
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Genet. , 03 February 2021
Sec. Statistical Genetics and Methodology
Volume 12 - 2021 | https://doi.org/10.3389/fgene.2021.579900
Bladder cancer is one of the most common urogenital malignancies in the world, and there are no adequate prognostic indicators. CNTD2 is one of the atypical cyclins, which may be related to the cell cycle and even the development of cancers. Early studies have shown that CNTD2 is closely related to the occurrence and development of many malignant tumors. However, the mechanism of CNTD2 in bladder cancer has not been reported. In our research, we explored the different expressions of CNTD2 between 411 bladder cancers and 19 normal bladder tissues based on the TCGA dataset. CNTD2-related signaling pathways were identified through the GSEA. We analyzed the associations of CNTD2 expression and bladder cancer progression and survival using GSE13507. Compared with 19 cases of normal bladder tissue, CNTD2 gene expression was increased in 411 cases of bladder cancer. The high expression of CNTD2 strongly correlated with grade (P < 0.0001), T classification (P = 0.0001), N classification (P = 0.00011), M classification (P = 0.044), age (P = 0.027), and gender (P = 0.0012). Bladder cancer patients with high CNTD2 expression had shorter overall survival (P < 0.001). In the meantime, univariate and multivariate analyses showed that the increased expression of CNTD2 was an independent factor for poor prognosis in bladder cancer patients (P < 0.001 and P < 0.001, respectively). CNTD2 expression is closely related to bladder cancer progression, and the high expression of CNTD2 may be an adverse biomarker in bladder cancer patients.
Bladder cancer is one of the most common genitourinary tract malignancies in the world (Torre et al., 2015) and the fourth most common carcinoma for males (Siegel et al., 2018). About 3.4 million people were affected by bladder cancer in the world, with 430,000 new cases each year since 2015 (Payton, 2013; Disease et al., 2016). The incidence and death rates of bladder cancer have also been rising rapidly in China in the past few decades (Pang et al., 2016; Liu et al., 2018). Despite the advancements in both local and systemic treatment, the progression and recurrence rates of non-invasive bladder cancer are still high, while survival rates for invasive and metastatic cancers are low (Jiang et al., 2019). Recurrence or metastasis of bladder cancer may refer to complex molecular regulatory mechanisms (Spiess et al., 2017; Wu et al., 2017). The biological characteristics and molecular regulatory mechanisms of bladder cancer have been studied extensively, and some progress has been made. However, the gene regulatory network of bladder cancer remains obscure (Jones et al., 2016; Yoshida et al., 2017). Therefore, it is urgent to study the pathogenesis and molecular regulation of bladder cancer. The exploration of suitable new therapeutic targets is a potential direction for the diagnosis and treatment of this disease.
The deregulation of cell cycle control is a common characteristic of cancer. Cyclin-dependent kinases (CDKs) were discovered in 1982, and were found to control the process of the cell cycle through interaction and activation of cyclins (Casimiro et al., 2012). CDKs are highly conserved serine/threonine kinases that must bind to cyclins to phosphorylate their substrates (Malumbres and Barbacid, 2009). Specific cyclin-CDK complexes are related to each important transition in the cell cycle. Many carcinomas exhibit inappropriate expression of typical cyclin cells. While most of the research conducted has focused on the function of canonical cyclins, the function of other proteins displaying the same characteristic “cyclin box,” which defines the CDK binding (Murray and Marks, 2001; Malumbres and Barbacid, 2005; Malumbres, 2014), remains largely unknown. This group of cyclins was called “atypical” due to their structural characteristics (Sánchez-Botet et al., 2018). CNTD2/CCNP is one of the atypical cyclins, and is known to overexpress in colon cancer and lung cancer. CNTD2 may be a prognostic biomarker and therapeutic target in these two cancers (Gasa et al., 2017; Sánchez-Botet et al., 2018). However, the prognostic role of CNTD2 in bladder cancer remains unclear.
Here, we investigated the relationship between CNTD2 expression and bladder cancer. In the first place, we investigated the different expressions of CNTD2 in 411 bladder cancers and 19 normal bladder tissues based on data obtained from The Cancer Genome Atlas (TCGA). Secondly, in order to detect the biological pathways involved in the CNTD2 regulatory network related to the pathogenesis of bladder cancer, we conducted Gene Set Enrichment Analysis (GSEA). Finally, we surveyed the relationship between CNTD2 expression and bladder cancer progression, stage, and survival through GSE13507. Our results suggest that high CNTD2 expression is an adverse indicator of bladder cancer and is associated with poor outcomes.
This study included 411 bladder cancers and 19 normal bladder tissues from the TCGA official website1. In order to investigate the role of CNTD2 expression in the development of bladder cancer, we chose GSE13507 because GSE13507 has detailed clinical data information that can be used for subsequent analysis. GSE13507 was downloaded from the Gene Expression Omnibus (GEO) database2, which was published on May 6, 2010. In GSE13507, the researchers used 165 primary bladder cancer samples, 23 recurrent non-muscle invasive tumor tissues, 58 normal-looking bladder mucosae surrounding cancer, and 10 normal bladder mucosae for microarray analysis. The gene expression data of all patients were assessed by the Illumina Human-6 v2.0 Expression BeadChip. The design, quality control, and data normalization of all chip experiments conform to the standard Illumina protocol. The clinical data of GSE13507 was also downloaded from the NCBI GEO databases (see text footnote 2).
