- Division of Immunobiology and Center for Inflammation and Tolerance, Cincinnati Children’s Hospital Medical Center and Department of Pediatrics, University of Cincinnati College of Medicine, Cincinnati, OH, United States
Domestic animals represent important resources for understanding shared mechanisms underlying complex natural diseases that arise due to both genetic and environmental factors. Intestinal inflammation, particularly inflammatory bowel disease (IBD), is a significant health challenge in humans and domestic animals. While the etiology of IBD is multifactorial, imbalance of symbiotic gut microbiota has been hypothesized to play a central role in disease pathophysiology. Advances in genomic sequencing and analytical pipelines have enabled researchers to decipher the composition of the intestinal microbiota during health and in the context of naturally occurring diseases. This review compiles microbiome genomic data across domestic species and highlights a common occurrence of gut microbiome dysbiosis during idiopathic intestinal inflammation in multiple species, including dogs, cats, horses, cows, and pigs. Current microbiome data obtained from animals with intestinal inflammation are mostly limited to taxonomical analyses in association with broad clinical phenotype. In general, a pathogen or pathosymbiont were not detected. Rather, functional potential of the altered microbiota has been suggested to be one of the key etiologic factors. Among the domestic species studied, canine analyses are currently the most advanced with incorporation of functional profiling of microbiota. Canine IBD parallels features of the disease in humans, thus canines represent a strong natural model for human IBD. While deeper analyses of metagenomic data, coupled with host molecular analyses are needed, comparative studies across domestic species can reveal shared microbial alterations and regulatory mechanisms that will improve our understanding of intestinal inflammation in both animals and humans.
Introduction
The mammalian intestine houses trillions of commensal microbes, collectively termed the microbiota. Host-microbiota symbiosis is supported by mounting studies with human subjects and lab animal models (Clemente et al., 2012). The importance of microbiota in the health of domestic animals has also been recognized for decades, yet largely supported by anecdotal evidence. In recent years, however, increasing studies on microbiota composition of companion and livestock animals using 16S based sequencing platforms have been conducted. In addition to advancing knowledge related to animal health, these studies help decipher pathophysiology of human diseases, as domestic animals naturally develop similar conditions. Microbiota findings across domestic species are summarized and compared below.
Dual Roles of Microbiota
Microbiota support mammalian health by promoting multiple physiologic processes, including nutrient metabolism, immune regulation, intestinal development, and antimicrobial protection (Jandhyala et al., 2015). On the other hand, alterations in the composition and function of the microbiota is implicated in many disorders such as inflammatory bowel disease (IBD) (Schirmer et al., 2019). IBD includes Crohn’s disease and ulcerative colitis and affects millions worldwide (Ng et al., 2017). The etiology of IBD is multifactorial including genetics, epigenetics, and environmental factors. Although IBD has been associated with alterations in the composition of microbiota in patients (Gevers et al., 2014; Schirmer et al., 2018; Lloyd-Price et al., 2019), the causative impact of altered microbiota is still under investigation.
Most studies aimed at understanding the underlying nature of IBD have relied heavily on experimental or genetic murine-models of intestinal inflammation in which altered microbiota composition also frequently occurs (Gkouskou et al., 2014). However, comparative studies in other mammalian species with intestinal inflammation is likely to yield critical clues into shared microbiome-based etiologies that occur in natural disease. Gastrointestinal disease is one of the leading health issues of domestic animals. IBD and other idiopathic enteropathies in animals pose challenges due to a lack of understanding of the cause. Microbiota disturbance has long been suggested, but more recent microbiome research has been undertaken to directly test this hypothesis in animal/veterinary science. In this review, we discuss these studies and highlight evidence supporting that gut dysbiosis and intestinal inflammation in domestic mammals are tightly linked, similar to observations in humans and murine models.
Gut Microbiota of Domestic Mammals
Dominance of two bacterial phyla, Firmicutes and Bacteroidetes, is unique to the mammalian hind gut ecosystem and not detected in fish intestine and plants (Mahowald et al., 2009; Ikeda-Ohtsubo et al., 2018). Microbiota of domestic animals generally follow this rule, with the exception that Fusobacteria can be one of the dominant phyla in canine and feline microbiota (Figure 1; Handl et al., 2011; Guard et al., 2015; Panasevich et al., 2015; Pilla and Suchodolski, 2020). Some symbiotic mechanisms provided by Firmicutes and Bacteroidetes include breaking down of dietary fibers and glycan to produce nutrients for colonocytes (Mahowald et al., 2009). Also, many indigenous Firmicutes and Bacteroidetes species are resistant to local antimicrobial peptides (Cullen et al., 2015). At lower taxonomic levels, microbiota compositions are distinct for each domestic species (Figure 1), possibly reflecting environment or host factors (Ley et al., 2008; Woo and Alenghat, 2017; Rothschild et al., 2018). Current knowledge of flora in domestic species during heath and inflammation are discussed below, focusing on data from 16S rRNA gene sequencing.
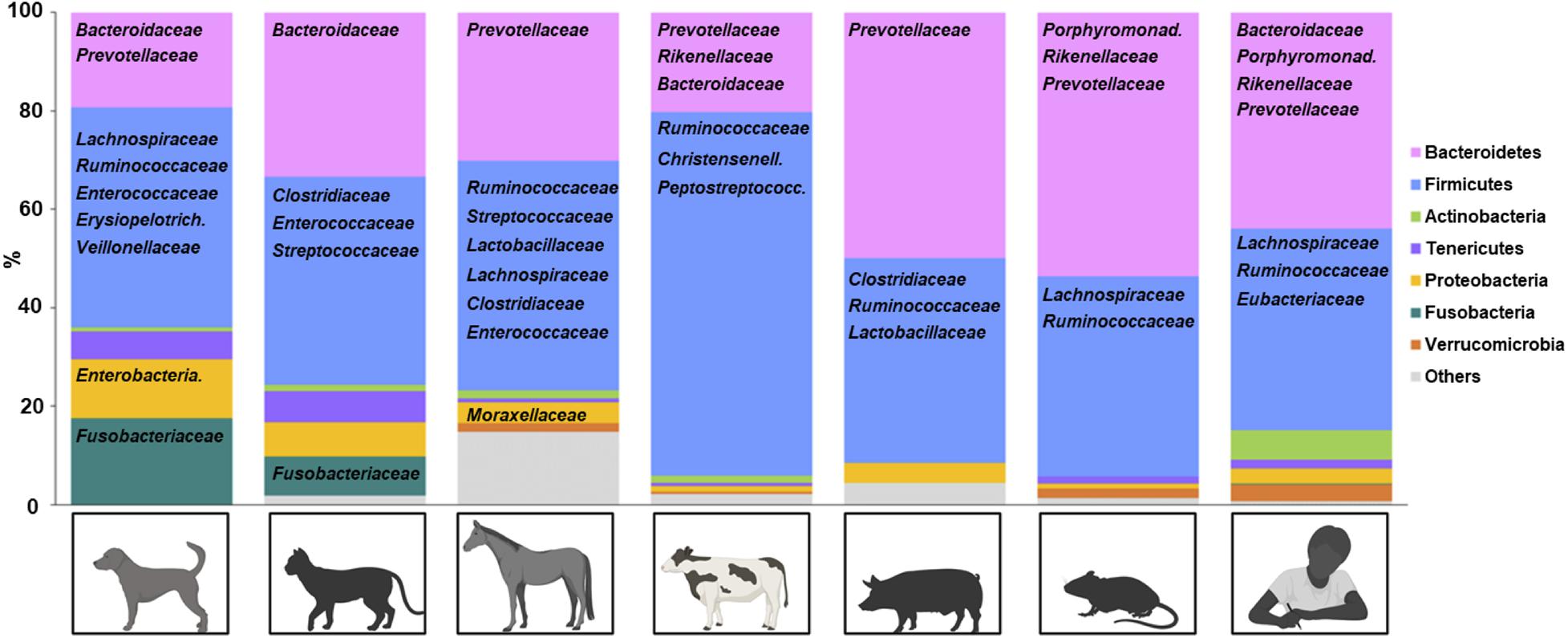
Figure 1. Representative fecal microbiota of healthy domestic animals. Phyla are represented by color and bacteria (family level) are represented below the corresponding phylum. Created with Biorender.com.
