
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Genet., 02 March 2021
Sec. RNA
Volume 12 - 2021 | https://doi.org/10.3389/fgene.2021.652129
This article is part of the Research TopicRNA Modifications and EpitranscriptomicsView all 30 articles
Small nuclear RNAs (snRNAs) are critical components of the spliceosome that catalyze the splicing of pre-mRNA. snRNAs are each complexed with many proteins to form RNA-protein complexes, termed as small nuclear ribonucleoproteins (snRNPs), in the cell nucleus. snRNPs participate in pre-mRNA splicing by recognizing the critical sequence elements present in the introns, thereby forming active spliceosomes. The recognition is achieved primarily by base-pairing interactions (or nucleotide-nucleotide contact) between snRNAs and pre-mRNA. Notably, snRNAs are extensively modified with different RNA modifications, which confer unique properties to the RNAs. Here, we review the current knowledge of the mechanisms and functions of snRNA modifications and their biological relevance in the splicing process.
Pre-mRNA splicing is, by definition, a co- or post-transcriptional RNA processing reaction by which introns are removed from mRNA precursors, and exons are precisely joined together to form functional mature mRNAs (Berget et al., 1977; Chow et al., 1977; Shi, 2017). The fidelity of this mechanism is critical for correct gene expression as proven by the fact that 10% of all disease-causing single-point mutations in humans generate splicing defects (Krawczak et al., 2007; Scotti and Swanson, 2016). Pre-mRNA splicing occurs via a two-step transesterification reaction pathway (Figure 1; Ruskin et al., 1984). In the first step, the 2'-hydroxyl group (2'-OH) of the branch point nucleotide (adenosine) attacks the phosphate at the 5' exon-intron junction (5' splice site), resulting in the cleavage of the phosphodiester bond between the 5' exon and intron, and the concurrent formation of a new 5'-2' phosphodiester bond between the 5' end of the intron and the branch point adenosine. Thus, a lariat-structured intermediate (lariat intron-3' exon) and a cut-off 5' exon intermediate are produced. In the second step, the 3'-OH group of the cut-off 5' exon attacks the phosphate at the intron-3' exon junction (3' splice site), releasing the lariat intron product and generating the spliced mature mRNA product.
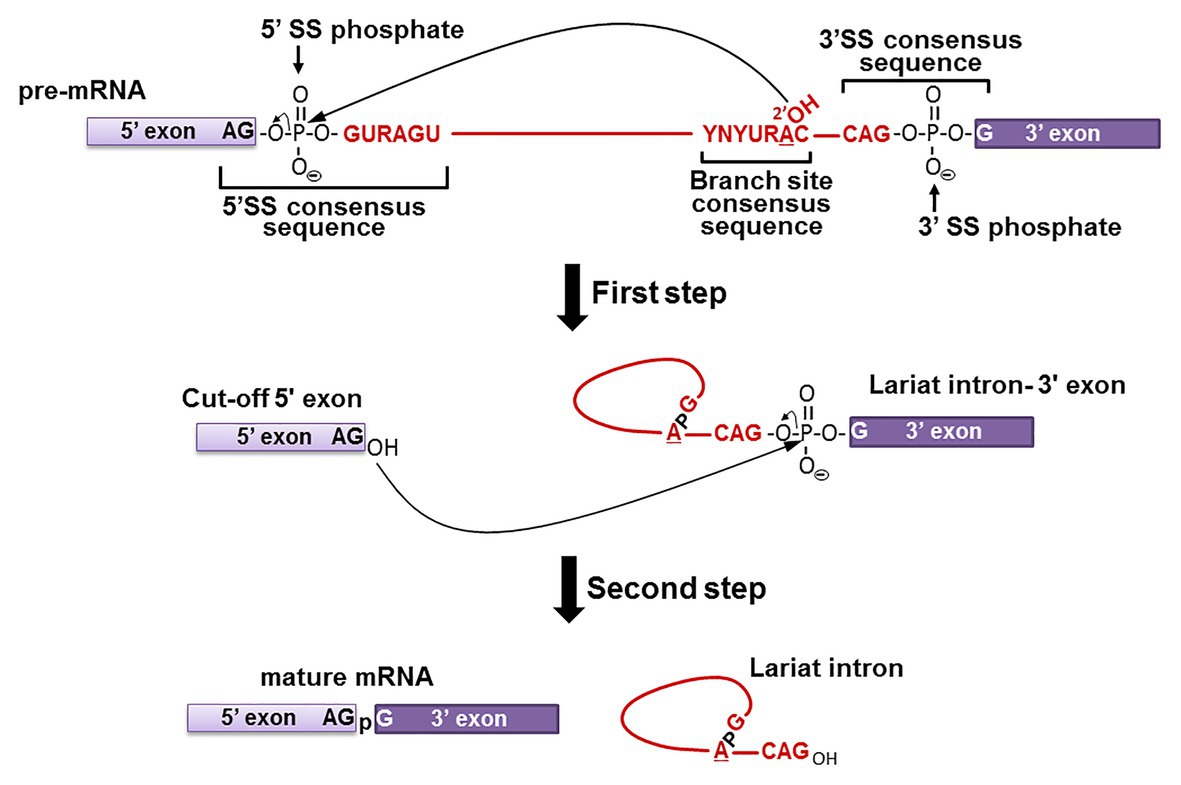
Figure 1. Pre-mRNA splicing pathway. Pre-mRNA splicing takes place via a two-step transesterification reaction pathway. In the first step, the 2'-OH group of the branch site adenosine attacks the phosphate at the 5' exon-intron junction (5' SS phosphate), generating the “Cut-off 5' exon” and the “Lariat intron-3' exon” intermediates. In the second step, the 3'-OH group of the “Cut-off 5' exon” attacks the phosphate at the intron-3' exon junction (3' SS phosphate), yielding the “mature mRNA” and “Lariat intron” products. The consensus sequences at the branch site and the 5' and 3' splice sites are shown (red letters, where Y is pyrimidine and R is purine). The 5' exon (light purple box), 3' exon (dark purple box), and the intron (red line) are also shown.
The two chemical reactions of pre-mRNA splicing occur only after the pre-mRNA is assembled into the functional spliceosome, a multi-component complex composed of five small nuclear RNAs (snRNAs U1, U2, U4, U5, and U6), which are present as small nuclear ribonucleoprotein particles (snRNPs, RNA-protein complexes) and a large number of splicing protein factors (Jurica and Moore, 2003).
During spliceosome assembly, spliceosomal snRNPs and splicing factors recognize and interact with the pre-mRNA consensus sequences, facilitating and specifying the transesterification reactions (Figure 2). Specifically, U1 snRNP recognizes the 5' splice site of a pre-mRNA to form a commitment complex (complex E) that commits the pre-mRNA to spliceosome assembly (Kondo et al., 2015). This recognition involves base-pairing interactions between the 10 highly conserved nucleotides at the 5' end of U1 snRNA and the intron sequences of the pre-mRNA at the 5' splice site (G/GUAUGU in yeast or G/GURAGU in vertebrates, where “/” represents the exon-intron junction and R stands for purine; Zhuang and Weiner, 1986). The U2 snRNP then recognizes the pre-mRNA branch site to form a pre-splicing complex called as complex A (Query et al., 1997). This recognition again involves a base-pairing interaction between a highly conserved sequence in U2 snRNA and the pre-mRNA branch site sequence (UACUA
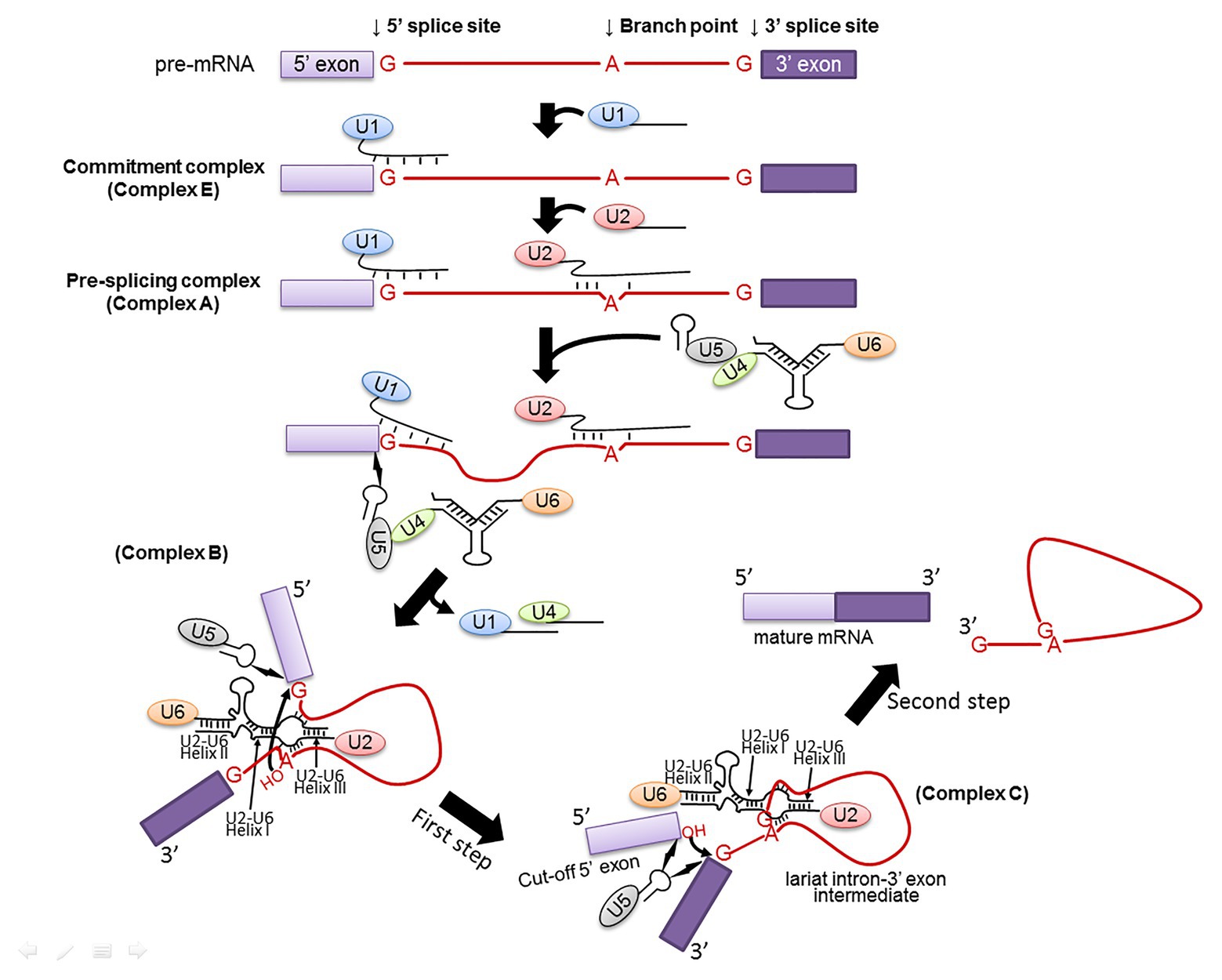
Figure 2. Spliceosome assembly. Spliceosome assembly is a dynamic multi-step process. Shown are several important steps resulting in Complexes E, A, B, and C. The branch site, the 5' and 3' splice sites are also shown. The 5' and 3' exons are in light and dark purple boxes, respectively, and the intron is in red line (and red letters). U1, U2, U4, U5, and U6 snRNPs are also depicted. RNA-RNA interactions, including U1–5' splice site, U2-branch site, U6–5' splice site, U4–U6, and U2–U6 (Helixes I, II, and III) are indicated as well. The two curved arrows indicate nucleophilic attacks (transesterification reactions). The lightning bolts indicate non-Watson-Crick nucleotide-nucleotide contacts.
