
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Genet., 03 June 2021
Sec. Computational Genomics
Volume 12 - 2021 | https://doi.org/10.3389/fgene.2021.671585
This article is part of the Research TopicComputational Approaches for Biomarker Detection and Precision Therapeutics in CancersView all 21 articles
Background: Protein-coding gene LIM Domain Kinase 1 (LIMK1) is upregulated in various tumors and reported to promote tumor invasion and metastasis. However, the prognostic values of LIMK1 and correlation with immune infiltrates in lung adenocarcinoma are still not understood. Therefore, we evaluated the prognostic role of LIMK1 and its correlation with immune infiltrates in lung adenocarcinoma.
Methods: Transcriptional expression profiles of LIMK1 between lung adenocarcinoma tissues and normal tissues were downloaded from the Cancer Genome Atlas (TCGA). The LIMK1 protein expression was assessed by the Clinical Proteomic Tumor Analysis Consortium (CPTAC) and the Human Protein Atlas. Receiver operating characteristic (ROC) curve was used to differentiate lung adenocarcinoma from adjacent normal tissues. Kaplan-Meier method was conducted to assess the effect of LIMK1 on survival. Protein-protein interaction (PPI) networks were constructed by the STRING. Functional enrichment analyses were performed using the “ClusterProfiler” package. The relationship between LIMK1 mRNA expression and immune infiltrates was determined by tumor immune estimation resource (TIMER) and tumor-immune system interaction database (TISIDB).
Results: The expression of LIMK1 in lung adenocarcinoma tissues was significantly upregulated than those in adjacent normal tissues. Increased LIMK1 mRNA expression was associated with lymph node metastases and high TNM stage. The ROC curve analysis showed that with a cutoff level of 4.908, the accuracy, sensitivity, and specificity for LIMK1 differentiate lung adenocarcinoma from adjacent controls were 69.5, 93.2, and 71.9%, respectively. Kaplan-Meier survival analysis showed lung adenocarcinoma patients with high- LIMK1 had a worse prognosis than those with low- LIMK1 (43.1 vs. 55.1 months, P = 0.028). Correlation analysis indicated LIMK1 mRNA expression was correlated with tumor purity and immune infiltrates.
Conclusion: Upregulated LIMK1 is significantly correlated with poor survival and immune infiltrates in lung adenocarcinoma. Our study suggests that LIMK1 can be used as a biomarker of poor prognosis and potential immune therapy target in lung adenocarcinoma.
Lung cancer is one of the most common malignant tumors around the world and the leading cause for cancer-related death (Jemal et al., 2011). The incidence of lung cancer has steadily increased over recent years. Lung cancer remains refractory and the 5-year survival rate continues to be the lowest among the major cancers. It is speculated that numerous people will be diagnosed with lung cancer in the future, which bring a heavy economic burden to our society (Torre et al., 2016; Albaba et al., 2017). In the subtypes of lung cancer, lung adenocarcinoma accounts for about 50% (Brustugun et al., 2018). Despite many therapeutic endeavors has been made in lung adenocarcinoma, such as targeted therapy and immunotherapy, the survival rate remains bleak and staggers at about 20% 5 years after treatment (Hirsch et al., 2017). Thus, it is imperative to search novel biomarkers for advancing the prognosis of lung adenocarcinoma.
LIM Domain Kinase 1 (LIMK1) is a protein known as a member of the LIM kinase protein family. LIMK1 is consisted of gene spans 39,499 base pairs with 16 exons and encoded by a gene located on human chromosome 7q11.23 (Scott and Olson, 2007). Through phosphorylation and inactivation to its downstream effector of cofilin, LIMK1 has been shown to be important in regulating the polymerization of actin (Liu et al., 2019). When LIMK1 is phosphorylated, cofilin loses the ability to bind to actin, leading to the accumulation of actin polymers dysregulation of actin-mediated cytoskeletal changes (Nishimura et al., 2006). The phosphorylation of LIMK1 has been implicated with many cellular functions including angiogenesis, proliferation, cell cycle, and metastasis progression (Foletta et al., 2004; Nishimura et al., 2006). Previous studies have confirmed that ectopic expression of LIMK1 was associated with the progression of several tumor types, such as colorectal cancer, gastric cancer, prostate cancer, and breast cancer (Davila et al., 2003; McConnell et al., 2011; You et al., 2015; Liao et al., 2017). A paper from Huang et al. (2020) indicated that the upregulation of LIMK1 is correlated with lymph node metastasis and poor biochemical-free survival in prostate cancer. In pancreatic cancer, Vlecken and Bagowski (2009) reported that knockdown of LIMK1 can lead to an inhibition of invasion and metastatic behavior, as well as suppression of pancreatic cancer cell-induced angiogenesis. Moreover, some recent findings suggested that downregulation of LIMK1 can inhibit lung cancer cell migration (Chen et al., 2013; Wan et al., 2014; Zhang et al., 2020). Thus, LIMK1 has great potential to be a biomarker of poor prognosis and therapeutic target for lung cancer.
The prognostic values and correlation with immune infiltrates of LIMK1 in lung adenocarcinoma are still not fully understood. Given the overexpression of LIMK1 in lung cancer and the downregulation of LIMK1 can inhibit lung cancer cell migration, we hypothesized that the level of LIMK1 is associated with survival in lung adenocarcinoma. To test this hypothesis, we evaluated the prognostic role of LIMK1 in lung adenocarcinoma based on data from The Cancer Genome Atlas (TCGA). In this study, we found that LIMK1 is upregulated in lung adenocarcinoma. Significantly, the upregulation of LIMK1 is correlated with poor clinical characteristics and risk factors. We further evaluated the diagnostic and prognostic values, the correlation with immune infiltrates of LIMK1 for lung adenocarcinoma. Our study links the overexpression of LIMK1 and poor survival in lung adenocarcinoma.
Transcriptional expression data of LIMK1 and corresponding clinical information were downloaded from TCGA official website1 (Tomczak et al., 2015). The 18 enrolled cancer types contained at least 5 samples in the normal group. Finally, the RNA-Seq gene expression data with workflow type of FPKM was transformed into TPM format and log2 conversion for further study. Since all the data were downloaded from TCGA, this study did not need approval from the Ethics Committee.
The RNA-Seq expression data of LIMK1 in lung adenocarcinoma was also downloaded from TCGA. Therefore, 535 lung adenocarcinoma and 59 adjacent normal tissue data were retained. The samples selected contained LIMK1 gene expression data and associated clinical information, including age, gender, smoker condition, T stage, N stage, M stage, and tumor location. The mRNA expression data were characterized by mean ± SD.
With the application of proteomic technologies, CPTAC2 analyzes tumor biospecimens using mass spectrometry, quantifying and identifying the constituent proteins and characterizing proteome of each tumor sample (Edwards et al., 2015). UALCAN3 is a user-friendly online web resource for analyzing publicly available cancer data (Chandrashekar et al., 2017). In this study, we performed UALCAN to present a throughout analysis of LIMK1 protein expression from CPTAC.
HPA4 contains normal tissues and tumor tissues information regarding the expression profiles of human genes on protein level (Uhlen et al., 2015, 2017). In this study, we conducted HPA to compare the protein expression of LIMK1 between normal lung tissue and lung adenocarcinoma tissue.
STRING is an online database for the retrieval of interacting genes (version 11.05; Szklarczyk et al., 2011). In this study, we conducted STRING to search co-expression genes and construct PPI networks with an interaction score >0.4. Gene ontology (GO) enrichment and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analyses of co-expression genes were performed by the “ClusterProfiler” package and visualized by the “ggplot2” package (Wickham, 2016; Yu et al., 2012).