The different expressions of CNTD2 between 411 bladder cancers and 19 normal bladder tissues from the TCGA were investigated. GSEA was used to identify CNTD2-related signaling pathways. GSE13507 was identified in GEO, patients with CNTD2 expression values above the median were considered as CNTD2high, and the others were classified to be CNTD2low in all patients with bladder cancer. P-value < 0.05 in unpaired t test analysis and fold change (FC, log2) > 0.5 or ≤0.5 were utilized to determine the differential expression of genes (DEGs) (Figure 1).
The experiment design, data normalization, and quality control conform to the standard scheme. The study was carried out in accordance with the International Conference and the Helsinki Declaration.
All statistical analysis was conducted by R software 3.6.2. The GSE13507 was divided into two groups (CNTD2high, n = 82; CNTD2low, n = 83) based on the median values of CNTD2 expression. The limma package was used to test differential gene expression (Ritchie et al., 2015). The Cox regression multivariate analysis and the Kaplan-Meier method were used to evaluate the survival analysis, with group comparisons made using the log-rank test. Gene Ontology (GO) enrichment terms and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways were conducted through the clusterProfiler package (Yu et al., 2012).
The gene expressions data of 411 bladder cancers and 19 normal bladder tissues were downloaded from the TCGA. The CNTD2 gene was highly expressed in bladder cancers compared with normal bladder tissues (Figure 2).
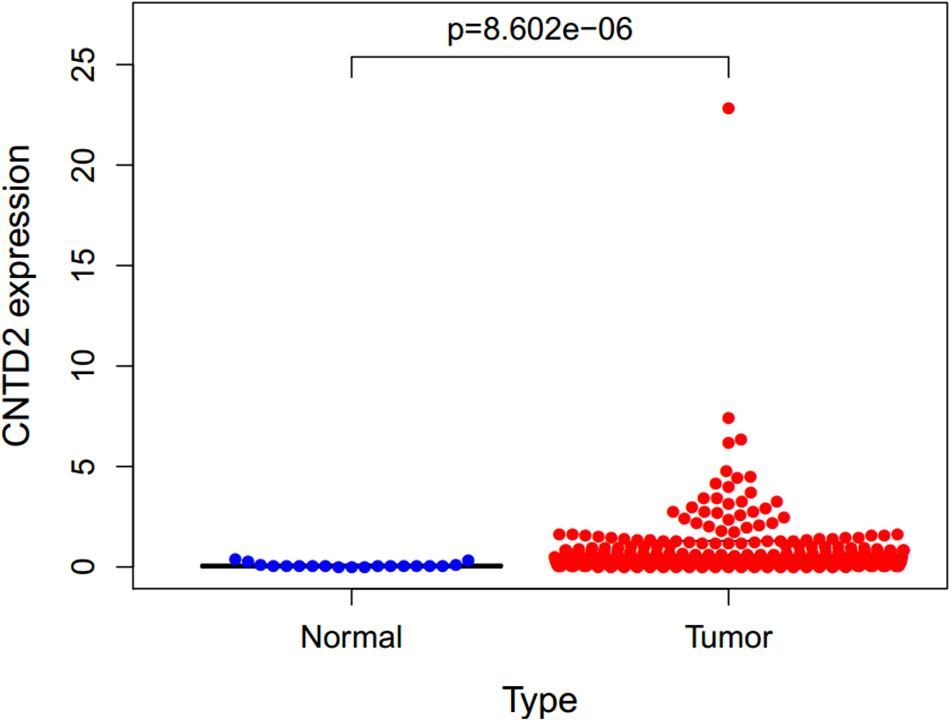
Figure 2. Expression of CNTD2 in 411 bladder cancers and 19 normal bladder tissues. DNA replication, RNA polymerase, base excision repair, and homologous recombination.
In order to investigate the different activation signaling pathways in bladder cancer, we performed GSEA between high and low CNTD2 expression datasets. GSEA was downloaded from https://www.gsea-msigdb.org/gsea/index.jsp. GSEA showed significant differences (NOM P-value < 0.05, FDR < 0.5) in the enrichment of the MSigDB collection (c2.cp.biocarta and h.all.v6.1.symbols). The most significant enrichment signaling pathways were selected according to the normalized enrichment score (NES) (Figure 3 and Table 1). Figure 3 demonstrates that DNA replication, RNA polymerase, base excision repair, and homologous recombination are differentially enriched in the CNTD2high expression phenotype.
