
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Genet. , 30 April 2021
Sec. RNA
Volume 12 - 2021 | https://doi.org/10.3389/fgene.2021.592669
 Xiangchun Pan1†
Xiangchun Pan1† Qingnan Li2†
Qingnan Li2† Danxia Chen1
Danxia Chen1 Wentao Gong1
Wentao Gong1 Nian Li1
Nian Li1 Yao Jiang1
Yao Jiang1 Hao Zhang1
Hao Zhang1 Yaosheng Chen2
Yaosheng Chen2 Xiaolong Yuan1,3*
Xiaolong Yuan1,3*The timing of puberty in mammals marks the point at which reproduction becomes possible. Abnormalities in the timing of puberty may exert a series of negative effects on subsequent health outcomes. Alternative splicing (AS) has not only emerged as a significant factor in the transcription of genes but it is also reported to play a role in the timing of puberty. However, to date, the changes and dynamics of AS during the onset of puberty is extremely seldom explored. In the present study, we used gilts as a research model to investigated the dynamics of AS and differentially expressed AS (DEAS) events within the hypothalamus–pituitary–ovary (HPO) axis across pre-, in-, and post-puberty. We detected 3,390, 6,098, and 9,085 DEAS events in the hypothalamus, pituitary, and ovary when compared across pre-, in-, and post-pubertal stages, respectively. Within the entire HPO axis, we also identified 22,889, 22,857, and 21,055 DEAS events in the pre-, in-, and post-pubertal stages, respectively. Further analysis revealed that the differentially spliced genes (DSGs) associated with staged DEAS events were likely to be enriched in the oxytocin signaling pathway, thyroid hormone signaling pathway, GnRH signaling pathway, and oocyte meiosis signaling pathway. The DSGs associated with DEAS events across the entire HPO axis were enriched in endocytosis signaling pathway, the MAPK signaling pathway, and the Rap1 signaling pathway. Moreover. the ASs of TAC1, TACR3, CYP19A1, ESR1, ESRRA, and FSHR were likely to regulate the functions of the certain HPO tissues during the onset of puberty. Collectively, the AS dynamics and DEAS events were comprehensively profiled in hypothalamus, pituitary, and ovary across the pre-, in-, and post-pubertal stages in pigs. These findings may enhance our knowledge of how puberty is regulated by AS and shed new light on the molecular mechanisms underlying the timing of puberty in mammals.
The timing of puberty in females relates to the first ovulation and the attainment of reproductive capability (Euling et al., 2008). Abnormalities in the timing of puberty exert a series of different diseases (Cesario and Hughes, 2007), including psychosocial disorders (Hankin et al., 2010), asthma (Al-Sahab et al., 2011), tumors of the reproductive system (Bodicoat et al., 2020), and hypogonadism (Abreu and Kaiser, 2016). Previous studies have demonstrated that the hypothalamic–pituitary–ovary (HPO) axis is responsible for controlling the timing of puberty in females (Zhu et al., 2019). During the onset of puberty, the hypothalamus releases gonadotropin-releasing hormone (GnRH) to the pituitary gland. This process promotes the release of follicle-stimulating hormone (FSH) and luteinizing hormone (LH) into the circulating system (Plant, 2015a). Subsequently, FSH and LH act to accelerate ovarian folliculogenesis and regulate the ovulation of mature follicles (Plant, 2015b; Avendaño et al., 2017; Zhu et al., 2019). Previous researchers have found that GnRH deficiency can cause delayed puberty (Balasubramanian et al., 2010). Furthermore, girls with abnormal levels of LH and FSH are associated to reduced ovarian weight and pubertal failure (Stagi et al., 2016). In rats, the disruption of follicular dynamics results in a delay in the onset of puberty (Lilienthal et al., 2006). These observations indicate that abnormalities in the HPO may lead to pubertal disorders, which in turn has a negative effect on health outcomes.
Alternative splicing (AS) is a post-transcriptional processing mechanism that assembles multiple transcripts with different functions from a single gene under the action of the spliceosome (Lee and Rio, 2015). AS determines whether exons are retained or skipped, excluded or included, and shortened or extended to form mature mRNA (Carazo et al., 2019). There are evidence reported that AS plays a vital role during the development of the hypothalamus (Hasin-Brumshtein et al., 2016), pituitary (Vazquez-Borrego et al., 2019), ovary (Wang et al., 2015), and estrus (Tang et al., 2018). For example, a previous study has shown that piRNAs regulate the pre-mRNA splicing of transcripts and may result in the production of a non-transposase-encoding mature mRNA isoform in germ cells (Teixeira et al., 2017). Also, other researchers have identified two alternative transcripts of the Lin28B gene (Lin28BS, encoding 247 amino acids (aa); and Lin28BL, encoding 261 aa) that regulate the timing of puberty in mammals (Cao et al., 2013). Similarly, another study has discovered that two Oct-2 transcripts (Oct-2a and Oct-2c) act as control mechanisms in the hypothalamus and regulate female puberty; these transcripts can act rapidly and alternatively, thus exerting control on the onset of puberty in the female mammal (Ojeda et al., 1999). Moreover, two isomers of the KISS1R gene (Ss kiss1r_v1 and Ss kiss1r_v2) have been shown to act as the gatekeeper for the onset of puberty, which also exhibit different functions during puberty (Mechaly et al., 2009). Collectively, these findings indicate that the different splicing types caused by AS may have an impact influence on the onset of puberty. However, little is known about the AS events that regulate the onset of puberty.
The rapid development of next-generation sequencing (NGS) technology has led to an increasing number of research studies on sequence-based AS. A previous study used RNA-sequencing (RNA-seq) to detect AS by accurately measuring the percent spliced-in (PSI) score (Hardwick et al., 2019). Another study also used RNA-seq to find that when a mutation occurred at a splice site, the proteins became non-functional (Mayerle et al., 2019). Furthermore, other researchers translated the sequences of new splice junctions, derived from RNA-seq into analogous polypeptide sequences, and created a database that can be used to discover new splice junction peptides that arise from AS (Sheynkman et al., 2013). In another study, researchers used long read sequencing to reveal the splicing status of introns in yeast (Oesterreich et al., 2016). Subsequently, other researchers developed an AS platform for regulating exons that undergo mutually exclusive exons (MXE) AS, ultimately identifying a functionally diverse range of RNA and protein isotypes (Mathur et al., 2019). In addition, the action of the splicing factors (SFs) as “scissors” can exert significant influence on AS. When the components of the SFs undergo alterations, it is likely to affect the occurrence of AS (Zhang et al., 2008). These results indicate that with the development of NGSs, the research on AS deserves more attention.
In the present study, we used gilts as a model and collected HPO tissues from pre-, in-, and post-pubertal animals. Then, we built strand-specific RNA libraries from the HPO tissues and investigated differentially expressed AS (DEAS) events and AS dynamics within the pubertal hypothalamus, pituitary, and ovary. Furthermore, we analyzed the comprehensive changes and dynamics of AS across pre-, in, and post-pubertal stages, as well as along the HPO axis. Finally, we predicted the interactions between several SFs and specifically spliced genes events might associated with puberty. This study may provide new insights into the mechanisms underlying the timing of puberty in female mammals, particularly with regards to AS.
Animal care and experiments were conducted in accordance with the regulations for the administration of affairs concerning experimental animals (Ministry of Science and Technology, China; revised in June 2004) and were approved by the Animal Care and Use Committee of the South China Agricultural University, Guangzhou, China (permit number SCAU#2013-10).
Pre-, in-, and post-pubertal stages were chosen during the onset of puberty; these different stages were identified by measured visually the reddening, swelling of the vulva, and by analyzing standing reflex with the back-pressure test and boar contact (Patterson et al., 2002). Nine Landrace × Yorkshire crossbred gilts with good body condition were used in this study. All gilts were raised in a clean, dry, and well-lit room with equal conditions and maintained on a standard normal diet (isoenergetic corn–barley and soybean meal diet) and water ad libitum. Three gilts without pubertal signs (no reddening, no swelling of the vulva, no standing reflex) were selected as the pre-pubertal gilts (age = 162 ± 3 days; weight = 81.38 ± 2.40 kg), and three gilts exhibiting the first pubertal signs (reddening, swelling of the vulva, standing reflex) were selected as the in-pubertal gilts (age = 212 ± 14 days; weight = 110 ± 2 kg). Gilts have regular estrus cycles, typically 18–23 days (Christenson and Ford, 1979). We used 14 days after in-puberty as the post-puberty, and this stage was diestrus in entire estrus cycles. Finally, three post-pubertal gilts were selected (age = 216 ± 17 days; weight = 122.82 ± 9.11 kg). All gilts were euthanized with an intravenous overdose of barbiturates (≥90 mg/kg). Subsequently, the hypothalamus, entire pituitary and ovary (left and right) were immediately removed. Immediately after decapitation, to dissect the hypothalamus regions, the cerebellum was removed to expose the junction of the brainstem and the cerebrum, so that the hypothalamus is visible, and the hypothalamus is cut along the root as well as the entire pituitary is taken from the pituitary fossa. Moreover, left and right ovaries were taken out. Meanwhile, the biological characteristics of these three stages were reconfirmed by the morphology of ovary (Supplementary Figure 1). All tissues were snap-frozen in liquid nitrogen and stored at −80°C to await subsequent sequencing.