Canine
Domestication of dogs is estimated to have occurred 12,000 years ago through acquisition of wild gray wolves (Larson and Fuller, 2014). Dogs have since shared environmental factors with humans. In particular, sharing food with human companions has impacted the microbiota composition of the dogs during evolution (Axelsson et al., 2013; Wang et al., 2013; Alessandri et al., 2019). Although large inter-individual variations were observed, five major phyla generally compose of dog microbiota including Firmicutes, Bacteroidetes, Fusobacteria, Actinobacteria and Proteobacteria. At the genus level, commonly isolated taxa in healthy dogs include Clostridium, Eubacterium, Lactobacillus, Faecalibacterium, Ruminococcus, Prevotella, Fusobacterium, Bacteroides, Lachnospira, and Megamonas (Handl et al., 2011; Guard et al., 2015; Panasevich et al., 2015; Pilla and Suchodolski, 2020).
Inflammatory bowel disease is a common non-infectious cause of chronic diarrhea and vomiting in dogs, with overrepresentation of certain breeds including German Shepherd, Norwegian Lundehund, Yorkshire terrier, Boxer and French bulldog (Dandrieux and Mansfield, 2019). While breed predisposition supports a genetic component, multiple studies have also shown association with dysbiosis in dogs. Some common microbiota findings were decreased Firmicutes, including Clostridia and Faecalibacterium, and decreased Fusobacterium, increase in Gammaproteobacteria including Escherichia. Alpha-diversity, which represents number of taxonomic groups, was decreased, and beta-diversity, which represents community similarity, was significantly different in IBD-microbiota compared to microbiota from non-IBD dogs (Table 1; Honneffer et al., 2014; Guard et al., 2015; Minamoto et al., 2015; Vázquez-Baeza et al., 2016; Niina et al., 2019). Aside from decreased Fusobacterium in canine IBD, these findings are similar to some human IBD studies (Pittayanon et al., 2020).
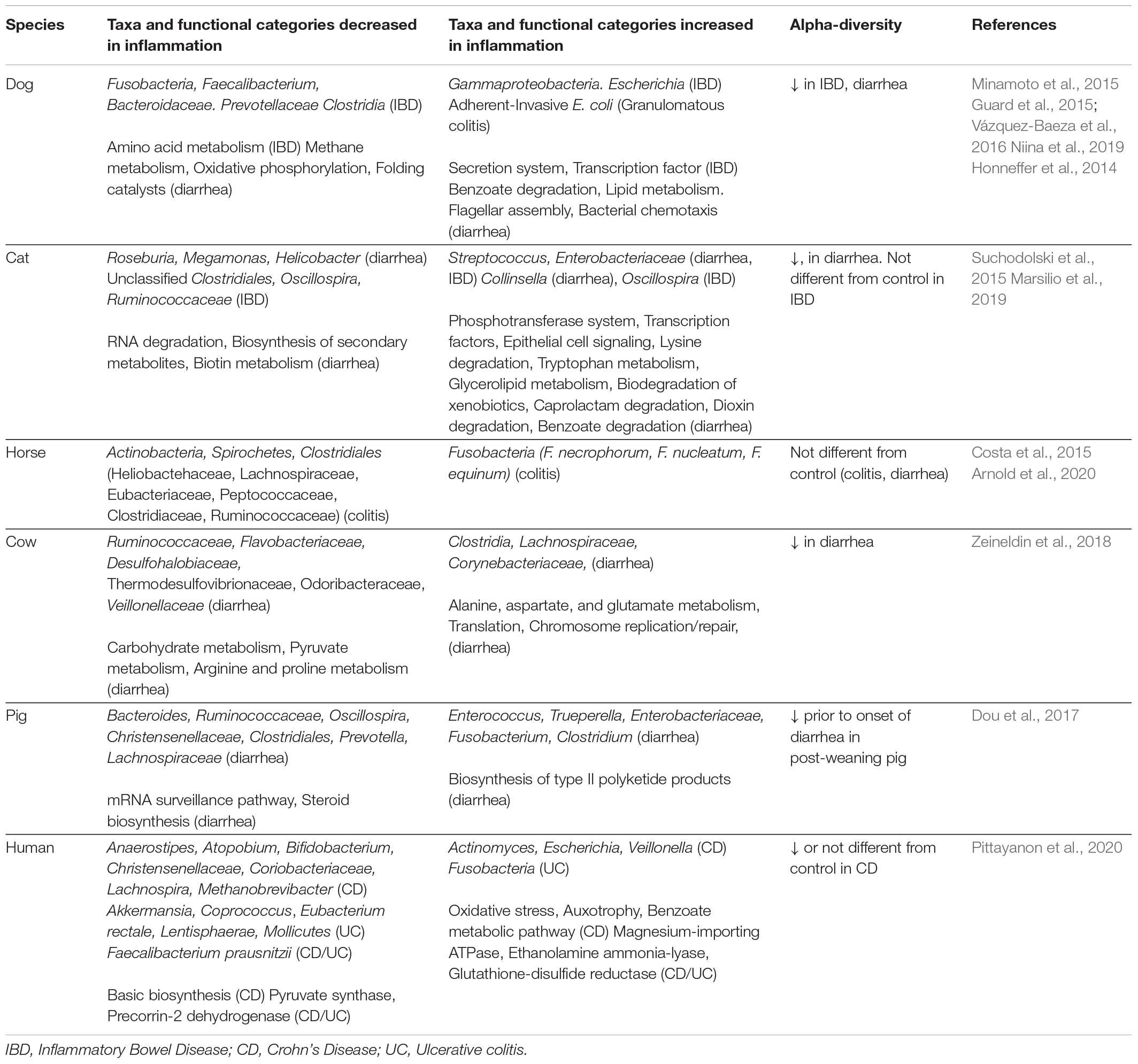
Table 1. Altered microbiota composition and function in intestinal inflammation across domestic animals.
