
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
CORRECTION article
Front. Genet. , 19 July 2021
Sec. Genomics of Plants and the Phytoecosystem
Volume 12 - 2021 | https://doi.org/10.3389/fgene.2021.632854
This article is part of the Research Topic Applied Genetics of Natural Fiber Plants View all 9 articles
This article is a correction to:
Genome-Wide Identification and Expression Pattern Analysis of the HAK/KUP/KT Gene Family of Cotton in Fiber Development and Under Stresses
 Xu Yang1,2
Xu Yang1,2 Jingjing Zhang2
Jingjing Zhang2 Aimin Wu2
Aimin Wu2 Hengling Wei2
Hengling Wei2 Xiaokang Fu2
Xiaokang Fu2 Miaomiao Tian2
Miaomiao Tian2 Liang Ma2
Liang Ma2 Jianhua Lu2
Jianhua Lu2 Hantao Wang2*
Hantao Wang2* Shuxun Yu2*
Shuxun Yu2*A Corrigendum on
Genome-Wide Identification and Expression Pattern Analysis of the HAK/KUP/KT Gene Family of Cotton in Fiber Development and Under Stresses
by Yang, X., Zhang, J., Wu, A., Wei, H., Fu, X., Tian, M., et al. (2020). Front. Genet. 11:566469. doi: 10.3389/fgene.2020.566469
In the original article, there were mistakes in Figures 8, 9, and 10 as published. Figures 8 & 10 and their captions were swapped Figure 9 had an incorrect caption. Also one of the Gene name was mislabeled in all the above mentioned figures. GhPOT1 should be GhPOT1-1. The corrected Figures 8, 9, and 10 appear below.
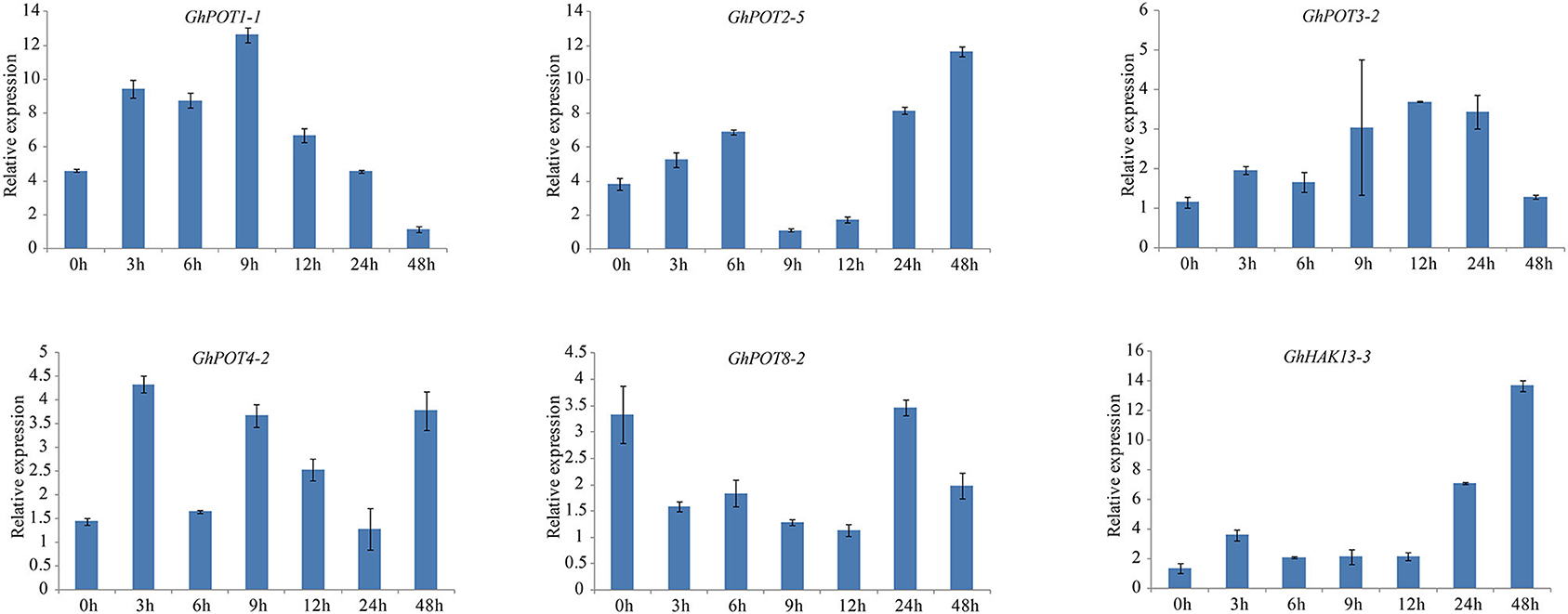
Figure 8. Expressions of GhPOT genes in response to potassium deficiency stress (0.03 mM KCl). The expressions of the GhPOT genes were determined by qRT-PCR using the total RNA isolated from TM-1 leaves at different time points (0, 3, 6, 9, 12, 24 and 48 h) of dehydration stress. The error bars indicate the standard error (SE) of three biological replicates.
