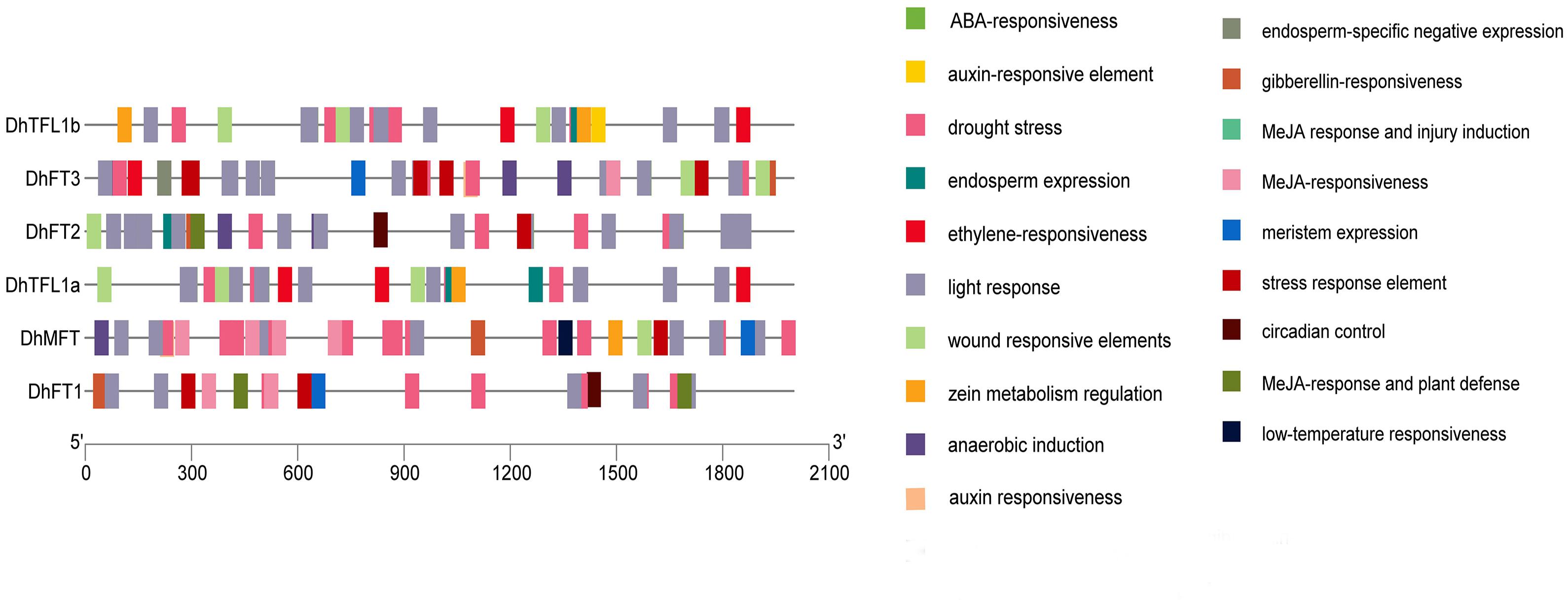
The PLANTCARE service was performed to analyze the upstream promoter sequences of six DhPEBP genes to discover hormone-responsive elements related to flowering regulation, such as GARE elements (Figure 7). The results showed that the promoter region of DhFT1, DhFT2, and DhMFT all contained a GARE-motif (TCTGTTG), while that of DhFT1 and DhFT3 both contained a P-box motif (CCTTTTG), implying that they may be involved in flowering regulation under the GA signaling pathway (Supplementary Table 1).
The Expression Profile of DhPEBPs Induced by GA
In the experiment, qRT-PCR was performed on Dendrobium leaves treated with gibberellin for 5 and 10 days and the CK group (Figure 8). The results showed that DhFT1 and DhFT3 were strongly induced by GA treatment at 10 days, and the expression levels were increased by 21.2 times and 6.2 times, respectively. The expression level of DhFT2 remained stable after 5 days of treatment. The expression of DhMFT was relatively stable, with a slight increase at 5 days. Due to the inhibition of negative feedback regulation, the expression of DhTFL1 decreased rapidly after GA treatment, especially DhTFL1b was hardly expressed at 10 days. The results indicated that DhFT and DhTFL1 may have different regulatory roles in the flowering regulation of D. huoshanense.
