- 1Department of Medical Genetics and Molecular Biology, School of Medicine, Iran University of Medical Sciences, Tehran, Iran
- 2Department of Medical Genetics, Ali-Asghar Children’s Hospital, Tehran, Iran
- 3Shahid Akbarabadi Clinical Research Development Unit (ShACRDU), Iran University of Medical Sciences, Tehran, Iran
Although it has been about 30 years since the discovery of circular RNAs (circRNAs) in mammalian cells, these subtypes of RNAs’ capabilities have come into focus in recent years. The unique structure and various functional roles of circRNAs in many cellular processes have aroused researchers’ interest and raised many questions about whether circRNAs can facilitate the diagnosis and treatment of diseases. To answer these questions, we will illustrate the main known functions and regulatory roles of circRNAs in the cell after presenting a brief history of the discovery of circRNAs and the main proposed theories of the biogenesis of circRNAs. Afterward, the practical application of circRNAs as biomarkers of different pathophysiological conditions will be discussed, mentioning some examples and challenges in this area. We also consider one of the main questions that human beings have always been faced, “the origin of life,” and its possible connection to circRNAs. Finally, focusing on the various capabilities of circRNAs, we discuss their potential therapeutic applications considering the immunity response toward exogenous circRNAs. However, there are still disputes about the exact immune system reaction, which we will discuss in detail.
Introduction
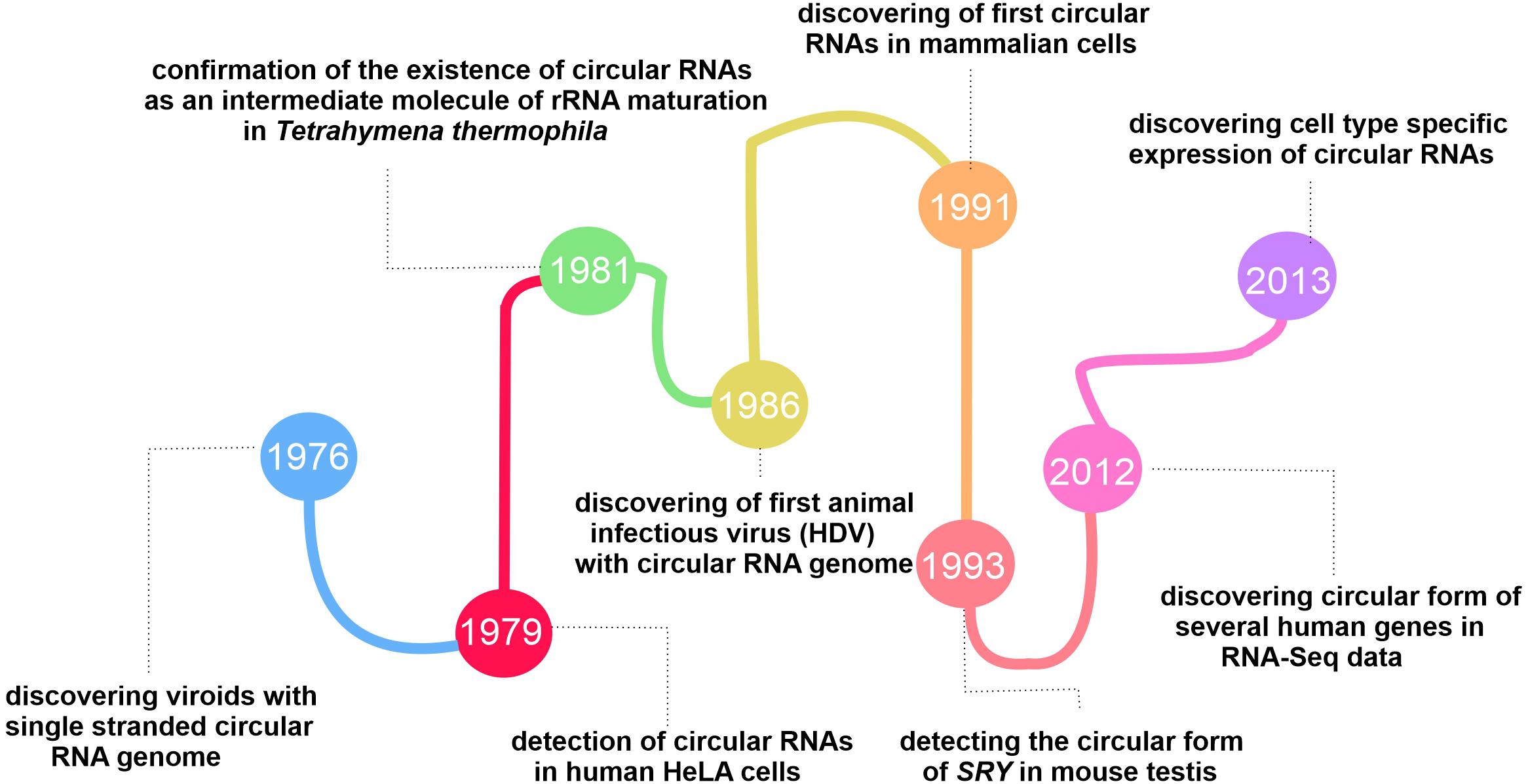
Our current knowledge of the role and importance of circRNAs in mammals, including humans, has come a long way. Since the first circular form of RNA was found in plant-infected viroids (uncoated RNA molecules that infect certain higher plants), scientist’s views on circRNAs have undergone tremendous changes, thanks to the development of new high-throughput sequencing techniques. Sanger et al. (1976) confirmed that viroids like PSTVd are single-stranded RNA molecules with a circular structure, based on the evidence from electron microscopic observations and biochemical investigation.
Nigro et al. (1991) announced the presence of circular forms of tumor suppressor gene, DCC (deleted in colorectal cancer). They referred to this finding merely as “scrambled exon” (Nigro et al., 1991).
The fluctuations in the circRNA discovery area continued until changes in the conventional algorithms and library preparation methods for RNA-seq in 2012, revealing the global expression of circRNAs in the mouse brain and different cell types (Salzman et al., 2012). The tissue-specificity and conserved expression of circRNAs mentioned in two separate published papers in 2013 revealed their functional role in cells (Jeck et al., 2013; Salzman et al., 2013). Since then, researchers have studied circRNAs from different angles to reveal this group of RNAs’ exact function and characteristics (More details on the history behind the evolution of circRNAs researches are presented in Figure 1).
Herein we summarize the known and unknown issues about circRNAs. The possible biogenesis models, roles in different pathologies, and the biological functions of circRNAs have been reviewed earlier (Salzman, 2016; Haddad and Lorenzen, 2019), so we overview these subjects briefly and from different perspectives. This review’s primary focus is on the potential applications of circRNAs, especially in the diagnosis and treatment of diseases, and the findings of studies on the possible immune system response toward the transfection of exogenous circRNAs. Although there are still many unknown issues in circRNAs’s field of research, it is likely that in the near future, given the characteristics of circRNAs and the spread of knowledge in this field of study, a number of the main challenges facing scientists may be addressed, especially in the treatment of diseases.
RNA Binding Proteins, the Main Contributors of circRNAs Biogenesis
The precise mechanism of circRNAs biogenesis is still a point of discussion. Although spliceosome appears to be the main actor of circRNA biogenesis, this process’s details remained unclear. The two highlighted mechanisms recommended by the researchers are direct backsplicing and exon skipping (lariat model). In the direct backsplicing model, there are inverted repeats in the vicinity of the exons. The base pairing of inverted repeats leads to forming a stem-loop structure, which brings circRNA-forming exons into proximity, thus enhancing the interaction between donor splice-site and upstream acceptor site (instead of downstream acceptor site in canonical splicing) (Figure 2A). Backsplicing seems to be a rare phenomenon due to the lower energy efficiency than canonical splicing and the need for complementary sequences that initiate the backsplicing process by bringing the splice sites close to each other (Zhang et al., 2016). Emerging shreds of evidence suggest that the complementary sequence is not enough for circularization, and backsplicing is assisted by many RNA binding proteins (RBPs).