Tissues of pre-, in-, and post-pubertal hypothalamus, pituitary, and ovary were homogenized separately in liquid nitrogen. Total RNAs were extracted from tissue extracts using Trizol (Invitrogen, Carlsbad, CA, United States). Quality control tests were carried out on the RNA using an Agilent Bioanalyzer 2100 system (Agilent, Palo Alto, CA, United States). Then, we used an Epicenter Ribo-zero rRNA Removal Kit (Epicenter, Madison, WI, United States) to remove rRNAs; total RNAs were subsequently quantified. After total RNA extraction and treatment with DNase I, we next used magnetic beads and oligo (dT) to extract mRNAs. Fragmentation buffer was then added to the purified mRNAs to cause fragmentation. Next, we used random hexamer primers and the mRNA fragments as templates to synthesize cDNAs. DNA fragments were then adenylated at the 3’-ends and then ligated to adapters. We then used cDNA fragments (100–200 bp in length) and PCR to generate cDNA libraries. A total of 5 μg of cDNA per sample was sequenced using the HiSeq 3000 Sequencer; this system was operated as recommended by the manufacturer (Illumina, San Diego, CA, United States) and 150 base paired-end reads were generated. The quality control of the raw and trimmed reads was performed using FastQC and Cutadapt software. FastQC software checked the quality of directional paired-end reads in the raw data (Ward et al., 2020). Cutadapt software discarded the low-quality reads (unknown bases >10%; low-mass bases > 50%) and the adapter contamination to generate the high-quality reads termed as clean data thereafter (Chen et al., 2014). Following quality control, the clean reads were mapped to Sus scrofa 11.1 by HISAT2 software (Kim et al., 2015).
The PSI value is an important index with which to identify AS events; significant pairwise differences can be identified using PSI values (| delta-PSI [ΔPSI]| ≥ 10%; FDR ≤ 5%) (Sandberg et al., 2008). The PSI metric represents the ratio of normalized read counts and indicates exon inclusion as a fraction of the total normalized reads for both exon inclusion and exon exclusion (Pervouchine et al., 2013). In addition, CASH software that can significantly improve the detection of DEAS events between samples1 (Wu W. et al., 2018). Moreover, CASH can directly identify different AS events by considering the value of ΔPSI as well as providing exon inclusion and exon exclusion reads (Wu W. et al., 2018). Therefore, in the present study, we primarily used CASH software to identify staged DEAS events across pre-, in-, and post-pubertal tissues, and to identify DEAS events across different tissues in the HPO axis. Then, we calculated the PSI using normalized reads while considering inclusion and exclusion data (FDR < 0.05). We used two specific modules in CASH software: SpliceCons and SpliceDiff. The first module detects eight different types of AS events that are classified according to splicing at different sites, including alternative 3’ splice site (A3SS), alternative 5′ splice site (A5SS), mutually exclusive exons (MXE), cassette exon (cassette, SE), multi-cassette exons (Cassette multi), intron retention (IR), alternative start exon (AltStart), and alternative end exon (AltEnd); these are depicted in Supplementary Figure 1. Then, we used the second module, SpliceDiff, to calculate the ΔPSI between each sample and thus identify DEAS events.
DEAS were identified using ΔPSI (FDR < 5%), and differentially spliced genes (DSGs) were defined as the parental genes from DEAS events. We also used R (version 3.5.1) to perform statistical analysis. Specifically, we used the functions of setdiff and intersect to analyze the number of genes undergoing AS. Next, we used the Venn Diagram package (Chen and Boutros, 2011) and the ggplot2 package (Villanueva and Chen, 2019) to visualize data. Upset plots were created using TBTools version 0.66836 software2. Stage-specific AS events were defined as those that only occurred in one pubertal stage with the screening condition of | △PSI| = 1. Similarly, tissue-specific AS events were defined as those that only occurred in one tissue with the screening condition of | △PSI| = 1 and FDR < 0.05. Finally, statistical differences among multiple groups were identified by ANOVA with Tukey’s multiple comparison test.
Next, DSGs were used for Gene Ontology (GO) analysis and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis using the FDR < 0.05 as a cutoff criterion. Co-existing DSGs were identified with KOBAS 3.0 online software. Furthermore, a list of splicing factors (SFs) was extracted from the SpliceAid-F database3; this database supplies the gene interactors of SFs (Giulietti et al., 2013). Finally, we used Cytoscape software (version 3.4.0) to generate a network of protein interactions between SFs and genes that are known to be associated with puberty.
Finally, we used RT-PCR to validate the reliability of the data of RNA-seq. Upstream and downstream primers were designed from the upper exon and lower exon of the target splicing region using NCBI primer designing tool. PCR amplification was performed using cDNA as a template. The PCR standard procedure was denaturation 94°C (3 min), 35 cycles at 94°C (30 s), 60°C (30 s), and 72°C (30 s), subsequently extending it for 10 min at 72°C. The PCR products were detected via running 2.5% agarose electrophoresis. All reactions were repeated three times.
In total, we generated 27 RNA-seq libraries consisting of nine hypothalamic tissues (611 million clean reads), nine pituitary tissues (673 billion clean reads), and nine ovarian tissues (597 million clean reads) obtained from gilts undergoing pubertal transition. These clean reads were subsequently mapped to the Sus scrofa reference genome by using HISAT2 with an average alignment rate of >90% (Supplementary Table 1).
In the present study, eight types of AS events were investigated in total, including A3SS, A5SS, MXE, Cassette, Cassette multi, IR, AltStart, and AltEnd events (Supplementary Figure 2). A total of 278,721 AS events were detected in pubertal HPO tissues across pre-, in-, and post-pubertal tissues (FDR < 0.05). In total, 33,535, 33,254, and 33,405 AS events were detected in the hypothalamus during pre-, in-, and post-pubertal stages, respectively (Figure 1 and Supplementary Tables 2, 3); 30,251, 30,135, and 29,767 AS events were identified in the pituitary during pre-, in-, and post-pubertal stages, respectively (Figure 1 and Supplementary Tables 2, 4); and 29,567, 28,684, and 30,123 AS events were detected in ovarian tissue during pre-, in-, and post-pubertal stages, respectively (Figure 1 and Supplementary Tables 2, 5). The most common AS event was Cassette events, while the least common was MXE events in all stages of HPO axis (Supplementary Table 6 and Supplementary Figure 3A, Tukey’s test). Collectively, these data indicate that we identified eight AS events during different pubertal stages in HPO tissues.
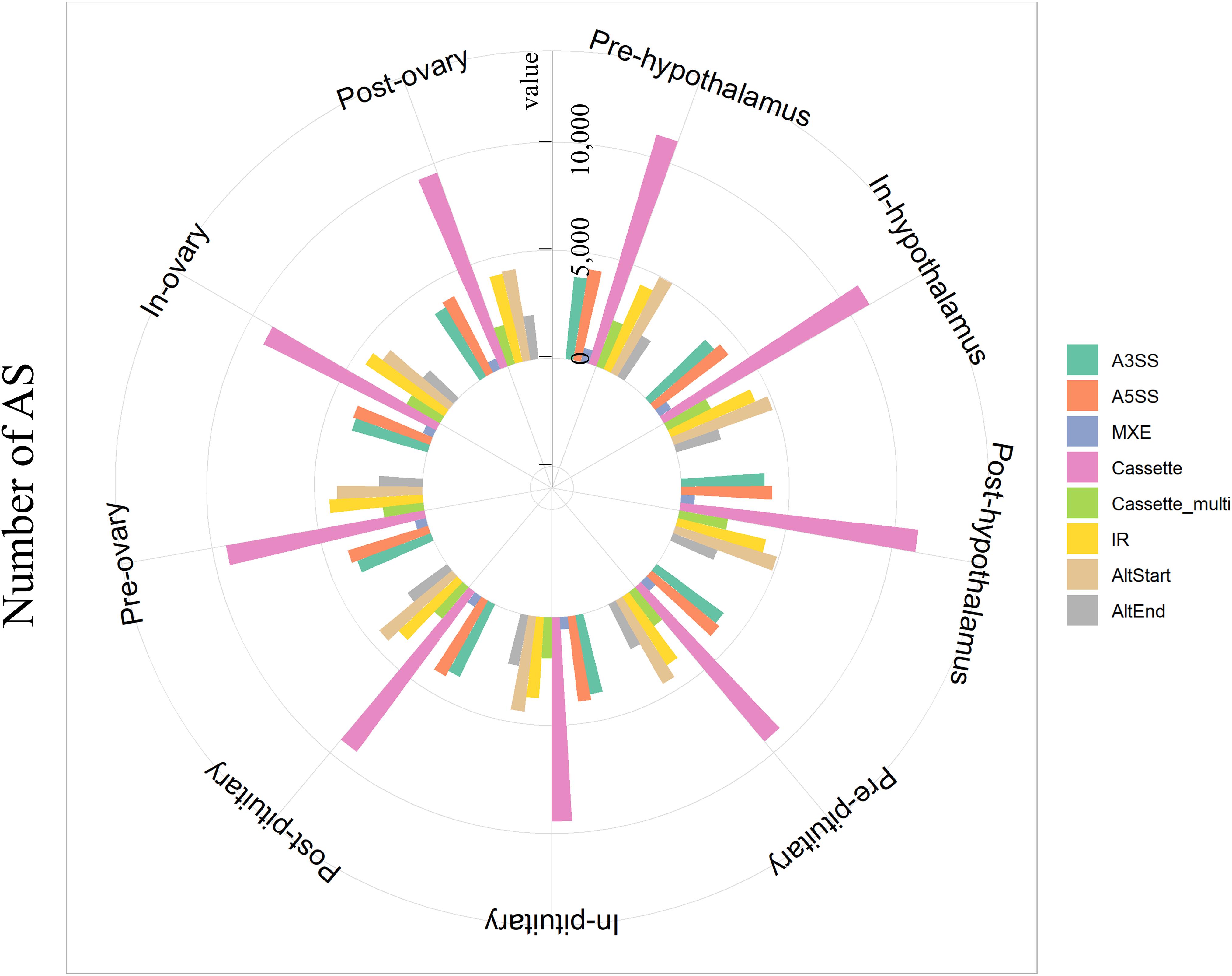
Figure 1. Overview of alternative splicing (AS) events in the pubertal hypothalamus–pituitary–ovary (HPO) axis. The number of AS events in the hypothalamus, pituitary, and ovary of gilts during pre-, in-, and post-pubertal stages.
