
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Genet. , 29 April 2021
Sec. Computational Genomics
Volume 12 - 2021 | https://doi.org/10.3389/fgene.2021.616110
This article is part of the Research Topic Computational tools in inferring cancer tissue-of-origin and molecular classification towards personalized cancer therapy, Volume II View all 13 articles
 Jie Huang1†
Jie Huang1† Xiang Wang1†
Xiang Wang1† Xue Zhang1
Xue Zhang1 Weijie Chen1
Weijie Chen1 Lijuan Luan1
Lijuan Luan1 Qi Song1
Qi Song1 Hao Wang2
Hao Wang2 Jia Liu1
Jia Liu1 Lei Xu1
Lei Xu1 Yifan Xu1
Yifan Xu1 Licheng Shen1
Licheng Shen1 Lijie Tan2
Lijie Tan2 Dongxian Jiang1
Dongxian Jiang1 Jieakesu Su1*
Jieakesu Su1* Yingyong Hou1,3*
Yingyong Hou1,3*In the present study, we aimed to investigate the clinical and prognostic values of CDK4 amplification and improve the risk stratification in patients with esophageal squamous cell carcinoma. CDK4 amplification was analyzed by fluorescence in situ hybridization using tissue microarray consisting of representative tissues of 520 patients with esophageal squamous cell carcinoma, and its correlation with clinicopathological features and clinical outcomes were evaluated. CDK4 amplification was found in 8.5% (44/520) of patients with esophageal squamous cell carcinoma. CDK4 amplification was negatively correlated with disease progression (P = 0.003) and death (P = 0.006). Patients with CDK4 amplification showed a significantly better disease-free survival (P = 0.016) and overall survival (P = 0.023) compared with those patients without CDK4 amplification. When patients were further stratified into I–II stage groups and III–IV stage groups, CDK4 amplification was significantly associated with both better disease-free survival (P = 0.023) and overall survival (P = 0.025) in the I–II stage group rather than the III–IV stage group. On univariate and multivariate analysis, invasive depth and CDK4 amplification were associated with disease-free survival and overall survival. Taken together, CDK4 amplification was identified as an independent prognostic factor for survival, which could be incorporated into the tumor–node–metastasis staging system to refine risk stratification of patients with esophageal squamous cell carcinoma.
Esophageal cancer (EC) is a lethal digestive tract malignancy with a poor prognosis, and an increasing incidence and mortality rate worldwide (Malhotra et al., 2017). There are two main histological types of EC: esophageal squamous cell cancer (ESCC) and esophageal adenocarcinoma (EAC), which have significant differences in pathogenesis, epidemiology, and risk factors (Rustgi and El-Serag, 2014; Arnold et al., 2015). ESCC usually occurs in flat cells lining the upper two thirds of the esophagus, predominantly in Africa and eastern Asia (especially in China), and smoking is the main risk factor (Lin et al., 2013; Rustgi and El-Serag, 2014), while EC mostly originates from the Barrett mucosa in the lower third of the esophagus and is prevalent in many developed countries (Edgren et al., 2013; Rustgi and El-Serag, 2014; Arnold et al., 2016). In addition to environmental and external factors, genetic factors may also contribute to the development of a specific type of EC. Recently, whole-genome sequencing and genome-wide association studies have been undertaken to identify EC-related genetic alterations (Gao et al., 2014; Lin et al., 2014; Cancer Genome Atlas Research Network, Analysis Working Group: Asan University, Bc Cancer Agency, Brigham and Women’s Hospital, Broad Institute, Brown University, et al., 2017).
Cell cycle dysregulation induced by abnormal genetic alterations (mutations, deletions, or amplifications) occur frequently in human malignancies (Hanahan and Weinberg, 2011; Gampenrieder et al., 2016). The deregulation of the cyclin D1–CDK4/6–Rb pathway, which will trigger loss of cell cycle control, is one of the hallmarks of carcinogenesis (Asghar et al., 2015). The cyclin-dependent kinase 4 (CDK4) gene located in the chromosomal region 12q14.1 might have oncogenic potential similar to other G1 regulatory genes (Haas et al., 1997). CDK4 is initially identified as a catalytic subunit present in the CDK/cyclin D complex in the G1 phase of the cell cycle (Matsushime et al., 1992). CDK4 coupled with cyclin D1 (CCND1) phosphorylates the retinoblastoma protein 1 (RB1), which leads to the release of the transcription factor EF2 and subsequently enables the cell cycle progress from G1 to S phase (Harbour et al., 1999). Alterations of these key components have been implicated in the pathogenesis of multiple tumor types (An et al., 1999). Overexpression of CDK4 could induce uncontrolled cell growth and eventually lead to tumorigenesis; moreover, amplification of the CDK4 gene, have been found in various cancers (Lee et al., 2014).
The perturbed cell cycle regulation pathway in ESCC mainly exhibited genetic alterations in the G1/S transition control, including mutations or deletions of TP53, RB1, CDKN2A, CHEK1, and CHEK2, and amplifications of CDK4, CCND1, CDK6, and MDM2 (Song et al., 2014). Alterations of these genes, such as inactivation of RB1 and CDKN2A and amplification of CCND1, CDK6, and MDM2 have been well documented in ESCC (Huang et al., 2007; Baba et al., 2014; Jiang et al., 2020). To date, the prognostic significance of CDK4 amplification in ESCC has not been described before. In this article, we describe CDK4 amplification in ESCC by fluorescence in situ hybridization (FISH) and meticulously investigated the clinical and prognostic values of CDK4 amplification in patients with ESCC to improve the risk stratification.
This study retrospectively enrolled 520 ESCC patients who had undergone surgical resection in the Department of Thorax Surgery, Zhongshan Hospital, Fudan University (Shanghai, China), between January 2007 and November 2010. Patients who received preoperative antitumor therapy, including neoadjuvant therapy, chemotherapy, and radiotherapy or died within 3 months were excluded from the current study. Ethical approval was granted by the Human Research Ethics Committee of Zhongshan Hospital, Fudan University. Signed informed consent for the acquisition and use of patient tissue specimens and clinical data was obtained from each patient.
All specimens were reassessed independently by two pathologists using hematoxylin and eosin (HE)-stained sections to determine the tumor grade, differentiation, invasion depth, lymph node metastasis, vessel and nerve involvement, and disease stage, according to the American Joint Committee on Cancer guidelines for tumor–node–metastasis (TNM) classification (eighth edition). Patients’ clinicopathological characteristics such as gender, age, smoking, tumor location, and clinical stage were collected from medical records. After surgery, patients were followed up with endoscopy and computed tomographic scan of the thorax and abdomen every 3 months for the first year, every 6 months for the second year, and every 6–12 months thereafter. Follow-up data of those patients who did not have themselves examined in our hospital were obtained by telephone.