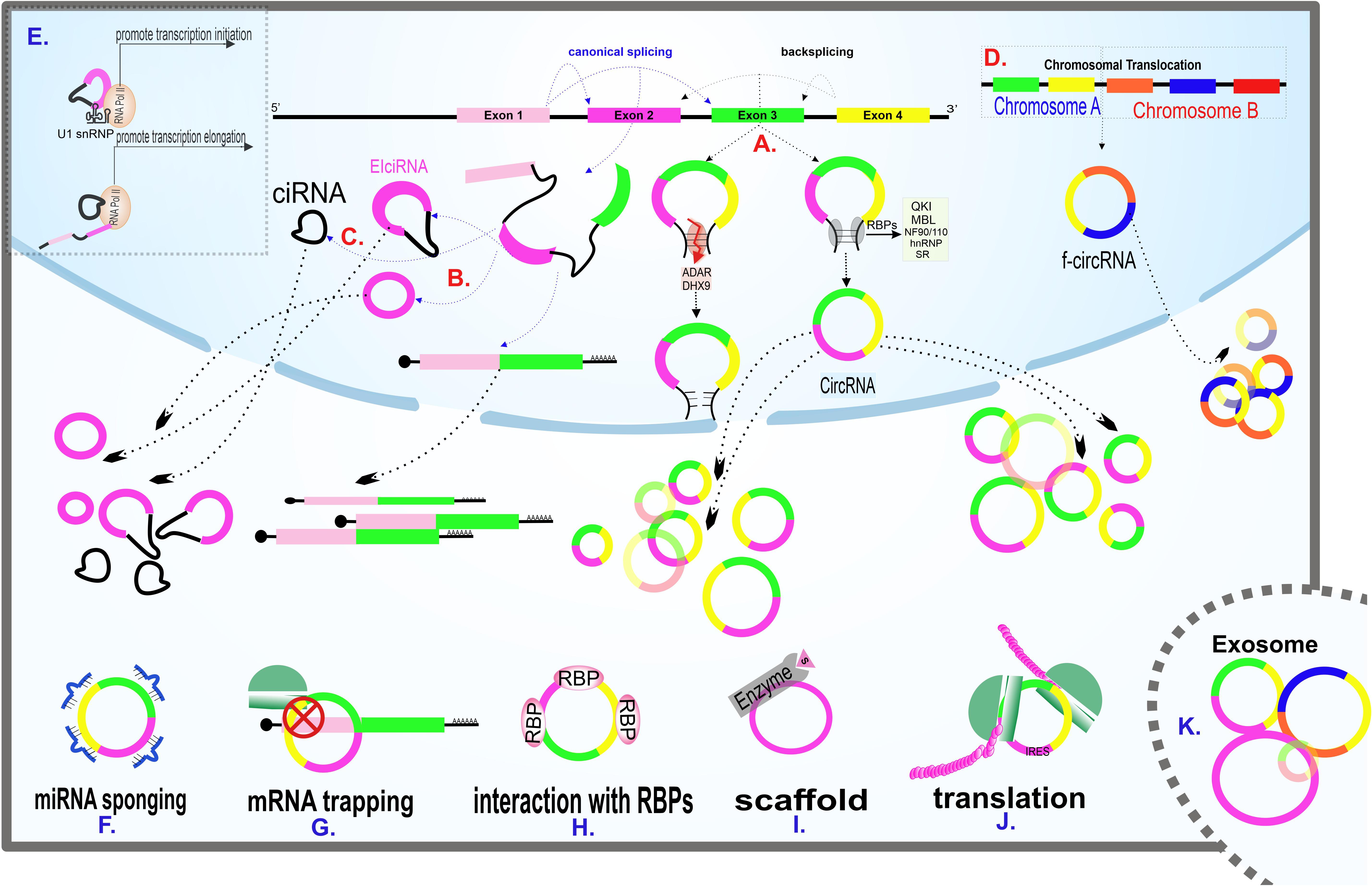
Figure 2. Scheme of main models of circRNAs biogenesis and circRNAs known cellular functions. (A) Generation of circRNAs via direct backsplicing and with the help of RBPs. ADAR and DHX9 inhibit circRNAs formation by unwinding RNA base pairings. (B) Exon skipping model of circRNAs biogenesis. The intermediate lariat formed in canonical splicing endures backsplicing. (C) The remained intron from splicing fails to debranch, leading to CIRNA formation. (D) F-circRNA formation following translocation of chromosomes. (E) Transcription regulation of circRNAs. EIciRNA interacts with the U1 snRNP subunit of spliceosome machinery at the beginning of transcription. CIRNA interacts with phosphorylated RNA polymerase II, facilitating elongation. (F) A circRNA sponging several miRNAs. (G) A circRNA traps its linear counterpart and inhibits translation by blocking the translation start site. (H) Interaction of a circRNA with RBPs. (I) CircRNA acting as a scaffold, facilitating enzyme and substrate reaction. (J) CircRNA containing IRES, coding protein. (K) Enrichment of various circRNAs in exosomes. Exosomal circRNAs are potential biomarkers for various diseases.
One of the most prominent RBPs contributing to the biogenesis of circRNAs is the splicing factor, namely Quaking (QKI). QKI, which is a member of the STAR family (a gene family involved in signal transduction and activation of RNA), binds to conserved binding sites in the introns adjacent to the circularization location and seems to catalysis the circularization process by dimer formation (Vernet and Artzt, 1997; Conn et al., 2015).
Muscleblind (MBL) -examined in Drosophila- is another splicing factor involved in circRNA’s biogenesis. MBL acts in a cis manner and binds to its conserved binding sites, and promotes the synthesis of circMbl, the most abundant circRNA in the Drosophila’s head. The feedback loop occurring in this process is remarkable. The MBL protein -synthesized from a linear Mbl transcript- helps create circular Mbl and prevents linear counterpart formation by competition with the canonical splicing (Ashwal-Fluss et al., 2014).
NF90/NF110, double-strand binding proteins originating from the ILF3 gene, also contribute to the backsplicing process. Studies indicated that circRNA’s overall expression decreases during viral infection, partly due to these antiviral operators’ nuclear export. Experiments such as the biotin-labeled RNA pull-down assays have shown that these proteins strongly bind to dsRNA and probably facilitate circRNA biogenesis by stabilizing RNA base pairing (Li et al., 2017).
Other facilitators of the backsplicing process include the two protein families called heterogeneous nuclear ribonucleoprotein (hnRNPs) and serine-arginine (SR). Previous studies have examined the role of hnRNPs in processes such as pre-mRNA maturation, cellular transport, and stabilizing of mRNA (Geuens et al., 2016). SR proteins also have multiple similar roles in regulating gene expression (Shepard and Hertel, 2009). SR and hnRNPs act together to circularize genes like Drosophila laccase2 (Kramer et al., 2015).
However, RBPs do not always accelerate the backsplicing process and circularization. Both ADAR (adenosine deaminases acting on RNA) and an RNA helicase called DHX9 can interact with inverted ALUs repeats and unwind RNA base pairing and, as a result, inhibit circularization (Ivanov et al., 2015; Aktaş et al., 2017).
As mentioned earlier, backsplicing is not the only possible mechanism of circRNA biogenesis. In the exon skipping (lariat) model, following canonical splicing, the intermediate lariat includes skipping exons endured backsplicing (Figure 2B). In this potential biogenesis model, repeated sequences surrounding the exons are unnecessary (Zaphiropoulos, 1997; Barrett et al., 2015).
Two other probable mechanisms of circRNA’s biogenesis are failure to debranch intron lariat (Figure 2C) and translocation (Figure 2D). Translocations mainly occur in cancers and are believed to be the potential biogenesis mechanisms (Guarnerio et al., 2016; Wilusz, 2017). In addition to the items mentioned earlier, the RNA pol II speed of synthesis also plays a decisive role in a transcript’s fate. The higher the RNA pol II elongation rate, the higher the ratio of circRNA to linear RNA synthesis. As the percentage of RNA synthesis increases, the pairing of complementary sequences is facilitated (Zhang et al., 2016).
circRNAs Play Fundamental Roles in Cell Operation
Discovering facts such as circRNAs tissue-specific expression and specific expression patterns during various development stages has led researchers to investigate the functions of circRNAs.
Herein, we will give a brief overview of some of the main functions that have been described for circRNAs.
miRNA Sponging, Important but Not General
MicroRNAs (miRNAs), RNA molecules with an approximate length of 22 nucleotides, are involved in post transcription regulation of genes by preventing their target mRNAs’ translation (Ambros, 2004). However, the regulatory network across the transcriptome does not end up with mRNAs and miRNAs. In 2013, an investigation showed an ability of circRNAs in miRNA sequestering, the phenomenon known as “miRNA sponging” (Hansen et al., 2013; Figure 2F). The miRNA sponge capability is not limited to circRNAs, and this feature is because of the presence of MREs (MiRNA Response Elements). CircRNAs are also referred to as competitive endogenous RNAs (ceRNAs) in some published data through their ability to inhibit miRNAs’ regulatory function on mRNAs (Mitra et al., 2018; Smillie et al., 2018). Although a significant number of published papers in the field of circRNA rely on this possible role, this phenomenon has been confirmed in just a small number of circRNAs (Dori and Bicciato, 2019). CiRS-7 (also known as CDR1as), circSRY, and circHIPK3 are three circRNAs involved in sponging miR-7, miR-138, and miR-124, respectively (Hansen et al., 2013; Zheng et al., 2016). CircRNAs indirectly affect extensive gene networks. Each circRNA could have several miRNA bindings sites, even for multiple different miRNAs, and each miRNA could inhibit numerous mRNAs. To attribute the sponging role to circRNAs, there must be evidence of miRNA-circRNA co-localization. Additionally, co-expression of circRNA and the related miRNA and the presence of MREs is necessary. Furthermore, the circRNA’s length and the adequacy of their expression abundance should be considered (Hansen et al., 2013; Dori and Bicciato, 2019).
Circular RNAs Regulate Gene Expression in Various Ways
CircRNAs are involved in several gene expression regulation stages, such as transcription initiation and elongation, splicing, and even translation. The three major known subtypes of the circRNAs, exonic circRNAs (EcircRNA), intronic circRNAs (CIRNA), and exon-intron circRNAs (EIciRNA), play roles in this area.
The existence of 2’–5’ Phosphodiester-linked in CIRNAs supports the hypothesis that the formation of this particular subgroup of circRNAs happened because of lariat debranching failure (Figure 2C). CIRNAs are enriched in the nucleus and positively interact with phosphorylated RNA pol II during elongation (Zhang et al., 2013; Figure 2E).
EIciRNAs, which contain both intron and exon in their structure, are also accumulated in the nucleus and indirectly regulate the RNA pol II by RNA-RNA interaction with the U1 snRNA subunit of spliceosome machinery and aid initiation of transcription (Li Z. et al., 2015; Figure 2E). Both CIRNAs and EIciRNAs apply for their regulatory role in a cis manner and enhance their parental gene transcription. The trans-regulatory role of these circRNAs is still a matter of debate.
CircRNAs are also involved in post-transcription regulation. Except for the sponge role of circRNAs described earlier, circular transcripts of a gene can prevent their linear counterparts’ efficient expression in other main ways. One way is the competition between the linear and the circular form of a gene in binding to translation-facilitating proteins. CircPABPN1, for example, appears to reduce the binding of HuR protein -which is known as translation promoter of PABPN1 mRNA- to its linear counterpart and thus reduces the efficiency of translation (Abdelmohsen et al., 2017). There is also a theory in which CircRNAs can prevent protein buildup in a phenomenon called “mRNA trapping.” CircRNAs like circFMN trap their linear mRNA counterparts and block the translation start site, thus protein synthesis (Chao et al., 1998; Figure 2G). Also, given that both linear and circular forms of a gene originating from the same splice site, the biogenesis of circRNAs decreases the final expression rate of linear RNA. It has been found that synthetic cirRNAs are not only transmissible to the cytoplasm but also the nucleus. Given that circRNAs are capable of regulating processes in the nucleus, it is possible to use circRNAs to regulate processes such as transcription and splicing (Jost et al., 2018).
Cell Directors in Close Contact With circRNAs
CircRNAs interact with proteins, the important cellular directors, in a variety of ways (Figures 2H,I). In addition to the above-mentioned RBPs-circRNAs interactions during circRNAs biogenesis, circRNAs can act as a protein transporter, subcellular localizer and scaffold, facilitating protein-protein interactions.
Although bioinformatic studies were done based on nucleotide sequences, the RBP chances of binding to circRNAs are no more than linear forms, but the interaction seems stronger (You et al., 2015).
Transfected cells with circAmotl1 showed increased AKT levels in the nucleus, supporting the fact that circAmotl1 may be involved in the nuclear translocation of AKT protein (Zeng et al., 2017). Previous research has investigated the potential interaction between circRNAs and proteins, denoted that circ-Foxo3 can potentially act as a scaffold and facilitate the interaction of CDK2 and its inhibitor, p21. The ternary complex formed of circ-Foxo3, p21, and CDK2, blocked the G1 to S phase transition of the cell cycle by inhibiting the CDK2 and cyclin A/cyclin E interactions (Du et al., 2016).