To explore the DEAS events in HPO tissues during different stages of pubertal transition, we carried out pairwise comparisons across the pre-, in-, and post-pubertal groups in three tissues from the HPO axis. The distribution of DEAS events is shown in Figure 2. When comparing pre- vs. in-puberty, in- vs. post-puberty, and pre- vs. post-pubertal groups, we detected 3,390 DEAS events in the hypothalamus (Figure 2A), 6,098 DEAS events in the pituitary (Figure 2B), and 9,085 DEAS events in the ovaries (Figure 2C). The most frequent DEAS in the hypothalamus and pituitary were Cassette events (Supplementary Table 7 and Supplementary Figures 3B,C, Tukey’s test). Pairwise comparisons of pre- and post-pubertal groups showed the highest frequency of DEAS events (Supplementary Figure 3D, Tukey’s test). In total, there were 18,573 DEAS involved in the pre-, in-, and post-pubertal HPO axis. Further analysis showed that the prevalence of staged DEAS was different when compared across the three tissues, thus indicating that different AS events might play various roles in the pubertal HPO axis.
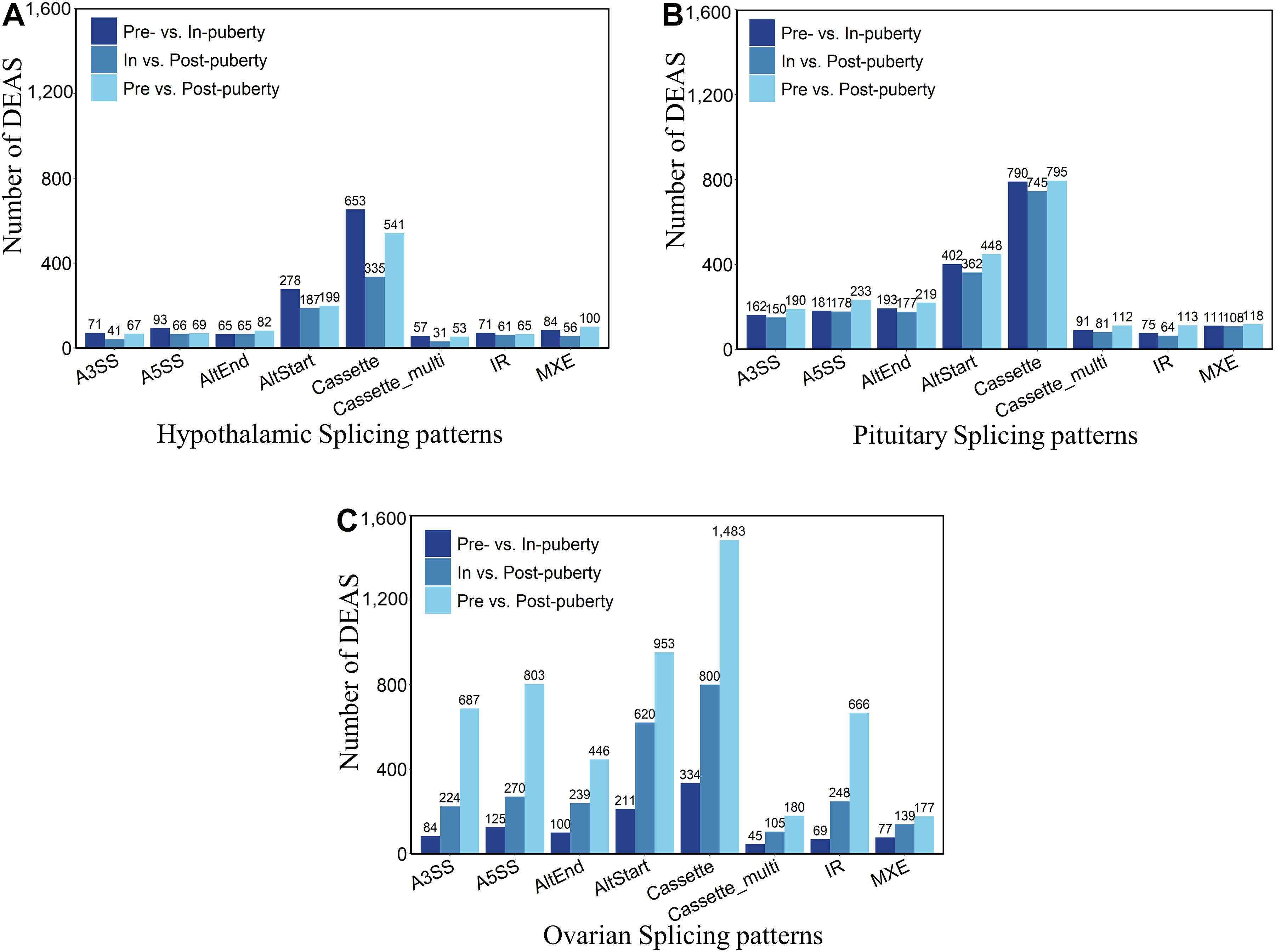
Figure 2. Staged differentially expressed alternative splicing (DEAS) events in the hypothalamus–pituitary–ovary (HPO) axis. Pairwise comparisons detected DEAS events in the hypothalamus (A), pituitary (B), and ovary (C) of pre-, in-, and post-pubertal tissues.
In total, we identified that 1,737 DSGs in the hypothalamus that were the parental genes of DEAS (Figure 3A), 3,088 DSGs in the pituitary (Figure 3B), and 3,780 DSGs in the ovaries (Figure 3C). It is noteworthy that several AS events might occur in the same gene. Specifically, an upset plot of multiple interactive sets demonstrated that 14.29% (Figure 3D), 22.72% (Figure 3E), and 33.54% (Figure 3F) of DSGs featured two or more DEAS events in the three tissues. In summary, these results showed that DEAS events might provide the possibility for improving diversity within the transcriptome.
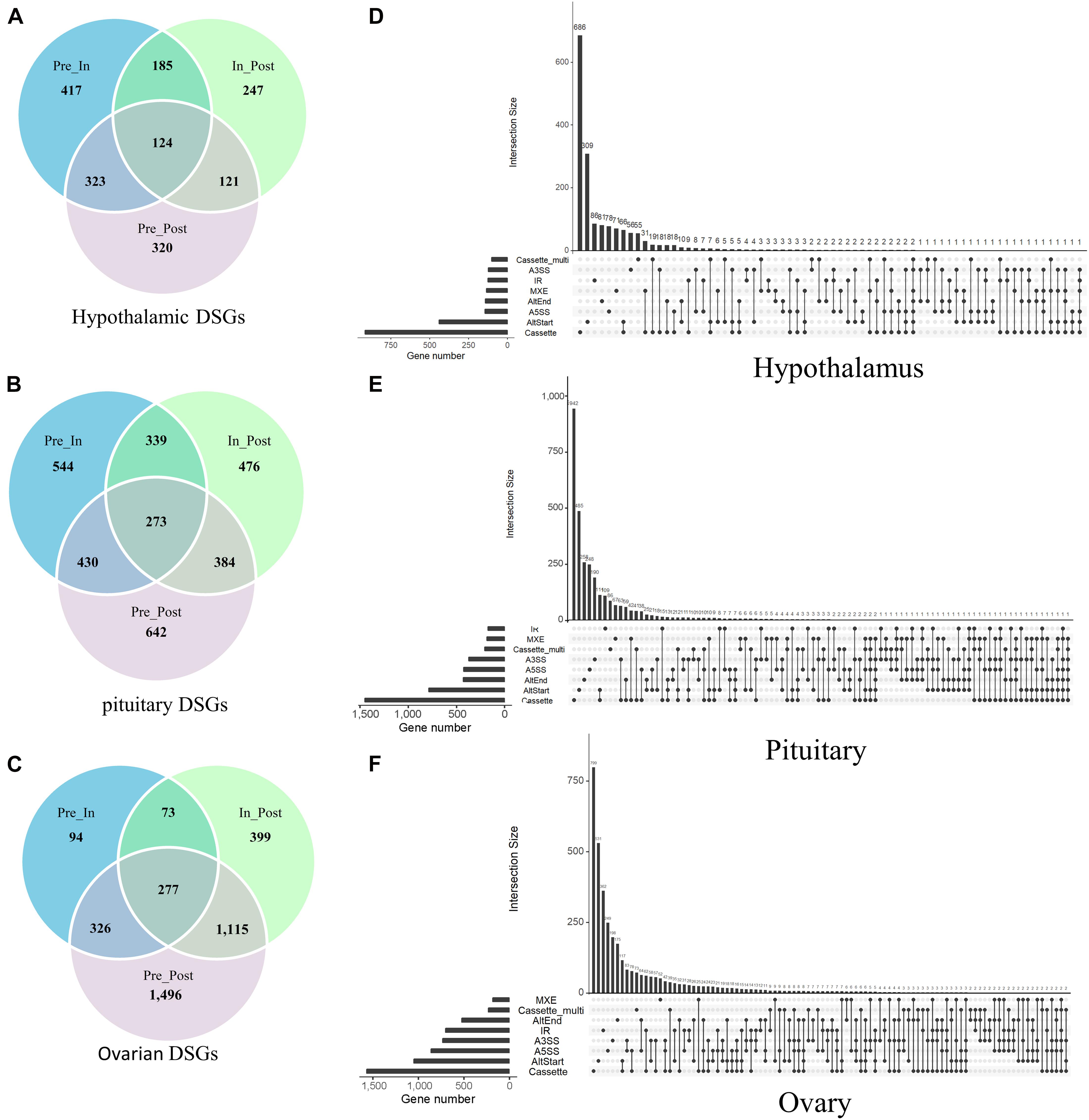
Figure 3. Characterization of the parental genes for staged differentially expressed alternative splicing (DEAS) events. The number of parental genes of the DEAS detected in the hypothalamus (A), pituitary (B), and ovary (C). Upset plot of different types of DEAS events, and their parent genes, in the hypothalamus (D), pituitary (E), and ovary (F). The abscissa is the number of genes that have occurred corresponding to AS, the ordinate represents the number of genes that occurred multiple AS. One gene may have up to six types of alternative splicing.
