
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Genet., 14 June 2021
Sec. Genetics of Aging
Volume 12 - 2021 | https://doi.org/10.3389/fgene.2021.675587
This article is part of the Research TopicOmics of Human Aging and Longevity in the Post Genome Era: From Single Biomarkers to Systems Biology ApproachesView all 7 articles
 Afsaneh Mohammadnejad1
Afsaneh Mohammadnejad1 Weilong Li1,2
Weilong Li1,2 Jesper Beltoft Lund1,3
Jesper Beltoft Lund1,3 Shuxia Li1
Shuxia Li1 Martin J. Larsen4,5
Martin J. Larsen4,5 Jonas Mengel-From1,4
Jonas Mengel-From1,4 Tanja Maria Michel6,7,8
Tanja Maria Michel6,7,8 Lene Christiansen1,9
Lene Christiansen1,9 Kaare Christensen1,4
Kaare Christensen1,4 Jacob Hjelmborg1
Jacob Hjelmborg1 Jan Baumbach10,11
Jan Baumbach10,11 Qihua Tan1,4*
Qihua Tan1,4*Cognitive aging is one of the major problems worldwide, especially as people get older. This study aimed to perform global gene expression profiling of cognitive function to identify associated genes and pathways and a novel transcriptional regulatory network analysis to identify important regulons. We performed single transcript analysis on 400 monozygotic twins using an assumption-free generalized correlation coefficient (GCC), linear mixed-effect model (LME) and kinship model and identified six probes (one significant at the standard FDR < 0.05 while the other results were suggestive with 0.18 ≤ FDR ≤ 0.28). We combined the GCC and linear model results to cover diverse patterns of relationships, and meaningful and novel genes like APOBEC3G, H6PD, SLC45A1, GRIN3B, and PDE4D were detected. Our exploratory study showed the downregulation of all these genes with increasing cognitive function or vice versa except the SLC45A1 gene, which was upregulated with increasing cognitive function. Linear models found only H6PD and SLC45A1, the other genes were captured by GCC. Significant functional pathways (FDR < 3.95e-10) such as focal adhesion, ribosome, cysteine and methionine metabolism, Huntington's disease, eukaryotic translation elongation, nervous system development, influenza infection, metabolism of RNA, and cell cycle were identified. A total of five regulons (FDR< 1.3e-4) were enriched in a transcriptional regulatory analysis in which CTCF and REST were activated and SP3, SRF, and XBP1 were repressed regulons. The genome-wide transcription analysis using both assumption-free GCC and linear models identified important genes and biological pathways implicated in cognitive performance, cognitive aging, and neurological diseases. Also, the regulatory network analysis revealed significant activated and repressed regulons on cognitive function.
Cognitive impairment is a global challenge that creates cost, social, and economic challenges for society in many populations, in particular, older populations worldwide. Although, some effort has been made to understand the genes and biological pathways involved in cognitive functioning through gene expression analysis, there is still a lack of knowledge. Harries et al. (2012) performed gene expression analysis on 691 subjects from the InCHIANTI study (mean age 72.6 years) and reported that the CCR2 gene was associated with cognitive function (FDR = 0.077). Also, no significant pathways were identified. Nygaard et al. (2019) performed gene expression analysis on 235 monozygotic (MZ) twin pairs and reported POU6F1 gene (FDR = 0.09) and significant pathways such as protein metabolism, translation, RNA metabolism, infectious disease, and the immune system.
The popular statistical models used in the analysis of gene expression data are usually linear models, which are controlled by multiple assumptions, including normality of phenotype and linear relations between expression level and the phenotype. In the case of having twins in the data, linear mixed-effect models are appropriate to deal with the correlation structure in the data. Imposing multiple assumptions in linear models might be the reason for having a smaller number of important markers in gene expression analysis. However, recently a couple of studies have shown the strength of generalize correlation coefficient (GCC) as a non-parametric method that is able to identify different patterns, deal with correlated twin samples as well as non-normality of the phenotype without imposing strict assumptions (Reshef et al., 2011; Speed, 2011; Murrell et al., 2016; Tan et al., 2017; Mohammadnejad et al., 2020, 2021). In fact, linear models are well-designed when there is a perfect linear relation; hence we applied both GCC and linear models.
Transcription factors (TFs) are specific DNA sequences that affect gene expression by promoting or repressing the target genes. Mutation in TFs and TF binding sites determine many human diseases. The group of genes that are controlled by TFs are called regulons (Lambert et al., 2018).
This study aimed to perform two analyses: (1) a global gene expression analysis of cognitive function measured in monozygotic (MZ) twins to identify significant genes and pathways associated with the phenotype by applying the assumption-free GCC and linear models, (2) investigate the significance of previously reported cognitive function-related TFs through a gene regulatory network analysis.
We used 400 MZ twins (220 males and 180 females) (Supplementary Table 1) recruited from the Danish Twin Registry from the Middle-Aged Danish Twin (MADT) study which were mainly healthy individuals. The whole blood samples were collected in the years 2008–2011 on a follow-up assessment. The general cognitive composite score consists of five cognitive tests, including verbal fluency, attention and working memory (digits forward and digits backward), and memory (immediate and delayed word recall) (McGue and Christensen, 2002). The cognitive test scores were standardized to mean 0 and standard deviation 1 and were summed to calculate the general cognitive composite scores (Petersen et al., 2016). The age ranged from 56 to 80 and the cognitive score ranged from 11.68 to 84.93. Informed consent was obtained from all participants and approved by The Regional Scientific Ethical Committees for Southern Denmark (S-VF-19980072). The study was conducted following the Helsinki II declaration. Blood cell counts for blood leukocyte subtypes (basophils, monocytes, eosinophils, lymphocytes, and neutrophils) were available for all the samples.
