- 1Division of Child and Adolescent Psychiatry, Department of Psychiatry, Severance Hospital, Institute of Behavioral Science in Medicine, Yonsei University College of Medicine, Seoul, South Korea
- 2Department of Psychiatry, Institute of Behavioral Science in Medicine, Yonsei University College of Medicine, Seoul, South Korea
- 3Department of Laboratory Medicine, Yonsei University College of Medicine, Seoul, South Korea
The clinical heterogeneity of autism spectrum disorder (ASD) is closely associated with the diversity of genes related to ASD pathogenesis. With their low effect size, it has been hard to define the role of common variants of genes in ASD phenotype. In this study, we reviewed genetic results and clinical scores widely used for ASD diagnosis to investigate the role of genes in ASD phenotype considering their functions in molecular pathways. Genetic data from next-generation sequencing (NGS) were collected from 94 participants with ASD. We analyzed enrichment of cellular processes and gene ontology using the Database for Annotation, Visualization, and Integrated Discovery (DAVID). We compared clinical characteristics according to genetic functional characteristics. We found 266 genes containing nonsense, frame shift, missense, and splice site mutations. Results from DAVID revealed significant enrichment for “ion channel” with an enrichment score of 8.84. Moreover, ASD participants carrying mutations in ion channel-related genes showed higher total IQ (p = 0.013) and lower repetitive, restricted behavior (RRB)-related scores (p = 0.003) and mannerism subscale of social responsiveness scale scores, compared to other participants. Individuals with variants in ion channel genes showed lower RRB scores, suggesting that ion channel genes might be relatively less associated with RRB pathogenesis. These results contribute to understanding of the role of common variants in ASD and could be important in the development of precision medicine of ASD.
Introduction
Autism spectrum disorder (ASD) is a neurodevelopmental disorder whose essential features include persistent impairment in reciprocal social communication and restricted, repetitive behaviors and interests (American Psychiatric Association and DSM-5 Task Force., 2013). Although ASD is now widely known to the public, with a reported prevalence of 18.5 per 1,000 children aged 8 years in the United States (Maenner et al., 2020), proper diagnosis and treatment are still major challenges for clinicians because of the heterogeneity of the disorder. Regardless of severity, most patients with ASD require suitable therapy considering their individual symptoms (Lord et al., 2018). Among various treatments applied for ASD patients, pharmacotherapy has proven effective in reducing behavioral problems associated with ASD (DeFilippis and Wagner, 2016). Previous studies have reported that over 30% of ASD patients used at least one antipsychotic drug and that the use of medication tends to be higher in ASD children (Rosenberg et al., 2010; Schubart et al., 2014). Pharmacologic treatment in ASD is usually focused on controlling restricted, repetitive behaviors (RRBs), irritability, and aggressive behaviors that are disruptive in behavioral therapy, in social situations, and in daily life (Williamson and Martin, 2012; Fung et al., 2016). Medications are more often prescribed if ASD patients are diagnosed of other comorbid psychiatric illnesses, and antipsychotics are the most frequently prescribed pharmacotherapy in ASD with intellectual disability (Houghton et al., 2017).
The diversity of genes related to ASD pathogenesis appears to be closely associated with the clinical heterogeneity of ASD (Persico and Napolioni, 2013). In addition to ASD-related syndromes or rare chromosomal abnormalities, additive effects from common genetic variants are also known to be related to ASD etiology (Lovato et al., 2019). Moreover, as rare gene variations with a high effect size account only for 10% of idiopathic autism (Persico and Napolioni, 2013), a cumulative effect for common genetic polymorphisms, such as single-nucleotide polymorphism (SNP), with a low effect size are thought to be important in explaining genetic components of ASD (Klei et al., 2012; Gaugler et al., 2014). Meanwhile, recent studies have indicated that common variants could be informative in identifying and diagnosing ASD (Wang and Avillach, 2021) and that cumulative dysfunction of genes by common variants could affect the severity of ASD manifestations (Toma, 2020). Additional research suggests that noncoding variants, as well as single-nucleotide variants and mosaic single-nucleotide variants, are implicated in autism susceptibility (Dias and Walsh, 2020).
However, defining the role of common variants in ASD still faces several obstacles. First, while common variants have been found to be related to ASD etiology in several studies, results have proven difficult to replicate, with an enormous number of genes suspected to be involved in ASD (Lovato et al., 2019). Also, it can be difficult to demonstrate the genetic contribution of a single common gene variant to ASD alone, because ASD shares genetic risks with other psychiatric illnesses, such as schizophrenia, and other neurodevelopmental disorders (Lee et al., 2013). Moreover, stochastic factors during gene expression and environmental factors can also affect the onset of ASD (Geschwind, 2011).
In recent years, next-generation sequencing (NGS), such as whole-genome sequencing, whole-exome sequencing, or clinical exome sequencing, has found use in identifying novel mutations in genes related to ASD (Jiang et al., 2013; Lovato et al., 2019). Most of the genes shown to be associated with ASD can be functionally classified into specific molecular pathways (Sahin and Sur, 2015; Parenti et al., 2020): the pathways include protein synthesis, transcriptional and epigenetic regulation, and synaptic signaling, affecting the functions of neurons and synapses important in neurodevelopment (De Rubeis et al., 2014; Sahin and Sur, 2015). Nevertheless, despite advancements in understanding of the molecular pathology of ASD, it is still unclear how molecular pathway alterations affect ASD phenotypes. For this reason, application of NGS in clinical settings remains limited. Understanding of the linkage between genotypes and ASD phenotypes, however, may help contribute to finally achieving proper diagnosis and predicting prognosis and individualized therapy.
In the present study, we investigated the role of genes in ASD phenotype in consideration of genetic functions in molecular pathways using NGS. We reviewed genetic results and clinical scores clinically used for ASD diagnosis. By excluding rare ASD-related syndromes and rare copy number variants, we only focused on common gene variants. To examine the relationship between ASD phenotype and genotype, we analyzed clinical scores for social function, RRB, and cognitive function in relation to the genetic results.
