
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Genet. , 17 June 2021
Sec. Evolutionary and Population Genetics
Volume 12 - 2021 | https://doi.org/10.3389/fgene.2021.665927
The environmental residue/sublethal doses of neonicotinoid insecticides are believed to generate a negative impact on pollinators, including honey bees. Here we report our recent investigation on how imidacloprid, one of the major neonicotinoids, affects worker bees by profiling the transcriptomes of various ages of bees exposed to different doses of imidacloprid during the larval stage. The results show that imidacloprid treatments during the larval stage severely altered the gene expression profiles and may induce precocious foraging. Differential expression of foraging regulators was found in 14-day-old treated adults. A high transcriptome similarity between larvae-treated 14-day-old adults and 20-day-old controls was also observed, and the similarity was positively correlated with the dose of imidacloprid. One parts per billion (ppb) of imidacloprid was sufficient to generate a long-term impact on the bee’s gene expression as severe as with 50 ppb imidacloprid. The disappearance of nurse bees may be driven not only by the hive member constitution but also by the neonicotinoid-induced precocious foraging behavior.
With high pollinating efficiency, workers of the honey bee, Apis mellifera, are the most important pollinators for agricultural crops as well as wild plants (Calderone, 2012; van der Zee et al., 2012; Garibaldi et al., 2013, 2016; Hung et al., 2018). Although honey bees may be exposed to different types of agricultural chemicals, previous studies evaluating the impacts of pesticides found that a group of insecticides called neonicotinoids may be particularly hazardous to honey bees (Goulson, 2013; Lu et al., 2014; Sandrock et al., 2014; Goñalongs and Farina, 2015). While foragers are more frequently in contact with the pesticides, the whole colony is also under the threat of sublethal neonicotinoids as neonicotinoid-contaminated nectar and pollen are delivered to the colony through foraging activities (Chauzat et al., 2006; Škerl et al., 2009; Codling et al., 2016; Mitchell et al., 2017; Böhme et al., 2018).
A sublethal dose of imidacloprid, one of the major neonicotinoids mostly used around the world, may not cause acute bee death, but it can generate chronic irreversible damage to the bee colony. Chronic exposure to sublethal doses of imidacloprid can generate abnormal behavior or physiological disorders in bees (Goulson, 2013; Lu et al., 2014; Sandrock et al., 2014; Goñalongs and Farina, 2015). A long-term investigation revealed that honey bee colony health and overwintering success are negatively affected by exposure to field doses [20–100 parts per billion (ppb)] of imidacloprid and that colony survival is correlated with the amount of imidacloprid (Dively et al., 2015; Woodcock et al., 2017; Wood et al., 2018).
For foragers, exposure to sublethal imidacloprid doses shortens their lifespan, permanently impairs their olfactory-associated leaning ability, and alters both their foraging behavior as well as foraging frequency (Decourtye et al., 2003, 2004; Faucon et al., 2005; Yang et al., 2008; Schneider et al., 2012; Zhang and Nieh, 2015). A higher rate of failure in homing results in a reduced number of foragers. To compensate for the shortage of foragers, nurse bees develop and reach an early behavioral maturation to generate young, precocious foragers (Beshers and Fewell, 2001; Leoncini et al., 2004; Sanchez-Bayo and Goka, 2014; Perry et al., 2015). Under these stressful conditions, the number of worker bees gradually decreases, resulting in colony collapse (Sanchez-Bayo and Goka, 2014; Perry et al., 2015; Ushitani et al., 2016).
Larvae and nurse bees are affected by consuming neonicotinoid-contaminated pollen or nectar. Honey bee larvae can tolerate higher doses of imidacloprid than nurse bees [LD50 = 4.17 μg, LC50 = 138.84 parts per million (ppm)] (Dai et al., 2017), but larval synapse development is affected by low doses. After exposure to 10 ppb imidacloprid, honey bee larvae pupation and eclosion rates are not affected. However, the developed adults show a reduced synaptic density in the brain region responsible for olfactory and visual function, and the olfaction-related learning behavior is permanently impaired (Yang et al., 2012; Peng and Yang, 2016). With continuous exposure, all these physiological and behavioral disorders induced by the sublethal dose of neonicotinoids can be sustained throughout the lifespan of workers and exhibit in different syndromes with the progress of bee development.
Due to difficulties in observation, only a few studies report the impact of imidacloprid on nurse bee behavior; they report that nurse bees show less activity and social interaction after being exposed to a low dose of imidacloprid (Medrzycki et al., 2003; Forfert and Moritz, 2017). From the molecular perspective, genes with functions related to metabolism, biosynthesis, and royal jelly synthesis are affected after exposure to sublethal doses of imidacloprid or other neonicotinoids (Aufauvre et al., 2014; Shi et al., 2017a,b; Wu et al., 2017; Christen et al., 2018; Wang et al., 2018). Most molecular studies suggest an immediate response after treatment; nevertheless, it is unclear whether the impact is long-lasting. In this report, we comprehensively evaluate the short- and long-term effects of sublethal doses of imidacloprid on honey bees of different ages following exposure during the larval stage. We used next-generation sequencing (NGS) to investigate the transcriptome of 9-day-old larvae and 0-, 7-, 14-, and 20-day-old adults. Different concentrations of imidacloprid solutions, including 1, 10, and 50 ppb, were applied to confirm the dosage effect and determine the lowest effective concentration of imidacloprid that could alter honey bee gene expression.
Honey bees were purchased from a local bee keeper at Hsinchu area and maintained at National Taiwan University and Miaoli District Agricultural Research and Extension Station (Council of Agriculture, Executive Yuan). Colonies contained at least eight honeycomb frames, a normal egg-laying queen, larvae, and pupae, as well as pollen and honey. Bee stability was continuously monitored. Pollen cake (45% sucrose, 30% pollens, 15% soybean powder, and 10% H2O) and 50% sucrose solution were provided during the non-flowering season. The area nearby the colonies was confirmed as not using neonicotinoid pesticides.
Imidacloprid powder (Sigma) was dissolved in 100% DMSO to a final concentration of 100 ppm and then diluted to 1, 10, and 50 ppm with DMSO. All solutions were aliquoted in 1.5-ml tubes and stored at −20°C until further use. Imidacloprid working solutions were freshly prepared every day before feeding. To prepare working solutions, 1, 10, and 50 ppm imidacloprid stock solutions were diluted with ddH2O to final concentrations of 1, 10, and 50 ppb (final concentration of DMSO is 0.1%).
The imidacloprid feeding assay and sample collection were performed during October to December 2016. For sample collection, two colonies were selected. For each colony, at least 300 larvae were used. The queen was restricted in one frame to lay eggs for 16 h to synchronize the age of experimental individuals. After the larvae hatched (defined as 1-day-old larvae), 1 μl of 1, 10, or 50 ppb of imidacloprid solution was added to each cell. This feeding procedure was performed daily for 4 days, from 1- to 4-day-old larvae, to mimic the number of days honey bee larvae consume bee bread. For each experimental group, a total of 4 μl of imidacloprid solution was applied into every cell, resulting in final amounts of imidacloprid consumed per larva of 0.004, 0.04, and 0.2 ng, respectively. Parallel controls were larvae fed with either ddH2O or 0.1% DMSO (see Supplementary Figure 1A for the experiment procedure). At 10 days after the cells were capped, pupae were pulled out and kept separately according to the treatments in 96-well plates at 37°C until eclosion. Bees from two different beehives were pooled as one group for the labeling and sampling procedure. Newly emerged adults were labeled on the thorax with different colors of acrylic paint based on the treatment and then randomly released into two beehives. Bees from different colonies were mixed and randomly assigned for the following experiment. The results represented in this report were the integrated effects from two hives of bee. Sampling was performed on 9-day-old larvae and 0- (newly emerged), 7-, 14-, and 20-day-old adults. The 0-, 7-, 14-, and 20-day-old adults conduct the general tasks of cell cleaning, brood rearing, pollen/nectar processing, and foraging, respectively (Seeley, 1982). For each age of worker bee, three biological replicates were collected, five individuals per replicate to final 15 individuals per age per treatment. Samples were stored in RNAlater (Invitrogen, Carlsbad, CA, United States) at −20°C for subsequent RNA extraction.