The emerging field of epitranscriptomics is continually unveiling additional unknown levels of complexity of the transcriptome (Nachtergaele and He, 2017; Roundtree et al., 2017). RNA modifications can have a major impact in RNA folding and function in all types of RNA, including snRNAs. All snRNAs (except for U6) have a 2,2,7-trimethylated 5' guanosine cap (U6 possesses a γ-monomethyl guanosine cap; Singh and Reddy, 1989). Further, numerous internal nucleotides are modified by pseudouridylation (5-ribosyl isomers of uridine), 2’-O-methylation, and in some cases, base methylation [e.g., N6-methyladenosine (m6A) and N2-methylguanosine (m2G)] (Reddy and Busch, 1988; Massenet et al., 1998; Bohnsack and Sloan, 2018). Notably, the modified nucleotides in spliceosomal snRNAs are remarkably conserved from species to species. For instance, various vertebrate snRNAs contain identical modified nucleotides at identical sites. Although a relatively small number of modified nucleotides have been identified in yeast snRNAs, those that are modified always have counterparts in higher eukaryotic snRNAs (Adachi and Yu, 2014). Furthermore, almost all the modified nucleotides are concentrated in regions that are functionally relevant to the splicing process (Adachi and Yu, 2014). Together, the conservation and the strategic location of these modified nucleotides strongly point to their importance in the process of spliceosome assembly and splicing. It should be pointed out that various post-transcriptional modifications, which are catalyzed by different types of machinery, generate diversity in the snRNAs that likely contribute to pre-mRNA splicing regulation.
In addition to U1, U2, U4, U5, and U6 snRNAs (major), there is also a set of minor spliceosomal snRNA species (U11, U12, U4atac, and U6atac) that participate in the splicing of a minor class of introns (Tarn and Steitz, 1997; Will and Lührmann, 2005; Turunen et al., 2013). Some of these minor snRNAs are also post-transcriptionally modified. This review will describe spliceosomal snRNA modifications (major and minor classes of snRNAs), focusing on the mechanisms and functions of these modifications.
The most abundant modified nucleotides in snRNAs are pseudouridine (Ψ) and 2'-O-methyl residues, whereas m6A and m2G are rarely present in only a few snRNA species (Epstein et al., 1980; Reddy et al., 1981b; Massenet et al., 1998; Bohnsack and Sloan, 2018). RNA modification can be catalyzed by either RNA-dependent or RNA-independent mechanism (De Zoysa and Yu, 2017; Meier, 2017; Wiener and Schwartz, 2020). While the RNA-independent mechanism depends on stand-alone protein enzymes capable of recognizing the substrates and catalyzing the chemical reaction, the RNA-dependent mechanism typically relies on RNA-protein enzyme complexes (RNPs), each of which is composed of one small RNA and several proteins. In each RNP, the RNA component functions as a guide recognizing the substrate RNA, and one of the protein components has enzymatic activity catalyzing the chemical reaction (Yu et al., 2005).
Both snRNA 2’-O-methylation and pseudouridylation are catalyzed by RNA-dependent mechanisms in high eukaryotes. Specifically, a family of box C/D RNPs is responsible for snRNA 2'-O-methylation (Balakin et al., 1996; Cavaillé et al., 1996; Kiss-László et al., 1996), and another family of RNPs, the box H/ACA RNP family, is accountable for snRNA pseudouridylation (Balakin et al., 1996; Ganot et al., 1997; Ni et al., 1997). Each member of the box C/D RNP family is composed of one unique box C/D RNA and four common core proteins (Fibrillarin, also known as Nop1, Nop56, Nop58, and Snu13). Likewise, members of the box H/ACA RNP family each consist of one unique box H/ACA RNA and a set of four common proteins (Dyskerin, also known as Nap57 or Cbf5, Nhp2, Nop10, and Gar1). Both box H/ACA and C/D snoRNAs are usually intron-encoded in mammals and are matured through splicing and processing (Tycowski et al., 1996; Hirose et al., 2003, 2006).
Mature box C/D RNAs and box H/ACA RNAs both fold into a unique secondary structure (Figure 3). The C/D RNAs, despite their sequence differences, form a signature structure with a terminal stem and two single-stranded sequences sandwiched between box C (RUGAUGA, where R is a purine) and box D' (CUGA) and between box C' (RUGAUGA) and box D (CUGA), respectively (Figure 3). It turns out that the single-stranded sequences serve as guides that base-pair with the substrate RNAs, forming a 10–21-nt duplex and specifying the target nucleotide that is precisely five nucleotides upstream from box D (or box D'). Once the target nucleotide is identified, fibrillarin (Nop1), one of the four box C/D RNP core proteins and a methyltransferase, delivers the methyl group to the target nucleotide at the 2'-O position. The “box D + 5” rule for box C/D RNA-guided snRNA 2'-O-methylation has been verified in various organisms, including Xenopus, mouse, and human, indicating that box C/D RNA-guided 2'-O-methylation of snRNA is universal in high eukaryotes (Kiss-László et al., 1996; Karijolich and Yu, 2010). Given that 2'-O-methylation is a sugar-ring modification, it can occur to any nucleotides. Interestingly, to date, no 2'-O-methylated residues have been identified in Saccharomyces cerevisiae snRNA.

Figure 3. RNA-guided snRNA modifications. (A) Box C/D RNA-guided 2'-O-methylation. Box C/D guide RNA (black line) and its substrate RNA (red line) are shown. Boxes C, D', C', and D of the box C/D RNA are indicated. Base-pairing interactions between the two RNAs (usually 10–21 base-pairs) are also depicted. The fifth nucleotide upstream from box D' is 2'-O-methylated (indicated), and so is the fifth nucleotide upstream from box D (also indicated). (B) Box H/ACA RNA-guided pseudouridylation. Both box H/ACA RNA (black line) and its substrate RNA (red line) are indicated. Boxes H, ACA, and CAB (a Cajal body localization signal sequence) of the box H/ACA RNA are also indicated. Base-pairing interactions between the guide and substrate RNAs in the pseudouridylation pockets (internal loops) are shown. The modified target uridines (Ψs) are indicated by two arrows. The 14–16 nt distance between the target uridine and box H (or box ACA) is indicated as well.
Similarly, despite their sequence differences, all members of the box H/ACA RNA family fold into a structure known as the “hairpin-hinge (H box, ANANNA, where N represents any nucleotide)-hairpin-tail (ACA box)” structure. In this structure, there are two independent hairpins, in each of which there exists an internal loop (single-stranded) that serves as a guide. In essence, the guide sequences are in two separate segments in the linear RNA that are brought together in internal loops within the hairpins. Base-pairing between the bipartite guide sequence and the snRNA, positions the target uridine at the base of the upper stem of the hairpin, leaving it unpaired within the internal loop (so-called “pseudouridylation pocket”) and located about 14–16 nucleotides upstream of box H or box ACA (Figure 3). When the target uridine is brought to the pocket, Dyskerin (NAP57 or Cbf5), one of the four box H/ACA RNP core proteins and also a pseudouridylase, converts the uridine to pseudouridine. The box H/ACA RNA-guided pseudouridylation mechanism has been tested and verified in various high eukaryotic systems (Terns and Terns, 2006). Recent analyses have further demonstrated that a minimum number of eight base-pairs between the guide and substrate in the pseudouridylation pocket is required for efficient pseudouridylation (Caton et al., 2018; De Zoysa et al., 2018).
In S. cerevisiae, spliceosomal snRNA pseudouridylation is more complex. Both RNA-dependent or RNA-independent mechanisms are used (Massenet et al., 1999; Ma et al., 2003, 2005; Yu et al., 2011). The yeast box H/ACA RNAs can be either encoded in introns of protein-coding genes or in independent transcripts of non-protein-coding genes. In S. cerevisiae, snRNA pseudouridylation can also be achieved via an RNA-independent (protein-only) mechanism.
Stand-alone (protein-only) pseudouridine synthases (Pus) can recognize the substrate and perform the uridine isomerization reaction in a site-specific manner. There are nine Pus enzymes in yeast (Pus1–9) and 11 human homologs (Pus1, Pus3, TruB1, TruB2, RusD1, RusD2, RusD3, RusD4, Pus7, Pus7L, and Pus10; Rintala-Dempsey and Kothe, 2017). In yeast, only Pus1 and Pus7 were identified as capable of RNA-independent pseudouridylation of snRNAs (Massenet et al., 1999; Ma et al., 2003; Basak and Query, 2014; Schwartz et al., 2014). Yeast Pus1 and Pus7 belong to the TruA and TruD families of pseudouridine synthases, respectively. Yeast Pus1 is localized in the nucleus and targets not only snRNAs but also other types of RNA, showing broad substrate specificity (Motorin et al., 1998). In addition to its pseudouridylation activity, Pus1 is also involved in tRNA biogenesis (Simos et al., 1996). Pus7 localizes in the nucleus and cytoplasm of cells and, like Pus1, can also target different RNAs. Pus7 is known for being able to recognize substrates relying on both the sequence (consensus UN
Other exotic base methylations, such as m6A and m2G, have also been identified in snRNAs. These base modifications are catalyzed by RNA-independent enzymatic machineries. Specifically, the m6A modification is catalyzed by the m6A methyltransferases, protein-only enzymes known as m6A writers (Frye et al., 2018). In this reaction, a methyl group is attached to N6 of an adenosine within a specific RNA motif, resulting in the production of m6A methylated RNA (RNA containing an N6-methyladenosine). m6A has been identified in human U2, U4, and U6 snRNAs, as well as in S. pombe U2 and U6 snRNAs. While m6A writers METTL4 and METTL16 are responsible for the formation of m6A in U2 and U6 snRNAs, respectively, the exact enzyme for the formation of m6A in human U4 snRNA (Reddy et al., 1981b) remains unknown.
The m2G modification results from the methylation of N2 of guanine (the guanosyl amino group at the position C2) and was initially identified in tRNAs (Grosjean et al., 1995). It is catalyzed by a class of enzymes known as guanine-(N2)-methyltransferases, which have been identified in several species (Sindhuphak et al., 1985; Sergiev et al., 2006) and seem to have a substrate consensus sequence (UG
It has long been known that mammalian major spliceosomal snRNAs contain a large number of modified nucleotides (Table 1). Specifically, there are two, fourteen, three, three, and three Ψs in mammalian U1, U2, U4, U5, and U6 snRNAs, respectively. There are also three, ten, four, five, and eight 2’-O-methylated residues in mammalian U1, U2, U4, U5, and U6 snRNAs, respectively. In addition, mammalian U2, U4, and U6 snRNAs each contain an m6A. Further, mammalian U6 snRNA contains an m2G as well. In contrast, there are a total of only six constitutively formed Ψs in S. cerevisiae snRNAs, including two in U1 snRNA, three in U2 snRNA, and one in U5 snRNA. In addition, S. cerevisiae U2 snRNA can be pseudouridylated at two novel sites under stress conditions. A set of minor class spliceosomal snRNAs also exists in mammals including U11, U12, U4atac, and U6atac. These snRNAs contain several Ψs and 2'-O-methylated residues as well. Over the years, the mechanisms (what enzymes are involved) and functions of these modifications have been studied, accumulating a wealth of knowledge.

Table 1. Yeast and human RNA modifications present in snRNAs with respective guide RNAs (when applicable) and catalyst machinery (Adachi and Yu, 2014).
