
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Genet. , 02 June 2021
Sec. Livestock Genomics
Volume 12 - 2021 | https://doi.org/10.3389/fgene.2021.649015
This article is part of the Research Topic Sheep and Goat Gene Exploration View all 19 articles
 Yuchun Xie1,2,3,4†
Yuchun Xie1,2,3,4† Zhihong Liu1,2,3,4†
Zhihong Liu1,2,3,4† Juntao Guo1,2,3,4†
Juntao Guo1,2,3,4† Xin Su1,2,3,4
Xin Su1,2,3,4 Cun Zhao1,2,3,4
Cun Zhao1,2,3,4 Chongyan Zhang1,2,3,4
Chongyan Zhang1,2,3,4 Qing Qin1,2,3,4
Qing Qin1,2,3,4 Dongliang Dai1,2,3,4
Dongliang Dai1,2,3,4 Yanhong Zhao1,2,3,4
Yanhong Zhao1,2,3,4 Zhiying Wang1,2,3,4
Zhiying Wang1,2,3,4 Ruijun Wang1,2,3,4
Ruijun Wang1,2,3,4 Yanjun Zhang1,2,3,4
Yanjun Zhang1,2,3,4 Rui Su1,2,3,4
Rui Su1,2,3,4 Zhixin Wang1,2,3,4*
Zhixin Wang1,2,3,4* Jinquan Li1,2,3,4*
Jinquan Li1,2,3,4*Fatty acid composition is an important aspect of meat quality in ruminants. Improving the beneficial fatty acid level in cashmere goat meat is important to its economic value. To investigate microRNAs (miRNAs) and mRNAs that regulate or coregulate polyunsaturated fatty acid (PUFA) synthesis and metabolism in the Inner Mongolia cashmere goat, we used longissimus dorsi muscle (WLM) and biceps femoris muscle (WBM) for transcript-level sequencing. RT-qPCR was used to evaluate the expression of mRNAs and miRNAs associated with PUFA synthesis and metabolism. The total PUFA content in the WBM was significantly higher than that in the WLM (P < 0.05). Our study is the first to systematically report miRNAs in cashmere goat meat. At the mRNA level, 20,375 genes were identified. ACSL1, CD36 and TECRL were at the center of a gene regulatory network and contributed significantly to the accumulation and metabolic regulation of fatty acids. At the miRNA level, 426 known miRNAs and 30 novel miRNAs were identified. KEGG analysis revealed that the miRNA target genes were involved mainly in the PPAR signaling pathway. The mRNA-miRNA coregulation analysis showed that ACSL1 was negatively targeted by nine miRNAs: chi-miR-10a-5p, chi-miR-10b-5p, chi-miR-130b-5p, chi-miR-15a-5p_R-1, chi-miR-15b-5p, chi-miR-16a-5p, chi-miR-16b-5p, chi-miR-181c-5p_R+1, and chi-miR-26b-5p. Finally, we speculated that the simultaneous silencing of ACSL1 by one or more of these nine miRNAs through PPAR signaling led to low ACSL1 expression in the WLM and, ultimately to high PUFA content in the WBM. Our study helps elucidate the metabolic regulation of fatty acids in Inner Mongolia cashmere goats.
The Inner Mongolian cashmere goat is a local breed that provides both cashmere and meat, but it is famous for its cashmere, which is known as “soft gold” (Su et al., 2015, 2020). The cashmere value of this goat has often caused its meat value to be ignored. Every year, nearly 700,000 cashmere goats are maintained, and the corresponding meat production is nearly 10,000 tons. Although the cashmere of this goat has been widely studied, its meat economic traits have not been reported. Fatty acid composition is a crucial aspect of meat quality in ruminants, as fatty acids often influence the flavor and juiciness of meat (Tvrzicka et al., 2011). In addition, fatty acids in meat are important for human health (Ladeira et al., 2018). Improving the quality of meat by increasing the concentrations of beneficial fatty acids to improve human health and reducing those of potentially detrimental fatty acids is an important undertaking (Scollan et al., 2014).
In the past few years, an increasing number of studies have demonstrated that microRNAs (miRNAs), once considered genomic “noise,” can mediate the epigenetic, transcriptional, and posttranscriptional regulation of genes involved in fatty acid metabolism by participating in the fatty acid regulatory network (Xu et al., 2010; Long et al., 2019; Zhang et al., 2019). Such studies have linked fatty acid metabolism with miRNA-mRNA relationships, which may provide new ideas and directions for exploration of the regulatory mechanisms of fatty acid metabolism in animals. Lu et al. (2020) found that miR-212 regulates fatty acid synthase (FASN) and sterol regulatory element binding factor 1 (SREBP1) expression by targeting the silent information regulator 2 (SIRT2) gene and repressing its expression, ultimately promoting fat deposition in mammary epithelial cells. In intramuscular preadipocytes, overexpression of miR-17-5p suppresses the expression of nuclear receptor coactivator 3 (NCOA3), fatty acid binding protein 4 (FABP4), and peroxisome proliferator-activated receptor gamma (PPARγ) and inhibits the differentiation of preadipocytes (Han et al., 2017). Depending on the degree of complementarity between miRNAs and the 3′-untranslated region (UTR) or 5′-UTR sequences of target mRNAs, miRNAs regulate mRNAs in two ways (Bartel, 2009): If the sequences are fully complementary, the miRNAs degrade the target mRNAs, whereas if the sequences are incompletely complementary, the miRNAs repress mRNA translation and thereby protein synthesis. Both ways accomplish gene regulation. Ma et al. (2018) found that overexpression of bta-miR-130a/b inhibits the expression of adipocyte differentiation-related genes, including peroxisome proliferator activated receptor gamma (PPARG), fatty acid binding protein 4 (FABP4), lipin 1 (LPIN1), and lipoprotein lipase (LPL). The aim of this study was to investigate polyunsaturated fatty acids (PUFAs) that are differentially present in different parts of the same goat, e.g., in the longissimus dorsi muscle (WLM) and biceps femoris muscle (WBM), and to use these two muscles as models to screen key miRNAs and mRNAs related to fatty acid synthesis and metabolism at the transcriptional level. Joint analysis of RNAs at both the mRNA and miRNA levels was further performed to construct a key miRNA-mRNA network associated with fatty acid metabolism and synthesis, which was further validated. The findings of this study help lay a foundation for the elucidation of the metabolic regulation of fatty acids in cashmere goats.
Samples were collected in accordance with the Guidelines for Experimental Animals of the Ministry of Science and Technology (Beijing, China) and were approved by the experimental animal ethics committee of Inner Mongolia Agricultural University (Approval No. [2020]056).
The animals were slaughtered under controlled conditions after being electrically stunned. WLM and WBM tissues were excised immediately between the 12th and 13th ribs (right half carcass) and rapidly stored in liquid nitrogen and subsequently stored at –80°C until analysis. Three replicates were provided by Inner Mongolia cashmere goats (Yiwei White Cashmere Goat Breeding Farm, Erdos, Inner Mongolia). In semidesert pastures, all goats are grazed year-round and feed freely. Two muscles from three goats (25.2 ± 0.16 kg, wether, 2 years old) were used for fatty acid composition analysis, miRNA sequencing (miRNA-Seq) and mRNA sequencing (mRNA-Seq) and RT-qPCR.
The extracted fatty acids were prepared with fatty acid methyl esters (Berton et al., 2016). The fatty acids were separated by an Agilent 5977B GC/MSD (Agilent, CA, United States) using a 100 m × 0.25 mm diameter SP-2560 capillary column of 0.02 mm thickness (Supelco, Bellefonte, PA) (Ebrahimi et al., 2014). Helium was used as a carrier gas with a spit ratio of 9:1. The experimental conditions for mass spectrometry were as follows: full scan mode, a solvent delay of 11 min, a gain factor of 10, an ion source temperature of 230°C, and a quadrupole temperature of 150°C. Qualitative Analysis B.07.00 software (Agilent) was used with the National Institute of Standards and Technology (NIST) database to identify the compound information for the reference standard (Sigma-Aldrich, United States) and samples. The results are expressed as a percentage of total fatty acids.
