- 1Department of Biology, University of Rochester, Rochester, NY, United States
- 2School of Biological Sciences, University of Nebraska, Lincoln, NE, United States
The three fruitfly species of the Drosophila simulans clade— D. simulans, D. mauritiana, and D. sechellia— have served as important models in speciation genetics for over 40 years. These species are reproductively isolated by geography, ecology, sexual signals, postmating-prezygotic interactions, and postzygotic genetic incompatibilities. All pairwise crosses between these species conform to Haldane’s rule, producing fertile F1 hybrid females and sterile F1 hybrid males. The close phylogenetic proximity of the D. simulans clade species to the model organism, D. melanogaster, has empowered genetic analyses of their species differences, including reproductive incompatibilities. But perhaps no phenotype has been subject to more continuous and intensive genetic scrutiny than hybrid male sterility. Here we review the history, progress, and current state of our understanding of hybrid male sterility among the D. simulans clade species. Our aim is to integrate the available information from experimental and population genetics analyses bearing on the causes and consequences of hybrid male sterility. We highlight numerous conclusions that have emerged as well as issues that remain unresolved. We focus on the special role of sex chromosomes, the fine-scale genetic architecture of hybrid male sterility, and the history of gene flow between species. The biggest surprises to emerge from this work are that (i) genetic conflicts may be an important general force in the evolution of hybrid incompatibility, (ii) hybrid male sterility is polygenic with contributions of complex epistasis, and (iii) speciation, even among these geographically allopatric taxa, has involved the interplay of gene flow, negative selection, and positive selection. These three conclusions are marked departures from the classical views of speciation that emerged from the modern evolutionary synthesis.
Introduction
From Darwin’s Origin of Species, to the modern evolutionary synthesis (Dobzhansky, 1937), and into the present era (Coyne and Orr, 2004), hybrid incompatibility— the intrinsic sterility or inviability of species hybrids— has held a central place in speciation research. The reason, most simply, is that hybrid incompatibility has contributed to reproductive isolation during the speciation histories of some species pairs, limiting gene flow and, on occasion, spurring the evolution of further reproductive isolation via reinforcement (Dobzhansky, 1940; Yukilevich, 2012). Conventional wisdom has it, however, that, during the time-course of speciation, hybrid incompatibility evolves late or, worse, after the fact (Mallet, 2006; Rabosky and Matute, 2013). Why then bother to study hybrid incompatibility? Apart from its sometimes role in speciation, there are at least two further reasons. One is that hybrid incompatibility is the manifestation of extreme species differences in genome function, structure, content, and regulation in which wildtype alleles from one species kill or sterilize when in the genomes of closely related species. Determining the molecular identities and forces involved in the evolution of these extreme species differences is therefore informative about the most rapidly evolving aspects of essential biological functions. The other reason is that hybrid incompatibility has presented a series of puzzles for evolutionary biology. For instance: as dead or sterile hybrid progeny are of no adaptive value, how could natural selection possibly explain the evolution of hybrid incompatibility? Darwin’s (1859) solution is that hybrid incompatibility is not itself adaptive but rather “incidental on other acquired differences” (p. 245). And: given Mendelian inheritance, how is the evolution of hybrid incompatibility permissible by natural selection at all? If heterozygous (Aa) hybrids are sterile because A and a alleles are incompatible, then the critical substitution (AA → Aa → aa) would be precluded by the sterile intermediate genotype. Bateson’s (1909) solution is that hybrid incompatibility can evolve readily, without passing through problematic intermediate genotypes, so long as substitutions occur at different interacting loci [AABB → aaBB → aabb, with, e.g., the A and b alleles being incompatible; see Orr (1996)]. The Darwin and Bateson solutions were later rediscovered and deepened by the modern synthesis thinkers (Dobzhansky, 1937; Muller, 1940, 1942) and today represent starting points for most genetic studies of hybrid incompatibility. As we explain below, modern genetic analyses have left us with several new puzzles concerning the evolution, biology, and consequences of hybrid incompatibilities.
Genetic incompatibility thinking was implicit in Sturtevant’s (1920) pioneering investigations of lethality in hybrids between Drosophila melanogaster and the newly discovered Drosophila simulans. Over the decades since, crosses between these two species have leveraged the ever-expanding genetic, molecular, and genomic resources of D. melanogaster to reveal, among other things, that F1 hybrid lethality involves sex chromosomes, autosomes, maternal factors (Sturtevant, 1920), protein-coding genes (Barbash et al., 2003; Brideau et al., 2006; Phadnis et al., 2015), and repetitive DNAs (Sawamura and Yamamoto, 1997; Ferree and Barbash, 2009, Satyaki et al., 2014). A major limitation of these species for genetic analysis of reproductive incompatibilities, however, is their age: as D. melanogaster diverged from D. simulans ∼3 Mya, all of their hybrids are dead or sterile, and genetic analyses involving D. melanogaster are limited, under most circumstances, to F1 hybrids [but see Sawamura et al. (2000); Masly et al. (2006))]. This problem has been circumvented in part by the discovery of “hybrid rescue mutations”— compatible alleles at otherwise incompatible loci that completely (or nearly so) reverse hybrid lethality (Watanabe, 1979; Hutter and Ashburner, 1987). The existence of these mutations shows that the genetic basis of hybrid lethality is sufficiently simple that hybrid rescue is possible by changing the genotype at any of a small number of loci. In contrast to hybrid lethality, however, F1 hybrid males from these crosses are sterile many times over so that no single-locus change in genotype can rescue their fertility (Sawamura, 2000; Sawamura et al., 2000). This implied faster accumulation of hybrid male sterility turns out to be a general feature of Drosophila speciation: among hundreds of species pairs, hybrid male sterility evolves earlier than other forms of hybrid incompatibility [e.g., hybrid lethality, hybrid female sterility (Wu, 1992; Wu and Davis, 1993, Coyne and Orr, 1997)]. Hybrid male sterility is therefore more likely to contribute to reproductive isolation, better reflects those biological functions that diverge fastest between species and, as discussed below, presents its own set of puzzles. To study hybrid incompatibility genes and phenotypes that evolve early during species divergence clearly requires studying younger species pairs than the D. melanogaster-D. simulans hybridization.
Here we review the history, progress, and major results of the genetics of HMS among the younger species of the D. simulans clade species— D. simulans (Sturtevant, 1919), Drosophila mauritiana (Tsacas and David, 1974), and D. sechellia (Tsacas and Bächli, 1981). These species diverged from one another ∼250 Kya [(Kliman et al., 2000; McDermott and Kliman, 2008, Garrigan et al., 2012); but see Schrider et al. (2018) for a younger estimated split time, ∼100 Kya], when a presumed D. simulans-like ancestor from Madagascar gave rise to D. sechellia on the Seychelles archipelago and D. mauritiana on Mauritius and Rodrigues islands. While morphologically similar [except for conspicuous differences in male genitalia (Ashburner et al., 2005)], the three simulans clade species are now partially reproductively isolated by geography, ecology (Lachaise et al., 1988), sexual isolation (Coyne, 1992a), postmating-prezygotic isolation (Price, 1997), and intrinsic hybrid incompatibility (Lachaise et al., 1986). During the past 40 years, these species have served as important models for the genetic analysis of speciation and species differences, producing many key breakthroughs that have influenced our thinking about speciation genetics. Here we focus on three major and, at the time, surprising observations to emerge from these efforts: the special role of sex chromosomes; the complex genetic architecture; and the interaction of gene flow and selection. Our review comes with two caveats. Rather than attempt a thorough survey of all hypotheses and alternative models, we focus on those that achieved some critical threshold of attention or otherwise guided the direction of research. And rather than present a taxonomically comprehensive review, we focus narrowly on lessons from the D. simulans clade species.
Sex Chromosomes and Hybrid Sterility
The sex chromosomes invariably play the largest role in hybrid sterility or inviability (Coyne and Orr, 1989a). The rapid evolution of hybrid male sterility (at least in male-heterogametic taxa) is arguably the most significant legacy from the last decade’s probing of Haldane’s rule (Wu et al., 1996).
Sex chromosomes feature prominently in three strong empirical patterns that characterize speciation. First, Haldane’s rule is the phenotypic observation from species crosses that if one F1 hybrid sex is dead, sterile or otherwise unfit, it tends to be the heterogametic (XY or ZW) sex (Haldane, 1922; Laurie, 1997, Orr, 1997; Schilthuizen et al., 2011, Delph and Demuth, 2016). Haldane’s rule is notable because it holds widely— in insects, birds, fish, mammals, and plants— and appears to be an obligate, intermediate phase in the gradual evolution of complete hybrid incompatibility (Coyne and Orr, 1989b). The fact that Haldane’s rule holds in male- (XY) and female-heterogametic (ZW) taxa implicates hybrid sex chromosome genotype rather than sex per se. Second, the large X-effect is the genetic observation in backcross analyses that the X chromosome has a disproportionately large effect on hybrid incompatibility, given its physical size and gene content (Dobzhansky, 1936; Coyne and Orr, 1989a; Coyne, 1992b; Presgraves, 2008a). To these “two rules of speciation” (Coyne and Orr, 1989a), a third population-genetic observation may be relevant: genetic differentiation between taxa is generally greater on the X chromosome than the autosomes, consistent with reduced interspecific gene flow on the X (Presgraves, 2018). All three patterns have attracted the attention of speciation geneticists as all three involve sex chromosomes, suggesting the possibility of a single, unitary explanation. If we can determine why the X (or Z) chromosome plays a special role in hybrid incompatibility, then we might have a ready explanation for three strong generalizations in speciation. Causes for the special role of sex chromosomes in speciation can be partitioned into (proximate) genetic causes and (ultimate) evolutionary causes which we discuss in turn.
Genetic Causes
Crosses between the three D. simulans clade species follow Haldane’s rule: F1 hybrid females are fertile but F1 hybrid males completely sterile (Lachaise et al., 1986). Backcross analyses of HMS between D. simulans (sim), D. mauritiana (mau), and D. sechellia (sech) show a large X-effect [Figure 1; (Coyne, 1984; Coyne and Kreitman, 1986)]. The X chromosome might have a disproportionately large effect on hybrid fitness problems for any of three genetic reasons: (1) the effect sizes of hybrid incompatibility factors on the X exceeds those of autosomal ones (Coyne and Orr, 1989a); (2) the density of hybrid incompatibility factors on the X exceeds that for the autosomes (Charlesworth et al., 1987; Coyne and Orr, 1989a; Turelli and Orr, 2000; Naveira, 2003); and/or (3) the negative effects of incompatibility alleles are on average recessive in hybrids (Muller, 1940, 1942, Turelli and Orr, 1995, 2000). Under this dominance theory, Haldane’s rule occurs because the XY sex, being hemizygous for the X, suffers the full effects of any X-linked hybrid incompatibilities whereas the XX sex, being heterozygous, is not. Similarly, the large X-effect occurs because foreign X-linked alleles are always hemizygous in backcross hybrid males whereas autosomal ones are always heterozygous (Wu and Davis, 1993).
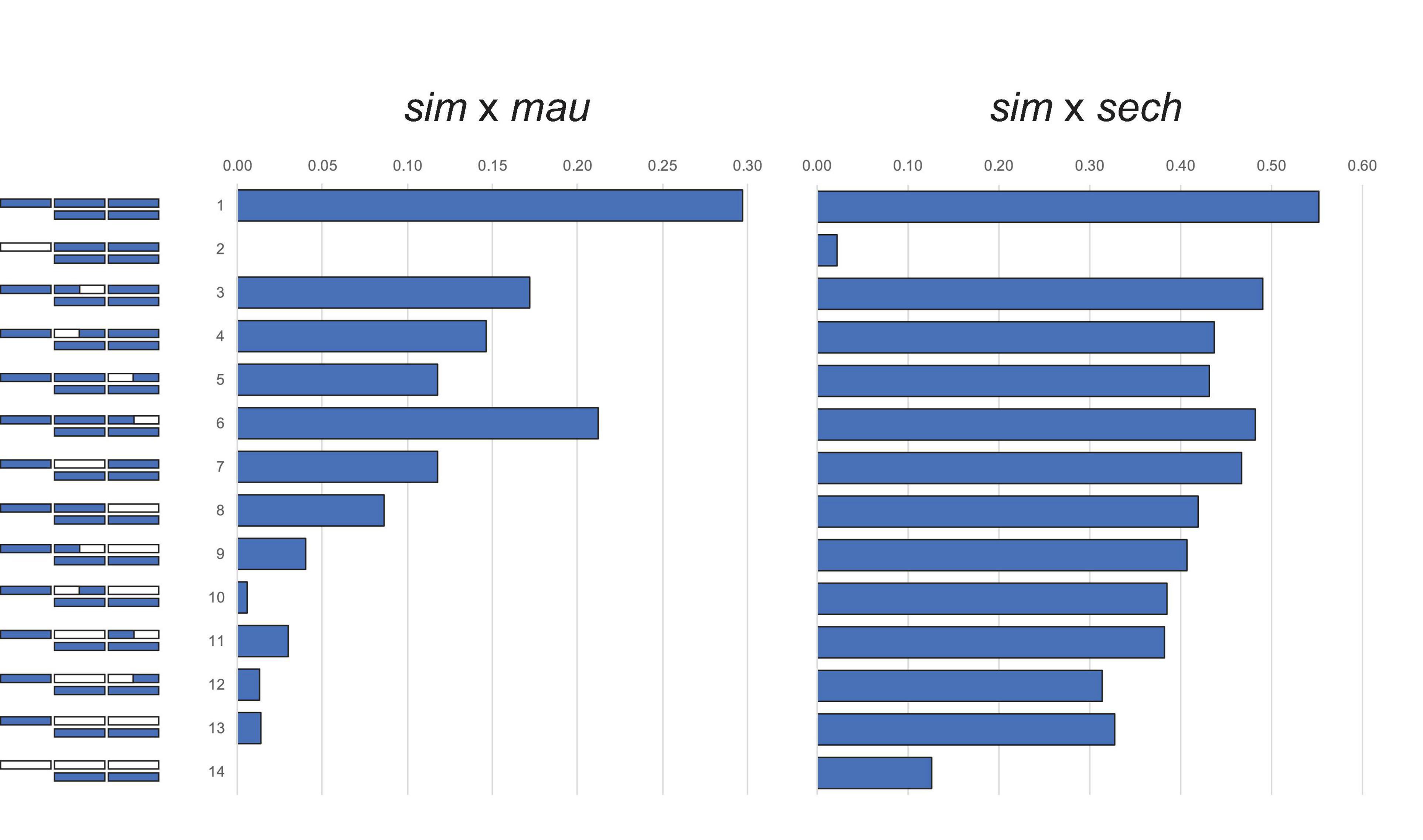
Figure 1. The large X-effect in backcross hybrid male sterility from crosses between D. simulans (sim) and D. mauritiana (mau) and between D. simulans and D. sechellia (sech). Bars represent the proportion males with motile sperm. D. simulans chromosome segments shown in purple, D. mauritiana or D. sechellia in white. Figures reproduced from data in Coyne (1984) and Coyne and Kreitman (1986).
