
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Genet. , 02 September 2021
Sec. Genomics of Plants and the Phytoecosystem
Volume 12 - 2021 | https://doi.org/10.3389/fgene.2021.680457
This article is part of the Research Topic Genetics and Genomics to Enhance Crop Production, Towards Food Security View all 36 articles
 Xirong Luo1,2†
Xirong Luo1,2† Shimei Yang1†
Shimei Yang1† Yong Luo2
Yong Luo2 Huarong Qiu2
Huarong Qiu2 Tangyan Li1,3
Tangyan Li1,3 Jing Li1
Jing Li1 Xiaocui Chen2
Xiaocui Chen2 Xue Zheng3
Xue Zheng3 Yongdui Chen3
Yongdui Chen3 Jie Zhang3
Jie Zhang3 Zhongkai Zhang3*
Zhongkai Zhang3* Cheng Qin1,3,4*
Cheng Qin1,3,4*The Na+/H+ exchangers (NHXs) are a class of transporters involved in ion balance during plant growth and abiotic stress. We performed systematic bioinformatic identification and expression-characteristic analysis of CaNHX genes in pepper to provide a theoretical basis for pepper breeding and practical production. At the whole-genome level, the members of the CaNHX gene family of cultivated and wild pepper were systematically identified using bioinformatics methods. Sequence alignment and phylogenetic tree construction were performed using MEGA X software, and the gene functional domain, conserved motif, and gene structure were analyzed and visualized. At the same time, the co-expression network of CaNHX genes was analyzed, and salt-stress analysis and fluorescence quantitative verification of the Zunla-1 cultivar under stress conditions were performed. A total of 9 CaNHX genes were identified, which have typical functional domains of the Na+/H+ exchanger gene. The physical and chemical properties of the protein showed that the protein was hydrophilic, with a size of 503–1146 amino acids. Analysis of the gene structure showed that Chr08 was the most localized chromosome, with 8–24 exons. Cis-acting element analysis showed that it mainly contains cis-acting elements such as light response, salicylic acid response, defense, and stress response. Transcriptom and co-expression network analysis showed that under stress, the co-expressed genes of CaNHX genes in roots and leaves were more obvious than those in the control group, including ABA, IAA, and salt. The transcriptome and co-expression were verified by qRT-PCR. In this study, the CaNHX genes were identified at the genome level of pepper, which provides a theoretical foundation for improving the stress resistance, production, development, and utilization of pepper in genetic breeding.
Pepper (Capsicum annuum L.), also known variously as capsicum, chili pepper, chile, and chili, is an annual or perennial plant belonging to the Solanaceae family. It is one of the most important vegetable crops in the world (Qin et al., 2015). Capsicum species were first introduced into China during the Ming Dynasty and today, China has the largest planting area and fresh yield in the world (FAO).1 It is an important cash crop with many varieties, and is considered also of ornamental value, with considerable genetic diversity for research purposes and breeding (Zhang et al., 2016). Many varieties—including Zunla-1, Yunnan Xiaomi Spicy, and Hainan Bell Pepper—are widely planted in China, and their market share is increasing every year.
Peppers contain many substances of nutritional value including vitamin C and vitamin A. The fruits are not only used for food seasoning, but also in the production of food pigments, medicine, and industrial chemicals (Kantar et al., 2016). In medicine, it is widely used for multiple functions, including antibiosis and the prevention and treatment of disease (Saleh et al., 2018). Three pepper genomes (Zunla-1, Chiltepin, and CM334) have been completely sequenced, and with continuing re-sequencing, transcriptome sequencing, and metabolomics based on the whole genome, an increasing amount of genetic data of various pepper varieties has been mined (Kim et al., 2014; Qin et al., 2014). A key component of peppers is their capsaicin. Peppers produced in northwest China contain higher capsaicin and heme levels, due to the dry climate, low rainfall, high solar radiation, and wide temperature difference between day and night (Liu et al., 2012). This study examines how the nutritional content and capsaicin levels in peppers change when Capsicum is stressed by its growing environment. Under drought conditions, the capsaicin content in pepper can be reduced; under certain salt conditions, a significantly higher concentration of salt can promote the yield of capsaicin compared with control and low-salt pepper growth, and photosynthetic efficiency does not necessarily increase with salt (Sarah et al., 2012; Khan et al., 2014).
Plant growth and development depend strongly on environmental factors, such as cold and heat, drought, soil salinity and alkalinity, and other abiotic stresses. When plants are under stress, including some major cash crops, the external environment directly affects plant production. Salt stress is one of the most serious abiotic stresses affecting plant productivity and causes significant crop loss worldwide (Zhang et al., 2018). When plants are in a saline-alkaline soil environment, their ion balance and water balance change significantly. A change in membrane permeability destroys the normal operation of transporters, causing plants to absorb additional sodium ions from the environment, affecting the absorption of other ions and causing nutritional imbalance. Na+/H+ antiporters play a key role in plant development and tolerance to salt stress (Akram et al., 2020). In response to the external influence on plants, the ions and water in plants are balanced through their own ion channels. In general, the cytoplasmic pH value is above neutral (pH 7.2–7.6), which is controlled by an array of regulating molecules such as Na+/K+ transporters, cation/proton exchangers like Ca2+/H+, sodium-proton antiporters (NHX), proton/nutrient transporters, and H+-translocating enzymes (Benèina et al., 2009). Studies indicate that NHX antiporters are involved in regulating the ion balance in plants under salt stress. Their primary physiological functions are the regulation of cytoplasmic pH and expulsion of H+ generated during metabolism, in exchange for transporting Na+ or K+ ions into the cytoplasm and vacuoles of plants and animals (Pedersen et al., 2006). This indicates that studying the salt-tolerance mechanism of plants can improve the growth of plants under salt and alkali stress.
Human HsNHE was the first eukaryotic sodium-hydrogen exchanger gene to be identified and cloned, and functions in transport, Na+/H+ exchange, and pH regulation (Sardet et al., 1989). The first NHX gene identified in plants was the AtNHX1 gene from Arabidopsis thaliana, and the expression of this gene can regulate NaCl in A. thaliana and is a salt-tolerance determiner (Gaxiola et al., 1999; Yokoi et al., 2002; Rodríguez-Rosales et al., 2009). Eight NHX genes were identified in A. thaliana, among which AtNHX1 and AtNHX2 were most common in the buds and roots of seedlings, while AtNHX5 mRNA was expressed in lower abundance in both buds and roots. AtNHX3 was detected in roots, while AtNHX4 and AtNHX6 mRNA were only detected by RT-PCR (Yokoi et al., 2002). To date, NHX genes of several plant species have been identified, such as Vitis vinifera (6 VvNHX genes) (Ayadi et al., 2020), Medicago truncatula (MtNHX1- MtNHX8) (Sandhu et al., 2018), Populus trichocarpa (PtNHX1- PtNHX8) (Tian et al., 2017), Populus euphratica (PeNHX1- PeNHX6) (Ye et al., 2009), Gossypium hirsutum (GhNHX1- GhNHX23) (Fu et al., 2020), Morus alba (MaNHX1- MaNHX7) (Cao et al., 2016), and Beta vulgaris (BvNHX1- BvNHX5) (Wu et al., 2019). This study performed different bioinformatic analyses of CaNHX genes in cultivated and wild peppers, and the CaNHX gene family of pepper was identified at the genome level, providing a theoretical basis for analyzing the function of the gene under salt stress.
In this study, the whole genome data of pepper (C. annuum L. Zunla-1 is hereinafter referred to as pepper) and C. annuum var. glabriusculum Chiltepin were taken as the research object.2 Zunla-1 pepper material was planted in the greenhouse of the Department of modern agriculture, Zunyi vocational and technical college (Zunyi, Guizhou, 107°045 ’E, 27°710’ N). The pepper was treated with 100 Mmol NaCl for 3, 6, 12, 24, and 72 h. The root material of pepper was stored in liquid nitrogen, and 3 samples were taken at each time point as biological replicates. RNA was extracted from the collected samples using the TianGene RNA Extraction Kit (DP432, Beijing, China). We then added root material with a weight of 50–100 ng for aseptic freezing grinding; 450 μL for oscillation mixing. This was transferred to the CS filter column and centrifuged for 3 min (12,000 rpm). The supernatant was transferred from the collection tube with a pipette gun to the Rnase-free centrifuge tube. Then, the supernatant was added and 0.5 times of anhydrous ethanol was mixed into the centrifuge tube and then transferred to the adsorption column CR3 for centrifugation for 30 s (12,000 rpm). A drop of 80 μl DNase I was added to the center of the collecting tube and left at room temperature for 15 min. 250 μL of protein-removing solution RW1 was added to the adsorption column CR3, and left to stand at room temperature for 2 min, before being centrifuged for 30 s (12,000 rpm) (this procedure was repeated once). We then took an enzyme-free centrifuge tube and placed the adsorption column in a new centrifuge tube for several minutes (until the rinsing solution RW was dried). 50 μL Rnase-free ddH2O was then vertically added to the adsorption column, and the obtained RNA was stored at –80°C for later use.