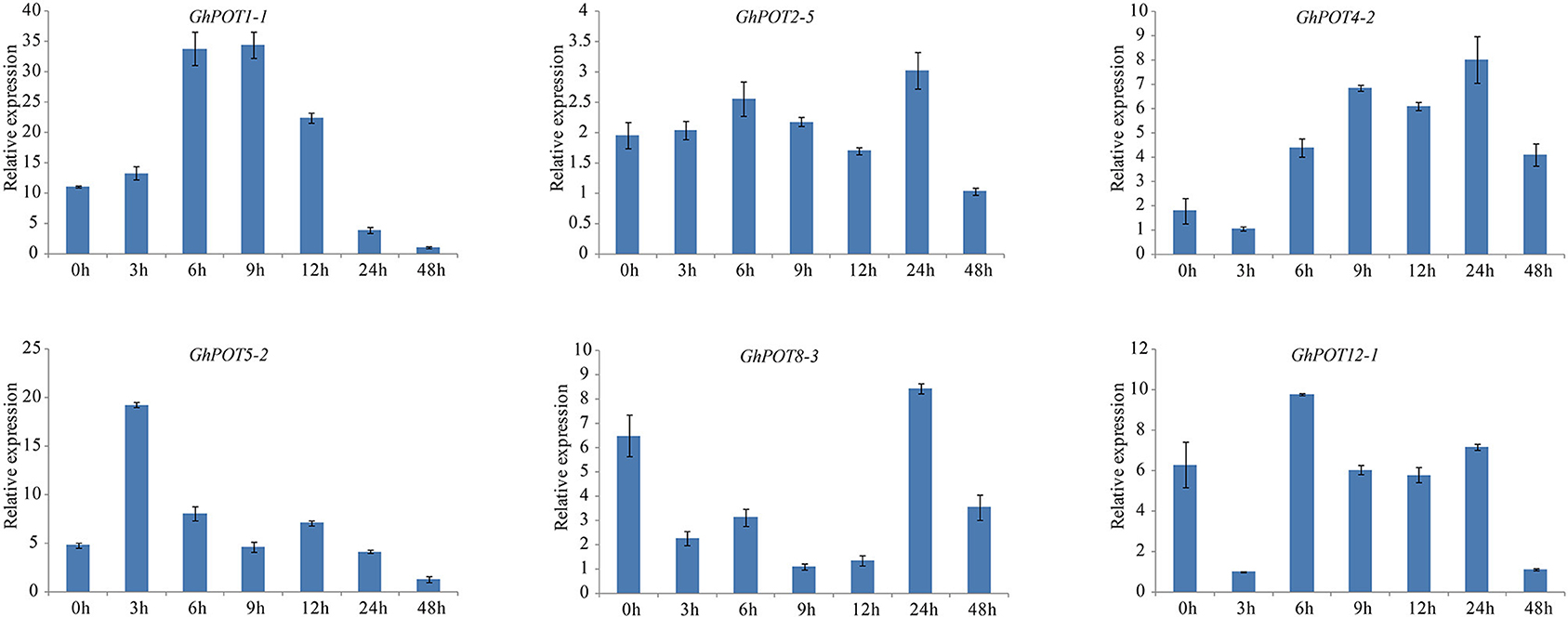
Figure 9. Expressions of GhPOT genes in response to dehydration stress (18% PEG6000). The expressions of GhPOT genes were determined by qRT-PCR using the total RNA isolated from TM-1 leaves at different time points (0, 3, 6, 9, 12, 24 and 48 h) of salt stress. The error bars indicate the standard error (SE) of three biological replicates.
