- 1Dipartimento Scienze Veterinarie, University of Messina, Messina, Italy
- 2Dipartimento di Scienze Agrarie Alimentari e Forestali, University of Palermo, Palermo, Italy
In the Mediterranean basin countries, the dairy sheep production is usually based on local breeds, which are very well-adapted to their production systems and environments and can indeed guarantee income, employment, and economic viability in areas where production alternatives are scarce or non-existent. Mastitis is still one of the greatest problems affecting commercial milk production. However, genetic evaluation of mastitis is particularly difficult because of its low heritability and the categorical nature of the trait. The aim of this study was to identify genomic regions putatively associated with somatic cells count (SCC) in the local economically important Valle del Belice sheep breed using of deregressed breeding values (DEBV) as response variables. All the samples were genotyped using the Illumina OvineSNP50K BeadChip. Genome-wide association analysis was carried out based on regression of DEBV. A total of eight markers were found to be significantly associated with log-transformed SCC. Several candidate genes associated with SCC were identified related to immunity system and udder conformation. The results can help improving the competitiveness of the local Valle del Belìce breed. Further studies considering a higher sample size or independent population will be needed to confirm our results.
Introduction
In the Mediterranean basin countries, the dairy sheep production is usually based on local breeds, which are very well-adapted to their production systems and environments and can indeed guarantee income, employment, and economic viability in areas where production alternatives are scarce or non-existent. Mastitis is the most important problem for the milk industry due to the decrease quality of milk and increased cost of flock regeneration due to early culling of ewes. It can be induced, for example, by a lack of hygiene, by pushed manual milking or feed disorder. In dairy sheep, generally, the most important agents involved in mastitis are the bacterial infections, and the most frequently isolated pathogens are coagulase-negative staphylococci (CNS) that are present on and around the udder skin (Leitner et al., 2008) with a different pathogenicity, causing clinical, and subclinical mastitis (Contreras et al., 2007; Riggio and Portolano, 2015). The udder infection determines the increase of the somatic cell count (SCC) in milk (Raynal-Ljutovac et al., 2007; Leitner et al., 2008) that causes significant damage of curd and cheese yields. Since the heritability of mastitis is low, genetic selection to improve mastitis by traditional selection is not very effective. SCC or log transformed SCC (i.e., somatic cell score, SCS) have relatively higher heritability compared to mastitis and is used as the first trait to improve mastitis resistance (Shook and Schutz, 1994). Kelly et al. (2000) found that an elevated SCC can alter the protein fractions distribution; decrease casein and lactose levels in milk; increase rennet clotting time, cheese moisture, and losses of fat and proteins in whey, and reduce curd firmness and cheese yielding. A study conducted by Sutera et al. (2018) confirmed that high levels of SCC in sheep milk are associated with milk yield losses and variations of fat and protein percentages. The estimated losses in milk yield ranged from 883 g for SCC ≤ 2,000 × 103 to 1,052 g for SCC ≤ 500 × 103 with an overall decrease of 16%, whereas fat and protein percentages increased to 0.06 and 0.29%, respectively. The negative effects of mastitis are provoked by a combination of animal characteristics (age, lactation stage, etc.), genetic (breed, inbreeding, etc.) and environmental factors (season, management, etc.) (Oget et al., 2019). Therefore, different individuals may have a different susceptibility to the disease, depending on their genetic heritage. In fact, there are several studies about the mastitis in dairy sheep confirming a genetic basis for mastitis resistance (Tolone et al., 2013; Oget et al., 2019), but no assumption had been made about the genes and the relative mechanisms.
The emergence of high-throughput genotyping technologies allowed routine genome-wide association studies (GWAS) to be performed in livestock populations. GWAS allows screening of the genome utilizing a large number of genetic markers spread across the entire genome to detect genetic variants associated with a particular disease or trait. The estimated breeding values (EBVs) were generally used to perform the GWAS. As an alternative, the EBVs can be “deregressed” (Garrick et al., 2009; Ostersen et al., 2011) to standardize the variance and influence of the individuals’ EBVs while still accounting for information from relatives. The use of deregressed EBVs (DEBVs) as dependent variables can improve the power of GWAS (Sell-Kubiak et al., 2015; Sevillano et al., 2015). An advantage of GWAS is that we can overcome the candidate gene approach through which sometimes significant results were not obtained due to the wrong or incomplete choice of candidate genes. In the last decades, several GWASs were conducted in sheep for milk production related traits (Sutera et al., 2019; Li et al., 2020), for fatty acids profile (Rovadoscki et al., 2018), for body weight (Ghasemi et al., 2019; Tao et al., 2020), for wool production (Wang et al., 2014), for nematode resistance (Becker et al., 2020) and ovine lentivirus resistance (White et al., 2012). To date, few GWASs have been conducted for SCC or SCS in dairy sheep (Oget et al., 2019), especially in local adapted breeds.
In Sicily, dairy sheep production represents an important resource for the local economy, and the Valle del Belice is the main local breed reared on the island for the production of traditional raw milk cheeses, at farm level by small local dairies. The breed is subjected to limited breeding selection programs for milk production traits, but shows excellent adaptability to local environments, sometimes with harsh conditions (Mastrangelo et al., 2017). Therefore, the aim of this study was to identify the genomic regions putatively associated with SCC in Valle del Belice sheep breed using of DEBVs as response variables.