TIMER is a comprehensive online resource for systematic analysis of immune infiltrates across various cancer types6 (Li et al., 2017). In this study, we performed TIMER to determine the relationship between LIMK1 expression in lung adenocarcinoma and six immune infiltrates (B cells, CD4+ T cells, CD8+ T cells, neutrophils, macrophages, and dendritic cells).
TISIDB7 is an online web integrated repository portal for tumor-immune system interaction (Ru et al., 2019). In this study, we performed TISIDB to determine the expression of LIMK1 and tumor-infiltrating lymphocytes (TILs) across human cancers. Based on the gene expression profile, the relative abundance of TILs was inferred by using gene set variation analysis. The correlations between LIMK1 and TILs were measured by Spearman’s test.
PrognoScan database8 is a powerful online platform to evaluate the correlation between gene expression and survival across various types of cancers (Mizuno et al., 2009). In this study, we performed PrognoScan database to analyze the correlation between LIMK1 expression and overall survival in lung adenocarcinoma with two different datasets (jacob-00182-CANDF, jacob-00182-MSK).
All statistical analyses were performed with R (V 3.6.3)9 and R package ggplot2 was used to visualize expression differences. Paired t-test and Mann-Whitney U-test were used to determine the differences between lung adenocarcinoma tissues and adjacent normal tissues. ROC curve was performed to detect the cutoff value of LIMK1 using the pROC package (Robin et al., 2011). Kaplan-Meier and log-rank tests were conducted with the survminer package10 to assess the effect of LIMK1 on survival.
To evaluate the mRNA expression pattern of LIMK1 across different cancer types, we excluded from the analysis the datasets from 15 cancer types that contained less than five samples in the normal group. The final working set refers to 18 cancer types. As shown in Figure 1, compared with normal tissues, LIMK1 was significantly upregulated in 16 of all 18 cancer types. This data indicated the mRNA expression of LIMK1 was abnormally expressed across different cancer types.
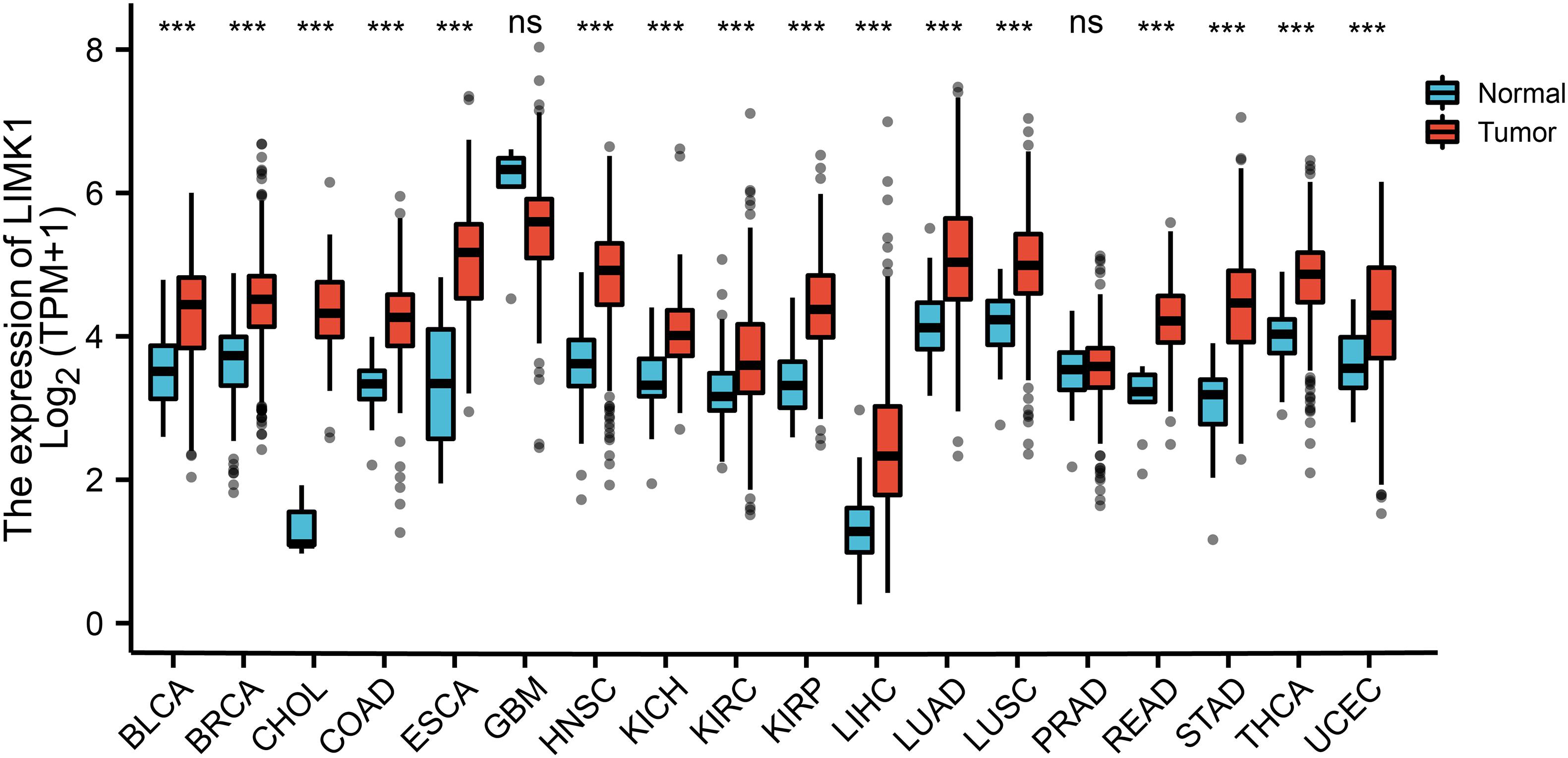
Figure 1. Expression pattern of LIMK1 in Pan-cancer perspective. The mRNA expression of LIMK1 was upregulated in 16 of 18 cancer types compared with normal tissues. (***P < 0.001). ns, no significance; BLCA, bladder urothelial carcinoma; BRCA, breast invasive carcinoma; CHOL, cholangiocarcinoma; COAD, colon adenocarcinoma; ESCA, esophageal carcinoma; GBM, glioblastoma mutiforme; HNSC, head and neck squamous cell carcinoma; KICH, kidney chromophobe; KIRC, kidney renal clear cell carcinoma; KIRP, kidney renal papillary cell carcinoma; LIHC, liver hepatocellular carcinoma; LUAD, lung adenocarcinoma; LUSC, lung squamous cell carcinoma; PRAD, prostate adenocarcinoma; READ, rectum adenocarcinoma; STAD, stomach adenocarcinoma; THCA, thyroid carcinoma; UCEC, uterine corpus endometrial carcinoma.
To determine the mRNA and protein expression of LIMK1 in lung adenocarcinoma, the LIMK1 expression data from TCGA and HPA were analyzed. The baseline characteristics of lung adenocarcinoma patients from TCGA were listed in Supplementary Table 1. As shown in Figure 2A, paired data analysis showed that the mRNA expression levels of LIMK1 in lung adenocarcinoma tissues (n = 57) were significantly higher than those in adjacent normal tissues (n = 57) (Figure 2A, 5.584 ± 0.747 vs. 4.320 ± 0.442, P < 0.001). Unpaired data analyses also showed that the mRNA expression levels of LIMK1 in lung adenocarcinoma tissues (n = 535) were significantly higher than those in adjacent normal tissues (n = 59) (Figure 2B, 5.314 ± 0.847 vs. 4.324 ± 0.437, Mann-Whitney U-test, P < 0.001). To present a throughout analysis of LIMK1 protein expression, we performed analysis on CPTAC with UALCAN. The result showed that the protein expression of LIMK1 in lung adenocarcinoma was significantly higher than those in normal tissues (Figure 2C). As shown in Figure 2D, immunohistochemical staining from HPA also revealed LIMK1 protein was upregulated in lung adenocarcinoma tissue. These results indicated that both mRNA and protein expression of LIMK1 are upregulated in lung adenocarcinoma tissues.