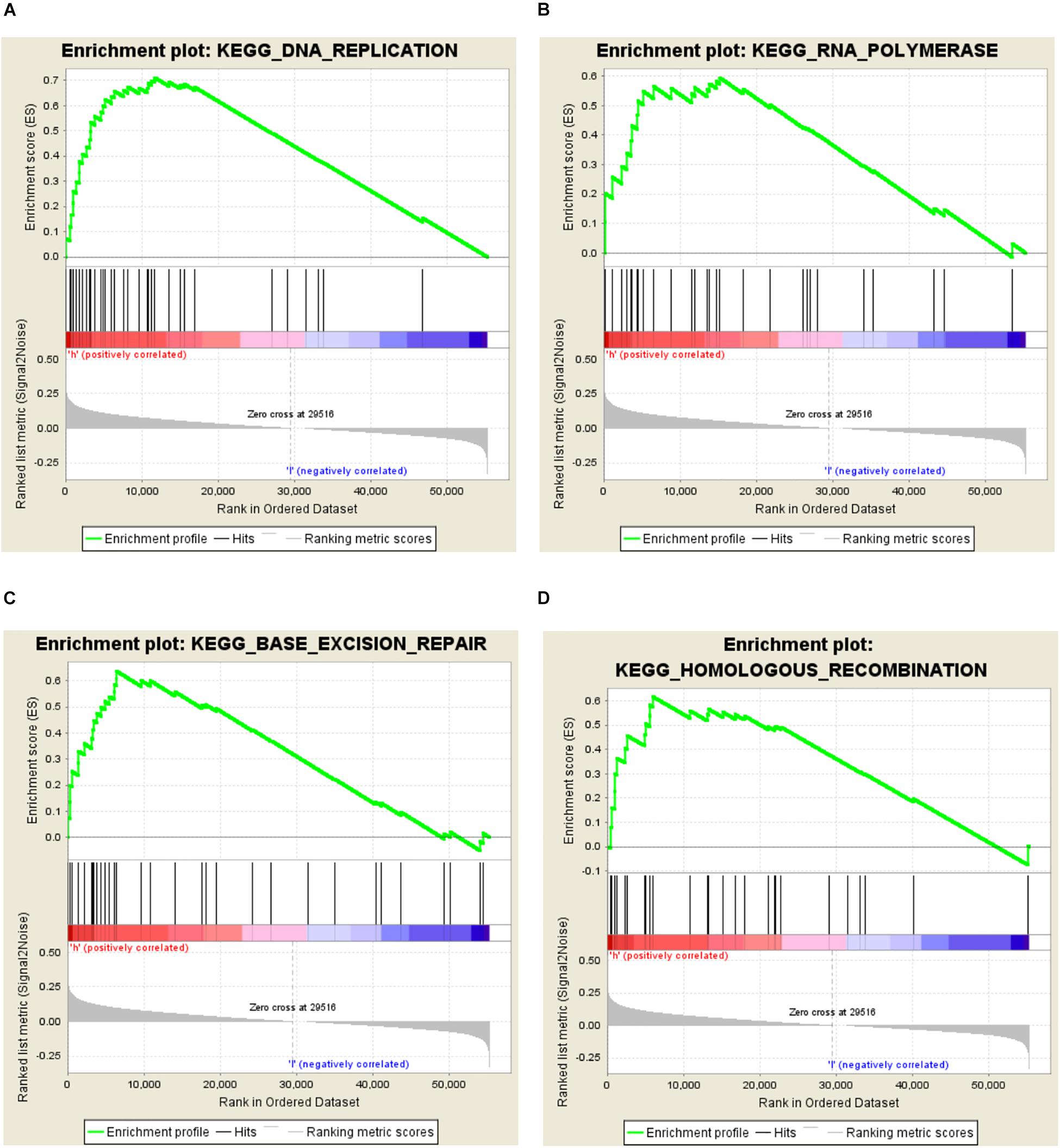
Figure 3. Enrichment plots from the Gene Set Enrichment Analysis (GSEA). GSEA results showing DNA replication (A), RNA polymerase (B), base excision repair (C), and homologous recombination (D) are differentially enriched in the CNTD2high expression phenotype.
We investigated the relationships between CNTD2 and clinicopathological characteristics using the GSE13507 dataset of 165 bladder cancer patients. Fuhrman grade is closely related to the differentiation of bladder cancer and the prognosis of patients. TNM stage is a very important classification of bladder cancer. T represents tumor size, N represents lymph node metastasis, and M represents distant metastasis. As shown in Figure 4, the high expression of CNTD2 strongly correlated with Fuhrman grade (P = 0.000), T classification (P = 0.000), N classification (P = 0.001), M classification (P = 0.044), gender (P = 0.001), and age (P = 0.027). The above results suggest that the high expression of CNTD2 is correlated with the prognosis of bladder cancer.
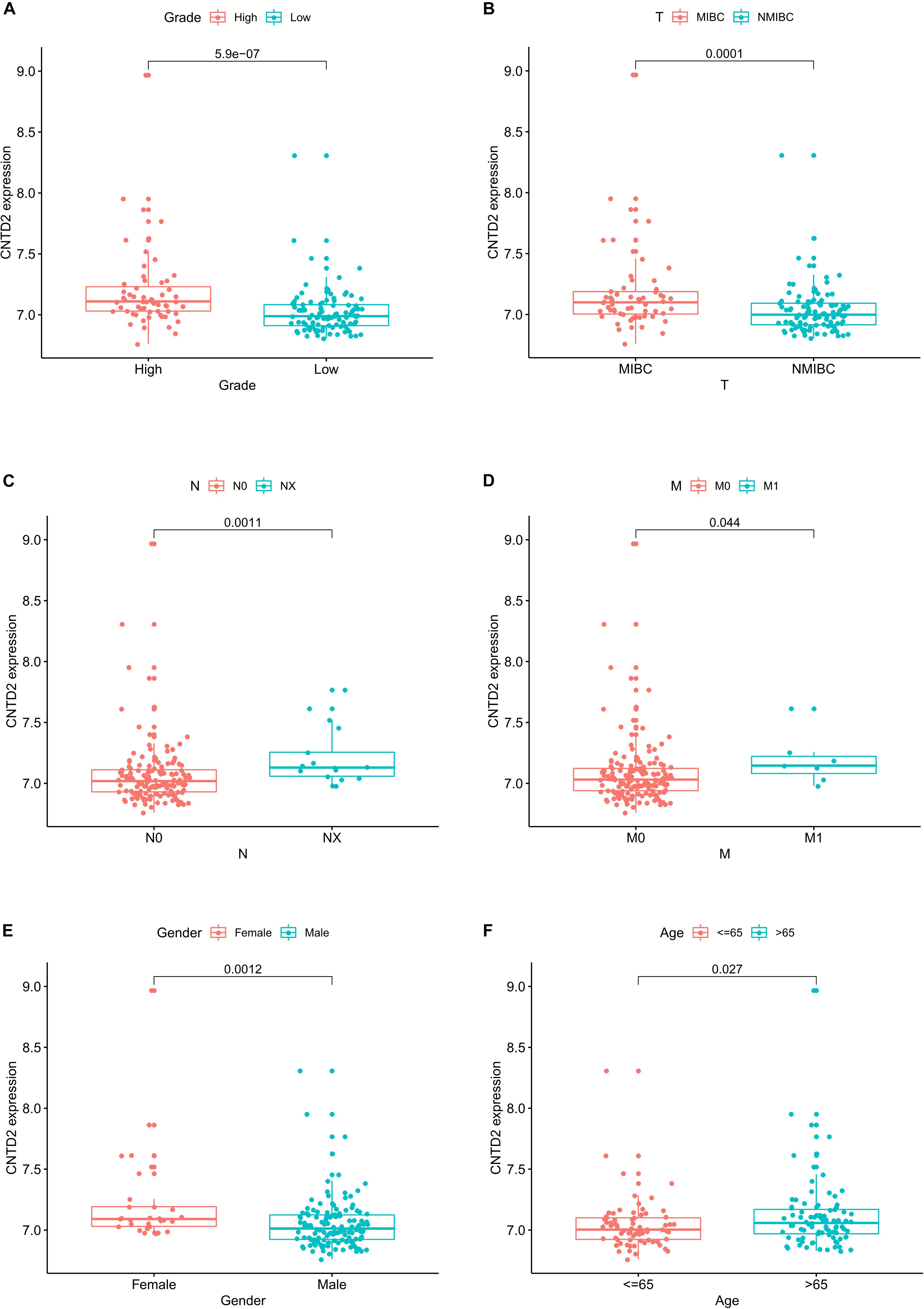
Figure 4. The relationships between CNTD2 expression and clinicopathological parameters in bladder cancer.
