
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Genet., 04 March 2021
Sec. Evolutionary and Population Genetics
Volume 12 - 2021 | https://doi.org/10.3389/fgene.2021.590830
This article is part of the Research TopicComparative Genomics and Functional Genomics Analyses in PlantsView all 37 articles
 Xiaochun Wei1†
Xiaochun Wei1† Md. Abdur Rahim2†
Md. Abdur Rahim2† Yanyan Zhao1
Yanyan Zhao1 Shuangjuan Yang1
Shuangjuan Yang1 Zhiyong Wang1
Zhiyong Wang1 Henan Su1
Henan Su1 Lin Li1
Lin Li1 Liujing Niu1
Liujing Niu1 Md. Harun-Ur-Rashid2
Md. Harun-Ur-Rashid2 Yuxiang Yuan1*
Yuxiang Yuan1* Xiaowei Zhang1*
Xiaowei Zhang1*Chinese cabbage is one of the most important and widely consumed vegetables in China. The developmental transition from the vegetative to reproductive phase is a crucial process in the life cycle of flowering plants. In spring-sown Chinese cabbage, late bolting is desirable over early bolting. In this study, we analyzed double haploid (DH) lines of late bolting (“Y410-1” and “SY2004”) heading Chinese cabbage (Brassica rapa var. pekinensis) and early-bolting Chinese cabbage (“CX14-1”) (B. rapa ssp. chinensis var. parachinensis) by comparative transcriptome profiling using the Illumina RNA-seq platform. We assembled 721.49 million clean high-quality paired-end reads into 47,363 transcripts and 47,363 genes, including 3,144 novel unigenes. There were 12,932, 4,732, and 4,732 differentially expressed genes (DEGs) in pairwise comparisons of Y410-1 vs. CX14-1, SY2004 vs. CX14-1, and Y410-1 vs. SY2004, respectively. The RNA-seq results were confirmed by reverse transcription quantitative real-time PCR (RT-qPCR). A Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis of DEGs revealed significant enrichment for plant hormone and signal transduction as well as starch and sucrose metabolism pathways. Among DEGs related to plant hormone and signal transduction, six unigenes encoding the indole-3-acetic acid-induced protein ARG7 (BraA02g009130), auxin-responsive protein SAUR41 (BraA09g058230), serine/threonine-protein kinase BSK11 (BraA07g032960), auxin-induced protein 15A (BraA10g019860), and abscisic acid receptor PYR1 (BraA08g012630 and BraA01g009450), were upregulated in both late bolting Chinese cabbage lines (Y410-1 and SY2004) and were identified as putative candidates for the trait. These results improve our understanding of the molecular mechanisms underlying flowering in Chinese cabbage and provide a foundation for studies of this key trait in related species.
Brassica belongs to the Brassicaceae family, which consists of several economically important vegetable crops consumed worldwide (Cheng et al., 2014). Vegetables in this genus exhibits various morphotypes, including leafy heads (cabbage, Brassica oleracea var. capitata; Chinese cabbage, Brassica rapa var. pekinensis), enlarged inflorescences (cauliflower, B. oleracea var. botrytis; broccoli, B. oleracea var. italica), enlarged axillary buds (Brussels sprouts, B. oleracea var. gemmifera), enlarged stems (kohlrabi, B. oleracea var. gongylodes), and enlarged roots (turnip, B. rapa subsp. rapa) (Zhao et al., 2005; Cheng et al., 2014, 2016). Furthermore, these vegetables contain various health-promoting secondary metabolites, including glucosinolates, phenolics, carotenoids, flavonoids, and anthocyanins, which have protective effects against inflammation, cardiovascular diseases, and age-related diseases (Manchali et al., 2012).
Brassica rapa subspecies show high morphological diversity (Zhao et al., 2005). In particular, the heading Chinese cabbage (B. rapa L. subsp. pekinensis) is one of the most important Brassica vegetables in Asian countries, especially in China, Korea, and Japan (Bong et al., 2012; Cheng et al., 2014). It is a major vegetable crop produced in China, where it is considered as a key source of mineral nutrition (Wu et al., 2008). Moreover, Chinese cabbage is the main ingredient of the most popular traditional Korean side dish kimchi (Bong et al., 2012; Lee et al., 2014). The flowering Chinese cabbage (B. rapa ssp. chinensis var. parachinensis) produces elongated, tender, and thick stalks with rapid bolting as edible organs (Cheng et al., 2014; Huang et al., 2017). Pak choi (B. rapa subsp. chinensis) produces smooth dark green leaves with a prominent white midrib instead of forming a leafy head (Zhao et al., 2005).
In angiosperms, flowering is the most important developmental transition in the plant life cycle (Song et al., 2015). This transition is controlled by endogenous and environmental signals (Song et al., 2015; Zhang et al., 2015). More than 180 genes identified by functional analyses are associated with flowering time in Arabidopsis (Fornara et al., 2010). Several of these genes form a complex regulatory network involving six key pathways, including photoperiod, vernalization, ambient temperature, age, autonomy, and gibberellin pathways (Fornara et al., 2010; Zhang et al., 2015; Huang et al., 2017). However, photoperiod and vernalization related to day length and low temperatures, respectively, have also been identified as the major pathways for the regulation of flowering time (reviewed in Song et al., 2013). Several homologs of Arabidopsis genes related to flowering in B. rapa and other plant species have been identified (Andersen et al., 2004; Greenup et al., 2009; Cheng et al., 2011; Duan et al., 2015; Xu et al., 2015). Mutations in the CONSTANS (CO), GIGANTEA (GI), and FLOWERING LOCUS T (FT), related to the photoperiod pathway, result in delayed flowering, but short days do not affect flowering time, unlike wild-type Arabidopsis (Suárez-López et al., 2001; Simpson and Dean, 2002; Moon et al., 2003). The CO gene encodes a zinc finger transcription factor that triggers the transcriptional upregulation of downstream floral integrator genes, including FT and SUPPRESSOR OF OVEREXPRESSION OF CO1 (SOC1), in leaves under long-day conditions (Samach et al., 2000; Srikanth and Schmid, 2011; Zhang et al., 2015). Furthermore, FT and SOC1 act as activators of floral meristem identity genes, such as LEAFY (LFY) and APETALA 1 (AP1) (Amasino, 2005). In Arabidopsis, FRIGIDA (FRI; encoding two coiled−coil motif-containing protein) and FLOWERING LOCUS C (FLC; encoding a MADS−box transcription factor) are involved in the annual winter habit (late flowering) (Michaels and Amasino, 1999; Johanson et al., 2000; Moon et al., 2003; Amasino, 2005). Mutations in these genes result in early flowering in Arabidopsis (Michaels and Amasino, 1999; Johanson et al., 2000; Choi et al., 2011). FRI positively regulates the transcription of the flowering repressor, FLC (Choi et al., 2011). In contrast, vernalization results in early flowering via suppressing the FLC (Michaels and Amasino, 1999; Sheldon et al., 1999). Three genes, VIN3 (VERNALIZATION INSENSITIVE 3), VRN2 (VERNALIZATION 2), and VRN1 (VERNALIZATION 1), are required for the cold-mediated repression of FLC (Kobayashi et al., 1999; Gendall et al., 2001; Sung and Amasino, 2004). Su et al. (2018) reported that BrVIN3.1 and BrFLC1 are the important genetic determinants of bolting time variation in B. rapa. Recent reports have revealed that there are various orthologs of FLC and FT for flowering time variation in B. rapa and B. oleracea (Schiessl et al., 2017). In B. rapa, there are two copies of FT on chromosomes A02 and A07 and four copies of FLC on chromosomes A02, A03, and A10 (reviewed in Schiessl et al., 2017). Moreover, Shu et al. (2018) reported a major quantitative trait locus (QTL) (Ef2.1) for early flowering in “broccoli × cabbage” and identified BolGRF6 as a putative candidate for early flowering in broccoli. BrSDG8 code for encoding a histone methyltransferase is associated with bolting in B. rapa ssp. pekinensis (Fu et al., 2020). Huang et al. (2020) found that histone methyltransferase CURLY LEAF (CLF; Bra032169) controls the expression of flowering-related genes, and mutation in the Bra032169 caused early bolting in Chinese cabbage.
Gibberellic acid (GA), a plant hormone, plays an important role in the regulation of flowering time (Bernier, 1988). In Arabidopsis, the application of exogenous GA promotes flowering under short-day conditions (Langridge, 1957; Chandler and Dean, 1994). Mutations in genes related to GA biosynthesis and signaling can alter flowering time in Arabidopsis (Wilson et al., 1992; Sun and Kamiya, 1994; Jacobsen et al., 1996). In Brassica napus, flower initiation, flowering time, and shoot elongation are regulated by endogenous GA (Dahanayake and Galwey, 1999). Exogenous GA can stimulate flower development, while an inhibitor of GA delays or inhibits flowering in Brassica (Rood et al., 1989; Zanewich et al., 1990).
In flowering Chinese cabbage, aerial vegetative parts and floral buds are consumed. The early-bolting flowering Chinese cabbage double haploid (DH) line “CX14-1” is characterized by rapid flower stalk development and early flowering. In contrast, late-bolting heading Chinese cabbage exhibits a long period of vegetative growth before flower bud initiation. In B. rapa, long-day conditions together with vernalization promote reproductive growth over vegetative growth; consequently, plants bolt before reaching the harvesting stage, which is a serious problem in this crop (Zhang et al., 2015). Therefore, late-bolting traits are desirable than early flowering for the cultivation of spring-sown Chinese cabbage.
In this study, we performed transcriptome sequencing of flower bud samples from early-bolting flowering Chinese cabbage (“CX14-1”) (B. rapa ssp. chinensis var. parachinensis) and late-bolting (“Y410-1” and “SY2004”) heading Chinese cabbage (B. rapa var. pekinensis) DH lines using the Illumina platform, with a focus on genes related to plant hormone signaling.
The late-bolting DH heading Chinese cabbage (B. rapa L. ssp. pekinensis) lines “Y410-1” and “SY2004” and a vernalization-independent early-bolting flowering Chinese cabbage (B. rapa ssp. chinensis var. parachinensis) DH line “CX14-1” were used (Figure 1). The plants were grown in the experimental field at Yuanyang (113° 97’ E and 35° 5’ N), Henan Academy of Agricultural Sciences, China. The flower primordia of these lines were used for RNA sequencing. The samples were collected, immediately frozen in liquid nitrogen, and stored at -80°C.

Figure 1. Phenotypes of early- and late-bolting Chinese cabbage double haploid lines used in the study.
The frozen flower bud samples were ground into a powder in liquid nitrogen. Thereafter, total RNA was isolated from 100 mg of powder using the RNeasy Mini Kit (Qiagen, Valencia, CA, United States) according to the manufacturer’s guidelines. The concentration, integrity, and purity of the RNA samples were determined using a NanoDrop spectrophotometer (NanoDrop Technologies, Wilmington, DE, United States) and Agilent 2100 Bio Analyzer (Agilent Technologies, Palo Alto, CA, United States). RNA samples with more than seven RNA integrity numbers (RINs) were used for RNA-seq library preparation with biological replications. Nine RNA-seq libraries were constructed using the NEBNext Ultra RNA Library Prep Kit for Illumina (New England Biolabs, Beverly, MA, United States) following the manufacturer’s recommendations. The clustering of the index-coded samples was performed on a cBot Cluster Generation System using the TruSeq PE Cluster Kit v4-cBot-HS (Illumina, San Diego, CA, United States) according to the manufacturer’s instructions. After cluster generation, the library preparations were sequenced on the Illumina high-throughput sequencing-by-synthesis technology platform to generate paired-end reads. The adapter sequences and low-quality reads were removed. The clean reads were then mapped to the B. rapa (Chiifu-401) v. 3.0 reference genome [Brassica database (BRAD)]1 using HISAT22.
