- 1Fisheries Research Institute of Fujian, Xiamen, China
- 2Fujian Key Laboratory of Genetics and Breeding of Marine Organisms, College of Ocean and Earth Sciences, Xiamen University, Xiamen, China
- 3Xiamen Key Laboratory of Urban Sea Ecological Conservation and Restoration, Xiamen, China
As a typical marine adaptive radiation species, most Takifugu species are widely distributed in East Asian offshore, which have diversified morphological characteristics and different ecological habits. The phylogenetic relationship and population structure of the Takifugu species was complicated because of incomplete lineage sorting, widespread hybridization and introgression. Therefore, to systematically clarify the phylogenetic relationships of Takifugu genus, explore the introgression and natural hybridization between different Takifugu species, and detect the selective signatures in the adaptive evolution of diversified traits, whole-genome resequencing was used in 122 Takifugu samples from 10 species. Phylogenetic analysis showed solid sister-group relationships between Takifugu bimaculatus and Takifugu flavidus, Takifugu oblongus, and Takifugu niphobles, Takifugu rubripes, and Takifugu obscurus, Takifugu xanthoptreus, and Takifugu ocellatus. Further admixture analysis indicated the divergence of T. obscurus population and the bidirectional gene flow between T. bimaculatus and T. flavidus. Using species-specific homozygous genetic variance sites, we detected the asymmetric introgression between T. bimaculatus and T. flavidus at China East sea and southern Taiwan Strait. By genome-scale genetic diversity scanning, we detected two copies of syt1, zar1 and tgfbr1 related to the semilunar reproduction rhythm in T. niphobles, involved in memory formation, embryo maturation and female reproduction. Furthermore, we also found lots of T. niphobles specific mutations in CDS region of circadian rhythm related genes and endocrine hormone genes. For Takifugu species, our research provides reliable genetic resources and results for the phylogeny, introgression, hybridization and adaptive evolution, and could be used as a guide for the formulation of the protection and proliferation release policies.
Introduction
Adaptive radiation leadsP to the evolution of different species with diverse ecological features and phenotypes within a rapidly multiplying lineage. Hybridization and gene introgression are the two main driving forces of adaptive radiation (Schluter, 2000) since they increase genetic diversity and enhance the ability of a species to quickly occupy a new ecological niche (Marsden et al., 2011; Todesco et al., 2016; Quilodrán et al., 2020). Conversely, hybridization is also considered as “genetic pollution,” which will accelerate the extinction of native species (Simon et al., 2020). Moreover, human activities are impacting the genetic makeup of an increasing number of native species (McFarlane et al., 2020). On the other hand, frequent gene exchange makes elucidating the lineage relationships between species complicated, and thus, it is difficult to identify their lineages and evolutionary history accurately through a few genes or the mitochondrial genome. Therefore, the phylogenetic relationships and evolutionary history of species that have arisen from adaptive radiation need to be explored on a genome-wide scale. Cichlid fish in East Africa have undergone one of the largest radiations and given rise to approximately 2,000 known species (Seehausen, 2006). The widespread incomplete lineage sorting (ILS), hybridization, gene introgression, and the complicated lineage relationships among cichlids were studied and elucidated on a genome-wide scale (Brawand et al., 2014; Meier et al., 2017; Malinsky et al., 2018). In addition, species that have arisen from adaptive radiation have high genetic similarity, exhibit diverse phenotypes, and are excellent specimens for the study of evolutionary developmental biology and evolutionary genomics (Macfadden and Hulbert, 1988). Using comparative genomics and population genetics, many studies have identified candidate genes underlying phenotypic differentiation among species are a product of adaptive radiation, such as dog (Akey et al., 2010), cichlid fishes (Torres-Dowdall et al., 2017) and finch (Tebbich et al., 2010).
Takifugu species are marine species that have arisen from adaptive radiation and are widely distributed in East Asian offshore. They belong to the family Tetraodontidae and the order Tetraodontiformes. Fatal toxicity and swelling body in danger are typical features of these species. In contrast to cichlid fish, only about 25 Takifugu species have undergone explosive speciation during Pliocene (Santini et al., 2013). However, they have diverse morphological characteristics, in terms of body size and coloration, and different ecological habits, such as spawning behavior and adaptability to freshwater (Kato et al., 2010; Motohashi et al., 2010). For example, Takifugu rubripes have a larger body size and faster growth rate than those of others (Hosoya et al., 2013). During the rising tides in the evening and around the days of the full moon, Takifugu niphobles spawn in intertidal beaches (Yamahira, 1994). Moreover, Takifugu obscurus and Takifugu ocellatus will migrate to the Yangtze River during the breeding season to spawn, and several differences in reproductive strategies between these two pufferfishes have been reported (Yang and Chen, 2008). Hence, Takifugu species are considered to be an excellent model for evolutionary studies of adaptive radiation in marine and estuarine environments.
Similar to cichlid fish, several natural and artificial hybridization studies of Takifugu species have been conducted and have reported the absence of absolute reproductive isolation (Takahashi et al., 2017). In addition, the hybridization of T. rubripes, T. obscurus, and T. flavidus, under artificial conditions, were reported to result in fertile offspring, which indicated that there were no reproductive isolation between these species (Jiang et al., 2016; Yoo et al., 2018). These types of hybridizations and introgression can contribute to phenotypic diversification, however, they also complicate the phylogenetic relationship between different Takifugu species. On the other hand, rapid speciation results in ILS, which is another obstacle in accurately describing phylogenetic relationships. Therefore, it is necessary to comprehensively explore phylogenetic relationships between the Takifugu species on a genome-wide scale. Compared with other vertebrate species, Takifugu species have the smallest genome of approximately 400 Mb, which is half the size of the cichlid genome and less than Gasterosteus aculeatus (Zhou Z. et al., 2019). This compact genome facilitates genetic research in Takifugu species. As the first sequenced teleost genome, the genome of T. rubripes was published in 2002 (Aparicio et al., 2002). To date, three other chromosome-level genomes have been published, including that of T. bimaculatus, T. flavidus, and T. obscurus (Zhou Y. et al., 2019; Zhou Z. et al., 2019; Kang et al., 2020). However, only a few studies involving phylogenetic and interspecific hybridization analysis in Takifugu species exist and are based on the mitochondrial genome and amplified fragment length polymorphisms (AFLP) (Yamanoue et al., 2009; Takahashi et al., 2017). In addition, there are only a few population genetic studies involving multiple Takifugu species to date (Zhang et al., 2020). The publication of high-quality genomes motivates population genetics and comparative genomic studies, which are crucial to the understanding of phylogenetic relationships, evolutionary histories, and adaptation strategies of the different Takifugu species during explosive speciation (Nielsen et al., 2009).
The primary aim of this study was to establish a phylogeographical relationship between 10 Takifugu species along the Chinese coast, including Yellow Sea, Eastern China sea, and Taiwan strait using the whole genome re-sequencing method for genotyping. Using this database, we identified reliable phylogenetic relationships and potential events of hybridization and introgression among these populations. In addition, we scanned the genome and detected several different genes in T. niphobles that are associated to the semilunar reproduction rhythm in the intertidal zone, and they are involved in memory formation, embryo development, and rhythm regulation, respectively. To the best of our knowledge, this is the first comprehensive phylogenetic and population genomics study of Takifugu species on a genome-wide scale. The results provide important clues for understanding the genetic divergence, hybridization, introgression, and speciation of marine species that have arisen from adaptive radiation in East Asia.
Materials and Methods
Sampling and DNA Extraction
Individuals from nine Takifugu species, consisting of T. obscurus (To), T. bimaculatus (Tb), T. rubripes (Tr), T. flavidus (Tf), T. oblongus (Tob), T. ocellatus (Toc), T. poechilonotus (Tp), T. alboplumbeus (Ta), and T. xanthoptreus (Tx), were sampled from marginal seas, straits, and estuaries along the Chinese coastal line during the summer and autumn in 2018, with two sites in the north Yellow sea (Tr, Tx), two sites in the estuary of the Yangzi river (To, Tf), one site in the China East sea (Tp), and four sites in the Taiwan Strait (Tob, Toc, Ta, Tb) (Figure 1A). In total, 70 fish were collected, and their tail fins were stored in 95% ethanol before genomic DNA extraction. All samples were collected following national legislation.
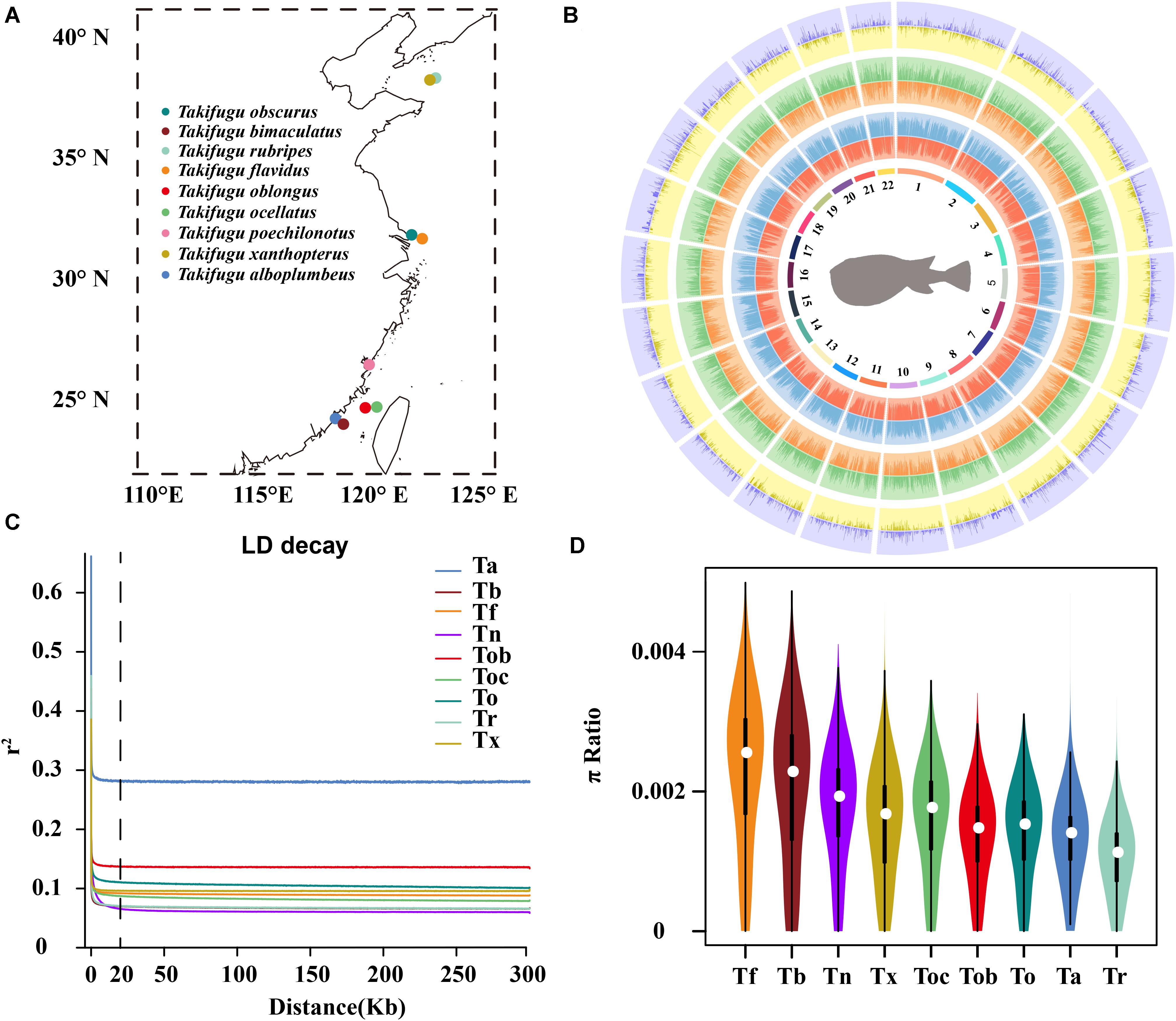
Figure 1. (A) The geographic locations of sample collection. (B) Circos histogram of the statistic of genetic variation calling. The tracks from inside to outside are 22 chromosomes, SNP number (red), indel number (blue), intergenic SNP number (orange), intergenic INDEL number (green), number of SNPs in CDS (yellow), and number of INDELs in CDS (purple). (C) The LD decay of 10 Takifugu species. (D) Genome-wide distribution of nucleotide diversity in a 50 kb non-overlapping window. The white dot was median, black rectangle represented the interquartile range, the black line represented the 95% confidence interval.