Materials and Methods
Data and Estimation of Breeding Value
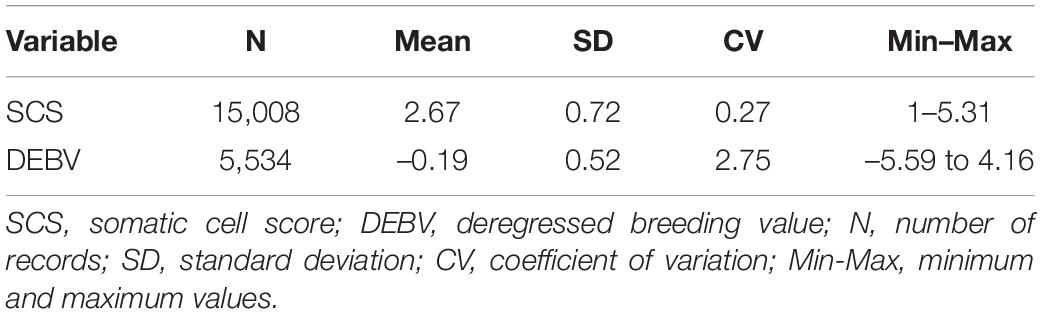
Between 2006 and 2016 the University of Palermo collected phenotypic data from 15 Valle del Belìce flocks, for a total of 1,813 individuals. The milk samples were collected aseptically from each individual from the two udder halves in sterile containers following an A4 recording procedure (ICAR, 2014), stored at 4°C and transferred to the laboratory to determine daily SCC using Fossomatic 6000 (Foss Electric Hillerød, Denmark) equipment. The phenotypic data set originated by these sampling works was composed of 15,008 observations. Animals with less than 3 test-day measurements within lactation were discarded. For each individual the following information were registered: order of parity, number of born lambs, lactation days, age, birth season and somatic cell count. Birth season was classified in three classes: 1 if the lambing was from August to November; 2 from December to March; 3 from April to July. SCC was normalized through a logarithmic transformation into somatic cell score (SCS) according to the formula of Ali and Shook (1980):
Preliminary analyses using the general linear model of ASReml R (Butler et al., 2009) were performed to determine the significance of the fixed effects where the Wald tests are implemented in the form of the ANOVA method. A single trait repeatability test day (TD) animal model was performed to estimate the breeding values (EBV) as follows:
where y is the observation vector for SCS TD; β is the vector of fixed effects that includes order of parity (op: 4 classes), age at first lambing (age: 4 classes, 1 when first lambing occurred at 10–14 months of age, 2 at 15–19 months of age, 3 at 20–24 months of age, and 4 at 25–29 months of age); birth season (bs: 3 classes), interaction between herd and birth season (hbs: 74 classes) and days of lactation (dim) modeled with a Legendre polynomial of order three. Htd is the vector of interaction between herd and test day random effect; a is the vector of direct additive genetic effects (breeding values); pe is the vectors of permanent environmental effect between lactations; e is the residual vector. X and Z are the corresponding incidence matrices relating records to fixed, animal, and permanent environmental between lactations effects, respectively. The pedigree file included 5,534 animals with 178 sires and 2,548 dams. The assumptions regarding the components of the model were:
and ; ; ; where A is the numerator relationship matrix based on pedigree and I are the identity matrix with orders equal to numbers of dams for htd and pe effects and equal to the records for residuals e. Variance components and breeding values for SCS were estimated based on REML method using ASReml R (Butler et al., 2009). In addition, EBVs were also deregressed according to Garrick et al. (2009) as follows:
where, EBV is the estimated breeding value and r2 is the reliability of that EBV.
Blood Sampling and DNA Extraction
A total of 476 sheep of Valle del Belice breed were sampled. About 10 mL of blood was collected from the jugular vein using vacutainer tubes containing EDTA as anticoagulant. The procedures involving animal sample collection followed the recommendation of directive 2010/63/EU. Sampling was carried out by trained veterinarians within the frame of vaccination Campaigns, hence no permission from the animal research ethics committee was necessary. Veterinarians adhered to standard procedures and relevant national guidelines to ensure appropriate animal care. Genomic DNA was extracted from each blood sample with a salting-out method (Miller et al., 1988). The DNA sample was quantified with a NanoDropND-1000 spectrophotometer (NanoDropTechnologies, Wilmington, DE, United States), diluted to a final concentration of 50 ng/mL (as required by the Illumina Infinium protocol), and stored at 4°C until use.
Genotyping and Quality Control
All the samples were genotyped using the Illumina OvineSNP50K BeadChip v2. Position and chromosomal coordinates for each SNP were obtained from the ovine genome sequence assembly (Oar 4.0)1. Quality control and association analyses were performed using GenABEL package (Aulchenko et al., 2007) in R environment2. Only SNPs located on autosomes were extracted and considered for further analyses. Animals and markers that fulfilled the following criteria were kept in the analysis: (i) call rate per individuals and per SNPs > 95%; (ii) minor allele frequency > 2%; (iii) no extreme deviation from Hardy-Weinberg equilibrium (P < 10–6).
GWAS Analyses
Genome-wide association analysis was carried out based on regression of DEBV with the genotypes of animals for one SNP at a time. We used the three-step approach referred to as genomic GRAMMAR-GC (Amin et al., 2007; Aulchenko et al., 2007). The advantage of this approach, especially in livestock, is that it accounts for cryptic population structure caused by the presence of closely related animals (Aulchenko et al., 2007) inferring relationships through genomic marker data. After Bonferroni correction, significant thresholds were P < 1.34 × 10–6 for genome-wide (P < 0.05) and P < 2.69 × 10–5 for suggestive (P < 0.10) (i.e., one false positive for genome scan), corresponding to −log10(P) equal to 5.87 and 4.56, respectively. Quantile-quantile (Q-Q) plots were used to analyze the extent to which the observed distribution of the statistic test followed the expected (null) distribution, in order to assess potential systematic bias due to population structure or analytical approach. Population substructure was explored using classical multidimensional scaling (MDS) in order to verify the genetic homogeneity of the sample before analysis using PLINK v1.9 (Purcell et al., 2007). The least square means of DEBV for the three genotypes affecting somatic cell count of significant SNP were also calculated by a general linear model (GLM) using R package lsmeans (Lenth and Lenth, 2018) and the significant threshold was set at P < 0.05.
Annotation
Genomic regions showing significant results were further explored to identify candidate genes underlying the loci. In particular, the gene contents located at ±250 kb distances from the significant SNP were annotated using Genome Data Viewer genome browser at the National Center for Biotechnology Information Database3. The presence of Quantitative Traits Loci (QTLs)4 related with the considered trait was also checked. Finally, to investigate the biological function and the phenotypes that are known to be affected by each annotated gene, we conducted a comprehensive literature search, including information from other species.
Results
Genetic Parameters and Estimated Breeding Values
Descriptive statistics and genetic merit for SCS in the sampled animals are presented in Table 1. About 15,000 TD observations for SCS were considered to estimate EBVs then, the DEBVs of 5,534 individuals were estimated. Heritability and repeatability estimates for SCS in the studied population were 0.045 (standard error = 0.02) and 0.40 (standard error = 0.01), respectively.
Quality Control for Genotyping Data
Among the 54,241 SNPs, 7,414 SNPs are located on sex chromosomes and thus were withdrawn from the analysis. A total of 3,999 SNPs were removed due to genotype rate <0.05, 2,037 SNPs due to minor allele frequency < 0.02 and 3,651 SNP due to Hardy-Weinberg disequilibrium (P < 10–6). Moreover, 12 individuals were also excluded for a low (<95%) call rate. Then, after quality control, we considered a total of 37,140 SNPs and 464 individuals for further analyses.