In contrast, a recent meta-analysis study comparing canine and human IBD metagenomes found distinct dysbiosis networks in canine versus human IBD microbiota (Vázquez-Baeza et al., 2016). Further studies are required to elucidate whether certain taxonomic groups found in multispecies have causative roles in IBD, or whether the imbalance of microbiota has collective functional effects. So far, a limited number of canine studies have conducted functional analyses of microbiota in the context of intestinal inflammation (Table 1; Guard et al., 2015; Minamoto et al., 2015). Interestingly, enrichment of the benzoate metabolism pathway was identified in the microbiota of dogs with diarrhea and this pathway has also been described as enriched in microbiota from IBD patients (Gevers et al., 2014; Guard et al., 2015; Lewis et al., 2015; Tataru and David, 2020).
One interesting exception in canine IBD is granulomatous colitis (GC) of Boxer and French bulldogs, in which the causative agent has been clearly associated with adherent-invasive E. coli (AIEC) (Simpson et al., 2006). Eradication of AIEC with antibiotics cures this disease (Manchester et al., 2013). Susceptibility to AIEC invasion was linked to polymorphism in SLAM family members/CD48 gene in the recent canine GWAS study (Hayward et al., 2016). Because AIEC has been isolated from subsets of human Crohn’s disease (CD) patients, and granulomatous lesions in canine GC mimic some CD lesions, microbiological comparisons between dog and human AIEC may elucidate a subset of CD pathophysiology (Dogan et al., 2020). Some canine studies have demonstrated beneficial effects of fecal microbiota transfer in IBD (Makielski et al., 2019; Niina et al., 2019), but factors that lead to improvement still need to be identified (Cammarota and Ianiro, 2019). Overall, canine microbiome analyses are the most advanced among domestic species and canine IBD may serve as a strong natural model for human IBD.
Feline
Unlike dogs that classify as omnivorous, cats are obligate carnivores and may not rely much on microbiome fermentation for energy. Nevertheless, commensal microbes inhabit the feline intestine at similar levels to those in the dog and human (Johnston et al., 2001; Sender et al., 2016; Pilla and Suchodolski, 2020), and bacterial fermentation occurs in the feline intestine (Whittemore et al., 2019; Lyu et al., 2020). Similar to dogs, the major bacterial phyla in the domestic cat intestine are Firmicutes, Bacteroidetes, Fusobacteria, Actinobacteria and Proteobacteria (Figure 1; Ramadan et al., 2014; Suchodolski et al., 2015; Lyu et al., 2020). Lower taxa commonly found include Clostridium, Enterococcus, Streptococcus, Bacteroide, Faecalibacterium, Fusobacterium, and Collinsella (Minamoto et al., 2012; Rochus et al., 2014; Lyu et al., 2020).
Idiopathic intestinal inflammation resulting in diarrhea is a common occurrence in domestic cats and differences in microbiota composition have been observed in cats with intestinal inflammation (Table 1; Suchodolski et al., 2015; Marsilio et al., 2019). For example, Enterobacteriaceae and Streptococcus were increased in affected animals in two studies. Alpha-diversity was decreased when feline patients exhibited diarrhea, but this parameter was not significantly different in IBD in the second study. Functional analysis of microbiota from the cats with diarrhea demonstrated enrichment of the following pathways: phosphotransferase system, tryptophan metabolism, dioxin degradation and xylene degradation (Table 1; Suchodolski et al., 2015). Interestingly, the benzoate degradation pathway found in dogs and humans also trended upwards in feline samples (adjusted p-value 0.07). In addition, microbiota composition has been associated with other feline conditions such as obesity and diabetes (Kieler et al., 2016, 2019). Although only a few studies have been conducted for cates, feline dysbiosis has been demonstrated with intestinal inflammation.
Equine
The horse is a monogastric herbivore that depends heavily on hind gut fermentation for its energy and nutritional requirements. Firmicutes, the major phylum fermenting dietary fibers in the equine gut, generally exhibits the largest relative abundance, followed by Bacteroidetes. Presence of Actinobacteria, Tenericutes, Proteobacteria, Verrucomicrobia, Fibrobacteres, and Spirochetes are also commonly reported in the equine intestine, albeit with much smaller abundances. Representative taxa at the genus level in healthy horses include Clostridium, Ruminococcus, Streptococcus, Butyrivibrio (Family Lachnospiraceae), Fibrobacter, Lactobacillus, Prevotella, and Enterococcus (Figure 1; Costa et al., 2012; Kauter et al., 2019; Stewart et al., 2019; Arnold et al., 2020). Balanced host-microbiota symbiosis in horses not only provides nutrition, but is also needed for healthy intestinal homeostasis. This is exemplified by the condition of antibiotic-induced equine colitis in which only a few doses of antibiotics can trigger fatal, non-infectious enterocolitis in horses (Cohen and Woods, 1999).
Inflammatory bowel disease in horses is less commonly reported partly due to difficulty obtaining biopsies for definitive diagnosis. Microbiota studies are also not extensively described specifically in equine IBD (Boshuizen et al., 2018). Instead, microbiota analyses in equine gut have been more commonly studied in idiopathic colitis, which can be caused by antibiotics, or stress-triggering factors such as transportation and dietary changes. Two studies found an increase in Fusobacteria in horses with idiopathic colitis, and a decrease in Clostridia (Table 1; Costa et al., 2012; Arroyo et al., 2020). However, alpha-diversity was not significantly different between healthy horses versus horses with colitis in these studies. Instead, antibiotics predictably reduced diversity of microbiota in asymptomatic horses (Costa et al., 2015; Arnold et al., 2020). It is likely that the microbiota shifts during disease are dynamic in the horse intestine. Among Fusobacteria species increased in equine colitis, Fusobacterium nucleatum has also been associated with human ulcerative colitis (Paramsothy et al., 2017; Tahara et al., 2017), and been found to exacerbate inflammation in a murine colitis model through IL-17F and NF-κB signaling. Thus F. nucleatum may represent an inflammation-triggering commensal that is shared across species (Chen et al., 2020). While association of dysbiosis and intestinal inflammation are demonstrated in multiple equine studies, functional microbiota analyses, coupled with molecular analyses of equine cells, will allow a better understanding for how equine dysbiosis triggers inflammation.
