
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Genet. , 20 May 2021
Sec. Cancer Genetics
Volume 12 - 2021 | https://doi.org/10.3389/fgene.2021.651882
 Guangjin Chen1,2,3†
Guangjin Chen1,2,3† Jiwei Sun1,2,3†
Jiwei Sun1,2,3† Mengru Xie1,2,3
Mengru Xie1,2,3 Shaoling Yu1,2,3
Shaoling Yu1,2,3 Qingming Tang1,2,3*
Qingming Tang1,2,3* Lili Chen1,2,3*
Lili Chen1,2,3*Plasminogen activator, urokinase (uPA) is a secreted serine protease whose Dysregulation is often accompanied by various cancers. However, the biological functions and potential mechanisms of PLAU in head and neck squamous cell carcinoma (HNSCC) remain undetermined. Here, the expression, prognosis, function, and coexpression genetic networks of PLAU in HNSCC were investigated by a series of public bioinformatics tools. A Higher PLAU level predicted a poorer clinical outcome. Meanwhile, functional network analysis implied that PLAU and associated genes mainly regulated cell-substrate adhesion, tissue migration, and extracellular matrix binding. The top 4 significantly associated genes are C10orf55, ITGA5, SERPINE1, and TNFRSF12A. Pathway enrichment analysis indicated that PLAU might activate the epithelial-to-mesenchymal transition (EMT) process, which could explain the poor prognosis in HNSCC. Besides, genes associated with PLAU were also enriched in EMT pathways. We further validated the bioinformatics analysis results by in vivo and in vitro experiments. Then, we found that much more PLAU was detected in HNSCC tissues, and the silencing of PLAU inhibit the proliferation, migration, and EMT process of CAL27 cell lines. Notably, the downregulation of PLAU decreased the expression of TNFRSF12A. Moreover, knockdown TNFRSF12A also inhibits cell proliferation and migration. In vivo experiment results indicated that PLAU inhibition could suppress tumor growth. Collectively, PLAU is necessary for tumor progression and can be a diagnostic and prognostic biomarker in HNSCC.
Head and neck cancer derives from the oral cavity, pharynx, and other upper aerodigestive tracts, in which HNSCC is the most common one (Mehanna et al., 2010). Carcinogenic factors of HNSCC contain smoking and alcohol as well as human papillomavirus (HPV) infection (Chow, 2020). Due to the high incidence of HNSCC and most HNCSS patients present a tendency of recurrence or distant metastasis, the prognosis of HNSCC patients is poor (Argiris et al., 2008). Certain mechanisms and pathways for tumor formation have been identified for targeted drug development, such as EGFR inhibitors. However, the benefit of existing drugs is dissatisfactory (Sacco and Cohen, 2015). Thus, the identification of new drug targets for the therapy and prognosis of HNSCC seems urgent.
PLAU encodes a secreted serine protease uPA (Mekkawy et al., 2014). uPA belongs to the plasminogen activator (PA) system and it serves to convert plasminogen to plasmin which is a broad-spectrum protease (Dano et al., 2005). Thus, uPA is involved in the basement membrane and extracellular matrix degradation process which counts for tumor invasion and metastasis (Mekkawy et al., 2014). Also, accumulating evidence indicated that the uPA system could participate in cell proliferation/apoptosis and EMT via other signaling pathways (Lester et al., 2007; Jo et al., 2009). Increasing PLAU expression was detected in various tumors, which is associated with patient survival, thus the uPA system could be a biomarker in malignancies (LeBeau et al., 2015; Su et al., 2016; Mahmood et al., 2018; Novak et al., 2019). However, the function of PLAU and specific mechanisms in HNSCC has not been determined till now.
We studied PLAU expression, mutation, and prognosis value in data from HNSCC patients with various open-access databases. We perform multi-dimensional analysis of genomic alteration and functional networks related to PLAU in HNSCC. Besides, we uncovered the biological function of PLAU in HNSCC through multiple experiments. Our results could confirm PLAU as a diagnosis and treatment target for HNSCC.
Oncomine1 is the largest cancer microarray database (Rhodes et al., 2004). We used Oncomine to analyze the mRNA levels of PLAU between HNSCC and adjacent normal tissues. We focused on several HNSCC studies, including Peng Head-Neck, Ginos Head-Neck, Estilo Head-Neck, Sengupta Head-Neck, Ye Head-Neck, Pyeon Multi-cancer (Ginos et al., 2004; Sengupta et al., 2006; Pyeon et al., 2007; Ye et al., 2008; Estilo et al., 2009; Peng et al., 2011).
UANLCAN2 is a web tool based on TCGA (The Cancer Genome Atlas) datasets (Chandrashekar et al., 2017). The analysis of PLAU expression in HNSCC and other cancer sub-groups including gender, ages, stages, or other features was conducted with UANLCAN. The survival probability was also analyzed by the UANLCAN database.
The genetic profile of PLAU in HNSCC was obtained by cBioPortal3 (Gao et al., 2013). The Oncoprint module showed the genetic alteration of PLAU in HNSCC.
GEPIA4 is a customized cancer database (Tang et al., 2017). We used GEPIA to visualize the analysis of specific genes from HNSC samples.
linkedOmics5 integrates multi-omics data from TCGA, which contains three analysis modules (Vasaikar et al., 2018). The PLAU association result in HNSCC (n = 520) was explored by the LinkFinder tab which used Pearson’s correlation coefficient. Then we used Gene Set Enrichment Analysis (GSEA) in the LinkInterpreter module to obtain the analysis of GO, KEGG pathways enrichment. The rank criteria were P-value, minimum genes size was 3 and simulations was 500.
The expression of PLAU in different cancers and immune infiltration levels were assessed by Time (cistrome. shinyapps.io/timer) (Li et al., 2017).
We investigated the single nucleotide variation, pathway activity, and drug sensitivity of query genes with TCGA HNSC samples by GSCALite6 (Liu et al., 2018).
Five matched pairs of oral squamous cell carcinoma tissues and matched adjacent non-tumorous tissues were obtained from the Department of Stomatology, Huazhong University of Science and Technology affiliated Union Hospital. This study protocol was approved by the Institutional Research Ethics Committee of Tongji Medical College (Wuhan, China). These specimens were further used in immunohistochemistry staining.
To establish subcutaneous xenograft, CAL-27 cells (1 × 107) were subcutaneously injected into 10 male BALB/c nude mice. One week after injection, 2 groups (n = 5 per group) mice were randomly distributed. uPA inhibitor UK-371804 (Selleck, S8457, dissolved in DMSO, 20 μg per mouse, treatment group) or DMSO (1 μL per mouse, control group) was twice weekly intratumorally injected. Vernier calipers were used for measuring the tumor (twice a week). Two weeks later, the subcutaneous tumors were harvested to calculate the volume by using the formula (length × width2)/2.
The human OSCC cell line CAL27 used for experiments was obtained from ATCC. CAL 27 cell lines were cultured in DMEM (Hyclone) supplemented with 10% fetal bovine serum (FBS), 100 μg/mL streptomycin, and 100 U/mL penicillin (Beyotime), at 37°C in 5% CO2.
Specific siRNA (GenePharma) and non-specific control were transfected into CAL27 cells with lipofectamine 3,000 (Invitrogen) reagents. After 4–6 h transfection, the medium was refreshed and the transfection effect was measured by real-time qPCR. The sequences of the siRNA are shown in Supplementary Table 1.
RNA was isolated from cells using TRIZOL (TAKARA) according to the manufacturer’s instructions and reverse-transcribed using HiScript II Q RT SuperMix for qPCR (Vazyme). Then, quantitative PCR was performed by the StepOne Plus PCR machine (Life Technologies, Carlsbad, CA). The PCR conditions are listed as follow: 5 min at 95°C,then 15 s at 95°C, 30 s at 60°C for 40 cycles, 15 s at 95°C, 60 s at 60°C and 15 s ay 90°C. GAPDH was used as internal control and the relative mRNA levels were analyzed by the 2–ΔΔCT method. The primers sequences are showed in Supplementary Table 2.
The proliferation and cytotoxicity of CAL-27 cells were measured by cell counting kit (CCK-8, Beyotime Institute of Biotechnology). In brief, CAL-27 cells were seeded into a 96-well plate (2,000 cells per well) and incubated for 48 h. After the CCK-8 reagent was added and incubated for 1 h, the optical density was detected at 450 nm wavelength. For cellular cytotoxicity assay, the CAL-27 cells were incubated with 0.34 nM bleomycin (Selleck, S1214) or 8.07 μM docetaxel (Selleck, S1148) for 48 h at 37°C.
CAL27 cell lines were transferred to a 12-well plate and a linear wound was formed by scratching with 200 μL micropipette tip, then cultured for 24 h with serum-free medium. The cells were photographed at 0 and 24 h and the wound distance was used to assess the cell migration rate.
The migration capacity of CAL27 cells was evaluated using the transwell chambers (Corning Life Sciences, MA, United States). CAL27 cells (5 × 104) with 200 μL serum-free medium were loaded into the upper chamber while medium supplemented with 10% serum were added to the lower well. After 24 h incubation, the top surface cells were removed and the rest were stained with crystal violet.