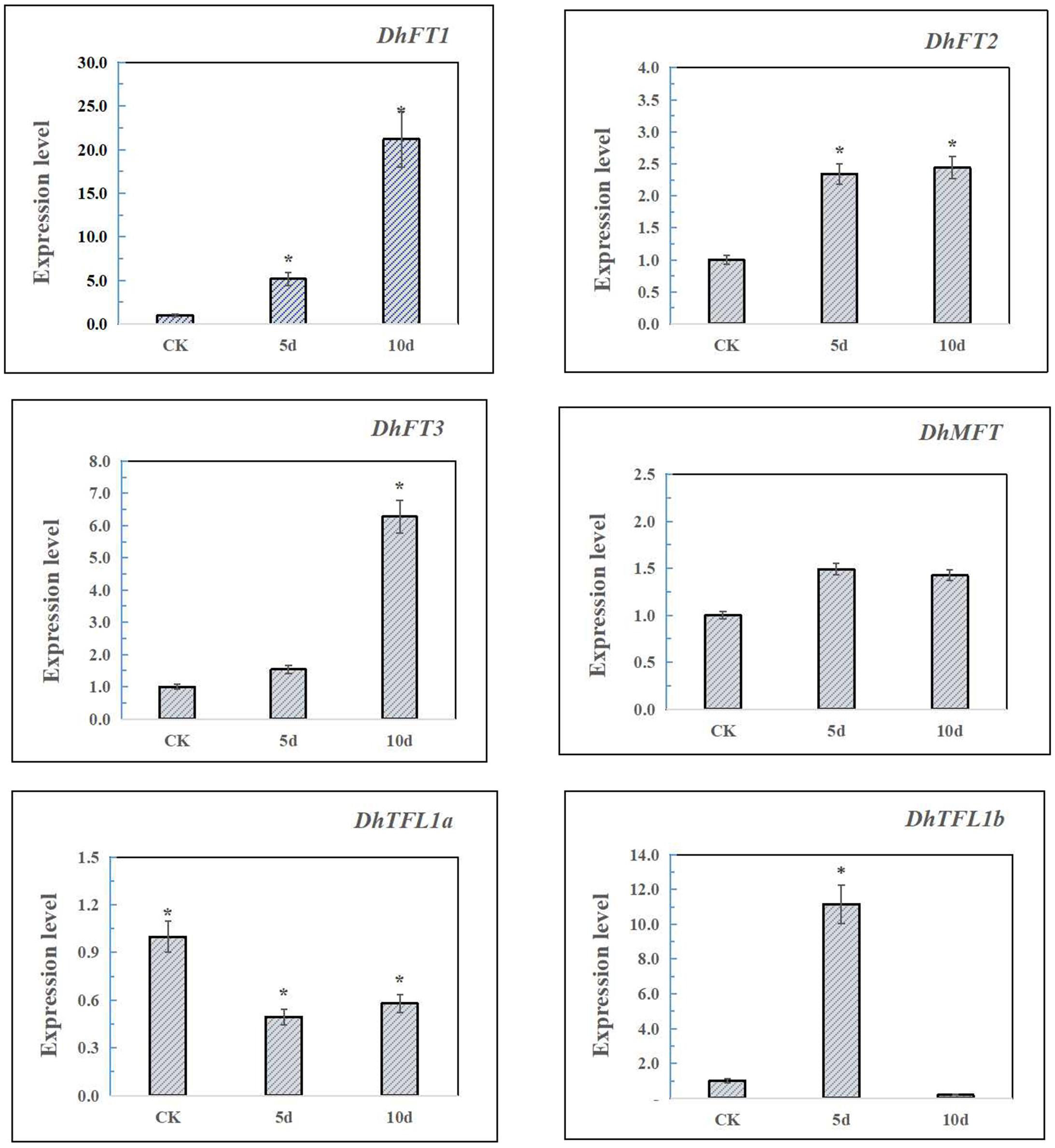
Figure 8. The qRT-PCR analysis of DhPEBPs. Asterisks mean that the expression levels have significant differences compared with CK.