Materials and Methods
Participants
In total, 197 children who underwent NGS for genetic evaluation were included in this study. All children were diagnosed with ASD by specialized child psychiatrists according to the diagnostic criteria suggested in the Diagnostic and Statistical Manual of Mental Disorders (DSM-5) (American Psychiatric Association and DSM-5 Task Force., 2013). The diagnosis of ASD was confirmed with Autism Diagnostic Interview-Revised (Lord et al., 1994) and Autism Diagnostic Observation Schedule-2 scores (Lord et al., 2000). Only children who showed severe autistic symptoms, morphologic problems, or other comorbidities were recommended for genetic evaluation in our clinical setting. Data were collected by retrospectively reviewing medical records for the children. Information on demographics, clinical symptom scores, genetics, comorbidities, and medications was collected. Among 197 children with ASD, we excluded children from analysis if any of the clinical symptom scores described below were missing. Ninety-six children were excluded before analysis due to insufficient clinical data. Afterward, seven children who were diagnosed with ASD-related syndromes (tuberous sclerosis or Rett syndrome) were additionally excluded from this study. This study was approved by the applicable institutional review boards for research with human subjects at Severance Hospital, Yonsei University College of Medicine, where this study was performed. Written informed consent agreeing to donation of human biologic materials was acquired from the participant’s legal guardian/next of kin.
Clinical Assessments
Autistic characteristics and intellectual function were assessed using the scales and tests listed below. For the assessment of intellectual function, the Korean-Wechsler Intelligence Scale for Children-IV (K-WISC-IV) (Gwak et al., 2011) or the Korean Wechsler Preschool and Primary Scales of Intelligence-IV (K-WPPSI-IV) (Park et al., 2016) was administered depending on the children’s age and ability to perform the test. The Korean-Bayley-III scale was also used for participants who were unable to perform intelligence tests. The Childhood Autism Rating Scale (CARS) was used to distinguish ASD from other developmental disorders and to assess the severity of ASD. A cutoff score of 30 points was applied, and the reliability and validity of the Korean version of CARS have been verified (Shin and Kim, 1998). The Social Communication Questionnaire (SCQ), which is based on the Autism Diagnostic Interview-Revised, was utilized to assess ASD symptoms (Anthony and Catherine, 2003). The Korean version of the SCQ was verified as a reliable and valid screening tool for autism in the Korean population (Kim et al., 2015).
The Social Responsiveness Scale (SRS) is a 65-item questionnaire of social interactions exhibited by children over the past 6 months (Constantino and Gruber, 2012). The test focuses on social impairments in naturalistic social settings and is measured by parents or teachers. It consists of five subscales (social awareness, social cognition, social communication, social motivation, and mannerisms). Each question is scored from 0 to 3 points, and the sum of scores for the social awareness, social cognition, social communication, and social motivation subscales is considered reflective of social communication, a core symptom of ASD in DSM-5. Similarly, the mannerisms subscale represents RRB symptoms, which is also a core symptom of ASD. We previously confirmed the clinical validity of the SRS in Korean children and suggested the relevance of SRS subscales to DSM-5 ASD diagnosis (Cheon et al., 2016). T-scores are used to resolve problems of differences in raw scores by sex or rater (parent or teacher). In this study, we used T-scores of each subscale, as well as total T-scores. T-scores over 75 indicate severe symptoms; T-scores between 60 and 75 are considered indicative of mid-to-moderate severity (Aldridge et al., 2012).
Next-Generation Sequencing
For exome sequencing, the xGen Inherited Diseases Panel (Integrated DNA Technologies, Coralville, IA, United States) including 4,503 candidate genes was used. The genes included in this panel are known to be related to ASD, intellectual disability, and other neurodevelopmental disorders.
The genomic DNA extracted from the children’s blood was used for library preparation and target capture using a custom panel targeting candidate genes. The NextSeq 550Dx System (Illumina, San Diego, CA, United States) was used to perform massively parallel sequencing. With our custom analysis pipeline, quality control and sequence analysis were proceeded, and copy number analysis was performed (Kim et al., 2019). The GRCh37 (hg19) built as the reference sequence was applied for mapping and variant calling while using the Burrows–Wheeler alignment (BWA) tool (version 0.7.12). HaplotypeCaller and MuTect2 in the GATK package (3.8-0) and VarScan2 (2.4.0) were used to identify single-nucleotide variations (SNVs) and insertion and deletions (indels). Online databases including the Human Gene Mutation Database (HGMD), Online Mendelian Inheritance in Man (OMIM), Clinvar, dbSNP, 1000 Genomes, the Exome Aggregation Consortium (ExAC), the Exome Sequencing Project (ESP), and the Korean Reference Genome Database (KRGDB) were used for analyses and variant annotation.
Classification of variants was conducted using a scoring algorithm implemented in the DxSeq Analyzer (Dxome, Seoul, Korea), based on the standards and guidelines established by the American College of Medical Genetics (ACMG) (Richards et al., 2015). We excluded genetic variants classified as benign or likely benign based on ACMG guidelines in NGS clinical reports by physicians in laboratory medicine. Afterward, variants were lined in order of higher probability of pathogenicity according to ACMG guidelines. Among various variants, we selected five variants with the greatest likelihood of being pathogenetic from each patient.
Gene Ontology
Using the result of NGS, we analyzed enrichment of cellular processes and gene ontology using the Database for Annotation, Visualization and Integrated Discovery (DAVID) (Huang et al., 2009; Sherman and Lempicki, 2009). Children were then classified according to genetic characteristics.
Statistical Analysis
Independent t-tests were used to estimate group differences in demographics and clinical scores. The chi-squared test was used for comparing categorical variables. Logistic regression analysis was applied to evaluate the relative risk of a group to another. Statistical significance was defined at p < 0.05. All analyses were performed using the Statistical Package for the Social Sciences software (version 25.0; SPSS Inc., Chicago, IL, United States).