The cells’ protein was collected by RIPA lysis buffer (Byotime, P0045), then the BCA protein assay kit (Biosharp, BL521A) normalizes the samples. The cell extracts were boiled with Sample Loading Buffer (Biosharp, BL529A) for 5 min. After separated by 10% SDS-PAGE gels and transferred to PVDF membrane (Millipore, Germany), the membranes were blocked in 5%BSA and incubated with primary antibodies at 4°C overnight. The primary antibodies are listed as follows: anti-PLAU (1:2,000, Proteintech, 17968-1-AP), anti-E-Cadherin (1:5,000, Proteintech, 20874-1-AP), anti-N-Cadherin (1:2,000, Proteintech, 22018-1-AP), anti-TWIST1 (1:1,000, Proteintech, 25465-1-AP), anti-SNAI1 (1:1,000, Proteintech, 13099-1-AP). Then the membranes were incubated with secondary antibody (Biosharp, BL003A) and the proteins were visualized by ECL enhanced chemiluminescence substrate (Millipore).
The oral cancer tissues and matched adjacent normal oral tissue were obtained from the Department of Stomatology, Huazhong University of Science and Technology affiliated Union Hospital. All the specimens were fixed with 10% buffered formalin, embedded in paraffin, and sectioned in sequence. The sections were deparaffinized by dimethyl benzene and boiled in alkaline Tris buffer for antigen repairing. UltraSensitiveTM SP IHC Kit (MXB, KIT-9710) was used to perform the immunostaining. After blocking, the slides were incubated with primary antibody for PLAU (1:200 dilution, Proteintech, 17968-1-AP) overnight at 4°C. After washing, the sections were immersed with biotin-conjugated IgG secondary antibody in IHC kit for 20 min, then with DAB for 2–5 min and stained with hematoxylin for 5 s. The sections were photographed with a Nikon microscope (Japan).
Statistical analysis was performed with GraphPad Prism 7.0 software and the significance was analyzed with Student’s t-test or ANOVA. In our study, P < 0.05 was considered statistically significant. All results were shown as Mean ± SD.
To identify the role of PLAU as a pan-cancer biomarker, a comparison between tumor and normal tissues was completed and multiple types of cancers, such as BLCA and BRCA, showed a high level of PLAU in tumor tissues among TCGA samples (Figure 1A). Then we evaluated expression levels of PLAU between HNSCC samples and normal samples in various studies from Oncomine. PLAU transcription levels were significantly higher in HNSCC tumor tissues among different studies. All the fold differences reached beyond 3 (Figures 1B–G). Then, we used the immunohistochemistry staining to confirmed that the PLAU expression was higher in OSCC tissues than in adjacent normal tissues (Figure 1H). The relative quantitive analyzing results showed that the ratio of PLAU-positive area in adjacent normal tissues was 17.58% while that in OSCC tissues was 44.35%. Further analysis of HNSCC tissues from TCGA based on clinical and pathological information revealed a higher abundance of PLAU in primary tumor patients than healthy ones based on gender, age, ethnicity as well as tumor stages, and grades (Figures 2A–F). Especially, PLAU expression of grade 1 was lowest while PLAU expression of grade 2 was highest in HNSCC. Taken together, PLAU act as a crucial diagnostic biomarker in HNSCC.
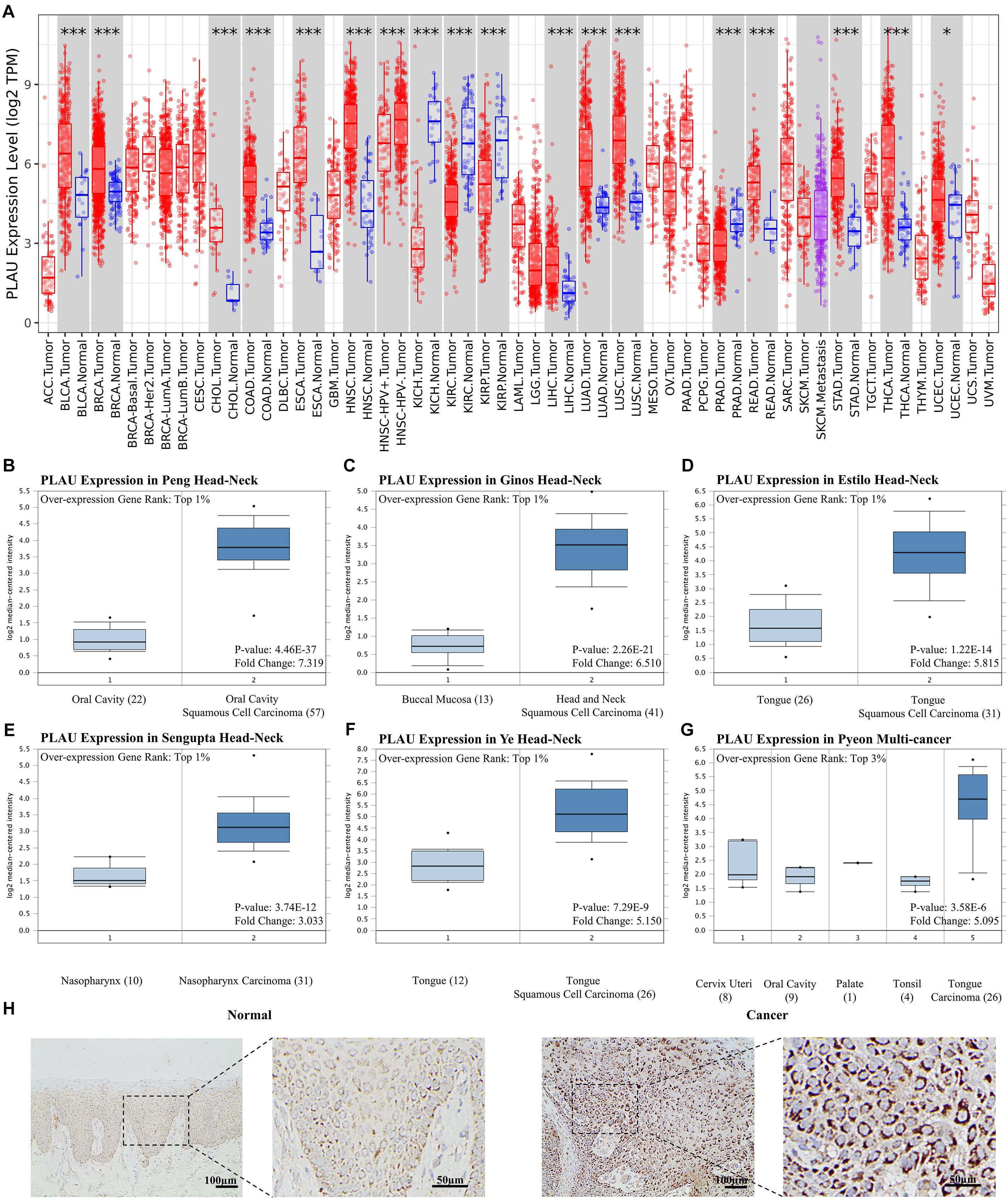
Figure 1. PLAU expression is upregulated in head and neck squamous cell carcinoma. (A) Relative PLAU expression levels in different cancers. (B–G) Boxplot showing PLAU mRNA levels in the Peng Head-Neck, Ginos Head-Neck, Estilo Head-Neck, Sengupta Head-Neck, Ye Head-Neck, Pyeon Multi-cancer, respectively. Levels of PLAU mRNA were significantly higher in tumor tissues than normal head and neck tissues. (H) Detection of PLAU expression levels in OSCC tissues and normal tissues with IHC.