We calculated multivariate hazard ratios for different variables using a Cox regression model in 165 patients with bladder cancer from the GSE13507 dataset. Univariate analysis results demonstrated that CNTD2 overexpression (P < 0.001), age (P < 0.001), grade (P < 0.001), T classification (P < 0.001), N classification (P < 0.001), and M classification (P < 0.001) were all closely correlated with a poor prognosis (Figure 5A). Furthermore, multivariate analyses revealed that CNTD2 overexpression (P < 0.001), age (P < 0.001), and T classification (P < 0.001) were all independent predictors of an unfavorable prognosis (Figure 5B). In summary, the results showed that overexpression of CNTD2 could be used as a potential marker in the prognosis of patients with bladder cancer.
According to the median expression of CNTD2, we separated the patients into two groups: CNTD2low (n = 83) and CNTD2high (n = 82). These results suggest that the high expression of CNTD2 is related to the poor prognosis of bladder cancer. Thus, we further investigated the level of survival in the GSE13507 dataset. The results demonstrated that the CNTD2high group had significantly shorter OS compared with the CNTD2low group (Figure 6, P < 0.001).
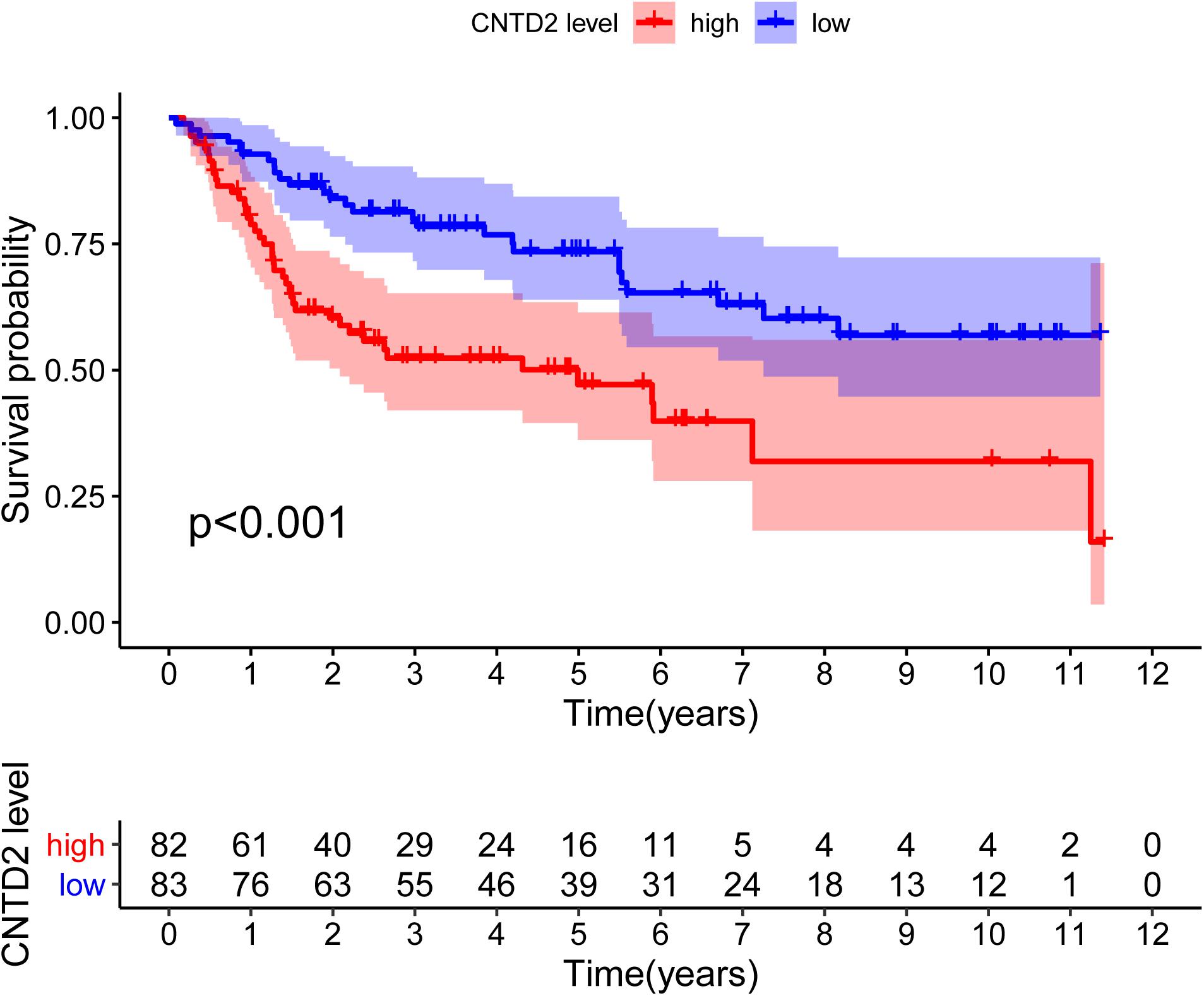
Figure 6. Overall survival curves of bladder cancer patients with high expression versus low expression of CNTD2.