Results
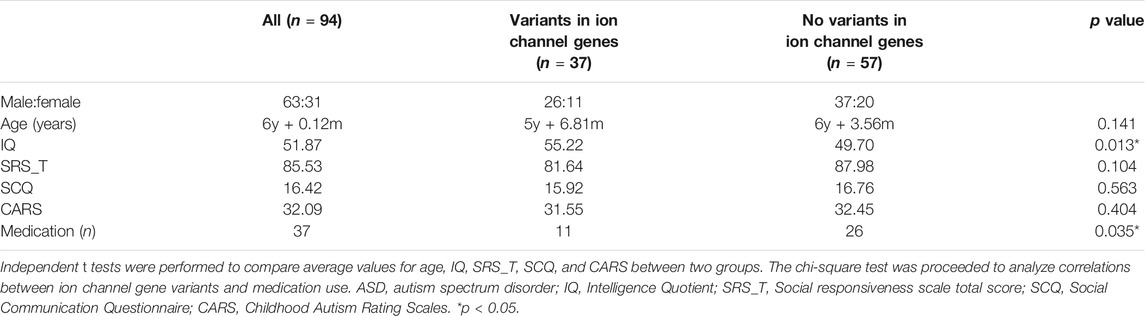
Applying the exclusion criteria, we excluded 94 children with ASD from final analysis (Figure 1). Children with ASD were about 6 years old on average (6.12 years, ranging from 3 years, 6 months to 15 years, 4 months). The male-to-female ratio was about 3:1, and 41 participants were using antipsychotics (aripiprazole or risperidone) because of excessive RRBs or irritability. The average IQ score was 51.87, ranging from 31 to 85. The T-score for total SRS was 85.53 on average, and all subscale T-scores in SRS exceeded 70 on average. CARS scores varied widely, from 21.00 to 51.50 (Table 1).
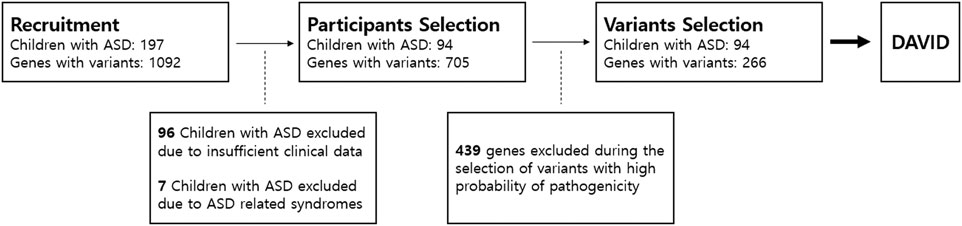
FIGURE 1. Participants and gene selection. We first recruited 197 children with ASD who underwent next-generation sequencing analysis. In total, 1,092 genes were detected with variants in NGS. We only included participants whose clinical assessment was complete with no missing data. After participant selection, we only selected five variants that were most likely pathogenic in each patient based on ACMG guidelines. A total of 439 genes with variants were excluded. Only 266 genes remained from participant and variant selection. The 266 genes were included in enrichment analysis using the Database for Annotation, Visualization and Integrated Discovery (DAVID). ASD, autism spectrum disorder.
Excluding known benign variants, we found that children carried 0 to 34 variants, with 14.44 variants per child on average. We collected up to five SNVs in children with ASD that had the highest probability of being pathogenic according to ACMG guidelines. In total, we collected 266 genes containing nonsense, frame shift, missense, and splice site mutations. More than one-third of genes (91 genes) overlapped at least twice. Variants in TSC2 (12 times), RAI1 (9 times), CHD7 (7 times), and RELN (7 times) were most frequently found among ASD children.
Results from DAVID highlighted significant enrichment for “ion channel” (UP_Keywords), with an enrichment score of 8.84 (corrected p = 1.9xe-13). In functional annotation clustering, 30 genes were involved in the ion channel cluster: CACNG2, CACNA1A, CACNA1C, CACNA1D, CACNA1G, and CACNA1H were associated with calcium voltage-gated channels; SCN1A, SCN10A, SCN2A, SCN3A, SCN7A, SCN9A, and SCN1B were involved in sodium voltage-gated channels; KCNMA1, KCNT1, KCNH2, KCNQ2, KCNQ4, HCN1, HCN2, and HCN4 were related to potassium channels; and CLCN2 and CHRNA4 were involved in chloride channel function. GABRB3, GABRG1, GABRR2, GRIN2B, P2RX7, RYR2, and RYR3 were also highlighted in ion channel functional cluster annotation (Figure 2). Details on variants of ion channel related genes are described in Supplementary Material S1.
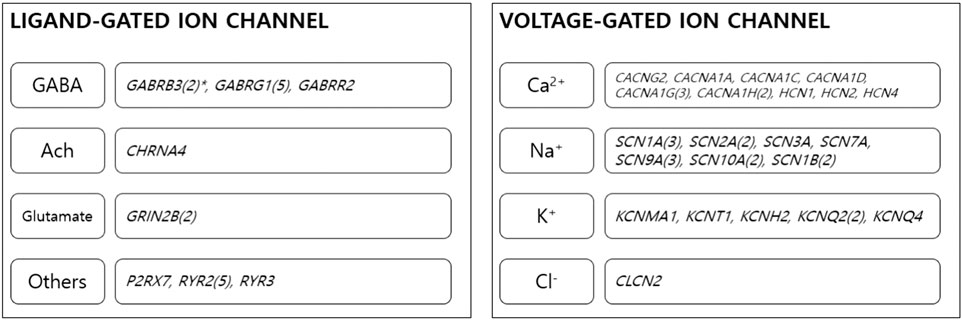
FIGURE 2. Ion channel genes. Thirty genes were classified as ion channel-related genes. The genes were involved in the function of several ion channels. *Numbers in brackets stand for number of overlapping genes among children with ASD. GABA, gamma aminobutyric acid; Ach, acetylcholine.