Total RNA was extracted using TRIzol reagent (Invitrogen, CA, United States) following the manufacturer’s procedure. The 6000 Nano LabChip Kit (Agilent, CA, United States) with the Agilent Bioanalyzer 2100 system (Agilent Technologies, CA, United States) was used to assess RNA integrity. Approximately 1 μg of total RNA with an RNA integrity number (RIN) > 7.0 was used to prepare a small RNA library according to the protocol of a TruSeq Small RNA Sample Prep Kit (Illumina, San Diego, United States). Then, we performed single-end sequencing (36 bp or 50 bp) on an Illumina HiSeq 2500. Approximately 10 μg of total RNA representing a specific adipose type was used to deplete ribosomal RNA with an Epicenter Ribo-Zero Gold Kit (Illumina). Following purification, the poly(A)- or poly(A)+ RNA fractions were fragmented into small pieces using divalent cations under elevated temperature. Then, the cleaved RNA fragments were reverse-transcribed to create the final cDNA library in accordance with the protocol for a mRNA-Seq sample preparation kit (Illumina), and the average insert size for the paired-end libraries was 300 bp (±50 bp). Then, we performed paired-end sequencing on an Illumina HiSeq 4000 (lc-bio, China) following the vendor’s recommended protocol.
First, Cutadapt (Martin, 2011) was used to remove the reads that contained adaptor sequences, contamination, low-quality bases and undetermined bases. Then, sequence quality was verified with FastQC1. We used Bowtie2 (Langmead and Salzberg, 2012) and TopHat2 (Kim et al., 2013) to map reads to the genome of Capra hircus. The mapped reads of each sample were assembled with StringTie (Pertea et al., 2015). Subsequently, the whole transcriptome of Capra hircus was obtained. The samples were merged to reconstruct a comprehensive transcriptome using Perl scripts. After the final transcriptome was generated, StringTie and Ballgown (Frazee et al., 2015) were used to estimate the expression levels of all transcripts.
The raw reads were subjected to an in-house program, ACGT101-miR (LC Sciences, Houston, Texas, United States), to remove adapter dimers, contamination, low-complexity sequences, sequences of common RNA families (rRNA, tRNA, small nuclear RNA (snRNA), snoRNA) and repeats. Subsequently, unique sequences with lengths of 18--26 nucleotides were mapped to specific species precursors in miRBase 22.0 with the Basic Local Alignment Search Tool (BLAST) to identify known miRNAs and novel 3p- and 5p-derived miRNAs. Length variation at both the 3′ and 5′ ends and one mismatch within the sequence were allowed in the alignment. The unique sequences that mapped to the hairpin arms of mature species-specific miRNAs were identified as known miRNAs. The unique sequences that mapped to the arms of known species-specific precursors opposite the annotated mature miRNA-containing arm were considered to be novel 5p- or 3p-derived miRNA candidates. The remaining sequences were mapped to precursors from other selected species (excluding species-specific precursors) in miRBase 22.0 through BLAST search, and the mapped premiRNAs were further BLASTed against the specific species genomes to determine their genomic locations. We defined the miRNAs obtained through the above two methods as known miRNAs. The unmapped sequences were BLASTed against the specific genomes, and the hairpin RNA structures containing sequences were predicted from the flanking 80 nt sequences using RNAfold software2.
StringTie was used to determine mRNA expression levels by calculating the fragments per kilobase of transcript per million mapped reads (FPKM) values. The differentially expressed mRNAs meeting the criteria of a log2 (fold change, FC) > 1 or a log2 (FC) < –1 and statistical significance (P < 0.05) were selected with the R package Ballgown.
The differential expression of miRNAs based on normalized deep-sequencing counts was analyzed through Student’s t-test based on the experimental design. The significance thresholds were 0.01 and 0.05 in each test. For the prediction of the target genes of the most abundant miRNAs, two computational target prediction algorithms (TargetScan 5.0 and miRanda 3.3a) were used to identify miRNA binding sites. When using the miRanda database, the species of Capra hircus was used. Finally, the genes predicted by both algorithms were combined, and the overlapping genes were identified. The Gene Ontology (GO) terms and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways of the most abundant miRNAs and their targets were also annotated.
Total RNA from six samples (three goats) was extracted with TRIzol (Invitrogen, CA) according to the manufacturer’s instructions. Total RNA was reverse-transcribed to cDNA using the PrimeScript RT reagent Kit (TaKaRa, Tokyo, Japan) and RT-qPCR was performed using SYBR Premix Ex Taq (TaKaRa, Tokyo, Japan). The mRNA primers were synthesized by Shanghai Shenggong, China (Supplementary Table 9). The relative mRNA levels were normalized to β-actin (Hou et al., 2016) expression for each sample; For miRNA detection, reverse transcription followed by stem-loop RT-qPCR was performed according to the manufacturer’s protocols using the Bulge-LoopTM miRNA RT-qPCR Primer (RiboBio, Guangzhou, China). Quantification of miRNA was performed with a stem-loop real-time PCR miRNA kit (RiboBio). The relative miRNA levels were normalized to U6 small nuclear RNA (Carvalho et al., 2019) expression for each sample.
The ct data from three goats and three technical replicates of all the candidate mRNA and miRNA obtained from the RT-qPCR experiments were evaluated with the 2–ΔΔCT method (Livak and Schmittgen, 2001). Student’s t-test was performed for WBM and WLM using SPSS (IBM SPSS Statistics). The results are presented as the mean ± standard deviation. A significance level of 0.05 was used.
To assess the fatty acid content, WLM and WBM tissues were collected from 2-year-old goats. A total of 34 fatty acids were found in the muscles of Inner Mongolian cashmere goats, including the omega-3 fatty acids C22:6 and C18:3n3 and the omega-6 fatty acids C18:2c9, C18:3n6, and C20:4 (Supplementary Table 1). Independent-sample t-tests were performed for fatty acids in different muscles (WBM and WLM). The results showed that the total PUFA content in WBM was significantly higher than that in WLM (P < 0.05) (Figure 1 and Supplementary Table 2), and the levels of the PUFAs methyl linoleate (C18:2c9), cis-13-16-docosadienoic acid methyl ester (C22:2), gamma-linolenic acid methyl ester (C18:3n6), cis-11,14,17-eicosatrienoic acid methyl ester (C20:3n3), methyl cis-5,8,11,11, and 14-eicosatetraenoic acid (C20:4) in WBM were significantly higher than those in WLM (P < 0.05). The levels of the monounsaturated fatty acids (MUFAs) cis-10-pentadecenoic acid methyl ester (C15:1) and methyl erucate (C22:1) in WBM were significantly higher than those in WLM (P < 0.05). In other words, WBM and WLM were representative and could be used as models for the study of PUFAs.