In a seminal paper, Coyne (1985) assayed the fertility of “unbalanced” F1 hybrid females that are homozygous for the sim X chromosome (Figure 2). If hybrid sterility in males results from exposure of recessive X-linked incompatibility alleles in hemizygous state, then genotypically equivalent hybrid females should be sterile too. They are not (Figure 2C). One possibility is that X-Y incompatibilities cause sterility in hybrid males but not in attached-X hybrid females, for which the Y is irrelevant for oogenesis. For sim and sech, however, direct tests for X-Y incompatibilities show that (1) Ysech does not cause HMS in a sim genetic background; and (2) the sterility of Xsech/Ysim males is caused by X-autosome, not X-Y, incompatibilities (Johnson et al., 1992, 1993, Zeng and Singh, 1993). Together, these findings falsify any model in which sex differences in hybrid fitness are solely attributable to sex differences in genotype, as the same genotype sterilizes males but not females— instead hybrid males are sterile because they suffer different hybrid incompatibilities than hybrid females. The results lead to two strong conclusions. Hybrid sterility factors are sex-specific, as might be expected given highly sex-specific gametogenic programs (Lindsley and Tokuyasu, 1980); and genetic factors causing hybrid male sterility accumulate faster between these species than those causing hybrid female sterility (Wu and Davis, 1993; Wu et al., 1996).
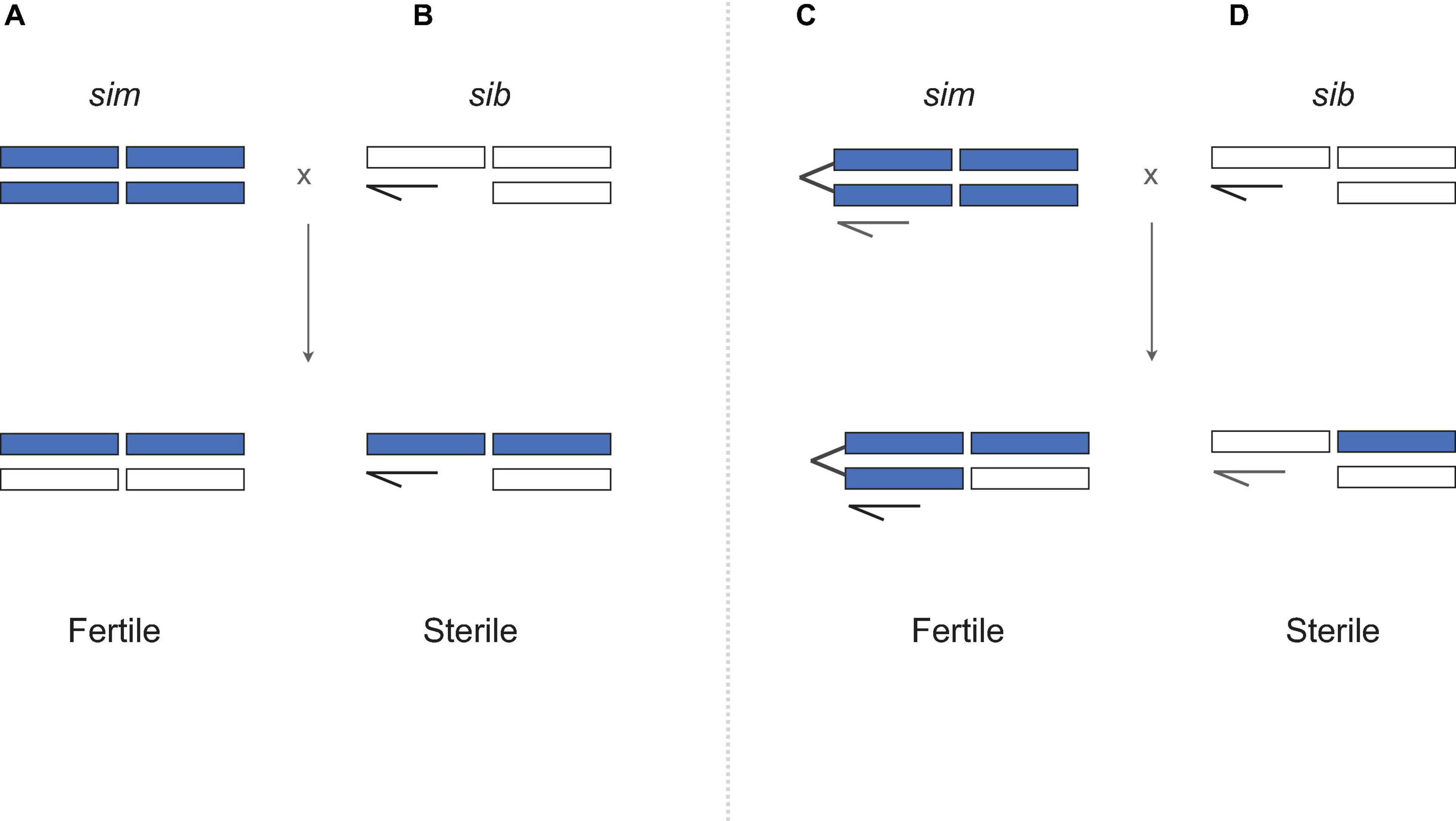
Figure 2. Querying the genetic basis of Haldane’s rule (A,B) using the attached-X test. (C,D) Attached-X chromosomes correspond to two X chromosomes that have been fused together and now segregate as a single chromosome. When D. simulans females carrying an attached-X are crossed to heterospecific males, the female F1 progeny inherit the sim attached-X and a heterospecific Y chromosome which has no effect in females (sex-determination in Drosophila is determined by the number of X chromosomes). Crosses involving attached-X chromosomes show that hybrid males are sterile (D) and hybrid females are fertile (C) despite having equivalent genotypes (B versus C). D. simulans (sim) material is shown in purple, D. mauritiana or D. sechellia (“sib”) material shown in white. Crosses based on Coyne (1985).
Fine-scale genetic analyses, in which small chromosomal segments generally comprising ≤2% of the genome are introgressed between species via recurrent backcrossing, confirm both conclusions: HMS factors accumulate at least ∼10 times faster than hybrid lethal or hybrid female sterility factors (Hollocher and Wu, 1996; True et al., 1996a, Tao et al., 2003a; Masly and Presgraves, 2007, Meiklejohn et al., 2018). These experiments also reveal a new, third conclusion— HMS accumulates between species at least 2.5- to 4-fold faster on the X chromosome than on the autosomes (True et al., 1996a; Tao et al., 2003a, Masly and Presgraves, 2007). This rapid accumulation of X-linked, male-specific hybrid sterility informs long-running debates about the genetic causes of Haldane’s rule and the large X-effect, and refutes the dominance explanation for hybrid male sterility. With these findings in mind, Turelli and Orr (Turelli and Orr, 1995, 2000) formulated a general “composite theory” for Haldane’s rule and the large X-effect. In male heterogametic taxa, Haldane’s rule is expected when
where τ is the ratio of the number of male- to female-specific incompatibilities, gX is the fraction of all incompatibilities that are X-linked, and d0 is the mean dominance of incompatibility alleles. (For brevity, we have suppressed some of the formal details of the model; see Turelli and Orr (2000) for the fuller treatment.) Data from the fine-scale genetic analyses suggest, conservatively, that τ ≥ 10 and gX ≥ 0.4 [(Tao and Hartl, 2003; Masly and Presgraves, 2007); see below]. Estimates of d0 are not available, but it does not matter: for all d0 ≤ 1, Haldane’s rule will be observed (R ≥ 6). Given the estimates of τ and gX, the large X-effect is similarly inevitable regardless of dominance (Naveira, 2003). Haldane’s rule among the simulans clade species can thus be explained entirely by the faster accumulation of factors causing hybrid male versus female sterility, whereas the large X-effect can be explained by the particularly fast accumulation of HMS factors on the X chromosome. The question now is why.
Evolutionary Causes
A simple mutagenic potential model cannot account for the rapid evolution of X-linked HMS: based on gene numbers and locations in Drosophila, viability presents a ∼10-fold larger mutational target for interspecific divergence than male fertility (Wu and Davis, 1993; Lindsley et al., 2013), and the X chromosome is neither enriched nor depleted for male fertility-essential or testis-expressed genes (Meiklejohn and Presgraves, 2012; Meisel et al., 2012, Lindsley et al., 2013). We therefore need evolutionary models that can account for the >10-fold excess of factors causing HMS versus hybrid lethality (or hybrid female sterility) and for the >2.5-fold excess of HMS factors on the X versus the autosomes.
Faster-Male Evolution
The faster evolution of HMS than hybrid female sterility might occur for one, or a combination, of two reasons. First, genes involved in male reproductive function might evolve faster as a consequence of sexual selection, giving rise to male-specific hybrid incompatibilities (Wu and Davis, 1993; Wu et al., 1996). There is no shortage of evidence for sexual selection on male reproductive signals (Anholt et al., 2020), morphology (e.g., genitalia, reproductive tract, and sperm [Miller and Pitnick, 2002; Frazee and Masly, 2015)], fertilization biology (Price, 1997), gene expression (Meiklejohn et al., 2003), and protein sequences (Swanson et al., 2001). Notably, however, the faster-male theory does not necessarily predict faster evolution for the X chromosome. Second, substitutions that affect spermatogenesis and oogenesis could in principle accumulate at similar rates, but spermatogenesis might be more sensitive to developmental disruption than oogenesis because, e.g., the elaborate postmeiotic stages of spermatogenesis proceed largely in the absence of transcription (Wu and Davis, 1993), leading to a greater proportion of those substitutions causing male-specific incompatibilities. As often noted (Wu and Davis, 1993; Laurie, 1997, Orr, 1997), a major weakness of both flavors of the faster-male theory— sexual selection and “spermatogenesis is special”— is that they are difficult to reconcile with the ubiquity of Haldane’s rule in female-heterogametic taxa where hybrid females are sterile but males are fertile (Presgraves, 2002; Price and Bouvier, 2002).
Faster-X Evolution
The faster-X theory shows that X-linked loci can experience higher rates of adaptive evolution than autosomal loci (Charlesworth et al., 1987). The original model assumes, critically, that adaptation proceeds via the fixation of unique beneficial mutations that increase heterozygous fitness by sh and hemi- or homozygous fitness by s (where s = the selection coefficient and h = the dominance coefficient); an equal sex ratio so that the effective population size of the X is 3/4 that of autosomes; and equal germline mutation rates for the two sexes. Then, the ratio of the rate of adaptive substitution on the autosomes and the X is
and faster-X evolution will occur when unique beneficial mutations are, on average, partially recessive (mean h < 1/2). (It is important here to distinguish the dominance of the beneficial effects of adaptive mutations within species versus the dominance of any incompatibility effects these mutations might have in species hybrids.) The magnitude of faster-X evolution can be enhanced for mutations that are more beneficial to males than females (Charlesworth et al., 1987). Empirical support the faster-X theory is found in multiple signatures of excess positive selection on the X relative to the autosomes, including more selective sweeps, more genes with histories of recurrent adaptive evolution, and a greater estimated proportion of adaptive substitutions (Meisel and Connallon, 2013).
If HMS arises as an incidental by-product of adaptive evolution within species, then faster-X evolution could help explain the faster evolution of HMS on the X (Charlesworth et al., 1987; Coyne and Orr, 1989a). More precisely, if adaptive substitutions on the X and autosomes have equal probabilities of causing HMS, then the X/A ratio of HMS factors is expected to be equal to the X/A ratio of substitution rates (Naveira, 2003). Instead, the observed X/A ratio for HMS factors between D. simulans and D. mauritiana (>2.5) cannot be explained by the observed X/A ratio for protein-coding sequence divergence [∼1.2; (Begun et al., 2007; Garrigan et al., 2014)]. The X/A ratio of interspecific divergence, however, confounds neutral, slightly deleterious, and adaptive substitutions. Theory shows that faster-X evolution occurs only when adaptation occurs via unique beneficial mutations, not via standing genetic variation or recurrent mutation (Orr and Betancourt, 2001). Therefore, to the extent that faster-X evolution contributes, HMS must be a by-product of the substitution of new rather than recurrent or segregating mutations. Despite these suggestions for a role for faster X evolution, there is an important caveat: while faster-X evolution can result in excess hybrid incompatibilities on the X, it does not on its own cause Haldane’s rule. If, in Eq. 1, τ = 1 and d0 = 0.5, then hybrid males and hybrid females will be equally fit despite any faster-X evolution (Orr, 1997). In principle, then, faster X evolution could contribute to the large X-effect and/or the high density of HMS factors on the X, but Haldane’s rule will require a separate explanation for the faster accumulation of HMS in general.
Misregulation of Gene Expression
Hybrid incompatibilities can result from misregulation of gene expression (Johnson and Porter, 2000; Mack and Nachman, 2016). Disproportionate misexpression of X-linked genes might arise in two ways. First, paralleling protein-coding sequence evolution, the expression levels of X-linked genes evolve faster than autosomal ones (Meisel et al., 2012). We might therefore expect greater misexpression of X-linked genes in hybrids. Second, and more narrowly, heteromorphic sex chromosomes often have chromosome-specific forms of regulation. In Drosophila, a dedicated sex chromosome dosage compensation complex (DCC) of proteins and RNAs distinguishes the single male X chromosome from the autosomes and enables hypertranscription of X-linked genes in male somatic cells (Gelbart and Kuroda, 2009). In some lineages, the components of the DCC evolve rapidly (Levine et al., 2007; Rodriguez et al., 2007), raising the possibility that hybrid incompatibilities that disrupt recognition and/or compensation could cause X-linked male-specific lethality [but not, it seems, between D. melanogaster and D. simulans; (Orr, 1989; Barbash, 2010)]. To explain HMS, we must consider sex chromosome regulation, and its possible disruption, in the male germline. Surprisingly, the regulation of the X in the male germline is still rather poorly understood in Drosophila. The facts are that X chromosome expression relative to autosomes is dynamic across stages of spermatogenesis but that, overall, X-linked genes are expressed, on average, at lower levels than autosomal ones. The lower expression of X-linked genes has been attributed to an absence of sex chromosome dosage compensation (Meiklejohn et al., 2011; Meiklejohn and Presgraves, 2012) and/or to a process similar to meiotic sex chromosome inactivation (Hense et al., 2007; Vibranovski et al., 2009, Meiklejohn et al., 2011; Landeen et al., 2016).
There is little indication that simulans clade hybrid males experience excess disruption of X chromosome regulation. Genome-wide gene expression analyses in testes from D. simulans-D. mauritiana F1 hybrids and from introgression hybrids that carry a single X-linked 500-kb region from D. mauritiana in a D. simulans background show that the proportion of X-linked genes that are misexpressed is roughly half that of autosomal genes that are misexpressed (Moehring et al., 2006b; Lu et al., 2010). If anything, then, the expression of autosomal genes appears more subject to disruption in hybrids. An outsized role of the X in hybrid sterility via gene misregulation therefore requires an excess of trans-acting X-linked factors that disrupt the expression of autosomal genes. But, even then, genome-wide studies of gene misexpression in hybrid males (or testes) can be challenging to interpret, for two reasons. First, it is difficult to distinguish gene misregulation that causes sterility versus misregulation that results from sterility. Second, and related, it is difficult to distinguish gene misexpression per se from the appearance of misexpression caused by the perturbed cell and/or tissue composition of hybrid versus parental species testes.
Gene Transposition
Dobzhansky (1937) and Muller (1942) both noted that evolutionary changes in the chromosomal locations of genes created the potential for recombinant hybrid genotypes that lack gene copies at both ancestral and transposed positions [see also (Stern, 1936; Werth and Windham, 1991, Lynch and Force, 2000)]. If the transposed gene is essential for male fertility then these double-null recombinant hybrid males would be sterile. The first evidence to support this model comes from certain F2-like hybrids between D. melanogaster and D. simulans: the fertility essential gene, JYalpha, is on chromosome 4 in D. melanogaster and on chromosome 3 in D. simulans; hybrid males homozygous for a D. simulans chromosome 4sim in an otherwise D. melanogaster background lack JYalpha entirely and are thus completely sterile (Muller and Pontecorvo, 1940; Orr, 1992, Masly et al., 2006). With proof of principle established, the question arises as to whether gene transposition might help explain Haldane’s rule and/or the large X-effect. Two genomic patterns characterizing gene transposition in Drosophila suggest a potential role (Moyle et al., 2010): there is excess gene transposition “traffic” involving the X chromosome, including both X → autosome and autosome → X gene movements; and transposed genes are disproportionately testis-expressed (Betran et al., 2002; Meisel et al., 2009, Han and Hahn, 2012). The absolute rate of X←→A gene traffic is, however, low relative to the rate of accumulation of HMS: among species lineages in the D. melanogaster group, the estimated mean rate of gene movement is ∼2 per million years (Meisel et al., 2009). This estimate suggests ∼0.5 species-specific gene movements in each of the ∼250-Ky old D. simulans clade lineages, far too few to account for the many X-linked HMS factors mapped between these species (True et al., 1996a; Tao et al., 2003a, Masly and Presgraves, 2007; Meiklejohn et al., 2018). The contribution of X-linked gene traffic to HMS in the D. simulans clade must therefore be negligible.