Tissue microarrays (TMAs) containing tumor tissues of the 520 patients under study were constructed as previously described (Shi et al., 2013). Briefly, the representative areas of 2 mm wide and 6 mm long with rich tumor cells were selected by two experienced pathologists according to HE-stained slides. The corresponding regions on archived formalin-fixed, paraffin-embedded (FFPE) tissue blocks were extracted, vertically planted into the recipient TMA blocks and then aggregated on the instrument.
Dual-color FISH assay was conducted on the TMA sections of 5 μm thickness using CDK4-specific probe (Spectrum orange) together with a centromere-specific probe (Spectrum green) for chromosome 12 (CEP12) (Empire Genomics, Buffalo, NY) for assessment of CDK4 amplification according to established laboratory protocol, as previously described (Zhang et al., 2014). FISH copy number evaluation was performed by two experienced pathologists blinded to patients’ clinicopathologic characteristics under a fluorescence microscope (BX43; Olympus, Tokyo, Japan) equipped with a DAPI/green/orange triple band pass filter and a Microscope Digital Camera (DP73; Olympus). At least 100 tumor cell nuclei of each ESCC sample were analyzed by counting orange signals for CDK4 and green signals for CEP12 under an oil microscope with a magnification of 1,000 times. Overlapping tumor nuclei were excluded from evaluation to avoid false-positive scoring. Then the average number of CDK4 and CEP12 signals and the ratio of CDK4/CEP12 were calculated for each case. Amplification of CDK4 was defined as a CDK4/CEP12 ratio ≥2.0 or an average copy number of CDK4 signals/tumor cell nucleus ≥5.0 or percentage of tumor cells containing large clusters of CDK4 signal ≥10%, respectively, based on previously reported modified scoring algorithms for HER2 and c-MYC (Wolff et al., 2007; Huang et al., 2019).
All the statistical analyses were carried out using SPSS 20.0 (SPSS Inc., Chicago, IL, United States). All P-values were two sided, and differences were considered statistically significant values of P < 0.05. Disease-free survival (DFS) was defined as the interval between surgical resection and recurrence, metastasis, or death from any cause. Overall survival (OS) was defined as the interval from date of curative surgery until death or last follow-up date. Correlations between CDK4 amplification and clinicopathologic variables were analyzed using the Fisher exact test or Pearson χ2 test. The Kaplan–Meier method with log-rank test was applied to calculate the cumulative survival proportion for OS and DFS by CDK4 amplification level and to determine if there were any significant differences between the survival curves. The Cox proportional hazard regression model was used to carry out the univariate and multivariate regression analyses, and the hazard ratio (HR) and 95% confidence intervals (CI) were determined.
Detailed clinicopathological characteristics of the study cohort including 520 ESCC specimens obtained for this study are summarized in Table 1. The median age of this cohort was 61 years (range, 34–83 years), of which 81.7% were men and 38.7% were smokers. By anatomic site, 44.0% of tumors were in the middle esophagus, whereas 51.2% of the tumors were in the upper and lower esophagus with a median tumor size of 3 cm (range, 0.3–10 cm). The tumor differentiation was defined as grade I in 20 (3.8%) patients, II in 292 (56.2%) patients, and III in 208 (40.0%) patients. Vessel and nerve invasions were presented in 111 (21.3%) and 177 (34.0%) tumors, respectively. Meanwhile, lymph node metastasis was observed in 238 (45.8%) of the patients. The depth of invasion was also evaluated. 15 (2.9%) cases were confined to the mucosa, 38 (7.3%) were in the submucosa, 115 (22.1%) were in the muscular layer, and 352 (67.7%) were beyond the muscular layer. Among these patients with ESCC, clinical stage was classified as I to II and III to IVb in 290 (55.8%) and 230 (44.2%) cases, respectively, according to the American Joint Committee on Cancer Staging Manual (eighth edition).
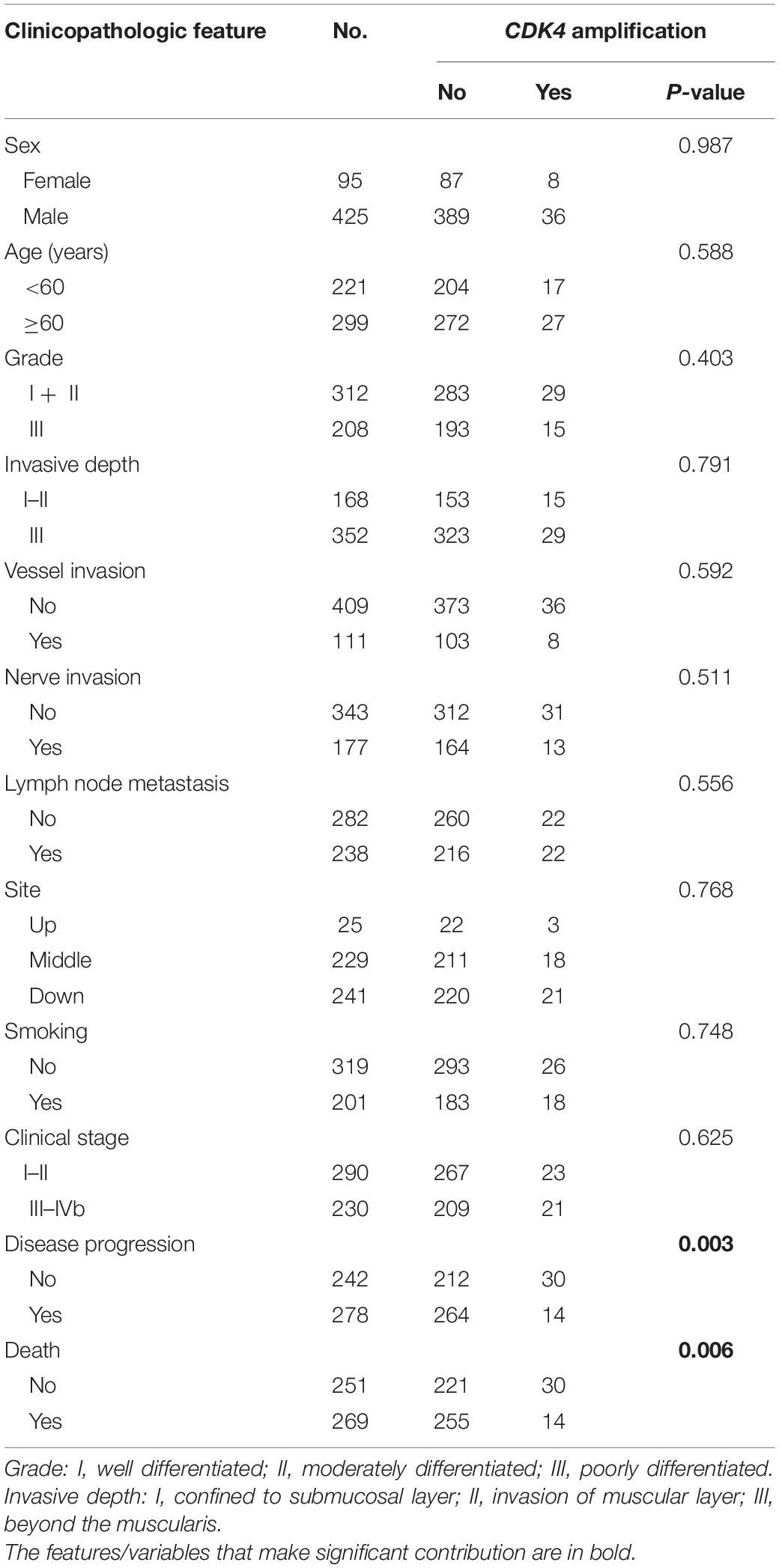
Table 1. Correlation between CDK4 amplification and clinicopathological features in full cohort of patients with ESCC.