RNA from whole larvae and adults’ heads was extracted using an RNeasy mini kit (Qiagen, Hilden, Germany) following the user manual. RNA quality was checked in an agarose gel, with no significant degradation of 28S and 18S bands. RNA quantity was measured using the NanoDrop 2000 UV-Vis spectrophotometer (Thermo Fisher Scientific, Waltham, MA, United States). For each sample, at least 10 μg of total RNA was used for RNA-seq library construction. The library construction was performed according to Zhong et al. (2011). Briefly, polyA RNA enrichment was performed using Oligo d(T)25 Magnetic Beads (NEB, Ipswich, MA, United States), then eluted and fragmented simultaneously in 2 × RT buffer in the presence of random hexamers (final concentration 6 μM) and d(T)23VN (final concentration 5 μM). First-strand cDNA was synthesized using a ProtoScript II First Strand cDNA synthesis kit (NEB). Second-strand cDNA was synthesized using DNA polymerase I (NEB) and RNase H (NEB) with the presence of dUTP mix (dUTP, dATP, dCTP, dGTP; final concentration 1 mM). After end repair and dA tailing, the product was ligated with an Illumina Y-shaped adapter. The final product was treated with UDG (NEB) then PCR amplified with an index primer set. Library size and quality were checked using a Bioanalyzer 2100 (Agilent, Santa Clara, CA, United States). Four lanes of 150-bp paired-end sequencing were performed using a NovaSeq 6000 sequencer (Illumina, San Diego, CA, United States).
FastQC was performed to check the quality of the reads, and seqtk1 was used to remove the reads that failed to achieve the quality score threshold (30). RNA-Seq reads were then aligned to the adaptor, tRNA, and ribosomal RNA reference sequences using Bowtie2 (v.2.3.4.1; doi: 10.1038/nmeth.1923) with default parameters. The remaining filtered reads were then mapped to the honey bee genome (GCA_000002195.1, Amel_4.5 assembly2) using HISAT2 (version 2.1.0) (Kim et al., 2015) with parameters [–max-intronlen 50000 -p 6 –dta-cufflinks]. Analysis of the differentially expressed genes (DEGs) was performed based on the number of mapped raw reads and identified using Bioconductor DEseq2 (v.1.12.4; Bioconductor 3.7) with default parameter and Wald test (Love et al., 2014). The DEGs with FDR (false discovery rate) < 0.05 and fold change ≥ 2 (log2 fold change ≥ 1 or ≤−1) were considered differentially expressed. The functional analysis and GO clustering of remaining DEGs were performed using the online gene functional classification tool, DAVID3 (Huang et al., 2009a; Huang et al., 2009b). Three tables were generated for each subset of DEGs, including “functional annotation clustering,” “functional annotation chart,” and “functional annotation table.”
To monitor the effect of sublethal doses of imidacloprid on honey bee gene expression, we collected larvae and adults of worker bee at five different ages. The sampling time was determined based on the relative probability of task performance as described in Seeley (1982). For each age, a total of three biological replicates were collected, with five bees per replicate to a final 15 bees per age per treatment. Honey bees were collected from two beehives and then randomly assigned for each replicate; thus, the results represented in this report may eliminate the colony effects. Four lanes of 150 bp Illumina paired-end sequencing were generated; read yields per sample are shown in Supplementary Table 1A. Read counts for each gene, as well as FPKM levels at various development stages, are listed in Supplementary Tables 1B–F, 2A–E, respectively. Imidacloprid was supplied in a feeding solution using 0.1% DMSO in ddH2O as a solvent; control bees were fed with 0.1% DMSO solution without insecticide.
For each age of worker bee, we compared the gene expression levels between bees fed with imidacloprid and the 0.1% DMSO control to examine the DEGs to understand the impact of imidacloprid on honey bee gene expression at various ages. For each age of bee, three comparisons against the solvent control (bees fed with 1 μl of 0.1% DMSO) were performed: (i) 1 μl of 1 ppb imidacloprid solution; (ii) 1 μl of 10 ppb imidacloprid solution; and (iii) 1 μl of 50 ppb imidacloprid solution (Supplementary Figure 1B). DEGs with FDR values < 5% were considered differentially expressed and selected for further analysis. The DEGs were then filtered based on their log2 fold change. DEGs with a log2 fold change < 1 or >−1 were discarded, while genes with a log2 fold change ≥ 1 or ≤−1 were considered twofold differentially expressed. DEGs with a log2 fold change ≤−1 were defined as “down-regulated,” and DEGs with a log2 fold change ≥ 1 were defined as “up-regulated” (Table 1 and Figure 1A). The list of DEGs is shown in Supplementary Tables 3A,B. The detailed analysis procedure is described in section “Materials and Methods.”
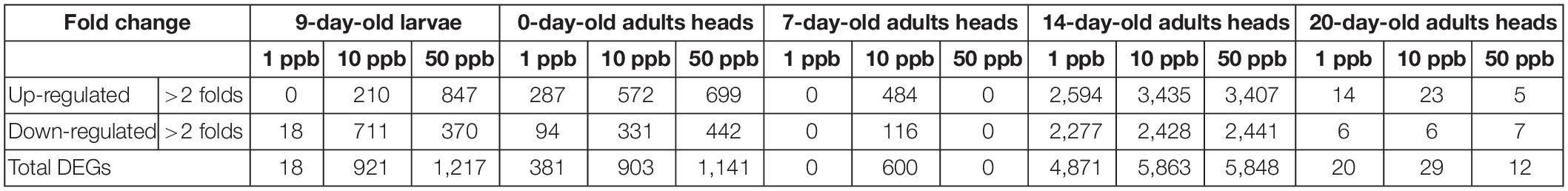
Table 1. DEG numbers in different stages of honey bee fed with different concentrations of imidacloprid.
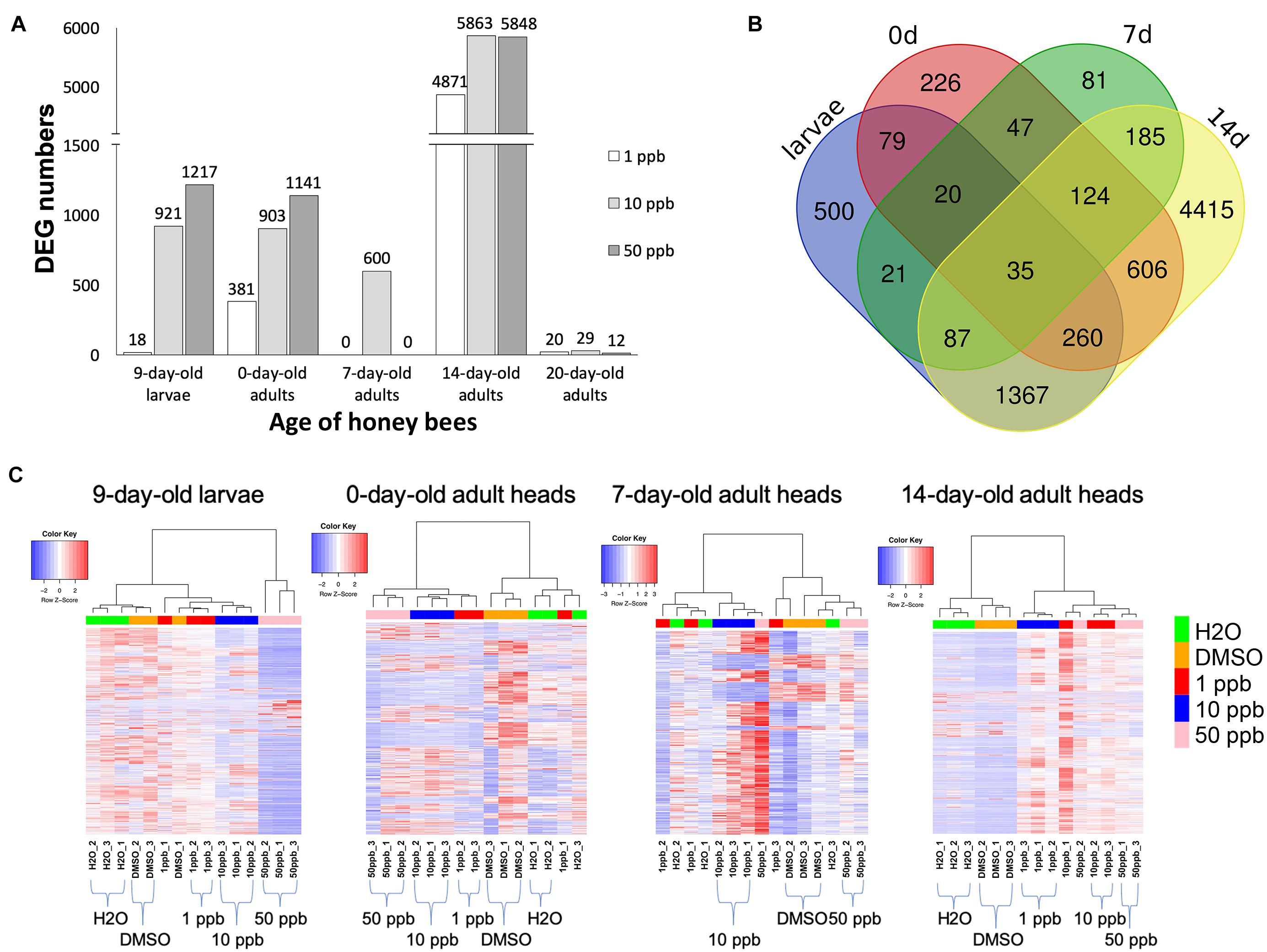
Figure 1. Numbers of differentially expressed genes at various developmental stages of worker bees. (A) Numbers of differentially expressed genes (DEGs) in 9-day-old larvae, as well as in 0-, 7-, 14-, and 20-day-old adults of honey bees exposed to 1-, 10-, or 50-ppb imidacloprid treatment for 4 days during the larval stage. Original numbers of DEGs are shown on top of each column. X-axis: the developmental stages of honey bees; Y-axis: the number of differentially expressed genes. (B) Venn diagram of shared DEGs of 9-day-old larvae, 0-, 7-, and 14-day-old adults among all concentrations of imidacloprid. The numbers of shared DEGs are shown in the overlapped region. Larvae: 9-day-old larvae; 0d: 0-day-old adults; 7d: 7-day-old adults; 14d: 14-day-old adults. (C) Cluster analysis of DEG expression patterns of 9-day-old honey bee larvae, as well as 0-, 7-, and 14-day-old adults. Read counts were used for cluster analysis. Complete normalization was applied, and the clustering was performed based on Pearson correlation coefficients. The expression trend of each replicate is shown. Controls (H2O and DMSO) and different concentrations of imidacloprid treatments (1, 10, and 50 ppb) are labeled in the bottom panel and marked with different colors in the panel of the dendrogram. Red: expression levels higher than average; blue: expression levels lower than average; white: expression levels close to average.