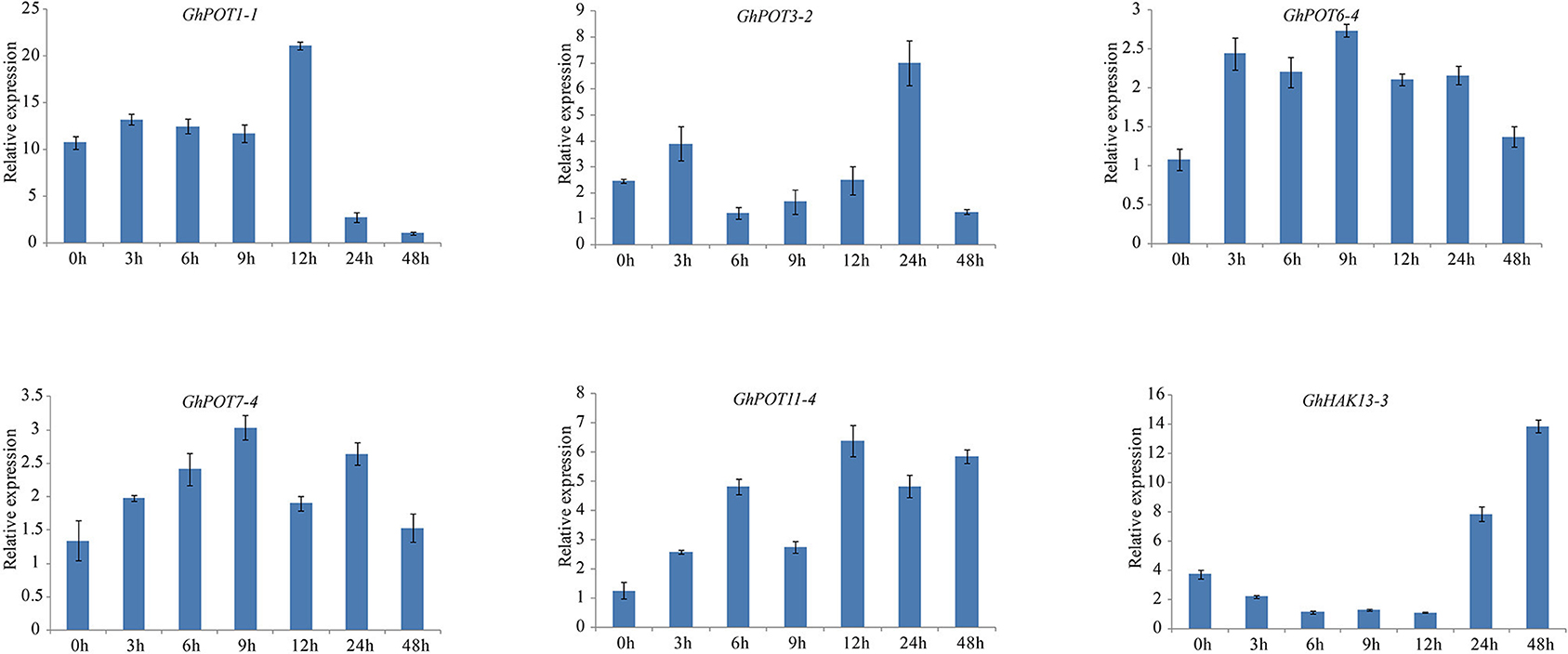
Figure 10. Expressions of GhPOT genes in response to salt stress (300 mM NaCl). The expressions of the GhPOT genes were determined by qRT-PCR using the total RNA isolated from TM-1 leaves at different time points (0, 3, 6, 9, 12, 24 and 48 h) of potassium deficiency stress. The error bars indicate the standard error (SE) of three biological replicates.
Additionally, there were mistakes in the below given sections: In the section: Results and sub-section: qRT-PCR Expression Analysis in Different Fiber Development Stages, the gene name was wrong, GhPOT12-1 should be as GHPOT12-2. The corrected paragraph appears below:
To investigate the possible functions of HAK/KUP/KT in fiber development, we selected 6 genes, which showed high expression levels in different fiber development stages in the transcriptome data (Figure 5) to examine their expression patterns. The results (Figure 7) showed that different genes had different expression patterns. The expression level of GhHAK4-3 was high in each period of fiber synthesis (especially 0 DPA ovule), except in 10 DPA fiber. GhPOT12-2 was significantly expressed in 0 DPA ovules, which is the initiation stage in fiber development. GhPOT11-3 and GhHAK13-1 were highly expressed in 5 and 10 DPA fibers, which are the expansion stages in fiber development. GhPOT2-4 and GhPOT8-2 were highly expressed in 20 and 25 DPA fibers, which are the secondary wall synthesis stages in fiber development.