The functions of assembled genes and novel gene discovery were performed by BLAST searches against the NR [National Center for Biotechnology Information (NCBI) non-redundant protein sequences3 ], Swiss-Prot4, gene ontology (GO)5, Clusters of Orthologous Groups of proteins (COG)6, Pfam7, and Kyoto Encyclopedia of Genes and Genomes (KEGG)8 databases. After prediction of the amino acid sequences of the new genes, HMMER (Eddy, 1998) was used for comparisons with the Pfam database to obtain annotation information.
Gene expression levels were estimated by fragments per kilobase of transcript per million fragments mapped (FPKM). The FPKM value for each gene was quantified according to the length of the gene and read count mapped to this gene. Differentially expressed genes (DEGs) between early-bolting and late-bolting Chinese cabbage lines were identified using the R package DEGseq (Wang et al., 2010). The resulting p-values were adjusted using the Benjamini and Hochberg method to control the false discovery rate. Genes with an adjusted p-value < 0.01 found by DEseq were identified as DEGs.
Gene ontology enrichment analysis of the DEGs was implemented by the GOseq R packages based on the Wallenius non-central hyper-geometric distribution (Young et al., 2010), which can adjust for gene length bias in DEGs. KEGG (Kanehisa et al., 2008) is a database resource for understanding high-level functions and utilities of the biological system from molecular-level information, especially large-scale molecular datasets generated by genome sequencing and other high-throughput experimental technologies (see text foot note 8). We used KOBAS (Mao et al., 2005) software to test the statistical enrichment of differential expression genes in KEGG pathways. A GO enrichment analysis of DEGs was performed using the GOseq R package based on the Wallenius non-central hyper-geometric distribution (Young et al., 2010), which can adjust for gene length bias in DEGs.
A total of 1 μg of RNA was used for cDNA synthesis using SuperScript III following the manufacturer’s protocol (Invitrogen, Gaithersburg, MD, United States). Then, the FPKM values for nine randomly selected genes were validated by reverse transcription quantitative real-time PCR (RT-qPCR) using the LightCycler 480II (Roche, Mannheim, Germany). A total of 45 ng of cDNA was used as a template for RT-qPCR with gene-specific primers (Supplementary Table 1) using 2 × SyGreen Mix (qPCRBIO Lo-ROX) (PCR Biosystems, London, United Kingdom). Thermocycling conditions were 95°C for 5 min, 45 cycles of 95°C for 10 s, 60°C for 10 s, and 72°C for 15 s. At the end of the PCR cycles, the Ct values were analyzed using LightCycler 480II software (Roche). The efficiency of each gene-specific primer was determined using pooled cDNA samples. The expression of each gene was normalized using the comparative 2–ΔΔCt method (Livak and Schmittgen, 2001) with BrActin as a reference gene.
We performed transcriptome analyses of two late-bolting heading Chinese cabbage (B. rapa L. ssp. pekinensis) DH lines (Y410-1 and SY2004) and one early-bolting flowering Chinese cabbage DH line (CX14-1) (B. rapa ssp. chinensis var. parachinensis) with three biological replications using Illumina sequencing technology. The raw reads generated by RNA-seq were deposited in the NCBI “Sequence Reads Archive” (SRA) under the accession number PRJNA605481. After removal of low-quality sequences, adapters, and ambiguous reads, a total of 721.49 million clean paired-end reads were obtained (Table 1). The clean reads for each sample totaled 8.81 Gb, and the Q30 base percentage was 92.71% or greater (Table 1). The clean reads were aligned with the B. rapa (Chiifu-401) v. 3.0 reference genome (BRAD, see text foot note 1), and the efficiency of the alignment ranged from 86.20 to 89.54%. Furthermore, variable splicing prediction, a gene structure analysis, and new gene discovery were performed based on the comparison. The clean reads were then assembled into 47,363 transcripts and 47,363 genes, of which 3,144 were predicted as novel genes (Supplementary Table 2).
A total of 30,375 DEGs (Supplementary Tables 3–5) were identified in pairwise comparisons (Y410-1 vs. SY2004, Y410-1 vs. CX14-1, and SY2004 vs. CX14-1) (Figure 2A). The most DEGs were found in Y410-1 vs. CX14-1 (12,932), with 7,913 upregulated and 5,019 downregulated genes (Figure 2). The fewest DEGs were found in Y410-1 vs. SY2004 (4732), with 2,925 and 1,807 up- and downregulated genes, respectively (Figure 2B). Overall, 1,445 DEGs were common to all comparisons. Volcano plots (Figure 3) and MA plots (Figure 4) were generated to summarize significant DEGs. The upregulated and downregulated genes in comparisons between each pair of early-bolting Chinese cabbage lines were determined by hierarchical clustering based on FPKM values (Figure 5).
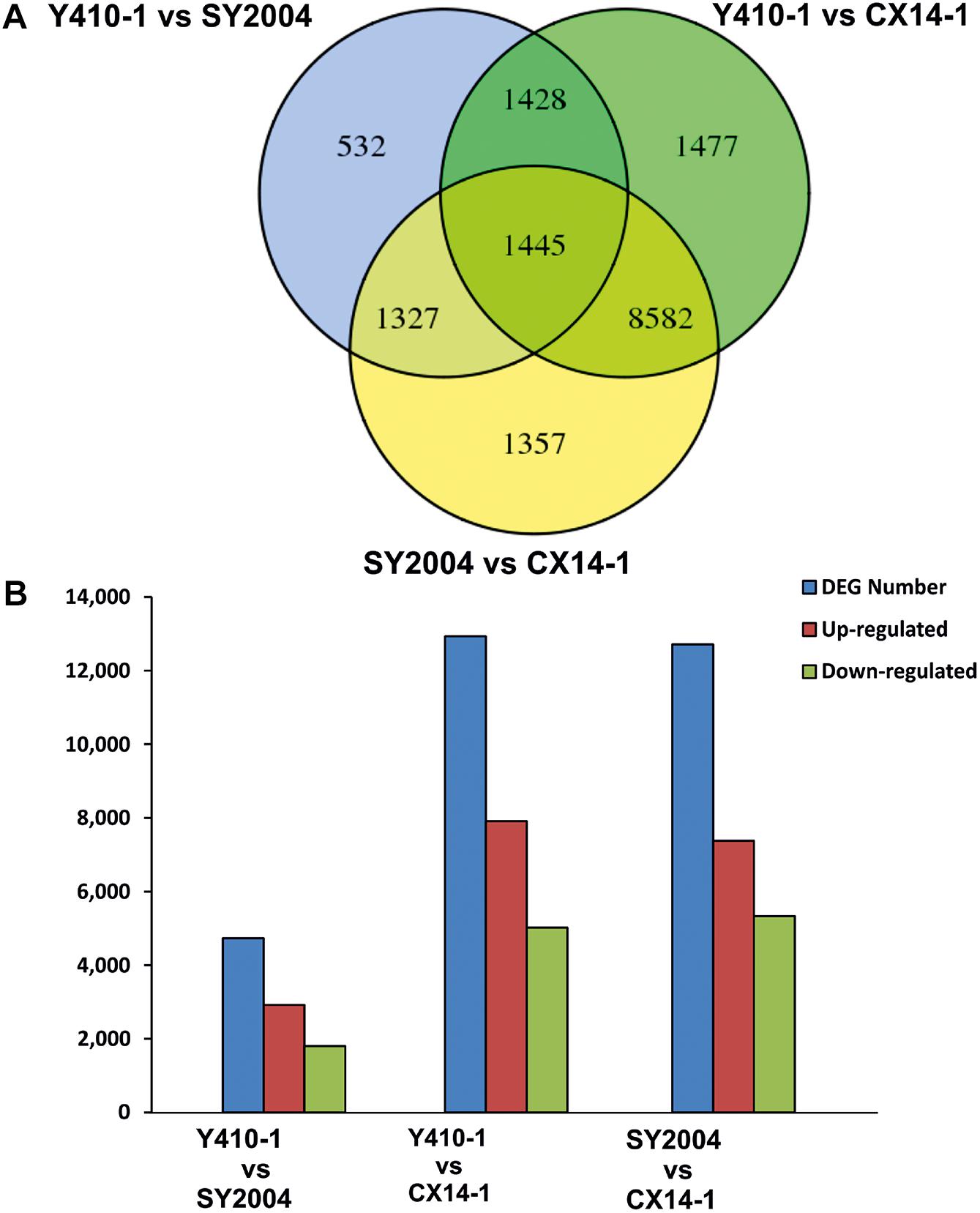
Figure 2. Differentially expressed genes (DEGs) between early- and late-bolting Chinese cabbage double haploid lines. (A) Venn diagram DEGs identified through pairwise comparisons. (B) Number of up- and downregulated genes in each comparison. Venn diagram was generated using the freely available VENNY 2.1 online tool (http://bioinfogp.cnb.csic.es/tools/venny/).
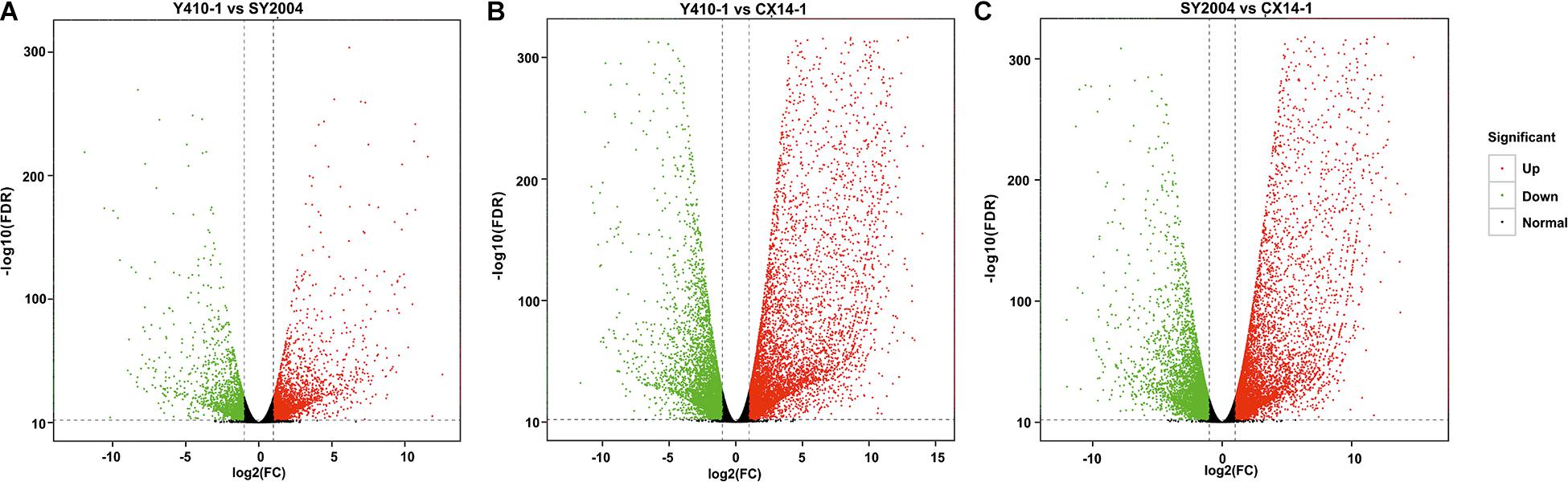
Figure 3. Volcano plots of differentially expressed genes (DEGs) for each comparison in flower bud of early- and late-bolting Chinese cabbage double haploid lines. The x and y axes indicate logarithm fold change [log2(FC)] of the difference in the expression of a gene between two samples and the negative logarithm of the statistical significance [log2(FDR)] of the change in gene expression, respectively. The red and green dots represent significantly up- and downregulated genes, respectively; while black dots represent non-differentiated genes. FC, fold change; FDR, false discovery rate.