C. annuum cv. CM334, C. chinense PI159236, C. annuum cv. ECW, C. annuum SF and C. baccatum PBC81 genomes come from PGP (Pepper Genome Platform).3 Cuneo, Corno di Carmagnola, Quadrato di Carmagnola, and Tumaticot genomes comes from RisEPP (Resequencing Piedmontese Pepper Ecotypes).4 According to the characteristics of the NHX gene family in Pfam5 data, there is an obvious conservative structure of the NHX gene family (PF00999) (El-Gebali et al., 2018). The genome-wide protein sequence of capsicum was searched by HMMER V3.3 software and verified with the Hmmer web server,6 and the sequences with the incomplete conservative structure were removed. meanwhile, the AtNHX gene of A. thaliana was used for blast comparison, and the E-value was maintained at 1e–20 for comparison. Selecting the intersection of HMMER identification and BLAST alignment, 9 candidate genes of CaNHX were finally identified for subsequent analysis (Potter et al., 2018). The physicochemical properties of the pepper CaNHX protein were analyzed using the online tool ExPASy7 (Artimo et al., 2012). Prediction of Plant-mPLoc by subcellular localization of the CaNHX gene in pepper was performed using online tools8 (Chou and Shen, 2010).
MEGA X was used to perform multiple sequence alignment analysis on the obtained 9 pepper NHX protein sequences obtained, and the phylogenetic tree (neighbor-joining, bootstrap = 1,000) was constructed, and other parameters were left at default settings (Kumar et al., 2018). The online tool Itol9 was used for the presentation and form of the pepper NHX phylogenetic tree (Letunic and Bork, 2019). The batch CD-search10 tool in NCBI was used to visually analyze the NHX gene structure of the NHX gene. The online tool GSDS11 was used for the visualization of pepper NHX gene structure (Hu et al., 2014). The online sequence analysis tool MEME Suite12 was used for motif analysis, with the motif number set at 10 (Bailey et al., 2009). Collinearity analysis of pepper was performed by BLAST for whole-genome protein levels, and the MCScanX tool was used for collinearity analysis (Wang et al., 2012). TBtools were used for the visualization of gene structure, motifs, and collinearity results (Chen et al., 2020).
Using BLAST to build a pepper comparison database, and the KaKs Calculator tool to calculate the synonymous substitution rate and nonsensical substitution rate of pepper CaNHX genes, the Ka/Ks ratio of genes was obtained, and evolutionary pressure was analyzed (Wang et al., 2010). The upstream 2,000 bp sequences of NHX genes were compared and extracted using the Bedtools genome analysis tool.13 The upstream 2,000 bp sequence was predicted and analyzed using the online tool PlantCARE.14 Visualization was performed with TBtools, the main action components were discussed (Lescot et al., 2002; Quinlan and Hall, 2010).
Transcription factors (TFs) in the C. annuum genome were identified using the online iTAK Plant Transcription factor and Protein Kinase Identifier and Classifier (Zheng et al., 2016). The expression data15 obtained from pepper informatic hub were analyzed using temporal and spatial expression patterns and co-expression network associations. The root and leaf tissues of the CM334 pepper cultivar were used for transcriptome and metabolomic analysis, with a total of 574 transcriptome data points. The co-expression results were visualized using Cytoscape 3.7.2 (Liu et al., 2017; Otasek et al., 2019).
The First Strand of RNA was synthesized using the Revertaid First Strand cDNA Synthesis Kit (K1622) from Thermo Field (RevertAid First Strand cDNA Synthesis Kit). The fluorescent quantitative primer Actin (GenBank: DQ832719) and Ubiquitin (GenBank: AY496112) were designed by Primer3plus software as the housekeeping gene (Supplementary Table 1). The fluorescence quantitative instrument for 15 samples was 96 Real-time qTOWER3.0 (Analytikjena, Germany), The fluorescence quantitative reaction system consisted of 10 μL SYBR Primix Ex Taq TM II (ZomBio, Beijing, China), and the upstream and downstream primers of each gene were 1 μL. And ddH2O to 20 μL. The PCR reaction procedure was 95°C for 30 s;95°C for 15 s;60°C for 30 s; and 72°C for 1 min, for 40 cycles. The quantitative RT-PCR results were analyzed using the 2–△△ Ct method (Livak and Schmittgen, 2001). GraphPad Prism v8 was used to visualize the fluorescence quantitative results.
According to the characteristics of the NHX gene family in the Pfam database, it contains Na_H_Exchanger (PF00999) functional domain. First, a total of 42 NHX genes were identified in pepper by the Hmmsearch identification method. Then, 8 AtNHX genes of A. thaliana were compared with the pepper genome by the BLASTP method. Combined with the two identification methods, the incomplete genes were removed by using the Hmmer online website. Nine NHX genes were obtained for subsequent analysis. These 9 CaNHX genes sequences were used for subsequent analysis and named CaNHX1-CaNHX9 in turn (Supplementary Table 2 and Table 1). The physical and chemical properties of the protein showed that the size was 360–1181 aa, the molecular weight was 398.06–129.91 kDa, the isoelectric value was 5.44–8.79, GRAVY was less than 1, and it was a hydrophilic protein. After subcellular localization of pepper CaNHX gene, it was found that the subcellular localization of CaNHX2 and CaNHX4 was in the Cell membrane, and the other 7 subcellular localization were all in Vacuole.
The 9 CaNHX genes in C. annuum identified were compared with 8 AtNHXs in A. thaliana, 12 GmNHXs in Glycine max, 8 PtNHXs in P. trichocarpa, 8 VvNHXs in V. vinifera, 7 OsNHXs in O. sativa, 7 ZmNHXs in Zea mays (Figure 1 and Supplementary Table 3). According to the phylogenetic tree, NHX genes were divided into three subgroups, among which subgroup I contained the most genes. Subgroup I and Subgroup III contain four genes, respectively, while subgroup II contains only one gene.