Evaluation of the role of circANRIL in atherosclerosis and ribosomal RNA (rRNA) maturation process using mass spectrometric and RNA immunoprecipitation (RIP) assays showed two proteins; PES1 and NOP14 interacted mostly with circANRIL, respectively (Holdt et al., 2016). Since the interaction between PES1 and rRNA is essential for rRNA’s maturation and ribosome formation, circANRIL binding to this member of the PeBoW complex blocks the rRNA binding site and consequently impair protein translation rate and cell growth (Holdt et al., 2016).
Cells perform their vital functions through proteins. Proteins interact with circRNAs for a variety of purposes. As can be deduced from the above examples, some of the most important proteins are transported in the cell with the help of circRNAs. Besides, proteins can interact effectively with other proteins and molecules by binding to circRNAs as a platform. That is one of the many reasons why the cell faces a crisis if mutations affect the expression levels or RNA binding sites of circRNAs.
CircRNAs, Blurring Boundary Between Coding and Non-coding World
CircRNAs are classified in the non-coding RNAs category. However, evidence from mass spectrometry, ribosomal footprinting, and CRISPR Cas9 based experiments indicated this subtype of RNAs’ potential to produce proteins (Legnini et al., 2017; Pamudurti et al., 2017; Yang Y.Y. et al., 2017). Two general mechanisms are known to initiate protein synthesis: cap-dependent and cap-independent [IRES (Internal Ribosome Entry Site)-mediated] translation.
Since there is no 5′ cap structure in circRNAs, the probable mechanism for circRNA translation is the cap-independent (Chen and Sarnow, 1995), and it is done with the help of a specific RNA element called IRES, which enables the interaction of the 40S ribosomal subunits to the transcript (Merrick, 2004; Figure 2J). The cap-independent translation is less effective than the cap-dependent mechanism, but is more commonly used during cellular stress like hypoxia and nutrient starvation, which are common in cancer cells (Walters and Thompson, 2016). The cap-independent mechanism promotes the translation of proteins, such as transcription and growth factors, required for cell survival (Walters and Thompson, 2016; Diallo et al., 2019; Zhao et al., 2019). Studies have shown that sequences containing methylated adenosine N6 (m6A) site at 5’ UTR can act as IRES (Yang Y. et al., 2017).
M6A modifications involvement in processes such as mRNA stability, splicing, and translation were approved, and now it seems that m6A is also effective at the beginning of circRNAs translation (Wang et al., 2014; Ma et al., 2016; Yang Y. et al., 2017; Zhou et al., 2019). CircRNA-m6A-seq revealed that m6A motifs are enriched in circRNAs. Yang Y. et al. (2017) showed that circRNAs containing even a single m6A motif are efficiently transcribed. Inserting m6a motifs before the start codon of circRNA reporter results in efficient GFP (green fluorescent protein) translation. The research revealed that the production of proteins from circRNAs containing m6A modification, which increased by heat shock, is similar to the translation of mRNAs containing m6A modifications. This study also demonstrated that the YTHDF3 plays an indispensable role in this process as an m6A reader, which interacts with the translation initiation factor eIF4G2 (Yang Y. et al., 2017).
Several studies have shown the potential role of circRNAs in protein production (Legnini et al., 2017; Zhang et al., 2018; Liang et al., 2019). Also, tools have been developed to predict the protein-coding potential of circRNAs. For example, CircCode is a python-based tool capable of predicting protein production in humans and Arabidopsis thaliana (Sun and Li, 2019).
It seems that by the advancement of technology in various fields of science and the elimination of technical problems in this area of research, more substantial evidence may be found to prove the ability of circRNAs to code protein.
As can be seen from the examples mentioned earlier, circRNAs are involved in different levels of cell metabolism. Understanding these functions and their effect on maintaining cell homeostasis can inspire new therapies. For example, targeting a circRNA, which is multidimensionally involved in the pathogenesis of a disease, can help effectively treat that disease. Even more, designing and transfecting a circRNA that is capable of adequately coding a tumor suppressor protein and also targeting multiple onco-miRs can be a valuable treatment method.
circRNA’s unique features nominated them as the origin of life
Features like development and tissue specificity, self-catalyzing capacity, and being conserve are among different species are mentioned for circRNAs. However, perhaps the most essential feature is the property acquired through their specific structure (Xia et al., 2017). The closed ring structure of circRNAs leads to the absence of a polyadenylated tail and 5’–3’ polarity. As a result, they are highly stable because of their resistance to exonucleases, including RNaseR (Suzuki et al., 2006). On average, the half-life of circRNAs is 2.5 times more than their linear counterpart, and this stability can last up to 48 h in cells (Jeck et al., 2013; Enuka et al., 2015). Considering this capability, circRNAs could answer the old question “where and how life begins?.” RNA molecules were introduced as a possible origin of life because they could have catalytic activity and store genetic information. However, one critique of the “RNA World Hypothesis” is that RNAs are not stable enough. Given the roles and properties mentioned for circRNAs, especially the super stability feature of these molecules, they can be considered as the potential candidate molecules in initiating life (Demongeot and Moreira, 2007; Neveu et al., 2013). However, answering such a comprehensive question is very challenging since there is no direct evidence of this process. Ideally, the primary circRNA should have a minimum number of nucleotides to minimize the possibility of breakage. It must also be capable of forming secondary structures such as hairpins to increase both the molecule’s stability and have the surface for interaction (Demongeot and Moreira, 2007). Putting aside questions about how early nucleotides formed on Earth and how nucleotides bind to each other in the “prebiotic pool,” this hypothesis seems to be worth further investigation.
CircRNAs, Potential Biomarkers, and the Challenges Ahead
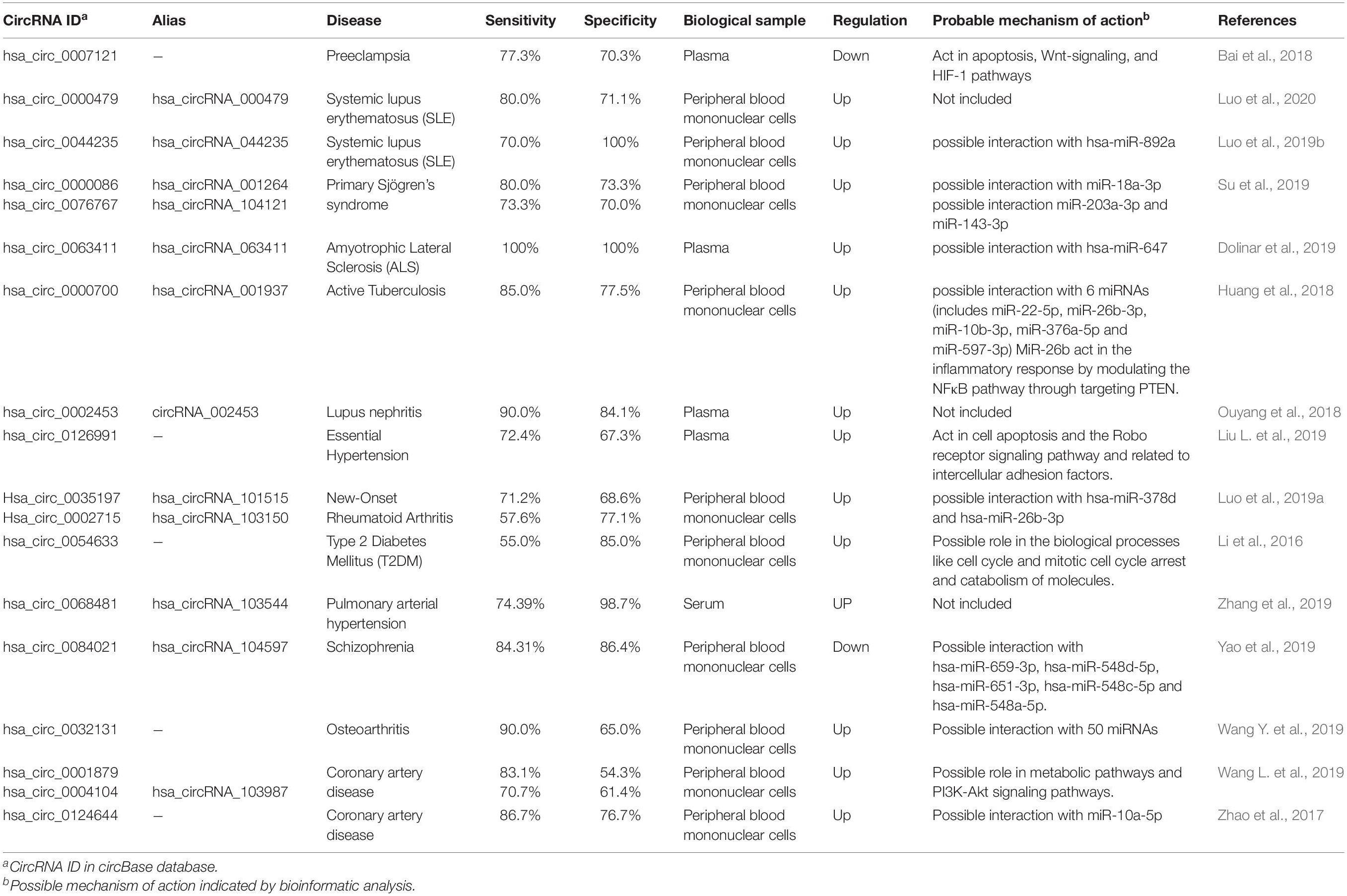
CircRNAs have been suggested as potential biomarkers, as their role in several intracellular molecular processes becomes apparent. Exceptional stability among all types of RNAs is perhaps the most important reason for using circRNAs as biomarkers. CircRNAs, especially those originated from exosomes (Figure 2K), can be detected in various body fluids, including saliva and serum, making it possible to diagnose and predict the prognosis of different kinds of diseases in a non-invasive or minimal-invasive manner (Li Y. et al., 2015). Some circRNAs suggested as the biomarker in various cancers and non-cancerous diseases with reported specificity and sensitivity are listed in Tables 1, 2.
