
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Genet. , 24 May 2021
Sec. Evolutionary and Population Genetics
Volume 12 - 2021 | https://doi.org/10.3389/fgene.2021.676917
This article is part of the Research Topic Role of Y Chromosome in Molecular Anthropology, Forensics, and Genetic Genealogy View all 5 articles
 Chunfang Luo1,2†
Chunfang Luo1,2† Lizhong Duan3†
Lizhong Duan3† Yanning Li1,4†
Yanning Li1,4† Qiqian Xie1†
Qiqian Xie1† Lingxiang Wang5
Lingxiang Wang5 Kai Ru5
Kai Ru5 Shahid Nazir6
Shahid Nazir6 Muhammad Jawad6
Muhammad Jawad6 Yifeng Zhao7
Yifeng Zhao7 Fenfen Wang8
Fenfen Wang8 Zhengming Du8
Zhengming Du8 Dehua Peng2
Dehua Peng2 Shao-Qing Wen5*‡
Shao-Qing Wen5*‡ Pingming Qiu1*‡
Pingming Qiu1*‡ Haoliang Fan1,5,9*‡
Haoliang Fan1,5,9*‡Guangdong province is situated in the south of China with a population size of 113.46 million. Hakka is officially recognized as a branch of Han Chinese, and She is the official minority group in mainland China. There are approximately 25 million Hakka people who mainly live in the East and North regions of China, while there are only 0.7 million She people. The genetic characterization and forensic parameters of these two groups are poorly defined (She) or still need to be explored (Hakka). In this study, we have genotyped 475 unrelated Guangdong males (260 Hakka and 215 She) with Promega PowerPlex® Y23 System. A total of 176 and 155 different alleles were observed across all 23 Y-STRs for Guangdong Hakka (with a range of allele frequencies from 0.0038 to 0.7423) and Guangdong She (0.0047–0.8605), respectively. The gene diversity ranged from 0.4877 to 0.9671 (Guangdong Hakka) and 0.3277–0.9526 (Guangdong She), while the haplotype diversities were 0.9994 and 0.9939 for Guangdong Hakka and Guangdong She, with discrimination capacity values of 0.8885 and 0.5674, respectively. With reference to geographical and linguistic scales, the phylogenetic analyses showed us that Guangdong Hakka has a close relationship with Southern Han, and the genetic pool of Guangdong Hakka was influenced by surrounding Han populations. The predominant haplogroups of the Guangdong She group were O2-M122 and O2a2a1a2-M7, while Guangdong She clustered with other Tibeto-Burman language-speaking populations (Guizhou Tujia and Hunan Tujia), which shows us that the Guangdong She group is one of the branches of Tibeto-Burman populations and the Huonie dialect of She languages may be a branch of Tibeto-Burman language families.
Hakka is one of the far-reaching ethnic groups that have a worldwide distribution and is officially recognized as a branch of Han Chinese in China. Hakka is a unique group that is not named after the region (Zhong, 2019). There are about 80 million Hakka people, and ∼50 million are situated in southern parts of China (mainly including Guangdong, Jiangxi, Fujian, Guangxi, Sichuan, Hainan, Hunan, Zhejiang, Taiwan, Hongkong, and Macao). Guangdong province is an important region for Lingnan culture, which has its unique styles of language, history, and culture and which lies in the southernmost part of mainland China. The Hakka population is mainly settled in the eastern and northern regions of Guangdong, comprising Meizhou, Heyuan, Huizhou, Shaoguan, and Qingyuan (Figure 1). The origin of Hakka has not been clearly defined yet. At present, there are two views about the origin of the Hakka population, either they belong to Northern Han (Luo, 1989) or they belong to Southern Han (Fang, 2007). According to a previous study (Li et al., 2003), majority of the Fujian Hakka gene pool (80.2%) came from Northern Han based on 14 Y-SNPs. On the other hand, the frequency of a 9-bp deletion in mitochondrial region V is 21.74% in Meizhou Hakka, which indicated that Meizhou Hakka had close relationships with Fujian Hakka (∼0.197) and other populations from South China (Cai et al., 2015). From the perspective of physical anthropology, Zheng et al. found that the physical characteristics of the Hakka population are a mixture of South-Asian and North-Asian ethnic populations, which is determined by 86 anthropologic characteristics in 650 male and 704 female Chinese Hakka adults living in Guangdong and Jiangxi (Zheng et al., 2013). Guangdong province is home to the Hakka population, but there is no comprehensive study available on the Y-chromosomal prospect of Guangdong Hakka.
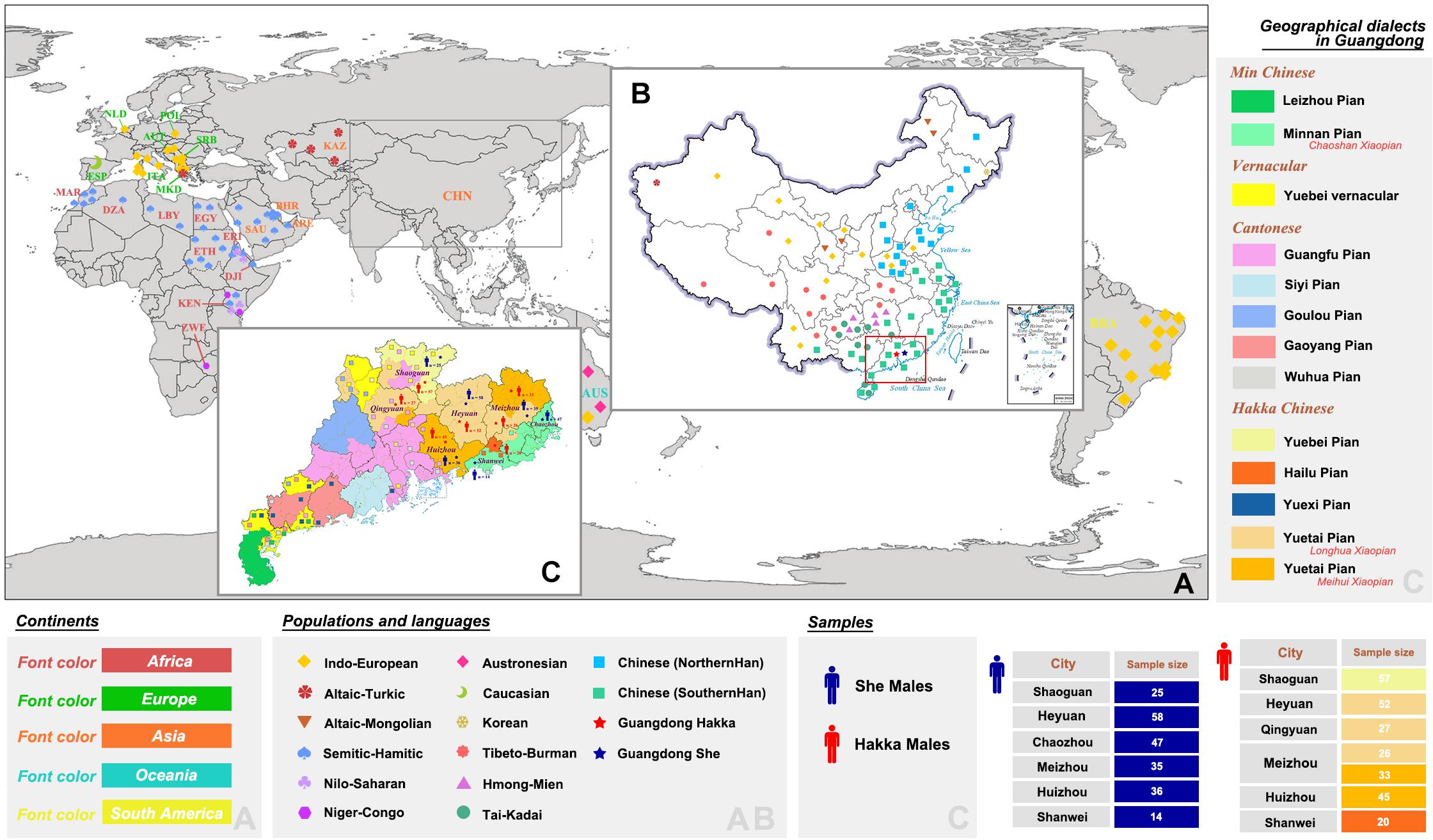
Figure 1. Locations of population distributions and sampling information. (A,B) Geographic positions of Guangdong Hakka, Guangdong She, and 159 worldwide populations with nine language families (48,637 individuals in total). (C) Distributions of geographical dialects in Guangdong province and detailed sampling information of Guangdong Hakka and Guangdong She groups.
She is one of the largest ethnic minorities in Jiangxi, Fujian, and Zhejiang provinces. Their presence is also reported in the provinces of Anhui and Guangdong. She people are shifting cultivation group in South China and had migrated from their primitive habitations in Fenghuang Mountain in Guangdong Chaozhou to Fujian, Zhejiang, Jiangxi, Anhui, and other provinces for more than a thousand years (Shi, 2013; Liu et al., 2017). The total population of the She group is 709,592 according to the 2010 census. Their presence in South Zhejiang is 25% and 53% in East Fujian. Their presence in Guangdong province is only 3.8% (27,000), although Guangdong Chaozhou Fenghuang Mountain is believed to be the definite headstream of She group’s culture and civilization (Liu et al., 2017). She people speak the She language with different dialects such as Shanke, Dongjia, and Huonie. She language is a branch of Sino-Tibetan language families and is the primary language for the She group (Lei, 2005). With reference to linguistics, the She language (mainly refers to Shanke and Dongjia dialects) belongs to the Hmong-Mien language branch (Lu and Wang, 2019). The Guangdong She population uses only the Huonie dialect. Interestingly, the divergences in languages may represent the differences in the genetic affinities of distinct She groups. Fujian She is more closely related to the Dongbei (Northern Han) and Hunan (Southern Han) populations (Liu et al., 2017). The genetic heterogeneities for the She population in the different regions indicate that the origins of Guangdong She and Fujian She may not be the same; Guangdong She may also not be a Hmong-Mien language-speaking group.
Y-chromosomal variant analysis for determining the patterns of present and past flow of genes between populations is very helpful (Oppenheimer, 2012). Y-chromosome short tandem repeats (Y-STRs) play an important role in forensic molecular biology (Prinz et al., 1997; Adnan et al., 2016). The use of Y-STRs also allows the simultaneous analysis of closely related and distantly related populations (Ballantyne and Kayser, 2012). Consequently, to address three main issues—(1) the forensic efficiencies of Promega PowerPlex® Y23 System (Promega Corporation, Fitchburg, WI, United States) in Guangdong Hakka and Guangdong She groups, (2) the population structures of the two groups, and (3) the language classification of Guangdong She (Huonie Dialect)—we used the Promega 5-dye multiplex system to genotype 23 Y-STRs in 260 Guangdong Hakka and 215 Guangdong She males and evaluated the system effectiveness of forensic applications in two groups. Furthermore, we conducted population genetics employing diversified analyses among globally dispersed human populations and regional closely related populations to make the issues clearer.
In this study, a total of 475 unrelated male individuals (260 Hakka and 215 She) were recruited from Guangdong province of China (Figure 1C). Blood samples of all volunteers were collected using the FTA cards (WhatmanTM, GE Healthcare, Chicago, IL, United States), with their written informed consent. This study and the procedures were approved by the Institutional Review Boards of Hainan Medical University and the Medical Ethics Committee of Hainan Medical University (no. HYLL-2020-012). All the experimental procedures were performed following the standards of the Declaration of Helsinki.