Genome-Wide Association Analyses
In total we detected eight significant SNPs for SCS, and among these, only one marker reached the genome-wide significant threshold (P < 4.72 × 10–7). The details of these SNPs including P-values, the positions on Ovis aries v4.0 genome assembly, the chromosomes and the closest known genes are given in Table 2. Manhattan plot, showing the profiles of the P-values [in terms of –log(P)] of all tested SNPs, is showed in Figure 1. The QQ-plot in Supplementary Figure S1 shows the observed and expected P-values of the GWAS for SCS. The genomic inflation factor (lambda) was lower than one indicating some population stratification. However, departure from this line is also expected for a really polygenic trait, as many causal SNPs may not yet have reached genome-wide significance owing to a lack of power (Power et al., 2017). The results for the MDS showed that the bulk of the samples were not separated by the first dimension, indicating a lack of substructure (Supplementary Figure S2). The eight SNPs were located on five different chromosomes: three SNPs on OAR1, one SNP on OAR3, one SNP on OAR7, two SNPs on OAR8 and one SNP on OAR10. Considering the range of ±250 kb surrounding each significant SNP, a total of 34 genes (Table 2) were found. The most significant SNP (rs161717499) was located within the coding region of the Stress Associated Endoplasmic Reticulum Protein 1 (SERP1) gene on OAR1.
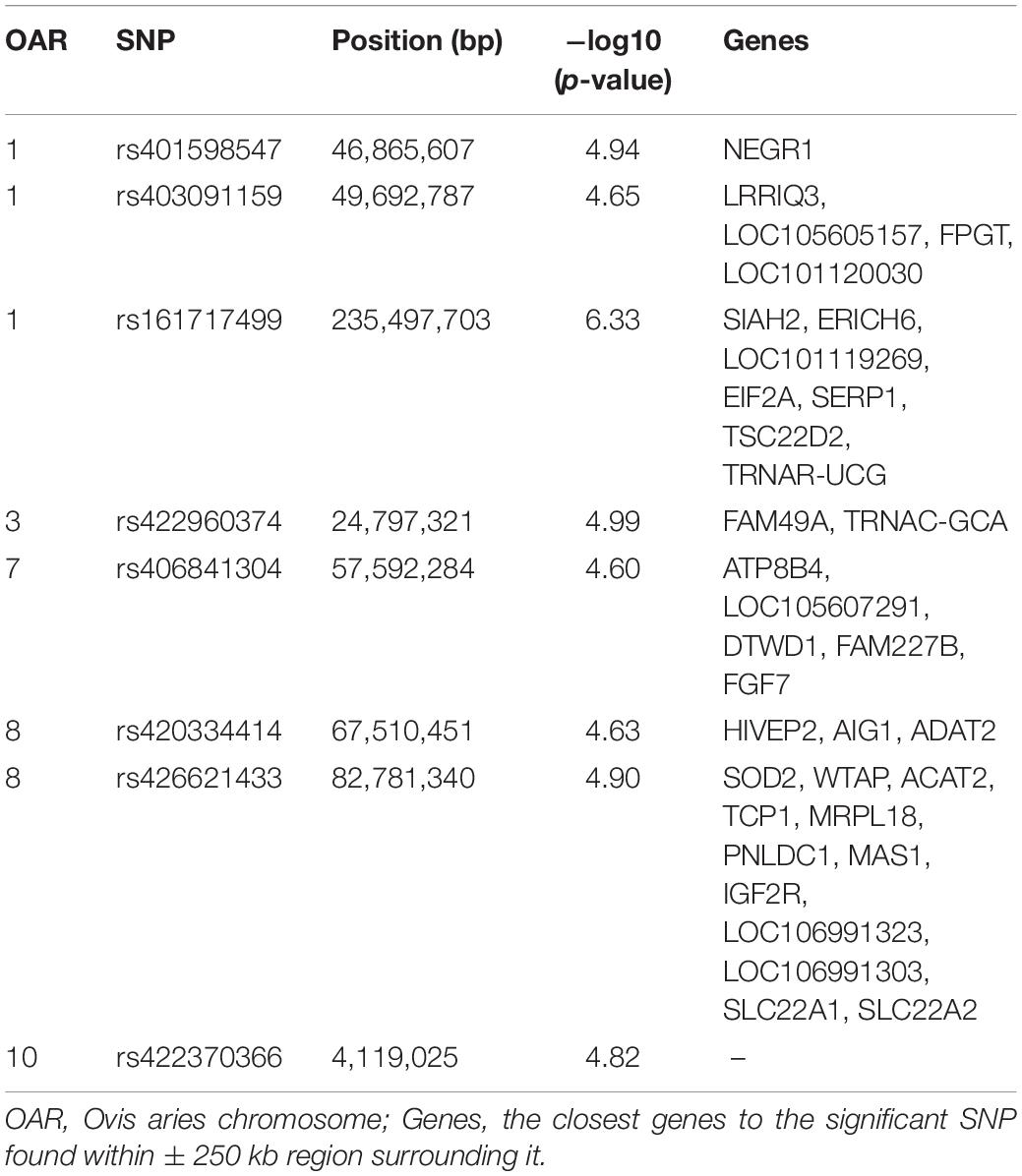
Table 2. Single nucleotide polymorphisms (SNPs) significantly associated with somatic cell score at genome-wide (P < 1.34 × 10–6) and suggestive (P < 2.69 × 10–5) thresholds.