All the patients were classified into two groups by using prespecified criteria for CDK4 amplification based on previous studies (Wolff et al., 2007; Huang et al., 2019). CDK4 amplification (a CDK4/CEP12 ratio ≥2.0 or an average copy number of CDK4 signals/tumor cell nucleus ≥5.0 or percentage of tumor cells containing large clusters of CDK4 signal ≥10%) was found in 8.5% (44 of 520) of patients (Figures 1A,B), and other patients (91.5%, 476 of 520) showed non-amplification (low polysomy or disomy) (Figures 1C,D). The correlations between CDK4 amplification and clinicopathological features are shown in Table 1. CDK4 amplification status significantly correlated with disease progression (P = 0.003) and death (P = 0.006). There was no significant difference between CDK4 amplification and CDK4 non-amplification group regarding sex (P = 0.987), age (P = 0.588), grade (P = 0.403), invasive depth (P = 0.791), vessel (P = 0.592) and nerve invasions (P = 0.511), lymph node metastasis (P = 0.556), anatomic site (P = 0.768), smoking (P = 0.748), and clinical stage (P = 0.625).
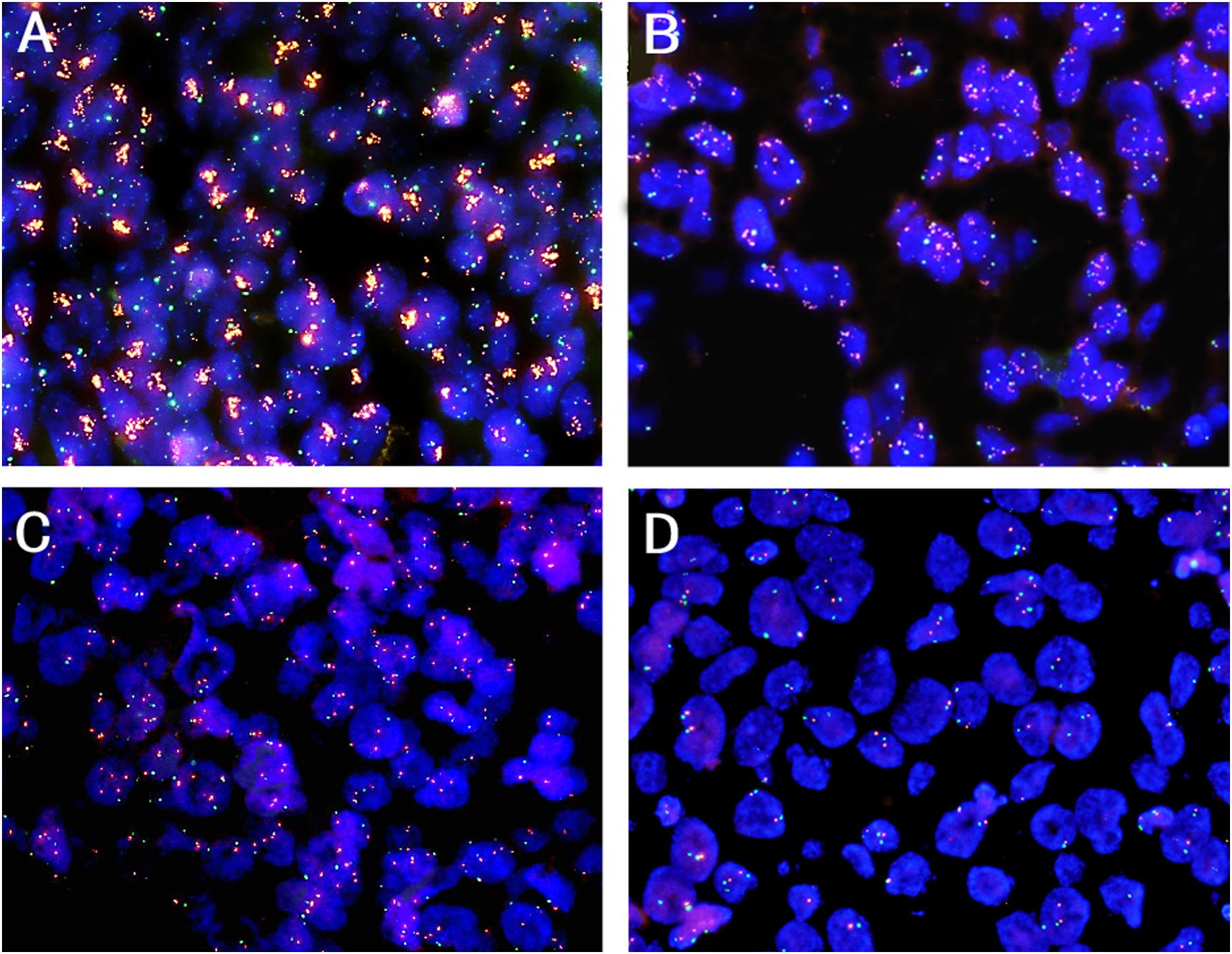
Figure 1. Representative patterns of CDK4 gene (orange color) and CEP12 (green color) copy number status by FISH (original magnification ×1,000). (A) CDK4 amplification, a CDK4/CEP12 ratio ≥2.0; (B) CDK4 amplification, an average copy number of CDK4 signals/tumor cell nucleus ≥5.0; (C) CDK4 non-amplification, low polysomy; (D) CDK4 non-amplification, disomy.