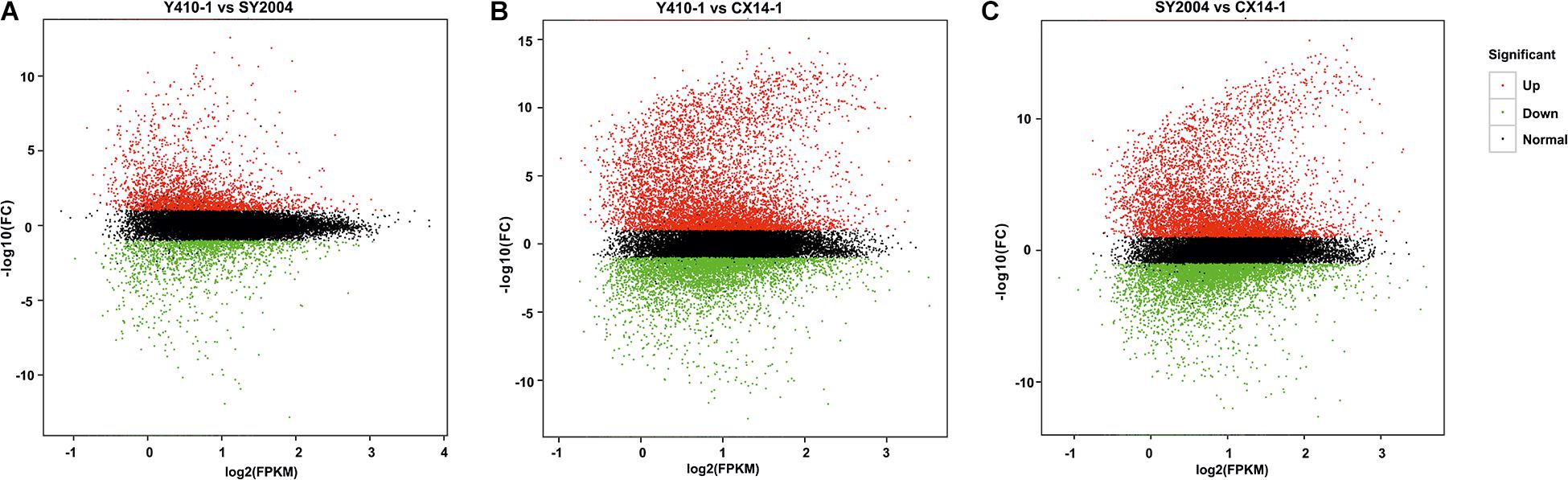
Figure 4. MA plot of differential expression for two samples. Each point represents the differentially expressed genes (DEGs). The x axis indicates log2(FPKM), and the y axis indicates log2(FC). The red and green dots represent significantly up- and downregulated genes, respectively; while black dots represent non-differentiated genes. FPKM, fragments per kilobase of transcript per million fragments mapped; FC, fold change.
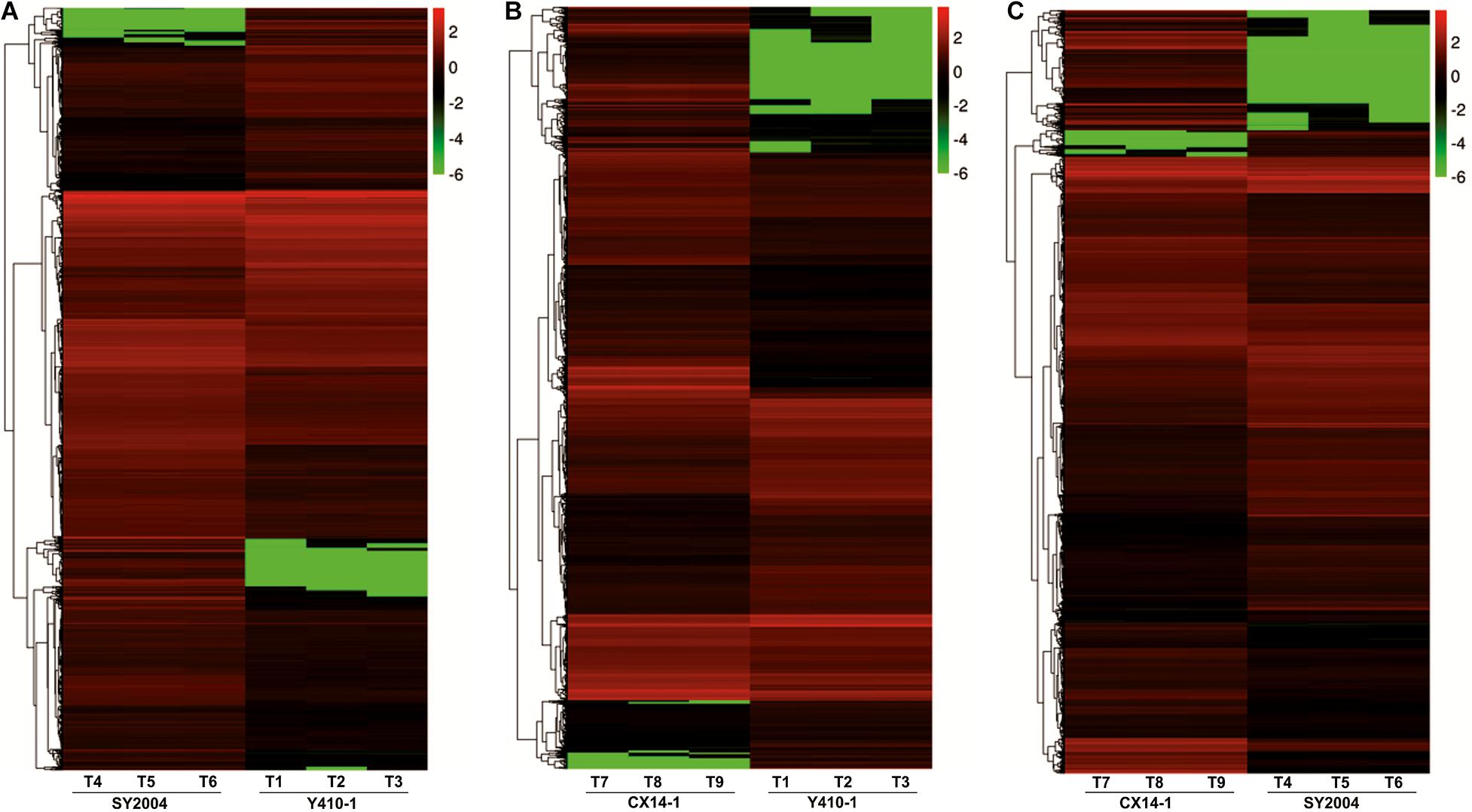
Figure 5. Heatmap representation of differentially expressed genes (DEGs) in the floral bud of early- and late-bolting Chinese cabbage double haploid lines. The expression patterns are based on the log2 of FPKM (fragments per kilobase of transcript per million fragments mapped for each gene) used for hierarchical clustering for each sample. The different columns represent different samples, with different rows representing different genes. Biological replicates: T1, T2, and T3 (Y410-1); T4, T5, and T6 (SY2004); and T7, T8, and T9 (CX14-1).
The putative functions of assembled genes were annotated by searches against public databases, including GO, COG, KEGG Orthology (KOG), KEGG, eggNOG, PFAM, NR, and SWISS-PROT. Among them, 47,214, 40,497, 35, 083, 32,944, and 24,390 genes were annotated to the NR, eggNOG, PFAM, SWISS-PROT, and KOG databases, respectively (Table 2). In addition, 2,564 of 3,144 novel genes were functionally annotated (Supplementary Table 2). Furthermore, we performed GO, COG, and KEGG pathway analyses to illustrate the biological functions of the Chinese cabbage floral bud transcriptomes. A GO term enrichment analysis was performed to identify terms in three general categories, biological process (BP), molecular function (MF), and cellular component (CC) (Berardini et al., 2004) (Figure 6A). A total of 37,394 genes were assigned to 54 main functional groups. The cellular process (GO:0009987), metabolic process (GO:0008152), and single-organism process (GO:0044699) were the most important in the BP category. Cell (GO:0005623), cell part (GO:0044464), and organelle (GO:0043226) were highly enriched in CC. Binding (GO:0005488) and catalytic activity (GO:0003824) were the most important GO terms in MF (Supplementary Table 6–8 and Figure 6A). These results indicated that early and late bolting might be associated with DEGs in these functional subgroups. According to a COG functional annotation analysis, 14,411 genes were classified into 25 COG categories (Figure 6B). The predominant COG categories represented in the Chinese cabbage floral bud transcriptomes were G (carbohydrate transport and metabolism), R (general function prediction only), and T (signal transduction mechanisms) (Figure 7B). Several genes in these categories were differentially expressed, which might contribute to flowering time differences between the early- and late-bolting Chinese cabbage DH lines.
