
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Genet. , 06 April 2021
Sec. Stem Cell Research
Volume 12 - 2021 | https://doi.org/10.3389/fgene.2021.625867
This article is part of the Research Topic The Role of Immunity Signaling in Stem Cell Therapy View all 8 articles
A correction has been applied to this article in:
Corrigendum: Characterization of Strip1 Expression in Mouse Cochlear Hair Cells
 Shasha Zhang1†
Shasha Zhang1† Ying Dong1†
Ying Dong1† Ruiying Qiang1†
Ruiying Qiang1† Yuan Zhang1†
Yuan Zhang1† Xiaoli Zhang2
Xiaoli Zhang2 Yin Chen2
Yin Chen2 Pei Jiang1
Pei Jiang1 Xiangyu Ma1
Xiangyu Ma1 Leilei Wu1
Leilei Wu1 Jingru Ai1
Jingru Ai1 Xia Gao2
Xia Gao2 Pengjun Wang3*
Pengjun Wang3* Jie Chen2*
Jie Chen2* Renjie Chai1,4,5,6*
Renjie Chai1,4,5,6*Striatin-interacting protein 1 (Strip1) is a core component of the striatin interacting phosphatase and kinase (STRIPAK) complex, which is involved in embryogenesis and development, circadian rhythms, type 2 diabetes, and cancer progression. However, the expression and role of Strip1 in the mammalian cochlea remains unclear. Here we studied the expression and function of Strip1 in the mouse cochlea by using Strip1 knockout mice. We first found that the mRNA and protein expression of Strip1 increases as mice age starting from postnatal day (P) 3 and reaches its highest expression level at P30 and that the expression of Strip1 can be detected by immunofluorescent staining starting from P14 only in cochlear HCs, and not in supporting cells (SCs). Next, we crossed Strip1 heterozygous knockout (Strip +/−) mice to obtain Strip1 homozygous knockout (Strip1−/−) mice for studying the role of Strip1 in cochlear HCs. However, no Strip1−/− mice were obtained and the ratio of Strip +/− to Strip1+/+ mice per litter was about 2:1, which suggested that homozygous Strip1 knockout is embryonic lethal. We measured hearing function and counted the HC number in P30 and P60 Strip +/− mice and found that they had normal hearing ability and HC numbers compared to Strip1+/+ mice. Our study suggested that Strip1 probably play important roles in HC development and maturation, which needs further study in the future.
Cochlear hair cells (HCs) are important cells in the inner ear for receiving sound signals and converting them into electrical signals to be transmitted to the brain (Hudspeth, 2014; Maoileidigh and Ricci, 2019). There are two types of HCs in the cochlea, inner HCs (IHCs) and outer HCs (OHCs), both of which develop in the embryo and mature after birth (Cotanche and Kaiser, 2010; Walters and Zuo, 2013; Fettiplace, 2017). Sensorineural hearing loss is mainly caused by HC loss, which results from many factors such as genetic factors, aging, noise exposure, and aminoglycosides (Dror and Avraham, 2009; Roth et al., 2011). Among these factors, mutations of many important genes involved in the development, maturation, structure, and function of cochlear HCs have been reported to cause HC loss and thus hearing loss (Venkatesh et al., 2015). However, there are still many more genes involved in HC loss that need further study in order to elucidate their roles and mechanisms.
Striatin-interacting protein 1 (Strip1), also called FAM40A, is one of the scaffold proteins in the striatin interacting phosphatase and kinase (STRIPAK) complex (Hwang and Pallas, 2014). The STRIPAK complex, an evolutionarily conserved supramolecular complex, is involved in many important physiological processes and diseases, including embryogenesis and development (Lant et al., 2015; Sakuma et al., 2015, 2016; Bazzi et al., 2017; Pal et al., 2017; Zheng et al., 2017), type 2 diabetes (Chursa et al., 2017), and cancer progression (Wong et al., 2014; Zhang et al., 2014; Madsen et al., 2015; Huang et al., 2017; Rodriguez-Cupello et al., 2020). The STRIPAK complex has several different formations depending on the combinations of different, mutually exclusive accessory proteins to the STRIPAK core components (Hwang and Pallas, 2014; Kuck et al., 2019; Rodriguez-Cupello et al., 2020; Seo et al., 2020; Stein et al., 2020; Xie et al., 2020). Several important proteins combine to form the core STRIPAK components, including a striatin family member, the PP2A A/C heterodimer, Mob3, Strip1 or Strip2, and a GCKIII kinase bound via Ccm3 (Goudreault et al., 2009). Loss of Strip1 and other scaffolding proteins leads to disassembly and thus to dysfunction of the STRIPAK complex (Madsen et al., 2015; Zheng et al., 2017; Bae et al., 2017; Tang et al., 2019).
The functions of Strip1 and its homologs in several eukaryotic organisms have been studied previously. In Neurospora crassa, the Strip1 homolog is required for hyphal fusion and cell-to-cell fusion (Xiang et al., 2002; Fu et al., 2011). In yeast, the Strip1 homolog is important for cell cycle and mitotic progression (Kemp and Sprague Jr., 2003; Frost et al., 2012), and it antagonizes mTORC2 signaling (Pracheil et al., 2012). In Drosophila, the Strip1 homolog regulates circadian rhythms by controlling daytime CLOCK dephosphorylation (Andreazza et al., 2015), plays roles in axon elongation by regulating early endosome organization (Sakuma et al., 2014), and antagonizes Hippo signaling for regulating cell proliferation (Ashton-Beaucage et al., 2014). In Caenorhabditis elegans, the Strip1 homolog is required for endoplasmic reticulum dynamics and function (Maheshwari et al., 2016). Cell migration and cytoskeleton dynamics are regulated by Strip1 in both Drosophila cells and human endothelial cells (Bai et al., 2011; Sakuma et al., 2015, 2016; Suryavanshi et al., 2018). In mouse embryo, knockout of Strip1 disrupts the migration of the mesoderm in the gastrula stage (Bazzi et al., 2017). Strip1 is also involved in cancer cell migration and metastasis (Madsen et al., 2015), and loss of Strip1 results in cell cycle arrest and subsequent reduced tumor growth by inducing the expression of cyclin-dependent kinase inhibitors (Rodriguez-Cupello et al., 2020). Strip2 (also called FAM40B), another Strip proteins, is also one of the scaffold proteins in the STRIPAK complex (Hwang and Pallas, 2014). Strip2 is also reported to play important roles in cell survival, growth, proliferation, differentiation and migration (Sabour et al., 2017; Dai et al., 2019; Pisciottano et al., 2019; Qiu et al., 2020).
In the mouse cochlea, the role of Strip1 remains unclear, while loss of Strip2 is reported to lead to a decrease in neural response amplitudes and a reduction in the number of afferent synapses (Pisciottano et al., 2019). Here we first studied the expression pattern of Strip1 in the cochlea as mice aged from neonates to adults. We then crossed Strip1 heterozygous knockout (Strip +/−) mice to obtain Strip1 homozygous knockout (Strip1−/−) mice but found that Strip1−/− mice were embryonic lethal. Finally, we tested the hearing function and counted the HC number in adult Strip +/− mice and found no difference compared to wild-type control mice. We suggest that future studies might use Strip1 conditional knockout mice to study the roles of Strip1 in cochlear HCs.