Whole blood was collected in PAXgene Blood RNA Tubes (PreAnalytiX GmbH, Hombrechtikon, Switzerland) and total RNA was extracted using the PAXgene Blood miRNA kit (QIAGEN) according to the manufacturer's protocol. The extracted RNA concentration was determined using a NanoDrop spectrophotometer ND-8000 (NanoDrop Technologies), and the quality was assessed by the Agilent 2100 Bioanalyzer. Gene expression profiling was performed using the Agilent SurePrint G3 Human GE v2 8×60K Microarray (Agilent Technologies). This array contains 62,976 60-mer probes. The array hybridization and sample labeling were done according to the “Two-Color Microarray-Based Gene Expression Analysis—Low Input Quick Amp Labeling” protocol. Samples were labeled Cy5 and the reference consisting of a pool of 16 samples was labeled Cy3. Hybridization, washing, scanning, and quantification were performed according to the array manufacturer's recommendations (Nygaard et al., 2019).
The R package limma was used for quality control (QC) of the data (Ritchie et al., 2007). Background correction using the normexp method was done on the raw intensity data, within-array normalization using loess normalization to intensity measurements of two colors (cy3/cy5) and between-array normalization based on the quantile normalization method to make data from the different arrays comparable. The missing expression values were imputed using the k-nearest neighbor algorithm and replicate probes were summarized calculating their median. All probes on sex chromosomes and long non-coding RNA (lncRNA) were excluded resulting in 27,734 mRNA probes. Prior to the statistical analysis, we calculated the coefficient of variation (CV) for each probe and excluded probes with CV < 0.1. This resulted in 27,647 mRNA probes.
First, we adjusted covariates age, sex, and cell composition on gene expression data. Next, we applied GCC, kinship, and LME models to investigate the association between mRNA expression level and cognitive function. In the linear models, both LME from the lme4 R package (Bates et al., 2015) and the kinship model from the kinship2 R package (Sinnwell et al., 2014) were applied. The LME model adjusts for correlation between twins in a pair by including twin pairing as a random effect in the model.
The kinship model calculates a kinship matrix and integrates it in the covariance matrix of the expression data. For GCC analysis, the Matie R package was applied (Murrell et al., 2016). Matie computes GCC by estimating a generalized R2, which is computed from the ratio of the likelihood of an alternative model (allowing dependence between variables) over the likelihood of a null model (that forces the variables to be independent). For each probe, we report the result from the model (linear or GCC) with the lowest p-value for statistical significance to ensure that the final results are based on the most proper model unlimited by linear assumption.
The adjustment for multiple testing was performed by the Benjamini & Hochberg false discovery rate (FDR) correction method (Benjamini and Hochberg, 1995). All analyses were carried out in R. We consider this an exploratory study, and to give an overview of the top findings we report findings with FDR ≤ 0.28.
A total number of 1,968 genes (p < 0.05) were used as input in the gene-set enrichment analysis (GSEA) website (https://www.gsea-msigdb.org/gsea/msigdb/index.jsp) to identify biological pathways over-represented by the list of genes for functional interpretation. Over-representation analysis is an enrichment test based on an overlap statistic (hypergeometric test) that uses a list of significant genes to identify significantly different pathways from what would be expected by chance.
We used the R package RTN (Castro et al., 2015) which constructs the transcriptional regulatory network and analysis of regulons. This package performs the analysis in two steps: (1) Transcriptional Network Inference (TNI): it checks the association between a given TF and all target genes using microarray transcriptome data. It uses the gene expression data and a list of all annotated target genes from the microarray (18,078 genes). Next, it computes mutual information (MI) between a regulator and all target genes, unstable interaction is removed by bootstrapping analysis and leads to a consensus network which is considered as a reference network. Then the ARACNe algorithm developed by Margolin et al. (2006) is applied to remove the redundant association between TFs and gene targets. (2) Transcriptional Network Analysis (TNA): it checks the enrichment of regulons by applying GSEA on the set of regulons. The two-tailed GSEA (GSEA-2T) is used to check if the regulon is positively or negatively associated with the gene expression and finally assesses their significance expression meaning that a large positive enrichment score (ES) represents an activated regulon, whereas a large negative ES represents a repressed regulon. We used a list of 17 TFs (CREB, MEF2, Npas4, SRF, CTCF, TCF4, DREAM, KChIP3, MeCP2, FOXP2, ZNF, SP3, ptf1a, REST, OTX2, XBP1, FOXO) which have already been discussed in relation to cognitive function (Manolopoulos et al., 2010; Wang and Konopka, 2013; Nonaka et al., 2014; Mozzi et al., 2017; Hwang and Zukin, 2018; Xiao et al., 2018; Badowska et al., 2020; Choi et al., 2021) and performed GSEA-2T with a default p-value cut-off set to 0.05 and using 10,000 permutations to identify significant regulons associated with cognitive function in our gene expression data (https://bioconductor.org/packages/devel/bioc/vignettes/RTN/inst/doc/RTN.html).