Genomic DNA was extracted from the tail fin as described previously (Zhou Z.X. et al., 2019). Briefly, approximately 10 mm2 of the fin samples were shredded and lysed at 56°C with 600 μL of a solution, consisting of STE buffer (10 mM of Tris-HCl, 10 mM of NaCl, 25 mM of EDTA), 10% SDS buffer, and 0.2 mg proteinase K. Subsequently, equal volumes of Tris-phenol and chloroform/isopropanol (24:1) were successively added to extract the DNA. The mixture was incubated for 1 h at −20°C for precipitation and the DNA was collected after centrifugation for 15 min. After washing twice with 70% ethanol, genomic DNA was resuspended in Milli-Q water, and RNase was added. The purified genomic DNA was run on a 1% agarose gel, using gel electrophoresis, and stained with GelGreen to confirm its genomic integrity. Next, DNA concentrations were quantified using a Qubit fluorometer 4.0 (Thermo Fisher Scientific, Waltham, MA, United States), and then diluted to 10 ng/μL for genotyping. Finally, all 70 samples were subjected to Whole-Genome Re-Sequencing.
Genome Resequencing and Genotyping
The sequencing libraries were constructed using the TruePrep DNA Library Prep Kit V2 for Illumina (Vazyme Biotec, Nanjing, China). Whole-genome re-sequencing was performed on the Illumina Hiseq X Ten platform at the Novogene Corporation, Beijing, China. Paired-end reads from each individual were aligned to the reference genome Tb (SWLE00000000) using the MEM algorithm of Burrows-Wheeler Aligner (BWA) (Li and Durbin, 2009). Samtools v1.8 and PICARD v2.18.9 were used to convert the binaries and sort files (Li et al., 2009). GATK v4.0.5.2 was employed to genotype all individuals, using standard procedures and preliminary filters for genetic variation sites with the parameters of “QD < 5.0 || FS > 35.0 || MQ < 55.0 || SOR > 3.0 || MQRankSum < −12.5 || ReadPosRankSum < −8.0 > “(McKenna et al., 2010). Finally, VCFTOOLS v0.1.06 was used to strictly filter low-quality sites with the parameters of “-mac 2 -min-alleles 2 -max-missing-count 2” (Danecek et al., 2011). To identify the sites of genetic variance in the mitochondrial genome, the same protocol and parameters were executed with the reference mitochondrial genome of T. bimaculatus (KP973944.1). Due to the insufficient sample size of Tp, two individuals were removed from evaluations of linkage disequilibrium (LD) and nucleotide diversity. LD decay was investigated for the nine species by PopLDdecay (Zhang et al., 2019).
Phylogenetics, Genetic Structure Analysis, and Identification of Hybridization and Introgression Events
Jmodeltest (2-2.1.10) was used to identify the best model for phylogenetic tree construction (Posada, 2008). Two maximum-likelihood trees were constructed using RAxML v8.2.12 with GTR + I + G model and 1000 bootstrap replicates, based on 236,794 single nucleotide polymorphisms (SNPs) on fourfold degenerate synonymous sites (4DTv) and 15,666 SNPs in the mitochondrial genome, respectively (Stamatakis, 2014). Principal component analysis (PCA) and structure analysis with all the SNPs were performed using GCTA v1.26.0 and ADMIXTURE v1.3.0, respectively (Alexander et al., 2009; Yang et al., 2011). The 3D plots and structure column charts were drawn using scatterplot3d in R.
After filtering the potential introgressive hybridized T. bimaculatus and T. flavidus individuals, we identified the Takifugu species-specific genetic variation sites among 10 species, which fixation index (Fst) were 1 between one Takifugu species and other individuals. ADMIXTURE was used to estimate the introgressive level of eight potential hybrid individuals from the Tb and Tf populations, and 851 species-specific genetic variation markers were identified between the remaining individuals from the Tb and Tf populations. To further estimate the levels of introgression of the remaining 28 Tb and Tf individuals, we used 31,131 highly differentiated markers (Fst > 0.8) between the Tb and Tf individuals. We scanned the whole genome for complete differentiation sites between Tob and Tx-Toc individuals, rather than using the sliding window scanning method for Fst. This is because weak gene exchange signals in a short region may be masked by numerous noise sites. The allele frequency was calculated using PLINK (v1.90b6.16) and all Manhattan scatterplots were drawn using R (Purcell et al., 2007).
Detecting Genome Regions and Candidate Genes Selective to the Semilunar Reproduction Rhythm
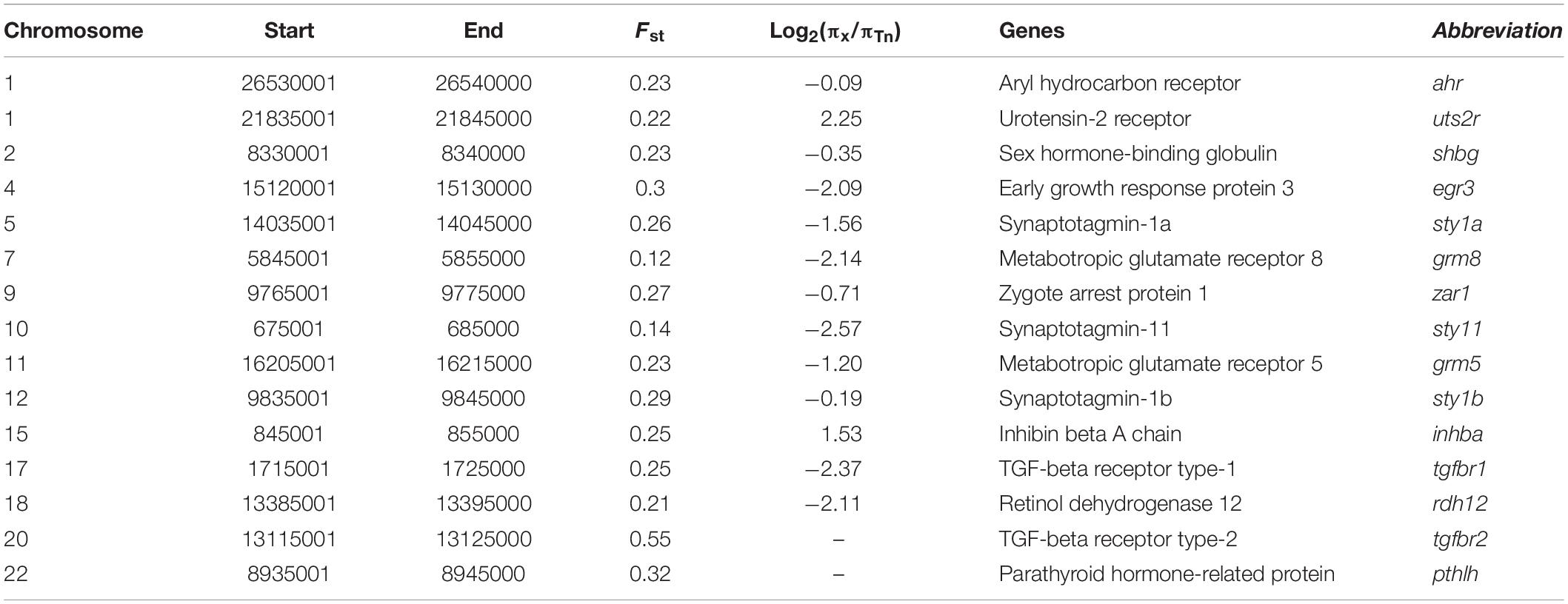
To estimate selective genes to the semilunar reproduction rhythm in Takifugu niphobles (Tn), we first scanned the genome using Fst and Nucleotide diversity (π) Ratio with a sliding window size of 50 Kb and step of 10 Kb. The average ratio of the π ratio of other Takifugu species and that of Tn [average(πx/πTn)] was used to represent the difference in nucleotide diversity. We identified the regions with the highest 1% of Fst values (Fst > 0.225) or a significant difference in nucleotide diversity (| Log2average(πx/πTn)| > 1.5). All candidate genes were annotated by BLAST in the NCBI database1. Fine map scanning was performed using Fst, π ratio, and Tajima’s D with a sliding window size of 1 kb and a step of 500 bp.
Results
Genetic Diversity Between Takifugu Species
We re-sequenced the genome of 70 wild Takifugu individuals and downloaded the re-sequencing data of an additional 52 individuals from NCBI, who belonged to 10 different Takifugu species, to estimate sites of genetic variation, phylogenetic relationships, potential hybridization and introgression, and the presence of selective signatures of adaptive evolution in the different species. A total of 663.7 GB of sequence data was collected, which is equivalent to 5.4 GB of data for each individual, with an average depth of approximately 13.5 (Supplementary Tables 2, 3). The data were mapped to their respective reference and mitochondrial genomes, and the average mapping ratio, genome coverage, and sequencing depth was 98.40%, 93.43%, and 13.21 X, respectively (Supplementary Table 4). Tb and Tf genomes showed the highest mapping ratio of 98.47%, 95.73%, and highest average genome coverage of 98.53% and 95.60%, for Tb and Tf, respectively (Supplementary Table 5). Furthermore, except for Toc12, the genome coverage of all other samples was greater than 90% (Supplementary Table 4). The interspecific universality of the reference genome in Takifugu species indicated similarity of genomes and a close phylogenetic relationship between them. After quality control, 13,598,656 SNPs and 1,629,677 insertion and deletion mutations (INDELs) were identified across 22 chromosomes (Figure 1B and Supplementary Table 6). The average length of INDELs was 5.7 bp and ranged from 1 bp to 259 bp, and the average distance between adjacent SNPs and INDELs was 27 bp and 225.7 bp, respectively (Supplementary Table 6). After careful classification of all sites of genetic variation, 55.21% SNPs and 53.52% INDELs were found to be in the intergenic region, and 7.74% SNPs and 1.18% INDELs were found to be in the coding sequence (CDS; Figure 1B and Supplementary Table 6). In the mitochondrial genome, 1,566 SNPs and 12 INDELs were identified, of which 8.23% SNPs and 75% INDELs were in the intergenic region (Supplementary Table 6).