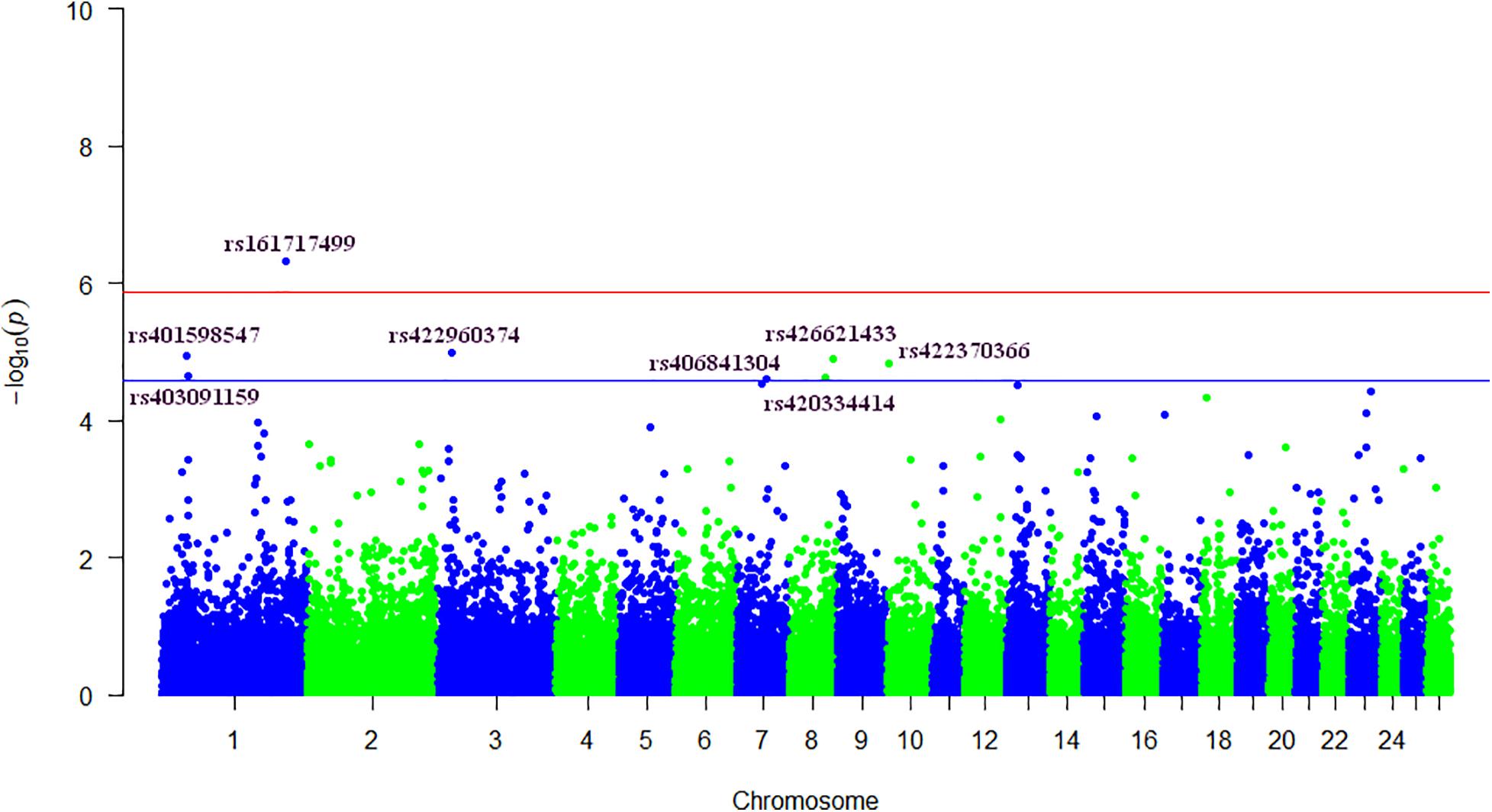
Figure 1. Genome-wide plot of −log10(P-values) for association of SNPs with somatic cell score. Blue and red lines represent suggestive [−log10(P-values) > 4.57] and genome-wide thresholds [−log10(P-values) > 5.87], respectively.
For each of the eight significant SNPs, we calculated the LSM of the DEBV for the three genotypes affecting the trait to investigate their genetic contribution (Figure 2). Five out of the eight above reported SNPs (rs401598547, rs403091159, rs161717499, rs422960374, rs426621433) reached the significance threshold (P < 0.05). Individuals with homozygous genotypes GG for rs401598547, CC for rs161717499 and AA for rs403091159, rs422960374, and rs426621433, showed lower somatic cells content among all three genotypes (Figure 2). After checking on SheepQTLdb tool, no one of the eight detected SNPs was located within a known QTLs related at SCS or mastitis.
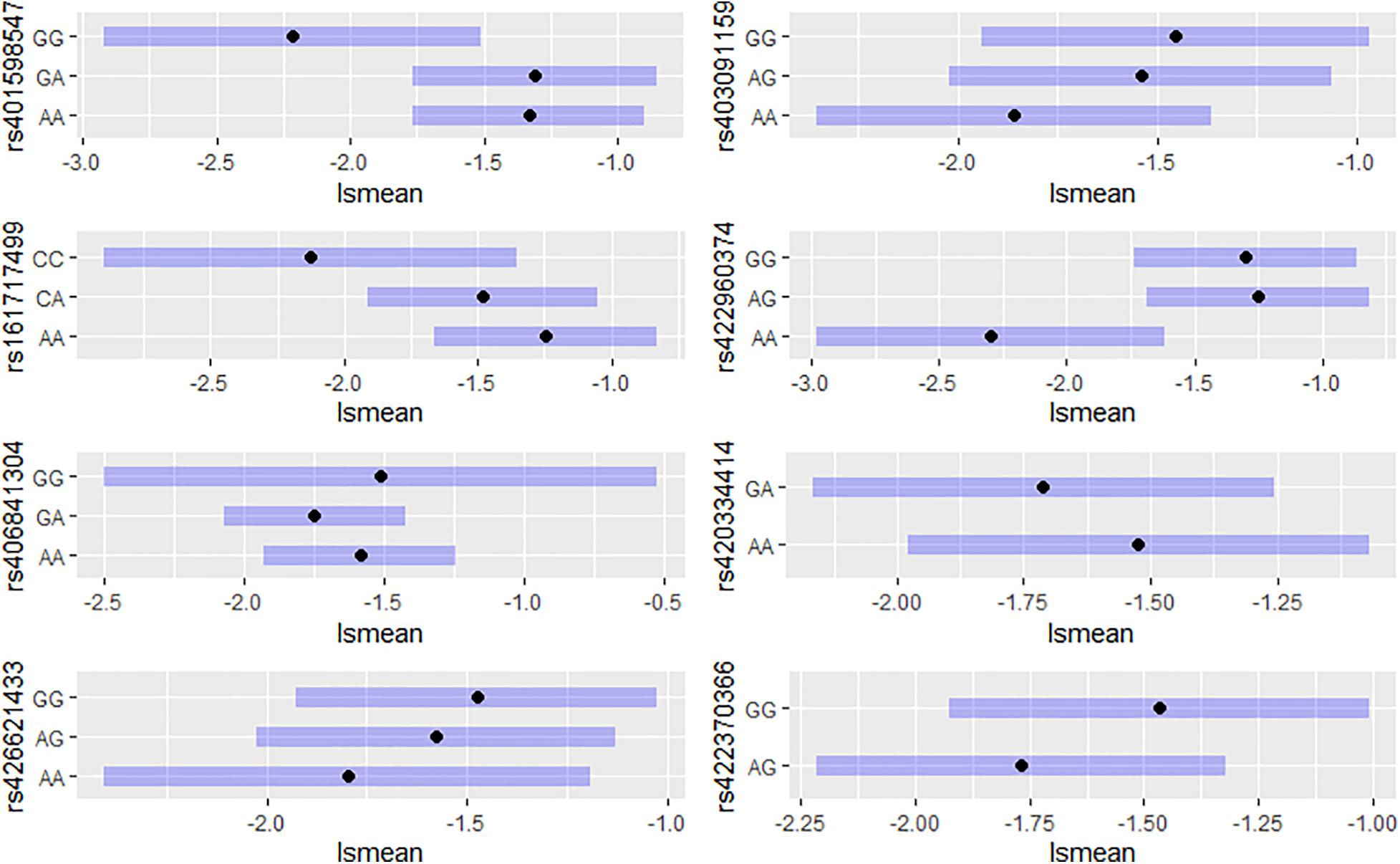
Figure 2. Least squares means (at 95% CI) of DEBV for the three genotypes affecting the trait of significant SNP detected from GWAS study.
