- 1Key Laboratory of Freshwater Fisheries and Germplasm Resources Utilization, Ministry of Agriculture, Freshwater Fisheries Research Center, Chinese Academy of Fishery Sciences, Wuxi, China
- 2Shenzhen Key Lab of Marine Genomics, Guangdong Provincial Key Lab of Molecular Breeding in Marine Economic Animals, BGI Academy of Marine Sciences, BGI Marine, Shenzhen, China
- 3BGI Education Center, University of Chinese Academy of Sciences, Shenzhen, China
- 4BGI Zhenjiang Institute of Hydrobiology, Zhenjiang, China
The mandarin fish, Siniperca chuatsi, is an economically important perciform species with widespread aquaculture practices in China. Its special feeding habit, acceptance of only live prey fishes, contributes to its delicious meat. However, little is currently known about related genetic mechanisms. Here, we performed whole-genome sequencing and assembled a 758.78 Mb genome assembly of the mandarin fish, with the scaffold and contig N50 values reaching 2.64 Mb and 46.11 Kb, respectively. Approximately 92.8% of the scaffolds were ordered onto 24 chromosomes (Chrs) with the assistance of a previously established genetic linkage map. The chromosome-level genome contained 19,904 protein-coding genes, of which 19,059 (95.75%) genes were functionally annotated. The special feeding behavior of mandarin fish could be attributable to the interaction of a variety of sense organs (such as vision, smell, and endocrine organs). Through comparative genomics analysis, some interesting results were found. For example, olfactory receptor (OR) genes (especially the beta and delta types) underwent a significant expansion, and endocrinology/vision related npy, spexin, and opsin genes presented various functional mutations. These may contribute to the special feeding habit of the mandarin fish by strengthening the olfactory and visual systems. Meanwhile, previously identified sex-related genes and quantitative trait locis (QTLs) were localized on the Chr14 and Chr17, respectively. 155 toxin proteins were predicted from mandarin fish genome. In summary, the high-quality genome assembly of the mandarin fish provides novel insights into the feeding habit of live prey and offers a valuable genetic resource for the quality improvement of this freshwater fish.
Introduction
The mandarin fish, Siniperca chuatsi, belonging to the family Percichthyidae and order Perciformes, has a relatively high market value and widespread aquaculture throughout China (Liang and Cui, 1982; Liu et al., 1998). It has a special feeding habit, accepting only live prey fishes and refusing dead food items in the wild (Chiang, 1959; Liu et al., 1998). The feeding behaviors of the mandarin fish require interactions of a variety of sense organs, such as eyes, mouth, lateral lines, and olfactory organs. Lateral-line may help alert the fish to vibrations that are made by nearby prey or approaching predators (Engelmann et al., 2000). Although the mandarin fish can feed properly on live prey fishes depending mainly on eyes and lateral-line, it can hunt prey fishes without these two organs (Liang et al., 1998). Researchers observed that the mandarin fish could recognize prey fishes using vision (Wu, 1988). A previous study (Liang et al., 1998) reported that the mandarin fish usually stayed more frequently near a perforated opaque cylinder containing live prey fishes rather than those without prey fishes, suggesting the importance of olfaction in searching for prey. However, this conclusion did not justify the food smells from other stimuli (such as hydromechanical stimulus).
Fish toxins have been poorly studied compared to venoms from other animals such as snakes, scorpions, spiders, and cone snails (Utkin, 2015). It is estimated that there are up to 2,900 venomous fishes (Xie et al., 2017) with venom systems convergently evolved 19 times (Harris and Jenner, 2019). Mandarin fish is one of those who can produce toxins in their hard spines to help them defense and prey, and cause pain and swelling at the site of the sting in human as well (Zhang F.-B. et al., 2019). However, apart from several antimicrobial peptides that can be regarded as toxins (Sun et al., 2007), there is no detailed report on venom genes and components of this fish yet.
In Mandarin fish species, females grow faster than males. Whether female mandarin fish have stronger predation ability is still unknown, so gender screening is of great significance to the cultivation of mandarin fish. So far, several gender-related molecular markers or functional genes have been screened, and even all-female mandarin fish have been bred (Guo et al., 2021; Liu et al., 2021). However, due to the lack of available genomic and transcriptome information, the mechanisms of sex differentiation remain poorly understood.
By far, genome data of the mandarin fish have been limited, which restricts genetic information for functional genomics studies. Therefore, in this study, we report a chromosome-level genome assembly of the mandarin fish using a combination of next-generation sequencing and previously reported genetic linkage map. The subsequent comparative genomic analysis provides novel insights into the feeding habit of live prey, toxin, and sex differentiation in the mandarin fish. This genome can not only serve as the genetic basis for in-depth investigations of fish evolution and biological functions but also offers a valuable genetic resource for quality improvement of this economically important fish.
Materials and Methods
Sample Collection, Library Construction, and Sequencing
We collected muscle samples and extracted genomic DNA from a mandarin fish (Figure 1), which was obtained from Freshwater Fisheries Research Center of Chinese Academy of Fishery Sciences, Wuxi City, Jiangsu Province, China. The extracted DNA was used to construct seven libraries, including three short-insert (270, 500, and 800 bp) and four long-insert (2, 5, 10, and 20 kb) libraries. Subsequently, applying the routine whole-genome shotgun sequencing strategy, we sequenced these libraries on a Hiseq2500 platform (Illumina, San Diego, CA, United States). Those raw reads with adapters or low-quality sequences were filtered by a SOAPfilter (v2.2) (Luo et al., 2015).
All experiments were carried out following the guidelines of the Animal Ethics Committee and were approved by the Institutional Review Board on Bioethics and Biosafety of BGI, China (No. FT 18134).
Estimation of the Genome Size and Generation of a Genome Assembly
We performed a 17-mer distribution analysis to estimate the target genome size using the clean reads from the short-insert libraries (Liu et al., 2013). The calculation of genome size was based on the following formula: G = knum/kdepth. Here, knum is the sequenced k-mer number and kdepth is the k-mer sequencing depth. We set optimized parameters (pregraph-K 41 –d 1; contig –M 1; scaff –b 1.5) for the SOAPdenovo2 software (v2.04) to generate contigs and original scaffolds (Luo et al., 2012). Subsequently, we employed GapCloser (v1.12; with parameter settings of −t 8 −l 150) to fill the gaps of intra-scaffolds (Li et al., 2009) using the clean reads from short-insert libraries (270, 500, and 800 bp). Finally, we used BUSCO (Benchmarking Universal Single-Copy Orthologs; v1.22) to assess genome integrity (Simao et al., 2015).
Repeat Annotation, Gene Prediction, and Functional Annotation
Repetitive sequences including tandem repeats and transposable elements (TEs) were predicted from the assembled genome. We used Tandem Repeats Finder (v4.07) to search for tandem repeats (Benson, 1999). RepeatMasker (v4.0.6) and RepeatProteinMask (v4.0.6) were employed to detect known TEs based on the Repbase TE library (Tarailo-Graovac and Chen, 2009; Bao et al., 2015). Besides, we used RepeatModeler (v1.0.8) and LTR_FINDER (v1.0.6) with default parameters to generate the de novo repeat library (Xu and Wang, 2007; Abrusan et al., 2009), and RepeatMasker was applied to search the repeat regions against the built repeat library.
We utilized three different approaches to annotate structures of predicted genes in our assembled genome, including de novo prediction, homology-based prediction, and transcriptome-based prediction. For the de novo prediction, we employed AUGUSTUS (v3.2.1) and GENSCAN (v1.0) to identify protein-coding genes within the mandarin fish genome, using the repeat-masked genome as the template (Burge and Karlin, 1997; Stanke et al., 2006). For the homology-based prediction, we aligned the homologous proteins of five other fish species, including zebrafish (Danio rerio), three-spined stickleback (Gasterosteus aculeatus), Nile tilapia (Oreochromis niloticus), medaka (Oryzias latipes), and fugu (Takifugu rubripes) (downloaded from Ensembl 83 release), to the repeat-masked genome using tblastn (v2.2.26) with an E-value ≤ 1e-5 (Mount, 2007; Cunningham et al., 2015). Subsequently, Solar (v0.9.6) and GeneWise (v2.4.1) (Birney and Durbin, 2000) were executed to define the potential gene structures for all alignments. The RNA-seq data from muscle tissues were aligned to the assembled genome also using tophat (v2.0.13) (Trapnell et al., 2009) and gene structures were predicted using cufflinks (v2.1.1) (Trapnell et al., 2012). Finally, we combined the above-mentioned three datasets to obtain a consistent and comprehensive gene set by GLEAN (v1.0) (Elsik et al., 2007).
These predicted coding proteins of the mandarin fish were aligned against public gene ontology (GO), Kyoto Encyclopedia of Genes and Genomes (KEGG), Swiss-Prot and TrEMBL databases for annotation of functions and pathways by using BLASTP (Kanehisa and Goto, 2000; Boeckmann et al., 2003; Harris et al., 2004). Subsequently, we applied InterProScan (v5.16−55.0) to identify functional motifs and domains through Pfam, PRINTS, ProDom, and SMART databases (Attwood, 2002; Letunic et al., 2004; Bru et al., 2005; Hunter et al., 2009; Finn et al., 2014).
Genome Evolution Analysis
Protein sequences of five ray-fin fishes, including spotted gar (Lepisosteus osseus), zebrafish, three-spined stickleback, medaka, and fugu were downloaded from the Ensembl (release-83) (Cunningham et al., 2015). Protein sequences of Asian arowana (Scleropages formosus, assembly fSclFor1.1) and giant-fin mudskipper (Periophthalmus magnuspinnatus, assembly fPerMag1.pri) were obtained from our recent works (You et al., 2014; Bian et al., 2016). Subsequently, OrthoMCL (v1.4) was executed to cluster the predicted mandarin fish genes into families (Li et al., 2003). We then identified and selected one-to-one orthologs from the above-mentioned eight teleost species, and finally used MUSCLE (v3.8.31) to perform multiple sequence alignment and PhyML (v3.0) to construct a phylogenetic tree (Edgar, 2004; Guindon et al., 2009, 2010).
Pseudo-Chromosome Construction
Single nucleotide polymorphisms (SNPs)-containing reads in the genetic linkage map of S. chuatsi (Sun et al., 2017) were mapped to our assembled mandarin fish genome, and the best hit reads were selected. Linkage groups (LGs) were assigned using the JoinMap4.1 software (Van Ooijen, 2006). Subsequently, a genetic linkage map of the mandarin fish was reconstructed, and SNPs in the genetic linkage map were used for assembling chromosomes. Based on genetic distances between these SNP markers, we determined the position and orientation of each scaffold and then anchored these scaffolds to construct pseudo-chromosomes.
To perform the genome synteny analysis, we downloaded genome sequences of European sea bass (Dicentrarchus labrax) from NCBI (Tine et al., 2014) as a reference. Genome-wide alignments were performed using lastz (Kurtz et al., 2004), and the best homology segments were selected using perl scripts. The final genomic synteny was visualized using the Circos software (Krzywinski et al., 2009).
Localization of Sex-Related Genes and Quantitative Trait Locis on Chromosomes
To identify candidate genes for underlying sex dimorphisms, we downloaded 81 putative sex-related genes from NCBI (Eshel et al., 2012, 2014; Zeng et al., 2016). The distribution of sex-related genes on chromosomes were determined by homologous sequence alignment.
Identification of leptin, Neuropeptide (npy), and spexin Genes From Teleost Fish Genomes
Protein sequences of three food intake genes, including leptin (NP_001122048.1), npy (AAI62071.1) and spexin (XP_010740053.1) protein sequences were downloaded from NCBI. We performed tblastn (v2.26) (Mount, 2007) with default parameters searching against the mandarin fish, large yellow croaker (Pseudosciaena crocea; assembly L_crocea_2.0), grass carp (Ctenopharyngodon idella; assembly CI01), yellowtail (Seriola dumerili; assembly Sdu_1.0), kingfish (Seriola lalandi; assembly Sedor1), orange-spotted grouper (Epinephelus coioides), sea bass (Lates calcarifer; assembly ASM164080v1), and juvenile ovate pompano (Trachinotus ovatus; Zhang D.-C. et al., 2019) genome sequences. Subsequently, Genewise (v2.2.0) was applied to extract the best alignment results (Birney et al., 2004).