Drive
Meiotic drive refers to the biased transmission of one allele—usually a selfish genetic element— over another from a heterozygous carrier. Drive in the male germline tends to involve two or more loci: a drive locus, with wildtype (D) and driving (d) alleles; a target-of-drive locus, with sensitive (S) and insensitive alleles (s); and a constellation of linked drive-enhancers and/or unlinked drive-suppressors. In heterozygous DS/ds males, spermatids bearing sensitive S alleles are killed or incapacitated by the action of the driving d allele, conferring a transmission advantage to the resistant ds haplotype. Whether d and s invade and spread in a population depends on the frequency of recombination: linkage enables transmission of d with s, whereas recombination yields “suicide” combinations of d with S. On autosomes, recombination therefore limits the opportunity for drive. On non-recombining sex chromosomes, however, there is no such limit—any factor on the X can drive against any target on the Y (and vice versa) without risk of suicide combinations. The resulting sex chromosome drive causes biased progeny sex ratios and reduced fertility, as well as many downstream knock-on consequences (Hamilton, 1967; Jaenike, 2001; Presgraves, 2008b; Meiklejohn and Tao, 2010). Most important among these, for our purposes, are molecular genetic arms races, as sex chromosome drive favors the evolution of resistant alleles at the target; suppressors at unlinked loci; and counter-resistance and/or suppressor-evasion at drive and linked enhancer loci. Sex chromosomes are thus subject to recurrent cycles of drive, resistance, suppression, counter-suppression, and so on (Carvalho et al., 1997; Hall, 2004), resulting in the potential accumulation of multiple, divergent, species-specific “cryptic drive” systems— drive loci that persist but in a silenced state.
From this seemingly fanciful premise, Frank (1991) and Hurst and Pomiankowski (1991) suggested that otherwise cryptic drivers might be released or aberrantly expressed in the naïve genetic backgrounds of species hybrids, with mutual destruction of X- and Y-bearing spermatids causing sterility that maps disproportionately to sex chromosomes (Frank, 1991; Hurst and Pomiankowski, 1991). Theirs was a radical suggestion, as most speciation geneticists at the time preferred to invoke classical neo-Darwinian phenomena, like genetic drift and ecological adaptation, and eschewed the seemingly exotic ones, like meiotic drive. Moreover, if meiotic drive causes Haldane’s rule and the large X-effect— the “two rules of speciation”— then drive must be far more ubiquitous than previously supposed. And, not least, some speciation geneticists had already looked for evidence of cryptic drive unleashed in hybrids but found none (Coyne, 1986). For these reasons, skeptics battered the drive theory for its implausibility and lack of evidence (Coyne et al., 1991; Johnson and Wu, 1992, Charlesworth et al., 1993; Coyne and Orr, 1993).
But the drive theory is enjoying a resurgence due to recent findings in mammals (Davis et al., 2015; Kruger et al., 2019, Rathje et al., 2019) and Drosophila (Orr and Irving, 2005; Phadnis and Orr, 2008, Zhang et al., 2015), including the D. simulans clade species. First, D. simulans harbors two well-characterized cryptic sex chromosome drive systems. The Paris drive system involves two co-drivers on the X chromosome, one a segmental duplication containing six genes and the other an allele of the HP1D2 gene, a rapidly evolving member of the HP1 heterochromatin protein family (Helleu et al., 2016). The Paris drivers are usually suppressed by resistant Y chromosomes and by multiple loci scattered across the autosomes (Courret et al., 2019; Helleu et al., 2019). The Winters drive system involves a new, lineage-restricted chimeric gene, Distorter on the X (Dox), that is usually suppressed by an autosomal retroduplicate, Not-much-yang (Nmy), which silences Dox expression by producing Dox-matching endogenous small interfering RNAs (Tao et al., 2007a,b, Lin et al., 2018). In a recent genetic analysis of X-linked HMS, we uncovered additional evidence for a novel cryptic X chromosome drive system in D. mauritiana (Meiklejohn et al., 2018). These discoveries confirm a key requirement of the drive theory, namely that closely related species accumulate divergent multilocus systems of cryptic drive.
A second requirement of the theory is that drive contributes to the evolution of HMS. Two observations are suggestive here. One is that genetic introgression of the Too much yin (Tmy) region on 3R of D. mauritiana into D. simulans releases cryptic drive and, along with other linked factors, contributes to HMS (Tao et al., 2001). Another is that the D. mauritiana allele of the X-linked gene, OdysseusH (OdsH) (Ting et al., 1998), contributes to HMS in a D. simulans genetic background, and the OdsHmau protein binds repetitive DNA sequences on the D. simulans Ysim chromosome but not its own D. mauritiana Ymau chromosome (Bayes and Malik, 2009). While there is no evidence that OdsH currently causes drive, it is easy to imagine a history in which Ymau evolved resistance to Odsmau-mediated drive by shedding target sequences, while the naïve Ysim retained them. Similar and more direct evidence links drive to HMS in two other Drosophila hybridizations (Orr and Irving, 2005; Phadnis and Orr, 2008, Zhang et al., 2015). These suggestive observations from the simulans clade species, and the more direct evidence from other species pairs, support the second key requirement that drive can contribute to HMS. The question now is not whether meiotic drive contributes to HMS but to what extent.
Satellite DNAs
Haldane’s rule and the large X-effect have never been short of competing explanatory hypotheses. We nevertheless hazard another, admittedly speculative, hypothesis here: the rapid evolution of HMS in the D. simulans clade could involve the rapid divergence of repetitive satellite DNA sequences (satDNAs), their regulation, and/or their functional effects. The notion that satDNAs contribute to hybrid incompatibility and speciation is not new (Rose and Doolittle, 1983; Ferree and Prasad, 2012, Sawamura, 2012; Gallach, 2014). But three recent findings from the D. simulans clade species and their close relative, D. melanogaster, are consistent with a role for satDNAs in the rapid evolution of HMS on sex chromosomes.
First, the sequences, copy numbers, genomic compositions, and chromosomal distributions of simple and complex satDNAs are strikingly different between the D. simulans clade species (Jagannathan et al., 2017; Sproul et al., 2020). Cytological analyses reveal that large blocks of [AACAAAC]n are detectable in D. mauritiana on chromosomes 2 and 3, in D. simulans on the Y chromosome, and in D. sechellia not at all (Jagannathan et al., 2017). Two complex satDNA families, in particular, show considerable turnover between the D. simulans clade species: the 1.688g/cm3 (Hsieh and Brutlag, 1979) and Rsp-like (Larracuente, 2014) satellites. In heterochromatic pericentromeric regions, large (Mb-scale) blocks of satDNAs reside in species-specific locations: 1.688 blocks are X-linked in D. sechellia but autosomal in D. simulans and D. mauritiana, whereas Rsp-like blocks are X-linked in D. simulans, autosomal in D. sechellia, and absent altogether from D. mauritiana centromeres (Sproul et al., 2020). High quality genome assemblies provide additional comprehensive, fine-scaled evidence for species-specific distributions of blocks of satDNA arrays in heterochromatic regions and small islands of satDNAs in euchromatic regions (Sproul et al., 2020; Chakraborty et al., 2021).
Second, euchromatic islands of satDNAs, including both 1.688 and Rsp-like, are significantly enriched on the X chromosome (Hsieh and Brutlag, 1979; Kuhn et al., 2012, Garrigan et al., 2014; Sproul et al., 2020). In the euchromatin, small (≤kb-scale) satDNA islands are enriched on the X relative to autosomes 15-fold in D. simulans, 29-fold in D. mauritiana, and 51-fold in D. sechellia (Chakraborty et al., 2021). This X chromosome enrichment results from expansion of satDNA islands on the X rather than loss from the autosomes. While some, presumably older, satDNA islands, like 1.688, are shared as homologous array loci among species, newer satDNA islands, like Rsp-like, tend to be species-specific (Sproul et al., 2020).
Third, and critically, satDNA-derived small RNAs are essential for male fertility in D. melanogaster (Mills et al., 2019). In particular, depletion of small RNAs from the highly abundant AAGAG tandem repeat results in defective histone-to-protamine transition during spermiogenesis (Mills et al., 2019). The exchange of canonical histones for sperm-specific protamines facilitates remodeling of sperm pronuclei into compact, non-nucleosomal structures that are ∼200-fold smaller in volume (Rathke et al., 2007). Processing of satDNAs during this radical chromatin remodeling appears to be susceptible to disruption. In D. melanogaster, for example, the autosomal meiotic drive gene complex Segregation Distorter (SD) achieves a >95% transmission advantage from heterozygous SD/ + males by disrupting the histone-to-protamine transition for + -bearing spermatids that have large blocks of Rsp satDNA (Wu et al., 1988; Gingell and McLean, 2020). In sterile F1 hybrid males between D. simulans and D. mauritiana, sperm nuclei show an age-dependent de-condensation phenotype (Kanippayoor et al., 2020), suggestive of compromised chromatin integrity, but whether specific misregulation of satDNAs is involved is unknown. [Spermatogenesis in sterile F1 hybrid males from the reciprocal cross, in contrast, arrests during premeiotic stages (Kulathinal and Singh, 1998)].
Overall, then, the satDNA hypothesis merits our attention because satDNA composition evolves quickly, satDNAs are enriched on the X, and disruption of satDNA regulation can disrupt male fertility. One obvious way to distinguish among these several hypotheses— faster male, faster X, drive, satDNAs, etc.— is to determine the molecular genetic basis of HMS. Our prospects for characterizing the molecular genetic basis of HMS depends on its genetic architecture, to which we turn next.
Genetic Architecture of Hybrid Male Sterility
When an introgressed segment that produces sterility is partitioned by recombination into shorter segments, sterility vanishes (Naveira and Maside, 1998). (H)ybrid sterility between incipient species is largely due to strong epistasis between genes of minor or no effect individually (Cabot et al., 1994).
The Dobzhansky-Muller (DM) model provides a simple, intuitively satisfying solution to the puzzle of how hybrid incompatibility might evolve, and it is supported by an abundance of genetic data from a variety of systems, including yeast, worms, flies, butterflies, fish, mouse, Arabidopsis, Mimulus, and more (Johnson, 2010; Presgraves, 2010). But the power of the DM model has not translated into routine identification of hybrid incompatibility genes. Aside from two notable successes (see below), the search for hybrid incompatibility genes has been hampered in several ways. First, reproductive isolation gets in the way, often prohibiting the creation and/or maintenance of desired genotypes. Second, hybrid incompatibilities are often polymorphic: the effect sizes, or even the existence, of genetic incompatibility alleles mapped between two particular strains may not hold for others. Third, despite the appeal of the simple two-locus DM model, hybrid incompatibilities almost always involve more than two factors— hybrids are sterile because they have the “wrong” genotype at ≥ 3 loci. Compounding these challenges, the “genetic architecture” of HMS loci appears to differ from that of other kinds of hybrid incompatibility.
Genetic Architecture
At the level of whole chromosomes, there are many HMS loci, few hybrid lethals, and even fewer hybrid female steriles (Hollocher and Wu, 1996; True et al., 1996a, Tao and Hartl, 2003; Masly and Presgraves, 2007, Meiklejohn et al., 2018). At the level of individual loci, the distribution of effect sizes for HMS and hybrid lethality differs as well. (We omit hybrid female sterility from this discussion because so little information is available). Numerous hybrid lethality factors of large effect have been identified between D. melanogaster and D. simulans (Sawamura and Yamamoto, 1997; Barbash et al., 2003, Presgraves et al., 2003; Tang and Presgraves, 2009, Phadnis et al., 2015) and several between D. mauritiana and its sister species (True et al., 1996a; Masly and Presgraves, 2007, Cattani and Presgraves, 2009). In all of these cases, hybrid lethality could be localized to an individual gene or repetitive DNA element. These findings, plus early genetic mapping results from the D. simulans clade species (Coyne and Charlesworth, 1986, 1989, Perez et al., 1993), nurtured expectations that HMS loci might also have large effects (Figures 3A–C).
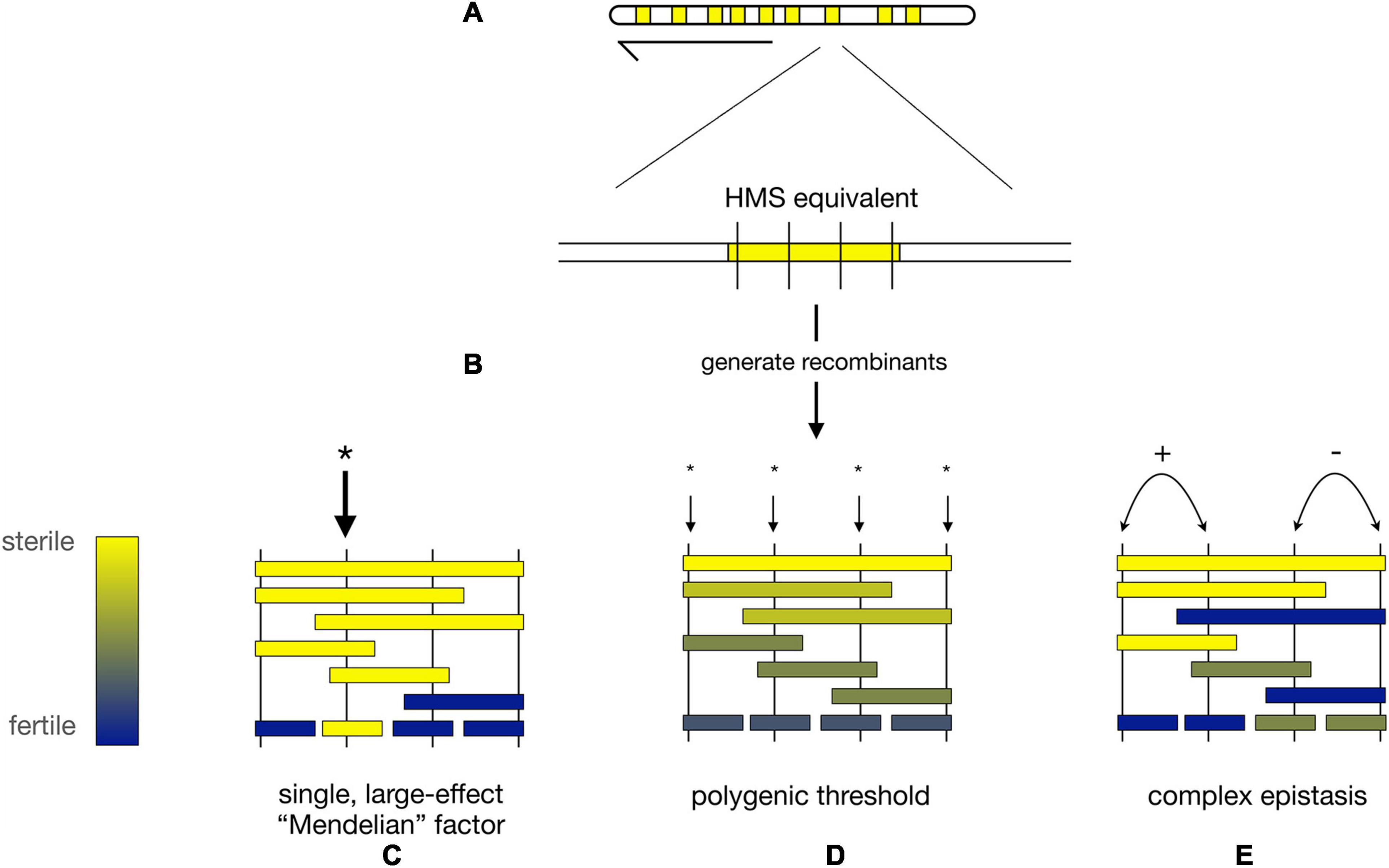
Figure 3. Alternative models for the genetic architecture of HMS. (A) Hypothetical map of nine X-linked regions from D. mauritiana (mau) that each cause strong HMS when introgressed into a D. simulans (sim) genetic background (“HMS equivalents,” yellow). (B) Fine-scale recombination mapping is used to dissect the genetic basis of each HMS equivalent region. (C) HMS regions may contain a single mau factor of large phenotypic effect (* = location of HMS factor). (D) Under the polygenic threshold model, multiple, interchangeable mau factors individually contribute to HMS; complete HMS occurs when a sufficient, threshold number of polygenic HMS factors is present simultaneously. (E) Under the complex epistasis model, two (or more) loci interact to determine the HMS phenotype. In the figure, there are two pairs of adjacent, epistatically interacting loci. The left pair of loci shows synergistic epistasis (+) in which two mau alleles interact to produce stronger HMS than the sum of their individual effects. The right pair of loci shows antagonistic epistasis (−) in which two mau alleles interact to produce weaker HMS than the sum of their individual effects.