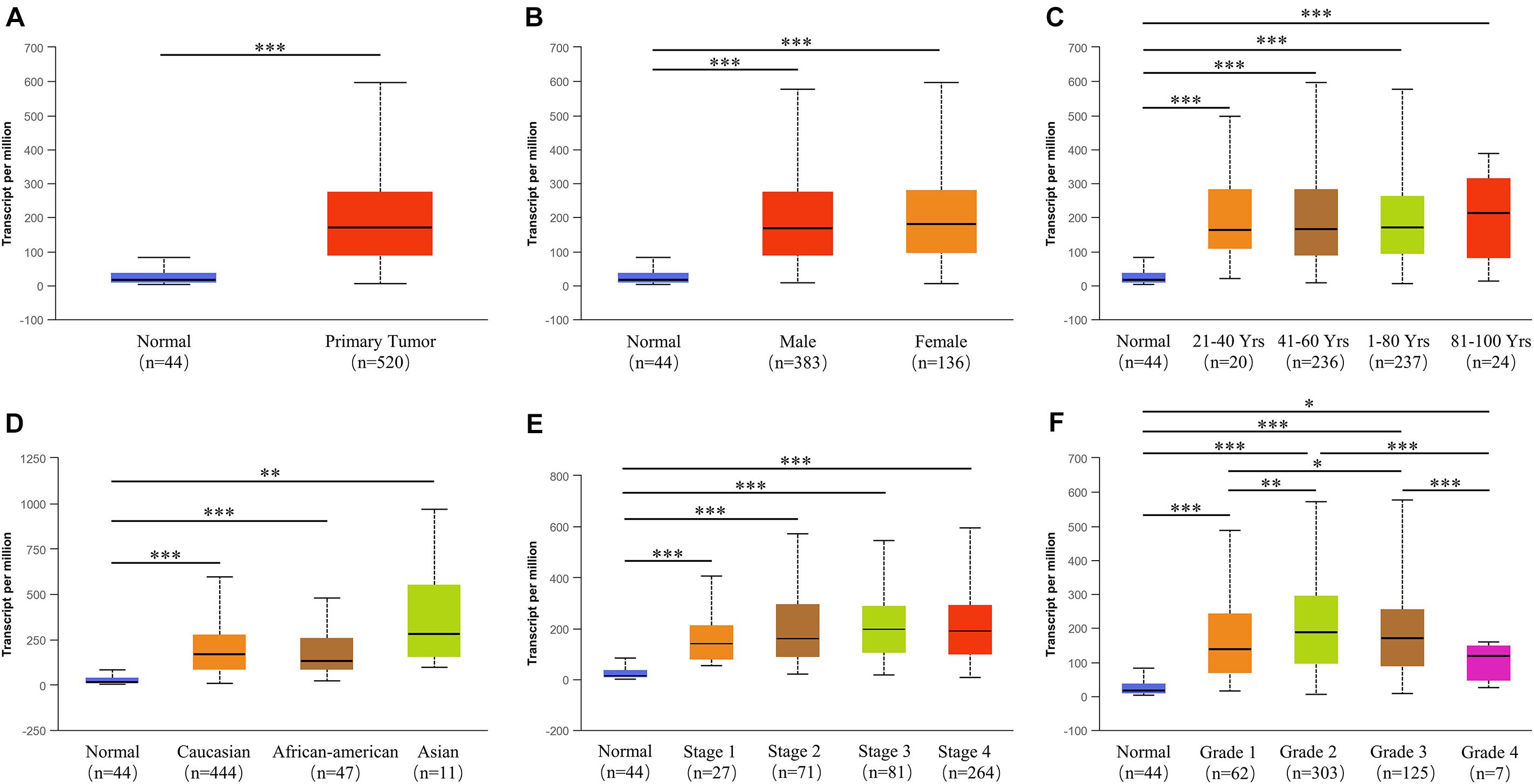
Figure 2. The relationship between PLAU expression and tumor subgroups in HNSCC. (A) Relative expression of PLAU in normal and HNSCC samples. (B) Relative expression of PLAU in normal individuals or HNSCC patients based on gender. (C) Relative expression of PLAU in normal individuals or HNSCC patients based on age. (D) Relative expression of PLAU in normal individuals or HNSCC patients based on race. (E) Relative expression of PLAU in normal individuals or HNSCC patients based on pathological stage. (F) Relative expression of PLAU in normal individuals or HNSCC patients based on tumor grade. *p < 0.05; **p < 0.01; ***p < 0.001.
CBioPortal was chosen to determine the PLAU alterations in HNSCC based on TCGA database. PLAU was altered in about 6% of the total patients. Most of them belonged to mRNA upregulation, with amplification and missense mutation taking part in as well (Figure 3A). We also analyzed the survival with the help of GEPIA web tools. The survival analysis results of PLAU in multiple cancer types demonstrated that PLAU was only negatively correlated with HNSCC, Brain Lower Grade Glioma (LGG), and Mesothelioma (MESO) (Figure 3B). HNSCC patients with a low PLAU level showed a better clinical outcome compared to the high expression group in HNSCC patients. Bothe OS and DFS showed a consistent relationship with PLAU expression (Figures 3C,D). Therefore, it is convinced that PLAU could be a prognostic biomarker for terrible clinical outcomes.
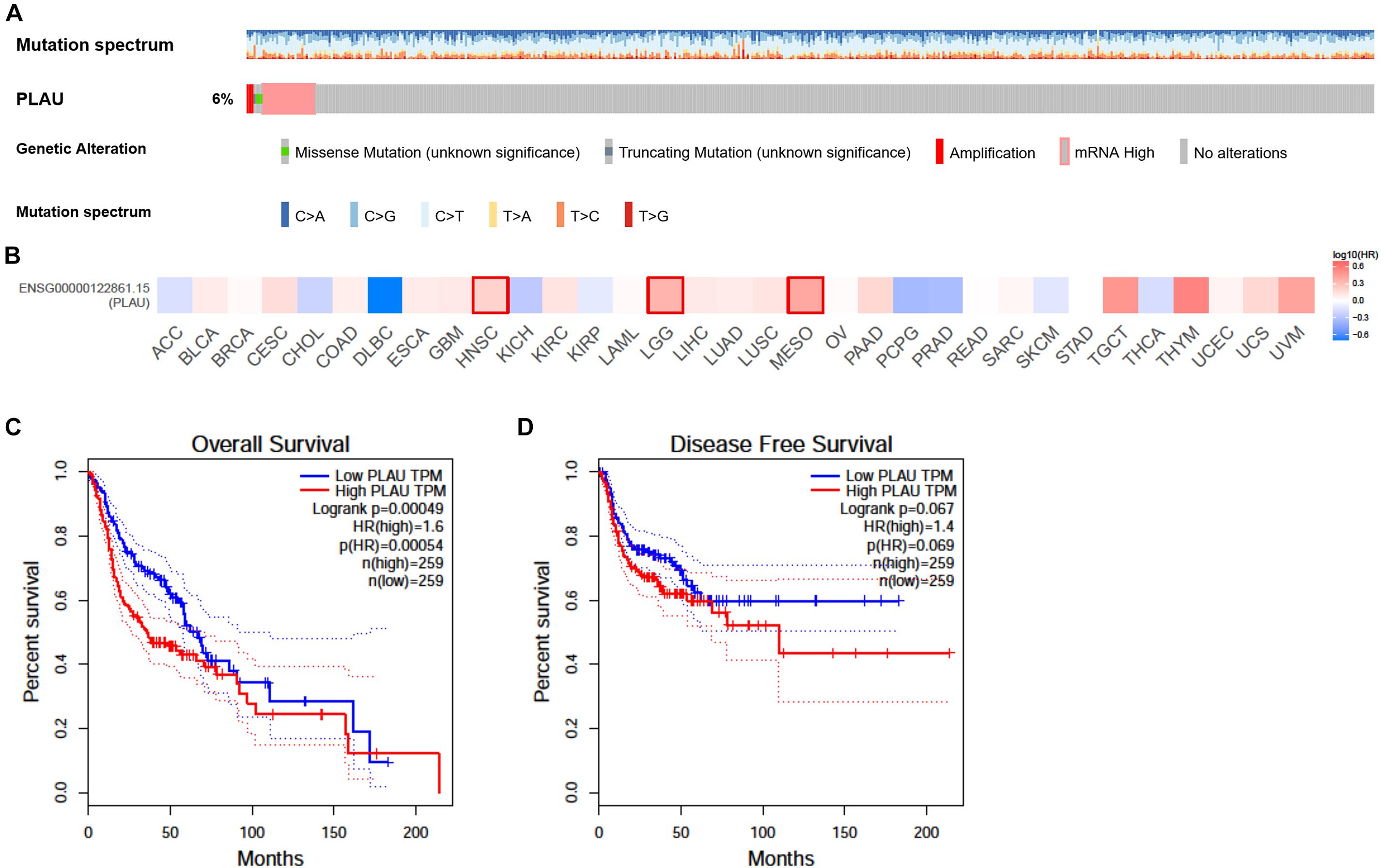
Figure 3. Alteration and prognostic value of PLAU in HNSCC. (A) The alterations of PLAU in HNSCC. (B) The association of PLAU expression and prognostic potential across different cancer types. (C) Overall survival of PLAU in HNSCC. (D) Disease-free survival of PLAU in HNSCC.
We next used LinkedOmics to analyze RNA sequencing data of HNSCC samples. The volcano plot depicted that 4,993 genes showed positive correlations with PLAU, while 7,385 showed negative correlations with PLAU significantly (Figure 4A). The top 50 significantly positively and negatively correlated co-expression genes were presented by heatmaps (Figures 4B,C). Four most significantly correlated genes, C10orf55 (cor = 0.7803, P = 1.133e-107), ITGA5 (cor = 0.6805, P = 649e-72), SERPINE1 (cor = 0.6592, P = 3.887e-66) and TNFRSF12A (cor = 0.6569, P = 1.554e-65), were selected according to the P-value and used to establish linear regression models (Supplementary Figure 1A). This result indicated that PLAU might function along with a large group of genes during HNSCC progress.
