
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
MINI REVIEW article
Front. Genet., 16 April 2021
Sec. Pharmacogenetics and Pharmacogenomics
Volume 12 - 2021 | https://doi.org/10.3389/fgene.2021.641905
This article is part of the Research TopicThe Immunology of Adverse Drug ReactionsView all 13 articles
 Yueran Li1
Yueran Li1 Pooja Deshpande1
Pooja Deshpande1 Rebecca J. Hertzman1
Rebecca J. Hertzman1 Amy M. Palubinsky2
Amy M. Palubinsky2 Andrew Gibson1*
Andrew Gibson1* Elizabeth J. Phillips1,2
Elizabeth J. Phillips1,2Adverse drug reactions (ADRs) remain associated with significant mortality. Delayed hypersensitivity reactions (DHRs) that occur greater than 6 h following drug administration are T-cell mediated with many severe DHRs now associated with human leukocyte antigen (HLA) risk alleles, opening pathways for clinical prediction and prevention. However, incomplete negative predictive value (NPV), low positive predictive value (PPV), and a large number needed to test (NNT) to prevent one case have practically prevented large-scale and cost-effective screening implementation. Additional factors outside of HLA contributing to risk of severe T-cell-mediated DHRs include variation in drug metabolism, T-cell receptor (TCR) specificity, and, most recently, HLA-presented immunopeptidome-processing efficiencies via endoplasmic reticulum aminopeptidase (ERAP). Active research continues toward identification of other highly polymorphic factors likely to impose risk. These include those previously associated with T-cell-mediated HLA-associated infectious or auto-immune disease such as Killer cell immunoglobulin-like receptors (KIR), epistatically linked with HLA class I to regulate NK- and T-cell-mediated cytotoxic degranulation, and co-inhibitory signaling pathways for which therapeutic blockade in cancer immunotherapy is now associated with an increased incidence of DHRs. As such, the field now recognizes that susceptibility is not simply a static product of genetics but that individuals may experience dynamic risk, skewed toward immune activation through therapeutic interventions and epigenetic modifications driven by ecological exposures. This review provides an updated overview of current and proposed genetic factors thought to predispose risk for severe T-cell-mediated DHRs.
Adverse drug reactions (ADRs) are estimated as the fourth to sixth leading cause of death (Dormann et al., 2000; Pouyanne et al., 2000; Miya et al., 2019). While the majority are classified as type A, predictable based on drug pharmacology, the remainder are off-target type B ADRs and inclusive of T-cell-mediated delayed drug hypersensitivity reactions (DHRs). While DHRs may elicit systemic effects, diverse clinical reactions also target specific organs including drug-induced liver injury (DILI), associated with nausea, fatigue, jaundice, and mortality up to 9.4% (Leise et al., 2014). However, most often they target skin, with presentation from mild rash (fixed drug eruption, maculopapular exanthema) to life-threatening severe cutaneous adverse reactions (SCARs) including Stevens-Johnson Syndrome/Toxic Epidermal Necrolysis (SJS/TEN) and drug reaction with eosinophilia and systemic symptoms (DRESS) (Peter et al., 2017). DRESS has a mortality up to 10% (Kardaun, 2019; Wolfson et al., 2019) and is characterized by widespread skin eruption, lymphadenopathy, fever, and multiple organ involvement (Choudhary et al., 2013; Kardaun, 2019). SJS and TEN are the same disease across a spectrum of severity with the higher end of mortality (TEN) associated with up to 50% death (Patel et al., 2013; Langley et al., 2018). SJS/TEN is characterized by blistering and involvement of at least two mucous membranes (Paulmann and Mockenhaupt, 2015; Miya et al., 2019; Zimmerman and Dang, 2020). Despite clinical distinction, lack of mechanistic delineation has precluded development of disease-specific treatment and prevention strategies (Pavlos et al., 2015; Redwood et al., 2018). In recent years many DHRs have been associated with strong human leukocyte antigen (HLA) class I associations opening pathways to prediction and prevention (Figure 1).
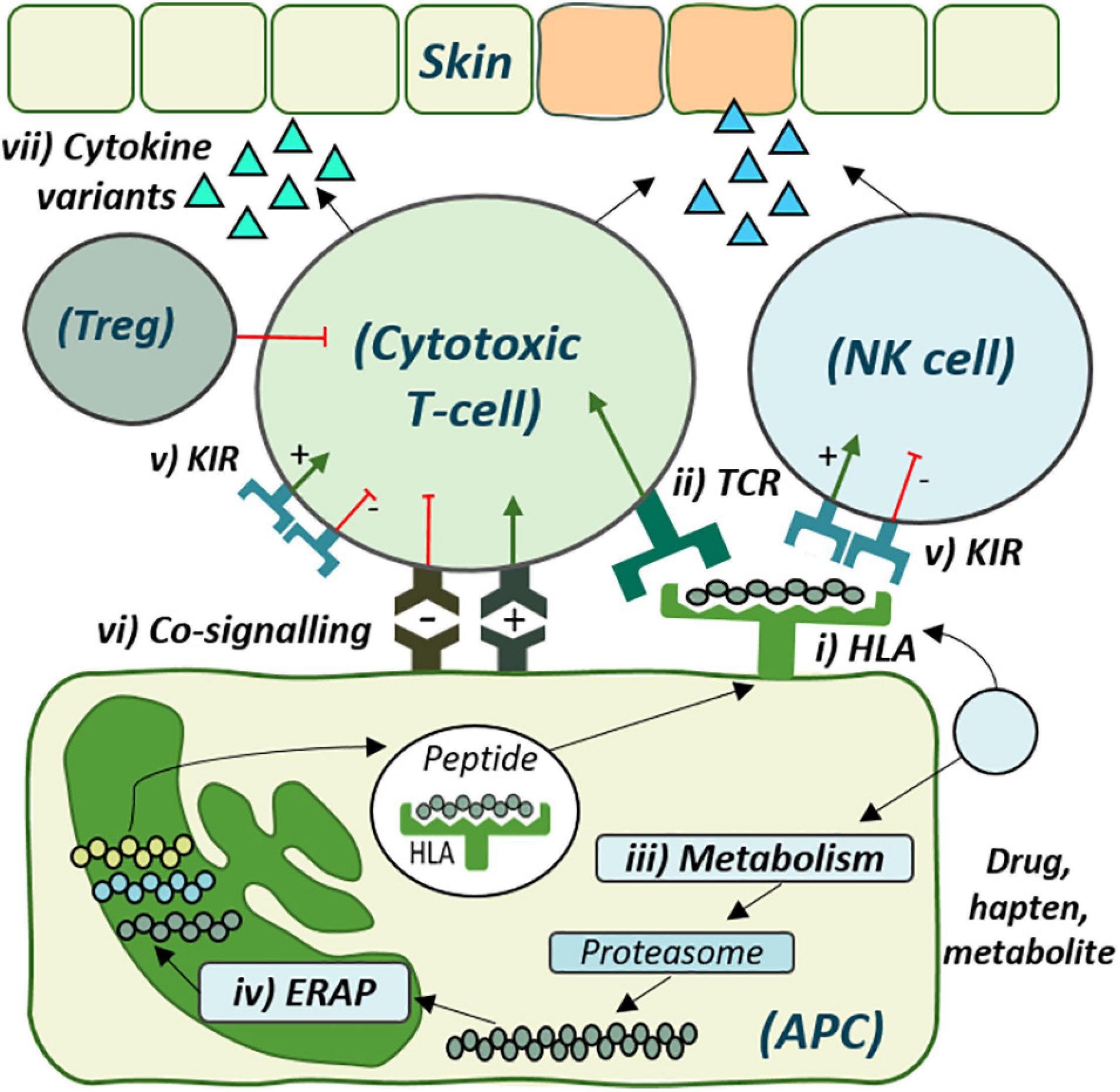
Figure 1. The cellular role of genetic risk factors defined and proposed for development of delayed drug hypersensitivity reactions. Drug antigen may bind directly to (i) specific human leukocyte antigen (HLA) molecules on the surface of antigen-presenting cells (APC) for recognition by corresponding (ii) T-cell receptors (TCRs). Alternatively, drug may undergo (iii) metabolism and cellular processing by (iv) endoplasmic reticulum aminopeptidase (ERAP), with the peptide products then presented by the risk HLA molecule for TCR recognition. (v) Distinct killer-cell immunoglobulin-like receptors (KIR), expressed on both T-cell and Natural Killer (NK) cells, also show specificity for HLA-antigen interaction, which may regulate cytotoxic degranulation. (vi) Co-signaling pathways also regulate T-cell activation, with overall co-inhibition or co-stimulation leading to (vii) cytokine release and respective tolerance with T-regulatory cell (Treg)-induced immunosuppression or inflammation and cytotoxic degranulation, respectively.
The HLA locus is highly polymorphic with >25,000 allelic variants annotated (HLA.alleles.org). In 2002, Mallal demonstrated carriage of HLA-B∗57:01 among 78% of HIV patients with abacavir hypersensitivity, which is a well-characterized systemic syndrome, opposed to just 2% of tolerant patients (Mallal et al., 2002). A randomized double blind clinical trial of real-time HLA-B∗57:01 screening versus abacavir treatment without real-time screening showed a negative predictive value (NPV) of 100% and a positive predictive value (PPV) of 55% (Mallal et al., 2008), demonstrating that HLA-B∗57:01 screening eliminates patch test positive abacavir hypersensitivity. This PREDICT-1 study was the licensing study upon which guideline-based HLA-B∗57:01 screening prior to abacavir prescription was established.
In 2004, association between HLA-B∗15:02 and carbamazepine (CBZ)-induced SJS/TEN in Taiwan was reported, which followed the translational roadmap provided by abacavir such that 0/4120 Taiwanese HLA-B∗15:02-negative patients developed SJS/TEN after CBZ exposure (Chung et al., 2004). Pre-prescription HLA-B∗15:02 screening for CBZ is now active in Hong Kong, Singapore, and Thailand where there is high allelic prevalence (FerrellJr., and McLeod, 2008). However, HLA-B∗15:02 is expressed in <1% of patients of European or African ancestry despite global disease burden, restricting universal screening and inferring that different HLA alleles drive reactions in different populations (Karnes et al., 2019). Indeed, multiple alleles are now associated with CBZ-SCAR in distinct populations, with HLA-A∗31:01 associated with DRESS in Europeans and Chinese, but not SJS/TEN (McCormack et al., 2011; Genin et al., 2014), highlighting propensity for distinct alleles to define risk for specific reactions. Most recently, Nicoletti reported HLA-A∗31:01 as a strong risk factor broadly across CBZ-induced SCAR and DILI in Europeans (Nicoletti et al., 2019) while Mockenhaupt described an HLA-B∗57:01 association for CBZ-SJS/TEN in Europeans (Mockenhaupt et al., 2019). These studies demonstrate that HLA restriction may be complex, with influence from multiple alleles restricted to antigen, reaction phenotype, and population (Table 1).
Other strong HLA associations have been described with near-complete NPV for WHO essential medicines, the most effective and safe drugs to meet the most important needs, such as allopurinol, dapsone, and vancomycin (WHO, 2021). Allopurinol is used for treatment of gout but is also the most prevalent drug cause of DRESS in the FDA Adverse event reporting system (Bluestein et al., 2021). In 2005, HLA-B∗58:01 was associated with allopurinol-induced SCAR with 100% NPV in Southeast Asians (Hung et al., 2005). Subsequent studies confirmed risk in cohorts from Europe (Lonjou et al., 2008), Japan (Kaniwa et al., 2008), Thailand (Yu et al., 2017), South Korea (Kang et al., 2011), and Portugal (Gonçalo et al., 2013), but, as with CBZ, comparative strength of association and allelic frequency is not replicated and is far lower in Europeans (Génin et al., 2011). Currently, where patients are known to be HLA-B∗58:01+, the European Medicines Agency advises clinicians to avoid allopurinol and screening is recommended in Korean, Thai, or Han Chinese patients (Ke et al., 2017). However, recent analysis in the UK defined the number needed to test (NNT) as 11,286, leading the panel to advise against routine screening (Plumpton et al., 2017).