RNA-seq and microarray are the two common high throughput methods for analyzing RNA expression profiles. Using the RNA-seq method seems to be more challenging and less efficient than microarray in the field of circRNAs. Using different library preparation methods, such as size selection or the RNA-seq reading depth, can lead to the loss of small-sized and low-abundant circRNAs data, respectively. Besides, although various algorithms have been developed for circRNA detection in RNA-seq data, none of them seems to be as efficient as they should be, and there is a slight overlap between the results of different algorithms (more details on Szabo and Salzman, 2016). It was demonstrated that microarrays are about 10 times more capable of detecting circRNAs than RNA-seq with an average of 20 million reads depth. However, it is impossible to detect unknown circRNAs using microarrays (Li et al., 2018).
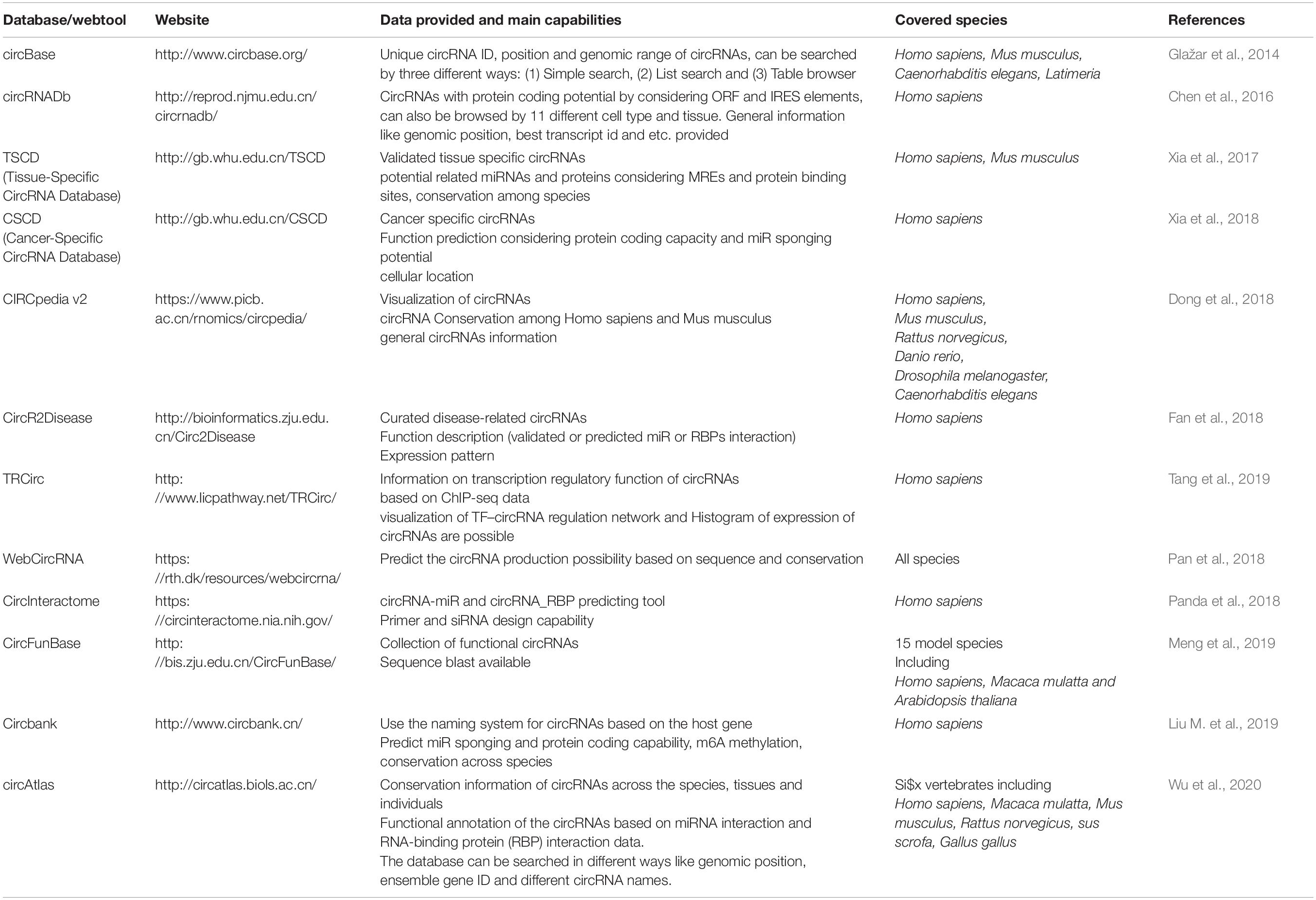
Another factor making the study of circRNAs demanding is the lack of a unit and standard nomenclature system (Franz et al., 2018). Various databases and articles use different naming methods, and converting names to each other is time-consuming and complicated; therefore, there is an urgent need for a single nomenclature system. Some of the most widely used databases for circRNAs and the information they provide is listed in Table 3.
Therapeutic Utility of CircRNAs
CircRNAs are molecules with the potential to treat a wide range of diseases. CircRNA-based treatments can be divided into two general categories. Not only can they be used as a therapeutic target, but it is also possible to take advantage of engineered circRNAs as medicinal operators (Figure 3 summarized some potential applications of circRNAs in the treatment of diseases).
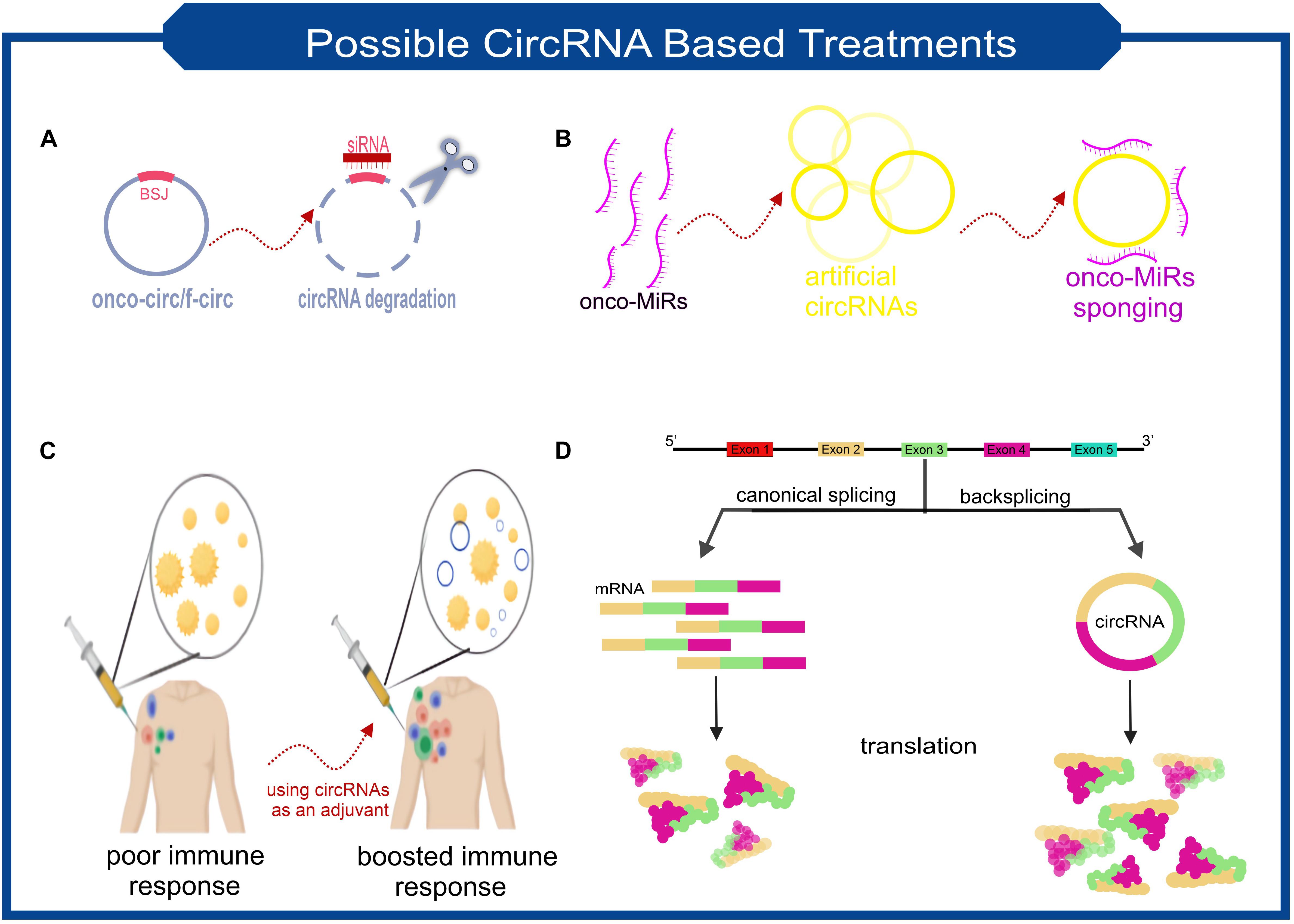
Figure 3. Scheme of some the possible treatment approaches based on the characteristics of circRNAs. (A) Targeting the backsplice junction of onco-circs or f-circs by siRNAs leads to the degradation of these harmful circRNAs. (B) Exclusive circRNAs designed to sequester/sponge oncogenic miRNAs. (C) Utilizing the possible immunogenic properties of circRNAs as an adjuvant. (D) Despite the lower biogenesis of circRNAs than linear RNAs, more stable translation can be achieved due to the high stability of circRNAs. This feature makes circRNAs ideal for protein replacement therapies.
Imagine a circRNA that is known to play a significant role in the exacerbation of a disease. One therapeutic approach can be to knockdown this circRNA with the help of RNA interference (RNAi) (Figure 3A). siRNA’s must be designed to target back-spliced junction; thus, the linear counterpart remains intact (Santer et al., 2019). Studies confirmed that using siRNAs-based knockdown of circRNAs involving various diseases’ pathogenesis will be a promising approach. In vitro assays demonstrated that circ-EIF4G3 and circ-0023642 knockdown would suppress cell proliferation and invasion of gastric cancer (Zhou et al., 2018; Wang Q. et al., 2019). Similar experiments in knockdown of circ-VAPA and circ_ANF609 have similar colorectal and nasopharyngeal carcinoma results, respectively (Yu, 2020; Zhang et al., 2020).