The 5-year DFS and OS rates for all patients were 32.1% and 32.9%, respectively, with a median follow-up period of 35.5 months (range, 3–102 months). Mean and median times to DFS were 41.6 and 31.0 months, while to OS were 44.9 and 35.5 months, respectively. Instances of disease progression (278), including 106 local recurrences and 172 lymph node or distant metastasis, were documented, and 277 patients (53.3%) died during the follow-up, in which 269 patients (51.7%) died of EC. To further explore the prognostic significance of CDK4 amplification and clinical outcomes, Kaplan–Meier analysis with log-rank test was used to compare differences between subgroups. The Kaplan–Meier curves revealed that the CDK4 amplification group with a median DFS and OS of 42.5 and 46.0 months, respectively, gained significant survival benefit compared with the group without CDK4 amplification (median DFS, 30.0 months, P = 0.016; median OS, 35.0 months, P = 0.023) (Figure 2). Univariate analysis of prognostic significance revealed that grade, invasive depth, vessel invasion, nerve invasion, lymph node metastasis, clinical stage, and CDK4 amplification were significantly associated with DFS and OS. In the multivariate analysis, invasive depth (P = 0.006, HR: 1.560, 95% CI: 1.133–2.149 for DFS; P = 0.008, HR: 1.542, 95% CI: 1.119–2.125 for OS) and CDK4 amplification (P = 0.015, HR: 0.512, 95% CI: 0.299–0.877 for DFS; P = 0.021, HR: 0.530, 95% CI: 0.309–0.908 for OS) were associated with DFS and OS (Table 2).
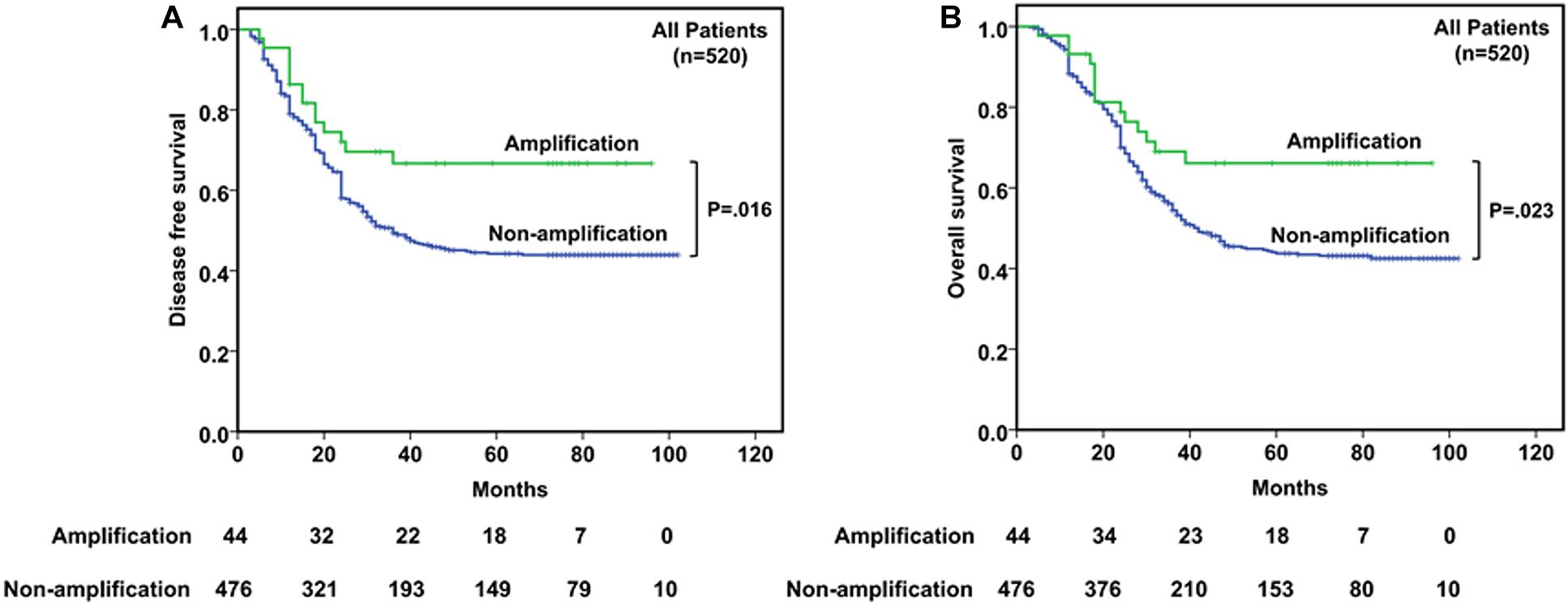
Figure 2. Kaplan–Meier curves of disease-free survival (DFS) (A) and overall survival (OS) (B) according to CDK4 amplification status in 520 esophageal squamous cell cancer (ESCC) patients.