Multiple linkage maps of the Takifugu genome showed a higher recombination rate compared with other vertebrates (Kai et al., 2005, 2011; Shi et al., 2020), and this was supported by our findings. In addition to Ta, all Takifugu species showed a very low degree (R2 < 0.2) of LD when the window of analysis was greater than 20 kb, indicating active recombination in the Takifugu genome (Figure 1C). Nucleotide diversity reflects the degree of polymorphism within a population. Using a 50 kb sliding window on a genome-wide scale, we found that Tf and Tb had the highest number of polymorphisms, which may be related to frequent introgressions between them (Figure 1D). In addition, Tr showed a reduced nucleotide diversity compared to that in other species. This reduction is likely the result of successive artificial inbreeding over generations and a decline in the wild population size (Nakajima et al., 2008).
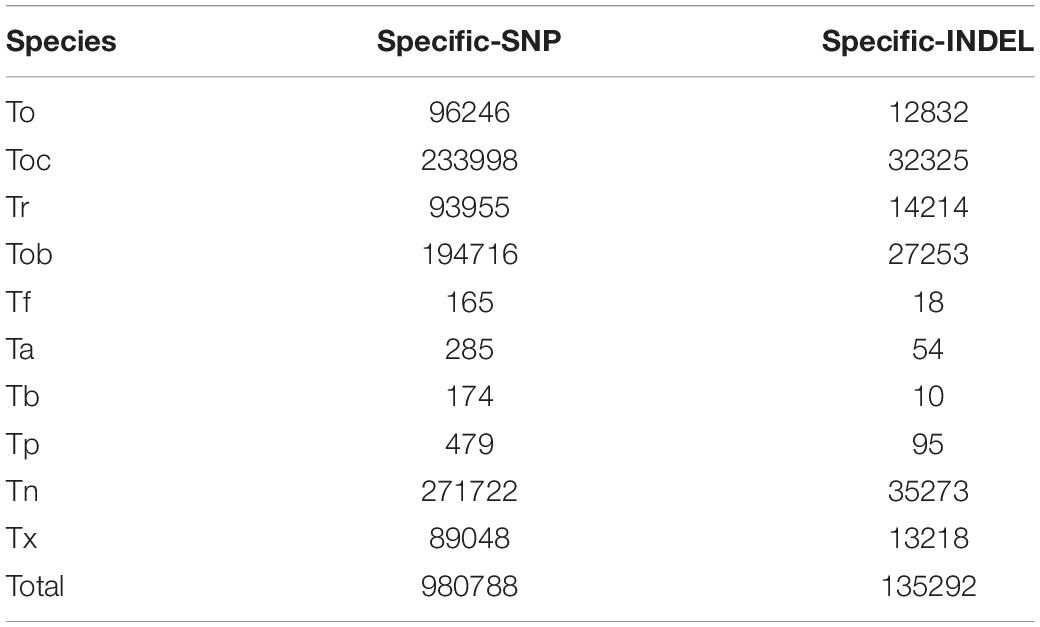
We identified the complete interspecific differentiation sites of genetic variation and thus removed potential hybrid individuals (Tb2, Tb3, Tb20, Tb22) (Figures 2A,B). Finally, we identified 980,788 species-specific SNPs and 135,292 species-specific INDELs. Tn exhibited the highest number of species-specific genetic variation sites, with 271,722 SNPs and 35,273 INDELs, and Tf and Tb exhibited the least number of species-specific genetic variation sites, with only 165 SNPs and 18 INDELs in Tf and 174 SNPs and 10 INDELs in Tb, indicating a possible close relationship during evolutionary history. Similarly, Ta and Tp exhibited a few numbers of species-specific genetic variation sites, with only 285 SNPs and 54 INDELs in Ta and 479 SNPs and 95 INDELs in Ta, which is due to the close relationship and small sample size between them. For other species, 96,246 SNPs and 12,832 INDELs were species-specific in To, 233,998 SNPs and 32,325 INDELs were species-specific in Toc, 93,955 SNPs and 14,214 INDELs were species-specific in Tr, 194,716 SNPs and 27,253 INDELs were species-specific in Tob, 89,048 SNPs and 13,218 INDELs were species-specific in Tx (Table 1).
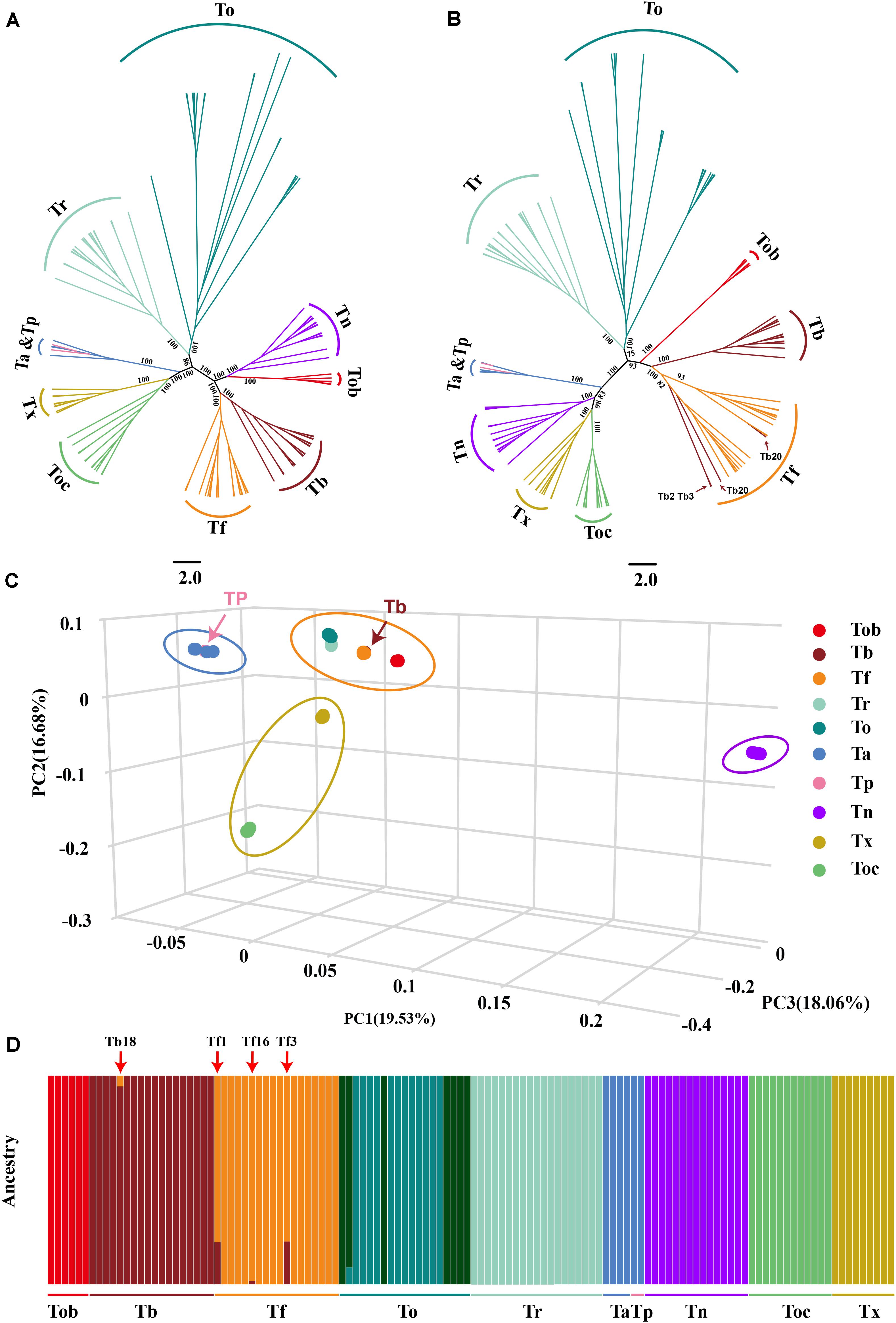
Figure 2. Population structure and relationships of 10 Takifugu species. (A) Maximum-likelihood tree of the relationships between the 10 Takifugu species (122 individuals) based on 4DTv sites. The bootstrap value of the main branches was listed on the branches. (B) Maximum-likelihood tree of the relationships between the 10 Takifugu species (122 individuals) based on mitochondrial genetic variation sites. The bootstrap value of the main branches was listed on the branches. (C) 3D plot visualizing the principal component (PC) analysis. (D) The proportion of ancestry for each individual assuming 10 ancestral population (K = 10). Colors in each columnar represent the likelihood proportion of a Takifugu genome assigned to a source population and each population is labeled at the bottom.
Phylogenetics and Structure Analysis
Considering that Takifugu is a product of adaptive radiation and is thus affected by natural selection, the accuracy of phylogenetic trees is reduced due to convergent evolution and ILS of the species (Jarvis et al., 2014; Chen et al., 2019). Hence, we used fourfold degenerate synonymous sites (4DTv) in the autosomes and SNPs in the mitochondrial genome to construct phylogenetic trees. In the 4DTv phylogenetic tree, sister-group relationships were confirmed between Tb-Tf, Tob-Tn, Tr-To, and Tx-Toc. In addition, four Ta individuals and two Tp individuals belonged to the same group, which indicated that these two species were closely linked (Figure 2A). In the wild, Tp and Ta individuals have very similar morphological patterns and almost overlapping habitats in the Northwest Pacific region (Supplementary Figure 1). Given our insufficient sample size, we cannot conclude whether these two species are synonymous to each other. Most topologies of the mitochondrial phylogenetic tree were consistent with that of the 4DTV phylogenetic tree. Interestingly, the Tn group was closer to the Tx-Toc sister-group, but Tn and Tob formed a reliable sister-group in the 4DTv phylogenetic tree. In previous studies, the close relationship between Tn and Tx-Toc sister-group was demonstrated in phylogenetic trees of the pufferfish based on the sequence of cytb, 12sRNA (Zhang and He, 2008) and the whole mitochondrial genome (Zhang and He, 2008; Yamanoue et al., 2009). In addition, four T. bimaculatus individuals were clustered in the Tf group (Figure 2B). The different phylogenetic relationships between mitochondria and autosomes indicated that hybridization played an important role in the evolutionary history of Takifugu species and hybrid progenies could survive in nature. PCA analysis further confirmed the phylogenetic relationship revealed by the two phylogenetic trees. Based on the three main principal components (percent cumulative eigenvalue variance = 54.27%; Figure 2C), the 10 Takifugu species were divided into four major groups, including the To-Tr-Tf-Tb-Tob group, the Ta-Tp group, the Tx-Toc group, and the Tn group.
To further understand the genetic structure in these Takifugu populations, we used the ADMIXTURE tool to analyze genetic variation sites with K values ranging from 3 to 18, wherein K is the assumed number of ancestral populations. The coefficient of variation suggested that K = 10 is the most likely number of genetically distinct groups within our samples (Supplementary Figure 2). Ta and TP showed the indistinguishable genetic group. To populations had two genetic groups that were not detected in other species, which indicated that there may be two populations with large genetic differences in To. In addition, we found four other potential hybrid individuals (Tb18, Tf1, Tf3, and Tf16), which indicated that a bidirectional gene flow existed between Tb and Tf (Figure 2D).