There were, however, early indications to the contrary. Genetic analyses between Drosophila buzzatti and Drosophila koepferae showed that interspecific introgressions on the autosomes were generally male-fertile when <30% the length of the chromosome but invariably male-sterile when >35% (Naveira and Fontdevila, 1985, 1986, 1991). No single locus caused HMS unless co-introgressed with some minimum number of additional HMS loci. The genetic basis for HMS appears similarly diffuse between D. mauritiana and D. simulans (Naveira, 1992). On the X chromosome, for instance, a ∼500-kb region from D. mauritiana containing OdsH causes strong HMS in a D. simulans genetic background only when co-introgressed with other (unknown) factors (Perez and Wu, 1995). On the autosomes, D. mauritiana introgressions into D. simulans uncovered ∼19 HMS loci on chromosome 3: all but one have modest effects on male fertility, and complete HMS requires the combined effects of multiple loci (Tao et al., 2001, 2003b).
Two models have been proposed to describe these observations. First, the polygenic threshold model posits that HMS results when a sufficient number of interchangeable, small-effect factors are co-introgressed together [Figure 3D; (Naveira and Maside, 1998)]. Under this model, the identity of the particular HMS loci involved matters less than the cumulative effects of the multiple, independent HMS factors. Second, the complex epistasis model posits that HMS is caused by synergistic epistatic interactions among co-introgressed conspecific alleles that are together incompatible with heterospecific factors (Figure 3E). Under this model, the genotypes at particular loci matters, as two (or more) weak alleles interact to produce an HMS effect greater than the sum of their individual effects (Cabot et al., 1994; Palopoli and Wu, 1994, Wu and Palopoli, 1994; Perez and Wu, 1995, Davis and Wu, 1996; Wu and Hollocher, 1998).
These models are not mutually exclusive, and indeed there is evidence consistent with both. We highlight examples gleaned from our recent X chromosome-wide introgression analysis of HMS. To map HMS factors, we assayed D. mauritiana introgressions in a D. simulans genetic background, delimited introgression size precisely using genotyping-by-sequencing, and measured fertility in replicate males for each genotype (Meiklejohn et al., 2018). Two features of the data are consistent with the polygenic threshold model. First, the polygenic threshold model predicts a correlation between penetrance (the proportion of males with a particular genotype that show the HMS phenotype) and the mean number of progeny for isogenic brothers that sired any progeny (Figure 4A). This prediction is supported by the introgression data (Figure 4B; Spearman’s ρ = −0.56, P < 0.0001). Second, within a 9 Mb interval in the middle of the X chromosome, male fertility appears to be a declining function of the amount of introgressed D. mauritiana sequence that is largely independent of chromosomal location, with a rough threshold length of ∼2 Mb, beyond which most introgressions are male-sterile (Figure 5A; Spearman’s ρ = −0.56, P < 0.0001). Of course, longer introgressions might cause HMS because they are more likely to capture a large-effect HMS factor. But with our high-coverage introgression map, we should observe at least some small introgressions that also capture large-effect HMS factors. We do not. Thus, for the medial ∼50% of the X chromosome, there appears to be no major effect factor that causes HMS on its own. These observations suggest that there must be functional divergence at a very large number of sites that each contribute, if weakly, to HMS.
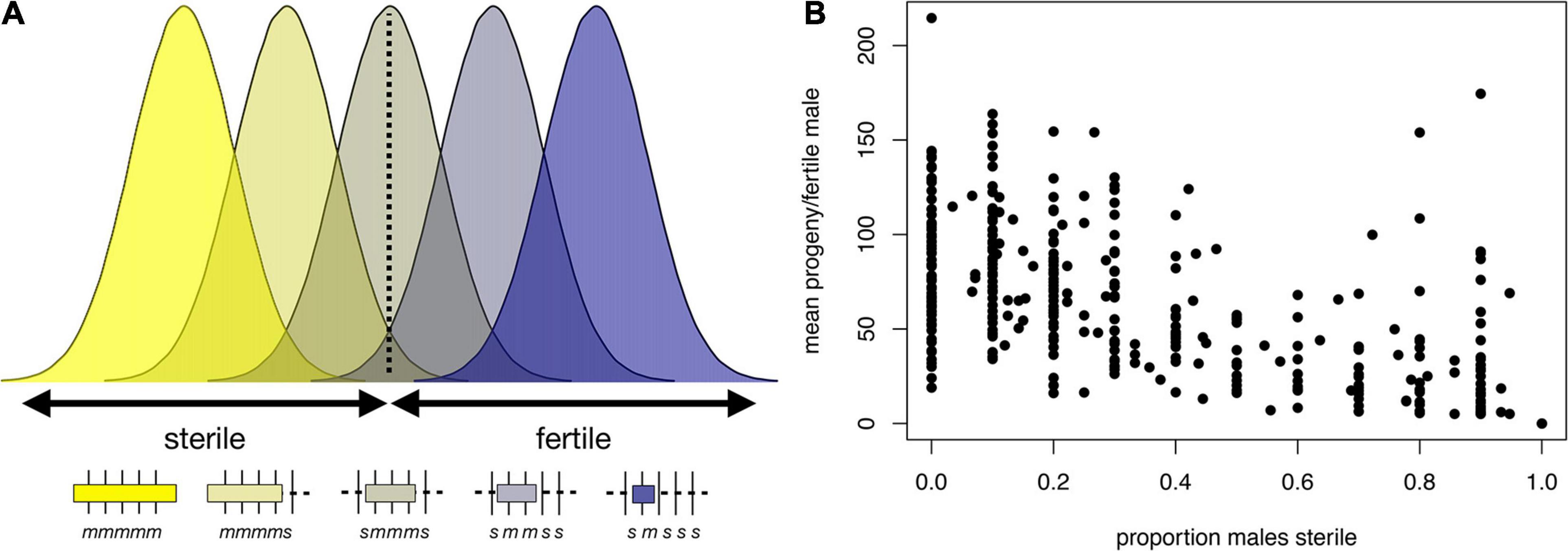
Figure 4. Polygenic threshold model of HMS (adapted from Lienard et al., 2016). (A) The plot shows the distribution of a hypothetical quantitative trait, fertility potential, for five hypothetical introgression genotypes shown beneath the x-axis. For each genotype, the length of a D. mauritiana introgression is represented by the length of the open bar; the genotypes at five markers (vertical tick marks) are indicated by m and s, for D. mauritiana and D. simulans, respectively; and the average fertility of males is indicated by the color of the bar. The largest introgression (mmmmm) is completely male-sterile, whereas the smallest introgression (smsss) is completely fertile. For intermediate introgression genotypes, some proportion of males produce no progeny (those falling below the threshold) whereas others produce >0 progeny. This polygenic threshold model suggests a correlation between the proportion of sterile males associated with a particular introgression genotype and the mean number of progeny produced by their fertile brothers. (B) Experimental data on D. mauritiana X-chromosome introgressions in a D. simulans genetic background show the predicted correlation (Spearman’s ρ = −0.91, P < 0.0001) between the proportion of sterile males and the mean number of progeny among fertile males (for data and details see Meiklejohn et al., 2018).
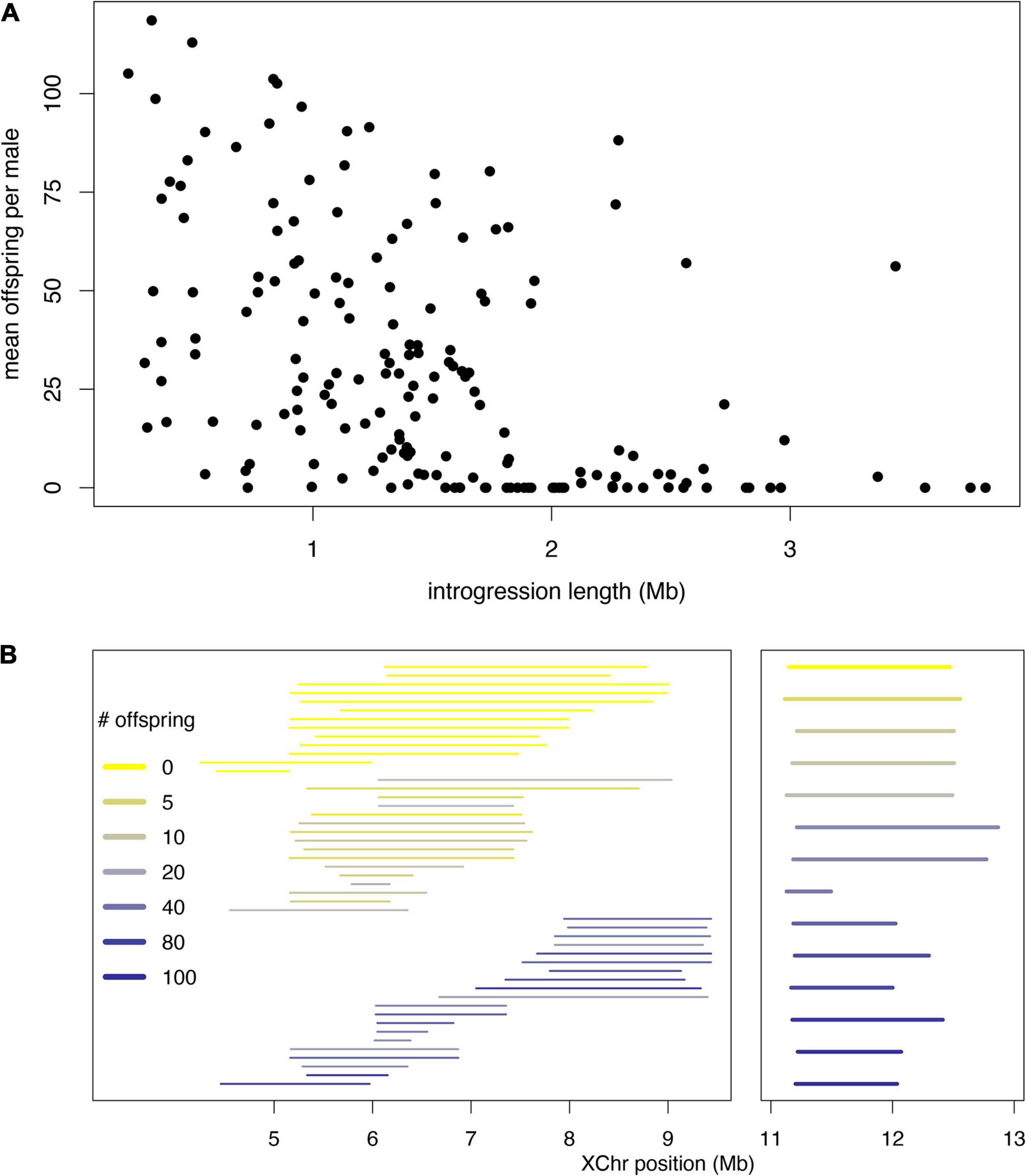
Figure 5. Data from fine-scale genomic analysis of HMS. X chromosome segments from D. mauritiana were introgressed via >25 generations of backcrossing into a D. simulans genetic background and assayed for fertility by crossing individual introgression males with three virgin D. simulans females for seven days (for data and details see Meiklejohn et al., 2018). For each introgression, ≥ 10 clonal males were phenotyped. (A) In genomic coordinates X:4–13 Mb, the fertility of all introgressions shows a strong negative correlation with length, with very few fertile introgressions >2 Mb. (B) Introgressions consistent with the polygenic threshold model. No single D. mauritiana locus between coordinates X:5–9 Mb is sufficient to cause HMS, but longer introgressions can. The median length of sterile introgressions (top group, yellow) is 1.7-fold greater than that for fertile introgression (bottom group, blue); Wilcoxon test P ∼ 0.0001. Between both X:4–6 Mb and X:11–12 Mb (right panel), small sterile introgressions can, however, fall within overlapping, larger fertile introgressions, consistent with complex epistasis.
At a smaller (sub-Mbp) scale, however, other features of the data implicate complex epistasis. In particular, sterile introgressions can be spanned by tiling paths of fertile introgressions or even completely overlapped by larger, fertile introgressions (Figure 5B). Similar observations obtain for autosomal introgressions between D. mauritiana and D. simulans (Tao et al., 2003b). These findings are difficult to reconcile with the polygenic threshold model: for any sterile introgression, a longer completely overlapping introgression should also be sterile. One explanation is that HMS alleles experience antagonistic interactions in which a small introgression causes HMS whereas co-introgression of additional factors suppresses HMS. Thus, evidence exists for both synergistic and antagonistic forms of complex epistasis (Figure 3E; Wu and Palopoli, 1994; Wu and Hollocher, 1998, Tao et al., 2003b). A polygenic architecture with two flavors of complex epistasis has two implications. The biological implication is that, in hybrids, HMS alleles do not behave as loss-of-function mutations at male fertility-essential genes. The practical implication is that individual HMS loci will be refractory to molecular identification.
HMS Genes
Despite the practical challenges, the molecular identities of HMS factors have been established in two cases in the D. simulans clade species. The first is the well-known, well-characterized X-linked HMS gene, OdsH (Ting et al., 1998). By itself, the D. mauritiana allele of OdsH causes sperm motility defects in ∼50% of introgression male carriers (Perez and Wu, 1995). Complete HMS (no sperm motility) occurs only when other factors are co-introgressed (Perez and Wu, 1995). Nevertheless, X chromosome-wide genetic analyses suggest that OdsH may be the HMS factor of the single largest individual effect (Meiklejohn et al., 2018). OdsH encodes a testes-expressed protein with a highly diverged DNA-binding homeodomain (Ting et al., 1998). While rapid evolution at OdsH was first hypothesized to result from sexual selection, its localization to Y chromosome satDNAs implicates genetic conflict [see above; (Bayes and Malik, 2009)].