The antibiotic dapsone is predominantly associated with treatment of leprosy (Wolf et al., 2002). In 2013, HLA-B∗13:01 was described with 99.8% NPV and 7.8% PPV as a risk factor among Chinese patients for dapsone hypersensitivity (Zhang et al., 2013). While prevalent in Chinese and Indian populations, HLA-B∗13:01 is comparatively absent among Europeans and Africans. HLA-B∗13:01 risk is now confirmed for dapsone-SCAR in Thailand (Tempark et al., 2017) and research has modeled drug interaction within the HLA binding site (Watanabe et al., 2017). Most recently, Chen expanded HLA-B∗13:01 risk to patients from Malaysia and Taiwan (Chen et al., 2018), and Zhao identified dapsone-responsive HLA-B∗13:01-restricted CD8+ T-cells in patients (Zhao et al., 2019).
Vancomycin, a front-line treatment for beta-lactam-resistant infections (Rybak et al., 2009; Frymoyer et al., 2013; Moore et al., 2020), is the most common antibiotic instigator of DRESS (Wolfson et al., 2019). In 2019, Konvinse published strong association between HLA-A∗32:01 and vancomycin-DRESS determining that 20% of HLA-A∗32:01+ patients would develop the disease (Konvinse et al., 2019). With a European prevalence of 6.8%, they predicted the NNT as just 75 and have since developed an HLA-A∗32:01-specific, cost-effective real-time PCR screen (Rwandamuriye et al., 2019). In 2020, Nakkam described cross-reactivity with an alternate glycopeptide antibiotic, teicoplanin, in 16% of HLA-A∗32:01+ vancomycin-DRESS patients predicted by a shared class II HLA haplotype (Nakkam et al., 2020). These data implicate risk alleles with influence not simply to dictate predisposition but with ramifications for ongoing treatment. Importantly, while predictive values defined by limited case-control studies may not be indicative of risk in the underlying population, warranting caution, in vitro assays have functionally confirmed that HLA risk restricted drug-specific T-cell activation for abacavir, CBZ, allopurinol, dapsone, and vancomycin (Chessman et al., 2008; Wei et al., 2012; Yun et al., 2014; Zhao et al., 2019; Nakkam et al., 2020).
Single HLA associations up until 2019 have been extensively reviewed (White et al., 2015; Karnes et al., 2019; Oussalah et al., 2020). Since then, further advancement in sequencing platforms has been providing increased resolution that has enabled discovery of novel HLA associations (LaHaye et al., 2016; van der Ven et al., 2018; Giannopoulou et al., 2019; Mimori et al., 2019). In 2019, Nakatani reported a Japanese association between SJS/TEN, HLA-A∗02:06:01, and cold medicines containing non-steroidal anti-inflammatories (Nakatani et al., 2019). Furthermore, Tangamornsuksan reported an association between methazolamide-induced SJS/TEN and HLA-B∗59:01 in Koreans and Han Chinese (Tangamornsuksan and Lohitnavy, 2019). In 2020, within a Thai HIV cohort, Sukasem reported an association between co-trimoxazole-induced DRESS with HLA-B∗13:01 and SJS/TEN with HLA-B∗15:02 and HLA-C∗08:01 (Sukasem et al., 2020). Furthermore, MPE and DRESS resulting from benznidazole was associated with HLA-A∗68, A∗11:01, and A∗29:02 in Bolivian patients with Chagas disease (Balas et al., 2020). Most recently, Zhao reported an association between oxcarbazepine-induced MPE and HLA-A∗03:01 and HLA-B∗07:02 in patients of Uighur Chinese ethnicity (Zhao et al., 2020). Moreover, HLA associations have also been reported for herbal medicines including green tea (Hoofnagle et al., 2020) and polygonum multiflorum with HLA-B∗35:01 (Li et al., 2019). These studies provide a glimpse into the recent progress toward risk prediction specific to populations, yet a significant hurdle remains risk discovery in minority groups for whom access to large cohorts for traditional population studies is nearly impossible. One strategy is to maximize utility of international SCAR registries where careful patient matching for drug, reaction phenotype, and ethnicity may provide means to explore shared risk (Somogyi et al., 2019). Indeed, Somogyi identified three patients of Australian Indigenous ethnicity with phenytoin-DRESS sharing HLA-B∗56:02 (Somogyi et al., 2019). Critically, HLA-B∗56:02 frequency ranges up to 19% in this population but is absent from the predominant Australian European populace, highlighting utility of detailed biobanking with functional validation of proposed risk alleles (Monshi et al., 2013; Pan et al., 2019). Another possibility is the likelihood that alleles with shared specificities drive response to the same drug, as for nevirapine (Chantarangsu et al., 2009; Carr et al., 2013). Here, association with HLA-C∗04 across ethnicities is driven by a unique F pocket motif that determines similar binding specificity for HLA-C∗04:01 with HLA-C∗05:01 and HLA-C∗18:01, dominant in Hispanics and Africans, respectively (Pavlos et al., 2017). The ability to design HLA crystal structures combined with HLA binding algorithms provides a functional bridge to understand whether proposed antigen binds to diverse alleles (Pavlos et al., 2017). Nonetheless, HLA is not the sole requirement for T-cell activation and other parameters are proposed to retain the HLA-restricted “positive predictive gap.”
Antigenic peptides bound to HLA must contact the T-cell receptor (TCR) to trigger T-cell activation (Figure 1). Each individual’s TCR repertoire comprises a diverse blend of public and private TCRs, which, through prior antigen exposure, may be uniquely distributed in tissues (Robins et al., 2010). A polyclonal response is well documented for abacavir (Redwood et al., 2019). This is in keeping with the altered peptide repertoire hypothesis suggesting that abacavir binds within the F pocket of the HLA-B∗57:01 peptide binding groove altering its peptide specificity and the repertoire of self-peptides recognized as immunogenic (Illing et al., 2012). Polyclonal response is also observed during CDR3 spectratyping after the in vitro priming of naïve T-cells to the immunogenic drug metabolite sulfamethoxazole-nitroso (SMX-NO) (Gibson et al., 2017). Here the authors implicate the high protein reactivity of SMX-NO thought to drive formation of multiple haptens, each with potential to produce a diverse array of antigenic peptides. However, early work by Nassif reports predominant expression of Vβ 13.1 and 14 on T-cells in the blister of such patients, suggesting that early response in tissue is driven by more select, dominant clonotypes (Nassif et al., 2002). In 2019, Pan reported dominant single, public “VFDNTDKLI” TCRα CDR3 and “ASSLAGELFF” TCRβ CDR3 in HLA-B∗15:02+ patients with CBZ hypersensitivity, rare in blood but dominantly expressed in blister (Pan et al., 2019). The dominant TCR was identified on T-cells expressing granulysin, a key cytotoxic mediator with precedent in eliciting tissue damage (Pan et al., 2019). Furthermore, the complete TCR blueprint provided by single-cell sequencing was synthetically reconstructed and shown to trigger T-cell activation specific to CBZ and HLA-B∗15:02. Preferential TCR expansion has also been described in blister during HLA-B∗58:01-associated allopurinol-SCAR (Chung et al., 2015). While further studies are warranted, those described begin to elucidate the specificity of a single dominantly expanded TCR to drive early response in the tissue of HLA-predisposed patients.
Although drug-protein conjugates are found at similar levels in allergic and tolerant patients (Park et al., 1998; Sullivan et al., 2015), the downstream impact of N-terminal peptide trimming that shapes the HLA-presented immunopeptidome has remained undefined. This process is performed by endoplasmic reticulum aminopeptidases (ERAPs) 1 and 2 (Serwold et al., 2001; Chang et al., 2005; Figure 1) for which polymorphic variants alter susceptibility and outcome to autoimmune disease and viral infections with HLA class I-restricted etiologies (Evans et al., 2011; Guerini et al., 2012; Biasin et al., 2013; Fruci et al., 2014; Reeves and James, 2015; Saulle et al., 2019; Vidal-Castiñeira et al., 2020). Specifically, distinct ERAP1 allotypes skew the HLA-class I-expressed immunopeptidome during infectious disease, where hypoactive allotypes result in longer sub-dominant peptides that impair CD8+ T-cell response (Kemming et al., 2019). Intriguingly, peptides with aromatic or hydrophobic C-terminal amino acids are favored by ERAP1 for efficient N-terminal trimming and treatment of cells with abacavir alters the self-peptide preference toward the same amino acids (Chang et al., 2005; Ostrov et al., 2012). In 2020, Pavlos identified ERAP1 as a novel predictor of abacavir tolerance among HLA-B∗57:01+ patients. Tolerant patients were significantly more likely to express ERAP1 hypoactive allotypes with reduced trimming efficiency compared to hypersensitive patients (Pavlos et al., 2020). While yet to transverse other drugs, the epistatic relationship between HLA and ERAP raises intrigue to the influence of other such genes. One such entity is the highly polymorphic Killer-cell Immunoglobulin-like receptors (KIRs) expressed on T-cells and Natural Killer (NK) cells (Mingari et al., 1997; LeMaoult et al., 2005), with both cell types reporting the predominant infiltrate of in SJS/TEN blister (Chung and Hung, 2010). HLA alleles are the distinct ligands for KIRs that regulate cytotoxic degranulation in a complex interaction with sensitivity to the presented peptide via overlapped TCR binding (Mandelboim et al., 1997; Boyington and Sun, 2002; Thananchai et al., 2007; Fadda et al., 2010; Figure 1). Notably, specific KIR have been associated with progression of HLA-restricted infectious disease (Bellón, 2019). Description by Fasbender of the induction of NK-activating ligands on hepatocytes after drug exposure, driving NK-mediated cytotoxicity, spurs interest given that T-cells in the blood of SJS/TEN patients overexpress KIR2DL2 and KIR2DL3 (Morel et al., 2010; Fasbender et al., 2020). With yet unreported genetic or functional assessment, studies are warranted to understand the combined influence of these interactions.
Drugs lacking protein reactivity may directly activate T-cells (Schnyder et al., 1997; Zanni et al., 1997; Naisbitt et al., 2003). However, metabolic detoxification pathways form protein-reactive metabolites, also reported to activate drug-specific T-cells (Naisbitt et al., 2001; Sullivan et al., 2015; Figure 1). Metabolism is highly varied due to polymorphic enzymes, with cytochrome P450 (CYP450) enzymes responsible for 90% of drug metabolism (Lynch and Price, 2007) and for which allelic variants are described from poor to ultrarapid metabolisers (Zanger and Schwab, 2013). While metabolic activity of skin is considered limited (Sharma et al., 2019), keratinocytes show capacity to metabolize and present drug-derived antigens (Reilly et al., 2000; Roychowdhury and Svensson, 2005). Several studies now investigate metabolic variants associated with DHR, most notably for phenytoin, predominantly oxidized to an inactive metabolite by CYP2C9 with minor contribution by CYP2C19. Genetic analyses show that CYP2C9∗2 and CYP2C9∗3 low function variants extend exposure to the immunogenic parent drug (Aynacioglu et al., 1999; Silvado et al., 2018). Specifically, CYP2C9∗3 is associated with SJS/TEN in both Han Chinese (Chung et al., 2014) and Thai (Suvichapanich et al., 2015; Tassaneeyakul et al., 2016). In addition, CYP2C19∗3 is associated with phenytoin-DRESS in Thai (Yampayon et al., 2017). In 2019, Su et al. (2019) published on the utility of combined risk HLA and CYP2C9∗3 genetic testing in Asian populations to prevent phenytoin hypersensitivity. It is now advised that physicians reduce the starting dose by 25% for patients classed as intermediate metabolizers, defined by CYP2C9∗1/∗3 and CYP2C9∗1/∗2 carriage (Caudle et al., 2014). Metabolic variation is also associated with DHR driven by nevirapine, hydroxylated by CYP2B6. Loss of functional alleles CYP2B6∗6 and CYP2B6∗18 are associated with increased susceptibility for nevirapine-SJS/TEN, with the ∗18 variant only observed in patients of African ancestry (Ciccacci et al., 2013; Carr et al., 2014). A handful of other associations are explored by Pirmohamed and were not significant upon multiple-testing correction (Pirmohamed et al., 2000); thus, most data to date implicate only a minor role for metabolic variation in DHR.