The U1 snRNA is one of the most abundant snRNAs in different species. Only two types of modifications, namely pseudouridylation and 2’-O-methylation, have been detected in mammalian U1 snRNA. Together, there is a total of five modified nucleotides, including Ψ5, Ψ6, Am1, Um2, and Am70 (Reddy et al., 1981a; Reddy and Busch, 1988; Massenet et al., 1999; Kiss et al., 2004; Gu et al., 2005; Krogh et al., 2017; Table 1). In yeast U1 snRNA, only Ψ5 and Ψ6 are identified (Massenet et al., 1999); no 2'-O-methylated residues have been detected. At present, it is still not clear whether the RNA-dependent or RNA-independent mechanism catalyzes pseudouridylation at positions 5 and 6 in yeast U1 snRNA. In human U1 snRNA, pseudouridylation at Ψ5, Ψ6 positions (Branlant et al., 1980) is catalyzed by H/ACA RNP machinery and guided by ACA47 (Kiss et al., 2004) and U109 (Gu et al., 2005), respectively. Mammalian U1 snRNA 2'-O-methylation at position 70 (Krogh et al., 2017) is likely catalyzed by an RNA-dependent mechanism, given that a box C/D RNA (SCARNA7, also known as U90) has been identified to target this site (Darzacq et al., 2002).
Notably, Ψ5 and Ψ6 are within the first 10-nucleotide sequence known to base-pair with the 5' splice site of pre-mRNA during splicing. Given that Ψ can affect local RNA structure and enhance base-pairing and base stacking (Ge and Yu, 2013), Ψ5 and Ψ6 are believed to be important in the recognition process of the 5' splice site. Indeed, an in vitro splicing assay performed to test competitive usage of two 5' splice sites suggested that the two Ψs in the U1 snRNA could provide a competitive advantage in the 5' splice site selection (Roca, 2005). In another study, it was shown that Ψ5 or Ψ6 can be bulged out in certain duplexes consisting of 5' splice sites and U1 snRNA (Roca et al., 2012). Thermodynamic analysis of these duplexes confirmed the stabilization properties of Ψs (and 2'-O-methylated residues at the first two positions) in this context, possibly by improving the base stacking of the helix. These results are consistent with the results of a previous work showing that a Ψ in a Ψ-G base pair strengthens the interaction between U1 snRNA and the 5' splice site (Freund, 2003). While these conclusions are exciting and make sense, they seem somewhat contradictory to an earlier work of Will et al. (1996), where the authors showed that U1 snRNA-depleted mammalian cell extracts could still be reconstituted for splicing when adding in vitro-transcribed (therefore unmodified) U1 snRNA. However, it could well be that the extracts could modify the in vitro-transcribed U1 snRNA upon addition. Alternatively, although the unmodified U1 snRNA could still support splicing, it may not be as active as the modified U1 snRNA. The reconstitution assay using unmodified U1 snRNA probably did not reflect the contributions of Ψ5 and Ψ6 in 5' splice site recognition. As for the function of 2’-O-methylated residue at position 70 (Am70) in mammalian U1 snRNA, not much is known.
U2 snRNA is the most extensively modified among all spliceosomal snRNAs (Shibata et al., 1975; Reddy and Busch, 1988). There are fourteen Ψs, ten 2'-O-methylated residues, and one m6Am residue in vertebrate U2 snRNA (Table 1). Given that a near-complete set of box H/ACA RNAs and a complete set of box C/D RNAs are identified and that they can account for almost all known pseudouridylation and 2'-O-methylation sites, it is believed that the RNA-dependent mechanisms are responsible for the formation of virtually all the Ψs and 2'-O-methylated residues (except for the first two 2'-O-methylated residues, Am1 and Um2) in vertebrate U2 snRNA (Hüttenhofer et al., 2001; Tycowski et al., 2004; Schattner et al., 2006; Deryusheva and Gall, 2017, 2018; Bohnsack and Sloan, 2018; Deryusheva et al., 2020). There are three Ψs and no 2'-O-methylated residues identified in S. cerevisiae U2 snRNA. The formation of Ψ at different positions within yeast U2 snRNA can be catalyzed by either RNA-dependent or RNA-independent mechanism (Massenet and Branlant, 1999; Ma et al., 2003, 2005; Schwartz et al., 2014).
Many of the U2 snRNA Ψs and 2'-O-methylated residues have been tested for function, and they are virtually all important for splicing. For example, using the Xenopus oocyte microinjection system, Yu et al. showed that only modified U2 snRNA (but not in vitro transcribed, unmodified U2 snRNA) was able to restore splicing in U2 snRNA-depleted oocytes, indicating that modified nucleotides of U2 snRNA are crucial for pre-mRNA splicing. Subsequently, they mapped the important modified nucleotides to the 5' end region. They further demonstrated that the modified nucleotides within the 5' end region are essential for the formation of functional U2 snRNP and splicing complexes (Yu et al., 1998). In a different study, Dönmez et al. tested these modified nucleotides individually and demonstrated that three Ψs (Ψ6, Ψ7, and Ψ15) and five 2'-O-methylated residues (Am1, Um2, Gm11, Gm12, and Gm19), located in the 5'-end region (first 24 nt) of human U2 snRNA, were required for efficient pre-mRNA splicing. While the Ψs have a cumulative effect in splicing, four of the five 2’-O-methylated residues (Am1, Um2, Gm12, and Gm19) were essential for activity (Dönmez et al., 2004). Soon after, it was shown that the Ψs in the branch site recognition region (BSRR, Ψ34, Ψ37, Ψ39, Ψ41, Ψ43, and Ψ44) are also essential for pre-mRNA splicing (Zhao and Yu, 2004).
Unlike vertebrate U2 snRNA, there are only three Ψs that are normally present in S. cerevisiae U2 snRNA. They are located at positions 35, 42, and 44 (equivalent to vertebrate U2 snRNA at positions 34, 41, and 43) in the BSRR (Figure 4; De Zoysa and Yu, 2017). Pseudouridylation at these positions, 35, 42, and 44, is catalyzed by Pus7, snR81 RNP, and Pus1, respectively. Among these pseudouridylation enzymes, Pus1 and Pus7 are stand-alone protein pseudouridylases, whereas snR81 RNP is a genuine box H/ACA RNP complex. Several lines of evidence indicate that all these Ψs contribute to branch site recognition during pre-mRNA splicing. For example, Yang et al. showed that a Pus7-deleted strain exhibited reduced levels of splicing and cell growth in certain conditions (Yang et al., 2005). By analyzing splicing in yeast strains deleted of any of the three pseudouridylases (in all combinations), Wu et al. (2016a) found that the three Ψs, in coordination with the ATPase Prp5, play an essential role in recognizing the branch site at an early stage during spliceosome assembly. Furthermore, structural studies of U2 snRNA showed the importance of the Ψ35 in splicing function. Specifically, it was proposed that Ψ35 affected the local RNA structure to expose the branch site adenosine 2'-OH group, making it available for nucleophilic attack on the 5' splice site – the first transesterification reaction or the first step of splicing (Newby and Greenbaum, 2001, 2002). However, recent work from Kielkopf’s lab (Kennedy et al., 2019) suggested that the role of Ψ35 could be indirect, perhaps more reliant on auxiliary factors.
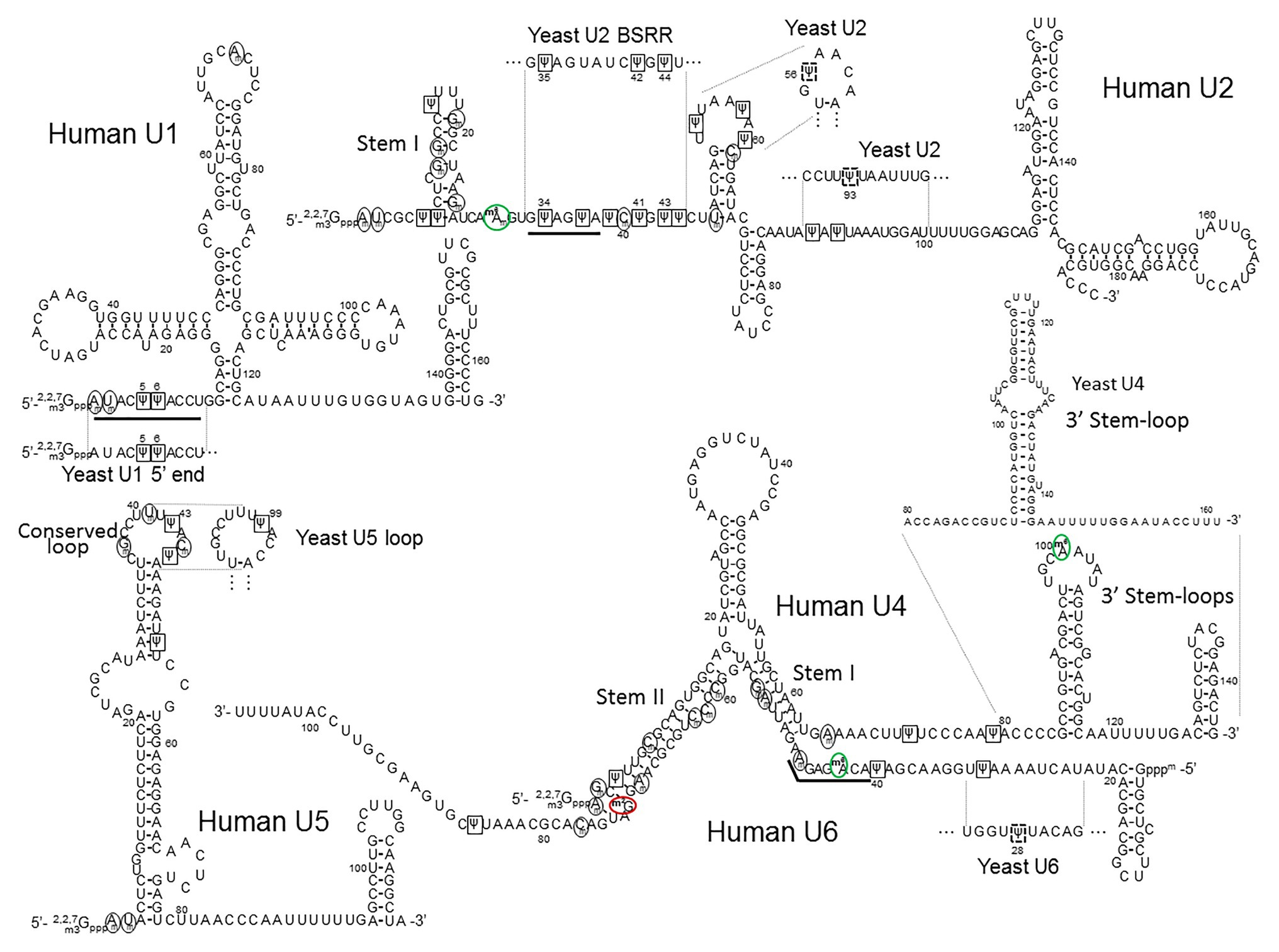
Figure 4. The primary sequences and the secondary structures of major snRNAs. The human U1, U2, U4/U6, and U5 snRNA sequences and structures are shown. The relevant yeast fragments are also depicted. The U2 Stem I, U4 3' Stem-loop, U5 Conserved loop, U4–U6 Stem I and Stem II, are indicated. The sequences that recognize the 5' splice site (the U1 5' end AmUmACΨΨACCU and the U6 ACm6AGAGAm sequences) and the branch site (the U2 branch site recognition sequence GΨAGΨA) are underlined. 2'-O-methylated nucleotides are circled (black circles), Ψs are boxed (black squares), inducible Ψs in yeast U2 and yeast U6 are boxed in dashed black squares, m6As (including m6Am) are circled in green (green circles), and m2G is circled in red (red circle).