The second identification of HMS factors comes from a 9-kb interval on chromosome arm 3R that contains just four protein-coding genes (Araripe et al., 2010). This HMS1 region has a very large effect (∼200 progeny for the D. simulans allele versus ∼2 for the D. mauritiana allele), making it a promising candidate for molecular characterization (Araripe et al., 2010). Transgenic experiments reveal, however, that even within this 9-kb region the genetic architecture of HMS is complex (Lienard et al., 2016). Transgenes carrying two different genes— agt and Taf1— each recover substantial (if not full) male fertility, implicating both in HMS. Both genes encode unrelated DNA binding and/or modifying proteins, but neither has signatures of recurrent positive selection. Chimeric transgenes that combine regulatory sequences from one species and coding sequences from the other at both genes similarly rescued fertility, further suggesting that multiple D. mauritiana substitutions distributed across coding and non-coding regions of both genes may be required for HMS.
Work on the genetic architecture of HMS— from large-scale high-resolution genetic analyses to the molecular identification of genes— supports a polygenic basis with additional evidence of complex (synergistic and antagonistic) epistasis. These inferences are necessarily based on analyses that seek to isolate individual HMS factor(s) in introgression hybrid male genotypes. The genetic basis of HMS in introgression hybrid males may of course differ from that in F1 hybrid males, as they have different genotypes. But if incompatibilities that conform to the polygenic threshold model are abundant, we may safely posit that F1 hybrid males are sterile due to the combined effects of very many, individually weak, HMS factors. If gene flow nevertheless occurs via fertile hybrid females, however, then many individually weak HMS factors will be exposed to selection in backcross or advanced backcross hybrid males. It is important to appreciate that many factors deemed to have “weak” phenotypic effects in the laboratory are readily detectable by natural selection.
Complex Speciation With Gene Flow
Species in sexual cross-fertilizing organisms are defined as groups of populations which are reproductively isolated to the extent that the exchange of genes between them is absent or so slow that the genetic differences are not diminished or swamped (Dobzhansky, 1944). (D)iverging genomes during (or even after) speciation can be quite “porous” with respect to gene flow at non-speciation loci (Wu, 2001).
Under simple allopatric speciation, populations isolated by geography eventually and incidentally evolve intrinsic reproductive incompatibility, a scenario that “appears so plausible that it hardly seems worth documenting” (Coyne and Orr, 2004). The three species of the D. simulans subcomplex would seem to be strong and obvious candidates for allopatric speciation via dispersal: they are believed to have originated on different Indian Ocean islands (Madagascar, the Seychelles, and Mauritius); D. simulans has never been collected on Mauritius (David et al., 1989); and, until recently, D. simulans had not been collected on the same islands of the Seychelles as D. sechellia (Lachaise et al., 1988). In geographic isolation, the three species have evolved ecological, sexual, postmating-prezygotic, and postzygotic barriers (Lachaise et al., 1986, 1988; R’Kha et al., 1991; Coyne, 1992a; Coyne and Charlesworth, 1997; Price, 1997). Early multi-locus population genetic analyses among the three species were, as expected, consistent with a simple model of isolation without gene flow (Kliman et al., 2000; Nunes et al., 2010).
There are now good reasons to doubt simple allopatric histories for these species. For D. sechellia and D. simulans, the evidence is direct: the two species now co-occur on a subset of the Seychelles (likely via human introductions), and hybrid males have been collected in the field (Matute and Ayroles, 2014; Navascues et al., 2014). For D. mauritiana and D. simulans, the first hints of gene flow came from mitochondria: ∼88% of D. mauritiana flies carry a D. simulans-like mitochondrial haplotype estimated to have introgressed ∼4,500 years ago (Solignac and Monnerot, 1986; Solignac et al., 1986, Satta and Takahata, 1990, Ballard, 2000a,b, Nunes et al., 2010). Genomic data have confirmed nuclear gene flow among all three species pairs. Simple (allopatric) speciation without gene flow predicts that the genealogical histories of all loci should be compatible with a single species divergence time (Figure 6A). The genomes of the three D. simulans clade species, however, present clear evidence for complex speciation with gene flow resulting in discrepant, reticulated genealogical histories (Figure 6B). Three different analyses, leveraging different (albeit overlapping) features of the data, estimate similar amounts of introgressed foreign material (2–5%) among the three species (Garrigan et al., 2012; Meiklejohn et al., 2018, Schrider et al., 2018). These findings underscore the limits of population genetic surveys at a small number of loci to detect introgression and contribute to the increasing evidence that gene flow is a common feature of divergence between closely related species, even for species pairs that are geographically allopatric (Mallet, 2005; Seehausen et al., 2014; Mallet et al., 2016).
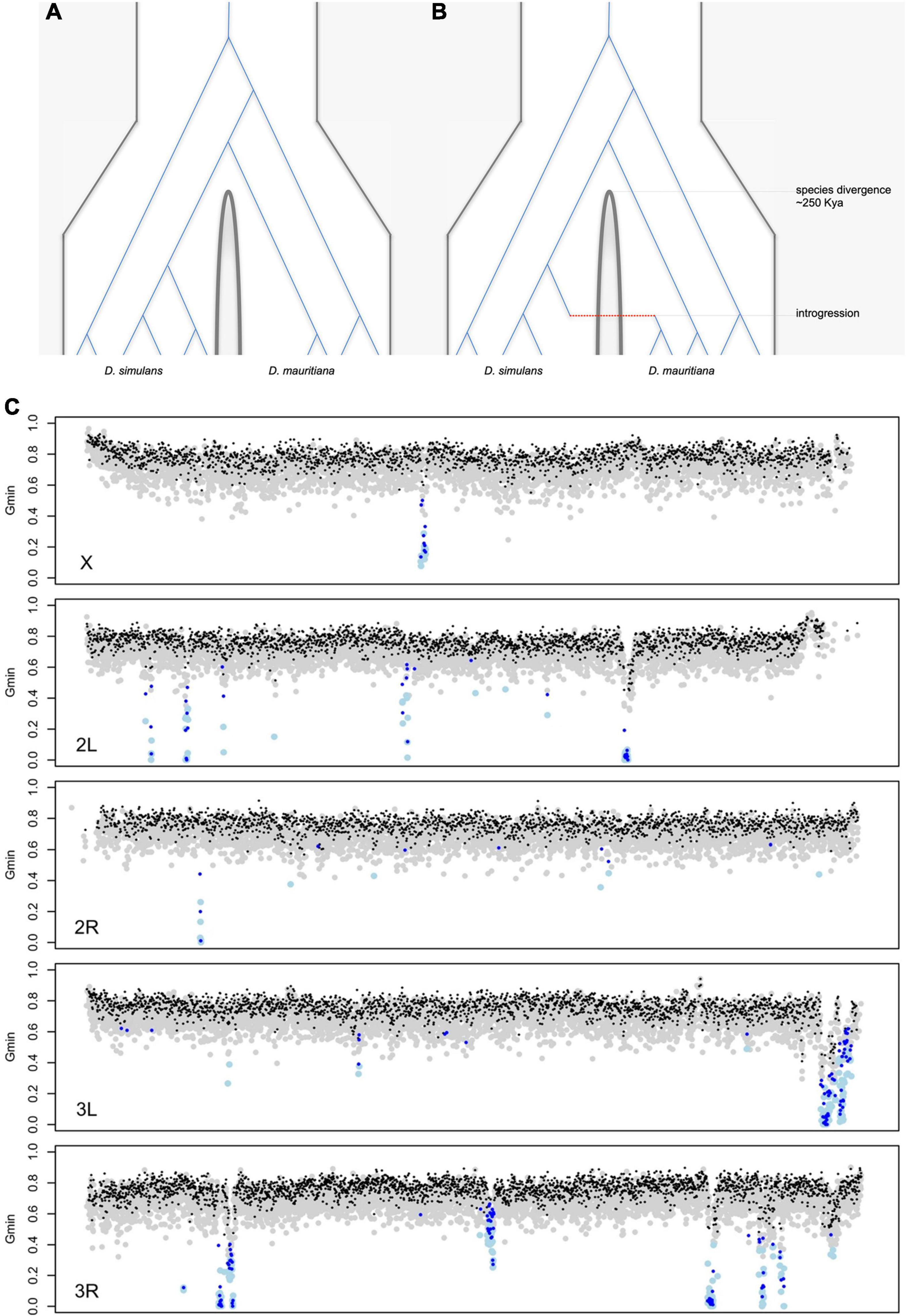
Figure 6. Hypothetical genealogical histories of multiple sequences sampled from two species under (A) simple allopatric speciation with no gene flow and (B) complex speciation with gene flow. Under simple allopatric speciation, all coalescent events among any two gene copies from the different species must predate the species divergence time. Under complex speciation, coalescent events between any two gene copies from the different species can postdate the species divergence time, as represented by the single introgression event (red). (C) A genome-wide scan for introgression in population samples from D. simulans (n = 20) and D. mauritiana (n = 10). The Gmin statistic was used to identify haplotypes with interspecific distances too low to be consistent with a simple allopatric speciation history (Geneva et al., 2016). Each gray (black) dot corresponds to a 5-kb (10-kb) genomic window consistent with a simple allopatric history, and each light blue (dark blue) dot corresponds to a 5-kb (10-kb) genomic window for which the simple null model is statistically rejected. Introgression is significantly underrepresented on the X chromosome (Meiklejohn et al., 2018).
Several features of the natural introgressions are informative. First, the introgressed haplotypes stand out from the genomic background for having aberrantly low interspecific sequence distances (Figure 6B). Second, the introgressed haplotypes show evidence of gradual erosion by recombination. The estimated lengths of introgressions depend both on local chromosomal recombination rate (e.g., longer introgressions tend to reside in low-recombination regions) and time-in-residence [e.g., older introgressions tend to be shorter; (Meiklejohn et al., 2018)]. Third, foreign introgressed material is two- to four-fold under-represented on the X chromosome [Figure 6C; (Garrigan et al., 2012; Meiklejohn et al., 2018)]. Between D. mauritiana and D. simulans, there is only one (∼130 kb-long) recent introgression on the X versus 47 on the autosomes [Figure 6C; (Meiklejohn et al., 2018)]. This X versus autosome difference in introgression density cannot be explained by, e.g., male-mediated admixture (F1 hybrid males are sterile so that all gene flow must be via fertile F1 hybrid females), nor by chromosomal differences in recombination rate (True et al., 1996b). The simplest interpretation is that X-linked material is less exchangeable between species. To introgress, compatible foreign alleles must first survive selection against genetically linked alleles that are incompatible (or otherwise locally maladaptive) and then escape from their deleterious chromosomal backgrounds by recombination (Bengtsson, 1985). Both are more difficult on the X chromosome, as the greater efficacy of selection on the X eliminates foreign material more quickly than on autosomes, and the higher density of hybrid incompatibilities on the X limits the opportunity to escape via recombination (Muirhead and Presgraves, 2016; Fraisse and Sachdeva, 2020).
The existence of interspecific introgression raises the question of what kinds of alleles do escape to persist in a foreign genetic background. Are most interspecific introgressions neutral (functionally equivalent) alleles? Or are interspecific introgressions enriched for globally adaptive alleles? At least three introgressions show signatures of positive selection. First, a ∼200-kb region on chromosome arm 3R has introgressed from D. simulans into D. sechellia, experienced a partial sweep in D. simulans and a complete sweep in D. sechellia (Garrigan et al., 2012; Brand et al., 2013, Schrider et al., 2018). The precise target of selection remains unclear. Second, the ∼130-kb haplotype on the X chromosome that has introgressed between D. simulans and D. mauritiana shows a large, partial sweep in D. simulans and a massive, ∼550-kb complete sweep in D. mauritiana (Nolte et al., 2013; Garrigan et al., 2014, Meiklejohn et al., 2018). Most intriguing, this introgressed interval spans the cryptic meiotic drive genes, Dox, and its parent gene, Mother of Dox (MDox) (Tao et al., 2007a,b). We hypothesize that Dox (and/or MDox) swept to high frequency in its native background before being suppressed, then migrated between species where, released from suppression in the new genetic background, it swept to high frequency again, resulting in parallel selective sweeps (Meiklejohn et al., 2018). Last, the introgression of a D. simulans mitochondrial haplotype into D. mauritiana appears to be non-neutral (Aubert and Solignac, 1990; Meany et al., 2019). These findings suggest that the most conspicuous signals of introgression correspond to loci favored globally by selection. However, the relative contributions of selection- versus drift-mediated introgression remains to be determined.
Overall, our findings imply that the interplay of gene flow and selection has shaped the genomic distribution of introgression. We do not know which form of reproductive isolation— geographic, ecological, sexual, postmating-prezygotic, or hybrid incompatibility— was most important during the history of speciation and admixture among these species. But there are compelling reasons to believe that HMS is among the important barriers to gene flow. For one, there is more HMS on the X chromosome and, consequently, less introgression on the X (Tao et al., 2003a; Masly and Presgraves, 2007, Garrigan et al., 2012; Meiklejohn et al., 2018). For another, the one region of the X chromosome where introgression has occurred is, conspicuously, where HMS is weak or absent (Meiklejohn et al., 2018). These two findings suggest that HMS has impeded X-linked introgression except for the one chromosomal region lacking HMS. Of course, not knowing the historical order of events, it is possible that the reverse is true: selection-driven introgressions may have shaped the genomic distribution of HMS. For instance, drive-mediated introgression of the MDox-Dox haplotype between species may have reduced local interspecific divergence and, incidentally, dampened the local accumulation of HMS (Meiklejohn et al., 2018). If true, it would imply that a drive-mediated trans-species sweep attenuated the evolution of HMS. This scenario highlights an implicit assumption of the drive theory, namely, that drive can contribute to divergence and HMS between strictly allopatric species. For species connected by gene flow, however, drive can introgress between species and erase local divergence. Furthermore, the introgression of a drive element creates additional pressure for any suppressors to follow (Crespi and Nosil, 2012). The role of drive in HMS is therefore contingent on the interplay of drive and gene flow.
Conclusion
The D. simulans clade species have been at the forefront of modern speciation genetics for over 40 years. While many puzzles remain unsolved, many of the successes have offered important lessons. From genetic analyses, we have learned that HMS in the D. simulans clade accumulates faster on the X chromosome. Why this is the case is still unresolved. The drive theory has been revived as a potential explanation, fueled by the discovery of multiple cryptic drive systems and by direct evidence for a role for drive in hybrid sterility (Tao et al., 2001; Orr and Irving, 2005, Phadnis and Orr, 2008; Zhang et al., 2015). But if HMS is primarily the result of drive, then the many HMS factors separating these species implies an extraordinary frequency of drive in the history of these species and/or an extraordinary proliferation of enhancer and suppressor loci associated with a smaller number of drive systems. We should also be clear that, with few exceptions (Moehring et al., 2006a; Good et al., 2008, Phadnis, 2011; Bi et al., 2015), genetic analyses have not yet established whether the X (or Z) has a relatively higher density of hybrid incompatibilities in other taxa. And, of course, Haldane’s rule and the large X effect may have different causes in different taxa. A composite model may well prevail (Wu and Davis, 1993), albeit one with different emphases than originally imagined. Explanations based on sexual selection, for instance, appear to have ceded ground to those based on genetic conflict.