Conclusion
Six PEBP genes were isolated from the D. huoshanense genome. The chromosome mapping suggested that the genes were distributed on chromosomes 6, 9, 15, and 17. The paralogs DhTFL1b and DhTFL1a that obtained by tandem duplication exhibited similar function. Gene structure and the conserved motif analysis further indicated 5 PEBP genes apart from DhMFT contained four exons and three introns entirely. The phylogeny showed that PEBP genes in A. thaliana, Z. mays, S. lycopersicum, O. sativa, and D. huoshanense can be classified into three subclades, FT, TFL, and MFT, which maintained a high homology with the same family in other species. The conserved domain of the amino acid demonstrated that two highly conserved short motifs DPDXP and GXHR embed in DhPEBPs. The 86th position of DhFTs was tyrosine (Y), while the 83th and 87th positions of DhTFL1s were both histidine (H), suggesting they had distinct functions in flowering regulation. The promoter analysis of six DhPEBPs revealed that several cis-elements related to hormone induction, light response and abiotic stress may be involved in the regulation of environmental stress and endogenous signaling pathways. The RT-PCR analysis in short-term days treated with GA3 indicated that the gene expressions of all DhFTs were gradually increased, whereas DhTFL1 were decreased. Taken together, DhPEBPs have various regulatory functions in modulating flowering.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author Contributions
CS and HD designed the research. CS, GL, and JD conducted the experiments. CS and GL analyzed the data. CS wrote the manuscript. HD and CS revised the manuscript, improved the English, and acquired the funding. All authors have read, reviewed, and approved the submitted version.
Funding
This work was supported by the Natural Science Foundation of Anhui Province (1808085MH307), High-Level Talents Research Initiation Funding Project (WGKQ202001011), and Quality Engineering Project in Anhui Province (2020jyxm2137).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2021.687689/full#supplementary-material
Supplementary Figure 1 | The phylogenic tree of five species by NJ method.
Supplementary Figure 2 | The orthologous clusters of five species and the main clusters involved in DhPEBPs.
Supplementary Figure 3 | Orthologous genes related to DhPEBPs in several clusters.
Footnotes
- ^ http://pfam.sanger.ac.uk/family/PF01161
- ^ http://pfam.xfam.org/search#tabview=tab1
- ^ http://smart.embl-heidelberg.de/
- ^ http://www.ebi.ac.uk/interpro/search/sequence/
- ^ http://plants.ensembl.org/index.html
- ^ https://www.ncbi.nlm.nih.gov/genome?LinkName=bioproject_genome&from_uid~$=$~192198
- ^ https://meme-suite.org/
- ^ https://orthovenn2.bioinfotoolkits.net/
- ^ https://www.psc.edu/biomed/genedoc
- ^ http://bioinformatics.psb.ugent.be/webtools/plantcare/html/
References
Adeyemo, O. S., Hyde, P. T., and Setter, T. L. (2019). Identification of FT family genes that respond to photoperiod, temperature and genotype in relation to flowering in cassava (Manihot esculenta, Crantz). Plant Reprod. 32, 181–191. doi: 10.1007/s00497-018-00354-5
Ahluwalia, J. S., and Hatsukami, D. (2015). FLOWERING LOCUS T4 (HvFT4) delays flowering and decreases floret fertility in barley. 4, 1–19.