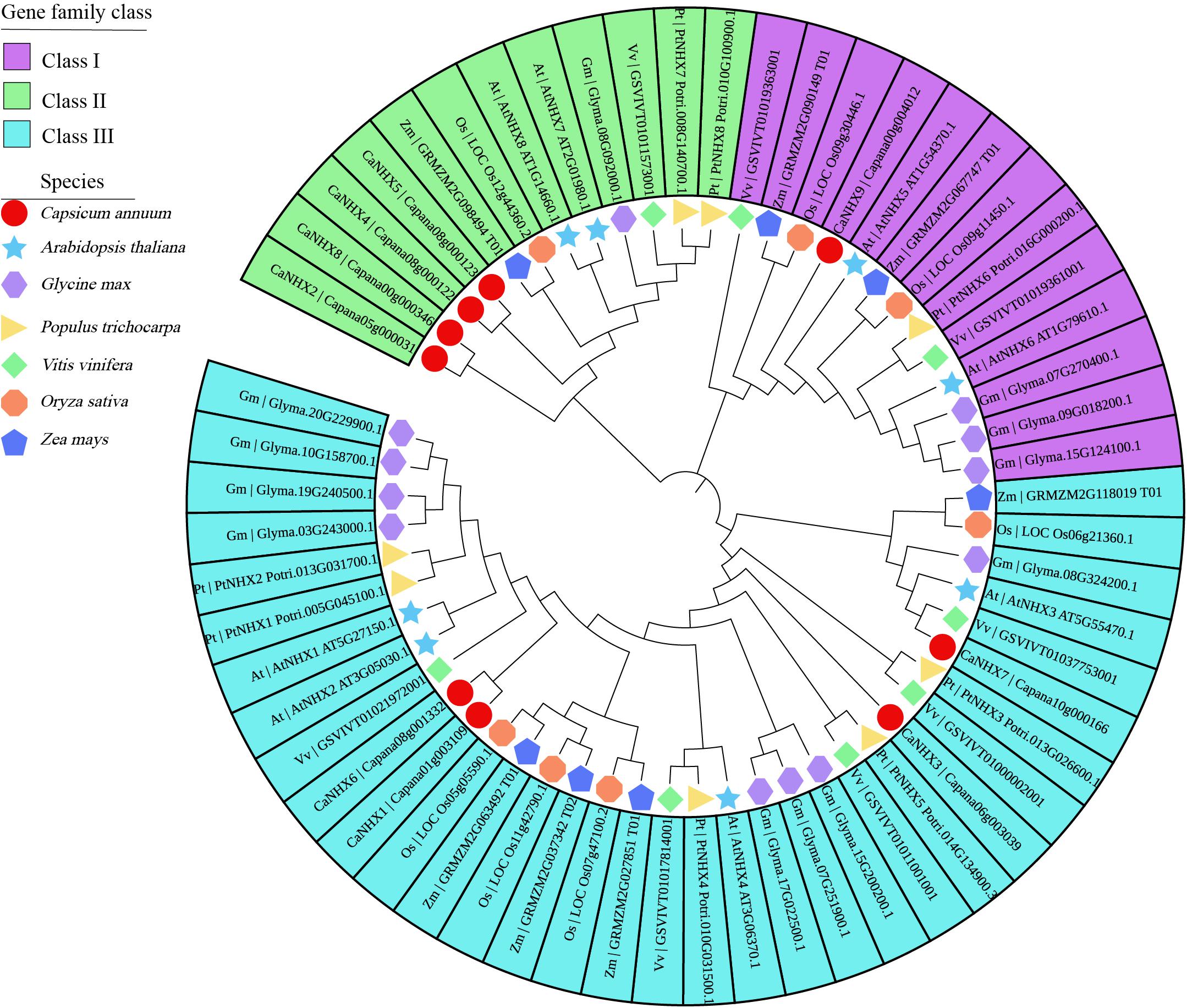
Figure 1. Phylogenetic analysis of CaNHX genes in C. annuum. Square: C. annuum; Star: A. thaliana; Horizontal hexagon: G. max; Right triangle: P. trichocarpa; Rhombus: V. vinifera; Octagon: O. sativa; Up pointing pentagram: Z. mays.
The NHX gene subfamily classification, gene structure, and motif analysis maps show the characteristics of A. thaliana and pepper gene families (Figure 2). Using TBtools to analyze the gene structure of 17 NHX genes family members in pepper and A. thaliana, the results showed that the exon number of the CaNHX gene family was mainly distributed between 8 and 24, while that of AtNHX gene family members in A. thaliana was between 12 and 23, among which the exon number of Class I subgroup was stable between 12 and 14, while the exon number of Class III was the largest. The NHX protein sequences of C. annuum and A. thaliana were analyzed by MEME web tool. According to the distribution of motif of CaNHX genes family members, the motif number is consistent with the phylogenetic tree. For example, in the Class I subfamily, there are 5 motif sequences, which are motif f1, motif2, motif5, motif6, and motif7. It is consistent with the phylogenetic tree classification of the Class I subgroup. The motif of the CaNHX gene in the Class II subfamily all contained motif8 and motif9, of which CaNHX8 was the one with the least motif number. In the Class III subfamily, the motif number of CaNHX1, CaNHX3, CaNHX6, and CaNHX7 remained at 7–8, of which motif8 and motif9 did not exist in the Class III subfamily. Motif7 is a typical amiloride-binding site (LFFIYLPPI), which is a motif contained in the genes of salt-tolerant plants and transgenic NHX plants, and contains the motif in CaNHX2, CaNHX3, CaNHX4, and CaNHX5 (Figures 2, 3).
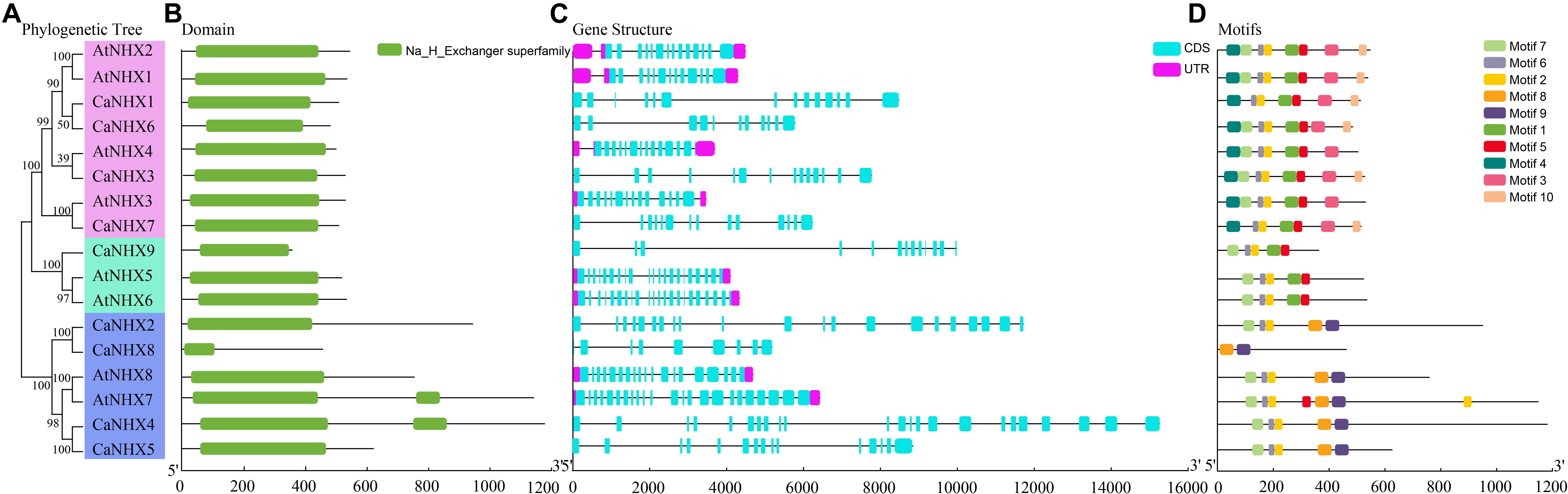
Figure 2. Gene structure and conserved sequence of C. annuum and A. thaliana. (A) Refined relationship between C annuum and A. thaliana. (B) Runctional structure domain. (B) NHX genes. (C) NHX genes structure. (D) Conservative motifs. (D) NHX genes. (E) Motifs.
Through identification, 9 were identified in cultivated pepper (Zunla-1) and 6 were identified in wild pepper (Chiltepin) (Supplementary Tables 5, 6). According to the gene sequence, 9 CaNHX gene sequences were mapped to 5 chromosomes, among which one was a simulated chromosome (not assembled to chromosome), 3 CaNHX genes were mapped to Chr08, but 2 genes were not mapped to the chromosome, and the other chromosomes Chr01, Chr05, Chr06, and Chr10 were all one CaNHX gene (Figure 4). To study the whole genome duplication (WGD) event, 42 cultivars were identified (Contains the confirmed 9 CaNHX genes) by Hmmsearch in cultivated pepper and 37 cultivars identified by hmmsearch in wild pepper were analyzed together (Supplementary Table 3). There are more collinearity relationships among the 9 pepper genes identified, among which CaNHX4 and CaNHX6 are on Chr08 of Zunla-1, while the collinearity block of this gene is on wild type Chr01. There is a collinearity block between CaNHX7 and wild-type Chr00 on Zunla-1 chromosome Chr10. Among them, CaNHX1, CaNHX5, and CaNHX8 have no collinearity block relationship, while CaNHX2, CaNHX3, and CaNHX9 also have collinearity, but their chromosomal positions do not change. The results indicated that the cultivated pepper Zunla-1 and the wild pepper were from the same ancestor, and there was a certain gene replication event. At the same time, we performed collinearity analysis on the genome of pepper varieties with Zunla-1and other pepper genomes, including Chiltepin, Corno, Cuneo, Quadrato, tumaticot, CM334, ECW, PBC81, and SF (Figure 5). The collinearity analysis between Zunla-1 and Corno, Cuneo, Quadrato, and Tumaticot showed that the chromosomal position relationship of Corno, Cuneo, Quadrato, and Tumaticot was the same. The results showed that the four pepper varieties were derived from the same ancestor and had less variation during the species evolution. However, the position relationship between the four pepper varieties and Zunla-1 changed greatly, which indicated that they had a far evolutionary relationship with more variation. Zunla-1 showed significant variation with Chiltepin, CM334, ECW, PBC81, and SF, and the changes of gene position were obvious, indicating that Zunla-1 was far related to the other five varieties of pepper. Three pairs of homologous loci were obtained by analyzing the Ka/Ks ratio of the CaNHX gene, and their Ka/Ks were less than one, indicating that the gene was mainly purified during the evolution of the CaNHX gene in pepper (Table 2).