One punched bloodstain paper (the size was 1.2 mm × 1.2 mm) was used as a PCR template directly for each sample without DNA extraction procedures. Y-STRs were co-amplified using Promega PowerPlex® Y23 System (Promega Corporation, Fitchburg, WI, United States), which is a five-dye multiplex kit that analyzes the 17 Yfiler Y-STRs (DYS19, DYS385a/b, DYS389I, DYS389II, DYS390, DYS391, DYS392, DYS393, DYS437, DYS438, DYS439, DYS448, DYS456, DYS458, DYS635, and Y_GATA_H4) together with six discriminating Y-STRs (DYS481, DYS533, DYS549, DYS570, DYS576, and DYS643) on a Veriti® 96 Well Thermal Cycler System (Thermo Fisher Scientific, Waltham, MA, United States) following the manufacturer’s instructions. The amplified products were separated by capillary electrophoresis on a 3500XL Genetic Analyzer (Thermo Fisher Scientific, Waltham, MA, United States), and data were analyzed using GeneMapper® ID-X Software v1.6 (Thermo Fisher Scientific, Waltham, MA, United States).
Haplotype and allele frequencies were calculated using Arlequin v3.5 (Excoffier and Lischer, 2010). Gene diversity (GD) was calculated as per the following formula:
where, n is the number of alleles at each Y-STR locus, and Pi is the frequency of the i-th allele. Haplotype diversity (HD) was estimated similarly as GD, while n and Pi were the total number of haplotypes and the frequency of the i-th haplotype, respectively. Discrimination capacity (DC) was determined as the ratio between the number of different haplotypes and the sample size. Match probability (MP) was defined as MP = , where Pi was the frequency of the i-th haplotype.
Genetic relationships between these populations and other reference populations were calculated employing RST (Excoffier et al., 1992; Excoffier and Smouse, 1994). Population pairwise genetic distances (RST) and corresponding p values between different populations were estimated by analysis of molecular variance (AMOVA) by the online tool available at YHRD1. The principal component analysis (PCA) and multidimensional scaling plot (MDS) were conducted based on allele frequencies using the R programming language. Additionally, phylogenetic relationships among different populations were drawn in Molecular Evolutionary Genetics Analysis 7.0 software (Kumar et al., 2016) by neighbor-joining phylogenetic tree (Saitou and Nei, 1987) based upon genetic distance matrices (RST matrices) and visualized by the Interactive Tree of Life v5 (Letunic and Bork, 2019).
The Y-STR typing experiments were performed strictly following the recommendations of the Chinese National Standards, Scientific Working Group on DNA Analysis Methods (SWGDAM, 2010) and the recommendations of the DNA Commission of the International Society of Forensic Genetics (Gusmao et al., 2006; Carracedo et al., 2013; Roewer et al., 2020). Control DNA 9948 and sdH2O were employed as positive and negative controls in each batch of PCR amplification and electrophoresis, respectively. In addition, our laboratory has passed the proficiency testing for Y-STR typing which is organized by YHRD and has been accredited under ISO/IEC 17025:2005 and China National Accreditation Service for Conformity Assessment.
The haplotype data of 475 unrelated male individuals from Guangdong Hakka and Guangdong She populations in the present study have been submitted to the YHRD database and received the accession number YA004707 (Guangdong Hakka, n = 260) and YA004708 (Guangdong She, n = 215). The Y-STR profiles, with variants (null alleles, off-ladder alleles, or copy number variants) for all samples, were re-amplified and re-genotyped by AmpFLSTR® Yfiler® PCR Amplification Kit (Thermo Fisher Scientific, Waltham, MA, United States) and Geno-ID Y41 Human Typing (Guangzhou Koalson Intelligent BioRobotics, Guangzhou, Guangdong, China) for confirmation.
The allele frequencies, allele numbers, and GD values of 23 Y-STR loci in the Guangdong Hakka and She groups are presented in Supplementary Tables 1, 2, respectively.
In Guangdong Hakka, 176 distinct alleles were observed at all loci, and the corresponding allelic frequencies ranged from 0.0038 to 0.7423 (DYS438). The number of different alleles varied from three (DYS437) to 40 (DYS385a/b). The GD values of six Y-STRs were greater than 0.9, and the highest (0.9671) and lowest (0.4877) estimates of GD corresponded to loci DYS385a/b and DYS438, respectively. We identified three off-ladders at DYS576 (19.1) and DYS458 (16.1) and the microvariant 19.1 at DYS576 which happened twice. All loci having intermediate values found in the Guangdong Hakka were re-amplified and re-genotyped for confirmation and were commonly observed in YHRD.
In the Guangdong She group, a total of 155 different alleles were obtained, and the number of diverse alleles ranged from four (at seven Y-STRs) to 36 (at DYS385a/b). The allele frequencies varied from 0.0047 to 0.8605 (DYS438). On one hand, the set of 23 Y-STR loci had a high level of genetic polymorphisms in the She group, DYS458 (0.9526), DYS385a/b (0.9180), DYS481 (0.9146), and DYS448 (0.9060), while the DYS438 (0.3277) and DYS391 (0.4875) showed extreme values of the GD distribution (with GD values < 0.5). In this study, no null allele, off-ladder, and copy number variant were found in the Guangdong She group.
The haplotypes and haplotype frequencies of 23 Y-STRs in the two groups are shown in Supplementary Tables 3, 4, respectively. A total of 231 different haplotypes were found in 260 Guangdong Hakka male individuals, of which 208 (90.04%) were unique. It was observed that 18 haplotypes (H006–H023) occurred twice, four haplotypes (H002–H005) occurred thrice, while only one haplotype (H001) was observed as being shared by four individuals (Supplementary Table 3). Genotyping with the 23 Y-STRs determined 122 distinct haplotypes in the Guangdong She group, and the fraction of unique haplotypes was only 53.28%. Additionally, 43 haplotypes (S015–S057) were detected twice, six haplotypes (S009–S014) and five haplotypes (S004–S008) were found three and four times, respectively, and the haplotypes S003, S002, S001 appeared five, six, and 15 times, respectively (Supplementary Table 4).
As shown in Table 1, the overall HD values of the Guangdong Hakka and She groups were 0.9994 and 0.9939, respectively. Moreover, it can be seen from Table 1 that the DC value of the Guangdong She group (0.5674) was much lower than those of the Guangdong Hakka group (0.8885) and Guangdong Han group (0.9706), which demonstrated that the discrimination power of Promega PowerPlex® Y23 System was not seemingly appropriate for the Guangdong She group. With the Guangdong She group being relatively isolated, there was limited gene flow, which led to the decrease in the discrimination capacity of the Y-STR system. In this regard, there is a dire need to get more knowledge about the mutability of currently known Y-STRs and to incorporate the rapidly mutating Y-STRs into forensic systems to enhance the capacity of discrimination, especially for isolated groups.
Dimensionality reduction analyses including PCA, MDS, linear discriminant analysis, Laplacian Eigenmaps, and locally linear embedding can accelerate the speed of algorithm execution, improve the performance of the analysis model, and reduce the complexity of data at the same time. To illustrate the genetic landscapes of globally dispersed human populations, especially for Guangdong Hakka and She groups, the dimensionality reduction analyses (PCA and MDS) were conducted based on the frequencies of 23 Y-STRs.
The PCA provides a method of visualizing the essential patterns of genetic relationships and allows us to identify and plot the major patterns within a multivariate dataset to indicate that the populations with closer geographical distances have more intimate relationships. We collected the 23 Y-STR frequency profiles of 48,162 individuals from 159 worldwide populations having nine language families within five continents (Supplementary Table 5) to conduct PCA with Guangdong Hakka and She groups (Yaju et al., 2014; Westen et al., 2015; Bai et al., 2016b; Garcia et al., 2016; Jung et al., 2016; Pickrahn et al., 2016; Wang L. et al., 2016; Wang Y. et al., 2016; Guo et al., 2017; Iacovacci et al., 2017; Jun et al., 2017; Lacerenza et al., 2017; Qiang et al., 2017; Spolnicka et al., 2017; Wang M. et al., 2017; Zgonjanin et al., 2017; Zhang et al., 2017; Cao et al., 2018; Fan et al., 2018a,b,c; Khubrani et al., 2018; Liu Y. et al., 2018; Juan et al., 2018; Liu Y. J. et al., 2018; Wang et al., 2018, 2020; Zhou et al., 2018; D’Atanasio et al., 2019; Fan et al., 2019; He et al., 2019; Henry et al., 2019; Ip et al., 2019; Jankova et al., 2019; Kai et al., 2019; Lang et al., 2019; Liu et al., 2019, 2020; Luo et al., 2019; Santos Stange et al., 2019; Tao et al., 2019a,b; Wang C. Z. et al., 2019; Wang X. et al., 2019; Wang Y. et al., 2019; Wu et al., 2019; Xie et al., 2019; Yaju et al., 2019, 2020; Zhabagin et al., 2019; Zhang et al., 2019; Al-Snan et al., 2020; Dezhi et al., 2020; Fan et al., 2020; Feng et al., 2020; Haiying et al., 2020; Hanguang et al., 2020; Jannuzzi et al., 2020; Kang et al., 2020; Shonhai et al., 2020; Song et al., 2020; Tang et al., 2020; Yin et al., 2020; Zeyad et al., 2020). As shown in Figure 2, the first and second components (PC1 and PC2) accounted for 6.85 and 5.69% of the total variances observed within these populations, respectively. From the perspective of geography, Guangdong Hakka was located in the cluster of south Chinese populations (Figure 2A). Additionally, Guangdong Hakka was observed close to the Southern Han populations (Figure 2B) from the linguistic point of view, while the Guangdong She group was situated in a relatively isolated location with no definite relationship with other Chinese populations from both geographic and linguistic scales (Figure 2).
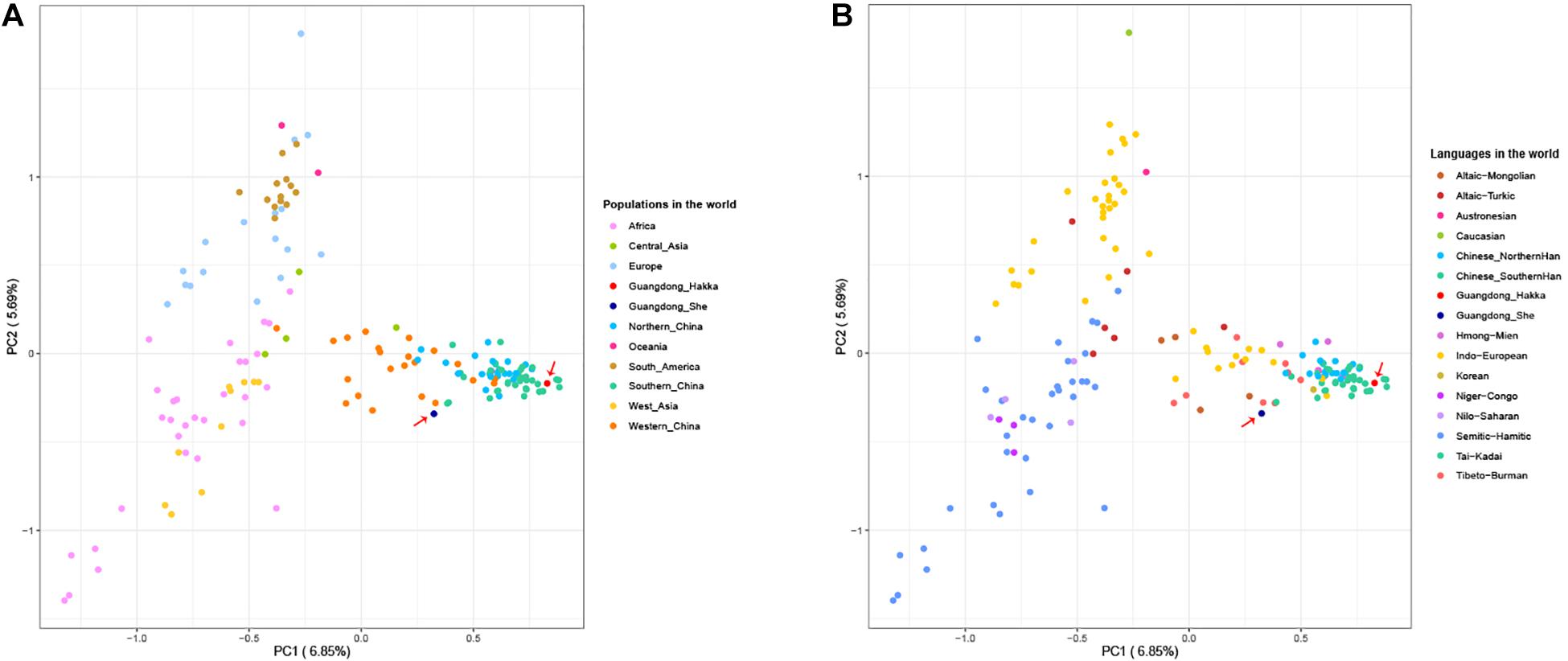
Figure 2. Principal component analysis (PCA) based on the frequencies of 23 Y-STRs among Guangdong Hakka, Guangdong She, and 159 worldwide human populations. (A) PCA from geographical scale. (B) PCA from linguistic scale.