A cluster analysis was performed using read counts to visualize patterns of DEGs at various developmental stages (Figure 1C). Complete normalization was applied, and the clustering was performed based on Pearson correlation coefficients. For 9-day-old larvae, honey bees fed with 50 ppb imidacloprid showed distinct expression patterns and were assigned as the outgroup, while patterns of DEGs, between 1 and 10 ppb imidacloprid, and between H2O and DMSO, were similar, with only slight differences between one replicate of the DMSO solvent control and one replicate of the 1-ppb treatment compared to the other replicates. For 0-day-old adults, a clearer grouping between controls (DMSO and H2O) and imidacloprid treatments (1, 10, and 50 ppb) was observed, although one replicate of the 1-ppb treatment had expression patterns closer to the H2O control rather than the other two 1-ppb replicates. For 7-day-old adults, the clustering was chaotic and correlations between controls and treatments could barely be identified. For 14-day-old adults, with vast numbers of DEGs identified at this stage, significantly distinct groups between controls and imidacloprid treatments were observed without ambiguous distributions. Cluster analysis for 20-day-old adults was not performed as less than 30 DEGs were identified.
After being exposed to a sublethal dose of imidacloprid, the gene expression of honey bee larvae and adults was significantly affected, and the effects were long-lasting (Table 1). The number of DEGs increased with the dosage of imidacloprid in 9-day-old larvae (18, 921, and 1,217 DEGs for 1, 10, and 50 ppb, respectively) and 0-day-old adults (381, 903, and 1,141 DEGs for 1, 10, and 50 ppb, respectively). For 7-day-old adults, 600 DEGs were identified in bees fed with 10 ppb imidacloprid, while no DEGs were identified in the other treatments (1 and 50 ppb imidacloprid). Surprisingly, more than 4,800 DEGs were observed in 14-day-old adults (4,871, 5,863, and 5,848 DEGs for 1, 10, and 50 ppb, respectively), and the number of DEGs seems to have increased slightly with the dosage of imidacloprid. For 20-day-old adults, fewer than 100 DEGs were identified for all treatments (Table 1) (20, 29, and 12 DEGs for 1, 10, and 50 ppb, respectively). For 9-day-old larvae and 0-day-old adults, there are fewer upregulated than downregulated genes. In contrast, for 7-, 14-, and 20-day-old adults, a greater number of DEGs were identified in the upregulated groups.
The numbers of unique and shared DEGs among different concentrations of imidacloprid treatment at every age of bee were examined to evaluate the effect of the dosage of imidacloprid (Supplementary Figure 2). DEGs were first divided based on the regulatory trend (i.e., upregulated or downregulated), and then the shared DEGs were examined. The numbers of shared DEGs among the different concentrations of imidacloprid treatments are shown in Supplementary Figures 2A–D. In 9-day-old larvae and 0- and 20-day-old adults, the percentage of shared DEGs was less than 30%; more DEGs were found to be uniquely identified in the different concentrations of imidacloprid treatments. In 14-day-old adults, the percentage of shared DEGs was higher than 50%. The shared DEGs in 9-day-old larvae and 0- to 14-day-old adults were then examined among all the imidacloprid treatments, disregarding the regulatory trend. Thirty-five genes were identified as shared among different aged bees and different concentrations of imidacloprid treatments, including gamma-aminobutyric acid receptor subunit beta (GB40975), DNA ligase 1-like (GB42264), cuticular protein (GB46297), RNA polymerase II elongation factor EII (GB48521), insulin-like growth factor 2 mRNA-binding protein 1 (GB52056), nicotinamide riboside kinase (Nrk1) (GB53410), trehalose transporter 1 (GB55302), and several other unknown-function-genes (Figure 1B and Table 2).
To further understand the biological meaning of the DEGs, Gene Ontology (GO) analysis was performed to predict their functions. DEGs of every treatment were first divided into two groups, i.e., upregulated and downregulated genes, based on their expression trend. An overview of the results of the “functional annotation chart” among all the treatments is shown in Supplementary Tables 3C,D. A GO term that could be identified in at least three ages of bee was considered as a constantly affected function. In the upregulated genes, GO terms identified in at least three ages of bees were as follows: Coiled coil, ion transport, nucleotide binding, Nucleotide-binding (alpha-beta plait), Ribosome, Ribonucleoprotein, Ribosomal protein, structural constituent of ribosome, Transport, and translation. Among the downregulated genes, most of the GO terms can be only identified in two ages of worker bee (Supplementary Tables 3C,D).
In general, the numbers of functional terms were correlated with the numbers of DEGs. For 9-day-old larvae, the number of downregulated clusters (79 and 35 for 10 and 50 ppb, respectively) was higher than that of upregulated clusters (7 and 14 for 10 and 50 ppb, respectively). The opposite pattern (i.e., a higher number of upregulated clusters) was observed in 0-, 7-, 14-, and 20-day-old adults. The numbers of functional terms of upregulated DEGs for 0-, 7-, 14-, and 20-day-old adults were as follows: 0-day-old adults, 16, 34, and 45 for 1, 10, and 50; 7-day-old adults, 23 for 10 ppb; and 14-day-old adults, 52, 91, and 81 for 1, 10, and 50 (Supplementary Table 3C). The numbers of GO cluster numbers of downregulated DEGs were as follows: 0-day-old adults, 8, 8, and 26 for 1, 10, and 50; 7-day-old adults, 10 for 10 ppb; and 14-day-old adults, 16, 27, and 37 for 1, 10, and 50 (Supplementary Table 3D). For upregulated terms, most of them were identified in 9-day-old larvae or 0-, 7-, and 20-day-old adults can be found in 14-day-old adults, while few functional clusters were only identified in 9-day-old larvae or 0-day-old adults. Terms such as ATPase/dynein-related/AAA domain, Dynein heavy chain, Frag1/DRAM/Sfk1, integral component of membrane, and transferase activity/transferring hexosyl groups were uniquely found in 9-day-old larvae, while 3′,5′-cyclic-nucleotide phosphodiesterase activity, 3′,5′-cyclic nucleotide phosphodiesterase/catalytic domain, Biotin/lipoyl attachment, Cyclic nucleotide-binding domain, Developmental protein, DnaJ domain, Groucho/transducin-like enhancer, Neuroactive ligand-receptor interaction, Nuclear hormone receptor/ligand-binding/core, Potassium channel, voltage-dependent, EAG/ELK/ERG, Ras-association, Serine-threonine/tyrosine-protein kinase catalytic domain, Steroid hormone receptor, Synaptotagmin, voltage-gated potassium channel activity, Zinc finger, and nuclear hormone receptor-type were found only in 0-day-old adults (Supplementary Table 3C). Terms of downregulated genes were found to be more diverse among different ages of bee, suggesting that the negatively affected genes may vary or change among different ages or tasks of worker bees. Higher numbers of DEGs were identified in 14-day-old adults than in 9-day-old larvae, yet fewer GO terms were identified in 14-day-old adults than in 9-day-old larvae. Nine-day-old honey bee larvae are transforming from larval to pupal, to finally adult stage, which may be involved in massive transcriptome reprogramming and diverse functional pathways for the differentiation and development of adults’ organs. High numbers of GO terms may suggest that various functional pathways could be affect after exposure to sublethal dose of imidacloprid.