There are three main aspects to consider for the impact of infectious disease on DHR. The first aspect is the effect of cumulative drug exposure in cohorts where long-term exposures are driven by repeat infection like antibiotic hypersensitivity in patients with cystic fibrosis (CF). Indeed, CF patients are far more likely to develop an allergy to beta-lactams than patients without (Burrows et al., 2007; Wright et al., 2018); thus, it is possible that repeat high dosing and antigen accumulation contributes to risk. Second is the potential for disease-associated immune dysregulation to heighten allergic susceptibility. An example is the reduced DHR incidence in HIV patients following initiation of successful highly active antiretroviral therapy, which controls viral progression, preventing deterioration of immune function (Coopman et al., 1993; Li et al., 1998). Similarly, studies show that CF patients have dysfunctional antiviral T-cell responses (Hubeau et al., 2004). Indeed, toll-like receptor 4, which mediates inflammatory cytokine expression, is reduced in CF airway cell lines (John et al., 2010; Keiser et al., 2015). Interestingly, cytokine variants predispose to DHRs such as liver injury: IL10-592 AA and IL10-819 TT are associated with docetaxel-induced liver injury, and polymorphism-380G/A in TNF−α is associated with hepatitis induced by antituberculosis drugs (Kim et al., 2011a; Liang et al., 2013; Figure 1). Evidence suggests that drug antigens may mount response in tissue through pre-existing antiviral T-cells in a heterologous immunity model (Descamps et al., 2003; Mitani et al., 2005). Functional evidence is based on work by Lucas who showed that all drug-naive HLA-B∗57:01+ individuals have T-cells responsive to abacavir (Lucas et al., 2015; Gibson et al., 2017). Such reactive promiscuity across all healthy donors implicates cross-reactivity with common broad-exposure pathogens (Smith et al., 2016).
It is now well established that epigenetic modifications to open or close the transcriptional template of genes impacts immunological processes (North and Ellis, 2011; Moggs et al., 2012). Epigenetic influence is environmental with documented effects from diet, viral exposures, and pollution driving distinguishable differences in immune status; thus, it may drive not only inter-individual but also intra-individual risk over time, proposing dynamic susceptibility. Indeed, Nadeau describes hypermethylation of the FOXP3 locus affecting Treg function and asthma severity in patients who live in areas with higher air pollution (Nadeau et al., 2010). Evidence now suggests that epigenetic effects may be multi-generational, with lead exposure and subsequent DNA methylation of fetal germ cells in grandparents traced through to grandchildren (Sen et al., 2015). While likely, epigenetic influence has yet to be directly inferred upon susceptibility to DHR, but there is some initial evidence. In 2018, Cheng published that risk of allopurinol-induced SCAR was attributed to variants of HCP5, PSORS1C1, TSHZ2, and NOTCH4. Although distinct polymorphisms and thus genetic variants, intriguingly NOTCH4 and TSHZ2, were included as genes that presented as highly differentially methylated, a form of epigenetic regulation (Cheng et al., 2017). Furthermore, Monroy-Arreola demonstrated upregulation of microRNA-21, -18, and -155 in drug-specific CD4+ T-cells from hypersensitive patients (Monroy-Arreola et al., 2018). While microRNA may regulate post-transcriptional gene expression, others bind to control regulators of epigenetic modification including DNA methyltransferases (Sato et al., 2011).
Immune checkpoints regulate T-cell activation to prevent uncontrolled activation. This complex process is the summation of varied co-stimulatory and opposingly co-inhibitory pathways (Figure 1). Intriguingly, polymorphic variants of checkpoints are linked to numerous autoimmune diseases including rheumatoid arthritis (Kong et al., 2005), multiple sclerosis (Kroner et al., 2005), and ankylosing spondylitis (Kantarci et al., 2003). While allelic influence is yet to be translated to risk for DHR, mechanistic studies have demonstrated the impact of blocking programmed death-1 (PD-1) or cytotoxic lymphocyte antigen-4 (CTLA4) axes to enhance naive T-cell priming to drug antigens (Gibson et al., 2014, 2017). Checkpoint inhibition is now widely adopted in cancer immunotherapy to re-invigorate anti-tumor T-cell responses, but dysregulation is not antigen-specific and immune-mediated ADR are common (Naidoo et al., 2015; Saw et al., 2017; Lomax et al., 2019). While reactions are varied and typically reported as enhanced immunogenicity to self (Mangan et al., 2020), emerging small cohort studies describe a high incidence of DHR in immune checkpoint inhibitor-treated patients (Imafuku et al., 2017; Ford et al., 2018). These studies remain only clinical observations and distinct checkpoint alleles have not been identified in genome-wide association studies; however, given the influence of multiple, counteracting co-signaling pathways, it may be that single variants have a low individual effect for which the previous studies have been underpowered. Further study is now warranted to define association with a greater risk of drug hypersensitivity reactions.
Given a lack of a single HLA allele to provide complete PPV, other risk factors must further restrict response and recent advances have detailed (i) application of single-cell sequencing to define the HLA-restricted dominant TCR driving early response in tissue and (ii) the impact of ERAP variants to skew immunodominant peptide presentation. Intriguingly, other proposed risk factors such as checkpoint receptors span genetic and epigenetic risk, with expression subject to environmental or therapeutic pressures, implicating highly dynamic risk. Strategies are now needed to identify risk alleles in minority populations where large clinical cohorts are impossible to obtain. The availability of multi-omic approaches offers opportunity to merge high-resolution genotyping with single-cell phenotyping to tease out more complex risk signatures that may also enable cost-effective patient screening.
YL, PD, RH, and AP contributed writing toward individual sections of the manuscript, led and majority authored by YL. AG and EP provided expert review, direction, and guidance. All authors contributed to the article and approved the submitted version.
EP reports grants from the National Institutes of Health (P50GM115305, R01HG010863, R01AI152183, R21AI139021, and U01AI154659) and from the National Health and Medical Research Council of Australia. EP was Drug Allergy Section Editor and receives royalties from Uptodate and consulting fees from Biocryst, Janssen and Vertex. She is co-director of IIID Pty Ltd. that holds a patent for HLA-B∗57:01 testing for abacavir hypersensitivity, and she holds a patent for Detection of Human Leukocyte Antigen-A∗32:01 in connection with Diagnosing Drug Reaction with Eosinophilia and Systemic Symptoms without any financial remuneration and not directly related to the submitted work. Funders played no role in any aspect of this Review.
The reviewer AC declared a past co-authorship with the authors AG and EP to the handling editor.
EP was Drug Allergy Section Editor and receives royalties from Uptodate and consulting fees from Biocryst, Janssen and Vertex. She is co-director of IIID Pty Ltd that holds a patent for HLA-B*57:01 testing for abacavir hypersensitivity, and she holds a patent for Detection of Human Leukocyte Antigen-A*32:01 in connection with Diagnosing Drug Reaction with Eosinophilia and Systemic Symptoms without any financial remuneration and not directly related to the submitted work. Funders played no role in any aspect of this Review.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Aynacioglu, A. S., Brockmöller, J., Bauer, S., Sachse, C., Güzelbey, P., Ongen, Z., et al. (1999). Frequency of cytochrome P450 CYP2C9 variants in a Turkish population and functional relevance for phenytoin. Br. J. Clin. Pharmacol. 48, 409–415. doi: 10.1046/j.1365-2125.1999.00012.x
Balas, A., Ramírez, E., Trigo, E., Cabañas, R., Fiandor, A., Arsuaga, M., et al. (2020). HLA-A* 68,-A* 11: 01, and-A* 29: 02 alleles are strongly associated with benznidazole-induced maculopapular exanthema (MPE)/DRESS. J. Allergy Clin. Immunol. Pract. 8, 3198–3200.e3.
Bellón, T. (2019). Mechanisms of severe cutaneous adverse reactions: recent advances. Drug Saf. 42, 973–992. doi: 10.1007/s40264-019-00825-2
Biasin, M., Sironi, M., Saulle, I., De Luca, M., La Rosa, F., Cagliani, R., et al. (2013). Endoplasmic reticulum aminopeptidase 2 haplotypes play a role in modulating susceptibility to HIV infection. AIDS 27, 1697–1706. doi: 10.1097/qad.0b013e3283601cee
Bluestein, S., Yu, R., Stone, C., and Phillips, E. (2021). A review of drug reaction with eosinophilia and systemic symptoms in the FDA adverse event reporting system (FAERS). J. Allergy Clin. Immunol. 147:AB12. doi: 10.1016/j.jaci.2020.12.085
Boyington, J. C., and Sun, P. D. (2002). A structural perspective on MHC class I recognition by killer cell immunoglobulin-like receptors. Mol. Immunol. 38, 1007–1021. doi: 10.1016/s0161-5890(02)00030-5
Burrows, J. A., Nissen, L. M., Kirkpatrick, C. M., and Bell, S. C. (2007). Beta-lactam allergy in adults with cystic fibrosis. J. Cystic Fibrosis 6, 297–303. doi: 10.1016/j.jcf.2006.11.001
Capule, F., Tragulpiankit, P., Mahasirimongkol, S., Jittikoon, J., Wichukchinda, N., Theresa Alentajan-Aleta, L., et al. (2020). Association of carbamazepine-induced Stevens-Johnson syndrome/toxic epidermal necrolysis with the HLA-B75 serotype or HLA-B∗15:21 allele in Filipino patients. Pharmacogenomics J. 20, 533–541. doi: 10.1038/s41397-019-0143-8
Carr, D. F., Bourgeois, S., Chaponda, M., Takeshita, L. Y., Morris, A. P., Castro, E. M. C., et al. (2017). Genome-wide association study of nevirapine hypersensitivity in a sub-Saharan African HIV-infected population. J. Antimicrob. Chemother. 72, 1152–1162.