From fine-scale genetic analyses, we have learned that the genetic architecture of HMS is best described by polygenic threshold and complex epistasis models with very few large-effect HMS factors separating the species. For both the X and the autosomes, apparently large-effect loci correspond to “HMS equivalents” (Tao et al., 2003b) that can be genetically decomposed into multiple factors with incomplete penetrance and/or sub-detectable phenotypic effects. This genetic architecture has important practical implications. First, the preponderance of individually weak-effect HMS factors hinders their genetic isolation and experimental validation (Wu and Palopoli, 1994). The HMS “success stories” (OdsH, JYalpha, and Overdrive) are not a random sample of HMS genes— they are large-effect outliers. In this sense, the genetics of speciation and the genetics of adaptation have, for similar practical reasons, both accumulated well-known, possibly unrepresentative, success stories involving large-effect, Mendelian factors (Rockman, 2012). Second, a polygenic architecture implies that the ∼15 HMS equivalents between D. simulans and D. mauritiana are underpinned by hundreds of substitutions with modest negative effects on male fertility. It is important to remember, however, that even “weak-effect” HMS factors, as determined by lab-based genetic analyses, are nonetheless readily detectable by natural selection in admixed populations and thus determine the level and genomic distribution of interspecific gene flow.
From population genomics analyses, we have learned that geographically allopatric species are not necessarily genetically allopatric. It appears that the inter-island dispersals of D. simulans-like ancestors to Mauritius and to the Seychelles ∼250,000 years ago were not unique events, as evidenced by recent nuclear and mitochondrial introgression. The resulting genomic distribution of introgression, however, is clearly shaped by the interplay of negative selection against incompatible and locally maladaptive alleles and positive selection for globally adaptive ones. Our findings reveal that selection against HMS disproportionately limits introgression on the X, whereas adaptation (Brand et al., 2013; Schrider et al., 2018) and drive (Meiklejohn et al., 2018) have enabled introgression. Now that we know that admixture has occurred, we can leverage the functional genetic and population genomics resources of the D. simulans clade species to further deconstruct the interaction of gene flow and natural selection during speciation.
Author Contributions
DP and CM contributed equally to the preparation of the manuscript. Both authors contributed to the article and approved the submitted version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Funding
Our work on hybrid incompatibilities in the D. simulans clade is supported by grants from the NIH (R01-GM123194 to CM and DP; and R01-GM135621-01 to DP).
References
Anholt, R. R. H., O’Grady, P., Wolfner, M. F., and Harbison, S. T. (2020). Evolution of reproductive behavior. Genetics 214, 49–73.
Araripe, L. O., Montenegro, H., Lemos, B., and Hartl, D. L. (2010). Fine-scale genetic mapping of a hybrid sterility factor between Drosophila simulans and D. mauritiana: the varied and elusive functions of speciation genes. BMC Evol. Biol. 10:385. doi: 10.1186/1471-2148-10-385
Ashburner, M., Golic, K. G., and Hawley, R. S. (2005). Drosophila: A Laboratory Handbook. Cold Spring Harbor. New York: Cold Spring Harbor Laboratory Press.
Aubert, J., and Solignac, M. (1990). Experimental evidence for mitochonrial DNA introgression between Drosophila species. Evolution 44, 1272–1282. doi: 10.1111/j.1558-5646.1990.tb05231.x
Ballard, J. W. O. (2000a). Comparative genomics of mitochondrial DNA in Drosophila simulans. J. Mol. Evol. 51, 64–75. doi: 10.1007/s002390010067
Ballard, J. W. O. (2000b). Comparative genomics of mitochondrial DNA in members of the Drosophila melanogaster subgroup. J. Mol. Evol. 51, 48–63. doi: 10.1007/s002390010066
Barbash, D. A. (2010). Genetic testing of the hypothesis that hybrid male lethality results from a failure in dosage compensation. Genetics 184, 313–316. doi: 10.1534/genetics.109.108100
Barbash, D. A., Siino, D. F., Tarone, A. M., and Roote, J. (2003). A rapidly evolving Myb-related protein causes species isolation in Drosophila. Proc. Natl. Acad. Sci. U.S.A. 100, 5302–5307. doi: 10.1073/pnas.0836927100
Bateson, W. (1909). “Heredity and variation in modern lights,” in Darwin and Modern Science, ed. A. C. Seward (Cambridge: Cambridge University Press), 85–101.
Bayes, J. J., and Malik, H. S. (2009). Altered heterochromatin binding by a hybrid sterility protein in Drosophila sibling species. Science 326, 1538–1541. doi: 10.1126/science.1181756
Begun, D. J., Holloway, A. K., Hillier, S. K. L. W., Poh, Y. P., Hahn, M. W., Nista, P. M., et al. (2007). Population genomics: whole-genome analysis of polymorphism and divergence in Drosophila simulans. Public Libr. Sci. Biol. 5, 2534–2559.
Bengtsson, B. O. (1985). “The flow of genes through a genetic barrier,” in Evolution: Essays in Honour of John Maynard Smith, eds P. J. Greenwood, P. H. Harvey, and M. Slatkin (Cambridge: Cambridge University Press), 31–42.
Betran, E., Thornton, K., and Long, M. (2002). Retroposed new genes out of the X in Drosophila. Genome Res. 12, 1854–1859. doi: 10.1101/gr.6049
Bi, Y., Ren, X., Yan, C., Shao, J., Xie, D., and Zhao, Z. (2015). A Genome-wide hybrid incompatibility landscape between Caenorhabditis briggsae and C. nigoni. PLoS Genet. 11:e1004993. doi: 10.1371/journal.pgen.1004993
Brand, C. L., Kingan, S. B., and Garrigan, D. (2013). A selective sweep across species boundaries in Drosophila. Mol. Biol. Evol. 30, 2177–2186. doi: 10.1093/molbev/mst123
Brideau, N. J., Flores, H. A., Wang, J., Maheshwari, S., Wang, X., and Barbash, D. A. (2006). Two dobzhansky-muller genes interact to cause hybrid lethality in Drosophila. Science 314, 1292–1295. doi: 10.1126/science.1133953
Cabot, E. L., Davis, A. W., Johnson, N. A., and Wu, C.-I. (1994). Genetics of reproductive isolation in the Drosophila simulans clade: complex epistasis underlying hybrid male sterility. Genetics 137, 175–189. doi: 10.1093/genetics/137.1.175
Carvalho, A. B., Vaz, S. C., and Klaczko, L. B. (1997). Polymorphism for Y-linked suppressors of sex-ratio in two natural populations of Drosophila mediopunctata. Genetics 146, 891–902. doi: 10.1093/genetics/146.3.891
Cattani, M. V., and Presgraves, D. C. (2009). Genetics and lineage-specific evolution of a lethal hybrid incompatibility between Drosophila mauritiana and its sibling species. Genetics 181, 1545–1555. doi: 10.1534/genetics.108.098392
Chakraborty, M., Chang, C.-H., Khost, D., Vedanayagam, J., Adrion, J. R., Liao, Y., et al. (2021). Evolution of genome structure in the Drosophila simulans species complex. Genome Res. 31, 380–396. doi: 10.1101/gr.263442.120
Charlesworth, B., Coyne, J. A., and Barton, N. H. (1987). The relative rates of evolution of sex-chromosomes and autosomes. Am. Nat. 130, 113–146. doi: 10.1086/284701
Charlesworth, B., Coyne, J. A., and Orr, H. A. (1993). Meiotic drive and unisexual hybrid sterility - a comment. Genetics 133, 421–424. doi: 10.1093/genetics/133.2.421
Courret, C., Gerard, P. R., Ogereau, D., Falque, M., Moreau, L., and Montchamp-Moreau, C. (2019). X-chromosome meiotic drive in Drosophila simulans: a QTL approach reveals the complex polygenic determinism of Paris drive suppression. Heredity 122, 906–915. doi: 10.1038/s41437-018-0163-1
Coyne, J. A. (1984). Genetic basis of male sterility in hybrids between two closely related species of Drosophila. Proc. Natl. Acad. Sci. U.S.A. 51, 4444–4447. doi: 10.1073/pnas.81.14.4444
Coyne, J. A. (1985). The genetic basis of Haldane’s rule. Nature 314, 736–738. doi: 10.1038/314736a0
Coyne, J. A. (1986). Meiotic segregation and male recombination in interspecific hybrids of Drosophila. Genetics 114, 485–494. doi: 10.1093/genetics/114.2.485
Coyne, J. A. (1992a). Genetics of sexual isolation in females of the Drosophila simulans species complex. Genet. Res. Camb. 60, 25–31. doi: 10.1017/s0016672300030639
Coyne, J. A., and Charlesworth, B. (1986). Location of an X-linked factor causing sterility in male hybrids of Drosophila-simulans and Drosophilia-mauritiana. Heredity 57, 243–246. doi: 10.1038/hdy.1986.114
Coyne, J. A., and Charlesworth, B. (1989). Genetic analysis of X-linked sterility in hybrids between three sibling species of Drosophila. Heredity 62, 97–106. doi: 10.1038/hdy.1989.13
Coyne, J. A., and Charlesworth, B. (1997). Genetics of a pheromonal difference affecting sexual isolation between Drosophila mauritiana and D-sechellia. Genetics 145, 1015–1030. doi: 10.1093/genetics/145.4.1015
Coyne, J. A., Charlesworth, B., and Orr, H. A. (1991). Haldane’s rule revisited. Evolution 45, 1710–1714.
Coyne, J. A., and Kreitman, M. (1986). Evolutionary genetics of two sibling species, Drosophila simulans and D. sechellia. Evolution 40, 673–691. doi: 10.2307/2408455
Coyne, J. A., and Orr, H. A. (1989a). “Two rules of speciation,” in Speciation and Its Consequences, eds D. Otte and J. Endler (Sunderland, MA: Sinauer Associates), 180–207.
Coyne, J. A., and Orr, H. A. (1989b). Patterns of speciation in Drosophila. Evolution 43, 362–381. doi: 10.2307/2409213
Coyne, J. A., and Orr, H. A. (1993). Further evidence against meiotic-drive models of hybrid sterility. Evolution 47, 685–687. doi: 10.2307/2410081
Coyne, J. A., and Orr, H. A. (1997). “Patterns of speciation in Drosophila” revisited. Evolution 51, 295–303. doi: 10.2307/2410984
Crespi, B. J., and Nosil, P. (2012). Conflictual speciation: species formation via genomic conflict. Trends Ecol. Evol. 28, 48–57. doi: 10.1016/j.tree.2012.08.015
David, J., McEvey, S. F., Solignac, M., and Tsacas, L. (1989). Drosophila communities on Mauritius and ecological niche of D. mauritiana (Diptera, Drosophilidae). J. Afr. Zool. 103, 107–116.
Davis, A. W., and Wu, C.-I. (1996). The broom of the sorcerer’s apprentice: the fine structure of a chromosomal region causing reproductive isolation between two sibling species of Drosophila. Genetics 143, 1287–1298. doi: 10.1093/genetics/143.3.1287
Davis, B. W., Seabury, C. M., Brashear, W. A., Li, G., Roelke-Parker, M., and Murphy, W. J. (2015). Mechanisms underlying mammalian hybrid sterility in two feline interspecies models. Mol. Biol. Evol. 32, 2534–2546. doi: 10.1093/molbev/msv124
Delph, L. F., and Demuth, J. P. (2016). Haldane’s rule: genetic bases and their empirical support. J. Heredity 107, 383–391. doi: 10.1093/jhered/esw026
Dobzhansky, T. (1936). Studies on hybrid sterility. II. Localization of sterility factors in Drosophila pseudoobscura hybrids. Genetics 21, 113–135. doi: 10.1093/genetics/21.2.113
Dobzhansky, T. (1944). On species and races of living and fossil man. Am. J. Phys. Anthropol. 2, 251–265. doi: 10.1002/ajpa.1330020303
Ferree, P. M., and Barbash, D. A. (2009). Species-specific heterochromatin prevents mitotic chromosome segregation to cause hybrid lethality in Drosophila. PLoS Biol. 7:e1000234. doi: 10.1371/journal.pbio.1000234
Ferree, P. M., and Prasad, S. (2012). How can satellite DNA divergence cause reproductive isolation? Let us count the chromosomal ways. Genet. Res. Int. 2012:430136.
Fraisse, C., and Sachdeva, H. (2020). The rates of introgression and barriers to genetic exchange between hybridizing species: sex chromosomes vs autosomes. Genetics 217:iyaa025.
Frank, S. A. (1991). Haldane’s rule: a defense of the meiotic drive theory. Evolution 45, 1714–1716. doi: 10.2307/2409793
Frazee, S. R., and Masly, J. P. (2015). Multiple sexual selection pressures drive the rapid evolution of complex morphology in a male secondary genital structure. Ecol. Evol. 5, 4437–4450. doi: 10.1002/ece3.1721
Gallach, M. (2014). Recurrent turnover of chromosome-specific satellites in Drosophila. Genome Biol. Evol. 6, 1279–1286. doi: 10.1093/gbe/evu104
Garrigan, D., Kingan, S. B., Geneva, A. J., Andolfatto, P., Clark, A. G., Thornton, K. R., et al. (2012). Genome sequencing reveals complex speciation in the Drosophila simulans clade. Genome Res. 22, 1499–1511. doi: 10.1101/gr.130922.111
Garrigan, D., Kingan, S. B., Geneva, A. J., Vedanayagam, J. P., and Presgraves, D. C. (2014). Genome diversity and divergence in Drosophila mauritiana: multiple signatures of faster X evolution. Genome Biol. Evol. 6, 2444–2458. doi: 10.1093/gbe/evu198
Gelbart, M. E., and Kuroda, M. I. (2009). Drosophila dosage compensation: a complex voyage to the X chromosome. Development 136, 1399–1410. doi: 10.1242/dev.029645
Geneva, A. J., Muirhead, C., Kingan, S. B., and Garrigan, D. (2016). A new method to scan genomes for introgression in a secondary contact model. PLoS One 10:e0118621. doi: 10.1371/journal.pone.0118621
Gingell, L. F., and McLean, J. R. (2020). A protamine knockdown mimics the function of Sd in Drosophila melanogaster. G3 10, 2111–2115. doi: 10.1534/g3.120.401307
Good, J. M., Dean, M. D., and Nachman, M. W. (2008). A complex genetic basis to X-Linked hybrid male sterility between two species of house mice. Genetics 179, 2213–2228. doi: 10.1534/genetics.107.085340
Haldane, J. B. S. (1922). Sex ratio and unisexual sterility in animal hybrids. J. Genet. 12, 101–109. doi: 10.1007/bf02983075
Hall, D. W. (2004). Meiotic drive and sex chromosome cycling. Evolution 58, 925–931. doi: 10.1554/03-440
Hamilton, W. D. (1967). Extraordinary sex ratios. Science 156, 477–488. doi: 10.1126/science.156.3774.477
Han, M. V., and Hahn, M. W. (2012). Inferring the history of interchromosomal gene transposition in Drosophila using n-dimensional parsimony. Genetics 190, 813–825. doi: 10.1534/genetics.111.135947
Helleu, Q., Courret, C., Ogereau, D., Burnham, K. L., Chaminade, N., Chakir, M., et al. (2019). Sex-ratio meiotic drive shapes the evolution of the Y chromosome in Drosophila simulans. Mol. Biol. Evol. 36, 2668–2681. doi: 10.1093/molbev/msz160
Helleu, Q., Gerard, P. R., Dubruille, R., Ogereau, D., Prud’homme, N., Loppin, B., et al. (2016). Rapid evolution of a Y-chromosome heterochromatin protein underlies sex chromosome meiotic drive. Proc. Natl. Acad. Sci. U.S.A. 113, 4110–4115. doi: 10.1073/pnas.1519332113
Hense, W., Baines, J. F., and Parsch, J. (2007). X chromosome inactivation during Drosophila spermatogenesis. Public Libr. Sci. Biol. 5, 2288–2295.