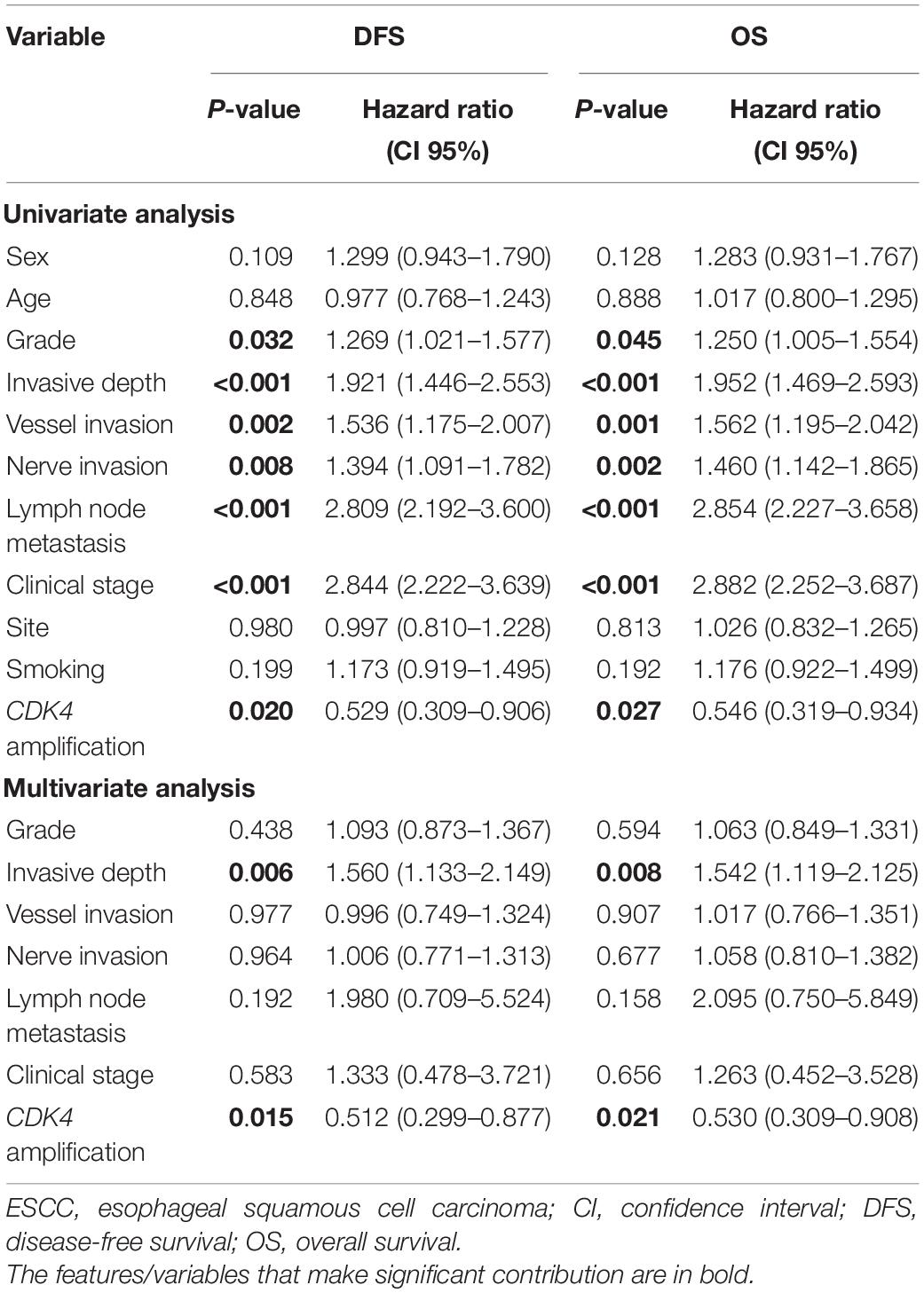
Table 2. Univariate and multivariate survival analyses for DFS and OS in full cohort of patients with ESCC.
In stages I–II patients (n = 290, Figures 3A,B), CDK4 amplification was significantly associated with better DFS (P = 0.023) and OS (P = 0.025). Among the 23 patients with CDK4 amplification, a better prognosis was observed, with a median DFS and OS being both 73.0 months compared with 45.0 and 47.0 months for 267 patients without CDK4 amplification. As to the stages III–IV patients (n = 230, Figures 3C,D), CDK4 amplification did not play the prognostic role whether in DFS (P = 0.144) or in OS (P = 0.211), since the median DFS and OS were 18.0 and 25.0 months, respectively, in 21 patients with CDK4 amplification, whereas it was 20.0 and 25.0 months, respectively, for 209 patients without CDK4 amplification.
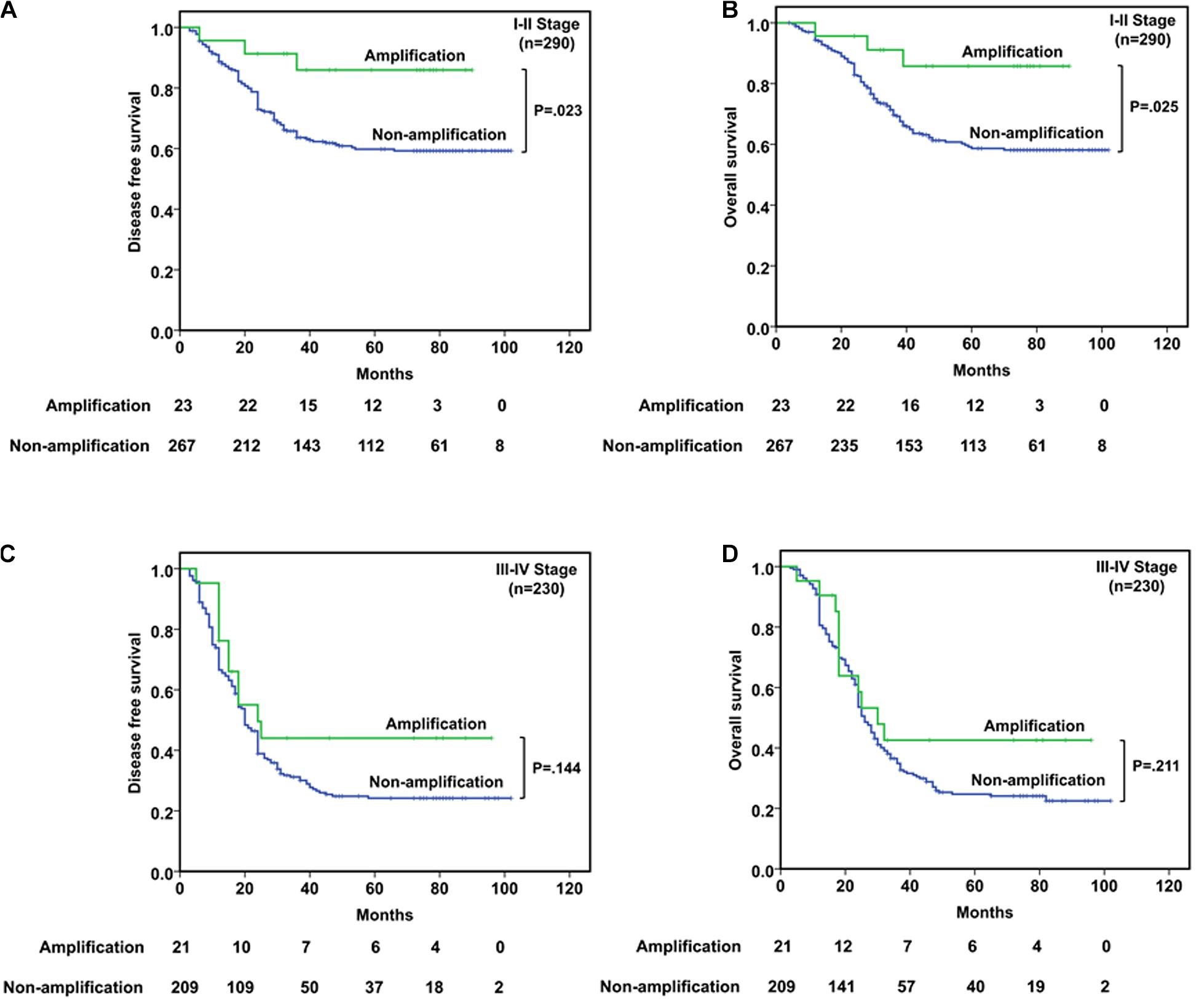
Figure 3. Survival analyses based on clinical stage of ESCC patients. (A,B) In stages I–II patients, CDK4 amplification was significantly associated with better DFS (P = 0.023) and OS (P = 0.025). (C,D) In stages III–IV patients, CDK4 amplification could not predict the prognosis in DFS (P = 0.144) or OS (P = 0.211).