For the sake of finding the genes related to CNTD2, we compared the differential expression values of CNTD2high versus CNTD2low. There were 246 downregulated genes and 211 upregulated genes [(FC, log2) > 0.5 or ≤0.5, P < 0.05, Figure 7A]. The first 20 downregulated and 20 upregulated genes were revealed in the heatmap (Figure 7B). We investigated the enriched GO terms and KEGG pathways using DEGs. In the biological process of GO, most DEGs are enriched in mitotic nuclear division (GO:0140014), nuclear division (GO:0000280), mitotic sister chromatid segregation (GO:0000070), regulation of mitotic nuclear division (GO:0007088), gland development (GO:0048732), regulation of nuclear division (GO:0051783), sister chromatid segregation (GO:0000819), organelle fission (GO:0048285), and embryonic organ development (GO:0048568) (Figure 7C). In the KEGG analysis results, cell cycle (hsa04110), small cell lung cancer (hsa05222), IL-17 signaling pathway (hsa04657), hepatocellular carcinoma (hsa05225), bladder cancer (hsa05219), and microRNAs in cancer (hsa05206) were the most enriched pathways (Figure 7D).
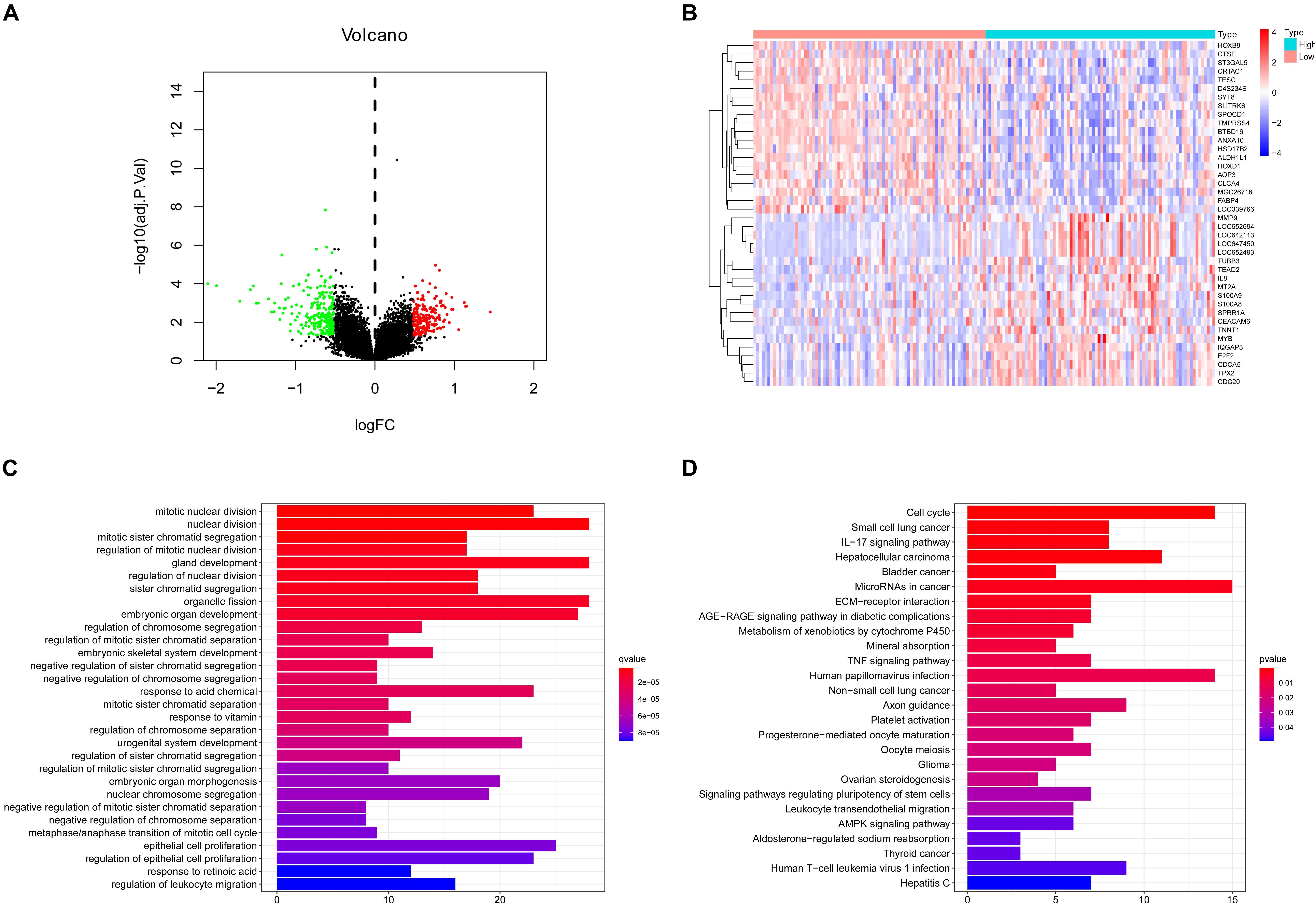
Figure 7. Different expression genes (DEGs), GO enrichment, and KEGG pathway analysis. (A) Volcano plot of the DEG expression between CNTD2high and CNTD2low. Cutoff criteria for DEG significance was P < 0.05 and the absolute value of the log2 fold change > 0.5. The red dots represent 211 upregulated genes, the green dots represent 246 downregulated genes, and black dots denote non-significant genes. (B) Heatmap demonstrates the top 20 upregulated genes and the top 20 downregulated genes. Red indicates high expression, white indicates intermediate expression, and blue represents low expression. (C,D) GO and KEGG analysis show different enriched pathways for differential expression genes.