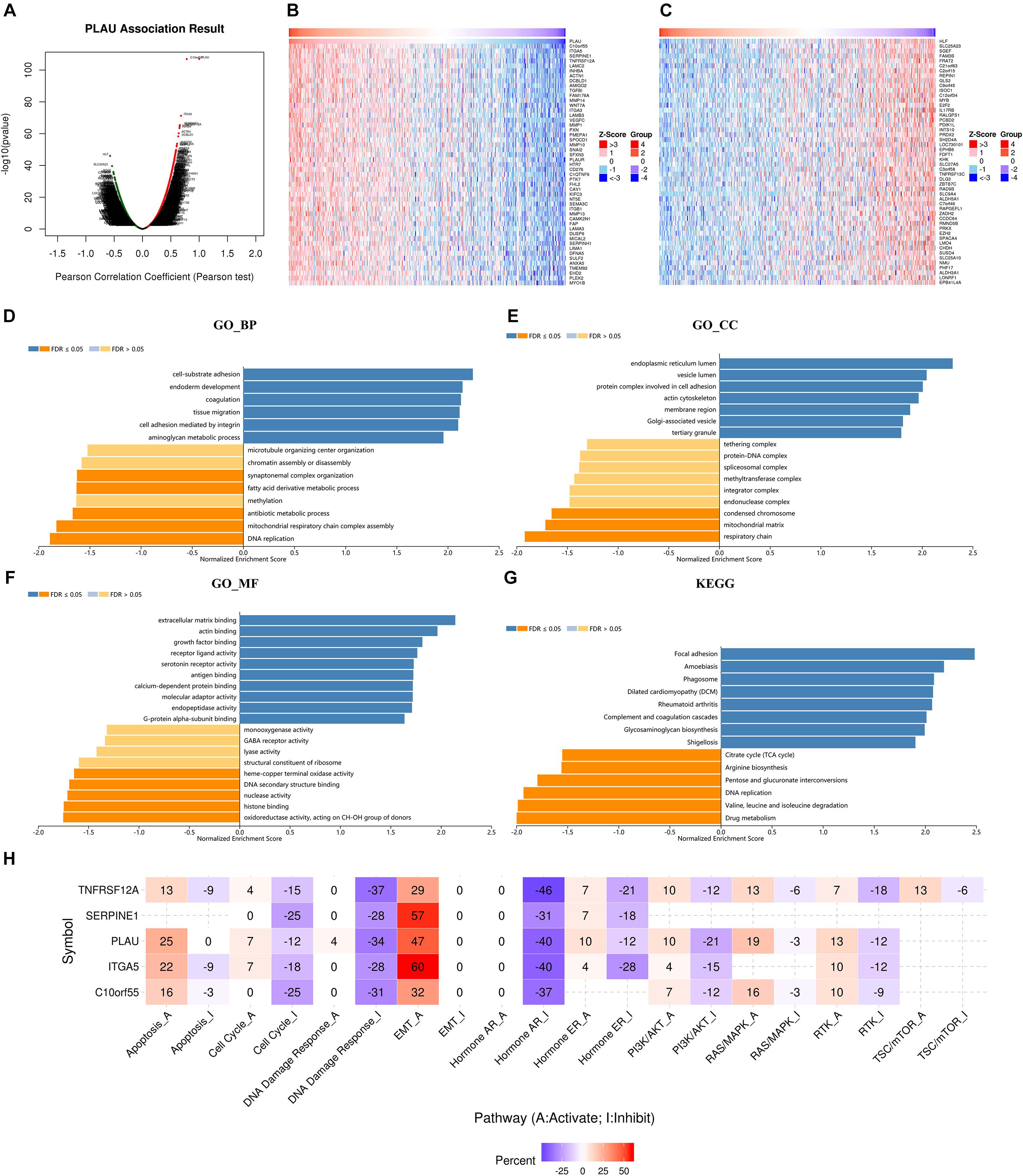
Figure 4. PLAU co-expression genes in HNSCC. (A) The genes correlated with PLAU were analyzed by Pearson test in HNSCC. (B) The top 50 genes positively correlated with PLAU in HNSCC. (C) The top 50 genes negatively correlated with PLAU in HNSCC. (D–G) Gene enrichment analysis of PLAU-associated genes in HNSCC. BP, biological process; CC, cellular components; MF, molecular functions. KEGG pathway analysis. (H) The relationships between hub genes expression and pathway activity groups.
Significant GO term by LinkedOmics revealed that PLAU along with its co-expressed genes were mainly enriched in cell-substrate adhesion and extracellular matrix binding, which indicated abnormality of cell membrane proteins and were correlated to the EMT process. This just coincided with the result that the most enriched cellular components were endoplasmic reticulum lumen, as endoplasmic reticulum lumen played a great part in protein synthesis (Figures 4D–F). KEGG pathway items suggested that PLAU along with its associated genes are most involved in the focal adhesion process (Figure 4G).
PLAU and the top 4 correlated genes, C10orf55, ITGA5, SERPINE1, and TNFRSF12A, were chosen as crucial mediators during HNSCC progress for further analysis. As genetic alteration analysis showed, 64% of HNSCC samples exhibited SERPINE1 genetic alteration, along with 27% samples showing ITGA5 and PLAU genetic alterations respectively. Among them, missense mutation was the most popular form of genetic mutations (Supplementary Figure 1B). The distribution of variants in HNSCC samples was described subsequently. SNP acted as the main form of variant types, in which C to T alteration counted the most (Supplementary Figure 1C).
Furthermore, we investigated the relationships between these genes and carcinogenesis-related pathways. In the perspective of PLAU as well as its most correlated genes, the EMT process was the most enriched pathway, further identifying the role of PLAU in tumor invasion and metastasis (Figure 4H).
Drug sensitivity analysis indicated that low ITGA5 showed its resistance to 14 types of drugs and small molecules consistently, while a high abundance of TNFRSF12A was resistant to multiple types of drugs. On the other hand, the relationship between the expression level of PLAU, SERPINE1, and drug resistance differed among drugs (Supplementary Figure 2). Importantly, high PLAU is sensitive to bleomycin and docetaxel in HNSCC. To validated the drug-sensitive data, we detected the effect of downregulating PLAU in bleomycin and docetaxel treated CAL-27 cells respectively. Suppressing PLAU led to tumor cells resistant to bleomycin and docetaxel (Supplementary Figure 3), which potentially helps the drug selection in future treatment.
Consequently, we presented the expression results of C10orf55, ITGA5, SERPINE1, and TNFRSF12A in HNSCC and normal samples from the TCGA database (Figures 5A–D). Even though the expression of C10orf55 had no statistical difference between normal and tumor tissues, the expression profiles of these four genes were consistent with PLAU. Then the prognosis of them in HNSCC patients was also established by GEPIA (Figures 5E–H). As the results showed, HNSCC patients with lower ITGA5, SERPINE1, and TNFRSF12A expression had a better OS rate while there was no connection between their expression levels and DFS. However, patients with high C10orf55 expression were correlated with poor DFS. Besides, the expression of co-expressed C10orf55, ITGA5, SERPINE1, and TNFRSF12A in Cal27 cells was detected by RT-PCR when PLAU has been downregulated. The results showed that only TNFRSF12A significantly decreased after downregulating PLAU, which implies TNFRSF12A is downstream of PLAU (Figures 5I–L). These results demonstrated that PLAU, as well as PLAU-associated genes, can be the potential prognostic markers of HNSCC patients.