In yeast U2 snRNA, two non-constitutive modifications (Ψ56 and Ψ93) can also be identified in stress conditions (Wu et al., 2011). Pseudouridylation at positions 56 and 93 is catalyzed, respectively, by Pus7, which normally catalyzes the formation of Ψ35, and snR81 RNP, which is responsible for the constitutive formation of Ψ42. The induction of Ψ formation at these two positions is through the Tor-signaling pathway under nutrient deprivation conditions (Wu et al., 2016b). The formation of Ψ56 can also be induced by heat shock. Although different types of machinery, stand-alone protein Pus7, and box H/ACA RNP snR81, catalyze the formation of Ψ56 and Ψ93, respectively, it appears that the imperfect sequences surrounding the inducible target sites (positions 56 and 93) vs. those flanking the constitutively modified sites (positions 35 and 42) could explain their inducibility (Wu et al., 2011). Induced formation of Ψ56 and Ψ93 plays a role in pre-mRNA splicing, perhaps by helping alter the U2 snRNA structure. Indeed, Ψ56- and Ψ93-mediated U2 snRNA structural change was observed in a structural study (van der Feltz et al., 2018).
Besides Ψs and 2'-O-methylated residues, one of the 2'-O-methylated adenosines (Am30) in human U2 snRNA is also base methylated in the N6-position (m6Am30; Figure 4). This modification is conserved through evolution, from yeast (S. pombe) to humans, in the same nucleotide position (Gu et al., 1996). Since m6Am30 is located almost immediately upstream of the branch site recognition sequence, it has recently drawn some attention. Chen et al. (2020) and Goh et al. (2020) have independently identified METTL4 as the methyltransferase responsible for the formation of m6Am of mammalian U2 snRNA at position 30. In their study, Chen et al. generated knocked-out METTL4 human cells and observed an effect on splicing in those cells when compared to wild-type cells. However, the direct link between the m6Am30 modification in U2 snRNA and splicing was not definitively established. Nonetheless, they demonstrated that METTL4 is the enzyme responsible for the m6Am30 modification. Specifically, using recombinant METTL4 and a fragment of U2 snRNA substrate, they carried out an in vitro biochemical assay and detected m6Am30 formation. However, the 2'-O-methylation of A30 is a prerequisite for the base methylation to occur. Thus, it appears that 2'-O-methylated adenosine (Am30), rather than unmodified adenosine (A30), is the true substrate. Additionally, the level of base methylation could also be severely reduced when changing the 5' and 3' nucleotides, pointing toward sequence recognition by the METTL4. In an independent study, Goh et al. (2020) confirmed METTL4 as the enzyme responsible for the m6Am30 modification (with Am being the true substrate). The authors also confirmed the base modification identity with HPLC-MS/MS (Goh et al., 2020). Using transcriptome-wide sequencing, they further showed that this modified nucleotide contributed to splicing regulation. As to the possible mechanism, the authors of this study hypothesized that the modified adenosine (m6Am30) could potentially be involved in the recruitment of U2 snRNA to the branch site by U2AF (Zamore et al., 1992; Zhang et al., 1992), a heterodimer that recognizes and binds to the 3' splice site at an early stage of spliceosome assembly (prior to complex A formation), thus affecting the pre-mRNA splicing process (Figure 2).
There is also a large number of modified nucleotides in mammalian U4, U5, and U6 snRNAs (Table 1). In total, human U4 snRNA has three Ψs (Ψ4, Ψ72, and Ψ79; Zerby and Patton, 1997), four 2'-O-methylated residues (Am1, Gm2, Cm8, and Am65; Krogh et al., 2017), and one m6A (m6A100; Reddy et al., 1981b). Human U5 contains several Ψs (Ψ43, Ψ46, and Ψ53; Shibata et al., 1975; Krol et al., 1981) and many 2'-O-methylated residues (Am1, Um2, Gm37, Um41, and Cm45; Krol et al., 1981; Krogh et al., 2017). The human U6 snRNA also has a large number of modified nucleotides, including three Ψs (31, 40, and 86), eight 2'-O-methylated residues (Am47, Am53, Gm54, Cm60, Cm62, Cm63, Am70, and Cm77), one m6A (m6A43), and one m2G (m2G72). Most of these modifications were identified decades ago (Epstein et al., 1980; Reddy and Busch, 1988). Similar to mammalian U2 snRNA modifications, pseudouridylation and 2'-O-methylation (except for the first two methylated residues) of human U4, U5, and U6 snRNAs are likely catalyzed by RNA-guided modification mechanisms (Tycowski et al., 1998; Ganot et al., 1999; Hüttenhofer et al., 2001; Jády and Kiss, 2001; Darzacq et al., 2002; Kiss et al., 2002, 2004; Vitali et al., 2003; Lestrade and Weber, 2006; Schattner et al., 2006; Bohnsack and Sloan, 2018). Interestingly, there is only one Ψ (Ψ99) and no 2'-O-methylated nucleotide in S. cerevisiae U5 snRNA (Massenet et al., 1999). No Ψ nor 2'-O-methylated residues were identified in yeast U4 and U6 snRNAs under normal growth conditions.
While the function of Ψs and 2'-O-methylated residues in yeast and mammalian U4, U5, and U6 snRNAs remains largely unclear, it is speculated that these modifications play a crucial role in splicing. Before participating in spliceosome assembly, U4, U5, and U6 snRNAs assemble into the U4/U6.U5 tri-snRNP particle, in which U4 and U6 snRNAs form an extensive base-pair interaction. The strength of this interaction was empirically determined as a stable one (Brow and Guthrie, 1988). Because of their presence in the base-paired region, Ψs and 2'-O-methylated residues seem to be particularly relevant (Figure 4). Given that Ψs and 2'-O-methylated residues are known to increase base-stacking and enhance base-pairing, it is possible that these modified nucleotides in the U4–U6 helixes contribute to stabilizing the interaction. However, the base-pairing between U4 and U6 snRNAs must eventually unwind for the catalytically active spliceosome to form after the U4/U6.U5 tri-snRNP particle enters complex A (pre-mRNA complexed with U1 and U2 snRNPs; see Figure 2). This unwinding event is performed by Brr2, an ATP-dependent helicase with two helicase cassettes in tandem, although only the N-terminal one has unwinding activity (Raghunathan and Guthrie, 1998). Nguyen et al. (2015) obtained the cryo-EM structure of U4/U6.U5 tri-snRNP in yeast and observed that the Brr2 active site is preloaded in the single-stranded region between the stem I of the U4-U6 duplex region and the 3' stem-loop of the U4 snRNA (Figure 4). It is speculated that, since the human U4 snRNA has two Ψs in this single-stranded region (Ψ72 and Ψ79), they could be involved in the recruitment of (or recognition by) the helicase. However, upon deciphering the structure of human U4/U6.U5 tri-snRNP (also by cryo-EM), Agafonov et al. were able to find that Brr2 is located in a different position within the human U4/U6.U5 tri-snRNP complex, approximately 8–10 nm away from the U4/U6 snRNA duplex (Agafonov et al., 2016). To understand the function of Ψ72 and Ψ79 of human U4 snRNA, further research is necessary.
In the course of activation of the spliceosome, or after the unwinding of the U4/U6 snRNA duplex, U1 and U4 snRNAs leave the spliceosome, and the U2, U5, and U6 snRNAs interact with pre-mRNA and with each other (see Figure 2). In particular, U2 and U6 snRNAs form three short base-paired duplexes (helixes I, II, and III; Datta and Weiner, 1991; Madhani and Guthrie, 1992; Sun and Manley, 1995; Burke et al., 2012), which are believed to be the catalytic center for splicing (transesterification) reactions. The dynamic formation of U2–U6 snRNA duplexes was also studied in a protein-free system, where U2–U6 snRNA interactions (likely related to the spliceosomal U2–U6 snRNA helixes) were detected (Burke et al., 2012; Chu et al., 2020). Notably, there are multiple modified nucleotides (Ψ and 2’-O-methylated residues) in the U2-U6 snRNA duplexes. Using single-molecule fluorescence, Karunatilaka and Rueda further investigated the role of these modified nucleotides in the dynamics of U2-U6 snRNA interactions. They concluded that the modifications present in the U2 snRNA stem I (Figure 4) contribute to the dynamics and conformation of the U2-U6 snRNA complex (Karunatilaka and Rueda, 2014). They also suggested that the modified nucleotides in the U2–U6 snRNA complex might also contribute to protein binding in addition to direct RNA structure stabilization.
In the spliceosome, U5 snRNA interacts, through its conserved loop, with the pre-mRNA by directly contacting (non-Watson-Crick pairing) the exon nucleotides at both the 5' and 3' splice sites. Notably, there are two Ψs and three 2'-O-methylated residues in this conserved loop sequence (GmCCUUmUΨACmΨ) of human U5 snRNA (Frank et al., 1994; Szkukalek et al., 1995). In S. cerevisiae U5, however, there is only one modified nucleotide (Ψ99), but it is located in the same conserved loop sequence (GCCUUUΨAC; Figure 4). However, it should be noted that despite the conservation, there is not yet direct evidence indicating that these modified nucleotides contribute to the U5-pre-mRNA interactions.
It was recently reported that METTL16 functions as the methyltransferase responsible for the formation of m6A at position 43 within mammalian U6 snRNA (Pendleton et al., 2017; Warda et al., 2017; Aoyama et al., 2020). This modification (m6A43; Shimba et al., 1995) could have a direct role in splicing regulation given that it is conserved from S. pombe (at position 37; Gu et al., 1996) to human and that it is located in the region which forms base-pairing interactions with the 5' splice site of pre-mRNA before the first step of splicing reaction (transesterification reaction) occurs (Figures 2, 4). In this regard, it has been shown that mutations in this region (AC
Another study was carried out in the Query lab, focusing on the inducible pseudouridylation of S. cerevisiae U6 snRNA at position 28 (Ψ28; Figure 4). They showed that the formation of Ψ28 occurred under certain filamentous growth conditions. This growth condition-induced pseudouridylation is catalyzed by Pus1. Subsequent analyses allowed the authors to conclude that Ψ28 in U6 snRNA directly contributes to filamentous formation (Basak and Query, 2014).
In addition to the major spliceosomal pathway (U2-dependent, described above), there is a less common (or minor) pathway required for splicing of rare class introns that contain different consensus sequences at the 5' and 3' splice sites and the branch site. Except for U5, which is common for both major and minor splicing pathways, a different set of spliceosomal snRNAs (U11, U12, U4atac, U6atac, and U5) is required for the minor splicing pathway (Figure 5). Because it depends on U12 (rather than U2), the minor splicing pathway is also known as the U12-dependent splicing pathway (Montzka and Steitz, 1988; Tarn and Steitz, 1996; Turunen et al., 2013). In the U12-dependent splicing pathway, U11, U12, U4atac, and U6atc snRNAs each play a role that is equivalent to the role of U1, U2, U4, and U6 snRNAs in the major spliceosome, respectively. Expectedly, the secondary structures of the minor class snRNAs are very similar to those of their major class snRNA counterparts (Figure 5). RNA-guided nucleotide modifications (pseudouridylation and 2'-O-methylation) in the minor-class snRNAs have also been studied (Jorjani et al., 2016).