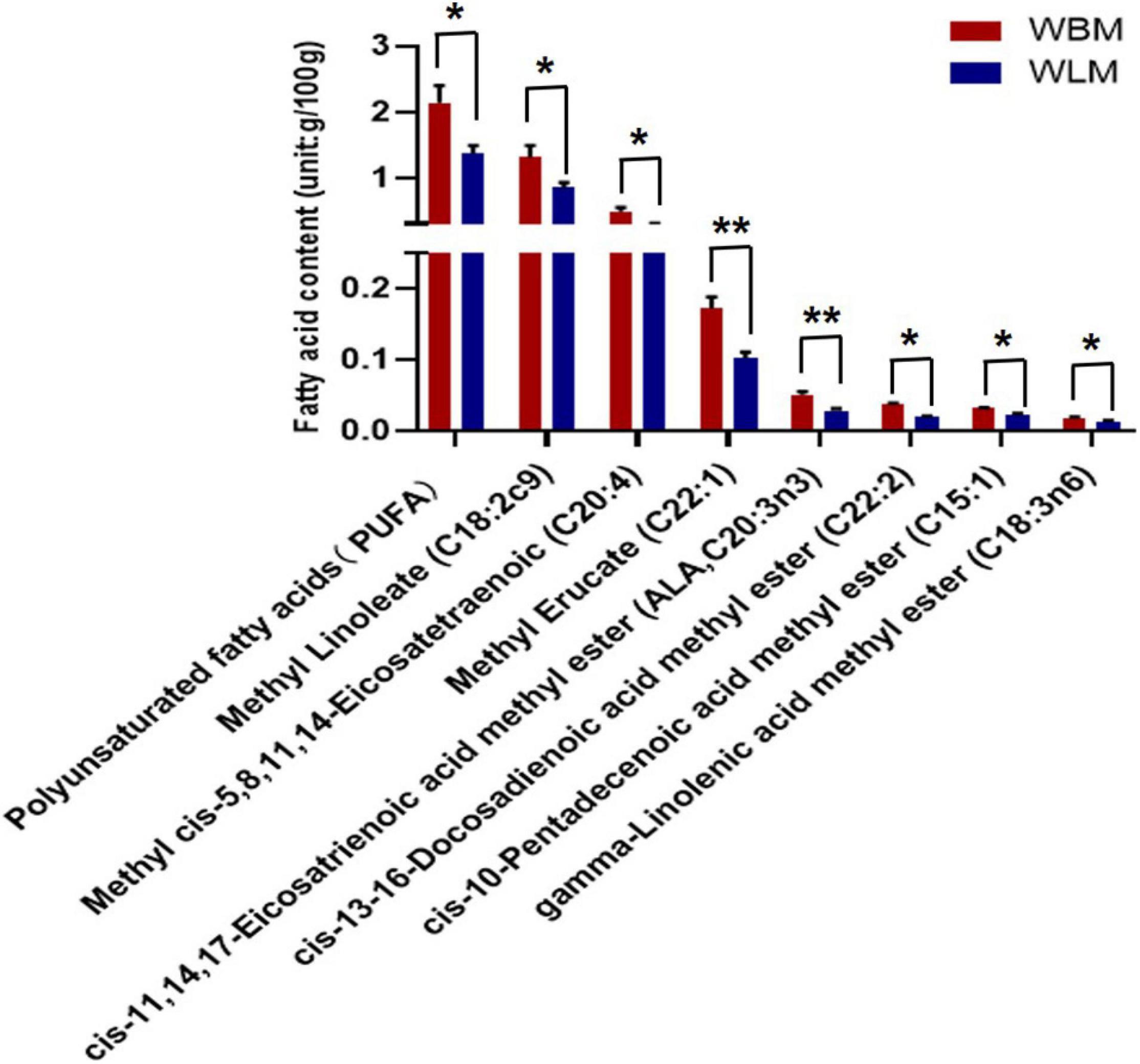
Figure 1. Fatty acid content of longissimus dorsi muscle (WLM) and biceps femoris muscle (WBM). Red represents the high-fatty acid group (the WBM tissue group), while blue represents the low-fatty acid group (the WLM tissue group). The asterisks represent the levels of significance (t-test: *P < 0.05, **P < 0.01).
To assess the mRNAs and miRNAs of the Inner Mongolia cashmere goat, we collected WLM tissues and WBM tissues for transcriptomic profiling of all mRNAs and miRNAs via high-throughput sequencing. For RNA-Seq library preparation, an average of 88,125,888 clean reads were obtained from the six samples tested, and 82.23–85.22% of these reads were uniquely aligned to the reference genome ARS1 in Ensembl. All 6 samples had at least 95.41% reads with quality equal to or exceeding Q30. An average of 20,375 genes were found in each sample of Inner Mongolia cashmere goat meat (Supplementary Table 3).
In addition, for the small RNA-Seq libraries, an average of 11,226,053 clean reads and 190,214 unique reads were obtained. An average of 426 known miRNAs and 30 novel miRNAs were obtained after a series of analyses (Supplementary Table 4).
There were 245 DEGs in WBM compared with WLM(Figure 2A and Supplementary Table 5), of which 85 were upregulated and 160 were downregulated in WBM. These DEGs included 18 DEGs related to the regulation of fatty acid synthesis and metabolism, including acyl-CoA synthetase long chain family member 1 (ACSL1) (FC = 2.23), lactate dehydrogenase B (LDHB, FC = 3.44), acyl-CoA dehydrogenase (ACADS, FC = 2.02), long-chain fatty acid acyl-CoA dehydrogenase (ACADVL, FC = 2.56), and trans-2,3-enoyl-CoA reductase like (TECRL) (FC = 2.67). These genes are important for the accumulation and metabolism of PUFAs.
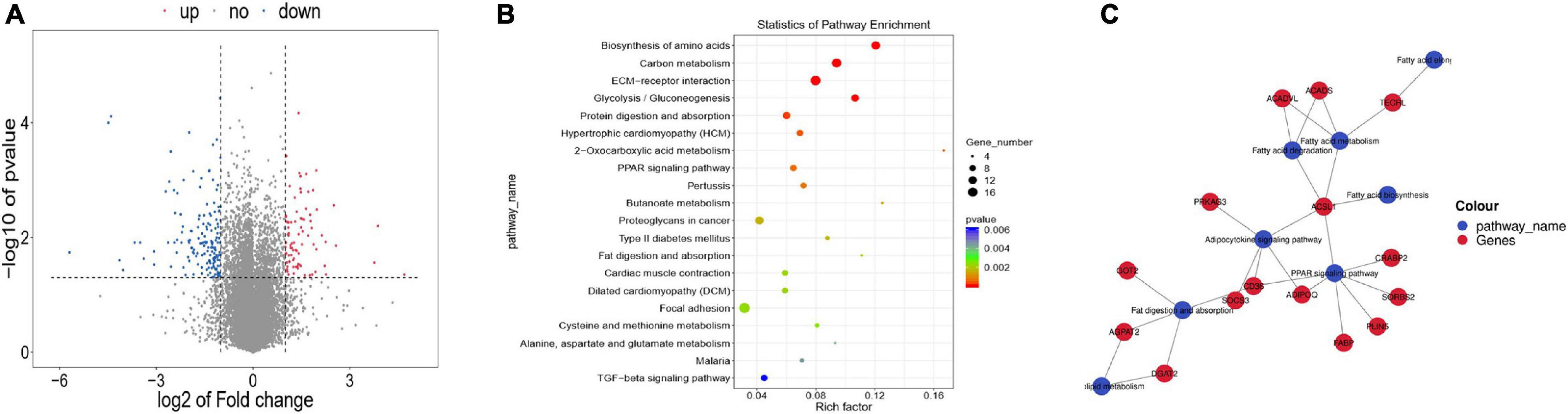
Figure 2. Differentially expressed genes (DEGs) in skeletal muscle in the high-fatty acid group and the low-fatty acid group. (A) DEGs. The red and blue dots represent upregulated and downregulated mRNAs, respectively. (B) KEGG pathway analysis for the DEGs. Only the top 20 enriched pathways are presented here. (C) KEGG-DEGs pairs and regulatory network.