Fusion circRNAs (f-circRNAs), primarily generated in chromosomal translocations during tumorigenesis, are potential cancer treatment targets. The formation of circRNAs can lead to drug resistance, cellular transformation, and other tumor-related hallmarks (Guarnerio et al., 2016). F-circRNAs knockdown would be a good choice for cancer cell-specific targeting (Figure 3A). Sometimes circRNAs knockdown can be used as a co-medication. A recently published study found that circRACGAP1 suppression could be used to reduce the toxic effects of apatinib (a drug that has anti-cancer effects by inhibiting VEGFR-2 tyrosine kinase). CircRACGAP1 enables autophagy-related gene 7 (ATG7) overexpression by sponging miR-3657. Inhibition of circRACGAP1 leads to cell apoptosis instead of autophagy, which sometimes causes cancer cells to survive under environmental stress (Yun and Lee, 2018; Ma et al., 2020).
Another role of circRNAs, which make them suitable candidates for disease treatment, lies in their ability to act as miRNA sponges (Figure 3B). A recently in vivo research result demonstrated that a small dose of unmodified circRNA could sponge two miRNAs involved in cardiac hypertrophy, thereby preventing heart failure (Lavenniah et al., 2020). It has also been shown that transfection of synthetic circRNA named scRNA21 increases the level of miR-21 target proteins such as Daxx by efficiently sponging miR-21 and inhibiting the proliferation of gastric cancer cells (Liu et al., 2018).
Perhaps the most revolutionary circRNA-based treatment lies in their stability and protein-coding capacity. Obtaining a stable expression of an exogenous protein is one of the main issues scientists have been facing. However, using mRNAs for this purpose has the advantage of being safer than DNA manipulation, but achieving adequate and stable expression has always been a challenge for researchers. Considering the potential ability of circRNAs to be translated and their high stability makes them applicable candidates. Researchers found an exogenous circRNA could produce much higher amounts of protein than unmodified linear mRNA and even modified linear mRNA counterparts (Figure 3D; Wesselhoeft et al., 2018). Unmodified circRNAs made from cloned self-splicing intron were transfected using a cationic lipid reagent. The protein production half-life of the highly purified circRNA was approximately two times higher than linear mRNA (Wesselhoeft et al., 2018).
Immune Response Toward Exogenous circRNAs
The ability of circRNAs in the prolonged production of proteins highly suites them as protein expression vectors and favorable in treating many diseases. Nevertheless, there are still fundamental questions to be answered. How does the immune system respond to exogenous circRNAs? Do circRNAs trigger the immune system as much as linear RNAs?
The truth is that researcher’s findings seem contradictory. A study done before investigating the ability of engineered circRNAs in protein production regarded exogenous circRNAs as an immunogenic agent. Chen Y.G. et al. (2017) showed that transfection of HeLa cells with RNAse R purified circRNAs results in overexpression of innate immunity genes like RIG-I (retinoic-acid-inducible gene-I) up to 500-fold. The role of RIG-I in controlling viral infections had been identified earlier. RIG-I triggers the innate immune system after recognizing dsRNA and 5′ triphosphate (Kell and Gale, 2015). CircRNAs lack 5′ triphosphate, so what triggers innate immunity, and how RIG-I sense endogenous circRNAs from exogenous ones? (Chen Y.G. et al., 2017) proved by an experiment that splicing of a circRNA with human introns in human cells does not induce RIG-I, and this result is independent of circRNA exon sequence. These results stated that RBPs associated with human-specific splicesome mark circRNAs as “self” or “non-self.” Nevertheless, two separate studies, both conducted by Wesselhoeft et al. (2018, 2019), assumed that the innate immune system provocation was only because of circRNAs contamination with other types of RNAs, and the use of ultra-pure circRNAs showed no significant sign of innate immune system provocation. Injection of RNaseR and phosphatase treated circRNAs, purified by HPLC into mice adipose tissue, did not show cytokine secretion. Also, the protein expression remained stable, supporting the ability of circRNAs to perform protein replacement therapy (Ma et al., 2016).
It is not the end of the story. In another research, Chen et al. (2019) confirmed their findings of circRNAs immunizing capacity. In this study, even though using HPLC purified circRNAs, immune system stimulation was still observed. Subcutaneous injection of exogenous circRNA results in dendritic cell activation and, as a result, indirect activation of T cells, which means not only the innate immune system but also the acquired immunity is triggered by exogenous circRNAs. These findings introduce another therapeutic potential of circRNAs to act as an adjuvant (Figure 3C). They also described the role of m6A modification patterns in distinguishing between endogenous and exogenous circRNAs. M6A modifications prevent RIG-I binding to circRNAs and, as a result, activation of the immune response. Experiments indicate that circRNAs from different origins (i.e., human vs. phage) have different m6A modification patterns at 3′ of back-splice junction and, therefore, different immunity response.
Differences in these researches findings may be attributed to details in the experiment’s design, such as cell types or the used reagents (Basavappa and Cherry, 2019). Further studies are needed to confirm each of these results and explain the underlying mechanisms of differences.
Discussion
For years, circRNAs were known as a “by-product of splicing” rather than a subspecies of RNAs with independent roles. However, in recent years series of functions have been defined for them.
This review summarized the current and potential applications of circRNAs based on their various functions and features. CircRNAs have multiple roles in biological processes, like gene expression regulation, interacting with proteins for different purposes, and coding proteins. Most of the published papers focused on the post transcription regulation of circRNAs, miR sponging. However, researchers should consider items such as sufficient circRNA amount, co-expression with the target miR, and the ratio between circRNA and target miRs in cases where the “sponging” capacity has not been investigated by experimental methods (Dori and Bicciato, 2019). In addition to the fact that circRNAs have various roles in the pathogenesis of various diseases, the stability and presence of circRNAs in body fluids like plasma and saliva have made them ideal diagnostic/prognostic biomarkers, and even several clinical trials are underway in this regard (ClinicalTrials.gov number: NCT04584996; NCT03170830). However, there are still challenges that need to be addressed to expedite and facilitate such studies. RNA-seq and microarray are among the high-throughput technologies that are used to screen differentially expressed circRNAs in different conditions. RNA-seq can evaluate the expression levels of unknown circRNAs. However, different analysis algorithms and even different library preparation methods lead to different results. To get the most out of this method, developing a gold standard for library preparation and bioinformatics analysis pipelines is highly recommended (Engström et al., 2013; Szabo and Salzman, 2016). Although microarray seems to be more efficient than RNA-seq, however, this method is limited to known circRNAs, and also more initial amount of RNA is needed (Li et al., 2018).
In addition to the role that circRNAs can play in the diagnosis and management of diseases, various features of circRNAs can also be used in the treatment of diseases. One possible treatment approach is to reduce the expression of circRNAs such as f-circRNAs and onco-circRNAs using siRNA designed for backspliced junction (Guarnerio et al., 2016). The alternative method for suppressing the circRNA expression is the use of CRISPR/CAS9. Adequate care must be taken in the use of the CRISPR/CAS9 system. Given that most circRNAs have a common origin with their linear counterpart, this method can only be used to remove the complementary sequences required for the biogenesis of circRNAs (Zhang et al., 2016). Moreover, engineered circRNAs can be used to sponge the disease-related miRNAs. Although anti-miRNA drugs such as Miravirsen (a locked-nucleic-acid (LNA)/DNA-mixmer oligonucleotide use to target HCV-related miRNA, miR22) have already been designed, LNA-based drugs show side effects related to the accumulation of unmetabolizable LNA nucleotides. Since the elimination cycle of circRNAs is naturally embedded in the cells, such complications are unlikely to occur (Janssen et al., 2013; Jost et al., 2018).
Another potential revolutionary circRNA-based treatment is achieving stable and efficient protein production in cells, regarding the resistance of circRNAs toward many ribonucleases. Nevertheless, further research needs to be done to establish the immune system’s precise mechanism of action toward exogenous circRNAs. If the immune system is not stimulated, stable protein expression is achieved. But if the activation of the immune system occurs, as happened in Chen et al. (2019) investigation, this event can be used positively, and circRNAs can be used as an adjuvant.
It is possible that by eliminating technical shortcomings and expanding knowledge in this area, circRNAs, once considered a by-product of splicing, shed new light on diagnosing and treating diseases.