To further clarify the genetic relationships on a relatively small scale, we performed PCA within Chinese populations (Figure 3). Populations from different regions of China clustered separately, and Guangdong Hakka and Southern Han clustered similarly on the left side. Guangdong She distributed on the middle bottom and clustered with south Chinese populations. However, from the perspective of linguistics, the language relationships between Guangdong She and the surrounding south Chinese populations which are illustrated in Figure 3B were still not distinct.
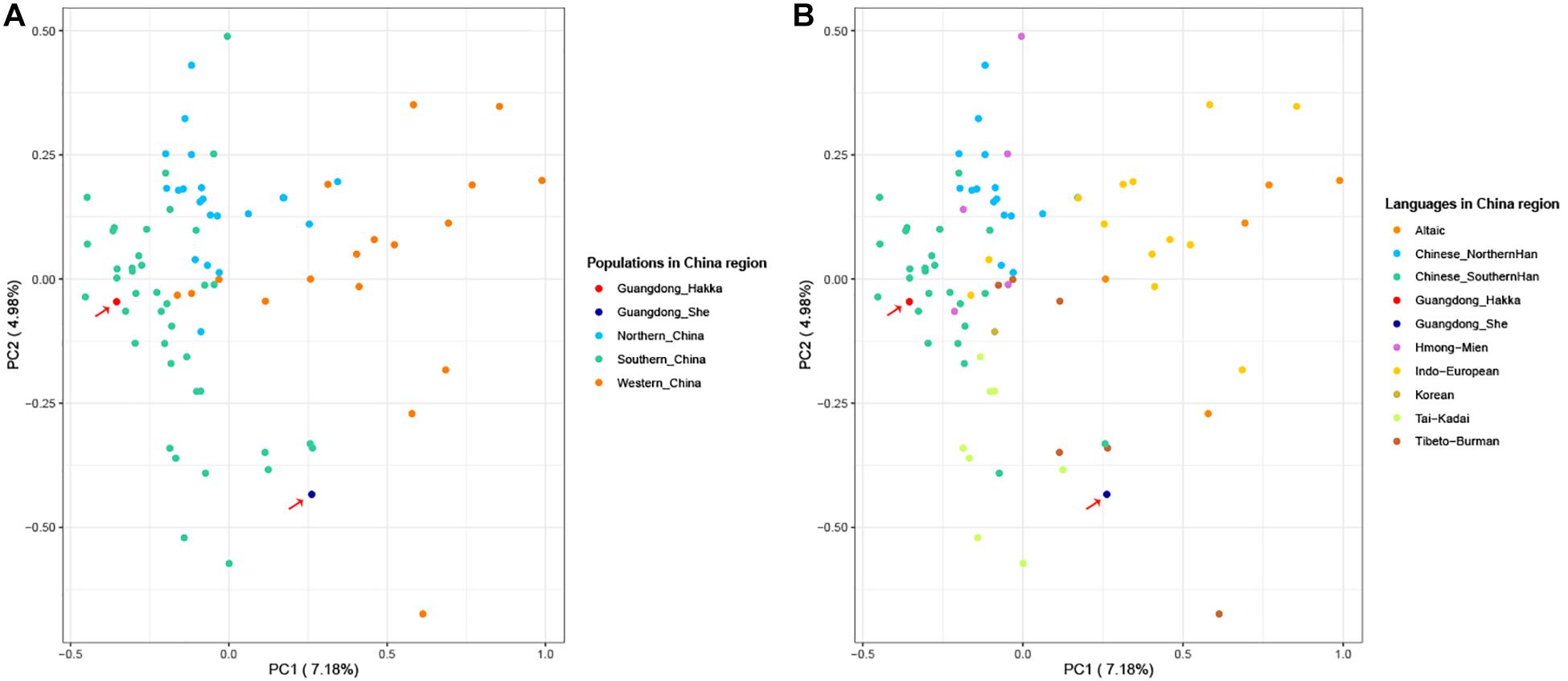
Figure 3. Principal component analysis (PCA) based on the 23 Y-STRs frequency profiles among Guangdong Hakka, Guangdong She, and 83 Chinese populations. (A) PCA from geographical scale. (B) PCA from linguistic scale.
MDS plots were conducted based on allele frequencies by Euclidean distance and Manhattan distance, respectively. As shown in Figure 4, each population was represented by a small dot with a different color in the multidimensional space, and the distances between the small dots showed the genetic relationships among the populations in distinct geographic areas or with different language families.
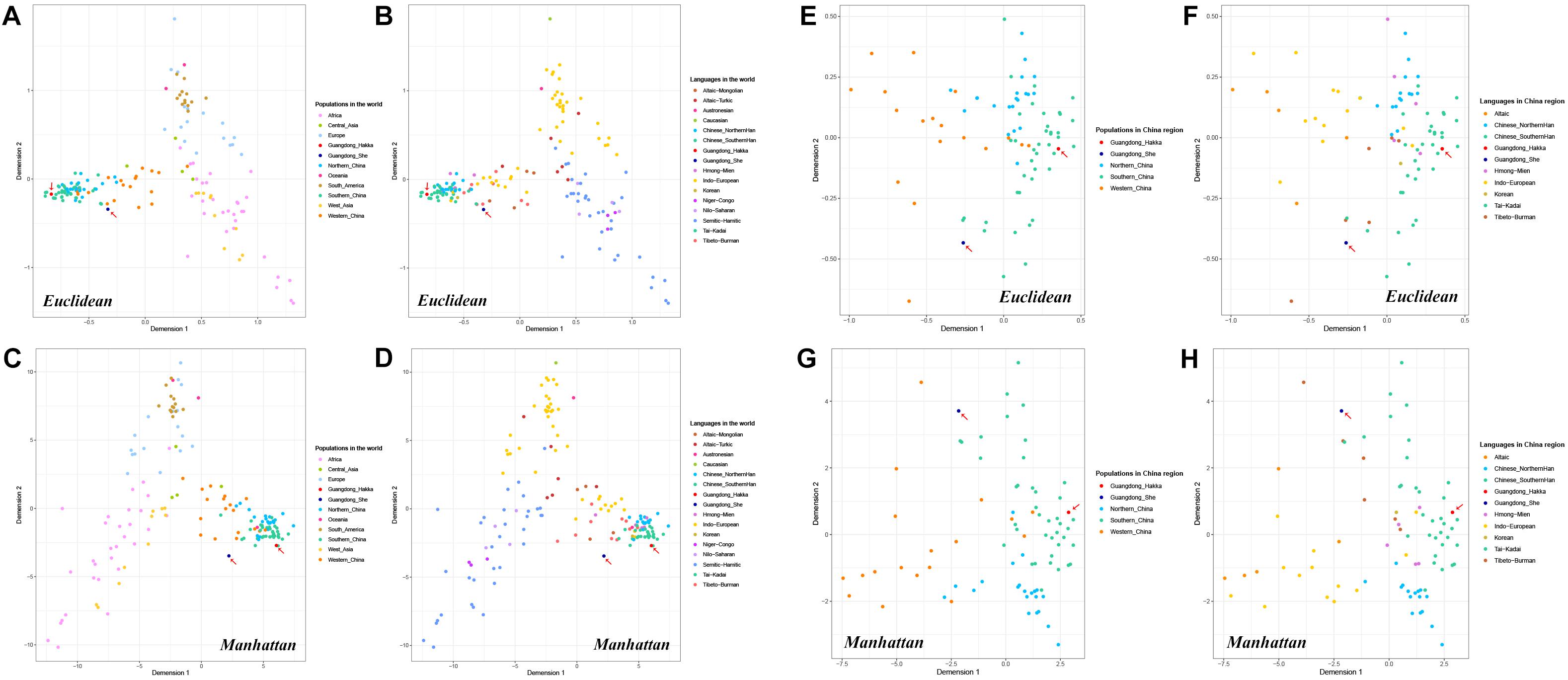
Figure 4. Multidimensional scaling (MDS) plots based on the frequencies of 23 Y-STRs among Guangdong Hakka, Guangdong She, and 159 worldwide human populations (including 83 Chinese populations). (A) Euclidean-based MDS plot in worldwide populations from geographical scale. (B) Euclidean-based MDS plot in worldwide populations from linguistic scale. (C) Manhattan-based MDS plot in worldwide populations from geographical scale. (D) Manhattan-based MDS plot in worldwide populations from linguistic scale. (E) Euclidean-based MDS plot in Chinese populations from geographical scale. (F) Euclidean-based MDS plot in Chinese populations from linguistic scale. (G) Manhattan-based MDS plot in Chinese populations from geographical scale. (H) Manhattan-based MDS plot in Chinese populations from the linguistic scale.
Whether from the perspective of geographical or linguistic scale, the results of MDS analysis by Euclidean distance (Figures 4A,B) had no apparent differences with the principal component analysis (Figures 2A,B). Moreover, in the Manhattan distance-based MDS plots (Figures 4C,D), Guangdong Hakka intertwining with the Southern Han cluster was in accordance with the performances in the MDS conducted by Euclidean distance (Figures 4A,B) and PCA plots (Figure 2). In addition, Guangdong She was located in a relatively isolated place.
Furthermore, to illustrate partial genetic relationships, we collected Guangdong Hakka, Guangdong She, and 83 other Chinese populations to perform MDS analyses with Euclidean distance and Manhattan distance (Figures 4E–H). The MDS results demonstrated that Guangdong Hakka had a close relationship with Southern Han populations, and Guangdong She was situated in a relatively isolated location from the perspectives of geography and linguistics, which were in line with the results of the PCA.