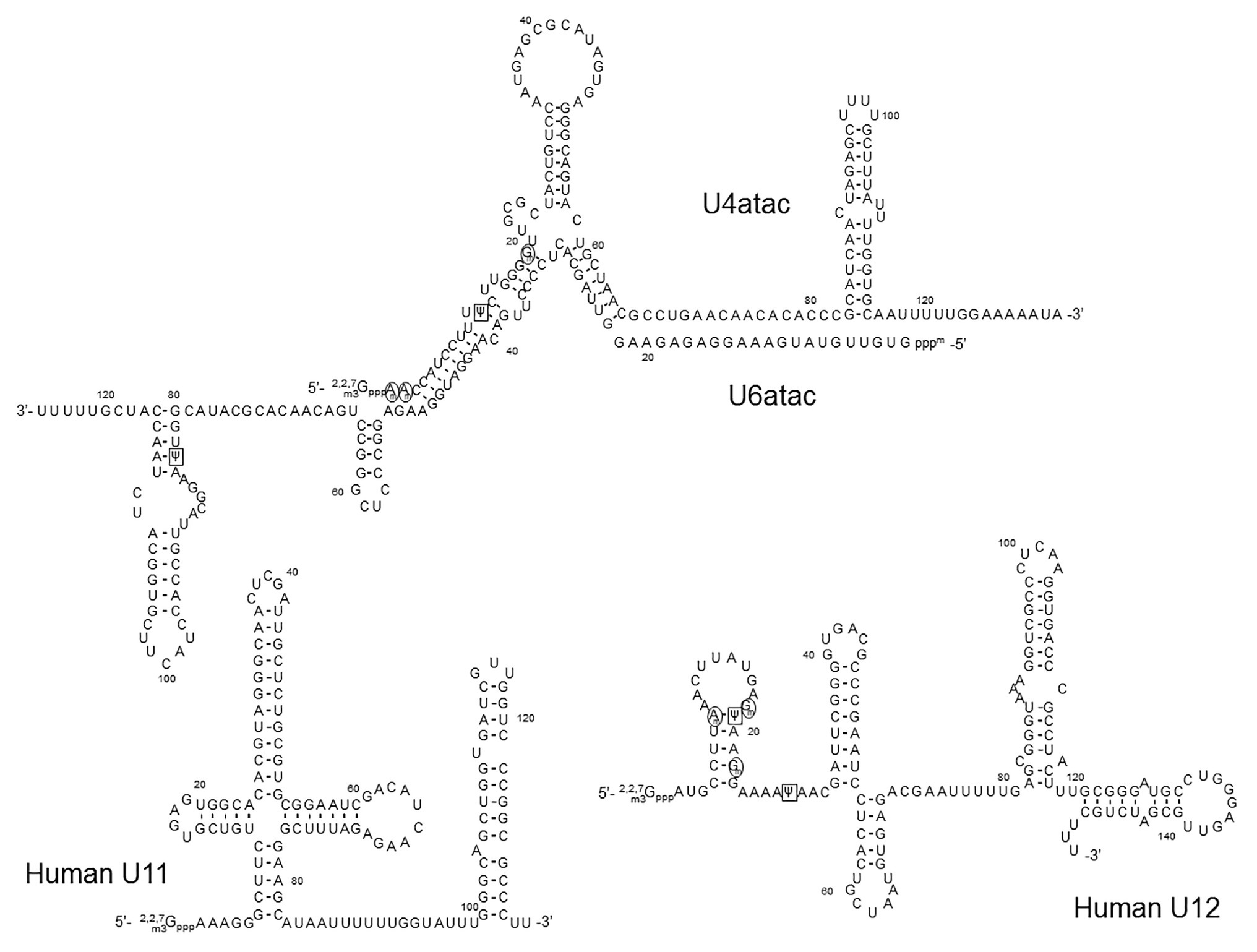
Figure 5. The primary sequences and the secondary structures of minor class snRNAs. The sequences and secondary structures of human U11, U12, U4atac, and U6atac snRNAs are shown. The sequences that recognize the 5' splice site of minor class introns (the U11 5' end and the U6atac sequences) and the branch site (the U12 branch site recognition sequence) are underlined. 2'-O-methylated nucleotides are circled (black circles), Ψs are boxed (black squares),
In an attempt to map Ψs in the minor spliceosome snRNAs in HeLa cells, Massenet and Branlant performed the pseudouridylation assay (CMC-modification followed by primer-extension) and identified Ψs in U12, U4atac, and U6atac snRNAs (Massenet and Branlant, 1999). Surprisingly, no Ψ was detected in U11 snRNA, although its major spliceosome counterpart U1 snRNA has two Ψs located in the 5'-end region that base-pairs with the 5'-splice site. It should be noted, however, that the 5' 10-nucleotide sequence (AAAAAGGGCU) of U11 snRNA (equivalent to the 5' 10-nucleotide sequence of U1 snRNA, AUACΨΨACCU) lacks the two U-residues (to be pseudouridylated) at the equivalent positions of U1 snRNA (Yu and Steitz, 1997). In the U12 snRNA, the authors detected only two Ψs at positions 19 and 28. Ψ19 is equivalent to Ψ34 of mammalian U2 snRNA that base-pairs with the nucleotide (A or G) within the branch site sequence that is immediately next to the bulged-out nucleotide (branch point adenosine), pointing toward a functional role of this Ψ. Ψ28 is located in a region that forms a base-pairing helix III with U6atac (equivalent to U2-U6 helix III), which has been shown to have a functional role in the splicing of U2-dependent introns (Sun and Manley, 1995). In the U4atac snRNA, a single Ψ was detected at position 12, located in the region that base-pairs with U6atac, equivalent to U4-U6 stem II in which there are several modified nucleotides in the U4 strand. Finally, Ψ83 was also identified in the U6atac snRNA 3'-end region. This Ψ could be functionally similar to Ψ86 in the U6 snRNA 3' terminal region. In a later study, these Ψs were all confirmed (Deryusheva et al., 2012). In addition, several 2'-O-methylations were also identified in U12 (at positions 8, 18, and 22) and U4atac snRNAs (at positions 1, 2, and 19, and potentially also position 8, although yet to be confirmed; Deryusheva et al., 2012). However, the exact function of these modified nucleotides remains unknown.
Understanding the splicing mechanisms at the molecular level is of critical importance not only to fully comprehend gene expression but also to develop new nucleic acid-based therapeutics, such as splice-switching oligonucleotides (Lim and Yokota, 2018), aimed at correcting splicing-associated mutations that lead to aberrant proteins and diseases. pre-mRNA splicing occurs in the spliceosome, an extremely large complex consisting of five snRNAs and a large number of proteins that interact with substrate pre-mRNA in a highly orchestrated manner. These snRNAs have a critical role in guiding the overall process via base-pairing interactions (and nucleotide-nucleotide contact) with the substrate pre-mRNA. Additionally, the snRNAs form dynamic structures that might be crucial for protein recruitment and catalysis. Post-transcriptionally modified nucleotides might contribute significantly in each of these steps during spliceosome assembly and splicing.
snRNA modifications, such as pseudouridine and 2'-O-methylation, have attracted a great deal of attention over the years, and extensive studies of these modifications have provided valuable insights into the mechanism of pre-mRNA splicing regulation. The continuous increase of knowledge of the fine-tuning and subtleties provided by RNA modifications in the spliceosome assembly and splicing processes are benefiting the development of better splicing modulation technologies. With the growing number of clinical trials based on splicing modulation therapies (exon-skipping or exon-inclusion) and the FDA-approved drugs based on this mechanism of action (Stein and Castanotto, 2017; Rüger et al., 2020), the interest in this field will certainly continue to grow. The novel deep sequencing chemical probing technologies and epitranscriptomics analytical techniques will help us to decipher the yet-to-be discovered code of spliceosomal RNA modifications.
PM, HA, and Y-TY wrote the manuscript and generated the figures. All authors read and approved the final manuscript.
The work performed in the Yu lab was supported by grants GM138387 and CA241111 from the US National Institutes of Health and grant CFF YU20G0 from the Cystic Fibrosis Foundation.
PM is a scientific director at ProQR Therapeutics.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We thank members of the Yu lab for insightful discussions.