Identification of Olfactory Receptor and Taste Receptor Genes From Genome Sequences
We used zebrafish and pufferfish olfactory receptor (OR) protein sequences (Supplementary Table 1) as the queries to extract the OR genes in the Mandarin fish, zebrafish, fugu, stickleback, medaka, giant-fin mudskipper, Asian arowana and spotted gar (following the method mentioned in the previous section).
Protein sequences of five different types of taste receptor (TR) genes, including sour TR genes (D. rerio: ENSDARP 00000119061), sweet TR genes (D. rerio: NP_001077325.1, NP_001034920.1, and T. rubripes: NP_001091094.1), umami TR genes (D. rerio: NP_001034614.2, NP_001034717.1 and T. rubripes: NP_001098687.1, NP_001072097.1), bitter TR genes (D. rerio: ENSDARG00000079880), salty TR genes (Homo sapiens: NP_001153048.1, NP_000327.2, and NP_001030.2), were downloaded from the NCBI or Ensembl database. We performed tblastn (Blast v2.26; Pevsner, 2005) with default parameters to search against these genome sequences. Subsequently, Genewise v2.2.0 (Birney et al., 2004) was employed to extract the best alignment results.
Proteins sequences were aligned by the MAFFT (v7.237) program (Katoh et al., 2002) with the einsi module. Phylogenetic trees were constructed using the PhyML (v3.0) program with bootstrap set to 1,000 (Guindon et al., 2010).
Identification of opsin Genes From Ray-Finned Fish Genomes
In this study, we chose eight teleost genomes to extract opsin protein sequences, including the mandarin fish, Asian arowana, mudskipper, spotted gar, medaka, stickleback, fugu, and zebrafish. Protein sequences of opsin genes (LWS-1: ENSDARP00000065940, LWS-2: ENSDARP00000149112, SWS-1: ENSDARP00000067159, SWS-2: ENSDARP00000144766, RH1: ENSDARP00000011562, RH2-1: ENSDARP00000001158, RH2-2: ENSDARP00000011837, RH2-3: ENSDARP00000001943, and RH2-4: ENSDARP000 00000979) from zebrafish were downloaded from the Ensembl database as the queries. We performed tblastn (v2.2.28) (Mount, 2007) to align these sequences. Finally, Exonerate (v2.2.0) (Slater and Birney, 2005) was employed to predict the perfect alignment results. Multiple sequence alignment of these predicted opsin genes was performed with the Muscle module in MEGA (v 7.0) (Kumar et al., 2016). They were then translated into protein sequences for phylogenetic analyses. Phylogenetic trees were constructed using the PhyML (v3.0) program with bootstrap set to 1,000 (Guindon et al., 2010).
Venom Proteins Prediction
Protein sequences of animal venoms and toxins (Supplementary Table 2) were downloaded from UniProtKB/Swiss-Prot (UniProt Consortium, 2018) via the Animal Toxin Annotation Project (Jungo et al., 2012). These protein sequences were then filtered and only reviewed references (7,093 in total) were maintained as the trust-worthy input queries for searching. Firstly, we blasted the reviewed toxins against the coding sequences (CDS) predicted from the mandarin fish genome assembly using blastp (Camacho et al., 2009) with an e-value of 1e-10. Subsequently, the mapped sequences of mostly partial or fragmented genes with aligning ratios less than 75% were discarded, and the remaining 195 hits were further filtered manually according to the constrained lengths of the venom sequences within the same family, conserved patterns (e.g., disulfide bonds) and other post-translational modifications (PTMs).
Results
Genome Sequencing and Assembly
Seven libraries including three short-insert (270, 500, and 800 bp) and four long-insert (2, 5, 10, and 20 kb) were constructed to generate a total of 327 Gb raw reads (Supplementary Table 3). Subsequently, these raw data were filtered, and 233 Gb clean data were obtained for subsequent genome assembly.
We calculated the genome size using the following formula: G = knum/kdepth. Here, the knum (i.e., k-mer number) was 43,888,350,480 and the kdepth (k-mer depth) was 59. Therefore, the estimated genome size of the mandarin fish is about 743.87 Mb (Supplementary Table 4 and Supplementary Figure 1).
We generated contigs and original scaffolds by paired-end reads to assemble the mandarin fish genome. After filling the gaps of intra-scaffolds, we obtained a 758.78-Mb genome assembly for the mandarin fish, with contig and scaffold N50 values of 46.11 Kb and 2.64 Mb, respectively (Table 1).
Using BUSCO analysis to determine the completeness of our assembly, it is found that the assembly contained 86.1% complete, 3.0% duplicated, 9.3% fragmented, and 3.7% missed BUSCOs. Besides, 74.6% of the clean reads of RNAseq could be mapped to the genome assembly. These results suggested that our genome assembly was relatively complete.
Chromosome-Level Genome Assembly
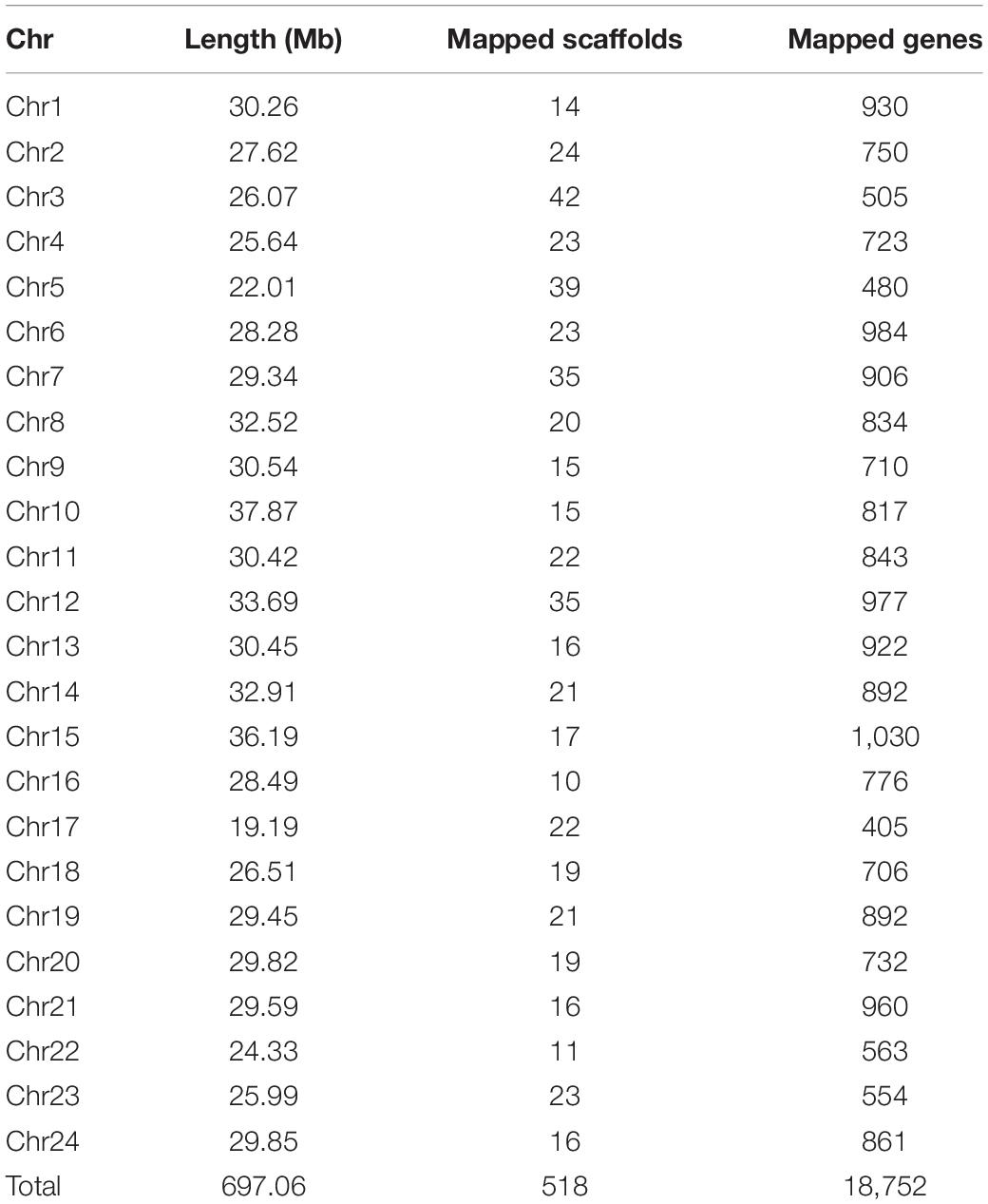
Based on the previously reported genetic linkage map of the mandarin fish (Sun et al., 2017), we anchored a total of 518 scaffolds into 24 chromosomes (Chr). A total of 697.06 Mb was assembled, corresponding to 92.8% of the assembled genome and 18,752 genes (from a total of 19,904 genes). The largest chromosome was Chr10 with 37.87 Mb in length containing 56 scaffolds, and the smallest was Chr17 with 19.19 Mb containing 22 scaffolds. The average chromosome length was 29 Mb with 21 scaffolds (Table 2 and Figure 2A).

Figure 2. Chromosome-level genome assembly of the mandarin fish. (A) A Circos atlas presents details of the pseudo-chromosome information from outside to inside: (I) the length of each pseudo-chromosome, (II) density of SNP distribution in each 100-kb genomic interval, (III) density of gene distribution in each 100-kb genomic interval, (IV) GC content of 100-kb genomic intervals, and (V) schematic presentation of major inter-chromosomal relationships in the mandarin fish genome. (B) A synteny comparison between mandarin fish and seabass genomes (chr: mandarin and LG: seabass) revealed high accuracy of our assembled genome for the mandarin fish.
There were 39,689 synteny blocks (>2 kb) between the assembled genomes of the mandarin fish and the reported European sea bass (Tine et al., 2014). We observed that almost all chromosomes showed the 1:1 synteny relationship, with an exception of Chr21 in the mandarin fish that aligned to two seabass chromosomes (Figure 2B). The results of collinearity between mandarin fish and European sea bass indicate that our chromosome assembly results are reliable.
Genome Annotation
Repeat sequences were identified based on homology search against the Repbase database and de novo prediction. We predicted that the mandarin fish genome contained 26.3% of repetitive elements. Compared with other perciforme fish, the mandarin fish was lower than red-spotted grouper (43.02%), giant grouper (45.1%), but much higher than large yellow croaker (18.1%) and golden pompano (20.25%) in the repeat sequence percentage. The most abundant TEs were long interspersed elements (13.96% of the genome), followed by DNA transposons (9.66%) and long terminal repeats (LTRs, 5.04%) (Supplementary Table 5).
Based on the genome with repeated elements masked, we integrated homology searching, de novo, and transcript methods to predict that the mandarin fish genome had 19,904 protein-coding genes (Supplementary Table 6), of which 19,059 (95.75%) genes were functionally annotated by at least one of the InterPro, GO, KEGG, Swiss-Prot, and TrEMBL protein databases (Supplementary Table 7). To estimate the completeness of our annotated genes, we determined that the annotated genes contained 88.9% complete, 3.2% duplicated, 6.5% fragmented, and 4.6% missed BUSCOs.