The results of the calculations of pairwise RST and the corresponding p values between Guangdong Hakka and 13 other Chinese Han populations from Southern and Northern Han regions (Kwak et al., 2005; Wu et al., 2011; Zhang et al., 2014; Shi et al., 2015; Shu et al., 2015; Bai et al., 2016b; Li et al., 2016; Wang L. et al., 2016; Wang Y. et al., 2016; Zhou et al., 2016, 2018; Nothnagel et al., 2017; Wang H. et al., 2017; Huang et al., 2019; Lang et al., 2019; Sun et al., 2019; Guan et al., 2020) based on 23 Y-STR haplotypes are listed in Supplementary Table 6. The population Guangdong Hakka has been found to be closely related to Guangdong Jieyang Han (RST = 0.0028) and Jiangxi Han (RST = 0.0029). Subsequently, the results of the phylogenetic analysis among Guangdong Hakka and Chinese Han populations are displayed in a phylogenetic tree based on the neighbor-joining method (Figure 5). Guangdong Hakka was first clustered with Guangdong Jieyang Han, followed by Jiangxi Han. Geographically, Jieyang is surrounded by Shanwei, Chaozhou, and Meizhou, whereas the habitations for Guangdong Hakka (Figure 1C) and Jiangxi bordering with Guangdong in the southwest were the dominating Hakka settlements. The phylogenetic tree showed the relationships between Guangdong Hakka and other Han populations in genetics, which were in accordance with the geographical relations. The paternal relationships revealed by Y-STRs indicate that Guangdong Hakka had close relationships with Southern Han, and there were extensive gene flows between Guangdong Hakka and the surrounding Han populations. Our results corroborated the findings of previous studies (Du et al., 2019; Han et al., 2020), which were conducted by using STRs and Y-STRs on the Guangdong Meizhou Hakka population.
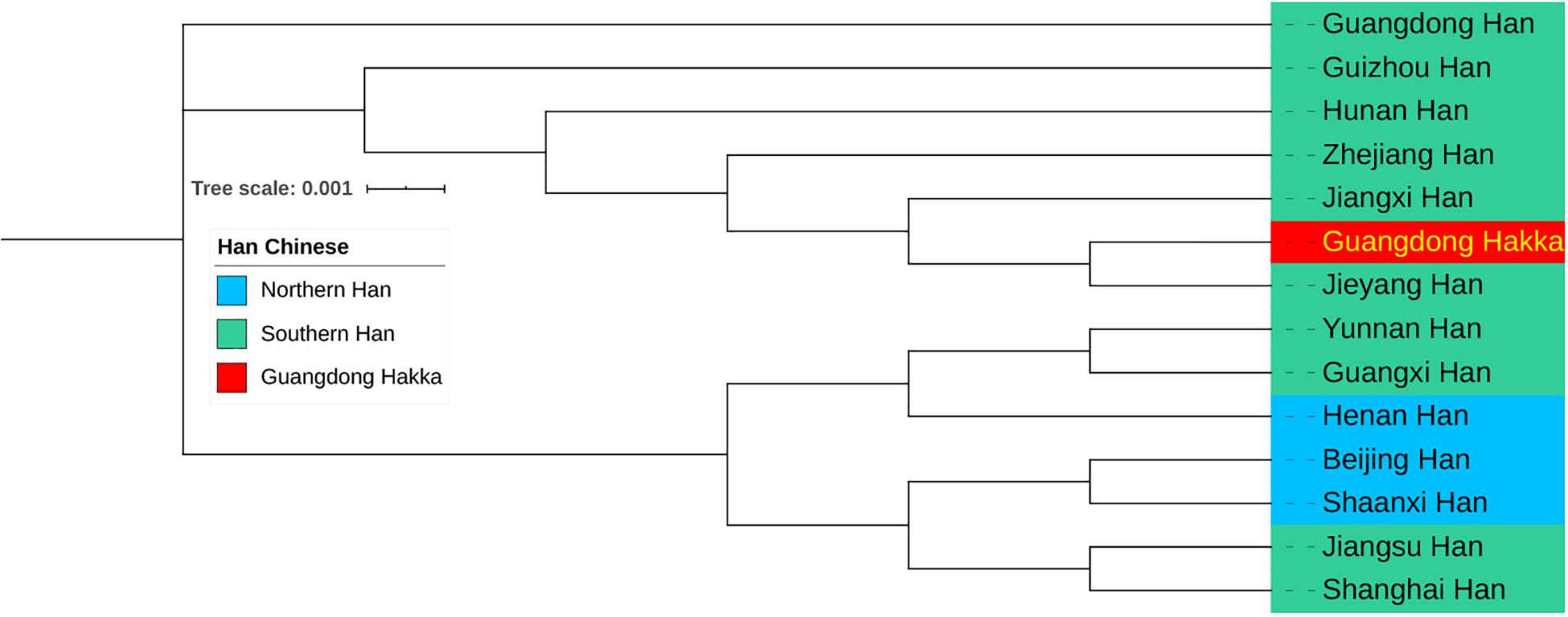
Figure 5. Neighbor-joining phylogenetic tree between Guangdong Hakka and 13 Han Chinese populations (including three Northern Han and 10 Southern Han) based on the matrix of pairwise RST values.
From the dimensionality reduction analyses discussed above, the genetic relationships between Guangdong She and other Chinese populations were not clarified. In addition, we employed the Y-STR haplotype profiles of 42 Chinese minorities and Guangdong She to assess the population pairwise genetic distances by AMOVA (Zhu et al., 2005, 2006a,b, 2008; He and Guo, 2013; Shan et al., 2014; Luo et al., 2015, 2019; Ou et al., 2015; Shu et al., 2015; Bai et al., 2016a; Bian et al., 2016; Fu et al., 2016; Gao et al., 2016; Yao et al., 2016; Guo et al., 2017; Ji et al., 2017; Zhao et al., 2017; Cao et al., 2018; Chen et al., 2018; Liu Y. J. et al., 2018; Atif et al., 2019; Liu et al., 2019; Xie et al., 2019; Dezhi et al., 2020; Feng et al., 2020; Song et al., 2020; Tang et al., 2020). The pairwise RST and the corresponding p values between Guangdong She and Chinese minority populations from five linguistically different families are displayed in Supplementary Table 7. No differences were observed between Guangdong She and Guizhou Tujia populations (RST = 0.0046, p = 0.0574), while significant genetic differences were observed between Guangdong She and all other Chinese minority populations (p < 0.05). The phylogenetic relationships between Guangdong She and 42 Chinese minorities were visualized in the neighbor-joining tree. As shown in Figure 6, the Tibeto-Burman language-speaking Tibetans and Altaic language-speaking Uighurs and Mongolians clustered together at the upper end, while the language relationships at the bottom, especially for Tai-Kadai, Hmong-Mien, and Tibeto-Burman, were ambiguous. The Guangdong She group clustered with two Tibeto-Burman populations, Guizhou Tujia and Hunan Tujia. However, the Fujian She group congregated with Guizhou Hmong-Mien language-speaking Miao populations.
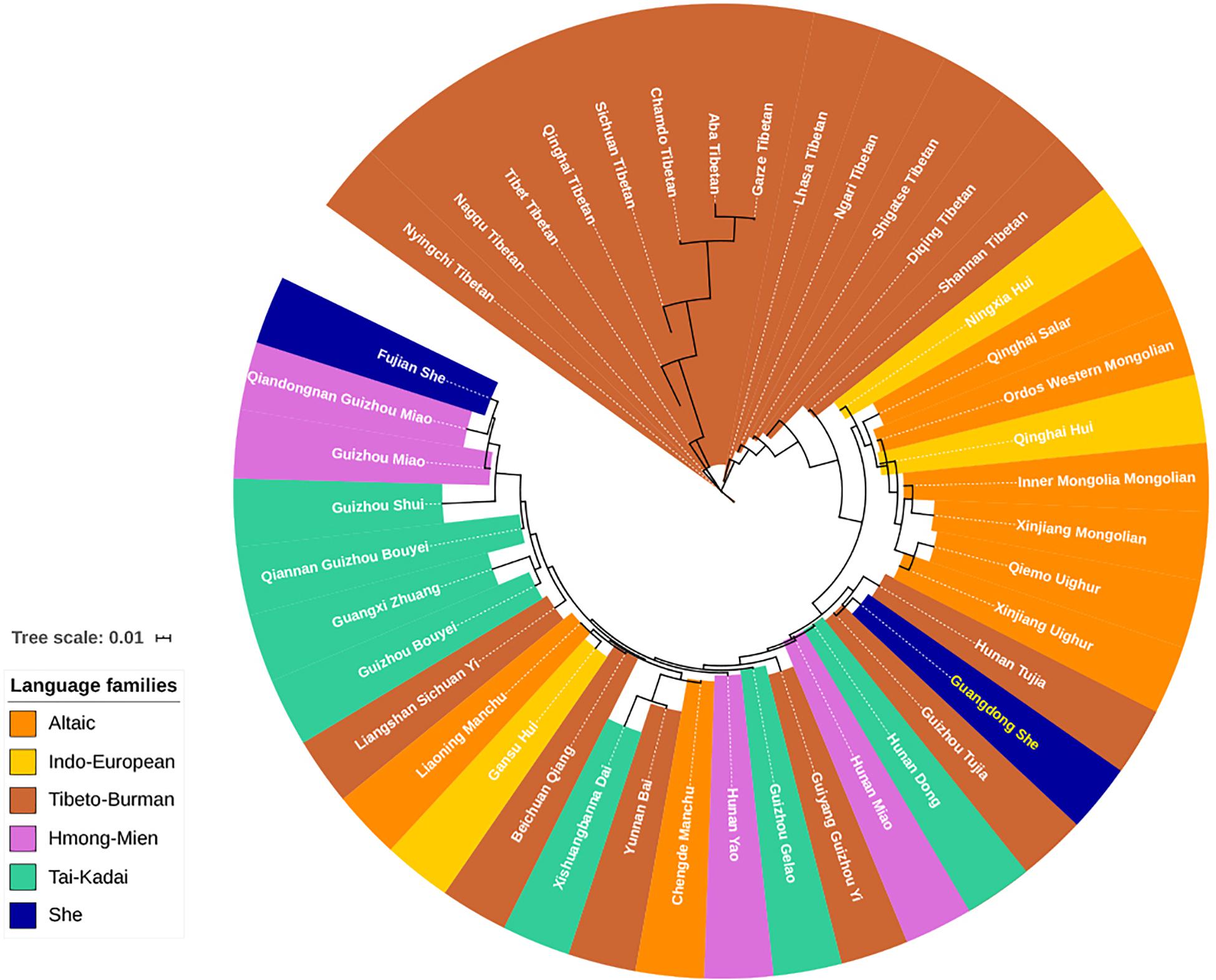
Figure 6. Neighbor-joining phylogenetic tree between Guangdong She and 42 other minorities with five different language families based on pairwise RST values.
The above-mentioned phylogenetic analysis hinted that Guangdong She clustered with Tibeto-Burman language-speaking Tujia populations in the same branch. To make further confirmation for the genetic relationships, we employed our in-house database which was composed of 37,754 pieces of Y-SNP/STR data and 109,142 Y-STR (Wang et al., 2015) mainly from East and Southeast Asia to make more precise predictions for 215 Guangdong She males in this study. Finally, 212 out of the 215 genotyped Y-STRs (98.60%) were observed, and a total of six Y-haplogroups were defined in Guangdong She which belong to major clades O2 and O1. The predominantly detected haplogroups were O2-M122 (45.75%), O2a2a1a2-M7 (25.47%), O1a-M119 (10.38%), O1b1a1a-M95 (8.02%), O-M175 (7.08%), and O2a2b1a1-M117 (3.30%), which were determined according to ISOGG, 20192. In addition, a PCA graph was performed among 77 populations (Wen et al., 2004; Gan et al., 2008; Li et al., 2008; Cai et al., 2011; Deng et al., 2013; Fan et al., 2018c; Rowold et al., 2019; Luo et al., 2020; Fan et al., 2021), which were composed of 4,195 individuals in total, including Tai-Kadai, Hmong-Mien, Tibeto-Burman, and Chinese (Southern and Northern Han) populations from Southeast and East Asia, which included three She groups (Fujian, Guangdong Chaoshan, and Guangdong She). From the diagram in Figure 7, the first and second principal components could explain 33.07% of the total variances. Moreover, the populations with different language branches were separated relatively, and the Guangdong She group had a close relationship with the Guangdong Chaoshan She group and clustered with Tibeto-Burman language-speaking populations (including Tujia, Tibetan, and Naxi minorities), while Fujian She located between Hmong-Mien and Han populations, especially Southern Han. The interpopulation comparison demonstrated that (1) different branches of She populations and Fujian and Guangdong She groups may have distinct origins from the perspectives of genetics and linguistics, especially from phylogenetic analyses based on Y-STRs and Y-SNPs, and (2) Guangdong She and Chaoshan She groups have close affinities with Tibeto-Burman language-speaking populations based on the evidence of Y-haplogroups which contained information about the subsequent colonization, differentiations, and migrations overlaid on recent population ranges.