Auge, G. A., Blair, L. K., Karediya, A., and Donohue, K. (2018). The autonomous flowering-time pathway pleiotropically regulates seed germination in Arabidopsis thaliana. Ann. Bot. 121, 183–191. doi: 10.1093/aob/mcx132
Bi, Z., Huang, H., and Hua, Y. (2019). Cloning and Characterization of Two FLOWERING LOCUS T-like Genes from Rubber Tree (Hevea brasiliensis). J. Plant Growth Regul. 38, 919–930. doi: 10.1007/s00344-018-9902-z
Carvalho, S. G., Guerra-Sá, R., de, C., and Merschmann, L. H. (2015). The impact of sequence length and number of sequences on promoter prediction performance. BMC Bioinformatics 16:S5. doi: 10.1186/1471-2105-16-S19-S5
Chen, C., Chen, H., Zhang, Y., Thomas, H. R., Frank, M. H., He, Y., et al. (2020). TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant. 8, 1194–1202. doi: 10.1016/j.molp.2020.06.009
Chen, M., and Penfield, S. (2018). Feedback regulation of COOLAIR expression controls seed dormancy and flowering time. Science 12, 1014–1017. doi: 10.1126/science.aar7361
Ge, W., Zhang, Y., Cheng, Z., Hou, D., Li, X., and Gao, J. (2017). Main regulatory pathways, key genes and microRNAs involved in flower formation and development of moso bamboo (Phyllostachys edulis). Plant Biotechnol. J. 15, 82–96. doi: 10.1111/pbi.12593
González-Suárez, P., Walker, C. H., and Bennett, T. (2020). Bloom and bust: understanding the nature and regulation of the end of flowering. Curr. Opin. Plant Biol. 57, 24–30. doi: 10.1016/j.pbi.2020.05.009
Guo, D., Li, C., Dong, R., Li, X., Xiao, X., and Huang, X. (2015). Molecular cloning and functional analysis of the FLOWERING LOCUS T (FT) homolog GhFT1 from Gossypium hirsutum. J. Integr. Plant Biol. 57, 522–533. doi: 10.1111/jipb.12316
Han, B., Jing, Y., Dai, J., Zheng, T., Gu, F., Zhao, Q., et al. (2020). A Chromosome-Level Genome Assembly of Dendrobium Huoshanense Using Long Reads and Hi-C Data. Genome Biol. Evol. 12, 2486–2490. doi: 10.1093/gbe/evaa215
Jiang, Z., Sun, L., Wei, Q., Ju, Y., Zou, X., Wan, X., et al. (2020). A new insight into flowering regulation: Molecular basis of flowering initiation in magnolia × soulangeana ‘changchun.’ Genes (Basel). 11, 1–18. doi: 10.3390/genes11010015
Jin, H., Tang, X., Xing, M., Zhu, H., Sui, J., Cai, C., et al. (2019). Molecular and transcriptional characterization of phosphatidyl ethanolamine-binding proteins in wild peanuts Arachis duranensis and Arachis ipaensis. BMC Plant Biol. 19:3. doi: 10.1186/s12870-019-2113-3
Jin, S., Nasim, Z., Susila, H., and Ahn, J. H. (2021). Evolution and functional diversification of FLOWERING LOCUS T/TERMINAL FLOWER 1 family genes in plants. Semin. Cell Dev. Biol. 109, 20–30. doi: 10.1016/j.semcdb.2020.05.007
Johnson, M. A., Harper, J. F., and Palanivelu, R. (2019). A Fruitful Journey: Pollen Tube Navigation from Germination to Fertilization. Annu. Rev. Plant Biol. 70, 809–837. doi: 10.1146/annurev-arplant-050718-100133
Li, C., Luo, L., Fu, Q., Niu, L., and Xu, Z. F. (2014). Isolation and functional characterization of JcFT, a FLOWERING LOCUS T (FT) homologous gene from the biofuel plant Jatropha curcas. BMC Plant Biol. 14:125. doi: 10.1186/1471-2229-14-125
Li, C., Luo, L., Fu, Q., Niu, L., and Xu, Z. F. (2015). Identification and Characterization of the FT/TFL1 Gene Family in the Biofuel Plant Jatropha curcas. Plant Mol. Biol. Report. 33, 326–333. doi: 10.1007/s11105-014-0747-8
Liu, H., Song, S., and Xing, Y. (2019). Beyond heading time: FT-like genes and spike development in cereals. J. Exp. Bot. 70, 1–3. doi: 10.1093/jxb/ery408
Luo, Y., Li, H., Xiang, Z., and He, N. (2018). Identification of Morus notabilis MADS-box genes and elucidation of the roles of MnMADS33 during endodormancy. Sci. Rep. 8, 1–16. doi: 10.1038/s41598-018-23985-0
Mackenzie, K. K., Coelho, L. L., Lütken, H., and Müller, R. (2019). Phylogenomic analysis of the PEBP gene family from Kalanchoë. Agronomy 9, 1–16. doi: 10.3390/agronomy9040171