To determine the biological functions of these staged DEAS events within the HPO axis, we performed KEGG enrichment analysis for the DSGs of DEAS events in the HPO axis during different stages of puberty. KEGG enrichment analysis showed that DSGs in the three tissues showed enrichment in different pathways (Supplementary Tables 8–10). DEGs were associated with the oxytocin signaling pathway, GnRH signaling pathway, insulin signaling pathway, thyroid hormone signaling pathway, and neurotrophin signaling pathway in the pituitary (Figure 4). Moreover, the oocyte meioses were enriched both in the hypothalamus and the pituitary. We also found that the tight junction was enriched in all three tissues (Figure 4).
Next, we investigated DEAS events in different HPO tissues during pubertal transition in gilts. AS events were compared between the three different tissues by pairwise comparisons during the three stages of puberty. When comparing the hypothalamus vs. pituitary, pituitary vs. ovary, and hypothalamus vs. ovary, we identified a total of 22,889, 22,857, and 21,055 DEAS events in the pre-, in-, and post-pubertal stages, respectively (Figures 5A–C). The most frequent DEAS events during pre- and post-puberty were Cassette events; however, this event was not significant in the in-pubertal stage (Supplementary Table 11 and Supplementary Figures 2E,F, Tukey’s test). In total, we detected 66,801 DEAS events in different HPO tissues. The differences between tissues with regards to AS patterns indicate that AS might play a different role in different HPO tissues during pubertal transition.
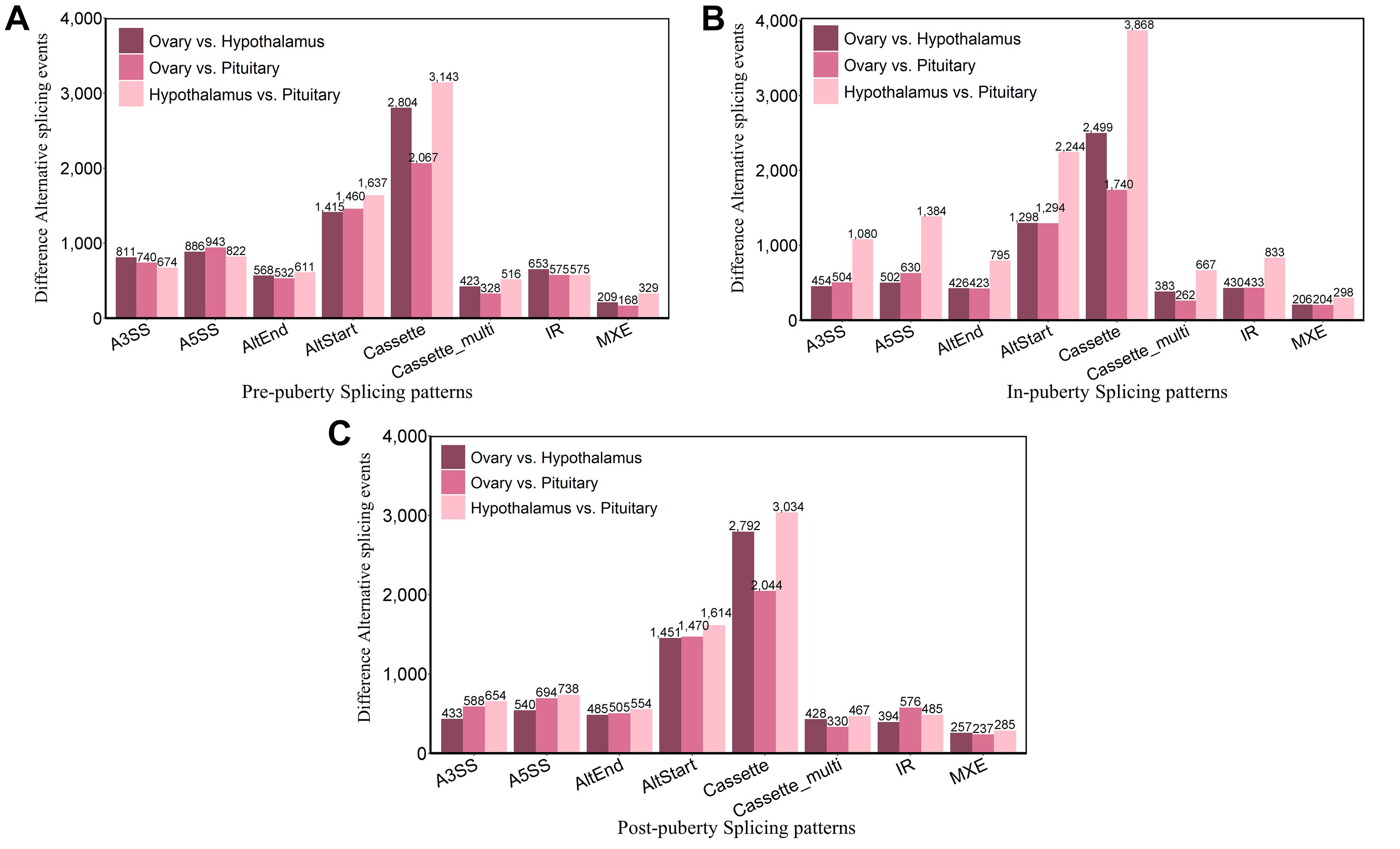
Figure 5. Tissular differentially expressed alternative splicing (DEAS) events during pubertal transition. Pairwise comparison–detected DEAS events were detected pre-puberty (A), in-puberty (B), and post-puberty (C) in the hypothalamus, pituitary, and ovary.
To characterize the biological functions of DEAS in different HPO tissues, we performed KEGG enrichment analysis for the DSGs of DEAS events in different HPO tissues across the three stages of puberty. We identified the top 30 significantly enriched pathways (Figure 6). Analysis of the three stages of puberty revealed that the DSGs were significantly enriched in endocytosis, the MAPK signaling pathway, the Rap1 signaling pathway, and axon guidance, in three stages (Figure 6). In particular, several tissular DSGs were enriched in the oocyte meiosis pathway during the pre-pubertal stage (Figure 6A and Supplementary Table 12). Some tissular DSGs were enriched in the spliceosome pathway during the in-puberty stage (Figure 6B and Supplementary Table 13). Finally, some tissular DSGs were enriched in the lysosome and peroxisome pathways during the post-pubertal phase (Figure 6C and Supplementary Table 14).
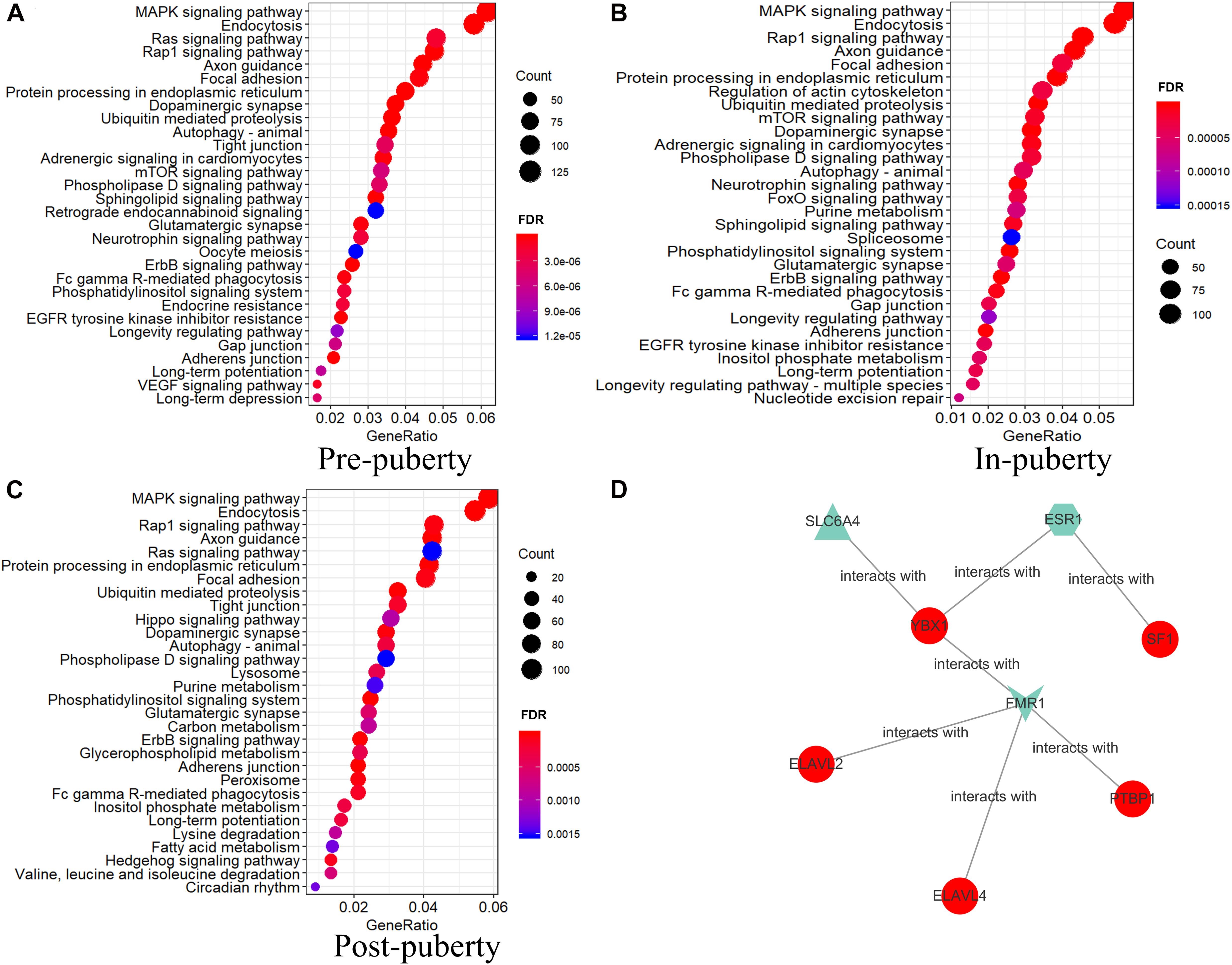
Figure 6. KEGG analysis of tissular differentially spliced genes and a network of splicing factors. KEGG analysis of DEAS in pre-puberty (A), in-puberty (B), and post-puberty (C) (FDR < 0.05). (D) Network of splicing factors (SFs) and pubertal genes of specific AS. Blue nodes represent stage-specific; purple nodes represent tissue-specific; and red nodes represent both stage- and tissue-specific AS.