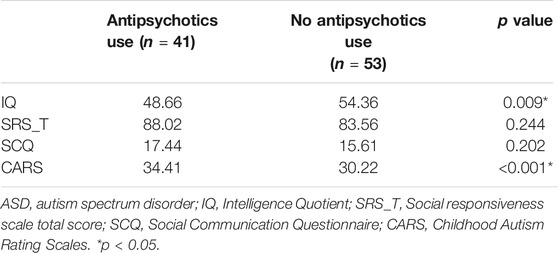
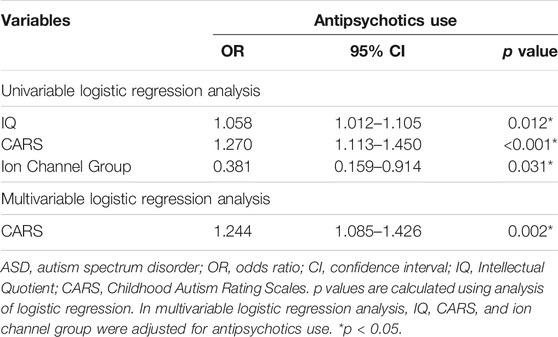
In our data, 37 children with ASD had at least one variant in a gene involved in ion channel function. ASD children carrying variants in genes related to ion channels (ion channel group) showed significantly higher IQ (p = 0.013) and mannerism subscale scores in SRS (p = 0.003) than other children that did not. Other clinical scores were not significantly different between groups (Table 1; Figure 3). Chi-square analysis indicated that the ion channel group and medication use were significantly related (p = 0.035). On the other hand, children with ASD who were on pharmacotherapy showed significantly lower IQ (p = 0.009) and higher CARS scores (p < 0.001). There were no differences in SRS total/subscale scores between participants with and without pharmacotherapy (Table 2). Univariate logistic regression analysis revealed that the ion channel group was at a lower risk of undergoing pharmacotherapy, compared to other children (odds ratio = 0.381, p = 0.031). Logistic regression also revealed that ASD children with lower IQ scores were more prone to use medication (odds ratio = 1.058, p = 0.012) and that higher CARS score were related to medication use (odds ratio = 1.270, p < 0.001). Multivariable logistic regression analysis for medication use in ASD children showed that only a high CARS score was predictive of a greater likelihood of receiving pharmacotherapy in ASD (odds ratio = 1.244, p = 0.002) (Table 3).
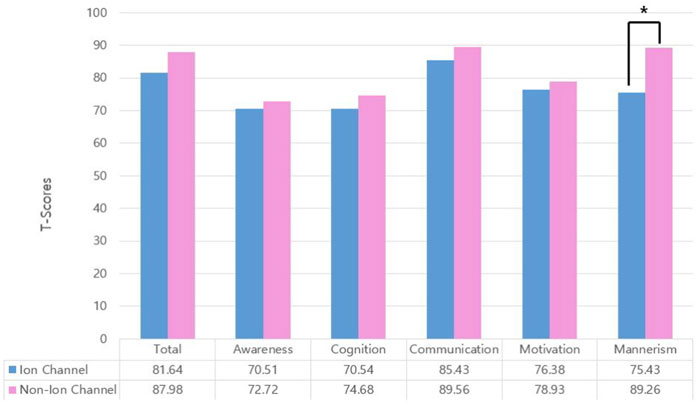
FIGURE 3. Group comparison of Social Responsiveness Scale (SRS) total T-scores and subscale T-scores. Comparison of children with variants in ion channel-related genes versus others. Only mannerism subscale scores were significantly different (*p = 0.003). Total: SRS total T-score; awareness: social awareness subscale T-score; cognition: social cognition subscale T-score; communication: social communication subscale T-score; motivation: social motivation subscale T-score; mannerism: mannerism subscale T-score.
Discussion
In this study, we examined the role of common genetic variants in ASD phenotype by comparing clinical scores in ASD children with different genetic characteristics. Functional cluster annotation revealed significant enrichment for genes involved in ion channels. ASD children with ion channel-related genetic variants presented with significantly higher IQ and less severe RRBs, leading to less exposure to antipsychotics. Our findings suggested that different molecular pathways regulated by related genes are associated with the different aspect of ASD phenotype. Finding the linkage between the molecular pathway and ASD characteristics may contribute to predict prognosis and precision medicine in ASD at the clinical site.
As NGS was proceeded for comparatively severe ASD patients in clinic, the average of IQ was 51.87, and the average of SRS total T-score was within severe criteria. Considering the result of genetic analysis that children with ASD possess more than 10 variants on average, additive genetic burdens of variants might have played a role in severe symptoms (Pizzo et al., 2019).
Impacting brain development in the prenatal period, defects in ion channels in the brain are critical not only in the pathogenesis of epilepsy but also in other neurodevelopmental disorders, including ASD (D'Adamo et al., 2020; Smith and Walsh, 2020). Mutations in ion channel-related genes seem to induce loss of function or gain of function of cell signaling (Imbrici et al., 2016), resulting in impairment of neuronal networks (Sanders et al., 2018). Although it would be difficult to discriminate the specific function of a particular ion channel in ASD pathogenesis, imbalances in excitation and inhibition have been emphasized in the development of neurodevelopmental disorders (Rubenstein and Merzenich, 2003). Additionally, both excitation and inhibition might play roles in complex neuronal circuits (Nelson and Valakh, 2015). In this study, genes associated with several ion channels were included in a functional cluster. Among 30 genes in the ion channel functional cluster, several genes appeared repeatedly in more than two children with ASD. GABRG1, one of the most frequently detected genes in our data, is a gamma-aminobutyric acid (GABA) receptor subunit gene. Although it is not yet clear whether mutated GABRG1 directly affects ASD pathogenesis, studies have highlighted GABA receptor genes as potentially important in ASD (Ma et al., 2005): for example, GABA receptor density was found to be reduced in ASD (Blatt et al., 2001). Interestingly, a GABA gene cluster on human chromosome 4 was shown to be related to vulnerability to social context in youth (Villafuerte et al., 2014; Trucco et al., 2020), suggesting that these genes affect social functioning. Another frequently detected gene in our data was RYR2, which is involved in calcium channel activation in the outer membrane of the endoplasmic reticulum (George et al., 2003). The gene is usually associated with dysregulation of cardiac muscles, but when expressed in the brain, the gene may take part in social functioning and delayed development (Lu and Cantor, 2012). Mutations in calcium voltage-gated channel-related genes (CACNA1A, CACNA1C, CACNA1D, CACNA1G, and CACNA1H) and sodium voltage-gated channel-related genes (SCN1A, SCN2A, SCN3A, SCN7A, SCN9A, SCN10A, and SCN1B) were also detected by our clinical exome sequencing, in line with previous studies reporting genetic associations with ASD (Schmunk and Gargus, 2013). Although it is unclear how much these genes contribute to actual ASD pathogenesis, they do, at the very least, appear to enhance susceptibility to ASD (Schmunk and Gargus, 2013).