Carr, D. F., Chaponda, M., Cornejo Castro, E. M., Jorgensen, A. L., Khoo, S., Van Oosterhout, J. J., et al. (2014). CYP2B6 c.983T>C polymorphism is associated with nevirapine hypersensitivity in Malawian and Ugandan HIV populations. J. Antimicrob. Chemother. 69, 3329–3334. doi: 10.1093/jac/dku315
Carr, D. F., Chaponda, M., Jorgensen, A. L., Castro, E. C., van Oosterhout, J. J., Khoo, S. H., et al. (2013). Association of human leukocyte antigen alleles and nevirapine hypersensitivity in a Malawian HIV-infected population. Clin. Infect. Dis. 56, 1330–1339. doi: 10.1093/cid/cit021
Caudle, K. E., Rettie, A. E., Whirl-Carrillo, M., Smith, L. H., Mintzer, S., Lee, M. T., et al. (2014). Clinical pharmacogenetics implementation consortium guidelines for CYP2C9 and HLA-B genotypes and phenytoin dosing. Clin. Pharmacol. Ther. 96, 542–548. doi: 10.1038/clpt.2014.159
Chang, C. C., Ng, C. C., Too, C. L., Choon, S. E., Lee, C. K., Chung, W. H., et al. (2017). Association of HLA-B∗15:13 and HLA-B∗15:02 with phenytoin-induced severe cutaneous adverse reactions in a Malay population. Pharmacogenomics J. 17, 170–173. doi: 10.1038/tpj.2016.10
Chang, S. C., Momburg, F., Bhutani, N., and Goldberg, A. L. (2005). The ER aminopeptidase, ERAP1, trims precursors to lengths of MHC class I peptides by a “molecular ruler” mechanism. Proc. Natl. Acad. Sci. U.S.A. 102, 17107–17112. doi: 10.1073/pnas.0500721102
Chantarangsu, S., Mushiroda, T., Mahasirimongkol, S., Kiertiburanakul, S., Sungkanuparph, S., Manosuthi, W., et al. (2009). HLA-B∗ 3505 allele is a strong predictor for nevirapine-induced skin adverse drug reactions in HIV-infected Thai patients. Pharmacogenetics Genomics 19, 139–146. doi: 10.1097/fpc.0b013e32831d0faf
Chen, W. T., Wang, C. W., Lu, C. W., Chen, C. B., Lee, H. E., Hung, S. I., et al. (2018). The function of HLA-B∗13:01 involved in the pathomechanism of dapsone-induced severe cutaneous adverse reactions. J. Invest. Dermatol. 138, 1546–1554. doi: 10.1016/j.jid.2018.02.004
Cheng, L., Sun, B., Xiong, Y., Hu, L., Gao, L., Lv, Q., et al. (2017). The minor allele HCP5 rs3099844 A, PSORS1C1 rs3131003 G are associated with allopurinol−induced severe cutaneous adverse reactions in Han Chinese: a multicentre retrospective case−control clinical study. Br. J. Dermatol. 178, e191–e193.
Chessman, D., Kostenko, L., Lethborg, T., Purcell, A. W., Williamson, N. A., Chen, Z., et al. (2008). Human leukocyte antigen class I-restricted activation of CD8+ T cells provides the immunogenetic basis of a systemic drug hypersensitivity. Immunity 28, 822–832. doi: 10.1016/j.immuni.2008.04.020
Cheung, Y.-K., Cheng, S.-H., Chan, E. J. M., Lo, S. V., Ng, M. H. L., and Kwan, P. (2013). HLA-B alleles associated with severe cutaneous reactions to antiepileptic drugs in Han Chinese. Epilepsia 54, 1307–1314. doi: 10.1111/epi.12217
Chiu, M. L. S., Hu, M., Ng, M. H. L., Yeung, C. K., Chan, J. C. Y., Chang, M. M., et al. (2012). Association between HLA−B58:01 allele and severe cutaneous adverse reactions with allopurinol in Han Chinese in Hong Kong. Br. J. Dermatol. 167, 44–49. doi: 10.1111/j.1365-2133.2012.10894.x
Choudhary, S., McLeod, M., Torchia, D., and Romanelli, P. (2013). Drug reaction with eosinophilia and systemic symptoms (DRESS) syndrome. J. Clin. Aesthet. Dermatol. 6:31.
Chung, W. H., Chang, W. C., Lee, Y. S., Wu, Y. Y., Yang, C. H., Ho, H. C., et al. (2014). Genetic variants associated with phenytoin-related severe cutaneous adverse reactions. JAMA 312, 525–534.
Chung, W. H., and Hung, S. I. (2010). Genetic markers and danger signals in stevens-johnson syndrome and toxic epidermal necrolysis. Allergol. Int. 59, 325–332. doi: 10.2332/allergolint.10-rai-0261
Chung, W.-H., Hung, S.-I., Hong, H.-S., Hsih, M.-S., Yang, L.-C., Ho, H.-C., et al. (2004). A marker for Stevens–Johnson syndrome. Nature 428:486.
Chung, W.-H., Pan, R.-Y., Chu, M.-T., Chin, S.-W., Huang, Y.-L., Wang, W.-C., et al. (2015). Oxypurinol-specific T cells possess preferential TCR clonotypes and express granulysin in allopurinol-induced severe cutaneous adverse reactions. J. Invest. Dermatol. 135, 2237–2248. doi: 10.1038/jid.2015.165
Ciccacci, C., Di Fusco, D., Marazzi, M. C., Zimba, I., Erba, F., Novelli, G., et al. (2013). Association between CYP2B6 polymorphisms and Nevirapine-induced SJS/TEN: a pharmacogenetics study. Eur. J. Clin. Pharmacol. 69, 1909–1916. doi: 10.1007/s00228-013-1549-x
Coopman, S. A., Johnson, R. A., Platt, R., and Stern, R. S. (1993). Cutaneous disease and drug reactions in HIV infection. New Engl. J. Med. 328, 1670–1674. doi: 10.1056/nejm199306103282304
Daly, A. K., Donaldson, P. T., Bhatnagar, P., Shen, Y., Pe’er, I., Floratos, A., et al. (2009). HLA-B5701 genotype is a major determinant of drug-induced liver injury due to flucloxacillin. Nat. Genet. 41, 816–819. doi: 10.1038/ng.379
Descamps, V., Mahe, E., Houhou, N., Abramowitz, L., Rozenberg, F., Ranger−Rogez, S., et al. (2003). Drug−induced hypersensitivity syndrome associated with Epstein–Barr virus infection. Br. J. Dermatol. 148, 1032–1034. doi: 10.1046/j.1365-2133.2003.05330.x
Dormann, H., Muth-Selbach, U., Krebs, S., Criegee-Rieck, M., Tegeder, I., Schneider, H. T., et al. (2000). Incidence and costs of adverse drug reactions during hospitalisation. Drug Saf. 22, 161–168. doi: 10.2165/00002018-200022020-00007
Evans, D. M., Spencer, C. C., Pointon, J. J., Su, Z., Harvey, D., Kochan, G., et al. (2011). Interaction between ERAP1 and HLA-B27 in ankylosing spondylitis implicates peptide handling in the mechanism for HLA-B27 in disease susceptibility. Nat. Genet. 43, 761–767.
Fadda, L., Borhis, G., Ahmed, P., Cheent, K., Pageon, S. V., Cazaly, A., et al. (2010). Peptide antagonism as a mechanism for NK cell activation. Proc. Natl. Acad. Sci. U.S.A. 107, 10160–10165. doi: 10.1073/pnas.0913745107
Fasbender, F., Obholzer, M., Metzler, S., Stöber, R., Hengstler, J. G., and Watzl, C. (2020). Enhanced activation of human NK cells by drug-exposed hepatocytes. Arch. Toxicol. 94, 439–448. doi: 10.1007/s00204-020-02668-8
Ferrell, P. B. Jr., and McLeod, H. L. (2008). Carbamazepine, HLA-B∗1502 and risk of Stevens-Johnson syndrome and toxic epidermal necrolysis: US FDA recommendations. Pharmacogenomics 9, 1543–1546. doi: 10.2217/14622416.9.10.1543
Fontana, R. J., Cirulli, E. T., Gu, J., Kleiner, D., Ostrov, D., Phillips, E., et al. (2018). The role of HLA-A∗33:01 in patients with cholestatic hepatitis attributed to terbinafine. J. Hepatol. 69, 1317–1325. doi: 10.1016/j.jhep.2018.08.004
Ford, M., Sahbudin, I., Filer, A., Steven, N., and Fisher, B. A. (2018). High proportion of drug hypersensitivity reactions to sulfasalazine following its use in anti-PD-1-associated inflammatory arthritis. Rheumatology 57, 2244–2246. doi: 10.1093/rheumatology/key234
Fruci, D., Romania, P., D’Alicandro, V., and Locatelli, F. (2014). Endoplasmic reticulum aminopeptidase 1 function and its pathogenic role in regulating innate and adaptive immunity in cancer and major histocompatibility complex class I−associated autoimmune diseases. Tissue Antigens 84, 177–186. doi: 10.1111/tan.12410
Frymoyer, A., Guglielmo, B. J., and Hersh, A. L. (2013). Desired vancomycin trough serum concentration for treating invasive methicillin-resistant Staphylococcal infections. Pediatr. Infect. Dis. J. 32, 1077–1079. doi: 10.1097/inf.0b013e318299f75c
Gao, S., Gui, X.-E., Liang, K., Liu, Z., Hu, J., and Dong, B. (2012). HLA-dependent hypersensitivity reaction to nevirapine in Chinese Han HIV-infected patients. AIDS Res. Hum. Retroviruses 28, 540–543. doi: 10.1089/aid.2011.0107
Gatanaga, H., Yazaki, H., Tanuma, J., Honda, M., Genka, I., Teruya, K., et al. (2007). HLA-Cw8 primarily associated with hypersensitivity to nevirapine. AIDS 21, 264–265. doi: 10.1097/qad.0b013e32801199d9
Genin, E., Chen, D., Hung, S., Sekula, P., Schumacher, M., Chang, P., et al. (2014). HLA-A∗ 31: 01 and different types of carbamazepine-induced severe cutaneous adverse reactions: an international study and meta-analysis. Pharmacogenomics J. 14, 281–288. doi: 10.1038/tpj.2013.40
Génin, E., Schumacher, M., Roujeau, J.-C., Naldi, L., Liss, Y., Kazma, R., et al. (2011). Genome-wide association study of Stevens-Johnson syndrome and toxic epidermal necrolysis in Europe. Orphanet J. Rare Dis. 6:52.
Giannopoulou, E., Katsila, T., Mitropoulou, C., Tsermpini, E. E., and Patrinos, G. P. (2019). Integrating next-generation sequencing in the clinical pharmacogenomics workflow. Front. Pharmacol. 10:384. doi: 10.3389/fphar.2019.00384
Gibson, A., Faulkner, L., Lichtenfels, M., Ogese, M., Al-Attar, Z., Alfirevic, A., et al. (2017). The effect of inhibitory signals on the priming of drug hapten-specific T cells that express distinct Vβ receptors. J. Immunol. 199, 1223–1237. doi: 10.4049/jimmunol.1602029
Gibson, A., Ogese, M., Sullivan, A., Wang, E., Saide, K., Whitaker, P., et al. (2014). Negative regulation by PD-L1 during drug-specific priming of IL-22-secreting T cells and the influence of PD-1 on effector T cell function. J. Immunol. 192, 2611–2621. doi: 10.4049/jimmunol.1302720
Gonçalo, M., Coutinho, I., Teixeira, V., Gameiro, A. R., Brites, M. M., Nunes, R., et al. (2013). HLA-B∗58:01 is a risk factor for allopurinol-induced DRESS and Stevens-Johnson syndrome/toxic epidermal necrolysis in a Portuguese population. Br. J. Dermatol. 169, 660–665. doi: 10.1111/bjd.12389
Guerini, F. R., Cagliani, R., Forni, D., Agliardi, C., Caputo, D., Cassinotti, A., et al. (2012). A functional variant in ERAP1 predisposes to multiple sclerosis. PLoS One 7:e29931. doi: 10.1371/journal.pone.0029931
Heap, G. A., Weedon, M. N., Bewshea, C. M., Singh, A., Chen, M., Satchwell, J. B., et al. (2014). HLA-DQA1–HLA-DRB1 variants confer susceptibility to pancreatitis induced by thiopurine immunosuppressants. Nat. Genet. 46, 1131–1134.