Next, we selected 36 pubertal genes and investigated how these genes underwent DEAS events. To do this, we screened literature and databases by hand. During the pubertal transition in pigs, as shown in Supplementary Table 15, AVP, BDNF, MSMB, and TAC1 genes respectively produce the A5SS, AltStart, and MXE of AS exclusively in the hypothalamus; FMR1, NCOA1, NOS1, PGR, TACR3, and TGFHR1 genes harbor the Cassette, AltStart, and A3SS of AS specially in the pituitary; and ADM, CYP19A1, DICER1, ENPP2, ESRRA, GDF9, PPARG, RIPK2, and STAT5B genes mainly have the IR, AltEnd, AltStart, A3SS, and Cassette of AS in the ovary (Supplementary Table 15). Besides, we found that 16 genes underwent only one AS event: ADM, APOE, APP, AVP, COMT, BDNF, FMR1, FSHR, MSMB, NOS1, NR5A1, PPARG, PTPN11, SLC6A4, STAT5A, and TACR3. These 16 genes were subjected to IR, Cassette, AltStart, A5SS, AltEnd, and MXE DEAS events, respectively. However, other genes exhibited complex DEAS events, such as ESR1 mediated by AltEnd, AltStart, A5SS, and Cassette events. In addition, 20 genes underwent DEAS only in certain tissues, including ADM, AVP, BDNF, COMT, CYP19A1, DICER1, ENPP2, ESR2, ESRRA, FMR1, GDF9, MSMB, NCOA1, NOS1, PGR, PPARG, PIPK2, STAT5B, TACR3, and TGFBR1. We also found that APOE, COMR, ESRRA, FSHR, NOS1, MSMB, SLC6A4, PGR, and NR2C2 only exhibited DEAS events in specific stages. These results demonstrated that diverse patterns of AS events occurred in a range of genes associated with puberty.
To investigate stage-specific and tissue-specific AS events in pubertal gilts, we screened our libraries for specific AS events (| △PSI| = 1, FDR < 0.05). We found that several stage-specific AS events were uniquely expressed in pre-, in-, and post-pubertal stages, of which specific MXE events repeatedly occurred (Supplementary Figure 4A). Similarly, some AS events were uniquely expressed in the three tissues, of which AltStart events repeatedly occurred (Supplementary Figure 4B). Notably, we found that the same gene could experience specific AS events at different sites (Supplementary Tables 16, 17).
To explore the relationship between SFs and specific AS, we went to screen the parental genes of specific AS that interacted with SFs by databases (Supplementary Table 18). We found that multiple SFs interact with specific splicing genes. For instance, the AS of ACTB in ovary is specific AltEnd, and SFs Sam68, hnRNP D, YB-1, etc. protein were predicted to interact with ACTB (Figure 6D and Supplementary Table 18). Moreover, the AS of RPS3 is exclusive A3SS, and SFs YB-1, hnRNP U, SLM-1 etc. protein were predicted to interact with RPS3 (Figure 6D and Supplementary Table 18). According to this analysis, several SFs were predicted to interact with specific spliced genes in specific stage or specific tissue. Future research should focus on investigating interactions between SFs and specific AS events.
To validate the reliability of the high-throughput RNA sequencing data, three puberty-related DEAS genes were randomly selected for validation experiments. RT-PCR was used to measure the expression of different transcripts that occurred in DEAS events. The primer from the upper and lower exon of the alternative exon was designed (Supplementary Table 19). Among them, TAC1 occurred different AltStart events of AS in pre-, in-, and post-puberty in hypothalamus, and the phenomenon of the first exon being jumped was most obvious in the pre-puberty (Figure 7A). TACR3 occurred in different Cassette events of AS in pre-pubertal hypothalamus and ovary, and the phenomenon of the exon being jumped was most obvious in the pre-pubertal ovary (Figure 7B). ESR1 occurred in different AltEnd events of AS in pre-pubertal ovary and post-pubertal ovary, and the phenomenon of the last exon being jumped was most obvious in the post-pubertal ovary (Figure 7C). The RT-PCR results showed that most of the AS events were in line with our prediction, confirming the reliability of sequencing. Further study should be focus on puberty-related DEAS.
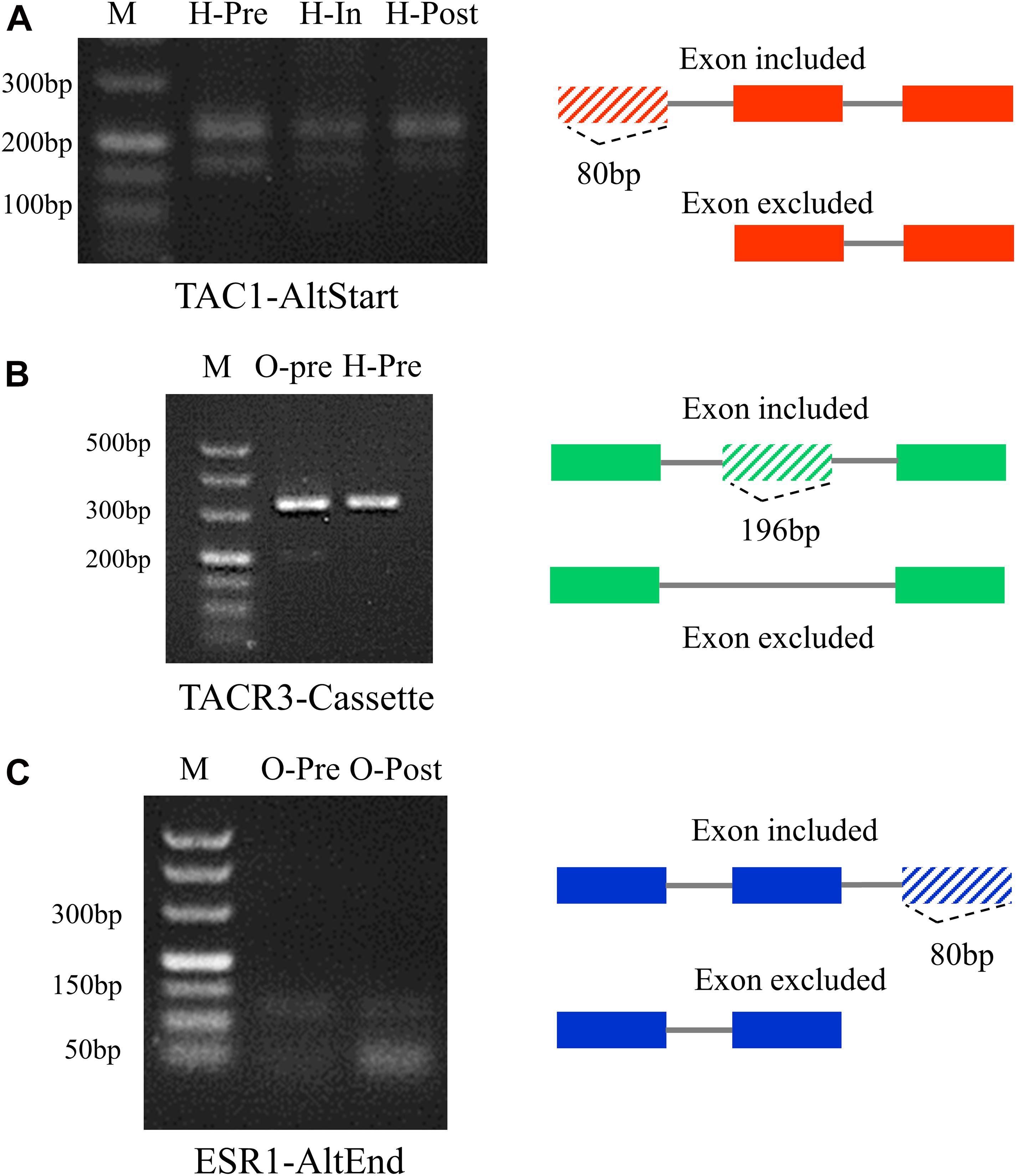
Figure 7. RT-PCR of alternative splicing (DEAS) events for validation. (A) DEAS of TAC1 in pre-, in-, and post-pubertal hypothalamus. (B) DEAS of TACR3 in pre-pubertal ovary and hypothalamus. (C) DEAS of ESR1 in pre- and post-pubertal ovary.
Puberty marks the achievement of sexual maturity and fertility and is mainly driven by the hypothalamic–pituitary–gonadal axis (Blakemore et al., 2010). Moreover, puberty is a very important biological process, irrespective of whether we consider reproduction, development, or disease (Nonneman et al., 2016; Vijayakumar et al., 2018). An increasing body of evidence now indicates that AS may be involved in regulating the function of HPO tissues and the onset of puberty in mammals (Ojeda et al., 1999; Mechaly et al., 2009; Cao et al., 2013). Previous studies have demonstrated that AS plays a crucial role in mammalian development, particularly AS events involving the ICE gene (Wang et al., 1998), the Bcl-2 gene family, the Ced-4 gene, and the family of caspase genes during the process of cellular apoptosis (Jiang and Wu, 1999). However, the AS profiles of tissues in the HPO axis have not been identified during pubertal transition in mammals. In this study, we systematically profiled AS events and analyzed DEAS events in pubertal HPO tissues from gilts. First, we found that the Cassette event was the most common form of AS in pubertal HPO tissues, which is in line with previous analysis of the human transcriptome showing that Cassette events were the most common form of AS (Sultan et al., 2008). Indeed, it has been reported that Cassette is the most common AS event due to the loss of functional domain/sites or the transfer of open reading framework, and the disruption of Cassette is one of the causes of mammalian disease (Kim et al., 2020). Next, we investigated AS events in different tissues of the HPO axis, and during different stages of puberty. We found that tissular DEAS events were much more than staged DEAS events, indicating that the degree of AS is greater between tissues. It is possible, therefore, that the underlying reason that the three tissues of HPO axis play different roles during puberty can be revealed by AS.