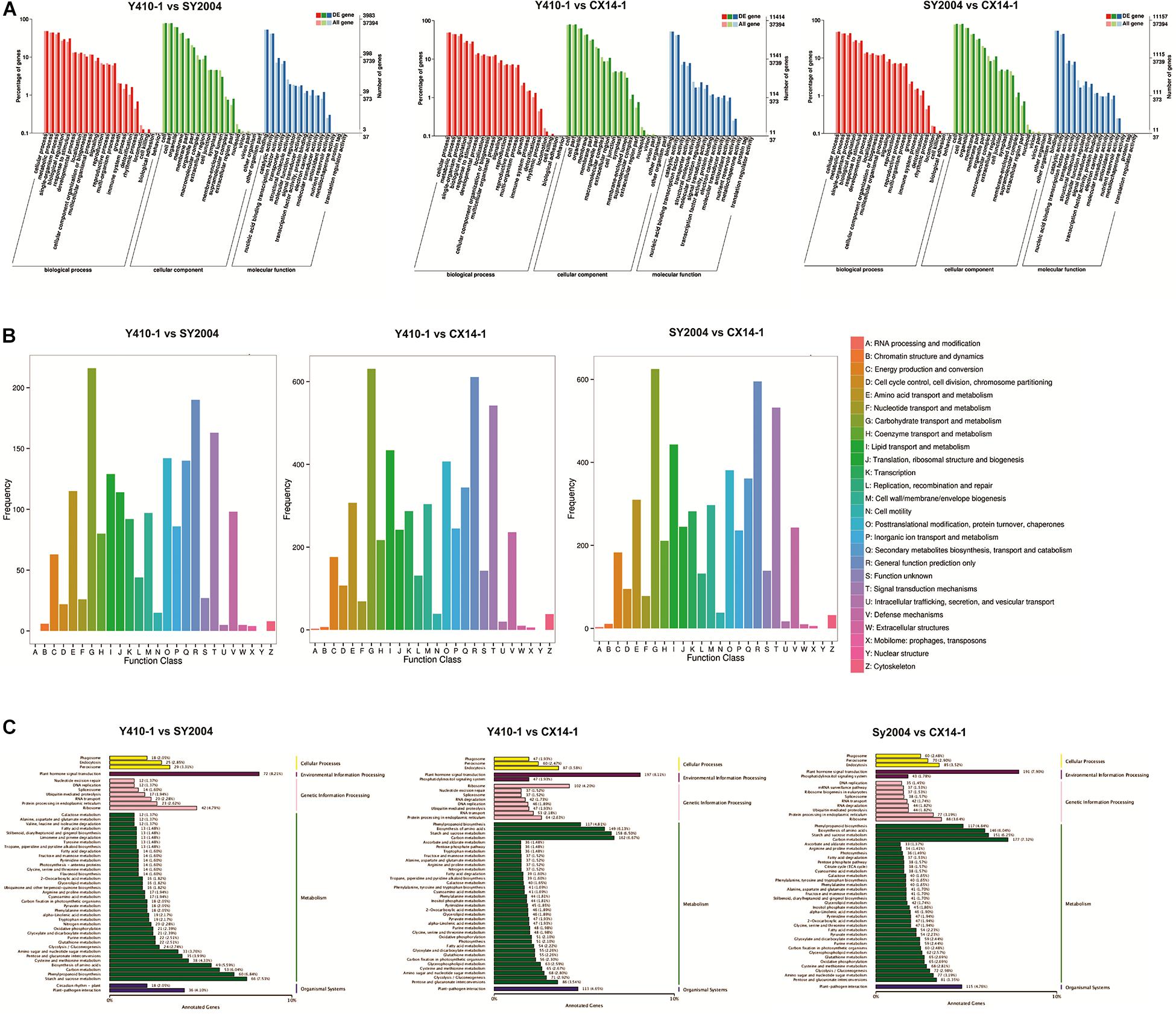
Figure 6. Functional classification of differentially expressed genes (DEGs) among early- and late-bolting Chinese cabbage double haploid lines. (A) Gene ontology (GO), (B) Clusters of Orthologous Groups of proteins (COG), and (C) Kyoto Encyclopedia of Genes and Genomes (KEGG) classification of DEGs.
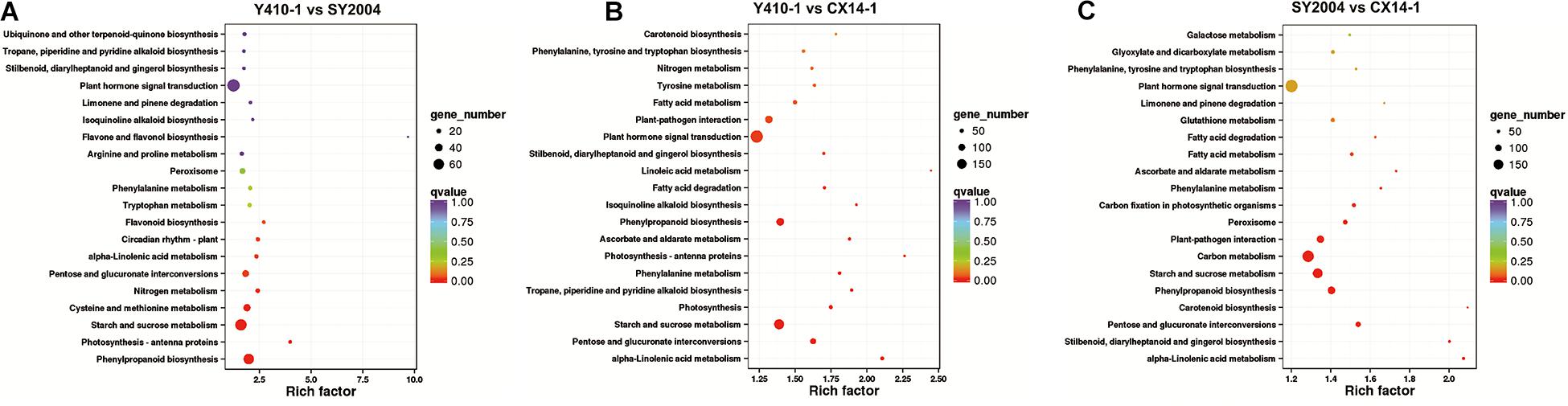
Figure 7. Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment scatter plot of differentially expressed genes. Rich factor is the ratio of the differentially expressed gene number to the total gene number in a certain pathway. The dot size and color indicate the number of genes and the range of the false discovery rate (FDR) value, respectively.
Different gene products interact with each other to exert biological functions, and pathway annotation analyses are therefore useful to predict the functions of gene. KEGG is a resource for the systematic analysis of gene function and genomic information databases, providing information about genes and expression patterns from a whole network perspective (Kanehisa et al., 2004). We therefore analyzed the metabolic pathways of the DEGs between early- and late-bolting Chinese cabbage lines using the KEGG database. Total 2,430 and 2,417 DEGs were assigned to 124, 128, and 127 pathways for Y410-1 vs. SY2004, Y410-1 vs. CX14-1, and SY2004 vs. CX14-1, respectively (Supplementary Table 9). Moreover, these pathways involved 15,408 genes, which were different from the DEGs assigned to pathways, indicating that some genes contribute to more than one KEGG pathway (Table 2 and Supplementary Table 9). For example, the novel gene predicted in this study “Brassica_rapa_newGene_2848” is involved in homologous recombination, mismatch repair, nucleotide excision repair, and DNA replication. However, six pathways, including plant–pathogen interaction, starch and sucrose metabolism, carbon metabolism, biosynthesis of amino acids, phenylpropanoid biosynthesis, and plant hormone signal transduction, contained over 100 DEGs between early- and late-bolting lines (SY410-1 vs. CX14-1 and SY2004 vs. CX14-1) (Figure 6C). Further, a KEGG pathway enrichment analysis of DEGs between early-and late-bolting lines revealed that plant hormone and signal transduction (197 genes) (Ko04075), starch and sucrose metabolism (158 genes) (Ko00500), and phenylalanine biosynthesis (117 genes) (Ko00940) are highly enriched for Y410-1 vs. CX14-1, while carbon metabolism (177 genes) (Ko01200), plant hormone and signal transduction (191 genes) (Ko04075), starch and sucrose metabolism (151 genes) (Ko00500), and phenylalanine biosynthesis (117 genes) (Ko00940) are highly enriched for SY2004 vs. CX14-1 (Figure 7 and Supplementary Table 8). Plant hormone and signal transduction as well as starch and sucrose metabolism were common to both comparisons. Among genes related to plant hormone and signal transduction, 98 and 99 genes were up- and downregulated in Y410-1 vs. CX14-1 (Supplementary Table 10), and 89 and 102 genes were up- and downregulated in SY2004 vs. CX14-1 (Supplementary Table 10).
The expression levels of most of the DEGs related to ABA receptors were downregulated, except PYL8 and PYL9, while most of the DEGs encoding ABA-insensitive five-like protein were upregulated in late-bolting Y410-1 compared with early-bolting flowering Chinese cabbage DH lines (Figure 8 and Supplementary Table 9). Similarly, the expression levels of most of the DEGs related to ABA receptors were downregulated, except for PYL8 and PYL9. In contrast, most of the DEGs related to ABA-insensitive 5-like proteins were upregulated in late-bolting compared with early-bolting flowering Chinese cabbage DH lines (Figure 8 and Supplementary Table 10).
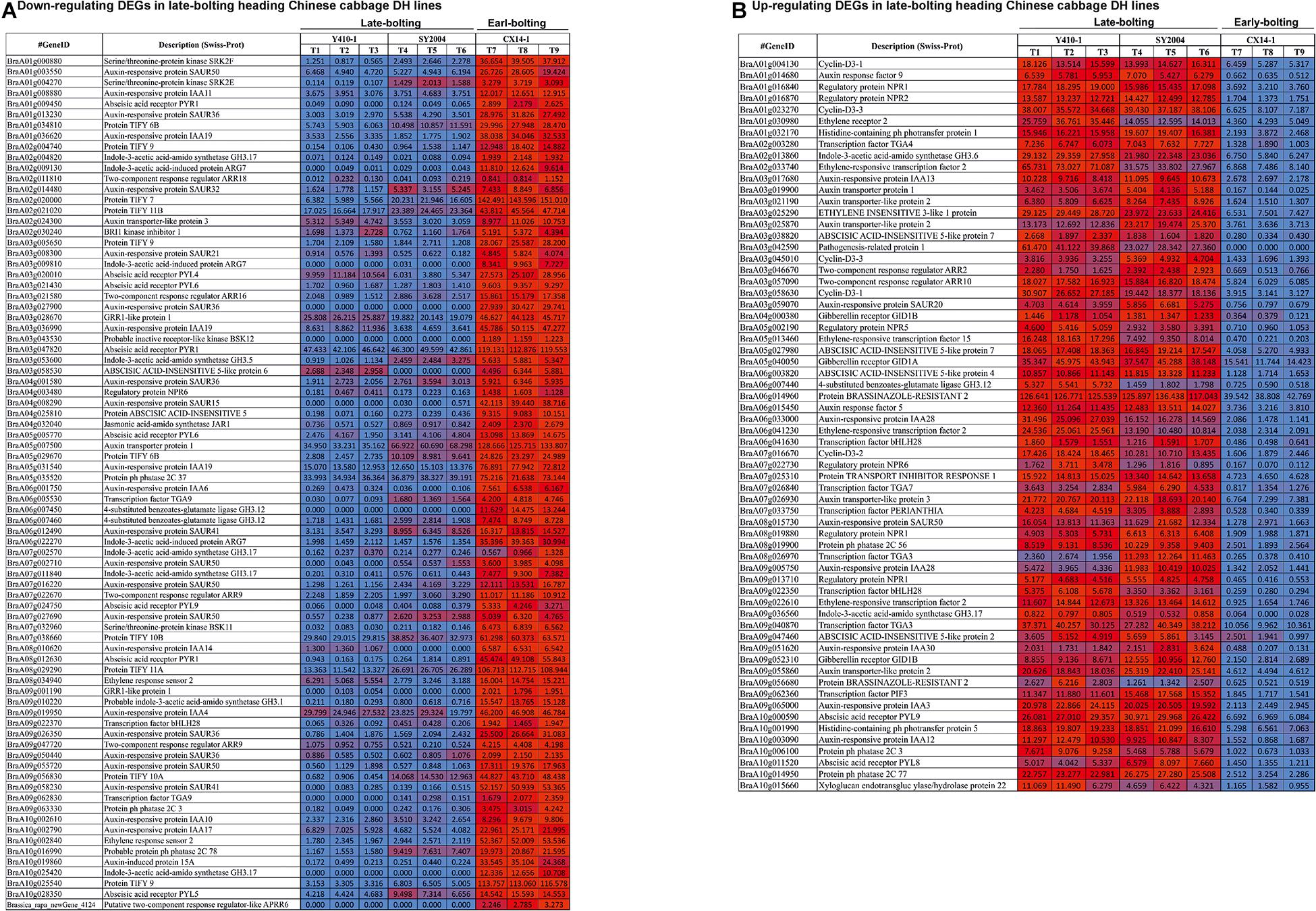
Figure 8. Heatmap representation of the expression pattern [fragments per kilobase of transcript per million fragments mapped (FPKM) values] of differentially expressed genes (DEGs) related to plant hormone and signal transduction. (A) Downregulated and (B) upregulated DEGs in both late-bolting heading Chinese cabbage double haploid (DH) lines.