Discussion
Mastitis is still one of the greatest problems affecting commercial milk production. However genetic evaluation of mastitis is particularly difficult because of the low heritability and the categorical nature of the trait. As a consequence, SCC has been promoted as an indirect method of predicting mammary infections due to the positive correlation between these two traits (Boettcher, 2005). It is worth to mention that collecting information on SCC is easier, cheaper, and less time demanding for farmers compared to use bacteriological status as direct measure of mastitis (Riggio, 2012). In this study we estimated the breeding value for SCS and identified the genomic regions putatively involved in mastitis resistance in the local economically important Valle del Belice dairy sheep breed.
The mean SCS (Table 1) was lower than those reported in previous studies (Riggio et al., 2010; Tolone et al., 2013) in the same breed, and by Ariznabarreta et al. (2002) in Churra sheep and Leitner et al. (2003) in Isaraeli-Assaf and Awassi sheep. These differences in SCS in the Valle del Belice breed were due to different sampled population. The heritability estimate for SCS in this study falls within the range (0.04–0.16) reported in literature for sheep (e.g., Barillet et al., 2001; Hamann et al., 2004; Tolone et al., 2013).
In this study, DEBVs of the SCS were used as trait scores for the association analysis. The estimated breeding values (EBVs) were generally used (like as pseudo-phenotypes) to perform the GWAS. Although EBVs have been used as dependent variables in GWAS (Johnston et al., 2011; Becker et al., 2013), this approach gave high false positive rate (Ekine et al., 2014). Consequences of using EBVs include varying levels of precision and “shrinkage effect” among the values used to represent phenotypes of different individuals, a reduction in the sample variance of the phenotypes, and double-counting of information from relatives (Garrick et al., 2009; Ostersen et al., 2011). The DEBV make good use of available information from genotyped animals as well as from their relatives, which can appropriately avoid bias introduced by simply pooling or averaging data information and account for heterogeneous variance (Garrick et al., 2009).
A total of eight SNPs were found to be significantly associated with SCS in Valle del Belice sheep. For five significant SNPs, results suggested that individuals with the GG genotypes at rs401598547, CC at rs161717499, and AA at rs403091159, rs422960374, and rs426621433, could be selected to reduce the somatic cells content in milk, although these genotypes had a low frequency in the breed. The lack of selection pressure in Valle del Belice dairy sheep may also contribute to the low frequency of the favorable alleles and genotypes. Therefore, the effect of these alleles for somatic cells content trait should be verified in a larger population or by testing them in an independent sample.
The most significant SNP associated with SCS was located in the intronic region of SERP1. This gene encodes the stress-associated endoplasmic reticulum protein 1 and was associated with immune system (Moravčíková et al., 2018). Another relevant gene close to the most significant SNP was SIAH2, involved to apoptosis and programmed cell death (Crisà et al., 2016). On the same chromosome, the other two markers were close to NEGR1, a gene involved with medium white blood cell count (a leukocyte trait) in Yak (Ma et al., 2019), LRRIQ3 related to the innate immune system upon recognition of pathogens (Pablo-Maiso et al., 2018) and FPGT, which is part of the L-fucose pathway, a key sugar in complex carbohydrates involved in cell-to-cell recognition, inflammation, and immune processes (Becker and Lowe, 2003). As above reported, mastitis is a persistent, inflammatory response of mammary tissue attributed to intramammary invasion of a mastitis-causing pathogen. Therefore, according to their role and function, these aforementioned genes can be considered as candidate involved in mastitis resistance and SCS. The SNP rs422960374 on OAR03, was close (∼70 Kb) to the FAM49A gene. Marete et al. (2018), in a GWAS for milking speed in French Holstein cows, reported the FAM49A as candidate gene for this trait. This gene was also associated to rear udder height in Holstein cattle (Gonzalez et al., 2020). The genetic correlation between the SCS and udder attachment in sheep was observed by Casu et al. (2010); De la Fuente et al. (1996) reported the indirect selection for subclinical mastitis resistance due to the inclusion of udder morphology traits in selection objectives. Moreover, Gutiérrez-Gil et al. (2018) suggested that sheep with udders and high degree of suspension or shallow udders close to the abdominal wall should be associated to lower SCS. Despite specific functions of this gene are not known yet, all the aforementioned aspects suggested a possible involving of FAM49A gene in our trait. Similarly, on OAR7, near to SNP rs406841304, two close genes are related with udder conformation (FAM227B) (Scienski et al., 2019) and with epithelial cell proliferation and differentiation (FGF7) (Bazer and Slayden, 2008; Yang et al., 2020), suggesting their possible role in the epithelial mammary cell proliferation. Moreover, the FGF7 has been reported as putative target gene in bovine mammary tissue infected with Streptococcus uberis (Naeem et al., 2012). Another significant SNP was located on OAR8 (rs426621433) at position 82,781,340 bp. This SNP mapped within a QTL for SCC (81.4–83.5 Mb) reported in a commercial French dairy sheep population (Rupp et al., 2015), and near a QTL for SCC (ID number 160869) on OAR 8 (80.5–80.6 Mb) in Churra sheep. Among the closest annotated genes in the region of ±250 kb surrounding it, the SOD2 gene seems to be the most plausible candidate affecting the SCS. In fact, the expression of SOD2 at mRNA and protein levels has been reported up-regulated in the mammary glands of ewes with clinical mastitis compared to healthy ewes (Gao et al., 2019). Mitterhuemer et al. (2010) showed an increase of SOD2 gene level in mammary tissue from mastitis cows inoculated with E. coli 24 h after infection as compared to controls. Finally, another candidate gene mapped near SNP rs426621433 on OAR8, was IGF2R, with a crucial role for the regulation of cell proliferation, growth, differentiation and survival, and associated with milk production traits. In fact, Dehoff et al. (1998) showed that lactation in the bovine mammary gland is associated with increased IGF2R concentration.