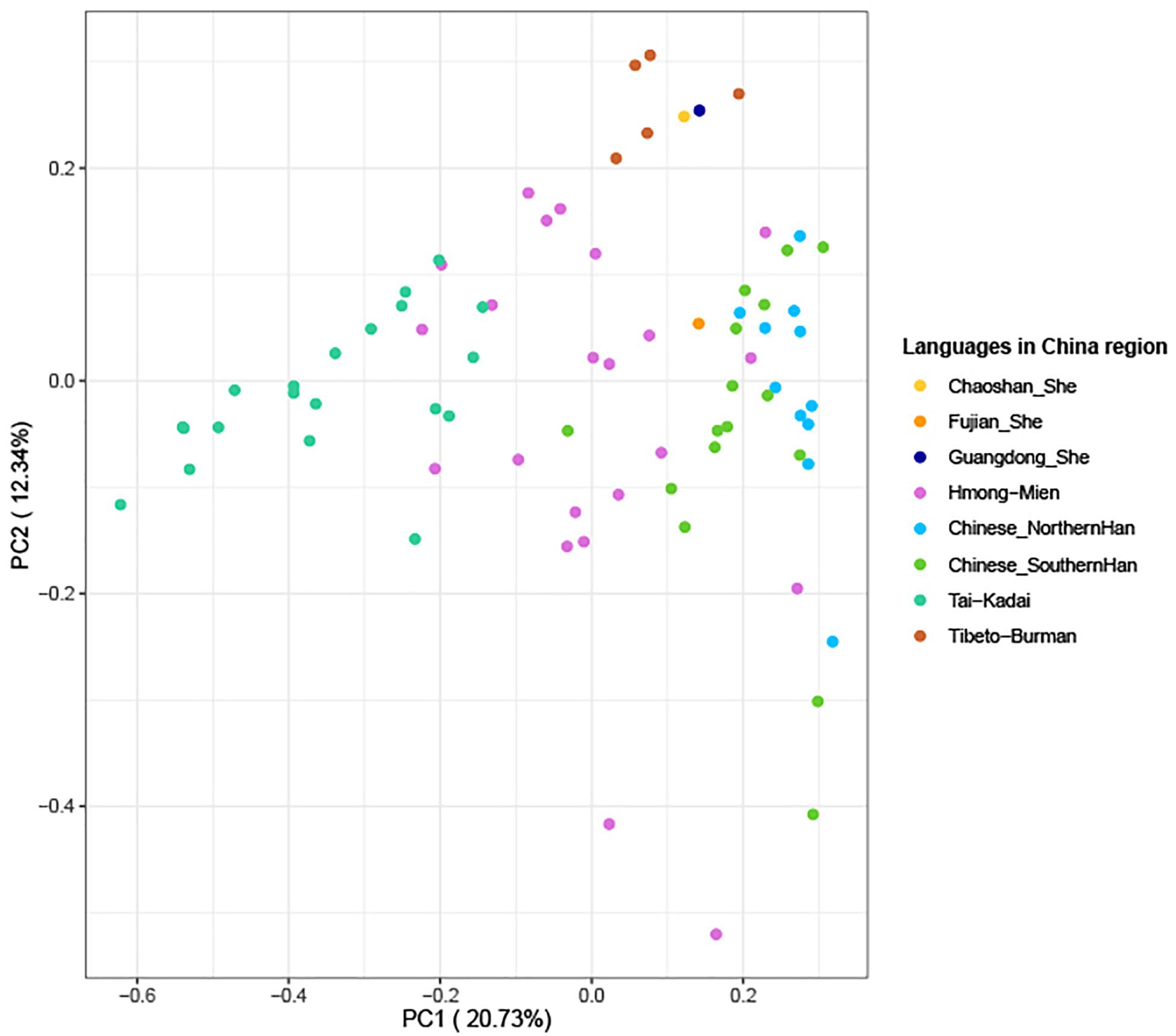
Figure 7. Principal component analysis based on Y-haplogroup frequencies between three She groups and 74 populations (4,195 individuals in total, which included 22 Hmong-Mien, 20 Tai-Kadai, 16 Southern Han, 11 Northern Han, and five Tibeto-Burman language-speaking populations mainly from Southeast and East Asia).
In the present study, a total of 475 unrelated Guangdong males (260 Hakka and 215 She) were genotyped by using 23 Y-STRs (including 17 Yfiler and 6 additional Y-STRs) by Promega PowerPlex® Y23 System. For Guangdong Hakka, a total of 176 different alleles were found, with corresponding allelic frequencies ranging from 0.0038 to 0.7423 and GD values that varied from 0.4877 to 0.9671, and the systematic effectiveness for Guangdong Hakka was observed to be sufficient (HD = 0.9994, DC = 0.8885). From the perspectives of geographical and linguistic scales, the phylogenetic analyses indicated that Guangdong Hakka had a close relationship with Southern Han, and there were extensive gene flows between Guangdong Hakka and the surrounding Han populations. For Guangdong She, we identified 155 distinct alleles with a range of allele frequencies from 0.0047 to 0.8605. The GD values for 23 Y-STRs ranged from 0.3277 to 0.9526, and the overall DC and HD were 0.5674 and 0.9939, respectively. The predominant haplogroups of the Guangdong She group were O2-M122 and O2a2a1a2-M7. Moreover, Guangdong She clustered with Tibeto-Burman language-speaking populations (Guizhou Tujia and Hunan Tujia), which demonstrates that the Guangdong She group seems to be one branch of Tibeto-Burman populations and the Huonie dialect of She languages may be a branch of Tibeto-Burman language families.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
The studies involving human participants were reviewed and approved by Medical Ethics Committee of Hainan Medical University. The patients/participants provided their written informed consent to participate in this study.
HF contributed to conceptualization, formal analysis, visualization, and writing—original draft preparation. CL and DP took charge of resources and contributed to project administration. HF, LW, and KR were in charge of software. LD, YL, QX, and YZ conducted the investigation. LD, YZ, FW, and ZD performed the validation. CL contributed to data curation. QX, SN, and MJ contributed to writing—review and editing. PQ and S-QW supervised the study. CL, HF, FW, and ZD took charge of funding acquisition. All authors reviewed the manuscript.
This study was supported by grants from the Program of Heyuan for Social Development and Technology Plans (No. 190620161503089), the Program of Hainan Association for Science and Technology Plans to Youth R&D Innovation (QCXM201705), and the National Undergraduate Innovation and Entrepreneurship Training Program (Nos. 201911810008 and 201911810023).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The handling editor AA declared a past collaboration with the author SN.
We would like to thank the donors who contributed samples for this study.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2021.676917/full#supplementary-material
Adnan, A., Ralf, A., Rakha, A., Kousouri, N., and Kayser, M. (2016). Improving empirical evidence on differentiating closely related men with RM Y-STRs: a comprehensive pedigree study from Pakistan. Forensic Sci. Int. Genet. 25, 45–51. doi: 10.1016/j.fsigen.2016.07.005
Al-Snan, N. R., Messaoudi, S. A., Khubrani, Y. M., Wetton, J. H., Jobling, M. A., and Bakhiet, M. (2020). Geographical structuring and low diversity of paternal lineages in Bahrain shown by analysis of 27 Y-STRs. Mol. Genet. Genom. 295, 1315–1324. doi: 10.1007/s00438-020-01696-4
Atif, A., Kasim, K., Rakha, A., Noor, A., Cheema, A. S., Hadi, S., et al. (2019). Population data of 23 Y STRs from Manchu population of Liaoning Province, Northeast China. Int. J. Legal. Med. 133, 785–788. doi: 10.1007/s00414-018-1957-7
Bai, R., Liu, Y., Lv, X., Shi, M., and Ma, S. (2016a). Genetic polymorphisms of 17 Y chromosomal STRs in She and Manchu ethnic populations from China. Forensic Sci. Int. Genet. 22, e12–e14. doi: 10.1016/j.fsigen.2016.01.011
Bai, R., Liu, Y., Zhang, J., Shi, M., Dong, H., Ma, S., et al. (2016b). Analysis of 27 Y-chromosomal STR haplotypes in a Han population of Henan province, Central China. Int. J. Legal. Med. 130, 1191–1194. doi: 10.1007/s00414-016-1326-3
Ballantyne, K. N., and Kayser, M. (2012). Additional Y-STRs in forensics: Why, Which, and When. Forens. Sci. Rev. 24, 63–78.
Bian, Y., Zhang, S., Zhou, W., Zhao, Q., Siqintuya, Z. R., and Li, C. (2016). Analysis of genetic admixture in Uyghur using the 26 Y-STR loci system. Sci. Rep. 6:19998. doi: 10.1038/srep19998
Cai, G. Q., Zhu, W. F., Wu, X. Y., Su, N., Yi, H., Chen, L. X., et al. (2015). Mitochondrial genetic analysis of the origin of Hakka in Meizhou, Guangdong. J. Sun Yat-sen Univ. 26, 129–131.
Cai, X., Qin, Z., Wen, B., Xu, S., Wang, Y., Lu, Y., et al. (2011). Human migration through bottlenecks from Southeast Asia into East Asia during Last Glacial Maximum revealed by Y chromosomes. PLoS One 6:e24282. doi: 10.1371/journal.pone.0024282
Cao, S., Bai, P., Zhu, W., Chen, D., Wang, H., Jin, B., et al. (2018). Genetic portrait of 27 Y-STR loci in the Tibetan ethnic population of the Qinghai province of China. Forensic Sci. Int. Genet. 34, e18–e19. doi: 10.1016/j.fsigen.2018.02.005
Carracedo, A., Butler, J. M., Gusmao, L., Linacre, A., Parson, W., Roewer, L., et al. (2013). New guidelines for the publication of genetic population data. Forensic Sci. Int. Genet. 7, 217–220. doi: 10.1016/j.fsigen.2013.01.001
Chen, P., He, G., Zou, X., Zhang, X., Li, J., Wang, Z., et al. (2018). Genetic diversities and phylogenetic analyses of three Chinese main ethnic groups in southwest China: a Y-chromosomal STR study. Sci. Rep. 8:15339. doi: 10.1038/s41598-018-33751-x
D’Atanasio, E., Iacovacci, G., Pistillo, R., Bonito, M., Dugoujon, J. M., Moral, P., et al. (2019). Rapidly mutating Y-STRs in rapidly expanding populations: discrimination power of the Yfiler Plus multiplex in northern Africa. Forensic Sci. Int. Genet. 38, 185–194. doi: 10.1016/j.fsigen.2018.11.002
Deng, Q.-Y., Wang, C.-C., Wang, X.-Q., Wang, L.-X., Wang, Z.-Y., Wu, W.-J., et al. (2013). Genetic affinity between the Kam-Sui speaking Chadong and Mulam people. J. Syst. Evol. 51, 263–270. doi: 10.1111/jse.12009
Dezhi, C., Meili, L., Yingjian, H., Yiping, H., Yu, T., and Weibo, L. (2020). Population genetics of 27 Y-STRs for the Yi population from Liangshan Yi Autonomous Prefecture, China. Int. J. Legal. Med. 135, 441–442. doi: 10.1007/s00414-020-02249-5
Du, W., Wu, W., Wu, Z., Guo, L., Wang, B., and Chen, L. (2019). Genetic polymorphisms of 32 Y-STR loci in Meizhou Hakka population. Int. J. Legal. Med. 133, 465–466. doi: 10.1007/s00414-018-1845-1
Excoffier, L., and Lischer, H. E. (2010). Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Mol. Eol. Resour. 10, 564–567. doi: 10.1111/j.1755-0998.2010.02847.x
Excoffier, L., and Smouse, P. E. (1994). Using allele frequencies and geographic subdivision to reconstruct gene trees within a species: molecular variance parsimony. Genetics 136, 343–359.