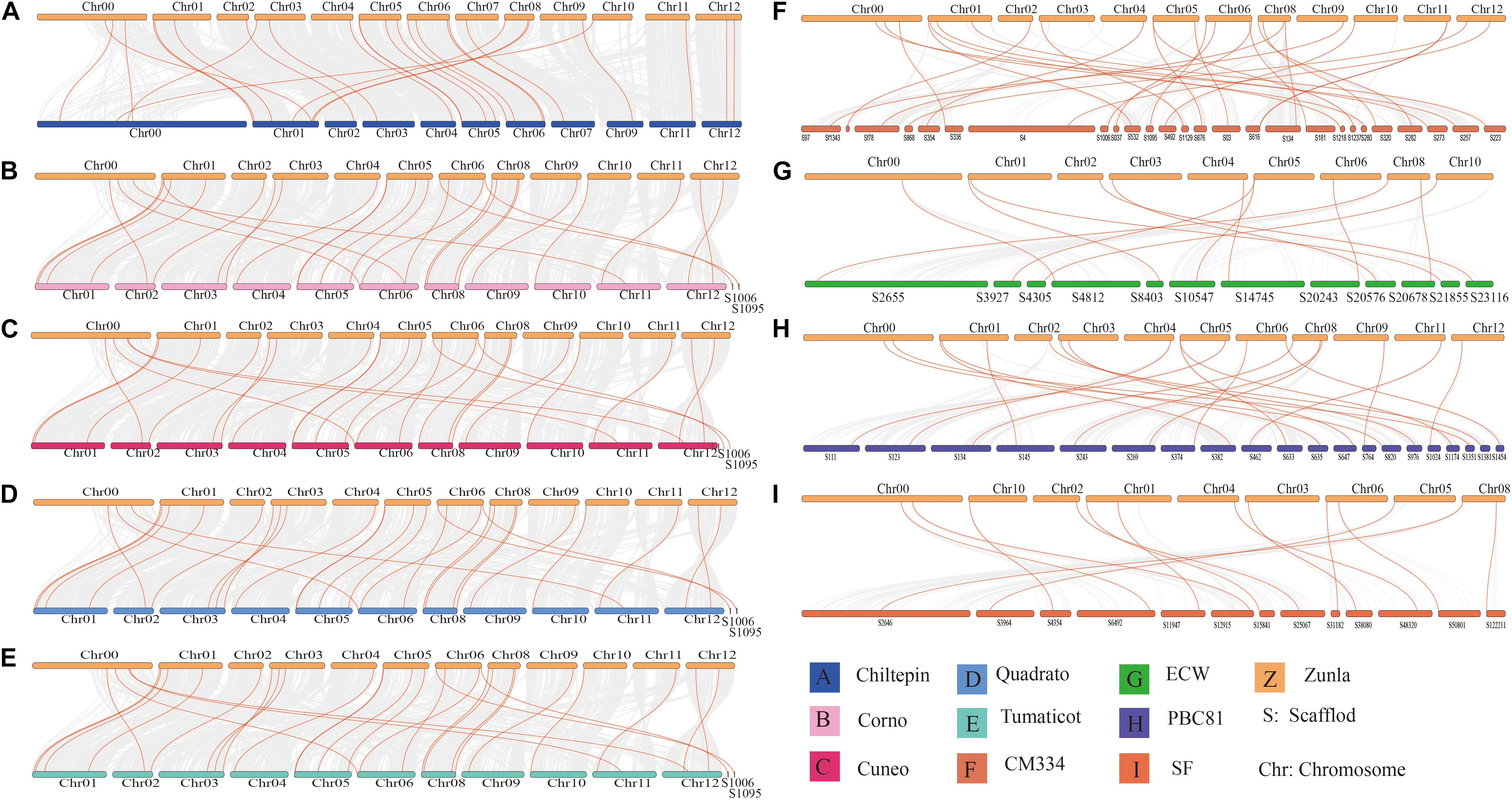
Figure 5. Collinearity analysis of CaNHX gene in pepper. (A) Chiltepin; (B) Corno; (C) Cuneo; (D) Quadrato; (E) Tumaticot; (F) CM334; (G) ECW; (H) PBC81; (I) SF. Z: Zunla-1.
By analyzing the upstream 2,000 bp sequence of the CaNHX genes, the cis-acting elements of the gene were predicted. In addition to a large number of basic elements—CAAT-box and TATA-box—there are also G-Box, GAG-motif, chs-CMA1a, TCT-motif, GATA-motif, GT1-motif, and AE-box in the CaNHX gene family, and TCA-elements in the salicylic acid reaction. Also present were cis-acting elements, TC-rich repeats, meristem expression elements, CAT-box, MYB binding site elements, MBS, MeJA response elements, GCTCA-motif, auxin response elements, TGA-elements, etc. (Figure 6 and Supplementary Table 7). The analysis showed that CaNHXs may be regulated by such things as light and salicylic acid, and may participate in defense mechanisms through these cis-acting elements, thus playing a role in protecting plant growth.
According to established methods, the co-expression network related to the CaNHX gene was extracted. Under stress, 10 groups of data were obtained: ABA, IAA, GA3, SA, JA, sodium chloride, mannitol, hydrogen peroxide, heat stress, cold stress, plus control groups. The co-expressed gene of the CaNHX gene was extracted by script, and the co-expressed gene related to the NHX gene under stress was obtained (Figure 7 and Supplementary Table 8). According to the co-expression network, in the control group, the genes co-expressed with the NHX gene contained fewer co-expression network genes than the other 10 groups, among which ABA, IAA, GA3, and mannitol were the most abundant. The number of co-expression genes was the highest under heat stress and cold stress, but other co-expression genes were also present under NaCl stress. Under salt stress, a total of 4 CaNHX genes were co-expressed with transcription factors, among which CaNHX9 co-expressed the most with 11 transcription factors, followed by CaNHX4 with 7 transcription factors, and CaNHX1 with only 1 NAC co-expressed with CaNHX1 was the least.
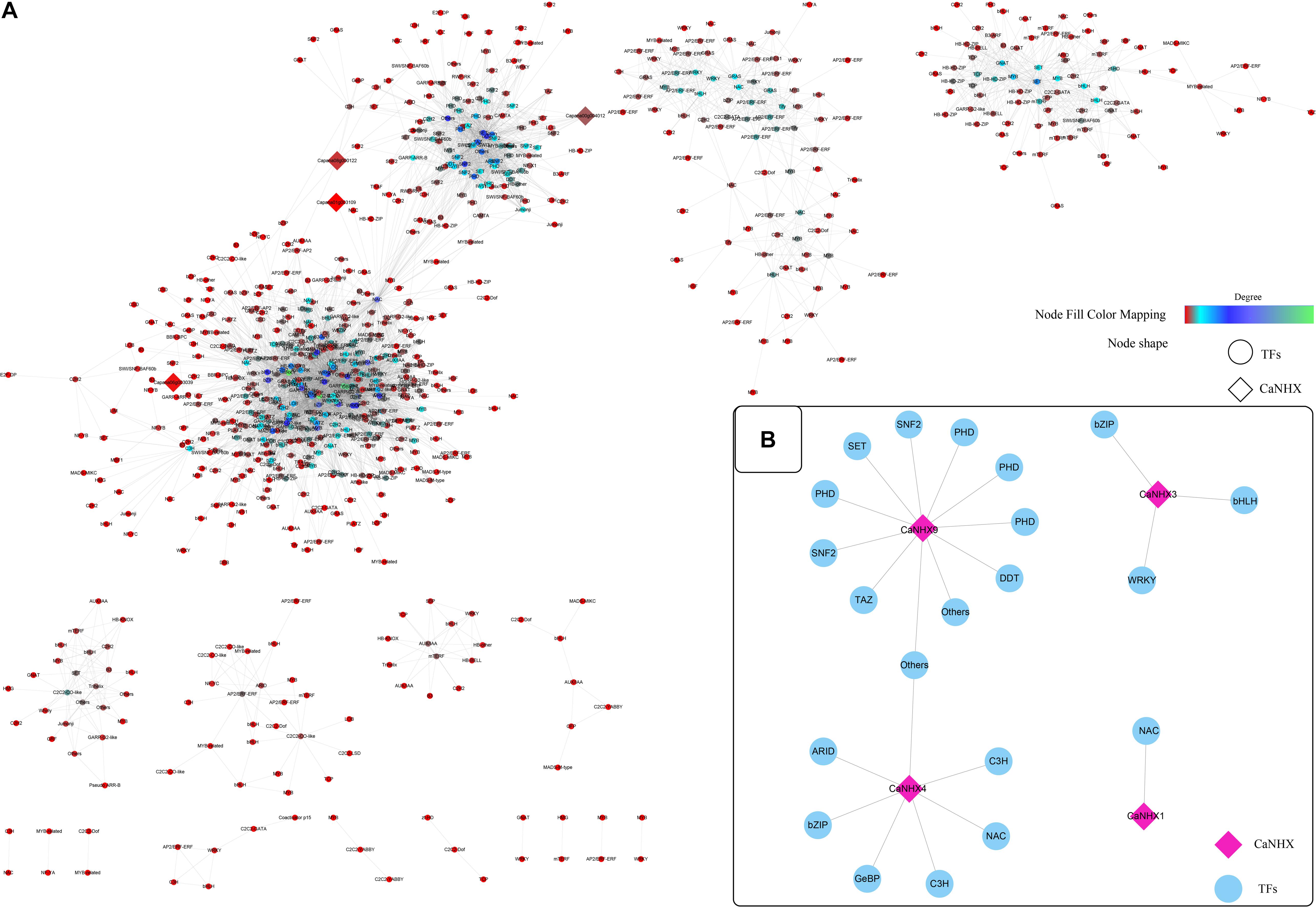
Figure 7. (A,B) Co-expression network of C. annuum CaNHX gene under NaCl stress (Diamond: CaNHX genes; Circular: TFs; Node Fill Color Mapping: Degree).