Interspecific Introgression and Hybridization
Two closely related species, Tb and Tf, coexisted across a vast area, from the southern East China Sea to the southern Yellow Sea (Supplementary Figure 3). Previous studies have shown that hybrid individuals of Tb and Tf existed in the East China Sea (Yin et al., 2016). However, there is no record of hybridization between Tb and Tf individuals in the southern Taiwan Strait. Through phylogenetic analysis of 4DTv and mitochondrial genome, we not only found the hybridization between female Tf and male Tb individuals was excited in nature but also extended the zone wherein hybridization occurs to the southern Taiwan Strait. In the phylogenetic analysis, we detected four potential hybrid individuals (Tb2, Tb3, Tb20, and Tb22). Except for them, we identified 143 SNPs in the mitochondrial genome and 15 SNPs in autosomes that Fst = 1 between Tb and Tf (Supplementary Figure 4A). In the mitochondrial genome, only Tb22 had one Tb-specific SNP, and all the remaining SNPs of four hybrid individuals were absolutely consistent with the genotype of Tf (Supplementary Figure 4B). In autosomes, Tb2 and Tb20 had a homozygous Tf-specific SNP (Supplementary Figure 4A). It suggested that both parents of Tb2 and Tb20 were Tf-specific homozygous or heterozygous on the respective loci.
To further evaluate the bidirectional gene flow in the Tb-Tf sister group, we re-detected sites of species-specific genetic variation between Tb and Tf in 28 “purebred” individuals, which were pedigrees in the phylogenetic and admixture analysis (excluding Tb2, Tb3, Tb18, Tb20, Tb22, Tf3 and Tf16). A total of 851 sites of genetic variation in the autosomes and 129 SNPs in the mitochondrial genome were identified and these were all homozygous and completely differentiated (Fst = 1) in the Tb-Tf sister group. Based on the 129 mitochondrial SNPs, four Tb individuals (Tb2, Tb3, Tb20, and Tb22) showed consistency genetic group with the Tf population, suggesting that these individuals had a hybrid background of female Tf and male Tb (Supplementary Figure 5). In contrast, all Tf individuals had pure mitochondrial genetic components. Based on the 851 SNPs and INDELs in the autosomes, three Tf individuals showed varying levels of introgression (14.33–41.34%) from Tb. Only two Tb individuals showed varying levels of introgressive (3.98% and 27.91%) from Tf (Figure 3A). In addition, we used 31,131 SNPs and INDELS on autosomes and 135 SNPs in the mitochondrial genome, which were highly differentiated in 28 “purebred” individuals of Tb and Tf (Fst > 0.8), to explore whether there was a weak introgression in these “purebred” individuals that showed no introgression signal by the phylogenetics and structure analysis in 122 Takifugu individuals. Results showed that there was low-level introgression in seven “purebred” Tf individuals with introgression rate ranging from 6.93 to 19.86%, but no introgressive signal was detected in the “purebred” Tb individuals (Figure 3B). Moreover, Tf14 showed a higher level of introgression than that of Tf16, and this was detected using ADMIXTURE on a genome-wide scale. For individuals with lower levels of introgression, a weak introgressive signal covered a large number of non-introgressive sites. With an increase in the frequency of backcrossing with the previous generation, the probability of detecting introgression declines. Hence, we did not rule out the possibility of genetic pollution in the remaining “purebred” individuals (Supplementary Figure 6). More importantly, as we relaxed the filtering standard of differentiated genetic variance sites between Tb and Tf, the impact of ILS on introgressive evaluation and the overshadowing effect of non-introgressive sites continued to expand. However, the significant difference between “purebred” Tb and Tf individuals indicated that there was widespread introgression from Tb to Tf at the junction of the Yellow Sea and the East China Sea. On the other hand, a reverse introgression existed at the southern Taiwan Strait.
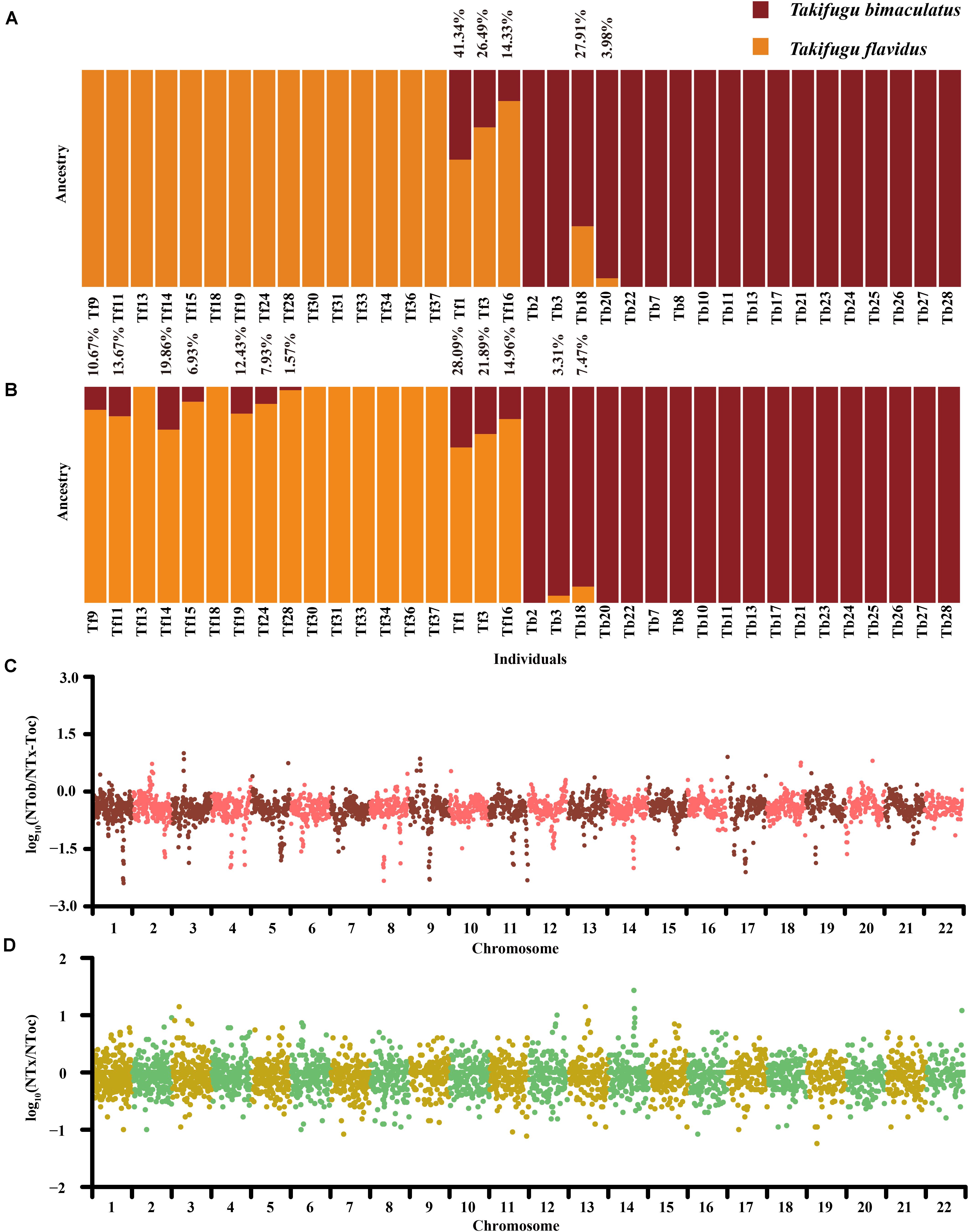
Figure 3. (A) Admixture analysis based on 851 homozygous genetic variation sites on autosomes between Tb and Tf. (B) Admixture analysis based on 31,131 homozygous genetic variation sites on autosomes between Tb and Tf. The orange represented the genetic group of Tf. The claret represented the genetic group of Tb. The introgressive ratio was listed on column. (C) The Genome scanning of genetic similarity between Tob and Tx-Toc in Tn. The ordinate is the Log10 of the ratio of Tob-specific homozygotes site number to Tx-Toc homozygotes site number. (D) The Genome scanning of genetic similarity between Tx and Toc in Tn. The ordinate is the Log10 of the ratio of Tx-specific homozygotes sites number to Toc homozygotes site number.
In the phylogenetic analysis, the phylogenetic difference between mitochondrial genomes and autosomes of Tn indicated that the ancestors of Tn may be influenced by asymmetric introgression from Tob and Tx-Toc (Figures 2A,B). We investigated the genotypes and allele frequencies of 563,658 SNPs and 60,752 INDELs in the autosomes and 254 SNPs in the mitochondrial genome in all Takifugu species; all of which were homozygous and completely differentiated (Fst = 1) between Tob and Tx-Toc group. In contrast to the largest number of Tx-Toc-specific mitochondrial SNPs, Tn had the largest number of homozygous sites of Tob-specific genetic variance in the autosomes, consisting of 132,295 SNPs and 12,008 INDELs, and the major allele in 53% of heterozygous loci was Tob-specific (Supplementary Figure 7 and Supplementary Table 7). A 100 kb sliding window was used to identify the conserved regions with low recombination frequencies after introgression by estimating the ratio of homozygous SNPs between Tob and Tx-Toc (Figure 3C). In the Tn population, several conserved regions were further confirmed as sites of potential introgression. For example, on chromosome 9, two regions between 4.36 and 4.59 Mb were conserved in Tob individuals, but the highly conserved regions in Tx-Toc individuals were between 7.23 and 7.38 Mb (Supplementary Figure 8). These numerous differences of genetic origin were more likely to be caused by frequent autosomal recombination after potential introgression, rather than selective sweep or convergent evolution. To further explore the genetic origin of Tn loci exchanged from the Tx-Toc group, we used 578,210 markers of genetic variance that exhibited complete differentiation (Fst = 1) between Tx and Toc. Further, these loci were homozygous in the Tob genome to reduce any statistical bias caused by genetic drift. Finally, 28,705 specific homozygous loci were identified in the Tx and the Tn genomes, and 33,469 specific homozygous loci were identified in the Toc and the Tn genomes. In addition, in the Tn population, almost all specific homozygous loci identified in Tx and Toc individuals were evenly scattered throughout the genome, and several regions showed higher homology between the Tn and Tx genomes (Figure 3D). For example, Tn and Tx individuals shared almost all specific homozygous loci from 10.2 Mb to 10.7 Mb on chromosome 14, which also showed higher homology with Tx-Toc-specific loci than Tob-specific loci (Supplementary Figure 9). The enrichment of Tx-specific homozygous loci in this region showed that ILS and gene draft may also play important roles in the speciation process of Takifugu.