The QQ plot and Manhattan plot are shown in Figures 1, 2. We saw more mRNA probes from GCC in the upper tail deviate from the diagonal line than those from the linear models as well as no indication of correlation structure which shows all models could perfectly deal with the correlation in the data. The summary of statistical information for all 27,647 mRNA probes is provided in Supplementary Table 2. The list of the top 20 mRNA probes from the analysis of both kinship and GCC is illustrated in Table 1. Among the list, 12 mRNA probes were identified by the GCC model and 8 mRNA probes by the linear model. The top six genes annotated from mRNA probes in a combined list of GCC and linear models were APOBEC3G (p = 1.569e-6, FDR= 0.04), H6PD (p =2.928e-5, FDR = 0.18), SLC45A1 (p = 3.027e-5, FDR = 0.18), GRIN3B (p = 3.197e-05, FDR = 0.18), PDE4D (p = 3.877e-5, FDR = 0.18), and PIGC (p = 0.0001, FDR = 0.28).
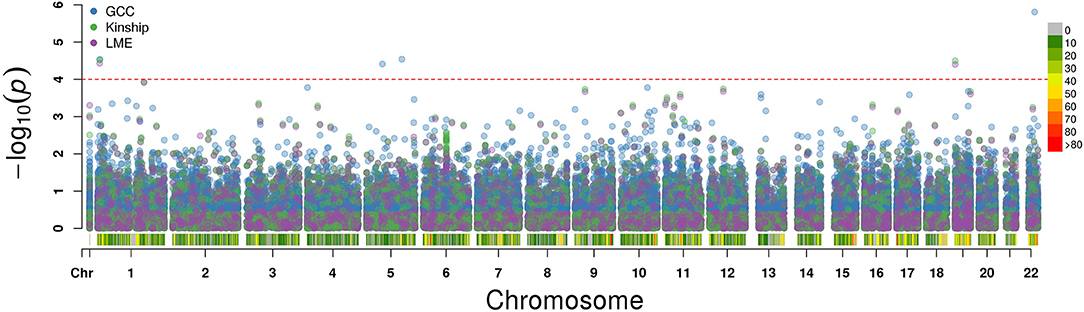
Figure 2. Manhattan plot for single mRNA probe analysis from GCC, kinship, and LME models along each chromosome.
The list of mRNA probes with p < 0.05 was used as input for over-representation analysis through the GSEA website. We identified significant functional KEGG and Reactome pathways with FDR < 3.95e-10 such as focal adhesion, ribosome, cysteine and methionine metabolism, Huntington's disease, eukaryotic translation elongation, nervous system development, influenza infection, metabolism of RNA, and cell cycle (Table 2).
We used 17 TFs as input and after the filtering and bootstrapping process, seven regulons remained, which among, five significant regulons from GSEA-2T analysis were identified with FDR < 1.3e-4. Among these significant regulons, two are positively associated with target genes, and three regulons are negatively associated with target genes. Table 3 shows the list of identified regulons with information about the number of TFs included in each regulon, a positive or negative score that gives information about the activating or repressing of the target genes, P-value, and adjusted P-value. The significant regulons were CTCF (ES = 0.89), REST (ES = 0.67), SP3 (ES = −1.24), SRF (ES = −0.87), and XBP1 (ES = −0.97) (Figure 3).
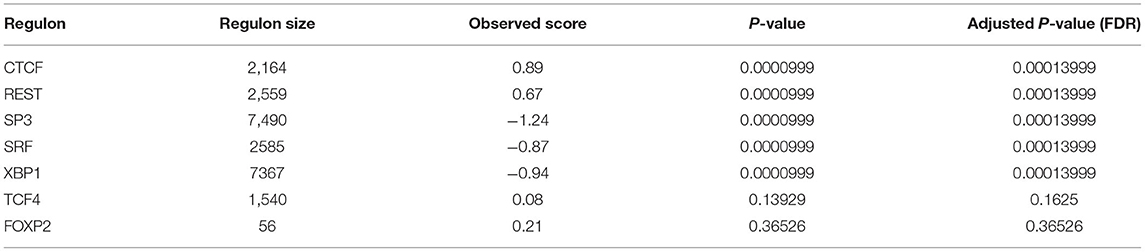
Table 3. List of seven regulons identified from GSEA-2T among which five significant regulons were identified with FDR < 1.3e-4.
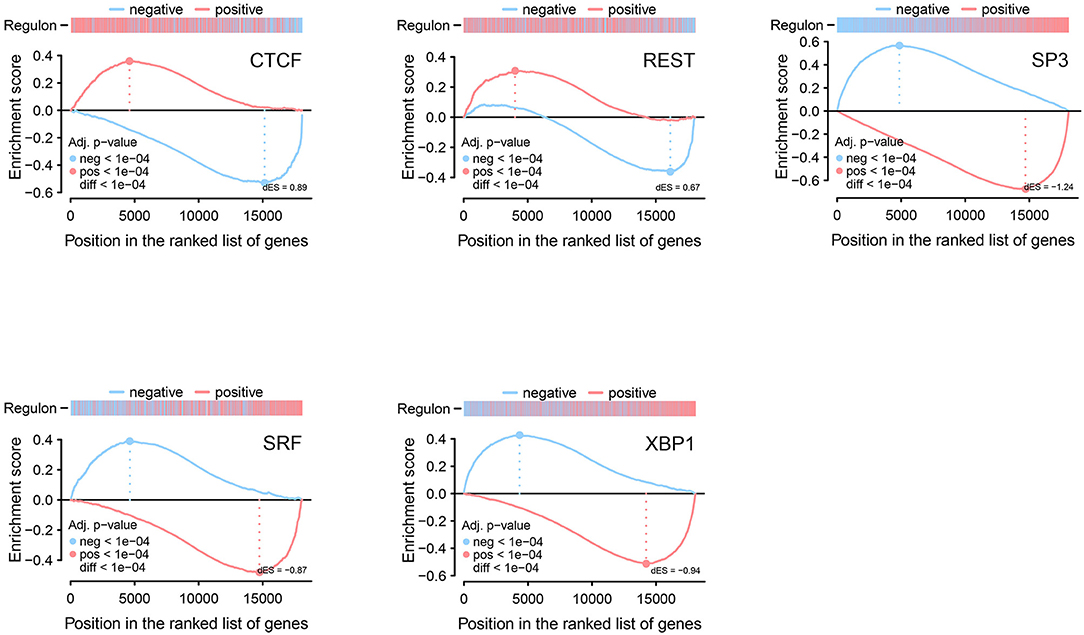
Figure 3. Two-tailed GSEA illustrating five significant identified regulons from regulatory network analysis. Regulons are split into positive and negative targets, and differential enrichment score (dES) is shown for positive (red line) and negative (blue line) targets.