Phylogenetic Analysis
To establish the phylogenetic position of the mandarin fish, we compared the genomes of the mandarin fish and seven other teleost fishes. We found that 16,922 orthologous gene families were shared among the eight teleost fishes, and identified 3,510 single-copy orthologs genes that were used to construct a phylogenetic tree (Figure 3). It appears that stickleback was most closely related to the mandarin fish. We selected the specific gene family in mandarin fish and they were functionally annotated by KEGG protein database (Supplementary Table 8). Finally, there were 74 specific families in mandarin fish, containing 194 genes. According to KEGG annotation, there were 45 genes associated to 205 pathways.
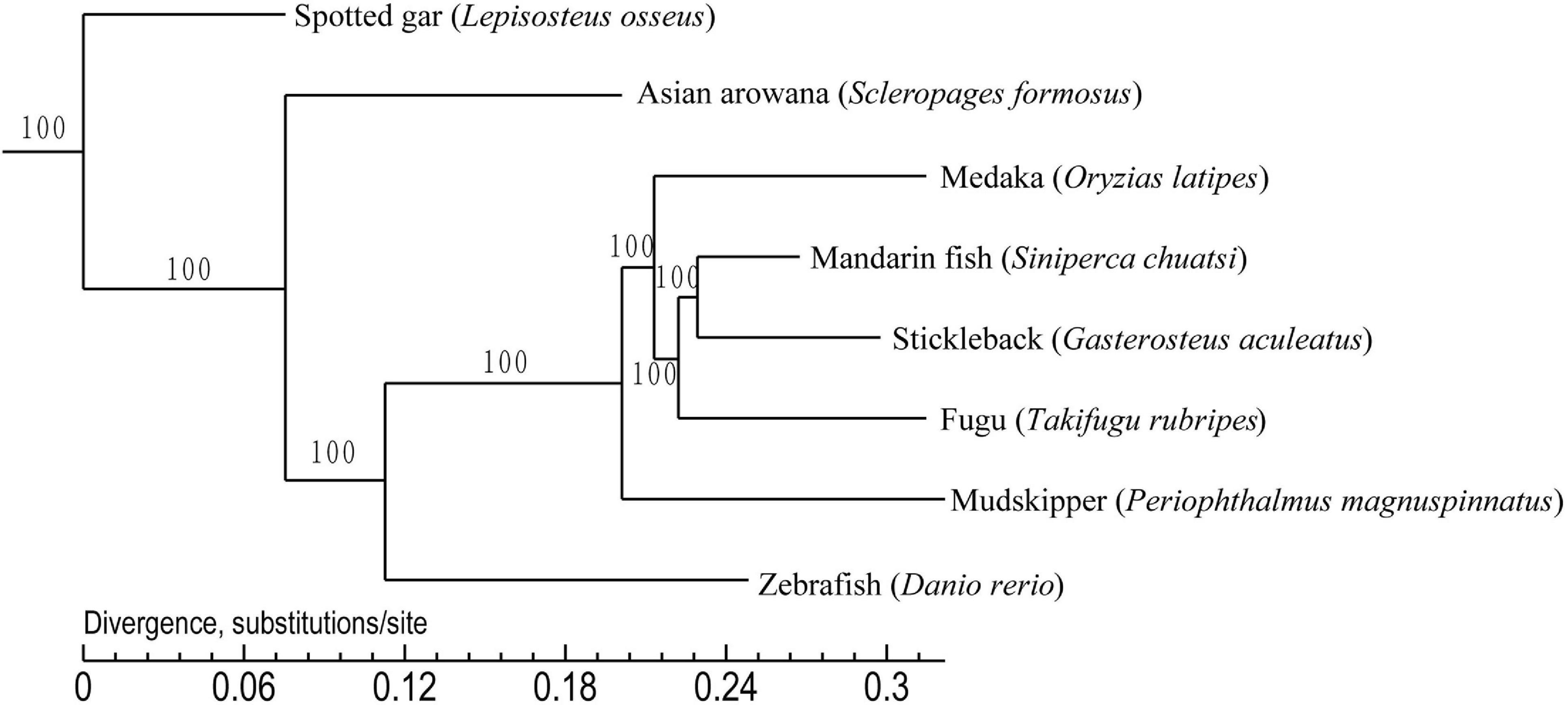
Figure 3. Phylogenetic tree of eight teleost fishes. It was constructed using the shared 3,510 single copy orthologous genes. Spotted gar was used as the outgroup.
Localization of Sex-Related Genes and QTLs on Chromosomes
Of the 81 sex-related genes, 19 genes were located on Chr14 (13 clustered in Figure 4A). In a previous study (Sun et al., 2017), five QTLs for sex determination (SD) were detected on LG23 (Sun et al., 2017) and thereby localized on Chr17 of the mandarin fish genome (clustered between 0 and 4 Mb; Figure 4B). Both r2_42410 and r2_237649 were located within the receptor-type tyrosine-protein phosphatase-like N (ptrpn) and SH3 domain-containing YSC84-like protein 1 (sh3yl1), respectively. The other three QTLs were located in the intergenic regions (Figure 4B). Genotypes of all the male and female fishes on r1_33008 were homozygous and heterozygous respectively, which was reported previously (Sun et al., 2017). Subsequently, we validated the marker r1_33008 (Sun et al., 2017) in another group (Figure 4B) and found that there was no difference between male and female, which may be unique in the genetic linkage map population.
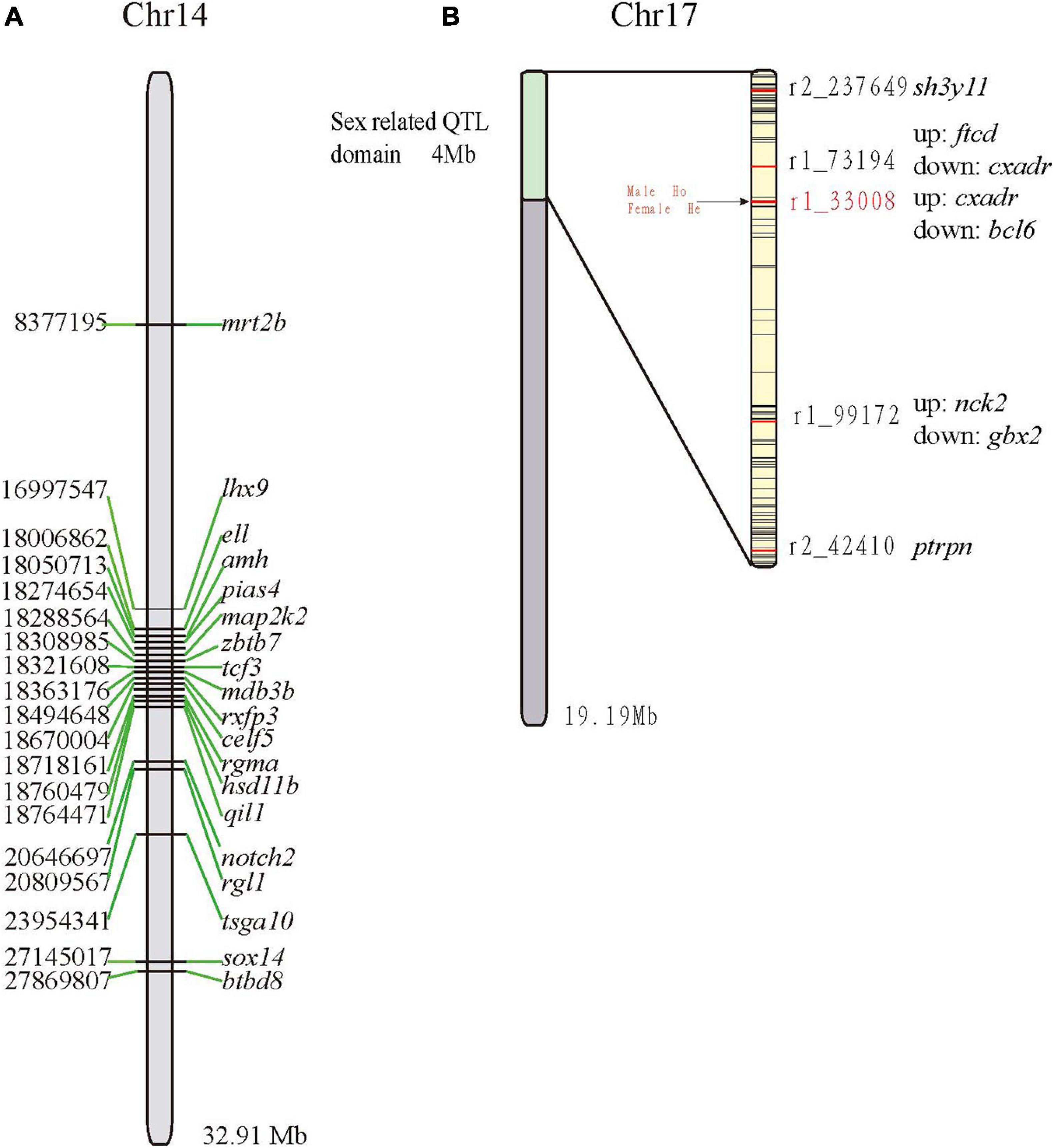
Figure 4. Localization of sex-related genes and QTLs on the Chr14 (A) and Chr17 (B), respectively. Abbreviations of genes: amh, anti-mullerian hormone; btbd8, BTB (POZ) domain containing 8; celf5, CUGBP Elav-like family member 5; dmrt2b, doublesex and mab-3 related transcription factor 2b; ell, RNA polymerase II elongation factor ELL; hsd11b, hydroxysteroid 11-beta-dehydrogenase 1-like protein; lhx9, LIM homeobox 9; map2k2, dual specificity mitogen-activated protein kinase kinase 2; notch2, notch homolog 2; pias4, Protein inhibitor of activated STAT 4; qil1, QIL1; rgl1, ral guanine nucleotide dissociation stimulator-like 1; rxfp3, relaxin family peptide receptor 3; tcf3, E2A-1 transcription factor; sox14, SRY-box transcription factor; zbtb7, zinc finger and BTB domain containing 7.
Analysis of Genes for Food Intake
In fishes, feeding behaviors are usually regulated by specific regions in the brain, the so-called feeding centers, which are under the influence of hormones produced by the brain and the periphery (Volkoff, 2016). The mandarin fish has a peculiar feeding habit of only accepting live prey fishes and refusing artificial diets or dead prey fishes. It is almost unknown about any genes for regulation of this unique food preference (Liu et al., 1998; Li et al., 2013). In our present study, several candidate genes for food intake were analyzed.
leptin is an important hormone involved in the regulation of food intake and energy balance (Kurokawa et al., 2005). Our synteny analysis of four representative fishes (mandarin, large yellow croaker, grouper and grass carp) (Figure 5) indicated that the upstream and downstream of the leptin genes are proline-rich transmembrane protein 4 (prrt4), transmembrane protein 53 (tmem53), RNA-binding protein 28 (rbm28) and Leucine-rich repeat-containing protein 4 (lrrc4), hepatocyte growth factor (hgf), voltage-dependent calcium channel subunit alpha (cacng-α) genes respectively, which is consistent with a previous report (Kurokawa et al., 2005). Compared with the other three species, the upstream genes {[F-actin]-monooxygenase MICAL3 (mical3) and zinc finger BED domain-containing protein 1 (zbed1)} of grass carp were not conserved (Figure 5). In a previous study (He et al., 2013), a typical leptin gene was reported to be composed of three exons and two introns, but the mandarin fish leptin gene consisted of two exons and one intron. However, our genomic results confirmed that the leptin structure of the mandarin fish is in fact consistent with the other three representative fishes (grouper, large yellow croaker, and grass carp) (Supplementary Figure 2). We thereby propose that the errors in the previous study are due to a shortage of whole genome sequence of the mandarin fish at that time.