AS events exhibit spatiotemporal specificity, which means that AS events vary across different tissues or different stages of development (Chakradhar, 2018). Our DEAS analysis showed that the Cassette event was the most frequent DEAS event in the hypothalamus and the pituitary, but not in the ovary, indicating that AS events might play different regulatory roles within the pubertal HPO axis. We found that the staged DEAS events were likely to occur on genes involving the signaling pathways that have all been reported to regulate the timing of puberty (Parent et al., 2008; Sun et al., 2009; Singh et al., 2015; Aliberti et al., 2019; Gioacchini et al., 2019). For example, previous studies have shown that tight junctions form in cells to create a major component of the blood–testis barrier during puberty in males (McCabe et al., 2012), and the blood–brain barrier also plays a crucial role in the microenvironment required to maintain neuronal function at puberty (Seker et al., 2016). Furthermore, growing ovaries that ingest ovoproteins from the blood stream were mediated by receptor-dependent endocytosis (Mizuta et al., 2013). These observations indicate that genes may be involved in regulating onset of puberty in certain different splicing patterns during mammalian puberty. According to our analysis of DEAS events within the entire HPO axis, one interesting finding is that the transcription of HPO tissues might undergo dramatic changes during the in-puberty stage. Furthermore, we found that DEAS events at the tissue level were more likely to occur on genes related to signaling pathways that have all been reported to affect cell apoptosis or the synthesis of the thyroid gland (Wu D.M. et al., 2018; Fan et al., 2019). It may be the case therefore that AS events affect the synthesis of the thyroid gland and, subsequently, the onset of puberty.
We also analyzed AS events in 36 genes that are known to be involved in puberty (Mechaly et al., 2009; Balasubramanian et al., 2010; Lomniczi et al., 2013; Plant, 2015b; Abreu and Kaiser, 2016; Chakradhar, 2018; Tang et al., 2018; Lents, 2019). Furthermore, the differential splicing of TAC1 can make it produce two isoforms (Page, 2005). The tachykinins encoded by these two isoforms have been shown to regulate the release of prolactin (PRL) in vivo, and these PRL secretagogues act on primary pituitary cells (Dupre et al., 2010). In the present study, we found that Cassette of TAC1 was more specifically identified in pituitary in in-pubertal stage, compared with that of hypothalamus (Supplementary Table 15). The AltStart in PGR gene, which has been shown to be affected by pituitary-specific Esr1 knockout (Sanchez-Criado et al., 2012), more exclusively occurred in pre-pubertal pituitary. The AltEnd of FSHR, which has been demonstrated to be involved in folliculogenesis (Candelaria et al., 2020), has been more specifically observed in ovaries in in-puberty, compared with hypothalamus. These finding and observations indicate that the ASs are expressed in a tissue-specific pattern based on their own functions during the onset of puberty. Moreover, each tissue is composed of different cells, and it is supposed that these ASs are cell specific in the cells of hypothalamus, pituitary, and ovary during the timing of puberty. A previous study has showed that cell-specific AS was essential for the function of cells (Lah et al., 2014; Gracida et al., 2016). Therefore, the more comprehensive profiles of AS changes and dynamics during the onset of puberty is supposed and proposed with the transcriptome data of single cells in HPO tissues.
In addition, the estrogen-related receptor alpha (ESRRA) gene encodes an orphan nuclear receptor that is involved in the release of GnRH in hypothalamus of marmoset monkey (Wahab et al., 2019). In the present study, the AltEnd event in ESRRA more exclusively occurred in pre-puberty, compared with post-puberty in ovary. It has been reported that TACR3 directly regulates the release of GnRH and the onset of puberty (Hietamaki et al., 2017). In the present study, we found that Cassette of TAC3R was more specifically identified in post-puberty in pituitary, compared with that of pre-puberty. The AltEnd and AltStart occurring in ESR1 gene, which is essential in response to normal onset of puberty and estrogen feedback (Cheong et al., 2015), more exclusively occurred in pre-puberty, compared with post-puberty in the ovary. These results suggest that ASs of genes exhibit in a specific pubertal stage during the timing of puberty in pigs.
Besides, we found that MXE and AltStart events highly occurred in HPO tissues during the pubertal transition of pigs. For example, COL3A1, which encodes the pro-α 1 chain of collagen III and is involved in the development of follicles (Yao et al., 2019), was identified as one of the genes that exclusively underwent MXE events in post-puberty. The CYP19A1 gene, which is involved in the estradiol biosynthesis (Sutherland et al., 2017), uniquely harbors AltStart in hypothalamus. Alternatively, a previous study shows that GH1 is involved in regulating onset of puberty (Hu et al., 2019), and we found that A5SS and MXE occur at different sites of GH1 by interacting with four SFs at the post-puberty in the present study. ACTB has been demonstrated to regulate the reproductive function of the ovary (Hassanpour et al., 2019), and the ACTB gene specially shows AltEnd in ovary by intersecting with five SFs at the post-puberty (Supplementary Tables 16–18). These results suggest that SFs might be involved in the onset of puberty by regulating specific splicing of genes. These results provide AS level data for a clearer understanding of the mechanisms underlying onset of puberty in the future.
Collectively, in this study, the AS dynamics and DEAS events were comprehensively profiled in hypothalamus, pituitary, and ovary across the pre-, in-, and post-pubertal stages in pigs. The related genes of DEAS were enriched in GnRH signaling pathway, thyroid hormone signaling pathway, oocyte meiosis, and oxytocin signaling pathway, which are all involved in the regulating and timing of puberty. Moreover, the ASs of TAC1, TACR3, CYP19A1, ESR1, ESRRA, and FSHR were likely to regulate the functions of certain HPO tissues during the onset of puberty. These findings may provide a step forward in our understanding of how molecular events, such as AS, can regulate the timing of puberty in mammals.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ebi.ac.uk/ena, PRJEB39754.
The animal study was reviewed and approved by the Animal Care and Use Committee of the South China Agricultural University, Guangzhou, China (permit number: SCAU#2013-10). Written informed consent was obtained from the owners for the participation of their animals in this study.
XP, QL, YC, and XY conceived and designed this research. XP, QL, and DC acquired the biological samples and analyzed the data. WG, NL, YJ, and HZ originally derived the data and helped in the analysis. XP and QL drafted the manuscript. XY and YC critically revised the first draft. All authors reviewed and approved the final manuscript.
This research was supported by the National Natural Science Foundation of China (Reference No. 31902131), the National Key R&D Program of China (Reference No. 2018YFD0501200), the earmarked fund for the China Agriculture Research System (Reference No. CARS-35), the Special Fund for Science and Technology Innovation of Guangdong Province (Reference No. 2018B020203003), and the National Natural Science Foundation of Guangdong Province (Reference No. 2019A1515010676).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We would like to thank the National Super computer Center in Guangzhou for its computing platform. Also, we are grateful to the editors and all the reviewers for their insightful comments and constructive suggestions that greatly improved our manuscript.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2021.592669/full#supplementary-material
Supplementary Figure 1 | The ovarian histology structure. (A) Pre-pubertal ovary. (B) In-pubertal ovary. (C) Post-pubertal ovary.
Supplementary Figure 2 | Representative model of eight different alternative splicing types. The black solid lines represent connections between exons that have undergone alternative splicing. The colored dotted line refers to the original connection.
Supplementary Figure 3 | Boxplots of number of AS and differentially expressed alternative splicing (DEAS). (A) AS in all pubertal tissue. (B) DEAS in hypothalamus. (C) DEAS in pituitary. (D) DEAS in ovary. (E) DEAS in pre-puberty. (F) DEAS in post-puberty. ∗∗FDR < 0.01, ∗∗∗FDR < 0.001.
Supplementary Figure 4 | Specific alternative splicing (AS) events in the pubertal hypothalamus–pituitary–ovary (HPO) axis. (A) Stage-specific AS. (B) Tissue-specific AS.
AS, alternative splicing; DSG, differential splicing gene; DEAS, differentially expressed alternative splicing; A3SS, alternative 3′ splice site; A5SS, alternative 5′ splice site; MXE, mutually exclusive exons; SE (Cassette), skipping exons; Cassette multi, multi-cassette exons; IR, intron retention; AltStart, alternative start exon; AltEnd, alternative end exon; HPO, hypothalamus–pituitary–ovary axis.