Her, Y., Kil, M. S., Park, J. H., Kim, C. W., and Kim, S. S. (2011). Stevens–Johnson syndrome induced by acetazolamide. J. Dermatol. 38, 272–275. doi: 10.1111/j.1346-8138.2010.00921.x
Hirata, K., Takagi, H., Yamamoto, M., Matsumoto, T., Nishiya, T., Mori, K., et al. (2008). Ticlopidine-induced hepatotoxicity is associated with specific human leukocyte antigen genomic subtypes in Japanese patients: a preliminary case–control study. Pharmacogenomics J. 8, 29–33. doi: 10.1038/sj.tpj.6500442
Hoofnagle, J. H., Bonkovsky, H. L., Phillips, E. J., Li, Y. J., Ahmad, J., Barnhart, H., et al. (2020). HLA−B35:01 and green tea induced liver injury. Hepatology doi: 10.1002/hep.31538 [Epub ahead of print]
Hubeau, C., Le Naour, R., Abély, M., Hinnrasky, J., Guenounou, M., Gaillard, D., et al. (2004). Dysregulation of IL-2 and IL-8 production in circulating T lymphocytes from young cystic fibrosis patients. Clin. Exp. Immunol. 135, 528–534. doi: 10.1111/j.1365-2249.2003.02385.x
Hung, S.-I., Chung, W.-H., Liou, L.-B., Chu, C.-C., Lin, M., Huang, H.-P., et al. (2005). HLA-B∗ 5801 allele as a genetic marker for severe cutaneous adverse reactions caused by allopurinol. Proc. Natl. Acad. Sci. U.S.A. 102, 4134–4139. doi: 10.1073/pnas.0409500102
Hung, S. I., Chung, W. H., Liu, Z. S., Chen, C. H., Hsih, M. S., Hui, R. C., et al. (2010). Common risk allele in aromatic antiepileptic-drug induced Stevens-Johnson syndrome and toxic epidermal necrolysis in Han Chinese. Pharmacogenomics 11, 349–356. doi: 10.2217/pgs.09.162
Illing, P. T., Vivian, J. P., Dudek, N. L., Kostenko, L., Chen, Z., Bharadwaj, M., et al. (2012). Immune self-reactivity triggered by drug-modified HLA-peptide repertoire. Nature 486, 554–558. doi: 10.1038/nature11147
Imafuku, K., Yoshino, K., Ymaguchi, K., Tsuboi, S., Ohara, K., and Hata, H. (2017). Nivolumab therapy before vemurafenib administration induces a severe skin rash. J. Eur. Acad. Dermatol. Venereol. 31, e169–e171.
Jarjour, S., Barrette, M., Normand, V., Rouleau, J. L., Dubé, M. P., and de Denus, S. (2015). Genetic markers associated with cutaneous adverse drug reactions to allopurinol: a systematic review. Pharmacogenomics 16, 755–767. doi: 10.2217/pgs.15.21
John, G., Yildirim, A. O., Rubin, B. K., Gruenert, D. C., and Henke, M. O. (2010). TLR-4–mediated innate immunity is reduced in cystic fibrosis airway cells. Am. J. Respir. Cell Mol. Biol. 42, 424–431. doi: 10.1165/rcmb.2008-0408oc
Kang, H.-R., Jee, Y. K., Kim, Y.-S., Lee, C. H., Jung, J.-W., Kim, S. H., et al. (2011). Positive and negative associations of HLA class I alleles with allopurinol-induced SCARs in Koreans. Pharmacogenet. Genomics 21, 303–307. doi: 10.1097/fpc.0b013e32834282b8
Kaniwa, N., Saito, Y., Aihara, M., Matsunaga, K., Tohkin, M., Kurose, K., et al. (2008). HLA-B locus in Japanese patients with anti-epileptics and allopurinol-related Stevens-Johnson syndrome and toxic epidermal necrolysis. Pharmacogenomics 9, 1617–1622. doi: 10.2217/14622416.9.11.1617
Kaniwa, N., Sugiyama, E., Saito, Y., Kurose, K., Maekawa, K., Hasegawa, R., et al. (2013). Specific HLA types are associated with antiepileptic drug-induced Stevens-Johnson syndrome and toxic epidermal necrolysis in Japanese subjects. Pharmacogenomics 14, 1821–1831. doi: 10.2217/pgs.13.180
Kantarci, O. H., Hebrink, D. D., Achenbach, S. J., Atkinson, E. J., Waliszewska, A., Buckle, G., et al. (2003). CTLA4 is associated with susceptibility to multiple sclerosis. J. Neuroimmunol. 134, 133–141. doi: 10.1016/s0165-5728(02)00395-8
Kardaun, S. H. (2019). “Drug reaction with eosinophilia and systemic symptoms (DRESS),” in Advances in Diagnosis and Management of Cutaneous Adverse Drug Reactions, eds N. H. Shear and R. P. Dodiuk-Gad (Cham: Springer), 87–104.
Karnes, J. H., Miller, M. A., White, K. D., Konvinse, K. C., Pavlos, R. K., Redwood, A. J., et al. (2019). Applications of immunopharmacogenomics: predicting, preventing, and understanding immune-mediated adverse drug reactions. Annu. Rev. Pharmacol. Toxicol. 59, 463–486. doi: 10.1146/annurev-pharmtox-010818-021818
Kazeem, G. R., Cox, C., Aponte, J., Messenheimer, J., Brazell, C., Nelsen, A. C., et al. (2009). High-resolution HLA genotyping and severe cutaneous adverse reactions in lamotrigine-treated patients. Pharmacogenet. Genomics 19, 661–665. doi: 10.1097/fpc.0b013e32832c347d
Ke, C. H., Chung, W. H., Wen, Y. H., Huang, Y. B., Chuang, H. Y., Tain, Y. L., et al. (2017). Cost-effectiveness analysis for genotyping before allopurinol treatment to prevent severe cutaneous adverse drug reactions. J. Rheumatol. 44, 835–843. doi: 10.3899/jrheum.151476
Keiser, N. W., Birket, S. E., Evans, I. A., Tyler, S. R., Crooke, A. K., Sun, X., et al. (2015). Defective innate immunity and hyperinflammation in newborn cystic fibrosis transmembrane conductance regulator–knockout ferret lungs. Am. J. Respir. Cell Mol. Biol. 52, 683–694. doi: 10.1165/rcmb.2014-0250oc
Kemming, J., Reeves, E., Nitschke, K., Widmeier, V., Emmerich, F., Hermle, T., et al. (2019). ERAP1 allotypes shape the epitope repertoire of virus-specific CD8+ T cell responses in acute hepatitis C virus infection. J. Hepatol. 70, 1072–1081. doi: 10.1016/j.jhep.2019.01.034
Kim, B. K., Jung, J. W., Kim, T. B., Chang, Y. S., Park, H. S., Moon, J., et al. (2017). HLA-A∗31:01 and lamotrigine-induced severe cutaneous adverse drug reactions in a Korean population. Ann. Allergy Asthma Immunol. 118, 629–630. doi: 10.1016/j.anai.2017.02.011
Kim, S.-H., Kim, S.-H., Yoon, H., Shin, D., Park, S.-S., Kim, Y.-S., et al. (2011a). TNF−α genetic polymorphism -308G/A and antituberculosis drug−induced hepatitis. Liver Int. 32, 809–814. doi: 10.1111/j.1478-3231.2011.02697.x
Kim, S.-H., Lee, K. W., Song, W.-J., Kim, S.-H., Jee, Y.-K., Lee, S.-M., et al. (2011b). Carbamazepine-induced severe cutaneous adverse reactions and HLA genotypes in Koreans. Epilepsy Res. 97, 190–197. doi: 10.1016/j.eplepsyres.2011.08.010
Kong, E. K., Prokunina-Olsson, L., Wong, W. H., Lau, C. S., Chan, T. M., Alarcón-Riquelme, M., et al. (2005). A new haplotype of PDCD1 is associated with rheumatoid arthritis in Hong Kong Chinese. Arthritis Rheum. 52, 1058–1062. doi: 10.1002/art.20966
Konvinse, K. C., Trubiano, J. A., Pavlos, R., James, I., Shaffer, C. M., Bejan, C. A., et al. (2019). HLA-A∗32:01 is strongly associated with vancomycin-induced drug reaction with eosinophilia and systemic symptoms. J. Allergy Clin. Immunol. 144, 183–192.
Koomdee, N., Pratoomwun, J., Jantararoungtong, T., Theeramoke, V., Tassaneeyakul, W., Klaewsongkram, J., et al. (2017). Association of HLA-A and HLA-B alleles with lamotrigine-induced cutaneous adverse drug reactions in the Thai population. Front. Pharmacol. 8:879. doi: 10.3389/fphar.2017.00879
Kroner, A., Mehling, M., Hemmer, B., Rieckmann, P., Toyka, K. V., Mäurer, M., et al. (2005). A PD-1 polymorphism is associated with disease progression in multiple sclerosis. Ann. Neurol. 58, 50–57. doi: 10.1002/ana.20514
LaHaye, S., Corsmeier, D., Basu, M., Bowman, J. L., Fitzgerald-Butt, S., Zender, G., et al. (2016). Utilization of whole exome sequencing to identify causative mutations in familial congenital heart disease. Circulation 9, 320–329. doi: 10.1161/circgenetics.115.001324
Langley, A., Worley, B., Pardo, J. P., Beecker, J., Ramsay, T., Saavedra, A., et al. (2018). Systemic interventions for treatment of Stevens−Johnson syndrome (SJS), toxic epidermal necrolysis (TEN), and SJS/TEN overlap syndrome. Cochrane Database Syst. Rev. 2018:CD013130.
Lee, H. Y., Shen, M. X., Lim, Y. L., Tay, Y. K., Chan, M. M. F., Pang, S. M., et al. (2016). Increased risk of strontium ranelate-related SJS/TEN is associated with HLA. Osteoporosis Int. 27, 2577–2583. doi: 10.1007/s00198-016-3568-9
Leise, M. D., Poterucha, J. J., and Talwalkar, J. A. (2014). Drug-induced Liver Injury. Mayo Clinic Proceedings. Amsterdam: Elsevier.
LeMaoult, J., Zafaranloo, K., Le Danff, C., and Carosella, E. D. (2005). HLA−G up−regulates ILT2, ILT3, ILT4, and KIR2DL4 in antigen presenting cells, NK cells, and T cells. FASEB J. 19, 1–23. doi: 10.1096/fj.04-1617fje
Li, C., Rao, T., Chen, X., Zou, Z., Wei, A., Tang, J., et al. (2019). HLA-B∗35:01 allele is a potential biomarker for predicting polygonum multiflorum-induced liver injury in humans. Hepatology 70, 346–357.
Li, L.-J., Hu, F.-Y., Wu, X.-T., An, D.-M., Yan, B., and Zhou, D. (2013). Predictive markers for carbamazepine and lamotrigine-induced maculopapular exanthema in Han Chinese. Epilepsy Res. 106, 296–300. doi: 10.1016/j.eplepsyres.2013.05.004
Li, T. S., Tubiana, R., Katlama, C., Calvez, V., Mohand, H. A., and Autran, B. (1998). Long-lasting recovery in CD4 T-cell function and viral-load reduction after highly active antiretroviral therapy in advanced HIV-1 disease. Lancet 351, 1682–1686. doi: 10.1016/s0140-6736(97)10291-4
Li, Y.-J., Phillips, E., Dellinger, A., Nicoletti, P., Schutte, R., Li, D., et al. (2020). HLA-B14:01 and HLA-B35:01 are associated with trimethoprim-sulfamethoxazole induced liver injury. Hepatology 73, 268–281.