One particularly noteworthy finding in this study is that children with variants in ion channel-related genes showed significantly lower RRB scores, suggesting that channelopathy is unlikely to be associated with RRB pathophysiology. In this study, lower RRB scores were significantly related to less use of antipsychotics, an important issue in managing ASD children. Genes related to RRB etiology have been shown to be highly heritable (Ronald et al., 2006) and to be independent of genes affecting social functioning (Ronald et al., 2005). Genetic differences in ASD core symptoms are also supported by differences in RRB symptom severity by sex (Szatmari et al., 2012). Although it remains difficult to explain the genetic differences between social impairment and RRBs, neurobiological factors, such as cortical–basal ganglia pathways, might be closely related (Turner et al., 2006). Considering it is the phenotypic heterogeneity and complex pathophysiology of RRBs in ASD, various genes may be involved (Lewis and Kim, 2009). In addition, neurotransmitter genes, such as dopamine (Lewis and Bodfish, 1998), glutamate (Purcell et al., 2001), serotonin (Di Giovanni et al., 2006), and GABA (Shao et al., 2003) genes, have been shown to be associated with RRBs (Lewis and Kim, 2009). While the cumulative burden of common genetic variants likely affects ASD phenotype the most, we assume that ion channel-related genes may be less connected to RRBs. Although the ion channel group was significantly associated with medication use in our study, the overall severity of ASD represented by CARS scores was more strongly associated with pharmacotherapy of ASD than the RRB score. This might be because RRBs are not the only reason for pharmacotherapy in ASD: emotional problems can also account for antipsychotic use (Stepanova et al., 2017).
There are several limitations to this study. First, the sample size was relatively small, compared to other genetic studies. As this study reviewed medical records retrospectively, we made an effort to include only ASD children with sufficient clinical scores. Also, NGS was only conducted for severe ASD patients at our clinic. As such, the average IQ score was 51.87, and the average total T-score for SRS fell within severe criteria. This limits the generalizability of our results to individuals with less severe ASD. Second, we used both K-WISC-IV and K-WPPSI-IV for intelligence tests because of differences in age at examination, resulting in un-unified subscales of IQ. Third, as reports on genetic results of NGS in Korean individuals with ASD are scarce, we were unable to compare our results within this population. Also, as we only reviewed medical records, we could not compare our results with a healthy control group. Similar studies including healthy controls should be followed to avoid false-positive results. Fourth, considering the preschool age and the low intellectual function, we did not evaluate the comorbidity of attention-deficit/hyperactivity disorder (ADHD) because of the diagnostic instability (Bunte et al., 2014). As ADHD could also possess genetic variants related to the ion channel pathway (Thapar et al., 2016), longitudinal follow-up of comorbidities should be followed. Fifth, we analyzed common variants in genes that seemed to have a low-to-moderate effect size, rather than rare variants. Our selection thereof may put into question if the genetic variants truly affect ASD etiology. Also, considering that genetic analysis has indicated that children with ASD possess more than 10 variants on average, we suspect that additive genetic burden from variants might have played a role in the more severe symptoms seen in our patients (Pizzo et al., 2019). In spite of these weaknesses, we present one possible way in which to interpret the meaning of numerous common variants in ASD. Also, we attempted to discriminate relatively pathogenic variants using ACMG guidelines.
In conclusion, we found several ion channel-related genes to be involved in ASD etiology. Although mutations in ion channel genes are expected to present low-to-moderate effect sizes, they might enhance susceptibility to ASD. Moreover, participants with variants in ion channel genes showed lower RRB scores, suggesting that ion channel genes might not be strongly associated with RRB pathogenesis. These results contribute to helping further understanding of the role of common variants in ASD and could prove to be important in the development of precision medicine for ASD.
Data Availability Statement
The datasets presented in this article are not readily available because it includes the patient’s genetic data for clinical purpose. Requests to access the datasets should be directed to a2FjaGVvbkB5dWhzLmFj.
Ethics Statement
The studies involving human participants were reviewed and approved by Institutional Review Boards for research with human subjects at Severance Hospital, Yonsei University College of Medicine. Written informed consent from the participant’s legal guardian/next of kin was not required to participate in this study in accordance with the national legislation and the institutional requirements.
Author Contributions
Conceptualization, JL, SH, JA, and KC. Data curation, SL, JC, and KC. Formal analysis, SL and JC. Methodology, JL, SH, JA. Supervision, KC. Writing—original draft, LS. Writing—review and editing, JS, SH, JA, SL, JC, and KC.
Funding
This research was supported by the Korea Health Technology R&D Project through the Korea Health Industry Development Institute (KHIDI) grant funded by the Korea government (Ministry Of Health and Welfare (MOHW)) (No. HI12C0021-A120029), the National Research Foundation of Korea (NRF) grant funded by the Korea government (Ministry of Science and Information and Communication Technology (MSIT)) (No. 2016R1A2B4006737 and No. 2021R1A2C2010913) and a faculty research grant of Yonsei University College of Medicine for 2020 (6-2020-0232).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2021.595934/full#supplementary-material
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Aldridge, F. J., Gibbs, V. M., Schmidhofer, K., and Williams, M. (2012). Investigating the Clinical Usefulness of the Social Responsiveness Scale (SRS) in a Tertiary Level, Autism Spectrum Disorder Specific Assessment Clinic. J. Autism Dev. Disord. 42 (2), 294–300. doi:10.1007/s10803-011-1242-9
American Psychiatric Association and DSM-5 Task Force (2013). Diagnostic and Statistical Manual of Mental Disorders : DSM-5. Washington, D.C.: American Psychiatric Association.