Strip +/− mice, bought from mouse bank of Cyagen company (Stock #KOCMP-229707-Strip1, Cyagen), were constructed by deleting exons 3–8 of the Strip1 gene, and mice of both sexes were used in the experiments. All animal experiments and procedures were approved by the Animal Care and Use Committee of Southeast University. All efforts were made to prevent animals’ suffering and minimize the number of animals used in the experiments.
Tail tips of mice were used to extract genomic DNA for genotyping PCR. A total volume of 180 μl 50 mM NaOH was added to the tail tips, and these were digested by incubating at 98°C for 1 h prior to adding 20 μl 1 M Tris-HCl (pH 7.0). The primers used in genotyping PCR were as follows: Strip1: (F1) 5′-GAC TGG CTG TTT TCC TAC TTA TTC CTA T-3′; (R1) 5′-AGA GCC AGT TCT TTC AAA CGT CAG-3′; and (F2) 5′-GTG GTC TGT TTC CTG AGG ATG TGT-3′.
At least 10 cochleae were dissected and used to extract total mRNA with TRIzol (Thermo, #15,596,026), and the mRNA was reverse transcribed into cDNA using the RevertAid First Strand CDNA Synthesis Kit (Thermo, #K1622). Real-time quantitative PCR (qPCR) was performed on a Bio-Rad C1000 Touch thermal cycler using the FastStart Universal SYBR Green Master (ROX) kit (Roche, #4,913,914,001) to quantify the gene expression levels. The real-time qPCR primers were as follows: Strip1: (F) 5′-AGT GGA GAA CCA TGC GAC AG-3′, (R) 5′-GGC AAA TGG CTC GTT GTT GT-3′; β-actin: (F) 5′-ACG GCC AGG TCA TCA CTA TTG-3′, (R) 5′-AGG GGC CGG ACT CAT CGT A-3′. The relative mRNA expression levels of Strip1 were calculated using 2–△△CT method relative to the house keeping gene β-actin.
At least 10 cochleae were dissected and homogenized in 100 μl ice-cold RIPA lysis buffer (Beyotime, #P0013B) using a tissue homogenizer (Shanghai Jingxin Industrial Development Co., Ltd., #JXFSTPRP-48). After centrifuging at 12,000 × g for 15 min at 4°C, the supernatant was boiled with 5 × SDS loading buffer, separated by 10% SDS-PAGE, and transferred to an Immobilon PVDF membrane (Millipore, #ISEQ00010). The membrane was first blocked for 1 h at room temperature with 5% non-fat dried milk in 0.1% PBS-Tween 20, incubated with the anti-Strip1 (Novusbio, # NBP2-45715) and anti-β-actin (Abcam, #ab119716) primary antibodies at 4°C overnight, and then incubated by HRP-conjugated secondary antibodies (Abmart, goat anti-mouse HRP, #M21001 and goat anti-rabbit HRP, #M21002) for 1 h at room temperature. The SupersignalTM West Femto Maximum Sensitivity Substrate (Thermo, #34,094) was used to detect the signals on a FluorChem M system (ProteinSimple, #FM0477).
The cochleae of neonatal mice (P0–P7) were directly dissected in cold HBSS with sharp forceps (WPI), and then fixed for 1 h at room temperature with 4% paraformaldehyde. The cochleae of mice older than P7 were fixed for 1 h at room temperature with 4% paraformaldehyde, decalcified at room temperature with 0.5 M EDTA for 1–3 days (depending on the mouse’s age), and then dissected. After blocking for 1 h at room temperature with blocking solution (5% donkey serum, 1% bovine serum albumin, 0.5% Triton X-100, and 0.02% sodium azide in pH 7.4 PBS), the cochleae were incubated with primary antibodies which are diluted in PBT1 (2.5% donkey serum, 1% bovine serum albumin, 0.1% Triton X-100, and 0.02% sodium azide in pH 7.4 PBS) at 4°C overnight. This was followed by incubation for 1 h at room temperature with Alexa FluorTM fluorescence-conjugated secondary antibody (Invitrogen) diluted 1:400 in PBT2 (1% bovine serum albumin and 0.1% Triton X-100 in pH 7.4 PBS). Fluorescence mounting medium (DAKO, #S3203) was used to mount the cochleae. The primary antibodies were anti-Myosin7a (Myo7a, Proteus Bioscience, #25-6790, 1:1,000 diluted in PBT1), anti-Sox2 (R&D Systems, #AF2018, 1:400 diluted in PBT1), and anti-Strip1 (Novusbio, #NBP2-45715, 1:400 diluted in PBT1). A Zeiss microscope (LSM 710) was used to scan the immunofluorescent images with the same hardware settings for all images.
P30 cochleae were isolated, fixed in 4% paraformaldehyde, and decalcified with 0.5 M EDTA at room temperature as mentioned above. Cochleae were then equilibrated with a series of ascending concentrations of sucrose (10–30%), and treated serially (1:1, 3:7, 9:1, and then 0:1) with a mixture of 30% sucrose and optimum cutting temperature (OCT) compound (Sakura Finetek). During the serial treatment, cochleae were put in each solution for at least 12 h at 4°C, and then vacuumed in a vacuum chamber for 1 h to remove bubbles from the tissue. Tissues were then frozen in OCT, sectioned (10 μm thick) with a freezing microtome (Leica CM1950) and processed for immunofluorecent staining.
A total of 100 mg per 1 kg mouse body weight pentobarbital sodium (0.01 g/ml) was intraperitoneally injected into P30 and P60 mice to achieve deep anesthesia, and the closed-field ABR thresholds of the mice were tested by using a TDT System III workstation (Tucker-Davis Technologies) as previously described (Chen et al., 2015). One fine needle electrode was inserted at the cranial vertex, one was inserted underneath the tested ear, and one was inserted in the back near the tail, and the mouse was placed on a thermostatic heating pad in a soundproof chamber. The ABR test was performed by generating 4, 8, 12, 16, 24, and 32 kHz tone pips and the sound intensities were decreased from 90 dB in 10 dB steps.
Data from at least three independent experiments were statistically analyzed using GraphPad Prism 6 software and are expressed as means± standard errors of the means. Unpaired, two-tailed Student’s t-tests were used to calculate P-values, and a P-value less than 0.05 was considered statistically significant.
We first studied the protein expression of Strip1 in the P3 mouse cochlea by western blotting and found that Strip1 is expressed at a low level in P3 cochlea but at a relatively high level in the HEI-OC1 HC cell line (Figure 1A). Considering that the expression of many genes increases with age, we next studied the expression of Strip1 at different ages by RT-PCR, qPCR, and western blotting. We found that both the mRNA and protein level of Strip1 increased as the mice aged and reached its highest level at P30 (Figures 1B–D). These results suggested that Strip1 might be involved in HC maturation in the mouse cochlea.