Bovine
The importance of microbiota composition in bovine health has long been recognized. The first compartment of cow’s forestomach, the rumen, holds 70–100 L of fluids composed of mutualistic flora and their metabolites, which are a major source of energy and nutrition of the cow. The cow’s diet is commonly formulated to maintain healthy rumen flora. Transfer of rumen fluid from a healthy cow to a cow with gastrointestinal disorders (transfaunation) has been practiced since the 18th century as a common therapeutic (DePeters and George, 2014). The microbiota of the cow intestine has received less attention than the rumen. Available data demonstrate a dominance of Firmicutes, suggesting there is continuous bacterial fermentation throughout the intestinal tract. Less abundant phyla in the cow intestines include Bacteroidetes, Actinobacteria, Proteobacteria, Tenericutes, and Verrucomicrobia. At the family/genus level, some of the representative taxa include Ruminococcaceae, Christensenellaceae, Peptostreptococcaceae, Clostridiaceae, Bacteroides, Prevotella, Rikenellaceae, Butyrivibrio, Bifidobacterium, Campylobacter and Acinetobacter (Figure 1; Mao et al., 2015; Fecteau et al., 2016; Li et al., 2020; Uchiyama et al., 2020). Functional analysis of the intestinal microbiota of cows has identified enriched pathways such as energy metabolism, replication and repair, amino acid metabolism, carbohydrate metabolism, and membrane transport. These indicate important roles of intestinal microbiota in nutrition and intestinal homeostasis.
Significant changes in microbiota composition in the intestine has been found in feedlot calves with hemorrhagic diarrhea with undefined etiology (Zeineldin et al., 2018). The calves with diarrhea had reduced Ruminococcaceae, Flavobacteriaceae, Desulfohalobiaceae, Thermodesulfovibrionaceae, and increased Clostridia, Lachnospiraceae. Some of the Clostridia in this data may have been pathogens, such as C. perfringens. Alpha-diversity of the microbiome was significantly decreased in the intestine of calves with diarrhea (Table 1; Zeineldin et al., 2018). IBD is not commonly diagnosed in cows, however, Johne’s disease, caused by Mycobacterium avium ssp. paratuberculosis (MAP) infection, shares many clinical and pathological features with Crohn’s disease (CD) of human (McNees et al., 2015). Because MAP infection has been controversially suspected as a cause of a subset of CD, a study was conducted to examine whether MAP infected, asymptomatic cows have dysbiosis (Fecteau et al., 2016). This work demonstrated dysbiosis in MAP-positive cows characterized by significantly decreased Firmicutes and Bacteroidetes and increased Actinobacteria. Because all animals were non-clinical in the study, the biological significance of this dysbiosis was unclear, though it was suggested as a potential predisposing factor for granulomatous enteritis and chronic enteropathy (Fecteau et al., 2016). Although the hind gut dysbiosis in bovine intestinal diseases are demonstrated in above studies, causal roles remain to be elucidated.
Porcine
Because of the similarity of intestinal anatomy of pigs and humans, pigs are often employed as experimental models for human intestinal diseases. Thus, their intestinal microbiota has been studied under more well-controlled conditions, relative to other large animal species. Firmicutes, Bacteroidetes and Proteobacteria are the major phyla most commonly found in pig distal intestine. At lower taxonomic levels, genera Clostridium, Blautia, Lactobacillus, Prevotella, Ruminococcus, Roseburia, and Subdoligranulum represent some of the core commensal bacteria of the domestic pig (Allen et al., 2011; Holman et al., 2017; Han et al., 2018). While several pig models of experimentally induced intestinal inflammation have been described, naturally occurring IBD is not reported in domestic pigs. A longitudinal study examining microbiota of post-weaning pigs in association of development of diarrhea revealed significant dysbiosis prior to onset of diarrhea (Table 1; Dou et al., 2017). Multiple taxa including Bacteroides, Ruminococcaceae, Oscillospira, Christensenellaceae, Prevotella, and Lachnospiraceae were decreased; and Enterococcus, Trueperella, Enterobacteriacea, Fusobacterium and Clostridium were increased in the affected group prior to onset of diarrhea, compared to the healthy control pigs that never developed diarrhea. Unlike the other clinical studies in which duration or presence of dysbiosis prior to onset of intestinal inflammation is unknown, significant dysbiosis prior to the onset of diarrhea in this study suggests a potential causative role. Causes of the dysbiosis itself were not determined in this study. This study highlighted an advantage of natural porcine models in which a large number of similar age pigs can be examined at the same time in a controlled environment.
Discussion and Future Directions
While most animal studies in this review did not employ statistical adjustment for confounding factors that could affect the intestinal flora, the findings support that dysbiosis plays a role in pathophysiology of intestinal inflammation across mammalian species. Aside from a subset of canine IBD and bovine MAP, the etiologies of IBD or non-infectious enteropathy in domestic animals are not attributed to a single pathogen or pathosymbiont. Rather, triggers may be associated with collective functional changes of microbiota and this effect on host cells, as proposed in human IBD (Ni et al., 2017). A recent systematic review of 48 human microbiome metagenomic studies did not identify any species that were universally associated with human IBD. However, multiple studies have identified common taxonomic perturbations in IBD, including increased Escherichia and decreased Faecalibacterium, which were also identified with IBD in dogs (Honneffer et al., 2014; Minamoto et al., 2015; Vázquez-Baeza et al., 2016; Pittayanon et al., 2020). A large human longitudinal study integrating metagenome, metatranscriptome and metabolome datasets did not identify taxa that commonly associate with IBD. Instead, functional profiling of microbiota, in combination with metabolite abundance, was more effective at separating IBD patients from controls (Lloyd-Price et al., 2019). While data examining functional alterations of microbiota in animal intestinal diseases are currently limited, a small number of dog and cat studies have described that the benzoate degradation pathway is increased in intestinal inflammation (Guard et al., 2015; Suchodolski et al., 2015). Interestingly, this pathway is similarly altered with human IBD (Gevers et al., 2014; Lewis et al., 2015; Tataru and David, 2020).