Anthony, B., and Catherine, L. (2003). Social Communication Questionnaire (SCQ). Los Angeles: Western Psychological Services.
Blatt, G. J., Fitzgerald, C. M., Guptill, J. T., Booker, A. B., Kemper, T. L., and Bauman, M. L. (2001). Density and Distribution of Hippocampal Neurotransmitter Receptors in Autism: an Autoradiographic Study. J. autism Dev. Disord. 31 (6), 537–543. doi:10.1023/a:1013238809666
Bunte, T. L., Schoemaker, K., Hessen, D. J., van der Heijden, P. G. M., and Matthys, W. (2014). Stability and Change of ODD, CD and ADHD Diagnosis in Referred Preschool Children. J. Abnorm Child. Psychol. 42 (7), 1213–1224. doi:10.1007/s10802-014-9869-6
Cheon, K.-A., Park, J.-I., Koh, Y.-J., Song, J., Hong, H.-J., Kim, Y.-K., et al. (2016). The Social Responsiveness Scale in Relation to DSM IV and DSM5 ASD in Korean Children. Autism Res. 9 (9), 970–980. doi:10.1002/aur.1671
Constantino, J. N., and Gruber, C. P. (2012). Social Responsiveness Scale. Second Edition. Torrance, CA: Western Psychological Services (WPS). (SRS-2): Manual.
D'Adamo, M. C., Liantonio, A., Conte, E., Pessia, M., and Imbrici, P. (2020). Ion Channels Involvement in Neurodevelopmental Disorders. Neuroscience 440, 337. doi:10.1016/j.neuroscience.2020.05.032
De Rubeis, S., He, X., Goldberg, A. P., Poultney, C. S., Samocha, K., Cicek, A. E., et al. (2014). Synaptic, Transcriptional and Chromatin Genes Disrupted in Autism. Nature 515 (7526), 209–215. doi:10.1038/nature13772
DeFilippis, M., and Wagner, K. D. (2016). Treatment of Autism Spectrum Disorder in Children and Adolescents. Psychopharmacol. Bull. 46 (2), 18–41.
Di Giovanni, G., Di Matteo, V., Pierucci, M., Benigno, A., and Esposito, E. (2006). Serotonin Involvement in the Basal Ganglia Pathophysiology: Could the 5-HT2C Receptor Be a New Target for Therapeutic Strategies? Curr. Med. Chem. 13 (25), 3069–3081. doi:10.2174/092986706778521805
Dias, C. M., and Walsh, C. A. (2020). Recent Advances in Understanding the Genetic Architecture of Autism. Annu. Rev. Genom. Hum. Genet. 21, 289–304. doi:10.1146/annurev-genom-121219-082309
Fung, L. K., Mahajan, R., Nozzolillo, A., Bernal, P., Krasner, A., Jo, B., et al. (2016). Pharmacologic Treatment of Severe Irritability and Problem Behaviors in Autism: a Systematic Review and Meta-Analysis. Pediatrics 137 (Suppl. 2), S124–S135. doi:10.1542/peds.2015-2851K
Gaugler, T., Klei, L., Sanders, S. J., Bodea, C. A., Goldberg, A. P., Lee, A. B., et al. (2014). Most Genetic Risk for Autism Resides with Common Variation. Nat. Genet. 46 (8), 881–885. doi:10.1038/ng.3039
George, C. H., Higgs, G. V., and Lai, F. A. (2003). Ryanodine Receptor Mutations Associated with Stress-Induced Ventricular Tachycardia Mediate Increased Calcium Release in Stimulated Cardiomyocytes. Circ. Res. 93 (6), 531–540. doi:10.1161/01.res.0000091335.07574.86
Geschwind, D. H. (2011). Genetics of Autism Spectrum Disorders. Trends Cognitive Sci. 15 (9), 409–416. doi:10.1016/j.tics.2011.07.003
Gwak, G., Oh, S., and Kim, C. (2011). K-WISC-IV (Korean Wechsler Intelligence Scale for Children-IV). Manual for Experts. Seoul: Hakjisa.
Houghton, R., Ong, R. C., and Bolognani, F. (2017). Psychiatric Comorbidities and Use of Psychotropic Medications in People with Autism Spectrum Disorder in the United States. Autism Res. 10 (12), 2037–2047. doi:10.1002/aur.1848
Huang, D. W., Sherman, B. T., and Lempicki, R. A. (2009). Bioinformatics Enrichment Tools: Paths toward the Comprehensive Functional Analysis of Large Gene Lists. Nucleic Acids Res. 37 (1), 1–13. doi:10.1093/nar/gkn923
Imbrici, P., Liantonio, A., Camerino, G. M., De Bellis, M., Camerino, C., Mele, A., et al. (2016). Therapeutic Approaches to Genetic Ion Channelopathies and Perspectives in Drug Discovery. Front. Pharmacol. 7, 121. doi:10.3389/fphar.2016.00121
Jiang, Y.-h., Yuen, R. K. C., Jin, X., Wang, M., Chen, N., Wu, X., et al. (2013). Detection of Clinically Relevant Genetic Variants in Autism Spectrum Disorder by Whole-Genome Sequencing. Am. J. Hum. Genet. 93 (2), 249–263. doi:10.1016/j.ajhg.2013.06.012
Kim, B., Lee, H., Jang, J., Kim, S.-J., Lee, S.-T., Cheong, J.-W., et al. (2019). Targeted Next Generation Sequencing Can Serve as an Alternative to Conventional Tests in Myeloid Neoplasms. PLoS One 14 (3), e0212228. doi:10.1371/journal.pone.0212228
Kim, J.-H., Sunwoo, H.-J., Park, S.-B., Noh, D.-H., Jung, Y. K., Cho, I.-H., et al. (2015). A Validation Study of the Korean Version of Social Communication Questionnaire. J. Korean Acad. Child Adolesc. Psychiatry 26 (3), 197–208. doi:10.5765/jkacap.2015.26.3.197