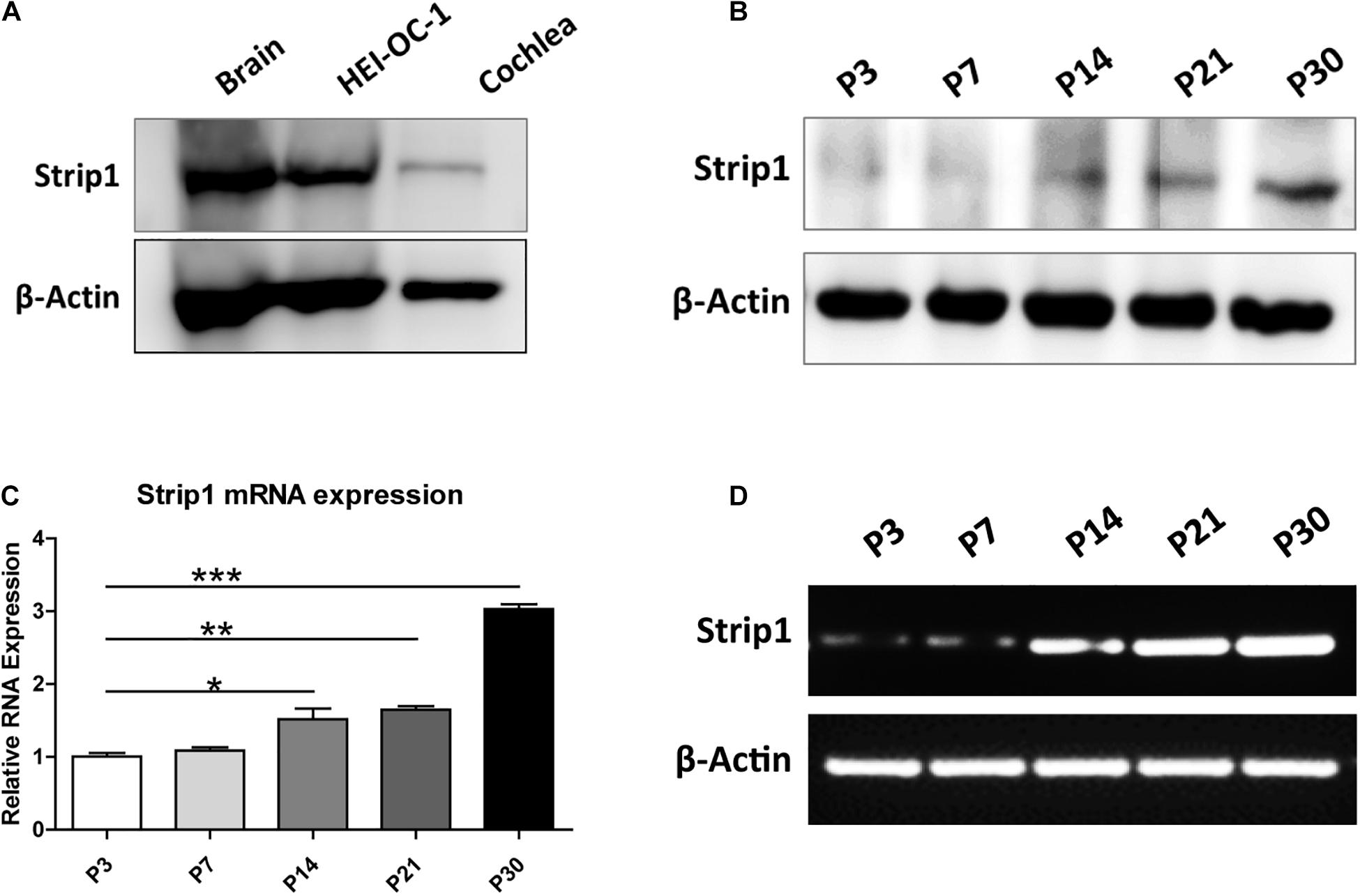
Figure 1. Protein and mRNA expression of Strip1 in the mouse cochlea. (A,B) Western blotting showing the protein expression of Strip1 in the mouse brain, the HEI-OC1 HC cell line, and in the mouse cochlea at P3 (A) and in the mouse cochlea at different ages (B). (C,D) mRNA expression of Strip1 in the mouse cochlea at different ages by qPCR (C) and RT-PCR (D). β-actin was used as the endogenous reference gene. Four independent qPCR were performed in (C). *p < 0.05, **p < 0.01, ***p < 0.001.
Next, we used the Strip1 antibody to study the specific expression pattern in the mouse cochlea by using immunofluorescent staining. We did not detect any Strip1 immunofluorescent signal in P3 or P7 cochlea in either the HC layer or SC layer (Figures 2A,B). At P14, we detected obvious Strip1 immunofluorescence in OHCs, but it was barely detectable in IHCs or the SC layer (Figure 2C). From P21 to P60, there was obvious Strip1 immunofluorescence in both OHCs and IHCs, but not in the SC layer (Figures 2D–F). We also immuno-stained frozen sections of P30 mice cochlea to confirm the limited expression of Strip1 in HCs (Figure 2G). These results also showed that Strip1 expression increased as mice age and were consistent with the mRNA and protein expression of Strip1.
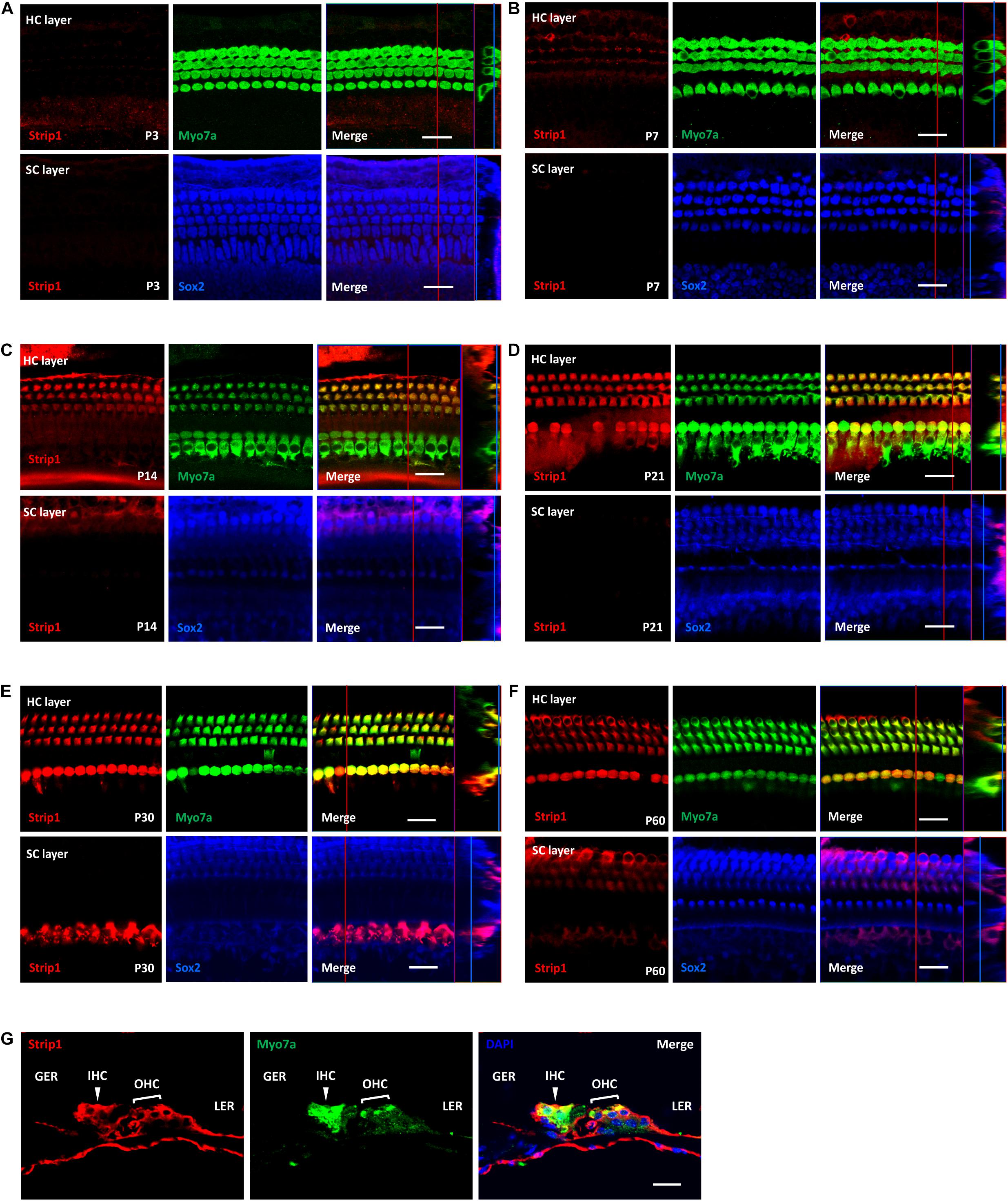
Figure 2. Immunofluorescent staining of Strip1 in the mouse cochlea at different ages. (A–F) The Strip1 antibody was used to stain Strip1 in mouse cochleae at P3 (A), P7 (B), P14 (C), P21 (D), P30 (E), and P60 (F). Because nucleus of SCs staining by Sox2 antibody are not always in the same layer, Z projection was performed with the ImageJ software in order to capture all the SCs in the images. (G) The immunofluorescent staining of P30 cochlear cryosections also showed the same expression pattern of Strip1. Myo7a and Sox2 were used as HC and SC markers, respectively. Scale bar, 20 μm.