Abreu, A. P., and Kaiser, U. B. (2016). Pubertal development and regulation. Lancet Diabetes Endocrinol. 4, 254–264. doi: 10.1016/S2213-8587(15)00418-0
Aliberti, P., Sethi, R., Belgorosky, A., Chandran, U. R., Plant, T. M., and Walker, W. H. (2019). Gonadotrophin-mediated miRNA expression in testis at onset of puberty in rhesus monkey: predictions on regulation of thyroid hormone activity and DLK1-DIO3 locus. Mol. Hum. Reprod. 25, 124–136. doi: 10.1093/molehr/gay054
Al-Sahab, B., Hamadeh, M. J., Ardern, C. I., and Tamim, H. (2011). Early menarche predicts incidence of asthma in early adulthood. Am. J. Epidemiol. 173, 64–70. doi: 10.1093/aje/kwq324
Avendaño, M. S., Vazquez, M. J., and Tena-Sempere, M. (2017). Disentangling puberty: novel neuroendocrine pathways and mechanisms for the control of mammalian puberty. Hum. Reprod. Update 23, 737–763. doi: 10.1093/humupd/dmx025
Balasubramanian, R., Dwyer, A., Seminara, S. B., Pitteloud, N., Kaiser, U. B., and Crowley, W. J. (2010). Human GnRH deficiency: a unique disease model to unravel the ontogeny of GnRH neurons. Neuroendocrinology 92, 81–99. doi: 10.1159/000314193
Blakemore, S. J., Burnett, S., and Dahl, R. E. (2010). The role of puberty in the developing adolescent brain. Hum. Brain Mapp. 31, 926–933. doi: 10.1002/hbm.21052
Bodicoat, D. H., Schoemaker, M. J., Jones, M. E., McFadden, E., Griffin, J., Ashworth, A., et al. (2020). Correction to: timing of pubertal stages and breast cancer risk: the breakthrough generations study. Breast Cancer Res. 22:19. doi: 10.1186/s13058-020-1257-2
Candelaria, J. I., Rabaglino, M. B., and Denicol, A. C. (2020). Ovarian preantral follicles are responsive to FSH as early as the primary stage of development. J. Endocrinol. 247, 153–168. doi: 10.1530/JOE-20-0126
Cao, G., Liu, Q., Chu, M., Di, R., Fang, L., Feng, T., et al. (2013). Analysis on cDNA sequence, alternative splicing and polymorphisms associated with timing of puberty of Lin28B gene in goats. Mol. Biol. Rep. 40, 4675–4683. doi: 10.1007/s11033-013-2562-y
Carazo, F., Romero, J. P., and Rubio, A. (2019). Upstream analysis of alternative splicing: a review of computational approaches to predict context-dependent splicing factors. Briefings Bioinform. 20, 1358–1375. doi: 10.1093/bib/bby005
Cesario, S. K., and Hughes, L. A. (2007). Precocious puberty: a comprehensive review of literature. J. Obstet Gynecol. Neonatal Nurs. 36, 263–274. doi: 10.1111/j.1552-6909.2007.00145.x
Chakradhar, S. (2018). Animals on the verge: what different species can teach us about human puberty. Nat. Med. 24, 114–117. doi: 10.1038/nm0218-114
Chen, C., Khaleel, S. S., Huang, H., and Wu, C. H. (2014). Software for pre-processing Illumina next-generation sequencing short read sequences. Source Code Biol. Med. 9:8. doi: 10.1186/1751-0473-9-8
Chen, H., and Boutros, P. C. (2011). VennDiagram: a package for the generation of highly-customizable Venn and Euler diagrams in R. BMC Bioinformatics 12:35. doi: 10.1186/1471-2105-12-35
Cheong, R. Y., Czieselsky, K., Porteous, R., and Herbison, A. E. (2015). Expression of ESR1 in glutamatergic and GABAergic neurons is essential for normal puberty onset, estrogen feedback, and fertility in female mice. J. Neurosci. 35, 14533–14543. doi: 10.1523/JNEUROSCI.1776-15.2015
Christenson, R. K., and Ford, J. J. (1979). Puberty and estrus in confinement-reared gilts. J. Anim. Sci. 49, 743–751. doi: 10.2527/jas1979.493743x
Dupre, S. M., Miedzinska, K., Duval, C. V., Yu, L., Goodman, R. L., Lincoln, G. A., et al. (2010). Identification of Eya3 and TAC1 as long-day signals in the sheep pituitary. Curr. Biol. 20, 829–835. doi: 10.1016/j.cub.2010.02.066
Euling, S. Y., Herman-Giddens, M. E., Lee, P. A., Selevan, S. G., Juul, A., Sorensen, T. I., et al. (2008). Examination of US puberty-timing data from 1940 to 1994 for secular trends: panel findings. Pediatrics 121(Suppl. 3), S172–S191. doi: 10.1542/peds.2007-1813D
Fan, Y., Li, J., Wei, W., Fang, H., Duan, Y., Li, N., et al. (2019). Ku80 gene knockdown by the CRISPR/Cas9 technique affects the biological functions of human thyroid carcinoma cells. Oncol. Rep. 42, 2486–2498. doi: 10.3892/or.2019.7348
Gioacchini, G., Marisaldi, L., Basili, D., Candelma, M., Pignalosa, P., Aiese, C. R., et al. (2019). A de novo transcriptome assembly approach elucidates the dynamics of ovarian maturation in the swordfish (Xiphias gladius). Sci. Rep. 9:7375. doi: 10.1038/s41598-019-43872-6
Gracida, X., Norris, A. D., and Calarco, J. A. (2016). Regulation of tissue-specific alternative splicing: C. elegans as a model system. Adv. Exp. Med. Biol. 907, 229–261. doi: 10.1007/978-3-319-29073-7_10
Giulietti, M., Piva, F., D’Antonio, M., D’Onorio, D. M. P., Paoletti, D., Castrignano, T., et al. (2013). SpliceAid-F: a database of human splicing factors and their RNA-binding sites. Nucleic Acids Res. 41, D125–D131. doi: 10.1093/nar/gks997
Hankin, B. L., Badanes, L. S., Abela, J. R., and Watamura, S. E. (2010). Hypothalamic-pituitary-adrenal axis dysregulation in dysphoric children and adolescents: cortisol reactivity to psychosocial stress from preschool through middle adolescence. Biol. Psychiatry 68, 484–490. doi: 10.1016/j.biopsych.2010.04.004
Hardwick, S. A., Joglekar, A., Flicek, P., Frankish, A., and Tilgner, H. U. (2019). Getting the entire message: progress in isoform sequencing. Front. Genet. 10:709. doi: 10.3389/fgene.2019.00709
Hasin-Brumshtein, Y., Khan, A. H., Hormozdiari, F., Pan, C., Parks, B. W., Petyuk, V. A., et al. (2016). Hypothalamic transcriptomes of 99 mouse strains reveal trans eQTL hotspots, splicing QTLs and novel non-coding genes. Elife 5:e15614. doi: 10.7554/eLife.15614
Hassanpour, H., Aghajani, Z., Bahadoran, S., Farhadi, N., Nazari, H., and Kaewduangta, W. (2019). Identification of reliable reference genes for quantitative real-time PCR in ovary and uterus of laying hens under heat stress. Stress 22, 387–394. doi: 10.1080/10253890.2019.1574294
Hietamaki, J., Hero, M., Holopainen, E., Kansakoski, J., Vaaralahti, K., Iivonen, A. P., et al. (2017). GnRH receptor gene mutations in adolescents and young adults presenting with signs of partial gonadotropin deficiency. PLoS One 12:e188750. doi: 10.1371/journal.pone.0188750
Hu, Z., Ai, N., Chen, W., Wong, Q. W., and Ge, W. (2019). Loss of growth hormone gene (gh1) in zebrafish arrests folliculogenesis in females and delays spermatogenesis in males. Endocrinology 160, 568–586. doi: 10.1210/en.2018-00878
Jiang, Z. H., and Wu, J. Y. (1999). Alternative splicing and programmed cell death. Proc. Soc. Exp. Biol. Med. 220, 64–72. doi: 10.1046/j.1525-1373.1999.d01-11.x
Kim, D., Langmead, B., and Salzberg, S. L. (2015). HISAT: a fast spliced aligner with low memory requirements. Nat. Methods 12, 357–360. doi: 10.1038/nmeth.3317
Kim, P., Yang, M., Yiya, K., Zhao, W., and Zhou, X. (2020). ExonSkipDB: functional annotation of exon skipping event in human. Nucleic Acids Res. 48, D896–D907. doi: 10.1093/nar/gkz917
Lah, G. J., Li, J. S., and Millard, S. S. (2014). Cell-specific alternative splicing of Drosophila Dscam2 is crucial for proper neuronal wiring. Neuron 83, 1376–1388. doi: 10.1016/j.neuron.2014.08.002
Lee, Y., and Rio, D. C. (2015). Mechanisms and regulation of alternative Pre-mRNA Splicing. Annu. Rev. Biochem. 84, 291–323. doi: 10.1146/annurev-biochem-060614-034316
Lents, C. A. (2019). Review: kisspeptin and reproduction in the pig. Animal 13, 2986–2999. doi: 10.1017/S1751731119001666
Lilienthal, H., Hack, A., Roth-Harer, A., Grande, S. W., and Talsness, C. E. (2006). Effects of developmental exposure to 2,2,4,4,5-pentabromodiphenyl ether (PBDE-99) on sex steroids, sexual development, and sexually dimorphic behavior in rats. Environ. Health Perspect 114, 194–201. doi: 10.1289/ehp.8391
Lomniczi, A., Loche, A., Castellano, J. M., Ronnekleiv, O. K., Bosch, M., Kaidar, G., et al. (2013). Epigenetic control of female puberty. Nat. Neurosci. 16, 281–289. doi: 10.1038/nn.3319
Mathur, M., Kim, C. M., Munro, S. A., Rudina, S. S., Sawyer, E. M., and Smolke, C. D. (2019). Programmable mutually exclusive alternative splicing for generating RNA and protein diversity. Nat. Commun. 10:2673. doi: 10.1038/s41467-019-10403-w
Mayerle, M., Yitiz, S., Soulette, C., Rogel, L. E., Ramirez, A., Ragle, J. M., et al. (2019). Prp8 impacts cryptic but not alternative splicing frequency. Proc. Natl. Acad. Sci. U.S.A. 116, 2193–2199. doi: 10.1073/pnas.1819020116
McCabe, M. J., Allan, C. M., Foo, C. F., Nicholls, P. K., McTavish, K. J., and Stanton, P. G. (2012). Androgen initiates Sertoli cell tight junction formation in the hypogonadal (hpg) mouse. Biol. Reprod. 87:38. doi: 10.1095/biolreprod.111.094318