Prognosis prediction and treatment guidance for ESCC are currently based on the TNM staging system, which provides prognostic information, and it will continue to be the most commonly applied approach for a fairly long time (Rustgi and El-Serag, 2014). However, patients with the same TNM stage may display different molecular phenotypes and prognoses. Many non-anatomic prognostic factors, especially genetic and molecular markers critical in carcinogenesis and cancer progression, are also found to have great significance in patient prognosis (Cao et al., 2014; Lin et al., 2017; Mei et al., 2017; Wang et al., 2017; Bi et al., 2020). Therefore, it is of great importance to identify accurate biological markers for the prognosis of ESCC, which may help subdivide patients at the same stage into different groups according to their prognosis. A better understanding of patient prognosis would help guide more personalized treatment for ESCC patients after curative surgery.
Aberrant CDK4 amplification in malignant tissues has been reported to be involved in the development and progression of various cancers including liposarcoma (Creytens et al., 2015), glioblastomas (Schmidt et al., 1994), breast cancer (Piezzo et al., 2020), ovarian cancer (Masciullo et al., 1997), and melanoma (Muthusamy et al., 2006) through the cyclin D1–CDK4/6–Rb pathway. Ricciotti et al. (2017) performed a cut point analysis of the prognostic significance of CDK4 amplification in patients with dedifferentiated liposarcoma by comparison of Kaplan–Meier survival curves using log rank tests. The study showed that CDK4 amplification was associated with decreased DFS (P = 0.0169) and disease-specific survival (DSS) (P = 0.0140). Saada-Bouzid et al. (2015) also demonstrated that CDK4 amplification was significantly associated with shorter recurrence-free survival, and overall survival in dedifferentiated liposarcoma patients. Altogether, the amplification of CDK4 appears to be a negative event in liposarcoma. In glioblastoma patients, Fischer et al. (2010) reported that lack of amplification of CDK4 was recognized to be associated with a significant longer survival time.
In the present study, we investigated CDK4 amplification and its value in the prediction of survival in patients with ESCC. The correlation between CDK4 amplification and the clinicopathological parameters of ESCC patients was also analyzed. Different from a singular criterion only using CDK4/CEP12 ratio or CDK4 copy numbers, we applied a more sophisticated CDK4 FISH criterion considering percentage of CDK4 clusters at the same time. Patients with CDK4 amplification and non-amplification account for 8.5% (n = 44) and 91.5% (n = 476) of all the 520 ESCC patients, respectively. CDK4 amplification rate (8.5%) determined by FISH analysis in our study is comparative with that of a previous study obtained by high-throughput sequencing methods (Song et al., 2014). There was no significant difference between CDK4 amplification and CDK4 non-amplification group regarding sex, age, grade, invasive depth, vessel and nerve invasions, lymph node metastasis, anatomic site, smoking, and clinical stage, which is in line with the conclusion that no significant associations were found between CDK4 gene amplification and patient’s age, tumor size, and lymph node status in breast cancer (An et al., 1999).
Although there was no statistical significance, CDK4 gene amplification was less common in tumors with higher histological grade. Moreover, it is worth noting that CDK4 amplification had a significant negative correlation with disease progression (P = 0.003) and death (P = 0.006) (Table 1). CDK4 seems to be negatively correlated with some indicators indicating poor prognosis in ESCC. Interestingly, different from the prognosis value of CDK4 amplification in dedifferentiated liposarcoma and glioblastoma patients, we demonstrated that CDK4 amplification was associated with a better DFS (P = 0.016) and OS (P = 0.023) (Figure 2). Furthermore, CDK4 amplification was not a common genetic alteration but proved to be an independent prognostic marker in patients with ESCC (Table 2). The results of this study seem to be opposite to the prognosis of other tumor types. This may be as a result of the complexity of the gene regulation process in ESCC. The occurrence and development of ESCC is a multistage and multifactor process, which involves the interaction of multiple oncogenes and tumor suppressor genes (Gao et al., 2014; Lin et al., 2014; Cancer Genome Atlas Research Network, Analysis Working Group: Asan University, Bc Cancer Agency, Brigham and Women’s Hospital, Broad Institute, Brown University, et al., 2017). In addition, we speculate that it may be due to the influence of cancer species, and geographical and environmental factors; the causes of different tumors are not the same, leading to the differences in research results. To the best of our knowledge, this study is the first to evaluate the value of CDK4 amplification as a novel candidate prognostic biomarker in patients with ESCC, so it is necessary to further investigate the upstream and downstream genes of CDK4 to clarify its role and elucidate the prognostic utility in ESCC.
Given that clinical stage is an important clinicopathological factor, the prognosis usually varies between patients with different stages. Therefore, we categorized the patients into the I–II stage group and III–IV stage group. In the I–II stage group, CDK4 amplification was significantly associated with both better DFS and OS compared with the non-amplification group. However, this significant correlation was not found in the III–IV stage patients implying that prognostic value of CDK4 amplification is relying on clinical stage (Figure 3). It is suggested that CDK4 may change in the early stage of ESCC and play an important role in the occurrence and development of the disease. With the increase in clinical stage, more and more genes in ESCC are changed (Sudo et al., 2019), and the interaction between genes becomes complex, which affects the role of CDK4.
In summary, we have first proved the prognostic significance of CDK4 amplification as a favorably independent prognostic factor for DFS and OS in Chinese patients with ESCC. Combining CDK4 amplification with the TNM staging system might add more information to better predict the prognosis of ESCC patients.