Considering the high expression level of Strip1 in the HCs of the adult mouse cochlea, we speculated that Strip1 might play important roles in HCs and thus in hearing function in the adult mouse cochlea. Therefore, we constructed Strip +/− mice and crossed them to get Strip1−/− mice to study the roles of Strip1 in cochlear HCs (Figures 3A,B). However, genotyping results showed that no Strip1−/− mice were born and that Strip +/− and Strip1+/+ mice were born in an approximately 2:1 ratio (Figure 3C). We also used Strip1 antibody to observe the protein expression level of Strip1 in Strip +/− and Strip1+/+ mice, and found that Strip1 protein level is decreased in Strip +/− mice compared to Strip1+/+ mice (Figure 3D). These results indicate that Strip1−/− are embryonic lethal, suggesting that Strip1 plays very important roles during embryo development.
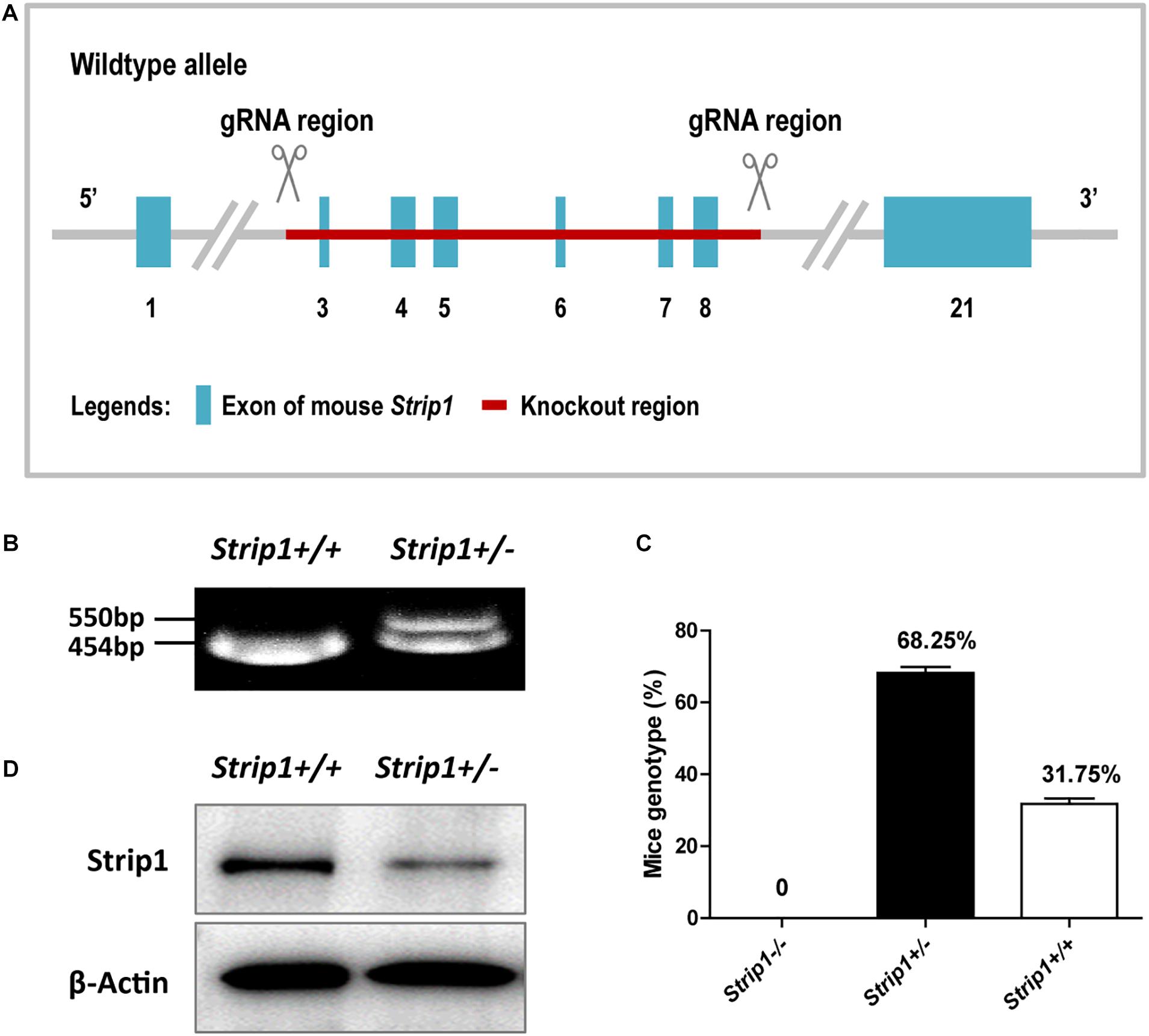
Figure 3. The construction and breeding of the Strip +/− mice. (A) Exons 3–8 of Strip1 were deleted to knock out the Strip1 gene. (B) Genotyping of Strip +/− mice and their offspring. The mutant allele is 550 bp, and the wild type allele is 454 bp. (C) The ratio of mice with different genotypes. There were no Strip1–/– mice, while the percentages of Strip +/− mice and Strip1+/+ mice were 68.25 and 32.75% per litter, respectively. Three litters of mice were counted. (D) Western blotting showing Strip1 protein expression level in Strip1+/+ and Strip +/− mice. β-actin was used as the endogenous reference gene.
Because we could not obtain Strip1−/− mice, we studied the hearing function of Strip +/− mice to see whether heterozygous knockout of Strip1 had any influence on hearing function. ABR tests were performed in P30 and P60 Strip +/− mice, and Strip1+/+ mice from the same litter were used as controls. The results showed that the hearing function of Strip +/− mice was normal at both P30 and P60 compared to the control mice (Figures 4A,B), which suggested that heterozygous knockout of Strip1 did not disrupt the hearing function of mice. We also examined the waveform of the 16 kHz ABR tests in P30 and P60 mice, and this showed no difference compared to controls (Figures 4C,D). Considering that HCs in the cochlea are necessary for sensing sound vibrations, we examined the HCs of P30 and P60 Strip +/− cochleae. Myo7a staining showed that the HCs had similar distributions in Strip +/− mice at both P30 and P60 compared to control mice (Figures 5A,B). We also quantified the HC number and found that there was no significant HC loss in Strip +/− mice at either P30 or P60 (Figures 5C,D). Taken together, these results suggest that Strip1 heterozygous knockout does not have any influence on the hearing function or HC number in the adult mouse cochlea. It is possible that only Strip1 homozygous knockout will have an effect on hearing function and HC number, but because Strip1−/− are embryonic lethal, Strip1 HC conditional knockout mice will be needed to further study the role of Strip1 in adult mouse cochlear HCs.