KEGG analysis revealed pathways associated with these DEGs (Figure 2B and Supplementary Table 6). Specifically, the DEGs were involved in 8 fatty acid-related signaling pathways, including the fat digestion and absorption (ko04975), fatty acid biosynthesis (ko00061), fatty acid metabolism (ko01212), fatty acid elongation (ko00062), PPAR signaling (ko03320), adipocytokine (ko04920) pathways, fatty acid degradation (ko00071), and glycerolipid metabolism(ko00561) pathways (Table 1). These eight signaling pathways are also important for the accumulation and metabolism of PUFAs. The mRNA-based KEGG network analysis of the above 18 DEGs and 8 signaling pathways revealed an indirect relationship among the 15 DEGs (Figure 2C). ACSL1 was associated with 5 fatty acid metabolic process terms: the adipocytokine signaling pathway, fatty acid biosynthesis, fatty acid metabolism, fatty acid degradation, and PPAR signaling pathway were thus indirectly linked to 11 genes. CD36 was found to participate in fat digestion and absorption, the PPAR signaling pathway and the adipocytokine signaling pathway and was indirectly linked to 11 genes. TECRL was revealed to be involved in fatty acid metabolism, fatty acid elongation and the biosynthesis of PUFAs and was indirectly linked to three genes. ACSL1, CD36 and TECRL were at the center of the gene regulatory network, indicating that they contribute significantly to the accumulation and metabolic regulation of fatty acids.
Venn analysis of the 409 miRNAs found in cashmere goat muscle showed that a total of 343 miRNAs were co-expressed in both WBM and WLM, 39 were expressed only in WLM, and 27 were expressed only in WBM (Supplementary Table 7). A total of 32 (P < 0.05) DEMs were found in different parts of the muscle (Figure 3A and Supplementary Table 8), of which 14 were upregulated and 18 were downregulated in WBM. PC-3p-32774_108 and hsa-miR-615-3p_R-1 were upregulated (P < 0.01), whereas chi-miR-10b-3p, pol-let-7a-5p_R+3_1ss17AG, chi-miR-362-5p, and bta-let-7a-5p_R+2 were downregulated (P < 0.01).
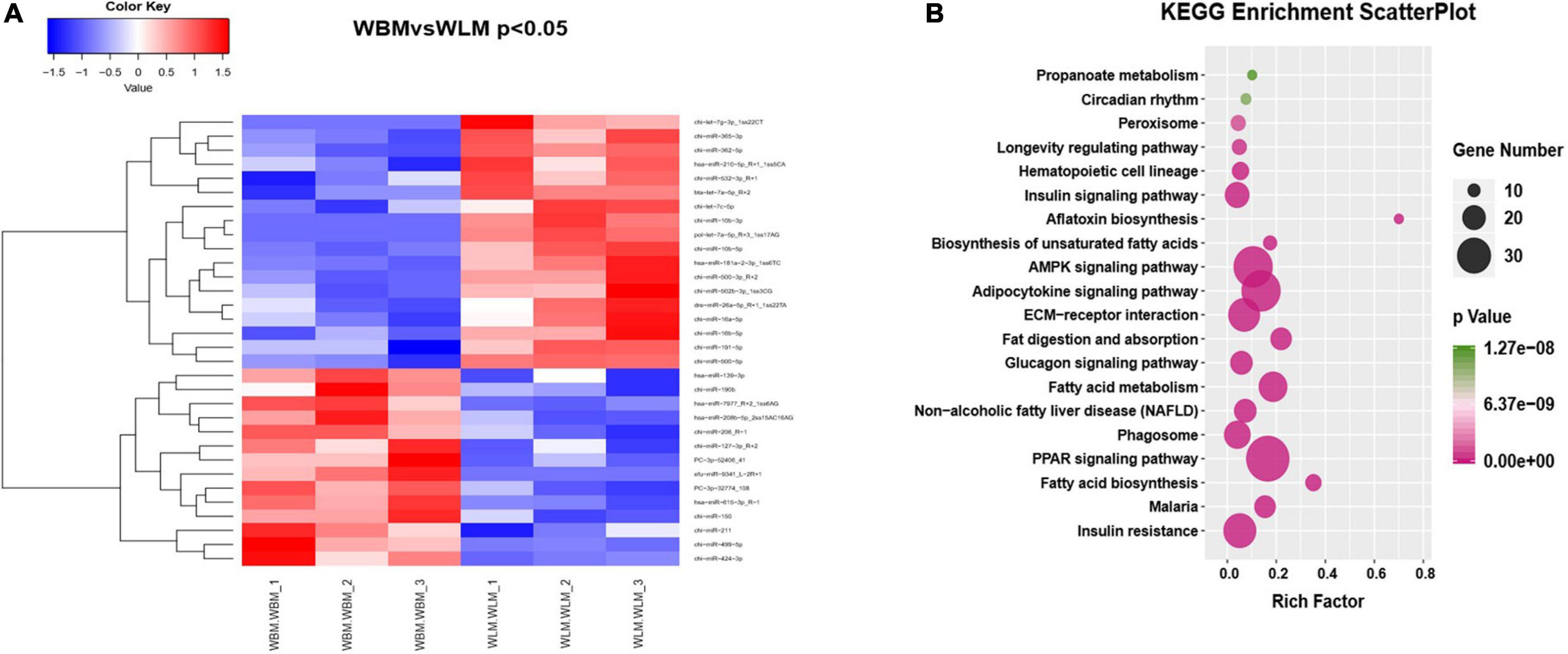
Figure 3. Differentially expressed miRNAs (DEMs) in skeletal muscle in the high-fatty acid group and the low-fatty acid group. (A) Heatmap of the significant DEMs. Red represents significant upregulation, while blue represents significant downregulation. (B) KEGG analysis of the targets of DEMs related to fatty acid metabolism.
KEGG analysis of the miRNA target genes (Figure 3B and Supplementary Table 8) revealed that they were associated mainly with biological processes such as fatty acid biosynthesis, the PPAR signaling pathway, and fatty acid degradation and absorption. The miRNA functional analysis results were consistent with the KEGG analysis results for the DEGs.
The miRNA target genes were predicted, and the transcriptome data were then used as a bridge to integrate the two sets of association data to obtain a miRNA-mRNA coregulatory network (Figure 4A). The constructed network contained 4 DEGs and 57 miRNAs. The network revealed that acetyl-CoA acyltransferase 2 (ACAA2) could be targeted by 8 miRNAs, while hydroxyacyl-CoA dehydrogenase trifunctional multienzyme complex subunit beta (HADHB) could be targeted by 11 miRNAs. ACSL1 could be targeted by 15 miRNAs, and enoyl-CoA hydratase short chain 1 (ECHS1) could be targeted by 23 miRNAs. These results suggest that ACAA2, HADHB, ACSL1, and ECHS1 may be the crucial genes mediated by miRNAs for the regulation of fatty acid metabolism.