The expression levels of DEGs encoding gibberellin receptors GID1A and GID1B were upregulated in both late-bolting (Y410-1 and SY2004) heading Chinese cabbage compared with the early-bolting flowering Chinese cabbage DH line (Figure 8 and Supplementary Table 10).
The expression levels of JA signaling genes, like jasmonate-amino synthetase (JAR1, a GH3 family of protein), were downregulated in both late-bolting heading Chinese cabbage DH lines (Y410-1 and SY2004) compared with the early-bolting flowering Chinese cabbage DH line (Figure 8 and Supplementary Table 10). Moreover, the expression levels of DEGs encoding TIFY proteins were upregulated in the early-bolting flowering Chinese cabbage DH line (CX14-1) compared with both late-bolting heading Chinese cabbage lines (Figure 8 and Supplementary Table 10).
The expression levels of DEGs encoding brassinazole-resistant 2 proteins (BraA09g056680, BraA06g014960, and BraA08g028240) and brassinosteroid-insensitive 1-associated receptor kinase 1 (BraA08g016610) were upregulated, whereas BRI1 kinase inhibitor 1 (BraA02g030240) was downregulated in both late-bolting heading Chinese cabbage lines compared with early-bolting flowering Chinese cabbage DH lines (Figure 8 and Supplementary Table 9). Furthermore, two DEGs related to DELLA protein RGA1 (BraA06g040430) and RGA2 (BraA09g023210) were upregulated in both late-bolting heading Chinese cabbage lines.
In case of ethylene signaling genes, DEGs encoding ethylene response sensors, such as ethylene response sensor 2 and ethylene-insensitive 3-like 3, were downregulated, while ethylene-responsive transcription factor 2 (BraA06g041230, BraA02g033740, and BraA09g022610), ethylene-responsive transcription factor 15 (BraA05g013460 and BraA04g022530), and ethylene receptors, such as ethylene-insensitive 3-like 1 (BraA03g025290), were upregulated in both late-bolting heading Chinese cabbage DH lines compared with the early-bolting flowering Chinese cabbage DH line (Figure 8 and Supplementary Table 10).
The unigenes related to auxin signal transduction, including auxin-responsive proteins, auxin transporter-like proteins, auxin responsive factor (ARF), and auxin-induced proteins, were differentially expressed between late-bolting heading Chinese cabbage and early-bolting flowering Chinese cabbage DH lines. The expression levels of 34 of these DEGs were upregulated in late-bolting heading Chinese cabbage Y410-1 compared with early-bolting flowering Chinese cabbage DH lines. Likewise, 27 and 33 DEGs were down- and upregulated, respectively, in another late-bolting DH line, SY2004 (Figure 8 and Supplementary Table 10). However, among these DEGs, 27 upregulated and 15 downregulated were common to both late-bolting lines (Supplementary Table 10).
Among the DEGs related to starch and sucrose metabolism, trehalose-phosphate phosphatase (A, B, G, and J), trehalase, alpha-amylase, beta-glucosidase, beta-fructofuranosidase, pectinesterase/pectinesterase inhibitor, sucrose-phosphate synthase, UDP-glucuronic acid decarboxylase, inactive beta-amylase, galacturonosyltransferase, UDP-glucuronate 4-epimerase, polygalacturonase, beta-D-xylosidase (2, 3, 5), alpha-glucosidase, UTP-glucose-1-phosphate uridylyltransferase 1, fructokinase (4, 5, 7), galacturonosyltransferase, polygalacturonate 4-alpha-galacturonosyltransferase, 1,4-alpha-glucan-branching enzyme, hexokinase, acid beta-fructofuranosidase, endoglucanase, alpha-trehalose-phosphate synthase, and exopolygalacturonase were downregulated in both late-bolting heading Chinese cabbage DH lines (Y410-1 and SY2004) compared with the early-bolting flowering Chinese cabbage (Figure 9). On the other hand, trehalose-phosphate phosphatase (I, H), glucan endo-1,3-beta-glucosidase, UTP-glucose-1-phosphate uridylyltransferase 2, alpha-glucan phosphorylase (1, 2), fructokinase-1, glucose-1-phosphate adenylyltransferase large subunit, beta-D-xylosidase (1, 4), starch synthase 1, and cellulose synthase-like protein D5 were upregulated in both late-bolting heading Chinese cabbage lines (Figure 9).
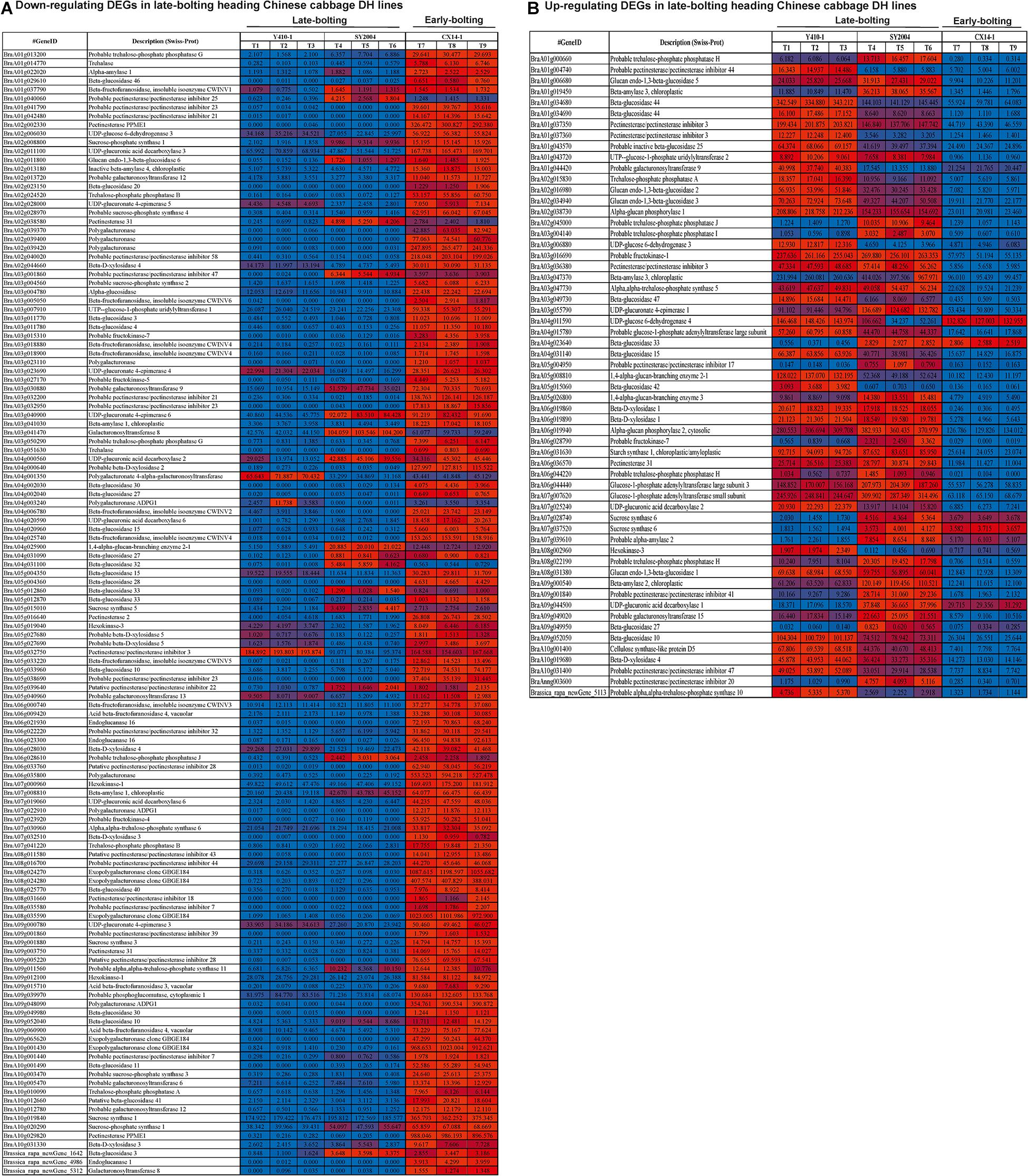
Figure 9. Heatmap representation of the expression pattern [fragments per kilobase of transcript per million fragments mapped (FPKM) values] of differentially expressed genes (DEGs) related to starch and sucrose metabolism. (A) Downregulated and (B) upregulated DEGs in both late-bolting heading Chinese cabbage double haploid (DH) lines.
We further tested the reliability of FPKM expression patterns (determined by RNA-seq) by RT-qPCR. Eight DEGs were randomly selected, and their relative expression levels were quantified using the same RNA samples extracted from early- and late-bolting Chinese cabbage lines for RNA sequencing. The results confirmed that the expression patterns of the analyzed unigenes were consistent with the FPKM expression pattern obtained by RNA-seq (Figure 10).