In the end, we calculated the correlativity between the top 40 DEGs of CNTD2high versus CNTD2low (Figure 8A). We also screened the protein–protein interaction (PPI) network in the STRING database using the top 40 DEGs and CNTD2. Most of the downregulated genes and the upregulated genes interact in the PPI network (Figure 8B), and CDC20, MYB, CEACAM6, and MMP9 were all reported to be associated with cancer.

Figure 8. The correlation analysis and PPI results of DEGs. (A) The correlation analysis of DEGs through the Pearson correlation coefficient; the green dots mean negative correlation, while the red dots mean positive correlation. (B) PPI network of the top DEGs; green indicates downregulated genes, while red represents upregulated genes.
Bladder cancer is one of the leading causes of cancer death in the world (Spiess et al., 2017). After the diagnosis of bladder cancer, most patients showed a significant decline in health-related quality of life scores over time (Smith et al., 2018). The patient often develops a new lesion with a more advanced stage when bladder cancer is discovered anywhere within the urothelial tract. Bladder cancer is one of the deadliest tumors in the world, but its intracellular mechanism is still unknown. Therefore, it is necessary to explore new oncogenes to help in the early diagnosis of bladder cancer and predict the prognosis of patients with bladder cancer.
CNTD2 is an atypical cyclin whose function is not well understood until now. It is currently speculated that CNTD2 may be related to the cell cycle and even the development of cancers. CNTD2 has been shown to be overexpressed in colon and lung cancers, and it may be a prognostic marker and therapeutic target in these two cancers (Gasa et al., 2017; Sánchez-Botet et al., 2018). However, the prognostic role of CNTD2 in bladder cancer remains unclear.
In order to investigate the relationship between CNTD2 and bladder cancer, we performed this research. Our results exhibited that the CNTD2 gene is highly expressed in bladder cancers compared with normal bladder tissues. The CNTD2high expression phenotype was associated with DNA replication, RNA polymerase, base excision repair, and homologous recombination by GSEA. Importantly, some studies have shown that DNA replication (Preston et al., 2010; Suzuki and Takahashi, 2013; Mughal et al., 2019), RNA polymerase (Marshall and White, 2008; Ferreira et al., 2020), base excision repair (Wallace et al., 2012; Marsden et al., 2017), and homologous recombination (Bishop and Schiestl, 2001) are all associated with the development of cancers.
In our research, we also found that the high level of CNTD2 expression was related to an upward trend in bladder cancer progression. The low expression of CNTD2 in bladder cancer patients was pertinent to favorable prognosis. Our results showed significant correlations between CNTD2 expression and Fuhrman grade, T classification, N classification, M classification, gender, and age, which suggest that CNTD2 expression might be important for the acquisition of malignant potential in bladder cancer. Furthermore, multivariate Cox regression analysis showed that CNTD2 expression was an independent prognostic factor affecting overall survival in bladder cancer patients. Thus, the results of our study suggested that CNTD2 could be a valuable biomarker for the prediction of bladder cancer prognosis, and to the best of our knowledge, this is the first report to address that CNTD2 is associated with poor prognosis in bladder cancer.
The GO analysis showed that most DEGs were enriched in mitotic nuclear division, nuclear division, mitotic sister chromatid segregation, regulation of mitotic nuclear division, gland development, regulation of nuclear division, sister chromatid segregation, organelle fission, and embryonic organ development. The KEGG pathways were mainly enriched in cell cycle, small cell lung cancer, IL-17 signaling pathway, hepatocellular carcinoma, bladder cancer, and microRNAs in cancer. All of these results show that CNTD2 may be a tumor oncogene in bladder cancer.