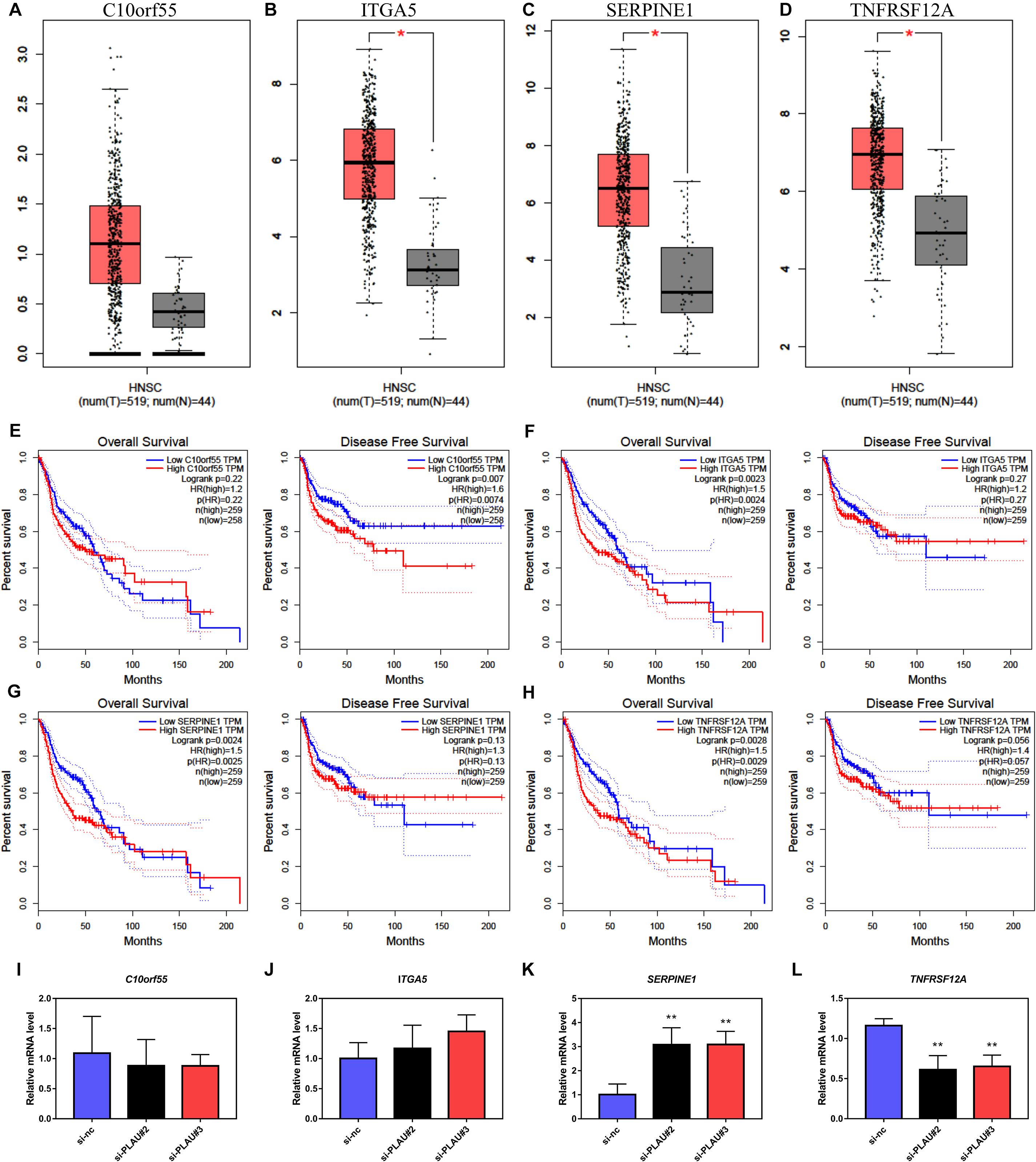
Figure 5. The expression levels and prognostic value of co-expressed genes in HNSCC. (A–D) The mRNA levels of C10orf55, ITGA5, SERPINE1, and TNFRSF12A in HNSCC and paired normal samples. (E–H) Prognostic role of C10orf55, ITGA5, SERPINE1, and TNFRSF12A in HNSCC. (I–L) The mRNA levels of C10orf55, ITGA5, SERPINE1, and TNFRSF12A in Cal27 cell lines under different si-RNA treatment.
Next, we evaluated the potential function of PLAU and associated genes in HNSCC cells. The PLAU-siRNA and TNFRSF12A-siRNA were transfected into the CAL27 cell line respectively and the transfected efficiency was validated by qRT-PCR (Figure 6A). Both silencings of PLAU and TNFRSF12A suppressed the proliferation of CAL-27 cells (Figure 6B). The in vitro experiments revealed that depletion of PLAU inhibited the migration of CAL27 cells (Figures 6C–F). To further elucidate the involvement of PLAU in the EMT pathway, we detected EMT-related markers. QRT-PCR demonstrated that E-cadherin was increased and N-cadherin, Fibronectin, SNAIL, TWIST1, TWIST2, ZEB1, ZEB2 were decreased in PLAU depletion CAL27 cell line (Figure 7A). The protein levels of N-cadherin, SNAIL, TWIST1 were also downregulated in the si-PLAU group (Figures 7B,C) what’s more, recovery of PLAU would increase migration rate and N-cadherin levels while decrease the SNAI1 levels in CAL-27 cells (Figures 7D–H).
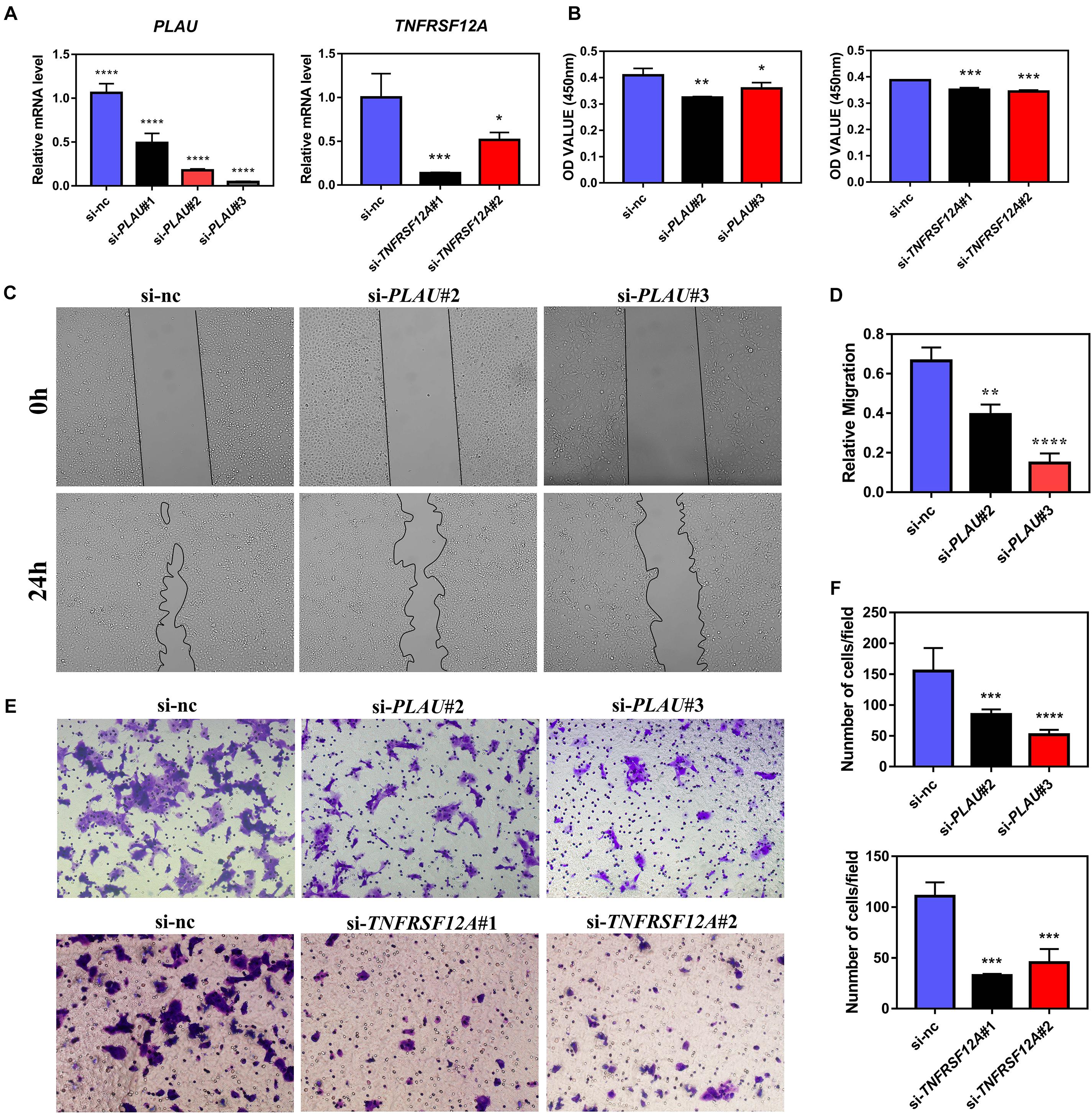
Figure 6. PLAU and TNFRSF12A regulate proliferation and migration in vitro. (A) Quantitative RT-PCR analysis of PLAU and TNFRSF12A expression. (B) Analysis of CCK8 assay. (C,D) Representative images and analysis of wound-healing assay. (E,F) Representative images and analysis of transwell assay.