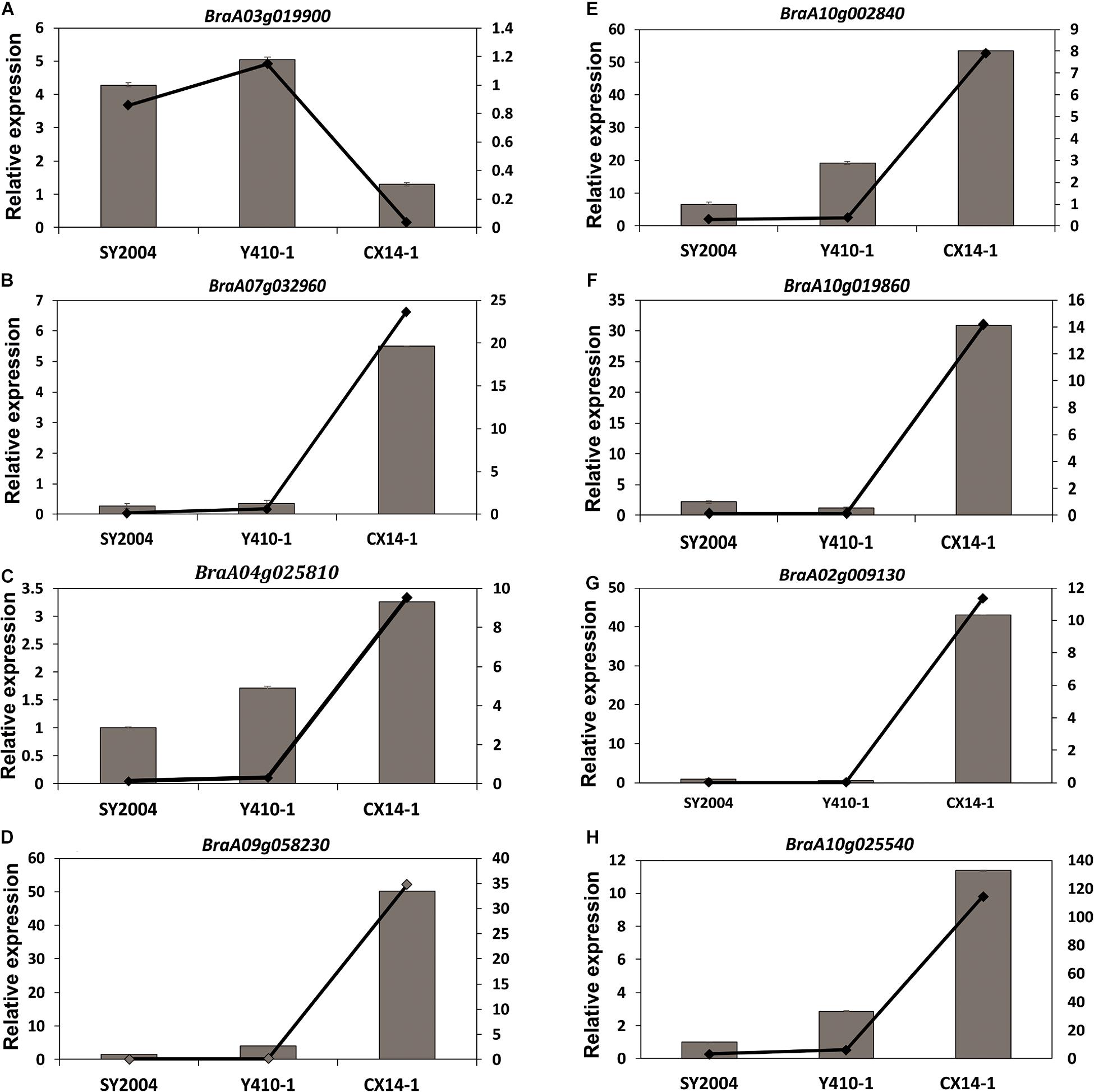
Figure 10. Relative expression of eight unigenes in the floral bud of early- and late-bolting Chinese cabbage double haploid lines. Error bar represents ± SE of the means of triplicates. The superimposed line graph represents the RNA-seq expression profiles [fragments per kilobase of transcript per million fragments mapped (FPKM)].
Late bolting is an important economic trait in spring-sown lines, and late-bolting varieties have been developed and characterized for off-season production to meet the annual demand for Chinese cabbage (Yang et al., 2007; Su et al., 2018). Nevertheless, little is known about the genes and pathways associated with flowering time in flowering Chinese cabbage (B. rapa ssp. chinensis var. parachinensis) and late-bolting heading Chinese cabbage (B. rapa var. pekinensis). RNA sequencing technology has been effectively utilized for transcriptome analyses in a wide range of texa (Wang et al., 2009; Lu et al., 2010). This high-throughput sequencing technology can be used to determine global gene expression differences between populations or species with phenotypic differences and responses to environmental stress (Mortazavi et al., 2008; Wang et al., 2009; Miao and Luo, 2013). In the present study, we performed a comparative transcriptome analysis of early-bolting Chinese cabbage DH lines by RNA-seq. We obtained 721.49 million clean paired-end reads assembled into 47,363 unigenes, including 3,144 genes predicted as novel (Table 1 and Supplementary Table 2). We detected 12,932 (Y410-1 vs. CX14-1) and 12,711 (SY2004 vs. CX14-1) DEGs between two sets of early- and late-bolting Chinese cabbage (Figure 2A and Supplementary Tables 4, 5).
A functional analysis of assembled unigenes revealed that the predominant COG categories were carbohydrate transport and metabolism, general function prediction only, and signal transduction mechanisms (Figure 6B). Several unigenes in these categories were differentially expressed and might explain the difference in flowering times in early-bolting Chinese cabbage DH lines. Moreover, we identified 3,144 novel unigenes, of which 2,564 were functionally annotated (Supplementary Table 2).
A KEGG pathway enrichment analysis of DEGs between early- and late-bolting DH lines indicated that plant hormone and signal transduction (Ko04075) and starch and sucrose metabolism (Ko00500) were the most enriched pathways in both comparisons (Supplementary Table 9). These results suggest that unigenes related to plant hormones, signal transduction, and sucrose metabolism are involved in the regulation of flowering time in these Chinese cabbage DH lines.
The “no hydrotropic response” (nhr1) Arabidopsis mutants show high levels of ABA, resulting in late flowering (Quiroz-Figueroa et al., 2010), whereas ABA-deficient (aba2 and aba3) or ABA-insensitive (abai4) mutants show early flowering than wild types (Martínez-Zapater et al., 1994; Matsoukas, 2014). Moreover, Zhu et al. (2013) demonstrated that ABA-insensitive 5 (ABI5, encoding a bZIP transcription factor) delayed flowering time in Arabidopsis under long-day conditions. Our results also indicated that the expression levels of most genes encoding ABI5-like proteins (ABA-insensitive 5-like protein 2, ABA-insensitive 5-like protein 4, ABA-insensitive 5-like protein 5, ABA-insensitive 5-like protein 6, and ABA-insensitive 5-like protein 7) were higher in late-bolting Chinese cabbage DH lines than in early-bolting flowering Chinese cabbage (Figure 8 and Supplementary Table 10).
Previous studies of mutants related to GA biosynthesis or signal transduction have revealed that GA can alter the flowering time (Wilson et al., 1992; Peng and Harberd, 1993; Sun and Kamiya, 1994; Jacobsen et al., 1996; Peng et al., 1997; Andres et al., 2014). Moreover, the DELLA domain protein RGA (repressor of ga1-3), GAI (GA insensitive), and RGA-like1 (RGL1 and RGL2) act as negative regulators of the GA signaling pathway (Yu et al., 2004; Silverstone et al., 2007). Our results revealed that two DEGs encoding DELLA proteins RGA1 (BraA06g040430) and RGA2 (BraA09g023210) are upregulated in late-bolting heading Chinese cabbage “Y410-1” (Figure 8 and Supplementary Table 10) and downregulated at the flowering stage compared with the vegetative stages in early-bolting flowering Chinese cabbage. These results suggest that the downregulation of negative regulators of the GA signaling pathway might increase GA levels, and the lack of function of these genes may trigger early bolting. Similar results have been reported by Huang et al. (2017), who reported that RGA1 and RGA2 are downregulated in flowering Chinese cabbage.
Several previous studies have indicated that the phytohormone jasmonic acid (JA) also regulates flowering time in Arabidopsis (Zhai et al., 2015). The F-box protein COI1 (coronatine insensitive 1) degrades JAZ (contains TIFY and Jas domains) repressors (Hoo and Howe, 2009). Transcript levels of NaJAZd and NaJAZh are upregulated in the early floral stages of NaJAZi-silenced plants due to the high JA content in the flowers (Li et al., 2017). Our results also showed that DEGs encoding TIFY proteins are more highly expressed in the early-bolting flowering Chinese cabbage DH line (CX14-1) than in both late-bolting heading Chinese cabbage lines (Figure 8 and Supplementary Table 10). These results suggest that the elevated expression of these genes in early flowering Chinese cabbage might be related to a high JA content.
Another steroidal phytohormone, brassinosteroid (BR), promotes flower induction in plants (Matsoukas, 2014). The BR signaling genes BZR1 (brassinazole-resistant1) and BES1 suppress the expression of important genes related to BR biosynthesis (CPD, constitutive photomorphogenesis, and dwarfism; DWF4, and DWARF4) by binding to their promoter regions in Arabidopsis (Wei et al., 2017). Moreover, BR-deficient/BR-insensitive mutants show delayed flowering time (Domagalska et al., 2007; Zhu et al., 2013). We also found that DEGs related to BZR1, BZR2, and BRI1 (brassinosteroid insensitive 1) were more highly expressed in both late-bolting heading Chinese cabbage DH lines than in early-bolting flowering Chinese cabbage (Figure 8 and Supplementary Table 10). These results suggest that the upregulation of BR signaling genes might affect BR biosynthesis and cause late bolting in late-bolting heading Chinese cabbage DH lines.
Starch and sucrose play important roles in flowering (Turnbull, 2011; Cho et al., 2018). Previous reports revealed that trehalose-6-phosphate acts as a signal molecule for flowering initiation in different plant species, including Arabidopsis thaliana (Sheen, 2014), grape (Caspari et al., 1998), and citrus (Shalom et al., 2014). Besides, Micallef et al. (1995) found that an increased level of endogenous sucrose promotes flowering in tomato. Wahl et al. (2013) reported that TREHALOSE-6-PHOSPHATE SYNTHASE 1 (TPS1) is required for the regulation of flowering time in A. thaliana. In the present study, alpha,alpha-trehalose-phosphate synthase 5 (BraA03g047730) was upregulated in both late-bolting heading Chinese cabbage lines compared with early-bolting flowering Chinese cabbage lines (Figure 9).
Razi et al. (2008) demonstrated that BoFLC alleles segregate independently from flowering time alleles in Brassica oleracea. However, Yuan et al. (2009) reported that variation in BrFLC1 is linked to flowering time in B. rapa. Moreover, Xie et al. (2015) performed a QTL analysis and showed that Br2 is the key determinant of BrFLC2 and a candidate flowering time locus in B. rapa. A transposon insertion in the coding sequence of BrFT2 located on a QTL on chromosome A07 (region Br5) causes late flowering (Zhang et al., 2015). Therefore, we further analyzed the flowering time-related genes, especially FLC and FT, based on FPKM expression values. Unigenes, such as FLK (BraA03g031700) and FLD (BraA03g034300), showed higher expression levels in both late-bolting lines than in early-bolting lines, while FLT (BraA02g016700) showed the opposite pattern (Supplementary Table 11). With regard to FT genes, the expression levels of three unigenes encoding proteins related to flowering time control, such as FCA (BraA01g020520), FY (BraA02g004930), and FPA (BraA09g036880), were higher in both late-bolting lines than in early-bolting lines (Supplementary Table 11). Nonetheless, none of these unigenes were differentially expressed between early- and late-bolting lines.
In late-bolting Chinese cabbage, 98 and 89 unigenes related to plant hormone and signal transduction were upregulated in Y410-1 vs. CX14-1 and SY2004 vs. CX14-1, respectively (Figure 8 and Supplementary Table 9). Among these, 19 and 12 unigenes showed log2 fold change values of > 5.0 in Y410-1 vs. CX14-1 and SY2004 vs. CX14-1, respectively; while six unigenes, including BraA02g009130 (indole-3-acetic acid-induced protein ARG7), BraA09g058230 (Auxin-responsive protein SAUR41), BraA07g032960 (serine/threonine-protein kinase BSK11), BraA10g019860 (auxin-induced protein 15A), BraA08g012630 (abscisic acid receptor PYR1), and BraA01g009450 (abscisic acid receptor PYR1), were common in both late-bolting heading Chinese cabbage lines (Y410-1 and SY2004). These unigenes are candidates for earl-bolting and late-bolting traits in these DH Chinese cabbage lines and could be useful for the development of molecular markers for the detection of early- and late-bolting cultivars.
Data is available at NCBI SRA accession: PRJNA605481.
XZ and YY conceived and designed the experiments. YZ, SY, and ZW performed the experiments. HS, LL, LN, and MH-U-R prepared the figures and tables. XW and MR drafted the work or revised it critically for important content. All authors contributed to the article and approved the submitted version.