Liang, X., Zhang, J., Zhu, Y., Lu, Y., Zhou, X., Wang, Z., et al. (2013). Specific genetic polymorphisms of IL10-592 AA and IL10-819 TT genotypes lead to the key role for inducing docetaxel-induced liver injury in breast cancer patients. Clin. Transl. Oncol. 15, 331–334. doi: 10.1007/s12094-012-0936-6
Littera, R., Carcassi, C., Masala, A., Piano, P., Serra, P., Ortu, F., et al. (2006). HLA-dependent hypersensitivity to nevirapine in Sardinian HIV patients. AIDS 20, 1621–1626. doi: 10.1097/01.aids.0000238408.82947.09
Locharernkul, C., Loplumlert, J., Limotai, C., Korkij, W., Desudchit, T., Tongkobpetch, S., et al. (2008). Carbamazepine and phenytoin induced Stevens−Johnson syndrome is associated with HLA−B∗ 1502 allele in Thai population. Epilepsia 49, 2087–2091. doi: 10.1111/j.1528-1167.2008.01719.x
Lomax, A. J., McQuillan, P. I. A., Hall, A., and McArthur, G. A. (2019). Acute toxic epidermal necrolysis reaction post single dose pembrolizumab with preceding cephalosporin exposure: successful rechallenge with anti-PD-1 therapy. Intern. Med. J. 49, 1051–1053. doi: 10.1111/imj.14388
Lonjou, C., Borot, N., Sekula, P., Ledger, N., Thomas, L., Halevy, S., et al. (2008). A European study of HLA-B in Stevens–Johnson syndrome and toxic epidermal necrolysis related to five high-risk drugs. Pharmacogenet. Genomics 18, 99–107. doi: 10.1097/fpc.0b013e3282f3ef9c
Lucas, A., Lucas, M., Strhyn, A., Keane, N. M., McKinnon, E., Pavlos, R., et al. (2015). Abacavir-reactive memory T cells are present in drug naïve individuals. PLoS One 10:e0117160. doi: 10.1371/journal.pone.0117160
Lucena, M. I., Molokhia, M., Shen, Y., Urban, T. J., Aithal, G. P., Andrade, R. J., et al. (2011). Susceptibility to Amoxicillin-Clavulanate-Induced Liver Injury Is Influenced by Multiple HLA Class I and II Alleles. Gastroenterology 141, 338–347. doi: 10.1053/j.gastro.2011.04.001
Lv, Y.-D., Min, F.-L., Liao, W.-P., He, N., Zeng, T., Ma, D.-H., et al. (2013). The association between oxcarbazepine-induced maculopapular eruption and HLA-B alleles in a Northern Han Chinese population. BMC Neurol. 13:75. doi: 10.1186/1471-2377-13-75
Lynch, T., and Price, A. (2007). The effect of cytochrome P450 metabolism on drug response, interactions, and adverse effects. Am. Fam. Phys. 76, 391–396.
Mallal, S., Nolan, D., Witt, C., Masel, G., Martin, A., Moore, C., et al. (2002). Association between presence of HLA-B∗ 5701, HLA-DR7, and HLA-DQ3 and hypersensitivity to HIV-1 reverse-transcriptase inhibitor abacavir. Lancet 359, 727–732. doi: 10.1016/s0140-6736(02)07873-x
Mallal, S., Phillips, E., Carosi, G., Molina, J.-M., Workman, C., Tomažiè, J., et al. (2008). HLA-B∗ 5701 screening for hypersensitivity to abacavir. New Engl. J. Med. 358, 568–579.
Mandelboim, O., Wilson, S. B., Valés-Gómez, M., Reyburn, H. T., and Strominger, J. L. (1997). Self and viral peptides can initiate lysis by autologous natural killer cells. Proc. Natl. Acad. Sci. U.S.A. 94, 4604–4609. doi: 10.1073/pnas.94.9.4604
Mangan, B. L., McAlister, R. K., Balko, J. M., Johnson, D. B., Moslehi, J. J., Gibson, A., et al. (2020). Evolving insights into the mechanisms of toxicity associated with immune checkpoint inhibitor therapy. Br. J. Clin. Pharmacol. 86, 1778–1789. doi: 10.1111/bcp.14433
Martin, A. M., Nolan, D., James, I., Cameron, P., Keller, J., Moore, C., et al. (2005). Predisposition to nevirapine hypersensitivity associated with HLA-DRB1∗0101 and abrogated by low CD4 T-cell counts. AIDS 19, 97–99. doi: 10.1097/00002030-200501030-00014
McCormack, M., Alfirevic, A., Bourgeois, S., Farrell, J. J., Kasperavièiûtë, D., Carrington, M., et al. (2011). HLA-A∗3101 and carbamazepine-induced hypersensitivity reactions in Europeans. N. Engl. J. Med. 364, 1134–1143.
Mehta, T. Y., Prajapati, L. M., Mittal, B., Joshi, C. G., Sheth, J. J., Patel, D. B., et al. (2009). Association of HLA-B∗ 1502 allele and carbamazepine-induced Stevens-Johnson syndrome among Indians. Indian J. Dermatol. Venereol. Leprol. 75:579.
Mimori, T., Yasuda, J., Kuroki, Y., Shibata, T. F., Katsuoka, F., Saito, S., et al. (2019). Construction of full-length Japanese reference panel of class I HLA genes with single-molecule, real-time sequencing. Pharmacogenomics J. 19, 136–146. doi: 10.1038/s41397-017-0010-4
Mingari, M. C., Ponte, M., Cantoni, C., Vitale, C., Schiavetti, F., Bertone, S., et al. (1997). HLA-class I-specific inhibitory receptors in human cytolytic T lymphocytes: molecular characterization, distribution in lymphoid tissues and co-expression by individual T cells. Int. Immunol. 9, 485–491. doi: 10.1093/intimm/9.4.485
Mitani, N., Aihara, M., Yamakawa, Y., Yamada, M., Itoh, N., Mizuki, N., et al. (2005). Drug−induced hypersensitivity syndrome due to cyanamide associated with multiple reactivation of human herpesviruses. J. Med. Virol. 75, 430–434. doi: 10.1002/jmv.20295
Miya, R., Malpani, A. K., Keri, S., and Panagaon, R. (2019). Drug induced Steven-Johnson syndrome (SJS). Indian J. Pharm. Pract. 12:133. doi: 10.5530/ijopp.12.2.28
Mockenhaupt, M., Wang, C. W., Hung, S. I., Sekula, P., Schmidt, A. H., Pan, R. Y., et al. (2019). HLA−B∗ 57: 01 confers genetic susceptibility to carbamazepine−induced SJS/TEN in Europeans. Allergy 74, 2227–2230. doi: 10.1111/all.13821
Moggs, J. G., Terranova, R., Kammüller, M. E., Chibout, S.-D., Chapman, V., Dearman, R. J., et al. (2012). Regulation of allergic responses to chemicals and drugs: possible roles of epigenetic mechanisms. Toxicol. Sci. 130, 60–69. doi: 10.1093/toxsci/kfs207
Monroy-Arreola, A., Durán-Figueroa, N. V., Méndez-Flores, S., Domínguez-Cherit, J., Watkinson, J., Badillo-Corona, J. A., et al. (2018). Up-regulation of T-cell activation microRNAs in drug-specific CD4(+) T-cells from hypersensitive patients. Chem. Res. Toxicol. 31, 454–461. doi: 10.1021/acs.chemrestox.7b00330
Monshi, M. M., Faulkner, L., Gibson, A., Jenkins, R. E., Farrell, J., Earnshaw, C. J., et al. (2013). Human leukocyte antigen (HLA)−B∗ 57: 01−restricted activation of drug−specific T cells provides the immunological basis for flucloxacillin−induced liver injury. Hepatology 57, 727–739. doi: 10.1002/hep.26077
Moon, J., Park, H. K., Chu, K., Sunwoo, J. S., Byun, J. I., Lim, J. A., et al. (2015). The HLA-A∗2402/Cw∗0102 haplotype is associated with lamotrigine-induced maculopapular eruption in the Korean population. Epilepsia 56, e161–e167.
Moore, J. A., Meakin, M., Earl, M. H., Kummer, T. M., McAleer, J. P., and Long, T. E. (2020). Effects of caspofungin, tolcapone, and other FDA-approved medications on MRSA susceptibility to vancomycin. J. Glob. Antimicrob. Resist. 22, 283–289. doi: 10.1016/j.jgar.2020.03.014
Morel, E., Escamochero, S., Cabañas, R., Díaz, R., Fiandor, A., and Bellón, T. (2010). CD94/NKG2C is a killer effector molecule in patients with Stevens-Johnson syndrome and toxic epidermal necrolysis. J. Allergy Clin. Immunol. 125, 703–710.e708. doi: 10.1016/j.jaci.2009.10.030
Nadeau, K., McDonald-Hyman, C., Noth, E. M., Pratt, B., Hammond, S. K., Balmes, J., et al. (2010). Ambient air pollution impairs regulatory T-cell function in asthma. J. Allergy Clin. Immunol. 126, 845–852.e10.
Naidoo, J., Page, D. B., Li, B. T., Connell, L. C., Schindler, K., Lacouture, M. E., et al. (2015). Toxicities of the anti-PD-1 and anti-PD-L1 immune checkpoint antibodies. Ann. Oncol. 26, 2375–2391. doi: 10.1093/annonc/mdv383
Naisbitt, D., Britschgi, M., Wong, G., Farrell, J., Depta, J., Chadwick, D., et al. (2003). Hypersensitivity reactions to carbamazepine: characterization of the specificity, phenotype, and cytokine profile of drug-specific T cell clones. Mol. Pharmacol. 63, 732–741. doi: 10.1124/mol.63.3.732
Naisbitt, D. J., Williams, D. P., Pirmohamed, M., Kitteringham, N. R., and Park, B. K. (2001). Reactive metabolites and their role in drug reactions. Curr. Opin. Allergy Clin. Immunol. 1, 317–325. doi: 10.1097/01.all.0000011033.64625.5a
Nakamura, R., Ozeki, T., Hirayama, N., Sekine, A., Yamashita, T., Mashimo, Y., et al. (2020). Association of HLA-A∗11:01 with sulfonamide-related severe cutaneous adverse reactions in Japanese patients. J. Invest. Dermatol. 140, 1659–1662.e6.
Nakatani, K., Ueta, M., Khor, S.-S., Hitomi, Y., Okudaira, Y., Masuya, A., et al. (2019). Identification of HLA-A∗ 0 2: 06: 01 as the primary disease susceptibility HLA allele in cold medicine-related Stevens-Johnson syndrome with severe ocular complications by high-resolution NGS-based HLA typing. Sci. Rep. 9, 1–8.
Nakkam, N., Gibson, A., Mouhtouris, E., Konvinse, K. C., Holmes, N., Chua, K. Y., et al. (2020). Cross-reactivity between vancomycin, teicoplanin, and telavancin in patients with HLA-A*32:01-positive vancomycin-induced DRESS sharing an HLA class II haplotype. J. Allergy Clin. Immunol. 147, 403–405. doi: 10.1016/j.jaci.2020.04.056
Nassif, A., Bensussan, A., Bachot, N., Bagot, M., Boumsell, L., Roujeau, J.-C., et al. (2002). Drug specific cytotoxic T-cells in the skin lesions of a patient with toxic epidermal necrolysis. J. Invest. Dermatol. 118, 728–733. doi: 10.1046/j.1523-1747.2002.01622.x
Nicoletti, P., Barrett, S., McEvoy, L., Daly, A. K., Aithal, G., Lucena, M. I., et al. (2019). Shared genetic risk factors across carbamazepine-induced hypersensitivity reactions. Clin. Pharmacol. Ther. 106, 1028–1036.