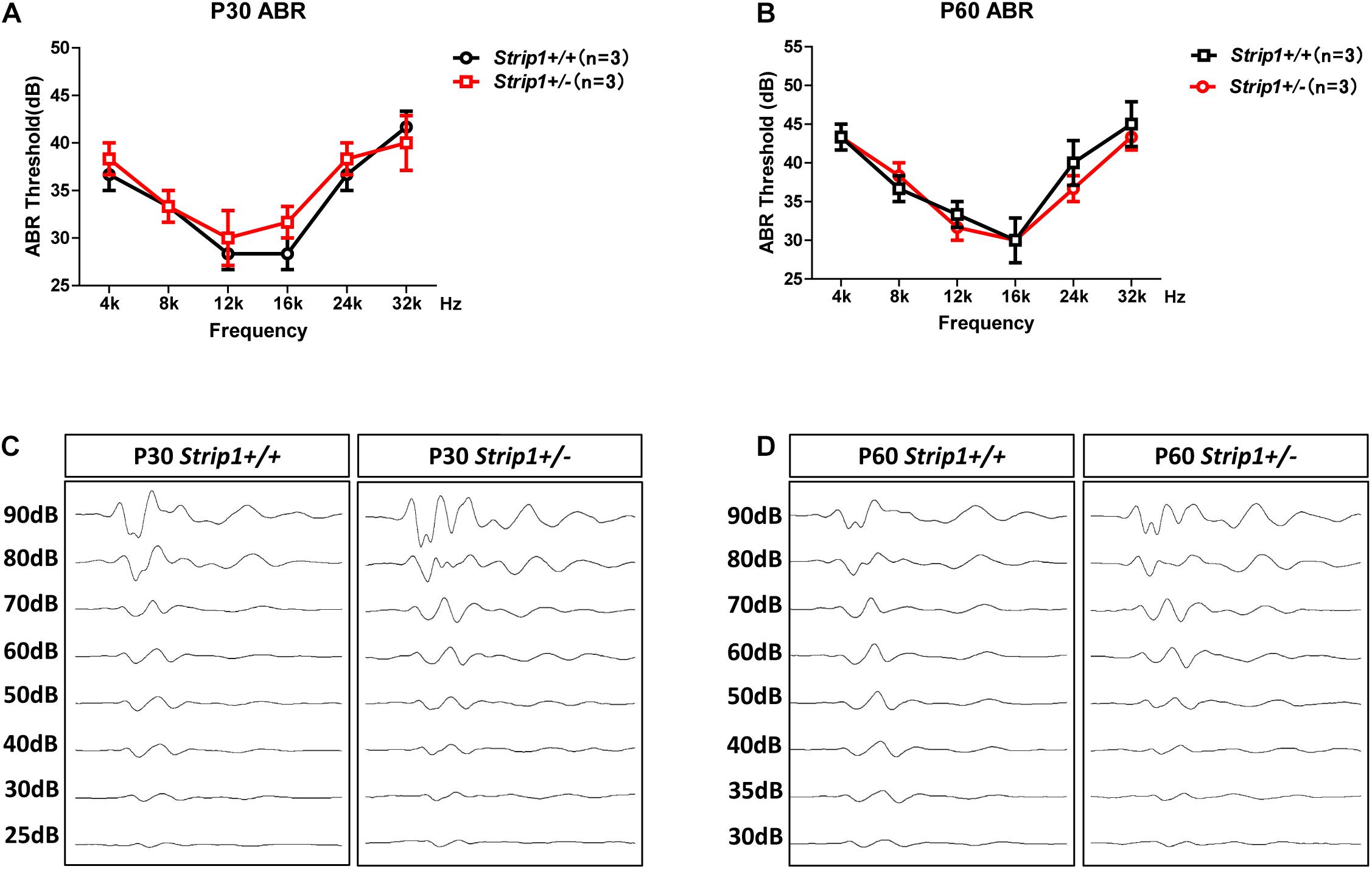
Figure 4. ABR tests of P30 and P60 mice. (A,B) The hearing function of P30 (A) and P60 (B) Strip +/− mice and Strip1+/+ mice was tested by ABR. n refers to the number of mice. (C,D) The ABR waveform at 16 kHz in P30 (C) and P60 (D) Strip +/− mice and Strip1+/+ mice.
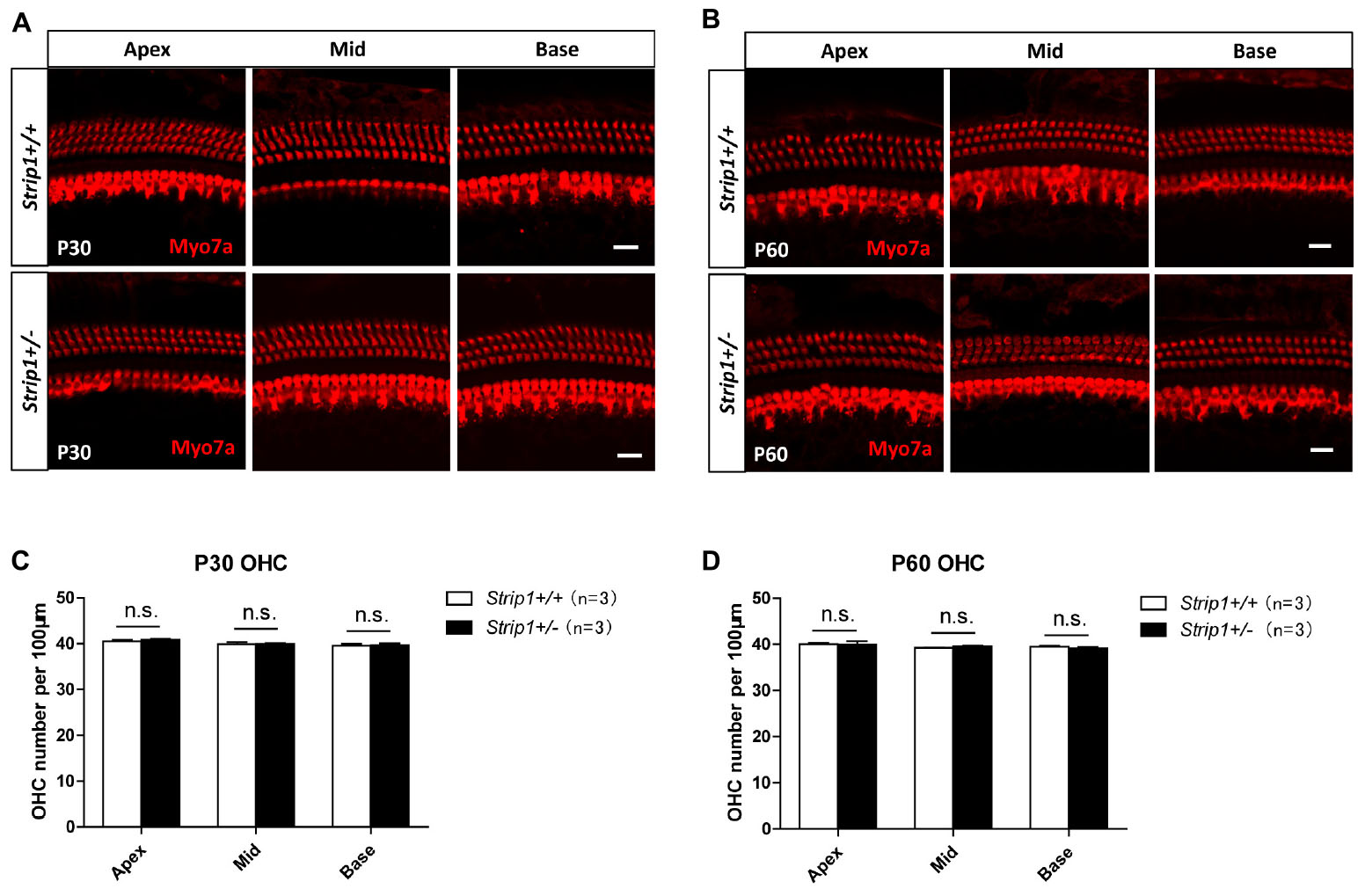
Figure 5. The cochlear HCs in adult Strip +/− mice are normal. (A,B) Immunofluorescent staining of P30 (A) and P60 (B) cochleae from Strip +/− mice and Strip1+/+ mice. Myo7a was used as the HC marker. Scale bar, 20 μm. (C,D) Quantification of the OHC number per 100 μm cochlear length in P30 (C) and P60 (D) Strip +/− mice and Strip1+/+ mice. n refers to the number of mice. n.s., not significant.