The complete GO analysis results generated using DAVID are shown in Supplementary Table 4 (9-day-old larvae, 1 ppb imidacloprid treatment), Supplementary Tables 5A–F (9-day-old larvae, 10 ppb imidacloprid treatment), Supplementary Tables 6A–E (9-day-old larvae, 50 ppb imidacloprid treatment), Supplementary Tables 7A–F (0-day-old adults, 1 ppb imidacloprid treatment), Supplementary Tables 8A–F (0-day-old adults, 10 ppb imidacloprid treatment), Supplementary Tables 9A–F (0-day-old adults, 50 ppb imidacloprid treatment), Supplementary Tables 10A–F (7-day-old adults, 10 ppb imidacloprid treatment), Supplementary Tables 11A–F (14-day-old adults, 1 ppb imidacloprid treatment), Supplementary Tables 12A–F (14-day-old adults, 10 ppb imidacloprid treatment), Supplementary Tables 13A–F (14-day-old adults, 50 ppb imidacloprid treatment), Supplementary Tables 14A–D (20-day-old adults, 1 ppb imidacloprid treatment), Supplementary Tables 15A–C (20-day-old adults, 10 ppb imidacloprid treatment), and Supplementary Tables 16A,B (20-day-old adults, 50 ppb imidacloprid treatment).
To examine whether the increasing amount of DEGs at 14-day-old adults is correlated with the cue for the switch of development of the worker bees, the expression trends of the key transcription factors that regulate the behavioral maturation in honey bees (Bloch et al., 2009) were examined. Among the 12 foraging regulators, eight including RXRA/RXRB (1, 10, and 50 ppb), CREB1 (10 and 50 ppb), DFD (1 and 10 ppb), DRI (1 and 50 ppb), EGR1/EGR2 (1, 10, and 50 ppb), PAX6 (10 ppb), HES1 (1, 10, and 50 ppb), and BHE40/BHE41 (10 and 50 ppb) were found upregulated in 14-day-old adults after imidacloprid treatment at the larval stage (Table 3; Supplementary Table 17A for read count of TF-related genes).
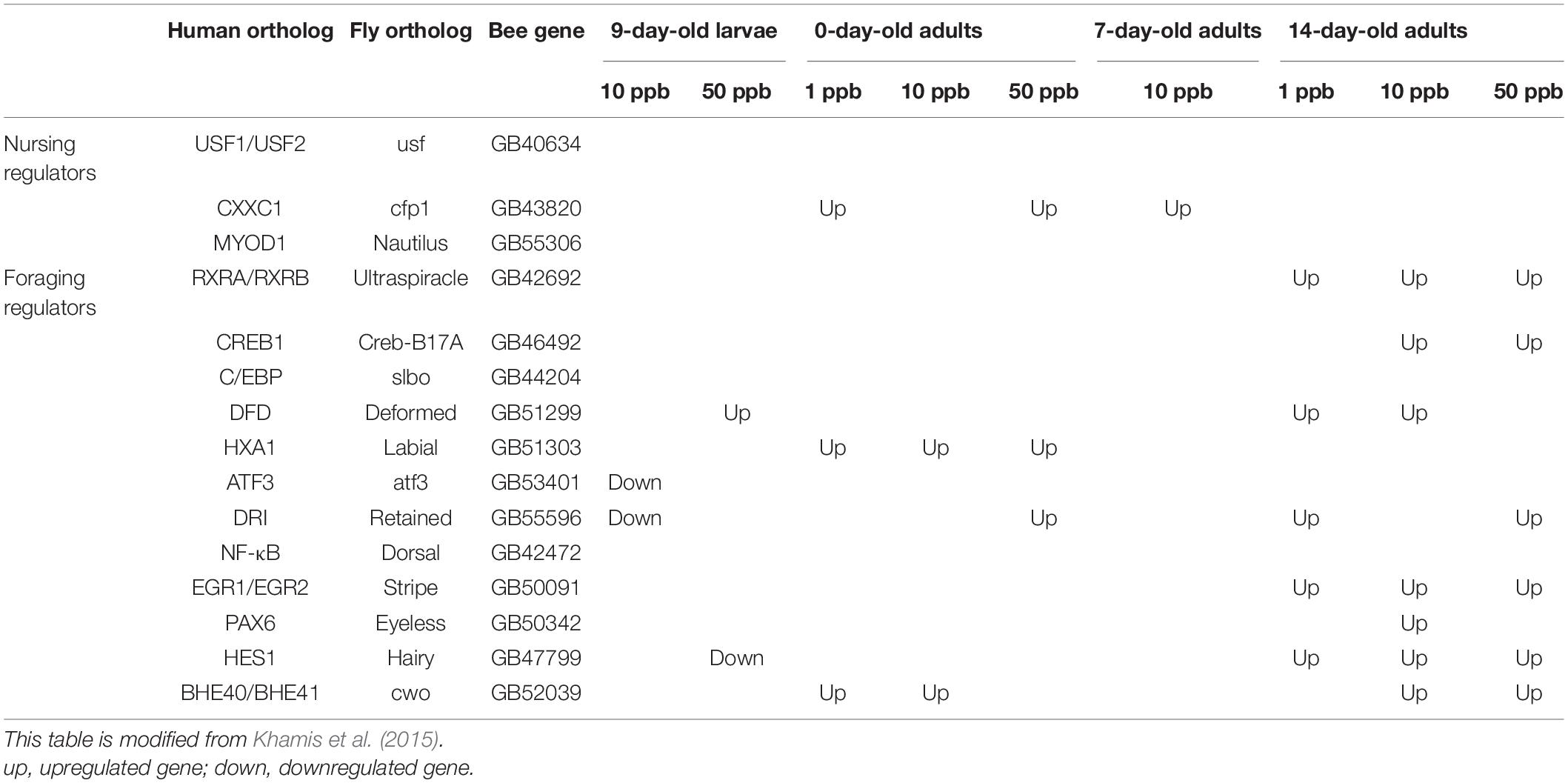
Table 3. Differentially expressed genes related to transcriptional regulators of honey bee behavioral state at different developmental ages after imidacloprid treatment.
The upregulation of foraging regulators in 14-day-old imidacloprid-treated adults suggests that the switch from nurse bee to forager was accelerated. To confirm this hypothesis, the transcriptome of 14-day-old bees treated with imidacloprid during larval stage was compared to that of 0-, 7-, and 20-day-old control bees (see Supplementary Figure 1C for a schematic diagram of comparison). A comparison between different ages of control bees was also performed (see Supplementary Figure 1D for a schematic diagram of comparison, Supplementary Figure 3 for results). Compared to 0- and 7-day-old controls, the total numbers of DEGs were 4,073 (14-day-old 1 ppb vs. 0-day-old control), 5,491 (14-day-old 10 ppb vs. 0-day-old control), 5,140 (14-day-old 50 ppb vs. 0-day-old control), 8,298 (14-day-old 1 ppb vs. 7-day-old control), 7,696 (14-day-old 10 ppb vs. 7-day-old control), and 7,436 (14-day-old 50 ppb vs. 7-day-old control) (Figures 2A,B). In the comparison of 14-day-old treatments to the 20-day-old control, the numbers of DEGs were negatively correlated with the concentration of imidacloprid. The numbers of DEGs identified from 14-day-old 1 ppb vs. 20-day-old control, 14-day-old 10 ppb vs. 20-day-old control, and 14-day-old 50 ppb vs. 20-day-old control were 4,743, 1,686, and 447, respectively (Figure 2C). The decreasing number of DEGs suggests the increase in transcriptome similarity, and the effect was dose-dependent. A cluster analysis of solvent controls and imidacloprid-treated workers in 9-day-old larvae and 0-, 7-, 14-, and 20-day-old adults was also performed to visualize expression patterns of whole transcriptome among different ages of bee. Fourteen-day-old workers with imidacloprid treatment during the larval stage were correlated with 20-day-old solvent control and treatments, suggesting the transcriptome similarity among them (Supplementary Figure 4).