Through applying both the assumption-free GCC and linear models, this exploratory study was able to capture diverse patterns of relations not limited to those from linear models. We were able to identify interesting genes and pathways implicated in cognitive function. Also, the novel transcription regulatory analysis paved the way for the detection of significant regulons associated with cognitive function.
The APOBEC3G gene belongs to a family of proteins grouped together due to their homology with the cytidine deaminase APOBEC1 (https://en.wikipedia.org/wiki/APOBEC3G). A very recent paper by Smith et al. (2020) reported that the APOBEC1 gene affected cognitive aging in animals. In addition, Cole et al. (2017) have reported that APOBEC1 is an essential regulatory mechanism of microglia (MG) function and is critical for overall brain homeostasis and healthy aging. The other gene SLC45A1, is highly expressed in the brain (https://gtexportal.org/home/). It is a protein-coding gene and associated with intellectual developmental disorder with neuropsychiatric features and autosomal recessive non-syndromic intellectual disability (https://www.genecards.org/cgi-bin/carddisp.pl?gene=SLC45A1). Srour et al. (2017) studied homozygous missense variants in SLC45A1 on four affected children from two unrelated consanguineous families with moderate to severe intellectual disability associated with epilepsy and variable neuropsychiatric features. They concluded that autosomal-recessive mutations in SLC45A1 result in intellectual disability, movement disorder, and epilepsy. SLC45A1 is thus the second cerebral glucose transporter, in addition to GLUT1, to be involved in human disease and implicated in neurodevelopmental disability. Also, SLC proteins are of great importance in the elucidation of neurodegenerative disorder mechanisms due to their important role in the synaptic regulation of neurotransmitters (Aykaç and Sehirli, 2020). Martínez-García and colleagues studied the association of rs6688832 and rs34603401 polymorphisms in the H6PD gene with obesity and polycystic ovary syndrome (PCOS) on 237 cases and 135 controls and they found that the variants in the H6PD gene were associated with obesity and PCOS (Martínez-García et al., 2012).
The other gene is PDE4D which is already reported to encode a cyclic AMP (cAMP) regulator which places PDE4D-related acrodysostosis in the same family of diseases as pseudohypoparathyroidism, pseudohypoparathyroidism, PRKAR1A-related acrodysostosis, and brachydactyly mental retardation syndrome, which are all characterized by cognitive impairment and short distal extremities (Lynch et al., 2013). Also, using gene knock-out and miRNA-induced gene knock-down in mice, the PDE4D gene and particularly its long-form isoforms has been shown to play a vital role in the mediation of memory and hippocampal neurogenesis which is mediated by cAMP signaling (Li et al., 2011; Michot et al., 2012). The gene GRIN3B is associated with depersonalization disorder and schizophrenia (https://www.genecards.org/cgi-bin/carddisp.pl?gene=GRIN3B). Additionally, in a study, Hornig et al. performed whole exome-sequencing in eight family members with a strong history of psychotic disorders over three generations and they found in all the affected family members frameshift mutation rs10666583 in the GRIN3B gene, which codes for the GluN3B subunit of the NMDA receptor (Hornig et al., 2017). NMDA receptors are ligand-gated cation channels that are blocked in the resting state by magnesium ions. They are involved in learning and memory, synaptic plasticity, and synaptogenesis (Harris et al., 1984; Normann et al., 2000; Lynch, 2004; Normann and Clark, 2005; Fan et al., 2014).
The PIGC gene is associated with diseases including glycosylphosphatidylinositol biosynthesis defect 16 and autosomal recessive non-syndromic intellectual disability (https://www.genecards.org/cgi-bin/carddisp.pl?gene=PIGC) which are both linked to intellectual disabilities. Moreover, Edvardson and colleagues studied to find disease-causing mutations in three patients from two unrelated families with severe intellectual disability, global developmental delay, and drug-responsive seizure disorder. They concluded that mutations in the PIGC gene were associated with epilepsy and intellectual disability (Edvardson et al., 2017).