Among the domestic species, canine research is currently the most advanced with incorporation of functional profiling of microbiota and stratification of patient populations by treatment (Minamoto et al., 2015; Niina et al., 2019). The genetics of dogs due to inbreeding may be advantageous for further dissection of genome-microbiota-disease analyses, as fewer samples may be required to identify disease polymorphisms (Hayward et al., 2016). Human IBD studies geared towards precision medicine employ multiomics to study interactions of genetics, epigenetics, microbiota and metabolites (Imhann et al., 2018; Lloyd-Price et al., 2019; Ryan et al., 2020). This type of work has shown connections between host genotype and microbiota in human IBD (Putignani et al., 2019; Benech and Sokol, 2020). Specifically, IBD risk polymorphisms of NOD2, AGL16L1 and FUT2 genes were associated with compositional and functional changes in the microbiota (Frank et al., 2011; Rausch et al., 2011; Knights et al., 2014). Interestingly, canine NOD2 genes also associated with IBD in German shepherds (Kathrani et al., 2014). Other analyses of human metagenomics and metabolomics have found that outcomes of fecal transplantation reflected both the donor and recipient microbiota composition and function (Paramsothy et al., 2019). Rodent studies suggest microbiota composition may also alter the epigenetic status of mammalian cells (Krautkramer et al., 2016; Kelly et al., 2018; Ansari et al., 2020). In line with this, epithelial DNA methylation profiles were found to correlate with both dysbiosis and inflammation in IBD patients (Ryan et al., 2020). In addition, modeling metagenomic data to predict network dynamics are expected to guide microbiota manipulation. For example, use of the Lotka-Volterra model led to an algorithm to calculate fecal transplant efficiency and design personalized probiotic cocktails (Xiao et al., 2020). Thus, the next steps for taking advantage of comparative animal models and developing microbiota-based strategies for domestic species will require integration of multiomic host data with microbiome and metabolite analyses.
Author Contributions
TA and SH-H planned and wrote the manuscript. Both authors contributed to the article and approved the submitted version.
Funding
The Alenghat laboratory is supported by the National Institutes of Health (DK114123, DK116868), Pew Charitable Trust, and a Kenneth Rainin Foundation award.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank members of the Alenghat lab for discussions and reading this work. TA holds an Investigator in the Pathogenesis of Infectious Disease Award from the Burroughs Wellcome Fund.
References
Alessandri, G., Milani, C., Mancabelli, L., Mangifesta, M., Lugli, G. A., Viappiani, A., et al. (2019). Metagenomic dissection of the canine gut microbiota: insights into taxonomic, metabolic and nutritional features. Environ. Microbiol. 21, 1331–1343. doi: 10.1111/1462-2920.14540
Allen, H. K., Looft, T., Bayles, D. O., Humphrey, S., Levine, U. Y., Alt, D., et al. (2011). Antibiotics in feed induce prophages in swine fecal microbiomes. MBio 2:e00260-11. doi: 10.1128/mBio.00260-11
Ansari, I., Raddatz, G., Gutekunst, J., Ridnik, M., Cohen, D., Abu-Remaileh, M., et al. (2020). The microbiota programs DNA methylation to control intestinal homeostasis and inflammation. Nat. Microbiol. 5, 610–619. doi: 10.1038/s41564-019-0659-3
Arnold, C. E., Isaiah, A., Pilla, R., Lidbury, J., Coverdale, J. S., Callaway, T. R., et al. (2020). The cecal and fecal microbiomes and metabolomes of horses before and after metronidazole administration. PLoS One 15:e0232905. doi: 10.1371/journal.pone.0232905
Arroyo, L. G., Rossi, L., Santos, B. P., Gomez, D. E., Surette, M. G., and Costa, M. C. (2020). Luminal and mucosal microbiota of the cecum and large colon of healthy and diarrheic horses. Animals 10:1403 doi: 10.3390/ani10081403
Axelsson, E., Ratnakumar, A., Arendt, M.-L., Maqbool, K., Webster, M. T., Perloski, M., et al. (2013). The genomic signature of dog domestication reveals adaptation to a starch-rich diet. Nature 495, 360–364. doi: 10.1038/nature11837
Benech, N., and Sokol, H. (2020). Fecal microbiota transplantation in gastrointestinal disorders: time for precision medicine. Genome Med. 12:58. doi: 10.1186/s13073-020-00757-y
Boshuizen, B., Ploeg, M., Dewulf, J., Klooster, S., Bruijn, M., de Picavet, M.-T., et al. (2018). Inflammatory bowel disease (IBD) in horses: a retrospective study exploring the value of different diagnostic approaches. BMC Vet. Res. 14:21. doi: 10.1186/s12917-018-1343-1
Cammarota, G., and Ianiro, G. (2019). FMT for ulcerative colitis: closer to the turning point. Nat. Rev. Gastroenterol. Hepatol. 16, 266–268. doi: 10.1038/s41575-019-0131-0
Chen, Y., Chen, Y., Cao, P., Su, W., Zhan, N., and Dong, W. (2020). Fusobacterium nucleatum facilitates ulcerative colitis through activating IL-17F signaling to NF-κB via the upregulation of CARD3 expression. J. Pathol. 250, 170–182. doi: 10.1002/path.5358
Clemente, J. C., Ursell, L. K., Parfrey, L. W., and Knight, R. (2012). The impact of the gut microbiota on human health: an integrative view. Cell 148, 1258–1270. doi: 10.1016/j.cell.2012.01.035
Cohen, N. D., and Woods, A. M. (1999). Characteristics and risk factors for failure of horses with acute diarrhea to survive: 122 cases (1990-1996). J. Am. Vet. Med. Assoc. 214, 382–390.