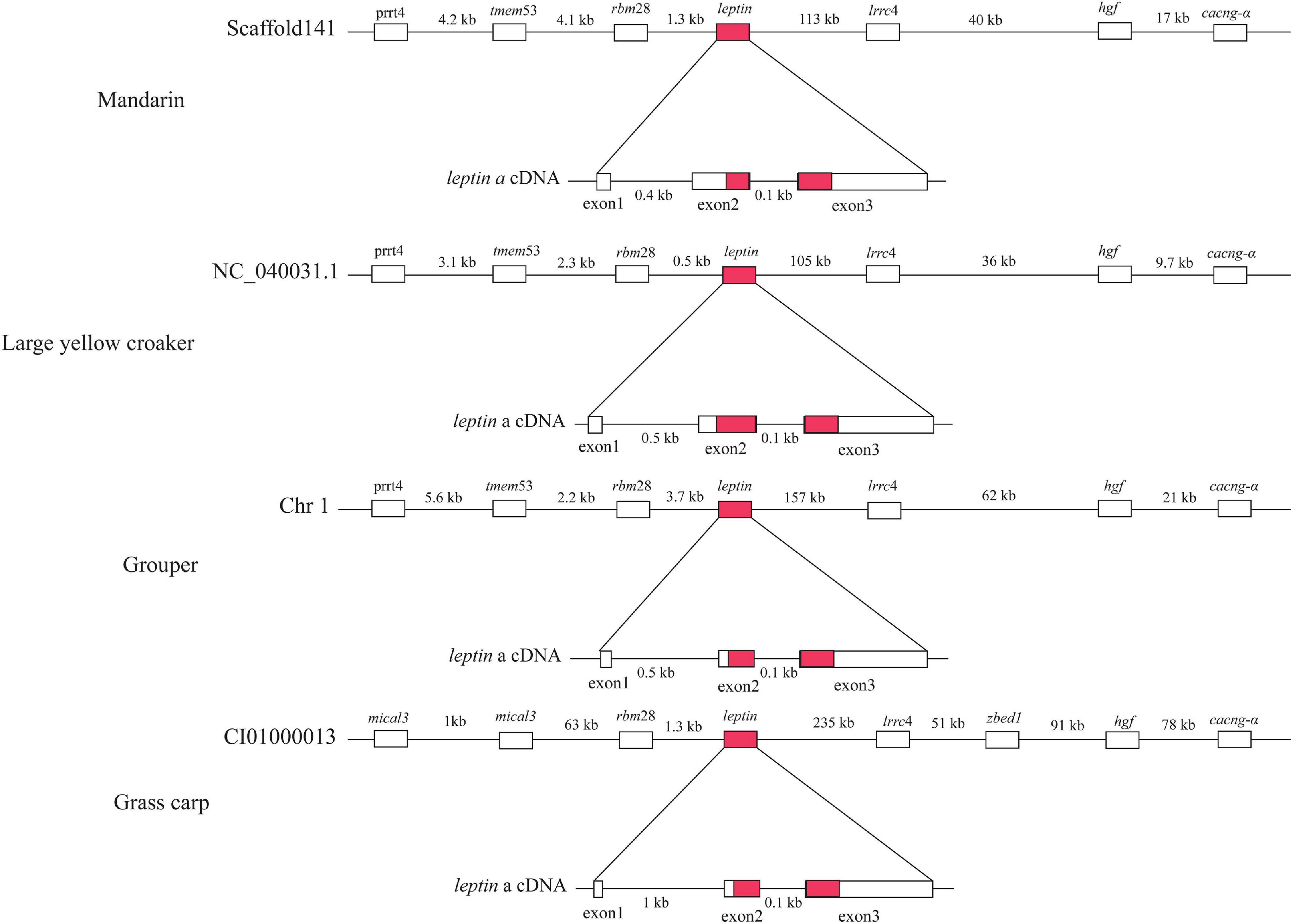
Figure 5. Comparison of leptin gene structures among mandarin fish, large yellow croaker, grouper and grass carp. Red marked areas represent open reading frames.
Neuropeptide Y (npy), belonging to the npy family, is abundant in the central nervous system (Volkoff, 2006; Holzer et al., 2012). npy has been implicated in several centrally mediated physiological functions, such as regulation of body temperature, sexual behavior, energy homeostasis, anxiety, mood, and neuroendocrine secretions (Holzer et al., 2012). Moreover, npy is one of the most abundant neuropeptides within the brain and has a major regulatory role in food intake (Yokobori et al., 2012; Zhou Y. et al., 2013). The npy gene of the mandarin fish on Chr 2 was comprised of three exons and two introns, which is consistent with other fishes. Mandarin fish compared to six perciforme fishes, NH2-terminal signal peptide (red box, Supplementary Figure 3) is variable, mature peptide (Black underline, Supplementary Figure 3) is highly conserved. However, its COOH-terminal domain had two significant variant sites (red asterisks in Supplementary Figure 3).
Spexin was identified in mammalian adipose tissue. It plays a significant role in the regulation of energy metabolism and food intake (Walewski et al., 2014; Zheng et al., 2017). Its expression is up-regulated in food deprivation and down-regulated in obese rats and humans, suggesting suppression of the orexin in the hypothalamus (Li et al., 2016). In the present study, we performed sequence analysis and found that the spexin gene is composed of six exons and five introns, and the amino acids of the mature peptide (spexin-14; Supplementary Figure 4a) in the mandarin fish are identical to that of the grouper (Li et al., 2016). Sequence alignment of mandarin fish spexin with the other six perciforme fishes reveals that the NH2-terminal signal peptide is highly variable, and its COOH-terminal domain had two significant variant sites (red asterisks). In contrast, the region covering the spexin mature peptide (black box) together with the dibasic processing sites flanking the two ends (RR and GRR, black triangle) is highly conserved (Supplementary Figure 4b).
Olfactory Receptor Genes in Teleost Fishes
We identified 133 OR genes in the mandarin fish genome (Figure 6A), including 119 functional genes and 13 pseudogenes. The numbers are different from those reported in zebrafish (102 functional genes and 35 pseudogenes) and pufferfish (44 functional genes and 54 pseudogenes) genomes (Niimura and Nei, 2005). We examined zebrafish (Ensembl version: GRCz11) and fugu genomes (Ensembl version: FUGU5) and identified 109 functional genes and 6 pseudogenes in the zebrafish and 72 functional genes and 12 pseudogenes in the fugu, respectively.
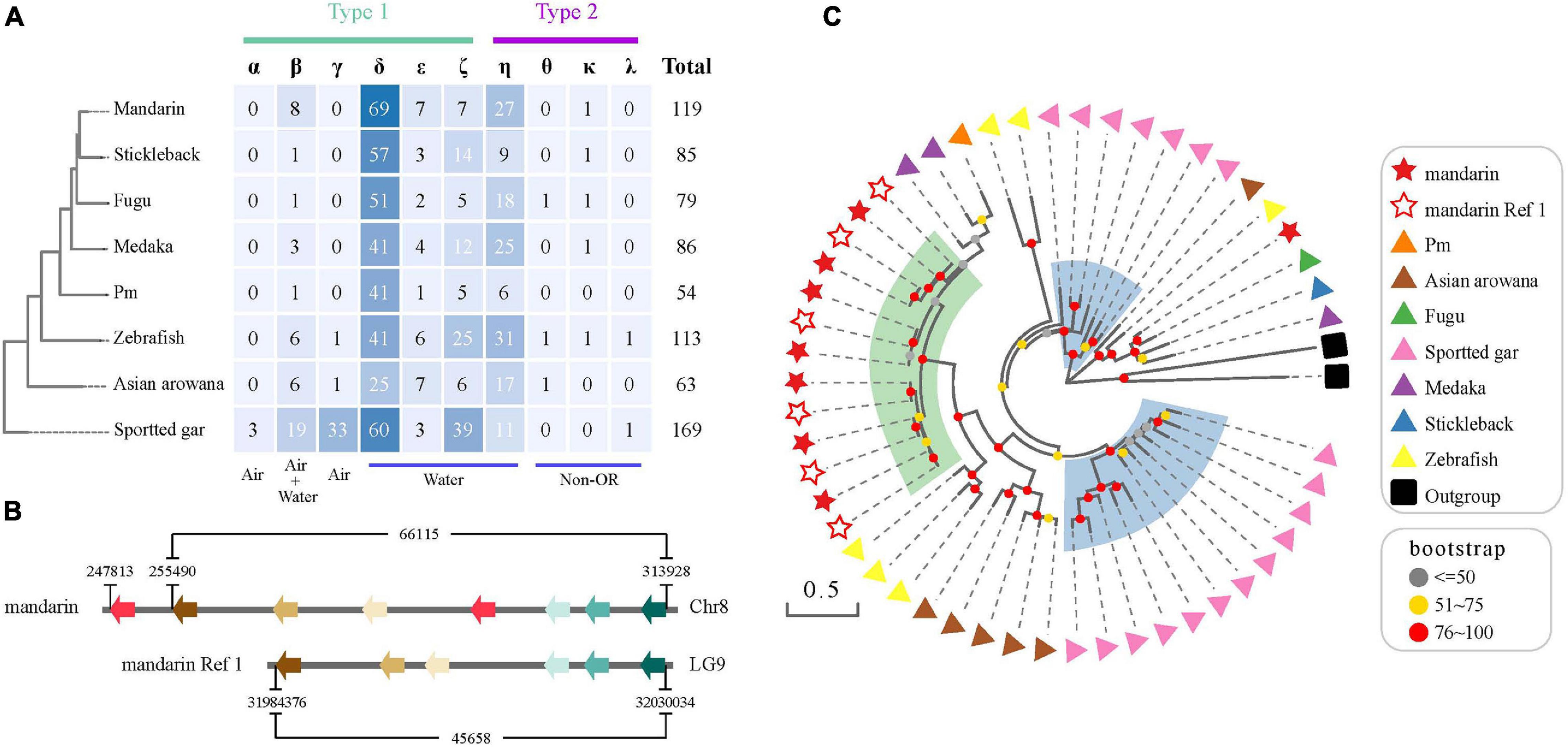
Figure 6. The OR gene family in the mandarin fish genome. (A) OR genes in the mandarin fish in comparison with seven other teleost fishes. “Air” and “water” represent the detection of airborne and water-soluble odorants, respectively. (B) A comparative genomic analysis of Group β OR functional genes in our genome assembly and the assembly of Ref 1. (C) A phylogenetic tree of Group β functional genes in the mandarin fish and seven other teleost fishes. Stars indicate the Group β OR functional genes of the mandarin fish [from our present work and Niimura and Nei (2005)]. Triangles with different colors represent various resources of the other seven teleost fishes. Green transparent rainbow indicates an expansion of Group β OR functional genes in the mandarin fish. Blue transparent rectangles indicate the expansion of Groupe β OR functional genes in spotted gar.
We found that the mandarin fish had more OR functional genes than other examined teleosts, except for the spotted gar that was diverged from teleosts before the teleost-specific genome duplication (TGD). Spotted gar has 36 functional genes ascribing to Groups α and γ, which were mostly absent in teleosts. The mandarin fish had the largest numbers of Group β (n = 8) and group δ (n = 69) functional genes among other teleosts. In a previous study (Lv et al., 2019), researchers indicated an expansion of Group β OR genes in the mandarin fish, which was confirmed in our present work (Figure 6C). We extracted more Group β OR genes (eight in Chr8) than a previous report (six in Reference (Niimura and Nei, 2005); see Figures 6B,C).