Niihara, H., Kakamu, T., Fujita, Y., Kaneko, S., and Morita, E. (2012). HLA−A31 strongly associates with carbamazepine−induced adverse drug reactions but not with carbamazepine−induced lymphocyte proliferation in a Japanese population. J. Dermatol. 39, 594–601. doi: 10.1111/j.1346-8138.2011.01457.x
North, M. L., and Ellis, A. K. (2011). The role of epigenetics in the developmental origins of allergic disease. Ann. Allergy Asthma Immunol. 106, 355–361; quiz62.
Ostrov, D. A., Grant, B. J., Pompeu, Y. A., Sidney, J., Harndahl, M., Southwood, S., et al. (2012). Drug hypersensitivity caused by alteration of the MHC-presented self-peptide repertoire. Proc. Natl. Acad. Sci. U.S.A. 109, 9959–9964. doi: 10.1073/pnas.1207934109
Oussalah, A., Yip, V., Mayorga, C., Blanca, M., Barbaud, A., Nakonechna, A., et al. (2020). Genetic variants associated with T cell-mediated cutaneous adverse drug reactions: a PRISMA-compliant systematic review-An EAACI position paper. Allergy 75, 1069–1098. doi: 10.1111/all.14174
Özkaya-Bayazit, E., and Akar, U. (2001). Fixed drug eruption induced by trimethoprim-sulfamethoxazole: evidence for a link to HLA-A30 B13 Cw6 haplotype. J. Am. Acad. Dermatol. 45, 712–717. doi: 10.1067/mjd.2001.117854
Pan, R.-Y., Chu, M.-T., Wang, C.-W., Lee, Y.-S., Lemonnier, F., Michels, A. W., et al. (2019). Identification of drug-specific public TCR driving severe cutaneous adverse reactions. Nat. Commun. 10, 1–13. doi: 10.1007/978-3-662-58713-3_36-1
Park, B. K., Pirmohamed, M., and Kitteringham, N. R. (1998). Role of drug disposition in drug hypersensitivity: a chemical, molecular, and clinical perspective. Chem. Res. Toxicol. 11, 969–988. doi: 10.1021/tx980058f
Patel, T. K., Barvaliya, M. J., Sharma, D., and Tripathi, C. (2013). A systematic review of the drug-induced Stevens-Johnson syndrome and toxic epidermal necrolysis in Indian population. Indian J. Dermatol. Venereol. Leprol. 79, 389. doi: 10.4103/0378-6323.110749
Paulmann, M., and Mockenhaupt, M. (2015). Severe drug-induced skin reactions: clinical features, diagnosis, etiology, and therapy. J. Dtsch. Dermatol. Ges. 13, 625–645. doi: 10.1111/ddg.12747
Pavlos, R., Deshpande, P., Chopra, A., Leary, S., Strautins, K., Nolan, D., et al. (2020). New genetic predictors for abacavir tolerance in HLA-B∗ 57: 01 positive individuals. Hum. Immunol. 81, 300–304. doi: 10.1016/j.humimm.2020.02.011
Pavlos, R., Mallal, S., Ostrov, D., Buus, S., Metushi, I., Peters, B., et al. (2015). T cell–mediated hypersensitivity reactions to drugs. Annu. Rev. Med. 66, 439–454. doi: 10.1146/annurev-med-050913-022745
Pavlos, R., McKinnon, E. J., Ostrov, D. A., Peters, B., Buus, S., Koelle, D., et al. (2017). Shared peptide binding of HLA Class I and II alleles associate with cutaneous nevirapine hypersensitivity and identify novel risk alleles. Sci. Rep. 7:8653.
Peter, J. G., Lehloenya, R., Dlamini, S., Risma, K., White, K. D., Konvinse, K. C., et al. (2017). Severe delayed cutaneous and systemic reactions to drugs: a global perspective on the science and art of current practice. J. Allergy Clin. Immunol. Pract. 5, 547–563. doi: 10.1016/j.jaip.2017.01.025
Pirmohamed, M., Alfirevic, A., Vilar, J., Stalford, A., Wilkins, E. G., Sim, E., et al. (2000). Association analysis of drug metabolizing enzyme gene polymorphisms in HIV-positive patients with co-trimoxazole hypersensitivity. Pharmacogenetics 10, 705–713. doi: 10.1097/00008571-200011000-00005
Plumpton, C. O., Alfirevic, A., Pirmohamed, M., and Hughes, D. A. (2017). Cost effectiveness analysis of HLA-B∗58:01 genotyping prior to initiation of allopurinol for gout. Rheumatology (Oxford) 56, 1729–1739. doi: 10.1093/rheumatology/kex253
Pouyanne, P., Haramburu, F., Imbs, J. L., and Bégaud, B. (2000). Admissions to hospital caused by adverse drug reactions: cross sectional incidence study. BMJ 320:1036. doi: 10.1136/bmj.320.7241.1036
Redwood, A. J., Pavlos, R. K., White, K. D., and Phillips, E. J. (2018). HLAs: Key regulators of T-cell-mediated drug hypersensitivity. HLA 91, 3–16. doi: 10.1111/tan.13183
Redwood, A. J., Rwandamuriye, F., Chopra, A., Leary, S., Ram, R., McDonnell, W., et al. (2019). Single-cell transcriptomics reveal polyclonal memory T-cell responses in skin with positive abacavir patch test results. J. Allergy Clin. Immunol. 144, 1413–1416.e7.
Reeves, E., and James, E. (2015). The role of endoplasmic reticulum aminopeptidase 1 biology in immune evasion by tumours. J. Vaccines Immunol. 1, 28–35.
Reilly, T. P., Lash, L. H., Doll, M. A., Hein, D. W., Woster, P. M., and Svensson, C. K. (2000). A role for bioactivation and covalent binding within epidermal keratinocytes in sulfonamide-induced cutaneous drug reactions. J. Invest. Dermatol. 114, 1164–1173. doi: 10.1046/j.1523-1747.2000.00985.x
Robins, H. S., Srivastava, S. K., Campregher, P. V., Turtle, C. J., Andriesen, J., Riddell, S. R., et al. (2010). Overlap and effective size of the human CD8+ T cell receptor repertoire. Sci. Transl. Med. 2:47ra64. doi: 10.1126/scitranslmed.3001442
Roujeau, J. C., Huynh, T. N., Bracq, C., Guillaume, J. C., Revuz, J., and Touraine, R. (1987). Genetic susceptibility to toxic epidermal necrolysis. Arch. Dermatol. 123, 1171–1173. doi: 10.1001/archderm.123.9.1171
Roychowdhury, S., and Svensson, C. K. (2005). Mechanisms of drug-induced delayed-type hypersensitivity reactions in the skin. AAPS J. 7, E834–E846.
Rwandamuriye, F. X., Chopra, A., Konvinse, K. C., Choo, L., Trubiano, J. A., Shaffer, C. M., et al. (2019). A rapid allele-specific assay for HLA-A∗ 32: 01 to identify patients at risk for vancomycin-induced Drug Reaction with Eosinophilia and systemic symptoms. J. Mol. Diagn. 21, 782–789. doi: 10.1016/j.jmoldx.2019.04.006
Rybak, M. J., Lomaestro, B. M., Rotscahfer, J. C., Moellering, R. C. Jr., Craig, W. A., Billeter, M., et al. (2009). Vancomycin therapeutic guidelines: a summary of consensus recommendations from the infectious diseases Society of America, the American society of health-system pharmacists, and the society of infectious diseases pharmacists. Clin. Infect. Dis. 49, 325–327. doi: 10.1086/600877
Saag, M., Balu, R., Phillips, E., Brachman, P., Martorell, C., Burman, W., et al. (2008). High sensitivity of human leukocyte antigen-b∗ 5701 as a marker for immunologically confirmed abacavir hypersensitivity in white and black patients. Clin. Infect. Dis. 46, 1111–1118. doi: 10.1086/529382
Sabourirad, S., Mortezaee, R., Mojarad, M., Eslahi, A., Shahrokhi, Y., Kiafar, B., et al. (2020). Investigating the association of Lamotrigine and Phenytoin-induced Stevens-Johnson syndrome/Toxic Epidermal Necrolysis with HLA-B*1502 in Iranian population. Exp. Dermatol. 30, 284–287. doi: 10.1111/exd.14240
Sato, F., Tsuchiya, S., Meltzer, S. J., and Shimizu, K. (2011). MicroRNAs and epigenetics. FEBS J. 278, 1598–1609. doi: 10.1111/j.1742-4658.2011.08089.x
Saulle, I., Ibba, S. V., Vittori, C., Fenizia, C., Piancone, F., Minisci, D., et al. (2019). Endoplasmic reticulum associated aminopeptidase 2 (ERAP2) is released in the secretome of activated MDMs and reduces in vitro HIV-1 infection. Front. Immunol. 10:1648. doi: 10.3389/fimmu.2019.01648
Saw, S., Lee, H. Y., and Ng, Q. S. (2017). Pembrolizumab-induced Stevens-Johnson syndrome in non-melanoma patients. Eur. J. Cancer 81, 237–239. doi: 10.1016/j.ejca.2017.03.026
Schnyder, B., Mauri-Hellweg, D., Zanni, M., Bettens, F., and Pichler, W. J. (1997). Direct, MHC-dependent presentation of the drug sulfamethoxazole to human alphabeta T cell clones. J. Clin. Invest. 100, 136–141. doi: 10.1172/jci119505
Sen, A., Heredia, N., Senut, M. C., Land, S., Hollocher, K., Lu, X., et al. (2015). Multigenerational epigenetic inheritance in humans: DNA methylation changes associated with maternal exposure to lead can be transmitted to the grandchildren. Sci. Rep. 5:14466.
Serwold, T., Gaw, S., and Shastri, N. (2001). ER aminopeptidases generate a unique pool of peptides for MHC class I molecules. Nat. Immunol. 2, 644–651. doi: 10.1038/89800
Sharma, A., Saito, Y., Hung, S.-I., Naisbitt, D., Uetrecht, J., and Bussiere, J. (2019). The skin as a metabolic and immune-competent organ: implications for drug-induced skin rash. J. Immunotoxicol. 16, 1–12. doi: 10.1080/1547691x.2018.1514444
Shi, Y.-W., Min, F.-L., Qin, B., Zou, X., Liu, X.-R., Gao, M.-M., et al. (2012). Association between HLA and Stevens–Johnson Syndrome Induced by Carbamazepine in Southern Han Chinese: genetic markers besides B*1502? Basic Clin. Pharmacol. Toxicol. 111, 58–64.
Silvado, C. E., Terra, V. C., and Twardowschy, C. A. (2018). CYP2C9 polymorphisms in epilepsy: influence on phenytoin treatment. Pharmgenomics Pers. Med. 11, 51–58. doi: 10.2147/pgpm.s108113
Smith, C. J., Quinn, M., and Snyder, C. M. (2016). CMV-specific CD8 T cell differentiation and localization: implications for adoptive therapies. Front. Immunol. 7:352. doi: 10.3389/fimmu.2016.00352
Somogyi, A. A., Barratt, D. T., Phillips, E. J., Moore, K., Ilyas, F., and Gabb, G. M. (2019). High and variable population prevalence of HLA−B∗ 56: 02 in indigenous Australians and relation to phenytoin−associated drug reaction with eosinophilia and systemic symptoms. Br. J. Clin. Pharmacol. 85, 2163–2169. doi: 10.1111/bcp.14025
Sousa-Pinto, B., Pinto-Ramos, J., Correia, C., Gonçalves-Costa, G., Gomes, L., Gil-Mata, S., et al. (2015). Pharmacogenetics of abacavir hypersensitivity: a systematic review and meta-analysis of the association with HLA-B∗57:01. J. Allergy Clin. Immunol. 136, 1092–1094.e3.