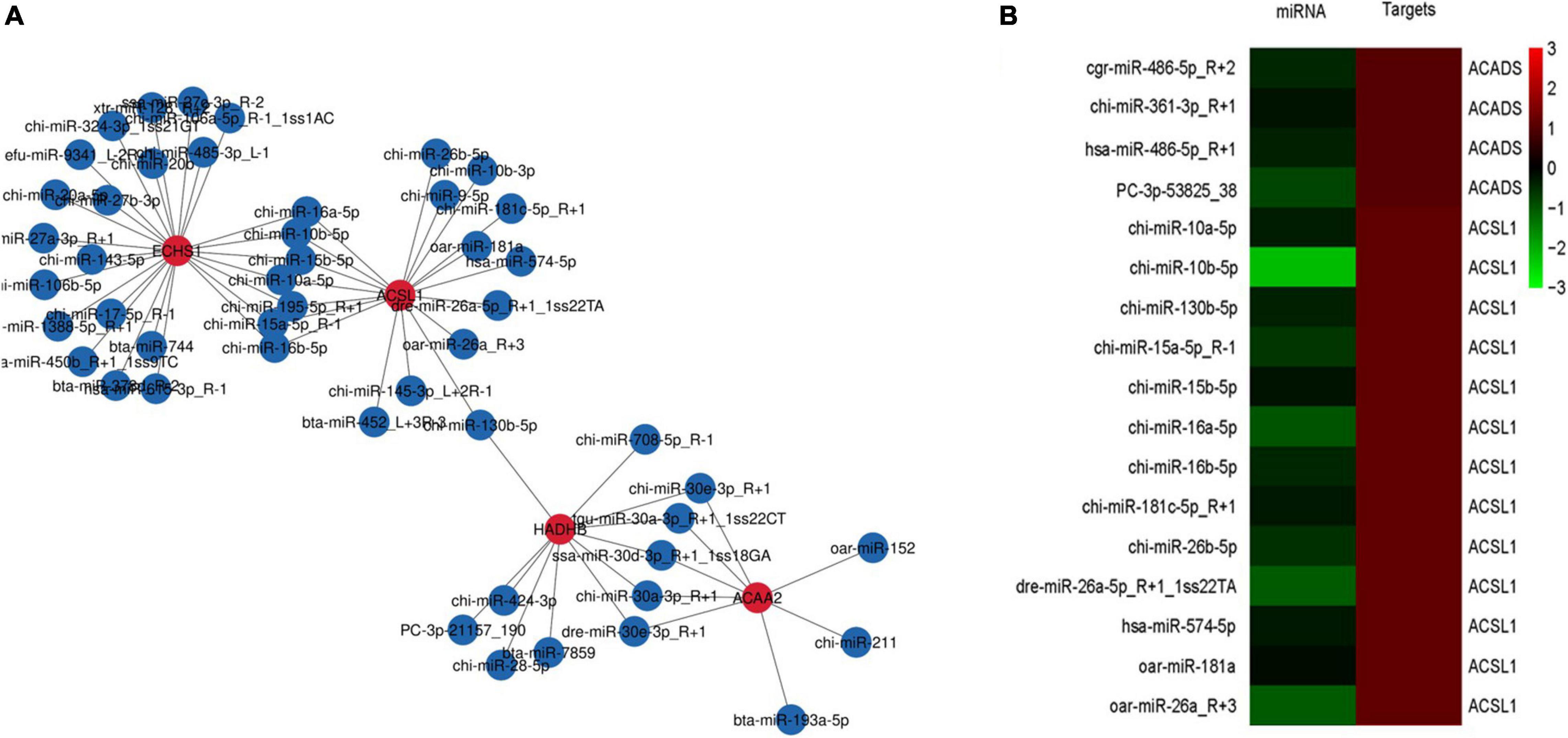
Figure 4. mRNA-microRNA coregulation analysis (A). miRNA-mRNA coregulatory network during fatty acid metabolism. The red and blue circles represent mRNAs and miRNAs, respectively. The solid lines indicate coregulation between miRNAs and mRNAs. (B) Clustering analysis of DEGs and miRNA expression patterns.
To determine which specific miRNAs regulate the DEGs related to fatty acid metabolism, we performed clustering analysis of the expression patterns of the target genes (Figure 4B). The analysis revealed the existence of 9 miRNAs that negatively regulate the ACSL1 gene in cashmere goats, including chi-miR-10a-5p, chi-miR-10b-5p, chi-miR-130b-5p, chi-miR-15a-5p_R-1, chi-miR-15b-5p, chi-miR-16a-5p, chi-miR-16b-5p, chi-miR-181c-5p_R + 1, and chi-miR-26b-5p. chi-miR-361-3p negatively regulates the ACADS gene in cashmere goats.
The expression of 6 DEGs related to unsaturated fatty acid synthesis and metabolism [TECRL, CD36, ACSL1, ACADS, ACADVL and diacylglycerol o-acyltransferase 2 (DGAT2)] was validated through RT-qPCR (Figure 5). The expression of these selected transcripts was significantly higher in the WBM than in the WLM tissues (P < 0.05), and the expression patterns were highly consistent with those obtained by the RNA-Seq method.
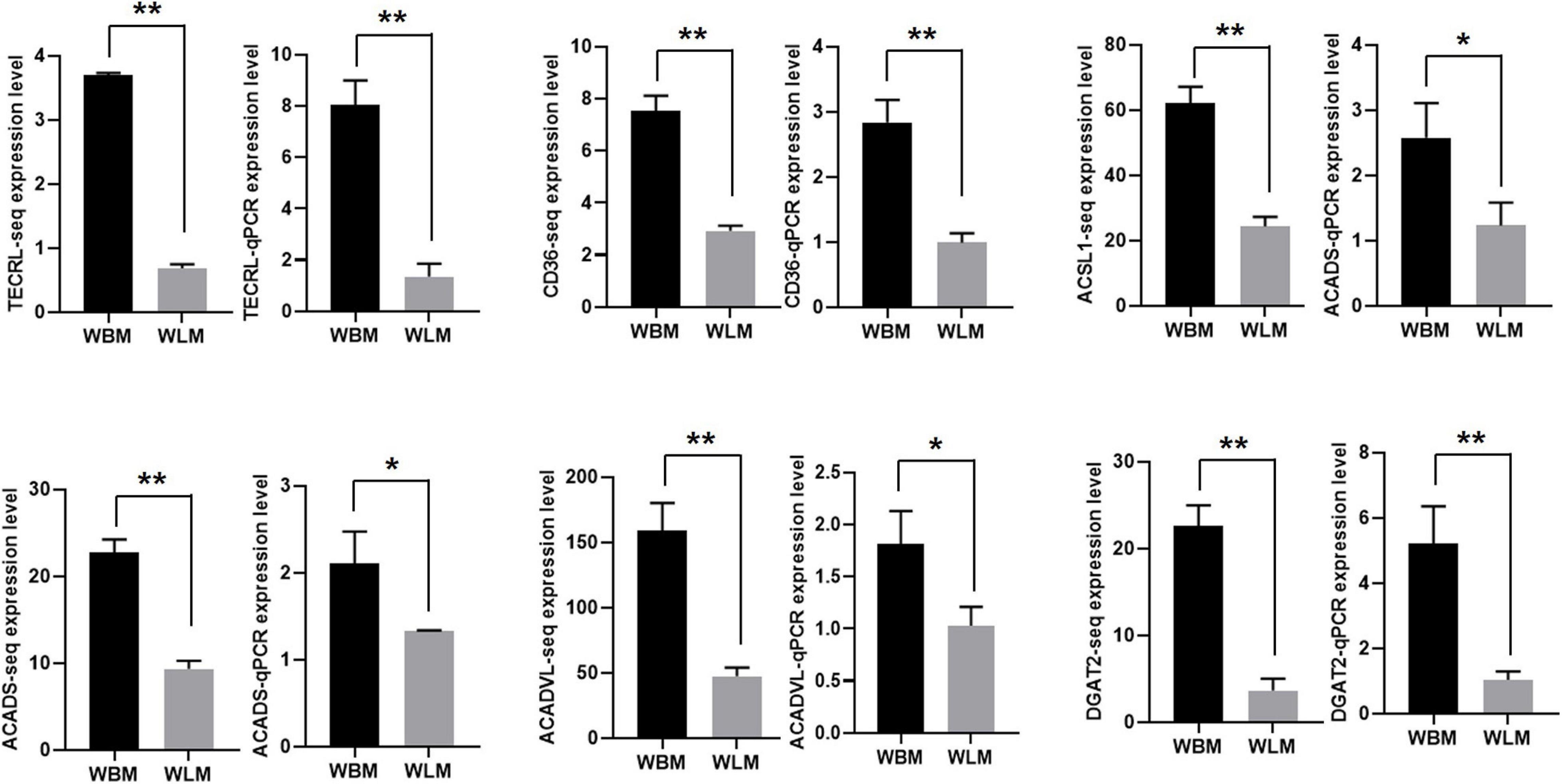
Figure 5. The expression of mRNA related to unsaturated fatty acid synthesis and metabolism. Biceps femoris muscle (WBM): high-fatty acid content group. Longissimus dorsi muscle (WLM): low-fatty acid content group. Expression of genes were normalized by β-actin. The asterisks represent the levels of significance (t-test: *P < 0.05, **P < 0.01).
The RNA-Seq results for chi-miR-16b-5p, chi-miR-10b-5p, chi-miR-365-3p, chi-miR-16a-5p, chi-miR-7c-5p, and chi-miR-191-5p were also validated through RT-qPCR (Figure 6). The expression of these selected miRNAs was significantly higher in the WLM than in the WBM tissues (P < 0.01), and the expression patterns were highly consistent with those obtained by the RNA-Seq method.