The expression profile of the CaNHX gene in pepper was analyzed using an online database (Figure 8). The results showed that the expression of CaNHX3, CaNHX4, and CaNHX5 in roots was upregulated compared with that in leaves during IR (IAA root stress) and GR (GA3 root stress), while the expression of CaNHX2, CaNHX6, CaNHX7, CaNHX8, and CaNHX9 was lower in leaves. Under hormone stress, CaNHX1 expression was upregulated. Under abiotic stress, CaNHX1 and CaNHX9 genes were upregulated in HR (heat root stress), while CaNHX2, CaNHX3, and CaNHX4 were upregulated in RR (H2O root stress), FR (cold root stress), and MR (mannitol root stress), whereas CaNHX5 was upregulated in FL (cold leaf stress), CaNHX6 was upregulated in HL (heat leaf stress), CaNHX7 was upregulated in RL (H2O leaf stress), CaNHX7 was upregulated in RL and CaNHX8 in FL. At the same time, expression analysis of the CaNHX gene family under salt stress showed that CaNHX1, CaNHX3, CaNHX4, CaNHX5, and CaNHX9 were upregulated in roots, and their expression tended to be consistent over time. CaNHX6, CaNHX7, and CaNHX8 were upregulated in NL (NaCl leaf stress), while the root expression of CaNHX2 was higher in the blank control than in the leaves.
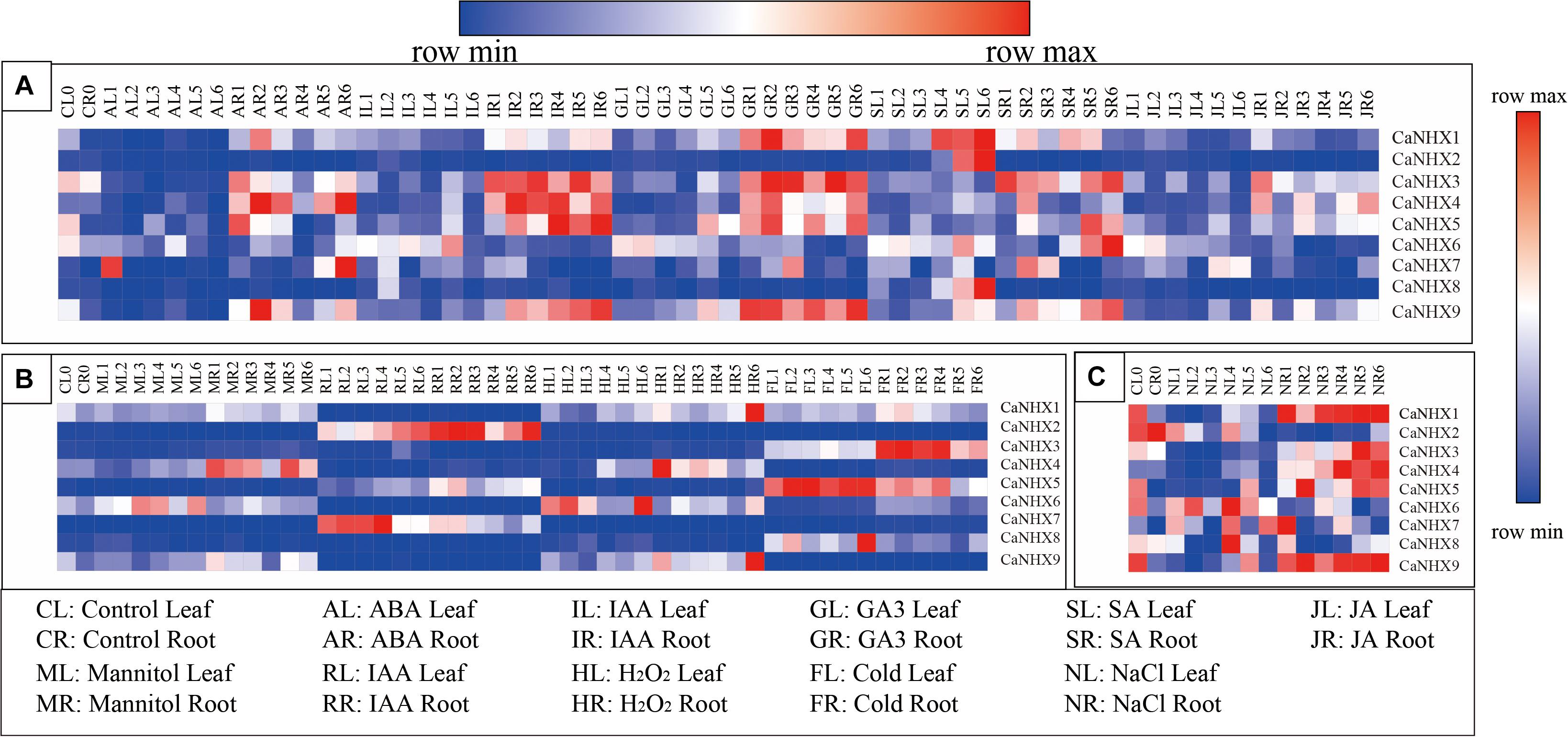
Figure 8. Expression pattern of CaNHX gene in pepper under different stress in root and leaf. (A) Hormonal stress. (B) Abiotic stress. (C) Salt stress.
The pepper at the 6-leaf growth stage was subjected to 100 Mmol salt stress, and the samples at different times (3, 6, 12, 24, and 72 h) were taken for fluorescence quantitative analysis (Figure 9). Results show that, under salt stress, when processing CaNHX1, CaNHX2, CaNHX6, CaNHX9 showed a trend of increased expression, the five time node, CaNHX1, CaNHX9 two genes in a state of relative balance, express no obvious floating. The expression of CaNHX2 and CaNHX6 began to be down-regulated over time after initial stress treatment and then began to be up-regulated after 24 h. The expression of CaNHX3, CaNHX4, CaNHX5, and CaNHX8 genes was less obvious than others. CaNHX1 with CaNHX9 fluorescence quantitative results agree with the transcriptome data, in response to salt stress were a higher expressed state, CaNHX2, CaNHX6, CaNHX7, CaNHX8 increase is not obvious in the transcriptome, which no expression in fluorescence quantitative CaNHX7 gene, do not make the same amount in the transcriptome. In conclusion, two genes, CaNHX1 and CaNHX9, were stably expressed in pepper under salt stress, which was consistent with transcriptome results. It was speculated that pepper could adjust its own ion balance by up-regulating the expression of CaNHX1 and CaNHX9 genes in the process of salt stress, so that pepper could adapt itself to the changes in the environment.