Adaptive Evolution of the Semilunar Reproduction Rhythm in T. niphobles
In contrast to other Takifugu species, Tn has specific reproductive habits, such as spawning in the intertidal zone during the rising tides, in the evening, around the days of the full moon (Yamahira, 1994). An experiment in an aquarium showed that Tn exhibited obvious aggregating behavior without tidal changes and also precisely maintained the semilunar reproductive rhythm during the spawning season (Motohashi et al., 2010). Hence, this reproductive rhythm is an innate social behavior in the Tn population. Moreover, different Tn populations prefer to spawn at particular beaches with an inclination angle of 8.84°, which is beneficial for gathering, stranding, and spawning (Yamahira, 1997). These social behaviors are closely related to the regulation of memory, rhythm, and levels of sex hormones. We compared the genetic differences between Tn and other Takifugu species to identify the genes specifically associated with spawning behavior. A total of 2,202 sliding windows showed selected signals, which were 17.16 Mb in size (Figures 4A,B and Supplementary Tables 8, 9). After functional annotation, we uncovered 15 candidate genes which are shown in Table 2. Among these, we found two copies of syt1, which encodes for Synaptotagmin-1, a calcium sensor that is responsible for calcium-triggered rapid release of neurotransmitters at the synapse (Geppert et al., 1994; Sudhof, 2013). syt1 plays a key role in α-amino-3-hydroxy-5-methyl-4-isoxazole-propionic acid receptor (AMPA) mediated long-term potentiation (LTP) and participates in the process of learning and memory formation (Wu et al., 2017). Upon fine scanning of Fst, π Ratio, and Tajima’s D, we detected sweep signals in the coding region of syt1a and syt1b (Figure 4C). In sty1a, 12 homozygous mutations were identified in the exons, consisting of two nonsynonymous mutations, two synonymous mutations, four mutations in the 3′UTR, and one mutation in the 5′UTR. The two nonsynonymous mutations changed proline to glutamine in exon 1 and aspartic acid to glutamic acid in exon 3. In sty1b, 16 homozygous mutations were identified in the exons, consisting of one nonsynonymous mutation, one synonymous mutation, three mutations in the 3′UTR, and eleven mutations in the 5′UTR. The nonsynonymous mutation on exon 1 changed aspartic acid to L-asparagine (Figure 4C and Supplementary Table 10). In addition, we also identified zygote arrest protein 1 (zar1), which is a critical maternal effect gene and participates in the process of oocyte-to-embryo transition (Wu et al., 2003). A study investigating embryo development in Tn reported shorter incubation periods and faster completion time in Tn than that of other Takifugu species (Sun et al., 2019). The shorter embryo maturation time of Tn may be due to a rapid change in tidelines around the days of full moon and new moon. In this study, we found significant differentiation signals around zar1 on chromosome 9 in Tn compared to that of other Takifugu species. Nine homozygous mutations were identified in the exons, consisting of one nonsynonymous mutation, three synonymous SNP mutations, three amino acid deletions, two SNPs, one INDEL mutation in the 3′UTR, and one mutation in the 5′UTR (Figure 4C and Supplementary Table 10). Moreover, TGF-β receptor type-1 (tgfbr1), which is an essential gene for the female reproductive system, had a positive selection signal on chromosome 17. Nine homozygous mutations were identified in the exons, consisting of two nonsynonymous mutations and seven synonymous mutations. In addition to the above genes, we found that many rhythm-related genes and endocrine hormone genes in Tn had specific mutations in the CDS (Supplementary Table 11). Many of these genes have been reported to play a key role in the lunar-associated spawning of Tn. Transcription studies have shown that three types of gonadotropin-releasing hormone exhibited different expression levels in the hypothalamus during the reproductive cycle (Shahjahan et al., 2010). In addition, the nocturnal secretion of melatonin from the pineal gland may be involved in the transmission of photoperiodic signals from the moonlight to the hypothalamus in Tn (Ando et al., 2013). In conclusion, our results showed that genes involved in LTP, maternal effect, rhythm regulation, and endocrine hormone secretion had species-specific mutations in the Tn genome, providing new insights into the genetic evolution of intertidal reproductive behavior and semilunar spawning rhythm.
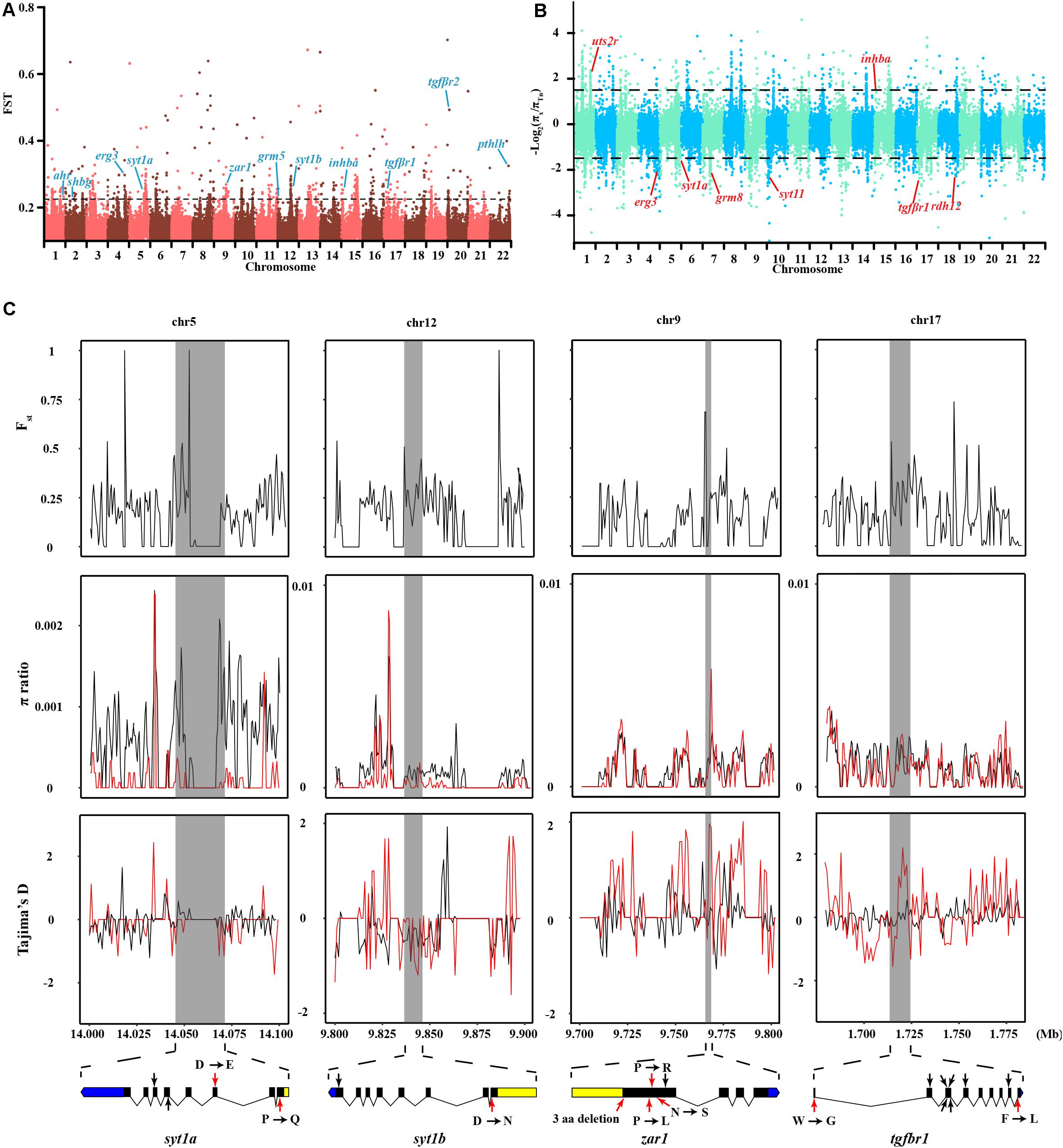
Figure 4. Genomic regions and candidate genes of T. niphobles under selected. (A) Selective scanning of Fst among Tn and other Takifugu species. ahr, aryl hydrocarbon receptor; shbg, sex hormone-binding globulin; erg3, early growth response protein 3; sty1a, synaptotagmin-1a; zar1, zygote arrest protein 1; grm5, metabotropic glutamate receptor 5; sty1b, synaptotagmin-1b; inhba, inhibin beta A chain; tgfβr1, TGF-beta receptor type-1; tgfβr2, TGF-beta receptor type-2; pthlh, parathyroid hormone-related protein. (B) Selective scanning of π ratio between Tn and other Takifugu species. uts2r, urotensin-2 receptor; grm8, metabotropic glutamate receptor 8; sty11, synaptotagmin-11; rdh12, retinol dehydrogenase 12. The candidate gene was marked in red on the Manhattan plot. (C) Selective scanning on four selection genes. The Fst, π ratios and Tajima’s D values were plotted using 500 bp sliding windows. D, aspartic acid; E, glutamic acid; P, proline; Q, glutamine; R, arginine; L, leucine; N, asparagine; S, serine; W, tryptophane; G, glycine; F, phenylalanine. In π ratio plotting and Tajima’s D plotting, the red curved lines represented the Tn individuals and the black curved lines represented the residual individuals. Genome annotations are shown at the bottom [black bar: coding sequences (CDS); blue bar: 5′UTR; yellow bar: 3′UTR]. The boundaries of syt1a, syt1b, zar1 and tgfbr1 are marked in gray shadow. The red arrows represented nonsynonymous mutations, the black arrows represented synonymous mutations.
Discussion
Asymmetric Hybridization Between T. bimaculatus and T. flavidus
Takifugu species had arisen from adaptive radiation in the coastal area of East Asia during the Pliocene epoch (1.8–5.3 million years ago) (Yamanoue et al., 2009). Similar to other species that have arisen from adaptive radiation, reproductive isolation is not absolute among the different Takifugu species, and several studies involving natural and artificial hybridization experiments have demonstrated the fertility of the hybrid offspring (Takahashi et al., 2017). Although female mate choice and prezygotic isolation (behavioral or ecological) in the different Takifugu species may reduce the frequency of hybridization, the rate of introgression is accelerated by changes in the environment and the population size (Saetre et al., 2001).
In this study, we detected bidirectional introgression between the Tf and Tb genomes by admixture analysis. Although we did not detect an F1 hybrid individual in the natural environment, there is sufficient evidence to show that Tb and Tf coexist in the region spanning the Taiwan Strait to the southern Yellow Sea, and their hybrid progeny is fertile. In the Taiwan Strait, four hybrid progeny, from matings between male Tb and female Tf, were identified based on phylogeny, but very low levels of introgression were detected in these individuals, indicating that there were multiple backcrossings between female hybrid individuals and male Tb individuals after primary hybridization. In the junction of the Yellow Sea and the East China Sea, many Tf individuals exhibited an introgression signal from the Tb genome in their autosomes, but no introgression signal was detected in the mitochondrial genome, indicating that matings between male Tb and female Tf were the most common hybrid combination in this region. These asymmetric hybridizations are widespread across Takifugu species and are often based on the extremely rare existence of F1 hybrids (Wirtz, 1999; Takahashi et al., 2017). For the first time, we report two opposite asymmetric hybridizations in Takifugu species between different geographical regions, thereby providing an excellent model to explore the influence of environment and geographical conditions on natural hybridization.