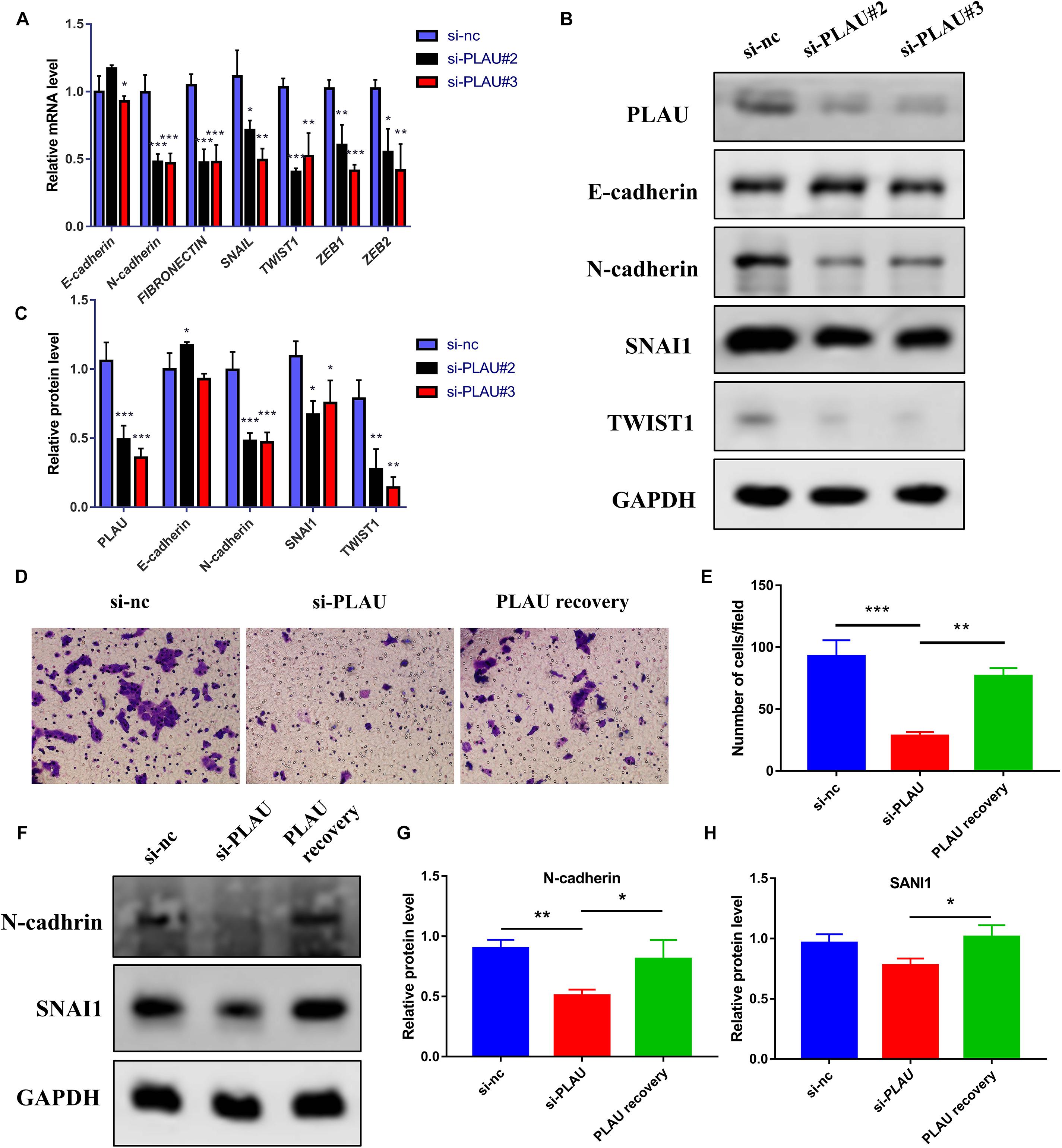
Figure 7. Loss of PLAU suppresses EMT process in CAL-27 cells. (A) Quantitative RT-PCR analysis of EMT-related markers in CAL27 cells under PLAU knockdown. (B,C) Western blot analysis of EMT-related proteins in CAL27 cells under PLAU knockdown. (D,E) Representative images and analysis of transwell assay. (F–H) Quantitative western blot analysis of N-cadherin and SNAI1 expression. *p < 0.05; **p < 0.01; ***p < 0.001.
To further reveal the function of PLAU on HNSCC progression in vivo, we construct xenografted tumor model and the mice were divided into the control group and UK-371804 (uPA inhibitor) treatment group. The UK-371804-treated group developed smaller tumor compared to the control group (Figures 8A,B). Consistent with these results, the levels of N-cadherin decreased and E-cadherin increased in UK-371804-treated group (Figures 8C–E). What’s more, the H&E staining show that the cancer cell density was lower in UK-371804-treated group (Figure 8F). Thus, intratumoral injection of UK-371804 can inhibit the progression of HNSCC.
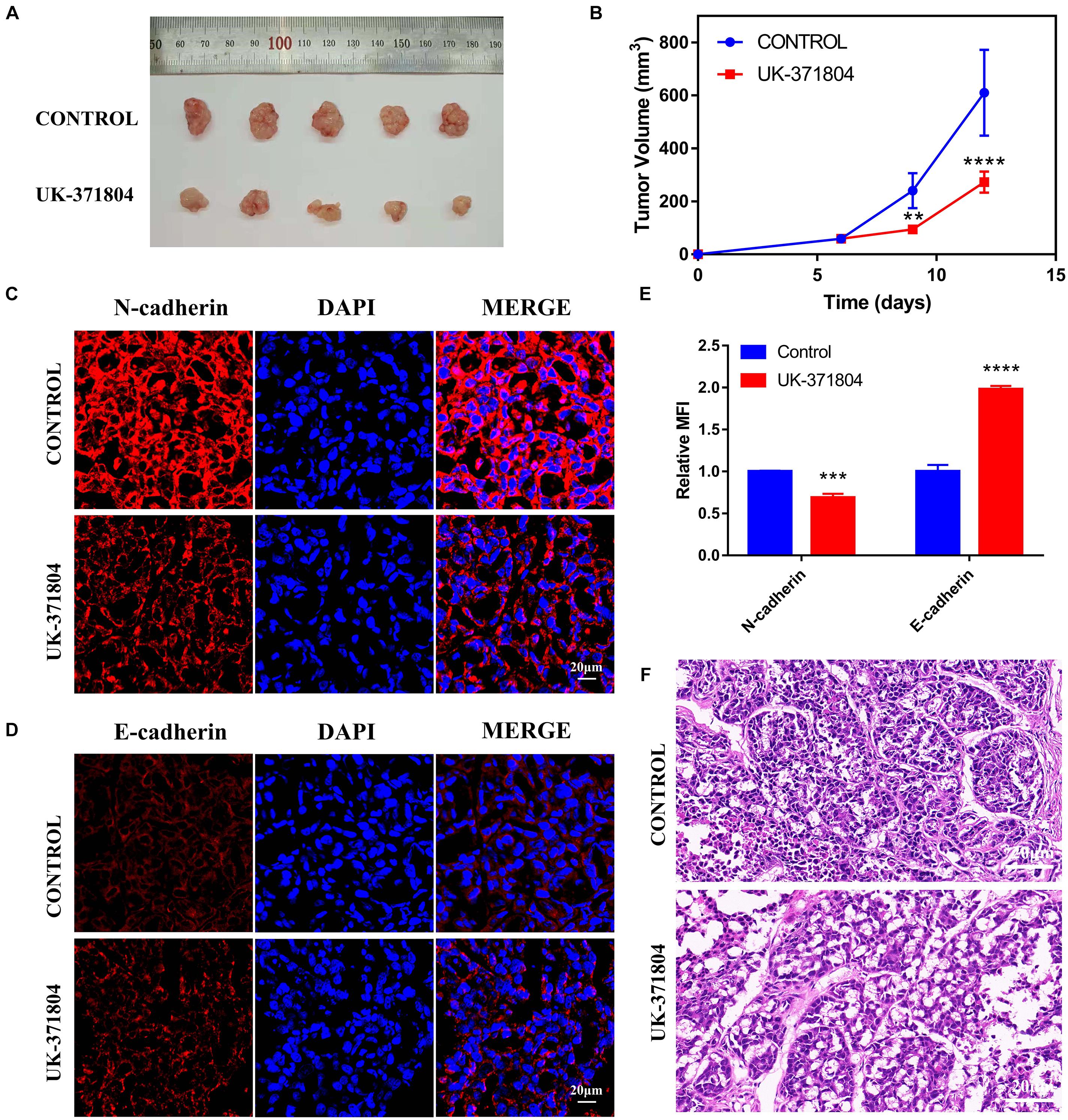
Figure 8. Loss of PLAU suppresses EMT and tumor growth. (A,B) Representative images of xenograft tumors and analysis of xenograft tumor volumes. (C–E) Immunofluorescent staining of N-cadherin and E-cadherin in xenograft tumors. *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001. (F) Images of H&E staining for xenograft tumors.
In this article, the expression level of PLAU was identified to increase in HNSCC among multiple Oncomine and TCGA datasets, suggesting its important oncogenic role in HNSCC. Subsequent immunohistologic staining results validated the upregulation of PLAU in OSCC tissues. Importantly, the PLAU expression level was significantly associated with tumor grades. The prognostic analysis results agree with the results of Li et al. (2021) who have constructed the cox regression model and validated the survival results from the GEO database. Multiple types of alterations, including mRNA upregulation, amplification, and missense mutation, could explain the overexpression of PLAU to a great extent. The significant prognostic value of PLAU in HNSCC ensures its role in tumor progression. The four associated genes identified in this study, TNFRSF12A was validated to be regulated by PLAU and TNFRSF12A downregulation restrains proliferation and migration of CAL-27 cells. These results implied that the PLAU-induced EMT process may need TNFRSF12A in HNSCC. Considering that TNFRSF12A plays an important role in immune with TWEAK (Burkly et al., 2011), we also explore Expression of PLAU showed a positive relationship with macrophage while a negative relationship with CD8+T cell infiltration in our study (Supplementary Figure 4). It has been proved macrophage and CD8+T cells act as pro-tumor and anti-tumor roles respectively. Thus, PLAU might promote tumor development by posing a pro-tumor immune microenvironment in HNSCC. However, the positive correlation between PLAU and CD4+T cell needs further study, as two subtypes of CD4+T cell, Th1 and Th2 cell, act opposite roles in carcinogenesis (Chandwaskar and Awasthi, 2019). The landscape of immunity influenced by PLAU deserved further exploration, as tumor immune response was regarded as a more and more important and complex part of HNSCC progress. Also, lots of genes were identified as co-expression profiles with PLAU. These co-expression genes construct a complex interacting network, together contributing to the progress of HNSCC. As known, PLAU affects the migration and invasion of the tumor (Blasi and Sidenius, 2010). The enrichment analysis of these associated genes also showed variations of many biological processes associated with cell adhesion and extracellular matrix. In consideration that the EMT process was significantly enriched by PLAU and its co-expression genes, it is expected that PLAU might play a role in tumor invasion. Drug sensitivity analysis of PLAU and its co-expression genes suggested that PLAU expression level could guide medical strategy for HNSCC.