Author Contributions
LY performed the literature search and drafted the manuscript. MA and MM critically revised the work. All authors read and approved the final manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
Abdelmohsen, K., Panda, A. C., Munk, R., Grammatikakis, I., Dudekula, D. B., De, S., et al. (2017). Identification of HuR target circular RNAs uncovers suppression of PABPN1 translation by CircPABPN1. RNA Biol. 14, 361–369. doi: 10.1080/15476286.2017.1279788
Aktaş, T., Ilik, I. A., Maticzka, D., Bhardwaj, V., Pessoa Rodrigues, C., Mittler, G., et al. (2017). DHX9 suppresses RNA processing defects originating from the Alu invasion of the human genome. Nature 544, 115–119. doi: 10.1038/nature21715
Ashwal-Fluss, R., Meyer, M., Pamudurti, N. R., Ivanov, A., Bartok, O., Hanan, M., et al. (2014). CircRNA biogenesis competes with Pre-mRNA splicing. Mol. Cell 56, 55–66. doi: 10.1016/j.molcel.2014.08.019
Bai, Y., Rao, H., Chen, W., Luo, X., Tong, C., and Qi, H. (2018). Profiles of circular RNAs in human placenta and their potential roles related to preeclampsia. Biol. Reprod. 98, 705–712. doi: 10.1093/biolre/ioy034
Barrett, S. P., Wang, P. L., and Salzman, J. (2015). Circular RNA biogenesis can proceed through an exon-containing lariat precursor. Elife 4:e07540. doi: 10.7554/eLife.07540
Basavappa, M. G., and Cherry, S. (2019). Going in circles: the black box of circular RNA immunogenicity. Mol. Cell 76, 3–5. doi: 10.1016/j.molcel.2019.08.027
Chao, C. W., Chan, D. C., Kuo, A., and Leder, P. (1998). The mouse formin (Fmn) gene: abundant circular RNA transcripts and gene- targeted deletion analysis. Mol. Med. 4, 614–628. doi: 10.1007/bf03401761
Chen, C. Y., and Sarnow, P. (1995). Initiation of protein synthesis by the eukaryotic translational apparatus on circular RNAs. Science 268, 415–417. doi: 10.1126/science.7536344
Chen, D., Zhang, C., Lin, J., Song, X., and Wang, H. (2018). Screening differential circular RNA expression profiles reveal that hsa_circ_0128298 is a biomarker in the diagnosis and prognosis of hepatocellular carcinoma. Cancer Manag. Res. 10, 1275–1283. doi: 10.2147/CMAR.S166740
Chen, S., Li, T., Zhao, Q., Xiao, B., and Guo, J. (2017). Using circular RNA hsa_circ_0000190 as a new biomarker in the diagnosis of gastric cancer. Clin. Chim. Acta 466, 167–171. doi: 10.1016/j.cca.2017.01.025
Chen, X., Han, P., Zhou, T., Guo, X., Song, X., and Li, Y. (2016). CircRNADb: a comprehensive database for human circular RNAs with protein-coding annotations. Sci. Rep. 6:34985. doi: 10.1038/srep34985
Chen, Y. G., Chen, R., Ahmad, S., Verma, R., Kasturi, S. P., Amaya, L., et al. (2019). N6-Methyladenosine modification controls circular RNA immunity. Mol. Cell 76, 96–109.e9. doi: 10.1016/j.molcel.2019.07.016
Chen, Y. G., Kim, M. V., Chen, X., Batista, P. J., Aoyama, S., Wilusz, J. E., et al. (2017). Sensing self and foreign circular RNAs by intron identity. Mol. Cell 67, 228–238.e5. doi: 10.1016/j.molcel.2017.05.022
Conn, S. J., Pillman, K. A., Gregory, P. A., Goodall, G. J., Conn, S. J., Pillman, K. A., et al. (2015). The RNA binding protein quaking regulates formation of circRNAs article the RNA binding protein quaking regulates formation of circRNAs. Cell 160, 1125–1134. doi: 10.1016/j.cell.2015.02.014
Demongeot, J., and Moreira, A. (2007). A possible circular RNA at the origin of life. J. Theor. Biol. 249, 314–324. doi: 10.1016/j.jtbi.2007.07.010
Diallo, L. H., Tatin, F., David, F., Godet, A. C., Zamora, A., Prats, A. C., et al. (2019). How are circRNAs translated by non-canonical initiation mechanisms? Biochimie 164, 45–52. doi: 10.1016/j.biochi.2019.06.015
Dolinar, A., Koritnik, B., Glavač, D., and Ravnik-Glavač, M. (2019). Circular RNAs as potential blood biomarkers in amyotrophic lateral sclerosis. Mol. Neurobiol. 56, 8052–8062. doi: 10.1007/s12035-019-1627-x
Dong, R., Ma, X. K., Li, G. W., and Yang, L. (2018). CIRCpedia v2: an updated database for comprehensive circular RNA annotation and expression comparison. Genomics Proteomics Bioinformatics 16, 226–233. doi: 10.1016/j.gpb.2018.08.001
Dori, M., and Bicciato, S. (2019). Integration of bioinformatic predictions and experimental data to identify circRNA-miRNA associations. Genes (Basel) 10, 26–32. doi: 10.3390/genes10090642
Dou, Z., Li, S., Ren, W., Wang, Q., Liu, J., Kong, X., et al. (2019). Decreased expression of hsa:circ_0072387 as a valuable predictor for oral squamous cell carcinoma. Oral Dis. 25, 1302–1308. doi: 10.1111/odi.13094
Du, W. W., Yang, W., Liu, E., Yang, Z., Dhaliwal, P., and Yang, B. B. (2016). Foxo3 circular RNA retards cell cycle progression via forming ternary complexes with p21 and CDK2. Nucleic Acids Res. 44, 2846–2858. doi: 10.1093/nar/gkw027
Engström, P. G., Steijger, T., Sipos, B., Grant, G. R., Kahles, A., Rätsch, G., et al. (2013). Systematic evaluation of spliced alignment programs for RNA-seq data. Nat. Methods 10, 1185–1191. doi: 10.1038/nmeth.2722
Enuka, Y., Lauriola, M., Feldman, M. E., Sas-chen, A., Ulitsky, I., and Yarden, Y. (2015). Circular RNAs are long-lived and display only minimal early alterations in response to a growth factor. Nucleic Acids Res. 44, 1370–1383. doi: 10.1093/nar/gkv1367
Fan, C., Lei, X., Fang, Z., Jiang, Q., and Wu, F. X. (2018). CircR2Disease: a manually curated database for experimentally supported circular RNAs associated with various diseases. Database 2018:bay044. doi: 10.1093/database/bay044
Fan, L., Cao, Q., Liu, J., Zhang, J., and Li, B. (2019). Correction to: circular RNA profiling and its potential for esophageal squamous cell cancer diagnosis and prognosis. Mol. Cancer 18:121. doi: 10.1186/s12943-019-1050-y
Franz, A., Rabien, A., Stephan, C., Ralla, B., Fuchs, S., Jung, K., et al. (2018). Circular RNAs: a new class of biomarkers as a rising interest in laboratory medicine. Clin. Chem. Lab. Med. 56, 1992–2003. doi: 10.1515/cclm-2018-0231
Fu, L., Yao, T., Chen, Q., Mo, X., Hu, Y., and Guo, J. (2017). Screening differential circular RNA expression profiles reveals hsa_circ_0004018 is associated with hepatocellular carcinoma. Oncotarget 8, 58405–58416. doi: 10.18632/oncotarget.16881
Geuens, T., Bouhy, D., and Timmerman, V. (2016). The hnRNP family: insights into their role in health and disease. Hum. Genet. 135, 851–867. doi: 10.1007/s00439-016-1683-5
Glažar, P., Papavasileiou, P., and Rajewsky, N. (2014). CircBase: a database for circular RNAs. RNA 20, 1666–1670. doi: 10.1261/rna.043687.113
Guarnerio, J., Bezzi, M., Jeong, J. C., Paffenholz, S. V., Berry, K., Naldini, M. M., et al. (2016). Oncogenic role of fusion-circRNAs derived from cancer-associated chromosomal translocations. Cell 165, 289–302. doi: 10.1016/j.cell.2016.03.020
Haddad, G., and Lorenzen, J. M. (2019). Biogenesis and function of circular RNAs in health and in disease. Front. Pharmacol. 10:428. doi: 10.3389/fphar.2019.00428
Hansen, T. B., Jensen, T. I., Clausen, B. H., Bramsen, J. B., Finsen, B., Damgaard, C. K., et al. (2013). Natural RNA circles function as efficient microRNA sponges. Nature 495, 384–388. doi: 10.1038/nature11993
Holdt, L. M., Stahringer, A., Sass, K., Pichler, G., Kulak, N. A., Wilfert, W., et al. (2016). Circular non-coding RNA ANRIL modulates ribosomal RNA maturation and atherosclerosis in humans. Nat. Commun. 7:12429. doi: 10.1038/ncomms12429
Huang, Z. K., Yao, F. Y., Xu, J. Q., Deng, Z., Su, R. G., Peng, Y. P., et al. (2018). Microarray expression profile of circular RNAs in peripheral blood mononuclear cells from active tuberculosis patients. Cell. Physiol. Biochem. 45, 1230–1240. doi: 10.1159/000487454
Ivanov, A., Memczak, S., Wyler, E., Torti, F., Porath, H. T., Orejuela, M. R., et al. (2015). Analysis of intron sequences reveals hallmarks of circular RNA biogenesis in animals. Cell Rep. 10, 170–177. doi: 10.1016/j.celrep.2014.12.019
Janssen, H. L. A., Reesink, H. W., Lawitz, E. J., Zeuzem, S., Rodriguez-Torres, M., Patel, K., et al. (2013). Treatment of HCV Infection by Targeting MicroRNA. N. Engl. J. Med. 368, 1685–1694. doi: 10.1056/nejmoa1209026
Jeck, W. R., Sorrentino, J. A., Wang, K., Slevin, M. K., Burd, C. E., Liu, J., et al. (2013). Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA 19, 141–157. doi: 10.1261/rna.035667.112
Pan, X., Xiong, K., Anthon, C., Hyttel, P., Freude, K. K., Jensen, L. J., et al. (2018). WebCircRNA: classifying the circular RNA potential of coding and noncoding RNA. Genes 9:536. doi: 10.3390/genes9110536
Jost, I., Shalamova, L. A., Gerresheim, G. K., Niepmann, M., Bindereif, A., and Rossbach, O. (2018). Functional sequestration of microRNA-122 from Hepatitis C Virus by circular RNA sponges. RNA Biol. 15, 1032–1039. doi: 10.1080/15476286.2018.1435248
Kramer, M. C., Liang, D., Tatomer, D. C., Gold, B., March, Z. M., Cherry, S., et al. (2015). Combinatorial control of Drosophila circular RNA expression by intronic repeats, hnRNPs, and SR proteins. Genes Dev. 29, 2168–2182. doi: 10.1101/gad.270421.115