Excoffier, L., Smouse, P. E., and Quattro, J. M. (1992). Analysis of molecular variance inferred from metric distances among DNA haplotypes: application to human mitochondrial DNA restriction data. Genetics 131, 479–491.
Fan, G. Y., An, Y. R., Peng, C. X., Deng, J. L., Pan, L. P., and Ye, Y. (2019). Forensic and phylogenetic analyses among three Yi populations in Southwest China with 27 Y chromosomal STR loci. Int. J. Legal. Med. 133, 795–797. doi: 10.1007/s00414-018-1984-4
Fan, H., Du, Z., Wang, F., Wang, X., Wen, S., Wang, L., et al. (2021). The forensic landscape and the population genetic analyses of Hainan Li based on massively parallel sequencing DNA profiling. Int. J. Legal. Med. doi: 10.1007/s00414-021-02590-3 [Epub ahead of print].
Fan, H., Wang, X., Chen, H., Long, R., Liang, A., Li, W., et al. (2018a). The evaluation of forensic characteristics and the phylogenetic analysis of the Ong Be language-speaking population based on Y-STR. Forensic Sci. Int. Genet. 37, e6–e11. doi: 10.1016/j.fsigen.2018.09.008
Fan, H., Wang, X., Chen, H., Zhang, X., Huang, P., Long, R., et al. (2018b). Population analysis of 27 Y-chromosomal STRs in the Li ethnic minority from Hainan province, southernmost China. Forensic Sci. Int. Genet. 34, e20–e22. doi: 10.1016/j.fsigen.2018.01.007
Fan, H., Zhang, X., Wang, X., Ren, Z., Li, W., Long, R., et al. (2018c). Genetic analysis of 27 Y-STR loci in Han population from Hainan province, southernmost China. Forensic Sci. Int. Genet. 33, e9–e10. doi: 10.1016/j.fsigen.2017.12.009
Fan, Z., Qunying, C., Xiaoxia, Z., Lei, J., and Meiman, D. (2020). Polymorphisms of 27 Y-STRs in Zhejiang Jinhua Han (in Chinese). J. Forensic Med. 2, 263–267.
Fang, X. J. (2007). Implications of Luo XiangLin’s Academic thoughts and practice of Hakka Studies. J. Jiaying Univ. 25, 11–16.
Feng, R., Zhao, Y., Chen, S., Li, Q., Fu, Y., Zhao, L., et al. (2020). Genetic analysis of 50 Y-STR loci in Dong, Miao, Tujia, and Yao populations from Hunan. Int. J. Legal. Med. 134, 981–983. doi: 10.1007/s00414-019-02115-z
Fu, X., Fu, Y., Liu, Y., Guo, J., Liu, Y., Guo, Y., et al. (2016). Genetic polymorphisms of 26 Y-STR loci in the Mongolian minority from Horqin district, China. Int. J. Legal. Med. 130, 941–946. doi: 10.1007/s00414-016-1387-3
Gan, R. J., Pan, S. L., Mustavich, L. F., Qin, Z. D., Cai, X. Y., Qian, J., et al. (2008). Pinghua population as an exception of Han Chinese’s coherent genetic structure. J. Hum. Genet. 53, 303–313. doi: 10.1007/s10038-008-0250-x
Gao, T., Yun, L., Gao, S., Gu, Y., He, W., Luo, H., et al. (2016). Population genetics of 23 Y-STR loci in the Mongolian minority population in Inner Mongolia of China. Int. J. Legal. Med. 130, 1509–1511. doi: 10.1007/s00414-016-1433-1
Garcia, O., Yurrebaso, I., Mancisidor, I. D., Lopez, S., Alonso, S., and Gusmao, L. (2016). Data for 27 Y-chromosome STR loci in the Basque Country autochthonous population. Forensic Sci. Int. Genet. 20, e10–e12. doi: 10.1016/j.fsigen.2015.09.010
Guan, T., Song, X., Xiao, C., Sun, H., Yang, X., Liu, C., et al. (2020). Analysis of 23 Y-STR loci in Chinese Jieyang Han population. Int. J. Legal. Med. 134, 505–507. doi: 10.1007/s00414-019-02019-y
Guo, F., Li, J., Chen, K., Tang, R., and Zhou, L. (2017). Population genetic data for 27 Y-STR loci in the Zhuang ethnic minority from Guangxi Zhuang Autonomous Region in the south of China. Forensic Sci. Int. Genet. 27, 182–183. doi: 10.1016/j.fsigen.2016.11.009
Gusmao, L., Butler, J. M., Carracedo, A., Gill, P., Kayser, M., Mayr, W. R., et al. (2006). DNA Commission of the International Society of Forensic Genetics (ISFG): an update of the recommendations on the use of Y-STRs in forensic analysis. Int. J. Legal. Med. 120, 191–200. doi: 10.1007/s00414-005-0026-1
Haiying, J., Kang, W., and Ke’er, Y. (2020). Investigation on Genetic Polymorphisms of 41 Y-STR Loci in the Zhuang Population in Guangxi (in Chinese). Biol. Chem. Eng. 6, 84–87.
Han, X., Shen, A., Yao, T., Wu, W., Wang, X., Sun, H., et al. (2020). Genetic diversity of 17 autosomal STR loci in Meizhou Hakka population. Int. J. Legal. Med. 135, 443–444. doi: 10.1007/s00414-020-02253-9
Hanguang, L., Peizhi, T., Qiansu, Y., Xin, Y., Tian, M., and Jianpin, T. (2020). Genetic Polymorphism of 36 Y-STR Loci among Han-ethnic Population in Guangxi Area (in Chinese). Forensic Sci. Technol. 45, 420–446. doi: 10.16467/j.1008-3650.2020.04.019
He, G., Wang, Z., Su, Y., Zou, X., Wang, M., Chen, X., et al. (2019). Genetic structure and forensic characteristics of Tibeto-Burman-speaking U-Tsang and Kham Tibetan Highlanders revealed by 27 Y-chromosomal STRs. Sci. Rep. 9:7739. doi: 10.1038/s41598-019-44230-2
He, J., and Guo, F. (2013). Population genetics of 17 Y-STR loci in Chinese Manchu population from Liaoning Province, Northeast China. Forensic Sci. Int. Genet. 7, e84–e85. doi: 10.1016/j.fsigen.2012.12.006
Henry, J., Dao, H., Scandrett, L., and Taylor, D. (2019). Population genetic analysis of Yfiler((R)) Plus haplotype data for three South Australian populations. Forensic Sci. Int. Genet. 41, e23–e25. doi: 10.1016/j.fsigen.2019.03.021
Huang, Y., Guo, L., Wang, M., Zhang, C., Kang, L., Wang, K., et al. (2019). Genetic analysis of 39 Y-STR loci in a Han population from Henan province, central China. Int. J. Legal. Med. 133, 95–97. doi: 10.1007/s00414-018-1852-2
Iacovacci, G., D’Atanasio, E., Marini, O., Coppa, A., Sellitto, D., Trombetta, B., et al. (2017). Forensic data and microvariant sequence characterization of 27 Y-STR loci analyzed in four Eastern African countries. Forensic Sci. Int. Genet. 27, 123–131. doi: 10.1016/j.fsigen.2016.12.015
Ip, S. C. Y., Lin, S. W., and Lam, T. T. (2019). Haplotype data of 27 Y-STR loci in Hong Kong Chinese. Forensic Sci. Int. Genet. 38, e14–e15. doi: 10.1016/j.fsigen.2018.11.001
Jankova, R., Seidel, M., Videtic Paska, A., Willuweit, S., and Roewer, L. (2019). Y-chromosome diversity of the three major ethno-linguistic groups in the Republic of North Macedonia. Forensic Sci. Int. Genet. 42, 165–170. doi: 10.1016/j.fsigen.2019.07.007
Jannuzzi, J., Ribeiro, J., Alho, C., de Oliveira Lazaro, E. A. G., Cicarelli, R., Simoes Dutra Correa, H., et al. (2020). Male lineages in Brazilian populations and performance of haplogroup prediction tools. Forensic Sci. Int. Genet. 44:102163. doi: 10.1016/j.fsigen.2019.102163
Ji, J., Ren, Z., Zhang, H., Wang, Q., Wang, J., Kong, Z., et al. (2017). Genetic profile of 23 Y chromosomal STR loci in Guizhou Shui population, southwest China. Forensic Sci. Int. Genet. 28, e16–e17. doi: 10.1016/j.fsigen.2017.01.010
Juan, M., Xang, W., Yanmei, H., Huiyong, J., Liwei, G., Qian, Z., et al. (2018). Genetic polymorphism of 27 Y-STR Loci among Xinxiang Han population in Henan Province (in Chinese). Med. Sci. 54, 452–457. doi: 10.13705/j.issn.1671-6825.2017.09.150
Jun, Y., Li, W., Jing, G., Jiaxin, X., Jinfeng, X., and Baojie, W. (2017). Polymorphisms of 27 Y-STRs in Liaoning Han (in Chinese). J. Forensic Med. 33, 666–668. doi: 10.3969/j.issn.1004-5619.2017.06.023
Jung, J. Y., Park, J. H., Oh, Y. L., Kwon, H. S., Park, H. C., Park, K. H., et al. (2016). Forensic genetic study of 29 Y-STRs in Korean population. Leg. Med. 23, 17–20. doi: 10.1016/j.legalmed.2016.09.001
Kai, Z., Xiaoye, J., Dajun, L., and Wuhu, G. (2019). Genetic polymorphic analysis of 27 Y-STR loci in Zhanjiang Han population (in Chinese). Chin. J. Forensic Med. 34, 449–453. doi: 10.13618/j.issn.1001-5728.2019.05.006
Kang, W., Hu, B., Bingjie, Z., Jiabin, H., and Peizhi, T. (2020). Investigation on the allele frequencies of 41 Y-STRLoci in Han Population in Jilin (in Chinese). Biol. Chem. Eng. 6, 103–107.