Asymmetric hybridization between Tb and Tf can only be attributed to prezygotic causes, since the bidirectional introgression implied there was no reproductive isolation among Tb, Tf, and their hybrid progenies. The most direct explanation for asymmetric hybridization is the sexual selection hypothesis which states that the females of a rare species are more likely to choose the males of a dominant species than the same species mating, because mate choice becomes weaker in the absence of males of the same species (Takahashi and Takata, 2000; Cahill et al., 2015). In the south Taiwan Strait, Tb is the dominant species, and estuaries of the Zhang River and the Jiulong River, located near our sample points, are the important spawning grounds (Fang and Du, 2008). Tf is the rare species of this area, and there are no reports of Tf spawning in this area. Hence, the primary hybrid offspring often arose from matings between female Tf and male Tb individuals, and the F1 hybrids likely backcrossed with Tb. Therefore, due to the independence of maternal inheritance, the introgression signal in the autosome was rapidly weakened after multiple backcrosses, but the introgression in the mitochondrial genome was completely preserved in a few individuals.
On the other hand, the situation is more complicated in the junction of the Yellow Sea and the East China Sea. First, this is an important fishing area in China, and fisheries in this region have been severely declining (Zhu et al., 2017). Further, Takifugu is an economically valuable species and thus they have a small population size (Zhang et al., 2010). Under huge fishing pressure, the delicate balance between interspecific hybridization and conservative mate choice based on sufficient population size is disturbed. Consequently, many female Tf frequently reproduced with males from other Takifugu species, and this accelerated the speed of interspecific introgression in the autosomes (Mai et al., 2014). Another explanation is the possible northward expansion of the Tb population. Migration is one of the most important ways to cause introgression, and the distribution of species is greatly impacted by environmental changes (Nogue et al., 2009). For example, with the increase in global warming, grizzly bears have become more common on the Arctic island in Canada, and hybridizations between grizzly bears and polar bears have been reported in several regions (Pongracz et al., 2017). In marine fishes, the impact of global warming is more significant, changing the distribution, spawning season, and even the offspring sex ratios of damselfish (Donelson and Munday, 2015), cod (McQueen and Marshall, 2017) and Argyrosomus coronus (Potts et al., 2014). As the two biggest marginal seas, east of China, the sea surface temperature (SST) of the Yellow Sea and East China Sea displayed a sharp incline from 1980 to 2016 (Pei et al., 2017). This change may have facilitated the invasion of Tb in the northern East China Sea and the southern Yellow Sea. Last but not least, individuals that have escaped from artificial breeding enclosures may also be one of the catalysts for introgression between Tb and Tf. In the past two decades, with the success of artificial breeding of Tb and Tf, the aquaculture industry has rapidly developed around the coast of the China East Sea and Yellow Sea (Zhou Z. et al., 2019). In particular, Tf is widely cultured in the central and northern Taiwan Straits, and the northern border of artificial Tb breeding grounds is in the Bohai Sea, which is not its traditional habitat (Zhang et al., 2020). In addition, breeding programs of Tb and Tf also involve their hybridization (Jiang et al., 2016). Inevitably, some purebred individuals and artificial hybrids escape into the wild and invade the local population. In summary, the huge pressure exerted by the fisheries at the junction of the East China Sea and Yellow Sea has impacted the ecological niche. Global warming and escaped artificial breeding individuals have accelerated the expansion of the Tb population to this region. To protect the pure genetic lines of Tf and Tb, more stringent culture and management practices need to be implemented, and a larger-scale fishery resource survey will help to further explore introgression and hybridization between Tb and Tf.
Diversified Reproductive Behaviors in Takifugu
Reproductive behavior includes any activity directed toward the perpetuation of a species, and the diversification of reproductive behavior will contribute to reproductive isolation and speciation (Chippindale, 2013). In species that have arisen from adaptive radiation due to the quick selection in different ecological environments, postzygotic isolation is not prevalent (Salzburger et al., 2002). Hence, prezygotic isolation caused by geographic isolation and different reproductive behaviors plays an important role in speciation during adaptive radiation. About 20 Takifugu species are widely distributed in the coastal of China, with only a few species exhibiting geographical isolation (Zhang and He, 2008). However, diversified reproductive behaviors, including mating period, spawning ground, and hatching conditions, accelerate speciation in Takifugu.
In this study, we identified several candidate genes, related to the semilunar reproduction rhythm, which had undergone selective sweep in Tn. As a widely distributed species, the geographical conditions promote natural hybridization of Tn with many Takifugu species, however, special reproductive behaviors of Tn lead to reproductive isolation. In addition to the specific occurrence of the mating period during the rising tides, Tn spawns only in the intertidal zone, which further reduces the possibility of hybridization with other species. Although we do not know how these specialized reproductive behaviors have developed, there is no doubt that they accelerates speciation in Takifugu. This rapid pattern of speciation also exists in Platichthys flesus (Momigliano et al., 2017) and stickleback (Berner et al., 2017). For example, Platichthys flesus in the Baltic Sea exhibit two reproductive behaviors: pelagic and demersal spawning. Since the low salinity of the Northern Baltic Sea cannot support the buoyancy of the egg, only demersal spawning flounders can thrive in this region. Molecular evidence showed that demersal and pelagic flounders were two ecological species that had arisen from a recent event of speciation, approximately 2,400 generations ago, and this speciation was faster than most known cases of ecological speciation in marine vertebrates (Momigliano et al., 2017). Migration during the mating period is another important reproductive behavior exhibited by Takifugu species. Toc migration has also been reported in the estuary along the Chinese coast from East China Sea to the Yangzi River for spawning during spring (Yang and Chen, 2008). Although other Takifugu species also migrate to the coastal area during the spawning season, they do not spawn in the freshwater river and rarely mate with To and Toc (Yasuji et al., 1991). On the other hand, a previous study showed that ecological shifts caused by colonization of and migration to estuarine habitats promote rapid adaptive divergence and reproductive isolation in incipient ecological species (Beheregaray and Sunnucks, 2001). Therefore, we speculate that the spawning behaviors of Takifugu species may be one of the driving forces of adaptive radiation, which is caused by changes in the estuarine environment during the glacial period. Hence, in future studies, we plan to reconstruct the demographic changes and historical population size of different Takifugu species, and then infer the geological events that lead to their differentiation, along with the period of occurrence, on a larger scale. In addition, we also plan to observe the reproductive behavior of more Takifugu species in the wild, which will provide more information for the study of speciation in Takifugu.
Conclusion
This study focused on the phylogenetic relationships, introgression, hybridization, adaptive evolution, and demographic history of Takifugu species in East Asia. We, except for the Tp-Ta sister group, reliably confirmed the sister-groups of Tb-Tf, Tob-Tn, Tr-To, Tx-Toc, based on 4DTv sites. Combined with the phylogenetic tree constructed on the basis of the mitochondrial genome sequence, we identified four hybridized individuals who are a product of a mating between male Tb and female Tf. Further, Tn may be a potential hybrid of To and the Tx-Toc sister group. Admixture analysis of all individuals showed a divergence of the To population from other species and a bidirectional gene flow between Tb and Tf. Using the species-specific highly differentiated sites of homozygous genetic variation between the “purebred” of Tb and Tf, we detected low levels of introgression in seven “purebred” Tf individuals, and no introgressive signal was detected in “purebred” Tb individuals, indicating that an asymmetric introgression existed at the East China Sea and southern Taiwan Strait. This phenomenon may be caused by the huge pressure exerted by fisheries, global warming, and individuals escaping from artificial breeding enclosures. Homozygous sites of complete differentiation (Fst = 1) between To and the Tx-Toc sister group revealed numerous differences in their genetic origin, which is a result of autosomal recombination after potential introgression. As a set of typical species that have arisen from adaptive radiation, Takifugu also exhibits diverse phenotypes and behaviors. On a genome-wide scale, we detected two copies of syt1, zar1 and tgfbr1, that are related to the semilunar reproduction rhythm in Tn and are involved in memory formation, embryo maturation, and the female reproductive system. Furthermore, we found that many genes regulating the reproductive rhythm and endocrine hormone levels in Tn had specific mutations in the CDS. Our research provides a reliable genetic resource of the phylogenetic relationships, evolutionary history, adaptive radiation, and hybridization in different Takifugu species. Additionally, our results can be used as a reference for the formulation of protection and release policies in artificial fisheries.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Ethics Statement
The animal study was reviewed and approved by Fujian Key Laboratory of Genetics and Breeding of Marine Organisms, College of Ocean and Earth Sciences, Xiamen University.
Author Contributions
PX and BL conceived the project. BL contributed to the funding acquisition. ZZ wrote the manuscript. ZZ, YB, and JY performed the analysis and designed the charts and tables. BL conducted the sampling collection. ZZ, FP, and YS conducted the Re-sequencing libraries. All authors have validated and approved the manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We acknowledge financial support from the Innovation and Industrialization Project of Takifugu breeding Industry (Grant No. 2017FJSCZY03), Science and Technology Major Project of Fujian Province (Grant No. 2020NZ08003), Industry-University Collaboration Project of Fujian Province (Grant No. 2019N5001), Fundamental and application foundation research funds for Guangdong Province (20201910240000762), and the Fundamental Research Funds for the Central Universities (Grant Nos. 20720180123 and 20720160110).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2021.625600/full#supplementary-material
Supplementary Figure 1 | The photo and distribution of T. poechilonotus, T. alboplumbeus. (A) The photo of T. poechilonotus. (B) The photo of T. alboplumbeus. (C) The distribution of T. poechilonotus. (D) The distribution of T. alboplumbeus.
Supplementary Figure 2 | The coefficient of variation of K in admixture from 3 to 18. The dotted line represents the best K value. CV, coefficient of variation; K, the number of groups.
Supplementary Figure 3 | The distribution of T. bimaculatus (Tb) and T. flavidus (Tf) in the coastal of China. Brown represents the habitat of Tb, orange represents the habitat of Tf, red represents the coexist region of Tb and Tf, gray represents the unknown region.
Supplementary Figure 4 | (A) The species-specific SNPs genotype of four hybrid individuals, Tb-specific and Tf-specific on mitochondrial genome. (B) The species-specific SNPs genotype of four hybrid individuals, Tb-specific and Tf-specific on autosomes.
Supplementary Figure 5 | (A) Admixture analysis based on 129 genetic variation sites on the mitochondrial genome between Tb and Tf. (B) Admixture analysis based on 135 genetic variation sites on the mitochondrial genome between Tb and Tf. The orange represented the genetic group of Tf, The claret represented the genetic group of Tb.
Supplementary Figure 6 | The hybrid diagram of Tb and Tf.
Supplementary Figure 7 | The distribution of homozygous and complete differentiation genetic variance sites between Tob and Tx-Toc group in seven Takifugu species. (A) The distribution of homozygous genetic variance sites in seven Takifugu species. (B) The distribution of heterozygous genetic variance sites in seven Takifugu species. (C) The distribution of genetic variance sites on the mitochondrial genome in seven Takifugu species.
Supplementary Figure 8 | The fine mapping of complete differentiation genetic variance sites between Tob and Tx-Toc group based on allele frequency of Tn. The dark blue plot represents the Tob-specific allele; the light blue plot represents the Tx-Toc specific allele.
Supplementary Figure 9 | The fine mapping of complete differentiation genetic variance sites between Tx and Toc group based on allele frequency of Tn. The dark blue plot represents the Tx-specific allele; the light blue plot represents the Toc specific allele.
Supplementary Table 1 | The statistics of sample size, collection location and accession number of sequencing data that download from NCBI database.
Supplementary Table 2 | The statistics of sequencing data of 70 wild Takifugu individuals.
Supplementary Table 3 | The statistics of sequencing data of 52 Takifugu individuals download from the NCBI database.