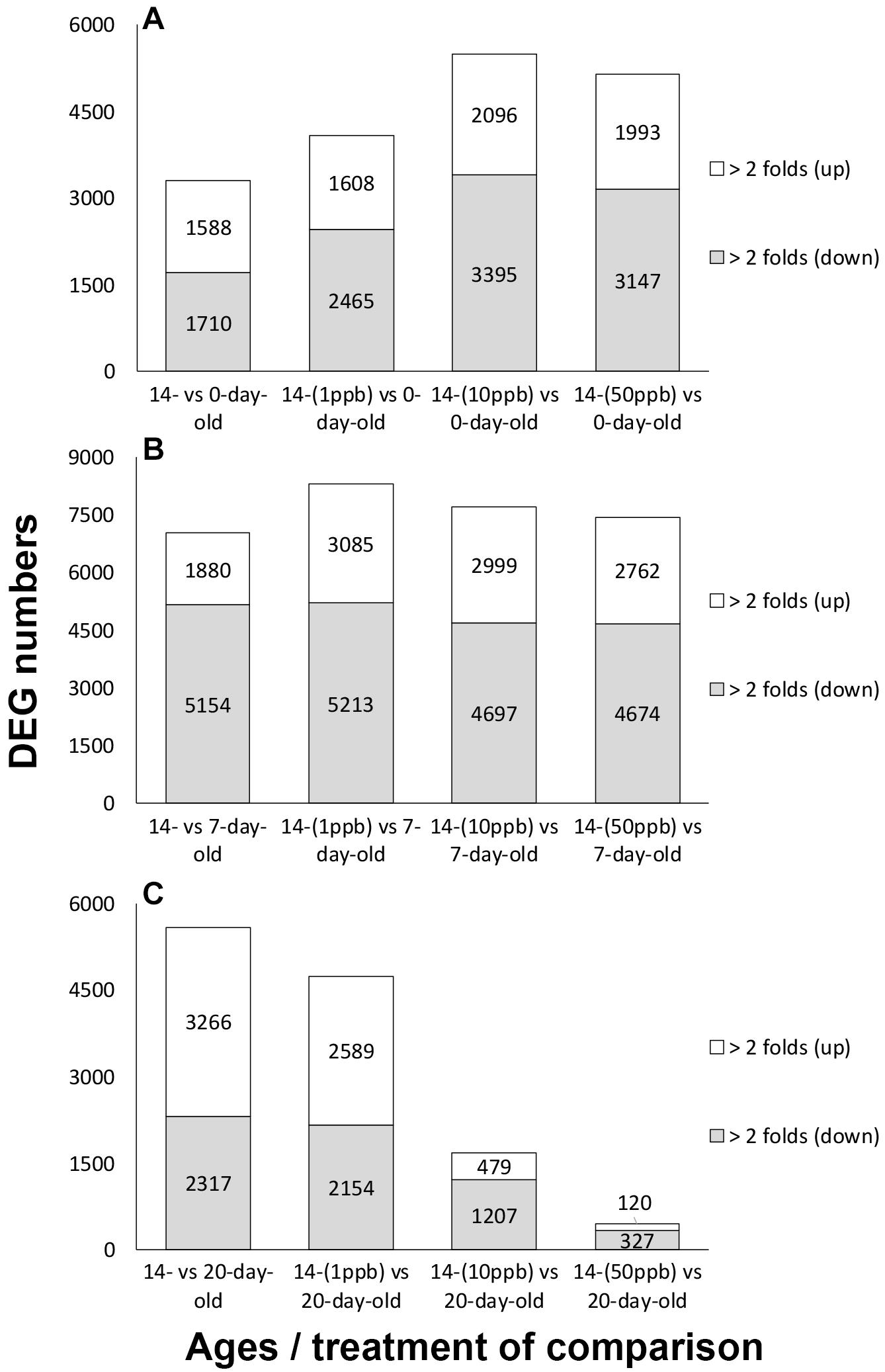
Figure 2. Numbers of differentially expressed genes (DEGs) identified from comparing 14-day-old imidacloprid treated bees with 0-, 7-, and 20-day-old control bees. (A) Fourteen-day-old imidacloprid treatments compared to 0-day-old adult control. (B) Fourteen-day-old imidacloprid treatments compared to 7-day-old adult control. (C) Fourteen-day-old imidacloprid treatments compared to 20-day-old adult control. DEGs were subdivided based on regulatory trends. Twofold (up): DEGs with a log2 fold change ≥ 1; twofold (down): DEGs with a log2 fold change ≤ –1. Numbers of DEGs are shown in each column.
The number of DEGs decreased with the progress of worker bee development from 9-day-old larva to 7-day-old adult, peaked in 14-day-old adults, and then dramatically decreased to less than 30 in 20-day-old adults (Table 1 and Figure 1A). The different concentrations of imidacloprid seem to have induced different effects, as the DEGs were diverse (Supplementary Figures 2A–D). The high similarity of expression patterns between biological replicates in every treatment excludes the possibility of individual variance or artifacts, and distinct DEG patterns were observed between controls and imidacloprid treatments (Figure 1C and Supplementary Figure 5 for sample correlation). The fluctuating DEG numbers suggest that the global gene expression in the honey bee changes with the progress of their development as well as task performance. As nurse bees and foragers contain distinct transcriptomes (Khamis et al., 2015), it is highly possible that the gene expression profile would change with the different tasks performed by the nurse bees. The upregulation of the nursing regulator at 0-day-old adults suggests that those DEGs could be correlated with the development of the nursing behavior (Table 3), which should be performed while the hive bees become 3-day-old or older. A similar trend was also observed for the foraging regulators for 14-day-old adults (Table 3). The DEGs identified at 9-day-old larvae and 0- and 7-day-old adults may reflect not only the damages but also the early maturation induced by sublethal imidacloprid treatment. The decrease in DEGs from 9-day-old larvae to 7-day-old adults may suggest that the task performance of nurse bees is attributed by a small subset of genes. Further experiments are needed to validate the behavioral change of newly emerged workers and the task-associated genes of nurse bees.
We compared our DEG results with previous studies (Forfert and Moritz, 2017; Shi et al., 2017a) and found that 329 DEGs were shared (Supplementary Table 18A). These genes are functionally related to lipid metabolism, pheromone/general odorant-binding protein, cytochrome P450, transmembrane transport, and ATP binding (Supplementary Tables 18B,C,D). Physiological experiments have confirmed that imidacloprid and clothianidin can impact energetic/nutrient homeostasis of honey bee (Cook, 2019). These genes could be toxification index genes expressed to inspect the existence of neonicotinoids in the honey bee. In bumble bee (Bombus terrestris) workers, exposure to field-realistic imidacloprid concentrations (10 ppb) can induce a total of 405 DEGs with functions related to energy reserve metabolism, synaptic transmission, apoptotic processes, xenobiotic transport, and complement activation, and including P450 genes (Manjon et al., 2018); exposure to a sublethal dose of clothianidin (6.47 ppb) induces the differential expression of 55 genes, including three putative cytochrome P450 genes (Bebane et al., 2019; Colgan et al., 2019). In Bradysia odoriphaga, exposure to the LC50 dose (4.08 mg/l) of imidacloprid for 24 h induces the differential expression of 674 unigenes with functions related to cytochrome P450, cell adhesion, axon guidance, and carbohydrate metabolic process (Chen et al., 2019). The effects of neonicotinoids on insect gene expression vary among different species of insects, but genes related to metabolism, detoxification (especially cytochrome P450), and neuron development/synaptic transmission seem to be commonly found. It is confirmed from molecular evidence that even at low concentrations, neonicotinoids can have an impact on the neuron development and detoxification of insects.
Honey bee division of labor correlates with the age of the adults, exposure to pheromones, ovary size, biogenic amines, and the proportion of members of the colony currently engaging in each task (Seeley, 1982; Huang and Robinson, 1992; Johnson, 2008, 2010; Bloch et al., 2009; Sagili et al., 2011; Wang et al., 2012; Reim and Scheiner, 2014). From a physiological perspective, a significant increase in the titers of JH coordinates the transformation from nurse bee to forager (Jaycox, 1967; Jaycox et al., 1974; Robinson, 1987; Fahrbach and Robinson, 1996; Bloch et al., 2009). From a molecular perspective, the switch from nurse bees to foragers is modulated by the differential expression of transcription factors and alternative transcription start sites, resulting in transcriptome changes followed by organized behavioral states (Whitfield et al., 2003; Khamis et al., 2015). In our study, it may not be easy to infer the trend of the JH titer based on the function of the identified DEGs, as the GO term related to JH synthesis/metabolite was not identified. Despite the expression of JH-related genes, the upregulation of the foraging regulators and the low transcriptomic difference between imidacloprid-treated 14-day-old adults and 20-day-old control adults suggest that the precocious worker bee may leave the beehive. Colin et al. (2019) examined the effect of a sublethal dose of imidacloprid by providing the beehive with a sucrose solution containing 5 ppb of the insecticide for 3 weeks and then monitored the foraging behavior of newly engaged foragers; foragers exposed to the insecticide started the orientation flight at an age 1.38 days younger than the control (Colin et al., 2019). Although the treatment methodology was different from the present study, it supports the relationship between the insecticide and precocious behavior. The occurrence of precocious foraging is believed to be triggered by a high mortality of foragers (Beshers and Fewell, 2001; Leoncini et al., 2004; Perry et al., 2015; Ushitani et al., 2016). The young foragers are found to be less efficient and have poor spatial memory and shorter lifespan (Chang et al., 2015; Perry et al., 2015; Ushitani et al., 2016) and cannot sustain the foraging task for long. This accelerated behavioral switch in our study is unlikely to be induced by the low number of foragers, as the experimental individuals were released into a healthy colony with task-performing foragers and food storage. We cannot exclude, however, that other unknown factors, such as pathogens or parasites, could have affected the task transition. Further validation and experiments are required to confirm if this phenomenon is a general effect, as two local colonies were used in this report.