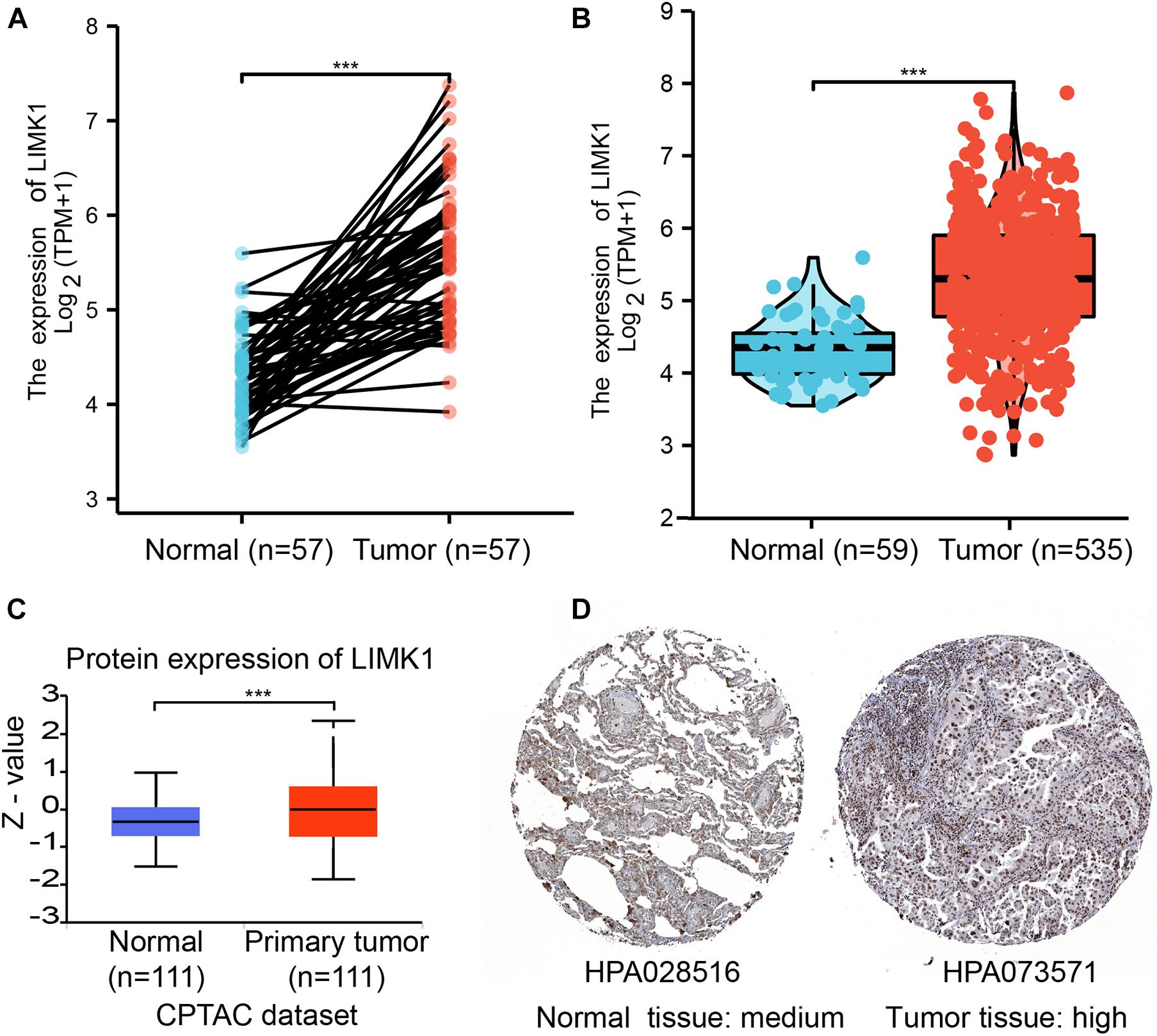
Figure 2. The mRNA and protein expression of LIMK1 in lung adenocarcinoma. (A) The mRNA expression levels of LIMK1 in 57 lung adenocarcinoma and matched-adjacent normal samples. (B) The mRNA expression levels of LIMK1 in 535 lung adenocarcinoma samples and 59 normal samples. (C) The protein expression levels of LIMK1 based on CPTAC. (D) The protein levels of LIMK1 based on Human Protein Atlas. Normal tissue, https://www.proteinatlas.org/ENSG00000106683-LIMK1/tissue/lung#img; Tumor tissue, https://www.proteinatlas.org/ENSG00000106683-LIMK1/pathology/lung+cancer#img (***P < 0.001).
To evaluate the association between the mRNA expression of LIMK1 and clinical pathological characteristics of lung adenocarcinoma samples, we performed Mann-Whitney U-test and logistic regression analysis. As shown in Table 1 and Figures 3A–I, higher expression levels of LIMK1 were observed in male patients (P = 0.004), patients with lymph node metastases (P = 0.022), and patients with high TNM stage (P = 0.048). However, no statistically significant correlation were found between the expression levels of LIMK1 and other clinical pathological characteristics, such as age (P = 0.113), smoker (P = 0.270), T stage (P = 0.129), M stage (P = 0.921), and anatomic subdivision (right vs. left, P = 0.959; peripheral vs. central, P = 0.562). Taken together, these results suggested that LIMK1 is correlated with lymph node metastases and high TNM stage, further suggesting LIMK1 may act as a biomarker of poor prognosis for lung adenocarcinoma.