Lan, X., Cao, J., Xu, J., Chen, C., Zheng, C., Wang, J., et al. (2018). Decreased expression of hsa_circ_0137287 predicts aggressive clinicopathologic characteristics in papillary thyroid carcinoma. J. Clin. Lab. Anal. 32:e22573. doi: 10.1002/jcla.22573
Lavenniah, A., Luu, T. D. A., Li, Y. P., Lim, T. B., Jiang, J., Ackers-Johnson, M., et al. (2020). Engineered circular RNA sponges act as miRNA Inhibitors to attenuate pressure overload-induced cardiac hypertrophy. Mol. Ther. 28, 1506–1517. doi: 10.1016/j.ymthe.2020.04.006
Legnini, I., Di Timoteo, G., Rossi, F., Morlando, M., Briganti, F., Sthandier, O., et al. (2017). Circ-ZNF609 Is a circular RNA that can be translated and functions in Myogenesis. Mol. Cell 66, 22–37.e9. doi: 10.1016/j.molcel.2017.02.017
Li, M., Zhao, Z., Li, X., Gao, C., Jian, D., Hao, P., et al. (2016). Hsa_circ_0054633 in peripheral blood can be used as a diagnostic biomarker of pre-diabetes and Type 2 diabetes mellitus. Acta Diabetol. 54, 237–245. doi: 10.20944/preprints201608.0225.v1
Li, P., Chen, S., Chen, H., Mo, X., Li, T., Shao, Y., et al. (2015). Using circular RNA as a novel type of biomarker in the screening of gastric cancer. Clin. Chim. Acta 444, 132–136. doi: 10.1016/j.cca.2015.02.018
Li, S., Teng, S., Xu, J., Su, G., Zhang, Y., Zhao, J., et al. (2018). Microarray is an efficient tool for circRNA profiling. Brief. Bioinform. 20, 1420–1433. doi: 10.1093/bib/bby006
Li, X., Liu, C. X., Xue, W., Zhang, Y., Jiang, S., Yin, Q. F., et al. (2017). Coordinated circRNA Biogenesis and function with NF90/NF110 in viral infection. Mol. Cell 67, 214–227.e7. doi: 10.1016/j.molcel.2017.05.023
Li, Y., Zheng, Q., Bao, C., Li, S., Guo, W., Zhao, J., et al. (2015). Circular RNA is enriched and stable in exosomes: a promising biomarker for cancer diagnosis. Cell Res. 25, 981–984. doi: 10.1038/cr.2015.82
Li, Z., Huang, C., Bao, C., Chen, L., Lin, M., Wang, X., et al. (2015). Exon-intron circular RNAs regulate transcription in the nucleus. Nat. Struct. Mol. Biol. 22, 256–264. doi: 10.1038/nsmb.2959
Liang, W. C., Wong, C. W., Liang, P. P., Shi, M., Cao, Y., Rao, S. T., et al. (2019). Translation of the circular RNA circβ-catenin promotes liver cancer cell growth through activation of the Wnt pathway. Genome Biol. 20:84. doi: 10.1186/s13059-019-1685-4
Liu, L., Gu, T., Bao, X., Zheng, S., Zhao, J., and Zhang, L. (2019). Microarray profiling of circular RNA identifies hsa-circ-0126991 as a potential risk factor for essential hypertension. Cytogenet. Genome Res. 157, 203–212. doi: 10.1159/000500063
Liu, M., Wang, Q., Shen, J., Yang, B. B., and Ding, X. (2019). Circbank: a comprehensive database for circRNA with standard nomenclature. RNA Biol. 16, 899–905. doi: 10.1080/15476286.2019.1600395
Liu, X., Abraham, J. M., Cheng, Y., Wang, Z., Wang, Z., Zhang, G., et al. (2018). Synthetic circular RNA Functions as a miR-21 sponge to suppress gastric carcinoma cell proliferation. Mol. Ther. Nucleic Acids 13, 312–321. doi: 10.1016/j.omtn.2018.09.010
Liu, X. X., Yang, Y. E., Liu, X., Zhang, M. Y., Li, R., Yin, Y. H., et al. (2019). A two-circular RNA signature as a non-invasive diagnostic biomarker for lung adenocarcinoma. J. Transl. Med. 17, 1–13. doi: 10.1186/s12967-019-1800-z
Lu, J., Zhang, P. Y., Xie, J. W., Wang, J. B., Lin, J. X., Chen, Q. Y., et al. (2019). Hsa_circ_0000467 promotes cancer progression and serves as a diagnostic and prognostic biomarker for gastric cancer. J. Clin. Lab. Anal 33:e22726. doi: 10.1002/jcla.22726
Lü, L., Sun, J., Shi, P., Kong, W., Xu, K., He, B., et al. (2017). Identification of circular RNAs as a promising new class of diagnostic biomarkers for human breast cancer. Oncotarget 8, 44096–44107. doi: 10.18632/oncotarget.17307
Luo, Q., Liu, J., Fu, B., Zhang, L., Guo, Y., Huang, Z., et al. (2019a). Circular RNAs Hsa_circ_0002715 and Hsa_circ_0035197 in peripheral blood are novel potential biomarkers for new-onset rheumatoid arthritis. Dis. Mark. 2019:2073139. doi: 10.1155/2019/2073139
Luo, Q., Zhang, L., Fang, L., Fu, B., Guo, Y., Huang, Z., et al. (2020). Circular RNAs hsa_circ_0000479 in peripheral blood mononuclear cells as novel biomarkers for systemic lupus erythematosus. Autoimmunity 53, 167–176. doi: 10.1080/08916934.2020.1728529
Luo, Q., Zhang, L., Li, X., Fu, B., Guo, Y., Huang, Z., et al. (2019b). Identification of circular RNAs hsa_circ_0044235 and hsa_circ_0068367 as novel biomarkers for systemic lupus erythematosus. Int. J. Mol. Med. 44, 1462–1472. doi: 10.3892/ijmm.2019.4302
Ma, H., Weng, X., Chen, K., Shi, H., He, C., and Street, E. (2016). N6 -methyladenosine modulates messenger RNA translation efficiency. Cell 161, 1388–1399. doi: 10.1016/j.cell.2015.05.014.N
Ma, L., Wang, Z., Xie, M., Quan, Y., Zhu, W., Yang, F., et al. (2020). Silencing of circRACGAP1 sensitizes gastric cancer cells to apatinib via modulating autophagy by targeting miR-3657 and ATG7. Cell Death Dis. 11:169. doi: 10.1038/s41419-020-2352-0
Meng, X., Hu, D., Zhang, P., Chen, Q., and Chen, M. (2019). CircFunBase: a database for functional circular RNAs. Database 2019:baz003. doi: 10.1093/database/baz003
Merrick, W. C. (2004). Cap-dependent and cap-independent translation in eukaryotic systems. Gene 332, 1–11. doi: 10.1016/j.gene.2004.02.051
Mitra, A., Pfeifer, K., and Park, K. S. (2018). Circular RNAs and competing endogenous RNA (ceRNA) networks. Transl. Cancer Res. 7, S624–S628. doi: 10.21037/tcr.2018.05.12
Neveu, M., Kim, H. J., and Benner, S. A. (2013). The “strong” RNA world hypothesis: fifty years old. Astrobiology 13, 391–403. doi: 10.1089/ast.2012.0868
Nigro, J. M., Cho, K. R., Fearon, E. R., Kern, S. E., Ruppert, J. M., Oliner, J. D., et al. (1991). Scrambled exons. Cell 64, 607–613. doi: 10.1016/0092-8674(91)90244-S
Ouyang, Q., Huang, Q., Jiang, Z., Zhao, J., Shi, G. P., and Yang, M. (2018). Using plasma circRNA_002453 as a novel biomarker in the diagnosis of lupus nephritis. Mol. Immunol. 101, 531–538. doi: 10.1016/j.molimm.2018.07.029
Pamudurti, N. R., Bartok, O., Jens, M., Ashwal-Fluss, R., Stottmeister, C., Ruhe, L., et al. (2017). Translation of CircRNAs. Mol. Cell 66, 9–21.e7. doi: 10.1016/j.molcel.2017.02.021
Panda, A. C., Dudekula, D. B., Abdelmohsen, K., and Gorospe, M. (2018). Analysis of circular RNAs using the web tool CircInteractome. Methods Mol. Biol. 1724, 43–56. doi: 10.1007/978-1-4939-7562-4_4
Qiao, G. L., Chen, L., Jiang, W. H., Yang, C., Yang, C. M., Song, L. N., et al. (2019). Hsa_circ_0003998 may be used as a new biomarker for the diagnosis and prognosis of hepatocellular carcinoma. Onco. Targets Ther. 12, 5849–5860. doi: 10.2147/OTT.S210363
Qin, M., Liu, G., Huo, X., Tao, X., Sun, X., Ge, Z., et al. (2016). Hsa-circ-0001649: a circular RNA and potential novel biomarker for hepatocellular carcinoma. Cancer Biomark. 16, 161–169. doi: 10.3233/CBM-150552
Salzman, J. (2016). Circular RNA expression: its potential regulation and function. Trends Genet. 32, 309–316. doi: 10.1016/j.tig.2016.03.002
Salzman, J., Chen, R. E., Olsen, M. N., Wang, P. L., and Brown, P. O. (2013). Cell-type specific features of circular RNA expression. PLoS Genet. 9:e1003777. doi: 10.1371/journal.pgen.1003777
Salzman, J., Gawad, C., Wang, P. L., Lacayo, N., and Brown, P. O. (2012). Circular RNAs are the predominant transcript isoform from hundreds of human genes in diverse cell types. PLoS One 7:e30733. doi: 10.1371/journal.pone.0030733
Sanger, H. L., Klotz, G., Riesner, D., Gross, H. J., and Kleinschmidt, A. K. (1976). Viroids are single stranded covalently closed circular RNA molecules existing as highly base paired rod like structures. Proc. Natl. Acad. Sci. U.S.A. 73, 3852–3856. doi: 10.1073/pnas.73.11.3852
Santer, L., Bär, C., and Thum, T. (2019). Circular RNAs: a novel class of functional RNA molecules with a therapeutic perspective. Mol. Ther. 27, 1350–1363. doi: 10.1016/j.ymthe.2019.07.001
Shao, Y., Tao, X., Lu, R., Zhang, H., Ge, J., Xiao, B., et al. (2019). Hsa_circ_0065149 is an indicator for early gastric cancer screening and prognosis prediction. Pathol. Oncol. Res. 26, 1475–1482. doi: 10.1007/s12253-019-00716-y
Shepard, P. J., and Hertel, K. J. (2009). The SR protein family. Genome Biol. 10:242. doi: 10.1186/gb-2009-10-10-242
Smillie, C. L., Sirey, T., and Ponting, C. P. (2018). Complexities of post-transcriptional regulation and the modeling of ceRNA crosstalk. Crit. Rev. Biochem. Mol. Biol. 53, 231–245. doi: 10.1080/10409238.2018.1447542
Su, L. C., Xu, W. D., Liu, X. Y., Fu, L., and Huang, A. F. (2019). Altered expression of circular RNA in primary Sjögren’s syndrome. Clin. Rheumatol. 38, 3425–3433. doi: 10.1007/s10067-019-04728-6
Sun, P., and Li, G. (2019). CircCode: a powerful tool for identifying circRNA coding ability. Front. Genet 10:981. doi: 10.3389/fgene.2019.00981