We found interesting and important pathways which might be implicated in cognitive impairment (Table 2). Ding et al. (2005) reported that in patients with mild cognitive impairment and Alzheimer's disease (AD), there is significant dysfunction in ribosome function that is not observed in the cerebellum of the same patients. Ribosome dysfunction is associated with a decline rate in protein synthesis, ribosomal RNA and tRNA levels, and increased RNA oxidation. Focal adhesion involves the integration of the adhesion, the communication between the extracellular matrix and the actin cytoskeleton, and the regulation of many cell types. Loss of cell adhesion can lead to cell death and altered focal signaling has been associated with synaptic loss, which may cause AD (Caltagarone et al., 2007). Wilson et al. (2002) reported that with increasing age peripheral cytokine dysregulation interacts with cognitive aging. Magaki et al. (2007) showed that alteration in cytokines through peripheral blood mononuclear cells (PBMCs) might be detected early in mild cognitive impairment. Additionally, other studies have been done on the role of cytokines in AD, cognitive impairment, and neurological disorders (Aarli, 2005; Nagae and Araki, 2016). Chang et al. (2012) discussed that p53 interacts with cellular factors, viral factors, and small RNAs, explaining its role in the development of neurodegenerative diseases. Previous studies point to evidence of the role and pathogenesis of Huntington's disease, endocytosis, eukaryotic translation elongation, nervous system development, influenza infection, metabolism of RNA, and cell cycle in relation to cognitive impairment, AD, and neurological diseases (Peavy et al., 2010; Jurgens et al., 2012; Mufson et al., 2012; Beckelman et al., 2016; Barbash et al., 2017; Zhu et al., 2020).
A total number of five significant regulons were enriched by GSEA-2T analysis of transcriptional regulation, in which CTCF and REST were identified as activated regulons and SP3, SRF, and XBP1 as repressed regulons in our study. A gene expression study on animal adult forebrain-restricted SRF deletion reported the decreased expression of several gene-containing serum response elements (SRE). And they proposed that these deficits in gene expression indicate SRF's role in the induction of genes necessary for long-term memory formation and the late phase of long-term potentiation-like (LTP) plasticity (Ramanan et al., 2005; Etkin et al., 2006). In our study, we also found the decreased expression of TF genes included in the SRF regulon with cognitive function. Yamakawa et al. (2017) discussed that SP3 tends to be a major negative regulator of synaptic gene expression and synaptic activity, which is also likely to play a significant role in cognitive decline in AD patients. They reported that SP3 and histone deacetylase HDAC2 negatively regulate synaptic function in neurons. Our study showed the downregulation of the SP3 regulon with cognitive function. Some studies have shown the role of XBP1 in endoplasmic reticulum stress, memory, and cognition (Valdés et al., 2014; Cissé et al., 2017a), rescuing hippocampal synaptic plasticity and memory through activating the Kalirin-7 (Kal7) pathway (Cissé et al., 2017b), XBP1's expression corrected age-associated changes in synaptic function (Cabral-Miranda et al., 2020). The other TF, CTCF has been studied by Choi et al. (2021) in adult CTCF cKO mice and concluded that deficiency in CTCF results in cognitive deficits. Some studies have reported the importance of REST due to its polymorphism role in cognitive function and the activation state of REST in the aging brain, which may differentiate neuroprotection from neurodegeneration (Lu et al., 2014; Warburton et al., 2016).
Overall, through applying GCC as a complementary method along with the linear models, this exploratory study was able to detect more important and meaningful differentially expressed genes and biological pathways implicated in cognitive function. Additionally, applying transcriptional analysis could reveal the link between significant regulons and cognition which further confirms that previously noted TFs are associated with cognitive function.
According to Danish and EU legislations, transfer and sharing of individual-level data require prior approval from the Danish Data Protection Agency and require that data sharing requests are dealt with on a case-by-case basis. However, we welcome any enquiries regarding collaboration and individual requests for data sharing. Requests can be directed to JH, amhqZWxtYm9yZ0BoZWFsdGguc2R1LmRr.
The studies involving human participants were reviewed and approved by The Regional Scientific Ethical Committees for Southern Denmark (S-VF-19980072). The patients/participants provided their written informed consent to participate in this study. Written informed consent was obtained from the individual(s) for the publication of any potentially identifiable images or data included in this article.