We identified 9 CaNHX genes in the pepper genome (zunla-1), 6 in C. annuum chiltepin, 9 in C. chinense. PI159236, and 9 in C. annnuum. Cv.CM334. There were 9 identified in C. nnuum. Cv. ECW, 10 identified in C. accatum. PBC81, 8 identified in C. nnuum. SF, 8 identified in Corno, 8 identified in Cuneo, 9 identified in Quadrato, and 8 identified in Tumaticot. The CaNHX gene was found to contain up to 9 genes in different varieties of pepper. So far, NHX genes have been identified in many species, with the most identified in Gossypium hirsutum and G. barbadense (23 GhNHX and 24 GbNHX, respectively) (Fu et al., 2020).
However, in subcellular localization, NHX genes can be classified into three subgroups according to their subcellular localization according to previous reports which were divided into three categories, namely Vac-Class, Endo-class, and PM-class, among which VAC-class is located in Vacuole, Endo-class in Endoretinal Reticulum, and PM-class in Plasma membrane (Wu et al., 2019). Other NHX gene species based on subcellular localization also include A. thaliana, P. trichocarpa, G. hirsutum, V. vinifera, Triticum aestivum, Oryza sativa, Sorghum bicolor, Cucurbita maxima, Solanum lycopersicum, Panicum virgatum, Eutrema halophilum,Spinacia oleracea, and Hordeum vulgare. According to the classification of grapes by Ayadi et al. (2020) the VvNHX genes of grapes are divided into two categories, namely Group I Vacuolar (VvNHX1–VvNHX5) and Group II Endosomal (VvNHX6). However, CaNHX genes in pepper are not classified according to their subcellular location. CaNHX2 and CaNHX4 are located in the Cell membrane, while the other subcellular locations of CaNHX gene family members are located in Vacuole. There is no PM-Class and Endo-class in pepper, which is quite different from previous studies.
The CaNHX gene in Zunla-1 can be divided into three subfamilies, namely Class I, Class II, and Class III. In the identified NHX gene families, NHX contains a complete functional domain. These CaNHX genes can be divided into 3 categories, which are the same as A. thaliana, beet, and other plants reported by predecessors (Wu et al., 2019). In addition to CaNHX7, CaNHX8, and CaNHX9, the other six genes also have the typical amilorid-binding site of the NHX gene (FFI/LY/FLLPPI), but this structure does not exist in the Class II subgroup. At the same time, the intermediate residues LF/AV/IY, LF in Class I, LA in Class II, and IY in Class III, PgNHX gene also had the same motif and residues in pomegranate. Not only pepper but also A. thaliana had an NHX gene without an amiloride-binding site (Counillon et al., 1993; Dong et al., 2021). In the wheat TaNHX gene, it was found that under salt stress, the expression of TaNHX2 and TaNHX3 genes were higher in leaves and roots, and the expression of TaNHX1 was higher only in roots. All three TaNHX genes contained an amiloride-binding site (LFFIYLLPPI) (Brini et al., 2005; Yu et al., 2007; Lu et al., 2014). It is speculated that the NHX gene containing an amiloride-binding site is more suitable for the growth of salinization conditions.
Plants are affected by the external environment during their growth, such as abiotic stress, drought, high temperature, salt, and alkali, etc. Transcription is particularly important in the response of plants to environmental changes. There are many cis-acting elements in the pepper CaNHX genes family, such as hormones and stress elements. Studies show that stress-related elements (such as high temperature, low temperature, drought, injury, and defense) and hormone-related elements (such as Auxin, Ethylene, GA, SA, MeJA, and ABA) are identified in the promoter of PtNHXs, and there are also cis-acting elements such as ABA and ABRE in sugar beet, which indicate that they can pass through during plant growth (Tian et al., 2017; Wu et al., 2019). In the transcriptional data of pepper, it was found that the co-expression networks of pepper under biotic and abiotic stress had higher gene network abundance than those under untreated conditions. In the co-expression network, CaNHX1, CaNHX3, CaNHX4, and CaNHX9 were found to be co-expressed with transcription factors, among which CaNHX3 was co-expressed with transcription factor WRKY, indicating that the cis-acting element of CaNHX3 was G-box, which has been found in studies. The G-box is an element associated with WRKY transcription factors under stress conditions. CaNHX3 was upregulated in several periods.
Up to now, the function of the CaNHX gene in pepper has not been analyzed, and no report on the CaNHX gene in pepper has been reported. With the transformation of salt-tolerant transgenic plants, the NHX gene will provide more benefits for agricultural development in soil salinization. Transgenic technology has become one of the important ways to obtain salt-tolerant plants and verify gene function (Dhankher and Foyer, 2018). NHX can improve the salt tolerance of transgenic plants, and the overexpression of AtNHX5 in rice can improve the salt tolerance and drought tolerance of transgenic rice, and the survival rate is higher (Li et al., 2011). SbNHXLP can improve the salt tolerance of tomatoe, and the Na+ level in tomato is lower, and the Ca2+ level is higher, compared with wild-type plants. SbNHXLP maintained ion homeostasis in tomato and alleviated NaCl stress (Kumari et al., 2017). When plants are subjected to salt stress, due to the lack of NHX expression to maintain homeostasis, the premature apoptosis of plants is caused, and the growth of plants is inhibited, thus affecting the yield of plants. Cao et al. (2011) through the overexpression of TaNHX2, improve the survival time of transgenic plants, showing salt tolerance. The number of flowering was more than that of the control group (Cao et al., 2011). In recent years, with the further exploration of the function of NHX, it has been found that NHX is resistant to cadmium, and the overexpression of GmNHX1 enhances the antioxidant capacity of plants and reduces the absorption of cadmium (Yang et al., 2017). It was also found that silencing genes in plants had a great influence on plant growth and development. Rodríguez-Rosales et al. (2008) found that tomato seedling growth, fruit, and seed yield had significant inhibitory effects by silencing tomato LeNHX2. Overexpression of LeNHX2 can enhance salt tolerance in plants (Rodríguez-Rosales et al., 2008; Baghour et al., 2019). In conclusion, overexpression of the NHX gene can improve ion homeostasis, osmotic regulation, reduce cell membrane damage, improve photosynthetic capacity, and play a role in plant protection and yield increase. The CaNHX gene in pepper has never been published and identified before. This study can provide theoretical support for research on the salt tolerance of pepper.
We found that the expression of CaNHXs was mainly concentrated in roots under hormone stress, while under abiotic stress, there were up-regulated expressions of CaNHX2, CaNHX3, and CaNHX4 genes in roots. In the cis-acting elements of the CaNHX gene family, it was found that CaNHX was expressed under various hormones and stress. However, under salt stress, most of the CaNHX genes were up-regulated, which indicated that CaNHX genes in Pepper could condition its ion balance by expressing NHX. The function of plant vacuole NHX antiporter has been identified and expressed in an exogenous system to enhance the salt tolerance of plants. Akram et al. (2020) found that the NHX gene was up-regulated under salt stress, and Yokoi et al. (2002) found that AtNHX1 had higher transcript abundance during salt stress (Yarra, 2019). The results indicated that the expression of NHX genes responded to salt stress during plant growth, which played a very important role in plant growth.
Transcriptome analysis found that the CaNHX gene had multiple expression patterns under single or multiple stress conditions. Meanwhile, the fluorescence quantitative verification in this study showed that the results were consistent with the transcriptome results, in which CaNHX1 and CaNHX9 were up-regulated under salt stress. These results indicated that the CaNHX gene provided a guarantee for the normal growth of pepper and the balance of ion channels of plant stress resistance.
In the pepper genome, 42 CaNHX genes were identified by a Hidden Markov Model database search (hmmsearch). Of these, 9 genes with complete functional domains were identified by BLASTP. We constructed a phylogenetic tree and found that the 9 CaNHX genes were divided into three categories: Class I, Class II, and Class III. The exon number of the Class I subgroup was relatively stable, and the genes were distributed on six chromosomes; these were for hydrophilic proteins. There was a motif amilorid-binding site of the NHX gene (FFI/LY/FLLPPI) associated with salt tolerance in the pepper CaNHX gene. There are many elements in the CaNHX gene, such as hormone stress, salt stress, and so on, and it was found that the CaNHX gene is associated with many genes in the co-expression process, and salt stress conditions are also associated with many genes. Transcriptome analysis showed that the CaNHX gene was up-regulated under various abiotic stresses, which was verified in combination with fluorescence quantification in this study and found to be consistent with transcriptome results. In this study, the whole gene of Pepper was identified at the genome level, which provided a theoretical basis for the genetic breeding of pepper under stress.