HC loss is the main cause of sensorial hearing loss. Although many genes have been reported to be involved in this process, the roles of many other genes remain unknown. Here we found that the expression of Strip1 in cochlear HCs increases as mice age, which suggests that Strip1 might play important roles in HC maturation and in maintaining normal hearing function. However, Strip1−/− mice die before birth, and we only tested the hearing function and counted the HC number in Strip +/− mice, both of which were normal compared to the control mice.
As mentioned above that core STRIPAK components include a striatin family member, the PP2A A/C heterodimer, Mob3, Strip1 or Strip2, and a GCKIII kinase bound via Ccm3 (Goudreault et al., 2009). Different combinations of these core proteins and accessory proteins results in the formation of different STRIPAK complexes. And it was reported that the STRIPAK complex plays many important roles in embryo development, cancer, diabetes, autism, and many other diseases, and regulates many important signaling pathways and cellular processes (Hwang and Pallas, 2014; Wong et al., 2014; Zhang et al., 2014; Lant et al., 2015; Madsen et al., 2015; Sakuma et al., 2015, 2016; Bazzi et al., 2017; Chursa et al., 2017; Huang et al., 2017; Pal et al., 2017; Zheng et al., 2017; Elramli et al., 2019; Kuck et al., 2019; Tang et al., 2019; Rodriguez-Cupello et al., 2020; Seo et al., 2020; Stein et al., 2020; Xie et al., 2020).
Strip1 is one of the scaffolding proteins in the STRIPAK complex, which is involved in embryogenesis and development (Lant et al., 2015; Sakuma et al., 2015, 2016; Bazzi et al., 2017; Pal et al., 2017; Zheng et al., 2017). Strip2, another scaffolding protein in the STRIPAK complex, is also reported to play roles in lung adenocarcinoma, smooth muscle, embryonic stem cells, and inner ear HCs (Sabour et al., 2017; Dai et al., 2019; Pisciottano et al., 2019; Qiu et al., 2020). In the cochlea, Strip2 is reported to be expressed in cochlear HCs, and mice lacking Strip2 have decreased neural response amplitudes and reduced numbers of afferent synapses (Pisciottano et al., 2019). Transcriptome analysis of cochlear IHCs and OHCs from adult mice also identified many differentially expressed genes between IHCs and OHCs, including Strip2 (Li et al., 2018). Considering the results from these studies, we suspected that Strip1 might also be expressed and play important roles in the mouse cochlea.
We used real-time qPCR and western blotting to study the mRNA and protein expression of Strip1 in the mouse cochlea and found that the expression of Strip1 increased as the mice aged and reached its highest expression level at P30. We also used immunofluorescent staining with a Strip1 antibody to study the expression pattern of Strip1 in the mouse cochlea. We found that Strip1 protein was barely detectable in any HCs at P3 and P7, that it was expressed in OHCs but only barely in IHCs at P14, and that it was expressed in both IHCs and OHCs starting from P21. We did not detect any Strip1 expression in SCs by immunofluorescent staining. Together, these results showed suggest that Strip1 is likely to be a very important gene for HC maturation and for normal hearing function.
The Strip +/− mice we obtained had knockout of one Strip1 allele by deleting exons 3–8 of the Strip1 gene. However, when we crossed Strip +/− mice we did not obtain any Strip1−/− mice. The Strip +/− mice were 68.25% of the litters and the Strip +/− mice were 31.75%, and their ratio was about 2:1, which was in accordance with Mendel’s law and suggested that the Strip1−/− mice died before birth. A recent study showed that Strip1 conditional knockout in Sox2-Cre mice experienced arrested development at mid-gestation because of serious disruptions of the mesoderm and its derivatives (Bazzi et al., 2017), and this might be the cause of death of the Strip1−/− mice in our study.
Because we could only obtain Strip +/− mice, we studied their hearing function and HC number at P30 and P60 and found that their hearing was normal at both P30 and P60 compared to the control mice. We also sacrificed the mice and dissected their cochleae and found that the HC number of Strip +/− mice was also normal compared to the control mice. In future studies, we plan to obtain Strip1-floxp mice and cross them with Atoh1-Cre mice to conditionally knockout Strip1 in HCs so as to study the roles of Strip1 in the mouse cochlea.
It was recently reported that Striatin is required for hearing and affects inner hair cells and ribbon synapses (Nadar-Ponniah et al., 2020). Striatin is found to be specifically expressed in the cell–cell junctions of the inner HCs, which is different from Strip1 expression as we shown. There are three proteins in the mammalian striatin family, striatin (STRN), S/G2 nuclear autoantigen (STRN3), and zinedin (STRN4) (Benoist et al., 2006). Thus we suspected that Strip1, expressed in inner and outer HCs, may interact with other two striatin family proteins which may have similar expression pattern as Strip1. This may be the reason why striatin expression is different from Strip1. And another Strip protein, Strip2, was also reported to be expressed in HCs (Scheffer et al., 2015; Pisciottano et al., 2019). It is possible that in cochlear HCs, both Strip1 and Strip2 interact with other two striatin family proteins, not with striatin, which needs further study in the future.
The original contributions presented in the study are included in the article/supplementary material, further inquiries can be directed to the corresponding author/s.
The animal study was reviewed and approved by the Animal Care and Use Committee of Southeast University.
SZ, YD, RQ, YZ, and RC conceived and designed the experiments. SZ, YD, RQ, YZ, XZ, YC, PJ, LW, and JA performed the experiments. XM, XG, PW, and JC analyzed the data. SZ, PW, JC, and RC wrote the manuscript. All authors read and approved the final manuscript.