This work was supported by the National Natural Science Foundation of China (31801874), the Programs for Science and Technology Development of Henan Province (212102110124 and 212102110126), the Modern Agro-Industry Technology Research System (CARS-23-G-16), and the Zhongyuan Scholar Program (202101510003).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2021.590830/full#supplementary-material
Amasino, R. M. (2005). Vernalization and flowering time. Curr. Opin. Biotech. 16, 154–158. doi: 10.1016/J.COPBIO.2005.02.004
Andersen, C. H., Jensen, C. S., and Petersen, K. (2004). Similar genetic switch systems might integrate the floral inductive pathways in dicots and monocots. Trends Plant Sci. 9, 105–107. doi: 10.1016/J.TPLANTS.2004.01.002
Andres, F., Porri, A., Torti, S., Mateos, J., Romera-Branchat, M., García-Martínez, J. L., et al. (2014). SHORT VEGETATIVE PHASE reduces gibberellin biosynthesis at the Arabidopsis shoot apex to regulate the floral transition Fernando Andrés. Proc. Natl. Acad. Sci. U.S.A. 111, E2760–E2769. doi: 10.1073/pnas.1409567111
Berardini, T. Z., Mundodi, S., Reiser, L., Huala, E., Garcia-Hernandez, M., Zhang, P., et al. (2004). Functional annotation of the Arabidopsis genome using controlled vocabularies. Plant Physiol. 135, 745–755. doi: 10.1104/pp.104.040071
Bernier, G. (1988). The control of floral evocation and morphogenesis. Ann. Rev. Plant Physiol. Plant Mol. Biol. 39, 175–219. doi: 10.1146/annurev.pp.39.060188.001135
Bong, Y. S., Shin, W. J., Gautam, M. K., Jeong, Y. J., Lee, A. R., Jang, C. S., et al. (2012). Determining the geographical origin of Chinese cabbages using multielement composition and strontium isotope ratio analyses. Food Chem. 135, 2666–2674. doi: 10.1016/J.FOODCHEM.2012.07.045
Caspari, H. W., Lang, A., and Alspach, P. (1998). Effects of girdling and leaf removal on fruit set and vegetative growth in grape. Am. J. Enol. Viti. 49, 359–366.
Chandler, J., and Dean, C. (1994). Factors influencing the vernalization response and flowering time of late flowering mutants of Arabidopsis thaliana (L.) Heynh. J. Exp. Bot. 45, 1279–1288. doi: 10.1093/jxb/45.9.1279
Cheng, F., Liu, S., Wu, J., Fang, L., Sun, S., Liu, B., et al. (2011). BRAD, the genetics and genomics database for Brassica plants. BMC Plant Biol. 11:136. doi: 10.1186/1471-2229-11-136
Cheng, F., Sun, R., Hou, X., Zheng, H., Zhang, F., Zhang, Y., et al. (2016). Subgenome parallel selection is associated with morphotype diversification and convergent crop domestication in Brassica rapa and Brassica oleracea. Nat. Genet. 48, 1218–1224. doi: 10.1038/ng.3634
Cheng, F., Wu, J., and Wang, X. (2014). Genome triplication drove the diversification of Brassica plants. Hort. Res. 1:14024. doi: 10.1038/hortres.2014.24
Cho, L. H., Pasriga, R., Yoon, J., Jeon, J. S., and An, G. (2018). Roles of sugars in controlling flowering time. J. Plant Biol. 61, 121–130. doi: 10.1007/s12374-018-0081-z
Choi, K., Kim, J., Hwang, H. J., Kim, S., Park, C., Kim, S. Y., et al. (2011). The FRIGIDA complex activates transcription of FLC, a strong flowering repressor in Arabidopsis, by recruiting chromatin modification factors. Plant Cell 23, 289–303. doi: 10.1105/tpc.110.075911
Dahanayake, S. R., and Galwey, N. W. (1999). Effects of Interactions between low-temperature treatments, gibberellin (GA3), and photoperiod on flowering and stem height of spring rape (Brassica napus var. annua). Ann. Bot. 84, 321–327. doi: 10.1006/ANBO.1999.0920
Domagalska, M. A., Schomburg, F. M., Amasino, R. M., Vierstra, R. D., Nagy, F., and Davis, S. J. (2007). Attenuation of brassinosteroid signaling enhances FLC expression and delays flowering. Development 134, 2841–2850. doi: 10.1242/dev.02866
Duan, W., Song, X., Liu, T., Huang, Z., Ren, J., Hou, X., et al. (2015). Genome-wide analysis of the MADS-box gene family in Brassica rapa (Chinese cabbage). Mol. Genet. Genom. 290, 239–255. doi: 10.1007/s00438-014-0912-7
Fornara, F., de Montaigu, A., and Coupland, G. (2010). Snapshot: control of flowering in Arabidopsis. Cell 141, 550–550. doi: 10.1016/j.cell.2010.04.024
Fu, W., Huang, S., Gao, Y., Zhang, M., Qu, G., Wang, N., et al. (2020). Role of BrSDG8 on bolting in Chinese cabbage (Brassica rapa). Theor. Appl. Genet. 133, 2937–2948. doi: 10.1007/s00122-020-03647-4
Gendall, A. R., Levy, Y. Y., Wilson, A., and Dean, C. (2001). The VERNALIZATION 2 gene mediates the epigenetic regulation of vernalization in Arabidopsis. Cell 107, 525–535.
Greenup, A., Peacock, W. J., Dennis, E. S., and Trevaskis, B. (2009). Molecular biology of seasonal flowering-responses in Arabidopsis and cereals. Ann. Bot. 103, 1165–1172. doi: 10.1093/aob/mcp063
Hoo, S. C., and Howe, G. A. (2009). A critical role for the TIFY motif in repression of jasmonate signaling by a stabilized splice variant of the JASMONATE ZIM-domain protein JAZ10 in Arabidopsis. Plant Cell 21, 131–145. doi: 10.1105/tpc.108.064097
Huang, S., Hou, L., Fu, W., Liu, Z., Li, C., Li, X., et al. (2020). An insertion mutation in Bra032169 encoding a histone methyltransferase is responsible for early bolting in chinese cabbage (Brassica rapa L. ssp. pekinensis). Front. Plant Sci. 11:547. doi: 10.3389/fpls.2020.00547
Huang, X., Lei, Y., Guan, H., Hao, Y., Liu, H., Sun, G., et al. (2017). Transcriptomic analysis of the regulation of stalk development in flowering Chinese cabbage (Brassica campestris) by RNA sequencing. Sci. Rep. 7:15517. doi: 10.1038/s41598-017-15699-6
Jacobsen, S. E., Binkowski, K. A., and Olszewski, N. E. (1996). SPINDLY, a tetratricopeptide repeat protein involved in gibberellin signal transduction in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 93, 9292–9296. doi: 10.1073/pnas.93.17.9292
Johanson, U., West, J., Lister, C., Michaels, S., Amasino, R., and Dean, C. (2000). Molecular analysis of FRIGIDA, a major determinant of natural variation in Arabidopsis flowering time. Science 290, 344–347.
Kanehisa, M., Araki, M., Goto, S., Hattori, M., Hirakawa, M., Itoh, M., et al. (2008). KEGG for linking genomes to life and the environment. Nucleic Acids Res. 36, D480–D484. doi: 10.1093/nar/gkm882
Kanehisa, M., Goto, S., Kawashima, S., Okuno, Y., and Hattori, M. (2004). The KEGG resource for deciphering the genome. Nucleic Acids Res. 32, 277–280. doi: 10.1093/nar/gkh063
Kobayashi, Y., Kaya, H., Goto, K., Iwabuchi, M., and Araki, T. (1999). A pair of related genes with antagonistic roles in mediating flowering signals. Science 286, 1960–1962. doi: 10.1126/science.286.5446.1960
Langridge, J. (1957). Effect of day-length and gibberellic acid on the flowering of Arabidopsis. Nature 180, 36–37. doi: 10.1038/180036a0
Lee, M. K., Chun, J. H., Byeon, D. H., Chung, S. O., Park, S. U., Park, S., et al. (2014). Variations in glucosinolates in 62 varieties of Chinese cabbage (Brassica rapa L. ssp. pekinensis) and their antioxidant activity. LWT Food Sci. Technol. 58, 93–101. doi: 10.1016/J.LWT.2014.03.001
Li, R., Wang, M., Wang, Y., Schuman, M. C., Weinhold, A., Schäfer, M., et al. (2017). Flower-specific jasmonate signaling regulates constitutive floral defenses in wild tobacco. Proc. Natl. Acad. Sci. U.S.A. 114, E7205–E7214. doi: 10.1073/pnas.1703463114
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Lu, T., Lu, G., Fan, D., Zhu, C., Li, W., Zhao, Q., et al. (2010). Functional annotation of the rice transcriptome at single-nucleotide resolution by RNA-seq. Genome Res. 20, 1238–1249. doi: 10.1101/gr.106120.110
Manchali, S., Chidambara Murthy, K. N., and Patil, B. S. (2012). Crucial facts about the health benefits of popular cruciferous vegetables. J. Funct. Foods 4, 94–106. doi: 10.1016/J.JFF.2011.08.004
Mao, X., Cai, T., Olyarchuk, J. G., and Wei, L. (2005). Automated genome annotation and pathway identification using KEGG Orthology (KO) as a controlled vocabulary. Bioinformatics 21, 3787–3793. doi: 10.1093/bioinformatics/bti430
Martínez-Zapater, J., Coupland, G., Dean, C., and Koornneef, M. (1994). Transition to flowering in Arabidopsis. Cold Spring Harb. Monogr. Arch. 27, 403–433.
Matsoukas, I. G. (2014). Interplay between sugar and hormone signaling pathways modulates floral signal transduction. Front. Genet. 5:218. doi: 10.3389/fgene.2014.00218
Miao, X., and Luo, Q. (2013). Genome-wide transcriptome analysis between small-tail Han sheep and the Surabaya fur sheep using high-throughput RNA sequencing. Reproduction 145, 587–596. doi: 10.1530/REP-12-0507
Micallef, B. J., Haskins, K. A., Vanderveer, P. J., Roh, K. S., Shewmaker, C. K., and Sharkey, T. D. (1995). Altered photosynthesis, flowering, and fruiting in transgenic tomato plants that have an increased capacity for sucrose synthesis. Planta 196, 327–334. doi: 10.1007/BF00201392
Michaels, S. D., and Amasino, R. M. (1999). FLOWERING LOCUS C encodes a novel MADS domain protein that acts as a repressor of flowering. Plant Cell 11, 949–956.