Khubrani, Y. M., Wetton, J. H., and Jobling, M. A. (2018). Extensive geographical and social structure in the paternal lineages of Saudi Arabia revealed by analysis of 27 Y-STRs. Forensic Sci. Int. Genet. 33, 98–105. doi: 10.1016/j.fsigen.2017.11.015
Kumar, S., Stecher, G., and Tamura, K. (2016). MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874. doi: 10.1093/molbev/msw054
Kwak, K. D., Jin, H. J., Shin, D. J., Kim, J. M., Roewer, L., Krawczak, M., et al. (2005). Y-chromosomal STR haplotypes and their applications to forensic and population studies in east Asia. Int. J. Legal. Med. 119, 195–201. doi: 10.1007/s00414-004-0518-4
Lacerenza, D., Aneli, S., Di Gaetano, C., Critelli, R., Piazza, A., Matullo, G., et al. (2017). Investigation of extended Y chromosome STR haplotypes in Sardinia. Forensic Sci. Int. Genet. 27, 172–174. doi: 10.1016/j.fsigen.2016.12.009
Lang, M., Liu, H., Song, F., Qiao, X., Ye, Y., Ren, H., et al. (2019). Forensic characteristics and genetic analysis of both 27 Y-STRs and 143 Y-SNPs in Eastern Han Chinese population. Forensic Sci. Int. Genet. 42, e13–e20. doi: 10.1016/j.fsigen.2019.07.011
Lei, F. Q. (2005). The she dialect language, widely-used in the she nationality. J. Lishui Univ. 27, 60–66.
Letunic, I., and Bork, P. (2019). Interactive Tree Of Life (iTOL) v4: recent updates and new developments. Nucleic Acids Res. 47, W256–W259. doi: 10.1093/nar/gkz239
Li, H., Pan, W. Y., Wen, B., Yang, N. N., Jin, J. Z., Jin, L., et al. (2003). Origin of Hakka and Hakkanese: a genetics analysis. Yi Chuan Xue Bao 30, 873–880.
Li, H., Wen, B., Chen, S. J., Su, B., Pramoonjago, P., Liu, Y., et al. (2008). Paternal genetic affinity between Western Austronesians and Daic populations. BMC Evol. Biol. 8:146. doi: 10.1186/1471-2148-8-146
Li, L., Yu, G., Li, S., Jin, L., and Yan, S. (2016). Genetic analysis of 17 Y-STR loci from 1019 individuals of six Han populations in East China. Forensic Sci. Int. Genet. 20, 101–102. doi: 10.1016/j.fsigen.2015.10.007
Liu, S., Chen, G., Huang, H., Lin, W., Guo, D., Zhao, S., et al. (2017). Patrilineal background of the She minority population from Chaoshan Fenghuang Mountain, an isolated mountain region, in China. Genomics 109, 284–289. doi: 10.1016/j.ygeno.2017.05.002
Liu, Y., Jin, X., Guo, Y., Zhang, X., Zhu, W., Zhang, W., et al. (2020). Haplotypic diversity and population genetic study of a population in Kashi region by 27 Y-chromosomal short tandem repeat loci. Mol. Genet. Genom. Med. 8:e1338. doi: 10.1002/mgg3.1338
Liu, Y., Wang, C., Zhou, W., Li, X. B., Shi, M., Bai, R., et al. (2019). Haplotypes of 27 Y-STRs analyzed in Gelao and Miao ethnic minorities from Guizhou Province, Southwest China. Forensic Sci. Int. Genet. 40, e264–e267. doi: 10.1016/j.fsigen.2019.03.002
Liu, Y., Wen, S., Guo, L., Bai, R., Shi, M., and Li, X. (2018). Haplotype data of 27 Y-STRs analyzed in the Hui and Tujia ethnic minorities from China. Forensic Sci. Int. Genet. 35, e7–e9. doi: 10.1016/j.fsigen.2018.04.006
Liu, Y. J., Guo, L. H., Li, J., Yue, J. T., and Shi, M. S. (2018). Genetic polymorphisms of 27 Y-STR Loci in Dongxiang population of Gansu province. Fa Yi Xue Za Zhi 34, 270–275. doi: 10.12116/j.issn.1004-5619.2018.03.011
Lu, J. N., and Wang, C. C. (2019). A new approach to the origin of she in the perspective of molecular anthropology. Philos. Soc. Sci. 145, 98–107.
Luo, H., Song, F., Zhang, L., and Hou, Y. (2015). Genetic polymorphism of 23 Y-STR loci in the Zhuang minority population in Guangxi of China. Int. J. Legal. Med. 129, 737–738. doi: 10.1007/s00414-015-1175-5
Luo, X. Q., Du, P. X., Wang, L. X., Zhou, B. Y., Li, Y. C., Zheng, H. X., et al. (2020). Uniparental Genetic analyses reveal the major origin of Fujian Tanka from ancient indigenous Daic populations. Hum. Biol. 91, 257–277. doi: 10.13110/humanbiology.91.4.05
Luo, Y., Wu, Y., Qian, E., Wang, Q., Wang, Q., Zhang, H., et al. (2019). Population genetic analysis of 36 Y-chromosomal STRs yields comprehensive insights into the forensic features and phylogenetic relationship of Chinese Tai-Kadai-speaking Bouyei. PLoS One 14:e0224601. doi: 10.1371/journal.pone.0224601
Nothnagel, M., Fan, G., Guo, F., He, Y., Hou, Y., Hu, S., et al. (2017). Revisiting the male genetic landscape of China: a multi-center study of almost 38,000 Y-STR haplotypes. Hum. Genet. 136, 485–497. doi: 10.1007/s00439-017-1759-x
Oppenheimer, S. (2012). Out-of-Africa, the peopling of continents and islands: tracing uniparental gene trees across the map. Philos. Trans. R. Soc. Lond. B Biol. Sci. 367, 770–784. doi: 10.1098/rstb.2011.0306
Ou, X., Wang, Y., Liu, C., Yang, D., Zhang, C., Deng, S., et al. (2015). Haplotype analysis of the polymorphic 40 Y-STR markers in Chinese populations. Forensic Sci. Int. Genet. 19, 255–262. doi: 10.1016/j.fsigen.2015.08.007
Pickrahn, I., Muller, E., Zahrer, W., Dunkelmann, B., Cemper-Kiesslich, J., Kreindl, G., et al. (2016). Yfiler((R)) Plus amplification kit validation and calculation of forensic parameters for two Austrian populations. Forensic Sci. Int. Genet. 21, 90–94. doi: 10.1016/j.fsigen.2015.12.014
Prinz, M., Boll, K., Baum, H., and Shaler, B. (1997). Multiplexing of Y chromosome specific STRs and performance for mixed samples. Forensic Sci. Int. 85, 209–218. doi: 10.1016/s0379-0738(96)02096-8
Qiang, L., Yue, X., Wei, Z., Dian, Z., Baowen, C., and Faming, Z. (2017). Genetic Polymorphism Analysis of 27 Y-STR Loci in the Hui Population from Weishan, Yunnan Province (in Chinese). J. Kunming Med. Univ. 38, 25–29.
Roewer, L., Andersen, M. M., Ballantyne, J., Butler, J. M., Caliebe, A., Corach, D., et al. (2020). DNA commission of the International Society of Forensic Genetics (ISFG): recommendations on the interpretation of Y-STR results in forensic analysis. Forensic Sci. Int. Genet. 48:102308. doi: 10.1016/j.fsigen.2020.102308
Rowold, D. J., Gayden, T., Luis, J. R., Alfonso-Sanchez, M. A., Garcia-Bertrand, R., and Herrera, R. J. (2019). Investigating the genetic diversity and affinities of historical populations of Tibet. Gene 682, 81–91. doi: 10.1016/j.gene.2018.09.043
Saitou, N., and Nei, M. (1987). The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4, 406–425. doi: 10.1093/oxfordjournals.molbev.a040454
Santos Stange, V., Silva Dos Reis, R., Mariano Garcia de Souza, R. F., Lima, L. M., Mayumi Vieira, C., de Paula, F., et al. (2019). Stratification among European descent and admixed Brazilian populations of Espirito Santo for 27 Y-STRs. Forensic Sci. Int. Genet. 41, e20–e22. doi: 10.1016/j.fsigen.2019.03.019
Shan, W., Ablimit, A., Zhou, W., Zhang, F., Ma, Z., and Zheng, X. (2014). Genetic polymorphism of 17 Y chromosomal STRs in Kazakh and Uighur populations from Xinjiang, China. Int. J. Legal. Med. 128, 743–744. doi: 10.1007/s00414-013-0948-y
Shi, M., Liu, Y., Zhang, J., Bai, R., Lv, X., and Ma, S. (2015). Analysis of 24 Y chromosomal STR haplotypes in a Chinese Han population sample from Henan Province, Central China. Forensic Sci. Int. Genet. 17, 83–86. doi: 10.1016/j.fsigen.2015.04.001
Shi, Z. J. (2013). An analysis of the origin and development of she culture. J. Mudanjiang Coll. Educ. 144, 122–124.
Shonhai, M., Nhiwatiwa, T., Nangammbi, T., and Mazando, S. (2020). Genetic analysis of 27 Y-chromosomal STR loci in a Zimbabwean Shona ethnic group. Leg. Med. 43:101660. doi: 10.1016/j.legalmed.2019.101660
Shu, L., Li, L., Yu, G., Yu, B., Liu, Y., Li, S., et al. (2015). Genetic analysis of 17 Y-STR loci in Han, Dong, Miao and Tujia populations from Hunan province, central-southern China. Forensic Sci. Int. Genet. 19, 250–251. doi: 10.1016/j.fsigen.2015.07.007
Song, F., Xie, M., Xie, B., Wang, S., Liao, M., and Luo, H. (2020). Genetic diversity and phylogenetic analysis of 29 Y-STR loci in the Tibetan population from Sichuan Province, Southwest China. Int. J. Legal. Med. 134, 513–516. doi: 10.1007/s00414-019-02043-y
Spolnicka, M., Dabrowska, J., Szablowska-Gnap, E., Paleczka, A., Jablonska, M., Zbiec-Piekarska, R., et al. (2017). Intra- and inter-population analysis of haplotype diversity in Yfiler((R)) Plus system using a wide set of representative data from Polish population. Forensic Sci. Int. Genet. 28, e22–e25. doi: 10.1016/j.fsigen.2017.01.014
Sun, H., Su, K., Fan, C., Long, F., Liu, Y., Sun, J., et al. (2019). Y-STRs’ genetic profiling of 1953 individuals from two Chinese Han populations (Guizhou and Shanxi). Forensic Sci. Int. Genet. 38, e8–e10. doi: 10.1016/j.fsigen.2018.10.011
SWGDAM (2010). Scientific Working Group on DNA Analysis Interpretation Guidelines for Autosomal STR Typing by Forensic DNA Testing Laboratories.