This work was supported by grants from the National Key RD Program of China (No. 2017YFA0103903), the Strategic Priority Research Program of the Chinese Academy of Sciences (No. XDA16010303), the National Natural Science Foundation of China (Nos. 81970892, 81970882, 81870721, 81900944, and 81700913), the Natural Science Foundation of Jiangsu Province (Nos. BK20190062 and BE2019711), the Shenzhen Fundamental Research Program (No. JCYJ20190814093401920), the Jiangsu Provincial Medical Youth Talent of the Project of Invigorating Health Care through Science, Technology and Education (No. QNRC2016002), and the Fundamental Research Funds for the Central Universities for the Support Program of Zhishan Youth Scholars of Southeast University (No. 2242021R41136).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Andreazza, S., Bouleau, S., Martin, B., Lamouroux, A., Ponien, P., Papin, C., et al. (2015). Daytime clock dephosphorylation is controlled by STRIPAK complexes in Drosophila. Cell Rep. 11, 1266–1279. doi: 10.1016/j.celrep.2015.04.033
Ashton-Beaucage, D., Udell, C. M., Gendron, P., Sahmi, M., Lefrancois, M., Baril, C., et al. (2014). A functional screen reveals an extensive layer of transcriptional and splicing control underlying RAS/MAPK signaling in Drosophila. PLoS Biol. 12:e1001809. doi: 10.1371/journal.pbio.1001809
Bae, S. J., Ni, L., Osinski, A., Tomchick, D. R., Brautigam, C. A., and Luo, X. (2017). SAV1 promotes Hippo kinase activation through antagonizing the PP2A phosphatase STRIPA. eLife 6:e30278.
Bai, S. W., Herrera-Abreu, M. T., Rohn, J. L., Racine, V., Tajadura, V., Suryavanshi, N., et al. (2011). Identification and characterization of a set of conserved and new regulators of cytoskeletal organization, cell morphology and migration. BMC Biol. 9:54. doi: 10.1186/1741-7007-9-54
Bazzi, H., Soroka, E., Alcorn, H. L., and Anderson, K. V. (2017). STRIP1, a core component of STRIPAK complexes, is essential for normal mesoderm migration in the mouse embryo. Proc. Natl. Acad. Sci. U S A. 114, E10928–E10936.
Benoist, M., Gaillard, S., and Castets, F. (2006). The striatin family: a new signaling platform in dendritic spines. J. Physiol. Paris 99, 146–153. doi: 10.1016/j.jphysparis.2005.12.006
Chen, Y., Li, L., Ni, W., Zhang, Y., Sun, S., Miao, D., et al. (2015). Bmi1 regulates auditory hair cell survival by maintaining redox balance. Cell Death Dis. 6:e1605. doi: 10.1038/cddis.2014.549
Chursa, U., Nunez-Duran, E., Cansby, E., Amrutkar, M., Sutt, S., Stahlman, M., et al. (2017). Overexpression of protein kinase STK25 in mice exacerbates ectopic lipid accumulation, mitochondrial dysfunction and insulin resistance in skeletal muscle. Diabetologia 60, 553–567. doi: 10.1007/s00125-016-4171-5
Cotanche, D. A., and Kaiser, C. L. (2010). Hair cell fate decisions in cochlear development and regeneration. Hear. Res. 266, 18–25. doi: 10.1016/j.heares.2010.04.012
Dai, L., Zhou, J., Li, T., Qian, Y., Jin, L., Zhu, C., et al. (2019). STRIP2 silencing inhibits vascular smooth muscle cell proliferation and migration via P38-AKT-MMP-2 signaling pathway. J. Cell Physiol. 234, 22463–22476. doi: 10.1002/jcp.28810
Dror, A. A., and Avraham, K. B. (2009). Hearing loss: mechanisms revealed by genetics and cell biology. Annu. Rev. Genet. 43, 411–437. doi: 10.1146/annurev-genet-102108-134135
Elramli, N., Karahoda, B., Sarikaya-Bayram, O., Frawley, D., Ulas, M., Oakley, C. E., et al. (2019). Assembly of a heptameric STRIPAK complex is required for coordination of light-dependent multicellular fungal development with secondary metabolism in Aspergillus nidulans. PLoS Genet. 15:e1008053. doi: 10.1371/journal.pgen.1008053
Fettiplace, R. (2017). Hair cell transduction. tuning, and synaptic transmission in the mammalian cochlea. Compr. Physiol. 7, 1197–1227. doi: 10.1002/cphy.c160049
Frost, A., Elgort, M. G., Brandman, O., Ives, C., Collins, S. R., Miller-Vedam, L., et al. (2012). Functional repurposing revealed by comparing S. pombe and S. cerevisiae genetic interactions. Cell 149, 1339–1352. doi: 10.1016/j.cell.2012.04.028
Fu, C., Iyer, P., Herkal, A., Abdullah, J., Stout, A., and Free, S. J. (2011). Identification and characterization of genes required for cell-to-cell fusion in Neurospora crassa. Eukaryot Cell 10, 1100–1109. doi: 10.1128/ec.05003-11
Goudreault, M., D’ambrosio, L. M., Kean, M. J., Mullin, M. J., Larsen, B. G., Sanchez, A., et al. (2009). A PP2A phosphatase high density interaction network identifies a novel striatin-interacting phosphatase and kinase complex linked to the cerebral cavernous malformation 3 (CCM3) protein. Mol. Cell Proteomics. 8, 157–171. doi: 10.1074/mcp.m800266-mcp200
Huang, T., Kim, C. K., Alvarez, A. A., Pangeni, R. P., Wan, X., Song, X., et al. (2017). MST4 phosphorylation of ATG4B regulates autophagic activity, tumorigenicity, and radioresistance in glioblastoma. Cancer Cell 32, 840–855 e8.
Hudspeth, A. J. (2014). Integrating the active process of hair cells with cochlear function. Nat. Rev. Neurosci. 15, 600–614. doi: 10.1038/nrn3786
Hwang, J., and Pallas, D. C. (2014). STRIPAK complexes: structure, biological function, and involvement in human diseases. Int. J. Biochem. Cell Biol. 47, 118–148. doi: 10.1016/j.biocel.2013.11.021
Kemp, H. A., and Sprague, G. F. Jr. (2003). Far3 and five interacting proteins prevent premature recovery from pheromone arrest in the budding yeast Saccharomyces cerevisiae. Mol. Cell Biol. 23, 1750–1763. doi: 10.1128/mcb.23.5.1750-1763.2003
Kuck, U., Radchenko, D., and Teichert, I. (2019). STRIPAK, a highly conserved signaling complex, controls multiple eukaryotic cellular and developmental processes and is linked with human diseases. Biol. Chem. Online ahead of print.
Lant, B., Yu, B., Goudreault, M., Holmyard, D., Knight, J. D., Xu, P., et al. (2015). CCM-3/STRIPAK promotes seamless tube extension through endocytic recycling. Nat. Commun. 6:6449.
Li, Y., Liu, H., Giffen, K. P., Chen, L., Beisel, K. W., and He, D. (2018). Transcriptomes of cochlear inner and outer hair cells from adult mice. Sci. Data 5:180199.