Costa, M. C., Arroyo, L. G., Allen-Vercoe, E., Stämpfli, H. R., Kim, P. T., Sturgeon, A., et al. (2012). Comparison of the fecal microbiota of healthy horses and horses with colitis by high throughput sequencing of the V3-V5 region of the 16S rRNA gene. PLoS One 7:e41484. doi: 10.1371/journal.pone.0041484
Costa, M. C., Stämpfli, H. R., Arroyo, L. G., Allen-Vercoe, E., Gomes, R. G., and Weese, J. S. (2015). Changes in the equine fecal microbiota associated with the use of systemic antimicrobial drugs. BMC Vet. Res. 11:19. doi: 10.1186/s12917-015-0335-7
Cullen, T. W., Schofield, W. B., Barry, N. A., Putnam, E. E., Rundell, E. A., Trent, M. S., et al. (2015). Antimicrobial peptide resistance mediates resilience of prominent gut commensals during inflammation. Science 347, 170–175. doi: 10.1126/science.1260580
Dandrieux, J. R. S., and Mansfield, C. S. (2019). Chronic enteropathy in canines: prevalence, impact and management strategies. Vet. Med. 10, 203–214. doi: 10.2147/VMRR.S162774
DePeters, E. J., and George, L. W. (2014). Rumen transfaunation. Immunol. Lett. 162, 69–76. doi: 10.1016/j.imlet.2014.05.009
Dogan, B., Zhang, S., Kalla, S. E., Dogan, E. I., Guo, C., Ang, C. R., et al. (2020). Molecular and phenotypic characterization of Escherichia coli associated with granulomatous colitis of boxer dogs. Antibiotics 9:540. doi: 10.3390/antibiotics9090540
Dou, S., Gadonna-Widehem, P., Rome, V., Hamoudi, D., Rhazi, L., Lakhal, L., et al. (2017). Characterisation of early-life fecal microbiota in susceptible and healthy pigs to post-weaning diarrhoea. PLoS One 12:e0169851. doi: 10.1371/journal.pone.0169851
Fecteau, M.-E., Pitta, D. W., Vecchiarelli, B., Indugu, N., Kumar, S., Gallagher, S. C., et al. (2016). Dysbiosis of the fecal microbiota in cattle infected with Mycobacterium avium subsp. paratuberculosis. PLoS One 11:e0160353. doi: 10.1371/journal.pone.0160353
Frank, D. N., Robertson, C. E., Hamm, C. M., Kpadeh, Z., Zhang, T., Chen, H., et al. (2011). Disease phenotype and genotype are associated with shifts in intestinal-associated microbiota in inflammatory bowel diseases. Inflamm. Bowel Dis. 17, 179–184. doi: 10.1002/ibd.21339
Gevers, D., Kugathasan, S., Denson, L. A., Vázquez-Baeza, Y., Van Treuren, W., Ren, B., et al. (2014). The treatment-naive microbiome in new-onset Crohn’s disease. Cell Host Microbe 15, 382–392. doi: 10.1016/j.chom.2014.02.005
Gkouskou, K., Deligianni, C., Tsatsanis, C., and Eliopoulos, A. (2014). The gut microbiota in mouse models of inflammatory bowel disease. Front. Cell. Infect. Microbiol. 4:28. doi: 10.3389/fcimb.2014.00028
Guard, B. C., Barr, J. W., Reddivari, L., Klemashevich, C., Jayaraman, A., Steiner, J. M., et al. (2015). Characterization of microbial dysbiosis and metabolomic changes in dogs with acute diarrhea. PLoS One 10:e0127259. doi: 10.1371/journal.pone.0127259
Han, G. G., Lee, J.-Y., Jin, G.-D., Park, J., Choi, Y. H., Kang, S.-K., et al. (2018). Tracing of the fecal microbiota of commercial pigs at five growth stages from birth to shipment. Sci. Rep. 8:6012. doi: 10.1038/s41598-018-24508-7
Handl, S., Dowd, S. E., Garcia-Mazcorro, J. F., Steiner, J. M., and Suchodolski, J. S. (2011). Massive parallel 16S rRNA gene pyrosequencing reveals highly diverse fecal bacterial and fungal communities in healthy dogs and cats. FEMS Microbiol. Ecol. 76, 301–310. doi: 10.1111/j.1574-6941.2011.01058.x
Hayward, J. J., Castelhano, M. G., Oliveira, K. C., Corey, E., Balkman, C., Baxter, T. L., et al. (2016). Complex disease and phenotype mapping in the domestic dog. Nat. Commun. 7:10460. doi: 10.1038/ncomms10460
Holman, D. B., Brunelle, B. W., Trachsel, J., and Allen, H. K. (2017). Meta-analysis to define a core microbiota in the swine gut. mSystems 2:e00004-17. doi: 10.1128/mSystems.00004-17
Honneffer, J. B., Minamoto, Y., and Suchodolski, J. S. (2014). Microbiota alterations in acute and chronic gastrointestinal inflammation of cats and dogs. World J. Gastroenterol. 20, 16489–16497. doi: 10.3748/wjg.v20.i44.16489
Ikeda-Ohtsubo, W., Brugman, S., Warden, C. H., Rebel, J. M. J., Folkerts, G., and Pieterse, C. M. J. (2018). How can we define “Optimal Microbiota?” A comparative review of structure and functions of microbiota of animals, fish, and plants in agriculture. Front. Nutr. 5:90. doi: 10.3389/fnut.2018.00090
Imhann, F., Vich Vila, A., Bonder, M. J., Fu, J., Gevers, D., Visschedijk, M. C., et al. (2018). Interplay of host genetics and gut microbiota underlying the onset and clinical presentation of inflammatory bowel disease. Gut 67, 108–119. doi: 10.1136/gutjnl-2016-312135
Jandhyala, S. M., Talukdar, R., Subramanyam, C., Vuyyuru, H., Sasikala, M., and Nageshwar Reddy, D. (2015). Role of the normal gut microbiota. World J. Gastroenterol. 21, 8787–8803. doi: 10.3748/wjg.v21.i29.8787
Johnston, K. L., Swift, N. C., Forster-van Hijfte, M., Rutgers, H. C., Lamport, A., Ballàvre, O., et al. (2001). Comparison of the bacterial flora of the duodenum in healthy cats and cats with signs of gastrointestinal tract disease. J. Am. Vet. Med. Assoc. 218, 48–51. doi: 10.2460/javma.2001.218.48
Kathrani, A., Lee, H., White, C., Catchpole, B., Murphy, A., German, A., et al. (2014). Association between nucleotide oligomerisation domain two (n.d.) gene polymorphisms and canine inflammatory bowel disease. Vet. Immunol. Immunopathol. 161, 32–41. doi: 10.1016/j.vetimm.2014.06.003
Kauter, A., Epping, L., Semmler, T., Antao, E.-M., Kannapin, D., Stoeckle, S. D., et al. (2019). The gut microbiome of horses: current research on equine enteral microbiota and future perspectives. Anim. Microbiome 1:14. doi: 10.1186/s42523-019-0013-3
Kelly, D., Denson, L. A., Alenghat, T., Kelly, D., Kotliar, M., Woo, V., et al. (2018). Microbiota-sensitive epigenetic signature predicts inflammation in Crohn’ s disease. J. Clin. Investig. Insight 3:e122104.