Adachi, H., and Yu, Y. -T. (2014). Insight into the mechanisms and functions of spliceosomal snRNA pseudouridylation. World J. Biol. Chem. 5, 398–408. doi: 10.4331/wjbc.v5.i4.398
Agafonov, D. E., Kastner, B., Dybkov, O., Hofele, R. V., Liu, W. -T., Urlaub, H., et al. (2016). Molecular architecture of the human U4/U6.U5 tri-snRNP. Science 351, 1416–1420. doi: 10.1126/science.aad2085
Aoyama, T., Yamashita, S., and Tomita, K. (2020). Mechanistic insights into m6A modification of U6 snRNA by human METTL16. Nucleic Acids Res. 48, 5157–5168. doi: 10.1093/nar/gkaa227
Balakin, A. G., Smith, L., and Fournier, M. J. (1996). The RNA world of the nucleolus: two major families of small RNAs defined by different box elements with related functions. Cell 86, 823–834. doi: 10.1016/S0092-8674(00)80156-7
Basak, A., and Query, C. C. (2014). A pseudouridine residue in the spliceosome core is part of the filamentous growth program in yeast. Cell Rep. 8, 966–973. doi: 10.1016/j.celrep.2014.07.004
Berget, S. M., Moore, C., and Sharp, P. A. (1977). Spliced segments at the 5' terminus of adenovirus 2 late mRNA. Proc. Natl. Acad. Sci. U. S. A. 74, 3171–3175. doi: 10.1073/pnas.74.8.3171
Boehringer, D., Makarov, E. M., Sander, B., Makarova, O. V., Kastner, B., Lührmann, R., et al. (2004). Three-dimensional structure of a pre-catalytic human spliceosomal complex B. Nat. Struct. Mol. Biol. 11, 463–468. doi: 10.1038/nsmb761
Bohnsack, M. T., and Sloan, K. E. (2018). Modifications in small nuclear RNAs and their roles in spliceosome assembly and function. Biol. Chem. 399, 1265–1276. doi: 10.1515/hsz-2018-0205
Branlant, C., Krol, A., Ebel, J. P., Lazar, E., Gallinaro, H., and Jacob, M., et al. (1980). Nucleotide sequences of nuclear U1A RNAs from chicken, rat and man. Nucleic Acids Res. 8, 4143–4154. doi: 10.1093/nar/8.18.4143
Brow, D. A., and Guthrie, C. (1988). Spliceosomal RNA U6 is remarkably conserved from yeast to mammals. Nature 334, 213–218. doi: 10.1038/334213a0
Burke, J. E., Sashital, D. G., Zuo, X., Wang, Y. -X., and Butcher, S. E. (2012). Structure of the yeast U2/U6 snRNA complex. RNA 18, 673–683. doi: 10.1261/rna.031138.111
Caton, E. A., Kelly, E. K., Kamalampeta, R., and Kothe, U. (2018). Efficient RNA pseudouridylation by eukaryotic H/ACA ribonucleoproteins requires high affinity binding and correct positioning of guide RNA. Nucleic Acids Res. 46, 905–916. doi: 10.1093/nar/gkx1167
Cavaillé, J., Nicoloso, M., and Bachellerie, J. P. (1996). Targeted ribose methylation of RNA in vivo directed by tailored antisense RNA guides. Nature 383, 732–735. doi: 10.1038/383732a0
Chen, H., Gu, L., Orellana, E. A., Wang, Y., Guo, J., Liu, Q., et al. (2020). METTL4 is an snRNA m6Am methyltransferase that regulates RNA splicing. Cell Res. 30, 544–547. doi: 10.1038/s41422-019-0270-4
Chow, L. T., Gelinas, R. E., Broker, T. R., and Roberts, R. J. (1977). An amazing sequence arrangement at the 5' ends of adenovirus 2 messenger RNA. Cell 12, 1–8. doi: 10.1016/0092-8674(77)90180-5
Chu, H., Perea, W., and Greenbaum, N. L. (2020). Role of the central junction in folding topology of the protein-free human U2-U6 snRNA complex. RNA 26, 836–850. doi: 10.1261/rna.073379.119
Darzacq, X., Jády, B. E., Verheggen, C., Kiss, A. M., Bertrand, E., and Kiss, T. (2002). Cajal body-specific small nuclear RNAs: a novel class of 2’-O-methylation and pseudouridylation guide RNAs. EMBO J. 21, 2746–2756. doi: 10.1093/emboj/21.11.2746
Datta, B., and Weiner, A. M. (1991). Genetic evidence for base pairing between U2 and U6 snRNA in mammalian mRNA splicing. Nature 352, 821–824. doi: 10.1038/352821a0
Deryusheva, S., Choleza, M., Barbarossa, A., Gall, J. G., and Bordonne, R. (2012). Post-transcriptional modification of spliceosomal RNAs is normal in SMN-deficient cells. RNA 18, 31–36. doi: 10.1261/rna.030106.111
Deryusheva, S., and Gall, J. G. (2017). Dual nature of pseudouridylation in U2 snRNA: Pus1p-dependent and Pus1p-independent activities in yeasts and higher eukaryotes. RNA 23, 1060–1067. doi: 10.1261/rna.061226.117
Deryusheva, S., and Gall, J. G. (2018). Orchestrated positioning of post-transcriptional modifications at the branch point recognition region of U2 snRNA. RNA 24, 30–42. doi: 10.1261/rna.063842.117
Deryusheva, S., Talhouarne, G. J. S., and Gall, J. G. (2020). “Lost and Found”: snoRNA annotation in the xenopus genome and implications for evolutionary studies. Mol. Biol. Evol. 37, 149–166. doi: 10.1093/molbev/msz209
De Zoysa, M. D., Wu, G., Katz, R., and Yu, Y. -T. (2018). Guide-substrate base-pairing requirement for box H/ACA RNA-guided RNA pseudouridylation. RNA 24, 1106–1117. doi: 10.1261/rna.066837.118
De Zoysa, M. D., and Yu, Y. -T. (2017). Posttranscriptional RNA pseudouridylation. The Enzymes 41, 151–167. doi: 10.1016/bs.enz.2017.02.001
Dönmez, G., Hartmuth, K., and Lührmann, R. (2004). Modified nucleotides at the 5' end of human U2 snRNA are required for spliceosomal E-complex formation. RNA 10, 1925–1933. doi: 10.1261/rna.7186504
Epstein, P., Reddy, R., Henning, D., and Busch, H. (1980). The nucleotide sequence of nuclear U6 (4.7 S) RNA. J. Biol. Chem. 255, 8901–8906. doi: 10.1016/S0021-9258(18)43587-9
Fica, S. M., Tuttle, N., Novak, T., Li, N. -S., Lu, J., Koodathingal, P., et al. (2013). RNA catalyses nuclear pre-mRNA splicing. Nature 503, 229–234. doi: 10.1038/nature12734
Frank, D. N., Roiha, H., and Guthrie, C. (1994). Architecture of the U5 small nuclear RNA. Mol. Cell. Biol. 14, 2180–2190. doi: 10.1128/MCB.14.3.2180
Freund, M. (2003). A novel approach to describe a U1 snRNA binding site. Nucleic Acids Res. 31, 6963–6975. doi: 10.1093/nar/gkg901
Frye, M., Harada, B. T., Behm, M., and He, C. (2018). RNA modifications modulate gene expression during development. Science 361, 1346–1349. doi: 10.1126/science.aau1646
Ganot, P., Bortolin, M. L., and Kiss, T. (1997). Site-specific pseudouridine formation in preribosomal RNA is guided by small nucleolar RNAs. Cell 89, 799–809. doi: 10.1016/S0092-8674(00)80263-9
Ganot, P., Jády, B. E., Bortolin, M. L., Darzacq, X., and Kiss, T. (1999). Nucleolar factors direct the 2’-O-ribose methylation and pseudouridylation of U6 spliceosomal RNA. Mol. Cell. Biol. 19, 6906–6917. doi: 10.1128/MCB.19.10.6906
Ge, J., and Yu, Y. -T. (2013). RNA pseudouridylation: new insights into an old modification. Trends Biochem. Sci. 38, 210–218. doi: 10.1016/j.tibs.2013.01.002
Goh, Y. T., Koh, C. W. Q., Sim, D. Y., Roca, X., and Goh, W. S. S. (2020). METTL4 catalyzes m6Am methylation in U2 snRNA to regulate pre-mRNA splicing. Nucleic Acids Res. 48, 9250–9261. doi: 10.1093/nar/gkaa684
Grosjean, H., Sprinzl, M., and Steinberg, S. (1995). Posttranscriptionally modified nucleosides in transfer RNA: their locations and frequencies. Biochimie 77, 139–141. doi: 10.1016/0300-9084(96)88117-X
Gu, J., Patton, J. R., Shimba, S., and Reddy, R. (1996). Localization of modified nucleotides in Schizosaccharomyces pombe spliceosomal small nuclear RNAs: modified nucleotides are clustered in functionally important regions. RNA 2, 909–918.
Gu, A. -D., Zhou, H., Yu, C. -H., and Qu, L. -H. (2005). A novel experimental approach for systematic identification of box H/ACA snoRNAs from eukaryotes. Nucleic Acids Res. 33:e194. doi: 10.1093/nar/gni185
Hirose, T., Ideue, T., Nagai, M., Hagiwara, M., Shu, M. -D., and Steitz, J. A. (2006). A spliceosomal intron binding protein, IBP160, links position-dependent assembly of intron-encoded box C/D snoRNP to pre-mRNA splicing. Mol. Cell 23, 673–684. doi: 10.1016/j.molcel.2006.07.011
Hirose, T., Shu, M. -D., and Steitz, J. A. (2003). Splicing-dependent and -independent modes of assembly for intron-encoded box C/D snoRNPs in mammalian cells. Mol. Cell 12, 113–123. doi: 10.1016/S1097-2765(03)00267-3
Hüttenhofer, A., Kiefmann, M., Meier-Ewert, S., O’Brien, J., Lehrach, H., and Bachellerie, J. P., et al. (2001). RNomics: an experimental approach that identifies 201 candidates for novel, small, non-messenger RNAs in mouse. EMBO J. 20, 2943–2953. doi: 10.1093/emboj/20.11.2943
Jády, B. E., and Kiss, T. (2001). A small nucleolar guide RNA functions both in 2’-O-ribose methylation and pseudouridylation of the U5 spliceosomal RNA. EMBO J. 20, 541–551. doi: 10.1093/emboj/20.3.541
Jorjani, H., Kehr, S., Jedlinski, D. J., Gumienny, R., Hertel, J., Stadler, P. F., et al. (2016). An updated human snoRNAome. Nucleic Acids Res. 44, 5068–5082. doi: 10.1093/nar/gkw386
Jurica, M. S., and Moore, M. J. (2003). Pre-mRNA splicing: awash in a sea of proteins. Mol. Cell 12, 5–14. doi: 10.1016/S1097-2765(03)00270-3
Jurica, M. S., Sousa, D., Moore, M. J., and Grigorieff, N. (2004). Three-dimensional structure of C complex spliceosomes by electron microscopy. Nat. Struct. Mol. Biol. 11, 265–269. doi: 10.1038/nsmb728
Karijolich, J., and Yu, Y. -T. (2010). Spliceosomal snRNA modifications and their function. RNA Biol. 7, 192–204. doi: 10.4161/rna.7.2.11207
Karunatilaka, K. S., and Rueda, D. (2014). Post-transcriptional modifications modulate conformational dynamics in human U2-U6 snRNA complex. RNA 20, 16–23. doi: 10.1261/rna.041806.113
Kennedy, S. D., Bauer, W. J., Wang, W., and Kielkopf, C. L. (2019). Dynamic stacking of an expected branch point adenosine in duplexes containing pseudouridine-modified or unmodified U2 snRNA sites. Biochem. Biophys. Res. Commun. 511, 416–421. doi: 10.1016/j.bbrc.2019.02.073
Kiss, A. M., Jády, B. E., Bertrand, E., and Kiss, T. (2004). Human box H/ACA pseudouridylation guide RNA machinery. Mol. Cell. Biol. 24, 5797–5807. doi: 10.1128/MCB.24.13.5797-5807.2004
Kiss, A. M., Jády, B. E., Darzacq, X., Verheggen, C., Bertrand, E., and Kiss, T. (2002). A Cajal body-specific pseudouridylation guide RNA is composed of two box H/ACA snoRNA-like domains. Nucleic Acids Res. 30, 4643–4649. doi: 10.1093/nar/gkf592
Kiss-László, Z., Henry, Y., Bachellerie, J. P., Caizergues-Ferrer, M., and Kiss, T. (1996). Site-specific ribose methylation of preribosomal RNA: a novel function for small nucleolar RNAs. Cell 85, 1077–1088. doi: 10.1016/S0092-8674(00)81308-2
Kondo, Y., Oubridge, C., van Roon, A. -M. M., and Nagai, K. (2015). Crystal structure of human U1 snRNP, a small nuclear ribonucleoprotein particle, reveals the mechanism of 5' splice site recognition. eLife 4:e04986. doi: 10.7554/eLife.04986
Krawczak, M., Thomas, N. S. T., Hundrieser, B., Mort, M., Wittig, M., Hampe, J., et al. (2007). Single base-pair substitutions in exon-intron junctions of human genes: nature, distribution, and consequences for mRNA splicing. Hum. Mutat. 28, 150–158. doi: 10.1002/humu.20400
Krogh, N., Kongsbak-Wismann, M., Geisler, C., and Nielsen, H. (2017). Substoichiometric ribose methylations in spliceosomal snRNAs. Org. Biomol. Chem. 15, 8872–8876. doi: 10.1039/C7OB02317K
Krol, A., Gallinaro, H., Lazar, E., Jacob, M., and Branlant, C. (1981). The nuclear 5S RNAs from chicken, rat and man. U5 RNAs are encoded by multiple genes. Nucleic Acids Res. 9, 769–787. doi: 10.1093/nar/9.4.769
Lestrade, L., and Weber, M. J. (2006). snoRNA-LBME-db, a comprehensive database of human H/ACA and C/D box snoRNAs. Nucleic Acids Res. 34, D158–D162. doi: 10.1093/nar/gkj002
Lim, K. R. Q., and Yokota, T. (2018). Invention and early history of exon skipping and splice modulation. Methods Mol. Biol. 1828, 3–30. doi: 10.1007/978-1-4939-8651-4_1
Ma, X., Yang, C., Alexandrov, A., Grayhack, E. J., Behm-Ansmant, I., and Yu, Y. -T. (2005). Pseudouridylation of yeast U2 snRNA is catalyzed by either an RNA-guided or RNA-independent mechanism. EMBO J. 24, 2403–2413. doi: 10.1038/sj.emboj.7600718
Ma, X., Zhao, X., and Yu, Y. -T. (2003). Pseudouridylation (psi) of U2 snRNA in S. cerevisiae is catalyzed by an RNA-independent mechanism. EMBO J. 22, 1889–1897. doi: 10.1093/emboj/cdg191
Madhani, H. D., Bordonné, R., and Guthrie, C. (1990). Multiple roles for U6 snRNA in the splicing pathway. Genes Dev. 4, 2264–2277. doi: 10.1101/gad.4.12b.2264
Madhani, H. D., and Guthrie, C. (1992). A novel base-pairing interaction between U2 and U6 snRNAs suggests a mechanism for the catalytic activation of the spliceosome. Cell 71, 803–817. doi: 10.1016/0092-8674(92)90556-R
Massenet, S., and Branlant, C. (1999). A limited number of pseudouridine residues in the human atac spliceosomal UsnRNAs as compared to human major spliceosomal UsnRNAs. RNA 5, 1495–1503. doi: 10.1017/S1355838299991537
Massenet, S., Motorin, Y., Lafontaine, D. L. J., Hurt, E. C., Grosjean, H., and Branlant, C. (1999). Pseudouridine mapping in the Saccharomyces cerevisiae spliceosomal U small nuclear RNAs (snRNAs) reveals that pseudouridine synthase Pus1p exhibits a dual substrate specificity for U2 snRNA and tRNA. Mol. Cell. Biol. 19, 2142–2154. doi: 10.1128/MCB.19.3.2142
Massenet, S., Mougin, A., and Branlant, C. (1998). “Posttranscriptional modifications in the U small nuclear RNAs” in Modification and Editingof RNA. ed. H. Grosjean (Washington, DC: ASM Press), 201–228.