Moon, J., Suh, S. S., Lee, H., Choi, K. R., Hong, C. B., Paek, N. C., et al. (2003). The SOC1 MADS-box gene integrates vernalization and gibberellin signals for flowering in Arabidopsis. Plant J. 35, 613–623. doi: 10.1046/j.1365-313X.2003.01833.x
Mortazavi, A., Williams, B. A., McCue, K., Schaeffer, L., and Wold, B. (2008). Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods 5, 621–628. doi: 10.1038/nmeth.1226
Peng, J., Carol, P., Richards, D. E., King, K. E., Cowling, R. J., and Murphy, G. P. (1997). The Arabidopsis GAI gene defines a signaling pathway that negatively regulates gibberellin responses. Genes Devel. 11, 3194–3205. doi: 10.1101/gad.11.23.3194
Peng, J., and Harberd, N. P. (1993). Derivative alleles of the Arabidopsis gibberellin-insensitive (gai) mutation confer a wild type phenotype. Plant Cell 5, 351–360. doi: 10.1105/tpc.5.3.351
Quiroz-Figueroa, F., Rodríguez-Acosta, A., Salazar-Blas, A., Hernández-Domínguez, E., Campos, M. E., Kitahata, N., et al. (2010). Accumulation of high levels of ABA regulates the pleiotropic response of the nhr1 Arabidopsis mutant. J. Plant Biol. 53, 32–44. doi: 10.1007/s12374-009-9083-1
Razi, H., Howell, E. C., Newbury, H. J., and Kearsey, M. J. (2008). Does sequence polymorphism of FLC paralogues underlie flowering time QTL in Brassica oleracea? Theor. Appl. Genet. 116, 179–192. doi: 10.1007/s00122-007-0657-3
Rood, S. B., Mandel, R., and Pharis, R. P. (1989). Endogenous gibberellins and shoot growth and development in Brassica napus. Plant Physiol. 89, 269–273. doi: 10.1104/PP.89.1.269
Samach, A., Onouchi, H., Gold, S. E., Ditta, G. S., Schwarz-Sommer, Z., Yanofsky, M. F., et al. (2000). Distinct roles of CONSTANS target genes in the reproductive development of Arabidopsis. Science 288, 1613–1616.
Schiessl, S. V., Huettel, B., Kuehn, D., Reinhardt, R., and Snowdon, R. J. (2017). Flowering time gene variation in Brassica species shows evolutionary principles. Front. Plant Sci. 8:1742. doi: 10.3389/fpls.2017.01742
Shalom, L., Samuels, S., Zur, N., Shlizerman, L., Doron-Faigenboim, A., Blumwald, E., et al. (2014). Fruit load induces changes in global gene expression and in abscisic acid (ABA) and indole acetic acid (IAA) homeostasis in citrus buds. J. Exp. Bot. 65, 3029–3044. doi: 10.1093/jxb/eru148
Sheen, J. (2014). Master regulators in plant glucose signaling networks. J. Plant Biol. 57, 67–79. doi: 10.1007/s12374-014-0902-7
Sheldon, C. C., Burn, J. E., Perez, P. P., Metzger, J., Edwards, J. A., Peacock, W. J., et al. (1999). The FLF MADS box gene: a repressor of flowering in Arabidopsis regulated by vernalization and methylation. Plant Cell 11, 445–458. doi: 10.1105/tpc.11.3.445
Shu, J., Liu, Y., Zhang, L., Li, Z., Fang, Z., Yang, L., et al. (2018). QTL-seq for rapid identification of candidate genes for flowering time in broccoli× cabbage. Theor. Appl. Genet. 131, 917–928. doi: 10.1007/s00122-017-3047-5
Silverstone, A. L., Tseng, T. S., Swain, S. M., Dill, A., Sun, Y. J., Olszewski, N. E., et al. (2007). Functional analysis of spindly in gibberellin signaling in Arabidopsis. Plant Physiol. 143, 987–1000. doi: 10.1104/pp.106.091025
Simpson, G. G., and Dean, C. (2002). Arabidopsis, the rosetta stone of flowering time? Science 296, 285–289. doi: 10.1126/science.296.5566.285
Song, X., Duan, W., Huang, Z., Liu, G., Wu, P., Liu, T., et al. (2015). Comprehensive analysis of the flowering genes in Chinese cabbage and examination of evolutionary patterns of CO-like genes in the plant kingdom. Sci. Rep. 5:14631. doi: 10.1038/srep14631
Song, Y. H., Ito, S., and Imaizumi, T. (2013). Flowering time regulation: photoperiod and temperature sensing in leaves. Trends Plant Sci. 18, 575–583. doi: 10.1016/J.TPLANTS.2013.05.003
Srikanth, A., and Schmid, M. (2011). Regulation of flowering time: all roads lead to Rome. Cell. Mol. Life Sci. 68, 2013–2037. doi: 10.1007/s00018-011-0673-y
Su, T., Wang, W., Li, P., Zhang, B., Li, P., Xin, X., et al. (2018). A genomic variation map provides insights into the genetic basis of spring Chinese cabbage (Brassica rapa ssp. pekinensis) selection. Mol. Plant 11, 1360–1376. doi: 10.1016/j.molp.2018.08.006
Suárez-López, P., Wheatley, K., Robson, F., Onouchi, H., Valverde, F., and Coupland, G. (2001). CONSTANS mediates between the circadian clock and control of flowering in Arabidopsis. Nature 410, 1116–1120. doi: 10.1038/35074138
Sun, T. P., and Kamiya, Y. (1994). The Arabidopsis GA1 locus encodes the cyclase ent-kaurene synthetase a of gibberellin biosynthesis. Plant Cell 6, 1509–1518. doi: 10.1105/TPC.6.10.1509
Sung, S., and Amasino, R. M. (2004). Vernalization in Arabidopsis thaliana is mediated by the PHD finger protein VIN3. Nature 427, 159–164. doi: 10.1038/nature02195
Turnbull, C. (2011). Long-distance regulation of flowering time. J. Exp. Bot. 62, 4399–4413. doi: 10.1093/jxb/err191
Wahl, V., Ponnu, J., Schlereth, A., Arrivault, S., Langenecker, T., Franke, A., et al. (2013). Regulation of flowering by trehalose-6-phosphate signaling in Arabidopsis thaliana. Science 339, 704–707. doi: 10.1126/science.1230406
Wang, L., Feng, Z., Wang, X., Wang, X., and Zhang, X. (2010). DEGseq: an R package for identifying differentially expressed genes from RNA-seq data. Bioinformatics 26, 136–138. doi: 10.1093/bioinformatics/btp612
Wang, Y., Li, L., Ye, T., Lu, Y., Chen, X., and Wu, Y. (2013). The inhibitory effect of ABA on floral transition is mediated by ABI5 in Arabidopsis. J. Exp. Bot. 64, 675–684. doi: 10.1093/jxb/ers361
Wang, Z., Gerstein, M., and Snyder, M. (2009). RNA-Seq: a revolutionary tool for transcriptomics Nat. Rev. Genet. 10, 57–63. doi: 10.1038/nrg2484
Wei, Z., Yuan, T., Tarkowská, D., Kim, J., Nam, H. G., Novák, O., et al. (2017). Brassinosteroid biosynthesis is modulated via a transcription factor cascade of COG1, PIF4, and PIF5. Plant Physiol. 174, 1260–1273. doi: 10.1104/pp.16.01778
Wilson, R. N., Heckman, J. W., and Somerville, C. R. (1992). Gibberellin is required for flowering in Arabidopsis thaliana under short days. Plant Physiol. 100, 403–408.
Wu, J., Yuan, Y. X., Zhang, X. W., Zhao, J., Song, X., Li, Y., et al. (2008). Mapping QTLs for mineral accumulation and shoot dry biomass under different Zn nutritional conditions in Chinese cabbage (Brassica rapa L. ssp. pekinensis). Plant Soil 310, 25–40. doi: 10.1007/s11104-008-9625-1
Xie, Q., Lou, P., Hermand, V., Aman, R., Park, H. J., Yun, D. J., et al. (2015). Allelic polymorphism of GIGANTEA is responsible for naturally occurring variations in circadian period in Brassica rapa. Proc. Natl. Acad. Sci. U.S.A. 112, 3829–3834. doi: 10.1073/pnas.1421803112
Xu, L., Hu, K., Zhang, Z., Guan, C., Chen, S., Hua, W., et al. (2015). A genome-wide association study revealed the genetic architecture of flowering time in rapeseed (Brassica napus L.). DNA Res. 23:dsv035. doi: 10.1093/dnares/dsv035
Yang, X., Yu, Y. J., Zhang, F. L., Zou, Z. R., Zhao, X. Y., Zhang, D. S., et al. (2007). Linkage map construction and quantitative trait loci analysis for bolting based on a double haploid population of Brassica rapa. J. Int. Plant Biol. 49, 664–671. doi: 10.1111/j.1744-7909.2007.00447.x
Young, M. D., Wakefield, M. J., Smyth, G. K., and Oshlack, A. (2010). Gene ontology analysis for RNA-seq: accounting for selection bias. Genome Biol. 11:R14. doi: 10.1186/gb-2010-11-2-r14
Yu, H., Ito, T., Zhao, Y., Peng, J., Kumar, P., and Meyerowitz, E. M. (2004). Floral homeotic genes are targets of gibberellin signaling in flower development. Proc. Natl. Acad. Sci. U.S.A. 101, 7827–7832. doi: 10.1073/pnas.0402377101
Yuan, Y. X., Wu, J., Sun, R. F., Zhang, X. W., Xu, D. H., Bonnema, G., et al. (2009). A naturally occurring splice site mutation in the Brassica rapa FLC1 gene is associated with variation in flowering time. J. Exp. Bot. 60, 1299–1308. doi: 10.1093/jxb/erp010
Zanewich, K. P., Rood, S. B., and Williams, P. H. (1990). Growth and development of Brassica genotypes differing in endogenous gibberellin content I. Leaf and reproductive development. Physiol. Plant 79, 673–678. doi: 10.1111/j.1399-3054.1990.tb00043.x
Zhai, Q., Zhang, X., Wu, F., Feng, H., Deng, L., Xu, L., et al. (2015). Transcriptional mechanism of the jasmonate receptor COI1-mediated delay of flowering time in Arabidopsis. Plant Cell 27, 2814–2828. doi: 10.1105/tpc.15.00619
Zhang, X., Meng, L., Liu, B., Hu, Y., Cheng, F., Liang, J., et al. (2015). A transposon insertion in FLOWERING LOCUS T is associated with delayed flowering in Brassica rapa. Plant Sci. 241, 211–220. doi: 10.1016/J.PLANTSCI.2015.10.007
Zhao, J., Wang, X., Deng, B., Lou, P., Wu, J., Sun, R., et al. (2005). Genetic relationships within Brassica rapa as inferred from AFLP fingerprints. Theor. Appl. Genet. 110, 1301–1314.
Keywords: comparative transcriptome, early bolting, late bolting, differentially expressed genes, Chinese cabbage
Citation: Wei X, Rahim MA, Zhao Y, Yang S, Wang Z, Su H, Li L, Niu L, Harun-Ur-Rashid M, Yuan Y and Zhang X (2021) Comparative Transcriptome Analysis of Early- and Late-Bolting Traits in Chinese Cabbage (Brassica rapa). Front. Genet. 12:590830. doi: 10.3389/fgene.2021.590830
Received: 03 August 2020; Accepted: 18 January 2021;
Published: 04 March 2021.
Edited by:
Zhiyong Liu, Shenyang Agricultural University, ChinaReviewed by:
Pengtao Ma, Yantai University, ChinaCopyright © 2021 Wei, Rahim, Zhao, Yang, Wang, Su, Li, Niu, Harun-Ur-Rashid, Yuan and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yuxiang Yuan, eXV4aWFuZ3l1YW4xMjZAMTI2LmNvbQ==; Xiaowei Zhang, eGlhb3dlaTU3MzdAMTYzLmNvbQ==
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.