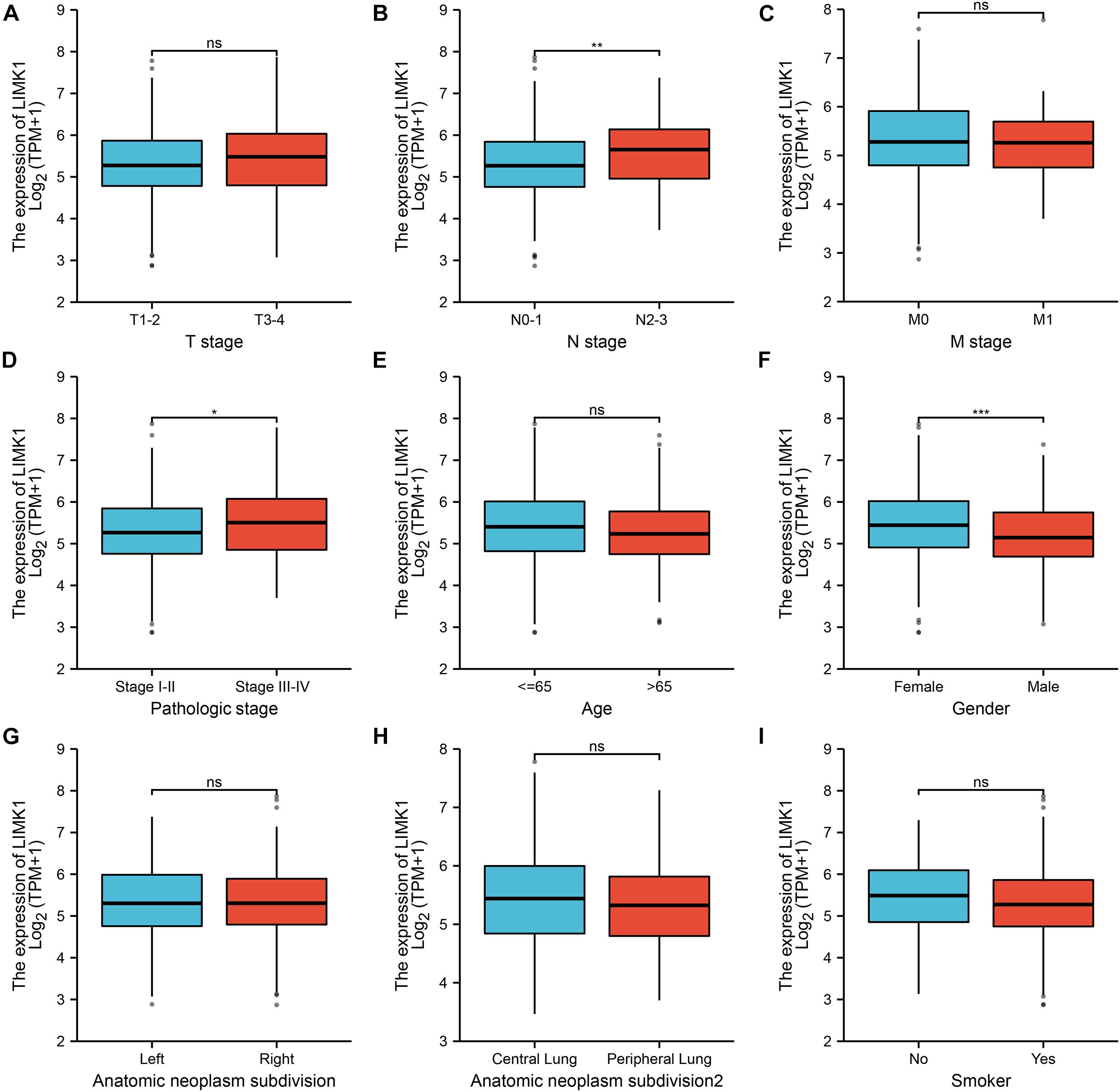
Figure 3. Relationships between LIMK1 mRNA levels and clinical pathological characteristics. LIMK1 mRNA expression was significantly correlated with lymph node metastases (B), high TNM stage (D) and gender was male (F). However, no statistically significant correlation were found between the expression levels of LIMK1 and T stage (A), M stage (C), age (E), anatomic neoplasm subdivision (G,H) and smoke condition (I) (ns, no significance, *P < 0.05, **P < 0.01, ***P < 0.001).
To investigate the value for LIMK1 to distinguish lung adenocarcinoma samples from normal smples, we performed a ROC curve analysis. As showed in Figure 4A, the ROC curve analysis showed LIMK1 had an AUC value of 0.851 (95% CI: 0.813–0.888). At a cutoff of 4.908, LIMK1 had a sensitivity, specificity, and accuracy of 69.5, 93.2, and 71.9%, respectively. The positive predictive value was 98.9% and the negative predictive value was 25.2%. These findings indicated that LIMK1 could be a promising biomarker to differentiate lung adenocarcinoma tissues from normal tissues.
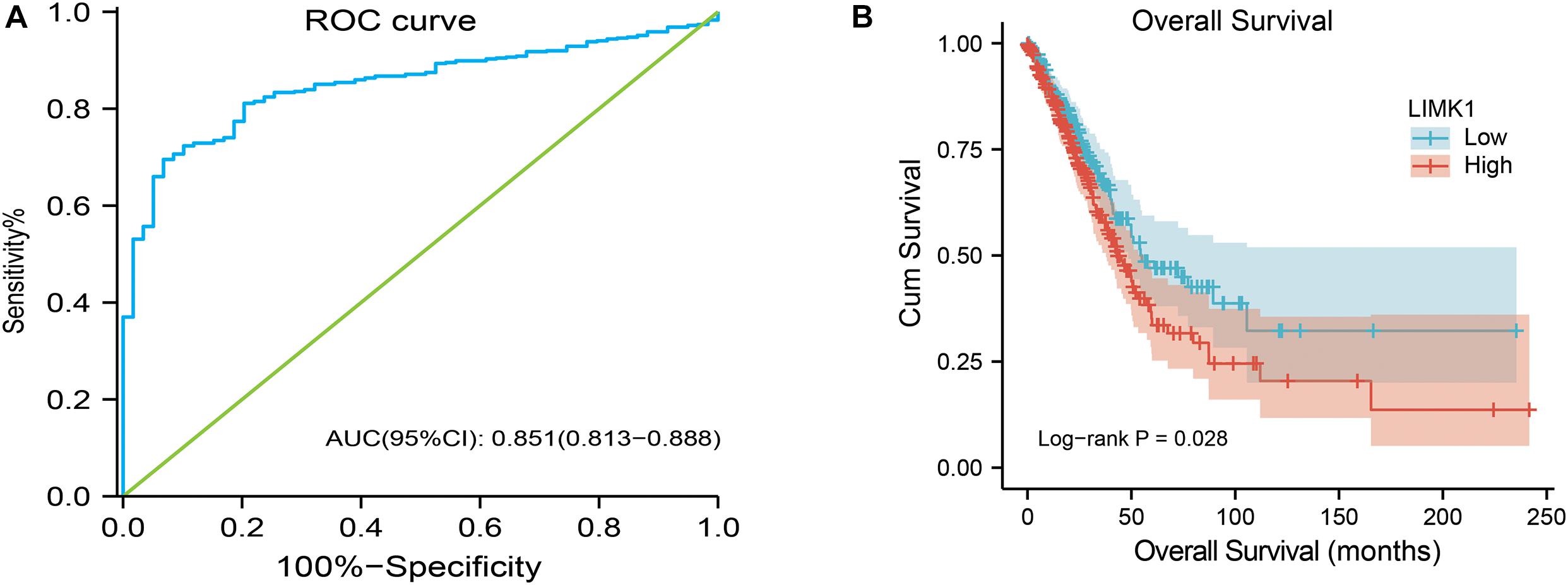
Figure 4. ROC and Kaplan-Meier curves for LIMK1. (A) ROC curve showed that LIMK1 had an AUC value of 0.851 to discriminate lung adenocarcinoma tissues from healthy controls. With a cutoff of 4.908, the sensitivity, specificity and accuracy were 93.2, 71.9, and 69.5%, respectively. (B) Kaplan-Meier survival curves indicated that lung adenocarcinoma patients with high LIMK1 mRNA expression had a shorter OS than those with low-level of LIMK1 (43.1 vs. 55.1 months, P = 0.028).
To explore the relationship between LIMK1 mRNA expression and OS in lung adenocarcinoma patients, Kaplan-Meier curves and PrognoScan database were performed. As shown in Figure 4B, the OS of lung adenocarcinoma patients with high-level of LIMK1 was significantly shorter than those with low-level of LIMK1 (43.1 vs. 55.1 months, P = 0.028). PrognoScan result with two different datasets (Supplementary Figure 1) also indicated that high expression of LIMK1 was correlated with poor overall survival in lung adenocarcinoma. These data indicated that high mRNA expression of LIMK1 is a biomarker of poor prognosis in lung adenocarcinoma.