Mauer, J., Sindelar, M., Despic, V., Guez, T., Hawley, B. R., Vasseur, J. -J., et al. (2019). FTO controls reversible m6Am RNA methylation during snRNA biogenesis. Nat. Chem. Biol. 15, 340–347. doi: 10.1038/s41589-019-0231-8
Meier, U. T. (2017). RNA modification in Cajal bodies. RNA Biol. 14, 693–700. doi: 10.1080/15476286.2016.1249091
Montzka, K. A., and Steitz, J. A. (1988). Additional low-abundance human small nuclear ribonucleoproteins: U11, U12, etc. Proc. Natl. Acad. Sci. 85, 8885–8889. doi: 10.1073/pnas.85.23.8885
Motorin, Y., Keith, G., Simon, C., Foiret, D., Simos, G., Hurt, E., et al. (1998). The yeast tRNA:pseudouridine synthase Pus1p displays a multisite substrate specificity. RNA 4, 856–869. doi: 10.1017/S1355838298980396
Nachtergaele, S., and He, C. (2017). The emerging biology of RNA post-transcriptional modifications. RNA Biol. 14, 156–163. doi: 10.1080/15476286.2016.1267096
Newby, M. I., and Greenbaum, N. L. (2001). A conserved pseudouridine modification in eukaryotic U2 snRNA induces a change in branch-site architecture. RNA N. Y. N 7, 833–845. doi: 10.1017/S1355838201002308
Newby, M. I., and Greenbaum, N. L. (2002). Sculpting of the spliceosomal branch site recognition motif by a conserved pseudouridine. Nat. Struct. Biol. 9, 958–965. doi: 10.1038/nsb873
Newman, A. J. (1997). The role of U5 snRNP in pre-mRNA splicing. EMBO J. 16, 5797–5800. doi: 10.1093/emboj/16.19.5797
Newman, A. J., and Norman, C. (1992). U5 snRNA interacts with exon sequences at 5' and 3' splice sites. Cell 68, 743–754. doi: 10.1016/0092-8674(92)90149-7
Newman, A. J., Teigelkamp, S., and Beggs, J. D. (1995). snRNA interactions at 5' and 3' splice sites monitored by photoactivated crosslinking in yeast spliceosomes. RNA 1, 968–980.
Nguyen, T. H. D., Galej, W. P., Bai, X. -C., Oubridge, C., Newman, A. J., Scheres, S. H. W., et al. (2016). Cryo-EM structure of the yeast U4/U6.U5 tri-snRNP at 3.7 Å resolution. Nature 530, 298–302. doi: 10.1038/nature16940
Nguyen, T. H. D., Galej, W. P., Bai, X., Savva, C. G., Newman, A. J., Scheres, S. H. W., et al. (2015). The architecture of the spliceosomal U4/U6.U5 tri-snRNP. Nature 523, 47–52. doi: 10.1038/nature14548
Ni, J., Samarsky, D. A., Liu, B., Ferbeyre, G., Cedergren, R., and Fournier, M. J. (1997). SnoRNAs as tools for RNA cleavage and modification. Nucleic Acids Symp. Ser. 36, 61–63.
Nilsen, T. W. (1994). RNA-RNA interactions in the spliceosome: unraveling the ties that bind. Cell 78, 1–4. doi: 10.1016/0092-8674(94)90563-0
Nilsen, T. W. (2003). The spliceosome: the most complex macromolecular machine in the cell? BioEssays 25, 1147–1149. doi: 10.1002/bies.10394.
Parker, R., Siliciano, P. G., and Guthrie, C. (1987). Recognition of the TACTAAC box during mRNA splicing in yeast involves base pairing to the U2-like snRNA. Cell 49, 229–239. doi: 10.1016/0092-8674(87)90564-2
Pendleton, K. E., Chen, B., Liu, K., Hunter, O. V., Xie, Y., and Tu, B. P., et al. (2017). The U6 snRNA m 6 a methyltransferase METTL16 regulates SAM synthetase intron retention. Cell 169, 824.e14–835.e14. doi: 10.1016/j.cell.2017.05.003
Plaschka, C., Lin, P. -C., and Nagai, K. (2017). Structure of a pre-catalytic spliceosome. Nature 546, 617–621. doi: 10.1038/nature22799
Query, C. C., McCaw, P. S., and Sharp, P. A. (1997). A minimal spliceosomal complex a recognizes the branch site and polypyrimidine tract. Mol. Cell. Biol. 17, 2944–2953. doi: 10.1128/MCB.17.5.2944
Raghunathan, P. L., and Guthrie, C. (1998). RNA unwinding in U4/U6 snRNPs requires ATP hydrolysis and the DEIH-box splicing factor Brr2. Curr. Biol. 8, 847–855. doi: 10.1016/S0960-9822(07)00345-4
Reddy, R., and Busch, H. (1988). “Small nuclear RNAs: RNA sequences, structure, and modifications” in Structure and function of major and minor small nuclear ribonucleoprotein particles. ed. M. L. Birnsteil (Heidelberg: Springer-Verlag Press), 1–37.
Reddy, R., Henning, D., and Busch, H. (1981a). Pseudouridine residues in the 5'-terminus of uridine-rich nuclear RNA I (U1 RNA). Biochem. Biophys. Res. Commun. 98, 1076–1083. doi: 10.1016/0006-291x(81)91221-3
Reddy, R., Henning, D., and Busch, H. (1981b). The primary nucleotide sequence of U4 RNA. J. Biol. Chem. 256, 3532–3538.
Rife, J. P., Cheng, C. S., Moore, P. B., and Strobel, S. A. (1998). N2-Methylguanosine is iso-energetic with guanosine in RNA duplexes and GNRA tetraloops. Nucleic Acids Res. 26, 3640–3644. doi: 10.1093/nar/26.16.3640
Rintala-Dempsey, A. C., and Kothe, U. (2017). Eukaryotic stand-alone pseudouridine synthases – RNA modifying enzymes and emerging regulators of gene expression? RNA Biol. 14, 1185–1196. doi: 10.1080/15476286.2016.1276150
Roca, X. (2005). Determinants of the inherent strength of human 5' splice sites. RNA 11, 683–698. doi: 10.1261/rna.2040605
Roca, X., Akerman, M., Gaus, H., Berdeja, A., Bennett, C. F., and Krainer, A. R. (2012). Widespread recognition of 5' splice sites by noncanonical base-pairing to U1 snRNA involving bulged nucleotides. Genes Dev. 26, 1098–1109. doi: 10.1101/gad.190173.112
Roundtree, I. A., Evans, M. E., Pan, T., and He, C. (2017). Dynamic RNA modifications in gene expression regulation. Cell 169, 1187–1200. doi: 10.1016/j.cell.2017.05.045
Rüger, J., Ioannou, S., Castanotto, D., and Stein, C. A. (2020). Oligonucleotides to the (gene) rescue: FDA approvals 2017-2019. Trends Pharmacol. Sci. 41, 27–41. doi: 10.1016/j.tips.2019.10.009
Ruskin, B., Krainer, A. R., Maniatis, T., and Green, M. R. (1984). Excision of an intact intron as a novel lariat structure during pre-mRNA splicing in vitro. Cell 38, 317–331. doi: 10.1016/0092-8674(84)90553-1
Ruskin, B., Zamore, P. D., and Green, M. R. (1988). A factor, U2AF, is required for U2 snRNP binding and splicing complex assembly. Cell 52, 207–219. doi: 10.1016/0092-8674(88)90509-0
Schattner, P., Barberan-Soler, S., and Lowe, T. M. (2006). A computational screen for mammalian pseudouridylation guide H/ACA RNAs. RNA 12, 15–25. doi: 10.1261/rna.2210406
Schwartz, S., Bernstein, D. A., Mumbach, M. R., Jovanovic, M., Herbst, R. H., León-Ricardo, B. X., et al. (2014). Transcriptome-wide mapping reveals widespread dynamic-regulated pseudouridylation of ncRNA and mRNA. Cell 159, 148–162. doi: 10.1016/j.cell.2014.08.028
Scotti, M. M., and Swanson, M. S. (2016). RNA mis-splicing in disease. Nat. Rev. Genet. 17, 19–32. doi: 10.1038/nrg.2015.3
Sergiev, P. V., Lesnyak, D. V., Bogdanov, A. A., and Dontsova, O. A. (2006). Identification of Escherichia coli m2G methyltransferases: II. The ygjO gene encodes a methyltransferase specific for G1835 of the 23 S rRNA. J. Mol. Biol. 364, 26–31. doi: 10.1016/j.jmb.2006.09.008
Shi, Y. (2017). Mechanistic insights into precursor messenger RNA splicing by the spliceosome. Nat. Rev. Mol. Cell Biol. 18, 655–670. doi: 10.1038/nrm.2017.86
Shibata, H., Ro-Choi, T. S., Reddy, R., Choi, Y. C., Henning, D., and Busch, H. (1975). The primary nucleotide sequence of nuclear U-2 ribonucleic acid. The 5'-terminal portion of the molecule. J. Biol. Chem. 250, 3909–3920. doi: 10.1016/S0021-9258(19)41485-3
Shimba, S., Bokar, J. A., Rottman, F., and Reddy, R. (1995). Accurate and efficient N-6-adenosine methylation in spliceosomal U6 small nuclear RNA by HeLa cell extract in vitro. Nucleic Acids Res. 23, 2421–2426. doi: 10.1093/nar/23.13.2421
Simos, G., Tekotte, H., Grosjean, H., Segref, A., Sharma, K., Tollervey, D., et al. (1996). Nuclear pore proteins are involved in the biogenesis of functional tRNA. EMBO J. 15, 2270–2284. doi: 10.1002/j.1460-2075.1996.tb00580.x
Sindhuphak, T., Hellman, U., and Svensson, I. (1985). Site specificities of three transfer RNA methyltransferases from yeast. Biochim. Biophys. Acta 824, 66–73. doi: 10.1016/0167-4781(85)90030-2
Singh, R., and Reddy, R. (1989). Gamma-monomethyl phosphate: a cap structure in spliceosomal U6 small nuclear RNA. Proc. Natl. Acad. Sci. U. S. A. 86, 8280–8283. doi: 10.1073/pnas.86.21.8280
Sontheimer, E. J., and Steitz, J. A. (1993). The U5 and U6 small nuclear RNAs as active site components of the spliceosome. Science 262, 1989–1996. doi: 10.1126/science.8266094
Staley, J. P., and Guthrie, C. (1998). Mechanical devices of the spliceosome: motors, clocks, springs, and things. Cell 92, 315–326. doi: 10.1016/S0092-8674(00)80925-3
Stein, C. A., and Castanotto, D. (2017). FDA-approved oligonucleotide therapies in 2017. Mol. Ther. J. Am. Soc. Gene Ther. 25, 1069–1075. doi: 10.1016/j.ymthe.2017.03.023