Accumulating bioinformatic studies indicate the crucial role of PLAU in cancer development. In gastric cancer, PLAU could act together with its co-expression gene to predict a worse survival status (Ai et al., 2020). Identification of PLAU as hub gene in pancreatic cancer through screening multiple datasets has also been completed recently (Chen Q. et al., 2019). In the perspective of HNSCC, an increase of PLAU was also observed by multiple bioinformatic analyses (Zhao et al., 2018; Yang et al., 2019). These results were just consistent with our observation, indicating that PLAU might be significant a pan-cancer oncogene. As is widely known, changes of cell-cell binding and EMT process were linked to cell migration both in epidermal cell and epithelial cell, which might lead to invasion and metastasis during tumor development (Xu et al., 2017; Aiello et al., 2018). PLAU was shown to participate in the migration and invasion of glioblastoma (Bernhart et al., 2013). In bladder cancer cells, stabilization of PLAU mRNA level might also contribute to tumor metastasis (Chen Z. et al., 2019). Considering that abnormality of cell adhesion, extracellular matrix, as well as enhancement of EMT process, were observed in our study by enrichment analysis, it might be speculated that upregulation of PLAU might promote tumor migration and invasion in HNSCC and have a clinical correlation with LN metastasis and distal metastasis. Importantly, PLAU was recommended for use as an indicator in breast cancer (Annecke et al., 2008; Sudol, 2011). We further explored the biological role of PLAU in vitro by manipulating the expression of PLAU. Our results demonstrated that depletion of PLAU could suppress the migration and invasion of OSCC cell line. Moreover, downregulation of PLAU reduced the EMT-related genes, indicating PLAU could induce the EMT process in the OSCC cell line. Besides, the expression and survival analysis of the top 4 associated genes of PLAU was conducted, which indicated that all these four genes and PLAU had a great predictive role in HNSCC. Drug resistance is an important factor influencing tumor prognosis (Adamska et al., 2018). Proper medical strategies were necessary for clinical treatment, which was hard to realize due to patient heterogeneity (Dagogo-Jack and Shaw, 2018). Due to drug sensitivity analysis, the association between drug sensitivity and PLAU as well as its co-expression genes was exhibited in our study. This result possesses huge clinical value, as a custom-tailored medical strategy in HNSCC could be figured out on the dependence of PLAU expression level.
Taken together, PLAU might act as an oncogene and effective biomarker in HNSCC. Enhancement of cell mobility and migration might account for its tumorigenesis. Indeed, we found that PLAU could regulate the EMT signal pathway in vitro. Adjustment of medical treatment based on PLAU expression might contribute to a better clinical outcome of HNSCC patients. The landscape of immunity influenced by PLAU deserved further exploration, as tumor immune response was regarded as a more and more important and complex part of HNSCC progress. In silico study combined experiments demonstrated that PLAU regulates the proliferation and migration via the EMT process in HNSCC.
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author/s.
The studies involving human participants were reviewed and approved by the Institutional Research Ethics Committee of Tongji Medical College (Wuhan, China). The patients/participants provided their written informed consent to participate in this study. The animal study was reviewed and approved by Institutional Animal Care and Use Committe of Tongji Medical College.
GC and JS performed the most analysis, experiments, and writing the manuscript. SY and MX helped to revised the manuscript. QT and LC designed this study and revised the manuscript. All authors contributed to the article and approved the submitted version.
This work was supported by the Young Elite Scientist Sponsorship Program by CAST (2018QNRC001 to QT).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2021.651882/full#supplementary-material
Supplementary Figure 1 | The single nucleotide variation (SNV) analysis of hub genes in HNSCC. (A) The Pearson correlation of PLAU expression with top 4 correlated genes, C10orf55, ITGA5, SERPINE1 and TNFRSF12A. (B) The SNV frequency and mutated types of hub genes in HNSCC. (C) The overview of the variation distribution of mutated genes and the classification of SNV types.
Supplementary Figure 2 | Drug sensitivity analysis from Genomics of Drug Sensitivity in Cancer (GDSC) data (GSCALite). The Spearman correlation of the genes expression and drug, The positive correlation indicated that high expression is resistant to the drug, vise verse.
Supplementary Figure 3 | Analysis of drug sensitivity of PLAU for bleomycin and docetaxel. ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001.
Supplementary Figure 4 | Correlations between PLAU expression and immune infiltration level in HNSCC (Timer). (A) The correlation between the abundance of immune cells and the level of PLAU. (B) The correlations between PLAU copy number variation (CNV) and infiltration level of immune cells. ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001.
Supplementary Figure 5 | Recovery of PLAU would increase N-cadherin levels and SNAI1 of CAL-27 cells. (A) The replicated western blots images of EMT-related proteins in CAL27 cells. (B) The replicated western blots images of N-cadherin and SNAI1 expression in CAL27 cells.
Supplementary Table 1 | Specific si-RNA used in this study.
Supplementary Table 2 | Genes and primers for PCR.
Adamska, A., Elaskalani, O., Emmanouilidi, A., Kim, M., Abdol Razak, N. B., Metharom, P., et al. (2018). Molecular and cellular mechanisms of chemoresistance in pancreatic cancer. Adv. Biol. Regul. 68, 77–87. doi: 10.1016/j.jbior.2017.11.007
Ai, C., Zhang, J., Lian, S., Ma, J., Gyõrffy, B., Qian, Z., et al. (2020). FOXM1 functions collaboratively with PLAU to promote gastric cancer progression. J. Cancer 11, 788–794. doi: 10.7150/jca.37323
Aiello, N. M., Maddipati, R., Norgard, R. J., Balli, D., Li, J., Yuan, S., et al. (2018). EMT subtype influences epithelial plasticity and mode of cell migration. Dev. Cell 45, 681–695.e4. doi: 10.1016/j.devcel.2018.05.027
Annecke, K., Schmitt, M., Euler, U., Zerm, M., Paepke, D., Paepke, S., et al. (2008). uPA and PAI-1 in breast cancer: review of their clinical utility and current validation in the prospective NNBC-3 trial. Adv. Clin. Chem. 45, 31–45. doi: 10.1016/S0065-2423(07)00002-9
Argiris, A., Karamouzis, M. V., Raben, D., and Ferris, R. L. (2008). Head and neck cancer. Lancet (Lond. Engl.) 371, 1695–1709. doi: 10.1016/S0140-6736(08)60728-X
Bernhart, E., Damm, S., Wintersperger, A., DeVaney, T., Zimmer, A., Raynham, T., et al. (2013). Protein kinase D2 regulates migration and invasion of U87MG glioblastoma cells in vitro. Exp. Cell Res. 319, 2037–2048. doi: 10.1016/j.yexcr.2013.03.029
Blasi, F., and Sidenius, N. (2010). The urokinase receptor: focused cell surface proteolysis, cell adhesion and signaling. FEBS Lett. 584, 1923–1930. doi: 10.1016/j.febslet.2009.12.039
Burkly, L. C., Michaelson, J. S., and Zheng, T. S. (2011). TWEAK/Fn14 pathway: an immunological switch for shaping tissue responses. Immunol. Rev. 244, 99–114. doi: 10.1111/j.1600-065X.2011.01054.x
Chandrashekar, D. S., Bashel, B., Balasubramanya, S. A. H., Creighton, C. J., Ponce-Rodriguez, I., Chakravarthi, B., et al. (2017). UALCAN: a portal for facilitating tumor subgroup gene expression and survival analyses. Neoplasia (New York, NY) 19, 649–658. doi: 10.1016/j.neo.2017.05.002