To construct PPI networks and functional annotations, we conducted STRING database, GO, and KEGG analyses. Figure 5A showed a network of LIMK1 and its 10 co-expression genes. As shown in Figure 5B, changes in the biological process of LIMK1 were associated with actin filament organization, regulation of actin filament-based process, and actin cytoskeleton organization. Functional annotations indicated that these genes were involved in purine ribonucleoside binding, GTP Binding, and GTPase activity. The correlation analyses between the expression of LIMK1 and co-expressed genes in lung adenocarcinoma from TCGA were shown in Figures 5C–I.
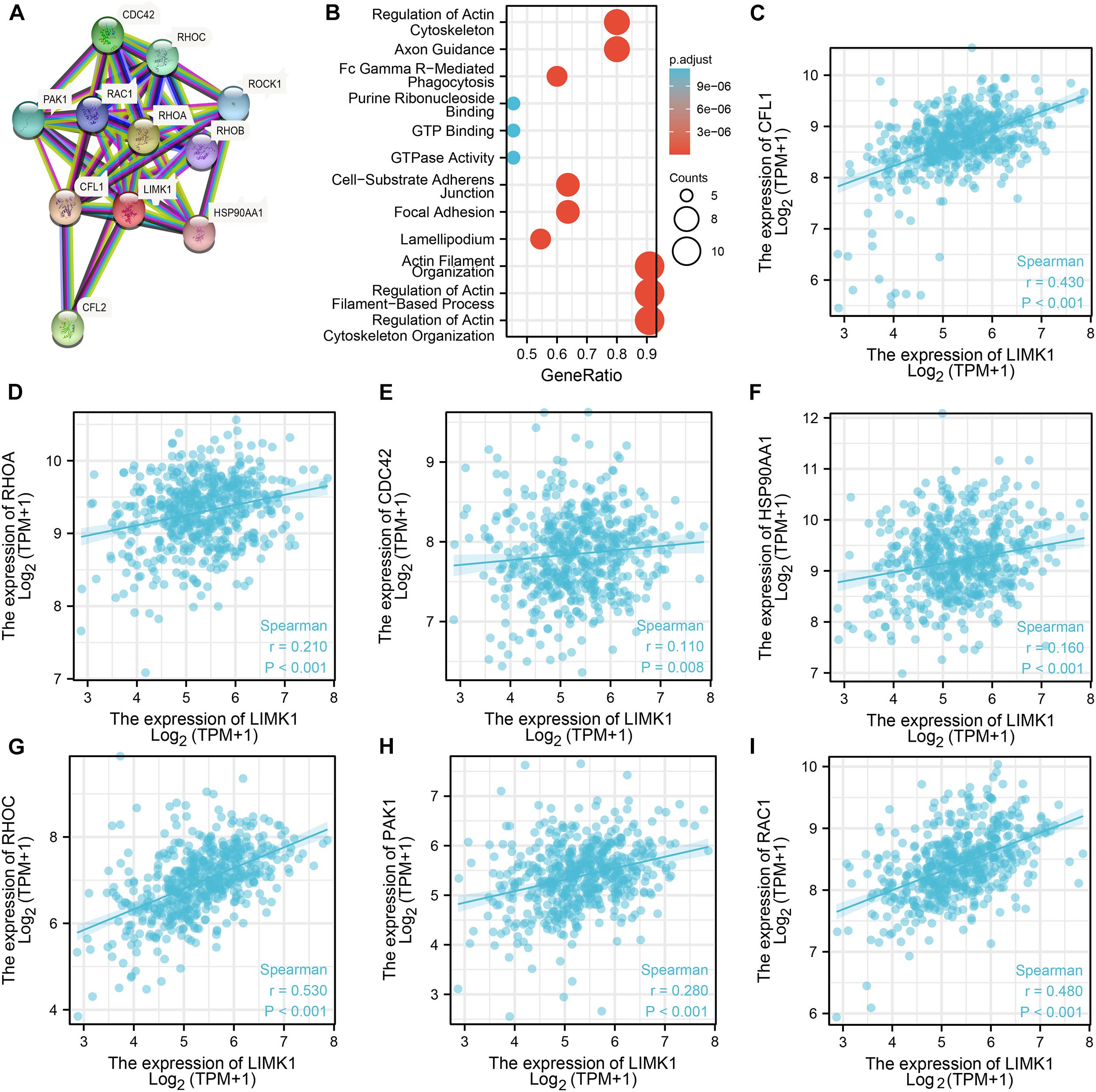
Figure 5. PPI networks and functional enrichment analyses. (A) A network of LIMK1 and its co-expression genes. (B) Functional enrichment analyses of 11 involved genes. LIMK1 was associated with actin filament organization, regulation of actin filament-based process, and actin cytoskeleton organization. These genes were involved in purine ribonucleoside binding, GTP Binding, and GTPase Activity. (C–I) The correlation analyses between the expression of LIMK1 and co-expressed genes in lung adenocarcinoma.CFL1, cofilin-1; RHOA, transforming protein RhoA; CFL2, cofilin-2; CDC42, cell division control protein 42 homolog; RHOC, Rho-related GTP-binding protein RhoC; PAK1, serine/threonine-protein kinase PAK 1; ROCK1, Rho-associated protein kinase 1; RAC1, Ras-related C3 botulinum toxin substrate 1; RHOB, Rho-related GTP-binding protein RhoB; HSP90AA1, heat shock protein HSP 90-alpha.
We analyzed the correlation between LIMK1 expression and the six types of tumor infiltrating immune cells in the TIMER database. As shown in Figure 6A, LIMK1 expression had correlations with tumor purity (r = −0.189, P = 2.37e-05), CD4+ T cell (r = 0.285, P = 1.65e-10), macrophage (r = 0.143, P = 1.64e-03), neutrophil (r = 0.263, P = 4.53e-09), dendritic cell (r = 0.363, P = 1.20e-16). We also evaluated the correlation between LIMK1 expression and 28 types of TILs in the TISIDB database. Figure 6B shown the relations between expression of LIMK1 and 28 types of TILs across human cancers. As shown in Figure 6C, the expression of LIMK1 was correlated with abundance of CD8+ T cells (r = 0.401, P = 2.2e-16), CD4+ T cells (r = 0.317, P = 1.92e-16), monocyte cells (r = 0.289, P = 2.71e-11), treg cells (r = 0.289, P = 4.41e-11), CD56dim cells (r = 0.275, P = 2.31e-10), and myeloid derived suppressor cells (MDSC, r = 0.275, P = 2.41e-10). These data indicated that LIMK1 may play a specific role in immune infiltration in lung adenocarcinoma.