Conclusion
In this study, we estimated the breeding value for SCS in Valle del Belice sheep. DEBVs of the SCS were used as trait scores for the association analysis. Several candidate genes associated with SCS were identified related to immunity system and udder conformation. These candidate genes provide valuable information for future functional characterization. Therefore, our results may contribute to increase knowledge on the role the genes play in the genetically determined mechanisms involved in mastitis in sheep. The results can help improving the competitiveness of the local Valle del Belìce breed, through the development of genetic improvement programs directed toward reducing the incidence of mastitis, also considering the udder conformation into selection objectives, with planned mating between subjects carrying favorable alleles. Anyway, further studies considering a higher sample size or independent population will be needed to confirm our results.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://figshare.com/ articles/dataset/GWAS_for_somatic_cell_counts_in_sheep/1379 7851.
Ethics Statement
Ethical review and approval was not required for the animal study because the procedures involving animal sample collection followed the recommendation of directive 2010/63/EU. Sampling was carried out by trained veterinarians within the frame of vaccination Campaigns, hence no permission from the animal research ethics committee was necessary.
Author Contributions
MT and AS: conception of the work. AM, RD, and MT: contributed to the data acquisition. MT, AS, and AM: data analysis. AS, MT, SM, and BP: results interpretation. MT, SM, and AM: drafting the article. MT, SM, MS, and AS: critical revision of the article. AS, AM, SM, MS, RD, BP, and MT: final approval of the version to be published. All authors contributed to the article and approved the submitted version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Funding
This research was financed by the LEO: Livestock Environment Opendata, PSRN 2014−2020−Sottomisura: 16.2, Project number PRJ−0185, CUP: J84I18000090007.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2021.643531/full#supplementary-material
Footnotes
- ^ https://www.ncbi.nlm.nih.gov/genome/?term=ovis+aries
- ^ http://www.r-project.org
- ^ https://www.ncbi.nlm.nih.gov/genome/gdv/browser/genome/?id=GCF_000298735.2
- ^ http://www.animalgenome.org/QTLdb
References
Ali, A., and Shook, G. E. (1980). An optimum transformation for somatic cell count in milk. J. Dairy Sci. 63, 487–490. doi: 10.3168/jds.S0022-0302(80)82959-6
Amin, N., Van Duijn, C. M., and Aulchenko, Y. S. A. (2007). Genomic background based method for association analysis in related individuals. PLoS One 2:e1274. doi: 10.1371/journal.pone.0001274
Ariznabarreta, A., Gonzalo, C., and San Primitivo, F. (2002). Microbiological quality and somatic cell count of ewe milk with special reference to staphylococci. J. Dairy Sci. 85, 1370–1375. doi: 10.3168/jds.S0022-0302(02)74203-3
Aulchenko, Y. S., de Koning, D. J., and Haley, C. (2007). Genome wide rapid association using mixed model and regression: a fast and simple method for genome wide pedigree-based quantitative trait loci as-sociation analysis. Genetics 177, 577–585. doi: 10.1534/genetics.107.075614
Barillet, F., Rupp, R., Mignon-Grasteau, S., Astruc, J. M., and Jacquin, M. (2001). Genetic analysis for mastitis resistance and milk somatic cell score in French Lacaune dairy sheep. Genet. Sel. Evol. 33, 397–415. doi: 10.1186/1297-9686-33-4-397
Bazer, F. W., and Slayden, O. D. (2008). Progesterone-induced gene expression in uterine epithelia: a myth perpetuated by conventional wisdom. Biol. Reprod. 79, 1008–1009. doi: 10.1095/biolreprod.108.072702
Becker, D., Wimmers, K., Luther, H., Hofer, A., and Leeb, T. (2013). A genome-wide association study to detect QTL for commercially important traits in Swiss Large White boars. PLoS One 8:e55951. doi: 10.1371/journal.pone.0055951
Becker, D. J., and Lowe, J. B. (2003). Fucose: biosynthesis and biological function in mammals. Glycobiology 13, 41R–53R. doi: 10.1093/glycob/cwg054
Becker, G. M., Davenport, K. M., Burke, J. M., Lewis, R. M., Miller, J. E., Morgan, J. L. M., et al. (2020). Genome−wide association study to identify genetic loci associated with gastrointestinal nematode resistance in Katahdin sheep. Anim. Genet. 51, 330–335. doi: 10.1111/age.12895
Boettcher, P. (2005). Breeding for improvement of functional traits in dairy cattle. Ital. J. Anim. Sci. 4, 7–16. doi: 10.4081/ijas.2005.3s.7
Butler, D. G., Cullis, B. R., Gilmour, A. R., and Gogel, B. J. (2009). ASReml-R Reference Manual. Technical Report. Brisbane, QLD: Queensland Department of Primary Industries.