Kieler, I. N., Mølbak, L., Hansen, L. L., Hermann-Bank, M. L., and Bjornvad, C. R. (2016). Overweight and the feline gut microbiome – a pilot study. J. Anim. Physiol. Anim. Nutr. 100, 478–484. doi: 10.1111/jpn.12409
Kieler, I. N., Osto, M., Hugentobler, L., Puetz, L., Gilbert, M. T. P., Hansen, T., et al. (2019). Diabetic cats have decreased gut microbial diversity and a lack of butyrate producing bacteria. Sci. Rep. 9:4822. doi: 10.1038/s41598-019-41195-0
Knights, D., Silverberg, M. S., Weersma, R. K., Gevers, D., Dijkstra, G., Huang, H., et al. (2014). Complex host genetics influence the microbiome in inflammatory bowel disease. Genome Med. 6:107. doi: 10.1186/s13073-014-0107-1
Krautkramer, K. A., Kreznar, J. H., Romano, K. A., Vivas, E. I., Barrett-Wilt, G. A., Rabaglia, M. E., et al. (2016). Diet-microbiota interactions mediate global epigenetic programming in multiple host tissues. Mol. Cell 64, 982–992. doi: 10.1016/j.molcel.2016.10.025
Larson, G., and Fuller, D. Q. (2014). The evolution of animal domestication. Annu. Rev. Ecol. Evol. Syst. 45, 115–136. doi: 10.1146/annurev-ecolsys-110512-135813
Lewis, J. D., Chen, E. Z., Baldassano, R. N., Otley, A. R., Griffiths, A. M., Lee, D., et al. (2015). Inflammation, antibiotics, and diet as environmental stressors of the gut microbiome in pediatric Crohn’s disease. Cell Host Microbe 18, 489–500. doi: 10.1016/j.chom.2015.09.008
Ley, R. E., Hamady, M., Lozupone, C., Turnbaugh, P. J., Ramey, R. R., Bircher, J. S., et al. (2008). Evolution of mammals and their gut microbes. Science 320, 1647–1651. doi: 10.1126/science.1155725
Li, H., Li, R., Chen, H., Gao, J., Wang, Y., Zhang, Y., et al. (2020). Effect of different seasons (spring vs summer) on the microbiota diversity in the feces of dairy cows. Int. J. Biometeorol. 64, 345–354. doi: 10.1007/s00484-019-01812-z
Lloyd-Price, J., Arze, C., Ananthakrishnan, A. N., Schirmer, M., Avila-Pacheco, J., Poon, T. W., et al. (2019). Multi-omics of the gut microbial ecosystem in inflammatory bowel diseases. Nature 569, 655–662. doi: 10.1038/s41586-019-1237-9
Lyu, Y., Su, C., Verbrugghe, A., Van de Wiele, T., Martos Martinez-Caja, A., and Hesta, M. (2020). Past, present, and future of gastrointestinal microbiota research in cats. Front. Microbiol. 11:1661 doi: 10.3389/fmicb.2020.01661
Mahowald, M. A., Rey, F. E., Seedorf, H., Turnbaugh, P. J., Fulton, R. S., Wollam, A., et al. (2009). Characterizing a model human gut microbiota composed of members of its two dominant bacterial phyla. Proc. Natl. Acad. Sci. U.S.A. 106, 5859–5864. doi: 10.1073/pnas.0901529106
Makielski, K., Cullen, J., O’Connor, A., and Jergens, A. E. (2019). Narrative review of therapies for chronic enteropathies in dogs and cats. J. Vet. Intern. Med. 33, 11–22. doi: 10.1111/jvim.15345
Manchester, A. C., Hill, S., Sabatino, B., Armentano, R., Carroll, M., Kessler, B., et al. (2013). Association between granulomatous colitis in french bulldogs and invasive Escherichia coli and response to fluoroquinolone antimicrobials. J. Vet. Intern. Med. 27, 56–61. doi: 10.1111/jvim.12020
Mao, S., Zhang, M., Liu, J., and Zhu, W. (2015). Characterising the bacterial microbiota across the gastrointestinal tracts of dairy cattle: membership and potential function. Sci. Rep. 5:16116. doi: 10.1038/srep16116
Marsilio, S., Pilla, R., Sarawichitr, B., Chow, B., Hill, S. L., Ackermann, M. R., et al. (2019). Characterization of the fecal microbiome in cats with inflammatory bowel disease or alimentary small cell lymphoma. Sci. Rep. 9:19208. doi: 10.1038/s41598-019-55691-w
McNees, A. L., Markesich, D., Zayyani, N. R., and Graham, D. Y. (2015). Mycobacterium paratuberculosis as a cause of Crohn’s disease. Expert Rev. Gastroenterol. Hepatol. 9, 1523–1534. doi: 10.1586/17474124.2015.1093931
Minamoto, Y., Hooda, S., Swanson, K. S., and Suchodolski, J. S. (2012). Feline gastrointestinal microbiota. Anim. Heal. Res. Rev. 13, 64–77. doi: 10.1017/S1466252312000060
Minamoto, Y., Otoni, C. C., Steelman, S. M., Büyükleblebici, O., Steiner, J. M., Jergens, A. E., et al. (2015). Alteration of the fecal microbiota and serum metabolite profiles in dogs with idiopathic inflammatory bowel disease. Gut Microbes 6, 33–47. doi: 10.1080/19490976.2014.997612
Ng, S. C., Shi, H. Y., Hamidi, N., Underwood, F. E., Tang, W., Benchimol, E. I., et al. (2017). Worldwide incidence and prevalence of inflammatory bowel disease in the 21st century: a systematic review of population-based studies. Lancet 390, 2769–2778. doi: 10.1016/S0140-6736(17)32448-0
Ni, J., Wu, G. D., Albenberg, L., and Tomov, V. T. (2017). Gut microbiota and IBD: causation or correlation? Nat. Rev. Gastroenterol. Hepatol. 14, 573–584. doi: 10.1038/nrgastro.2017.88
Niina, A., Kibe, R., Suzuki, R., Yuchi, Y., Teshima, T., Matsumoto, H., et al. (2019). Improvement in clinical symptoms and fecal microbiome after fecal microbiota transplantation in a dog with inflammatory bowel disease. Vet. Med. 10, 197–201. doi: 10.2147/VMRR.S230862
Panasevich, M. R., Kerr, K. R., Dilger, R. N., Fahey, G. C., Guérin-Deremaux, L., Lynch, G. L., et al. (2015). Modulation of the faecal microbiome of healthy adult dogs by inclusion of potato fibre in the diet. Br. J. Nutr. 113, 125–133. doi: 10.1017/S0007114514003274
Paramsothy, S., Kamm, M. A., Kaakoush, N. O., Walsh, A. J., van den Bogaerde, J., Samuel, D., et al. (2017). Multidonor intensive faecal microbiota transplantation for active ulcerative colitis: a randomised placebo-controlled trial. Lancet 389, 1218–1228. doi: 10.1016/S0140-6736(17)30182-4
Paramsothy, S., Nielsen, S., Kamm, M. A., Deshpande, N. P., Faith, J. J., Clemente, J. C., et al. (2019). Specific bacteria and metabolites associated with response to fecal microbiota transplantation in patients with ulcerative colitis. Gastroenterology 156, 1440–1454.e2. doi: 10.1053/j.gastro.2018.12.001