Supplementary Table 4 | Statistics of genotyping of 122 Takifugu species.
Supplementary Table 5 | Statistics of genotyping of 10 Takifugu species.
Supplementary Table 6 | Statistics of genetic variance sites.
Supplementary Table 7 | Statistics of the number of homozygous and complete differentiation genetic variance sites between Tob and Tx-Toc group in seven Takifugu species.
Supplementary Table 8 | The significant selected regions of Tn based on the Fst threshold (Fst > 0.225).
Supplementary Table 9 | The significant selected regions of Tn based on the π Ratio threshold (| Log2()| > 1.5).
Supplementary Table 10 | Mutations that in the exon of four candidate genes of Tn.
Supplementary Table 11 | Mutations that in the exon of rhythm and sex hormones related genes of Tn.
Footnotes
References
Akey, J. M., Ruhe, A. L., Akey, D. T., Wong, A. K., Connelly, C. F., Madeoy, J., et al. (2010). Tracking footprints of artificial selection in the dog genome. Proc. Natl. Acad. Sci. U.S.A. 107, 1160–1165. doi: 10.1073/pnas.0909918107
Alexander, D. H., Novembre, J., and Lange, K. (2009). Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 19, 1655–1664. doi: 10.1101/gr.094052.109
Ando, H., Shahjahan, M., and Hattori, A. (2013). Molecular neuroendocrine basis of lunar-related spawning in grass puffer. Gen. Compar. Endocrinol. 181, 211–214. doi: 10.1016/j.ygcen.2012.07.027
Aparicio, S., Chapman, J., Stupka, E., Putnam, N., Chia, J. M., Dehal, P., et al. (2002). Whole-genome shotgun assembly and analysis of the genome of Fugu rubripes. Science 297, 1301–1310. doi: 10.1126/science.1072104
Beheregaray, L. B., and Sunnucks, P. (2001). Fine-scale genetic structure, estuarine colonization and incipient speciation in the marine silverside fish Odontesthes argentinensis. Mol. Ecol. 10, 2849–2866. doi: 10.1046/j.1365-294x.2001.t01-1-01406.x
Berner, D., Ammann, M., Spencer, E., Ruegg, A., Luscher, D., and Moser, D. (2017). Sexual isolation promotes divergence between parapatric lake and stream stickleback. J. Evol. Biol. 30, 401–411. doi: 10.1111/jeb.13016
Brawand, D., Wagner, C. E., Li, Y. I., Malinsky, M., Keller, I., Fan, S., et al. (2014). The genomic substrate for adaptive radiation in African cichlid fish. Nature 513, 375–381. doi: 10.1038/nature13726
Cahill, J. A., Stirling, I., Kistler, L., Salamzade, R., Ersmark, E., Fulton, T. L., et al. (2015). Genomic evidence of geographically widespread effect of gene flow from polar bears into brown bears. Mol. Ecol. 24, 1205–1217. doi: 10.1111/mec.13038
Chen, L., Qin, Q., Jiang, Y., Wang, K., Lin, Z. S., Li, Z. P., et al. (2019). Large-scale ruminant genome sequencing provides insights into their evolution and distinct traits. Science 364, 1152. doi: 10.1126/science.aav6202
Chippindale, A. K. (2013). Evolution: sperm, cryptic choice, and the origin of species. Curr. Biol. 23, R885–R887. doi: 10.1016/j.cub.2013.08.024
Danecek, P., Auton, A., Abecasis, G., Albers, C. A., Banks, E., Depristo, M. A., et al. (2011). The variant call format and VCFtools. Bioinformatics 27, 2156–2158. doi: 10.1093/bioinformatics/btr330
Donelson, J. M., and Munday, P. L. (2015). Transgenerational plasticity mitigates the impact of global warming to offspring sex ratios. Glob Chang. Biol. 21, 2954–2962. doi: 10.1111/gcb.12912
Fang, M., and Du, Q. (2008). Preliminary study on technique of tagged and released Takifugu bimaculatus in Jiulongjiang Estuary. J. Oceanogr. Taiwan Strait 27, 326–328.
Geppert, M., Goda, Y., Hammer, R. E., Li, C., Rosahl, T. W., Stevens, C. F., et al. (1994). Synaptotagmin-I – a major Ca2+ sensor for transmitter release at a central synapse. Cell 79, 717–727. doi: 10.1016/0092-8674(94)90556-8
Hosoya, S., Kai, W., Fujita, M., Miyaki, K., Suetake, H., Suzuki, Y., et al. (2013). The genetic architecture of growth rate in juvenile Takifugu species. Evolution 67, 590–598. doi: 10.1111/j.1558-5646.2012.01781.x
Jarvis, E. D., Mirarab, S., Aberer, A. J., Li, B., Houde, P., Li, C., et al. (2014). Whole-genome analyses resolve early branches in the tree of life of modern birds. Science 346, 1320–1331. doi: 10.1126/science.1253451
Jiang, H. B., Bao, J., and Han, Y. (2016). Mitochondrial DNA sequence of the hybrid of Takifugu flavidus (♀) x Takifugu rubripes (♂). Mitochondr. DNA A DNA Mapp. Seq. Anal. 27, 2117–2118. doi: 10.3109/19401736.2014.982583
Kai, W., Kikuchi, K., Fujita, M., Suetake, H., Fujiwara, A., Yoshiura, Y., et al. (2005). A genetic linkage map for the tiger pufferfish, Takifugu rubripes. Genetics 171, 227–238. doi: 10.1534/genetics.105.042051
Kai, W., Kikuchi, K., Tohari, S., Chew, A. K., Tay, A., Fujiwara, A., et al. (2011). Integration of the genetic map and genome assembly of fugu facilitates insights into distinct features of genome evolution in teleosts and mammals. Genom. Biol. Evol. 3, 424–442. doi: 10.1093/gbe/evr041
Kang, S., Kim, J. H., Jo, E., Lee, S. J., Jung, J., Kim, B. M., et al. (2020). Chromosomal-level assembly of Takifugu obscurus (Abe, 1949) genome using third-generation DNA sequencing and Hi-C analysis. Mol. Ecol. Resour. 20, 520–530. doi: 10.1111/1755-0998.13132
Kato, A., Maeno, Y., and Hirose, S. (2010). Brief migration of the grass puffer, Takifugu niphobles, to fresh water from salt water. Ichthyol. Res. 57, 298–304. doi: 10.1007/s10228-009-0150-x
Li, H., and Durbin, R. (2009). Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25, 1754–1760. doi: 10.1093/bioinformatics/btp324
Li, H., Handsaker, B., Wysoker, A., Fennell, T., Ruan, J., Homer, N., et al. (2009). The sequence alignment/map format and SAMTOOLS. Bioinformatics 25, 2078–2079. doi: 10.1093/bioinformatics/btp352
Macfadden, B. J., and Hulbert, R. C. (1988). Explosive speciation at the base of the adaptive radiation of miocene grazing horses. Nature 336, 466–468. doi: 10.1038/336466a0
Mai, A. C. G., Condini, M. V., Albuquerque, C. Q., Loebmann, D., St Pierre, T. D., Miekeley, N., et al. (2014). High plasticity in habitat use of Lycengraulis grossidens (Clupeiformes, Engraulididae). Estuar. Coast. Shelf Sci. 141, 17–25. doi: 10.1016/j.ecss.2014.01.014
Malinsky, M., Svardal, H., Tyers, A. M., Miska, E. A., Genner, M. J., Turner, G. F., et al. (2018). Whole-genome sequences of Malawi cichlids reveal multiple radiations interconnected by gene flow. Nat. Ecol. Evol. 2, 1940–1955. doi: 10.1038/s41559-018-0717-x
Marsden, C. D., Lee, Y., Nieman, C. C., Sanford, M. R., Dinis, J., Martins, C., et al. (2011). Asymmetric introgression between the M and S forms of the malaria vector, Anopheles gambiae, maintains divergence despite extensive hybridization. Mol. Ecol. 20, 4983–4994. doi: 10.1111/j.1365-294X.2011.05339.x
McFarlane, S. E., Hunter, D. C., Senn, H. V., Smith, S. L., Holland, R., Huisman, J., et al. (2020). Increased genetic marker density reveals high levels of admixture between red deer and introduced Japanese sika in Kintyre, Scotland. Evol. Appl. 13, 432–441. doi: 10.1111/eva.12880
McKenna, A., Hanna, M., Banks, E., Sivachenko, A., Cibulskis, K., Kernytsky, A., et al. (2010). The genome analysis toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genom. Res. 20, 1297–1303. doi: 10.1101/gr.107524.110
McQueen, K., and Marshall, C. T. (2017). Shifts in spawning phenology of cod linked to rising sea temperatures. Ices J. Mar. Sci. 74, 1561–1573. doi: 10.1093/icesjms/fsx025
Meier, J. I., Marques, D. A., Mwaiko, S., Wagner, C. E., Excoffier, L., and Seehausen, O. (2017). Ancient hybridization fuels rapid cichlid fish adaptive radiations. Nat. Commun. 8:14363. doi: 10.1038/ncomms14363
Momigliano, P., Jokinen, H., Fraimout, A., Florin, A. B., Norkko, A., and Merila, J. (2017). Extraordinarily rapid speciation in a marine fish. Proc. Natl. Acad. Sci. U.S.A. 114, 6074–6079. doi: 10.1073/pnas.1615109114
Motohashi, E., Yoshihara, T., Doi, H., and Ando, H. (2010). Aggregating behavior of the grass puffer, takifugu niphobles, observed in aquarium during the spawning period. Zool. Sci. 27, 559–564. doi: 10.2108/zsj.27.559
Nakajima, H., Kai, M., Koizumi, K., Tanaka, T., and Machida, M. (2008). Optimal release locations of juvenile ocellate puffer Takifugu rubripes identified by tag and release experiments. Rev. Fisher. Sci. 16, 228–234. doi: 10.1080/10641260701684106
Nielsen, E. E., Hemmer-Hansen, J., Larsen, P. F., and Bekkevold, D. (2009). Population genomics of marine fishes: identifying adaptive variation in space and time. Mol. Ecol. 18, 3128–3150. doi: 10.1111/j.1365-294X.2009.04272.x
Nogue, S., Rull, V., and Vegas-Vilarrubia, T. (2009). Modeling biodiversity loss by global warming on Pantepui, northern South America: projected upward migration and potential habitat loss. Clim. Change 94, 77–85. doi: 10.1007/s10584-009-9554-x
Pei, Y. H., Liu, X. H., and He, H. L. (2017). Interpreting the sea surface temperature warming trend in the Yellow Sea and East China Sea. Sci. China Earth Sci. 60, 1558–1568. doi: 10.1007/s11430-017-9054-5
Pongracz, J. D., Paetkau, D., Branigan, M., and Richardson, E. (2017). Recent Hybridization between a Polar Bear and Grizzly Bears in the Canadian Arctic. Arctic 70, 151–160. doi: 10.14430/arctic4643
Posada, D. (2008). jModelTest: phylogenetic model averaging. Mol. Biol. Evol. 25, 1253–1256. doi: 10.1093/molbev/msn083