Klei, L., Sanders, S. J., Murtha, M. T., Hus, V., Lowe, J. K., Willsey, A. J., et al. (2012). Common Genetic Variants, Acting Additively, Are a Major Source of Risk for Autism. Mol. Autism 3 (1), 9–13. doi:10.1186/2040-2392-3-9
Lee, S. H., Ripke, S., Neale, B. M., Faraone, S. V., Purcell, S. M., Perlis, R. H., et al. (2013). Genetic Relationship between Five Psychiatric Disorders Estimated from Genome-wide SNPs. Nat. Genet. 45 (9), 984–994. doi:10.1038/ng.2711
Lewis, M. H., and Bodfish, J. W. (1998). Repetitive Behavior Disorders in Autism. Ment. Retard. Dev. Disabil. Res. Rev. 4 (2), 80–89. doi:10.1002/(sici)1098-2779(1998)4:2<80::aid-mrdd4>3.0.co;2-0
Lewis, M., and Kim, S.-J. (2009). The Pathophysiology of Restricted Repetitive Behavior. J. Neurodev. Disord. 1 (2), 114–132. doi:10.1007/s11689-009-9019-6
Lord, C., Elsabbagh, M., Baird, G., and Veenstra-Vanderweele, J. (2018). Autism Spectrum Disorder. Lancet 392 (10146), 508–520. doi:10.1016/s0140-6736(18)31129-2
Lord, C., Risi, S., Lambrecht, L., Cook, Jr., E. H., Leventhal, B. L., DiLavore, P. C., et al. (2000). The Autism Diagnostic Observation Schedule—Generic: A Standard Measure of Social and Communication Deficits Associated with the Spectrum of Autism. J. Autism Dev. Disord. 30 (3), 205–223. doi:10.1023/a:1005592401947
Lord, C., Rutter, M., and Le Couteur, A. (1994). Autism Diagnostic Interview-Revised: a Revised Version of a Diagnostic Interview for Caregivers of Individuals with Possible Pervasive Developmental Disorders. J. Autism Dev. Disord. 24 (5), 659–685. doi:10.1007/bf02172145
Lovato, D. V., Herai, R. R., Pignatari, G. C., and Beltrão-Braga, P. C. B. (2019). The Relevance of Variants with Unknown Significance for Autism Spectrum Disorder Considering the Genotype-Phenotype Interrelationship. Front. Psychiatry 10, 409. doi:10.3389/fpsyt.2019.00409
Lu, A. T.-H., and Cantor, R. M. (2012). Allowing for Sex Differences Increases Power in a GWAS of Multiplex Autism Families. Mol. Psychiatry 17 (2), 215–222. doi:10.1038/mp.2010.127
Ma, D. Q., Whitehead, P. L., Menold, M. M., Martin, E. R., Ashley-Koch, A. E., Mei, H., et al. (2005). Identification of Significant Association and Gene-Gene Interaction of GABA Receptor Subunit Genes in Autism. Am. J. Hum. Genet. 77 (3), 377–388. doi:10.1086/433195
Maenner, M. J., Shaw, K. A., and Baio, J. (2020). Prevalence of Autism Spectrum Disorder Among Children Aged 8 Years—Autism and Developmental Disabilities Monitoring Network, 11 Sites, United States, 2016. MMWR Surveill. Summ. 69 (4), 1–21. doi:10.15585/mmwr.ss6904a1
Nelson, S. B., and Valakh, V. (2015). Excitatory/inhibitory Balance and Circuit Homeostasis in Autism Spectrum Disorders. Neuron 87 (4), 684–698. doi:10.1016/j.neuron.2015.07.033
Parenti, I., Rabaneda, L. G., Schoen, H., and Novarino, G. (2020). Neurodevelopmental Disorders: From Genetics to Functional Pathways. Trends Neurosci. 43, 608. doi:10.1016/j.tins.2020.05.004
Park, H., Lee, K., Lee, S.-H., and Park, M. (2016). A Study on Standardization of K-WPPSI-IV: Analyses of Reliability and Validity. Korean J. Childcare Educ. 12 (4), 111–130. doi:10.14698/jkcce.2016.12.04.111
Persico, A. M., and Napolioni, V. (2013). Autism Genetics. Behav. Brain Res. 251, 95–112. doi:10.1016/j.bbr.2013.06.012
Pizzo, L., Jensen, M., Polyak, A., Rosenfeld, J. A., Mannik, K., Krishnan, A., et al. (2019). Rare Variants in the Genetic Background Modulate Cognitive and Developmental Phenotypes in Individuals Carrying Disease-Associated Variants. Genet. Med. 21 (4), 816–825. doi:10.1038/s41436-018-0266-3
Purcell, A. E., Jeon, O. H., Zimmerman, A. W., Blue, M. E., and Pevsner, J. (2001). Postmortem Brain Abnormalities of the Glutamate Neurotransmitter System in Autism. Neurology 57 (9), 1618–1628. doi:10.1212/wnl.57.9.1618
Richards, S., Aziz, N., Aziz, N., Bale, S., Bick, D., Das, S., et al. (2015). Standards and Guidelines for the Interpretation of Sequence Variants: a Joint Consensus Recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet. Med. 17 (5), 405–423. doi:10.1038/gim.2015.30
Ronald, A., Happé, F., Bolton, P., Butcher, L. M., Price, T. S., Wheelwright, S., et al. (2006). Genetic Heterogeneity between the Three Components of the Autism Spectrum: a Twin Study. J. Am. Acad. Child Adolesc. Psychiatry 45 (6), 691–699. doi:10.1097/01.chi.0000215325.13058.9d
Ronald, A., Happé, F., and Plomin, R. (2005). The Genetic Relationship between Individual Differences in Social and Nonsocial Behaviours Characteristic of Autism. Dev. Sci. 8 (5), 444–458. doi:10.1111/j.1467-7687.2005.00433.x
Rosenberg, R. E., Mandell, D. S., Farmer, J. E., Law, J. K., Marvin, A. R., and Law, P. A. (2010). Psychotropic Medication Use Among Children with Autism Spectrum Disorders Enrolled in a National Registry, 2007-2008. J. Autism Dev. Disord. 40 (3), 342–351. doi:10.1007/s10803-009-0878-1