We found that many cancer-related genes interact with CNTD2 through the PPI network. Gayyed et al. (2016) reported that high expression of CDC20 was closely related with advanced tumor stage in carcinoma of the colon, breast, endometrium, and prostate. Choi et al. (2013) demonstrated that elevated CDC20 expression is associated with poor prognosis in bladder cancer. CEACAM6 is a member of the CEA family. Abnormal overexpression of CEACAM6 plays a role in several characteristics of cancer, including uncontrolled proliferation, anoikis resistance, neovascularization, immune avoidance, invasion, and metastasis (Johnson and Mahadevan, 2015; Lee et al., 2015; Rizeq et al., 2018). MYB has now been investigated as an oncogene that is involved in some human leukemias. In addition, recent research suggests that MYB is activated in colon and breast cancers (Ramsay and Gonda, 2008).
In summary, the expression of CNTD2 may be a potential prognostic molecular marker for predicting the survival of bladder cancer. Combined with the above GO and KEGG analysis results, CNTD2 might interact with other bladder cancer-associated genes and was mainly involved in mitotic nuclear division, nuclear division, mitotic sister chromatid segregation, regulation of mitotic nuclear division, gland development, regulation of nuclear division, sister chromatid segregation, organelle fission, and embryonic organ development. Further experimental validation should be performed to prove the biologic impact of CNTD2. Nevertheless, our study is mainly based on the GSE13507 dataset, and further studies are still needed for verification.
The original contributions presented in the study are included in the article/supplementary material, further inquiries can be directed to the corresponding author/s.
The studies involving human participants were reviewed and approved by Ethics Committee of Zhongshan People’s Hospital. The patients/participants provided their written informed consent to participate in this study. Written informed consent was obtained from the individual(s) for the publication of any potentially identifiable images or data included in this article.
WD and RY conceived and designed the study. MG data collected and performed the analyses. MG, ES, and GH drafted the manuscript. MG, WN, and WD prepared the figures and table. All authors reviewed and revised the manuscript.
This study was supported by the Science and Technology Plan Project of Zhongshan City 2019B1064 and the Major Science and Technology Plan Project of Zhongshan City 2016B 1003.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Bishop, A. J., and Schiestl, R. H. (2001). Homologous recombination as a mechanism of carcinogenesis. Biochim. Biophys. Acta 1471, M109–M121. doi: 10.1016/s0304-419x(01)00018-x
Casimiro, M. C., Crosariol, M., Loro, E., Li, Z., and Pestell, R. G. (2012). Cyclins and cell cycle control in cancer and disease. Genes Cancer 3, 649–657. doi: 10.1177/1947601913479022
Choi, J. W., Kim, Y., Lee, J. H., and Kim, Y. S. (2013). High expression of spindle assembly checkpoint proteins CDC20 and MAD2 is associated with poor prognosis in urothelial bladder cancer. Virchows Arch. 463, 681–687. doi: 10.1007/s00428-013-1473-6
Disease, G. B. D., Injury, I., and Prevalence, C. (2016). Global, regional, and national incidence, prevalence, and years lived with disability for 310 diseases and injuries,1990-2015: a systematic analysis for the global burden of disease study. Lancet 388, 1545–1602. doi: 10.1007/s00586-017-5432-9
Ferreira, R., Schneekloth, J. S., Panov, K. I., Hannan, K. M., and Hannan, R. D. (2020). Targeting the RNA polymerase I transcription for cancer therapy comes of age. Cells 9:266. doi: 10.3390/cells9020266
Gasa, L., Sanchez-Botet, A., Quandt, E., Hernández-Ortega, S., Jiménez, J., and Carrasco-García, M. A. (2017). A systematic analysis of orphan cyclins reveals CNTD2 as a new oncogenic driver in lung cancer. Sci. Rep. 7:10228. doi: 10.1038/s41598-017-10770-8
Gayyed, M. F., El-Maqsoud, N. M., Tawfiek, E. R., El Gelany, S. A., and Rahman, M. F. (2016). A comprehensive analysis of CDC20 overexpression in common malignant tumors from multiple organs its correlation with tumor grade and stage. Tumour. Biol. 37, 749–762. doi: 10.1007/s13277-015-3808-1
Jiang, F., Qi, W., Wang, Y., Wang, W. H., and Fan, L. (2019). lncRNA PEG10 promotes cell survival, invasion and migration by sponging miR-134 in human bladder cancer. Biomed. Pharmacother. 114:108814. doi: 10.1016/j.biopha.2019.108814
Johnson, B., and Mahadevan, D. (2015). Emerging role and targeting of carcinoembryonic antigen-related cell adhesion molecule 6 (CEACAM6) in human malignancies. Clin. Cancer Drugs 2, 100–111. doi: 10.2174/2212697X02666150602215823
Jones, R. T., Felsenstein, K. M., and Theodorescu, D. (2016). Pharmacogenomics: biomarker-directed therapy for bladder cancer. Urol. Clin. North Am. 43, 77–86. doi: 10.1016/j.ucl.2015.08.007