JH and QT contributed to the conception and design. AM performed the data analysis and wrote the manuscript. All authors read and approved the final manuscript.
This study was jointly supported by the Independent Research Fund Denmark (Grant Number DFF–6110-00114), Ph.D. scholarship Grant from the Faculty of Health Science, University of Southern Denmark, and Lundbeck Foundation (grant number R170-2014-1353). JB was grateful for financial support by the VILLUM Young Investigator Grant no. 13154 and H2020 Grant REPOTRIAL no. 777111.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2021.675587/full#supplementary-material
Aarli, J. (2005). Role of cytokines in neurological disorders. Curr. Med. Chem. 10, 1931–1937. doi: 10.2174/0929867033456918
Aykaç, A., and Sehirli, A. Ö. (2020). The role of the SLC transporters protein in the neurodegenerative disorders. Clin. Psychopharmacol. Neurosci. 18, 174–187. doi: 10.9758/cpn.2020.18.2.174
Badowska, D. M., Brzózka, M. M., Kannaiyan, N., Thomas, C., Dibaj, P., Chowdhury, A., et al. (2020). Modulation of cognition and neuronal plasticity in gain- and loss-of-function mouse models of the schizophrenia risk gene Tcf4. Transl. Psychiatry. 10:343. doi: 10.1038/s41398-020-01026-7
Barbash, S., Garfinkel, B. P., Maoz, R., Simchovitz, A., Nadorp, B., Guffanti, A., et al. (2017). Alzheimer's brains show inter-related changes in RNA and lipid metabolism. Neurobiol. Dis. 106, 1–13. doi: 10.1016/j.nbd.2017.06.008
Bates, D., Mächler, M., Bolker, B. M., and Walker, S. C. (2015). Fitting linear mixed-effects models using lme4. J. Stat. Softw. 67, 1–48. doi: 10.18637/jss.v067.i01
Beckelman, B. C., Zhou, X., Keene, C. D., and Ma, T. (2016). Impaired eukaryotic elongation factor 1A expression in Alzheimer's disease. Neurodegen. Dis. 16, 39–43. doi: 10.1159/000438925
Benjamini, Y., and Hochberg, Y. (1995). Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B 57, 289–300. doi: 10.1111/j.2517-6161.1995.tb02031.x
Cabral-Miranda, F., Tamburini, G., Martinez, G., Medinas, D., Gerakis, Y., Miedema, T., et al. (2020). Control of mammalian brain aging by the unfolded protein response (UPR). bioRxiv, 2020.04.13.039172. doi: 10.1101/2020.04.13.039172
Caltagarone, J., Jing, Z., and Bowser, R. (2007). Focal adhesions regulate Aβ signaling and cell death in Alzheimer's disease. Biochim. Biophys. Acta - Mol. Basis Dis. 1772, 438–445. doi: 10.1016/j.bbadis.2006.11.007
Castro, M. A. A., De Santiago, I., Campbell, T. M., Vaughn, C., Hickey, T. E., Ross, E., et al. (2015). Regulators of genetic risk of breast cancer identified by integrative network analysis. Nat. Genet. 48, 12–21. doi: 10.1038/ng.3458
Chang, J. R., Ghafouri, M., Mukerjee, R., Bagashev, A., Chabrashvili, T., and Sawaya, B. E. (2012). Role of p53 in neurodegenerative diseases. Neurodegener. Dis. 9, 68–80. doi: 10.1159/000329999
Choi, D., Il, K.im, M., Kim, S., Yu, N. K., and Kwak, C., Seo, H., et al. (2021). Conditional knock out of transcription factor CTCF in excitatory neurons induces cognitive deficiency. Mol. Brain. 14:1. doi: 10.1186/s13041-020-00716-z
Cissé, M., Duplan, E., and Checler, F. (2017a). The transcription factor XBP1 in memory and cognition: implications in Alzheimer's disease. Mol. Med. 22, 905–917. doi: 10.2119/molmed.2016.00229
Cissé, M., Duplan, E., Lorivel, T., Dunys, J., Bauer, C., Meckler, X., et al. (2017b). The transcription factor XBP1s restores hippocampal synaptic plasticity and memory by control of the Kalirin-7 pathway in Alzheimer model. Mol. Psychiatry 22, 1562–1575. doi: 10.1038/mp.2016.152
Cole, D. C., Chung, Y., Gagnidze, K., Hajdarovic, K. H., Rayon-Estrada, V., Harjanto, D., et al. (2017). Loss of APOBEC1 RNA-editing function in microglia exacerbates age-related CNS pathophysiology. Proc. Natl. Acad. Sci. U.S.A. 114, 13272–13277. doi: 10.1073/pnas.1710493114
Ding, Q., Markesbery, W. R., Chen, Q., Li, F., and Keller, J. N. (2005). Ribosome dysfunction is an early event in Alzheimer's disease. J. Neurosci. 25, 9171–9175. doi: 10.1523/JNEUROSCI.3040-05.2005
Edvardson, S., Murakami, Y., Nguyen, T. T. M., Shahrour, M., St-Denis, A., Shaag, A., et al. (2017). Mutations in the phosphatidylinositol glycan C (PIGC) gene are associated with epilepsy and intellectual disability. J. Med. Genet. 54, 196–201. doi: 10.1136/jmedgenet-2016-104202
Etkin, A., Alarcón, J. M., Weisberg, S. P., Touzani, K., Huang, Y. Y., Nordheim, A., et al. (2006). A Role in Learning for SRF: deletion in the adult forebrain disrupts Ltd and the formation of an immediate memory of a novel context. Neuron 50, 127–143. doi: 10.1016/j.neuron.2006.03.013
Fan, X., Jin, W. Y., and Wang, Y. T. (2014). The NMDA receptor complex: a multifunctional machine at the glutamatergic synapse. Front. Cell. Neurosci. 8:160. doi: 10.3389/fncel.2014.00160
Harries, L. W., Bradley-Smith, R. M., Llewellyn, D. J., Pilling, L. C., Fellows, A., Henley, W., et al. (2012). Leukocyte CCR2 expression is associated with mini-mental state examination score in older adults. Rejuvenation Res. 15, 395–404. doi: 10.1089/rej.2011.1302