Rubenstein, J. L. R., and Merzenich, M. M. (2003). Model of Autism: Increased Ratio of Excitation/inhibition in Key Neural Systems. Genes Brain Behav. 2 (5), 255–267. doi:10.1034/j.1601-183x.2003.00037.x
Sahin, M., and Sur, M. (2015). Genes, Circuits, and Precision Therapies for Autism and Related Neurodevelopmental Disorders. Science 350 (6263), aa3897. doi:10.1126/science.aab3897
Sanders, S. J., Campbell, A. J., Cottrell, J. R., Moller, R. S., Wagner, F. F., Auldridge, A. L., et al. (2018). Progress in Understanding and Treating SCN2A-Mediated Disorders. Trends Neurosci. 41 (7), 442–456. doi:10.1016/j.tins.2018.03.011
Schmunk, G., and Gargus, J. J. (2013). Channelopathy Pathogenesis in Autism Spectrum Disorders. Front. Genet. 4, 222. doi:10.3389/fgene.2013.00222
Schubart, J. R., Camacho, F., and Leslie, D. (2014). Psychotropic Medication Trends Among Children and Adolescents with Autism Spectrum Disorder in the Medicaid Program. Autism 18 (6), 631–637. doi:10.1177/1362361313497537
Shao, Y., Cuccaro, M. L., Hauser, E. R., Raiford, K. L., Menold, M. M., Wolpert, C. M., et al. (2003). Fine Mapping of Autistic Disorder to Chromosome 15q11-Q13 by Use of Phenotypic Subtypes. Am. J. Hum. Genet. 72 (3), 539–548. doi:10.1086/367846
Sherman, B. T., and Lempicki, R. A. (2009). Systematic and Integrative Analysis of Large Gene Lists Using DAVID Bioinformatics Resources. Nat. Protoc. 4 (1), 44. doi:10.1038/nprot.2008.211
Shin, M. S., and Kim, Y.-H. (1998). Standardization Study for the Korean Version of the Childhood Autism Rating Scale: Reliability, Validity and Cut-Off Score. Korean J. Clin. Psychol. 17, 1–15.
Smith, R. S., and Walsh, C. A. (2020). Ion Channel Functions in Early Brain Development. Trends Neurosci. 43 (2), 103–114. doi:10.1016/j.tins.2019.12.004
Stepanova, E., Dowling, S., Phelps, M., and Findling, R. L. (2017). Pharmacotherapy of Emotional and Behavioral Symptoms Associated with Autism Spectrum Disorder in Children and Adolescents. Dialogues Clin. Neurosci. 19 (4), 395–402. doi:10.31887/dcns.2017.19.4/rfindling
Szatmari, P., Liu, X. Q., Goldberg, J., Zwaigenbaum, L., Paterson, A. D., Woodbury‐Smith, M., et al. (2012). Sex Differences in Repetitive Stereotyped Behaviors in Autism: Implications for Genetic Liability. Am. J. Med. Genet. 159B (1), 5–12. doi:10.1002/ajmg.b.31238
Thapar, A., Martin, J., Martin, J., Mick, E., Arias Vásquez, A., Langley, K., et al. (2016). Psychiatric Gene Discoveries Shape Evidence on ADHD's Biology. Mol. Psychiatry 21 (9), 1202–1207. doi:10.1038/mp.2015.163
Toma, C. (2020). Genetic Variation across Phenotypic Severity of Autism. Trends Genet. 36 (4), 228–231. doi:10.1016/j.tig.2020.01.005
Trucco, E. M., Yang, S., Yang, J. J., Zucker, R. A., Li, R., and Buu, A. (2020). Time-varying Effects of GABRG1 and Maladaptive Peer Behavior on Externalizing Behavior from Childhood to Adulthood: Testing Gene × Environment × Development Effects. J. Youth Adolescence 49 (7), 1351–1364. doi:10.1007/s10964-019-01171-3
Turner, K. C., Frost, L., Linsenbardt, D., McIlroy, J. R., and Müller, R.-A. (2006). Atypically Diffuse Functional Connectivity between Caudate Nuclei and Cerebral Cortex in Autism. Behav. Brain Funct. 2 (1), 34. doi:10.1186/1744-9081-2-34
Villafuerte, S., Trucco, E. M., Heitzeg, M. M., Burmeister, M., and Zucker, R. A. (2014). Genetic Variation in GABRA 2 Moderates Peer Influence on Externalizing Behavior in Adolescents. Brain Behav. 4 (6), 833–840. doi:10.1002/brb3.291
Wang, H., and Avillach, P. (2021). Diagnostic Classification and Prognostic Prediction Using Common Genetic Variants in Autism Spectrum Disorder: Genotype-Based Deep Learning. JMIR Med. Inform. 9 (4), e24754. doi:10.2196/24754
Keywords: autism spectrum disorder, next-generation sequencing, ion channel genes, common variants, restricted repetitive behavior
Citation: Lee J, Ha S, Ahn J, Lee S-T, Choi JR and Cheon K-A (2021) The Role of Ion Channel-Related Genes in Autism Spectrum Disorder: A Study Using Next-Generation Sequencing. Front. Genet. 12:595934. doi: 10.3389/fgene.2021.595934
Received: 18 August 2020; Accepted: 21 September 2021;
Published: 12 October 2021.
Edited by:
Devanand Sadanand Manoli, University of California, San Francisco, United StatesReviewed by:
Rezvan Noroozi, Jagiellonian University, PolandClaudia Lattig, University of Los Andes, Colombia
Copyright © 2021 Lee, Ha, Ahn, Lee, Choi and Cheon. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Keun-Ah Cheon, a2FjaGVvbkB5dWhzLmFj
 Junghan Lee
Junghan Lee Sungji Ha
Sungji Ha Jaeun Ahn1
Jaeun Ahn1 Jong Rak Choi
Jong Rak Choi Keun-Ah Cheon
Keun-Ah Cheon