Tang, J., Yang, M., Wang, X., Wang, Q., Wang, Q., Zhang, H., et al. (2020). Genetic structure and forensic characterisation of 36 Y-chromosomal STR loci in Hmong-Mien-speaking Miao population. Ann. Hum. Biol. 47, 541–548. doi: 10.1080/03014460.2020.1788159
Tao, R., Jin, M., Ji, G., Zhang, J., Zhang, J., Yang, Z., et al. (2019a). Forensic characteristics of 36 Y-STR loci in a Changzhou Han population and genetic distance analysis among several Chinese populations. Forensic Sci. Int. Genet. 40, e268–e270. doi: 10.1016/j.fsigen.2019.03.011
Tao, R., Wang, S., Zhang, J., Zhang, J., Yang, Z., Zhang, S., et al. (2019b). Genetic characterization of 27 Y-STR loci analyzed in the Nantong Han population residing along the Yangtze Basin. Forensic Sci. Int. Genet. 39, e10–e13. doi: 10.1016/j.fsigen.2018.11.015
Wang, C. C., Wang, L. X., Shrestha, R., Wen, S., Zhang, M., Tong, X., et al. (2015). Convergence of Y Chromosome STR haplotypes from different SNP haplogroups compromises accuracy of haplogroup prediction. J. Genet. Genom. 42, 403–407. doi: 10.1016/j.jgg.2015.03.008
Wang, C. Z., Su, M. J., Li, Y., Chen, L., Jin, X., Wen, S. Q., et al. (2019). Genetic polymorphisms of 27 Yfiler((R)) Plus loci in the Daur and Mongolian ethnic minorities from Hulunbuir of Inner Mongolia Autonomous Region, China. Forensic Sci. Int. Genet. 40, e252–e255. doi: 10.1016/j.fsigen.2019.02.003
Wang, C. Z., Zhang, J. S., Li, X. B., Bai, R. F., Shi, M. S., and Wang, C. C. (2020). Haplotype analysis of 36 Y-STR loci in a Chinese Han population from Anhui Province, Eastern China. Int. J. Legal. Med. 134, 2063–2065. doi: 10.1007/s00414-020-02321-0
Wang, H., Ba, H., Yang, C., Zhang, J., and Tai, Y. (2017). Inner and inter population structure construction of Chinese Jiangsu Han population based on Y23 STR system. PLoS One 12:e0180921. doi: 10.1371/journal.pone.0180921
Wang, J., Wen, S., Shi, M., Liu, Y., Zhang, J., Bai, R., et al. (2018). Haplotype structure of 27 Yfiler((R))Plus loci in Chinese Dongxiang ethnic group and its genetic relationships with other populations. Forensic Sci. Int. Genet. 33, e13–e16. doi: 10.1016/j.fsigen.2017.12.014
Wang, L., Chen, F., Kang, B., Zheng, H., Zhao, Y., Li, L., et al. (2016). Genetic population data of Yfiler Plus kit from 1434 unrelated Hans in Henan Province (Central China). Forensic Sci. Int. Genet. 22, e25–e27. doi: 10.1016/j.fsigen.2016.02.009
Wang, M., Wang, Z., Zhang, Y., He, G., Liu, J., and Hou, Y. (2017). Forensic characteristics and phylogenetic analysis of two Han populations from the southern coastal regions of China using 27 Y-STR loci. Forensic Sci. Int. Genet. 31, e17–e23. doi: 10.1016/j.fsigen.2017.10.009
Wang, X., Li, Y., and Fan, H. (2019). The associations between screen time-based sedentary behavior and depression: a systematic review and meta-analysis. BMC Public Health 19:1524. doi: 10.1186/s12889-019-7904-9
Wang, Y., Li, S., Dang, Z., Kong, X., Zhang, Y., Ma, L., et al. (2019). Genetic diversity and haplotype structure of 27 Y-STR loci in a Yanbian Korean population from Jilin Province, Northeast China. Leg. Med. 36, 110–112. doi: 10.1016/j.legalmed.2018.11.010
Wang, Y., Zhang, Y. J., Zhang, C. C., Li, R., Yang, Y., Ou, X. L., et al. (2016). Genetic polymorphisms and mutation rates of 27 Y-chromosomal STRs in a Han population from Guangdong Province, Southern China. Forensic Sci. Int. Genet. 21, 5–9. doi: 10.1016/j.fsigen.2015.09.013
Wen, B., Li, H., Lu, D., Song, X., Zhang, F., He, Y., et al. (2004). Genetic evidence supports demic diffusion of Han culture. Nature 431, 302–305. doi: 10.1038/nature02878
Westen, A. A., Kraaijenbrink, T., Clarisse, L., Grol, L. J., Willemse, P., Zuniga, S. B., et al. (2015). Analysis of 36 Y-STR marker units including a concordance study among 2085 Dutch males. Forensic Sci. Int. Genet. 14, 174–181. doi: 10.1016/j.fsigen.2014.10.012
Wu, W., Pan, L., Hao, H., Zheng, X., Lin, J., and Lu, D. (2011). Population genetics of 17 Y-STR loci in a large Chinese Han population from Zhejiang Province, Eastern China. Forensic Sci. Int. Genet. 5, e11–e13. doi: 10.1016/j.fsigen.2009.12.005
Wu, Z., Chen, T. F., Zeng, Z. F., Zhang, Y. W., Tang, Z., Su, K. Y., et al. (2019). Genetic Structure Analysis of Y-Chromosome STR and SNP in Population of Wujiang Area, Suzhou City. Fa Yi Xue Za Zhi 35, 448–454. doi: 10.12116/j.issn.1004-5619.2019.04.014
Xie, M., Song, F., Li, J., Lang, M., Luo, H., Wang, Z., et al. (2019). Genetic substructure and forensic characteristics of Chinese Hui populations using 157 Y-SNPs and 27 Y-STRs. Forensic Sci. Int. Genet. 41, 11–18. doi: 10.1016/j.fsigen.2019.03.022
Yaju, L., Jian, M., Chuanhong, Z., Xuebo, L., and Meisen, S. (2019). Genetic polymorphisms of 27 Y-STR loci in Tujia and Hui population and the cluster analysis of 13 ethnic groups (in Chinese). Basic Clin. Med. 39, 314–320.
Yaju, L., Jun-tao, Z., Shaoxing, S., Hai, L., Lihong, G., Yi, Z., et al. (2014). Genetic polymorphisms of 27Y-STR loci in Henna Han population. Forensic Sci. Technol. 4, 18–20.
Yaju, L., Lin, G., Juntao, Y., Jin, L., Meisen, S., and Xuebo, L. (2020). Genetic polymorphisms of 27 Y-STR in 3 Chinese populations and the cluster analysis of 13 ethnic groups (in Chinese). Lab. Med. Clin. 5, 577–585. doi: 10.3969/j.issn.1672-9455.2020.05.001
Yao, H. B., Wang, C. C., Tao, X., Shang, L., Wen, S. Q., Zhu, B., et al. (2016). Genetic evidence for an East Asian origin of Chinese Muslim populations Dongxiang and Hui. Sci. Rep. 6:38656. doi: 10.1038/srep38656
Yin, C., Su, K., He, Z., Zhai, D., Guo, K., Chen, X., et al. (2020). Genetic Reconstruction and Forensic Analysis of Chinese Shandong and Yunnan Han Populations by Co-Analyzing Y Chromosomal STRs and SNPs. Genes 11:743. doi: 10.3390/genes11070743
Zeyad, T., Adam, A., Alghafri, R., and Iratni, R. (2020). Study of 27 Y-STR markers in United Arab Emirates population. Forensic Sci. Intern. Rep. 2:100057.
Zgonjanin, D., Alghafri, R., Antov, M., Stojiljkovic, G., Petkovic, S., Vukovic, R., et al. (2017). Genetic characterization of 27 Y-STR loci with the Yfiler((R)) Plus kit in the population of Serbia. Forensic Sci. Int. Genet. 31, e48–e49. doi: 10.1016/j.fsigen.2017.07.013
Zhabagin, M., Sarkytbayeva, A., Tazhigulova, I., Yerezhepov, D., Li, S., Akilzhanov, R., et al. (2019). Development of the Kazakhstan Y-chromosome haplotype reference database: analysis of 27 Y-STR in Kazakh population. Int. J. Legal. Med. 133, 1029–1032. doi: 10.1007/s00414-018-1859-8
Zhang, J., Tao, R., Zhong, J., Sun, D., Qiao, L., Shan, S., et al. (2019). Genetic polymorphisms of 27 Y-STR loci in the Dezhou Han population from Shandong province, Eastern China. Forensic Sci. Int. Genet. 39, e26–e28. doi: 10.1016/j.fsigen.2018.11.021
Zhang, J., Wang, J., Liu, Y., Shi, M., Bai, R., and Ma, S. (2017). Haplotype data for 27 Y-chromosomal STR loci in the Chaoshan Han population, South China. Forensic Sci. Int. Genet. 31, e54–e56. doi: 10.1016/j.fsigen.2017.08.003
Zhang, S., Tian, H., Wang, Z., Zhao, S., Hu, Z., Li, C., et al. (2014). Development of a new 26plex Y-STRs typing system for forensic application. Forensic Sci. Int. Genet. 13, 112–120. doi: 10.1016/j.fsigen.2014.06.015
Zhao, Q., Bian, Y., Zhang, S., Zhu, R., Zhou, W., Gao, Y., et al. (2017). Population genetics study using 26 Y-chromosomal STR loci in the Hui ethnic group in China. Forensic Sci. Int. Genet. 28, e26–e27. doi: 10.1016/j.fsigen.2017.01.018
Zheng, L., Li, Y., Lu, S., Bao, J., Wang, Y., Zhang, X., et al. (2013). Physical characteristics of Chinese Hakka. Sci. China Life Sci. 56, 541–551. doi: 10.1007/s11427-013-4471-7
Zhong, J. K. (2019). The review and prospect of the study in Hakka historical origin. Local Cult. Res. 02, 101–122.
Zhou, H., Ren, Z., Zhang, H., Wang, J., and Huang, J. (2016). Genetic profile of 17 Y chromosome STRs in the Guizhou Han population of southwestern China. Forensic Sci. Int. Genet. 25, e6–e7. doi: 10.1016/j.fsigen.2016.05.010
Zhou, Y., Shao, C., Li, L., Zhang, Y., Liu, B., Yang, Q., et al. (2018). Genetic analysis of 29 Y-STR loci in the Chinese Han population from Shanghai. Forensic Sci. Int. Genet. 32, e1–e4. doi: 10.1016/j.fsigen.2017.11.003
Zhu, B., Deng, Y., Zhang, F., Wei, W., Chen, L., Zhao, J., et al. (2006a). Genetic analysis for Y chromosome short tandem repeat haplotypes of Chinese Han population residing in the Ningxia province of China. J. Forensic Sci. 51, 1417–1420. doi: 10.1111/j.1556-4029.2006.00282.x
Zhu, B., Liu, S., Ci, D., Huang, J., Wang, Y., Chen, L., et al. (2006b). Population genetics for Y-chromosomal STRs haplotypes of Chinese Tibetan ethnic minority group in Tibet. Forensic Sci. Int. 161, 78–83. doi: 10.1016/j.forsciint.2005.09.003
Zhu, B., Li, X., Wang, Z., Wu, H., He, Y., Zhao, J., et al. (2005). Y-STRs haplotypes of Chinese Mongol ethnic group using Y-PLEX 12. Forensic Sci. Int. 153, 260–263. doi: 10.1016/j.forsciint.2004.11.002
Keywords: Guangdong Hakka, Guangdong She, Y-STR, forensic characteristics, phylogenetic analyses
Citation: Luo C, Duan L, Li Y, Xie Q, Wang L, Ru K, Nazir S, Jawad M, Zhao Y, Wang F, Du Z, Peng D, Wen S-Q, Qiu P and Fan H (2021) Insights From Y-STRs: Forensic Characteristics, Genetic Affinities, and Linguistic Classifications of Guangdong Hakka and She Groups. Front. Genet. 12:676917. doi: 10.3389/fgene.2021.676917
Received: 06 March 2021; Accepted: 06 April 2021;
Published: 24 May 2021.
Edited by:
Atif Adnan, China Medical University, ChinaReviewed by:
Guanglin He, Sichuan University, ChinaCopyright © 2021 Luo, Duan, Li, Xie, Wang, Ru, Nazir, Jawad, Zhao, Wang, Du, Peng, Wen, Qiu and Fan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shao-Qing Wen, d2Vuc2hhb3FpbmdAZnVkYW4uZWR1LmNu; Pingming Qiu, cWl1cG1meUAxMjYuY29t; Haoliang Fan, ZmFuaGFvbGlhbmcxOTg5MzFAMTYzLmNvbQ==
†These authors have contributed equally to this work
‡ORCID: Shao-Qing Wen, orcid.org/0000-0003-1223-4720; Pingming Qiu, orcid.org/0000-0002-5579-1124; Haoliang Fan, orcid.org/0000-0002-3214-0177
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.