Casu, S., Sechi, S., Salaris, S. L., and Carta, A. (2010). Phenotypic and genetic relationships between udder morphology and udder health in dairy ewes. Small Rumin. Res. 88, 77–83. doi: 10.1016/j.smallrumres.2009.12.013
Contreras, A., Sierra, D., Sánchez, A., Corrales, J. C., Marco, J. C., and Paape, M. J. (2007). Mastitis in small ruminants. Small Rumin. Res. 68, 145–153. doi: 10.1016/j.smallrumres.2006.09.011
Crisà, A., Ferrè, F., Chillemi, G., and Moioli, B. (2016). RNA-Sequencing for profiling goat milk transcriptome in colostrum and mature milk. BMC Vet. Res. 12:264. doi: 10.1186/s12917-016-0881-7
De la Fuente, L. F., Fernandez, G., and San Primitivo, F. (1996). A linear evaluation system for udder traits of dairy ewes. Livest. Prod. Sci. 45, 171–178. doi: 10.1016/0301-6226(96)00003-6
Dehoff, M. H., Elgin, R. G., Collier, R. J., and Clemmons, D. R. (1998). Both type I and II insulin-like growth factor receptor binding increase during lactogenesis in bovine mammary tissue. Endocrinology 122, 2412–2417. doi: 10.1210/endo-122-6-2412
Ekine, C. C., Rowe, S. J., Bishop, S. C., and de Koning, D. J. (2014). Why breeding values estimated using familial data should not be used for genome-wide association studies. G3 Genes Genom. Genet. 4, 341–347. doi: 10.1534/g3.113.008706
Gao, J., Li, T., Lu, Z., Wang, X., Zhao, X., and Ma, Y. (2019). Proteomic analyses of mammary glands provide insight into the immunity and metabolism pathways associated with clinical mastitis in meat sheep. Animals 9:309. doi: 10.3390/ani9060309
Garrick, D. J., Taylor, J. F., and Fernando, R. L. (2009). Deregressing estimated breeding values and weighting information for genomic regression analyses. Genet. Sel. Evol. 41:55. doi: 10.1186/1297-9686-41-55
Ghasemi, M., Zamani, P., Vatankhah, M., and Abdoli, R. (2019). Genome-wide association study of birth weight in sheep. Animal 13, 1797–1803. doi: 10.1017/S1751731118003610
Gonzalez, M., Villa, R., Villa, C., Gonzalez, V., Montano, M., Medina, G., et al. (2020). Inspection of real and imputed genotypes reveled 76 SNPs associated to rear udder height in Holstein cattle. Adv. Vet. Anim. Res. 7, 234–241.
Gutiérrez-Gil, B., Esteban-Blanco, C., Suarez-Vega, A., and Arranz, J. J. (2018). Detection of quantitative trait loci and putative causal variants affecting somatic cell score in dairy sheep by using a 50K SNP chip and whole-genome sequencing. J. Dairy Sci. 101, 9072–9088. doi: 10.3168/jds.2018-14736
Hamann, H., Horstick, A., Wessels, A., and Distl, O. (2004). Estimation of genetic parameters for test day milk production, somatic cell score and litter size at birth in East Friesian ewes. Livest. Prod. Sci. 87, 153–160. doi: 10.1016/j.livprodsci.2003.09.015
ICAR (2014). International Agreement of Recording Practices. International Committee for Animal Recording. Available online at: https://www.icar.org/index.php/icar-recording-guidelines/ (accessed September 10, 2020).
Johnston, S. E., McEwan, J. C., Pickering, N. K., Kijas, J. W., Beraldi, D., Pilkington, J. G., et al. (2011). Genome-wide association mapping identifies the genetic basis of discrete and quantitative variation in sexual weaponry in a wild sheep population. Mol. Ecol. 20, 2555–2566. doi: 10.1111/j.1365-294X.2011.05076.x
Kelly, A. L., Tiernan, D., O’sullivan, C., and Joyce, P. (2000). Correlation between bovine milk somatic cell count and polymorphonuclear leukocyte level for samples of bulk milk and milk from individual cows. J. Dairy Sci. 83, 300–304. doi: 10.3168/jds.S0022-0302(00)74878-8
Leitner, G., Chaffer, M., Caraso, Y., Ezra, E., Kababea, D., Winkler, M., et al. (2003). Udder infection and milk somatic cell count, NAGase activity and milk composition—fat, protein and lactose—in Israeli-Assaf and Awassi sheep. Small Rumin. Res. 49, 157–164. doi: 10.1016/S0921-4488(03)00079-8
Leitner, G., Silanikove, N., and Merin, U. (2008). Estimate of milk and curd yield loss of sheep and goats with intrammamary infection and its relation to somatic cell count. Small Rumin. Res. 74, 221–225. doi: 10.1016/j.smallrumres.2007.02.009
Li, H., Wu, X. L., Tait, R. G. Jr., Bauck, S., Thomas, D. L., Murphy, T. W., et al. (2020). Genome−wide association study of milk production traits in a crossbred dairy sheep population using three statistical models. Anim. Genet. 51, 624–628. doi: 10.1111/age.12956
Ma, X., Jia, C., Fu, D., Chu, M., Ding, X., Wu, X., et al. (2019). Analysis of hematological traits in polled yak by genome-wide association studies using individual SNPs and haplotypes. Genes 10:463. doi: 10.3390/genes10060463
Marete, A., Sahana, G., Fritz, S., Lefebvre, R., Barbat, A., Lund, M. S., et al. (2018). Genome-wide association study for milking speed in French Holstein cows. J. Dairy Sci. 101, 6205–6219. doi: 10.3168/jds.2017-14067
Mastrangelo, S., Tolone, M., Sardina, M. T., Sottile, G., Sutera, A. M., Di Gerlando, R., et al. (2017). Genome-wide scan for runs of homozygosity identifies potential candidate genes associated with local adaptation in Valle del Belice sheep. Genet. Sel. Evol. 49, 84. doi: 10.1186/s12711-017-0360-z
Miller, S. A., Dykes, D. D., and Polesky, H. F. (1988). A salting out method for DNA preparation. Nucleic Acids Res. 16:1215. doi: 10.1093/nar/16.3.1215
Moravčíková, N., Simčič, M., Mészáros, G., Sölkner, J., Kukučková, V., Vlček, M., et al. (2018). Genomic response to natural selection within alpine cattle breeds. Czech J. Anim. Sci. 63, 136–143. doi: 10.17221/62/2017-CJAS
Naeem, A., Zhong, K., Moisá, S. J., Drackley, J. K., Moyes, K. M., and Loor, J. J. (2012). Bioinformatics analysis of microRNA and putative target genes in bovine mammary tissue infected with Streptococcus uberis. J. Dairy Sci. 95, 6397–6408. doi: 10.3168/jds.2011-5173
Oget, C., Tosser-Klopp, G., and Rupp, R. (2019). Genetic and genomic studies in ovine mastitis. Small Rumin. Res. 176, 55–64. doi: 10.1016/j.smallrumres.2019.05.011
Ostersen, T., Christensen, O. F., Henryon, M., Nielsen, B., Su, G., and Madsen, P. (2011). Deregressed EBV as the response variable yield more reliable genomic predictions than traditional EBV in pure-bred pigs. Genet. Sel. Evol. 43:38. doi: 10.1186/1297-9686-43-38
Pablo-Maiso, D., Doménech, A., Echeverría, I., Gómez-Arrebola, C., De Andrés, D., Rosati, S., et al. (2018). Prospects in innate immune responses as potential control strategies against non-primate Lentiviruses. Viruses 10:435. doi: 10.3390/v10080435
Power, R. A., Parkhill, J., and de Oliveira, T. (2017). Microbial genome-wide association studies: lessons from human GWAS. Nat Rev Genet 18:41. doi: 10.1038/nrg.2016.132
Purcell, S., Neale, B., Todd-Brown, K., Thomas, L., Ferreira, M. A., Bender, D., et al. (2007). PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 81, 559–575. doi: 10.1086/519795
Raynal-Ljutovac, K., Pirisi, A., de Crémoux, R., and Gonzalo, C. (2007). Somatic cells of goat and sheep milk analytical, sanitary, productive and technological aspects. Small Rumin. Res. 68, 126–144. doi: 10.1016/j.smallrumres.2006.09.012
Riggio, V. (2012). Genetic Aspects of Somatic Cell Count and Udder Health in the Italian Valle del Belice Dairy Sheep. Gelderland: Wageningen University.