In the Section: Results, Sub-section: qRT-PCR Expression Analysis in Response to Multiple Stress Treatments, Paragraph 2, there was an incorrect citation of Figures 6, 8, it should be Figures 8, 6. The corrected paragraph appears below:
As shown in Figures 8, 6, random selected genes were examined under potassium deficiency stress, and all showed basically the same expression pattern—after potassium deficiency treatment, the expression was upregulated for a few hours and then downregulated. GhPOT1-1 and GhPOT3-2 precisely followed this pattern, arriving at their peaks at 9 h and 12 h, respectively. GhHAK13-3 and GhPOT2-5, showed upregulated expressions after the basic pattern and reached their maximum expression levels at 48 h. Although GhPOT8-2 was downregulated after treatment, the maximum value was reached at 24 h after treatment, and its change trend followed the pattern.
Lastly, in the Section: Discussion, Sub section: Salt Stress, there was an incorrect citation of Figures 6, 9, it should be Figures 6, 10. The corrected paragraph appears below:
Salt stress is an abiotic stress relevant to modern agricultural production. In this study, transcriptome analysis indicated that most GhPOT genes were responsive to salt stress, and the qRT-PCR results showed that all the selected genes were upregulated after treatment with high concentrations of NaCl. There have been some reports of HAK/KUP/KT genes in other species that could relieve salt stress in plants. Salt stress significantly decreased the root net K+ uptake rate in WT rice and almost completely blocked net K+ uptake in Oshak1 mutants when the K+ concentration was below 0.05 mm. However, plants overexpressing OsHAK1 were more tolerant of salt stress compared to the wild type. The same results were shown in HvHAK1, LeHAK5, and CaHAK1 (Martínez-Cordero et al., 2004; Nieves-Cordones et al., 2007; Fulgenzi et al., 2008). GhPOT5, which is a homolog of OsHAK1, showed higher expression after treatment than did other genes (Figure 6). AtHKT1 provides a key mechanism for protecting leaves from salt stress (Hamamoto et al., 2015), and GhPOT1, which is homologous to AtHKT1, showed significantly increased expression after salt treatment (Figures 6, 10).
The authors apologize for this error and state that this does not change the scientific conclusions of the article in any way. The original article has been updated.
Fulgenzi, F. R., Peralta, M. L., Mangano, S., Dann, C. H., Vallejo, A. J., Puigdomenech, P., et al. (2008). The ionic environment controls the contribution of the barley HvHAK1 transporter to potassium acquisition. Plant Physiol. 147, 252–262. doi: 10.1104/pp.107.114546
Hamamoto, S., Horie, T., Hauser, F., Deinlein, U., Schroeder, J. I., and Uozumi, N. (2015). HKT transporters mediate salt stress resistance in plants: from structure and function to the field. Curr. Opin. Biotechnol. 32, 113–120. doi: 10.1016/j.copbio.2014.11.025
Martínez-Cordero, M. A., Martínez, V., and Rubio, F. (2004). Cloning and functional characterization of the high-affinity K+ transporter HAK1 of pepper. Plant Mol. Biol. 56, 413–421. doi: 10.1007/s11103-004-3845-4
Keywords: HAK/KUP/KT, cotton, expression patterns, fiber development, stress
Citation: Yang X, Zhang J, Wu A, Wei H, Fu X, Tian M, Ma L, Lu J, Wang H and Yu S (2021) Corrigendum: Genome-Wide Identification and Expression Pattern Analysis of the HAK/KUP/KT Gene Family of Cotton in Fiber Development and Under Stresses. Front. Genet. 12:632854. doi: 10.3389/fgene.2021.632854
Received: 24 November 2020; Accepted: 22 April 2021;
Published: 19 July 2021.
Edited and reviewed by: Andrew H. Paterson, University of Georgia, United States
Copyright © 2021 Yang, Zhang, Wu, Wei, Fu, Tian, Ma, Lu, Wang and Yu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hantao Wang, dy53YW5naGFudGFvQDE2My5jb20=; Shuxun Yu, eXN4MTk1MzExQDE2My5jb20=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.