Taste Receptor Genes in Teleost Fishes
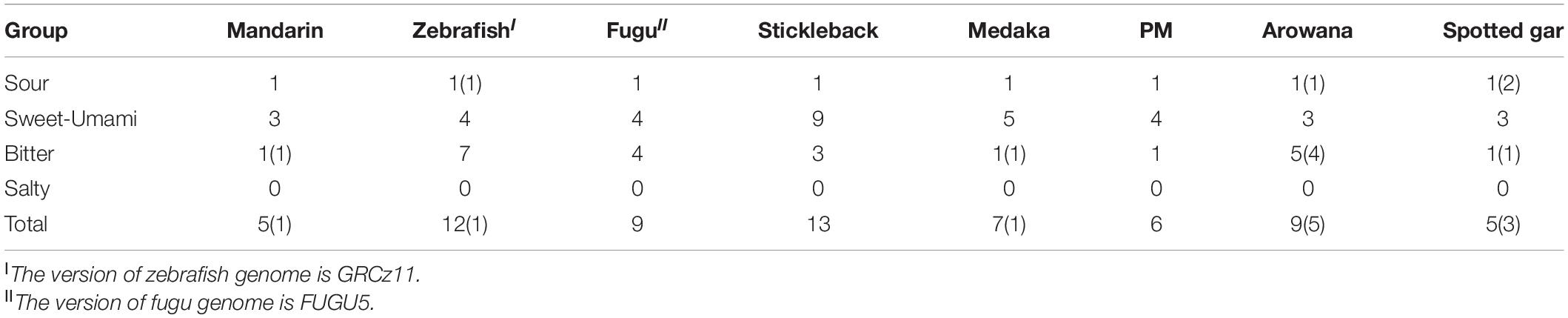
Taste receptor type 1family (tas1r), belonging to the G protein-coupled receptor (gpcr), plays a central role in the reception of sweet and umami taste in many vertebrates. A tas1r2 + 3 heterodimer was identified as the sweet TR (Nelson et al., 2001; Li et al., 2002). Tas1r3 may serve as a receptor for high sucrose concentrations (Zhao et al., 2003). A tas1r1 + 3 heterodimer and multiple combinations of tas1r2 with tas1r3 were identified as a tuned L-amino acid TR in fish (Oike et al., 2007). We extracted three tas1r genes in the mandarin fish, Asian arowana and spotted gar (Table 3 and Figure 7). It seems that the gene numbers responding to sweet and umami tastes in the mandarin fish is more primitive since they are much closer to ancient fishes (such as arowana and gars).
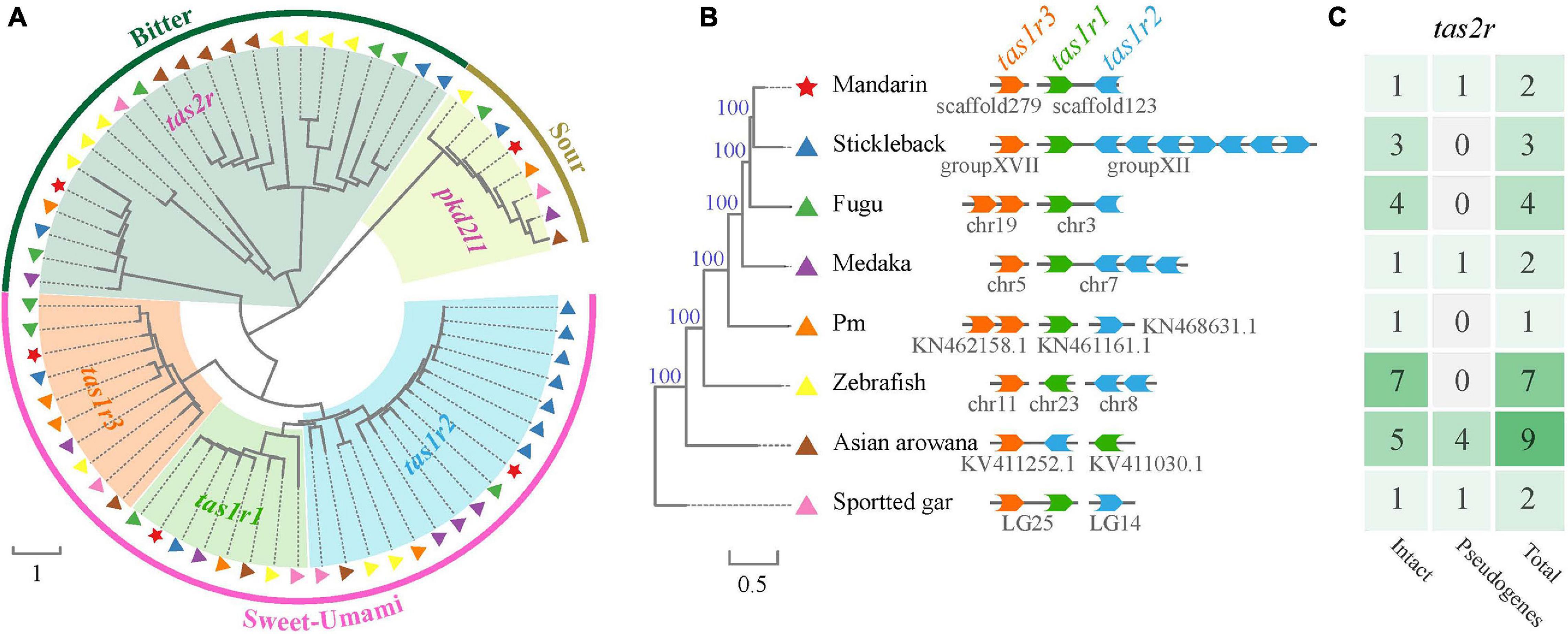
Figure 7. A phylogenetic tree of taste-related genes in the mandarin fish and seven other teleost fishes. (A) Comparisons of taste-related genes in the mandarin fish and seven other teleost fishes. Red stars indicate the gens of the mandarin fish. Other symbols indicate the sources of other species. (B) Genomic locations of tas1r1, tas1r2, and tas1r3 genes in our genome assembly. (C) Summary of the copy number of tas2r gene in different species [corresponding to those fishes in panel (B)].
Bitter taste preference was likely recognized as a mechanism for avoiding toxic foods. Bitter foods evoke innate aversive behaviors in many animals. Taste receptor type 2 (tas2r) family was identified as bitter TRs in mammals. Most vertebrate species have several tas2r genes, and their copy numbers varied among various species. In our present study, we identified only one intact tas2r gene in the mandarin fish, giant-fin mudskipper, and spotted gar (Figure 7C), suggesting that these three species possibly have a low ability to distinguish the bitter foods. However, compared with the other seven teleost fishes, the mandarin fish showed no difference in number of genes for responding to salty and sour tastes (Table 3).
Comparison of Mandarin Fish opsin Genes With Other Teleost Fish
A total of 50 opsin nucleotide sequences, including 14 RH1 (rhodopsin), 14 RH2 (green-sensitive), 4 SWS1 (short wavelength-sensitive 1), 7 SWS2 (short wavelength-sensitive 2), and 11 LWS (long wavelength-sensitive), were successfully derived from eight teleost fishes (Figure 8A). To understand the evolutionary relationships among these opsins in teleosts, we constructed a phylogenetic tree (Figure 8B) using spotted gar LWS as the outgroup. Obviously, all opsin could be divided into five main clades (RH1, RH2, SWS1, SWS2, and LWS) in the eight examined teleost species.
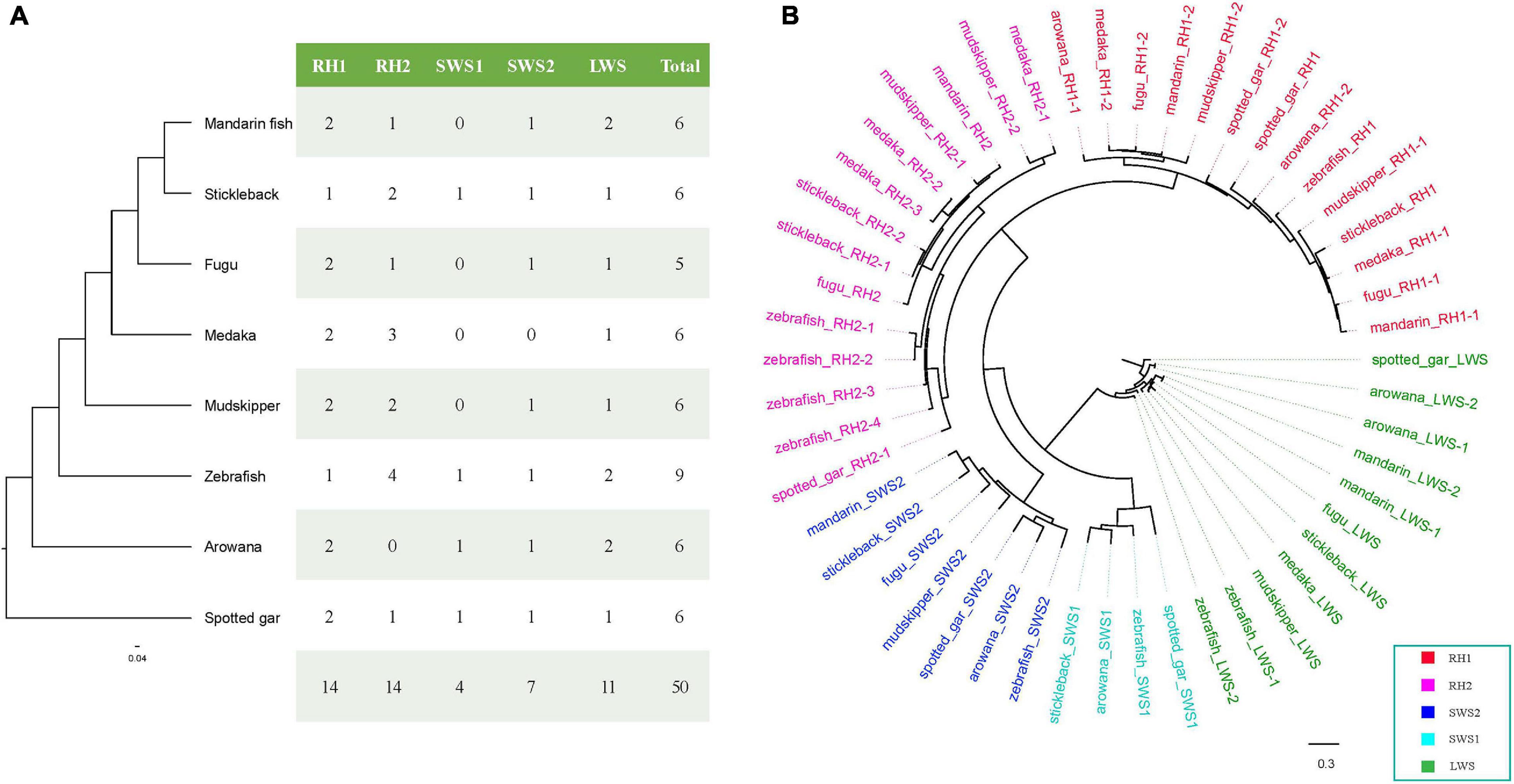
Figure 8. opsin genes in the mandarin fish genome. (A) opsin genes in the mandarin fish and seven other teleost fishes. (B) A phylogenetic tree of opsin genes in the mandarin fish and seven other teleost fishes. LWS, long wavelength-sensitive; RH1, rhodopsin; RH2, green-sensitive; SWS1, short wavelength-sensitive 1; SWS2, short wavelength-sensitive 2.
In this study, we identified six opsin genes in the mandarin fish genome (Figure 8A), including two RH1, one RH2, one SWS2, and one LWS. A previous study (Neafsey and Hartl, 2005) reported loss of SWS1 genes in fugu and mudskipper (You et al., 2014) genome. We tried to extract SWS1 sequences in the examined teleosts, but could not find SWS1 in the mandarin fish, medaka, fugu, and giant-fin mudskipper either. This loss of SWS1 could be an adaptation to minimize retinal damage from ultraviolet. We identified two LWS genes in the mandarin fish, zebrafish and arowana, while only one LWS in other fishes. In zebrafish, two LWS genes locating in tandem encode different protein sequences (Chinen et al., 2003). However, in the mandarin fish and arowana, two LWS genes were also in tandem but encoded the identical protein sequences. According to Figure 8B and Supplementary Figure 4, m and arin LWS-1 and LWS-2 were completely same in amino acid sequences. And arowana LWS-1 and LWS-2 had same sequences but they were different in zebrafish.