Chandwaskar, R., and Awasthi, A. (2019). Emerging roles of Th9 cells as an anti-tumor helper T cells. Int. Rev. Immunol. 38, 204–211. doi: 10.1080/08830185.2019.1648453
Chen, Q., Yu, D., Zhao, Y., Qiu, J., Xie, Y., and Tao, M. (2019). Screening and identification of hub genes in pancreatic cancer by integrated bioinformatics analysis. J. Cell. Biochem. 120, 19496–19508. doi: 10.1002/jcb.29253
Chen, Z., Chen, X., Xie, R., Huang, M., Dong, W., Han, J., et al. (2019). DANCR promotes metastasis and proliferation in bladder cancer cells by enhancing IL-11-STAT3 signaling and CCND1 expression. Mol. Ther. J. Am. Soc. Gene Ther. 27, 326–341. doi: 10.1016/j.ymthe.2018.12.015
Chow, L. Q. M. (2020). Head and neck cancer. N. Engl. J. Med. 382, 60–72. doi: 10.1056/NEJMra1715715
Dagogo-Jack, I., and Shaw, A. T. (2018). Tumour heterogeneity and resistance to cancer therapies. Nat. Rev. Clin. Oncol. 15, 81–94. doi: 10.1038/nrclinonc.2017.166
Dano, K., Behrendt, N., Hoyer-Hansen, G., Johnsen, M., Lund, L. R., Ploug, M., et al. (2005). Plasminogen activation and cancer. Thromb. Haemostasis 93, 676–681. doi: 10.1160/TH05-01-0054
Estilo, C. L., Oc, P., Talbot, S., Socci, N. D., Carlson, D. L., Ghossein, R., et al. (2009). Oral tongue cancer gene expression profiling: identification of novel potential prognosticators by oligonucleotide microarray analysis. BMC Cancer 9:11. doi: 10.1186/1471-2407-9-11
Gao, J., Aksoy, B. A., Dogrusoz, U., Dresdner, G., Gross, B., Sumer, S. O., et al. (2013). Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal. 6:l1. doi: 10.1126/scisignal.2004088
Ginos, M. A., Page, G. P., Michalowicz, B. S., Patel, K. J., Volker, S. E., Pambuccian, S. E., et al. (2004). Identification of a gene expression signature associated with recurrent disease in squamous cell carcinoma of the head and neck. Cancer Res. 64, 55–63. doi: 10.1158/0008-5472.CAN-03-2144
Jo, M., Lester, R. D., Montel, V., Eastman, B., Takimoto, S., and Gonias, S. L. (2009). Reversibility of epithelial-mesenchymal transition (EMT) induced in breast cancer cells by activation of urokinase receptor-dependent cell signaling. J. Biol. Chem. 284, 22825–22833. doi: 10.1074/jbc.M109.023960
LeBeau, A. M., Sevillano, N., Markham, K., Winter, M. B., Murphy, S. T., Hostetter, D. R., et al. (2015). Imaging active urokinase plasminogen activator in prostate cancer. Cancer Res. 75, 1225–1235. doi: 10.1158/0008-5472.CAN-14-2185
Lester, R. D., Jo, M., Montel, V., Takimoto, S., and Gonias, S. L. (2007). uPAR induces epithelial-mesenchymal transition in hypoxic breast cancer cells. J. Cell Biol. 178, 425–436. doi: 10.1083/jcb.200701092
Li, T., Fan, J., Wang, B., Traugh, N., Chen, Q., Liu, J. S., et al. (2017). TIMER: a web server for comprehensive analysis of tumor-infiltrating immune cells. Cancer Res. 77, e108–e110. doi: 10.1158/0008-5472.CAN-17-0307
Li, Z., Chen, C., Wang, J., Wei, M., Liu, G., Qin, Y., et al. (2021). Overexpressed PLAU and its potential prognostic value in head and neck squamous cell carcinoma. PeerJ 9:e10746. doi: 10.7717/peerj.10746
Liu, C. J., Hu, F. F., Xia, M. X., Han, L., Zhang, Q., and Guo, A. Y. (2018). GSCALite: a web server for gene set cancer analysis. Bioinformatics (Oxford Engl.) 34, 3771–3772. doi: 10.1093/bioinformatics/bty411
Mahmood, N., Mihalcioiu, C., and Rabbani, S. A. (2018). Multifaceted role of the urokinase-type plasminogen activator (uPA) and its receptor (uPAR): diagnostic, prognostic, and therapeutic applications. Front. Oncol. 8:24. doi: 10.3389/fonc.2018.00024
Mehanna, H., Paleri, V., West, C. M., and Nutting, C. (2010). Head and neck cancer–Part 1: epidemiology, presentation, and prevention. BMJ (Clin. Res. ed) 341:c4684. doi: 10.1136/bmj.c4684
Mekkawy, A. H., Pourgholami, M. H., and Morris, D. L. (2014). Involvement of urokinase-type plasminogen activator system in cancer: an overview. Med. Res. Rev. 34, 918–956. doi: 10.1002/med.21308
Novak, C. M., Horst, E. N., Taylor, C. C., Liu, C. Z., and Mehta, G. (2019). Fluid shear stress stimulates breast cancer cells to display invasive and chemoresistant phenotypes while upregulating PLAU in a 3D bioreactor. Biotechnol. Bioeng. 116, 3084–3097. doi: 10.1002/bit.27119
Peng, C. H., Liao, C. T., Peng, S. C., Chen, Y. J., Cheng, A. J., Juang, J. L., et al. (2011). A novel molecular signature identified by systems genetics approach predicts prognosis in oral squamous cell carcinoma. PLoS One 6:e23452. doi: 10.1371/journal.pone.0023452
Pyeon, D., Newton, M. A., Lambert, P. F., den Boon, J. A., Sengupta, S., Marsit, C. J., et al. (2007). Fundamental differences in cell cycle deregulation in human papillomavirus-positive and human papillomavirus-negative head/neck and cervical cancers. Cancer Res. 67, 4605–4619. doi: 10.1158/0008-5472.CAN-06-3619
Rhodes, D. R., Yu, J., Shanker, K., Deshpande, N., Varambally, R., Ghosh, D., et al. (2004). ONCOMINE: a cancer microarray database and integrated data-mining platform. Neoplasia (New York, NY) 6, 1–6. doi: 10.1016/S1476-5586(04)80047-2
Sacco, A. G., and Cohen, E. E. (2015). Current treatment options for recurrent or metastatic head and neck squamous cell carcinoma. J. Clin. Oncol. 33, 3305–3313. doi: 10.1200/JCO.2015.62.0963
Sengupta, S., den Boon, J. A., Chen, I. H., Newton, M. A., Dahl, D. B., Chen, M., et al. (2006). Genome-wide expression profiling reveals EBV-associated inhibition of MHC class I expression in nasopharyngeal carcinoma. Cancer Res. 66, 7999–8006. doi: 10.1158/0008-5472.CAN-05-4399
Su, S. C., Lin, C. W., Yang, W. E., Fan, W. L., and Yang, S. F. (2016). The urokinase-type plasminogen activator (uPA) system as a biomarker and therapeutic target in human malignancies. Expert Opin. Ther. Targets 20, 551–566. doi: 10.1517/14728222.2016.1113260
Sudol, M. (2011). From Rous sarcoma virus to plasminogen activator, src oncogene and cancer management. Oncogene 30, 3003–3010. doi: 10.1038/onc.2011.38
Tang, Z., Li, C., Kang, B., Gao, G., Li, C., and Zhang, Z. (2017). GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 45, W98–W102. doi: 10.1093/nar/gkx247
Vasaikar, S. V., Straub, P., Wang, J., and Zhang, B. (2018). LinkedOmics: analyzing multi-omics data within and across 32 cancer types. Nucleic Acids Res. 46, D956–D963. doi: 10.1093/nar/gkx1090
Xu, F., Zhang, J., Hu, G., Liu, L., and Liang, W. (2017). Hypoxia and TGF-β1 induced PLOD2 expression improve the migration and invasion of cervical cancer cells by promoting epithelial-to-mesenchymal transition (EMT) and focal adhesion formation. Cancer Cell Int. 17:54. doi: 10.1186/s12935-017-0420-z
Yang, K., Zhang, S., Zhang, D., Tao, Q., Zhang, T., Liu, G., et al. (2019). Identification of SERPINE1, PLAU and ACTA1 as biomarkers of head and neck squamous cell carcinoma based on integrated bioinformatics analysis. Int. J. Clin. Oncol. 24, 1030–1041. doi: 10.1007/s10147-019-01435-9
Ye, H., Yu, T., Temam, S., Ziober, B. L., Wang, J., Schwartz, J. L., et al. (2008). Transcriptomic dissection of tongue squamous cell carcinoma. BMC Genomics. 9:69. doi: 10.1186/1471-2164-9-69
Keywords: prognostic biomarker, EMT, PLAU, TNFRSF12A, head and neck squamous cell carcinoma
Citation: Chen G, Sun J, Xie M, Yu S, Tang Q and Chen L (2021) PLAU Promotes Cell Proliferation and Epithelial-Mesenchymal Transition in Head and Neck Squamous Cell Carcinoma. Front. Genet. 12:651882. doi: 10.3389/fgene.2021.651882
Received: 11 January 2021; Accepted: 23 April 2021;
Published: 20 May 2021.
Edited by:
Manoj Kumar Kashyap, Amity University Gurgaon, IndiaReviewed by:
Yashwanth Subbannayya, Norwegian University of Science and Technology, NorwayCopyright © 2021 Chen, Sun, Xie, Yu, Tang and Chen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Qingming Tang, aHVzdF90YW5nQGh1c3QuZWR1LmNu; Lili Chen, Y2hlbmxpbGkxMDMwQGh1c3QuZWR1LmNu
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.