Imidacloprid acts as an agonist of the insect neuronal nicotinic acetylcholine receptors and would consequently affect the expression of nicotinic acetylcholine receptor genes (nAChRs) (Déglise et al., 2002; Eiri and Nieh, 2016). nAChRs mediate insect synaptic neurotransmission as well as many cognitive processes, including vision and olfactory learning (Breer and Sattelle, 1987). In the GO cluster analysis of DEGs, Nicotinic acetylcholine-gated receptor, transmembrane domain, was identified (Supplementary Tables 12A, 13A), although the numbers of nAChRs-related DEGs were related low [five out of 3,435 (10 ppb) and 3,407 (50 ppb)] and resulted in a high p-value. We examined the expression of nAChRs-related genes and found five of them that are differentially expressed in 14-day-old treated adults. These genes include nAChRa6 (GB43416; 1, 10, and 50 ppb), nAChRa8 (GB40923; 10 and 50 ppb), nAChRa9 (GB53427; 1 ppb), nAChRb1 (GB53055; 50 ppb), and nAChRa9 (GB53427; 10 and 50 ppb), which were upregulated, and nAChRb2 (GB53428, 10 and 50 ppb) which was downregulated (Table 4; Supplementary Table 17B for read counts). Among 11 nicotinic acetylcholine receptors subunit genes (Jones et al., 2006), four genes, including nAChRa2, nAChRa7, nAChRa8, and nAChRb1, are expressed in the Kenyon cells, the ordered neuropils in mushroom bodies that are crucial for olfactory learning and memory (Hammer and Menzel, 1995). Most of the differentially expressed nAChRs were found in 14-day-old treated adults regardless of the dose of imidacloprid. This might be correlated with the development of foraging behavior in adults over 10 days of age (Khamis et al., 2015). Food reward-induced olfactory stimuli are required to perform food collection and foraging behavior (Menzel, 1999; Dupuis et al., 2011; Giurfa and Sandoz, 2012). It is also possible for this differential expression to be associated with the development of precocious foraging behavior in the 14-day-old treated adults, as previously described due to the transcriptome similarity between these adults and the 20-day-old control. In the 35 shared DEGs among the four ages, we also found nicotinamide riboside kinase (Nrk1) (GB53410). This gene is induced in eukaryotes by the presence of nerve damage to protect damaged neurons from degradation (Belenky et al., 2007). Differential expression of Nrk1 among all ages of treated bees suggests that the neurons were damaged after the sublethal imidacloprid treatment. These results support previous findings that the exposure to sublethal doses of imidacloprid during the larval stage can induce neuron damage and consequently impair honey bee adults’ olfaction and related learning behavior (Medrzycki et al., 2003; Forfert and Moritz, 2017).
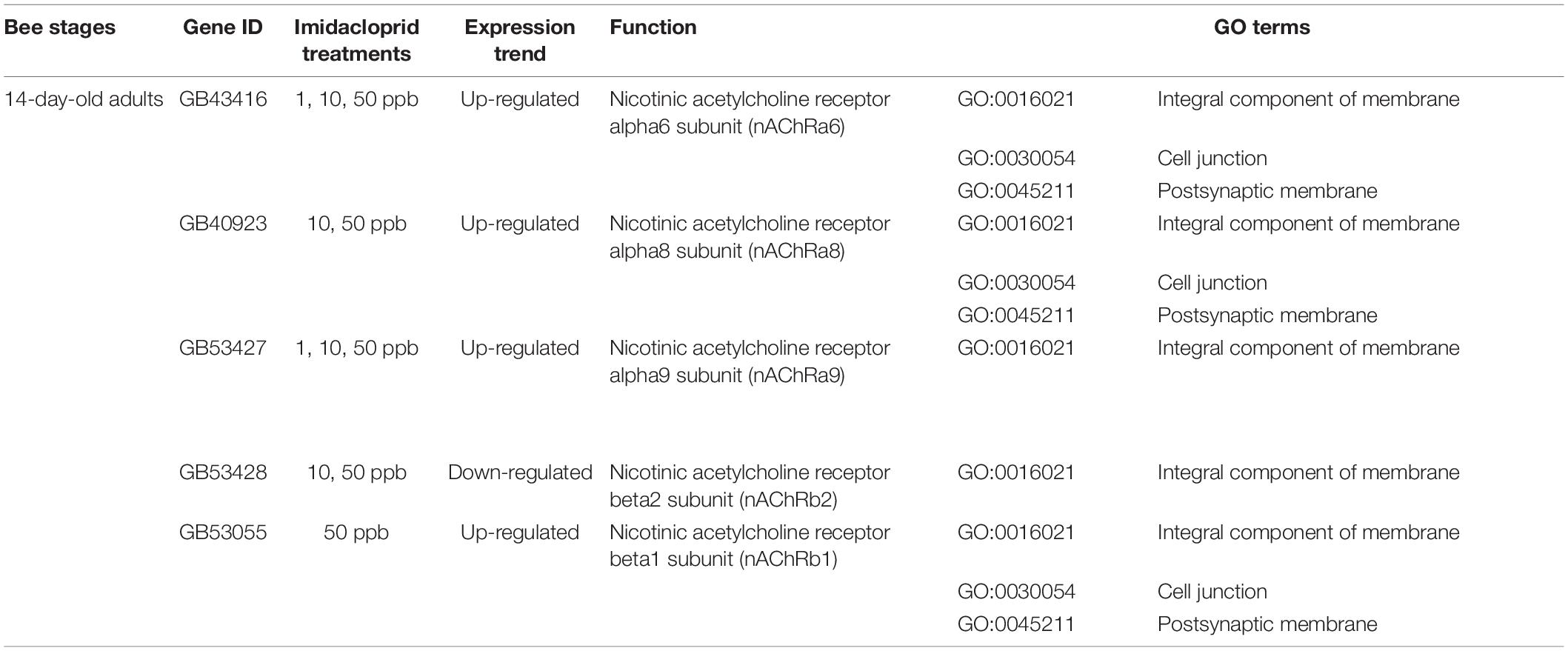
Table 4. Differentially expressed nAChRs family subunit genes in 14-day-old adults with 1-, 10-, and 50-ppb imidacloprid treatments.
Honey bee RNA-seq raw reads are available at https://www.ncbi.nlm.nih.gov/sra/PRJNA521949.
E-CY and Y-RC designed the experiments. Y-RC, CT, P-SH, and T-HW performed the imidacloprid treatment and honey bee sampling. Y-RC prepared the RNA-Seq library. DT performed the data analysis. Y-RC, DT, E-CY, and SZ wrote the manuscript. All authors contributed to the article and approved the submitted version.
This work was supported by grants from the Taiwan Ministry of Science and Technology, 104-2313-B-002-019-MY3 and 107-2313-B-002-038-MY3; 107-2811-B-002-596, Postdoc funding to Y-RC; and Hong Kong AoE (AoE/M-403/16) for SZ.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2021.665927/full#supplementary-material
Aufauvre, J., Misme-Aucouturier, B., Viguès, B., Texier, C., Delbac, F., and Blot, N. (2014). Transcriptome analyses of the honeybee response to Nosema ceranae and insecticides. PLoS One 9:e91686. doi: 10.1371/journal.pone.0091686
Bebane, P. S. A., Hunt, J. H., Pegoraro, M., Jones, A. R. C., Marchall, H., Rosato, E., et al. (2019). The effects of the neonicotinoid imidacloprid on gene expression and DNA methylation in the biff-tailed bumblebee Bumbus terrestris. bioRxiv [Preprint] 590091. doi: 10.1098/rspb.2019.0718
Belenky, P., Racette, F. G., Bogan, K. L., McClure, J. M., Smith, J. S., and Brenner, C. (2007). Nicotinamide riboside promotes sir2 silencing and extends lifespan via Nrk and Urh1/Pnp1/Meu1 pathways to NAD(+). Cell 129, 473–484. doi: 10.1016/j.cell.2007.03.024
Beshers, S. N., and Fewell, J. H. (2001). Models of division of labor in social insects. Ann. Rev. Entomol. 46, 413–440. doi: 10.1146/annurev.ento.46.1.413
Bloch, G., Shpigler, H., Wheeler, D. E., and Robinson, G. E. (2009). “Chapter 30 Endocrine influences on the organization of insect societies,” in Hormones, Brain and Behavior, 2nd Edn, eds D. W. Pfaff, A. P. Arnold, A. M. Etgen, S. E. Fahrbach, and R. T. Rubin, (San Diego, CA: Academic Press), 1027–1068.