The primary amino acid sequence is very important for opsin molecular properties. In this study, we compared their sequences and identify the differences among the eight examined fishes. We identified some amino acid changes in the mandarin fish that are probably critical for wavelength absorption. Here, we observed five specific sites in the mandarin fish, with significant differences from the other seven fish species (see more details in Supplementary Figure 5). We identified the transmembrane domains of LWS with TMHMM Server (Version 2.0)1 (Supplementary Figure 5). In the mandarin fish, two specific sites are in the transmembrane domains, L98 and T219, which are potentially important for light absorption. LWS is often used for red vision, and shallow water receives more red light. The freshwater fishes have more LWS genes than those in seawater (Lin et al., 2017). The more LWS genes and several specific sites in transmembrane domains help fishes be more sensitive to light and live prey.
Toxin Genes Were Identified in the Mandarin Fish Genome
Fish toxins have been poorly studied compared to venoms from other animals such as snakes, scorpions, spiders, and cone snails (Utkin, 2015). It is estimated that there are up to 2,900 venomous fishes (Xie et al., 2017) with venom systems convergently evolved 19 times (Harris and Jenner, 2019). Mandarin fish is one of those who can produce toxins in their hard spines to help them defense and prey, and cause pain and swelling at the site of the sting in human as well (Zhang F.-B. et al., 2019). However, apart from several antimicrobial peptides that can be regarded as toxins (Sun et al., 2007), there is no detailed report on venom genes and components of this fish yet.
In this study, a total of 155 toxin proteins were predicted from the mandarin fish genome assembly. They ranged from 87 to 1,895 amino acids (aa), with more than half of them less than 300 aa (Supplementary Figure 6). Unlike a vast number (125) of short-length “fragmented” venoms (less than 100 aa) in the Chinese yellow catfish genome (Zhang et al., 2018), there were only two short-length venoms in the mandarin genome, with a length of 87 and 98 aa, respectively. Consistent with the common findings that most toxins are short peptides, the majority (96; 62%) of our predicted toxin proteins had an entire length between 100 and 300 aa.
Among the 155 putative venom proteins, 144 were classified into 37 families, with 11 unclassified toxins (Supplementary Figure 7). The top four biggest groups in these toxins included peptidase S1, venom metalloproteinase (M12B), Type-B carboxylesterase, and calmodulin, consisting of 27, 13, 10, and 9 toxin genes respectively. Interestingly, several fish-specific toxins were identified, including SC_GLEAN_10016806 and SC_GLEAN_10016808 that belonged to the stonustoxin (SNTX-a), SC_GLEAN_10016805, and SC_GLEAN_10016807 annotated as SNTX-β. SNTX is a soluble heterodimeric assembly of α and β subunits that share a sequence identity of ∼50% (Ellisdon et al., 2015). It was firstly isolated from the stonefish (Poh et al., 1991) and has been proved to induce platelet aggregation and hemolytic activity (Khoo et al., 1992) and also function as a neurotoxin (Low et al., 1994). The existence of two copies of both SNTX-α and SNTX-β suggesting the probability of the forming of active and functional toxins in the mandarin fish. SNTXs, along with all other toxins identified in this assembled genome, showed the great potential of discovering new drugs.
Discussion
Our high-quality genome assembly of the mandarin fish could provide opportunities to understand SD, special food intake, or other biological processes at the genome level in this economically important fish. The final assembled genome was 758.78 Mb, and approximately 92.8% of the scaffolds were ordered onto 24 chromosomes. The mandarin fish has been widely cultivated in China, with a special feeding habit of accepting only live prey fishes for its delicious meat. However, little is currently known about related genetic mechanisms. Uncovering the molecular mechanisms for regulation of feeding behaviors may not only lead to specific adjustments in fish culture conditions and feeding strategies but also gradually instruct us to develop new technologies to improve feeding, food conversion efficiency and the growth of aquaculture fishes (Volkoff et al., 2010; Zhou Y. et al., 2013).
In fact, feeding is a complex of behaviors, including at least food intake itself and foraging or appetite behavior. Eating is ultimately regulated by the central feeding center in the brain (Keen-Rhinehart et al., 2013; Woods et al., 2014). It also processes information from endocrine signals from the brain and the surrounding environments. These endocrine signals include various hormones. For example, npy and spexin are two important hormones involved in the regulation of food intake and energy balance (Zheng et al., 2017; Zhou Y. et al., 2013). The latest research suggests that smell might regulate appetite through npy in yellowtail (Senzui et al., 2020). npy as a neuromodulator in the olfactory epithelium and intensified the activity of OR neurons and olfaction (Negroni et al., 2012; Senzui et al., 2020). In our present study, we observed that the amino acid sequence of npy and spexin showed a high level of conservation, when compared with the other six examined Perciformes fishes. It seems that npy and spexin are conserved neuropeptides in fish evolution with important physiological functions. However, the npy and spexin genes of the mandarin fish had significant variations at the C-termini of the protein sequences (Supplementary Figures 2, 3), which may be related to the special diet of the mandarin fish.
Olfaction is also crucial for animals to find foods and to judge whether potential foods are edible or not (Chandrashekar et al., 2000; Nei et al., 2008). It is controlled by a large family of OR genes. Fishes also have this gene family, but the number of genes is much less than mammals (Niimura and Nei, 2006). Previous studies have demonstrated that the beta type OR genes are presented in both aquatic and terrestrial vertebrates, indicating that these receptors detect both water-soluble and airborne odorants (Niimura, 2009); however, delta type OR genes are only in aquatic organisms (You et al., 2014). In the present study, we determined that the mandarin fish had the largest numbers of Group β (n = 8) and group δ (n = 69) functional genes than the other teleost fishes (Figure 6A), which might contribute to its particular carnivorous diet.
Vision is very important for animals because it plays important roles in foraging, mating, information transmission, and escaping from predators (Yokoyama, 2000). Based on their amino acid compositions, opsin genes are classified into five common clusters: RH1 (rhodopsin), RH2 (rhodopsin-like or the green light-sensitive pigments), SWS1 (short wavelength−, or the UV or violet light-sensitive pigments), SWS2 (SWS1-like or the blue light-sensitive pigments); LWS/MWS (long wavelength- or middle wavelength-sensitive, or the red- and green-sensitive pigments) (Yokoyama, 2000; Shichida and Matsuyama, 2009). opsin diversity is usually generated by gene duplication and/or accumulation of mutations. MWS/LWS opsins have peak values of light absorption (Terai et al., 2002). The light sensitivity of a visual pigment is determined not only by the chromophore itself, but also by its interaction with the amino acid residues lining the pocket of the opsin (Yokoyama, 1995). In this study, compared with other closely related species, the mandarin fish was identified with more LWS genes. LWS1/2 had five specific sites in the mandarin fish with remarkable differences from the other seven fish species (Supplementary Figure 5). Certain mutations of the transmembrane domains, L98 and T219 in the LWS genes might be expected to contribute to the special feeding habit of live prey.
Many fish species exhibit sexual dimorphisms, such as Japanese flounder (Paralichthys olivaceus) (Shao et al., 2015), half-smooth tongue sole (Cynoglossus semilaevis) (Song et al., 2012), displaying significant differences in growth rates or sizes between male and female individuals. Females of mandarin fish present higher growth rates (by 10–20% in body weight) than males (Sun et al., 2017). Therefore, screening of sex-related genes or markers is important for the development of the mandarin industry, which will be helpful for the elucidation of the SD mechanisms in the mandarin fish. Nineteen sex-related genes, localized on the Chr14, were previously reported to be involved in spermatogenesis, SD, and testicular determination. Some studies support that SD is controlled by many major genetic factors that may interact with minor genetic factors, thereby implying that SD should be analyzed as a quantitative trait (Eshel et al., 2012). Five sex-related QTLs in the mandarin fish were previously detected on the Chr17. Therefore, we speculate that both the Chr14 and Chr17 are the potential to be related to SD in the mandarin fish. These results suggest the involvement of multiple chromosomes in sex relation, and provide supportive evidence to the polygenic SD in fishes (Zhang S. et al., 2019). In the coming future, the development of unisex male populations will be necessary for rapid improvement of the quality and quantity of the mandarin fish.
Conclusion
In our present study, we generated a chromosomal-level genome assembly for the mandarin fish, which has been an economically important fish in China. Our genome assembly is high in quality, completeness, and accuracy based on multiple evaluations. Gene prediction, functional annotation, and evolutionary analysis provided novel insights into the genomic structure and mechanisms underlying food intake, SD, and prediction of new toxins. Our genome sequences will also offer a valuable genetic resource to support extensive fisheries and artificial breeding programs, and thereby allows for effective disease management, growth improvement, and discovering new drugs in the mandarin fish.
Data Availability Statement
This Whole Genome Shotgun project of mandarin fish has been deposited in CNGBdb with accession number CNA0013732. Raw reads from Illumina sequencing are deposited in the CNGBdb with accession number CNS0204384. The genome assembly of mandarin fish has been deposited in the CNGB Nucleotide Sequence Archive (https://db.cngb.org/cnsa/) under the Project ID CNP0000961.
Ethics Statement
The animal study was reviewed and approved by the Institutional Review Board on Bioethics and Biosafety of BGI, China (No. FT 18134). Written informed consent was obtained from the owners for the participation of their animals in this study.
Author Contributions
XB, XY, and WD conceived and designed the research. WD and XZ performed the genome sequencing. XZ, JL, and YH performed data analyses and wrote the manuscript. WJ, ZC, and MW performed sample preparation. WD, XZ, XB, QS, and XY revised the manuscript. All authors approved submission of the manuscript for publication.
Funding
This work was supported by the Jiangsu Agriculture Science and Technology Innovation Fund [No. CX(17)3005]; the Central Public-Interest Scientific Institution Basal Research Fund (2017JBFZ02); the Shenzhen Special Program for Development of Emerging Strategic Industries (No. JSGG20170412153411369); the Ministry of Agriculture and Rural Affairs Project for Conservation of Species Resources (No. 17190412); and the Natural Science Foundation for Fundamental Research in Shenzhen (JCYJ20190812105801661).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2021.671650/full#supplementary-material
Footnotes
References
Abrusan, G., Grundmann, N., Demester, L., and Makalowski, W. (2009). TEclass–a tool for automated classification of unknown eukaryotic transposable elements. Bioinformatics 25, 1329–1330. doi: 10.1093/bioinformatics/btp084
Attwood, T. K. (2002). The PRINTS database: a resource for identification of protein families. Brief Bioinform. 3, 252–263. doi: 10.1093/bib/3.3.252
Bao, W., Kojima, K. K., and Kohany, O. (2015). Repbase update, a database of repetitive elements in eukaryotic genomes. Mob. DNA 6:11.
Benson, G. (1999). Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res. 27, 573–580. doi: 10.1093/nar/27.2.573
Bian, C., Hu, Y., Ravi, V., Kuznetsova, I. S., Shen, X., Mu, X., et al. (2016). The Asian arowana (Scleropages formosus) genome provides new insights into the evolution of an early lineage of teleosts. Sci. Rep. 6:24501.
Birney, E., Clamp, M., and Durbin, R. (2004). GeneWise and Genomewise. Genome Res. 14, 988–995. doi: 10.1101/gr.1865504
Birney, E., and Durbin, R. (2000). Using GeneWise in the Drosophila annotation experiment. Genome Res. 10, 547–548. doi: 10.1101/gr.10.4.547
Boeckmann, B., Bairoch, A., Apweiler, R., Blatter, M. C., Estreicher, A., Gasteiger, E., et al. (2003). The SWISS-PROT protein knowledgebase and its supplement TrEMBL in 2003. Nucleic Acids Res. 31, 365–370. doi: 10.1093/nar/gkg095
Bru, C., Courcelle, E., Carrere, S., Beausse, Y., Dalmar, S., and Kahn, D. (2005). The ProDom database of protein domain families: more emphasis on 3D. Nucleic Acids Res. 33, D212–D215.