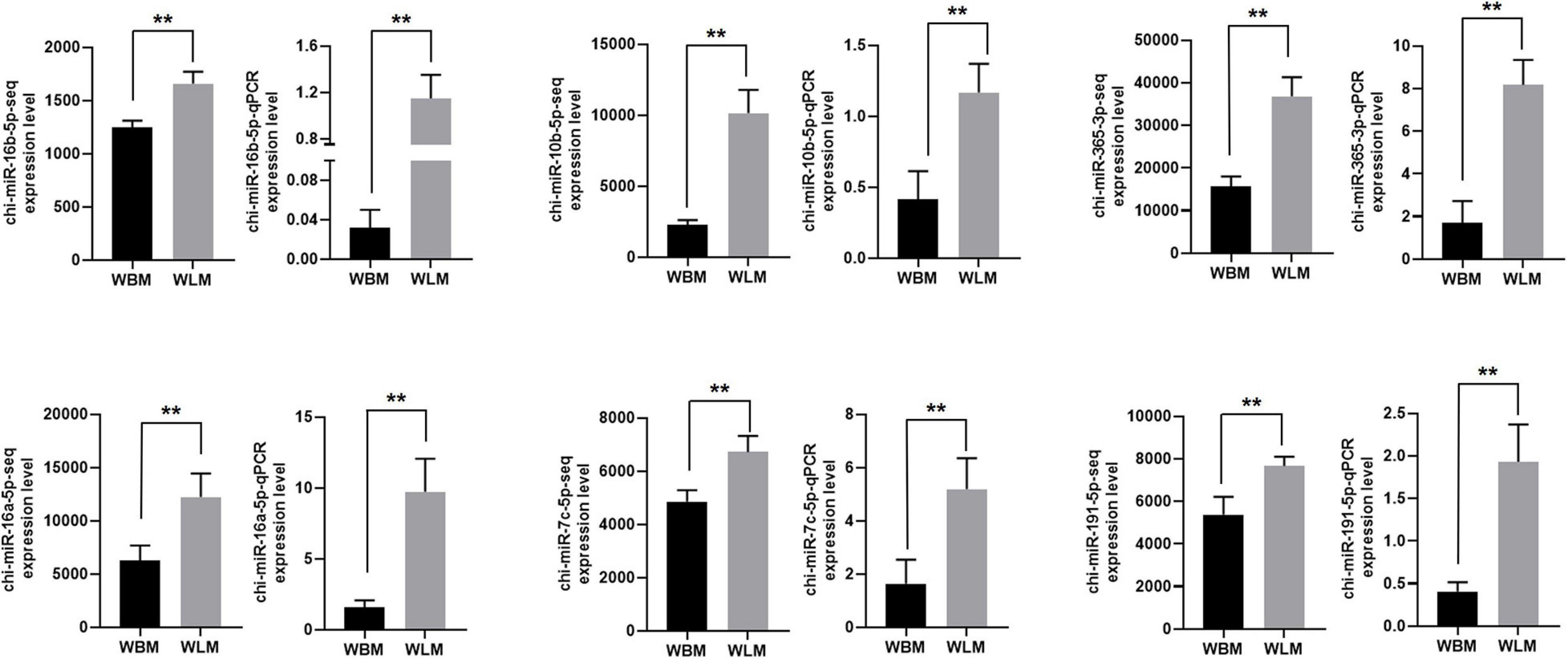
Figure 6. Validation of the miRNA-Seq results. Biceps femoris muscle (WBM): high-fatty acid group. Longissimus dorsi muscle (WLM): low-fatty acid group. Expression of miRNAs were normalized by U6 snRNA. The asterisks represent the levels of significance (t-test: **P < 0.01).
Our results show that Inner Mongolian cashmere goat meat contains a variety of omega-3 and omega-6 fatty acids that are beneficial to humans, such as C18:3n3 and C18:2c9, which have protective effects against cardiovascular and inflammatory diseases via the downregulation of proinflammatory genes (De Caterina et al., 2006; Yalcintan et al., 2018). There are two essential types of fatty acids in human nutrition: omega-3 fatty acids (alpha-linolenic acid) and omega-6 fatty acids (linoleic acid). The human body cannot synthesize essential fatty acids and thus must obtain them from food. Although seafood is the major dietary source of omega-3 fatty acids, a recent fatty acid intake survey indicated that red meat also serves as a significant source of omega-3 fatty acids for some populations (Sinclair et al., 1994). Therefore, understanding the molecular mechanisms related to fatty acids in the meat of the Inner Mongolian cashmere goat is vital. Improving our understanding of the sensory characteristics and nutritional value of goat meat is also key. Fatty acid composition is species- and tissue-specific. In our study, the PUFA content was significantly higher in WBM than in WLM. The findings indicate that the WBM and WLM of Inner Mongolia cashmere goats are effective models for research on the accumulation and metabolism of PUFAs.
Improving the beneficial fatty acid content of cashmere goat meat is important to improving the economic traits of cashmere goats. Thus, it is necessary to understand the molecules and molecular mechanisms that affect the accumulation and metabolism of fatty acids. To investigate which genes and miRNAs coregulate PUFA expression in Inner Mongolia cashmere goats, we selected WBM and WLM tissues with significantly different PUFA contents for miRNA and mRNA sequencing. We identified 20,375 genes and 426 known miRNAs and 30 novel miRNAs in the meat of Inner Mongolia cashmere goats. At the mRNA level, 18 DEGs and 8 signaling pathways were related to fatty acid metabolism. The KEGG network analysis of the above 18 DEGs and 8 signaling pathways revealed that ACSL1, CD36, and TECRL are at the center of the gene regulatory network. ACSL1 exists in fat cells and is considered to play an important role in activating the synthesis of triglycerides from fatty acids (Li et al., 2009). High expression of ACSL1 reduces fatty acid β-oxidation through the PPARγ pathway, thereby increasing triglyceride levels (Li T. et al., 2020). In addition, CD36 can activate fatty acid β-oxidation (Shi and Burn, 2004; Kennedy and Kashyap, 2011). CD36+CD44bright cells express relatively high levels of three key enzymes involved in fatty acid β-oxidation (ACADVL, ACADM, and HADHA) (Pascual et al., 2017). CD36 recognizes a number of lipid ligands, binds native and oxidized lipoproteins and then coordinates fat metabolism (Pepino et al., 2014). Furthermore, through GeneCards, we found that TECRL is a protein-coding gene. Among its related pathways are metabolism and fatty acyl-CoA biosynthesis. However, there have been no reports about TECRL in muscle tissue. In conclusion, the ACSL1, CD36, and TECRL genes are important participants in molecular mechanisms related to fatty acids in the Inner Mongolia cashmere goat.
At the miRNA level, our study is the first to systematically report that miRNAs regulate PUFA metabolism in the meat of Inner Mongolia cashmere goats. There were 214, 222, and 207 mature miRNAs in skeletal muscles of Landrace, Tongcheng, and Wuzhishan pigs, respectively (Hou et al., 2016). There were 767 known miRNAs in the WLM of Xinjiang brown cattle and Kazakh cattle (Li N. et al., 2020). In our study, we identified 426 known miRNAs and 30 novel miRNAs in the meat of cashmere goats. The number of miRNAs identified in the meat of cashmere goats was less than that in beef, but more than that in pork. Our results provide data to elucidate the molecular mechanisms affecting PUFA metabolism in Inner Mongolia cashmere goats. KEGG analysis showed that the miRNA target genes were mainly involved in fatty acid biosynthesis, fatty acid elongation, fatty acid degradation and absorption, and the PPAR signaling pathway. These results are consistent with those of the DEG KEGG analysis. Fatty acid uptake and decreased lipolysis are associated with increased intramuscular fat (IMF) deposition. Specifically, changes in the balance between synthesis and degradation can increase or decrease fatty acids (Teixeira et al., 2017). PPARs are a family of nuclear receptors that bind to fatty acids and perform significant functions in the regulation of nutrient metabolism and energy homeostasis (Lemay and Hwang, 2006). PUFAs can bind to PPARα at physiological concentrations and induce the expression of several genes involved in fatty acid metabolism, including fatty acid transport, synthesis and β oxidation (Rodríguez-Cruz and Serna, 2017). In conclusion, the PPAR pathway is important for fatty acid metabolism in the Inner Mongolia cashmere goat.