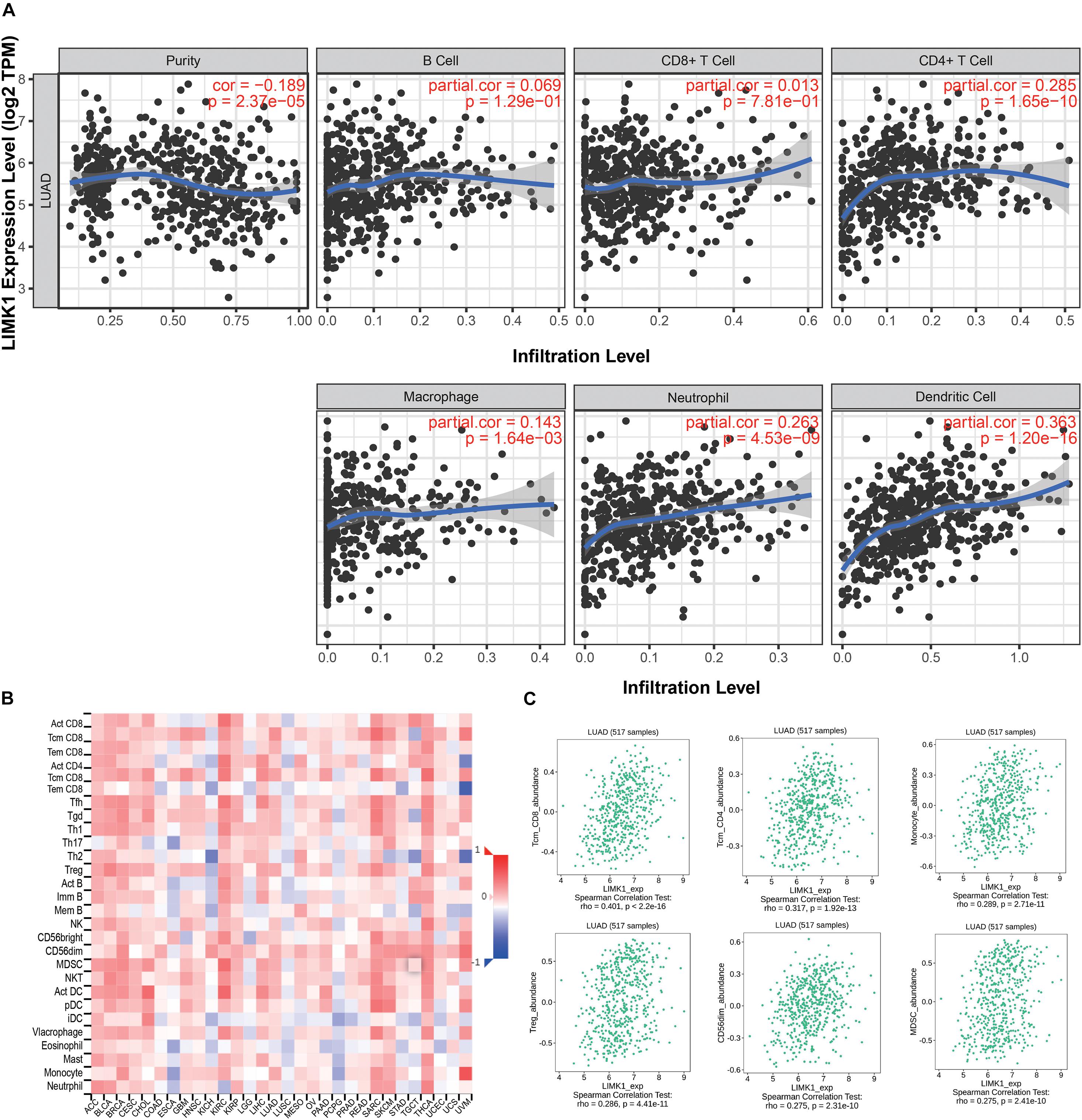
Figure 6. Correlations of LIMK1 expression with immune infiltration level. (A) LIMK1 expression is negatively related to tumor purity and has correlations with dendritic cell, CD4+ T cell, neutrophil, and macrophage in lung adenocarcinoma. (B) Relations between the expression of LIMK1 and 28 types of TILs across human cancers. (C) LIMK1 was correlated with abundance of CD8+ T cells, CD4+ T cells, monocyte cells, Treg cells, CD56dim cells, and MDSC cells.
In this study, we found that the mRNA expression of LIMK1 is upregulated in lung adenocarcinoma tissues. The upregulated mRNA expression of LIMK1 is positively correlated with lymph node metastases and high TNM stage. ROC curve analysis indicated that LIMK1 could be a promising diagnostic biomarker to differentiate lung adenocarcinoma from normal tissues. In light of Kaplan-Meier curves and univariate analysis, we confirmed that high mRNA expression of LIMK1 is associated with short OS and LIMK1 can be used as a potential biomarker of poor prognosis for lung adenocarcinoma. Moreover, LIMK1 may play a specific role in immune infiltration in lung adenocarcinoma.
LIMK1 is one of the members of the LIM kinase family and has been reported to play a significant role in promoting cell invasion and metastasis (Scott and Olson, 2007). Many studies about the oncogenic role of LIMK1 in several human cancers have been emerged in recent years, including gastric cancer, pancreatic cancer, as well as lung cancer (McConnell et al., 2011; Chen et al., 2013). Furthermore, it is reported that LIMK1 is upregulated in various cancers and associates with an unfavorable prognosis (Huang et al., 2020). However, the expression of LIMK1 and its prognostic value has not been fully investigated in lung adenocarcinoma. Here, in this study, based on pan-cancer analysis, our results are consistent with those reports that LIMK1 mRNA is abnormally expressed in various cancers. We also confirmed that LIMK1 is significantly upregulated in lung adenocarcinoma. High mRNA expression of LIMK1 is positively associated with lymph node metastases and high TNM stage, our finding agrees with the previous report by Chen et al. (2013). These findings suggest that LIMK1 might act as a potential biomarker of poor prognosis to identify lung adenocarcinoma with poor clinical outcome.
Currently, the function of LIMK1 in tumors had not been fully reported. Previous trials suggest that LIMK1 may be a target of dasatinib which can inhibit LIMK1 to suppress lung cancer cell proliferation and growth (Zhang et al., 2020). Other studies have shown LIMK1 acts as a direct target of miRNA-27-3p and miRNA-128-3p (Chen et al., 2017; Zhao et al., 2019), both miRNA-27-3p and miRNA-128-3p can suppress cancer cell proliferation, migration, and invasion. The underlying mechanism analysis showed that the LIMK1-cofilin signaling pathway plays an important role in tumor progression (Nishimura et al., 2006). All these results suggest that LIMK1 could be regarded as a promising biomarker or emerging target for cancer therapy. Given the condition that mRNA expression of LIMK1 is significantly higher in lung adenocarcinoma than in normal lung tissues, we speculate LIMK1 can act as a biomarker to differentiate lung adenocarcinoma from normal controls. In order to validate the clinical value of LIMK1 in the diagnosis of lung adenocarcinoma, we conducted ROC curve analysis. Our results showed that LIMK1 had a significantly high AUC value in the detection of lung adenocarcinoma, with 69.5% in sensitivity, 93.2% in specificity, and 71.9% in accuracy. On the basis of our finding, we conclude that LIMK1 might act as a potential diagnostic biomarker to differentiate lung adenocarcinoma from normal controls.
Recent studies have characterized LIMK1 as an important biomarker for poor prognosis and associated upregulated mRNA expression of LIMK1 with poor overall survival in many cancers. In prostate cancer, it is reported that elevated LIMK1 is positively associated with higher Gleason Scores and incidence of metastasis, as well as poor clinical outcome and reduced survival (Davila et al., 2007; Mardilovich et al., 2015; Huang et al., 2020). In ovarian cancer, Zhang et al. (2012) demonstrated that overexpression of LIMK1 is significant correlated with severity and poor differentiation level of ovarian cancer. A paper from Zhang et al. (2011) suggested that upregulation of LIMK1 can promote the invasion and metastasis in drug-resistant osteosarcoma and in turn LIMK1 can act as a potential novel therapeutic target. In glioblastoma, Chen et al. (2020) reported that LIMK1 is increased and the overexpression of LIMK1 is associated with high grade and poor prognosis. In contrast, suppression of LIMK1 can prolong survival time. However, the prognostic value of LIMK1 has not been investigated in lung adenocarcinoma. Given the upregulation of LIMK1 is positively correlated with lymph node metastases and high TNM stage, we speculated LIMK1 is involved in the development of lung adenocarcinoma. Moreover, since lymph node metastases and high TNM stage are correlated with poor survival, we speculated that the upregulation of LIMK1 is a biomarker of poor prognosis. Furthermore, in light of Kaplan-Meier curves and log-rank test, lung adenocarcinoma patients with high mRNA expression of LIMK1 are associated with a decreased survival rate than those with low LIMK1 levels. On the basis of our data, we concluded that LIMK1 can be used as a biomarker of poor prognosis for determining prognosis in lung adenocarcinoma.