Madsen, C. D., Hooper, S., Tozluoglu, M., Bruckbauer, A., Fletcher, G., Erler, J. T., et al. (2015). STRIPAK components determine mode of cancer cell migration and metastasis. Nat. Cell Biol. 17, 68–80. doi: 10.1038/ncb3083
Maheshwari, R., Pushpa, K., and Subramaniam, K. (2016). A role for post-transcriptional control of endoplasmic reticulum dynamics and function in C. elegans germline stem cell maintenance. Development 143, 3097–3108. doi: 10.1242/dev.134056
Maoileidigh, O., and Ricci, D. A. J. (2019). A bundle of mechanisms: inner-ear hair-cell mechanotransduction. Trends Neurosci. 42, 221–236. doi: 10.1016/j.tins.2018.12.006
Nadar-Ponniah, P. T., Taiber, S., Caspi, M., Koffler-Brill, T., Dror, A. A., Siman-Tov, R., et al. (2020). Striatin is required for hearing and affects inner hair cells and ribbon synapses. Front. Cell Dev. Biol. 8:615. doi: 10.3389/fcell.2020.00615
Pal, S., Lant, B., Yu, B., Tian, R., Tong, J., Krieger, J. R., et al. (2017). CCM-3 Promotes C. elegans germline development by regulating vesicle trafficking cytokinesis and polarity. Curr. Biol. 27, 868–876. doi: 10.1016/j.cub.2017.02.028
Pisciottano, F., Cinalli, A. R., Stopiello, J. M., Castagna, V. C., Elgoyhen, A. B., Rubinstein, M., et al. (2019). Inner ear genes underwent positive selection and adaptation in the mammalian lineage. Mol. Biol. Evol. 36, 1653–1670. doi: 10.1093/molbev/msz077
Pracheil, T., Thornton, J., and Liu, Z. (2012). TORC2 signaling is antagonized by protein phosphatase 2A and the Far complex in Saccharomyces cerevisiae. Genetics 190, 1325–1339. doi: 10.1534/genetics.111.138305
Qiu, L. M., Sun, Y. H., Chen, T. T., Chen, J. J., and Ma, H. T. (2020). STRIP2, a member of the striatin-interacting phosphatase and kinase complex, is implicated in lung adenocarcinoma cell growth and migration. FEBS Open Biol. 10, 351–361. doi: 10.1002/2211-5463.12785
Rodriguez-Cupello, C., Dam, M., Serini, L., Wang, S., Lindgren, D., Englund, E., et al. (2020). The STRIPAK complex regulates response to chemotherapy through p21 and p27. Front. Cell Dev. Biol. 8:146. doi: 10.3389/fcell.2020.00146
Roth, T. N., Hanebuth, D., and Probst, R. (2011). Prevalence of age-related hearing loss in Europe: a review. Eur. Arch. Otorhinolaryngol. 268, 1101–1107. doi: 10.1007/s00405-011-1597-8
Sabour, D., Srinivasan, S. P., Rohani, S., Wagh, V., Gaspar, J. A., Panek, D., et al. (2017). STRIP2 is indispensable for the onset of embryonic stem cell differentiation. Mol. Ther. Methods Clin. Dev. 5, 116–129. doi: 10.1016/j.omtm.2017.04.001
Sakuma, C., Kawauchi, T., Haraguchi, S., Shikanai, M., Yamaguchi, Y., Gelfand, V. I., et al. (2014). Drosophila Strip serves as a platform for early endosome organization during axon elongation. Nat. Commun. 5:5180.
Sakuma, C., Okumura, M., Umehara, T., Miura, M., and Chihara, T. (2015). A STRIPAK component Strip regulates neuronal morphogenesis by affecting microtubule stability. Sci. Rep. 5:17769.
Sakuma, C., Saito, Y., Umehara, T., Kamimura, K., Maeda, N., Mosca, T. J., et al. (2016). The Strip-hippo pathway regulates synaptic terminal formation by modulating actin organization at the Drosophila neuromuscular synapses. Cell Rep. 16, 2289–2297. doi: 10.1016/j.celrep.2016.07.066
Scheffer, D. I., Shen, J., Corey, D. P., and Chen, Z. Y. (2015). Gene expression by mouse inner ear hair cells during development. J. Neurosci. 35, 6366–6380. doi: 10.1523/jneurosci.5126-14.2015
Seo, G., Han, H., Vargas, R. E., Yang, B., Li, X., and Wang, W. (2020). MAP4K interactome reveals STRN4 as a key STRIPAK complex component in hippo pathway regulation. Cell Rep. 32:107860. doi: 10.1016/j.celrep.2020.107860
Stein, V., Blank-Landeshammer, B., Muntjes, K., Marker, R., Teichert, I., Feldbrugge, M., et al. (2020). The STRIPAK signaling complex regulates dephosphorylation of GUL1, an RNA-binding protein that shuttles on endosomes. PLoS Genet. 16:e1008819. doi: 10.1371/journal.pgen.1008819
Suryavanshi, N., Furmston, J., and Ridley, A. J. (2018). The STRIPAK complex components FAM40A and FAM40B regulate endothelial cell contractility via ROCKs. BMC Cell Biol. 19:26. doi: 10.1186/s12860-018-0175-y
Tang, Y., Chen, M., Zhou, L., Ma, J., Li, Y., Zhang, H., et al. (2019). Architecture, substructures, and dynamic assembly of STRIPAK complexes in Hippo signaling. Cell Discov. 5:3.
Venkatesh, M. D., Moorchung, N., and Puri, B. (2015). Genetics of non syndromic hearing loss. Med. J. Armed Forces India 71, 363–368.
Walters, B. J., and Zuo, J. (2013). Postnatal development, maturation and aging in the mouse cochlea and their effects on hair cell regeneration. Hear. Res. 297, 68–83. doi: 10.1016/j.heares.2012.11.009
Wong, M., Hyodo, T., Asano, E., Funasaka, K., Miyahara, R., Hirooka, Y., et al. (2014). Silencing of STRN4 suppresses the malignant characteristics of cancer cells. Cancer Sci. 105, 1526–1532. doi: 10.1111/cas.12541
Xiang, Q., Rasmussen, C., and Glass, N. L. (2002). The ham-2 locus, encoding a putative transmembrane protein, is required for hyphal fusion in Neurospora crassa. Genetics 160, 169–180.
Xie, R., Wen, F., and Qin, Y. (2020). The dysregulation and prognostic analysis of STRIPAK complex across cancers. Front. Cell Dev. Biol. 8:625. doi: 10.3389/fcell.2020.00625
Zhang, H., Ma, X., Peng, S., Nan, X., and Zhao, H. (2014). Differential expression of MST4, STK25 and PDCD10 between benign prostatic hyperplasia and prostate cancer. Int. J. Clin. Exp. Pathol. 7, 8105–8111.
Keywords: hair cell, cochlea, inner ear, expression, Strip1
Citation: Zhang S, Dong Y, Qiang R, Zhang Y, Zhang X, Chen Y, Jiang P, Ma X, Wu L, Ai J, Gao X, Wang P, Chen J and Chai R (2021) Characterization of Strip1 Expression in Mouse Cochlear Hair Cells. Front. Genet. 12:625867. doi: 10.3389/fgene.2021.625867
Received: 04 November 2020; Accepted: 12 March 2021;
Published: 06 April 2021.
Edited by:
Chen Zhang, Capital Medical University, ChinaCopyright © 2021 Zhang, Dong, Qiang, Zhang, Zhang, Chen, Jiang, Ma, Wu, Ai, Gao, Wang, Chen and Chai. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Renjie Chai, cmVuamllY0BzZXUuZWR1LmNu; Jie Chen, bmpqaWVjaGVuQDE2My5jb20=; Pengjun Wang, d2FuZ3BqQHNqdHUuZWR1LmNu
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.