The mRNA-miRNA coregulation analysis showed that only ACSL1 was negatively regulated by miRNAs in cashmere goats. ACSL1 was negatively targeted by 9 miRNAs: chi-miR-10a-5p, chi-miR-10b-5p, chi-miR-130b-5p, chi-miR-15a-5p_R-1, chi-miR-15b-5p, chi-miR-16a-5p, chi-miR-16b-5p, chi-miR-181c-5p_R + 1, and chi-miR-26b-5p. ACSL1 is involved in 5 fatty acid metabolic pathways: the adipocytokine signaling pathway, fatty acid biosynthesis, fatty acid metabolism, fatty acid degradation, and the PPAR signaling pathway; all pathways are related to PPAR signaling. Thus, we speculated that the simultaneous silencing of ACSL1 by one or more of these nine negatively regulated miRNAs through PPAR signaling led to the low expression of the ACSL1 gene in the WLM and finally to the high PUFA content in the WBM. A study by Lian showed that bta-miR-181a may contribute to the negative regulation of lipid synthesis in mammary cells by targeting ACSL1 (Lian et al., 2016). In addition, miR-34a-5p can increase intracellular lipid content by reducing the ACSL1 protein level (Tian et al., 2019). miR-126-3 might be involved in lipid metabolism in the mammary gland (Chu et al., 2017). However, there were no prior reports on the 9 miRNAs identified in our study and their association with fatty acids in cashmere goat meat. Furthermore, which miRNAs interact with ACSL1 through PPAR signaling pathways to ultimately affect fatty acid synthesis or breakdown in the Inner Mongolia cashmere goat requires further cellular functional validation.
Our study is the first to systematically report miRNAs in cashmere goat meat and 426 known miRNAs and 30 novel miRNAs were identified. KEGG analysis revealed that the miRNA target genes were involved mainly in the PPAR signaling pathway. At the mRNA level, 20375 genes were identified. The mRNA-miRNA coregulation analysis showed that ACSL1 was negatively targeted by nine miRNAs. We speculated that the simultaneous silencing of ACSL1 by one or more of these nine miRNAs through PPAR signaling led to low ACSL1 expression in the WLM and, ultimately, to high PUFA content in the WBM.
The data presented in the study are deposited in the SRA database (https://www.ncbi.nlm.nih.gov/sra) repository, accession number (SRA, PRJNA689238).
The animal study was reviewed and approved by the experimental animal ethics committee of Inner Mongolia Agricultural University [Approval No. (2020)056].
YX, ZL, and JG made substantial contributions to the conception and design of the experiments. YX, ZL, JG, XS, CYZ, and CZ conceived and designed the experiments YX, RS, QQ, and DD performed the experiments. YX, ZL, ZYW, YJZ, and RW analyzed the data. YX, ZL, YHZ, and ZYW wrote the manuscript. JL and ZXW critically revised the manuscript. All authors read and approved the final manuscript.
This work received funding from the National Natural Science Foundation of China (31660640 and 32060742), the Major Science and Technology Projects of the Inner Mongolia Autonomous Region of China (2020ZD0004), the Key Technology Project of the Inner Mongolia Autonomous Region (2020GG0030), and National Key Research and Development Program of China (2018YFD0502000). The funding bodies played no role in the design of the study, collection, analysis, and interpretation of data or in the writing of the manuscript.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We are thankful for the samples provided by the Aerbasi White Cashmere Goat Breeding Farm. JL provided the test platform, and the authors of this article, ZL helped in designing and conducting the experiments and in analyzing, evaluating and interpreting the results.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2021.649015/full#supplementary-material
Supplementary Table 1 | Fatty acid content of cashmere goat muscle.
Supplementary Table 2 | Fatty acid classification.
Supplementary Table 3 | Overview of the data for RNA sequencing.
Supplementary Table 4 | Overview of the data for small RNA sequencing.
Supplementary Table 5 | Differentially regulated mRNAs.
Supplementary Table 6 | KEGG enrichment of DEGs.
Supplementary Table 7 | Detected miRNA.
Supplementary Table 8 | Differently expressed miRNAs.
Supplementary Table 9 | The primer information of mRNA.
Bartel, D. P. (2009). MicroRNAs: target recognition and regulatory functions. Cell 136, 215–233. doi: 10.1016/j.cell.2009.01.002
Berton, M. P., Fonseca, L. F. S., Gimenez, D. F. J., Utembergue, B. L., Cesar, A. S., Coutinho, L. L., et al. (2016). Gene expression profile of intramuscular muscle in Nellore cattle with extreme values of fatty acid. Bmc Genomics 17:972. doi: 10.1186/s12864-016-3232-y
Carvalho, E. B., Gionbelli, M. P., Rodrigues, R. T. S., Bonilha, S. F. M., Newbold, C. J., Guimarães, S. E. F., et al. (2019). Differentially expressed mRNAs, proteins and miRNAs associated to energy metabolism in skeletal muscle of beef cattle identified for low and high residual feed intake. BMC Genomics 20:501. doi: 10.1186/s12864-019-5890-z
Chu, M., Zhao, Y., Feng, Y., Zhang, H., Liu, J., Cheng, M., et al. (2017). MicroRNA-126 participates in lipid metabolism in mammary epithelial cells. Mol. Cell. Endocrinol. 15, 77–86. doi: 10.1016/j.mce.2017.05.039
De Caterina, R., Zampolli, A., Del Turco, S., Madonna, R., and Massaro, M. (2006). Nutritional mechanisms that influence cardiovascular disease. Am. J. Clin. Nutr. 83, 421S–426S.
Ebrahimi, M., Rajion, M., and Goh, Y. (2014). Effects of oils rich in linoleic and α-linolenic acids on fatty acid profile and gene expression in goat meat. Nutrients 6, 3913–3928. doi: 10.3390/nu6093913
Frazee, A. C., Pertea, G., Jaffe, A. E., Langmead, B., Salzberg, S. L., and Leek, J. T. (2015). Ballgown bridges the gap between transcriptome assembly and expression analysis. Nat. Biotechnol. 33:243. doi: 10.1038/nbt.3172
Han, H. Y., Gu, S. H., Chu, W., Sun, W., Wei, W., Dang, X., et al. (2017). miR-17–5p Regulates differential expression of NCOA3 in pig intramuscular and subcutaneous adipose tissue. Lipids 52, 939–949.