Burge, C., and Karlin, S. (1997). Prediction of complete gene structures in human genomic DNA. J. Mol. Biol. 268, 78–94. doi: 10.1006/jmbi.1997.0951
Camacho, C., Coulouris, G., Avagyan, V., Ma, N., Papadopoulos, J., Bealer, K., et al. (2009). BLAST+: architecture and applications. BMC Bioinform. 10:421. doi: 10.1186/1471-2105-10-421
Chandrashekar, J., Mueller, K. L., Hoon, M. A., Adler, E., Feng, L., Guo, W., et al. (2000). T2Rs function as bitter taste receptors. Cell 100, 703–711. doi: 10.1016/s0092-8674(00)80706-0
Chiang, L. (1959). On the biology of mandarinfish, Siniperca chuatsi, of Liangtze Lake. Acta Hydrobiol. Sin 3, 365–385.
Chinen, A., Hamaoka, T., Yamada, Y., and Kawamura, S. (2003). Gene duplication and spectral diversification of cone visual pigments of zebrafish. Genetics 163, 663–675. doi: 10.1093/genetics/163.2.663
UniProt Consortium, (2018). UniProt: a worldwide hub of protein knowledge. Nucleic Acids Res. 47, D506–D515.
Cunningham, F., Amode, M. R., Barrell, D., Beal, K., Billis, K., Brent, S., et al. (2015). Ensembl 2015. Nucleic Acids Res. 43, D662–D669.
Edgar, R. C. (2004). MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797. doi: 10.1093/nar/gkh340
Ellisdon, A. M., Reboul, C. F., Panjikar, S., Huynh, K., Oellig, C. A., Winter, K. L., et al. (2015). Stonefish toxin defines an ancient branch of the perforin-like superfamily. Proc. Natl. Acad. Sci. U.S.A. 112, 15360–15365. doi: 10.1073/pnas.1507622112
Elsik, C. G., Mackey, A. J., Reese, J. T., Milshina, N. V., Roos, D. S., and Weinstock, G. M. (2007). Creating a honey bee consensus gene set. Genome Biol. 8:R13.
Engelmann, J., Hanke, W., Mogdans, J., and Bleckmann, H. (2000). Hydrodynamic stimuli and the fish lateral line. Nature 408, 51–52. doi: 10.1038/35040706
Eshel, O., Shirak, A., Dor, L., Band, M., Zak, T., Markovich-Gordon, M., et al. (2014). Identification of male-specific amh duplication, sexually differentially expressed genes and microRNAs at early embryonic development of Nile tilapia (Oreochromis niloticus). BMC Genom. 15:774. doi: 10.1186/1471-2164-15-774
Eshel, O., Shirak, A., Weller, J. I., Hulata, G., and Ron, M. (2012). Linkage and physical mapping of sex region on LG23 of Nile Tilapia (Oreochromis niloticus). G3 2, 35–42. doi: 10.1534/g3.111.001545
Finn, R. D., Bateman, A., Clements, J., Coggill, P., Eberhardt, R. Y., Eddy, S. R., et al. (2014). Pfam: the protein families database. Nucleic Acids Res. 42, D222–D230.
Guindon, S., Delsuc, F., Dufayard, J. F., and Gascuel, O. (2009). Estimating maximum likelihood phylogenies with PhyML. Methods Mol. Biol. 537, 113–137. doi: 10.1007/978-1-59745-251-9_6
Guindon, S., Dufayard, J. F., Lefort, V., Anisimova, M., Hordijk, W., and Gascuel, O. (2010). New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst. Biol. 59, 307–321. doi: 10.1093/sysbio/syq010
Guo, W., He, S., Liang, X. T., Changxu, D., Yaqi, L., and Liyuan, M. (2021). A high-density genetic linkage map for Chinese perch (Siniperca chuatsi) using 2.3K genotyping-by-sequencing SNPs. Anim. Genet. 52, 311–320. doi: 10.1111/age.13046
Harris, M. A., Clark, J., Ireland, A., Lomax, J., Ashburner, M., Foulger, R., et al. (2004). The Gene Ontology (GO) database and informatics resource. Nucleic Acids Res. 32, D258–D261.
Harris, R. J., and Jenner, R. A. (2019). Evolutionary ecology of fish venom: adaptations and consequences of evolving a venom system. Toxins 11:60. doi: 10.3390/toxins11020060
He, S., Liang, X. F., Li, L., Huang, W., Shen, D., and Tao, Y. X. (2013). Gene structure and expression of leptin in Chinese perch. Gen. Comp. Endocrinol. 194, 183–188. doi: 10.1016/j.ygcen.2013.09.008
Holzer, P., Reichmann, F., and Farzi, A. (2012). Neuropeptide Y, peptide YY and pancreatic polypeptide in the gut-brain axis. Neuropeptides 46, 261–274. doi: 10.1016/j.npep.2012.08.005
Hunter, S., Apweiler, R., Attwood, T. K., Bairoch, A., Bateman, A., Binns, D., et al. (2009). InterPro: the integrative protein signature database. Nucleic Acids Res. 37, D211–D215.
Jungo, F., Bougueleret, L., Xenarios, I., and Poux, S. (2012). The UniProtKB/Swiss-Prot Tox-Prot program: a central hub of integrated venom protein data. Toxicon 60, 551–557. doi: 10.1016/j.toxicon.2012.03.010
Kanehisa, M., and Goto, S. (2000). KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 28, 27–30.
Katoh, K., Misawa, K., Kuma, K., and Miyata, T. (2002). MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 30, 3059–3066. doi: 10.1093/nar/gkf436
Keen-Rhinehart, E., Ondek, K., and Schneider, J. E. (2013). Neuroendocrine regulation of appetitive ingestive behavior. Front. Neurosci. 7:213. doi: 10.3389/fnins.2013.00213
Khoo, H. E., Yuen, R., Poh, C. H., and Tan, C. H. (1992). Biological activities of Synanceja horrida (stonefish) venom. Nat. Toxins 1, 54–60.
Krzywinski, M., Schein, J., Birol, I., Connors, J., Gascoyne, R., Horsman, D., et al. (2009). Circos: an information aesthetic for comparative genomics. Genome Res. 19, 1639–1645. doi: 10.1101/gr.092759.109
Kumar, S., Stecher, G., and Tamura, K. (2016). MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874. doi: 10.1093/molbev/msw054
Kurokawa, T., Uji, S., and Suzuki, T. (2005). Identification of cDNA coding for a homologue to mammalian leptin from pufferfish, Takifugu rubripes. Peptides 26, 745–750. doi: 10.1016/j.peptides.2004.12.017
Kurtz, S., Phillippy, A., Delcher, A. L., Smoot, M., Shumway, M., Antonescu, C., et al. (2004). Versatile and open software for comparing large genomes. Genome Biol. 5:R12.
Letunic, I., Copley, R. R., Schmidt, S., Ciccarelli, F. D., Doerks, T., Schultz, J., et al. (2004). SMART 4.0: towards genomic data integration. Nucleic Acids Res. 32, D142–D144.
Li, L., Stoeckert, C. J. Jr., and Roos, D. S. (2003). OrthoMCL: identification of ortholog groups for eukaryotic genomes. Genome Res. 13, 2178–2189. doi: 10.1101/gr.1224503
Li, R., Yu, C., Li, Y., Lam, T. W., Yiu, S. M., Kristiansen, K., et al. (2009). SOAP2: an improved ultrafast tool for short read alignment. Bioinformatics 25, 1966–1967. doi: 10.1093/bioinformatics/btp336
Li, S., Liu, Q., Xiao, L., Chen, H., Li, G., Zhang, Y., et al. (2016). Molecular cloning and functional characterization of spexin in orange-spotted grouper (Epinephelus coioides). Comp. Biochem. Physiol. B Biochem. Mol. Biol. 196–197, 85–91. doi: 10.1016/j.cbpb.2016.02.009
Li, W., Zhang, T., Ye, S., Liu, J., and Li, Z. (2013). Feeding habits and predator-prey size relationships of mandarin fish Siniperca chuatsi (Basilewsky) in a shallow lake, central China. J. Appl. Ichthyol. 29, 56–63. doi: 10.1111/j.1439-0426.2012.02044.x
Li, X., Staszewski, L., Xu, H., Durick, K., Zoller, M., and Adler, E. (2002). Human receptors for sweet and umami taste. Proc. Natl. Acad. Sci. U.S.A. 99, 4692–4696.
Liang, X., Kiu, J., and Huang, B. (1998). The role of sense organs in the feeding behaviour of Chinese perch. J. Fish Biol. 52, 1058–1067. doi: 10.1111/j.1095-8649.1998.tb00603.x
Liang, Y., and Cui, X. (1982). The eco-physiological characteristics of artificial propagation in mandarin fish (Siniperca chuatsi). Acta Hydrobiol. Sin. 16, 90–92.
Lin, J.-J., Wang, F.-Y., Li, W.-H., and Wang, T.-Y. (2017). The rises and falls of opsin genes in 59 ray-finned fish genomes and their implications for environmental adaptation. Sci. Rep. 7:15568.
Liu, B., Shi, Y., Yuan, J., Hu, X., Zhang, H., Li, N., et al. (2013). Estimation of genomic characteristics by analyzing k-mer frequency in de novo genome projects. arXiv preprint. arXiv:13082012.
Liu, J., Cui, Y., and Liu, J. (1998). Food consumption and growth of two piscivorous fishes, the mandarin fish and the Chinese snakehead. J. Fish Biol. 53, 1071–1083. doi: 10.1111/j.1095-8649.1998.tb00464.x
Liu, S., Xu, P., Liu, X., Guo, D., Chen, X., Bi, S., et al. (2021). Production of neo-male mandarin fish Siniperca chuatsi by masculinization with orally administered 17 alpha-methyltestosterone. Aquaculture 530:680.
Low, K. S., Gwee, M. C., Yuen, R., Gopalakrishnakone, P., and Khoo, H. E. (1994). Stonustoxin: effects on neuromuscular function in vitro and in vivo. Toxicon 32, 573–581. doi: 10.1016/0041-0101(94)90205-4
Luo, R., Liu, B., Xie, Y., Li, Z., Huang, W., Yuan, J., et al. (2012). SOAPdenovo2: an empirically improved memory-efficient short-read de novo assembler. Gigascience 1:18.
Luo, R., Liu, B., Xie, Y., Li, Z., Huang, W., Yuan, J., et al. (2015). Erratum: SOAPdenovo2: an empirically improved memory-efficient short-read de novo assembler. Gigascience 4:30.
Lv, L.-Y., Liang, X.-F., and He, S. (2019). Genome-wide identification and characterization of olfactory receptor genes in chinese perch, Siniperca chuatsi. Genes 10:178. doi: 10.3390/genes10020178
Mount, D. W. (2007). Using the basic local alignment search tool (BLAST). Cold Spring Harb. Protoc. 2007:pdb. top17.