Riggio, V., and Portolano, B. (2015). Genetic selection for reduced somatic cell counts in sheep milk: a review. Small Rumin. Res. 126, 33–42. doi: 10.1016/j.smallrumres.2015.01.020
Riggio, V., Portolano, B., Bovenhuis, H., and Bishop, S. C. (2010). Genetic parameters for somatic cell score according to udder infection status in Valle del Belìce dairy sheep and impact of imperfect diagnosis of infection. Genet. Sel. Evol. 42:30. doi: 10.1186/1297-9686-42-30
Rovadoscki, G. A., Pertile, S. F. N., Alvarenga, A. B., Cesar, A. S. M., Pértille, F., Petrini, J., et al. (2018). Estimates of genomic heritability and genome-wide association study for fatty acids profile in Santa Inês sheep. BMC Genomics 19:375. doi: 10.1186/s12864-018-4777-8
Rupp, R., Senin, P., Sarry, J., Allain, C., Tasca, C., Ligat, L., et al. (2015). A point mutation in suppressor of cytokine signalling 2 (Socs2) increases the susceptibility to inflammation of the mammary gland while associated with higher body weight and size and higher milk production in a sheep model. PLoS Genet 11:e1005629. doi: 10.1371/journal.pgen.1005629
Scienski, K., Ialacci, A., Bagnato, A., Reginelli, D., Durán-Aguilar, M., and Strillacci, M. G. (2019). Genetic variability in a Holstein population using SNP markers and their use for monitoring mating strategies. Rev. Mex. Cienc. Pecu. 10, 643–663. doi: 10.22319/rmcp.v10i3.4842
Sell-Kubiak, E., Duijvesteijn, N., Lopes, M. S., Janss, L. L. G., Knol, E. F., Bijma, P., et al. (2015). Genome-wide association study reveals novel loci for litter size and its variability in a Large White pig population. BMC Genomics 16:1049. doi: 10.1186/s12864-015-2273-y
Sevillano, C. A., Lopes, M. S., Harlizius, B., Hanenberg, E. H., Knol, E. F., and Bastiaansen, J. W. M. (2015). Genome-wide association study using deregressed breeding values for cryptorchidism and scrotal/inguinal hernia in two pig lines. Genet. Sel. Evol. 47:18. doi: 10.1186/s12711-015-0096-6
Shook, G. E., and Schutz, M. M. (1994). Selection on somatic cell score to improve resistance to mastitis in the United States. J. Dairy Sci. 77, 648–658. doi: 10.3168/jds.S0022-0302(94)76995-2
Sutera, A. M., Portolano, B., Di Gerlando, R., Sardina, M. T., Mastrangelo, S., and Tolone, M. (2018). Determination of milk production losses and variations of fat and protein percentages according to different levels of somatic cell count in Valle del BeliceBelìce dairy sheep. Small Rumin. Res. 162, 39–42. doi: 10.1016/j.smallrumres.2018.03.002
Sutera, A. M., Riggio, V., Mastrangelo, S., Di Gerlando, R., Sardina, M. T., Pong−Wong, R., et al. (2019). Genome−wide association studies for milk production traits in Valle del Belìce sheep using repeated measures. Anim. Genet. 50, 311–314. doi: 10.1111/age.12789
Tao, L., He, X. Y., Pan, L. X., Wang, J. W., Gan, S. Q., and Chu, M. X. (2020). Genome−wide association study of body weight and conformation traits in neonatal sheep. Anim. Genet. 51, 336–340. doi: 10.1111/age.12904
Tolone, M., Riggio, V., and Portolano, B. (2013). Estimation of genetic and phenotypic parameters for bacteriological status of the udder, somatic cell score, and milk yield in dairy sheep using a threshold animal model. Livest. Sci. 151, 134–139. doi: 10.1016/j.livsci.2012.11.014
Wang, Z., Zhang, H., Yang, H., Wang, S., Rong, E., Pei, W., et al. (2014). Genome-wide association study for wool production traits in a Chinese Merino sheep population. PLoS One 9:e107101. doi: 10.1371/journal.pone.0107101
White, S. N., Mousel, M. R., Herrmann-Hoesing, L. M., Reynolds, J. O., Leymaster, K. A., Neibergs, H. L., et al. (2012). Genome-wide association identifies multiple genomic regions associated with susceptibility to and control of ovine lentivirus. PLoS One 7:e47829. doi: 10.1371/journal.pone.0047829
Keywords: mastitis, local dairy sheep, GWAS, SNPs arrays, candidate genes
Citation: Sutera AM, Moscarelli A, Mastrangelo S, Sardina MT, Di Gerlando R, Portolano B and Tolone M (2021) Genome-Wide Association Study Identifies New Candidate Markers for Somatic Cells Score in a Local Dairy Sheep. Front. Genet. 12:643531. doi: 10.3389/fgene.2021.643531
Received: 18 December 2020; Accepted: 01 March 2021;
Published: 22 March 2021.
Edited by:
Mohammed Ali Al Abri, Sultan Qaboos University, OmanCopyright © 2021 Sutera, Moscarelli, Mastrangelo, Sardina, Di Gerlando, Portolano and Tolone. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Marco Tolone, bWFyY28udG9sb25lQHVuaXBhLml0
 Anna Maria Sutera
Anna Maria Sutera Angelo Moscarelli
Angelo Moscarelli Salvatore Mastrangelo
Salvatore Mastrangelo Maria Teresa Sardina2
Maria Teresa Sardina2 Marco Tolone
Marco Tolone