The original contributions presented in the study are included in the article/supplementary material, further inquiries can be directed to the corresponding author/s.
The studies involving human participants were reviewed and approved by the Human Research Ethics Committee of Zhongshan Hospital, Fudan University. The patients/participants provided their written informed consent to participate in this study.
JH wrote the draft of the manuscript. XW and YH evaluated CDK4 copy numbers. XZ, WC, and LL analyzed the results of the experiment. QS, HW, and JL constructed Tissue microarrays (TMA). LX, YX, and LS were involved in the picture editing. LT and DJ put forward suggestions for improvement. YH and JS revised the manuscript. All authors contributed to the article and approved the submitted version.
This work was supported by the Shanghai Natural Science Foundation of China (No. 18ZR1406800), Xiamen Science and Technology Project of Fujian Province, China (No. 3502Z20184003), National Natural Science Foundation of China (No. 81702372), Shanghai Municipal Commission of Science and Technology (No. 19441904000), Shanghai Municipal Key Clinical Specialty (No. shslczdzk01302), and Shanghai Science and Technology Development Fund (No. 19MC1911000).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
An, H., Beckmann, M., Reifenberger, G., Bender, H., and Niederacher, D. (1999). Gene amplification and overexpression of CDK4 in sporadic breast carcinomas is associated with high tumor cell proliferation. Am. J. Pathol. 154, 113–118. doi: 10.1016/s0002-9440(10)65257-1
Arnold, M., Colquhoun, A., Cook, M. B., Ferlay, J., Forman, D., and Soerjomataram, I. (2016). Obesity and the incidence of upper gastrointestinal cancers: an ecological approach to examine differences across age and sex. Cancer Epidemiol. Biomarkers Prev. 25, 90–97. doi: 10.1158/1055-9965.epi-15-0753
Arnold, M., Soerjomataram, I., Ferlay, J., and Forman, D. (2015). Global incidence of oesophageal cancer by histological subtype in 2012. Gut 64, 381–387. doi: 10.1136/gutjnl-2014-308124
Asghar, U., Witkiewicz, A. K., Turner, N. C., and Knudsen, E. S. (2015). The history and future of targeting cyclin-dependent kinases in cancer therapy. Nat. Rev. Drug Discov. 14, 130–146.
Baba, Y., Watanabe, M., Murata, A., Shigaki, H., Miyake, K., Ishimoto, T., et al. (2014). LINE-1 hypomethylation, DNA copy number alterations, and CDK6 amplification in esophageal squamous cell carcinoma. Clin. Cancer Res. 20, 1114–1124. doi: 10.1158/1078-0432.ccr-13-1645
Bi, Y., Guo, S., Xu, X., Kong, P., Cui, H., Yan, T., et al. (2020). Decreased ZNF750 promotes angiogenesis in a paracrine manner via activating DANCR/miR-4707-3p/FOXC2 axis in esophageal squamous cell carcinoma. Cell Death Dis. 11:296.
Cancer Genome Atlas Research Network, Analysis Working Group: Asan University, Bc Cancer Agency, Brigham and Women’s Hospital, Broad Institute, Brown University, et al. (2017). Integrated genomic characterization of oesophageal carcinoma. Nature 541, 169–175. doi: 10.1038/nature20805
Cao, F., Han, H., Zhang, F., Wang, B., Ma, W., Wang, Y., et al. (2014). HPV infection in esophageal squamous cell carcinoma and its relationship to the prognosis of patients in northern China. Sci. World J. 2014:804738.
Creytens, D., van Gorp, J., Ferdinande, L., Speel, E. J., and Libbrecht, L. (2015). Detection of MDM2/CDK4 amplification in lipomatous soft tissue tumors from formalin-fixed, paraffin-embedded tissue: comparison of multiplex ligation-dependent probe amplification (MLPA) and fluorescence in situ hybridization (FISH). Appl. Immunohistochem. Mol. Morphol. 23, 126–133. doi: 10.1097/pdm.0000000000000041
Edgren, G., Adami, H. O., Weiderpass, E., and Nyren, O. (2013). A global assessment of the oesophageal adenocarcinoma epidemic. Gut 62, 1406–1414. doi: 10.1136/gutjnl-2012-302412
Fischer, U., Leidinger, P., Keller, A., Folarin, A., Ketter, R., Graf, N., et al. (2010). Amplicons on chromosome 12q13-21 in glioblastoma recurrences. Int. J. Cancer 126, 2594–2602.
Gampenrieder, S. P., Rinnerthaler, G., and Greil, R. (2016). CDK4/6 inhibition in luminal breast cancer. Memo 9, 76–81. doi: 10.1007/s12254-016-0268-2
Gao, Y. B., Chen, Z. L., Li, J. G., Hu, X. D., Shi, X. J., Sun, Z. M., et al. (2014). Genetic landscape of esophageal squamous cell carcinoma. Nat. Genet. 46, 1097–1102.
Haas, K., Staller, P., Geisen, C., Bartek, J., Eilers, M., and Moroy, T. (1997). Mutual requirement of CDK4 and Myc in malignant transformation: evidence for cyclin D1/CDK4 and p16INK4A as upstream regulators of Myc. Oncogene 15, 179–192. doi: 10.1038/sj.onc.1201171
Hanahan, D., and Weinberg, R. A. (2011). Hallmarks of cancer: the next generation. Cell 144, 646–674. doi: 10.1016/j.cell.2011.02.013
Harbour, J. W., Luo, R. X., Dei Santi, A., Postigo, A. A., and Dean, D. C. (1999). Cdk phosphorylation triggers sequential intramolecular interactions that progressively block Rb functions as cells move through G1. Cell 98, 859–869. doi: 10.1016/s0092-8674(00)81519-6
Huang, C., Yang, L., Li, Z., Yang, J., Zhao, J., Dehui, X., et al. (2007). Detection of CCND1 amplification using laser capture microdissection coupled with real-time polymerase chain reaction in human esophageal squamous cell carcinoma. Cancer Genet. Cytogenet. 175, 19–25. doi: 10.1016/j.cancergencyto.2007.01.003
Huang, J., Jiang, D., Zhu, T., Wang, Y., Wang, H., Wang, Q., et al. (2019). Prognostic significance of c-MYC amplification in esophageal squamous cell carcinoma. Ann. Thorac. Surg. 107, 436–443. doi: 10.1016/j.athoracsur.2018.07.077
Jiang, D., Chen, L., Huang, J., Wang, H., Song, Q., Shi, P., et al. (2020). Mouse double minute 2 amplification in oesophageal squamous cell carcinoma is associated with better outcome. Histopathology 77, 963–973. doi: 10.1111/his.14208
Lee, S., Park, H., Ha, S. Y., Paik, K. Y., Lee, S. E., Kim, J. M., et al. (2014). CDK4 amplification predicts recurrence of well-differentiated liposarcoma of the abdomen. PLoS One 9:e99452. doi: 10.1371/journal.pone.0099452
Lin, D. C., Hao, J. J., Nagata, Y., Xu, L., Shang, L., Meng, X., et al. (2014). Genomic and molecular characterization of esophageal squamous cell carcinoma. Nat. Genet. 46, 467–473.