Sun, J. S., and Manley, J. L. (1995). A novel U2-U6 snRNA structure is necessary for mammalian mRNA splicing. Genes Dev. 9, 843–854. doi: 10.1101/gad.9.7.843
Szkukalek, A., Myslinski, E., Mougin, A., Luhrmann, R., and Branlant, C. (1995). Phylogenetic conservation of modified nucleotides in the terminal loop 1 of the spliceosomal U5 snRNA. Biochimie 77, 16–21. doi: 10.1016/0300-9084(96)88099-0
Tarn, W. Y., and Steitz, J. A. (1996). A novel spliceosome containing U11, U12, and U5 snRNPs excises a minor class (AT-AC) intron in vitro. Cell 84, 801–811. doi: 10.1016/S0092-8674(00)81057-0
Tarn, W. Y., and Steitz, J. A. (1997). Pre-mRNA splicing: the discovery of a new spliceosome doubles the challenge. Trends Biochem. Sci. 22, 132–137. doi: 10.1016/S0968-0004(97)01018-9
Terns, M., and Terns, R. (2006). Noncoding RNAs of the H/ACA family. Cold Spring Harb. Symp. Quant. Biol. 71, 395–405. doi: 10.1101/sqb.2006.71.034
Turunen, J. J., Niemelä, E. H., Verma, B., and Frilander, M. J. (2013). The significant other: splicing by the minor spliceosome. Wiley Interdiscip. Rev. RNA 4, 61–76. doi: 10.1002/wrna.1141
Tycowski, K. T., Aab, A., and Steitz, J. A. (2004). Guide RNAs with 5' caps and novel box C/D snoRNA-like domains for modification of snRNAs in metazoa. Curr. Biol. CB 14, 1985–1995. doi: 10.1016/j.cub.2004.11.003
Tycowski, K. T., Shu, M. D., and Steitz, J. A. (1996). A mammalian gene with introns instead of exons generating stable RNA products. Nature 379, 464–466. doi: 10.1038/379464a0
Tycowski, K. T., You, Z. H., Graham, P. J., and Steitz, J. A. (1998). Modification of U6 spliceosomal RNA is guided by other small RNAs. Mol. Cell 2, 629–638. doi: 10.1016/S1097-2765(00)80161-6
Urban, A., Behm-Ansmant, I., Branlant, C., and Motorin, Y. (2009). RNA sequence and two-dimensional structure features required for efficient substrate modification by the Saccharomyces cerevisiae RNA:{psi}-synthase Pus7p. J. Biol. Chem. 284, 5845–5858. doi: 10.1074/jbc.M807986200
van der Feltz, C., DeHaven, A. C., and Hoskins, A. A. (2018). Stress-induced pseudouridylation alters the structural equilibrium of yeast U2 snRNA stem II. J. Mol. Biol. 430, 524–536. doi: 10.1016/j.jmb.2017.10.021
Vitali, P., Royo, H., Seitz, H., Bachellerie, J. -P., Hüttenhofer, A., and Cavaillé, J. (2003). Identification of 13 novel human modification guide RNAs. Nucleic Acids Res. 31, 6543–6551. doi: 10.1093/nar/gkg849
Wahl, M. C., Will, C. L., and Lührmann, R. (2009). The spliceosome: design principles of a dynamic RNP machine. Cell 136, 701–718. doi: 10.1016/j.cell.2009.02.009
Wan, R., Yan, C., Bai, R., Wang, L., Huang, M., Wong, C. C. L., et al. (2016). The 3.8 Å structure of the U4/U6.U5 tri-snRNP: insights into spliceosome assembly and catalysis. Science 351, 466–475. doi: 10.1126/science.aad6466
Warda, A. S., Kretschmer, J., Hackert, P., Lenz, C., Urlaub, H., and Höbartner, C., et al. (2017). Human METTL16 is a N6-methyladenosine (m6A) methyltransferase that targets pre-mRNAs and various non-coding RNAs. EMBO Rep. 18, 2004–2014. doi: 10.15252/embr.201744940
Wassarman, D. A., and Steitz, J. A. (1992). Interactions of small nuclear RNA’s with precursor messenger RNA during in vitro splicing. Science 257, 1918–1925. doi: 10.1126/science.1411506
Wiener, D., and Schwartz, S. (2020). The epitranscriptome beyond m6A. Nat. Rev. Genet. 22, 119–131. doi: 10.1038/s41576-020-00295-8
Will, C. L., and Lührmann, R. (2005). Splicing of a rare class of introns by the U12-dependent spliceosome. Biol. Chem. 386, 713–724. doi: 10.1515/BC.2005.084
Will, C. L., and Lührmann, R. (2011). Spliceosome structure and function. Cold Spring Harb. Perspect. Biol. 3:a003707. doi: 10.1101/cshperspect.a003707
Will, C. L., Rümpler, S., and Klein Gunnewiek, J., van Venrooij, W. J., and Lührmann, R. (1996). In vitro reconstitution of mammalian U1 snRNPs active in splicing: the U1-C protein enhances the formation of early (E) spliceosomal complexes. Nucleic Acids Res. 24, 4614–4623. doi: 10.1093/nar/24.23.4614
Wu, G., Adachi, H., Ge, J., Stephenson, D., Query, C. C., and Yu, Y. (2016a). Pseudouridines in U2 snRNA stimulate the ATPase activity of Prp5 during spliceosome assembly. EMBO J. 35, 654–667. doi: 10.15252/embj.201593113
Wu, J. A., and Manley, J. L. (1991). Base pairing between U2 and U6 snRNAs is necessary for splicing of a mammalian pre-mRNA. Nature 352, 818–821. doi: 10.1038/352818a0
Wu, G., Radwan, M. K., Xiao, M., Adachi, H., Fan, J., and Yu, Y. -T. (2016b). The TOR signaling pathway regulates starvation-induced pseudouridylation of yeast U2 snRNA. RNA 22, 1146–1152. doi: 10.1261/rna.056796.116
Wu, S., Romfo, C. M., Nilsen, T. W., and Green, M. R. (1999). Functional recognition of the 3' splice site AG by the splicing factor U2AF35. Nature 402, 832–835. doi: 10.1038/45590
Wu, G., Xiao, M., Yang, C., and Yu, Y. -T. (2011). U2 snRNA is inducibly pseudouridylated at novel sites by Pus7p and snR81 RNP. EMBO J. 30, 79–89. doi: 10.1038/emboj.2010.316
Wyatt, J. R., Sontheimer, E. J., and Steitz, J. A. (1992). Site-specific cross-linking of mammalian U5 snRNP to the 5' splice site before the first step of pre-mRNA splicing. Genes Dev. 6, 2542–2553. doi: 10.1101/gad.6.12b.2542
Yan, C., Wan, R., Bai, R., Huang, G., and Shi, Y. (2017). Structure of a yeast step II catalytically activated spliceosome. Science 355, 149–155. doi: 10.1126/science.aak9979
Yang, C., McPheeters, D. S., and Yu, Y. -T. (2005). ψ35 in the branch site recognition region of U2 small nuclear RNA is important for pre-mRNA splicing in Saccharomyces cerevisiae. J. Biol. Chem. 280, 6655–6662. doi: 10.1074/jbc.M413288200
Yean, S. L., Wuenschell, G., Termini, J., and Lin, R. J. (2000). Metal-ion coordination by U6 small nuclear RNA contributes to catalysis in the spliceosome. Nature 408, 881–884. doi: 10.1038/35048617
Yu, A. T., Ge, J., and Yu, Y. -T. (2011). Pseudouridines in spliceosomal snRNAs. Protein Cell 2, 712–725. doi: 10.1007/s13238-011-1087-1
Yu, Y. T., Shu, M. D., and Steitz, J. A. (1998). Modifications of U2 snRNA are required for snRNP assembly and pre-mRNA splicing. EMBO J. 17, 5783–5795. doi: 10.1093/emboj/17.19.5783
Yu, Y. T., and Steitz, J. A. (1997). Site-specific crosslinking of mammalian U11 and u6atac to the 5' splice site of an AT-AC intron. Proc. Natl. Acad. Sci. U. S. A. 94, 6030–6035. doi: 10.1073/pnas.94.12.6030
Yu, Y. -T., Terns, R. M., and Terns, M. P. (2005). “Mechanisms and functions of RNA-guided RNA modification” in Fine-tuning of RNA functions by modification and editing. ed. H. Grosjean (New York: Springer-Verlag Press), 223–262.
Zamore, P. D., Patton, J. G., and Green, M. R. (1992). Cloning and domain structure of the mammalian splicing factor U2AF. Nature 355, 609–614. doi: 10.1038/355609a0
Zerby, D. B., and Patton, J. R. (1997). Modification of human U4 RNA requires U6 RNA and multiple pseudouridine synthases. Nucleic Acids Res. 25, 4808–4815. doi: 10.1093/nar/25.23.4808
Zhang, X., Yan, C., Zhan, X., Li, L., Lei, J., and Shi, Y. (2018). Structure of the human activated spliceosome in three conformational states. Cell Res. 28, 307–322. doi: 10.1038/cr.2018.14
Zhang, M., Zamore, P. D., Carmo-Fonseca, M., Lamond, A. I., and Green, M. R. (1992). Cloning and intracellular localization of the U2 small nuclear ribonucleoprotein auxiliary factor small subunit. Proc. Natl. Acad. Sci. U. S. A. 89, 8769–8773. doi: 10.1073/pnas.89.18.8769
Zhao, X., and Yu, Y. -T. (2004). Detection and quantitation of RNA base modifications. RNA 10, 996–1002. doi: 10.1261/rna.7110804
Zhuang, Y. A., Goldstein, A. M., and Weiner, A. M. (1989). UACUAAC is the preferred branch site for mammalian mRNA splicing. Proc. Natl. Acad. Sci. U. S. A. 86, 2752–2756. doi: 10.1073/pnas.86.8.2752
Keywords: pre-mRNA splicing, small nuclear RNA, RNA modifications, epitranscriptomics, pseudouridine, 2'-O-methylation, N6-methyladenosine, N2-methylation
Citation: Morais P, Adachi H and Yu Y-T (2021) Spliceosomal snRNA Epitranscriptomics. Front. Genet. 12:652129. doi: 10.3389/fgene.2021.652129
Received: 11 January 2021; Accepted: 08 February 2021;
Published: 02 March 2021.
Edited by:
Jia Meng, Xi’an Jiaotong-Liverpool University, ChinaReviewed by:
Piyush Khandelia, Birla Institute of Technology and Science, India Olivier Bensaude, École Normale Supérieure, FranceCopyright © 2021 Morais, Adachi and Yu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yi-Tao Yu, eWl0YW9feXVAdXJtYy5yb2NoZXN0ZXIuZWR1; Pedro Morais, cG1vcmFpc0Bwcm9xci5jb20=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.