Harris, E. W., Ganong, A. H., and Cotman, C. W. (1984). Long-term potentiation in the hippocampus involves activation of N-methyl-D-aspartate receptors. Brain Res. 323, 132–137. doi: 10.1016/0006-8993(84)90275-0
Hornig, T., Grüning, B., Kundu, K., Houwaart, T., Backofen, R., Biber, K., et al. (2017). GRIN3B missense mutation as an inherited risk factor for schizophrenia: Whole-exome sequencing in a family with a familiar history of psychotic disorders. Genet. Res. (Camb). 99:e1. doi: 10.1017/S0016672316000148
Hwang, J. Y., and Zukin, R. S. (2018). REST, a master transcriptional regulator in neurodegenerative disease. Curr. Opin. Neurobiol. 48, 193–200. doi: 10.1016/j.conb.2017.12.008
Jurgens, H. A., Amancherla, K., and Johnson, R. W. (2012). Influenza infection induces neuroinflammation, alters hippocampal neuron morphology, and impairs cognition in adult mice. J. Neurosci. 32:3958–3968. doi: 10.1523/JNEUROSCI.6389-11.2012
Lambert, S. A., Jolma, A., Campitelli, L. F., Das, P. K., Yin, Y., Albu, M., et al. (2018). The human transcription factors. Cell 172, 650–665. doi: 10.1016/j.cell.2018.01.029
Li, Y. F., Cheng, Y. F., Huang, Y., Conti, M., Wilson, S. P., O'Donnell, J. M., et al. (2011). Phosphodiesterase-4D knock-out and RNA interference-mediated knock-down enhance memory and increase hippocampal neurogenesis via increased cAMP signaling. J. Neurosci. 31, 172–183. doi: 10.1523/JNEUROSCI.5236-10.2011
Lu, T., Aron, L., Zullo, J., Pan, Y., Kim, H., Chen, Y., et al. (2014). REST and stress resistance in ageing and Alzheimer's disease. Nature 507, 448–454. doi: 10.1038/nature13163
Lynch, D. C., Dyment, D. A., Huang, L., Nikkel, S. M., Lacombe, D., Campeau, P. M., et al. (2013). Identification of novel mutations confirms Pde4d as a major gene causing acrodysostosis. Hum. Mutat. 34, 97–102. doi: 10.1002/humu.22222
Lynch, M. A. (2004). Long-term potentiation and memory. Physiol. Rev. 84, 87–136. doi: 10.1152/physrev.00014.2003
Magaki, S., Mueller, C., Dickson, C., and Kirsch, W. (2007). Increased production of inflammatory cytokines in mild cognitive impairment. Exp. Gerontol. 42, 233–240. doi: 10.1016/j.exger.2006.09.015
Manolopoulos, K. N., Klotz, L. O., Korsten, P., Bornstein, S. R., and Barthel, A. (2010). Linking Alzheimer's disease to insulin resistance: the FoxO response to oxidative stress. Mol. Psychiatry 15, 1046–1052. doi: 10.1038/mp.2010.17
Margolin, A. A., Nemenman, I., Basso, K., Wiggins, C., Stolovitzky, G., Favera, R. D., et al. (2006). ARACNE: an algorithm for the reconstruction of gene regulatory networks in a mammalian cellular context. BMC Bioinform. 7:S7. doi: 10.1186/1471-2105-7-S1-S7
Martínez-García, M. A., San-Millán, J. L., and Escobar-Morreale, H. F. (2012). The R453Q and D151A polymorphisms of Hexose-6-Phosphate Dehydrogenase Gene (H6PD) influence the polycystic ovary syndrome (PCOS) and obesity. Gene. 497, 38–44. doi: 10.1016/j.gene.2012.01.047
McGue, M., and Christensen, K. (2002). The heritability of level and rate-of-change in cognitive functioning in Danish twins aged 70 years and older. Exp. Aging Res. 28, 435–451. doi: 10.1080/03610730290080416
Michot, C., Le Goff, C., Goldenberg, A., Abhyankar, A., Klein, C., Kinning, E., et al. (2012). Exome sequencing identifies PDE4D mutations as another cause of acrodysostosis. Am. J. Hum. Genet. 90, 740–745. doi: 10.1016/j.ajhg.2012.03.003
Mohammadnejad, A., Nygaard, M., Li, S., Zhang, D., Xu, C., Li, W., et al. (2020). Generalized correlation coefficient for genome-wide association analysis of cognitive ability in twins. Aging (Albany. NY). 12, 22457–22494. doi: 10.18632/aging.104198
Mohammadnejad, A., Soerensen, M., Baumbach, J., Mengel-From, J., Li, W., Lund, J., et al. (2021). Novel DNA methylation marker discovery by assumption-free genome-wide association analysis of cognitive function in twins. Aging Cell. 20:e13293. doi: 10.1111/acel.13293
Mozzi, A., Guerini, F. R., Forni, D.i., Costa, A. S., Nemni, R., Baglio, F., et al. (2017). REST, a master regulator of neurogenesis, evolved under strong positive selection in humans and in non human primates. Sci. Rep. 7:9530. doi: 10.1038/s41598-017-10245-w
Mufson, E. J., Binder, L., Counts, S. E., Dekosky, S. T., Detoledo-Morrell, L., Ginsberg, S. D., et al. (2012). Mild cognitive impairment: pathology and mechanisms. Acta Neuropathol. 123, 13–30. doi: 10.1007/s00401-011-0884-1
Murrell, B., Murrell, D., and Murrell, H. (2016). Discovering general multidimensional associations. PLoS ONE 11:e0151551. doi: 10.1371/journal.pone.0151551
Nagae, T., and Araki, K. (2016). Cytokines and cytokine receptors involved in the pathogenesis of Alzheimers disease. J. Clin. Cell. Immunol. 7:441. doi: 10.4172/2155-9899.1000441
Nonaka, M., Kim, R., Sharry, S., Matsushima, A., Takemoto-Kimura, S., and Bito, H. (2014). Towards a better understanding of cognitive behaviors regulated by gene expression downstream of activity-dependent transcription factors. Neurobiol. Learn. Mem. 115, 21–29. doi: 10.1016/j.nlm.2014.08.010
Normann, C., and Clark, K. (2005). Selective modulation of Ca(2+) influx pathways by 5-HT regulates synaptic long-term plasticity in the hippocampus. Brain Res. 1037, 187–193. doi: 10.1016/j.brainres.2005.01.001