Lin, Y., Shen, L. Y., Fu, H., Dong, B., Yang, H. L., Yan, W. P., et al. (2017). P21, COX-2, and E-cadherin are potential prognostic factors for esophageal squamous cell carcinoma. Dis. Esophagus 30, 1–10. doi: 10.1007/978-981-15-4190-2_1
Lin, Y., Totsuka, Y., He, Y., Kikuchi, S., Qiao, Y., Ueda, J., et al. (2013). Epidemiology of esophageal cancer in Japan and China. J. Epidemiol. 23, 233–242. doi: 10.2188/jea.je20120162
Malhotra, G. K., Yanala, U., Ravipati, A., Follet, M., Vijayakumar, M., and Are, C. (2017). Global trends in esophageal cancer. J. Surg. Oncol. 115, 564–579. doi: 10.1002/jso.24592
Masciullo, V., Scambia, G., Marone, M., Giannitelli, C., Ferrandina, G., Bellacosa, A., et al. (1997). Altered expression of cyclin D1 and CDK4 genes in ovarian carcinomas. Int. J. Cancer 74, 390–395. doi: 10.1002/(sici)1097-0215(19970822)74:4<390::aid-ijc5>3.0.co;2-q
Matsushime, H., Ewen, M. E., Strom, D. K., Kato, J. Y., Hanks, S. K., Roussel, M. F., et al. (1992). Identification and properties of an atypical catalytic subunit (p34PSK-J3/cdk4) for mammalian D type G1 cyclins. Cell 71, 323–334. doi: 10.1016/0092-8674(92)90360-o
Mei, L. L., Qiu, Y. T., Zhang, B., and Shi, Z. Z. (2017). MicroRNAs in esophageal squamous cell carcinoma: potential biomarkers and therapeutic targets. Cancer Biomark. 19, 1–9. doi: 10.3233/cbm-160240
Muthusamy, V., Hobbs, C., Nogueira, C., Cordon-Cardo, C., McKee, P. H., Chin, L., et al. (2006). Amplification of CDK4 and MDM2 in malignant melanoma. Genes Chromosomes Cancer 45, 447–454.
Piezzo, M., Cocco, S., Caputo, R., Cianniello, D., Gioia, G. D., Lauro, V. D., et al. (2020). Targeting cell cycle in breast cancer: CDK4/6 inhibitors. Int. J. Mol. Sci. 21:6479.
Ricciotti, R. W., Baraff, A. J., Jour, G., Kyriss, M., Wu, Y., Liu, Y., et al. (2017). High amplification levels of MDM2 and CDK4 correlate with poor outcome in patients with dedifferentiated liposarcoma: a cytogenomic microarray analysis of 47 cases. Cancer Genet. 218-219, 69–80. doi: 10.1016/j.cancergen.2017.09.005
Saada-Bouzid, E., Burel-Vandenbos, F., Ranchere-Vince, D., Birtwisle-Peyrottes, I., Chetaille, B., Bouvier, C., et al. (2015). Prognostic value of HMGA2, CDK4, and JUN amplification in well-differentiated and dedifferentiated liposarcomas. Mod. Pathol. 28, 1404–1414. doi: 10.1038/modpathol.2015.96
Schmidt, E. E., Ichimura, K., Reifenberger, G., and Collins, V. P. (1994). CDKN2 (p16/MTS1) gene deletion or CDK4 amplification occurs in the majority of glioblastomas. Cancer Res. 54, 6321–6324.
Shi, Y., He, D., Hou, Y., Hu, Q., Xu, C., Liu, Y., et al. (2013). An alternative high output tissue microarray technique. Diagn. Pathol. 8:9.
Song, Y., Li, L., Ou, Y., Gao, Z., Li, E., Li, X., et al. (2014). Identification of genomic alterations in oesophageal squamous cell cancer. Nature 509, 91–95.
Sudo, K., Kato, K., Matsuzaki, J., Boku, N., Abe, S., Saito, Y., et al. (2019). Development and validation of an esophageal squamous cell carcinoma detection model by large-scale microRNA profiling. JAMA Netw. Open 2:e194573. doi: 10.1001/jamanetworkopen.2019.4573
Wang, C., Wang, J., Chen, Z., Gao, Y., and He, J. (2017). Immunohistochemical prognostic markers of esophageal squamous cell carcinoma: a systematic review. Chin. J. Cancer 36:65.
Wolff, A. C., Hammond, M. E., Schwartz, J. N., Hagerty, K. L., Allred, D. C., Cote, R. J., et al. (2007). American society of clinical oncology/college of American pathologists guideline recommendations for human epidermal growth factor receptor 2 testing in breast cancer. J. Clin. Oncol. 25, 118–145.
Keywords: esophageal squamous cell carcinoma, CDK4 amplification, clinical stage, prognostic value, fluorescence in situ hybridization
Citation: Huang J, Wang X, Zhang X, Chen W, Luan L, Song Q, Wang H, Liu J, Xu L, Xu Y, Shen L, Tan L, Jiang D, Su J and Hou Y (2021) CDK4 Amplification in Esophageal Squamous Cell Carcinoma Associated With Better Patient Outcome. Front. Genet. 12:616110. doi: 10.3389/fgene.2021.616110
Received: 11 October 2020; Accepted: 26 February 2021;
Published: 29 April 2021.
Edited by:
Ling Kui, Harvard Medical School, United StatesReviewed by:
Yulin Zhang, Shandong University, ChinaCopyright © 2021 Huang, Wang, Zhang, Chen, Luan, Song, Wang, Liu, Xu, Xu, Shen, Tan, Jiang, Su and Hou. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yingyong Hou, aG91eWluZ3lvbmdAYWxpeXVuLmNvbQ==; Jieakesu Su, MTM4MTYzMjcxMzZAMTI2LmNvbQ==
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.