Hou, X., Yang, Y., Zhu, S., Hua, C., Zhou, R., Mu, Y., et al. (2016). Comparison of skeletal muscle miRNA and mRNA profiles among three pig breeds. Mol. Genet. Genomics 291, 559–573. doi: 10.1007/s00438-015-1126-3
Kennedy, D. J., and Kashyap, S. R. (2011). Pathogenic role of scavenger receptor CD36 in the metabolic syndrome and diabetes. Metab. Syndr. Relat. Disord. 9, 239–245. doi: 10.1089/met.2011.0003
Kim, D., Pertea, G., Trapnell, C., Pimentel, H., Kelley, R., and Salzberg, S. L. (2013). TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 14:R36. doi: 10.1186/gb-2013-14-4-r36
Ladeira, M. M., Schoonmaker, J. P., Swanson, K. C., Duckett, S. K., Gionbelli, M. P., Rodrigues, L. M., et al. (2018). Review: nutrigenomics of marbling and fatty acid profile in ruminant meat. Animal 12, s282–s294. doi: 10.1017/S1751731118001933
Langmead, B., and Salzberg, S. L. (2012). Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359. doi: 10.1038/nmeth.1923
Lemay, D. G., and Hwang, D. H. (2006). Genome-wide identification of peroxisome proliferator response elements using integrated computational genomics. J. Lipid Res. 47, 1583–1587. doi: 10.1194/jlr.M500504-JLR200
Li, L. O., Ellis, J. M., Paich, H. A., Wang, S., Gong, N., Altshuller, G., et al. (2009). Liver-specific loss of long chain acyl-CoA synthetase-1 decreases triacylglycerol synthesis and beta-oxidation and alters phospholipid fatty acid composition. J. Biol. Chem. 284, 27816–27826. doi: 10.1074/jbc.M109.022467
Li, N., Yu, Q. L., Yan, X. M., Li, H. B., and Zhang, Y. (2020). Sequencing and characterization of miRNAs and mRNAs from the longissimus dorsi of Xinjiang brown cattle and Kazakh cattle. Gene 741:144537. doi: 10.1016/j.gene.2020.144537
Li, T., Li, X., Meng, H., Chen, L., and Meng, F. (2020). ACSL1 affects triglyceride levels through the PPARγ Pathway. Int. J. Med. Sci. 17, 720–727. doi: 10.7150/ijms.42248
Lian, S., Guo, J. R., Nan, X. M., Ma, L., Loor, J. J., and Bu, D. P. (2016). MicroRNA Bta-miR-181a regulates the biosynthesis of bovine milk fat by targeting ACSL1. J. Dairy Sci. 99, 3916–3924. doi: 10.3168/jds.2015-10484
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Long, J. K., Dai, W., Zheng, Y. W., and Zhao, S. P. (2019). miR-122 promotes hepatic lipogenesis via inhibiting the LKB1/AMPK pathway by targeting Sirt1 in non-alcoholic fatty liver disease. Mol. Med. 25:26. doi: 10.1186/s10020-019-0085-2
Lu, X., Xia, H., Jiang, J., Xu, X., Li, M., Chen, Z., et al. (2020). MicroRNA-212 targets SIRT2 to influence lipogenesis in bovine mammary epithelial cell line. J. Dairy Res. 87, 232–238. doi: 10.1017/S0022029920000229
Ma, X., Wei, D., Cheng, G., Li, S., Wang, L., Wang, Y., et al. (2018). Bta-miR-130a/b regulates preadipocyte differentiation by targeting PPARG and CYP2U1 in beef cattle. Mol. Cell. Probes 42, 10–17. doi: 10.1016/j.mcp
Martin, M. (2011). Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 24, 1138–1143. doi: 10.1089/cmb.2017.0096
Pascual, G., Avgustinova, A., Mejetta, S., Martín, M., Castellanos, A., Attolini, C. S., et al. (2017). Targeting metastasis-initiating cells through the fatty acid receptor CD36. Nature 541, 41–45. doi: 10.1038/nature20791
Pepino, M. Y., Kuda, O., Samovski, D., and Abumrad, N. A. (2014). Structure-function of CD36 and importance of fatty acid signal transduction in fat metabolism. Annu. Rev. Nutr. 34, 281–303. doi: 10.1146/annurev-nutr-071812-161220
Pertea, M., Pertea, G. M., Antonescu, C. M., Chang, T. C., Mendell, J. T., and Salzberg, S. L. (2015). StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 33, 290–295. doi: 10.1038/nbt.3122
Rodríguez-Cruz, M., and Serna, D. S. (2017). Nutrigenomics of n-3 fatty acids: regulators of the master transcription factors. Nutrition 41, 90–96. doi: 10.1016/j.nut.2017.04.012
Scollan, N. D., Dannenberger, D., Nuernberg, K., Richardson, I., MacKintosh, S., Hocquette, J. F., et al. (2014). Enhancing the nutritional and health value of beef lipids and their relationship with meat quality. Meat Sci. 97, 384–394. doi: 10.1016/j.meatsci.2014.02.015
Shi, Y., and Burn, P. (2004). Lipid metabolic enzymes: emerging drug targets for the treatment of obesity. Nat. Rev. Drug Discov. 3, 695–710.
Sinclair, A. J., Johnson, L., O’Dea, K., and Holman, R. T. (1994). Diets rich in lean beef increase arachidonic acid and long-chain omega 3 polyunsaturated fatty acid levels in plasma phospholipids. Lipids 29, 337–343. doi: 10.1007/BF02537187
Su, R., Fu, S., Zhang, Y., Wang, R., Zhou, Y., Li, J., et al. (2015). Comparative genomic approach reveals novel conserved microRNAs in Inner Mongolia cashmere goat skin and longissimus dorsi. Mol. Biol. Rep. 42, 989–995. doi: 10.1007/s11033-014-3835-9
Su, R., Gong, G., Zhang, L., Yan, X., Wang, F., Zhang, L., et al. (2020). Screening the key genes of hair follicle growth cycle in Inner Mongolian Cashmere goat based on RNA sequencing. Arch. Anim. Breed. 63, 155–164. doi: 10.5194/aab-63-155-2020
Teixeira, P. D., Oliveira, D. M., Chizzotti, M. L., Chalfun-Junior, A., Coelho, T. C., Gionbelli, M., et al. (2017). Subspecies and diet affect the expression of genes involved in lipid metabolism and chemical composition of muscle in beef cattle. Meat Sci. 133, 110–118. doi: 10.1016/j.meatsci.2017.06.009
Tian, W. H., Wang, Z., Yue, Y. X., Li, H., Li, Z. J., Han, R. L., et al. (2019). miR-34a-5p increases hepatic triglycerides and total cholesterol levels by regulating ACSL1 protein expression in laying hens. Int. J. Mol. Sci. 20:4420. doi: 10.3390/ijms20184420
Tvrzicka, E., Kremmyda, L. S., Stankova, B., and Zak, A. (2011). Fatty acids as biocompounds: their role in human metabolism, health and disease–a review. Part 1: classification, dietary sources and biological functions. Biomed. Pap. Med. Fac. Univ. Palacky Olomouc Czech. Repub. 155, 117–130.
Xu, B., Gerin, I., Miao, H. Z., Vu-Phan, D., Johnson, C. N., Xu, R., et al. (2010). Multiple roles for the non-coding RNA SRA in regulation of adipogenesis and insulin sensitivity. PLoS ONE 5:e14199. doi: 10.1371/journal.pone.0014199
Yalcintan, H., Ekiz, B., and Ozcan, M. (2018). Comparison of meat quality characteristics and fatty acid composition of finished goat kids from indigenous and dairy breeds. Trop. Anim. Health Prod. 50, 1261–1269.
Keywords: microRNA, mRNA, cashmere goat, ACSL1, fatty acid
Citation: Xie Y, Liu Z, Guo J, Su X, Zhao C, Zhang C, Qin Q, Dai D, Zhao Y, Wang Z, Wang R, Zhang Y, Su R, Wang Z and Li J (2021) MicroRNA-mRNA Regulatory Networking Fine-Tunes Polyunsaturated Fatty Acid Synthesis and Metabolism in the Inner Mongolia Cashmere Goat. Front. Genet. 12:649015. doi: 10.3389/fgene.2021.649015
Received: 15 February 2021; Accepted: 29 April 2021;
Published: 02 June 2021.
Edited by:
Sonia Andrade, University of São Paulo, BrazilReviewed by:
Siyuan Zhan, Sichuan Agricultural University, ChinaCopyright © 2021 Xie, Liu, Guo, Su, Zhao, Zhang, Qin, Dai, Zhao, Wang, Wang, Zhang, Su, Wang and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhixin Wang, aGhodHd6eEAxMjYuY29t; Jinquan Li, bGlqaW5xdWFuX25kQDEyNi5jb20=
†These authors have contributed equally to this work and share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.