Su, S. C., Chen, C. B., Chang, W. C., Wang, C. W., Fan, W. L., Lu, L. Y., et al. (2019). HLA Alleles and CYP2C9∗3 as predictors of phenytoin hypersensitivity in East Asians. Clin. Pharmacol. Ther. 105, 476–485. doi: 10.1002/cpt.1190
Sukasem, C., Chaichan, C., Nakkrut, T., Satapornpong, P., Jaruthamsophon, K., Jantararoungtong, T., et al. (2018). Association between HLA-B alleles and carbamazepine-induced maculopapular exanthema and severe cutaneous reactions in Thai Patients. J. Immunol. Res. 2018, 1–11. doi: 10.1155/2018/2780272
Sukasem, C., Jantararoungtong, T., Kuntawong, P., Puangpetch, A., Koomdee, N., Satapornpong, P., et al. (2016). HLA-B (∗) 58:01 for allopurinol-induced cutaneous adverse drug reactions: implication for clinical interpretation in Thailand. Front. Pharmacol. 7:186. doi: 10.3389/fphar.2016.00186
Sukasem, C., Pratoomwun, J., Satapornpong, P., Klaewsongkram, J., Rerkpattanapipat, T., Rerknimitr, P., et al. (2020). Genetic association of co-trimoxazole-induced severe cutaneous adverse reactions is phenotype-specific: HLA class I genotypes and haplotypes. Clin. Pharmacol. Therap. 108, 1078–1089. doi: 10.1002/cpt.1915
Sullivan, A., Gibson, A., Park, B. K., and Naisbitt, D. J. (2015). Are drug metabolites able to cause T-cell-mediated hypersensitivity reactions? Expert Opin. Drug Metab. Toxicol. 11, 357–368. doi: 10.1517/17425255.2015.992780
Suvichapanich, S., Jittikoon, J., Wichukchinda, N., Kamchaisatian, W., Visudtibhan, A., Benjapopitak, S., et al. (2015). Association analysis of CYP2C9∗3 and phenytoin-induced severe cutaneous adverse reactions (SCARs) in Thai epilepsy children. J. Hum. Genet. 60, 413–417. doi: 10.1038/jhg.2015.47
Tangamornsuksan, W., Chaiyakunapruk, N., Somkrua, R., Lohitnavy, M., and Tassaneeyakul, W. (2013). Relationship between the HLA-B∗1502 Allele and Carbamazepine-induced stevens-johnson syndrome and toxic epidermal necrolysis: a systematic review and meta-analysis. JAMA Dermatol. 149, 1025–1032. doi: 10.1001/jamadermatol.2013.4114
Tangamornsuksan, W., and Lohitnavy, M. (2019). Association between HLA-B∗5901 and methazolamide-induced Stevens-Johnson syndrome/toxic epidermal necrolysis: a systematic review and meta-analysis. Pharmacogenomics J. 19, 286–294. doi: 10.1038/s41397-018-0052-2
Tassaneeyakul, W., Prabmeechai, N., Sukasem, C., Kongpan, T., Konyoung, P., Chumworathayi, P., et al. (2016). Associations between HLA class I and cytochrome P450 2C9 genetic polymorphisms and phenytoin-related severe cutaneous adverse reactions in a Thai population. Pharmacogenet. Genomics 26, 225–234. doi: 10.1097/fpc.0000000000000211
Tempark, T., Satapornpong, P., Rerknimitr, P., Nakkam, N., Saksit, N., Wattanakrai, P., et al. (2017). Dapsone-induced severe cutaneous adverse drug reactions are strongly linked with HLA-B∗13: 01 allele in the Thai population. Pharmacogenet. Genomics 27, 429–437. doi: 10.1097/fpc.0000000000000306
Thananchai, H., Gillespie, G., Martin, M. P., Bashirova, A., Yawata, N., Yawata, M., et al. (2007). Cutting Edge: Allele-specific and peptide-dependent interactions between KIR3DL1 and HLA-A and HLA-B. J. Immunol. 178, 33–37. doi: 10.4049/jimmunol.178.1.33
Thomas, M., Hopkins, C., Duffy, E., Lee, D., Loulergue, P., Ripamonti, D., et al. (2017). Association of the HLA-B∗53:01 Allele with drug reaction with eosinophilia and systemic symptoms (DRESS) syndrome during treatment of HIV infection with raltegravir. Clin. Infect. Dis. 64, 1198–1203. doi: 10.1093/cid/cix096
Urban, T. J., Nicoletti, P., Chalasani, N., Serrano, J., Stolz, A., Daly, A. K., et al. (2017). Minocycline hepatotoxicity: clinical characterization and identification of HLA-B*35:02 as a risk factor. J. Hepatol. 67, 137–144. doi: 10.1016/j.jhep.2017.03.010
van der Ven, A. T., Connaughton, D. M., Ityel, H., Mann, N., Nakayama, M., Chen, J., et al. (2018). Whole-exome sequencing identifies causative mutations in families with congenital anomalies of the kidney and urinary tract. J. Am. Soc. Nephrol. 29, 2348–2361.
Vidal-Castiñeira, J. R., López-Vázquez, A., Diaz-Bulnes, P., Díaz-Coto, S., Márquez-Kisinousky, L., Martínez-Borra, J., et al. (2020). Genetic contribution of endoplasmic reticulum aminopeptidase 1 polymorphisms to liver fibrosis progression in patients with HCV infection. J. Mol. Med. 98, 1245–1254. doi: 10.1007/s00109-020-01948-1
Wang, C.-W., Tassaneeyakul, W., Chen, C.-B., Chen, W.-T., Teng, Y.-C., Huang, C.-Y., et al. (2020). Whole genome sequencing identifies genetic variants associated with co-trimoxazole hypersensitivity in Asians. J. Allergy Clin. Immunol. [Epub ahead of print].
Wang, Q., Sun, S., Xie, M., Zhao, K., Li, X., and Zhao, Z. (2017). Association between the HLA-B alleles and carbamazepine-induced SJS/TEN: a meta-analysis. Epilepsy Res. 135, 19–28. doi: 10.1016/j.eplepsyres.2017.05.015
Watanabe, H., Watanabe, Y., Tashiro, Y., Mushiroda, T., Ozeki, T., Hashizume, H., et al. (2017). A docking model of dapsone bound to HLA-B∗13:01 explains the risk of dapsone hypersensitivity syndrome. J. Dermatol. Sci. 88, 320–329. doi: 10.1016/j.jdermsci.2017.08.007
Wei, C.-Y., Chung, W.-H., Huang, H.-W., Chen, Y.-T., and Hung, S.-I. (2012). Direct interaction between HLA-B and carbamazepine activates T cells in patients with Stevens-Johnson syndrome. J. Allergy Clin. Immunol. 129, 1562–1569.e5.
White, K. D., Chung, W.-H., Hung, S.-I., Mallal, S., and Phillips, E. J. (2015). Evolving models of the immunopathogenesis of T cell-mediated drug allergy: the role of host, pathogens, and drug response. J. Allergy Clin. Immunol. 136, 219–235. doi: 10.1016/j.jaci.2015.05.050
Wolfson, A. R., Zhou, L., Li, Y., Phadke, N. A., Chow, O. A., and Blumenthal, K. G. (2019). Drug reaction with eosinophilia and systemic symptoms (DRESS) syndrome identified in the electronic health record allergy module. J. Allergy Clin. Immunol. Pract. 7, 633–640. doi: 10.1016/j.jaip.2018.08.013
Wright, M. F., Bush, A., and Carr, S. B. (2018). Hypersensitivity reactions to intravenous antibiotics in cystic fibrosis. Paediatr. Respir. Rev. 27, 9–12. doi: 10.1016/j.prrv.2018.01.003
Yampayon, K., Sukasem, C., Limwongse, C., Chinvarun, Y., Tempark, T., Rerkpattanapipat, T., et al. (2017). Influence of genetic and non-genetic factors on phenytoin-induced severe cutaneous adverse drug reactions. Eur. J. Clin. Pharmacol. 73, 855–865. doi: 10.1007/s00228-017-2250-2
Yang, F., Gu, B., Zhang, L., Xuan, J., Luo, H., Zhou, P., et al. (2014). HLA-B*13:01 is associated with salazosulfapyridine-induced drug rash with eosinophilia and systemic symptoms in Chinese Han population. Pharmacogenomics 15, 1461–1469. doi: 10.2217/pgs.14.69
Yang, F., Xuan, J., Chen, J., Zhong, H., Luo, H., Zhou, P., et al. (2015). HLA-B*59:01: a marker for Stevens-Johnson syndrome/toxic epidermal necrolysis caused by methazolamide in Han Chinese. Pharmacogenomics J. 16, 83–87. doi: 10.1038/tpj.2015.25
Yu, K. H., Yu, C. Y., and Fang, Y. F. (2017). Diagnostic utility of HLA−B* 5801 screening in severe allopurinol hypersensitivity syndrome: an updated systematic review and meta−analysis. Int. J. Rheum. Dis. 20, 1057–1071. doi: 10.1111/1756-185x.13143
Yun, J., Marcaida, M. J., Eriksson, K. K., Jamin, H., Fontana, S., Pichler, W. J., et al. (2014). Oxypurinol directly and immediately activates the drug-specific T cells via the preferential use of HLA-B∗ 58: 01. J. Immunol. 192, 2984–2993. doi: 10.4049/jimmunol.1302306
Zanger, U. M., and Schwab, M. (2013). Cytochrome P450 enzymes in drug metabolism: regulation of gene expression, enzyme activities, and impact of genetic variation. Pharmacol. Therap. 138, 103–141. doi: 10.1016/j.pharmthera.2012.12.007
Zanni, M. P., Mauri-Hellweg, D., Brander, C., Wendland, T., Schnyder, B., Frei, E., et al. (1997). Characterization of lidocaine-specific T cells. J. Immunol. 158, 1139–1148.
Zhang, F.-R., Liu, H., Irwanto, A., Fu, X.-A., Li, Y., Yu, G.-Q., et al. (2013). HLA-B∗ 13: 01 and the dapsone hypersensitivity syndrome. New Engl. J. Med. 369, 1620–1628.
Zhao, Q., Alhilali, K., Alzahrani, A., Almutairi, M., Amjad, J., Liu, H., et al. (2019). Dapsone−and nitroso dapsone−specific activation of T cells from hypersensitive patients expressing the risk allele HLA−B∗ 13: 01. Allergy 74, 1533–1548.
Zhao, T., Wang, T.-T., Jia, L., Wang, F., Bahatibieke, M., and Liu, W-l, et al. (2020). The association between HLA-A∗03:01 and HLA-B∗07:02 alleles and oxcarbazepine-induced maculopapular eruption in the Uighur Chinese Population. Seizure 81, 43–46. doi: 10.1016/j.seizure.2020.05.006
Keywords: delayed hypersensitivity, human leukocyte antigen, T-cell receptor, endoplasmic reticulum aminopeptidase, genetic risk, immune checkpoint
Citation: Li Y, Deshpande P, Hertzman RJ, Palubinsky AM, Gibson A and Phillips EJ (2021) Genomic Risk Factors Driving Immune-Mediated Delayed Drug Hypersensitivity Reactions. Front. Genet. 12:641905. doi: 10.3389/fgene.2021.641905
Received: 15 December 2020; Accepted: 08 March 2021;
Published: 16 April 2021.
Edited by:
Jason Trubiano, University of Melbourne, AustraliaReviewed by:
Ana Copaescu, McGill University Health Centre, CanadaCopyright © 2021 Li, Deshpande, Hertzman, Palubinsky, Gibson and Phillips. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Andrew Gibson, YS5naWJzb25AaWlpZC5jb20uYXU=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.