Normann, C., Peckys, D., Schulze, C. H., Walden, J., Jonas, P., and Bischofberger, J. (2000). Associative long-term depression in the hippocampus is dependent on postsynaptic N-type Ca2+ channels. J. Neurosci. 20, 8290–8297. doi: 10.1523/JNEUROSCI.20-22-08290.2000
Nygaard, M., Larsen, M. J., Thomassen, M., McGue, M., Christensen, K., Tan, Q., et al. (2019). Global expression profiling of cognitive level and decline in middle-aged monozygotic twins. Neurobiol. Aging 84, 141–147. doi: 10.1016/j.neurobiolaging.2019.08.019
Peavy, G. M., Jacobson, M. W., Goldstein, J. L., Hamilton, J. M., Kane, A., Gamst, A. C., et al. (2010). Cognitive and functional decline in Huntington's disease: dementia criteria revisited. Mov. Disord. 25, 1163–1169. doi: 10.1002/mds.22953
Petersen, I., McGue, M., Tan, Q., Christensen, K., and Christiansen, L. (2016). Change in depression symptomatology and cognitive function in twins: a 10-year follow-up study. Twin Res. Hum. Genet. Off. J. Int. Soc. Twin Stud. 19, 104–111. doi: 10.1017/thg.2016.3
Ramanan, N., Shen, Y., Sarsfield, S., Lemberger, T., Schütz, G., Linden, D. J., et al. (2005). SRF mediates activity-induced gene expression and synaptic plasticity but not neuronal viability. Nat. Neurosci. 8, 759–767. doi: 10.1038/nn1462
Reshef, D. N., Reshef, Y. A., Finucane, H. K., Grossman, S. R., McVean, G., Turnbaugh, P. J., et al. (2011). Detecting novel associations in large data sets. Science 334, 1518–1524. doi: 10.1126/science.1205438
Ritchie, M. E., Silver, J., Oshlack, A., Holmes, M., Diyagama, D., Holloway, A., et al. (2007). A comparison of background correction methods for two-colour microarrays. Bioinformatics 23, 2700–2707. doi: 10.1093/bioinformatics/btm412
Sinnwell, J. P., Therneau, T. M., and Schaid, D. J. (2014). The kinship2 R package for pedigree data. Hum. Hered. 78, 91–93. doi: 10.1159/000363105
Smith, G., Rani, A., Kumar, A., Barter, J., and Foster, T. C. (2020). Hippocampal subregion transcriptomic profiles reflect strategy selection during cognitive aging. J. Neurosci. 40, 4888–4899. doi: 10.1523/JNEUROSCI.2944-19.2020
Speed, T. (2011). A correlation for the 21st century. Science 334, 1502–1503. doi: 10.1126/science.1215894
Srour, M., Shimokawa, N., Hamdan, F. F., Nassif, C., Poulin, C., Al Gazali, L., et al. (2017). Dysfunction of the cerebral glucose transporter SLC45A1 in individuals with intellectual disability and epilepsy. Am. J. Hum. Genet. 100, 824–830. doi: 10.1016/j.ajhg.2017.03.009
Tan, Q., Thomassen, M., Burton, M., Mose, K. F., Andersen, K. E., Hjelmborg, J., et al. (2017). Generalized correlation coefficient for non-parametric analysis of microarray time-course data. J. Integr. Bioinform. 14. doi: 10.1515/jib-2017-0011
Valdés, P., Mercado, G., Vidal, R. L., Molina, C., Parsons, G., Court, F. A., et al. (2014). Control of dopaminergic neuron survival by the unfolded protein response transcription factor XBP1. Proc. Natl. Acad. Sci. U. S. A. 111, 6804–6809. doi: 10.1073/pnas.1321845111
Wang, G.-Z., and Konopka, G. (2013). Decoding human gene expression signatures in the brain. Transcription 4, 102–108. doi: 10.4161/trns.24885
Warburton, A., Miyajima, F., Shazadi, K., Crossley, J., Johnson, M. R., Marson, A. G., et al. (2016). NRSF and BDNF polymorphisms as biomarkers of cognitive dysfunction in adults with newly diagnosed epilepsy. Epilepsy Behav. 54, 117–127. doi: 10.1016/j.yebeh.2015.11.013
Wilson, C. J., Finch, C. E., and Cohen, H. J. (2002). Cytokines and cognition - The case for a head-to-toe inflammatory paradigm. J. Am. Geriatr. Soc. 50, 2041–2056. doi: 10.1046/j.1532-5415.2002.50619.x
Xiao, D., Liu, X., Zhang, M., Zou, M., Deng, Q., Sun, D., et al. (2018). Direct reprogramming of fibroblasts into neural stem cells by single non-neural progenitor transcription factor Ptf1a. Nat. Commun. 9, 2865. doi: 10.1038/s41467-018-05209-1
Yamakawa, H., Cheng, J., Penney, J., Gao, F., Rueda, R., Wang, J., et al. (2017). The transcription factor Sp3 cooperates with HDAC2 to regulate synaptic function and Plasticity in neurons. Cell Rep. 20, 1319–1334. doi: 10.1016/j.celrep.2017.07.044
Keywords: cognitive aging, transcript, generalized correlation coefficient, linear regression, twins, regulons
Citation: Mohammadnejad A, Li W, Lund JB, Li S, Larsen MJ, Mengel-From J, Michel TM, Christiansen L, Christensen K, Hjelmborg J, Baumbach J and Tan Q (2021) Global Gene Expression Profiling and Transcription Factor Network Analysis of Cognitive Aging in Monozygotic Twins. Front. Genet. 12:675587. doi: 10.3389/fgene.2021.675587
Received: 03 March 2021; Accepted: 26 April 2021;
Published: 14 June 2021.
Edited by:
Ignazio Piras, Translational Genomics Research Institute, United StatesReviewed by:
Mauro Pala, National Research Council (CNR), ItalyCopyright © 2021 Mohammadnejad, Li, Lund, Li, Larsen, Mengel-From, Michel, Christiansen, Christensen, Hjelmborg, Baumbach and Tan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Qihua Tan, cXRhbkBoZWFsdGguc2R1LmRr
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.