Mechaly, A. S., Vinas, J., and Piferrer, F. (2009). Identification of two isoforms of the Kisspeptin-1 receptor (kiss1r) generated by alternative splicing in a modern teleost, the Senegalese sole (Solea senegalensis). Biol. Reprod. 80, 60–69. doi: 10.1095/biolreprod.108.072173
Mizuta, H., Luo, W., Ito, Y., Mushirobira, Y., Todo, T., Hara, A., et al. (2013). Ovarian expression and localization of a vitellogenin receptor with eight ligand binding repeats in the cutthroat trout (Oncorhynchus clarki). Comp. Biochem. Physiol. B Biochem. Mol. Biol. 166, 81–90. doi: 10.1016/j.cbpb.2013.07.005
Nonneman, D. J., Schneider, J. F., Lents, C. A., Wiedmann, R. T., Vallet, J. L., and Rohrer, G. A. (2016). Genome-wide association and identification of candidate genes for age at puberty in swine. BMC Genet. 17:50. doi: 10.1186/s12863-016-0352-y
Oesterreich, F. C., Herzel, L., Straube, K., Hujer, K., Howard, J., and Neugebauer, K. M. (2016). Splicing of nascent RNA coincides with intron exit from RNA polymerase II. Cell 165, 372–381. doi: 10.1016/j.cell.2016.02.045
Ojeda, S. R., Hill, J., Hill, D. F., Costa, M. E., Tapia, V., Cornea, A., et al. (1999). The Oct-2 POU domain gene in the neuroendocrine brain: a transcriptional regulator of mammalian puberty. Endocrinology 140, 3774–3789. doi: 10.1210/endo.140.8.6941
Page, N. M. (2005). New challenges in the study of the mammalian tachykinins. Peptides 26, 1356–1368. doi: 10.1016/j.peptides.2005.03.030
Parent, A. S., Rasier, G., Matagne, V., Lomniczi, A., Lebrethon, M. C., Gerard, A., et al. (2008). Oxytocin facilitates female sexual maturation through a glia-to-neuron signaling pathway. Endocrinology 149, 1358–1365. doi: 10.1210/en.2007-1054
Patterson, J. L., Willis, H. J., Kirkwood, R. N., and Foxcroft, G. R. (2002). Impact of boar exposure on puberty attainment and breeding outcomes in gilts. Theriogenology 57, 2015–2025. doi: 10.1016/s0093-691x(02)00674-x
Pervouchine, D. D., Knowles, D. G., and Guigo, R. (2013). Intron-centric estimation of alternative splicing from RNA-seq data. Bioinformatics 29, 273–274. doi: 10.1093/bioinformatics/bts678
Plant, T. M. (2015a). Neuroendocrine control of the onset of puberty. Front. Neuroendocrinol. 38:73–88. doi: 10.1016/j.yfrne.2015.04.002
Plant, T. M. (2015b). 60 Years of Neuroendocrinology: the hypothalamo-pituitary–gonadal axis. J. Endocrinol. 226, T41–T54. doi: 10.1530/JOE-15-0113
Sanchez-Criado, J. E., Trudgen, K., Millan, Y., Blanco, A., Monterde, J., Garrido-Gracia, J. C., et al. (2012). Estrogen receptor (ESR) 2 partially offsets the absence of ESR1 in gonadotropes of pituitary-specific Esr1 knockout female mice. Reproduction 143, 549–558. doi: 10.1530/REP-11-0214
Sandberg, R., Zhang, L., Mayr, C., Kingsmore, S. F., Wang, E. T., Luo, S., et al. (2008). Alternative isoform regulation in human tissue transcriptomes. Nature 456, 470–476.
Seker, F. B., Yorulmaz, H., Kaptan, E., Caglayan, B., and Oztas, B. (2016). Gestational treatment of folic acid attenuates blood-brain barrier leakage in pregnant- and prepubertal rats after pentylenetetrazole-induced seizure. Nutr. Neurosci. 19, 55–62. doi: 10.1179/1476830514Y.0000000154
Sheynkman, G. M., Shortreed, M. R., Frey, B. L., and Smith, L. M. (2013). Discovery and mass spectrometric analysis of novel splice-junction peptides using RNA-Seq. Mol. Cell Proteomics 12, 2341–2353. doi: 10.1074/mcp.O113.028142
Singh, G. K., Balzer, B. W., Desai, R., Jimenez, M., Steinbeck, K. S., and Handelsman, D. J. (2015). Requirement for specific gravity and creatinine adjustments for urinary steroids and luteinizing hormone concentrations in adolescents. Ann. Clin. Biochem. 52(Pt 6), 665–671. doi: 10.1177/0004563215580385
Stagi, S., di Tommaso, M., Scalini, P., Lapi, E., Losi, S., Bencini, E., et al. (2016). Triple X syndrome and puberty: focus on the hypothalamus-hypophysis-gonad axis. Fertil Steril 105, 1547–1553. doi: 10.1016/j.fertnstert.2016.02.019
Sultan, M., Schulz, M. H., Richard, H., Magen, A., Klingenhoff, A., Scherf, M., et al. (2008). A global view of gene activity and alternative splicing by deep sequencing of the human transcriptome. Science 321, 956–960. doi: 10.1126/science.1160342
Sun, Q. Y., Miao, Y. L., and Schatten, H. (2009). Towards a new understanding on the regulation of mammalian oocyte meiosis resumption. Cell Cycle 8, 2741–2747. doi: 10.4161/cc.8.17.9471
Sutherland, H. G., Champion, M., Plays, A., Stuart, S., Haupt, L. M., Frith, A., et al. (2017). Investigation of polymorphisms in genes involved in estrogen metabolism in menstrual migraine. Gene 607, 36–40. doi: 10.1016/j.gene.2017.01.008
Tang, L. T., Ran, X. Q., Mao, N., Zhang, F. P., Niu, X., Ruan, Y. Q., et al. (2018). Analysis of alternative splicing events by RNA sequencing in the ovaries of Xiang pig at estrous and diestrous. Theriogenology 119, 60–68. doi: 10.1016/j.theriogenology.2018.06.022
Teixeira, F. K., Okuniewska, M., Malone, C. D., Coux, R., Rio, D. C., and Lehmann, R. (2017). piRNA-mediated regulation of transposon alternative splicing in the soma and germ line. Nature 552, 268–272. doi: 10.1038/nature25018
Vazquez-Borrego, M. C., Fuentes-Fayos, A. C., Venegas-Moreno, E., Rivero-Cortes, E., Dios, E., Moreno-Moreno, P., et al. (2019). Splicing machinery is dysregulated in pituitary neuroendocrine tumors and is associated with aggressiveness features. Cancers (Basel) 11:1439. doi: 10.3390/cancers11101439
Vijayakumar, N., Op, D. M. Z., Shirtcliff, E. A., and Pfeifer, J. H. (2018). Puberty and the human brain: insights into adolescent development. Neurosci. Biobehav. Rev. 92, 417–436. doi: 10.1016/j.neubiorev.2018.06.004
Villanueva, R. A. M., and Chen, Z. J. (2019). “ggplot2: elegant graphics for data analysis,”in Measurement: Interdisciplinary Research and Perspectives, 2nd Edn, 17, 160–167. doi: 10.1080/15366367.2019.1565254
Wahab, F., Khan, I. U., Polo, I. R., Zubair, H., Drummer, C., Shahab, M., et al. (2019). Irisin in the primate hypothalamus and its effect on GnRH in vitro. J. Endocrinol. 241, 175–187. doi: 10.1530/JOE-18-0574
Wang, F., Pan, J., Liu, Y., Meng, Q., Lv, P., Qu, F., et al. (2015). Alternative splicing of the androgen receptor in polycystic ovary syndrome. Proc. Natl. Acad. Sci. U.S.A. 112, 4743–4748. doi: 10.1073/pnas.1418216112
Wang, S., Miura, M., Jung, Y. K., Zhu, H., Li, E., and Yuan, J. (1998). Murine caspase-11, an ICE-interacting protease, is essential for the activation of ICE. Cell 92, 501–509. doi: 10.1016/s0092-8674(00)80943-5
Ward, C. M., To, T., and Pederson, S. M. (2020). ngsReports: a Bioconductor package for managing FastQC reports and other NGS related log files. Bioinformatics 36, 2587–2588. doi: 10.1093/bioinformatics/btz937
Wu, D. M., Wang, Y. J., Han, X. R., Wen, X., Wang, S., Shen, M., et al. (2018). LncRNA LINC00880 promotes cell proliferation, migration, and invasion while inhibiting apoptosis by targeting CACNG5 through the MAPK signaling pathway in spinal cord ependymoma. J. Cell Physiol. 233, 6689–6704. doi: 10.1002/jcp.26329
Wu, W., Zong, J., Wei, N., Cheng, J., Zhou, X., Cheng, Y., et al. (2018). CASH: a constructing comprehensive splice site method for detecting alternative splicing events. Brief Bioinform. 19, 905–917. doi: 10.1093/bib/bbx034
Yao, G., He, J., Kong, Y., Zhai, J., Xu, Y., Yang, G., et al. (2019). Transcriptional profiling of long noncoding RNAs and their target transcripts in ovarian cortical tissues from women with normal menstrual cycles and primary ovarian insufficiency. Mol. Reprod. Dev. 86, 847–861. doi: 10.1002/mrd.23158
Zhang, Z., Lotti, F., Dittmar, K., Younis, I., Wan, L., Kasim, M., et al. (2008). SMN deficiency causes tissue-specific perturbations in the repertoire of snRNAs and widespread defects in splicing. Cell 133, 585–600. doi: 10.1016/j.cell.2008.03.031
Keywords: alternative splicing, differentially spliced genes, hypothalamus-pituitary-ovary axis, gilts, puberty
Citation: Pan X, Li Q, Chen D, Gong W, Li N, Jiang Y, Zhang H, Chen Y and Yuan X (2021) Alternative Splicing Dynamics of the Hypothalamus–Pituitary–Ovary Axis During Pubertal Transition in Gilts. Front. Genet. 12:592669. doi: 10.3389/fgene.2021.592669
Received: 07 August 2020; Accepted: 24 March 2021;
Published: 30 April 2021.
Edited by:
Naoyuki Kataoka, The University of Tokyo, JapanReviewed by:
Yinghui Ling, Anhui Agricultural University, ChinaCopyright © 2021 Pan, Li, Chen, Gong, Li, Jiang, Zhang, Chen and Yuan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiaolong Yuan, eXhsQHNjYXUuZWR1LmNu
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.