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author/s.
XL, SY, ZZ, and CQ conceived and designed the experiments, and drafted the manuscript. YL, HQ, TL, JL, XC, XZ, YC, and JZ performed the experiments. SY and CQ analyzed the data. All authors read and approved the manuscript.
This work was supported by the Special Financial Grant from the China Postdoctoral Science Foundation (No. 2017T100718), the National Natural Science Foundation of China (Nos. 31660575 and 31860498), the Guizhou Province Science and Technology Plan Program of China (Qian Kehe Support Nos. [2019]2258, [2020]1Y088, and [2019]2413), the Government Special Funds for Guiding Local Science and Technology Development of China (Qianke Zhongyindi No. 20174003), the Zunyi Innovative Talent Team Training Project of China (Zunshi Kehe Rencai No. 201904), and the Zunyi Excellent Young Scientific and Technological Innovative Talents Training Project (Zunyouqingke No. 201806).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2021.680457/full#supplementary-material
Akram, U., Song, Y., Liang, C., Abid, M. A., Askari, M., Myat, A. A., et al. (2020). Genome-wide characterization and expression analysis of NHX Gene family under salinity stress in gossypium barbadense and its comparison with Gossypium hirsutum. Genes 11:803. doi: 10.3390/genes11070803
Artimo, P., Jonnalagedda, M., Arnold, K., Baratin, D., Csardi, G., de Castro, E., et al. (2012). ExPASy: SIB bioinformatics resource portal. Nucleic Acids Res. 40, W597–W603. doi: 10.1093/nar/gks400
Ayadi, M., Martins, V., Ben Ayed, R., Jbir, R., Feki, M., Mzid, R., et al. (2020). Genome wide identification, molecular characterization, and gene expression analyses of grapevine NHX antiporters suggest their involvement in growth, ripening, seed dormancy, and stress response. Biochem. Genet. 58, 102–128. doi: 10.1007/s10528-019-09930-4
Baghour, M., Gálvez, F. J., Sánchez, M. E., Aranda, M. N., Venema, K., and Rodríguez-Rosales, M. P. (2019). Overexpression of LeNHX2 and SlSOS2 increases salt tolerance and fruit production in double transgenic tomato plants. Plant Physiol. Biochem. 135, 77–86. doi: 10.1016/j.plaphy.2018.11.028
Bailey, T. L., Boden, M., Buske, F. A., Frith, M., Grant, C. E., Clementi, L., et al. (2009). MEME Suite: tools for motif discovery and searching. Nucleic Acids Res. 37, W202–W208. doi: 10.1093/nar/gkp335
Benèina, M., Bagar, T., Lah, L., and Kraševec, N. (2009). A comparative genomic analysis of calcium and proton signaling/homeostasis in Aspergillus species. Fungal Genet. Biol. 46, S93–S104. doi: 10.1016/j.fgb.2008.07.019
Brini, F., Gaxiola, R. A., Berkowitz, G. A., and Masmoudi, K. (2005). Cloning and characterization of a wheat vacuolar cation/proton antiporter and pyrophosphatase proton pump. Plant Physiol. Bioch. 43, 347–354. doi: 10.1016/j.plaphy.2005.02.010
Cao, B., Long, D., Zhang, M., Liu, C., Xiang, Z., and Zhao, A. (2016). Molecular characterization and expression analysis of the mulberry Na+/H+ exchanger gene family. Plant Physiol. Biochem. 99, 49–58. doi: 10.1016/j.plaphy.2015.12.010
Cao, D., Hou, W., Liu, W., Yao, W., Wu, C., Liu, X., et al. (2011). Overexpression of TaNHX2 enhances salt tolerance of ‘composite’ and whole transgenic soybean plants. Plant Cell Tiss. Org. 10, 541–552. doi: 10.1007/s11240-011-0005-9
Chen, C., Chen, H., Zhang, Y., Thomas, H. R., Frank, M. H., He, Y., et al. (2020). TBtools: an integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 13, 1194–1202. doi: 10.1016/j.molp.2020.06.009
Chou, K. C., and Shen, H. B. (2010). Plant-mPLoc: a top-down strategy to augment the power for predicting plant protein subcellular localization. PLoS One 5:e11335. doi: 10.1371/journal.pone.0011335
Counillon, L., Franchi, A., and Pouyssegur, J. (1993). A point mutation of the Na+/H+ exchanger gene (NHE1) and amplification of the mutated allele confer amiloride resistance upon chronic acidosis. Proc. Natl. Acad. Sci. U.S.A. 90, 4508–4512.
Dhankher, O. P., and Foyer, C. H. (2018). Climate resilient crops for improving global food security and safety. Plant Cell Environ. 41, 877–884. doi: 10.1111/pce.13207
Dong, J., Liu, C., Wang, Y., Zhao, Y., Ge, D., and Yuan, Z. (2021). Genome-wide identification of the NHX gene family in Punica granatum L. and their expressional patterns under salt stress. Agronomy 11:264. doi: 10.3390/agronomy11020264
El-Gebali, S., Mistry, J., Bateman, A., Eddy, S. R., Luciani, A., Potter, S. C., et al. (2018). The Pfam protein families database in 2019. Nucleic Acids Res. 47, D427–D432. doi: 10.1093/nar/gky995
Fu, X., Lu, Z., Wei, H., Zhang, J., Yang, X., Wu, A., et al. (2020). Genome-wide identification and expression analysis of the NHX (Sodium/Hydrogen Antiporter) gene family in cotton. Front. Genet. 11:964. doi: 10.3389/fgene.2020.00964
Gaxiola, R. A., Rao, R., Sherman, A., Grisafi, P., Alper, S. L., and Fink, G. R. (1999). The Arabidopsis thaliana proton transporters, AtNhx1 and Avp1, can function in cation detoxification in yeast. Proc. Natl. Acad. Sci. U.S.A. 96:1480.
Hu, B., Jin, J., Guo, A. Y., Zhang, H., Luo, J., and Gao, G. (2014). GSDS 2.0: an upgraded gene feature visualization server. Bioinformatics 31, 1296–1297. doi: 10.1093/bioinformatics/btu817
Kantar, M. B., Anderson, J. E., Lucht, S. A., Mercer, K., Bernau, V., Case, K. A., et al. (2016). Vitamin variation in Capsicum Spp. Provides opportunities to improve nutritional value of human diets. PLoS One 11:e0161464. doi: 10.1371/journal.pone.0161464
Khan, A. L., Shin, J. H., Jung, H. Y., and Lee, I. J. (2014). Regulations of capsaicin synthesis in Capsicum annuum L. by Penicillium resedanum LK6 during drought conditions. Sci. Hortic. 175, 167–173. doi: 10.1016/j.scienta.2014.06.008
Kim, S., Park, M., Yeom, S. I., Kim, Y. M., Lee, J. M., Lee, H. A., et al. (2014). Genome sequence of the hot pepper provides insights into the evolution of pungency in Capsicum species. Nat. Genet. 46, 270–278. doi: 10.1038/ng.2877
Kumar, S., Stecher, G., Li, M., Knyaz, C., and Tamura, K. (2018). MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 35, 1547–1549. doi: 10.1093/molbev/msy096
Kumari, P. H., Kumar, S. A., Sivan, P., Katam, R., Suravajhala, P., Rao, K. S., et al. (2017). Overexpression of a plasma membrane bound Na+/H+ antiporter-like protein (SbNHXLP) confers salt tolerance and improves fruit yield in tomato by maintaining ion homeostasis. Front. Plant Sci. 7:2027. doi: 10.3389/fpls.2016.02027
Lescot, M., Déhais, P., Thijs, G., Marchal, K., Moreau, Y., Van de Peer, Y., et al. (2002). PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 30, 325–327. doi: 10.1093/nar/30.1.325
Letunic, I., and Bork, P. (2019). Interactive Tree Of Life (iTOL) v4: recent updates and new developments. Nucleic Acids Res. 47, W256–W259. doi: 10.1093/nar/gkz239
Li, M., Lin, X., Li, H., Pan, X., and Wu, G. (2011). Overexpression of AtNHX5 improves tolerance to both salt and water stress in rice (Oryza sativa L.). Plant Cell Tiss. Org. 107, 283–293. doi: 10.1007/s11240-011-9979-6