Pilla, R., and Suchodolski, J. S. (2020). The role of the canine gut microbiome and metabolome in health and Gastrointestinal disease. Front. Vet. Sci. 6:498. doi: 10.3389/fvets.2019.00498
Pittayanon, R., Lau, J. T., Leontiadis, G. I., Tse, F., Yuan, Y., Surette, M., et al. (2020). Differences in gut microbiota in patients with vs without inflammatory bowel diseases: a systematic review. Gastroenterology 158, 930–946.e1. doi: 10.1053/j.gastro.2019.11.294
Putignani, L., Gasbarrini, A., and Dallapiccola, B. (2019). Potential of multiomics technology in precision medicine. Curr. Opin. Gastroenterol. 35, 491–498. doi: 10.1097/MOG.0000000000000589
Ramadan, Z., Xu, H., Laflamme, D., Czarnecki-Maulden, G., Li, Q. J., Labuda, J., et al. (2014). Fecal microbiota of cats with naturally occurring chronic diarrhea assessed using 16S rRNA gene 454-pyrosequencing before and after dietary treatment. J. Vet. Intern. Med. 28, 59–65. doi: 10.1111/jvim.12261
Rausch, P., Rehman, A., Künzel, S., Häsler, R., Ott, S. J., Schreiber, S., et al. (2011). Colonic mucosa-associated microbiota is influenced by an interaction of Crohn disease and FUT2 (Secretor) genotype. Proc. Natl. Acad. Sci. U.S.A. 108, 19030–19035. doi: 10.1073/pnas.1106408108
Rochus, K., Janssens, G. P. J., and Hesta, M. (2014). Dietary fibre and the importance of the gut microbiota in feline nutrition: a review. Nutr. Res. Rev. 27, 295–307. doi: 10.1017/S0954422414000213
Rothschild, D., Weissbrod, O., Barkan, E., Kurilshikov, A., Korem, T., Zeevi, D., et al. (2018). Environment dominates over host genetics in shaping human gut microbiota. Nature 555, 210–215. doi: 10.1038/nature25973
Ryan, F. J., Ahern, A. M., Fitzgerald, R. S., Laserna-Mendieta, E. J., Power, E. M., Clooney, A. G., et al. (2020). Colonic microbiota is associated with inflammation and host epigenomic alterations in inflammatory bowel disease. Nat. Commun. 11:1512. doi: 10.1038/s41467-020-15342-5
Schirmer, M., Denson, L., Vlamakis, H., Franzosa, E. A., Thomas, S., Gotman, N. M., et al. (2018). Compositional and temporal changes in the gut microbiome of pediatric ulcerative colitis patients are linked to disease course. Cell Host Microbe 24, 600–610.e4. doi: 10.1016/j.chom.2018.09.009
Schirmer, M., Garner, A., Vlamakis, H., and Xavier, R. J. (2019). Microbial genes and pathways in inflammatory bowel disease. Nat. Rev. Microbiol. 17, 497–511. doi: 10.1038/s41579-019-0213-6
Sender, R., Fuchs, S., and Milo, R. (2016). Revised estimates for the number of human and bacteria cells in the body. PLoS Biol. 14:e1002533. doi: 10.1371/journal.pbio.1002533
Simpson, K. W., Dogan, B., Rishniw, M., Goldstein, R. E., Klaessig, S., McDonough, P. L., et al. (2006). Adherent and invasive Escherichia coli is associated with granulomatous colitis in boxer dogs. Infect. Immun. 74, 4778–4792. doi: 10.1128/IAI.00067-06
Stewart, H. L., Southwood, L. L., Indugu, N., Vecchiarelli, B., Engiles, J. B., and Pitta, D. (2019). Differences in the equine faecal microbiota between horses presenting to a tertiary referral hospital for colic compared with an elective surgical procedure. Equine Vet. J. 51, 336–342. doi: 10.1111/evj.13010
Suchodolski, J. S., Foster, M. L., Sohail, M. U., Leutenegger, C., Queen, E. V., Steiner, J. M., et al. (2015). The fecal microbiome in cats with diarrhea. PLoS One 10:e0127378. doi: 10.1371/journal.pone.0127378
Tahara, T., Hirata, I., Nakano, N., Tahara, S., Horiguchi, N., Kawamura, T., et al. (2017). Potential link between Fusobacterium enrichment and DNA methylation accumulation in the inflammatory colonic mucosa in ulcerative colitis. Oncotarget 8, 61917–61926. doi: 10.18632/oncotarget.18716
Tataru, C. A., and David, M. M. (2020). Decoding the language of microbiomes using word-embedding techniques, and applications in inflammatory bowel disease. PLoS Comput. Biol. 16:e1007859 doi: 10.1371/journal.pcbi.1007859
Uchiyama, J., Murakami, H., Sato, R., Mizukami, K., Suzuki, T., Shima, A., et al. (2020). Examination of the fecal microbiota in dairy cows infected with bovine leukemia virus. Vet. Microbiol. 240:108547. doi: 10.1016/j.vetmic.2019.108547
Vázquez-Baeza, Y., Hyde, E. R., Suchodolski, J. S., and Knight, R. (2016). Dog and human inflammatory bowel disease rely on overlapping yet distinct dysbiosis networks. Nat. Microbiol. 1:16177. doi: 10.1038/nmicrobiol.2016.177
Wang, G., Zhai, W., Yang, H., Fan, R., Cao, X., Zhong, L., et al. (2013). The genomics of selection in dogs and the parallel evolution between dogs and humans. Nat. Commun. 4:1860. doi: 10.1038/ncomms2814
Whittemore, J. C., Stokes, J. E., Price, J. M., and Suchodolski, J. S. (2019). Effects of a synbiotic on the fecal microbiome and metabolomic profiles of healthy research cats administered clindamycin: a randomized, controlled trial. Gut Microbes 10, 521–539. doi: 10.1080/19490976.2018.1560754
Woo, V., and Alenghat, T. (2017). Host-microbiota interactions: epigenomic regulation. Curr. Opin. Immunol. 44, 52–60. doi: 10.1016/j.coi.2016.12.001
Xiao, Y., Angulo, M. T., Lao, S., Weiss, S. T., and Liu, Y.-Y. (2020). An ecological framework to understand the efficacy of fecal microbiota transplantation. Nat. Commun. 11:3329. doi: 10.1038/s41467-020-17180-x
Keywords: microbiota, IBD, canine, feline, equine, bovine, porcine
Citation: Hashimoto-Hill S and Alenghat T (2021) Inflammation-Associated Microbiota Composition Across Domestic Animals. Front. Genet. 12:649599. doi: 10.3389/fgene.2021.649599
Received: 06 January 2021; Accepted: 27 May 2021;
Published: 21 June 2021.
Edited by:
Xiaojiang Xu, National Institute of Environmental Health Sciences (NIEHS), United StatesReviewed by:
Scott H. Harrison, North Carolina Agricultural and Technical State University, United StatesAli Salehzadeh-Yazdi, University of Rostock, Germany
Copyright © 2021 Hashimoto-Hill and Alenghat. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Theresa Alenghat, dGhlcmVzYS5hbGVuZ2hhdEBjY2htYy5vcmc=
 Seika Hashimoto-Hill
Seika Hashimoto-Hill Theresa Alenghat
Theresa Alenghat