Lee, O. J., Son, S. M., Hong, K. P., Lee, Y. M., Kim, M. Y., Choi, J. W., et al. (2015). CEACAM6 as detected by the AP11 antibody is a marker notable for mucin-producing adenocarcinomas. Virchows Arch. 466, 151–159. doi: 10.1007/s00428-014-1688-1
Liu, X., Cheng, X., Liu, X., He, L., Zhang, W., and Wang, Y. (2018). Investigation of the urinary metabolic variations and the application in bladder cancer biomarker discovery. Int. J. Cancer 143, 408–418. doi: 10.1002/ijc.31323
Malumbres, M., and Barbacid, M. (2005). Mammalian cyclin-dependent kinases. Trends Biochem. Sci. 30, 630–641. doi: 10.1016/j.tibs.2005.09.005
Malumbres, M., and Barbacid, M. (2009). Cell cycle, CDKs and cancer: a changing paradigm. Nat. Rev. Cancer 9, 153–166. doi: 10.1038/nrc2602
Marsden, C. G., Dragon, J. A., Wallace, S. S., and Sweasy, J. B. (2017). Base excision repair variants in cancer. Methods Enzymol. 591, 119–157. doi: 10.1016/bs.mie.2017.03.003
Marshall, L., and White, R. J. (2008). Non-coding RNA production by RNA polymerase III is implicated in cancer. Nat. Rev. Cancer 8, 911–914. doi: 10.1038/nrc2539
Mughal, M. J., Mahadevappa, R., and Kwok, H. F. (2019). DNA replication licensing proteins Saints and sinners in cancer. Semin. Cancer Biol. 58, 11–21. doi: 10.1016/j.semcancer.2018.11.009
Murray, A. W., and Marks, D. (2001). Can sequencing shed light on cell cycling? Nature 409, 844–846. doi: 10.1038/35057033
Pang, C., Guan, Y., Li, H., Chen, W., and Zhu, G. (2016). Urologic cancer in China. Jpn. J. Clin. Oncol. 46, 497–501. doi: 10.1093/jjco/hyw034
Payton, S. (2013). Bladder cancer: biomarker panel predicts recurrence after radical cystectomy. Nat. Rev. Urol. 10:309. doi: 10.1038/nrurol.2013.95
Preston, B. D., Albertson, T. M., and Herr, A. J. (2010). DNA replication fidelity and cancer. Semin. Cancer Biol. 20, 281–293. doi: 10.1016/j.semcancer.2010.10.009
Ramsay, R. G., and Gonda, T. J. (2008). MYB function in normal and cancer cells. Nat. Rev. Cancer 8, 523–534. doi: 10.1038/nrc2439
Ritchie, M. E., Phipson, B., Wu, D., Hu, Y. F., Law, C. W., Shi, W., et al. (2015). limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 43:e47. doi: 10.1093/nar/gkv007
Rizeq, B., Zakaria, Z., and Ouhtit, A. (2018). Towards understanding the mechanisms of actions of carcinoembryonic antigen-related cell adhesion molecule 6 in cancer progression. Cancer Sci. 109, 33–42. doi: 10.1111/cas.13437
Sánchez-Botet, A., Gasa, L., Quandt, E., Hernández-Ortega, S., Jiménez, J., Mezquita, P., et al. (2018). The atypical cyclin CNTD2 promotes colon cancer cell proliferation and migration. Sci. Rep. 8:11797. doi: 10.1038/s41598-018-30307-x
Siegel, R. L., Miller, K. D., and Jemal, A. (2018). Cancer statistics, 2018. CA Cancer J. Clin. 68, 7–30. doi: 10.3322/caac.21442
Smith, A. B., Jaeger, B., Pinheiro, L. C., Edwards, L. J., Tan, H. J., Nielsen, M. E., et al. (2018). Impact of bladder cancer on health-related quality of life. BJU Int. 121, 549–557. doi: 10.1111/bju.14047
Spiess, P. E., Agarwal, N., Bangs, R., Boorjian, S. A., Buyyounouski, M. K., Clark, P. E., et al. (2017). Bladder Cancer, Version 5.2017, NCCN Clinical Practice Guidelines in Oncology. J. Natl. Compr. Canc. Netw. 15, 1240–1267. doi: 10.6004/jnccn.2017.0156
Suzuki, M., and Takahashi, T. (2013). Aberrant DNA replication in cancer. Mutat. Res. 743-744, 111–117. doi: 10.1016/j.mrfmmm.2012.07.003
Torre, L. A., Bray, F., Siegel, R. L., Ferlay, J., Lortet-Tieulent, J., and Jemal, A. (2015). Global cancer statistics, 2012. CA Cancer J. Clin. 65, 87–108. doi: 10.3322/caac.21262
Wallace, S. S., Murphy, D. L., and Sweasy, J. B. (2012). Base excision repair and cancer. Cancer Lett. 327, 73–89. doi: 10.1016/j.canlet.2011.12.038
Wu, S., Zheng, J., Li, Y., Yu, H., Shi, S. Y., Xie, W. B., et al. (2017). A radiomics nomogram for the preoperative prediction of lymph node metastasis in bladder cancer. Clin. Cancer Res. 23, 6904–6911. doi: 10.1158/1078-0432.CCR-17-1510
Yoshida, S., Takahara, T., Kwee, T. C., Waseda, Y., Kobayashi, S., and Fujii, Y. (2017). DWI as an imaging biomarker for bladder cancer. AJR Am. J. Roentgenol. 208, 1218–1228. doi: 10.2214/AJR.17.17798
Keywords: CNTD2, bladder cancer, bioinformatics analysis, prognosis, survival
Citation: Gong M, Song E, Huang G, Ni W, Dong W and Yuan R (2021) Enhanced Expression of CNTD2/CCNP Predicts Poor Prognosis in Bladder Cancer Based on the GSE13507. Front. Genet. 12:579900. doi: 10.3389/fgene.2021.579900
Received: 14 July 2020; Accepted: 08 January 2021;
Published: 03 February 2021.
Edited by:
Ni Zhao, Johns Hopkins University, United StatesReviewed by:
Hong Zhang, Merck (United States), United StatesCopyright © 2021 Gong, Song, Huang, Ni, Dong and Yuan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Wenjing Dong, dmFsZXJpZTAwNjdAMTYzLmNvbQ==; Runqiang Yuan, eXVhbnJ1bnFpYW5nMjAxM0AxNjMuY29t
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.