LIMK1 is a crucial component of Rac1/PAK1/LIMK1/cofilin signaling pathway, which is involved in several cancers. For example, in cervical cancer, miR-509-3p can regulate this pathway to enhance the apoptosis and chemo-sensitivity of cervical cancer cells (Xu et al., 2012). In gastric cancer, the inhibition of Rho GDP dissociation inhibitor 2 can suppress tumor cell migration and invasion via signaling pathway (Zeng et al., 2020). In this study, co-expression analyses indicated that the expression of LIMK1 is significantly correlated to that of Rac1, PAK1, and CLF1. On the basis of our finding, we speculate that the upregulation of LIMK1 expression would affect the entire pathway. However, this should be tested in other experiments.
Many studies about the possible role of LIMK1 in human TILs have emerged in recent years. Xu et al. (2012) reported that LIMK1 may be involved in spontaneous actin polarization in transformed CD4 T cells. However, the correlation analysis between LIMK1 expression and immune cell infiltration in lung adenocarcinoma has not been investigated. In this study, we found that several tumor infiltrating immune cells (CD4+ T cell, macrophage, neutrophil, dendritic cell) were correlated with the expression of LIMK1 in lung adenocarcinoma by using TIMER. We also found that positive correlation were indicated between LIMK1 expression and CD8+ T cells, CD4+ T cells, monocyte cells, treg cells, CD56dim cells, and myeloid derived suppressor cells. These findings suggest that there is a potential correlation between LIMK1 and immune infiltration in lung adenocarcinoma. However, further research should be designed to confirm this correlation.
There are several limitations in this study. First, the expression and prognostic implication of LIMK1 were conducted with online public databases, further study with clinical samples is required to validate these results. Second, to further examine the detailed mechanism of the impact of LIMK1 on immune infiltration in lung adenocarcinoma, in vivo/vitro experiments should be designed.
In conclusion, in this study, we showed for the first time that mRNA expression of LIMK1 is upregulated in lung adenocarcinoma and positively correlated with lymph node metastases and high TNM stage. Our research suggests that LIMK1 could be regarded as a potential biomarker of poor prognosis to identify lung adenocarcinoma patients with poor clinical outcomes and may play a specific role in immune infiltration.
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
YuZ conceived and designed the study. GL performed data analysis and wrote the manuscript. YiZ and CZ contributed analysis tools. All authors reviewed the manuscript.
This study was supported by the Science and Technology Development Fund of Nanjing Medical University (NMUB2019112), “The 13TH Five-Year Plan” Major Program of Nanjing Medical Science and Technique Development Foundation (ZDX16012).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2021.671585/full#supplementary-material
Supplementary Figure 1 | The correlation between LIMK1 and overall survival in two different datasets analyzed with PrognoScan. High expression of LIMK1 was correlated with poor overall survival in dataset jacob-00182-CANDF (A) and jacob-00182-MSK (B).
Albaba, H., Lim, C., and Leighl, N. B. (2017). Economic Considerations in the Use of Novel Targeted Therapies for Lung Cancer: review of Current Literature. Pharmacoeconomics 35, 1195–1209. doi: 10.1007/s40273-017-0563-8
Brustugun, O. T., Gronberg, B. H., Fjellbirkeland, L., Helbekkmo, N., Aanerud, M., Grimsrud, T. K., et al. (2018). Substantial nation-wide improvement in lung cancer relative survival in Norway from 2000 to 2016. Lung Cancer 122, 138–145. doi: 10.1016/j.lungcan.2018.06.003
Chandrashekar, D. S., Bashel, B., Balasubramanya, S. A. H., Creighton, C. J., Ponce-Rodriguez, I., Chakravarthi, B., et al. (2017). UALCAN: a Portal for Facilitating Tumor Subgroup Gene Expression and Survival Analyses. Neoplasia 19, 649–658. doi: 10.1016/j.neo.2017.05.002
Chen, J., Ananthanarayanan, B., Springer, K. S., Wolf, K. J., Sheyman, S. M., Tran, V. D., et al. (2020). Suppression of LIM Kinase 1 and LIM Kinase 2 Limits Glioblastoma Invasion. Cancer Res. 80, 69–78. doi: 10.1158/0008-5472.CAN-19-1237
Chen, Q., Jiao, D., Hu, H., Song, J., Yan, J., Wu, L., et al. (2013). Downregulation of LIMK1 level inhibits migration of lung cancer cells and enhances sensitivity to chemotherapy drugs. Oncol. Res. 20, 491–498. doi: 10.3727/096504013x13657689382699
Chen, Y., Chen, G., Zhang, B., Liu, C., Yu, Y., and Jin, Y. (2017). miR-27b-3p suppresses cell proliferation, migration and invasion by targeting LIMK1 in colorectal cancer. Int. J. Clin. Exp. Pathol. 10, 9251–9261.
Davila, M., Frost, A. R., Grizzle, W. E., and Chakrabarti, R. (2003). LIM kinase 1 is essential for the invasive growth of prostate epithelial cells: implications in prostate cancer. J. Biol. Chem. 278, 36868–36875. doi: 10.1074/jbc.M306196200
Davila, M., Jhala, D., Ghosh, D., Grizzle, W. E., and Chakrabarti, R. (2007). Expression of LIM kinase 1 is associated with reversible G1/S phase arrest, chromosomal instability and prostate cancer. Mol. Cancer 6:40. doi: 10.1186/1476-4598-6-40
Edwards, N. J., Oberti, M., Thangudu, R. R., Cai, S., McGarvey, P. B., Jacob, S., et al. (2015). The CPTAC Data Portal: a Resource for Cancer Proteomics Research. J. Proteome. Res. 14, 2707–2713. doi: 10.1021/pr501254j
Foletta, V. C., Moussi, N., Sarmiere, P. D., Bamburg, J. R., and Bernard, O. (2004). LIM kinase 1, a key regulator of actin dynamics, is widely expressed in embryonic and adult tissues. Exp. Cell. Res. 294, 392–405. doi: 10.1016/j.yexcr.2003.11.024
Hirsch, F. R., Scagliotti, G. V., Mulshine, J. L., Kwon, R., Curran, W. J., Wu, Y. L., et al. (2017). Lung cancer: current therapies and new targeted treatments. Lancet 389, 299–311. doi: 10.1016/s0140-6736(16)30958-8
Huang, J. B., Wu, Y. P., Lin, Y. Z., Cai, H., Chen, S. H., Sun, X. L., et al. (2020). Up-regulation of LIMK1 expression in prostate cancer is correlated with poor pathological features, lymph node metastases and biochemical recurrence. J. Cell. Mol. Med. 24, 4698–4706. doi: 10.1111/jcmm.15138