Potts, W. M., Henriques, R., Santos, C. V., Munnik, K., Ansorge, I., Dufois, F., et al. (2014). Ocean warming, a rapid distributional shift, and the hybridization of a coastal fish species. Glob. Change Biol. 20, 2765–2777. doi: 10.1111/gcb.12612
Purcell, S., Neale, B., Todd-Brown, K., Thomas, L., Ferreira, M. A., Bender, D., et al. (2007). PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 81, 559–575. doi: 10.1086/519795
Quilodrán, C. S., Nussberger, B., Macdonald, D. W., Montoya-Burgos, J. I., and Currat, M. (2020). Projecting introgression from domestic cats into European wildcats in the Swiss Jura. Evol. Appl. 13, 1–12. doi: 10.1111/eva.12968
Saetre, G. P., Borge, T., Lindell, J., Moum, T., Primmer, C. R., Sheldon, B. C., et al. (2001). Speciation, introgressive hybridization and nonlinear rate of molecular evolution in flycatchers. Mol. Ecol. 10, 737–749. doi: 10.1046/j.1365-294x.2001.01208.x
Salzburger, W., Baric, S., and Sturmbauer, C. (2002). Speciation via introgressive hybridization in East African cichlids? Mol. Ecol. 11, 619–625. doi: 10.1046/j.0962-1083.2001.01438.x
Santini, F., Nguyen, M. T. T., Sorenson, L., Waltzek, T. B., Alfaro, J. W. L., Eastman, J. M., et al. (2013). Do habitat shifts drive diversification in teleost fishes? An example from the pufferfishes (Tetraodontidae). J. Evol. Biol. 26, 1003–1018. doi: 10.1111/jeb.12112
Seehausen, O. (2006). African cichlid fish: a model system in adaptive radiation research. Proc. R. Soc. B Biol. Sci. 273, 1987–1998. doi: 10.1098/rspb.2006.3539
Shahjahan, M., Hamabata, T., Motohashi, E., Doi, H., and Ando, H. (2010). Differential expression of three types of gonadotropin-releasing hormone genes during the spawning season in grass puffer, Takifugu niphobles. Gen. Compar. Endocrinol. 167, 153–163. doi: 10.1016/j.ygcen.2010.01.018
Shi, Y., Zhou, Z., Liu, B., Kong, S., Chen, B., Bai, H., et al. (2020). Construction of a High-density genetic linkage map and QTL mapping for growth-related traits in Takifugu bimaculatus. Mar. Biotechnol. 22, 130–144. doi: 10.1007/s10126-019-09938-2
Simon, A., Arbiol, C., Nielsen, E. E., Couteau, J., Sussarellu, R., Burgeot, T., et al. (2020). Replicated anthropogenic hybridisations reveal parallel patterns of admixture in marine mussels. Evol. Appl. 13:12879. doi: 10.1111/eva.12879
Stamatakis, A. (2014). RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30, 1312–1313. doi: 10.1093/bioinformatics/btu033
Sudhof, T. C. (2013). A molecular machine for neurotransmitter release: synaptotagmin and beyond. Nat. Med. 19, 1227–1231. doi: 10.1038/nm.3338
Sun, Z., Jiang, X., Zhang, H., Zhao, Y., Ren, Y., and Zhou, Q. (2019). Observation on embryonic development of Takifugu niphobles. Mar. Fisher. 41, 65–72. doi: 10.13233/j.cnki.mar.fish.2019.01.008
Takahashi, H., and Takata, K. (2000). Multiple lineages of the mitochondrial DNA introgression from Pungitius pungitius (L.) to Pungitius tymensis (Nikolsky). Canad. J. Fisher. Aquatic Sci. 57, 1814–1833. doi: 10.1139/cjfas-57-9-1814
Takahashi, H., Toyoda, A., Yamazaki, T., Narita, S., Mashiko, T., and Yamazaki, Y. (2017). Asymmetric hybridization and introgression between sibling species of the pufferfish Takifugu that have undergone explosive speciation. Mar. Biol. 158, 1815–1827. doi: 10.1007/s00227-017-3120-2
Tebbich, S., Sterelny, K., and Teschke, I. (2010). The tale of the finch: adaptive radiation and behavioural flexibility. Philos. Transact. R. Soc. Lond. Ser. B Biol. Sci. 365, 1099–1109. doi: 10.1098/rstb.2009.0291
Todesco, M., Pascual, M. A., Owens, G. L., Ostevik, K. L., Moyers, B. T., Hubner, S., et al. (2016). Hybridization and extinction. Evol. Appl. 9, 892–908. doi: 10.1111/eva.12367
Torres-Dowdall, J., Pierotti, M. E. R., Harer, A., Karagic, N., Woltering, J. M., Henning, F., et al. (2017). Rapid and parallel adaptive evolution of the visual system of neotropical midas cichlid fishes. Mol. Biol. Evol. 34, 2469–2485. doi: 10.1093/molbev/msx143
Wirtz, P. (1999). Mother species-father species: unidirectional hybridization in animals with female choice. Anim. Behav. 58, 1–12. doi: 10.1006/anbe.1999.1144
Wu, D., Bacaj, T., Morishita, W., Goswami, D., Arendt, K. L., Xu, W., et al. (2017). Postsynaptic synaptotagmins mediate AMPA receptor exocytosis during LTP. Nature 544, 316–321. doi: 10.1038/nature21720
Wu, X. M., Viveiros, M. M., Eppig, J. J., Bai, Y. C., Fitzpatrick, S. L., and Matzuk, M. M. (2003). Zygote arrest 1 (Zar1) is a novel maternal-effect gene critical for the oocyte-to-embryo transition. Nat. Genet. 33, 187–191. doi: 10.1038/ng1079
Yamahira, K. (1994). Combined effects of tidal and diurnal cycles on spawning of the puffer, Takifugu niphobles (Tetraodontidae). Environ. Biol. Fishes 40, 255–261. doi: 10.1007/Bf00002513
Yamahira, K. (1997). Proximate factors influencing spawning site specificity of the puffer fish Takifugu niphobles. Mar. Ecol. Progr. Ser. 147, 11–19. doi: 10.3354/meps147011
Yamanoue, Y., Miya, M., Matsuura, K., Miyazawa, S., Tsukamoto, N., Doi, H., et al. (2009). Explosive speciation of Takifugu: another use of fugu as a model system for evolutionary biology. Mol. Biol. Evol. 26, 623–629. doi: 10.1093/molbev/msn283
Yang, J., Lee, S. H., Goddard, M. E., and Visscher, P. M. (2011). GCTA: a tool for genome-wide complex trait analysis. Am. J. Hum. Genet. 88, 76–82. doi: 10.1016/j.ajhg.2010.11.011
Yang, Z., and Chen, Y. F. (2008). Differences in reproductive strategies between obscure puffer Takifugu obscurus and ocellated puffer Takifugu ocellatus during their spawning migration. J. Appl. Ichthyol. 24, 569–573. doi: 10.1111/j.1439-0426.2008.01071.x
Yasuji, M., Nobuo, S., Yasuhiro, T., Osame, T., and Keiichi, M. (1991). Occurrence of natural hybrid between pufferfishes, Takifugu xanthopterus and T. vermicularis, in Ariake Bay, Kyushu, Japan. Nipp. Suis. Gakkai. 57, 1247–1255. doi: 10.2331/suisan.57.1247
Yin, L., Gong, X., and Bao, B. (2016). Occurrence of hybrid among Takifugu in East China Sea. J. Shang. Ocean Univ. 25, 359–366. doi: 10.12024/jsou.20150401409
Yoo, G. Y., Lee, T. H., Gil, H. W., Lim, S. G., and Park, I. S. (2018). Cytogenetic analysis of hybrids and hybrid triploids between the river puffer, Takifugu obscurus, and the tiger puffer, Takifugu rubripes. Aquacult. Res. 49, 637–650. doi: 10.1111/are.13493
Zhang, C., Dong, S. S., Xu, J. Y., He, W. M., and Yang, T. L. (2019). PopLDdecay: a fast and effective tool for linkage disequilibrium decay analysis based on variant call format files. Bioinformatics 35, 1786–1788. doi: 10.1093/bioinformatics/bty875
Zhang, G. Y., Shi, Y. H., Zhu, Y. Z., Liu, J. Z., and Zang, W. L. (2010). Effects of salinity on embryos and larvae of tawny puffer Takifugu flavidus. Aquaculture 302, 71–75. doi: 10.1016/j.aquaculture.2010.02.005
Zhang, H., Hou, J., Liu, H., Zhu, H., Xu, G., and Xu, J. (2020). Adaptive evolution of low-salinity tolerance and hypoosmotic regulation in a euryhaline teleost, Takifugu obscurus. Mar. Biol. 167:90. doi: 10.1007/s00227-020-03705-x
Zhang, Y. B., and He, S. P. (2008). Investigations into the perplexing interrelationship of the Genus Takifugu Abe, 1949 (Tetraodontiformes, Tetraodontidae). Chin. Sci. Bul. 53, 233–244. doi: 10.1007/s11434-008-0066-2
Zhou, Y., Xiao, S., Lin, G., Chen, D., Cen, W., Xue, T., et al. (2019). Chromosome genome assembly and annotation of the yellowbelly pufferfish with PacBio and Hi-C sequencing data. Sci. Data 6:267. doi: 10.1038/s41597-019-0279-z
Zhou, Z., Liu, B., Chen, B., Shi, Y., Pu, F., Bai, H., et al. (2019). The sequence and de novo assembly of Takifugu bimaculatus genome using PacBio and Hi-C technologies. Sci. Data 6:187. doi: 10.1038/s41597-019-0195-2
Zhou, Z. X., Han, K. H., Wu, Y. D., Bai, H. Q., Ke, Q. Z., Pu, F., et al. (2019). Genome-Wide association study of growth and body-shape-related traits in large Yellow croaker (Larimichthys crocea) using ddRAD sequencing. Mar. Biotechnol. 21, 655–670. doi: 10.1007/s10126-019-09910-0
Zhu, M. Y., Yamakawa, T., Yoda, M., Yasuda, T., Kurota, H., Ohshimo, S., et al. (2017). Using a multivariate auto-regressive state-space (MARSS) model to evaluate fishery resources abundance in the East China Sea, based on spatial distributional information. Fisher. Sci. 83, 499–513. doi: 10.1007/s12562-017-1089-x
Keywords: Takifugu species, phylogenetic, introgression, hybridization, adaptive evolution
Citation: Liu B, Zhou Z, Bai Y, Yang J, Shi Y, Pu F and Xu P (2021) Genome-Scale Phylogenetic and Population Genetic Studies Provide Insight Into Introgression and Adaptive Evolution of Takifugu Species in East Asia. Front. Genet. 12:625600. doi: 10.3389/fgene.2021.625600
Received: 03 November 2020; Accepted: 07 January 2021;
Published: 22 February 2021.
Edited by:
Xu Wang, Auburn University, United StatesReviewed by:
Baofeng Su, Auburn University, United StatesHuitong Shi, Stanford University, United States
Copyright © 2021 Liu, Zhou, Bai, Yang, Shi, Pu and Xu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Peng Xu, eHVwZW5nNzdAeG11LmVkdS5jbg==; eHVwZW5nNzdAZ21haWwuY29t
†These authors have contributed equally to this work
 Bo Liu1†
Bo Liu1† Zhixiong Zhou
Zhixiong Zhou Fei Pu
Fei Pu Peng Xu
Peng Xu