Böhme, F., Bischoff, G., Zebitz, C. P. W., Rosenkranz, P., and Wallner, K. (2018). Pesticide residue survey of pollen loads collected honeybees (Apis mellifera) in daily internals at three agricultural sites in South Germany. PLoS One 13:e0199995. doi: 10.1371/journal.pone.0199995
Breer, H., and Sattelle, D. B. (1987). Molecular properties and functions of insect acetylcholine receptors. J. Insect Physiol. 33, 771–790. doi: 10.1016/0022-1910(87)90025-4
Calderone, N. W. (2012). Insect pollinated crops, insect pollinators and US agriculture: trend analysis of aggregate data for the period 1992 – 2009. PLoS One 7:e37235. doi: 10.1371/journal.pone.0037235
Chang, L. H., Barron, A. B., and Cheng, K. (2015). Effects of the juvenile hormone analogue methoprene on rate of behavioral development, foraging performance and navigation in honey bee (Apis mellifera). J. Exp. Biol. 218, 1715–1724. doi: 10.1242/jeb.119198
Chauzat, M. P., Faucon, J. P., Martel, A. C., Lachaize, J., Cougoule, N., and Aubert, M. (2006). A survey of pesticide residues in pollen loads collected by honey bees in France. J. Econ. Entomol. 99, 253–262. doi: 10.1603/0022-0493-99.2.253
Chen, C., Shan, T., Liu, Y., Shi, X., and Gao, X. (2019). Identification of a novel cytochrome P450 CYP3356A1 linker with insecticide detoxification in Bradysia odoriphaga. Pest Manag. Sci. 75, 1006–1013. doi: 10.1002/ps.5208
Christen, V., Shirrmann, M., Frey, J. E., and Fent, K. (2018). Global transcriptomic effects of environmentally relevant concentrations of the neonicotinoids clothianidin, imidacloprid, and thiamethoxam in the brain of honey bees (Apis mellifera). Environ. Sci. Technol. 52, 7534–7544. doi: 10.1021/acs.est.8b01801
Codling, G., Naggar, Y. A., Giesy, J. P., and Robertson, A. J. (2016). Concentrations of neonicotinoid insecticides in honey, pollen and honey bees (Apis mellifera) in central Saskatchewan, Canada. Chemosphere 144, 2321–2328. doi: 10.1016/j.chemosphere.2015.10.135
Colgan, T. J., Fletcher, I. K., Arce, A. N., Gill, R. J., Rodrigues, A. R., Stolle, E., et al. (2019). Caste- and pesticide-specific effects of neonicotinoid pesticide exposure on gene expression in bumblebees. Mol. Ecol. 28, 1964–1974. doi: 10.1111/mec.15047
Colin, T., Meikle, W. G., Wu, X., and Barron, A. B. (2019). Traces of a neonicotinoid induce precocious foraging and reduce foraging performance in honey bees. Environ. Sci. Technol. 53, 8252–8261. doi: 10.1021/acs.est.9b02452
Cook, S. C. (2019). Compound and dose-dependent effects of two neonicotinoids pesticides on honey bee (Apis mellifera) metabolic physiology. Insects 10:18. doi: 10.3390/insects10010018
Dai, P., Jack, C. J., Mortensen, A. N., and Ellis, J. D. (2017). Acute toxicity of five pesticides to Apis mellifera larvae reared in vitro. Pest Manag. Sci. 73, 2282–2286. doi: 10.1002/ps.4608
Decourtye, A., Devillers, J., Cluzeau, S., Charreton, M., and Pham-DeleÌgue, M. H. (2004). Effects of imidacloprid and deltamethrin on associative learning in honeybees under semi-field and laboratory conditions. Ecotoxicol. Environ. Saf. 57, 410–419. doi: 10.1016/j.ecoenv.2003.08.001
Decourtye, A., Lacassie, E., and Pham-Delègue, M. H. (2003). Learning performances of honeybees (Apis mellifera) are differentially affected by imidacloprid according to the season. Pest Manag. Sci. 59, 269–278. doi: 10.1002/ps.631
Déglise, P., Grünewald, B., and Gauthier, M. (2002). The insecticide imidacloprid is a partial agonist of the nicotinic receptor of honeybee Kenyon cells. Neurosci. Lett. 321, 13–16. doi: 10.1016/S0304-3940(01)02400-4
Dively, G. P., Embrey, M. S., Kamel, A., Hawthorne, D. J., and Pettis, J. S. (2015). Assessment of chronic sublethal effects of imidacloprid on honey bee colony health. PLoS One 10:e0118748. doi: 10.1371/journal.pone.0118748
Dupuis, J. P., Gauthier, M., and Raymond-Delpech, V. (2011). Expression patterns of nicotinic subunits alpha 2, alpha 7, alpha 8, and beta 1 affect the kinetics and pharmacology of ACh-induced currents in adult bee olfactory neuropiles. J. Neurophysiol. 106, 1604–1613. doi: 10.1152/jn.00126.2011
Eiri, D. M., and Nieh, J. C. (2016). A nicotinic acetylcholine receptor agonist affects honey bee sucrose responsiveness and decrease waggle dancing. J. Exp. Biol. 219(Pt 13):2081. doi: 10.1242/jeb.143727
Fahrbach, S. E., and Robinson, G. E. (1996). Juvenile hormone, behavior maturation, and brain structure in the honey bee. Dev. Neurosci. 18, 102–114. doi: 10.1159/000111474
Faucon, J. P., Aurières, C., Drajnudel, P., Mathieu, L., Ribière, M., Martel, A. C., et al. (2005). Experimental study on the toxicity of imidacloprid given in syrup to honey bee (Apis mellifera) colonies. Pest Manag. Sci. 6, 111–125. doi: 10.1002/ps.957
Forfert, N., and Moritz, R. F. A. (2017). Thiacloprid alters social interactions among honey bee workers (Apis mellifera). J. Apic. Res. 56, 467–474. doi: 10.1080/00218839.2017.1332542
Garibaldi, L. A., Carvalheiro, L. G., Vaissière, B. E., Gemmill-Herren, B., Hipólito, J., Freitas, B. M., et al. (2016). Mutually beneficial pollinator diversity and crop yield outcomes in small and large farms. Science 351, 388–391. doi: 10.1126/science.aac7287
Garibaldi, L. A., Sreffan-Dewenter, I., Winfree, R., Aizen, M. A., Bommarco, R., Cunningham, S. A., et al. (2013). Wild pollinators enhance fruit set of crops regardless of honey bee abundance. Science 339, 1608–1611. doi: 10.1126/science.1230200
Giurfa, M., and Sandoz, J. C. (2012). Invertebrate learning and memory: fifty years of olfactory conditioning of the proboscis extension response in honeybees. Learn. Mem. 19, 54–66. doi: 10.1101/lm.024711.111
Goñalongs, C. M., and Farina, W. M. (2015). Effects of sublethal doses of imidacloprid on young adult honeybee behaviour. PLoS One 10:e0140814. doi: 10.1371/journal.pone.0140814
Goulson, D. (2013). Review: an overview of the environmental risks posed by neonicotinoid insectcides. J. Appl. Ecol. 50, 977–987. doi: 10.1111/1365-2664.12111
Hammer, M., and Menzel, R. (1995). Learning and memory in the honeybee. J. Neurosci. 15, 1617–1630. doi: 10.1523/JNEUROSCI.15-03-01617.1995
Huang, D. W., Sherman, B. T., and Lempicki, R. A. (2009a). Bioinformatics enrichment tools: paths towards the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 37, 1–13. doi: 10.1093/nar/gkn923
Huang, D. W., Sherman, B. T., and Lempicki, R. A. (2009b). Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4, 44–57. doi: 10.1038/nprot.2008.211
Huang, Z. Y., and Robinson, G. E. (1992). Honeybee colony integration: worker-worker interactions mediate hormonally regulated plasticity in division of labor. Proc. Natl. Acad. Sci. U.S.A. 89, 11726–11729. doi: 10.1073/pnas.89.24.11726
Hung, K. L. J., Kingston, J. M., Albrecht, M., Holway, D. A., and Kohn, J. R. (2018). The worldwide importance of honey bees as pollinators in natural habitats. Proc. R. Soc. B 285:20172140. doi: 10.1098/rspb.2017.2140
Jaycox, E. R. (1967). Behavioral changes in worker honey bees (Apis mellifera L.) after injection with synthetic juvenile hormone (Hymenoptera: Apidae). J. Kans. Entomol. Soc. 49, 165–170.
Jaycox, E. R., Skowronek, W., and Gwynn, G. (1974). Behavioral changes in a worker honey bee (Apis mellifera) induced by injections of a juvenile hormone mimic. Ann. Entomol. Soc. Am. 67, 529–534. doi: 10.1093/aesa/67.4.529
Johnson, B. R. (2008). Within-nest temporal polyethism in the honey bee. Behav. Ecol. Sociobiol. 62, 777–784. doi: 10.1007/s00265-007-0503-2
Johnson, B. R. (2010). Division of labor in honeybees: form, functions, and proximate mechanisms. Behav. Ecol. Sociobiol. 64, 305–316. doi: 10.1007/s00265-009-0874-7
Jones, A. K., Raymond-Delpech, V., Thany, S. H., Gauthier, M., and Sattelle, D. B. (2006). The nicotinic acetylcholine receptor gene family of the honey bee, Apis mellifera. Genome Res. 16, 1422–1430. doi: 10.1101/gr.4549206
Khamis, A. M., Hamilton, A. R., Medvedeca, Y. A., Alam, T., Alam, I., Essack, M., et al. (2015). Insights into the transcriptional architecture of behavioral plasticity in the honey bee Apis mellifera. Sci. Rep. 5:11136. doi: 10.1038/srep11136
Kim, D., Langmead, B., and Salzberg, S. L. (2015). HISAT: a fast spliced aligner with low memory requirements. Nat. Methods 12, 357–360. doi: 10.1038/nmeth.3317
Leoncini, I., Crauser, D., Robinson, G. E., and Conte, Y. L. (2004). Worker–worker inhibition of honey bee behavioural development independent of queen and brood. Insect. Soc. 51, 392–394. doi: 10.1007/s00040-004-0757-x
Love, M. I., Huber, W., and Anders, S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DEseq2. Genome Biol. 15:550. doi: 10.1186/s13059-014-0550-8
Lu, C., Warchol, K. M., and Callahan, R. A. (2014). Sub-lethal exposure to neonicotinoids impaired honey bees winterization before proceeding to colony collapse disorder. Bull. Insectol. 67, 125–130.