Jemal, A., Bray, F., Center, M. M., Ferlay, J., Ward, E., and Forman, D. (2011). Global cancer statistics. CA Cancer J. Clin. 61, 69–90. doi: 10.3322/caac.20107
Li, T., Fan, J., Wang, B., Traugh, N., Chen, Q., Liu, J. S., et al. (2017). TIMER: a Web Server for Comprehensive Analysis of Tumor-Infiltrating Immune Cells. Cancer Res. 77, e108–e110. doi: 10.1158/0008-5472.can-17-0307
Liao, Q., Li, R., Zhou, R., Pan, Z., Xu, L., Ding, Y., et al. (2017). LIM kinase 1 interacts with myosin-9 and alpha-actinin-4 and promotes colorectal cancer progression. Br. J. Cancer 117, 563–571. doi: 10.1038/bjc.2017.193
Liu, J., Zhang, Z., Liu, J., and Wang, D. (2019). LIM Kinase 1 Mediates Estradiol Effects on the Phosphorylation of Cofilin1 in Eutopic Endometrial Stromal Cells During the Invasion and Proliferation of Endometriosis. Reprod. Sci. 26, 1499–1505. doi: 10.1177/1933719119828076
Mardilovich, K., Gabrielsen, M., McGarry, L., Orange, C., Patel, R., Shanks, E., et al. (2015). Elevated LIM kinase 1 in nonmetastatic prostate cancer reflects its role in facilitating androgen receptor nuclear translocation. Mol. Cancer Ther. 14, 246–258. doi: 10.1158/1535-7163.MCT-14-0447
McConnell, B. V., Koto, K., and Gutierrez-Hartmann, A. (2011). Nuclear and cytoplasmic LIMK1 enhances human breast cancer progression. Mol. Cancer 10:75. doi: 10.1186/1476-4598-10-75
Mizuno, H., Kitada, K., Nakai, K., and Sarai, A. (2009). PrognoScan: a new database for meta-analysis of the prognostic value of genes. BMC Med. Genom. 2:18. doi: 10.1186/1755-8794-2-18
Nishimura, Y., Yoshioka, K., Bernard, O., Bereczky, B., and Itoh, K. (2006). A role of LIM kinase 1/cofilin pathway in regulating endocytic trafficking of EGF receptor in human breast cancer cells. Histochem. Cell. Biol. 126, 627–638. doi: 10.1007/s00418-006-0198-x
Robin, X., Turck, N., Hainard, A., Tiberti, N., Lisacek, F., Sanchez, J. C., et al. (2011). pROC: an open-source package for R and S+ to analyze and compare ROC curves. BMC Bioinform. 12:77. doi: 10.1186/1471-2105-12-77
Ru, B., Wong, C. N., Tong, Y., Zhong, J. Y., Zhong, S. S. W., Wu, W. C., et al. (2019). TISIDB: an integrated repository portal for tumor-immune system interactions. Bioinformatics 35, 4200–4202. doi: 10.1093/bioinformatics/btz210
Scott, R. W., and Olson, M. F. (2007). LIM kinases: function, regulation and association with human disease. J. Mol. Med. 85, 555–568. doi: 10.1007/s00109-007-0165-6
Szklarczyk, D., Franceschini, A., Kuhn, M., Simonovic, M., Roth, A., Minguez, P., et al. (2011). The STRING database in 2011: functional interaction networks of proteins, globally integrated and scored. Nucleic Acids Res. 39, D561–8. doi: 10.1093/nar/gkq973
Tomczak, K., Czerwińska, P., and Wiznerowicz, M. (2015). The Cancer Genome Atlas (TCGA): an immeasurable source of knowledge. Contemp. Oncol. 19, A68–A77. doi: 10.5114/wo.2014.47136
Torre, L. A., Siegel, R. L., and Jemal, A. (2016). Lung Cancer Statistics. Adv. Exp. Med Biol. 893, 1–19. doi: 10.1007/978-3-319-24223-1_1
Uhlen, M., Fagerberg, L., Hallstrom, B. M., Lindskog, C., Oksvold, P., Mardinoglu, A., et al. (2015). Proteomics. Science 347:1260419. doi: 10.1126/science.1260419
Uhlen, M., Zhang, C., Lee, S., Sjostedt, E., Fagerberg, L., Bidkhori, G., et al. (2017). A pathology atlas of the human cancer transcriptome. Science 357:eaan2507. doi: 10.1126/science.aan2507
Vlecken, D. H., and Bagowski, C. P. (2009). LIMK1 and LIMK2 are important for metastatic behavior and tumor cell-induced angiogenesis of pancreatic cancer cells. Zebrafish 6, 433–439. doi: 10.1089/zeb.2009.0602
Wan, L., Zhang, L., Fan, K., and Wang, J. (2014). MiR-27b targets LIMK1 to inhibit growth and invasion of NSCLC cells. Mol. Cell. Biochem. 390, 85–91. doi: 10.1007/s11010-013-1959-1
Wickham, H. (2016). ggplot2 Elegant Graphics for Data Analysis. Germany: Springer International Publishing. doi: 10.1007/978-3-319-24277-4
Xu, X., Guo, J., Vorster, P., and Wu, Y. (2012). Involvement of LIM kinase 1 in actin polarization in human CD4 T cells. Commun. Integr. Biol. 5, 381–383. doi: 10.4161/cib.20165
You, T., Gao, W., Wei, J., Jin, X., Zhao, Z., Wang, C., et al. (2015). Overexpression of LIMK1 promotes tumor growth and metastasis in gastric cancer. Biomed. Pharmacother. 69, 96–101. doi: 10.1016/j.biopha.2014.11.011
Yu, G., Wang, L. G., Han, Y., and He, Q. Y. (2012). clusterProfiler: an R package for comparing biological themes among gene clusters. Omics 16, 284–287. doi: 10.1089/omi.2011.0118
Zeng, Y., Ren, M., Li, Y., Liu, Y., Chen, C., Su, J., et al. (2020). Knockdown of RhoGDI2 represses human gastric cancer cell proliferation, invasion and drug resistance via the Rac1/Pak1/LIMK1 pathway. Cancer Lett. 492, 136–146. doi: 10.1016/j.canlet.2020.07.013
Zhang, H., Wang, Y., Xing, F., Wang, J., Wang, Y., Wang, H., et al. (2011). Overexpression of LIMK1 promotes migration ability of multidrug-resistant osteosarcoma cells. Oncol. Res. 19, 501–509. doi: 10.3727/096504012x13286534482511
Zhang, M., Tian, J., Wang, R., Song, M., Zhao, R., Chen, H., et al. (2020). Dasatinib Inhibits Lung Cancer Cell Growth and Patient Derived Tumor Growth in Mice by Targeting LIMK1. Front. Cell. Dev. Biol. 8:556532. doi: 10.3389/fcell.2020.556532
Zhang, W., Gan, N., and Zhou, J. (2012). Immunohistochemical Investigation of the Correlation between LIM Kinase 1 Expression and Development and Progression of Human Ovarian Carcinoma. J. Int. Med. Res. 40, 1067–1073. doi: 10.1177/147323001204000325
Keywords: lung adenocarcinoma, LIMK1, LIM domain kinase1, biomarker, prognosis, immune infiltrates
Citation: Lu G, Zhou Y, Zhang C and Zhang Y (2021) Upregulation of LIMK1 Is Correlated With Poor Prognosis and Immune Infiltrates in Lung Adenocarcinoma. Front. Genet. 12:671585. doi: 10.3389/fgene.2021.671585
Received: 24 February 2021; Accepted: 06 May 2021;
Published: 03 June 2021.
Edited by:
Suman Ghosal, National Institutes of Health (NIH), United StatesReviewed by:
Enrique Medina-Acosta, State University of the North Fluminense Darcy Ribeiro, BrazilCopyright © 2021 Lu, Zhou, Zhang and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yu Zhang, emhhbmd5dTIxMTNfbmpAMTYzLmNvbQ==
†These authors have contributed equally to this work and share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.