Neafsey, D. E., and Hartl, D. L. (2005). Convergent loss of an anciently duplicated, functionally divergent RH2 opsin gene in the fugu and Tetraodon pufferfish lineages. Gene 350, 161–171. doi: 10.1016/j.gene.2005.02.011
Negroni, J., Meunier, N., Monnerie, R., Salesse, R., Baly, C., Caillol, M., et al. (2012). Neuropeptide Y enhances olfactory mucosa responses to odorant in hungry rats. PLoS One 7:e45266. doi: 10.1371/journal.pone.0045266
Nei, M., Niimura, Y., and Nozawa, M. (2008). The evolution of animal chemosensory receptor gene repertoires: roles of chance and necessity. Nat. Rev. Genet. 9, 951–963. doi: 10.1038/nrg2480
Nelson, G., Hoon, M. A., Chandrashekar, J., Zhang, Y., Ryba, N. J., and Zuker, C. S. (2001). Mammalian sweet taste receptors. Cell 106, 381–390. doi: 10.1016/s0092-8674(01)00451-2
Niimura, Y. (2009). On the origin and evolution of vertebrate olfactory receptor genes: comparative genome analysis among 23 chordate species. Genome Biol. Evol. 1, 34–44. doi: 10.1093/gbe/evp003
Niimura, Y., and Nei, M. (2005). Evolutionary dynamics of olfactory receptor genes in fishes and tetrapods. Proc. Natl. Acad. Sci. U.S.A. 102, 6039–6044. doi: 10.1073/pnas.0501922102
Niimura, Y., and Nei, M. (2006). Evolutionary dynamics of olfactory and other chemosensory receptor genes in vertebrates. J. Hum. Genet. 51, 505–517. doi: 10.1007/s10038-006-0391-8
Oike, H., Nagai, T., Furuyama, A., Okada, S., Aihara, Y., Ishimaru, Y., et al. (2007). Characterization of ligands for fish taste receptors. J. Neurosci. 27, 5584–5592. doi: 10.1523/jneurosci.0651-07.2007
Poh, C. H., Yuen, R., Khoo, H. E., Chung, M., Gwee, M., and Gopalakrishnakone, P. (1991). Purification and partial characterization of stonustoxin (lethal factor) from Synanceja horrida venom. Comp. Biochem. Physiol. B 99, 793–798. doi: 10.1016/0305-0491(91)90143-2
Senzui, A., Masumoto, T., and Fukada, H. (2020). Neuropeptide Y expression in response to sensory organ-detected fish meal soluble components and orally fed fish meal-based diet in yellowtail Seriola quinqueradiata. Aquaculture 514:734512. doi: 10.1016/j.aquaculture.2019.734512
Shao, C., Niu, Y., Rastas, P., Liu, Y., Xie, Z., Li, H., et al. (2015). Genome-wide SNP identification for the construction of a high-resolution genetic map of Japanese flounder (Paralichthys olivaceus): applications to QTL mapping of Vibrio anguillarum disease resistance and comparative genomic analysis. DNA Res. 22, 161–170. doi: 10.1093/dnares/dsv001
Shichida, Y., and Matsuyama, T. (2009). Evolution of opsins and phototransduction. Philos. Trans. R. Soc. Lond. B Biol. Sci. 364, 2881–2895. doi: 10.1098/rstb.2009.0051
Simao, F. A., Waterhouse, R. M., Ioannidis, P., Kriventseva, E. V., and Zdobnov, E. M. (2015). BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 31, 3210–3212. doi: 10.1093/bioinformatics/btv351
Slater, G. S., and Birney, E. (2005). Automated generation of heuristics for biological sequence comparison. BMC Bioinform. 6:31. doi: 10.1186/1471-2105-6-31
Song, W., Li, Y., Zhao, Y., Liu, Y., Niu, Y., Pang, R., et al. (2012). Construction of a High-density microsatellite genetic linkage map and mapping of sexual and growth-related traits in half-smooth tongue sole (Cynoglossus semilaevis). PLoS One 7:e52097. doi: 10.1371/journal.pone.0052097
Stanke, M., Keller, O., Gunduz, I., Hayes, A., Waack, S., and Morgenstern, B. (2006). AUGUSTUS: ab initio prediction of alternative transcripts. Nucleic Acids Res. 34, W435–W439.
Sun, B. J., Xie, H. X., Song, Y., and Nie, P. (2007). Gene structure of an antimicrobial peptide from mandarin fish, Siniperca chuatsi (Basilewsky), suggests that moronecidins and pleurocidins belong in one family: the piscidins. J. Fish Dis. 30, 335–343. doi: 10.1111/j.1365-2761.2007.00789.x
Sun, C., Niu, Y., Ye, X., Dong, J., Hu, W., Zeng, Q., et al. (2017). Construction of a high-density linkage map and mapping of sex determination and growth-related loci in the mandarin fish (Siniperca chuatsi). BMC Genom. 18:446. doi: 10.1186/s12864-017-3830-3
Tarailo-Graovac, M., and Chen, N. (2009). Using RepeatMasker to identify repetitive elements in genomic sequences. Curr. Protoc. Bioinform. 25, 4.10.11–14.10.14.
Terai, Y., Mayer, W. E., Klein, J., Tichy, H., and Okada, N. (2002). The effect of selection on a long wavelength-sensitive (LWS) opsin gene of Lake Victoria cichlid fishes. Proc. Natl. Acad. Sci. U.S.A. 99, 15501–15506. doi: 10.1073/pnas.232561099
Tine, M., Kuhl, H., Gagnaire, P. A., Louro, B., Desmarais, E., Martins, R. S., et al. (2014). European sea bass genome and its variation provide insights into adaptation to euryhalinity and speciation. Nat. Commun. 5:5770.
Trapnell, C., Pachter, L., and Salzberg, S. L. (2009). TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25, 1105–1111. doi: 10.1093/bioinformatics/btp120
Trapnell, C., Roberts, A., Goff, L., Pertea, G., Kim, D., Kelley, D. R., et al. (2012). Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and cufflinks. Nat. Protoc. 7, 562–578. doi: 10.1038/nprot.2012.016
Utkin, Y. N. (2015). Animal venom studies: current benefits and future developments. World J. Biol. Chem. 6, 28–33. doi: 10.4331/wjbc.v6.i2.28
Van Ooijen, J. (2006). JoinMap§4, Software for the Calculation of Genetic Linkage Maps in Experimental Populations. Wageningen: Kyazma BV.
Volkoff, H. (2006). The role of neuropeptide Y, orexins, cocaine and amphetamine-related transcript, cholecystokinin, amylin and leptin in the regulation of feeding in fish. Comp. Biochem. Physiol. A Mol. Integr. Physiol. 144, 325–331. doi: 10.1016/j.cbpa.2005.10.026
Volkoff, H. (2016). The neuroendocrine regulation of food intake in fish: a review of current knowledge. Front. Neurosci. 10:540. doi: 10.3389/fnins.2016.00540
Volkoff, H., Hoskins, L. J., and Tuziak, S. M. (2010). Influence of intrinsic signals and environmental cues on the endocrine control of feeding in fish: potential application in aquaculture. Gen. Comp. Endocrinol. 167, 352–359. doi: 10.1016/j.ygcen.2009.09.001
Walewski, J. L., Ge, F., Lobdell, H. T., Levin, N., Schwartz, G. J., Vasselli, J. R., et al. (2014). Spexin is a novel human peptide that reduces adipocyte uptake of long chain fatty acids and causes weight loss in rodents with diet-induced obesity. Obesity 22, 1643–1652. doi: 10.1002/oby.20725
Woods, I. G., Schoppik, D., Shi, V. J., Zimmerman, S., Coleman, H. A., Greenwood, J., et al. (2014). Neuropeptidergic signaling partitions arousal behaviors in zebrafish. J. Neurosci. 34, 3142–3160. doi: 10.1523/jneurosci.3529-13.2014
Wu, Z. (1988). A preliminary ethological analysis on the feeding behavior of mandarin fish. Freshw. Fish 5, 18–21.
Xie, B., Huang, Y., Baumann, K., Fry, B. G., and Shi, Q. (2017). From marine venoms to drugs: efficiently supported by a combination of transcriptomics and proteomics. Mar. Drugs 15, 1–10. doi: 10.3390/md15040103
Xu, Z., and Wang, H. (2007). LTR_FINDER: an efficient tool for the prediction of full-length LTR retrotransposons. Nucleic Acids Res. 35, W265–W268.
Yokobori, E., Azuma, M., Nishiguchi, R., Kang, K. S., Kamijo, M., Uchiyama, M., et al. (2012). Neuropeptide Y stimulates food intake in the Zebrafish, Danio rerio. J. Neuroendocrinol. 24, 766–773. doi: 10.1111/j.1365-2826.2012.02281.x
Yokoyama, S. (1995). Amino acid replacements and wavelength absorption of visual pigments in vertebrates. Mol. Biol. Evol. 12, 53–61. doi: 10.1093/oxfordjournals.molbev.a040190
Yokoyama, S. (2000). Molecular evolution of vertebrate visual pigments. Prog. Retin. Eye Res. 19, 385–419. doi: 10.1016/s1350-9462(00)00002-1
You, X., Bian, C., Zan, Q., Xu, X., Liu, X., Chen, J., et al. (2014). Mudskipper genomes provide insights into the terrestrial adaptation of amphibious fishes. Nat. Commun. 5:5594.
Zeng, Q., Liu, S., Yao, J., Zhang, Y., Yuan, Z., Jiang, C., et al. (2016). Transcriptome display during testicular differentiation of channel catfish (Ictalurus punctatus) as revealed by RNA-Seq analysis. Biol. Reprod. 95:19. doi: 10.1095/biolreprod.116.138818
Zhang, F.-B., Wang, Y., Xiao, J., and Zeng, Y. (2019). Study on distribution of Ichthyotoxic fish in the Jialing River. Resourc. Environ. Yangtze Basin 28, 2901–2909.
Zhang, S., Li, J., Qin, Q., Liu, W., Bian, C., Yi, Y., et al. (2018). Whole-genome sequencing of chinese yellow catfish provides a valuable genetic resource for high-throughput identification of toxin genes. Toxins 10:488. doi: 10.3390/toxins10120488
Zhang, D.-C., Guo, L., Guo, H.-Y., Zhu, K.-C., Li, S.-Q., Zhang, Y., et al. (2019). Chromosome-level genome assembly of golden pompano (Trachinotus ovatus) in the family Carangidae. Sci. Data. 6, 1–11.
Zhang, S., Zhang, X., Chen, X., Xu, T., Wang, M., Qin, Q., et al. (2019). Construction of a High-density linkage map and QTL fine mapping for growth- and sex-related traits in channel catfish (Ictalurus punctatus). Front. Genet. 10:251. doi: 10.3389/fgene.2019.00251
Zhao, G. Q., Zhang, Y., Hoon, M. A., Chandrashekar, J., Erlenbach, I., Ryba, N. J., et al. (2003). The receptors for mammalian sweet and umami taste. Cell 115, 255–266. doi: 10.1016/s0092-8674(03)00844-4
Zheng, B., Li, S., Liu, Y., Li, Y., Chen, H., Tang, H., et al. (2017). Spexin suppress food intake in zebrafish: evidence from gene knockout study. Sci. Rep. 7:14643.
Keywords: the mandarin fish (Siniperca chuatsi), whole-genome sequencing, feeding habit, molecular mechanism, chromosome-level genome assembly
Citation: Ding W, Zhang X, Zhao X, Jing W, Cao Z, Li J, Huang Y, You X, Wang M, Shi Q and Bing X (2021) A Chromosome-Level Genome Assembly of the Mandarin Fish (Siniperca chuatsi). Front. Genet. 12:671650. doi: 10.3389/fgene.2021.671650
Received: 24 February 2021; Accepted: 07 May 2021;
Published: 23 June 2021.
Edited by:
Ka Yan Ma, Sun Yat-sen University, ChinaReviewed by:
Xinhai Ye, Zhejiang University, ChinaChangwei Shao, Chinese Academy of Fishery Sciences, China
Copyright © 2021 Ding, Zhang, Zhao, Jing, Cao, Li, Huang, You, Wang, Shi and Bing. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xuwen Bing, YmluZ3h3QGZmcmMuY24=; Xinxin You, eW91eGlueGluQGdlbm9taWNzLmNu
†These authors have contributed equally to this work
 Weidong Ding
Weidong Ding Xinhui Zhang
Xinhui Zhang Xiaomeng Zhao2,3†
Xiaomeng Zhao2,3† Xuwen Bing
Xuwen Bing