Manjon, C., Troczka, B. J., Zaworra, M., Beadle, K., Randall, E., Hertlein, G., et al. (2018). Unravelling the molecular determinants of bee sensitivity to neonicotinoid insecticides. Curr. Biol. 28, 1137–1143. doi: 10.1016/j.cub.2018.02.045
Medrzycki, P., Montanari, R., Bortolotti, L., Maini, S., and Porrini, C. (2003). Effects of imidacloprid administered in sub-lethal doses on honey bee behaviour. Laboratory tests. Bull. Insectol. 56, 59–62.
Menzel, R. (1999). Memory dynamics in the honeybee. J. Comp. Physiol. A 185, 323–340. doi: 10.1007/s003590050392
Mitchell, E. A. D., Mulhauser, B., Mulot, M., Mutabazi, A., Glauser, G., and Aebi, A. (2017). A worldwide survey of neonicotinoids in honey. Science 358, 109–111. doi: 10.1126/science.aan3684
Peng, Y. C., and Yang, E. C. (2016). Sublethal dosage of imidacloprid reduces the microglomerular density of honey bee mushroom bodies. Sci. Rep. 6:19298. doi: 10.1038/srep19298
Perry, C. J., Søvik, E., Myerscough, M. R., and Barron, A. B. (2015). Rapid behavioral maturation accelerates failure of stressed honey bee colonies. Proc. Natl. Acad. Sci. U.S.A. 112, 3427–3432. doi: 10.1073/pnas.1422089112
Reim, T., and Scheiner, R. (2014). Division of labour in honey bees: age- and task- related changes in the expression of octopamine receptor genes. Insect Mol. Biol. 23, 833–841. doi: 10.1111/imb.12130
Robinson, G. E. (1987). Regulation of honey bee age polyethism by juvenile hormone. Behav. Ecol. Sociobiol. 20, 329–338.
Sagili, R. R., Pankiw, T., and Metz, B. N. (2011). Division of labor associated with brood rearing in the honey bee: how does it translate to colony fitness? PLoS One 6:e16785. doi: 10.1371/journal.pone.0016785
Sanchez-Bayo, F., and Goka, K. (2014). Pesticide residues and bees – a risk assessment. PLoS One 9:e94482. doi: 10.1371/journal.pone.0094482
Sandrock, C., Tanadini, M., Tanadini, L. G., Fauser-Misslin, A., Potts, S. G., and Neumann, P. (2014). Impact of chronic neonicotinoid exposure on honeybee colony performance and queen supersedure. PLoS One 9:e103592. doi: 10.1371/journal.pone.0103592
Schneider, C. W., Tautz, J., Grünewald, B., and Fuchs, S. (2012). RFID tracking of sublethal effects of two neonicotinoid insecticides on the foraging behavior of Apis mellifera. PLoS One 7:e30023. doi: 10.1371/journal.pone.0030023
Seeley, T. D. (1982). Adaptive significance of the age polyethism schedule in honeybee colonies. Behav. Ecol. Sociobiol. 11, 287–293. doi: 10.1007/BF00299306
Shi, T. F., Wang, Y. F., Liu, F., Qi, L., and Yu, L. S. (2017a). Influence of the neonicotinoid insecticide thiamethoxam on miRNA expression in the honey bee (Hymenoptera: Apidae). J. Insect Sci. 17:96. doi: 10.1093/jisesa/iex074
Shi, T. F., Wang, Y. F., Liu, F., Qi, L., and Yu, L. S. (2017b). Sublethal effects of the neonicotinoid insectice thiamethoxam on the transcriptome of the honey bees (Hymenoptera: Apidae). J. Econ. Entomol. 110, 2283–2289. doi: 10.1093/jee/tox262
Škerl, M. I. S., Bolta, ŠV., Česnik, H. B., and Gregorc, A. (2009). Residues of pesticides in honeybee (Apis mellifera carnica) bee bread and in pollen loads from treated apple orchards. Bull. Environ. Contam. Toxicol. 83, 374–377. doi: 10.1007/s00128-009-9762-0
Ushitani, T., Perry, C. J., Cheng, K., and Barron, A. B. (2016). Accelerate behavioral development changes fine-scale search behaviour and spatial memory in honey bees (Apis mellifera L). J. Exp. Biol. 219, 412–418. doi: 10.1242/jeb.126920
van der Zee, R., Pisa, L., Andonov, S., Brodschneider, R., Charriére, J. D., Chlebo, R., et al. (2012). Managed honey bee colony losses in Canada, China, Europe, Israel and Turkey, for the winters of 2008-9 and 2009-10. J. Apic. Res. 51, 100–114. doi: 10.3896/IBRA.1.51.1.12
Wang, K., Fan, R. L., Ji, W. N., Zhang, W. W., Chen, X. M., Wang, S., et al. (2018). Transcriptome analysis of newly emerged honeybees exposure to sublethal carbendazim during larval stage. Front. Genet. 9:426. doi: 10.3389/fgene.2018.00426
Wang, Y., Kocher, S. D., Lunksvayer, T. A., Grozinger, C. M., Page, R. E. Jr., and Amdam, G. V. (2012). Regulation of behaviorally associated gene networks in worker honey bee ovaries. J. Exp. Biol. 215(Pt 1), 124–134. doi: 10.1242/jeb.060889
Whitfield, C. W., Cziko, A. M., and Robinson, G. E. (2003). Gene expression profiles in the brain predict behavior in individual honey bees. Science 302, 296–299. doi: 10.1126/science.1086807
Wood, S. C., Kozii, I. V., Koziy, R. V., Epp, T., and Simko, E. (2018). Comparative chronic toxicity of three neonicotinoids on New Zealand packaged honey bees. PLoS One 13:e0190517. doi: 10.1371/journal.pone.0190517
Woodcock, B. A., Bullock, J. M., Shore, R. F., Heard, M. S., Pereira, M. G., Redhead, J., et al. (2017). Country- specific effects of neonicotinoid pesticides on honey bees and wild bees. Science 356, 1393–1395. doi: 10.1126/science.aaa1190
Wu, M. C., Chang, Y. W., Lu, K. H., and Yang, E. C. (2017). Gene expression changes in honey bees induced by sublethal imidacloprid exposure during the larval stage. Insect Biochem. Mol. Biol. 88, 12–20. doi: 10.1016/j.ibmb.2017.06.016
Yang, E. C., Chang, H. C., We, W. Y., and Chen, Y. W. (2012). Impaired olfactory associative behavior of honeybee workers due to contamination of imidacloprid in the larval stage. PLoS One 7:e49472. doi: 10.1371/journal.pone.0049472
Yang, E. C., Chuang, Y. C., Chen, Y. L., and Chang, L. H. (2008). Abnormal foraging behavior induced by sublethal dosage of imidacloprid in the honey bee. J. Econ. Entomol. 101, 1743–1748. doi: 10.1603/0022-0493-101.6.1743
Zhang, E., and Nieh, J. C. (2015). The neonicotinoid imidacloprid impairs honey bee aversive learning of simulated predation. J. Exp. Biol. 218(Pt 20), 3199–3205. doi: 10.1242/jeb.127472
Keywords: honey bee, imidacloprid, sublethal dosage, transcriptome, precocious foraging
Citation: Chen Y-R, Tzeng DTW, Ting C, Hsu P-S, Wu T-H, Zhong S and Yang E-C (2021) Missing Nurse Bees—Early Transcriptomic Switch From Nurse Bee to Forager Induced by Sublethal Imidacloprid. Front. Genet. 12:665927. doi: 10.3389/fgene.2021.665927
Received: 09 February 2021; Accepted: 12 May 2021;
Published: 17 June 2021.
Edited by:
Serena Aceto, University of Naples Federico II, ItalyReviewed by:
Sonia Andrade, University of São Paulo, BrazilCopyright © 2021 Chen, Tzeng, Ting, Hsu, Wu, Zhong and Yang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Silin Zhong, c2lsaW4uemhvbmdAY3Voay5lZHUuaGs=; En-Cheng Yang, ZWN5YW5nQG50dS5lZHUudHc=
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.