Melzer, R. (2017). Regulation of flowering time: A splicy business. J. Exp. Bot. 68, 5017–5020. doi: 10.1093/jxb/erx353
Nguyen, L. T., Schmidt, H. A., Von Haeseler, A., and Minh, B. Q. (2015). IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 32, 268–274. doi: 10.1093/molbev/msu300
Ospina-Zapata, D. A., Madrigal, Y., Alzate, J. F., and Pabón-Mora, N. (2020). Evolution and Expression of Reproductive Transition Regulatory Genes FT/TFL1 With Emphasis in Selected Neotropical Orchids. Front. Plant Sci. 11:469. doi: 10.3389/fpls.2020.00469
Plackett, A. R. G., Powers, S. J., Phillips, A. L., Wilson, Z. A., Hedden, P., and Thomas, S. G. (2018). The early inflorescence of Arabidopsis thaliana demonstrates positional effects in floral organ growth and meristem patterning. Plant Reprod. 31, 171–191. doi: 10.1007/s00497-017-0320-3
Schiessl, S. V., Huettel, B., Kuehn, D., Reinhardt, R., and Snowdon, R. J. (2017). Flowering time gene variation in Brassica species shows evolutionary principles. Front. Plant Sci. 8:01742. doi: 10.3389/fpls.2017.01742
Tamura, K., Stecher, G., Peterson, D., Filipski, A., and Kumar, S. (2013). MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol. 12, 2725–2729. doi: 10.1093/molbev/mst197
Wang, M., Tan, Y., Cai, C., and Zhang, B. (2019). Identification and expression analysis of phosphatidy ethanolamine-binding protein (PEBP) gene family in cotton. Genomics 111, 1373–1380. doi: 10.1016/j.ygeno.2018.09.009
Wang, Z., Yang, R., Devisetty, U. K., Maloof, J. N., Zuo, Y., Li, J., et al. (2017). The divergence of flowering time modulated by FT/TFL1 is independent to their interaction and binding activities. Front. Plant Sci. 8:697. doi: 10.3389/fpls.2017.00697
Wang, Z., Zhou, Z., Liu, Y., Liua, T., Li, Q., Ji, Y., et al. (2015). Functional evolution of phosphatidylethanolamine binding proteins in soybean and arabidopsis. Plant Cell 27, 323–336. doi: 10.1105/tpc.114.135103
Xiao, G., Li, B., Chen, H., Chen, W., Wang, Z., Mao, B., et al. (2018). Overexpression of PvCO1, a bamboo CONSTANS-LIKE gene, delays flowering by reducing expression of the FT gene in transgenic Arabidopsis. BMC Plant Biol. 18:1469. doi: 10.1186/s12870-018-1469-0
Yang, Z., Chen, L., Kohnen, M. V., Xiong, B., Zhen, X., Liao, J., et al. (2019). Identification and Characterization of the PEBP Family Genes in Moso Bamboo (Phyllostachys heterocycla). Sci. Rep. 9, 1–12. doi: 10.1038/s41598-019-51278-7
Yu, X., Liu, H., Sang, N., Li, Y., Zhang, T., Sun, J., et al. (2019). Identification of cotton mother of FT and TFL1 homologs, GhMFT1 and GhMFT2, involved in seed germination. PLoS One 14:e0215771. doi: 10.1371/journal.pone.0215771
Zhang, X., Wang, C., Pang, C., Wei, H., Wang, H., Song, M., et al. (2016). Characterization and functional analysis of PEBP Family genes in upland cotton (Gossypium hirsutum L.). PLoS One 11:e0161080. doi: 10.1371/journal.pone.0161080
Keywords: Dendrobium huoshanense, flowering regulation, flowering locus T, homology, collinearity analysis, PEBP genes
Citation: Song C, Li G, Dai J and Deng H (2021) Genome-Wide Analysis of PEBP Genes in Dendrobium huoshanense: Unveiling the Antagonistic Functions of FT/TFL1 in Flowering Time. Front. Genet. 12:687689. doi: 10.3389/fgene.2021.687689
Received: 29 March 2021; Accepted: 18 May 2021;
Published: 09 July 2021.
Edited by:
Yunpeng Cao, Central South University of Forestry and Technology, ChinaReviewed by:
Quan Gan, Anhui Academy of Agricultural Sciences, Chinese Academy of Agricultural Sciences (CAAS), ChinaIrfan Ali Sabir, Shanghai Jiao Tong University, China
Cheng Qin, Zunyi Vocational and Technical College, China
Copyright © 2021 Song, Li, Dai and Deng. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hui Deng, ZGh1cEBxcS5jb20=
 Cheng Song
Cheng Song Guohui Li3
Guohui Li3