Suzuki, H., Zuo, Y., Wang, J., Zhang, M. Q., Malhotra, A., and Mayeda, A. (2006). Characterization of RNase R-digested cellular RNA source that consists of lariat and circular RNAs from pre-mRNA splicing. Nucleic Acids Res. 34:e63. doi: 10.1093/nar/gkl151
Szabo, L., and Salzman, J. (2016). Detecting circular RNAs: bioinformatic and experimental challenges. Nat. Rev. Genet. 17, 679–692. doi: 10.1038/nrg.2016.114
Tang, Z., Li, X., Zhao, J., Qian, F., Feng, C., Li, Y., et al. (2019). TRCirc: a resource for transcriptional regulation information of circRNAs. Brief. Bioinform. 20, 2327–2333. doi: 10.1093/bib/bby083
Tian, J., Xi, X., Wang, J., Yu, J., Huang, Q., Ma, R., et al. (2019). CircRNA hsa_circ_0004585 as a potential biomarker for colorectal cancer. Cancer Manag. Res. 11, 5413–5423. doi: 10.2147/CMAR.S199436
Vernet, C., and Artzt, K. (1997). STAR, a gene family involved in signal transduction and activation of RNA. Trends Genet. 13, 479–484. doi: 10.1016/S0168-9525(97)01269-9
Walters, B., and Thompson, S. R. (2016). Cap-independent translational control of carcinogenesis. Front. Oncol. 6:128. doi: 10.3389/fonc.2016.00128
Wang, J., Li, X., Lu, L., He, L., Hu, H., and Xu, Z. (2018). Circular RNA hsa_circ_0000567 can be used as a promising diagnostic biomarker for human colorectal cancer. J. Clin. Lab. Anal. 32:e22379. doi: 10.1002/jcla.22379
Wang, L., Shen, C., Wang, Y., Zou, T., Zhu, H., Lu, X., et al. (2019). Identification of circular RNA Hsa_circ_0001879 and Hsa_circ_0004104 as novel biomarkers for coronary artery disease. Atherosclerosis 286, 88–96. doi: 10.1016/j.atherosclerosis.2019.05.006
Wang, Q., Wang, T., Hu, Y., Jiang, W., Lu, C., Zheng, W., et al. (2019). Circ-EIF4G3 promotes the development of gastric cancer by sponging miR-335. Pathol. Res. Pract. 215:152507. doi: 10.1016/j.prp.2019.152507
Wang, X., Lu, Z., Gomez, A., Hon, G. C., Yue, Y., Han, D., et al. (2014). N 6-methyladenosine-dependent regulation of messenger RNA stability. Nature 505, 117–120. doi: 10.1038/nature12730
Wang, Y., Wu, C., Yang, Y., Ren, Z., Lammi, M. J., and Guo, X. (2019). Preliminary exploration of hsa-circ-0032131 levels in peripheral blood as a potential diagnostic biomarker of osteoarthritis. Genet. Test. Mol. Biomark. 23, 717–721. doi: 10.1089/gtmb.2019.0036
Wesselhoeft, R. A., Kowalski, P. S., and Anderson, D. G. (2018). Engineering circular RNA for potent and stable translation in eukaryotic cells. Nat. Commun. 9:2629. doi: 10.1038/s41467-018-05096-6
Wesselhoeft, R. A., Kowalski, P. S., Parker-Hale, F. C., Huang, Y., Bisaria, N., and Anderson, D. G. (2019). RNA circularization diminishes immunogenicity and can extend translation duration in vivo. Mol. Cell 74, 508–520.e4. doi: 10.1016/j.molcel.2019.02.015
Wilusz, J. E. (2017). Circular RNAs: unexpected outputs of many protein-coding genes Circular RNAs: unexpected outputs of many protein-coding genes. RNA Biol. 14, 1007–1017. doi: 10.1080/15476286.2016.1227905
Wu, W., Ji, P., and Zhao, F. (2020). CircAtlas: an integrated resource of one million highly accurate circular RNAs from 1070 vertebrate transcriptomes. Genome Biol. 21:101. doi: 10.1186/s13059-020-02018-y
Xia, S., Feng, J., Chen, K., Ma, Y., Gong, J., Cai, F., et al. (2018). CSCD: a database for cancer-specific circular RNAs. Nucleic Acids Res. 46, D925–D929. doi: 10.1093/nar/gkx863
Xia, S., Feng, J., Lei, L., Hu, J., Xia, L., Wang, J., et al. (2017). Comprehensive characterization of tissue-specific circular RNAs in the human and mouse genomes. Brief. Bioinform. 18, 984–992. doi: 10.1093/bib/bbw081
Yang, F., Liu, D. Y., Guo, J. T., Ge, N., Zhu, P., Liu, X., et al. (2017). Circular RNA circ-LDLRAD3 as a biomarker in diagnosis of pancreatic cancer. World J. Gastroenterol. 23, 8345–8354. doi: 10.3748/wjg.v23.i47.8345
Yang, L., Yu, Y., Yu, X., Zhou, J., Zhang, Z., Ying, S., et al. (2019). Downregulated expression of hsa-circ-0005556 in gastric cancer and its clinical significance. Dis. Markers 2019:2624586. doi: 10.1155/2019/2624586
Yang, Y., Fan, X., Mao, M., Song, X., Wu, P., Zhang, Y., et al. (2017). Extensive translation of circular RNAs driven by N 6 -methyladenosine. Cell Res. 27, 626–641.
Yang, Y. Y., Fan, X., Mao, M., Song, X., Wu, P., Zhang, Y., et al. (2017). Extensive translation of circular RNAs driven by N 6 -methyladenosine. Cell Res. 27, 626–641. doi: 10.1038/cr.2017.31
Yao, G., Niu, W., Zhu, X., He, M., Kong, L., Chen, S., et al. (2019). Hsa-circRNA-104597: a novel potential diagnostic and therapeutic biomarker for schizophrenia. Biomark. Med. 13, 331–340. doi: 10.2217/bmm-2018-0447
You, X., Vlatkovic, I., Babic, A., Will, T., Epstein, I., Tushev, G., et al. (2015). Neural circular RNAs are derived from synaptic genes and regulated by development and plasticity. Nat. Neurosci. 18, 603–610. doi: 10.1038/nn.3975
Yu, M. (2020). CircRNA ZNF609 Knockdown suppresses cell growth via modulating miR-188 / ELF2 Axis in nasopharyngeal carcinoma. Onco Targets Ther. 13, 2399–2409.
Yu, X., Ding, H., Yang, L., Yu, Y., Zhou, J., Yan, Z., et al. (2019). Reduced expression of circRNA hsa_circ_0067582 in human gastric cancer and its potential diagnostic values. J. Clin. Lab. Anal. 34:e23080. doi: 10.1002/jcla.23080
Yun, C. W., and Lee, S. H. (2018). The roles of autophagy in cancer. Int. J. Mol. Sci. 19:3466. doi: 10.3390/ijms19113466
Zaphiropoulos, P. G. (1997). Exon skipping and circular RNA formation in transcripts of the human cytochrome P-450 2C18 gene in epidermis and of the rat androgen binding protein gene in testis. Mol. Cell. Biol. 17, 2985–2993. doi: 10.1128/mcb.17.6.2985
Zeng, Y., Du, W. W., Wu, Y., Yang, Z., Awan, F. M., Li, X., et al. (2017). A circular RNA binds to and activates AKT phosphorylation and nuclear localization reducing apoptosis and enhancing cardiac repair. Theranostics 7, 3842–3855. doi: 10.7150/thno.19764
Zhang, M., Huang, N., Yang, X., Luo, J., Yan, S., Xiao, F., et al. (2018). A novel protein encoded by the circular form of the SHPRH gene suppresses glioma tumorigenesis. Oncogene 37, 1805–1814. doi: 10.1038/s41388-017-0019-9
Zhang, X., Xu, Y., Yamaguchi, K., Hu, J., Zhang, L., Wang, J., et al. (2020). Circular RNA circVAPA knockdown suppresses colorectal cancer cell growth process by regulating miR – 125a / CREB5 axis. Cancer Cell Int. 20:103. doi: 10.1186/s12935-020-01178-y
Zhang, Y., Chen, Y., Yao, H., Lie, Z., Chen, G., Tan, H., et al. (2019). Elevated serum circ_0068481 levels as a potential diagnostic and prognostic indicator in idiopathic pulmonary arterial hypertension. Pulm. Circ. 9, 1–9. doi: 10.1177/2045894019888416
Zhang, Y., Xue, W., Li, X., Zhang, J., Chen, S., Zhang, J. L., et al. (2016). The biogenesis of nascent circular RNAs. Cell Rep. 15, 611–624. doi: 10.1016/j.celrep.2016.03.058
Zhang, Y., Zhang, X., Chen, T., Xiang, J., Yin, Q., Xing, Y., et al. (2013). Circular intronic long noncoding RNAs. Mol. Cell 51, 792–806. doi: 10.1016/j.molcel.2013.08.017
Zhao, W., Qi, X., Liu, L., Liu, Z., Liu, J., and Wu, J. (2019). Epigenetic regulation of m6A modifications in human cancer. Mol. Ther. Nucleic Acid 19, 405–412. doi: 10.1016/j.omtn.2019.11.022
Zhao, Z., Li, X., Gao, C., Jian, D., Hao, P., Rao, L., et al. (2017). Peripheral blood circular RNA hsa-circ-0124644 can be used as a diagnostic biomarker of coronary artery disease. Sci. Rep. 7:39918. doi: 10.1038/srep39918
Zheng, Q., Bao, C., Guo, W., Li, S., Chen, J., Chen, B., et al. (2016). Circular RNA profiling reveals an abundant circHIPK3 that regulates cell growth by sponging multiple miRNAs. Nat. Commun. 7:11215. doi: 10.1038/ncomms11215
Zhou, K. I., Shi, H., Lyu, R., Wylder, A. C., Matuszek, Ż, Pan, J. N., et al. (2019). Regulation of Co-transcriptional Pre-mRNA Splicing by m6A through the Low-Complexity Protein hnRNPG. Mol. Cell 76, 70–81.e9.
Zhou, L.-H., Yang, Y.-C., Zhang, R.-Y., Wang, P., Pang, M.-H., and Liang, L.-Q. (2018). CircRNA_0023642 promotes migration and invasion of gastric cancer cells by regulating EMT. Eur. Rev. Med. Pharmacol. Sci. 22, 2297–2303. doi: 10.26355/eurrev_201804_14818
Keywords: circRNA, biomarker, biological function, molecular therapy, protein-coding, immunity response, origin of life
Citation: Yesharim L, Mojbafan M and Abiri M (2021) Hints From the Cellular Functions to the Practical Outlook of Circular RNAs. Front. Genet. 12:679446. doi: 10.3389/fgene.2021.679446
Received: 11 March 2021; Accepted: 25 May 2021;
Published: 17 June 2021.
Edited by:
Kathleen Boris-Lawrie, University of Minnesota Twin Cities, United StatesReviewed by:
Alper Yilmaz, Yıldız Technical University, TurkeyMalgorzata Kloc, Houston Methodist Research Institute, United States
Gatikrushna Singh, University of Minnesota Twin Cities, United States
Copyright © 2021 Yesharim, Mojbafan and Abiri. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Maryam Abiri, YWJpcmkubUBpdW1zLmFjLmly
 Liora Yesharim
Liora Yesharim Marzieh Mojbafan
Marzieh Mojbafan Maryam Abiri
Maryam Abiri