Hollocher, H., and Wu, C.-I. (1996). The genetics of reproductive isolation in the Drosophila simulans clade: X vs. autosomal effects and male vs. female effects. Genetics 143, 1243–1255. doi: 10.1093/genetics/143.3.1243
Hsieh, T., and Brutlag, D. (1979). Sequence and sequence variation within the 1.688 g/cm3 satellite DNA of Drosophila melanogaster. J. Mol. Biol. 135, 465–481. doi: 10.1016/0022-2836(79)90447-9
Hurst, L. D., and Pomiankowski, A. (1991). Causes of sex ratio bias may account for unisexual sterility in hybrids: a new explanation of Haldane’s rule and related phenomena. Genetics 128, 841–858. doi: 10.1093/genetics/128.4.841
Hutter, P., and Ashburner, M. (1987). Genetic rescue of inviable hybrids between Drosophila melanogaster and its sibling species. Nature 327, 331–333. doi: 10.1038/327331a0
Jagannathan, M., Warsinger-Pepe, N., Watase, G. J., and Yamashita, Y. (2017). Comparative analysis of satellite DNA in the Drosophila melanogaster species complex. G3 3, 693–704. doi: 10.1534/g3.116.035352
Johnson, N. (2010). Hybrid incompatibility genes: remnants of a genomic battlefield? Trends Genet. 26, 317–325. doi: 10.1016/j.tig.2010.04.005
Johnson, N. A., Hollocher, H., Noonburg, E., and Wu, C.-I. (1993). The effects of interspecific Y chromosome replacements on hybrid sterility within the Drosophila simulans clade. Genetics 135, 443–453. doi: 10.1093/genetics/135.2.443
Johnson, N. A., Perez, D. E., Cabot, E. L., Hollocher, H., and Wu, C.-I. (1992). A test of reciprocal X-Y interactions as a cause of hybrid sterility in Drosophila. Nature 358, 751–753. doi: 10.1038/358751a0
Johnson, N. A., and Porter, A. H. (2000). Rapid speciation via parallel, directional selection on regulatory genetic pathways. J. Theor. Biol. 205, 527–542. doi: 10.1006/jtbi.2000.2070
Johnson, N. A., and Wu, C.-I. (1992). An empirical test of the meiotic drive models of hybrid sterility: sex ratio data from hybrids between Drosophila simulans and Drosophila sechellia. Genetics 130, 507–511. doi: 10.1093/genetics/130.3.507
Kanippayoor, R. L., Alpern, J. H. M., and Moehring, A. J. (2020). A common suite of cellular abnormalities and spermatogenetic errors in sterile hybrid males in Drosophila. Proc. R. Soc. B Biol. Sci. 287:20192291. doi: 10.1098/rspb.2019.2291
Kliman, R. M., Andolfatto, P., Coyne, J. A., Depaulis, F., Kreitman, M., Berry, A. J., et al. (2000). The population genetics of the origin and divergence of the Drosophila simulans complex species. Genetics 156, 1913–1931.
Kruger, A. N., Brogley, M. A., Huizinga, J. L., Kidd, J. M., de Rooij, D. G., Hu, Y. C., et al. (2019). A neofunctionalized X-linked ampliconic gene family is essential for male fertility and equal sex ratio in mice. Curr. Biol. 29, 3699.e5–3706.e5.
Kuhn, G. C., Kuttler, H., Moreira-Filho, O., and Heslop-Harrison, J. S. (2012). The 1.688 repetitive DNA of Drosophila: concerted evolution at different genomic scales and association with genes. Mol. Biol. Evol. 29, 7–11. doi: 10.1093/molbev/msr173
Kulathinal, R. J., and Singh, R. S. (1998). Cytological characterization of premeiotic versus postmeiotic defects producing hybrid male sterility among sibling species of the Drosophila melanogaster complex. Evolution 52, 1067–1079. doi: 10.2307/2411237
Lachaise, D., Cariou, M.-L., David, J. R., Lemeunier, F., Tsacas, L., and Ashburner, M. (1988). Historical biogeography of the Drosophila melanogaster species subgroup. Evol. Biol. 22, 159–225. doi: 10.1007/978-1-4613-0931-4_4
Lachaise, D., David, J. R., Lemeunier, F., Tsacas, L., and Ashburner, M. (1986). The reproductive relationships of Drosophila sechellia with D. mauritiana, D. simulans, and D. melanogaster from the Afrotropical region. Evolution 1986, 262–271. doi: 10.1111/j.1558-5646.1986.tb00468.x
Landeen, E. L., Muirhead, C., Wright, L., Meiklejohn, C. D., and Presgraves, D. C. (2016). Sex chromosome-wide transcriptional suppression and compensatory cis-regulatory evolution mediate gene expression in the Drosophila male germline. PLoS Biol. 14:e1002499. doi: 10.1371/journal.pbio.1002499
Larracuente, A. M. (2014). The organization and evolution of the Responder satellite in species of the Drosophila melanogaster group: dynamic evolution of a target of meiotic drive. BMC Evol. Biol. 14:233. doi: 10.1186/s12862-014-0233-9
Laurie, C. C. (1997). The weaker sex is heterogametic: 75 years of Haldane’s rule. Genetics 147, 937–951. doi: 10.1093/genetics/147.3.937
Levine, M. T., Holloway, A. K., Arshad, U., and Begun, D. J. (2007). Pervasive and largely lineage-specific adaptive protein evolution in the dosage compensation complex of Drosophila melanogaster. Genetics 177, 1959–1962. doi: 10.1534/genetics.107.079459
Lienard, M. A., Araripe, L. O., and Hartl, D. L. (2016). Neighboring genes for DNA-binding proteins rescue male sterility in Drosophila hybrids. Proc. Natl. Acad. Sci. U.S.A. 113, E4200–E4207.
Lin, C. J., Hu, F., Dubruille, R., Vedanayagam, J., Wen, J., Smibert, P., et al. (2018). The hpRNA/RNAi pathway is essential to resolve intragenomic conflict in the Drosophila male germline. Dev. Cell. 46, 316.e5–326.e5.
Lindsley, D. L., Roote, J., and Kennison, J. A. (2013). Anent the genomics of spermatogenesis in Drosophila melanogaster. PLoS One 8:e55915. doi: 10.1371/journal.pone.0055915
Lindsley, D. L., and Tokuyasu, K. T. (1980). “Spermatogenesis,” in The Genetics and Biology of Drosophila, eds M. Ashburner and T. R. F. Wright (New York: Academic Press), 226–294.
Lu, X., Shapiro, J. A., Ting, C.-T., Li, Y., Li, C., Xu, J., et al. (2010). Genome-wide misexpression of X-linked versus autosomal genes associated with hybrid male sterility. Genome Res. 20, 1097–1102. doi: 10.1101/gr.076620.108
Lynch, M., and Force, A. G. (2000). The origin of interspecific genomic incompatibility via gene duplication. Am. Nat. 156, 590–605. doi: 10.2307/3079065
Mack, K. L., and Nachman, M. W. (2016). Gene regulation and speciation. Trends Genet. 33, 68–80. doi: 10.1016/j.tig.2016.11.003
Mallet, J. (2005). Hybridization as an invasion of the genome. Trends Ecol. Evol. 20, 229–237. doi: 10.1016/j.tree.2005.02.010
Mallet, J. (2006). What does Drosophila genetics tell us about speciation? Trends Ecol. Evol. 21, 386–393. doi: 10.1016/j.tree.2006.05.004
Mallet, J., Besansky, N., and Hahn, M. W. (2016). How reticulated are species? BioEssays 38, 140–149. doi: 10.1002/bies.201500149
Masly, J. P., Jones, C. D., Noor, M. A. F., Locke, J., and Orr, H. A. (2006). Gene transposition as a novel cause of hybrid male sterility. Science 313, 1448–1450. doi: 10.1126/science.1128721
Masly, J. P., and Presgraves, D. C. (2007). High-resolution genome-wide dissection of the two rules of speciation in Drosophila. Public Libr. Sci. Biol. 5, 1890–1898.
Matute, D. R., and Ayroles, J. F. (2014). Hybridization occurs between Drosophila simulans and D. sechellia in the Seychelles archipelago. J. Evol. Biol. 27, 1057–1068. doi: 10.1111/jeb.12391
McDermott, S. R., and Kliman, R. M. (2008). Estimation of isolation times of the island species in the Drosophila simulans complex from multilocus sequence data. PLoS One 3:e2442. doi: 10.1371/journal.pone.0002442
Meany, M. K., Conner, W. R., Richter, S. V., Bailey, J. A., Turelli, M., and Cooper, B. S. (2019). Loss of cytoplasmic incompatibility and minimal fecundity effects explain relatively low Wolbachia frequencies in Drosophila mauritiana. Evolution 73, 1278–1295. doi: 10.1111/evo.13745
Meiklejohn, C. D., Landeen, E. L., Cook, J. M., Kingan, S. B., and Presgraves, D. C. (2011). Sex chromosome-specific regulation in the Drosophila male germline but little evidence for chromosomal dosage compensation or meiotic inactivation. PLoS Biol. 9:e1001126. doi: 10.1371/journal.pbio.1001126
Meiklejohn, C. D., Landeen, E. L., Gordon, K. E., Rzatkiewicz, T., Kingan, S. B., Geneva, A. J., et al. (2018). Gene flow mediates the role of sex chromosome meiotic drive during complex speciation. eLife 7:e35468.
Meiklejohn, C. D., Parsch, J., Ranz, J. M., and Hartl, D. L. (2003). Rapid evolution of male-biased gene expression in Drosophila. Proc. Natl. Acad. Sci. U.S.A. 100, 9894–9899. doi: 10.1073/pnas.1630690100
Meiklejohn, C. D., and Presgraves, D. C. (2012). Little evidence for demasculinization of the Drosophila X chromosome among genes expressed in the male germline. Genome Biol. Evol. 4, 895–904.
Meiklejohn, C. D., and Tao, Y. (2010). Genetic conflict and sex chromosome evolution. Trends Ecol. Evol. 25, 215–223. doi: 10.1016/j.tree.2009.10.005
Meisel, R. P., and Connallon, T. (2013). The faster-X effect: integrating theory and data. Trends Genet. 29, 537–544. doi: 10.1016/j.tig.2013.05.009
Meisel, R. P., Han, M. V., and Hahn, M. W. (2009). A complex suite of forces drives gene traffic from Drosophila X chromosomes. Genome Biol. Evol. 1, 176–188. doi: 10.1093/gbe/evp018
Meisel, R. P., Malone, J. H., and Clark, A. G. (2012). Faster-X evolution of gene expression in Drosophila. PLoS Genet. 8:e1003013. doi: 10.1371/journal.pgen.1003013
Miller, G. T., and Pitnick, S. (2002). Sperm-female coevolution in Drosophila. Science 298, 1230–1233. doi: 10.1126/science.1076968
Mills, W. K., Lee, Y. C. G., Kochendoerfer, A. M., Dunleavy, E. M., and Karpen, G. H. (2019). RNA from a simple-tandem repeat is required for sperm maturation and male fertility in Drosophila melanogaster. eLife 8:e48940.
Moehring, A. J., Llopart, A., Elwyn, S., Coyne, J. A., and Mackay, T. F. (2006a). The genetic basis of postzygotic reproductive isolation between Drosophila santomea and D. yakuba due to hybrid male sterility. Genetics 173, 225–233. doi: 10.1534/genetics.105.052985
Moehring, A. J., Teeter, K. C., and Noor, M. A. F. (2006b). Genome-wide patterns of expression in Drosophila pure species and hybrid males. II. Examination of multiple-species hybridizations, platforms, and life cycle stages. Mol. Biol. Evol. 24, 137–145. doi: 10.1093/molbev/msl142
Moyle, L. C., Muir, C. D., Han, M. V., and Hahn, M. W. (2010). The contribution of gene movement to the two rules of speciation. Evolution 64, 1541–1557. doi: 10.1111/j.1558-5646.2010.00990.x
Muirhead, C., and Presgraves, D. C. (2016). Hybrid incompatibilities, local adaptation, and the genomic distribution of natural introgression between species. Am. Nat. 187, 249–261. doi: 10.1086/684583
Muller, H. J. (1940). “Bearing of the Drosophila work on systematics,” in The New Systematics, ed. J. S. Huxley (Oxford: Clarendon Press), 185–268.
Muller, H. J., and Pontecorvo, G. (1940). Recombinants between Drosophila species, the F1 hybrids of which are sterile. Nature 146:199. doi: 10.1038/146199b0
Navascues, M., Legrand, D., Campagne, C., Cariou, M. L., and Depaulis, F. (2014). Distinguishing migration from isolation using genes with intragenic recombination: detecting introgression in the Drosophila simulans species complex. BMC Evol. Biol. 14:89. doi: 10.1186/1471-2148-14-89
Naveira, H., and Fontdevila, A. (1985). The evolutionary history of Drosophila buzzattii. IX. High frequencies of chromosome rearrangements induced by introgressive hybridization. Chromosoma 91, 87–94. doi: 10.1007/bf00294050
Naveira, H., and Fontdevila, A. (1986). The evolutionary history of Drosophila buzzatii. XII. The genetic basis of sterility in hybrids between D. buzzatii and its sibling D. serido from Argentina. Genetics 114, 841–857. doi: 10.1093/genetics/114.3.841
Naveira, H., and Fontdevila, A. (1991). The evolutionary history of Drosophila buzzatii. XXI. Cumulative action of multiple sterility factors on spermatogenesis in hybrids of D. buzzatii and D. koepferae. Heredity 67, 57–72. doi: 10.1038/hdy.1991.65
Naveira, H. F. (1992). Location of X-linked polygenic effects causing sterility in male hybrids of Drosophila simulans and D. mauritiana. Heredity 68, 211–217. doi: 10.1038/hdy.1992.34
Naveira, H. F. (2003). On the relative roles of faster-X evolution and dominance in the establishment of intrinsic postzygotic isolating barriers. Genetica 118, 41–50.
Naveira, H. F., and Maside, X. R. (1998). “The genetics of hybrid male sterility in Drosophila,” in Endless Forms, eds D. J. Howard and S. H. Berlocher (Oxford: Oxford University Press), 330–338.