Liu, F., Yu, H., Deng, Y., Zheng, J., Liu, M., Ou, L., et al. (2017). PepperHub, an informatics hub for the chili pepper research community. Mol. Plant 10, 1129–1132. doi: 10.1016/j.molp.2017.03.005
Liu, H., Yang, H., Zheng, J., Jia, D., Wang, J., Li, Y., et al. (2012). Irrigation scheduling strategies based on soil matric potential on yield and fruit quality of mulched-drip irrigated chili pepper in Northwest China. Agric. Water Manage. 115, 232–241. doi: 10.1016/j.agwat.2012.09.009
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Lu, W., Guo, C., Li, X., Duan, W., Ma, C., Zhao, M., et al. (2014). Overexpression of TaNHX3, a vacuolar Na+/H+ antiporter gene in wheat, enhances salt stress tolerance in tobacco by improving related physiological processes. Plant Physiol. Biochem. 76, 17–28. doi: 10.1016/j.plaphy.2013.12.013
Otasek, D., Morris, J. H., Bouças, J., Pico, A. R., and Demchak, B. (2019). Cytoscape automation: empowering workflow-based network analysis. Genome Biol. 20, 1–15. doi: 10.1186/s13059-019-1758-4
Pedersen, S. F., O’Donnell, M. E., Anderson, S. E., and Cala, P. M. (2006). Physiology and pathophysiology of Na+/H+ exchange and Na+-K+-2Cl- cotransport in the heart, brain, and blood. Am. J. Physiol. Reg. I 291, R1–R25. doi: 10.1152/ajpregu.00782.2005
Potter, S. C., Luciani, A., Eddy, S. R., Park, Y., Lopez, R., and Finn, R. D. (2018). HMMER web server: 2018 update. Nucleic Acids Res. 46, W200–W204. doi: 10.1093/nar/gky448
Qin, C., Cheng, J., Wu, Z., Luo, X., and Hu, K. (2015). Research advances in the genomics of pepper. J. Agric. Biotechem. 23, 953–966. doi: 10.1111/j.1475-6773.2012.01428.x
Qin, C., Yu, C., Shen, Y., Fang, X., Min, J., Cheng, J., et al. (2014). Whole-genome sequencing of cultivated and wild peppers provides insights into Capsicum domestication and specialization. Proc. Natl. Acad. Sci. U.S.A. 111:5135. doi: 10.1073/pnas.1400975111
Quinlan, A. R., and Hall, I. M. (2010). BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26, 841–842. doi: 10.1093/bioinformatics/btq033
Rodríguez-Rosales, M. P., Gálvez, F. J., Huertas, R., Aranda, M. N., Baghour, M., Cagnac, O., et al. (2009). Plant NHX cation/proton antiporters. Plant Signal. Behav. 4, 265–276. doi: 10.4161/psb.4.4.7919
Rodríguez-Rosales, M. P., Jiang, X., Gálvez, F. J., Aranda, M. N., Cubero, B., and Venema, K. (2008). Overexpression of the tomato K+/H+ antiporter LeNHX2 confers salt tolerance by improving potassium compartmentalization. New Phytol. 179, 366–377. doi: 10.1111/j.1469-8137.2008.02461.x
Saleh, B., Omer, A., and Teweldemedhin, B. (2018). Medicinal uses and health benefits of chili pepper (Capsicum spp.): a review. MOJ Food Proc. Technol. 6, 325–328. doi: 10.15406/mojfpt.2018.06.00183
Sandhu, D., Pudussery, M. V., Kaundal, R., Suarez, D. L., Kaundal, A., and Sekhon, R. S. (2018). Molecular characterization and expression analysis of the Na+/H+ exchanger gene family in Medicago truncatula. Funct. Integr. Genomic. 18, 141–153. doi: 10.1007/s10142-017-0581-9
Sarah, A., Todd, P. E., Meekins, J. F., Dale, P., and Marcia, M. (2012). Research article: effects of salt stress on capsaicin content, growth, and fluorescence in a Jalapeño cultivar of Capsicum annuum (Solanaceae). BIOS 83, 1–7. doi: 10.1893/0005-3155-83.1.1
Sardet, C., Franchi, A., and Pouysségur, J. (1989). Molecular cloning, primary structure, and expression of the human growth factor-activatable Na+/H+ antiporter. Cell 56, 271–280. doi: 10.1016/0092-8674(89)90901-X
Tian, F., Chang, E., Li, Y., Sun, P., Hu, J., and Zhang, J. (2017). Expression and integrated network analyses revealed functional divergence of NHX-type Na+/H+ exchanger genes in poplar. Sci. Rep. UK 7:2607. doi: 10.1038/s41598-017-02894-8
Wang, D., Zhang, Y., Zhang, Z., Zhu, J., and Yu, J. (2010). KaKs_calculator 2.0: a toolkit incorporating gamma-series methods and sliding window strategies. Genom. Proteom. Bioinf. 8, 77–80. doi: 10.1016/S1672-0229(10)60008-3
Wang, Y., Tang, H., DeBarry, J. D., Tan, X., Li, J., Wang, X., et al. (2012). MCScanX: a toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 40:e49. doi: 10.1093/nar/gkr1293
Wu, G. Q., Wang, J. L., and Li, S. J. (2019). Genome-wide identification of Na+/H+ antiporter (NHX) genes in sugar beet (Beta vulgaris L.) and their regulated expression under salt stress. Genes 10:401. doi: 10.3390/genes10050401
Yang, L., Han, Y., Wu, D., Yong, W., Liu, M., Wang, S., et al. (2017). Salt and cadmium stress tolerance caused by overexpression of the Glycine Max Na+/H+ Antiporter (GmNHX1) gene in duckweed (Lemna turionifera 5511). Aquat. Toxicol. 192, 127–135. doi: 10.1016/j.aquatox.2017.08.010
Yarra, R. (2019). The wheat NHX gene family: potential role in improving salinity stress tolerance of plants. Plant Gene 18:100178. doi: 10.1016/j.plgene.2019.100178
Ye, C. Y., Zhang, H. C., Chen, J. H., Xia, X. L., and Yin, W. L. (2009). Molecular characterization of putative vacuolar NHX-type Na+/H+ exchanger genes from the salt-resistant tree Populus euphratica. Physiol. Plantarum 137, 166–174. doi: 10.1111/j.1399-3054.2009.01269.x
Yokoi, S., Quintero, F. J., Cubero, B., Ruiz, M. T., Bressan, R. A., Hasegawa, P. M., et al. (2002). Differential expression and function of Arabidopsis thaliana NHX Na+/H+ antiporters in the salt stress response. Plant J. 30, 529–539. doi: 10.1046/j.1365-313X.2002.01309.x
Yu, J. N., Huang, J., Wang, Z. N., Zhang, J. S., and Chen, S. Y. (2007). An Na+/H+ antiporter gene from wheat plays an important role in stress tolerance. J. Biosci. 32, 1153–1161. doi: 10.1007/s12038-007-0117-x
Zhang, X. M., Zhang, Z. H., Gu, X. Z., Mao, S. L., Li, X. X., Chadśuf, J., et al. (2016). Genetic diversity of pepper (Capsicum spp.) germplasm resources in China reflects selection for cultivar types and spatial distribution. J. Integr. Agric. 15, 1991–2001. doi: 10.1016/S2095-3119(16)61364-3
Zhang, Y., Fang, J., Wu, X., and Dong, L. (2018). Na+/K+ balance and transport regulatory mechanisms in weedy and cultivated rice (Oryza sativa L.) under salt stress. BMC Plant Biol. 18:375. doi: 10.1186/s12870-018-1586-9
Keywords: Capsicum annuum, CaNHX, phylogenetic tree, co-expression, abiotic stress
Citation: Luo X, Yang S, Luo Y, Qiu H, Li T, Li J, Chen X, Zheng X, Chen Y, Zhang J, Zhang Z and Qin C (2021) Molecular Characterization and Expression Analysis of the Na+/H+ Exchanger Gene Family in Capsicum annuum L.. Front. Genet. 12:680457. doi: 10.3389/fgene.2021.680457
Received: 14 March 2021; Accepted: 02 August 2021;
Published: 02 September 2021.
Edited by:
Deepmala Sehgal, International Maize and Wheat Improvement Center Mexico, MexicoReviewed by:
Ratan Chopra, University of Minnesota Twin Cities, United StatesCopyright © 2021 Luo, Yang, Luo, Qiu, Li, Li, Chen, Zheng, Chen, Zhang, Zhang and Qin. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhongkai Zhang, emhvbmdrYWk5OUBzaW5hLmNvbQ==; Cheng Qin, cWluY2hlbmcxMDAxQDE2My5jb20=; orcid.org/0000-0002-8519-4697
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.