Nolte, V., Pandey, R. V., Kofler, R., and Schlotterer, C. (2013). Genome-wide patterns of natural variation reveal strong selective sweeps and ongoing genomic conflict in Drosophila mauritiana. Genome Res. 23, 99–110. doi: 10.1101/gr.139873.112
Nunes, M. D., Wengel, P. O., Kreissl, M., and Schlotterer, C. (2010). Multiple hybridization events between Drosophila simulans and Drosophila mauritiana are supported by mtDNA introgression. Mol. Ecol. 19, 4695–4707. doi: 10.1111/j.1365-294x.2010.04838.x
Orr, H. A. (1989). Does postzygotic isolation result from improper dosage compensation? Genetics 122, 891–894. doi: 10.1093/genetics/122.4.891
Orr, H. A. (1992). Mapping and characterization of a speciation gene in Drosophila. Genet. Res. 59, 73–80. doi: 10.1017/s0016672300030275
Orr, H. A. (1996). Dobzhansky, Bateson, and the genetics of speciation. Genetics 144, 1331–1335. doi: 10.1093/genetics/144.4.1331
Orr, H. A., and Betancourt, A. J. (2001). Haldane’s sieve and adaptation from the standing genetic variation. Genetics 157, 875–884. doi: 10.1093/genetics/157.2.875
Orr, H. A., and Irving, S. (2005). Segregation distortion in hybrids between the bogota and USA subspecies of Drosophila pseudoobscura. Genetics 169, 671–682. doi: 10.1534/genetics.104.033274
Palopoli, M. F., and Wu, C.-I. (1994). Genetics of hybrid male sterility between Drosophila sibling species: a complex web of epistasis is revealed in interspecific studies. Genetics 138, 329–341. doi: 10.1093/genetics/138.2.329
Perez, D. E., and Wu, C.-I. (1995). Further characterization of the Odysseus locus of hybrid sterility in Drosophila: one gene is not enough. Genetics 140, 201–206. doi: 10.1093/genetics/140.1.201
Perez, D. E., Wu, C.-I., Johnson, N. A., and Wu, M.-L. (1993). Genetics of reproductive isolation in the Drosophila simulans clade: DNA-marker assisted mapping and characterization of a hybrid-male sterility gene, Odysseus (Ods). Genetics 134, 261–275. doi: 10.1093/genetics/134.1.261
Phadnis, N. (2011). Genetic architecture of male sterility and segregation distortion in Drosophila pseudoobscura Bogota-USA hybrids. Genetics 189, 1001–1009. doi: 10.1534/genetics.111.132324
Phadnis, N., Baker, E. C. P., Cooper, J. C., Frizzell, K. A., Hsieh, E., de la Cruz, A. F. A., et al. (2015). An essential cell cycle regulation gene causes hybrid inviability in Drosophila. Science 350, 1552–1555. doi: 10.1126/science.aac7504
Phadnis, N., and Orr, H. A. (2008). A single gene causes both male sterility and segregation distortion in Drosophila hybrids. Science 323, 376–379. doi: 10.1126/science.1163934
Presgraves, D. C. (2002). Patterns of postzygotic isolation in Lepidoptera. Evolution 56, 1168–1183. doi: 10.1554/0014-3820(2002)056[1168:popiil]2.0.co;2
Presgraves, D. C. (2008a). Sex chromosomes and speciation in Drosophila. Trends Genet. 24, 336–343. doi: 10.1016/j.tig.2008.04.007
Presgraves, D. C. (2008b). “Drive and sperm: evolution and genetics of male meiotic drive,” in Sperm Biology: An Evolutionary Perspective, eds T. R. Birkhead, D. J. Hosken, and S. Pitnick (Amsterdam: Elsevier Press).
Presgraves, D. C. (2010). The molecular evolutionary basis of species formation. Nat. Rev. Genet. 11, 175–180. doi: 10.1038/nrg2718
Presgraves, D. C. (2018). Evaluating genomic signatures of the large X-effect during complex speciation. Mol. Ecol. 27, 3822–3830. doi: 10.1111/mec.14777
Presgraves, D. C., Balagopalan, L., Abmayr, S. M., and Orr, H. A. (2003). Adaptive evolution drives divergence of a hybrid inviability gene between two species of Drosophila. Nature 423, 715–719. doi: 10.1038/nature01679
Price, C. S. C. (1997). Conspecific sperm precedence in Drosophila. Nature 388, 663–666. doi: 10.1038/41753
Price, T. D., and Bouvier, M. M. (2002). The evolution of F1 postzygotic incompatibilities in birds. Evolution 56, 2083–2089. doi: 10.1554/0014-3820(2002)056[2083:teofpi]2.0.co;2
Rabosky, D. L., and Matute, D. R. (2013). Macroevolutionary speciation rates are decoupled from the evolution of intrinsic reproductive isolation in Drosophila and birds. Proc. Natl. Acad. Sci. U.S.A. 110, 15354–15359. doi: 10.1073/pnas.1305529110
Rathje, C. C., Johnson, E. E. P., Drage, D., Patinioti, C. G., Silvestri, G., Affara, N. A., et al. (2019). Differential sperm motility mediates the sex ratio drive shaping mouse sex chromosome evolution. Curr. Biol. 29, 3692–3698. doi: 10.1016/j.cub.2019.09.031
Rathke, C., Baarends, W. M., Jayaramaiah-Raja, S., Bartkuhn, M., Renkawitz, R., and Renkawitz-Pohl, R. (2007). Transition from a nucleosome-based to a protamine-based chromatin configuration during spermiogenesis in Drosophila. J. Cell Sci. 120, 1689–1700. doi: 10.1242/jcs.004663
R’Kha, S., Capy, P., and David, J. R. (1991). Host-plant specialization in the Drosophila melanogaster species complex: a physiological, behavioral, and genetical analysis. Proc. Natl. Acad. Sci. U.S.A. 88, 1835–1839. doi: 10.1073/pnas.88.5.1835
Rockman, M. V. (2012). The QTN program and the alleles that matter for evolution: all that’s gold does not glitter. Evolution 66, 1–17. doi: 10.1111/j.1558-5646.2011.01486.x
Rodriguez, M. A., Vermaak, D., Bayes, J. J., and Malik, H. S. (2007). Species-specific positive selection of the male-specific lethal complex that participates in dosage compensation in Drosophila. Proc. Natl. Acad. Sci. U.S.A. 104, 15412–15417. doi: 10.1073/pnas.0707445104
Rose, M., and Doolittle, W. F. (1983). Molecular biological mechanisms of speciation. Science 220, 157–162. doi: 10.1126/science.220.4593.157
Satta, Y., and Takahata, N. (1990). Evolution of Drosophila mitochondrial DNA and the history of the melanogaster subgroup. Proc. Natl. Acad. Sci. U.S.A. 87, 9558–9562. doi: 10.1073/pnas.87.24.9558
Satyaki, P. R., Cuykendall, T. N., Wei, K. H., Brideau, N. J., Kwak, H., Aruna, S., et al. (2014). The Hmr and Lhr hybrid incompatibility genes suppress a broad range of heterochromatic repeats. PLoS Genet. 10:e1004240. doi: 10.1371/journal.pgen.1004240
Sawamura, K. (2000). Genetics of hybrid inviability and sterility in Drosophila: the Drosophila melanogaster-Drosophila simulans case. Plant Species Biol. 15, 237–247. doi: 10.1046/j.1442-1984.2000.00043.x
Sawamura, K. (2012). Chromatin evolution and molecular drive in speciation. Int. J. Evol. Biol. 2012:301894.
Sawamura, K., Davis, A. W., and Wu, C.-I. (2000). Genetic analysis of speciation by means of introgression into Drosophila melanogaster. Proc. Natl. Acad. Sci. U.S.A. 97, 2652–2655. doi: 10.1073/pnas.050558597
Sawamura, K., and Yamamoto, M. T. (1997). Characterization of a reproductive isolation gene, Zygotic hybrid rescue, of Drosophila melanogaster by using minichromosomes. Heredity 79, 97–103. doi: 10.1038/hdy.1997.127
Schilthuizen, M., Giesbers, M. C. W. G., and Beukeboom, L. W. (2011). Haldane’s rule in the 21st century. Heredity 107, 95–102.
Schrider, D. R., Ayroles, J., Matute, D. R., and Kern, A. D. (2018). Supervised machine learning reveals introgressed loci in the genomes of Drosophila simulans and D. sechellia. PLoS Genet. 14:e1007341. doi: 10.1371/journal.pgen.1007341
Seehausen, O., Butlin, R. K., Keller, I., Wagner, C. E., Boughman, J. W., Hohenlohe, P. A., et al. (2014). Genomics and the origin of species. Nat. Rev. Genet. 15, 176–192.
Solignac, M., and Monnerot, M. (1986). Race formation, speciation, and introgression within Drosophila simulans, D. mauritiana, and D. sechellia inferred from mitonchondrial DNA analysis. Evolution 40, 531–539. doi: 10.1111/j.1558-5646.1986.tb00505.x
Solignac, M., Monnerot, M., and Mounolou, J.-C. (1986). Mitochondrial DNA evolution in the melanogaster species subgroup of Drosophila. J. Mol. Evol. 23, 31–40.
Sproul, J. S., Khost, D. E., Eickbush, D. G., Negm, S., Wei, X., Wong, I., et al. (2020). Dynamic evolution of euchromatic satellites on the X Chromosome in Drosophila melanogaster and the simulans Clade. Mol. Biol. Evol. 37, 2241–2256. doi: 10.1093/molbev/msaa078
Sturtevant, A. H. (1919). A new species closely resembling Drosophila melanogaster. Psyche 26, 153–155. doi: 10.1155/1919/97402
Sturtevant, A. H. (1920). Genetic studies on Drosophila simulans. I. Introduction. Hybrids with Drosophila melanogaster. Genetics 5, 488–500. doi: 10.1093/genetics/5.5.488
Swanson, W. J., Clark, A. G., Waldrip-Dail, H. M., Wolfner, M. F., and Aquadro, C. F. (2001). Evolutionary EST analysis identifies rapidly evolving male reproductive proteins in Drosophila. Proc. Natl. Acad. Sci. U.S.A. 98, 7375–7379. doi: 10.1073/pnas.131568198
Tang, S., and Presgraves, D. C. (2009). Evolution of the Drosophila nuclear pore complex results in multiple hybrid incompatibilities. Science 323, 779–782. doi: 10.1126/science.1169123
Tao, Y., Araripe, L., Kingan, S. B., Ke, Y., Xiao, H., and Hartl, D. L. (2007a). A sex-ratio meiotic drive system in Drosophila simulans. II: an X-linked distorter. Public Libr. Sci. Biol. 5:e293. doi: 10.1371/journal.pbio.0050293
Tao, Y., Chen, S., Hartl, D. L., and Laurie, C. C. (2003a). Genetic dissection of hybrid incompatibilities between Drosophila simulans and D. mauritiana. I. Differential accumulation of hybrid male sterility effects on the X and autosomes. Genetics 164, 1383–1397. doi: 10.1093/genetics/164.4.1383
Tao, Y., Masly, J. P., Araripe, L., Ke, Y., and Hartl, D. L. (2007b). A sex-ratio meiotic drive system in Drosophila simulans. I: an autosomal suppressor. Public Libr. Sci. Biol. 5:e292. doi: 10.1371/journal.pbio.0050292
Tao, Y., and Hartl, D. L. (2003). Genetic dissection of hybrid incompatibilities between Drosophila simulans and D. mauritiana. III. Heterogeneous accumulation of hybrid incompatibilities, degree of dominance, and implications for Haldane’s rule. Evolution 57, 2580–2589. doi: 10.1554/03-094
Tao, Y., Hartl, D. L., and Laurie, C. C. (2001). Sex-ratio segregation distortion associated with reproductive isolation in Drosophila. Proc. Natl. Acad. Sci. U.S.A. 98, 13183–13188. doi: 10.1073/pnas.231478798
Tao, Y., Zeng, Z. B., Hartl, D. L., and Laurie, C. C. (2003b). Genetic dissection of hybrid incompatibilities between Drosophila simulans and D. mauritiana. II. Mapping hybrid male sterility loci on the third chromosome. Genetics 164, 1399–1418. doi: 10.1093/genetics/164.4.1399
Ting, C.-T., Tsaur, S.-C., Wu, M.-L., and Wu, C.-I. (1998). A rapidly evolving homeobox at the site of a hybrid sterility gene. Science 282, 1501–1504. doi: 10.1126/science.282.5393.1501
True, J. R., Mercer, J. M., and Laurie, C. C. (1996b). Differences in crossover frequency and distribution among three sibling species of Drosophila. Genetics 142, 507–523. doi: 10.1093/genetics/142.2.507
True, J. R., Weir, B. S., and Laurie, C. C. (1996a). A genome-wide survey of hybrid incompatibility factors by the introgression of marked segments of Drosophila mauritiana chromosomes into Drosophila simulans. Genetics 142, 819–837. doi: 10.1093/genetics/142.3.819
Tsacas, L., and Bächli, G. (1981). Drosophila sechellia, n.sp., huitieme espece du sous-goupe melanogaster des Iles Sechelles [Diptera, Drosophilidae]. Rev. Franc. D’entomol. Nouvelle Ser. 3, 146–150.
Tsacas, L., and David, J. R. (1974). Drosophila mauritiana n.sp. du groupe melanogaster de l’Ile Maurice. Bull. Soc. Entomol. France 79, 42–46.
Turelli, M., and Orr, H. A. (1995). The dominance theory of Haldane’s rule. Genetics 140, 389–402. doi: 10.1093/genetics/140.1.389
Turelli, M., and Orr, H. A. (2000). Dominance, epistasis and the genetics of postzygotic isolation. Genetics 154, 1663–1679. doi: 10.1093/genetics/154.4.1663
Vibranovski, M. D., Lopes, H. F., Karr, T. L., and Long, M. (2009). Stage-specific expression profiling of Drosophila spermatogenesis suggests that meiotic sex chromosome inactivation drives genomic relocation of testis-expressed genes. PLoS Genet. 5:e1000731. doi: 10.1371/journal.pgen.1000731
Watanabe, T. K. (1979). A gene that rescues the lethal hybrids between Drosophila melanogaster and D. simulans. Jpn. J. Genet. 54, 325–331. doi: 10.1266/jjg.54.325
Werth, C. R., and Windham, M. D. (1991). A model for divergent, allopatric speciation of polyploid pteridophytes resulting from silencing of duplicate-gene expression. Am. Nat. 137, 515–526. doi: 10.1086/285180
Wu, C.-I. (1992). A note on Haldane’s rule: hybrid inviability versus hybrid sterility. Evolution 46, 1584–1587. doi: 10.2307/2409965
Wu, C.-I. (2001). The genic view of the process of speciation. J. Evol. Biol. 14, 851–865. doi: 10.1046/j.1420-9101.2001.00335.x
Wu, C.-I., and Davis, A. W. (1993). Evolution of postmating reproductive isolation: the composite nature of Haldane’s rule and its genetic bases. Am. Nat. 142, 187–212. doi: 10.1086/285534
Wu, C.-I., and Hollocher, H. (1998). “Subtle is Nature: the genetics of species differentiation and speciation,” in Endless Forms, eds D. J. Howard and S. H. Berlocher (Oxford: Oxford University Press), 339–351.
Wu, C.-I., Johnson, N. A., and Palopoli, M. F. (1996). Haldane’s rule and its legacy: why are there so many sterile males? Trends Ecol. Evol. 11, 281–284. doi: 10.1016/0169-5347(96)10033-1
Wu, C.-I., Lyttle, T. W., Wu, M.-L., and Lin, G. F. (1988). Association between DNA satellite sequences and the responder of segregation distortion in D. melanogaster. Cell 54, 179–189. doi: 10.1016/0092-8674(88)90550-8
Wu, C.-I., and Palopoli, M. F. (1994). Genetics of postmating reproductive isolation in animals. Ann. Rev. Genet. 27, 283–308. doi: 10.1146/annurev.ge.28.120194.001435
Yukilevich, R. (2012). Asymmetrical patterns of speciation uniquely support reinforcement in Drosophila. Evolution 66, 1430–1446. doi: 10.1111/j.1558-5646.2011.01534.x
Zeng, L.-W., and Singh, R. S. (1993). The genetic basis of Haldane’s rule and the nature of asymmetric hybrid male sterility among Drosophila simulans, Drosophila mauritiana and Drosophila sechellia. Genetics 134, 251–260. doi: 10.1093/genetics/134.1.251
Keywords: speciation, Haldane’s rule, large X-effect, genetic conflict, gene flow
Citation: Presgraves DC and Meiklejohn CD (2021) Hybrid Sterility, Genetic Conflict and Complex Speciation: Lessons From the Drosophila simulans Clade Species. Front. Genet. 12:669045. doi: 10.3389/fgene.2021.669045
Received: 17 February 2021; Accepted: 20 May 2021;
Published: 23 June 2021.
Edited by:
Igor V. Sharakhov, Virginia Tech, United StatesReviewed by:
Axel Imhof, Ludwig-Maximilians University of Munich, GermanyAaron Arthur Comeault, Bangor University, United Kingdom
Copyright © 2021 Presgraves and Meiklejohn. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Daven C. Presgraves, ZGF2ZW4ucHJlc2dyYXZlc0Byb2NoZXN0ZXIuZWR1
 Daven C. Presgraves
Daven C. Presgraves Colin D. Meiklejohn
Colin D. Meiklejohn